Abstract
Background and aims
Cardiomyocyte hypertrophy and interstitial fibrosis are key components of myocardial remodeling in Heart Failure (HF) with preserved (HFpEF) or reduced ejection fraction (HFrEF). MicroRNAs (miRNAs) are non-coding, evolutionarily conserved RNA molecules that may offer novel insights into myocardial remodeling. This study aimed to characterize miRNA expression in HFpEF (LVEF 45%) and HFrEF (LVEF < 45%) and its association with myocardial remodeling.
Methods
Prospectively enrolled symptomatic HF patients (HFpEF:n = 36; HFrEF:n = 31) and controls (n = 23) underwent cardiac magnetic resonance imaging with T1-mapping and circulating miRNA expression (OpenArray system).
Results
13 of 188 miRNAs were differentially expressed between HF groups (11 downregulated in HFpEF). Myocardial extracellular volume (ECV) was increased in both HF groups (HFpEF 30 ± 5%; HFrEF 30 ± 3%; controls 26 ± 2%, p < 0.001). miR-128a-3p, linked to cardiac hypertrophy, fibrosis, and dysfunction, correlated positively with ECV in HFpEF (r = 0.60, p = 0.01) and negatively in HFrEF (r = 0.51, p = 0.04). miR-423-5p overexpression, previously associated HF mortality, was inversely associated with LVEF (r = 0.29, p = 0.04) and intracellular water lifetime (ic) (r = 0.45, p < 0.05) in both HF groups, and with NT-proBNP in HFpEF (r = 0.63, p < 0.01).
Conclusions
miRNA expression profiles differed between HF phenotypes. The differential expression and association of miR-128a-3p with ECV may reflect the distinct vascular, interstitial, and cellular etiologies of HF phenotypes.
Keywords: Heart failure, Biomarkers, MicroRNA, Interstitial fibrosis, Myocardial remodeling
1. Introduction
Heart failure (HF) is a clinical syndrome characterized by changes in cardiac muscle structure and function [1] which affects more than 60 million people worldwide and is strongly associated with high morbidity and mortality rates [2]. While several therapeutic interventions can improve symptoms and prognosis in HF with reduced ejection fraction (HFrEF) [3,4], only sodium-glucose co-transporter-2 (SGLT2) inhibitors have proven effective in reducing cardiovascular events in both HFrEF [5,6] and HF with preserved ejection fraction (HFpEF) [7]. Left ventricular (LV) hypertrophy and increased interstitial myocardial fibrosis are key pathophysiological features of HF that may offer valuable information to improve our current understanding of HFpEF, enabling its better characterization and differentiation from HFrEF [8,9].
MicroRNAs (miRNAs) are a group of small non-coding, evolutionarily conserved RNA molecules (22 nucleotides) that resemble messenger RNA (mRNA) sequences of target genes and cause the repression of protein transcription [10]. Although the HFrEF and HFpEF phenotypes share certain similarities, differences regarding clinical characteristics and response to medical therapy support the exploratory analysis of miRNAs as a complementary approach to better understand the existing singularities of HFrEF and HFpEF [11] in terms of myocardial tissue characteristics. Cardiac magnetic resonance (CMR) T1 mapping techniques [9,12] allow for the simultaneous quantification of two central components of myocardial remodeling among HF patients: the myocardial extracellular volume fraction (ECV) [13,14] which is a biomarker of interstitial fibrosis, and the intracellular water-lifetime (), a novel marker of cardiomyocyte diameter [12].
This prospective study aimed to investigate the expression of circulating miRNAs in symptomatic patients with HFrEF and with HFpEF, and to establish the correlation of such miRNAs with markers of global LV and myocardial remodeling, such as and ECV, in both HF phenotypes.
2. Methods
2.1. Subject selection and clinical assessment
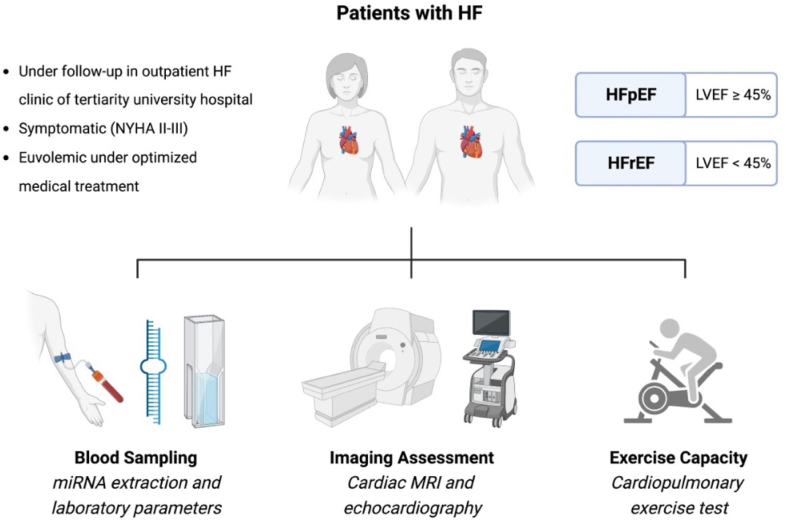
We prospectively enrolled 67 patients with symptomatic HF (NYHA class ≥ II, 36 with HFpEF, and thirty-one with HFrEF) from an outpatient HF clinic of a tertiary university hospital (Clinical Hospital of the State University of Campinas, UNICAMP, Campinas, São Paulo, Brazil) from February/2017 until February/2020. All patients with HF fulfilled the established Framingham criteria [15] as well as the recently published universal definition [16] for HF diagnosis. All enrolled HF patients had a documented medical history of hospitalization and/or an urgent visit to the emergency department related to HF within 12 months. They were carefully evaluated to ensure euvolemia and were receiving optimal medical management for both HF [17] and any accompanying comorbidities. Based on a transthoracic echocardiogram performed at the time of enrollment, patients were classified as having either HFrEF (left ventricular ejection fraction [LVEF] < 45%) or HFpEF (LVEF ≥ 45%). Patients with Chagas disease, hypertrophic cardiomyopathy, cardiac amyloidosis, or any sign of infiltrative and/or storage heart disease were excluded. In addition, patients with any preexisting genetic disorder involving cardiac involvement were also excluded. Detailed inclusion and exclusion criteria are displayed in Supplemental Table 1 (Supplemental Table 1, online-only Data Supplement), which highlights that only symptomatic HF patients with exercise limitation and dyspnea (NYHA ≥ II), as well as no contraindication to CMR or cardiopulmonary exercise testing (CPET), were eligible to participate. The H2FPEF score, a recently validated diagnostic score for HFpEF based on six clinical and echocardiographic criteria, was used to assess the likelihood of a HFpEF diagnosis [18]. All recruited patients, blinded to the patient group and to any clinical information relevant for sub-group classification, underwent the following procedures: CPET, CMR, and laboratory analyses, including high-sensitivity troponin T, NT-proBNP, and miRNAs (Fig. 1). Laboratory analyses were performed using blood samples obtained after 12-h fasting immediately before CMR scanning and comprised hematocrit, renal function, total cholesterol and fractions, glucose, and glycosylated hemoglobin in addition to high-sensitivity troponin T, NT-proBNP, and miRNAs. Laboratory parameters and biomarkers were quantified using standard methodology from commercially available supplies. A group of healthy volunteers was studied for comparison and were also subjected to laboratory analyses, echocardiogram, CPET, and CMR.
Fig. 1.
Study design including all procedures performed. Briefly, HF patients with reduced and preserved EF under optimized medical therapy were prospectively enrolled and subject to imaging, biomarkers, and functional assessment.
The current study was conducted in accordance with the Helsinki Declaration. The Institutional Review Board at UNICAMP approved the current protocol (CAA: 63097016.5.2011.5404), and all participants provided written informed consent.
