Abstract
Defective interfering viral particles are readily produced in cell culture after a high multiplicity of infection with many animal RNA viruses. Due to defects that they carry in their genomes, their life cycle needs to be complemented by the helper functions provided by a parental virus which makes them both dependent on and competitive with the parental virus. In many instances, this may cause the abrogation of a lytic cycle of the parental virus, leading to a persistent infection. In this paper, we describe for the first time the presence of truncated or defective interfering viral RNAs produced in Vero cells persistently infected with the flavivirus Murray Valley encephalitis virus. While these RNAs have not been detected in acutely infected Vero cells, their appearance coincided with the establishment of persistent infection. We also show for the first time that the defective viral RNAs replicate well in both cell culture and cell-free virus replication systems, indicating that they may interfere with the replication of parental virus at the level of viral RNA synthesis. Significantly, structural analyses of these RNA species including nucleotide sequencing have revealed that they carry similar nucleotide deletions encompassing the genes coding for the prM and E proteins and various gene segments coding for the N terminus of the NS1 protein. These deletions are in frame, allowing the synthesis of truncated NS1 proteins to occur in persistently infected cells. This may have further implications for the interference with the parental virus at the level of viral RNA synthesis in addition to a major one at the level of virion assembly and release.
Viral persistence in vivo and in vitro has been reported for a number of animal RNA and DNA viruses. Two major strategies have been employed by these viruses to establish and maintain a persistent infection: one which involves the evasion of the host immune surveillance by the virus by using a variety of tactics to either remove recognition molecules on infected cells or abrogate lymphocyte and/or macrophage functions and the other which alters viral replication and transcription, resulting in nonlytic infection with the production of incomplete or defective viruses (3, 23, 24).
Defective interfering (DI) viruses were initially described for influenza virus by von Magnus (29), who observed their production in embryonated chicken eggs after infection with a large inoculum. They have been also reported for other RNA virus families, including picornaviruses, alphaviruses, and flaviviruses (1, 18, 26). Structural analyses of DI viral genomes revealed that the majority of them are deletion mutants which originate from the genome of parental virus. Although essential replication and encapsidation signals are preserved, DI viruses usually lack part or most of their protein-encoding sequences, which makes them highly dependent upon the parental virus for both replication and encapsidation factors. The competition for these factors between defective and parental viruses affects the replication of parental virus, which eventually leads to a reduction in the titers of infectious parental virus (16). It has also been proposed that DI viruses may play a significant role in the establishment and maintenance of persistent infection (17).
The ability to establish persistent infection in vivo and in vitro has been previously reported for many flaviviruses, including yellow fever (21), St. Louis encephalitis (27), Japanese encephalitis (26), tick-borne encephalitis (13), West Nile (3, 11), and Murray Valley encephalitis (MVE) viruses (24). The cause of persistence in some of the persistent flavivirus infections described so far has been suggested to be the production of DI viral particles, which interfered with the replication of parental virus (3, 24, 26). It has recently been reported that the persistent infection with the Japanese encephalitis virus may also be attributed to the presence of a truncated viral protein NS1 in the absence of any detectable DI virus (6).
Flaviviruses are positive single-stranded RNA viruses with linear RNA genomes of approximately 11 kb. They have a single open reading frame, the 5′ portion of which encodes three structural proteins, i.e., the capsid (C), membrane (M), and envelope (E) proteins, and the 3′ portion of which encodes seven nonstructural proteins, designated NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. While the role of all structural proteins is known and several nonstructural proteins such as NS2B, NS3, and NS5 have been shown to be involved in the virus polyprotein processing and RNA synthesis (2, 4), the functions of the remaining nonstructural proteins, including the NS1 protein, have not been fully characterized. These viruses use a unique strategy of replication involving double-stranded RNAs such as a replicative form (RF), which serves as the template for the synthesis of virion RNA, and a replicative intermediate (RI), which carries nascent genomic RNA (7, 9). It has been shown that similar viral RNA species are produced during acute infection regardless of the system used to study flavivirus replication, whether in cell culture, in a cell-free system of an RNA-dependent RNA polymerase (RDRP) assay, or in mouse brains in vivo (2, 7–9, 28). In this study, we sought to characterize the viral RNA species produced in persistently infected Vero cells with MVE virus by an RDRP assay, Northern blot analysis, and reverse transcription (RT)-PCR. Here we describe for the first time the presence of truncated viral RNAs in Vero cells persistently infected with MVE virus. These defective viral RNAs may be responsible for the establishment of persistent infection in vitro by interfering with the lytic cycle of the parental virus.
MATERIALS AND METHODS
Cells and media.
African green monkey kidney (Vero) cells were maintained in growth medium consisting of M199 medium supplemented with 10% fetal calf serum (FCS) and l-glutamine. The cells were subcultured weekly by standard trypsinization techniques. Vero cells used for virus propagation were maintained in a minimal growth medium supplemented with 2% FCS and l-glutamine.
Virus.
A standard virus stock of the Australian flavivirus MVE virus, strain OR2 (19), was obtained from the departmental collection as a 10% homogenate of infected suckling mouse brain and was propagated twice in Vero cell monolayers at a multiplicity of infection (MOI) of 0.1. The cell monolayers were incubated at 37°C for 1 h with the virus, and then the inoculum was replaced with fresh medium and the cultures were incubated for an additional 72 h or until a cytopathic effect (CPE) was clearly visible. The cell culture medium containing the infectious virus was then harvested, clarified by centrifugation at 1,000 × g, and divided into aliquots before being stored at −70°C. Acute infection of Vero cells was similarly performed at an MOI of 0.5 to 1.0.
Virus titration.
The titration of the infectious virus in the cell culture medium was performed by 50% tissue culture infective dose (TCID50) assay in serial 10-fold dilutions in Vero cells as described previously (25).
Establishment of a persistent infection.
A confluent Vero cell monolayer was infected with MVE virus strain OR2 at an MOI of 10 for 1 h at 37°C. After infection, the medium was replaced, and the cells were maintained in culture until extensive CPE occurred. Lysed cells and medium were removed and replaced with fresh M199 medium supplemented with 10% FCS and l-glutamine. The remaining viable cells grew back, resulting in persistently infected, confluent monolayers. The cells were passaged by trypsinization every 4 to 5 days. The cell culture medium from Vero cells persistently infected with the MVE virus was collected at every passage from passages 1 to 10 (P1 to P10), clarified by centrifugation as described above, aliquoted, and stored at −70°C. To study the effect of dilution of the viral stocks obtained after harvesting the medium from the persistently infected Vero cell cultures, 10−2 and 10−4 dilutions were made in M199 medium supplemented with 2% FCS and l-glutamine and together with undiluted stocks were used to infect the fresh confluent monolayers of Vero cells for 1 h at 37°C. The amount of virus used in this set of experiments was estimated to be 50 times lower than that standardly used in the TCID50 assay. Three to five days later, when the CPE was evident, the culture medium was collected, clarified, and titrated for the presence of infectious virus by TCID50 assay, and it was also used to infect fresh monolayers of Vero cells for further use in a cell-free virus replication assay.
