Abstract
Purpose
Hypertoxigenic Streptococcus pyogenes emm1 lineage M1UK has recently been associated with upsurges of invasive infections and scarlet fever in several countries, but whole-genome sequencing surveillance data of lineages circulating in Germany is lacking. In this study, we investigated recent iGAS isolates from our laboratory at a German tertiary care center for the presence of the M1UK lineage.
Methods
Whole-genome sequencing was employed to characterize a collection of 47 consecutive non-copy isolates recovered from blood cultures (21) and tissue samples (26) in our laboratory between October 2022 and April 2023.
Results
M protein gene (emm) typing distinguished 14 different emm types, with emm1 (17) being the dominant type. Single-nucleotide polymorphism (SNP) analysis confirmed the presence of all 27 SNPs characteristic for the M1UK lineage in 14 of 17 emm1 isolates.
Conclusion
This study has shown for the first time that M1UK is present in Germany and might constitute a driving force in the observed surge of GAS infections. This observation mirrors developments in the UK and other countries and underscores the importance of WGS surveillance to understand the epidemiology of GAS.
Supplementary Information
The online version contains supplementary material available at 10.1007/s15010-023-02137-1.
Keywords: Streptococcus pyogenes, Epidemiology, Emm type, M1UK
Introduction
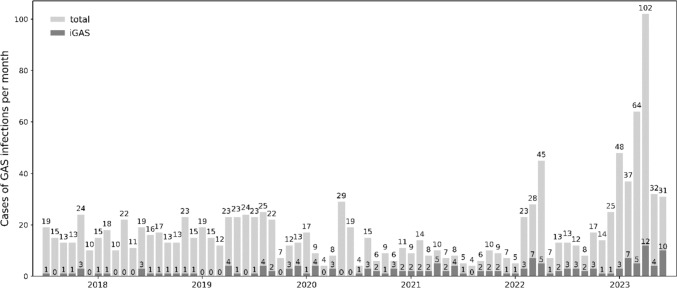
Streptococcus pyogenes, also referred to as Group A Streptococcus (GAS), is an important human pathogen that causes non-invasive infections such as scarlet fever, pharyngitis and impetigo, and also life-threatening invasive infections (iGAS) such as necrotising fasciitis, pneumonia, meningitis and puerperal sepsis [1]. In 2022 and 2023, several European countries (including Denmark, Ireland, France, the Netherlands, Sweden, Spain, and the UK) have reported a marked increase in scarlet fever and iGAS [2–6]. The observed increase followed a period of low incidence during the COVID-19 pandemic, but has now exceeded pre-pandemic levels [5, 6]. This phenomenon is likely attributable to reduced exposure at the population level and an associated so-called immunity gap [7], which may have led to widespread dissemination in the population after the lifting of COVID-19-related restrictions. In addition, the current high activity of viral respiratory infections might have contributed to an increase in iGAS cases with a respiratory focus [6, 8]. On the other hand, an increase of scarlet fever and iGAS had been observed in the UK some years before the pandemic and was associated with the emergence and spread of a new lineage of S. pyogenes designated M1UK [9]. The M1UK lineage is a variant of the highly successful, contemporary epidemic M1global strain [10]. M1UK is differentiated from M1global by 27 chromosomal single nucleotide polymorphisms (SNPs) and exhibits enhanced expression of the superantigenic scarlet fever toxin SpeA (streptococcal pyrogenic exotoxin A) in vitro [9, 11]. The M1UK lineage has subsequently been identified in several other countries (Australia, Belgium, Canada, Netherlands, Portugal, Scotland, USA) [3, 8, 11–15], where in some cases (Australia, Belgium, Netherlands, Portugal) it has also expanded and displaced the M1global lineage [11, 12, 16, 17]. Recently, the emergence and spread of two more new clones designated M1DK (emm1) and M4NL22 (emm4) were reported in Denmark and the Netherlands, respectively [4, 18]. In Germany, the Robert Koch Institute (RKI) also reported a strong increase in iGAS infections for the fourth quarter of 2022 [19]. RKI surveillance data also show a marked increase in the number of reported scarlet fever cases from two federated German states with mandatory scarlet fever reporting (https://survstat.rki.de/; accessed 2023/08/25). This trend was reflected in the number of GAS/iGAS isolates recovered from clinical samples at our laboratory (Fig. 1). Currently, whole-genome surveillance data of circulating S. pyogenes strains from Germany is not yet available. To explore the GAS population and to elucidate whether any of the epidemic clones recently described in neighbouring countries might have contributed to the reported increase in iGAS infections, we analysed a collection of 47 S. pyogenes isolates recovered from blood and tissue samples from October 2022 to April 2023 by whole-genome sequencing.