2.2. Echocardiogram
Standard and tissue Doppler measurements were conducted in accordance with the current American Society of Echocardiography (ASE) guidelines [19] using a 3.5 MHz transducer (Vivid-S70, GE Healthcare, Milwaukee, Wisconsin, USA). The E/A ratio was acquired on the apical axis of four chambers by pulsed Doppler. Tissue Doppler myocardial velocities were obtained in early diastole (E’) at the lateral and septal aspects of the mitral annulus in the apical four-chamber view. Two-dimensional myocardial deformation (strain 2D) was acquired in apical axial images at four-, three-, and two-chamber view, to obtain the global longitudinal strain (GLS) (%).
2.3. cardiac magnetic resonance
CMR was performed using a 3 T system (Achieva, Philips Medical Systems, Best, the Netherlands) with a 6-element phased-array surface coil and image acquisition gated to the ECG. The CMR protocol included morphology and function assessment by parallel-receiver-accelerated cine SSFP imaging (8-mm thickness, repetition time: 3.4 ms, echo time: 1.2 ms, and in-plane spatial resolution 1.5 mm, SENSE factor = 2), scar assessment by phase-sensitive inversion-recovery-prepared (PSIR) gradient-echo imaging 10 min after a cumulative gadolinium dose of 0.2 mmol/kg (Gadoterate-meglumine; Dotarem, Guerbet, Aulnay-sous-Bois, France), and Modified Look-Locker Imaging (MOLLI) for T1 mapping before and after contrast administration for native T1, ECV, and [12]. Late gadolinium enhancement (LGE) was quantified as a percentage of total LV mass using a 5SD criterion. MOLLI T1 mapping for ECV and was performed in a mid-ventricular LV short-axis slice using the following parameters: field of view 240 mm; echo time 1.7 ms; repetition time 2.1 ms; flip angle 50°; 256 × 196 matrix; 8-mm slice, acceleration factor 2. The segmental T1* in 6 LV segments was determined by a nonlinear least-squares fitting to an analytic expression for the inversion recovery. T1 was obtained from T1* by correction for the effects of the image-readouts during the inversion recovery. Then, myocardial R1 (R1myocardial) in each wall segment was fit as a function of blood R1 (R1blood) with a 2-compartment model of transcytolemmal water exchange to determine ECV and , as previously described [12]. Segments with LGE were excluded from the quantification of mean LV ECV and . The MASS CMR software (Mass Research, Leiden University Medical Center, Leiden, the Netherlands, version 2022-EXP) was used for all imaging analyses.
2.4. Cardiopulmonary exercise testing
CPET was performed with breath-to-breath technique at rest and throughout the exercise period using a respiratory gas analysis interface with a validated cycle ergometer (Med Graphics, St. Paul, MN, USA), as previously described [20]. Oxygen (VO2) and carbon dioxide (VCO2) uptake, as well as the respiratory exchange ratio, were calculated. Peak VO2 was defined as the highest consumption of O2, obtained by averaging data from the last 30 s of peak effort [21]. VO2 was measured at the anaerobic threshold using the V-slope method. In addition, oxygen pulse (VO2/heart rate), ventilatory efficiency (VE/VCO2), oscillatory exercise (EOV, a recognized prognostic marker in HF), and aerobic efficiency (VO2/watt, a marker of efficiency and cardiac metabolism) were assessed.
2.5. RNA isolation and circulating miRNA expression analysis using OpenArray
miRNA extraction was performed using the miRneasy serum/plasma kit (Qiagen, Valencia, CA, USA). A NanoDrop ND-2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) was used to measure miRNA quantity and quality [22]. For the miRNA analysis, the TaqMan OpenArray Human System was employed, which is based on quantitative real-time PCR (qRT-PCR) and contains 754 miRNAs on a microfluidic platform. Assessment of both transcription and complementary DNA synthesis reaction (cDNA) was performed using a reagent mix containing two primer pools A and B for each sample (Life Technologies, Carlsbad, CA, USA). The miRNA panels were loaded using the AccuFill system and the qRT-PCR was run on the QuantStudio 12 K Flex platform (Applied Biosystems, Waltham, MA USA). Data analyses were performed using the Expression Suite Software (version 1.0.4; Thermo Fisher Scientific, Waltham, MA, USA), and data were excluded from analyses with an amplification score <1.2 and a cycle threshold (Ct) value > 35. For the differential analysis, only miRNAs that were expressed in at least 50% of the participants within each study group were considered. Global normalization [23] was used as recommended by the manufacturer (Thermo Fisher Scientific, Wilmington, DE, USA).
2.6. Gene set enrichment analysis
For gene enrichment analyses, the miRWalk 2.0 database was used [24]. mRNAs predicted in at least four of the five tools (miRDB, miRWalk, RNA22, miRanda and TargetScan) were considered as possible miRNA targets. To understand the biological relevance of differentially expressed miRNAs, we used the Database for Annotation, Visualization, and Integrated Discovery (DAVID), Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis to predict potential target genes and determine the enriched pathways. Only pathways with >1.5-fold enrichment and p-values ≤0.05 were considered.
2.7. Statistical analysis
Continuous variables are summarized as mean ± standard deviation (SD) or median with interquartile range (IQR), and categorical variables as proportions. Variables were compared using Mann-Whitney, T-test, or Chi-square test as appropriate. The differences in miRNA expression among the groups with a p-value <0.05 were considered significant. Only miRNAs expressed in at least 50% of the cohort were taken into consideration. Correlations between the expression of circulating miRNAs (log-transformed), imaging data (echocardiogram and CMR), and NT-proBNP were calculated using Spearman's correlation. Mann-Whitney testing was employed to compare the expressions of miRNAs between groups. All analyses were performed with SPSS (IBM SPSS Statistics 2.0 Inc. Chicago, Illinois, USA) and the R environment (version 4.1.2, https://www.R-project.org/).
3. Results
3.1. Patients’ clinical and laboratory characteristics
Patients’ clinical and demographic characteristics are shown in Table 1. HFpEF patients were mostly female (58% vs. 48%; p = 0.569) and older (60.81 ± 11.92 years vs. 53.32 ± 11.10years, p = 0.010) and presented higher systolic blood pressure (144.18 ± 20.51 mmHg vs. 127.74 ± 20.12 mmHg, p = 0.002) compared to HFrEF patients. With regard to laboratory parameters, HFpEF patients presented higher median glycated hemoglobin (HFpEF: 7.0 mg/dL [IQR 6.0 to 8.0] vs. HFrEF: 6.0 mg/dL [IQR 5.0 to 7.0], p = 0.044, Table 1), while HFrEF patients had greater NT-proBNP values (HFpEF: 198.0 ng/L [IQR 50.0 to 680.0] vs. HFrEF: 660.0 ng/L [IQR 132.0 to 2415.0], p = 0.011, Table 1). All recruited patients were symptomatic (NYHA ≥ II) despite optimal medical treatment for HF and for comorbidities. As expected, and in accordance with the latest guideline recommendations [17], most HFrEF patients were under prognosis-modifying HF therapy (β-blockers [100%], angiotensin-converting enzyme inhibitor (ACEi) or angiotensin receptor blocker or sacubitril-valsartan [100%], spironolactone [74.2%] and diuretics [100%]).
Table 1.