Immunofluorescence studies.
Acutely and persistently infected cultures of Vero cells were fixed with acetone at −20°C and probed with a mixture of monoclonal antibodies directed against the NS1, prM, and E proteins of the MVE virus (15). The cells were then incubated with a goat anti-mouse antibody conjugated to fluorescein (TAGO) for 1 h at room temperature. Fluorescence was observed by a Leitz fluorescence microscope.
Preparation of cell extracts.
Cell extracts were prepared from both acutely and persistently infected Vero cells with MVE virus strain OR2 at 24 h or 5 to 6 days postinfection (p.i.), respectively, as described previously (7). Cell extracts were harvested over several passages of persistently infected Vero cells, from P1 to P10. Briefly, cell monolayers were harvested and washed in 1× phosphate-buffered saline and then resuspended in 10 mM Tris-HCl (pH 8.0) containing 10 mM sodium acetate (TN buffer) at a concentration of 4 × 107 cells/ml. Cell membranes were disrupted by passaging the cells 20 times through a 21-gauge needle and then 20 times through a 26-gauge needle. The cell lysates were centrifuged at 800 × g for 5 min, and the supernatants (cytoplasmic fraction) were collected and stored in aliquots at −70°C. The cell extracts were either used in cell-free virus replication assays, i.e., the RDRP assays, or extracted by phenol to isolate RNA for Northern blot analysis. Protein concentrations in the cell extracts were determined by using a Micro BCA protein assay reagent kit (Pierce).
RDRP assay.
RDRP assays were carried out by using the extracts of infected cells which had been stored at −70°C. The standard assay contained 50 mM Tris-HCl (pH 8.5), 10 mM Mg acetate, 7.5 mM K acetate, 6 mg of actinomycin D per ml, 10 mM 2-mercaptoethanol, 5 mM phosphoenolpyruvate, 3 U of pyruvate kinase (Boehringer Mannheim) per μl, 0.5 mM each CTP, UTP, and ATP, 25 μM GTP, 5 μCi of [α-33P]GTP, and the cellular extract containing 5 to 8 mg of cellular proteins per ml. The assays were carried out at 37°C for 2 h or as indicated in the figure legends. The RNA products were extracted with phenol-sodium dodecyl sulfate (phenol-SDS) in TNE buffer (TN + EDTA), resuspended in 10 μl of double-distilled water, and separated by agarose gel electrophoresis.
Nondenaturing agarose gel electrophoresis.
RNA products were separated on nondenaturing 0.8% agarose gels (15 by 15 by 0.5 cm) at 100 V for 2.5 to 3 h. All solutions were prepared with double-distilled water. Gel electrophoresis equipment was pretreated by soaking in 3% H2O2 solution for 30 min. The agarose gels were prepared with 1× TBE buffer (89 mM Tris base, 89 mM boric acid, 2 mM EDTA), which also served as the electrode buffer. Samples were mixed with 3 to 5 μl of gel loading buffer (0.04% bromophenol blue, 0.04% xylene cyanol, 5% glycerol) and loaded onto the gel without heating.
Northern blot analyses.
Electrophoretically separated viral RNAs were transferred to a nylon membrane (Hybond N+; Amersham) for 2 h by vacuum with 50 mM NaOH as the transfer buffer. The RNA was fixed to the membrane by baking at 80°C for 15 min and then subjected to hybridization with either cDNA or oligonucleotide probes.
(i) cDNA hybridization.
The viral cDNA clone MVE CL1/1/12 (10) was used as a probe to detect viral RNAs. This clone was digested with EcoRI, and a 6.5-kb DNA fragment which covers the 5′ half of the MVE genome was obtained, gel purified, and randomly labelled with [α-32P]dCTP by using the GIGAprime random labelling kit (Bresatec, Adelaide, Australia). After being prehybridized for 3 h at 65°C, the membranes were hybridized to the labelled viral cDNA probe at the same temperature overnight in the same buffer (7% SDS, 0.5 M sodium phosphate buffer [pH 7.0], 1 mM EDTA, 1% bovine serum albumin). The membranes were washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature for 5 min followed by two washes with 2× SSC–1% SDS at 65°C for 30 min and finally two washes with 0.1× SSC at room temperature for 30 min. The filters were air dried and exposed to X-ray film (Kodak X-Omat AR or Fuji X-ray) overnight at −70°C in the presence of an intensifying screen.
(ii) Oligonucleotide hybridization.
Oligonucleotide probes were designed to hybridize to the specific regions of the MVE genome by using the Oligo design program and used for either mapping of the deletion in defective viral RNA or the detection of positive- and negative-sense viral RNAs (see Table 2). The oligonucleotides were synthesized on an Oligo 1000M DNA synthesizer (Beckman) and were either 5′ end labelled with [γ-32P]ATP or 3′ end labelled with [α-32P]Cordycepin 5′ triphosphate (NEN). Hybridizations were carried out with each of the oligonucleotide probes by incubation at 42°C overnight in hybridization buffer containing 5× SSC, 20 mM NaH2PO4, 0.5% SDS, and 10× Denhardt solution. Membranes were washed twice with 2× SSC for 5 min at room temperature, followed by 2 washes with 2× SSC–1% SDS at 42°C for 15 min. The membranes were exposed to X-ray film (Kodak X-Omat AR or Fuji X-ray) for 1 to 2 days in the presence of an intensifying screen at −70°C.
TABLE 2.
Oligonucleotide primers used in Northern blot hybridization and RT-PCR
| Primera | 5′–3′ sequence | Nucleotide positions in the virus genome |
|---|---|---|
| 13as | TCGGAAGCTCACGCAG | 13–28 |
| 138as | CCGCGTTTTAGCATATTGA | 138–156 |
| 440as | CCAATCAGCATGAAAATGAC | 440–459 |
| 949s | GCTCCTCGTTGCTCCTGCCTAC | 949–970 |
| 2,021s | ACGCCTGTCGGGAGAATGGTG | 2021–2041 |
| 3,018as | TGCGGTCCCAATAATGGTGC | 3018–3037 |
| 3,994as | CCGGAGCAAGCATTGCTAGC | 3994–4013 |
| 4,927s | AAACGTGCAGACCAAGCCCG | 4927–4946 |
| 6,031s | TGGAGGCGGAACCAGTGAGG | 6031–6050 |
| 7,017as | AGCAGGCTTCAAATCCAGGG | 7017–7036 |
| 8,001as | TTCTTCATGGCCTGGCCCAC | 8001–8020 |
| 9,061s | ACGCGAGAAAAAACCCGGAG | 9061–9080 |
| 9,991s | TAGGGCAGCTACCTTCATGT | 9991–10010 |
| 10,006s | GGGATCTGCGCCTGATGGCC | 10006–10025 |
| 10,849as | GGGTCTCCTCTAACCTCTAGTCC | 10849–10871 |
| K1Ls | GGCAGAGGGCCAATACGG | 215–232 |
| K2Ls | AGACGTTCATCTGCGTGAGC | 1–20 |
as and s, antisense or sense orientation of the oligonucleotide primer, respectively.
