Inulin fiber diet induces microbial dysbiosis, production of bile acids, and inflammatory ILC2s responses, triggering IL-5 production, eosinophilia, and intestinal inflammation. These studies highlight that diet and microbiota-derived metabolites are significant determinants of the balance between inflammatory versus tissue-protective ILC2s with implications for chronic inflammation.
Abstract
Innate lymphoid cells (ILCs) can promote host defense, chronic inflammation, or tissue protection and are regulated by cytokines and neuropeptides. However, their regulation by diet and microbiota-derived signals remains unclear. We show that an inulin fiber diet promotes Tph1-expressing inflammatory ILC2s (ILC2INFLAM) in the colon, which produce IL-5 but not tissue-protective amphiregulin (AREG), resulting in the accumulation of eosinophils. This exacerbates inflammation in a murine model of intestinal damage and inflammation in an ILC2- and eosinophil-dependent manner. Mechanistically, the inulin fiber diet elevated microbiota-derived bile acids, including cholic acid (CA) that induced expression of ILC2-activating IL-33. In IBD patients, bile acids, their receptor farnesoid X receptor (FXR), IL-33, and eosinophils were all upregulated compared with controls, implicating this diet–microbiota–ILC2 axis in human IBD pathogenesis. Together, these data reveal that dietary fiber–induced changes in microbial metabolites operate as a rheostat that governs protective versus pathologic ILC2 responses with relevance to precision nutrition for inflammatory diseases.
Introduction
Mammalian barrier immunity is influenced by a variety of environmental factors including diet and signals derived from the microbiota (Alexander and Turnbaugh, 2020; Blander et al., 2017; Lee and Dixit, 2020; Nobs et al., 2020; Rooks and Garrett, 2016). Diet-regulated metabolites of host or microbial origin can be sensed by diverse cell types including epithelial, stromal, neuronal, and immune cells, all of which contribute to a balanced state of immunity, inflammation, and tissue protection (Blander et al., 2017; McCarville et al., 2020; Rooks and Garrett, 2016; Zheng et al., 2020). Innate lymphoid cells are enriched at barrier surfaces, including intestine, lung, and skin, and can regulate barrier function, immune cell homeostasis, and tissue protection (Ebbo et al., 2017; Eberl et al., 2015; Klose and Artis, 2016, 2020; Ricardo-Gonzalez et al., 2022; Vivier et al., 2018). In this context, group 2 innate lymphoid cells (ILC2s) can exert proinflammatory or tissue-protective effects in a tissue- and context-dependent manner. For example, a subset of proinflammatory ILC2s (ILC2INFLAM) marked by elevated expression of Tph1 produces type 2 inflammatory cytokines IL-5 and IL-13. These cytokines promote recruitment of eosinophils which protect against helminth infection and promote allergic diseases (Flamar et al., 2020; Huang et al., 2015; Kim et al., 2013). In contrast, some ILC2s express the growth factor amphiregulin (AREG) that contributes to tissue protection and repair (Monticelli et al., 2011, 2012, 2015). ILC2s can be activated by a variety of cytokines and neuropeptides including IL-25, IL-33, thymic stromal lymphopoietin, neuromedin U, vasoactive intestinal peptide, and calcitonin gene–related peptide (CGRP) (Halim et al., 2012; Klose et al., 2017; Nagashima et al., 2019; Neill et al., 2010; Nussbaum et al., 2013; Saenz et al., 2010). However, the pathways that balance proinflammatory versus tissue-protective ILC2 responses as well as the impact of diet, the microbiota, and their metabolites on their functional potential remain poorly understood.
In this study, we employed an inulin fiber dietary intervention to perturb intestinal microbiota and interrogate their potential roles in the activation and functional potential of proinflammatory versus tissue-protective ILC2s. We observed that fiber diet–induced dysregulation of the microbiota and their associated metabolites elicits activation of a distinct population of ILC2INFLAM in the murine colon. While ILC2s have been shown to be tissue-protective during intestinal damage and inflammation in some contexts, the inulin fiber diet–activated ILC2INFLAM worsens the clinical outcomes via eosinophilic inflammation in multiple murine models of intestinal damage and inflammation. Collectively, these data highlight that environmental triggers including diet, microbiota, and their metabolites are significant environmental determinants of the balance between homeostatic versus pathologic functions of ILC2s with implications for multiple inflammatory diseases at barrier surfaces.
Results
Inulin fiber diet activates ILC2INFLAM and exacerbates intestinal damage
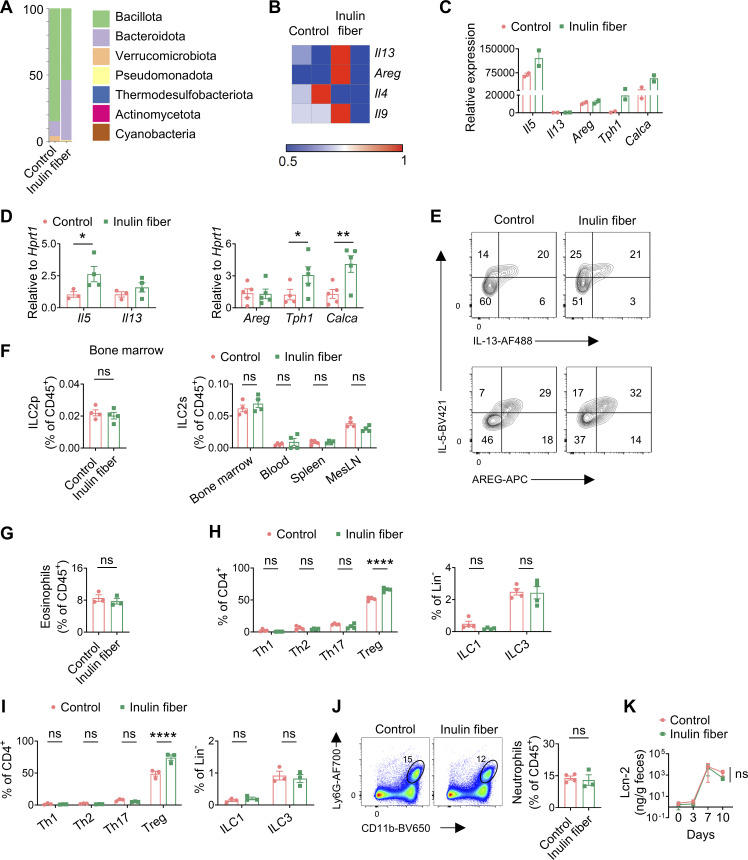
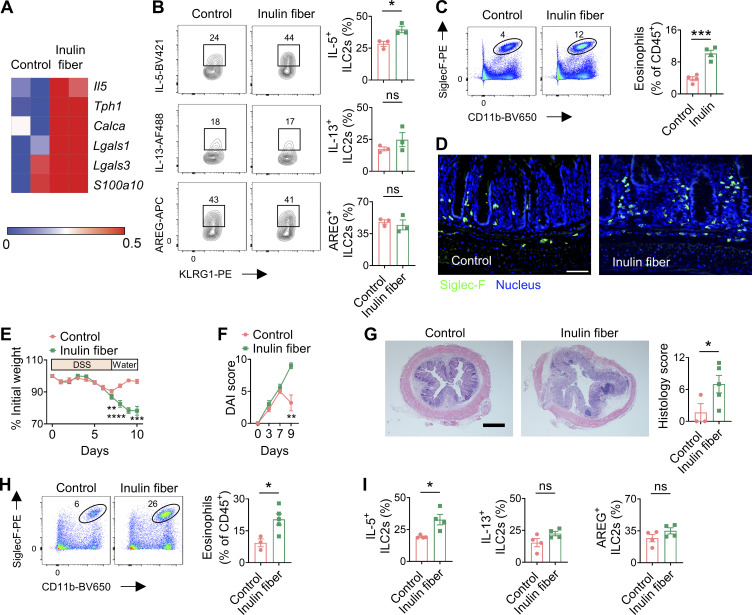
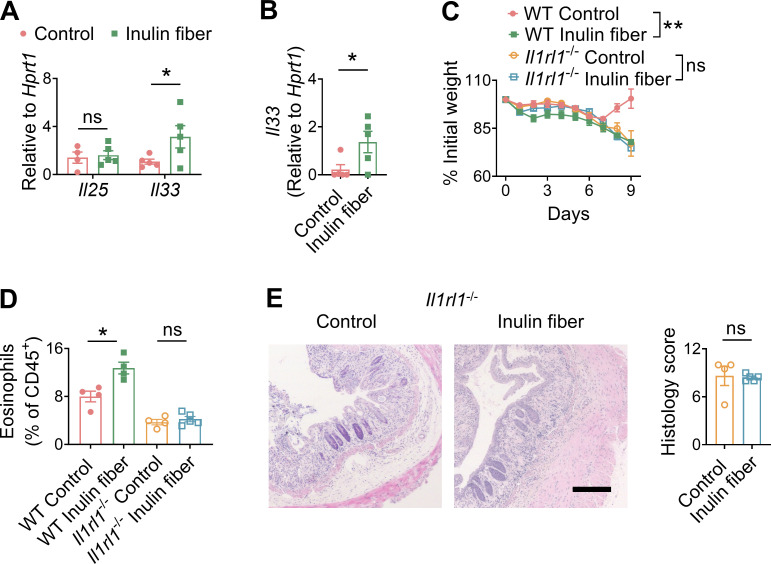
To study the influence of microbiota on the functional potential of ILC2s, we employed an inulin fiber diet that induces significant shifts in the composition of the microbiota including an increase in Bacteroidota (previously known as Bacteroidetes) and a decrease in Bacillota (previously known as Firmicutes) (Arifuzzaman et al., 2022; Chambers et al., 2019; Chijiiwa et al., 2020) (Fig. S1 A). Wild-type (WT) mice were exposed to a control diet or an inulin fiber diet for 2 wk, and flow cytometry–sorted colonic ILC2s were analyzed by RNA sequencing (RNA-seq). We observed that inulin fiber diet significantly altered the transcriptome of colonic ILC2s (Fig. 1 A). Inulin fiber diet significantly upregulated inflammatory Il5 but not Il13 or tissue-protective Areg (Fig. 1 A and Fig. S1, B–D), indicating a unique inflammatory signature of this ILC2 population. These colonic ILC2INFLAM expressed high levels of Calca (Fig. 1 A; and Fig. S1, C and D) that encodes CGRP, which is known to promote IL-5 production in ILC2s (Nagashima et al., 2019; Xu et al., 2019). The inulin fiber diet–induced colonic ILC2INFLAM also expressed high levels of Tph1 (Fig. 1 A and Fig. S1, C and D) which encodes serotonin-producing enzyme tryptophan hydroxylase 1 and has previously been associated with small intestinal ILC2INFLAM cells (Flamar et al., 2020). Further, we observed upregulation of several additional inflammatory genes in the inulin fiber diet group, which were previously described in ILC2INFLAM in lung tissue including Lgals1, Lgals3, and S100a10 (Nagashima et al., 2019) (Fig. 1 A). At the protein level, IL-5+ ILC2s also produce IL-13 and AREG; however, the inulin fiber diet specifically promoted the production of IL-5 but not IL-13 or AREG (Fig. 1 B and Fig. S1 E). The frequencies of ILC2s in the lymphoid tissues or their progenitors (ILC2p) in the bone marrow were unaffected by the inulin fiber diet (Fig. S1 F), suggesting that the generation of colonic ILC2INFLAM is due to an alteration of ILC2 phenotype within the tissue rather than an increase of ILC2 population. ILC2-secreted IL-5 recruits eosinophils (Klose and Artis, 2020; Klose et al., 2017; Nussbaum et al., 2013), and consistent with this finding, we also observed a significant increase in the number of eosinophils in the colon of inulin fiber diet–fed mice compared with control diet–fed mice (Fig. 1, C and D). The eosinophilia was reversed upon withdrawal from the inulin fiber diet, suggesting that a continuous supply of inulin is required to maintain the elevated eosinophil level (Fig. S1 G). Together, these data suggest that the inulin fiber diet induces ILC2INFLAM with distinct inflammatory features and associated colonic eosinophilia.
Figure S1.
Microbial and immunological parameters in mice upon exposure to inulin fiber diet and DSS. (A) Representative taxonomic classification of 16S rRNA genes in fecal suspension from mice fed with control or inulin fiber diet for 2 wk. (B) Heatmap showing gene expression of colonic ILC2s in mice fed the control or inulin fiber diet (n = 2 mice). (C) Expression levels of various genes determined by RNA-seq of sorted ILC2s (n = 2 mice). (D) Expression levels of various genes determined by RT-qPCR of sorted ILC2s (n = 3–5 mice). (E) Representative flow cytometry plots showing co-expression of IL-5 and IL-13 (top panels) and IL-5 and AREG (bottom panels) in the colonic ILC2s. (F) Frequency of ILC2 progenitor cells (ILC2p) in the bone marrow and ILC2s in various tissues of mice fed with control or inulin fiber diet for 2 wk (n = 4 mice). (G) Frequency of colonic eosinophils in mice fed with control or inulin fiber diet for 2 wk and then regular chow for 4 wk (n = 3 mice). (H) Frequency of various T cells and ILCs in the colons of mice fed with control or inulin fiber diet for 2 wk (n = 4 mice). (I) Frequency of various T cells and ILCs in the colons of mice at the endpoint after DSS treatment (n = 3 mice). (J) Representative flow cytometry plots and frequency of CD11b+Ly6G+ neutrophils in colons of mice treated with DSS (n = 3–4 mice). (K) Levels of fecal lipocalin-2 (Lcn-2) in DSS-treated mice measured by ELISA (n = 5 mice). Data are representative of two independent experiments (A and D–K). Data are means ± SEM. Statistics were calculated by unpaired two-tailed t test (F, G, J, and K) or two-way ANOVA with uncorrected Fisher’s LSD test (D, H, and I). *P < 0.05, **P < 0.01, ****P < 0.0001, ns, not significant.
Figure 1.
