Abstract
Chemokine receptor CXCR4 (also known as LESTR and fusin) has been shown to function as a coreceptor for T-cell-tropic strains of human immunodeficiency virus type 1 (HIV-1). We have developed a binding assay to show that HIV envelope (Env) can interact with CXCR4 independently of CD4 but that this binding is markedly enhanced by the previous interaction of Env with soluble CD4. We also show that nonglycosylated HIV-1SF-2 gp120 or sodium metaperiodate-treated oligomeric gp160 from HIV-1451 bound much more readily to CXCR4 than their counterparts with intact carbohydrate residues did.
In the recent past, several members of the family of chemokine receptors have been identified as cofactors for human immunodeficiency virus type 1 (HIV-1) entry (1, 6, 8, 10). Specifically, CCR5 (as well as CCR3 and CCR2b in some instances) has been shown to mediate entry of viruses characterized as macrophage tropic or dual tropic (1, 5–8), while CXCR4 has been shown to mediate entry of T-cell-tropic or dual-tropic strains (7, 10). While several ligands have been found for CCR5, CXC chemokine stromal derivative factor (SDF1) remains the only known ligand for CXCR4 (4, 24). Coimmunoprecipitation studies have shown that HIV-1 Env from T-cell-tropic strains forms a complex with CD4 and CXCR4 (18), but the nature of the binding events leading to the formation of this complex and the possibility of a direct interaction between HIV Env and CXCR4 remained speculative. Data from Hesselgesser et al. (15) have more recently shown that gp120 from the T-cell-tropic strains IIIB or BRU was able to compete with SDF1 for binding to CXCR4 in hNT cells (a neuronal CD4-negative cell line), indicating the possibility of a direct interaction between CXCR4 and gp120, but no information was presented on the relevance of the interaction with CD4. Other data have shown that gp120 from macrophage-tropic strains of HIV might be able to bind directly to CCR5 and that the affinity for binding between the two molecules can be increased significantly by the presence of soluble CD4 (sCD4) (34), although this effect could not be reproduced by a different group (32).
We have performed the following studies to determine if HIV Env binds to CXCR4 independently of CD4 and, if so, what would be the effect of previous binding of HIV Env to sCD4.
CD4-independent binding of HIV Env to CXCR4.
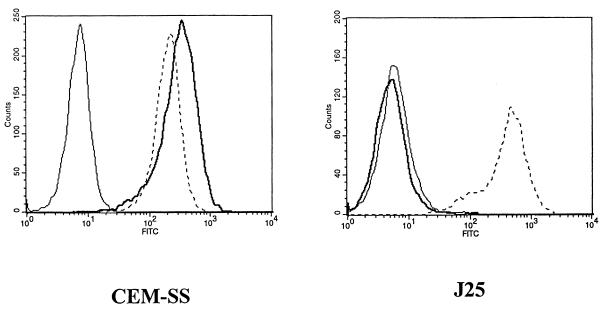
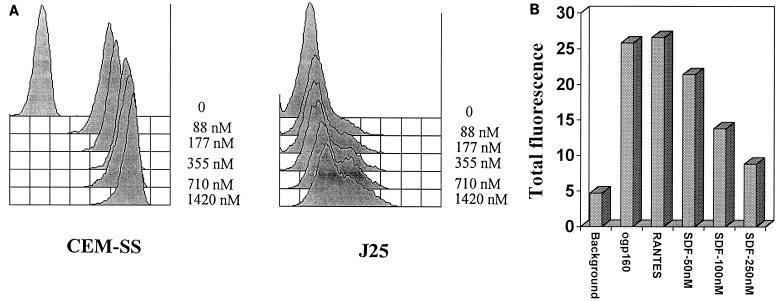
The phenotypes of the T-cell lines CEM-SS and Jurkat 25 (J25) were evaluated with respect to surface expression of both CD4 and CXCR4. J25 clone 22F6 cells (3, 21) were grown in complete medium (RPMI 1640, 2% penicillin-streptomycin, 2% l-glutamine; BioWhittaker, Walkersville, Md.) containing heat-inactivated 10% fetal calf serum at 37°C in a 5% CO2 atmosphere. CEM-SS is a T-cell line that was obtained from the AIDS Research and Reference Reagent Program and maintained in complete medium. CEM-SS cells were derived from a human lymphoblastoid tumor (22, 23). Commercial monoclonal antibody (MAb) to CD4 (mouse immunoglobulin G2a [IgG2a], clone S3.5), fluorescein isothiocyanate (FITC) labeled, and the necessary isotypic controls were obtained from Caltag Laboratories (San Francisco, Calif.). Mouse MAb 12G5 against CXCR4 was raised in BALB/c mice and has been described previously (9). Goat anti-mouse IgG–FITC was purchased from Becton Dickinson (San Jose, Calif.). Flow cytometric analysis was performed on a Becton Dickinson FACScan cytometer equipped with a 15-mW argon laser emitting at 488 nm. Dead cells were detected on the basis of their scatter and eliminated from the analysis. Live cells (10,000) were analyzed