RT-PCR.
Phenol-extracted samples from persistently infected Vero cells were used as a template for RT-PCR analysis using the Titan one-tube RT-PCR system (Boehringer Mannheim). The same strain of MVE virus free of any DI virus particles was propagated in laboratory mice. Viral RNA was isolated from mouse brains as described by Urosevic et al. (28) and used as a positive control. The RT-PCR mix consisted of 0.2 mM each deoxynucleoside triphosphate, 100 ng of forward primer (see Table 2) (primer K2Ls), 200 ng of reverse primer (see Table 2) (primer 3018as), 5 mM dithiothreitol, 10 ml of 5× RT-PCR buffer, 1.5 mM MgCl2, 1 μl of avian myeloblastosis virus, and Expand high-fidelity enzyme blend (Boehringer Mannheim) in a total reaction volume of 50 μl. The template and reverse primer were incubated at 70°C for 10 min and chilled on ice prior to the addition of the reaction mix. RT was conducted at 42°C for 2 h. This was followed by an initial 3-min denaturation at 95°C and 35 subsequent cycles of 1 min of denaturation at 95°C, 1 min of annealing at 55°C, and 5 min of extension at 72°C. PCR products were analyzed on nondenaturing agarose gels as described previously.
Cloning and sequencing.
RT-PCR products were excised from the agarose gels and purified with a JETquick gel extraction kit (Genomed). Purified RT-PCR products were either used directly for sequencing or cloned into a pGEM-T vector (Promega) and used to transform competent XL1 Blue cells. Positive clones were confirmed by PCR by using the same conditions described above and prepared for sequencing. Both plasmid DNA obtained by a modified mini-alkaline lysis-polyethylene glycol precipitation procedure (Applied Biosystems) and RT-PCR products obtained as described above were subjected to sequencing with an ABI PRISM dye terminator cycle sequencing kit (Perkin-Elmer) in the presence of either primer K2Ls, K1Ls, or 3,018as (Table 2) and analyzed by using an ABI 373A automated DNA sequencer (Applied Biosystems).
Western blot analysis.
Polyacrylamide gel electrophoresis (PAGE) was used to separate 15 to 20 μg of protein present in cytoplasmic lysates of Vero cells persistently infected with MVE virus. This was performed in parallel with lysates from cells acutely infected with the same virus and mock-infected cells. Samples were either boiled for 5 min or not boiled in nonreducing sample buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 0.025% bromophenol blue) before loading. Following PAGE, the proteins were transferred to nitrocellulose membranes and treated with blocking buffer as described elsewhere (14). The membranes were incubated with a cocktail of monoclonal antibodies to NS1 (1:1 [vol/vol] dilution in culture supernatant), M2-10C6, M-E6, M2-8C4, and M2-9A2 (15), overnight at 4°C. Peroxidase-labelled antibodies to mouse immunoglobulin G (goat anti-mouse immunoglobulin G; Bio-Rad) were used as a secondary antibody at 1/1,000. Blots were stained with diaminobenzidine (0.05% in phosphate-buffered saline with 0.018% H2O2) until the desired intensity was obtained and then rinsed in distilled water before being photographed (15).
RESULTS
Establishment of persistent infection in Vero cells with MVE virus.
A persistent infection of Vero cells with MVE virus strain OR2 was established by infecting the cells at an MOI of 10. Three to five days later, 80 to 90% of the cells showed severe CPE and were detached from the monolayer, while the surviving cells continued to grow to confluence and were routinely passaged up to 10 times. The cell culture medium from each passage was collected and titrated for infectious virus by TCID50 assay (Table 1). As shown in Table 1, the viral titers in persistently infected cells from P1 to P10 were constantly 10- to 100-fold lower than the viral titer in acutely infected cells. In addition, it has also been observed that the lower dilutions (10−1 and 10−2) of the cell supernatants from the persistently infected cells did not produce CPE in indicator cell monolayers during the TCID50 assay (data not shown). This has been assumed to be a result of interference with the defective virus at the higher titers since the same cell supernatants at dilutions higher than 10−2 produced an obvious CPE (data not shown).
TABLE 1.
Virus titers in acute and persistent infections of Vero cells with MVE virus
| Infection | Passage or virus | Log10TCID50/100 ml |
|---|---|---|
| Persistent | P1 | 5.77 |
| P2 | 6.45 | |
| P3 | 5.5 | |
| P4 | 6.12 | |
| P5 | 5.2 | |
| P6 | 5.1 | |
| P7 | 4.3 | |
| P8 | 5.5 | |
| P9 | 4.9 | |
| P10 | 5.46 | |
| Acute | Parental virus | 7.16 |
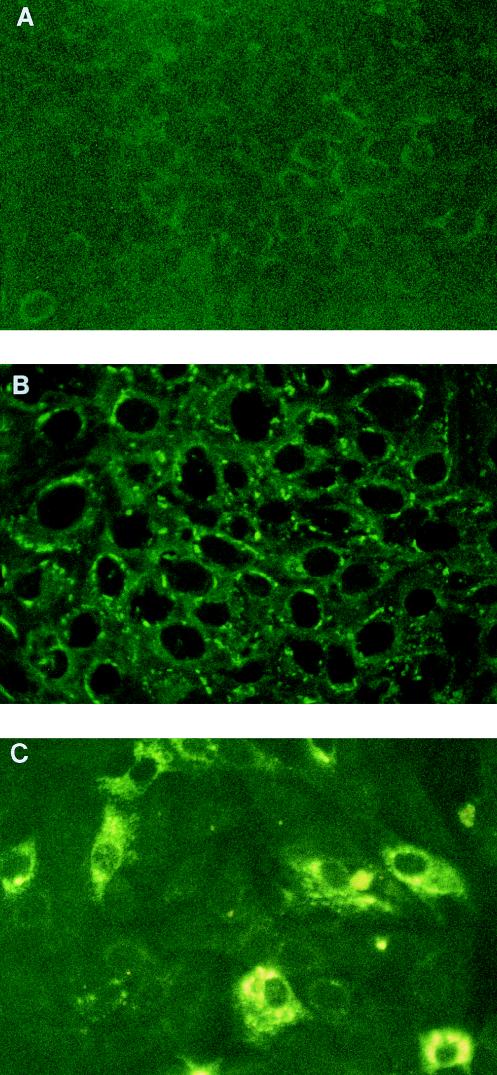
To confirm the presence of virus in persistently infected cells, an indirect immunofluorescence assay using a mixture of virus-specific monoclonal antibodies to the viral proteins NS1, preM, and E (15) was carried out. This assay revealed the presence of MVE virus antigens in approximately 40 to 60% of the cells in persistently infected cultures at P1 compared to 100% of the cells acutely infected with parental virus at a lower input dose (MOI of 1) (Fig. 1). Although the acutely infected cells showed an extensive presence of viral proteins, the intensity of the positive signal in those cells was much lower than that in persistently infected ones, indicating the presence of smaller amounts of viral proteins (Fig. 1). This may have resulted from the smaller amount of template available in acutely infected cells due to both the lower input dose of the virus (MOI of 1 versus 10) and shorter duration of replication, which was only 24 h for acutely infected cells compared to a minimum of 4 to 5 days for persistently infected ones.