Fig. 1.
Monthly cases of GAS infections identified by our laboratory between 7/2017 and 6/2023. Total number of cases and cases of invasive infections (iGAS) are represented by light and dark grey bars, respectively. Numbers above the light grey bars indicate the number of all GAS detections in the respective period, and numbers above the dark grey bars indicate the number of iGAS
Results and discussion
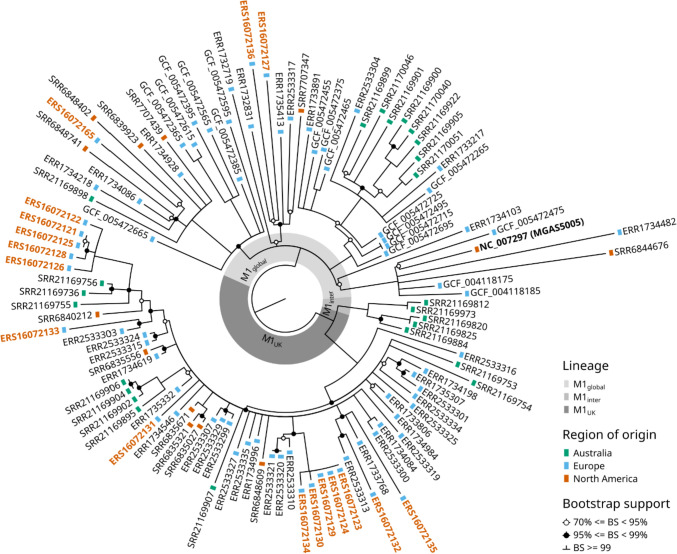
The S. pyogenes M1UK lineage has emerged and rapidly spread in several countries worldwide. Owing to the lack of nationwide whole-genome surveillance data for S. pyogenes, information on the presence of the M1UK lineage in Germany is not yet available. This prompted us to characterize the population structure of contemporary S. pyogenes isolated during routine diagnostic workup of blood cultures and tissue samples at the microbiology laboratory of the University Medical Center Hamburg-Eppendorf, a 1600-bed tertiary care center. Between October 2022 and April 2023, a total of 53 non-copy S. pyogenes isolates were recovered from eligible specimens. Of those, 47 (21 blood culture isolates and 26 isolates from tissue specimens) were available for whole-genome sequencing and subsequent delineation of the recently described new M1UK, M1DK and M4NL22 lineages (supplemental file 1). M protein gene (emm) typing distinguished 14 different emm types, with emm1 [17] being the dominant type, followed by emm89 [7] (Table 1). Single-nucleotide polymorphism (SNP) analysis confirmed the presence of all 27 SNPs characteristic for the M1UK lineage [9] in 14 of 17 emm1 isolates. The remaining three emm1 isolates lacked any of the M1UK-defining SNPs. SNP-based phylogenetic analysis grouped our M1UK isolates together with representative M1UK isolates recovered from iGAS in the UK, Australia, Canada and the USA and separated them from the M1global and M1inter lineages. M1inter lineages carry subsets of the SNPs that define M1UK, but failed to spread as successful as M1UK [9, 11] (Fig. 2). Analysis of virulence and resistance gene content of our 14 M1UK isolates revealed no striking differences as compared to other M1UK or local M1global strains (supplemental file 1) [9]. The emm1 clone M1DK, reported to be highly prevalent in Denmark [4], was not found, and only one isolate of emm4, a prevalent genotype in invasive infections in the Netherlands [18], was identified in our collection.
Table 1.