for each marker. CXCR4 surface expression was determined by washing the cells taken in logarithmic growth phase with phosphate-buffered saline (PBS) containing 1% horse serum and incubating them with 10 μl of 12G5 antibody/100 μl (0.16 mg/ml) at 4°C for 30 min. The cells were then washed again in PBS, and a secondary goat anti-mouse IgG–FITC (Becton Dickinson) was incubated with the cells for another 30 min at 4°C. Finally, the cells were washed with PBS and fixed with 2% paraformaldehyde. As a control, equal amounts of mouse IgG2a (the same isotype as 12G5) were used. Both cell lines expressed significant levels of CXCR4 on their surfaces (Fig. 1), but only CEM-SS had measurable levels of surface CD4. This characteristic of the phenotype of J25 cells, with respect to CD4 expression, has been reported before (3). To assess binding of HIV Env to CXCR4, the following binding assay was developed. Oligomeric gp160 (ogp160) was purified from cell cultures (obtained from T. C. Van Cott (Henry M. Jackson Foundation, Rockville, Md.) infected with HIV451 (17). The cells were washed once with PBS and then incubated with ogp160 for 1 h at 37°C in RPMI medium. The cells were washed again in PBS and incubated with 10 μg of human MAb 1331A [IgG3(λ)]/ml, which is specific for the C terminus of gp120 (i.e., amino acids 510 to 516 of HIVLAI), or with a human MAb against p24 (MAb 71-31) as a control (12) for 30 min at 4°C. The secondary antibody was a goat anti-human IgG phycoerythrin labeled (Caltag). The cells were fixed in 2% paraformaldehyde, and the fluorescence intensity was determined by flow cytometry. Background was obtained by adding MAb 1331 and goat anti-human IgG, phycoerythrin labeled, to the cells in the absence of ogp160. The results of the binding assay with ogp160 from HIV451 and both cell lines are shown in Fig. 2A. By using the high-affinity human MAb 1331A against the C-terminal region of gp120, our assay was able to detect significant binding of the ogp160 molecule to the surfaces of both cell lines even at concentrations of only 88 nM. The very high relative affinity of MAb 1331A for the gp120 molecule appears to be critical to demonstrate this interaction, as other antibodies with lower relative affinities for gp120 were incapable of detecting this low-level binding (data not shown). The binding of ogp160 to the CD4-expressing CEM-SS cells was several orders of magnitude higher than that to the J25 cells. To prove the specificity of the binding assay for CXCR4, a synthetic form of SDF1 was produced and tested for its ability to block infection by the HIV-1 strain NL4-3 in HeLa CD4-positive long terminal repeat (LTR)-LacZ cells. These data have been published elsewhere (2). SDF1 synthesis and composition have been described previously (24). Exposure of J25 cells to SDF1 was shown to produce a dose-dependent blockage of the binding of ogp160 to the surfaces of the J25 cells (Fig. 2B), indicating the specific nature of the assay.
FIG. 1.
Phenotype analysis of CEM-SS and J25 cell lines. Thin solid line, background; thick solid line, CD4; dashed line, CXCR4.
FIG. 2.
(A) Binding of ogp160 from HIV451 to the surfaces of CEM-SS or J25 cells. Fluorescence intensity is expressed on a logarithmic scale on the x axis, with each line representing one-half log. Concentrations of ogp160 are shown at the right of each graph. The experiments were done in duplicate to ensure consistency of results. (B) Effect of RANTES (250 nM) or increasing amounts of SDF1 (up to 250 nM) on binding of ogp160 (355 nM) to J25 cells. The results are expressed as mean channel fluorescence. Experiments were repeated twice to ensure consistency of results.