FIG. 1.
Indirect immunofluorescence of Vero cells infected with MVE virus. A mixture of monoclonal antibodies to the NS1, prM, and E proteins of MVE virus was used to detect the virus in mock-infected Vero cells (A), in Vero cells acutely infected with MVE virus strain OR2 at an MOI of 1 at 24 h p.i. (B), and in persistently infected Vero cells which survived an acute infection with the same virus at an MOI of 10 and were passaged once (P1) without further addition of the virus (C).
Replication of defective viral RNA in persistently infected cells.
To study the viral RNA species produced during persistent infection of Vero cells with MVE virus, cell extracts were harvested from persistently infected cells at P1 to P10 and analyzed by an RDRP assay. Initially, the RDRP assay was performed in the absence of label and the viral RNA products were analyzed by Northern blot hybridization after separation on an agarose gel under nondenaturing conditions (Fig. 2A). By this approach, the total viral RNAs produced in both cell culture during virus infection and RDRP assay during cell-free virus replication in either persistently or acutely infected Vero cells were determined. As shown before by using a similar approach to study total viral RNA produced in the mouse brain (28), Northern blot analysis revealed the presence of three standard viral RNA species in the acutely infected cells: virion RNA (vRNA), RF RNA, and RI RNA (Fig. 2A, lane 2). However, persistently infected cells showed two additional RNA species which migrated faster than the standard RF and vRNA species (Fig. 2A, lane 1). As will be shown later, these faster-migrating RNAs contain large internal deletions. Also, at this stage, we were not able to distinguish how many individual defective viral RNA species were present in the broad bands shown in Fig. 2A due to resolution limitations of agarose gel electrophoresis. It will be shown later by using RT-PCR and nucleotide sequencing that there are at least four defective viral RNA species which comigrated in the gels shown in Fig. 2 and 3. Hence, the RNAs present only in persistently infected cells are labelled defective RF (dRF) and defective vRNA (dvRNA) in Fig. 2 and subsequent figures. The identity of the RF and vRNA forms, both standard and defective, was confirmed by oligonucleotide hybridization in which the oligonucleotide probe complementary to the viral negative-strand RNA (Table 2; primer 10,006s) hybridized to RI and both standard and defective RFs, while the oligonucleotide probe complementary to the viral positive-strand RNA (Table 2; primer 440as) hybridized to all viral RNA species, including RI, standard and defective RFs, and standard and defective vRNAs (data not shown).
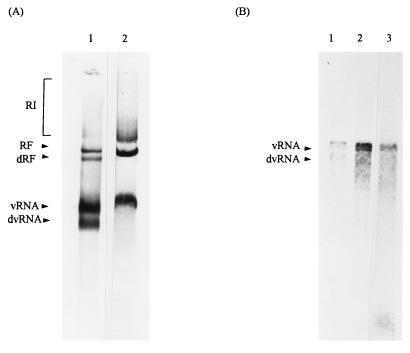
FIG. 2.
RDRP assay using cell extracts of persistently infected Vero cells with the MVE virus. Viral RNA products of the standard RDRP assay were separated on an agarose gel under either nondenaturing (A) or denaturing conditions (B), transferred to Hybond N+ membrane, and hybridized to a 32P-labelled viral cDNA probe, MVE CL1/1/12. Cellular extracts from persistently (lanes 1) and acutely (lanes 2) infected Vero cells were prepared, assayed by the RDRP assay, and electrophoresed in parallel. Migration of the control vRNA, isolated from viral particles collected by polyethylene glycol precipitation, is presented (panel B, lane 3).
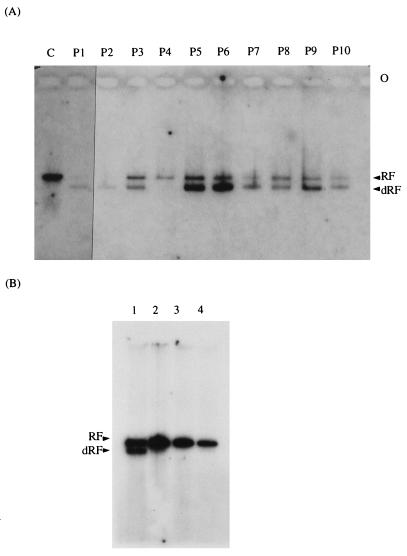
FIG. 3.
Effect of passaging and dilution on the presence of defective viral RNA. (A) Cellular extracts from P1 to P10 of Vero cells persistently infected with MVE virus were subjected to the RDRP assay in the presence of [α-32P]GTP, and viral RNA was separated on an agarose gel under nondenaturing conditions. The extracts of cells acutely infected with MVE virus at an MOI of 1 are shown in lane C. (B) Cellular extracts of Vero cells infected with the second-round culture medium (M2) collected from cells acutely infected with either neat (lane 1) or diluted (10−2 dilution, lane 2; 10−4 dilution, lane 3) cell culture medium (M1) obtained from persistently infected cells at P1. Vero cells acutely infected with MVE virus at an MOI of 1 are also shown (lane 4).
The standard RDRP assay was also performed in the presence of [α-32P]GTP with cellular extracts of either acutely or persistently infected cells from all passages to detect viral RNA species which are replicated de novo under the cell-free conditions. Under such conditions, the dRF species was observed only in persistently infected cells (Fig. 3A, lanes 2 to 11). The appearance of the respective standard and dvRNAs was observed after a longer exposure (data not shown).