Inulin fiber diet activates ILC2INFLAM and exacerbates intestinal damage and inflammation. (A) Heatmap showing differential gene expression of colonic ILC2s in mice fed the control or inulin fiber diet for 2 wk (n = 2 mice). (B) Frequency of IL-5–expressing, IL-13–expressing, or AREG-expressing KLRG1+ ILC2s in colonic lamina propria, determined by intracellular cytokine staining (n = 3 mice). Flow cytometry plots were gated from CD45+Lin−CD90.2+CD127+ cells. (C) Representative flow cytometry plots and frequency of CD11b+SiglecF+ eosinophils in the CD45+ population (n = 4 mice). (D) Representative immunofluorescence staining showing Siglec-F+ eosinophils in the colons. Scale bar = 50 μm. (E and F) Disease and recovery of DSS-treated control diet– and inulin fiber diet–fed mice were monitored by daily weight loss (E) and DAI (F). Mice were fed control or inulin fiber diet from 2 wk prior to the DSS challenge to the endpoint of the experiment (n = 5 mice). (G) Representative H&E staining of distal colons at the endpoint of the DSS experiment. Scale bar = 500 μm. Graph shows histological scores (n = 3–5 mice). (H) Representative flow cytometry plots and frequency of CD11b+SiglecF+ eosinophils in the CD45+ population (n = 3–5 mice). (I) Frequency of IL-5–expressing, IL-13–expressing, or AREG-expressing KLRG1+ ILC2s in colonic lamina propria, determined by intracellular cytokine staining (n = 4 mice). Data are representative of three (C and E–G) or two (B, D, H, and I) independent experiments. Data are means ± SEM. Statistics were calculated by unpaired two-tailed t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant.
Dietary fiber promotes a group of microbiota-derived metabolites called short-chain fatty acids (SCFA) that have been shown to exert anti-inflammatory effects by promoting regulatory T cells (Tregs). Indeed, we observed that in addition to eosinophils, Tregs were also increased in the colon following exposure to an inulin fiber diet (Fig. S1 H). Therefore, to investigate the impact of inulin fiber diet–induced type 2 inflammation and eosinophil accumulation on intestinal inflammation and tissue repair, we next employed a chemically induced murine model of acute intestinal damage and inflammation (Monticelli et al., 2015; Tsou et al., 2022; Zhang et al., 2022). WT mice were exposed to a control diet or an inulin fiber diet for 2 wk and then exposed to dextran sulfate sodium (DSS) for 7 days (Fig. 1 E). While mice fed with a control diet began to recover after withdrawal of DSS on day 7, mice exposed to an inulin fiber diet continued to lose weight (Fig. 1 E) and exhibited significantly elevated clinical disease activity index (DAI), including general morbidity, rectal bleeding, and diarrhea (Fig. 1 F). Increased immune cell infiltration, intestinal edema, and loss of epithelial crypt structure were also observed in the colons isolated from inulin fiber diet–fed mice (Fig. 1 G), indicating that dietary inulin fiber increases inflammation and impairs tissue repair processes following colonic injury despite elevated Tregs (Fig. S1 I). Notably, the dominant immune cell populations associated with type 1/type 3 inflammatory signatures of intestinal inflammation, including neutrophil influx in the colon or fecal lipocalin levels, were not significantly different between control or inulin fiber diet–fed groups (Fig. S1, I–K). Conversely, the colons of inulin fiber diet–fed mice displayed a significant increase in eosinophils compared with mice fed control diet (Fig. 1 H). Furthermore, IL-5+ ILC2s remained elevated in the inulin fiber diet–fed mice compared with the control diet–fed mice after the DSS challenge (Fig. 1 I). Together, these data indicate that the inulin fiber diet–mediated exacerbation of disease severity in the chemically induced acute model of intestinal damage and inflammation could be driven by an ILC2INFLAM-triggered and eosinophil-mediated pathologic type 2 inflammatory response.
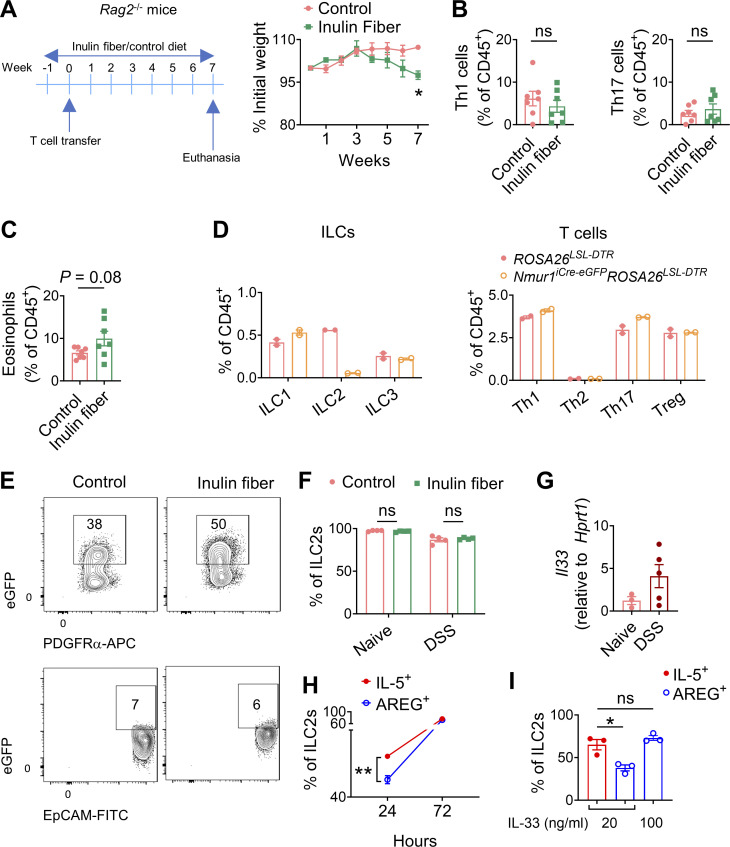
To determine the role of the inulin fiber diet in chronic intestinal inflammation, we employed a T cell transfer model. Rag2−/− mice were fed a control or inulin fiber diet for a week and then administered with CD4+CD45RBhiCD25lo naive T cells. We observed that weight loss in the inulin fiber group was significantly higher than in the control diet group (Fig. S2 A). While levels of Th1 and Th17 cells, the major drivers of intestinal inflammation in this model (Leppkes et al., 2009), were comparable between control and inulin fiber diet groups (Fig. S2 B), increased accumulation of eosinophils was observed in the colons of the inulin fiber group compared with control (Fig. S2 C). These data suggest that eosinophils could be the driver of inulin fiber diet–induced weight loss during chronic intestinal inflammation.
Figure S2.
Immune profiling related to T cell transfer colitis, ILC2 depletion and IL-33 treatment. (A) Experimental schematic and body weights in a T cell transfer model of colitis (n = 3–4 mice). (B and C) Frequency of T-bet+ Th1 and RORγt+ Th17 cells (B) and eosinophils (C) in the colon in a T cell transfer model of colitis (n = 7). (D) Colonic levels of various immune cells in DT-treated mice (n = 2). (E) Representative flow cytometry plots showing IL33-eGFP expression in epithelial (EpCAM+) and stromal (PDGFRα+) cell subsets in the colons of mice fed with control or inulin fiber diet for 2 wk. (F) Frequency of ST2+ ILC2s in the colons of mice after 2 wk of diets (naïve) or after 2 wk of diets followed by DSS treatment and recovery (DSS) (n = 4 mice). (G) Expression of Il33 in the colons of naïve or DSS-treated mice on a control diet (n = 3–5 mice). (H and I) ILC2s sorted from mouse colons were plated at a density of 5,000 cells per well in a 96-well round-bottom plate in complete medium with 100 ng/ml IL-2, 100 ng/ml IL-7, and various concentrations of IL-33. Graphs show frequencies of IL-5+ ILC2s and AREG+ ILC2s at various time points after incubation with 20 ng/ml IL-33 (H) and at 72 h after incubation with various concentrations of IL-33 (I). Data are representative of A and D–I or pooled from B and C two independent experiments. Data are means ± SEM. Statistics were calculated by unpaired two-tailed t test. *P < 0.05, **P < 0.01, ns, not significant.
ILC2s are required for inulin fiber diet–induced colonic eosinophilia
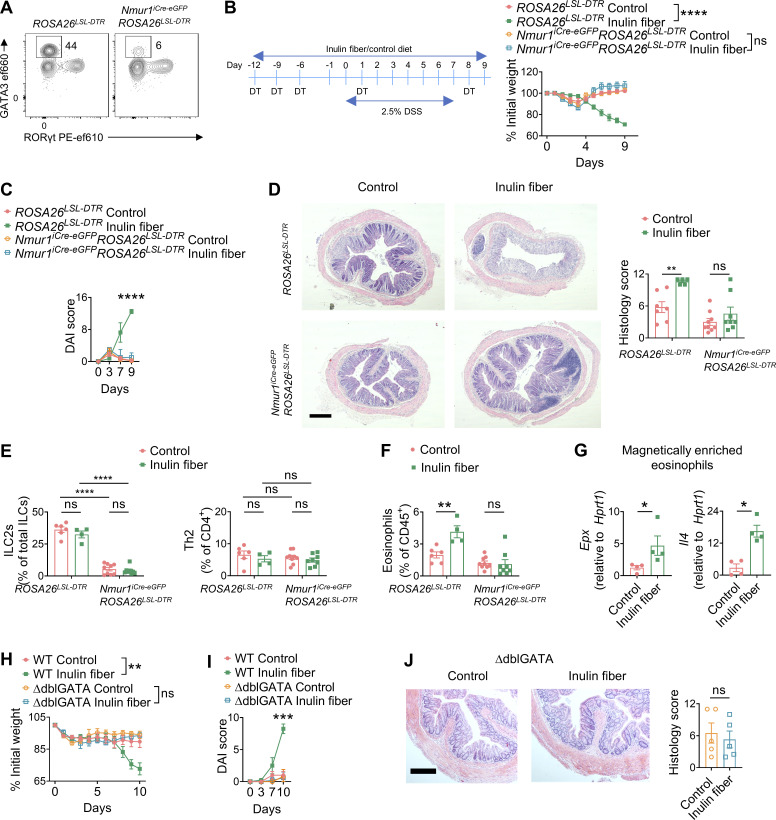
Next, we sought to determine the role of ILC2s in inulin fiber diet–mediated intestinal inflammation. We previously reported that among various colonic immune cells, ILC2s selectively express the G protein–coupled receptor NMUR1 (Jarick et al., 2022; Tsou et al., 2022). Therefore, next, we employed a new mouse model that we recently developed for specific targeting of ILC2s (Nmur1iCre-eGFP) (Jarick et al., 2022; Tsou et al., 2022). Employing repeated injection of diphtherial toxin (DT) in Nmur1iCre-eGFPROSA26LSL-DTR mice or ROSA26LSL-DTR littermates, we validated that ILC2s can be selectively depleted for at least up to 4 wk (Fig. 2 A and Fig. S2 D). We observed that depletion of ILC2s abrogated the ability of the inulin fiber diet to exacerbate weight loss (Fig. 2 B), disease activity (Fig. 2 C), and histopathology, including crypt loss (Fig. 2 D). Furthermore, inulin fiber diet did not induce colonic eosinophilia in the Nmur1iCre-eGFPROSA26LSL-DTR mice in which ILC2s were specifically depleted (Fig. 2, E and F). Together, these data indicate that ILC2s are required for inulin fiber diet–induced inflammation and exacerbation of disease severity.
Figure 2.
ILC2s and eosinophils mediate the exacerbation of intestinal damage by the inulin fiber diet. (A) Representative flow cytometry plots showing depletion of ILC2s 4 wk after intraperitoneal injections with 100 ng/mouse DT on days 0, 3, 6, 14, and 21. (B and C) Experimental schematic and body weights (B) and DAI (C) of DSS-treated mice (n = 4–5 mice). (D) Representative H&E staining of distal colons of DSS-treated mice (scale bar = 500 μm), and histology scores at the endpoint (day 9) (n = 5–9 mice). (E and F) Frequency of ILC2s and GATA3+ Th2 cells (E) and eosinophils (F) in the colon of the DSS-treated mice at the endpoint (n = 4–9 mice). (G) Gene expression in magnetically enriched colonic eosinophils determined by RT-qPCR from mice fed with control or inulin fiber diet for 2 wk (n = 4 mice). (H–J) Body weights (H) and DAI (I) on various days and colon histology at the endpoint (J) in DSS-treated mice fed with control or inulin fiber diet (n = 4–9 mice). Scale bar = 200 μm. Data are representative of (A–C and G) or pooled from (D–F and H) two independent experiments. Data are means ± SEM. Statistics were calculated by unpaired two-tailed t test (C, I, and J) or two-way ANOVA with Tukey’s multiple comparison test (B and H) or uncorrected Fisher’s LSD test (D–F) or Mann–Whitney U test (G). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant.
Eosinophils mediate the exacerbation of intestinal damage by an inulin fiber diet in mice
While inulin fiber diet triggers colonic eosinophilia (Arifuzzaman et al., 2022) (Fig. 1 D), the effects of inulin fiber diet on the activation of eosinophils remain to be explored. Therefore, we magnetically enriched colonic eosinophils from control or inulin fiber diet–fed mice and analyzed gene expression of eosinophil-intrinsic inflammatory and tissue-damaging factors. We observed that inulin fiber diet significantly upregulated the genes encoding IL-4 (Il4) and eosinophil peroxidase (Epx) (Fig. 2 G), both of which have previously been shown to contribute to the pathogenesis in the DSS model of intestinal damage and inflammation (Forbes et al., 2004; Hertati et al., 2020; Pushparaj et al., 2013; Stevceva et al., 2001). Therefore, to test the requirement of eosinophils in inulin-induced inflammation, WT and eosinophil-deficient (ΔdblGATA) mice were given the control or inulin fiber diet followed by exposure to DSS. We observed that in contrast to WT mice, ΔdblGATA mice fed with an inulin fiber diet did not exhibit higher weight loss compared with the control diet (Fig. 2 H). Disease activity and crypt loss were also comparable between the two diet groups in the absence of eosinophils (Fig. 2, I and J), supporting that eosinophils are required for inulin fiber diet–induced exacerbation of disease severity. In summary, these findings demonstrate that an inulin fiber diet triggers an ILC2-dependent accumulation of colonic eosinophils which contributes to intestinal inflammatory disease.