Patients’ clinical and demographic characteristics.
| Variables | All patients N = 67 | HFpEF N = 36 | HFrEF N = 31 | p-value (HFpEF vs. HFrEF) |
|---|---|---|---|---|
| Female, % (n) | 53.7% (36) | 58.33% (21) | 48.39% (15) | 0.569 |
| Age, years | 57.34 ± 12.01 | 60.81 ± 11.92 | 53.32 ± 11.10 | 0.010 |
| Body mass index, kg/m2 | 30.70 ± 5.53 | 30.92 ± 5.60 | 30.45 ± 5.53 | 0.731 |
| Hemodynamic Data: | ||||
| Resting HR, bpm | 65.23 ± 13.07 | 65.09 ± 9.89 | 65.52 ± 16.18 | 0.897 |
| Systolic blood pressure, mmHg | 136.22 ± 21.79 | 144.18 ± 20.51 | 127.74 ± 20.12 | 0.002 |
| Diastolic blood pressure, mmHg | 84.91 ± 13.43 | 88.91 ± 12.11 | 80.65 ± 13.64 | 0.013 |
| NYHA functional class | ||||
| Class II, % (n) | 61.2% (41) | 52.8% (19) | 71.0% (22) | 0.203 |
| Class III, % (n) | 38.8% (26) | 47.2% (17) | 29.0% (9) | 0,203 |
| Comorbidities | ||||
| Hypertension, % (n) | 80.6% (54) | 86.1% (31) | 74.2% (23) | 0.203 |
| Diabetes, % (n) | 46.2% (31) | 55.5% (20) | 35.5% (11) | 0.162 |
| History of alcohol abuse, % (n) | 11.9% (8) | 5.6% (2) | 19.4% (6) | 0.174 |
| Dyslipidemia, % (n) | 73.2% (49) | 77.7% (28) | 67.7% (21) | 0.517 |
| Smoking, % | 22.4% (15) | 19.4% (7) | 25.8% (8) | 0.742 |
| Atrial fibrillation, % (n) | 26.9% (18) | 38.9% (14) | 12.9% (4) | 0.034 |
| Previous Myocardial Infarction, % | 8.9% (6) | 5.6% (2) | 12.9% (4) | 0.534 |
| Previous PCI or CABG, % | 10.4% (7) | 13.9% (5) | 6.5% (2) | 0.554 |
| HF Etiology | ||||
| Ischemic, % (n) | 14.9% (10) | 19.4% (7) | 9.7% (3) | 0.438 |
| Hypertensive, % (n) | 49.3% (33) | 61.1% (22) | 35.5% (11) | 0.065 |
| Idiopathic, % (n) | 35.8% (24) | 19.4% (7) | 54.8% (17) | 0.006 |
| Laboratory parameters | ||||
| Hemoglobin, mg/dL | 13.80 ± 1.11 | 13.68 ± 1.18 | 13.94 ± 1.28 | 0.357 |
| Hematocrit, mg/dL | 41.03 ± 2.80 | 40.82 ± 2.95 | 41.28 ± 2.63 | 0.516 |
| Urea, mg/dL | 38 (31–46) | 39 (32–45) | 38 (30–46) | 0.608 |
| Creatinine, mg/dL | 0.99 ± 0.31 | 0.95 ± 0.32 | 1.03 ± 0.29 | 0.304 |
| Glycated hemoglobin, mg/dL | 6 (5–8) | 7 (6–8) | 6 (5–7) | 0.044 |
| Glucose, mg/dL | 107 (93–160) | 113 (98–171) | 101 (60–124) | 0.247 |
| LDL cholesterol, mg/dL | 92.55 ± 38.81 | 90.87 ± 39.49 | 94.40 ± 38.63 | 0.722 |
| HDL cholesterol, mg/dL | 41.31 ± 11.83 | 41.47 ± 10.48 | 41.13 ± 13.38 | 0.912 |
| Triglycerides, mg/dL | 139 (91–211) | 141 (110–217) | 133 (89–194) | 0.353 |
| Biomarkers | ||||
| Hight-sensitivity troponin T, ng/dL | 10 (6–18) | 10 (6–16) | 13 (6–19) | 0.648 |
| NT-proBNP, pg/ml | 293 (84–1710) | 198 (50–680) | 660 (132–2415) | 0.011 |
| Heart Failure Pharmacotherapy | ||||
| ACEi, % (n) | 25.4% (17) | 33.3% (12) | 16.1% (5) | 0.183 |
| Angiotensin receptor blocker, % (n) | 43.3% (29) | 52.8% (19) | 32.3% (10) | 0.150 |
| ACEi/ARB, % (n) | 68.7% (46) | 86.1% (31) | 48.4% (15) | 0.002 |
| Sacubitril/Valsartan, % (n) | 25.4% (17) | 2.8% (1) | 51.6% (16) | <0.001 |
| ACEi/ARB/Sacubitril/Valsartan, %(n) | 94.0% (63) | 88.9% (32) | 100% (31) | 0.162 |
| Beta-blockers, % (n) | 94.0% (63) | 88.9% (32) | 100% (31) | 0.162 |
| Spironolactone, % (n) | 59.7% (40) | 47.2% (17) | 74.2% (23) | 0.046 |
| Diuretics, % (n) | 89.5% (60) | 80.5% (29) | 100% (31) | 0.028 |
| Amiodarone, % (n) | 4.5% (3) | 2.8% (1) | 6.5% (2) | 0.894 |
| Aspirin, % (n) | 27.3% (18) | 31.4% (11) | 22.6% (7) | 0.647 |
| Warfarin, % (n) | 23.9% (16) | 30.6% (11) | 16.1% (5) | 0.274 |
| Calcium channel blocker, % (n) | 32.8% (22) | 52.8% (19) | 9.7% (3) | <0.001 |
| Statin, % (n) | 73.2% (49) | 77.8% (28) | 67.7% (21) | 0.517 |
Variables are summarized as mean ± SD if normally distributed, median (IQR) if not normally distributed, of as number (percentage) for categorical variables. NYHA: New York Heart Association functional classification; HFpEF: heart failure preserved ejection fraction; HFrEF: heart failure reduced ejection fraction; NT-proBNP: brain natriuretic peptide. ACEi,- angiotensin-converting enzyme inhibitor; ARB- angiotensin receptor blocker.
3.2. LV parameters assessed by echocardiogram
The HF groups differed importantly in cardiac remodeling patterns as assessed by echocardiogram, with HFpEF patients presenting a smaller mean LV volume index than HFrEF patients. The left ventricular end-diastolic volume (LVEDV) to LV mass ratio was significantly higher in patients with HFpEF (HFpEF: 2.18 ± 0.62 vs. HFrEF: 1.49 ± 0.40, p = 0.034, Table 2), confirming that these patients had a more concentrically shaped LV's than those with HFrEF. As expected, the HFrEF group had a lower mean LVEF (HFpEF: 58.41% ± 9.32 vs. HFrEF: 34.08% ± 9.72, p < 0.001, Table 2) and a markedly reduced GLS (HFpEF: −14.99% ± 5.37 vs. HFrEF: −8.84% ± 3.63, p < 0.001, Table 2). Mean GLS in HFpEF patients was lower compared to both previously reported healthy controls [25] and the prospectively recruited group of healthy controls (Supplemental Table 2, online-only Data Supplement). In addition, HFpEF patients showed an elevated early diastolic transmitral flow velocity (E) to early diastolic mitral annular tissue velocity (e’) ratio (HFpEF: 12.73 ± 5.80 vs. HFrEF: 16.56 ± 5.77, p = 0.010), which confirmed the presence of a higher LV filling pressure in the HFpEF group. The recently validated H2FPEF score resulted in overall concordance with the HFpEF diagnosis (63.63% ± 25.67; 5.0 points [IQR 3.0 to 6.0]), corroborating the diagnosis by standard clinical criteria [18].
Table 2.
Echocardiographic imaging parameters.
| Variables | All patients N = 67 | HFpEF N = 36 | HFrEF N = 31 | p-value (HFpEF vs. HFrEF) |
|---|---|---|---|---|
| LV mass index, g/m2 | 131.85 ± 58.24 | 107.51 ± 36.21 | 157.76 ± 66.19 | <0.001 |
| Relative wall thickness | 0.31 ± 0.07 | 0.34 ± 0.05 | 0.27 ± 0.06 | <0.001 |
| LVDV/LV mass, ratio | 1.85 ± 0.63 | 2.18 ± 0.62 | 1.49 ± 0.40 | 0.034 |
| LA volume index, ml | 41.18 ± 16.52 | 39.6 ± 11.2 | 42.74 ± 16.60 | 0.389 |
| LV diastolic volume, ml | 146.24 ± 94.41 | 92.91 ± 35.41 | 206.46 ± 104.05 | <0.001 |
| LV systolic volume, ml | 87.92 ± 81.44 | 40.09 ± 21.73 | 141.91 ± 90.30 | <0.001 |
| LV ejection fraction, % | 46.98 ± 15.45 | 58.41 ± 9.32 | 34.08 ± 9.72 | <0.001 |
| Global longitudinal strain, % | −12.10 ± 5.54 | −14.99 ± 5.37 | −8.84 ± 3.63 | <0.001 |
| E/A, ratio | 1.34 ± 0.84 | 1.35 ± 0.83 | 1.32 ± 0.86 | 0.913 |
| E/e', ratio | 14.50 ± 6.05 | 12.73 ± 5.80 | 16.56 ± 5.77 | 0.010 |
Variables are summarized as mean ± SD if normally distributed, as median [IQR] if not normally distributed, or as number (percentage) for categorical variables. LA: left atrium; LV: left ventricle; DV: diastolic volume; E/A. e’- early diastole.