If the defective viral RNA described above has accumulated as a result of the high MOI of the virus used to produce a persistent infection in Vero cells (MOI of 10), then diluting the medium collected from the persistently infected cells containing the defective virus should have the opposite effect on the accumulation of defective RNA. This hypothesis was tested by the following experiment in which neat cell culture medium and two dilutions (10−2 and 10−4) of it obtained from persistently infected cells at P1 (medium 1 [M1]) were used to acutely infect the fresh monolayers of Vero cells. To achieve the acute infection of Vero cells with viral stocks containing defective virus, neat, 10−2-diluted, and 10−4-diluted M1 was used in aliquots 50 times smaller than those previously employed in the TCID50 assay (Table 1). The second round of cell culture medium (M2) was collected from acutely infected cells 3 to 5 days p.i. with either neat, 10−2-diluted, or 10−4-diluted M1 at the time of the CPE appearance and used to either titrate infectious virus by TCID50 or infect fresh monolayers of Vero cells for the preparation of cellular extracts for the RDRP assay. Although virus titration revealed similar amounts of infectious virus in M2 medium regardless of the dilution of the M1 stock used initially to infect the cells (log10TCID50s after infection with neat, 10−2-diluted, and 10−4-diluted M1 were 6.75, 6.71, and 6.75, respectively), the pattern of interference in the TCID50 assay was markedly different. No interference was observed at any dilution of M2 collected from the cells infected with a 10−4 dilution of M1. Very low interference was found with the 10−1 dilution of M2 collected from cells infected with the 10−2 dilution of M1. Interference was observed with the 10−1 and 10−2 dilutions of M2 obtained from cells infected with the neat M1, as described above (data not shown). The cell extracts obtained 24 h p.i. with M2 collected from the cell culture infected with the neat M1, as described above, was used in the RDRP assay and, as shown in Fig. 3B (lane 1), displayed de novo synthesis of both RF and dRF species. In contrast, when the cell extracts prepared from Vero cells infected with M2 of cells infected with either the 10−2 (Fig. 3B, lane 2) or 10−4 dilution of M1 (Fig. 3B, lane 3) were used in the RDRP assay, no production of dRF was observed. In fact, they resembled the cell extract obtained from the acutely infected cells with the low MOI (MOI of 1) of the parental virus (Fig. 3B, lane 4). A small amount of dRF species could be seen in the cell extract of cells infected with M2 obtained from the cells infected with 10−2-diluted M1 after a longer exposure (data not shown).
Structural analysis of defective viral RNA.
As shown above, for the first time we have detected fast-migrating viral RNA species in persistently infected cells by use of the RDRP assay in combination with nondenaturing agarose gel electrophoresis and Northern blot hybridization analysis (Fig. 2A, lane 1). To confirm that the viral RNAs identified by this procedure are not artifacts of either the virus RNA isolation procedure or the cell-free replication assay, we have used either the RDRP assay alone (Fig. 3) or direct Northern blot analysis without prior submission to RDRP assay to study total viral RNA in the cell extracts of both acutely and persistently infected cells. By using either approach, similar viral RNA species were revealed to either preexist in cellular extracts (data not shown) or incorporate the label during the RDRP assay (Fig. 3), strongly suggesting that the faster-migrating viral RNA species are genuine products of virus replication. To characterize them further, we decided to use agarose gel electrophoresis under denaturing conditions in combination with Northern blot analysis. These conditions were supposed to resolve whether the fast-migrating, defective viral RNAs were truncated or had an unusually compact conformation. Since the cell-free virus replication assay in the absence of label in combination with Northern blot analysis proved to increase the detection of viral RNAs in the sample (Fig. 2A), we subjected those samples to agarose gel electrophoresis under denaturing conditions (Fig. 2B). Northern blot hybridization with a virus-specific cDNA probe revealed a single RNA species in both persistently and acutely infected cells of 11 kb (Fig. 2B, lanes 1 and 2, respectively), which corresponds to the size of packaged vRNA of the parental virus (Fig. 2B, lane 3), while in persistently infected cells, an additional RNA species of approximately 8 to 9 kb was observed (Fig. 2B, lane 1). This RNA species is likely to be a truncated viral RNA which carries a deletion of approximately 2 to 3 kb.
To determine the position of the deletion in the defective RNA, total RNA isolated from persistently infected cells, containing both parental and defective vRNAs, was hybridized to a panel of oligonucleotide probes that were complementary to the viral genomic sequences and distributed at 1-kb intervals along the MVE genome (Table 2). The results of this hybridization revealed that the parental RNA hybridized to all oligonucleotide probes used in this experiment. In contrast, the defective viral RNA contained sequences complementary to the majority of the oligonucleotides used, including those complementary to the conserved elements at the 5′ and 3′ noncoding regions of the viral genome but not those corresponding to the region containing oligonucleotides at positions 949 and 2012 (data not shown). This indicated that the deletion in the defective RNA was somewhere between positions 440 and 3037 in the virus genome.
RT-PCR and nucleotide sequencing.
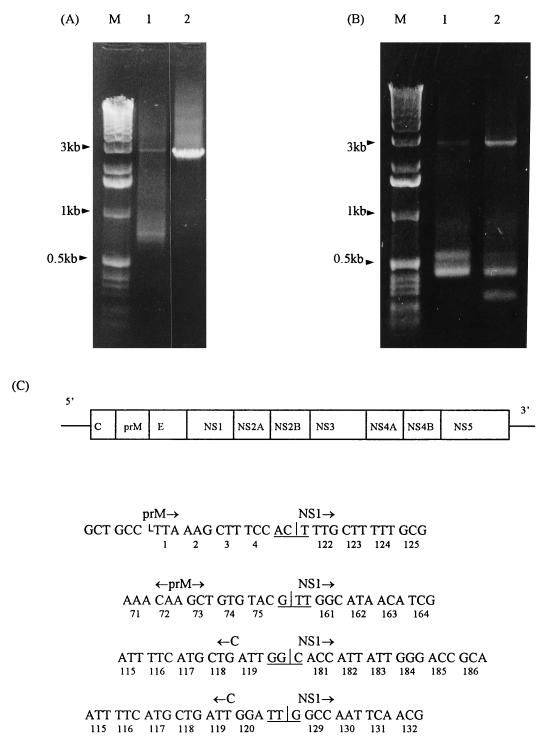
The precise location of the deletion in the defective viral RNAs was determined and the defective viral RNA in persistently infected cells was further characterized by nucleotide sequencing of the cDNA corresponding to the dvRNA, which was obtained by RT-PCR (Fig. 4). In the initial attempt, total RNA purified from persistently infected Vero cells at P7 and full-length vRNA isolated from infected mouse brain were used as templates in RT-PCR. Given the approximate location of the deletion determined by oligonucleotide hybridization, a primer pair encompassing positions 1 to 3037 of the MVE virus genome was used in RT-PCR. As predicted, a 3-kb DNA product was obtained from both the total viral RNA extracted from the brain of a flavivirus-susceptible C3H/HeJ mouse and the total viral RNA extracted from persistently infected Vero cells (Fig. 4A, lanes 1 and 2). In contrast, only the RNA extract of persistently infected cells supported the amplification of a smaller viral PCR product of approximately 0.7 kb (Fig. 4A, lane 1). This product was gel purified and cloned into pGEM-T vector (Promega), and a single positive clone was isolated and sequenced. When these sequence data were compared to the published sequence of the MVE virus prototype, strain 1-51 (10), a large gap of 2,353 nucleotides between positions 486 and 2839 was detected in the smaller PCR product obtained from the defective RNA of MVE virus strain OR2 (Fig. 4C). In the second attempt, a new primer designated K1Ls (Table 2) was used in RT-PCR together with the previously used 3018 primer to amplify a smaller region of the virus genome encompassing the deletion, i.e., from positions 215 to 3037. By using this new set of primers, viral RNAs isolated from persistently infected Vero cells at P7 and P8 were subjected to RT-PCR in parallel under the same conditions, and the resulting cDNA products were analyzed by agarose gel electrophoresis (Fig. 4B). As shown in Fig. 4B, a new round of RT-PCR using the new set of primers to amplify viral RNA present in P7 of persistently infected cells has produced a cDNA fragment of 0.55 kb in addition to the expected one of 0.46 kb (lane 1). A parallel RT-PCR performed on the subsequent passage (P8) of persistently infected cells revealed another cDNA product of approximately 0.3 kb in addition to a very strong band of the expected 0.46-kb product and a very faint band of a 0.55-kb fragment (Fig. 4B, lane 2). Four additional cDNA fragments, corresponding to 0.55- and 0.46-kb bands (Fig. 4B, lane 1) and 0.46- and 0.3-kb bands (Fig. 4B, lane 2), were cut out of the gel and subjected to either direct DNA sequencing or cloning into the pGEM-T vector. The cloning resulted in three additional positive clones which, when sequenced, revealed the presence of additional deletions (Fig. 4C). Sequencing of single positive clones for 0.55-, 0.46-, and 0.3-kb products revealed three additional deletions spanning nucleotides 698 to 2955, 462 to 2860, and 459 to 3019 of the MVE virus genome, respectively (Fig. 4C). Direct sequencing of the 0.55- and 0.3-kb RT-PCR products confirmed the presence of the same deletions as those in the corresponding cloned cDNA fragments (data not shown). However, direct sequencing of the 0.46-kb RT-PCR product was not conclusive in the vicinity of the deletion, indicating heterogeneity of the cDNA products (data not shown). This may suggest the presence of additional truncated viral RNA species which carry deletions with a similar size although at slightly different positions. It could be expected from the data described above that the positions of the other deletions may vary from nucleotides 459 to 698 on the left to nucleotides 2839 to 3019 on the right, although the number and sequence of additional defective viral RNA species would be difficult to predict. This may further be complicated by the variability of their occurrence from passage to passage, as already shown for the previously characterized ones (Fig. 4B).