IL-33 is required for inulin fiber diet–induced colonic eosinophilia
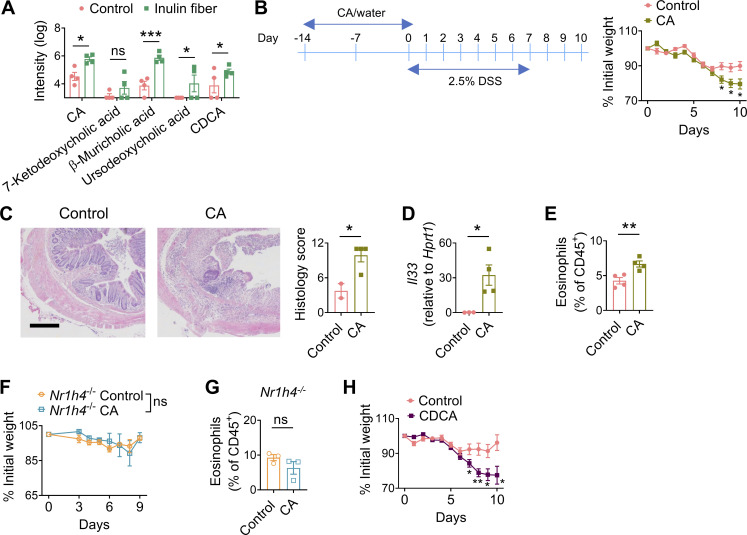
Since both IL-33 and IL-25 can promote ILC2INFLAM induction (Flamar et al., 2020; Huang et al., 2015), we next sought to determine their involvement in the inulin fiber diet–induced eosinophilia. We observed that an inulin fiber diet significantly increased the levels of Il33 but not Il25 in the colon (Fig. 3 A). The primary source of inulin fiber–induced IL-33 was mesenchymal stromal cells in the colon (Fig. S2 E). The inulin fiber diet also upregulated Il33 during DSS-induced intestinal damage (Fig. 3 B). Notably, the levels of the IL-33 receptor in colonic ILC2s were not altered by inulin fiber diet (Fig. S2 F). We and others have previously shown that IL-33 can promote ILC2-intrinsic AREG, Tregs, and tissue repair during intestinal damage (Lopetuso et al., 2018; Monticelli et al., 2015; Ngo Thi Phuong et al., 2021; Schiering et al., 2014; Sedhom et al., 2013). Indeed, when we employed Il1rl1−/− mice, which are deficient in IL-33 receptor T1/ST2, we observed that Il1rl1−/− mice fed with control diet lost more weight than WT mice fed with control diet (Fig. 3 C), supporting that IL-33/ST2 signaling in ILC2s or other cells including Tregs has a protective role during intestinal inflammatory disease (Lopetuso et al., 2018; Schiering et al., 2014; Sedhom et al., 2013). However, while WT mice fed an inulin fiber diet exhibited greater loss of body weight compared with that on the control diet, Il1rl1−/− mice did not display a difference in weight loss between control and inulin fiber diets (Fig. 3 C). In addition, the levels of eosinophils and crypt loss in the colons of Il1rl1−/− mice were also comparable between the control and inulin fiber diet (Fig. 3, D and E). Together, these data indicate that the inulin fiber diet–induced eosinophilia and exacerbated inflammation and disease severity require IL-33.
Figure 3.
IL-33 is required for inulin fiber diet–induced colonic eosinophilia. (A) Gene expression in the distal colon was determined by RT-qPCR analysis of mice fed with control or inulin fiber diet for 2 wk (n = 4–5 mice). (B) Expression of Il33 in the distal colon of mice at the endpoint (day 9) after challenge with DSS. (C–E) Body weights (C) and colonic eosinophil levels (D) and histology scores (E) at the endpoint in DSS-treated mice fed with control or inulin fiber diet (n = 4–5 mice). Scale bar = 200 μm. Data are representative of two independent experiments. Data are means ± SEM. Statistics were calculated by unpaired two-tailed t test (A, B, D, and E) or two-way ANOVA with Tukey’s multiple comparison test (C). *P < 0.05, **P < 0.01, ns, not significant.
Since we observed that the exposure of naïve mice to an inulin fiber diet led to the production of stromal IL-33, which activated ILC2s to produce IL-5 but not AREG (Fig. 1 B), and that the colons of naïve mice express lower Il33 than DSS-challenged mice (Fig. S2 G), we further investigated the mechanism of IL-33–mediated induction of AREG. Our in vitro studies showed that while IL-33 could promote both IL-5 and AREG, the kinetics of AREG production is slower than IL-5 (Fig. S2 H), and a higher concentration of IL-33 is required for the production of AREG that is comparable with levels of IL-5 production (Fig. S2 I). Together, these data suggest that a low concentration of IL-33 is sufficient to trigger IL-5 production, and the IL-5–mediated accumulation of eosinophils prior to DSS challenge supersedes any beneficial effects of IL-33 during DSS-induced tissue damage.
Bile acids mimic inulin-induced exacerbation of intestinal damage and inflammation
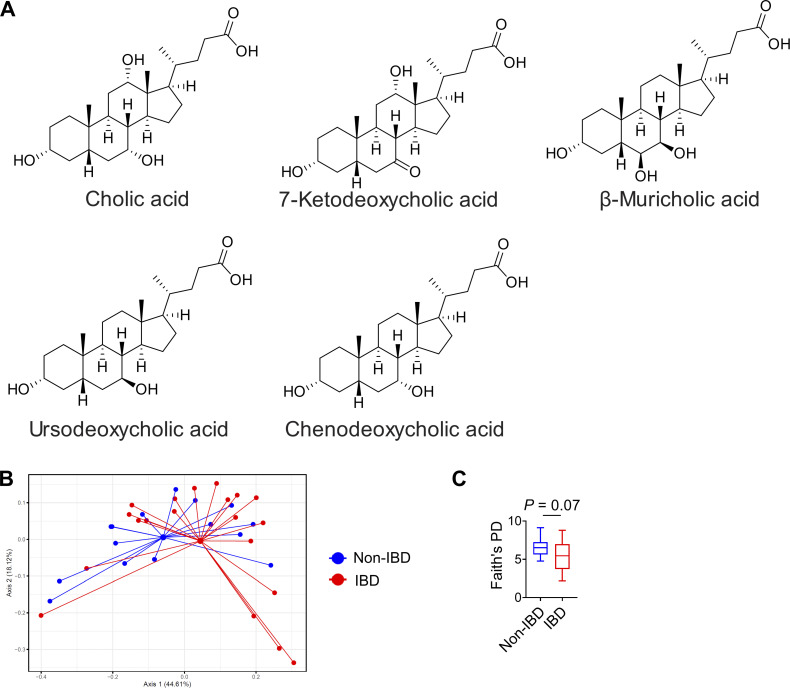
We previously reported that an inulin fiber diet increases not only the level of Bacteroidetes population but also their capacity to hydrolyze bile salts, leading to elevated systemic levels of several unconjugated bile acids including cholic acid (CA) and chenodeoxycholic acid (CDCA) (Arifuzzaman et al., 2022) (Fig. 4 A and Fig. S3 A). Therefore, we investigated whether direct administration of bile acids also triggers type 2 inflammation and exacerbates tissue damage. WT mice were exposed to control drinking water or CA-supplemented drinking water for 2 wk and then exposed to DSS. Similar to exposure to inulin fiber diet, mice that received CA exhibited significantly more severe loss of body weight (Fig. 4 B). Histological analyses revealed more crypt loss during the recovery period, and immunologic analyses revealed increased Il33 and eosinophils in the colon of CA-treated mice compared with control mice (Fig. 4, C–E). When mice deficient in the bile acid receptor farnesoid X receptor (FXR) (Nr1h4−/−) were exposed to CA and then challenged with DSS, we did not observe any increase in the frequency of colonic eosinophils or any alteration in the severity of disease (Fig. 4, F and G), suggesting that CA exerts its immunomodulatory effects via FXR. Since CDCA is the most abundant bile acid in humans (Thakare et al., 2018), we also employed CDCA and observed that CDCA-treated mice exposed to DSS exhibited significantly greater weight loss compared with control mice exposed to DSS (Fig. 4 H). Together, these data demonstrate that these primary bile acids, either administered directly or induced by diet and microbiota, can have profound pathological effects during intestinal inflammation.
Figure 4.
Administration of bile acids worsens DSS-induced disease severity. (A) Bile acid levels were measured by LC-MS in the serum of mice after feeding with a control or inulin fiber diet for 2 wk (n = 4 mice). Peak integration data from HPLC-MS analysis were log-transformed (Karpievitch et al., 2012) prior to statistical analysis. (B) Experimental schematic and body weights of DSS-treated mice under control (water) and CA treatment groups (n = 7–9 mice). (C) Representative H&E staining of distal colons at the endpoint of DSS experiment (day 10), Scale bar = 200 μm. Graph shows histological scores (n = 2–4 mice). (D–E) Expression of Il33 (D) and frequency of eosinophils (E) in the colons of DSS-treated mice at the endpoint (n = 3–4 mice). (F) Body weights of DSS-treated Nr1h4−/− mice under control (water) and CA-treated groups (n = 3 mice). (G) Frequency of eosinophils in the colons of mice at the endpoint of DSS experiment (n = 3 mice). (H) Body weights of DSS-treated mice under control (water) and CDCA treatment groups (n = 8–9 mice). Data are pooled from (B and H) or representative of (A and C–G) two independent experiments. Data are means ± SEM. Statistics were calculated by unpaired two-tailed t test. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant.
Figure S3.
Chemical structures of bile acids and analyses of human fecal microbial composition. (A) Chemical structures of differential unconjugated bile acids identified in mouse serum. (B and C) Weighted UniFrac PCoA of 16S rRNA (B) and Faith’s phylogenic diversity (PD) (C) in fecal samples from non-IBD controls and IBD patients (n = 15–20 humans). For PCoA plot PERMANOVA: F = 12.6, Df = 1, P = 0.09. Statistics were calculated by unpaired two-tailed t test with Welch’s correction (C).
Human fecal microbiota transfer (FMT) can reproduce inulin fiber diet–induced eosinophilia and intestinal damage in mice
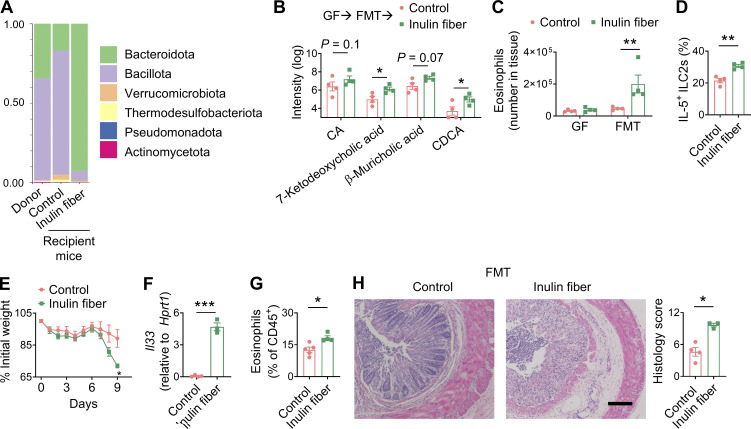
To determine whether the inulin fiber diet–induced and bile acid–mediated type 2 inflammation is relevant in the context of the human microbiota, we performed human FMT from healthy individuals into germ-free (GF) mice. Upon receiving human FMT, mice received either a control or inulin fiber diet for 2 wk. As we and others have shown previously (Arifuzzaman et al., 2022; Chijiiwa et al., 2020), administration of inulin increased the abundance of Bacteroidota (Fig. 5 A) in the human FMT mice and upregulated serum bile acids including CDCA (Fig. 5 B). While GF mice did not exhibit any increase in eosinophils upon administration of the inulin fiber diet, human FMT restored the inulin fiber diet–induced eosinophilia in GF mice (Fig. 5 C). The eosinophilia in the human FMT mice was associated with an increased number of colonic IL-5+ ILC2s (Fig. 5 D). Upon exposure to DSS, mice receiving human FMT and an inulin fiber diet exhibited significantly greater body weight loss (Fig. 5 E), which was accompanied by increased expression of Il33, accumulation of eosinophils, and enhanced tissue damage in the colon (Fig. 5, F–H). Together, these data demonstrate that the human microbiota can mediate the inulin fiber diet–induced elevation of bile acids and subsequent type 2 inflammation, further supporting that this diet microbiota pathway could contribute to inflammation in human inflammatory bowel disease (IBD).
Figure 5.
Human FMT can reproduce inulin fiber diet–induced eosinophilia and intestinal damage. (A) Representative taxonomic classification of 16S rRNA genes in fecal suspension from individual human donor or stool pellets collected from recipient mice with human microbiota on control or inulin fiber diet for 2 wk. (B–D) Serum bile acids (B), colonic eosinophils (C), and colonic IL-5+ ILC2s (D) in the recipient mice (n = 4) after 2 wk of diet. (E) Body weights of DSS-treated control diet– and inulin fiber diet–fed human FMT mice (n = 4 mice). (F and G) Expression of Il33 (F) and frequency of eosinophils (G) in the colon of DSS-treated human FMT mice at the endpoint (day 9) (n = 3–5 mice). (H) Representative H&E staining and histology scores of distal colons at the endpoint (day 9) of the DSS experiment. Scale bar = 100 μm. Data are representative of two independent experiments. Data are means ± SEM. Statistics were calculated by unpaired two-tailed t test (D–H) or two-way ANOVA with uncorrected Fisher’s LSD test (B and C). *P < 0.05, **P < 0.01, ***P < 0.001.