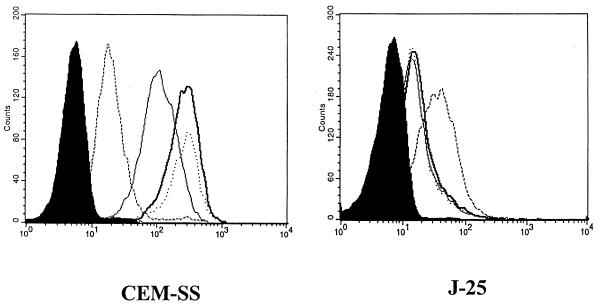
To further test the fact that HIV Env binding to CXCR4 could occur independently of CD4, and to evaluate the effect of prior binding of Env to sCD4, the following experiments were performed. We preexposed CEM-SS as well as J25 cells to either the anti-CD4 antibody Leu3a (Becton Dickinson), which blocks the CD4 binding domain of HIV Env, or OKT4 (Ortho Diagnostics, Costa Mesa, Calif.), which does not block binding of HIV Env to CD4. The cells were then tested for their ability to bind ogp160 to their surfaces. As shown in Fig. 3, OKT4 had no significant effect on the binding of ogp160 to either CEM-SS or J25 cells while Leu3a readily inhibited binding of ogp160 to CEM-SS cells but had no such effect on J25 cells. Furthermore, when ogp160 was allowed to react in advance with recombinant sCD4 produced in CHO cells (Intracel, Issaquah, Wash.) for 30 min at 4°C at a concentration of 1 μg/ml, we were able to show a clear decrease in the surface binding of ogp160 to CEM-SS cells while the opposite, an obvious enhancement in surface binding, was demonstrated for J25 cells (Fig. 3).
FIG. 3.
Binding of ogp160 to CEM-SS or J25 cells after exposure of the cells to the anti-CD4 antibodies Leu3a (thin solid lines), OKT4 (dotted lines), or a combination of ogp160 with sCD4 (dashed lines). The shaded areas represent background. The thick solid lines represent binding in the absence of antibodies or sCD4. The experiments were performed in quadruplicate with similar results. Mean channel fluorescence is represented on the x axis.
Taken together, these data indicate that HIV Env can bind to CXCR4 independently of CD4. On the other hand, prior interaction of HIV Env with CD4 results in a clear increase in the binding of HIV Env to CXCR4.
Relevance of the glycosylation state of HIV Env in binding to CXCR4.
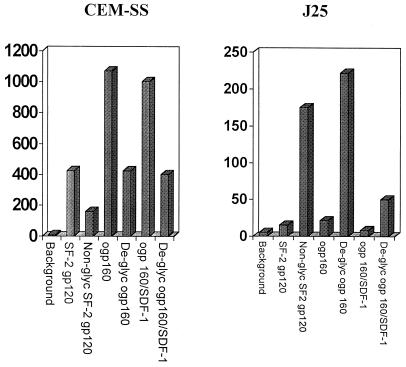
The binding of HIV Env to CD4 is dependent on the appropriate conformation of the Env molecule (27), which can be significantly altered by changes in its carbohydrate content. We next tested the hypothesis that alterations in the carbohydrate moieties of Env would affect its binding to CXCR4. To do so, we used the gp120 molecule from HIVSF2, produced in CHO cells, and its counterpart, nonglycosylated HIVSF2 Env 2-3, produced in yeast strain 2150, and tested both in the binding assay with CEM-SS or J25 cells. HIVSF-2 gp120 and its nonglycosylated counterpart, Env 2-3, were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, from Kathelyn Steimer, Chiron Corp. (13, 14, 19, 26, 29–31). The results are shown in Fig. 4. As expected, nonglycosylated HIVSF2 Env 2-3 bound to the surfaces of the CEM-SS cells to a lesser extent than did HIVSF2 gp120. On the other hand, and unexpectedly, nonglycosylated HIVSF2 Env 2-3 bound much more readily to the surfaces of the J25 cells than its glycosylated counterpart, HIVSF-2 gp120, even when used at equal molar concentrations. To determine whether these findings could be generalized to other Env molecules that lacked intact carbohydrate molecules, we treated ogp160 with sodium metaperiodate. ogp160 from HIV451 at 1.25 μg/ml was treated with sodium metaperiodate (Sigma, St. Louis, Mo.) in acetate buffer for 2 h at 4°C in the dark (33). The cells to be tested had been treated previously with 1% glycine (Sigma) for 30 min at 37°C. Such treatment results in the oxidation and cleavage of the carbohydrate hydroxyl groups without affecting the structure of the polypeptide chains (33). Nonspecific binding by the resulting aldehyde groups was prevented by blocking the target cells beforehand with 1% glycine. The results are shown in Fig. 4. Sodium metaperiodate treatment of ogp160 resulted in a marked inhibition of the