FIG. 4.
RT-PCR and sequence analyses of defective viral RNA. (A) RT-PCR analysis of viral RNA present in either persistently infected Vero cells at P7 (lane 1) or a mouse brain after an intracerebral challenge with MVE virus (lane 2). The oligonucleotide primers K2Ls and 3018as were used to amplify the region of the viral genomic RNA from position 1 to 3037. (B) RT-PCR analysis of viral RNA present in two successive passages, P7 and P8, of persistently infected Vero cells (lanes 1 and 2, respectively) by using the oligonucleotide primers K1Ls and 3018as to amplify the region of the viral genome from position 215 to 3037. (C) Diagram showing the sequences of four defective viral RNAs in the vicinity of deletions. The viral genome is depicted at the top. Vertical lines denote the sites of deletions; horizontal lines denote in-frame codons formed by joining nucleotides left of the deletion of the prM- and C-coding units to various nucleotides following the deletion in the NS1-coding unit. The numbers below the sequence denote the positions of codons (amino acids) in the prM-, NS1-, and C-coding units (proteins), respectively.
When the sequence data were summarized, four large deletions were revealed which encompass the region of the viral genome from the C terminus of the C protein to the middle of NS1, eliminating a region coding for viral proteins prM and E and the N-terminal portion of NS1 (Fig. 4C). This suggests the presence of at least four distinct although similar defective viral RNAs in persistently infected cells. General features of all four of these deletions are that they are in frame and that their presence and position may not interfere with the synthesis and processing of the viral polyprotein. As an outcome, both shorter polyprotein and truncated NS1 protein may be expected to be produced in persistently infected cells.
NS1 protein expression in persistently infected Vero cells.
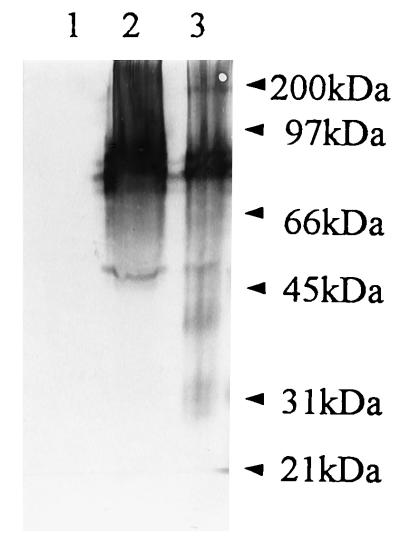
To test the hypothesis described above about the expression and processing of defective polyprotein and expression of different forms of NS1 protein, we have subjected cell lysates from persistently infected Vero cells to Western blot analysis. The samples of mock-infected and acutely and persistently infected Vero cells were separated by 10% PAGE before (Fig. 5) and after boiling in nonreducing sample buffer to allow detection of both dimeric and monomeric forms of NS1 (15). A cocktail of monoclonal antibodies to NS1, M2-10C6, M-E6, M2-8C4, and M2-9A2 (15), has been used in immunoblotting, revealing the presence of two truncated forms of NS1 of approximately 39 and 29 kDa in persistently infected cells, two dimeric forms of 80 and 100 kDa, and one monomeric form of 45 kDa (Fig. 5, lane 3). No additional bands were visible in the sample of persistently infected cells, indicating that truncated NS1 did not form dimers (Fig. 5, lane 3). The same dimeric forms of 80 and 100 kDa and the monomeric form of 45 kDa as those seen in the persistently infected sample were detected in acutely infected cells at 48 h p.i., although no truncated NS1 protein was observed (Fig. 5, lane 2). No reactivity with the monoclonal antibodies was detected in the mock-infected sample (Fig. 5, lane 1).
FIG. 5.
Western blot analysis of NS1 protein produced during persistent infection of Vero cells with MVE virus. Monoclonal antibodies to NS1, M2-10C6, M-E6, M2-8C4, and M2-9A2 (15), were used in a cocktail to probe for the presence of viral NS1 forms in mock-infected (lane 1), acutely infected (48 h p.i.) (lane 2), and persistently infected (lane 3) Vero cells at P8. Samples (15 to 20 μg of protein) were separated by 10% PAGE in the absence of 2-mercaptoethanol without boiling in parallel with prestained protein molecular size standards (Bio-Rad; 6.5 to 200 kDa).