Bile acid metabolites are dysregulated in IBD patients
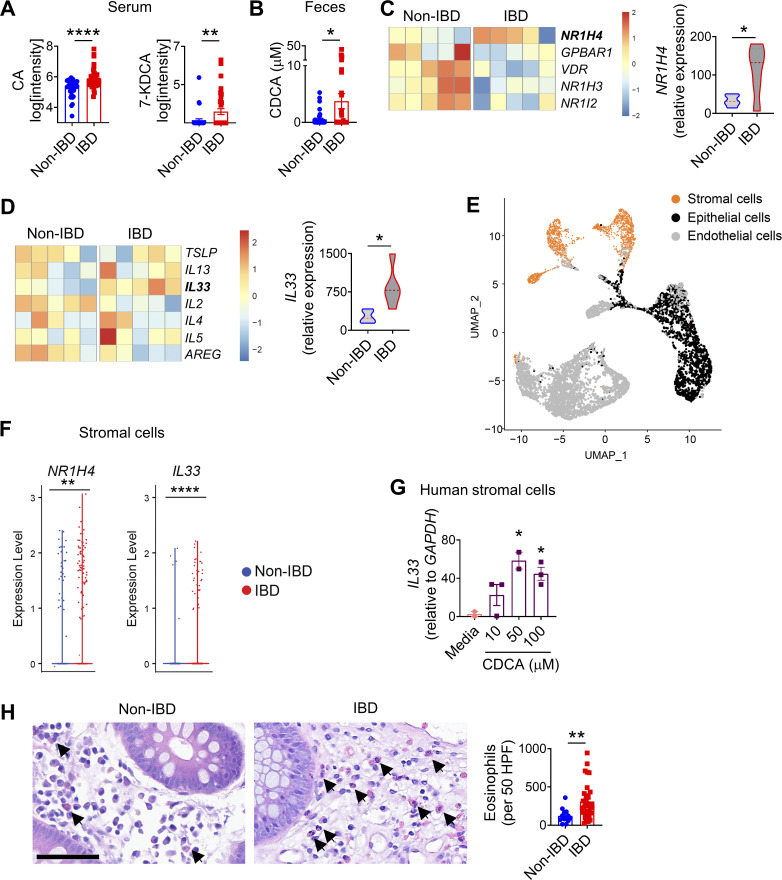
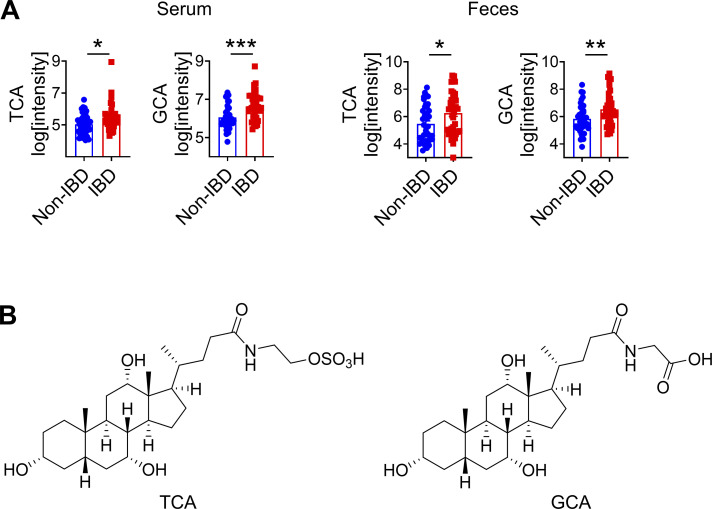
To interrogate the relationship between intestinal microbiota and bile acids in the context of human IBD, we recruited a cohort of adult IBD patients with ulcerative colitis or pancolitis (n = 40) and a comparable cohort of non-IBD controls (n = 40). We employed sequencing of 16S ribosomal RNA (rRNA) to determine the composition of the fecal microbiota. We observed that the microbial community in IBD patients has a trend to be distinct and with decreased taxonomic diversity compared with non-IBD controls (Fig. S3, B and C), consistent with other clinical studies (Franzosa et al., 2019). Next, we performed untargeted comparative metabolomic analyses of serum and fecal samples obtained from IBD patients and non-IBD controls using liquid chromatography coupled with high-resolution mass spectrometry. Among a total of >40,000 detected features, we detected bile acids as the most significantly differential metabolites between IBD and non-IBD samples. Levels of unconjugated bile acids, including CA and 7-ketodeoxycholic acid (7-KDCA), were significantly increased in the serum of IBD patients compared with non-IBD controls (Fig. 6 A). We also detected increased CDCA in the feces of IBD patients compared with non-IBD controls (Fig. 6 B). Levels of taurine- and glycine-conjugated bile acids were significantly increased in both the serum and feces of IBD patients compared with non-IBD controls (Fig. S4, A and B). Collectively, our data demonstrate that active IBD is associated with significant changes in bile acid profiles.
Figure 6.
The bile acid–eosinophil axis is dysregulated in IBD. (A and B) Serum (A) and fecal (B) levels of unconjugated bile acids (n = 37–40 humans). (C) Heatmap showing bile acid receptor genes and the violin plot showing NR1H4 expression in the colon biopsies of non-IBD controls and IBD patients (n = 5 humans). (D) Heatmap showing type 2 cytokine genes and the violin plot showing IL33 expression in the colons of non-IBD controls and IBD patients (n = 5 humans). (E) UMAP plot of scRNA-seq data of non-IBD control and IBD patient colons. (F) Violin plots showing expression of NR1H4 and IL33 in the stromal cells shown in E. (G) IL33 gene expression in human colonic cell line CCD-18Co determined by RT-qPCR after 4 h incubation with increasing concentrations of CDCA (n = 2–3 replicates). Data are representative of two independent experiments. (H) Representative H&E staining of human colorectal tissues (scale bar = 60 μm). Bar plot shows the number of eosinophils in 50 high power fields (HPF) in the colons of non-IBD controls (n = 13) and IBD patients (n = 35). Data are means ± SEM. Statistics were calculated by unpaired two-tailed t test (A–E, F, and H) or one-way ANOVA with Dunnett’s multiple comparisons test (G). *P < 0.05, **P < 0.01, ****P < 0.0001.
Figure S4.
Analyses of metabolites in human serum and fecal samples. (A) Serum and fecal levels of conjugated bile acids (n = 37–40 humans). (B) Chemical structures of TCA and GCA. Data are means ± SEM. Statistics were calculated by unpaired two-tailed t test (A). *P < 0.05, **P < 0.01, ***P < 0.001.
A bile acid type 2 inflammatory gene signature is activated in IBD patients
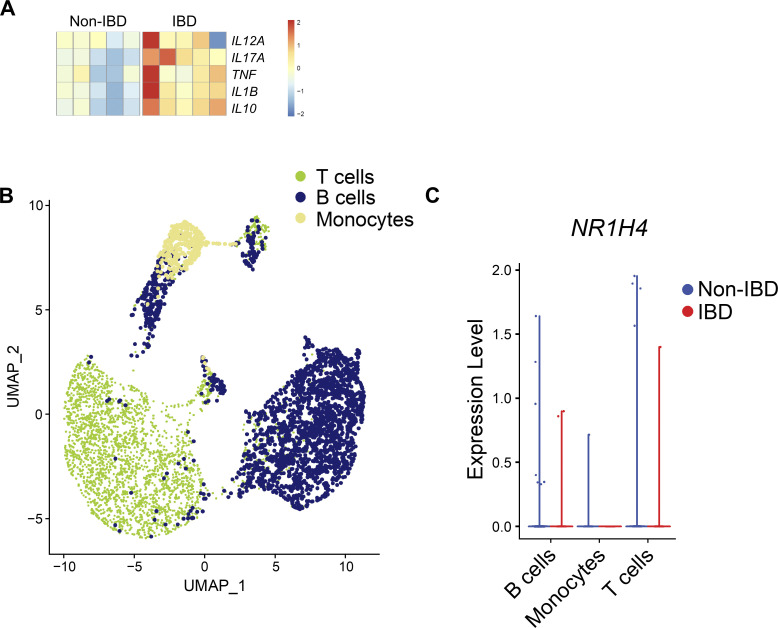
Since bile acids were the most significantly altered metabolites in active IBD patients, we next sought to determine the potential impact of altered bile acid levels in the intestine of IBD patients. First, by employing bulk RNA-seq, we compared the gene expression levels of known bile acid receptors in the colon tissue biopsies isolated from IBD patients and healthy controls. We observed that while most bile acid receptor genes were downregulated in IBD patients, NR1H4, the gene encoding bile acid receptor FXR, was significantly upregulated in IBD patients compared with non-IBD controls (Fig. 6 C), suggesting that FXR could be a mediator of bile acid signaling in the inflamed intestine. Next, to identify a potential functional link between bile acids and the immune response, we analyzed the expression of inflammatory cytokine genes in the same dataset. As expected, all major type 1/type 3 inflammatory genes known to be associated with IBD (Friedrich et al., 2019) were upregulated in the colons of IBD patients (Fig. S5 A). However, when we analyzed type 2 cytokines, IL33 was significantly upregulated in IBD patients compared with non-IBD controls (Fig. 6 D). We also observed trends for increased levels of IL5 and decreased levels of AREG in IBD patients compared with non-IBD controls (Fig. 6 D).
Figure S5.
Analyses of human intestinal transcriptional profile. (A) Heatmap showing type 1/3 cytokine genes in non-IBD controls and IBD patients (n = 5 humans). (B) UMAP plot of scRNA-seq data of non-IBD control and IBD patient colons. (C) Violin plot showing expression of NR1H4 in various immune cell clusters in B.
To determine the cellular sources of NR1H4 and IL33 in the human intestine, we performed single-cell RNA-seq (scRNA-seq) analyses to directly compare cell lineage–specific gene expression profiles in colonic biopsies isolated from IBD patients versus non-IBD controls. First, we analyzed major immune cell types including T cells and monocytes, where we did not observe any differential expression of NR1H4 between IBD and non-IBD biopsies (Fig. S5, B and C). Next, we focused on nonhematopoietic cell types, primarily epithelial and mesenchymal stromal cells which have previously been shown to undergo extensive remodeling and display inflammatory signatures associated with IBD (Friedrich et al., 2021; Kinchen et al., 2018; Parikh et al., 2019; Smillie et al., 2019). Unbiased clustering identified several nonhematopoietic cell types, including epithelial, stromal, and endothelial cell subsets (Fig. 6 E). We observed that NR1H4 was upregulated in the stromal cell subset of the IBD colon tissue compared with non-IBD control tissue (Fig. 6 F). Notably, more stromal cells expressed IL33 in the IBD colon compared with the control (Fig. 6 F). Together, these data demonstrate that expression of both bile acid receptor FXR and type 2 inflammatory cytokine IL-33 are increased in the colonic stromal cells isolated from IBD patients, suggesting that a bile acid–IL-33 axis in these cell types is associated with inflammation in IBD patients. To test this hypothesis, we incubated a human colonic stromal cell line with CDCA. We observed that exposure to CDCA triggered IL33 expression in a concentration-dependent manner (Fig. 6 G), further confirming that bile acids can promote stromal cell-intrinsic IL-33 production in the human colon.
IL-33 can activate multiple immune cell types associated with type 2 inflammation, including type 2 helper T cell (Th2), ILC2s, and eosinophils (Cayrol and Girard, 2018). In mice, IL-33 has been shown to play a critical regulatory role in the development, homeostasis, and activation of eosinophils (Johnston and Bryce, 2017; Johnston et al., 2016). In humans, IL-33 can directly activate eosinophils as well as promote the expression of eosinophil-recruiting IL-5 in ILC2s (Cherry et al., 2008; Nussbaum et al., 2013; Salimi et al., 2013). Therefore, we next compared the numbers of eosinophils in the colorectal biopsies from IBD patients and healthy non-IBD controls. We observed significantly higher infiltration of eosinophils in the lamina propria and intercryptic regions in tissues isolated from IBD patients compared with non-IBD controls (Fig. 6 H). Together, these data support the existence of a dysregulated bile acid–IL-33–eosinophil axis in IBD patients.
Discussion
In this study, we identify that dietary inulin fiber–induced alterations in the composition of the microbiota trigger the accumulation of ILC2INFLAM in the colon, promoting tissue eosinophilia that exacerbates disease outcomes in two murine models of intestinal damage and inflammation. Mechanistically, inulin fiber diet–induced and microbiota-derived bile acids, including CA and CDCA, upregulate the expression of IL-33 to trigger IL-5–producing ILC2INFLAM. Consistent with this model, elevated levels of bile acids (including CA and CDCA) and higher accumulation of eosinophils were observed in the colon of IBD patients compared to healthy individuals. Additional studies are needed to determine the casual link between the IBD microbiota and IL-5–producing ILC2s.
These new data demonstrate that diet and microbiota function as determinants of pathologic versus protective ILC2 function by the induction of ILC2INFLAM, which produce inflammatory IL-5 but not tissue-protective AREG. While IL-33 and ILC2s have been shown to have specific tissue-protective roles in murine models of colitis (Lopetuso et al., 2018; Monticelli et al., 2015; Ngo Thi Phuong et al., 2021; Tsou et al., 2022), they have also been implicated in promoting the disease (Qiu et al., 2020; Sedhom et al., 2013). This study shows that while IL-33 has a tissue-protective role during intestinal damage following exposure to a normal diet, the inulin fiber diet–mediated production of IL-33 in the colon can be detrimental due to the activation of ILC2INFLAM and subsequent eosinophilia that exacerbates inflammation. While we observed that the inflammatory effects of the inulin fiber diet were IL-33 receptor–dependent, the role of ILC2-intrinsic IL-33 receptor signaling requires further investigation.
Elevation of fecal CDCA and colonic eosinophils in IBD patients have previously been reported by other studies (Ahrens et al., 2008; Franzosa et al., 2019; Raab et al., 1998); however, transcriptomic analyses of human colons identified a potential link between these two observations. We found that expression of both the bile acid receptor FXR and type 2 cytokine IL-33 are upregulated in IBD patients compared with non-IBD controls, suggesting that elevated bile acid levels may be the underlying cause of increased eosinophils in human IBD. Several recent scRNA-seq studies of the human colon found that colonic stromal cells undergo extensive remodeling in IBD, including activation of inflammatory gene signatures, leading to the emergence of activated stromal cell subsets including inflammatory fibroblasts (Friedrich et al., 2021; Kinchen et al., 2018; Smillie et al., 2019). The scRNA-seq analysis of human colon tissues shown here revealed that upregulation of FXR and IL-33 expression is restricted to specific stromal cell subsets, suggesting that bile acid signaling functions upstream of cytokine production by these cells, which was further confirmed by in vitro experiments with human colonic stromal cell lines. Similar regulatory signaling could exist between microbiota-derived metabolites and other inflammatory cytokines in the stromal cell subsets (Friedrich et al., 2021; Kinchen et al., 2018; Smillie et al., 2019).
Dietary fibers including inulin promote the production of SCFAs that have anti-inflammatory effects (Furusawa et al., 2013; Maslowski et al., 2009; Smith et al., 2013). However, IBD patients are frequently advised to refrain from a fiber-rich diet since dietary fiber often exacerbates the disease (Ananthakrishnan, 2015; Cohen et al., 2013; Forbes et al., 2017). Although SCFAs have been shown to reduce intestinal inflammation in multiple murine models (Furusawa et al., 2013; Maslowski et al., 2009; Smith et al., 2013), similar studies using various high-fiber diets have reported conflicting results. For example, while some galactose-based fermentable fibers such as pectin and guar gum had protective effects (Macia et al., 2015; Silveira et al., 2017), fructan-based fibers such as inulin or fructose oligosaccharides exacerbated inflammation (Goto et al., 2010; Miles et al., 2017). While we observed increased bile acids in our cohort of IBD patients compared with non-IBD controls, it was not possible to associate these data with fiber intake due to the lack of sufficient dietary information. However, a recent randomized control trial demonstrated that administration of inulin to UC patients increased the colonic levels of inflammatory cytokines including IL-5 as well as symptomatic relapse in patients (Armstrong et al., 2022), supporting the capacity of inulin to trigger type 2 inflammation and tissue damage in humans. However, how specific fiber types exacerbate inflammation in human and mouse models was not clear. Our findings demonstrate that inulin fiber can be proinflammatory by acting as a critical environmental determinant of inflammatory ILC2 responses.