binding of ogp160 to the surfaces of the CEM-SS cells. In contrast, sodium metaperiodate treatment of ogp160 resulted in a very clear increase in the binding of HIV Env to the surfaces of the J25 cells. The preexposure of CEM-SS cells to SDF1 did not significantly affect the binding of ogp160 or sodium metaperiodate-treated ogp160. On the other hand, preexposure of J25 cells to 250 nM SDF1 resulted in a marked decrease in binding of both ogp160 and sodium metaperiodate-treated ogp160. These data indicate the specificity of the interaction of the deglycosylated form of ogp160 with CXCR4. The results of these experiments suggest that the alteration in the carbohydrate content of the HIV Env molecules resulted in a better exposure of the epitopes involved in gp120 binding to CXCR4.
FIG. 4.
Binding of HIVSF-2 gp120 or the nonglycosylated form, HIVSF-2 Env 2-3 (Non-glyc SF-2 gp120), to CEM-SS or J25 cells. The concentration was 355 nM for both. The binding of ogp160 and sodium metaperiodate-treated ogp160 (De-glyc ogp160), each at a concentration of 355 nM, to CEM-SS or J25 cells is also shown. The two right-hand bars in each graph show results for cells preexposed to SDF1 at 150 nM. The results are expressed as mean channel fluorescence. The experiments were performed in duplicate with similar results.
The understanding of the underlying mechanisms by which HIV Env, CD4, and the newly discovered HIV coreceptors interact to mediate viral entry remains a very significant issue. The way that HIV Env and CD4 interact is well established (28), and some information exists about the interaction between HIV Env, CCR5, and CD4 (34). In this paper we have shown that HIV Env is able to interact in a CD4-independent manner with CXCR4. Still, the extent of such interaction was clearly lower than that of the sCD4-HIV Env complex and CXCR4. This effect of sCD4 seems to be consistent with the observation that the complexing of this molecule with HIV Env from the strains JRFL or BAL resulted in a significant increase in the affinity of HIV Env for CCR5 (34). We speculate that this interaction between sCD4 and HIV Env results in a conformational change that exposes the binding epitopes in HIV Env relevant for binding to CXCR4, as it does with other gp120 epitopes (16). A different scenario would involve a change in both molecules, resulting in a newly formed common binding epitope. This second alternative seems less likely given our data showing CD4-independent binding of HIV Env to CXCR4, as well as previous data showing the existence of HIV strains capable of CD4-independent entry into target cells (9, 15).
The gp120 molecule from HIV contains 20 potential N-linked glycosylation sites, with N-linked glycans representing at least 50% of the molecular mass. Their role in CD4 binding has been studied extensively, although some of the results remain somewhat controversial. Most of the available data seem to indicate that complete lack of glycosylation completely (20), or at least partially (25), inhibits HIV Env binding to CD4. Also, enzymatic manipulation of the carbohydrate residues results in a significant decrease but not in complete abrogation of the binding of HIV Env to CD4 (11, 20, 25). It was therefore somewhat unexpected to find that the nonglycosylated form, as well as the sodium metaperiodate-treated form, of HIV Env was able to bind in such an enhanced way to CXCR4. This would appear to reinforce the concept of the existence of a binding epitope for CXCR4 within HIV Env which is different from the one for CD4. It also suggests that the changes occurring as a consequence of the manipulation of the carbohydrate residues likely result in a better exposure of the CXCR4 binding epitope(s) within the HIV Env molecule.