DISCUSSION
In this study, we have analyzed viral RNA molecules involved in the establishment and maintenance of persistent infection with MVE virus strain OR2 in Vero cells. We have used similar experimental conditions to establish persistent infection with MVE virus as previously described (24), although an alternative approach including Northern blot analysis, cell-free virus replication assay, and RT-PCR has been applied to analyze the viral RNA present in persistently infected cells. Using this approach, we were able to identify at least four defective viral RNA species with similar sizes, averaging 8,600 nucleotides. Due to the similarity in their sizes (8,453, 8,615, 8,660, and 8,756 nucleotides), it was not possible to resolve them under the standard agarose gel conditions, either nondenaturing or denaturing, used in our study (Fig. 2 and 3). Their existence and precise position and the size of the deletion they carry were determined by RT-PCR and nucleotide sequencing (Fig. 4). However, their relative abundance has been shown to vary from passage to passage, indicating variability in their occurrence (Fig. 4B). These RNAs may either be responsible for or result from the establishment of persistent infection with the normally cytocidal MVE virus. Although some earlier reports suggested the presence of truncated viral RNAs and DI viruses in a cell culture affiliated with persistent flavivirus infection (3, 26), the present study is the first to demonstrate the presence of discrete defective flaviviral RNA species in persistently infected cells. We have also demonstrated for the first time that some of these defective flaviviral RNAs can replicate in both infected cells and a cell-free virus replication system, as revealed by the appearance of their RF(s) in the RDRP assay and Northern blot analysis (Fig. 2 and 3). According to these data, the level of RNA synthesis of the DI viruses is equal to or slightly higher than that of parental virus, indicating that interference may occur at the level of RNA synthesis since it is likely that both defective and parental RNAs use the same replicative machinery. The ability of the defective RNAs to be released from the infected cells together with the parental virus and to infect new cells has also been demonstrated in the set of experiments in which the cell culture medium collected from the persistently infected cells was passaged and used to infect the fresh monolayers of Vero cells. These experiments indicated that the DI viruses present in the cell culture medium of persistently infected cells were able to infect fresh monolayers of Vero cells during an acute infection, replicate and be released into the second-round cell culture medium (M2), preserve their interference capacity in the same dilution range, and persist to the second-round acute infection in which their replication was revealed by the RDRP assay (Fig. 3B, lane 1). With the same set of experiments, we were able to show that the DI viruses could be reduced to undetectable levels by the use of increasing dilutions (10−2 and 10−4) of the same cell culture medium (Fig. 3B, lanes 2 and 3). These results are in agreement with the notion that the enrichment of the DI viruses is usually dependent on a high multiplicity of infection, which provides both more DI and more parental viruses with the helper functions needed in the multiplication of the DI viruses, whereas a low multiplicity of infection reduces the amount of both DI and parental viruses, which eventually limits the production of DI viruses (12).
We have shown only indirectly the capacity of the defective viral RNAs to interfere with the replication of the parental virus. This has been revealed by the ability of these RNAs to prevent the appearance of CPE in indicator cells at the lower dilutions (10−1 and 10−2) of the cell culture medium in the TCID50 assay, as described in this paper. The presence of DI viruses in persistently infected cell cultures affected both the titer of the infectious virus in all passages studied (Table 1) and the percentage of cells infected with the parental virus, as shown by immunofluorescence (Fig. 1). The levels of parental and DI viruses in different passages of persistently infected cells showed similar cyclic patterns in both the TCID50 (Fig. 1) and RDRP assays (Fig. 3A), further indicating the close interrelationship between these two viral entities.
Structural analyses of the defective viral RNAs have revealed 2,257-, 2,353-, 2,398-, and 2,560-nucleotide deletions, which represent 20 to 23% loss of the viral genome encompassing the coding units for the structural proteins prM and E and various segments of the gene coding for the N terminus of the NS1 protein (Fig. 4C). The 2,398- and 2,560-nucleotide deletions apparently affect the sequence coding for the carboxy terminus of the C protein (10). This region is a membrane-associated domain not present in the mature virion form of the C protein and has been implicated in the expression of the prM protein (5). The observed deletions are in frame, and some of them were shown to allow the synthesis and processing of the truncated viral polyprotein, which in turn resulted in the production of truncated forms of viral NS1 protein of 39 and 29 kDa (Fig. 5, lane 3). This indicates that the defective viral RNAs are expressed at the protein level and that, due to the deletions they carry, are incapable of supporting the production of the structural proteins prM and E. This makes defective virus highly dependent on the parental virus for the virion assembly and release from the cell and probably represents the major point of interference with the parental virus. The major feature of the defective viral RNA expression is, as shown in Fig. 5, the appearance of truncated NS1 protein forms, which is in agreement with the expression of the similarly truncated form of the NS1 protein of 39 kDa recently reported to be strongly associated with the persistent infection of a variety of cell types, including Vero cells, with the Japanese encephalitis virus (6). There is also evidence of the existence of similarly truncated NS1 protein in Vero cells acutely infected with the higher dose (MOI of 10) of MVE virus prototype strain 1-51 (15a). The viral NS1 protein has been recently suggested to have an important role in viral RNA replication; this suggestion was based on both its colocalization with the viral RF RNA form in dengue virus- or Kunjin virus-infected cells (20, 30) and the impaired replication of the viral ts mutants of the yellow fever virus carrying the lesions in the NS1 gene (22). The truncation of the NS1 protein may create two different scenarios for the establishment of persistent infection, one in which the DI RNA incapable of producing its own NS1 protein competes with the parental viral RNA for the full-length NS1 and the other in which the truncated form of the NS1 protein has a direct effect on the virus infection as observed by Chen et al. (6), providing an additional, yet-unknown drive to the flavivirus-persistent state. At this stage, we are unable to distinguish between these two scenarios, although evidence presented in this paper strongly supports the hypothesis that the truncated forms of NS1 protein observed in persistently infected cells originate from and are encoded by the defective viral RNAs produced during persistent infection.
The structural defects in flaviviral DI RNAs, as described in this paper, partially resemble those previously detected in a poliovirus system in which naturally occurring deletions were always found to be in frame, which affected the synthesis of structural but not of nonstructural proteins (18). The incorporation of an out-of-frame mutation into the poliovirus DI RNA has been shown to abolish the replication of this RNA even in the presence of parental helper virus (14). The ability of the defective viral RNA to be replicated in the presence or absence of parental virus is restored when an in-frame mutation is introduced into the poliovirus genome, suggesting that a cis-acting protein or coupling between translation and replication may be required for poliovirus RNA replication (14). It is our further aim to study the replicon-like properties of the defective viral RNAs described in this paper and the relationship between translation and replication during the replication of flaviviruses.
ACKNOWLEDGMENTS
This work was supported by the Australian Research Council.
We thank the staff of the Centre of Molecular and Cellular Biology, University of Western Australia, for the technical assistance with the oligonucleotide synthesis and DNA sequencing. We are especially indebted to R. A. Hall for the monoclonal antibodies to the MVE virus proteins prM, E, and NS1. We also thank R. A. Hall, M. Poidinger, and A. Scalzo for critical comments on the manuscript.