In summary, this study identifies a previously unrecognized pathway through which the inulin fiber diet regulates the composition and metabolic output of the intestinal microbiota to trigger type 2 inflammation in the context of ongoing intestinal damage and inflammation. The identification of a microbiota-dependent immunomodulatory role for dietary fiber could have implications for the development of microbiota- and metabolite-based precision nutrition that could influence the efficacy of therapeutic intervention strategies for IBD and other chronic inflammatory diseases.
Materials and methods
Mice and diets
C57BL/6 (Jax 000664), Il33fl/fl-eGFP (Jax 030619) (Han et al., 2018), Nr1h4−/− (Jax 007214) (Sinal et al., 2000), ROSA26LSL-DTR (ROSA26iDTR; Jax 007900), and ΔdblGATA (Jax 005653) (Yu et al., 2002) mice were originally purchased from The Jackson Laboratories. Rag2−/− (TAC no. RAGN12) mice were originally purchased from Taconic Farms. The transgenic Nmur1iCre-eGFP reporter mouse was generated by Cyagen and was described previously (Tsou et al., 2022). These strains, as well as Il1rl1−/− (Hsu et al., 2010) (provided by Andrew N.J. McKenzie, MRC Laboratory of Molecular Biology, Cambridge, UK) mice on a C57BL/6 background, were bred at Weill Cornell Medicine (WCM). Germ-free C57BL/6J mice and gnotobiotic mice were bred and maintained in flexible PVC isolators (Park Bioservices) at WCM. All other mice were maintained under specific pathogen–free conditions. Both female and male mice were included for all strains used in this study. All mice used were between 6 and 12 wk old, and age- and sex-matched for each experiment. All mice were maintained in facilities with a 12-h light–dark cycle, an average ambient temperature of 21°C, an average humidity of 48%, and were provided food and water ad libitum. When studying the effects of dietary fiber, mice were given an inulin fiber diet (D16052309; Research Diets, Inc.) supplemented with 30% fiber (26% inulin and 4% cellulose) or a calorie-matched control diet (D12450J-1.5; Research Diets, Inc.) containing 4.7% cellulose, which is comparable to the crude fiber content of standard chow. The duration of dietary intervention was 2 wk unless otherwise stated. Double-irradiated sterile diets were used for all GF and FMT mice. All mouse experiments were approved by and performed in accordance with the Institutional Animal Care and Use Committee guidelines at WCM.
Human samples
Feces, serum, and colon tissue biopsies of IBD patients and age- and sex-matched non-IBD healthy controls were obtained from the JRI Live Cell Bank at WCM following protocols approved by the Institutional Review Board (protocol number 1503015958). The non-IBD controls did not have any major inflammatory diseases, cancer, or obesity. However, they might have other common conditions including diabetes and cardiovascular diseases. Informed consent was obtained from all subjects. All IBD patients used for various analyses were diagnosed as UC patients, except one patient whose colon tissues used for scRNA-seq was pan-colitic. H&E slides were generated from paraffinized colorectal biopsies. Biopsies used for RNA-seq were cryopreserved in 90% FBS and 10% dimethylsulfoxide for future side-by-side comparison. For FMT and metabolomic studies, donor fecal samples were resuspended in PBS (Goc et al., 2021).
Isolation of cells from various tissues
Mouse colons were removed, cleaned, and washed in ice-cold PBS (Sigma-Aldrich). They were then opened longitudinally and washed again in ice-cold PBS. Dissociation of epithelial cells was performed by shaking at 37°C in HBSS (Sigma-Aldrich) containing 10 mM HEPES and 5 mM EDTA (Thermo Fisher Scientific) two times for 15 min. After each step, samples were vortexed, and the supernatant containing the epithelial fraction was removed. The remaining tissue was chopped into small pieces and enzymatic digestion was performed using collagenase III (1 mg/ml; Worthington), dispase (0.4 U/ml; Thermo Fisher Scientific), DNase I (20 μg/ml; Sigma-Aldrich), and 4% FBS for 40 min in a shaker at 37°C. The single-cell suspension was then filtered through a 70-μm cell strainer and centrifuged through a Percoll (Sigma-Aldrich) gradient and washed. Eosinophils were enriched magnetically using PE anti-SiglecF antibody and anti-PE microbeads (Milteny), according to the manufacturer’s protocol. For human colorectal samples, tissues were incubated in 0.5 mg ml−1 collagenase D and 20 mg ml−1 DNase I for 1 h at 37°C with shaking. After digestion, the remaining tissues were further dissociated mechanically by a syringe plunger. Cells were filtered through a 70-mm cell strainer. Mesenteric lymph nodes and spleens were minced and incubated in RPMI 1640 medium (Sigma-Aldrich) supplemented with 1% BSA (Sigma-Aldrich), Collagenase II (1 mg/ml; Sigma-Aldrich), and DNaseI (20 μg/ml) for 20 min in a shaker at 37°C. Cells were then dissociated using a Pasteur pipette and filtered through a 70-μm cell strainer. For the bone marrow, the ends of the bones were cut, then the marrow tissue was flushed out using a 25-gauge needle and ice-cold RPMI 1640 medium with 5% FBS. The collected bone marrow was then processed through a 40-μm cell strainer with gentle agitation with a syringe plunger. For the spleen, bone barrow, and blood, red blood cells were lysed with ACK lysing buffer (Lonza) and washed with ice-cold RPMI 1640 medium supplemented with 5% FBS.
Flow cytometry and cell sorting
Mouse single-cell suspensions were pretreated with anti-CD16/32 and then incubated on ice with conjugated antibodies in PBS. Dead cells were routinely excluded with Fixable Aqua Dead Cell Stain (Thermo Fisher Scientific). Lineage (Lin) markers used were as follows: Lin1: CD3ε (145-2C11), CD5 (53-7.3), CD8a (53-6.7), Lin2: CD19 (1D3), and CD11c (N418). FcεRIα (MAR-1) was included in either Lin1 or Lin2. NK1.1 (PK136) was included in Lin2 for intracellular cytokine staining. CD45 (30-F11), CD127 (A7R34), CD90.2 (30-H12), KLRG1 (2F1), CD11b (M1/70), SiglecF (E50-2440), Ly6G (1A8), c-Kit (CD117) (2B8), Ter119 (TER119), EpCAM (G8.8), CD31 (390), Sca-1 (D7), PDGFRα (CD140a) (APA5), CD135 (Flt3), and α4β7 were used for surface staining. Transcription factors were detected by intranuclear staining using T-bet (4B10), GATA3 (TWAJ), RORγt (B2D), and FoxP3 (FJK-16s). CD45+CD11b+SiglecF+CD11c−SSChi cells were defined as eosinophils. ILC2s were defined as CD45+Lin−CD90.2+CD127+GATA3+ for phenotyping and as CD45+Lin−CD90.2+CD127+KLRG1+ for sorting and intracellular cytokine assay. ST2+ cells were detected by a biotinylated anti-ST2 (IL33R) antibody (RMST2-2) followed by streptavidin-PE. CD45+Lin–CD127+Flt3–α4β7+c-Kit–ST2+ cells were defined as ILC2p. CD45–Ter119–EpCAM–CD31–Sca-1+PDGFRα+ cells were defined as mesenchymal stromal cells (Arifuzzaman et al., 2022). For the detection of intracellular cytokines, isolated cells were stimulated with 100 ng/ml phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich) and 1 μg/ml ionomycin (Sigma-Aldrich) for 3–4 h in the presence of 10 μg/ml brefeldin A (Sigma-Aldrich) in complete RPMI-1640 medium (containing 10% FBS, 50 mM 2-mercaptoethanol, 1 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin) to evaluate their cytokine production. IL-5+ and IL-13+ cells were detected by anti-IL-5 (TRFK5) and anti-IL-13 (eBio13A) antibodies, respectively. AREG+ cells were detected by biotinylated anti-AREG antibody (BAF989; R&D Systems) followed by streptavidin-APC. All antibodies used in flow cytometry were purchased from Thermo Fisher Scientific, BioLegend, or BD. All antibodies above were used at 1:200 dilutions, except CD127-BV421, IL-5-BV421, and IL-13-AF488, which were used at 1:100 dilutions, and anti-AREG antibody was used at 1:400 dilution. BD Cytofix/Cytoperm Fixation and Permeabilization Solution (BD Bioscience) and eBioscience Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific) were used for cytokine staining and intranuclear staining, respectively. Stained cells were analyzed on a 5-laser, 18-color custom-configuration BD LSRFortessa (BD) or sorted on a 5-laser, 18-color custom-configuration FACSAria III. Data were collected using BD FACSDiva version 8.0.1 or 9.0 and analyzed using FlowJo (version 10.6.1 or 10.7.1, Tree Star).
Quantitative real-time PCR (RT-qPCR)
Total RNA was isolated from tissues or magnetically enriched cells using the RNeasy Plus mini kit (Qiagen) and from cell cultures using TRIzol reagent (Thermo Fisher Scientific), according to the protocol provided by the manufacturer. cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit with Multiscribe Reverse Transcriptase (Thermo Fisher Scientific). When the number of sorted cells was low, 10X lysis buffer (Takara) or TRIzol reagent was used for RNA isolation, and Superscript II Reverse Transcriptase or the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific) was used for cDNA synthesis. qPCR reactions were set up using the Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) and run on a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) using QuantStudio Real-Time PCR software v1.0. Gene expression was normalized to Hprt1 for mouse tissues and GAPDH for the human cell line and displayed as a fold change compared with controls. The following primers were used: murine Il5 (QT00099715; Qiagen), murine Il13 (QT02423449; Qiagen), murine Areg (QT00112217; Qiagen), murine Tph1 (Mm01202614_m1; Thermo Fisher Scientific), murine Il33 (QT00135170; Qiagen), murine Calca (forward: 5′-GAGCAGATCAGGAGGTGTGG-3′ and reverse: 5′-GGCTGTTATCTGTTCAGGCCT-3′, murine Epx (forward: 5′-CTGTCTCCTGACTAACCGCTCT-3′ and reverse: 5′-TCAGCGGCTAGGCGATTGTGTT-3′, murine Il4 (QT00160678), murine Hprt1 (forward: 5′-CTTGCTGGTGAAAAGGACCTCTC-3′ and reverse: 5′-GAAGTACTCATTATAGTCAAGGGCA-3′, human IL33 (QT00041559; Qiagen), and human GAPDH (QT00040894; Qiagen).
Immunofluorescence microscopy
Colons were coiled into a “Swiss roll” and fixed in 4% paraformaldehyde (BioWorld) in PBS for 4 h at 4°C. Tissues were then placed in 30% sucrose dissolved in PBS at 4°C overnight. Tissues were then embedded in optimal cutting temperature (OCT) medium (Tissue-Tek, Sakura) and stored at −80°C until sectioning. Frozen tissues were sectioned at 10 μm using a cryotome (Leica Instruments), immobilized on Superfrost Plus slides (VWR), and stored at −20°C until immunostaining. For immunostaining, slides were allowed to come to room temperature, and any excess OCT medium was washed off using PBS. Tissue sections were then blocked in PBS with 5% normal goat serum (Jackson Immunoresearch), 5% normal donkey serum (Jackson Immunoresearch), and 0.2% Triton X-100 (Sigma-Aldrich) for 30 min. Tissue sections were then stained with rat anti-mouse Siglec-F Clone E50-2440 (Clone E50-2440, 552125; BD) 1:200 diluted in blocking buffer overnight at 4°C. Sections were washed three times with PBS and then incubated with anti-rat IgG-A488, diluted 1:500 in blocking buffer for 1 h at room temperature. Tissue sections were then washed three times in PBS and mounted with Prolong Diamond antifade reagent with DAPI (Invitrogen). Stained sections were imaged on an inverted Nikon Eclipse Ti microscope (Nikon) and analyzed by Fiji v1.53g.
Lipocalin ELISA
For fecal lipocalin-2 detection, fecal samples were collected and weighted for data normalization, and then homogenized in PBS and centrifuged at 12,000 rpm to remove aggregates, and the resulting supernatant was collected. Afterward, a sandwich ELISA was performed using mouse lipocalin-2/NGAL DuoSet ELISA (R&D Systems), according to the manufacturer’s instructions.
Administration of metabolites
For in vivo studies, CA or CDCA (sodium salt form, VWR) was administered in drinking water at a 6 mM concentration. All metabolites were administered for a duration of 2 wk. For in vitro studies, human colonic stromal cell line CCD-18Co (ATCC) (Friedrich et al., 2021) was cultured to a monolayer and then subjected to incubation with various concentrations of CDCA for 4 h.
Fecal microbiota transplantation
For FMT studies, fecal suspensions from individual donors were administered to recipient GF mice by oral gavage (100 μl per mouse). Transplanted animals were maintained in sterile isocages for 4–6 wk before administration of diets. Animals were evaluated for successful transplantation by comparing 16S sequencing between human donors and recipient mice.
Models of intestinal damage and inflammation
For the chemically induced model, mice were administered with 2.5–3.0% colitis-grade DSS with an average molecular weight of 36,000–50,000 Da (MP Biomedicals) in drinking water for 6–7 days, followed by drinking water for 2–3 days. Mice were monitored daily for morbidity (piloerection, lethargy), weight loss, and rectal bleeding. The severity of disease (DAI) was scored as follows: weight loss (no change, 0; <5%, 1; 6–10%, 2; 11–20%, 3; >20%, 4); feces (normal, 0; pasty, semi-formed, 2; liquid, sticky, or unable to defecate after 5 min, 4), rectal bleeding (no blood, 0; visible blood in the rectum, 1; visible blood on fur, 2); general appearance (normal, 0; piloerect, 1; lethargic and piloerect, 2; hunched, 3; motionless, 4). For a subset of experiments related to intracellular cytokine staining, mice were treated with DSS for 4–5 days, followed by drinking water for 2–3 days. For the T cell transfer model, Rag2−/− mice were retro-orbitally injected with 105 naïve CD4+CD25−CD45RBhi cells and body weight was monitored for 6 wk.