3.3. LV parameters assessed by CMR imaging
CMR confirmed that HFrEF patients had more dilated ventricles (LVEDV index: HFpEF: 75.30 ± 28.01, ml/m2 vs. HFrEF: 152.54 ± 73.87 ml/m2, p < 0.001, Table 3) and a greater LV mass index (HFpEF: 68.65 ± 25.73 g/m2 vs. 85.59 ± 40.62 g/m2, p = 0.047, Table 3) compared to HFpEF patients. The LV mass/LVEDV ratio was indicative of a concentric geometric hypertrophy pattern in HFpEF patients. HFrEF patients were more likely to present focal myocardial fibrosis revealed by LGE (HFpEF: 44.4% vs. HFrEF: 74.0%, p = 0.013, Table 3) with an atypical pattern (HFpEF: 30.5% vs. HFrEF: 61.3%, p = 0.023, Table 3). Native T1 and ECV were similar between the HF groups (native T1: HFpEF 1252.63 ± 55.59 ms vs. HFrEF: 1263.81 ± 72.51 ms, p = 0.533, Table 3; ECV: HFpEF 0.30 ± 0.05 vs. HFrEF 0.30 ± 0.03, p = 0.617, Table 3). , a surrogate of cardiomyocyte diameter, was significantly higher in HFpEF patients compared to HFrEF patients (HFpEF: 0.15 ± 0.05 s vs. HFrEF: 0.11 ± 0.04 s, p = 0.004, Table 3).
Table 3.
Cardiac magnetic resonance imaging parameters.
| Variables | All patients N = 67 | HFpEF N = 36 | HFrEF N = 31 | p-value (HFpEF vs. HFrEF) |
|---|---|---|---|---|
| LVDV index, ml/m2 | 112.14 ± 66.89 | 75.30 ± 28.01 | 152.54 ± 73.87 | <0.001 |
| LVSV index, ml/m2 | 70.28 ± 63.64 | 31.11 ± 19.05 | 113.24 ± 67.78 | <0.001 |
| LV ejection fraction, % | 44.94 ± 18.80 | 60.25 ± 10.89 | 28.15 ± 8.18 | <0.001 |
| LV mass index, g/m2 | 76.73 ± 34.46 | 68.65 ± 25.73 | 85.59 ± 40.62 | 0.047 |
| LV mass/LV DV, g/ml | 0.75 ± 0.26 | 0.90 ± 0.25 | 0.58 ± 0.16 | <0.001 |
| LA volume index, ml/m2 | 39.64 ± 20.16 | 37.00 ± 19.24 | 42.56 ± 21.06 | 0.315 |
| Presence of LGE, % (n) | 58.2% (39) | 44.4% (16) | 74.0% (23) | 0.013 |
| LGE with atypical pattern for CAD, % (n) | 44.8% (30) | 30.5% (11) | 61.3% (19) | 0.023 |
| LGE with typical pattern for CAD, % (n) | 8.9% (6) | 8.4% (3) | 9.7% (3) | 0.847 |
| Intracellular lifetime of water, s | 0.13 ± 0.05 | 0.15 ± 0.05 | 0.11 ± 0.04 | 0.004 |
| Native T1, ms | 1257.81 ± 63.60 | 1252.63 ± 55.59 | 1263.81 ± 72.51 | 0.533 |
| ECV, % | 0.30 ± 0.04 | 0.30 ± 0.05 | 0.30 ± 0.03 | 0.617 |
| LGE mass, % per gram of tissue | 15.78 ± 11.21 | 11.68 ± 7.67 | 19.20 ± 12.69 | 0.017 |
Variables are summarized as mean ± SD if normally distributed, median (IQR) if not normally distributed, of as number (percentage) for categorical variables. LA: left atrium.
HF patients contrasted importantly with healthy controls in terms of laboratory, echocardiographic, CPET, and CMR parameters (Supplemental Table 2, online-only Data Supplement). Briefly, NT-proBNP levels, GLS, LVEF, LV mass, native T1 and ECV differed significantly between controls and HF patients, and was similarly elevated in both HF groups. As expected, the functional parameters assessed by CPET confirmed that both HF groups had markedly reduced functional capacity as compared to controls (Supplemental Table 2, online-only Data Supplement).
3.4. Circulating microRNA expression analysis
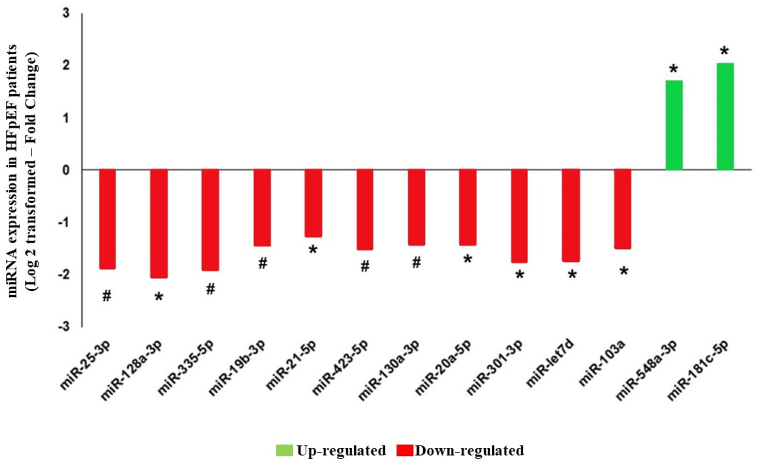
Among a total of 754 expressed miRNAs, one hundred and eighty-eight were detectable in all HF patients. Thirteen of these miRNAs were differentially expressed in HFpEF versus HFrEF: miR-181c-5p and miR-548a-3p had an increased expression, and miR-21- 5p, miR-20a-5p, miR-130a-3p, miR-103a-3p, miR-423-5p, miR-19b-3p, miR-301-3p, let-7d-5p, miR-335-5p, miR-128a-3p, and miR-25-3p had a reduced expression in HFpEF compared to HFrEF, as shown in Fig. 2.
Fig. 2.
Serum expression of microRNAs (miRNAs) in heart failure patients with preserved ejection fraction (HFpEF) compared with reduced ejection fraction (HFrEF) as a reference. Data are expressed as fold change (Log2 transformed). Positive values mean upregulation (green), and negative values mean downregulation (red). *p < 0.05; #p < 0.005.