Distribution of emm types
| emm type | Number [n] |
|---|---|
| 1 | 17 (14 M1UK) |
| 12 | 6 |
| 89 | 6 |
| 27 | 3 |
| 49 | 3 |
| 53 | 2 |
| 66 | 2 |
| 87 | 2 |
| 4 | 1 |
| 11 | 1 |
| 77 | 1 |
| 102 | 1 |
| 106 | 1 |
| 218 | 1 |
| Total | 47 |
Fig. 2.
Maximum likelihood phylogenetic tree from core SNPs (excluding prophage regions). Isolates are labelled with accession numbers; isolates sequenced for the present study are shown in orange boldface print. M1 lineage is indicated by shading of the inner doughnut plot. Region of strain origin is indicated by coloured rectangles. Nodes with bootstrap support values lower than 70% have been collapsed, and bootstrap support values between 70 and 99% are marked with circles
In conclusion, this study has shown for the first time that M1UK is present in Germany and might constitute a driving force in the observed surge of GAS infections. We concede that our study is only a snapshot of a regional S. pyogenes population, which may not be representative of the German S. pyogenes population. In addition, due to the lack of patient travel information, acquisition of M1UK GAS outside of Germany cannot be excluded in all cases. Our study population also did not encompass commensal isolates or isolates from cases of uncomplicated pharyngitis and might thus not reflect the overall composition of our local S. pyogenes population. Nevertheless, our preliminary data underscore the need for further studies of larger strain collections to reconstruct the spread of M1UK in Germany and elucidate its role in the current surge of GAS infections [19].
Methods
S. pyogenes study isolates (n = 47) were thawed from a − 80 °C cryo-collection of contemporary isolates and underwent WGS. In brief, DNA was extracted using QiaSymphony mericon extraction kits (Qiagen) on a QiaSymphony SP instrument according to the manufacturer’s instructions. Libraries for paired end sequencing were prepared using the NEB NextUltra II DNA library Prep Kit for Illumina (NEB) and sequenced with 2 × 150 cycles on an Illumina MiSeq instrument. Reads or nucleotide sequences from additional isolates were obtained from the National Center for Biotechnology Information (NCBI) sequence read archive, the NCBI reference sequence database and the European Nucleotide Archive (ENA). Reads were assembled with shovill 1.1 employing spades 3.15.5 [20], annotated with bakta 1.8.1 [21] and subjected to pan genome analysis with roary 3.13.0 [21]. Emm types were assigned from bakta assemblies using the Centers for Disease Control Streptococcus Laboratory GAS bioinformatic pipeline (https://github.com/BenJamesMetcalf/GAS_Scripts_Reference). Resistance genes where detected using AMRFinderPlus 3.11.14 with the NCBI reference gene database version 2023–08-08.2 [22]. Known virulence markers were identified with abricate 1.0.1 (https://github.com/tseemann/abricate) and the virulence factor database version 2022–08-26 [23]. Additional allelic profiling was performed with chewBBACA 3.3.0 [24] and a S. pyogenes wgMLST schema from Chewie-NS [25]. Core SNPs were identified with snippy 4.6.0 (https://github.com/tseemann/snippy) using S. pyogenes MGAS5005 (GenBank accession NC_007297.2) as a reference. Maximum likelihood phylogenies from concatenated core SNPs were constructed using gubbins 3.3.0 [26] with IQTree [27] and visualized with TreeViewer 2.1.0 (https://github.com/arklumpus/TreeViewer/tree/v2.1.0).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank the technical staff of the Institute of Medical Microbiology, Virology and Hygiene for their excellent support.
Author contributions
M.W. Designed the study, analyzed data, wrote the manuscript. B.B. Performed experiments, analyzed data, edited the manuscript. N.D-B. Performed experiments, analyzed data, edited the manuscript. A.H. Performed experiments, prepared figure 1, edited the manuscript R.B. Performed experiments, edited the manuscript M.A. Provided ressources, edited the manuscript. M.C. Performed experiments, analyzed data, prepared figures 1 and 2, wrote the manuscript. H.R. Designed the study, analyzed data, wrote the manuscript
Funding
Open Access funding enabled and organized by Projekt DEAL. No funds, grants or other support was received.