Histological analysis
Distal colon tissues from mice were fixed with 4% paraformaldehyde, followed by embedment in paraffin. 5–6 μm sections were used for staining with H&E by IDEXX BioResearch. H&E staining of human colorectal biopsies was performed by the Center for Translational Pathology at WCM. Histological scoring of H&E-stained tissues was performed by a blinded scorer based on epithelial hyperplasia, depletion of goblet cells, inflammation in the lamina propria, and submucosal inflammation on a point scale of 0–3, with 0 being no pathology and 3 being the most severe pathology. Eosinophils were counted by a blinded scorer, and eosinophils were counted per field of view (250 µm2) and then normalized to the number of eosinophils per 50 fields of view.
16S amplicon sequencing of intestinal microbiota
DNA extraction
Library preparation, sequencing, and operational taxonomic unit table generation were performed by the Microbiome Core Lab at WCM. Each fecal pellet was deposited into a Qiagen PowerBead tube with 0.1 mm glass beads (13118-50). Using a Promega Maxwell RSC PureFood GMO and Authentication Kit (AS1600), 1 ml of cetyltrimethylammonium bromide buffer and 20 μl of RNAse A solution were added to the PowerBead tube that contained the sample. Then the sample/buffer was mixed for 10 s on a Vortex Genie2 and then incubated at 95°C for 5 min on an Eppendorf ThermoMixer F2.0, shaking at 1,500 rpm. The tube was removed and clipped to a horizontal microtube attachment on a Vortex Genie2 and vortexed at high-speed for 20 min. The sample was then removed from the Vortex and centrifuged at 40°C and 12,700 rpm for 10 min. Upon completion, the sample was centrifuged again for an additional 10 min to eliminate foam. The sample tube cap was removed, and the sample was checked for foam and particulates. If foam or particulates were found in the sample, they were carefully removed using a P1000 pipette. The opened tube was then added to a Promega MaxPrep Liquid Handler tube rack. The Liquid Handler instrument was loaded with proteinase K tubes, lysis buffer, elution buffer, 1,000 ml tips, 50 ml tips, 96-sample deep-well plate, and Promega Maxwell RSC 48 plunger tips. The Promega MaxPrep Liquid Handler instrument was programmed to use 300 μl of the sample and transfer all sample lysate into a Promega Maxwell RSC 48 extraction cartridge for DNA extraction. Upon completion, the extraction cartridge was loaded into a Promega Maxwell RSC 48 for DNA extraction and elution. DNA was eluted in 100 μl and transferred to a standard 96-well plate. DNA was quantified using a Quant-iT dsDNA High Sensitivity Assay Kit using a Promega GloMax plate reader on a microplate.
16S library generation
Library generation follows the protocol from the Earth Microbiome Project (https://earthmicrobiome.org/protocols-and-standards/16s/).
Library verification, quality check, and pooling
Amplicon libraries were washed using Beckman Coulter AMPure XP magnetic beads. Library quality and size verification was performed using PerkinElmer LabChip GXII instrument with DNA 1K Reagent Kit (CLS760673). Library concentrations were quantified using Quant-iT dsDNA High Sensitivity Assay Kit using Promega GloMax plate reader on a microplate (655087). Library molarity was calculated based on library peak size and concentration. Libraries were normalized to 2 nM using the PerkinElmer Zephyr G3 NGS Workstation (133750) and pooled together using the same volume across all normalized libraries into a 1.5 ml Eppendorf DNA tube (022431021).
Sequencing
Pooled libraries were sequenced on the Illumina MiSeq instrument at a loading concentration of 7pM with 15% PhiX, paired-end 250 using MiSeq Reagent Kit v2, and 500 cycles (MS-102-2003).
Data processing
Demultiplexed raw reads were processed using the Nextflow (Di Tommaso et al., 2017) nf-core (Ewels et al., 2020) ampliseq pipeline (Straub et al., 2020), version 2.3.1, with the following parameters: -profile singularity --FW_primer GTGYCAGCMGCCGCGGTAA --RV_primer CCGYCAATTYMTTTRAGTTT --dada_ref_taxonomy silva --ignore_empty_input_files --ignore_failed_trimming --min_frequency 10 --retain_untrimmed --trunclenf 240 --trunclenr 160. Specifically, reads were trimmed with cutadapt (Martin, 2011), PhiX and quality filtering, read pair merging, and amplicon sequence variant resolution was performed with DADA2 (Callahan et al., 2016). Subsequent taxonomic assignment was also performed with DADA2 using the Silva reference database (Quast et al., 2013), version 138. Diversity analysis was performed with QIIME2 (Bolyen et al., 2019).
RNA-seq of ILC2s
RNA-seq libraries were prepared from 1,000 sorted colonic lamina propria ILC2s by the Epigenomics Core at WCM using the Clontech SMARTer Ultra Low Input RNA Kit V4 (Clontech Laboratories). Libraries were sequenced on an Illumina HiSeq 2500, generating 50 bp single-end reads. Two samples from two control diet–fed WT SPF mice and two samples from two inulin fiber diet–fed WT SPF mice were used. Reads were demultiplexed using Illumina’s CASAVA (v1.8.2), adapter-trimmed using FLEXBAR v2.4 (Dodt et al., 2012), and aligned to the mouse genome (NCBI GRCm38/mm10) using STAR aligner v2.3.0 (Dobin et al., 2013) with default parameters. Read counts per gene (RefSeq annotation) were determined using the Rsubread R package v3.10 I (Liao et al., 2019). Normalization and differential expression analysis were performed using DESeq2 version 1.26.0 (Love et al., 2014). Prior to differential expression testing, genes were prefiltered to only include genes that had a minimum of 50 raw reads in at least two samples. Tests were corrected for multiple comparisons using a false discovery rate (FDR) of 10% to determine significance.
Bulk RNA-seq of human colon tissues
Colonic biopsies were digested into single-cell suspensions and checked for quality control. RNA-seq libraries were prepared from the single-cell suspensions by Bristol Myers Squibb. Libraries were sequenced on an Illumina HiSeq 2500, generating 2 × 50 bp paired-end reads. Reads were demultiplexed using Illumina’s CASAVA (v1.8.2), adapter-trimmed using FLEXBAR v2.4 (Dodt et al., 2012), and aligned to the human reference genome (GRC GRCg38-95; NCBI) using STAR aligner v2.3.0 (Dobin et al., 2013) with default parameters. Read counts per gene (RefSeq annotation) were determined using the Rsubread R package v3.10 I (Liao et al., 2019). Normalization and differential expression analysis were performed using DESeq2 version 1.38.3 (Love et al., 2014). Tests were corrected for multiple comparisons, using an FDR of 5% to determine significance. Heatmap visualization of select genes of interest was performed using the pheatmap R package (https://cran.r-project.org/package=pheatmap), version 1.0.12, using normalized counts and scaling by row.
scRNA-seq of human colon tissues
Single-cell suspensions generated from colon biopsies from a non-IBD healthy control and a pan-colitic IBD patient were used for scRNA-seq. Tissue samples were thawed into warm media with FBS, and preservation media was removed by washing. Samples were treated with EDTA predigestion with rotation to remove dead/damaged epithelial cells. Samples were then dissociated enzymatically into a single-cell suspension with rotation. Cells were washed, strained, and counted for viability using acridine orange/propidium iodide, and 10,000 cells were loaded onto 10X channels. The GEX 3′ V3 protocol was followed throughout sequencing. Libraries were sequenced on an Illumina NovaSeq instrument. Raw scRNA-seq data was obtained from Celsius Therapeutics and the reads were processed by 10X’s Cell Ranger version 3.1.0 using the GRCg38-95 reference genome, resulting in filtered matrix, barcode, and feature files for both samples. scRNA-seq data were further processed and analyzed using R version 4.2.2 (R Core Team 2022) and the Seurat package version 4.3.0. Specifically, Cell Ranger output was imported using the Read10X function. Seurat objects were created using only genes appearing in at least three cells. Cells were further filtered to exclude those with <200 genes detected or >7,000 genes detected. Read counts were then normalized within each sample using the SCTransform function with the glmGamPoi method, percent mitochondrial reads as the vars.to.regress argument, and vst.flavor v2. For each sample, integration features were selected using the SelectIntegrationFeatures function with nfeatures set to 3,000. Integration of both samples was then performed with the PrepSCTIntegration, FindIntegrationAnchors, and IntegrateData functions. The RunPCA function was then run with npcs set to 30. The graph representing cells with similar expression patterns was generated with the FindNeighbors function using the 30 largest principal components. Cell clusters were generated using the Louvain algorithm implemented by the FindClusters function with a resolution parameter equal to 0.6. Marker genes for each cluster were determined using the Wilcoxon test on the SCT normalized counts after running the PrepSCTFindMarkers function using the function FindAllMarkers, and including only genes with log fold changes >0.25 and Bonferroni-corrected P values <0.01. Cluster names were determined initially by SingleR to annotate the clusters using the Human Primary Cell Atlas reference data followed by manual inspection of the lists of cluster marker genes. Cells were reclustered after subsetting compartments of interest (stromal, epithelial, endothelial, and immune) based on the selection of variable genes, dimensionality reduction, and graph clustering. Dimensionality reduction by Uniform Manifold Approximation and Projection was performed using the RunUMAP function with the 30 largest principal components. All visualizations of scRNA-seq data were generated using the Seurat package as well as ggplot2. Statistics for the violin plots were calculated by unpaired two-tailed t test.
Metabolomic analysis of serum and feces
Metabolite extraction
Metabolites were extracted from serum and fecal samples with methanol. 4 ml of methanol was added to 1 ml of serum in 4-ml glass vials. The vials were then vortexed for 1 min. 300 mg of dried fecal pellet content was vortexed and sonicated in 6 ml of 100% methanol for 1 min. Extracts from serum and fecal samples were pelleted at 5,000 g for 10 min at 4°C and supernatants were transferred to 2-ml high-performance liquid chromatography (HPLC) vials. Samples were then dried in a SpeedVac (Thermo Fisher Scientific) vacuum concentrator. Dried materials were resuspended in 1 ml of methanol and vortexed for 1 min. Samples were pelleted at 5,000 g for 5 min at 22°C and supernatants were transferred to HPLC vials and dried in a SpeedVac vacuum concentrator. Samples were then resuspended in 150 μl of methanol and centrifuged at 5,000 g for 10 min at 22°C. Clarified extracts were transferred to fresh HPLC vials and stored at −20°C until analysis.
Mass spectrometry
Liquid chromatography–mass spectrometry (LC-MS) was performed on a Thermo Fisher Scientific Vanquish ultra-high performance liquid chromatography (UHPLC) system coupled with a Thermo Q-Exactive HF hybrid quadrupole-orbitrap high-resolution mass spectrometer equipped with a heated electrospray ionization ion source. Metabolites were separated using a water−acetonitrile gradient on an Agilent Zorbax Eclipse XDB-C18 column (150 × 2.1 mm, particle size = 1.8 μm) maintained at 40°C; solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile. The A/B gradient started at 1% B for 3 min after injection and increased linearly to 100% B at 20 min, then 100% B for 5 min, and down to 1% B for 3 min using a flow rate of 0.5 ml/min. Mass spectrometer parameters were as follows: spray voltage, 3.5 kV; capillary temperature, 380°C; probe heater temperature, 400°C; 60, sheath flow rate; 20, auxiliary flow rate, and one spare gas; S-lens RF level, 50; resolution, 240,000; and AGC target, 3 × 106. The instrument was calibrated weekly with positive and negative ion calibration solutions (Thermo Fisher Scientific). Each sample was analyzed in negative and positive ionization modes using an m/z range of 100–800. Data were collected using Thermo Fisher Scientific Xcalibur software version 4.1.31.9 and quantified via integration in Excalibur Quan Browser version 4.1.31.9.
Feature detection and characterization
LC-MS RAW files from biospecimens were converted to mzXML format (centroid mode) using MSconvert v3.0.20315-7da487568 (ProteoWizard), followed by analysis using the XCMS analysis feature in Metaboseek v0.9.7 (http://metaboseek.com) (Helf et al., 2022) based on the centWave XCMS algorithm to extract features (Tautenhahn et al., 2008). Peak detection values were set as follows: 4 ppm, 3–20 peakwidth, 3 snthresh, 3 and 100 prefilter, FALSE fitgauss, 1 integrate, TRUE firstBaselineCheck, 0 noise, wMean mzCenterFun, and −0.005 mzdiff. XCMS feature grouping values were set as follows: 0.2 minfrac, 2 bw, 0.002 mzwid, 500 max, 1 minsamp, and FALSE usegroup. Metaboseek peak filling values were set as follows: 5 ppm_m, 5 rtw, and TRUE rtrange. The resulting tables of all detected features were then processed with the Metaboseek data explorer. To select differential features, we applied a filter that only retained entries with peak area ratios <0.2 (down in IBD patients) or >5 (up in IBD patients), with a retention time window of 1–20 min, >20,000 Maxint, and >0.95 peak quality as calculated by Metaboseek. We manually curated the resulting list to remove false positive entries (i.e., features that upon manual inspection of raw data were not differential). For verified differential features, we examined elution profiles, isotope patterns, and MS1 spectra to find molecular ions and remove adducts, fragments, and isotope peaks. The remaining masses were put on the inclusion list for MS/MS (ddMS2) characterization. Positive and negative ionization mode data were processed separately. To acquire MS2 spectra, we ran a top 10 data-dependent MS2 method on a Thermo Q-exactive-HF mass spectrometer with MS1 resolution, 60,000; AGC target, 1 × 106; maximum injection time (IT), 50 ms; MS2 resolution, 45,000; AGC target, 5 × 105; maximum IT, 80 ms; isolation window, 1.0 m/z; stepped normalized collision energy, 10 and 30 for positive ion mode and 40 and 60 for negative ion mode; and dynamic exclusion, 3 s.
Bile acids analysis
The identities of free bile acids, taurocholic acid (TCA), and glycocholic acid (GCA) were confirmed by coinjection with authentic standards (Schymanski annotation level one, according to previously published standards [Schymanski et al., 2014] as previously described [Arifuzzaman et al., 2022]). A methanol stock solution of standard bile acids was prepared and diluted at five different concentrations for the construction of calibration curves. The resulting mixtures and sample extracts were analyzed using the LC-MS conditions described above. For the quantification of bile acids in fecal samples, the amounts were normalized based on the weight of the lyophilized samples prior to extraction.
Statistical tests were performed with GraphPad Prism version 9.4.1 or R 3.6.3. P values of datasets were determined by unpaired, or paired (where applicable), two-tailed t test with a 95% confidence interval. Where appropriate, Mann–Whitney U test or one or two-way ANOVA followed by post-hoc tests were performed. P values for RNA-seq datasets were determined by Wilcoxon test. P values of <0.05 were considered to be significant. The entity n represents biologically independent samples and not technical replicates unless specified otherwise. Error bars depict the standard error of the mean (SEM).