In summary, we have shown that HIV Env can interact with CXCR4 in a CD4-independent manner. We have also shown how the interaction of CD4 with HIV Env results in a significant increase in the binding of the latter to CXCR4 and how the alterations in the carbohydrate composition of the HIV Env molecule affect its binding to CXCR4. The complete definition of these interactions may result in novel approaches to protect against cell infection by HIV.
Acknowledgments
This work was supported by funds from the Research Center for AIDS and HIV Infection at the Manhattan VA Medical Center, New York, N.Y., and CFAR-NIAID grant P30 AI27742.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP1 alpha, MIP 1 beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Amara A, Le Gall S, Schwartz O, Salamero J, Montes M, Loetscher P, Baggiolini M, Virelizier J-L, Arenzana-Seisdedos F. HIV coreceptor downregulation as antiviral principle: SDF-1 alpha-dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J Exp Med. 1997;186:139–146. doi: 10.1084/jem.186.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandres J C, Shaw A S, Ratner L. HIV-1 Nef protein downregulation of CD4 surface expression: relevance of the lck binding domain of CD4. Virology. 1995;207:338–341. doi: 10.1006/viro.1995.1089. [DOI] [PubMed] [Google Scholar]
- 4.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 5.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 6.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 7.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 8.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 9.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Broder C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven transmembrane G protein coupled receptor. Science. 1996;272:872–876. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 11.Fenouillet E, Miquelis R, Drillien R. Biological properties of recombinant HIV envelope synthesized in CHO glycosylation-mutant cell lines. Virology. 1996;218:224–231. doi: 10.1006/viro.1996.0182. [DOI] [PubMed] [Google Scholar]
- 12.Gorny M K, Gianakakos L, Sharpe S, Zolla-Pazner S. Generation of human monoclonal antibodies to human immunodeficiency virus. Proc Natl Acad Sci USA. 1989;86:1624–1628. doi: 10.1073/pnas.86.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haigwood N L, Barker C B, Higgins K W, Skiles P V, Moore G K, Mann K A, Lee D R, Eichberg J W, Steimer K S. Evidence for neutralizing antibodies directed against conformational epitopes of HIV-1 gp120. In: Ginsberg H, Chanock R, editors. Vaccines Ninety, modern approaches to new vaccines including prevention of AIDS. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1990. pp. 313–320. [Google Scholar]
- 14.Haigwood N L, Schuster J R, Moore G K, Lee H, Skiles P V, Higgins K H, Barr P J, George-Nascimiento C, Steimer K S. Importance of hypervariable regions of HIV-1 gp120 in generation of virus neutralizing antibodies. AIDS Res Hum Retroviruses. 1990;6:855–869. doi: 10.1089/aid.1990.6.855. [DOI] [PubMed] [Google Scholar]
- 15.Hesselgesser J, Halks-Miller M, DelVecchio V, Peiper S C, Hoxie J A, Kolson D L, Taub D, Horuk R. CD4-independent association between HIV-1 gp120 and CXCR4: functional chemokine receptors are expressed in human neurons. Curr Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- 16.Kang C-Y, Hariharan K, Nara P L, Sodroski J, Moore J P. Immunization with a soluble CD4-gp120 complex preferentially induces neutralizing anti-human immunodeficiency virus type 1 antibodies directed to conformation-dependent epitopes of gp120. J Virol. 1994;68:5854–5862. doi: 10.1128/jvi.68.9.5854-5862.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kayanamaran V S, Pal R, Gallo R C, Sarngadharan M G. A unique HIV culture secreting soluble gp160. AIDS Res Hum Retroviruses. 1988;4:319. doi: 10.1089/aid.1988.4.319. [DOI] [PubMed] [Google Scholar]