Data availability
Whole genome sequences generated for this study are available in the European Nucleotide Archive at https://www.ebi.ac.uk/ena/browser/home and can be accessed with the project number PRJEB64404. Isolate metadata are available in the online supplementary material.
Declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
References
- 1.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. Disease manifestations and pathogenic mechanisms of group A streptococcus. Clin Microbiol Rev. 2014;27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO . Increase in invasive group A streptococcal infections among children in Europe, including fatalities. WHO; 2022. [Google Scholar]
- 3.van der Putten BCL, Vlaminckx BJM, de Gier B, Freudenburg-de Graaf W, van Sorge NM. Group A streptococcal meningitis with the M1UK variant in the Netherlands. JAMA. 2023;329:1791–1792. doi: 10.1001/jama.2023.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johannesen TB, Munkstrup C, Edslev SM, Baig S, Nielsen S, Funk T, Kristensen DK, Jacobsen LH, Ravn SF, Bindslev N, Gubbels S, Voldstedlund M, Jokelainen P, Hallstrom S, Rasmussen A, Kristinsson KG, Fuglsang-Damgaard D, Dessau RB, Olsen AB, Jensen CS, Skovby A, Ellermann-Eriksen S, Jensen TG, Dzajic E, Ostergaard C, Lomborg Andersen S, Hoffmann S, Andersen PH, Stegger M. Increase in invasive group A streptococcal infections and emergence of novel, rapidly expanding sub-lineage of the virulent Streptococcus pyogenes M1 clone, Denmark, 2023. Euro Surveill. 2023 doi: 10.2807/1560-7917.ES.2023.28.26.2300291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cobo-Vazquez E, Aguilera-Alonso D, Carrasco-Colom J, Calvo C, Saavedra-Lozano J, Ped GASNWG. Increasing incidence and severity of invasive group A streptococcal disease in Spanish children in 2019–2022. Lancet Reg Health Europe. 2023;27:100597. doi: 10.1016/j.lanepe.2023.100597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lassoued Y, Assad Z, Ouldali N, Caseris M, Mariani P, Birgy A, Bonacorsi S, Bidet P, Faye A. Unexpected increase in invasive group A streptococcal infections in children after respiratory viruses outbreak in france: a 15-year time-series analysis. Open Forum Infect Dis. 2023;10:188. doi: 10.1093/ofid/ofad188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messacar K, Baker RE, Park SW, Nguyen-Tran H, Cataldi JR, Grenfell B. Preparing for uncertainty: endemic paediatric viral illnesses after COVID-19 pandemic disruption. Lancet. 2022;400:1663–1665. doi: 10.1016/S0140-6736(22)01277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies PJB, Russell CD, Morgan AR, Taori SK, Lindsay D, Ure R, Brown D, Smith A. Increase of severe pulmonary infections in adults caused by M1(UK) Streptococcus pyogenes, Central Scotland UK. Emerg Infect Dis. 2023;29:1638. doi: 10.3201/eid2908.230569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynskey NN, Jauneikaite E, Li HK, Zhi X, Turner CE, Mosavie M, Pearson M, Asai M, Lobkowicz L, Chow JY, Parkhill J, Lamagni T, Chalker VJ, Sriskandan S. Emergence of dominant toxigenic M1T1 Streptococcus pyogenes clone during increased scarlet fever activity in England: a population-based molecular epidemiological study. Lancet Infect Dis. 2019;19:1209–1218. doi: 10.1016/S1473-3099(19)30446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, Kristinsson KG, Gottfredsson M, Vuopio J, Raisanen K, Caugant DA, Steinbakk M, Low DE, McGeer A, Darenberg J, Henriques-Normark B, Van Beneden CA, Hoffmann S, Musser JM. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci USA. 2014;111:E1768–1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies MR, Keller N, Brouwer S, Jespersen MG, Cork AJ, Hayes AJ, Pitt ME, De Oliveira DMP, Harbison-Price N, Bertolla OM, Mediati DG, Curren BF, Taiaroa G, Lacey JA, Smith HV, Fang NX, Coin LJM, Stevens K, Tong SYC, Sanderson-Smith M, Tree JJ, Irwin AD, Grimwood K, Howden BP, Jennison AV, Walker MJ. Detection of Streptococcus pyogenes M1(UK) in Australia and characterization of the mutation driving enhanced expression of superantigen SpeA. Nat Commun. 