Online supplemental material
Fig. S1 shows the intestinal microbiota composition of the control diet– and inulin fiber diet–fed mice, the gene and protein expression patterns of different cytokines in ILC2s, and immune profiling of the colons from naïve and DSS-treated mice. Fig. S2 shows data from a T cell transfer model of colitis, immune phenotyping of the colons from ILC2-depleted mice, IL-33 and ST2 expression in the colon, and IL-33–mediated activation of ILC2s in vitro. Fig. S3 shows chemical structures of unconjugated bile acids and the microbiota composition in IBD patient and non-IBD controls. Fig. S4 shows chemical structures of conjugated bile acids and their concentration in IBD patient and non-IBD controls. Fig. S5 shows bulk and scRNA-seq data from the colonic biopsies of in IBD patient and non-IBD controls.
Acknowledgments
We thank members of the Artis laboratory for discussion and critical reading of the manuscript. We thank all contributing members of the JRI IBD Live Cell Bank consortium, which is supported by the Jill Roberts Institute for Research in IBD, the Jill Roberts Center for IBD, Cure for IBD, the Rosanne H. Silbermann Foundation, the Sanders Family, and WCM Division of Pediatric Gastroenterology and Nutrition. We thank Bristol Meyers Squibb and Celsius Therapeutics for their help with bulk and scRNA-seq of human samples, respectively.
This work was supported by the Crohn’s & Colitis Foundation (851136 to M. Arifuzzaman, 937437 to H. Yano, 901000 to W. Zhang), AGA Research Foundation, WCM Research Assistance for Primary Parents Initiative, The. W.M. Keck Foundation (all to C.-J. Guo), the Howard Hughes Medical Institute (to F.C. Schroeder), CURE for IBD, the Jill Roberts Institute for Research in IBD, Kenneth Rainin Foundation, the Sanders Family Foundation, Rosanne H. Silbermann Foundation, Linda and Glenn Greenberg, the Allen Discovery Center Program, a Paul G. Allen Frontiers Group advised program of the Paul G. Allen Family Foundation (all to D. Artis), and the National Institutes of Health (K99AI173660 to M. Arifuzzaman, DP2 HD101401-01 to C.-J. Guo, R35 GM131877 to F.C. Schroeder, and DK126871, AI151599, AI095466, AI095608, AR070116, AI172027, DK132244 to D. Artis).
Author contributions: M. Arifuzzaman carried out most of the experiments and analyzed the data. T.H. Won and F.C. Schroeder performed human and mouse metabolomic data generation and analysis. T.-T. Li, W.-B. Jin, and C.-J. Guo helped with LC-MS of human and mouse fecal samples. H. Yano, E. Hu, J. Uddin, S. Kashyap, and E.R. Emanuel helped with various experiments with mice. J. Uddin and A. Grier performed RNA-seq data analyses. The JRI Live Cell Bank Consortia contributed to human sample acquisition, processing, and storage. M. Arifuzzaman and D. Artis conceived the project, analyzed data, and wrote the manuscript with input from all coauthors.
Footnotes
JRI Live Cell Bank members: David Artis, Randy Longman, Gregory F. Sonnenberg, Ellen Scherl, Robbyn Sockolow, Dana Lukin, Vinita Jacob, Laura Sahyoun, Michael Mintz, Lasha Gogokhia, Thomas Ciecierega, Aliza Solomon, Arielle Bergman, Kimberley Chein, Elliott Gordon, Michelle Ramos, Kenny Joselin Castro Ochoa, Victoria Ribeiro de Godoy, Adriana Brcic-Susak, Seun Oguntunmibi, Dario Garone, and Caitlin Mason.
Contributor Information
JRI Live Cell Bank:
David Artis, Randy Longman, Gregory F. Sonnenberg, Ellen Scherl, Robbyn Sockolow, Dana Lukin, Vinita Jacob, Laura Sahyoun, Michael Mintz, Lasha Gogokhia, Thomas Ciecierega, Aliza Solomon, Arielle Bergman, Kimberley Chein, Elliott Gordon, Michelle Ramos, Kenny Joselin Castro Ochoa, Victoria Ribeiro de Godoy, Adriana Brcic-Susak, Seun Oguntunmibi, Dario Garone, and Caitlin Mason
Data availability
All data necessary to understand and evaluate the conclusions of this paper are provided in this published article, the accompanying source data, or the supplemental material. The 16S rRNA-seq data are available at the NCBI Sequence Read Archive under accession number BioProject PRJNA1074072. The mouse ILC2 RNA-seq data are available at Gene Expression Omnibus (GEO) under accession number GSE183443. The human RNA-seq and scRNA-seq data are available at GEO under accession numbers GSE255720 and GSE255243, respectively. The MS1/MS2 data for mouse and human samples analyzed in this study are available at the GNPS website (http://massive.ucsd.edu) under MassIVE ID numbers MSV000086890 and MSV000092648, respectively.
Data availability
All data necessary to understand and evaluate the conclusions of this paper are provided in this published article, the accompanying source data, or the supplemental material. The 16S rRNA-seq data are available at the NCBI Sequence Read Archive under accession number BioProject PRJNA1074072. The mouse ILC2 RNA-seq data are available at Gene Expression Omnibus (GEO) under accession number GSE183443. The human RNA-seq and scRNA-seq data are available at GEO under accession numbers GSE255720 and GSE255243, respectively. The MS1/MS2 data for mouse and human samples analyzed in this study are available at the GNPS website (http://massive.ucsd.edu) under MassIVE ID numbers MSV000086890 and MSV000092648, respectively.
References
- Ahrens, R., Waddell A., Seidu L., Blanchard C., Carey R., Forbes E., Lampinen M., Wilson T., Cohen E., Stringer K., et al. 2008. Intestinal macrophage/epithelial cell-derived CCL11/eotaxin-1 mediates eosinophil recruitment and function in pediatric ulcerative colitis. J. Immunol. 181:7390–7399. 10.4049/jimmunol.181.10.7390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, M., and Turnbaugh P.J.. 2020. Deconstructing mechanisms of diet-microbiome-immune interactions. Immunity. 53:264–276. 10.1016/j.immuni.2020.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthakrishnan, A.N. 2015. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 12:205–217. 10.1038/nrgastro.2015.34 [DOI] [PubMed] [Google Scholar]
- Arifuzzaman, M., Won T.H., Li T.T., Yano H., Digumarthi S., Heras A.F., Zhang W., Parkhurst C.N., Kashyap S., Jin W.B., et al. 2022. Inulin fibre promotes microbiota-derived bile acids and type 2 inflammation. Nature. 611:578–584. 10.1038/s41586-022-05380-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, H.K., Bording-Jorgensen M., Santer D.M., Zhang Z., Valcheva R., Rieger A.M., Sung-Ho Kim J., Dijk S.I., Mahmood R., Ogungbola O., et al. 2022. Unfermented beta-fructan fibers fuel inflammation in select inflammatory bowel disease patients. Gastroenterology. 164:228–240. 10.1053/j.gastro.2022.09.034 [DOI] [PubMed] [Google Scholar]
- Blander, J.M., Longman R.S., Iliev I.D., Sonnenberg G.F., and Artis D.. 2017. Regulation of inflammation by microbiota interactions with the host. Nat. Immunol. 18:851–860. 10.1038/ni.3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen, E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37:852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., and Holmes S.P.. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 13:581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrol, C., and Girard J.P.. 2018. Interleukin-33 (IL-33): A nuclear cytokine from the IL-1 family. Immunol. Rev. 281:154–168. 10.1111/imr.12619 [DOI] [PubMed] [Google Scholar]
- Chambers, E.S., Byrne C.S., Morrison D.J., Murphy K.G., Preston T., Tedford C., Garcia-Perez I., Fountana S., Serrano-Contreras J.I., Holmes E., et al. 2019. Dietary supplementation with inulin-propionate ester or inulin improves insulin sensitivity in adults with overweight and obesity with distinct effects on the gut microbiota, plasma metabolome and systemic inflammatory responses: A randomised cross-over trial. Gut. 68:1430–1438. 10.1136/gutjnl-2019-318424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, W.B., Yoon J., Bartemes K.R., Iijima K., and Kita H.. 2008. A novel IL-1 family cytokine, IL-33, potently activates human eosinophils. J. Allergy Clin. Immunol. 121:1484–1490. 10.1016/j.jaci.2008.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chijiiwa, R., Hosokawa M., Kogawa M., Nishikawa Y., Ide K., Sakanashi C., Takahashi K., and Takeyama H.. 2020. Single-cell genomics of uncultured bacteria reveals dietary fiber responders in the mouse gut microbiota. Microbiome. 8:5. 10.1186/s40168-019-0779-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, A.B., Lee D., Long M.D., Kappelman M.D., Martin C.F., Sandler R.S., and Lewis J.D.. 2013. Dietary patterns and self-reported associations of diet with symptoms of inflammatory bowel disease. Dig. Dis. Sci. 58:1322–1328. 10.1007/s10620-012-2373-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso, P., Chatzou M., Floden E.W., Barja P.P., Palumbo E., and Notredame C.. 2017. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35:316–319. 10.1038/nbt.3820 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt, M., Roehr J.T., Ahmed R., and Dieterich C.. 2012. FLEXBAR-flexible barcode and adapter processing for next-generation sequencing platforms. Biology. 1:895–905. 10.3390/biology1030895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbo, M., Crinier A., Vély F., and Vivier E.. 2017. Innate lymphoid cells: Major players in inflammatory diseases. Nat. Rev. Immunol. 17:665–678. 10.1038/nri.2017.86 [DOI] [PubMed] [Google Scholar]
- Eberl, G., Colonna M., Di Santo J.P., and McKenzie A.N.. 2015. Innate lymphoid cells. Innate lymphoid cells: A new paradigm in immunology. Science. 348:aaa6566. 10.1126/science.aaa6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels, P.A., Peltzer A., Fillinger S., Patel H., Alneberg J., Wilm A., Garcia M.U., Di Tommaso P., and Nahnsen S.. 2020. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38:276–278. 10.1038/s41587-020-0439-x [DOI] [PubMed] [Google Scholar]
- Flamar, A.L., Klose C.S.N., Moeller J.B., Mahlakõiv T., Bessman N.J., Zhang W., Moriyama S., Stokic-Trtica V., Rankin L.C., Putzel G.G., et al. 2020. Interleukin-33 induces the enzyme tryptophan hydroxylase 1 to promote inflammatory group 2 innate lymphoid cell-mediated immunity. Immunity. 52:606–619.e6. 10.1016/j.immuni.2020.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes, A., Escher J., Hébuterne X., Kłęk S., Krznaric Z., Schneider S., Shamir R., Stardelova K., Wierdsma N., Wiskin A.E., and Bischoff S.C.. 2017. ESPEN guideline: Clinical nutrition in inflammatory bowel disease. Clin. Nutr. 36:321–347. 10.1016/j.clnu.2016.12.027 [DOI] [PubMed] [Google Scholar]
- Forbes, E., Murase T., Yang M., Matthaei K.I., Lee J.J., Lee N.A., Foster P.S., and Hogan S.P.. 2004. Immunopathogenesis of experimental ulcerative colitis is mediated by eosinophil peroxidase. J. Immunol. 172:5664–5675. 10.4049/jimmunol.172.9.5664 [DOI] [PubMed] [Google Scholar]
- Franzosa, E.A., Sirota-Madi A., Avila-Pacheco J., Fornelos N., Haiser H.J., Reinker S., Vatanen T., Hall A.B., Mallick H., McIver L.J., et al. 2019. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 4:293–305. 10.1038/s41564-018-0306-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich, M., Pohin M., Jackson M.A., Korsunsky I., Bullers S.J., Rue-Albrecht K., Christoforidou Z., Sathananthan D., Thomas T., Ravindran R., et al. 2021. IL-1-driven stromal-neutrophil interactions define a subset of patients with inflammatory bowel disease that does not respond to therapies. Nat. Med. 27:1970–1981. 10.1038/s41591-021-01520-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich, M., Pohin M., and Powrie F.. 2019. Cytokine networks in the pathophysiology of inflammatory bowel disease. Immunity. 50:992–1006. 10.1016/j.immuni.2019.03.017 [DOI] [PubMed] [Google Scholar]
- Furusawa, Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., et al. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 504:446–450. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- Goc, J., Lv M., Bessman N.J., Flamar A.L., Sahota S., Suzuki H., Teng F., Putzel G.G., Eberl G., Withers D.R., et al. 2021. Dysregulation of ILC3s unleashes progression and immunotherapy resistance in colon cancer. Cell. 184:5015–5030.e16. 10.1016/j.cell.2021.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, H., Takemura N., Ogasawara T., Sasajima N., Watanabe J., Ito H., Morita T., and Sonoyama K.. 2010. Effects of fructo-oligosaccharide on DSS-induced colitis differ in mice fed nonpurified and purified diets. J. Nutr. 140:2121–2127. 10.3945/jn.110.125948 [DOI] [PubMed] [Google Scholar]
- Halim, T.Y., Krauss R.H., Sun A.C., and Takei F.. 2012. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity. 36:451–463. 10.1016/j.immuni.2011.12.020 [DOI] [PubMed] [Google Scholar]
- Han, H., Roan F., Johnston L.K., Smith D.E., Bryce P.J., and Ziegler S.F.. 2018. IL-33 promotes gastrointestinal allergy in a TSLP-independent manner. Mucosal Immunol. 11:394–403. 10.1038/mi.2017.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helf, M.J., Fox B.W., Artyukhin A.B., Zhang Y.K., and Schroeder F.C.. 2022. Comparative metabolomics with Metaboseek reveals functions of a conserved fat metabolism pathway in C. elegans. Nat. Commun. 13:782. 10.1038/s41467-022-28391-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertati, A., Hayashi S., Ogawa Y., Yamamoto T., and Kadowaki M.. 2020. Interleukin-4 receptor α subunit deficiency alleviates murine intestinal inflammation in vivo through the enhancement of intestinal mucosal barrier function. Front. Pharmacol. 11:573470. 10.3389/fphar.2020.573470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, C.L., Neilsen C.V., and Bryce P.J.. 2010. IL-33 is produced by mast cells and regulates IgE-dependent inflammation. PLoS One. 5:e11944. 10.1371/journal.pone.0011944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Guo L., Qiu J., Chen X., Hu-Li J., Siebenlist U., Williamson P.R., Urban J.F. Jr., and Paul W.E.. 2015. IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential “inflammatory” type 2 innate lymphoid cells. Nat. Immunol. 16:161–169. 10.1038/ni.3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarick, K.J., Topczewska P.M., Jakob M.O., Yano H., Arifuzzaman M., Gao X., Boulekou S., Stokic-Trtica V., Leclère P.S., Preußer A., et al. 2022. Non-redundant functions of group 2 innate lymphoid cells. Nature. 611:794–800. 10.1038/s41586-022-05395-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, L.K., and Bryce P.J.. 2017. Understanding interleukin 33 and its roles in eosinophil development. Front. Med. 4:51. 10.3389/fmed.2017.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, L.K., Hsu C.L., Krier-Burris R.A., Chhiba K.D., Chien K.B., McKenzie A., Berdnikovs S., and Bryce P.J.. 2016. IL-33 precedes IL-5 in regulating eosinophil commitment and is required for eosinophil homeostasis. J. Immunol. 197:3445–3453. 10.4049/jimmunol.1600611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpievitch, Y.V., Dabney A.R., and Smith R.D.. 2012. Normalization and missing value imputation for label-free LC-MS analysis. BMC Bioinformatics. 13:S5. 10.1186/1471-2105-13-S16-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B.S., Wojno E.D., and Artis D.. 2013. Innate lymphoid cells and allergic inflammation. Curr. Opin. Immunol. 25:738–744. 10.1016/j.coi.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen, J., Chen H.H., Parikh K., Antanaviciute A., Jagielowicz M., Fawkner-Corbett D., Ashley N., Cubitt L., Mellado-Gomez E., Attar M., et al. 2018. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell. 175:372–386.e17. 10.1016/j.cell.2018.08.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, C.S., and Artis D.. 2016. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 17:765–774. 10.1038/ni.3489 [DOI] [PubMed] [Google Scholar]