3.5. Association of differentially expressed miRNAs with myocardial remodeling and functional capacity in the entire HF cohort
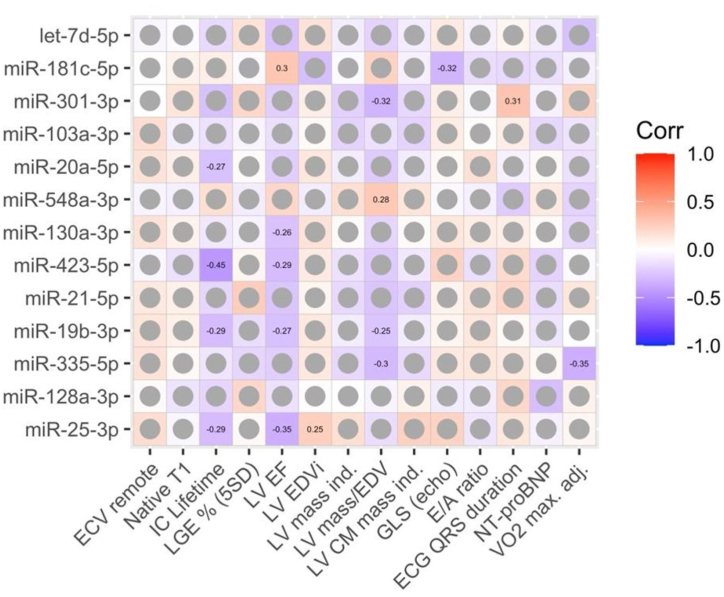
The association between the thirteen miRNAs differentially expressed between HF phenotypes among the entire HF cohort with echocardiographic, CMR, and laboratory parameters are shown in Fig. 5. LVEF was associated with the highest number of miRNAs, presenting a negative correlation with miR-130a-3p (r = −0.26, p = 0.039), miR-423-5p (r = −0.29, p = 0.040), miR-19b-3p (r = −0.27, p = 0.029), and miR-25-3p (r = −0.35, p = 0.005), in addition to a positive correlation with miR-181c-5p (r = 0.30, p = 0.048). miR-128a-3p correlated positively with ECV in HFpEF (r = 0.60, p = 0.01) and negatively in HFrEF (r = 0.51, p = 0.04). τic was inversely associated with four miRNAs (miR-20a-5p [r = −0.27, p = 0.047], miR-423-5p [r = −0.45, p = 0.003], miR-19b-3p [r = −0.29, p = 0.035], and miR-25-3p [r = −0.29, p = 0.035]). LV mass/EDV was negatively associated with other three miRNAs (miR-301-3p [r = −0.32, p = 0.022], miR-19b-3p [r = −0.25, p = 0.044], and miR-335-5p [r = −0.30, p = 0.025]) and positively with miR-548a-3p (r = 0.28, p = 0.025). Other parameters correlated only with one miRNA each: LV EDV index showed a direct correlation with miR-25-3p (r = 0.25, p = 0.047); GLS, an inverse correlation with miR-181c-5p (r = −0.32, p = 0.037); ECG QRS duration, a direct correlation with miR-301-3p (r = 0.31, p = 0.027); and peak VO2, adjusted for body mass, a negative correlation with miR-335-5p (r = −0.35, p = 0.012).
Fig. 5.
Association of miRNA expressions with clinical and laboratory features. The numeric values in cells represent spearman's correlation coefficients with significance levels <0.05. Non-significant correlations are shown as grey circles.
3.6. Circulating microRNA expression and NT-proBNP analysis
In the exploratory analysis considering the HF phenotypes separately (Supplemental Fig. 1, online-only Data Supplement), it is possible to observe that the HFpEF group presented a negative correlation between NT-proBNP and miR-103a-3p (r = −0.40, p = 0.048), miR-423-5p (r = −0.63, p = 0.001), miR-21-5p (r = −0.52, p = 0.002), miR-19b-3p (r = −0.35, p = 0.036), miR-25-3p (r = −0.49, p = 0.003), as well as a positive correlation with miR-548a-3p (r = 0.34, p = 0.041), while the HFrEF group showed only a negative association between NT-proBNP and miR-128a-3p (r = −0.69, p = 0.001).
3.7. Correlation between circulating miRNAs levels and structural and functional parameters by echocardiogram and CMR
With respect to echocardiographic parameters, GLS correlated negatively with miR-128a-3p (r = −0.63, p = 0.002), while LV mass index was negatively associated with miR-335-5p (r = −0.39, p = 0.046) and miR-301-3p (r = −0.51, p = 0.008) in the HFrEF group (Supplemental Fig. 1, online-only Data Supplement). LVEF, in turn, showed a direct correlation with miR-103a-3p (r = 0.58, p = 0.004) in the HFpEF group and with miR-128a-3p (r = 0.50, p = 0.02) in the HFrEF group.
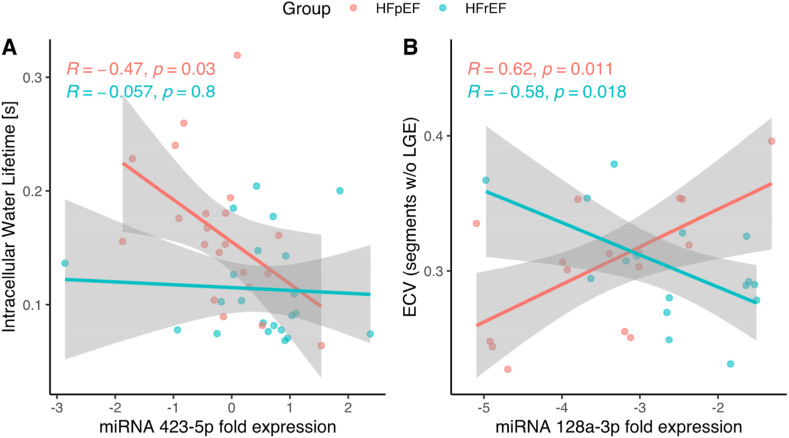
Regarding CMR parameters, τic was inversely associated with miR-423-5p (r = −0.49, p = 0.023) in the HFpEF group (Fig. 3A and Supplemental Fig. 1, online-only Data Supplement), and ECV showed a positive correlation in the HFpEF group (r = 0.60, p = 0.013) and an inverse correlation in the HFrEF group (r = −0.51, p = 0.043) with miR-128a-3p (Fig. 3B and Supplemental Fig. 1, online-only Data Supplement).
Fig. 3.
Correlation analysis of miRNA expression and Cardiac Magnetic Resonance in HF patients. A- Correlation of intracellular lifetime of water (seconds) and expression of miR-423-5p. B- Correlation of extracellular volume (ECV) with expression of miR-128a-3p. Values related to heart failure with preserved ejection fraction (HFpEF) and heart failure with reduced ejection fraction (HFrEF) patients are presented in orange and in green, respectively. (The continuous lines and shaded areas represent linear regression fits with confidence ranges for the HFrEF and HFpEF subgroups, respectively. Pearson's R′ and the p-values are shown in colors corresponding to the two groups).
Concerning CPET, peak VO2 adjusted for body mass was inversely associated with miR-335-5p (r = −0.56, p = 0.002) and with miR-548a-3p (r = −0.45, p = 0.01) and positively associated with miR-301-3p (r = 0.50, p = 0.01) in the HFpEF group, while only miR-128a-3p showed a positive correlation with peak VO2 adjusted for body mass (r = 0.46, p = 0.044) in the HFrEF group (Supplemental Fig. 1, online-only Data Supplement). Supplemental Fig. 2 highlights the correlations of miR-128a-3p and miR-485-5p with LV mass/EDV and LVEF.