2023;14:1051. doi: 10.1038/s41467-023-36717-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rumke LW, de Gier B, Vestjens SMT, van der Ende A, van Sorge NM, Vlaminckx BJM, Witteveen S, van Santen M, Schouls LM, Kuijper EJ. Dominance of M1(UK) clade among Dutch M1 Streptococcus pyogenes. Lancet Infect Dis. 2020;20:539–540. doi: 10.1016/S1473-3099(20)30278-4. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Nanduri SA, Van Beneden CA, Beall BW. M1(UK) lineage in invasive group A streptococcus isolates from the USA. Lancet Infect Dis. 2020;20:538–539. doi: 10.1016/S1473-3099(20)30279-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demczuk W, Martin I, Domingo FR, MacDonald D, Mulvey MR. Identification of Streptococcus pyogenes M1(UK) clone in Canada. Lancet Infect Dis. 2019;19:1284–1285. doi: 10.1016/S1473-3099(19)30622-X. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Rivers J, Mathis S, Li Z, Chochua S, Metcalf BJ, Beall B, Onukwube J, Gregory CJ, McGee L. Expansion of invasive group A streptococcus M1(UK) lineage in active bacterial core surveillance, United States, 2019–2021. Emerg Infect Dis. 2023;29:2116–2120. doi: 10.3201/eid2910.230675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-Ruiz JP, Lin Q, Lammens C, Smeesters PR, van Kleef-van Koeveringe S, Matheeussen V, Malhotra-Kumar S. Increase in bloodstream infections caused by emm1 group A streptococcus correlates with emergence of toxigenic M1(UK), Belgium, May 2022 to August 2023. Euro Surveill. 2023;28:2300422. doi: 10.2807/1560-7917.ES.2023.28.36.2300422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouveia C, Bajanca-Lavado MP, Mamede R, Araujo Carvalho A, Rodrigues F, Melo-Cristino J, Ramirez M, Friaes A, Portuguese Study Group of Pediatric Invasive Streptococcal, D., and Portuguese Study Group of Paediatric Invasive Streptococcal, D Sustained increase of paediatric invasive Streptococcus pyogenes infections dominated by M1(UK) and diverse emm12 isolates, Portugal, September 2022 to May 2023. Euro Surveill. 2023;28:2300427. doi: 10.2807/1560-7917.ES.2023.28.36.2300427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Putten BCL, Bril-Keijzers WCM, Rumke LW, Vestjens SMT, Koster LAM, Willemsen M, van Houten MA, Rots NY, Vlaminckx BJM, de Gier B, van Sorge NM. Novel emm4 lineage associated with an upsurge in invasive group A streptococcal disease in the Netherlands, 2022. Microb Genom. 2023 doi: 10.1099/mgen.0.001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.RKI. Epidemiologisches Bulletin. 2023.
- 20.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes De Novo assembler. Curr Protoc Bioinform. 2020;70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 21.Schwengers O, Jelonek L, Dieckmann MA, Beyvers S, Blom J, Goesmann A. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb Genom. 2021 doi: 10.1099/mgen.0.000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, Hoffmann M, Pettengill JB, Prasad AB, Tillman GE, Tyson GH, Klimke W. AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep. 2021;11:12728. doi: 10.1038/s41598-021-91456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 2016;44:D694–697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva M, Machado MP, Silva DN, Rossi M, Moran-Gilad J, Santos S, Ramirez M, Carrico JA. chewBBACA: a complete suite for gene-by-gene schema creation and strain identification. Microb Genom. 2018 doi: 10.1099/mgen.0.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friaes A, Mamede R, Ferreira M, Melo-Cristino J, Ramirez M. Annotated whole-genome multilocus sequence typing schema for scalable high-resolution typing of Streptococcus pyogenes. J Clin Microbiol. 2022;60:e0031522. doi: 10.1128/jcm.00315-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole genome sequences generated for this study are available in the European Nucleotide Archive at https://www.ebi.ac.uk/ena/browser/home and can be accessed with the project number PRJEB64404. Isolate metadata are available in the online supplementary material.