- 18.Lapham C K, Ouyang J, Chandrasekar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 19.Levy J A, Hoffman A D, Kramer S M, Landis J A, Shimabukuro J M, Oshiro L S. Isolation of lymphocytopathic retroviruses from San Francisco patients with AIDS. Science. 1984;225:840–842. doi: 10.1126/science.6206563. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Luo L, Rasool N, Kang C Y. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J Virol. 1993;67:584–588. doi: 10.1128/jvi.67.1.584-588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luria S, Chambers I, Berg P. Expression of the HIV-1 Nef protein in T cells prevents antigen receptor-mediated induction of interleukin-2 mRNA. Proc Natl Acad Sci USA. 1991;88:5326–5330. doi: 10.1073/pnas.88.12.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nara P L, Hatch W C, Dunlop N M, Robey W G, Arthur L O, Gonda M A, Fischinger P J. Simple, rapid, quantitative syncitium forming microassay for the detection of HIV neutralization antibody. AIDS Res Hum Retroviruses. 1987;3:283–302. doi: 10.1089/aid.1987.3.283. [DOI] [PubMed] [Google Scholar]
- 23.Nara P L, Fischinger P J. Quantitative infectivity assay for HIV-1 and -2. Nature. 1988;332:469–470. doi: 10.1038/332469a0. [DOI] [PubMed] [Google Scholar]
- 24.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Hayami M. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 25.Papandreou M J, Idziorek T, Miquelis R, Fenouillet E. Glycosylation and stability of mature HIV envelope glycoprotein conformation under various conditions. FEBS Lett. 1996;379:171–176. doi: 10.1016/0014-5793(95)01505-1. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez-Pescador R, Power M D, Barr P J, Steimer K S, Stempien M M, Brown-Shimer S L, Gee W W, Renard A, Randolph A, Levy J A, Dina D, Luciw P A. Nucleotide sequence and expression of an AIDS-associated retrovirus (ARV-2) Science. 1985;227:484–492. doi: 10.1126/science.2578227. [DOI] [PubMed] [Google Scholar]
- 27.Sattentau Q J, Weiss R A. The CD4 antigen: physiological ligand and HIV receptor. Cell. 1988;52:631–633. doi: 10.1016/0092-8674(88)90397-2. [DOI] [PubMed] [Google Scholar]
- 28.Sattentau Q J, Clapham P R, Weiss R A, Beverley P C, Montagnier L, Alhalabi M F, Gluckmann J C, Klatzmann D. The human and simian immunodeficiency viruses HIV-1, HIV-2 and SIV interact with similar epitopes on their cellular receptor, the CD4 molecule. AIDS. 1988;2:101–105. doi: 10.1097/00002030-198804000-00005. [DOI] [PubMed] [Google Scholar]
- 29.Scandella C J, Kilpatrick J, Lidster W, Parker C, Moore J P, Moore G K, Mann K A, Brown P, Coates S, Chapman B, Masiarz F R, Steimer K S, Haigwood N L. Nonaffinity purification of recombinant gp120 for use in AIDS vaccine development. AIDS Res Hum Retroviruses. 1993;9:1233–1244. doi: 10.1089/aid.1993.9.1233. [DOI] [PubMed] [Google Scholar]
- 30.Steimer K S, Puma J P, Power M D, Powers M A, George-Nascimiento C, Stephans J C, Levy J A, Sanchez-Pescador R, Luciw P A, Barr P J, Hallewell R A. Differential antibody responses of individuals infected with AIDS-associated retroviruses surveyed using the viral core antigen p25 gag expressed in bacteria. Virology. 1986;150:283–290. doi: 10.1016/0042-6822(86)90289-8. [DOI] [PubMed] [Google Scholar]
- 31.Steimer K S, Van Nest G A, Haigwood N L, Tillson E M, George-Nascimiento C, Barr P J, Dina D. Recombinant gag and env polypeptides in characterizing HIV-1 neutralizing antibodies. In: Ginsberg H, Brown F, Lerner R A, Chanock R, editors. Vaccines 88, new chemical and genetic approaches to vaccination: prevention of AIDS and other viral, bacterial and parasitic diseases. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. pp. 347–355. [Google Scholar]
- 32.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Meyer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 33.Woodward M P, Young W W, Jr, Bloodgood R A. Detection of monoclonal antibodies specific for carbohydrate epitopes using periodate oxidation. J Immunol Methods. 1985;78:143–153. doi: 10.1016/0022-1759(85)90337-0. [DOI] [PubMed] [Google Scholar]
- 34.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV gp120 glycoproteins with the chemokine receptor CCR5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]