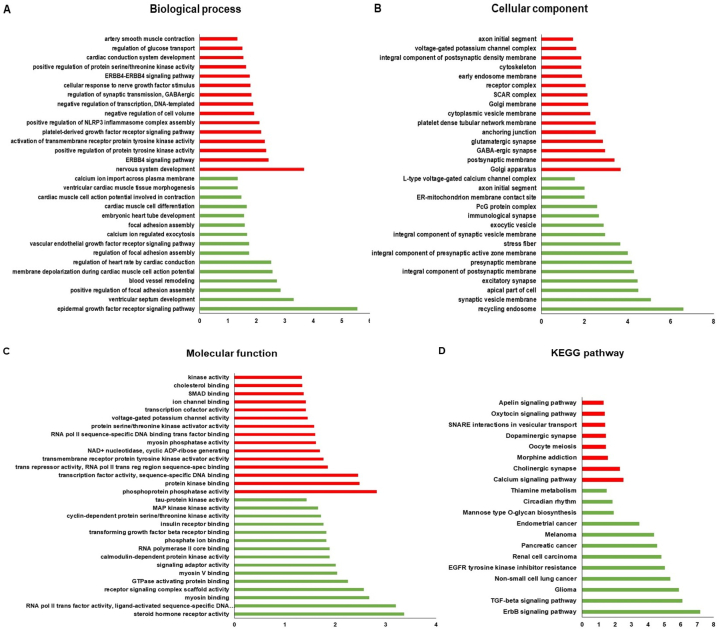
3.8. Target gene enrichment analyses
The miRwalk database was used for gene enrichment analyses, through which it was possible to demonstrate the pathways and percentages of genes involved with the differentially expressed miRNAs. As displayed in Fig. 4(A–D), the pathways involved with the thirteen miRNAs were found to be differentially expressed between the HFpEF and HFrEF groups. These pathways show enrichment of genes associated with cardiovascular development characterized in the database by terms such as “blood vessel remodeling,” “regulation of heart rate by cardiac conduction”, and “artery smooth muscle contraction”, as well as “protein kinase binding” and “tau-protein kinase activity” in molecular function. The cellular components are mainly composed of “recycling endosome”, “Golgi apparatus”, “L-type voltage-gated calcium channel complex”, and “voltage-gated potassium channel activity”, in addition to “ErbB signaling”, “TGF-beta signaling”, and “Calcium signaling” pathways on KEGG pathways.
Fig. 4.
Functional and enrichment analysis of upregulated (Red) and downregulated (Green) miRNA target gene pathways targeted by differentially expressed miRNAs in HF. The pathways were classified based on Biological Processes (A), Cellular Components (B), Molecular Function (C) and Enriched Genes in the analysis of the KEGG pathway (D). The abscissa is the value of −log 10 (p-value).
4. Discussion
miRNAs participate in important physiological and pathological processes in the human body, including those involved in cardiovascular diseases such as stroke, coronary artery disease, HF, and hypertension [26]. In the current study, we identified thirteen miRNAs that were differentially expressed in patients with HFpEF compared to those with HFrEF, two of which were up-regulated (miR-181c-5p and miR-548a-3p) and eleven were down-regulated (miR-21- 5p, miR-20a-5p, miR-130a-3p, miR-103a-3p, miR-423-5p, miR-19b-3p, miR-301-3p, let-7d-5p, miR-335-5p, miR-128a-3p, and miR-25-3p). Wong et al. [27] presented a study with the differential expression of miRNAs in HFpEF patients compared to HFrEF patients, and several of them were also differentially expressed in our study, corroborating the relevance of these miRNAs in HF.
The miRNAs expressed differentially in this study between the HF phenotypes correlated with key cardiac features on both echocardiogram and CMR. ECV, a validated non-invasive marker of extracellular matrix expansion and diffuse interstitial fibrosis [28,29] was significantly associated with four miRNAs (miR-335-5p, miR-20a-5p, miR-181c-5p, and miR-128a-3p), with associations of opposite sign in the case of miR-128a-3p, though ECV was similarly elevated in both HF groups, compared to controls. We also observed an increase in miR-181c-5p expression in patients with HFpEF compared to HFrEF, in addition to an inverse correlation between the levels of this miRNA and ECV in patients with HFrEF. Das et al. [30] demonstrated that miR-181a offers a protective myocardial response to oxidative stress, while an increase in miR-181c levels may have detrimental effects, indicating that members of the same family of miRNA have considerably different phenotypic effects on the cardiovascular system. Moreover, Wang et al. [31] have shown that overexpression of miR-181c-5p promotes cardiomyocyte injury and apoptosis by prompting hypoxia/reoxygenation, suggesting that increased levels of this miRNA can be used as a biomarker for risk assessment in cardiac diseases.
In our study, HFrEF patients showed a higher expression of miR-128a-3p compared to HFpEF patients, and such overexpression may influence cardiomyocyte size, myocardial fibrosis, and reduce LV systolic performance [32]. miR-128 is a miRNA that stimulates cardiomyocyte apoptosis during ischemia/reperfusion injury and has been found to be upregulated in both patients and animal models with HF [32]. In addition, there is evidence that miR-128 plays an important role in hypertension-induced myocardial injury, which can possibly explain the positive association between its expression and an increase in ECV among patients with HFpEF in which hypertension is not only prevalent but an essential comorbidity associated with myocardial remodeling and fibrosis [33]. Modulation of miR-128 has also been implicated in cardiomyocyte proliferation and vascular smooth muscle cell (VSMC) physiology [34]. Interestingly, miR-128 expression is reduced in sick/impaired vessels, and controlling miR-128 in animal models demonstrated promising results. In HFpEF, microvascular dysfunction consolidates an emergent pathophysiological mechanism and is associated with adverse clinical outcomes [35,36]. The positive association of ECV with miR-128a-3p among patients with HFpEF indicates that this pathway plays an essential role in the myocardial remodeling progression in this HF phenotype.
MiR-335-5p was downregulated in HFpEF patients. This miRNA had an inverse correlation with LV mass in the HFrEF group and a direct correlation with ECV in the HFpEF group. miR-335-5p is involved in cardiomyocyte differentiation, which can activate the Wnt and transforming growth factor (TGF-β) signaling pathways by increasing the expression of GATA4 and NKX2-5, as well as mesoderm, progenitor, and cardiac differentiation markers [37]. Another interesting cell culture study showed that overexpression of miR-335-5p attenuated cell growth as well as increased apoptosis in cells treated with 25 nM glucose and down-regulated the expression of the glucose transporter 4 gene SLC2A4 [38], supporting the notion that this miRNA is associated with myocardial remodeling.
The intracellular water lifetime (τic), a CMR marker of cardiomyocyte size and diameter [12], was significantly increased in patients with HFpEF. While LV mass index was increased in patients with HFrEF, the reduced τic indicates that cardiomyocytes had a smaller diameter, reflecting a more eccentric global remodeling and larger end-diastolic volume index seen in this phenotype compared to HFpEF. Furthermore, miR-423-5p expression was reduced in patients with HFpEF compared to those with HFrEF and inversely correlated with τic and NT-proBNP, suggesting an association between reduced expression of this miRNA and cardiomyocyte stretch in patients with HFpEF. More recently, Kanagalal et al. [39] suggested that while inflammatory/fibrotic/renal abnormalities were comparable in both HF phenotypes, markers of cardiomyocyte stretch/damage were more pronounced altered in HFrEF compared to HFpEF. In a recent meta-analysis including 967 HF patients, reduced expression levels of miR-423-5p were associated with increased mortality, suggesting that this miRNA may serve as a useful prognostic biomarker of HF [40]. In addition, Park et al. [41] observed reduced levels of miR-423-5p in patients with atrial fibrillation, while Luo et al. [42] demonstrated that overexpression of this miRNA could exert a cardioprotective effect against ischemia–reperfusion injury, corroborating the role of this miRNA in our study.
Analysis of the thirteen miRNAs differentially expressed between patients with HFpEF and HFrEF in the miRwalk database identified several regulatory pathways associated with cardiovascular development, such as “blood vessel remodeling”, “regulation of heart rate by cardiac conduction” and “artery smooth muscle contraction” in biological processes, “protein kinase binding” and “tau-protein kinase activity” in molecular function, in addition to “ErbB signaling pathway” and “calcium signaling pathway” in KEGG pathways.
Our prospective design, echocardiographic and CMR quantification, together with miRNA analysis by qRT-PCR in a TaqMan-based system, reinforce the importance of our study. However, we recognize that our study has several limitations. First, our study sample was limited. Second, as this was an exploratory study, it was not possible to adjust for multiple comparisons for all confounding factors, including different medical treatments and subgroups of interests according to imaging characteristics, to avoid type II errors. Third, we did not validate differentially expressed miRNAs in another cohort, and the results should be validated in larger, independent cohorts.