REFERENCES
- 1.Barrett A D T, Dimmock N J. Variation in homotypic and heterotypic interference by defective interfering viruses derived from different strains of Semliki Forest virus and from Sindbis virus. J Gen Virol. 1984;65:1119–1122. doi: 10.1099/0022-1317-65-6-1119. [DOI] [PubMed] [Google Scholar]
- 2.Bartholomeusz A I, Wright P J. Synthesis of dengue virus RNA in vitro: initiation and the involvement of proteins NS3 and NS5. Arch Virol. 1993;128:111–121. doi: 10.1007/BF01309792. [DOI] [PubMed] [Google Scholar]
- 3.Brinton M A. Characterisation of West Nile virus persistent infections in genetically resistent and susceptible mouse cells. I. Generation of defective non-plaquing virus particles. Virology. 1982;116:84–98. doi: 10.1016/0042-6822(82)90405-6. [DOI] [PubMed] [Google Scholar]
- 4.Chambers T J, Weir R C, Grakoui A, McCourt D W, Bazan J F, Fletterick R J, Rice C M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci USA. 1990;87:8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers T J, Hahn C S, Galler R, Rice C M. Flavivirus genome organization, expression and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 6.Chen L K, Liao C L, Lin C G, Lai S C, Liu C I, Ma S H, Huang Y Y, Lin Y L. Persistence of Japanase encephalitis virus is associated with abnormal expression of the non-structural protein NS1 in host cell. Virology. 1996;217:220–229. doi: 10.1006/viro.1996.0109. [DOI] [PubMed] [Google Scholar]
- 7.Chu P W G, Westaway E G. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology. 1985;140:68–79. doi: 10.1016/0042-6822(85)90446-5. [DOI] [PubMed] [Google Scholar]
- 8.Chu P W G, Westaway E G. Characterization of Kunjin virus RNA-dependent RNA polymerase: reinitiation of synthesis in vitro. Virology. 1987;157:330–337. doi: 10.1016/0042-6822(87)90275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleaves G R, Ryan T E, Schlesinger R W. Identification and characterisation of type 2 dengue virus replicative intermediate and replicative form RNAs. Virology. 1981;111:73–83. doi: 10.1016/0042-6822(81)90654-1. [DOI] [PubMed] [Google Scholar]
- 10.Dalgarno L, Trent D W, Strauss J H, Rice C M. Partial nucleotide sequence of the Murray Valley encephalitis virus genome. J Mol Biol. 1986;187:309–323. doi: 10.1016/0022-2836(86)90435-3. [DOI] [PubMed] [Google Scholar]
- 11.Debnath N C, Tiernery R, Sil B K, Wills M R, Barrett A D T. In vitro homotypic interference by defective interfering particles of West Nile virus. J Gen Virol. 1991;72:2705–2711. doi: 10.1099/0022-1317-72-11-2705. [DOI] [PubMed] [Google Scholar]
- 12.Dimmock N J. Defective interfering viruses: modulators of infection. Microbiol Sci. 1985;2:1–7. [PubMed] [Google Scholar]
- 13.Fokina G I, Malenko G V, Levina L S, Koreshkova G V, Rzhakhova O E, Mamanenko L L, Pogodina V V, Frolova M P. Persistence of tick-borne encephalitis virus in monkeys. V. Virus localization after subcutaneous inoculations. Acta Virol. 1982;26:369–375. [PubMed] [Google Scholar]
- 14.Hagino-Yamagishi K, Nomoto A. In vitro construction of poliovirus defective interfering particles. J Virol. 1989;63:5386–5392. doi: 10.1128/jvi.63.12.5386-5392.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall R A, Kay B H, Burgess G W, Clancy P, Fanning I D. Epitope analysis of the envelope and non-structural glycoproteins of Murray Valley encephalitis virus. J Gen Virol. 1990;71:2923–2930. doi: 10.1099/0022-1317-71-12-2923. [DOI] [PubMed] [Google Scholar]
- 15a.Hall, R. A. Personal communication.
- 16.Holland J J. Defective viral genomes. In: Fields B N, Knipe D M, et al., editors. Virology. New York, N.Y: Raven Press, Ltd.; 1990. pp. 151–165. [Google Scholar]
- 17.Huang A S, Baltimore D. Defective viral particles and viral disease processes. Nature. 1970;226:325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- 18.Kajigaya S, Arakawa H, Kuge S, Koi T, Imura N, Nomoto A. Isolation and characterization of defective-interfering particles of poliovirus Sabin 1 strain. Virology. 1985;142:307–316. doi: 10.1016/0042-6822(85)90339-3. [DOI] [PubMed] [Google Scholar]
- 19.Liehne P F S, Anderson S, Stanley N F, Liehne C G, Wright A E, Chan K H, Leivers S, Britten D K, Hamilton N P. Isolation of Murray Valley encephalitis virus and other arboviruses in the Ord river valley 1972–1976. Aust J Exp Biol Med Sci. 1981;59:347–356. doi: 10.1038/icb.1981.29. [DOI] [PubMed] [Google Scholar]
- 20.Mackenzie J M, Jones M K, Young P R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 21.Monath T P C. Neutralizing antibody responses in the major immunoglobulin classes to yellow fever 17D vaccination of humans. Am J Epidemiol. 1971;93:123–129. doi: 10.1093/oxfordjournals.aje.a121232. [DOI] [PubMed] [Google Scholar]
- 22.Muylaert I R, Galler R, Rice C M. Genetic analysis of the yellow fever virus NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J Virol. 1997;71:291–298. doi: 10.1128/jvi.71.1.291-298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oldstone M B A. Viral persistence. Cell. 1989;56:517–520. doi: 10.1016/0092-8674(89)90573-4. [DOI] [PubMed] [Google Scholar]
- 24.Poidinger M, Coelen R J, Mackenzie J S. Persistent infection of Vero cells by the flavivirus Murray Valley encephalitis virus. J Gen Virol. 1991;72:573–578. doi: 10.1099/0022-1317-72-3-573. [DOI] [PubMed] [Google Scholar]
- 25.Rosenbaum M J, Sullivan E J, Edwards E A. Techniques for cell cultivation in plastic microtitration plates and their application in biological assays. In: Wasky G O, editor. Animal tissue culture advances in techniques. London, United Kingdom: Butterworths; 1972. pp. 49–81. [Google Scholar]
- 26.Schmaljohn C, Blair C P. Persistent infection of cultured mammalian cells by Japanese encephalitis virus. J Virol. 1977;24:580–589. doi: 10.1128/jvi.24.2.580-589.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slavin H B. Persistence of the virus of St. Louis encephalitis in the central nervous system of mice for over five months. J Bacteriol. 1943;46:113–116. doi: 10.1128/jb.46.2.113-116.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urosevic N, van Maanen M, Mansfield J P, Mackenzie J S, Shellam G S. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and -resistant mice with Murray Valley encephalitis virus. J Gen Virol. 1997;78:23–29. doi: 10.1099/0022-1317-78-1-23. [DOI] [PubMed] [Google Scholar]
- 29.von Magnus Propagation of the PR8 strain of influenza virus in chick embryos. III. Properties of the incomplete virus produced in serial passages of undiluted virus. Acta Pathol Microbiol Scand. 1951;29:157–181. doi: 10.1111/j.1699-0463.1951.tb00114.x. [DOI] [PubMed] [Google Scholar]
- 30.Westaway E G, Mackenzie J M, Kenney M T, Jones M K, Khromykh A A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]