- Klose, C.S.N., and Artis D.. 2020. Innate lymphoid cells control signaling circuits to regulate tissue-specific immunity. Cell Res. 30:475–491. 10.1038/s41422-020-0323-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, C.S.N., Mahlakõiv T., Moeller J.B., Rankin L.C., Flamar A.L., Kabata H., Monticelli L.A., Moriyama S., Putzel G.G., Rakhilin N., et al. 2017. The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature. 549:282–286. 10.1038/nature23676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, A.H., and Dixit V.D.. 2020. Dietary regulation of immunity. Immunity. 53:510–523. 10.1016/j.immuni.2020.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppkes, M., Becker C., Ivanov I.I., Hirth S., Wirtz S., Neufert C., Pouly S., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., et al. 2009. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 136:257–267. 10.1053/j.gastro.2008.10.018 [DOI] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2019. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47:e47. 10.1093/nar/gkz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopetuso, L.R., De Salvo C., Pastorelli L., Rana N., Senkfor H.N., Petito V., Di Martino L., Scaldaferri F., Gasbarrini A., Cominelli F., et al. 2018. IL-33 promotes recovery from acute colitis by inducing miR-320 to stimulate epithelial restitution and repair. Proc. Natl. Acad. Sci. USA. 115:E9362–E9370. 10.1073/pnas.1803613115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia, L., Tan J., Vieira A.T., Leach K., Stanley D., Luong S., Maruya M., Ian McKenzie C., Hijikata A., Wong C., et al. 2015. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 6:6734. 10.1038/ncomms7734 [DOI] [PubMed] [Google Scholar]
- Martin, M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 17:10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Maslowski, K.M., Vieira A.T., Ng A., Kranich J., Sierro F., Yu D., Schilter H.C., Rolph M.S., Mackay F., Artis D., et al. 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 461:1282–1286. 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarville, J.L., Chen G.Y., Cuevas V.D., Troha K., and Ayres J.S.. 2020. Microbiota metabolites in health and disease. Annu. Rev. Immunol. 38:147–170. 10.1146/annurev-immunol-071219-125715 [DOI] [PubMed] [Google Scholar]
- Miles, J.P., Zou J., Kumar M.V., Pellizzon M., Ulman E., Ricci M., Gewirtz A.T., and Chassaing B.. 2017. Supplementation of low- and high-fat diets with fermentable fiber exacerbates severity of DSS-induced acute colitis. Inflamm. Bowel Dis. 23:1133–1143. 10.1097/MIB.0000000000001155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli, L.A., Osborne L.C., Noti M., Tran S.V., Zaiss D.M., and Artis D.. 2015. IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc. Natl. Acad. Sci. USA. 112:10762–10767. 10.1073/pnas.1509070112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli, L.A., Sonnenberg G.F., Abt M.C., Alenghat T., Ziegler C.G., Doering T.A., Angelosanto J.M., Laidlaw B.J., Yang C.Y., Sathaliyawala T., et al. 2011. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12:1045–1054. 10.1038/ni.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli, L.A., Sonnenberg G.F., and Artis D.. 2012. Innate lymphoid cells: Critical regulators of allergic inflammation and tissue repair in the lung. Curr. Opin. Immunol. 24:284–289. 10.1016/j.coi.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima, H., Mahlakõiv T., Shih H.Y., Davis F.P., Meylan F., Huang Y., Harrison O.J., Yao C., Mikami Y., Urban J.F. Jr., et al. 2019. Neuropeptide CGRP limits group 2 innate lymphoid cell responses and constrains type 2 inflammation. Immunity. 51:682–695.e6. 10.1016/j.immuni.2019.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill, D.R., Wong S.H., Bellosi A., Flynn R.J., Daly M., Langford T.K., Bucks C., Kane C.M., Fallon P.G., Pannell R., et al. 2010. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 464:1367–1370. 10.1038/nature08900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo Thi Phuong, N., Palmieri V., Adamczyk A., Klopfleisch R., Langhorst J., Hansen W., Westendorf A.M., and Pastille E.. 2021. IL-33 drives expansion of type 2 innate lymphoid cells and regulatory T cells and protects mice from severe, acute colitis. Front. Immunol. 12:669787. 10.3389/fimmu.2021.669787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobs, S.P., Zmora N., and Elinav E.. 2020. Nutrition regulates innate immunity in health and disease. Annu. Rev. Nutr. 40:189–219. 10.1146/annurev-nutr-120919-094440 [DOI] [PubMed] [Google Scholar]
- Nussbaum, J.C., Van Dyken S.J., von Moltke J., Cheng L.E., Mohapatra A., Molofsky A.B., Thornton E.E., Krummel M.F., Chawla A., Liang H.E., and Locksley R.M.. 2013. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 502:245–248. 10.1038/nature12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh, K., Antanaviciute A., Fawkner-Corbett D., Jagielowicz M., Aulicino A., Lagerholm C., Davis S., Kinchen J., Chen H.H., Alham N.K., et al. 2019. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 567:49–55. 10.1038/s41586-019-0992-y [DOI] [PubMed] [Google Scholar]
- Pushparaj, P.N., Li D., Komai-Koma M., Guabiraba R., Alexander J., McSharry C., and Xu D.. 2013. Interleukin-33 exacerbates acute colitis via interleukin-4 in mice. Immunology. 140:70–77. 10.1111/imm.12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, X., Qi C., Li X., Fang D., and Fang M.. 2020. IL-33 deficiency protects mice from DSS-induced experimental colitis by suppressing ILC2 and Th17 cell responses. Inflamm. Res. 69:1111–1122. 10.1007/s00011-020-01384-4 [DOI] [PubMed] [Google Scholar]
- Quast, C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., and Glöckner F.O.. 2013. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab, Y., Fredens K., Gerdin B., and Hällgren R.. 1998. Eosinophil activation in ulcerative colitis: Studies on mucosal release and localization of eosinophil granule constituents. Dig. Dis. Sci. 43:1061–1070. 10.1023/A:1018843104511 [DOI] [PubMed] [Google Scholar]
- Ricardo-Gonzalez, R.R., Molofsky A.B., and Locksley R.M.. 2022. ILC2s - development, divergence, dispersal. Curr. Opin. Immunol. 75:102168. 10.1016/j.coi.2022.102168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks, M.G., and Garrett W.S.. 2016. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16:341–352. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz, S.A., Siracusa M.C., Perrigoue J.G., Spencer S.P., Urban J.F. Jr., Tocker J.E., Budelsky A.L., Kleinschek M.A., Kastelein R.A., Kambayashi T., et al. 2010. IL25 elicits a multipotent progenitor cell population that promotes T(H)2 cytokine responses. Nature. 464:1362–1366. 10.1038/nature08901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi, M., Barlow J.L., Saunders S.P., Xue L., Gutowska-Owsiak D., Wang X., Huang L.C., Johnson D., Scanlon S.T., McKenzie A.N., et al. 2013. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J. Exp. Med. 210:2939–2950. 10.1084/jem.20130351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiering, C., Krausgruber T., Chomka A., Fröhlich A., Adelmann K., Wohlfert E.A., Pott J., Griseri T., Bollrath J., Hegazy A.N., et al. 2014. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 513:564–568. 10.1038/nature13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymanski, E.L., Jeon J., Gulde R., Fenner K., Ruff M., Singer H.P., and Hollender J.. 2014. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 48:2097–2098. 10.1021/es5002105 [DOI] [PubMed] [Google Scholar]
- Sedhom, M.A., Pichery M., Murdoch J.R., Foligné B., Ortega N., Normand S., Mertz K., Sanmugalingam D., Brault L., Grandjean T., et al. 2013. Neutralisation of the interleukin-33/ST2 pathway ameliorates experimental colitis through enhancement of mucosal healing in mice. Gut. 62:1714–1723. 10.1136/gutjnl-2011-301785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira, A.L.M., Ferreira A.V.M., de Oliveira M.C., Rachid M.A., da Cunha Sousa L.F., Dos Santos Martins F., Gomes-Santos A.C., Vieira A.T., and Teixeira M.M.. 2017. Preventive rather than therapeutic treatment with high fiber diet attenuates clinical and inflammatory markers of acute and chronic DSS-induced colitis in mice. Eur. J. Nutr. 56:179–191. 10.1007/s00394-015-1068-x [DOI] [PubMed] [Google Scholar]
- Sinal, C.J., Tohkin M., Miyata M., Ward J.M., Lambert G., and Gonzalez F.J.. 2000. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 102:731–744. 10.1016/S0092-8674(00)00062-3 [DOI] [PubMed] [Google Scholar]
- Smillie, C.S., Biton M., Ordovas-Montanes J., Sullivan K.M., Burgin G., Graham D.B., Herbst R.H., Rogel N., Slyper M., Waldman J., et al. 2019. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 178:714–730.e22. 10.1016/j.cell.2019.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, P.M., Howitt M.R., Panikov N., Michaud M., Gallini C.A., Bohlooly-Y M., Glickman J.N., and Garrett W.S.. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 341:569–573. 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevceva, L., Pavli P., Husband A., Ramsay A., and Doe W.F.. 2001. Dextran sulphate sodium-induced colitis is ameliorated in interleukin 4 deficient mice. Genes Immun. 2:309–316. 10.1038/sj.gene.6363782 [DOI] [PubMed] [Google Scholar]
- Straub, D., Blackwell N., Langarica-Fuentes A., Peltzer A., Nahnsen S., and Kleindienst S.. 2020. Interpretations of environmental microbial community studies are biased by the selected 16S rRNA (gene) amplicon sequencing pipeline. Front. Microbiol. 11:550420. 10.3389/fmicb.2020.550420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn, R., Böttcher C., and Neumann S.. 2008. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics. 9:504. 10.1186/1471-2105-9-504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakare, R., Alamoudi J.A., Gautam N., Rodrigues A.D., and Alnouti Y.. 2018. Species differences in bile acids I. Plasma and urine bile acid composition. J. Appl. Toxicol. 38:1323–1335. 10.1002/jat.3644 [DOI] [PubMed] [Google Scholar]
- Tsou, A.M., Yano H., Parkhurst C.N., Mahlakõiv T., Chu C., Zhang W., He Z., Jarick K.J., Zhong C., Putzel G.G., et al. 2022. Neuropeptide regulation of non-redundant ILC2 responses at barrier surfaces. Nature. 611:787–793. 10.1038/s41586-022-05297-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier, E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. 2018. Innate lymphoid cells: 10 Years on. Cell. 174:1054–1066. 10.1016/j.cell.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Xu, H., Ding J., Porter C.B.M., Wallrapp A., Tabaka M., Ma S., Fu S., Guo X., Riesenfeld S.J., Su C., et al. 2019. Transcriptional Atlas of intestinal immune cells reveals that neuropeptide α-CGRP modulates group 2 innate lymphoid cell responses. Immunity. 51:696–708.e9. 10.1016/j.immuni.2019.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C., Cantor A.B., Yang H., Browne C., Wells R.A., Fujiwara Y., and Orkin S.H.. 2002. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J. Exp. Med. 195:1387–1395. 10.1084/jem.20020656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Lyu M., Bessman N.J., Xie Z., Arifuzzaman M., Yano H., Parkhurst C.N., Chu C., Zhou L., Putzel G.G., et al. 2022. Gut-innervating nociceptors regulate the intestinal microbiota to promote tissue protection. Cell. 185:4170–4189.e20. 10.1016/j.cell.2022.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, D., Liwinski T., and Elinav E.. 2020. Interaction between microbiota and immunity in health and disease. Cell Res. 30:492–506. 10.1038/s41422-020-0332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data necessary to understand and evaluate the conclusions of this paper are provided in this published article, the accompanying source data, or the supplemental material. The 16S rRNA-seq data are available at the NCBI Sequence Read Archive under accession number BioProject PRJNA1074072. The mouse ILC2 RNA-seq data are available at Gene Expression Omnibus (GEO) under accession number GSE183443. The human RNA-seq and scRNA-seq data are available at GEO under accession numbers GSE255720 and GSE255243, respectively. The MS1/MS2 data for mouse and human samples analyzed in this study are available at the GNPS website (http://massive.ucsd.edu) under MassIVE ID numbers MSV000086890 and MSV000092648, respectively.
All data necessary to understand and evaluate the conclusions of this paper are provided in this published article, the accompanying source data, or the supplemental material. The 16S rRNA-seq data are available at the NCBI Sequence Read Archive under accession number BioProject PRJNA1074072. The mouse ILC2 RNA-seq data are available at Gene Expression Omnibus (GEO) under accession number GSE183443. The human RNA-seq and scRNA-seq data are available at GEO under accession numbers GSE255720 and GSE255243, respectively. The MS1/MS2 data for mouse and human samples analyzed in this study are available at the GNPS website (http://massive.ucsd.edu) under MassIVE ID numbers MSV000086890 and MSV000092648, respectively.