In conclusion, from the analysis of 754 miRNAs on the OpenArray system, we observed thirteen miRNAs expressed differentially in HFpEF patients compared to HFrEF patients, most of which have been previously shown to be involved in pathways that could help explain the differences between HF phenotypes. Correlation analysis of miRNA expression with several echocardiographic and CMR variables identified miRNAs related to myocardial structural characteristics, suggesting that these miRNAs are implicated in cardiac remodeling. Target gene enrichment analyses also indicated potential metabolic and signaling pathways associated with cardiac remodeling. Taken together, our results demonstrate an important association between the serum expression of miRNAs and the components of myocardial remodeling in patients with HF, suggesting potential targets for further studies and, ultimately, for therapeutic interventions.
Funding
The National Council for Scientific and Technological Development (CNPq) (grant number 303366/2015-0) to O.R.C.F.; and São Paulo Research Foundation (FAPESP) (grant numbers 2015/15402-2, 2016/26209-1, and 2017/03708-5) to O.R.C.F.
Grahic element of structured astract
Study design and summary of its main findings. In this prospective observational study, thirty-six patients with heart failure with preserved ejection fraction and thirty-one patients with heart failure with reduced ejection fraction were subjected to cardiac magnetic resonance (CMR) imaging, echocardiography, cardiopulm onary exercise testing (CPET), and laboratory analyses, including high-sensitivity troponin T, NT-proBNP, and microRNA (miRNA) expression profiling. Of the one hundred and eighty-eight miRNAs found in both HF phenotypes, thirteen were expressed differentially between the HF groups and correlated with clinical and imaging parameters associated with myocardial remodeling. Of note, miR-423-5p (overexpressed in HFrEF) was inversely associated with cardiomyocyte diameter (expressed by CMR intracellular lifetime of water) in the HFpEF group, and miR-128-a-3p (under-expressed in HFpEF) was directly associated with diffuse myocardial fibrosis (indicated by CMR extracellular volume fraction) in the HFpEF group. Abbreviations: HF heart failure, HFpEF heart failure with preserved ejection fraction, HFrEF heart failure with reduced ejection fraction, LVEF left ventricular ejection fraction, NYHA New York Heart Association, miRNA microRNA, MRI magnetic resonance imaging, corr correlation, ECV extracellular volume fraction, IC intracellular, LGE late gadolinium enhancement, 5SD five standard deviation, LVEDVi left ventricular end-diastolic volume index, ind index, LVCM left ventricular cardiomyocyte mass, GLS global longitudinal strain, E/A early to late diastolic transmitral flow velocity, VO2 oxygen uptake.
Data availability statement
The data underlying this article cannot be shared publicly due to ethical and legal restrictions on sharing sensitive patient information. The data will be shared on reasonable request to the corresponding author. The datasets generated and/or analyzed during the current study are available in the ArrayExpress repository; Access code: https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-13180?key=e5e0a577-810c-49d8-8c77-5ae918bf013b.
CRediT authorship contribution statement
Layde Rosane Paim: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Luis Miguel da Silva: Writing – review & editing, Writing – original draft, Formal analysis, Data curation. Lígia M. Antunes-Correa: Writing – review & editing, Writing – original draft, Methodology, Formal analysis. Vinicius Citelli Ribeiro: Writing – review & editing, Visualization, Validation, Methodology, Formal analysis. Roberto Schreiber: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation, Formal analysis, Conceptualization. Eduarda O.Z. Minin: Writing – review & editing, Methodology, Formal analysis. Larissa C.M. Bueno: Writing – review & editing, Methodology, Formal analysis. Elisangela C.P. Lopes: Writing – review & editing, Methodology, Formal analysis. Renan Yamaguti: Writing – review & editing, Methodology, Formal analysis. Andréa Coy-Canguçu: Writing – review & editing, Writing – original draft, Visualization, Methodology, Formal analysis. Sergio San Juan Dertkigil: Writing – review & editing, Visualization, Validation, Supervision, Methodology. Andrei Sposito: Writing – review & editing, Methodology, Formal analysis. Jose Roberto Matos-Souza: Writing – review & editing, Methodology, Formal analysis. Thiago Quinaglia: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation, Formal analysis, Conceptualization. Tomas G. Neilan: Writing – review & editing, Visualization, Methodology, Formal analysis. Licio A. Velloso: Writing – review & editing, Visualization, Validation, Methodology. Wilson Nadruz: Writing – review & editing, Validation, Supervision, Methodology, Formal analysis. Michael Jerosch-Herold: Writing – review & editing, Validation, Supervision, Methodology, Formal analysis. Otavio R. Coelho-Filho: Writing – review & editing, Writing – original draft, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Otavio R Coelho-Filho reports financial support was provided by State of Sao Paulo Research Foundation. Otavio R Coelho-Filho reports was provided by. Otavio R Coelho-Filho reports a relationship with AstraZeneca Pharmaceuticals LP that includes: funding grants and speaking and lecture fees. Otavio R Coelho-Filho reports a relationship with Amgen Inc that includes: funding grants. Otavio R Coelho-Filho reports a relationship with Pfizer Inc that includes: funding grants and speaking and lecture fees. Otavio R Coelho-Filho reports a relationship with Novartis AG that includes: speaking and lecture fees. Otavio R Coelho-Filho reports a relationship with Bayer AG that includes: speaking and lecture fees. Otavio R Coelho-Filho reports a relationship with Boehringer Ingelheim Ltd that includes: speaking and lecture fees. Otavio R Coelho-Filho reports a relationship with Takeda Pharmaceutical Company Limited that includes: funding grants and speaking and lecture fees. Tomas G Neilan reports a relationship with H3 Biomedicine Inc that includes: funding grants. Tomas G Neilan reports a relationship with Amgen Inc that includes: funding grants. Tomas G Neilan reports a relationship with AbbVie Inc that includes: funding grants. Tomas G Neilan reports a relationship with AstraZeneca that includes: funding grants. T.G.N. has received support from AstraZeneca, Bristol Myer Squibb, AbbVie, Amgen, and H3 Biomedicine. O.R.C.F. has received research grants and/or speaking honoraria from Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Novartis, Takeda, and Pfizer. The remaining authors declare no conflict of interest.
Acknowledgements
The authors would like to thank the CMR biomedical technologists for their efforts in the CMR scanning of the patients in this study.
Footnotes
The work described in the following manuscript was performed at the State University of Campinas Faculty of Medical Sciences, Campinas, Brazil.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27206.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.McMurray J.J., Pfeffer M.A. Heart failure. Lancet. 2005;365(9474):1877–1889. doi: 10.1016/S0140-6736(05)66621-4. [DOI] [PubMed] [Google Scholar]
- 2.Disease G.B.D., Injury I., Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1789–1858. doi: 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcondes-Braga F.G., et al. Emerging topics update of the Brazilian heart failure guideline - 2021. Arq. Bras. Cardiol. 2021;116(6):1174–1212. doi: 10.36660/abc.20210367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonagh T.A., et al. ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 2021;42(36):3599–3726. doi: 10.1093/eurheartj/ehab368. 2021. [DOI] [PubMed] [Google Scholar]
- 5.McMurray J.J.V., et al. Dapagliflozin in patients with heart failure and reduced ejection fraction. N. Engl. J. Med. 2019;381(21):1995–2008. doi: 10.1056/NEJMoa1911303. [DOI] [PubMed] [Google Scholar]
- 6.Packer M., et al. Effect of empagliflozin on worsening heart failure events in patients with heart failure and preserved ejection fraction: EMPEROR-preserved trial. Circulation. 2021;144(16):1284–1294. doi: 10.1161/CIRCULATIONAHA.121.056824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anker S.D., et al. Empagliflozin in heart failure with a preserved ejection fraction. N. Engl. J. Med. 2021;385(16):1451–1461. doi: 10.1056/NEJMoa2107038. [DOI] [PubMed] [Google Scholar]
- 8.van Heerebeek L., et al. Myocardial structure and function differ in systolic and diastolic heart failure. Circulation. 2006;113(16):1966–1973. doi: 10.1161/CIRCULATIONAHA.105.587519. [DOI] [PubMed] [Google Scholar]
- 9.Shah R.V., et al. Myocardial tissue remodeling in adolescent obesity. J. Am. Heart Assoc. 2013;2(4) doi: 10.1161/JAHA.113.000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammond S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitter D., Cotter G., Voors A.A. Clinical use of novel biomarkers in heart failure: towards personalized medicine. Heart Fail. Rev. 2014;19(3):369–381. doi: 10.1007/s10741-013-9396-5. [DOI] [PubMed] [Google Scholar]
- 12.Coelho-Filho O.R., et al. Quantification of cardiomyocyte hypertrophy by cardiac magnetic resonance: implications for early cardiac remodeling. Circulation. 2013;128(11):1225–1233. doi: 10.1161/CIRCULATIONAHA.112.000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flett A.S., et al. Equilibrium contrast cardiovascular magnetic resonance for the measurement of diffuse myocardial fibrosis: preliminary validation in humans. Circulation. 2010;122(2):138–144. doi: 10.1161/CIRCULATIONAHA.109.930636. [DOI] [PubMed] [Google Scholar]
- 14.White S.K., et al. T1 mapping for myocardial extracellular volume measurement by CMR: bolus only versus primed infusion technique. JACC Cardiovasc. Imag. 2013;6(9):955–962. doi: 10.1016/j.jcmg.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 15.McKee P.A., et al. The natural history of congestive heart failure: the Framingham study. N. Engl. J. Med. 1971;285(26):1441–1446. doi: 10.1056/NEJM197112232852601. [DOI] [PubMed] [Google Scholar]
- 16.Bozkurt B., et al. Universal definition and classification of heart failure: a report of the heart failure society of America, heart failure association of the European Society of cardiology, Japanese heart failure society and writing committee of the universal definition of heart failure. J. Card Fail. 2021 doi: 10.1016/j.cardfail.2021.01.022. [DOI] [PubMed] [Google Scholar]
- 17.McDonagh T.A., et al. ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 2021;42(36):3599–3726. doi: 10.1093/eurheartj/ehab368. 2021. [DOI] [PubMed] [Google Scholar]
- 18.Reddy Y.N.V., et al. A simple, evidence-based approach to help guide diagnosis of heart failure with preserved ejection fraction. Circulation. 2018;138(9):861–870. doi: 10.1161/CIRCULATIONAHA.118.034646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang R.M., et al. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J. Am. Soc. Echocardiogr. 2015;28(1):1–39.e14. doi: 10.1016/j.echo.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Corra U., et al. Role of cardiopulmonary exercise testing in clinical stratification in heart failure. A position paper from the committee on exercise physiology and training of the heart failure association of the European society of cardiology. Eur. J. Heart Fail. 2018;20(1):3–15. doi: 10.1002/ejhf.979. [DOI] [PubMed] [Google Scholar]
- 21.Lewis G.D., et al. Sildenafil improves exercise capacity and quality of life in patients with systolic heart failure and secondary pulmonary hypertension. Circulation. 2007;116(14):1555–1562. doi: 10.1161/CIRCULATIONAHA.107.716373. [DOI] [PubMed] [Google Scholar]
- 22.Li Y., Kowdley K.V. Method for microRNA isolation from clinical serum samples. Anal. Biochem. 2012;431(1):69–75. doi: 10.1016/j.ab.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mestdagh P., et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dweep H., et al. miRWalk--database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J. Biomed. Inf. 2011;44(5):839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Yingchoncharoen T., et al. Normal ranges of left ventricular strain: a meta-analysis. J. Am. Soc. Echocardiogr. 2013;26(2):185–191. doi: 10.1016/j.echo.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Rao P.K., et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ. Res. 2009;105(6):585–594. doi: 10.1161/CIRCRESAHA.109.200451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong L.L., et al. Combining circulating MicroRNA and NT-proBNP to detect and categorize heart failure subtypes. J. Am. Coll. Cardiol. 2019;73(11):1300–1313. doi: 10.1016/j.jacc.2018.11.060. [DOI] [PubMed] [Google Scholar]
- 28.Coelho-Filho O.R., et al. Role of transcytolemmal water-exchange in magnetic resonance measurements of diffuse myocardial fibrosis in hypertensive heart disease. Circ Cardiovasc Imaging. 2013;6(1):134–141. doi: 10.1161/CIRCIMAGING.112.979815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coelho-Filho O.R., et al. Cardiac magnetic resonance assessment of interstitial myocardial fibrosis and cardiomyocyte hypertrophy in hypertensive mice treated with spironolactone. J. Am. Heart Assoc. 2014;3(3) doi: 10.1161/JAHA.114.000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das S., et al. Divergent effects of miR-181 family members on myocardial function through protective cytosolic and detrimental mitochondrial microRNA targets. J. Am. Heart Assoc. 2017;6(3) doi: 10.1161/JAHA.116.004694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S., et al. MiR-181c-5p promotes inflammatory response during hypoxia/reoxygenation injury by downregulating protein tyrosine phosphatase nonreceptor type 4 in H9C2 cardiomyocytes. Oxid. Med. Cell. Longev. 2020;2020 doi: 10.1155/2020/7913418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz-Velasco A., et al. Targeting mir128-3p alleviates myocardial insulin resistance and prevents ischemia-induced heart failure. Elife. 2020;9 doi: 10.7554/eLife.54298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin J., et al. Role of miR-128 in hypertension-induced myocardial injury. Exp. Ther. Med. 2017;14(4):2751–2756. doi: 10.3892/etm.2017.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farina F.M., et al. miR-128-3p is a novel regulator of vascular smooth muscle cell phenotypic switch and vascular diseases. Circ. Res. 2020;126(12):e120–e135. doi: 10.1161/CIRCRESAHA.120.316489. [DOI] [PubMed] [Google Scholar]
- 35.Paulus W.J., Tschope C. A novel paradigm for heart failure with preserved ejection fraction: comorbidities drive myocardial dysfunction and remodeling through coronary microvascular endothelial inflammation. J. Am. Coll. Cardiol. 2013;62(4):263–271. doi: 10.1016/j.jacc.2013.02.092. [DOI] [PubMed] [Google Scholar]
- 36.Sinha A., et al. Untangling the pathophysiologic link between coronary microvascular dysfunction and heart failure with preserved ejection fraction. Eur. Heart J. 2021;42(43):4431–4441. doi: 10.1093/eurheartj/ehab653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kay M., et al. Hsa-miR-335 regulates cardiac mesoderm and progenitor cell differentiation. Stem Cell Res. Ther. 2019;10(1):191. doi: 10.1186/s13287-019-1249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G., Zhang L. miR-335-5p aggravates type 2 diabetes by inhibiting SLC2A4 expression. Biochem. Biophys. Res. Commun. 2021;558:71–78. doi: 10.1016/j.bbrc.2021.04.011. [DOI] [PubMed] [Google Scholar]
- 39.Kanagala P., et al. Characterizing heart failure with preserved and reduced ejection fraction: an imaging and plasma biomarker approach. PLoS One. 2020;15(4) doi: 10.1371/journal.pone.0232280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J., et al. Prognostic value of microRNAs in heart failure: a meta-analysis. Medicine (Baltim.) 2021;100(46) doi: 10.1097/MD.0000000000027744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park H., Park H., Park J. Circulating microRNA-423 attenuates the phosphorylation of calcium handling proteins in atrial fibrillation. Mol. Med. Rep. 2022;25(5) doi: 10.3892/mmr.2022.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo H., et al. microRNA-423-3p exosomes derived from cardiac fibroblasts mediates the cardioprotective effects of ischaemic post-conditioning. Cardiovasc. Res. 2019;115(7):1189–1204. doi: 10.1093/cvr/cvy231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly due to ethical and legal restrictions on sharing sensitive patient information. The data will be shared on reasonable request to the corresponding author. The datasets generated and/or analyzed during the current study are available in the ArrayExpress repository; Access code: https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-13180?key=e5e0a577-810c-49d8-8c77-5ae918bf013b.