Abstract
Traditionally, public health surveillance relied on individual-level data but recently wastewater-based epidemiology (WBE) for the detection of infectious diseases including COVID-19 became a valuable tool in the public health arsenal. Here, we use WBE to follow the course of the COVID-19 pandemic in Rochester, Minnesota (population 121,395 at the 2020 census), from February 2021 to December 2022. We monitored the impact of SARS-CoV-2 infections on public health by comparing three sets of data: quantitative measurements of viral RNA in wastewater as an unbiased reporter of virus level in the community, positive results of viral RNA or antigen tests from nasal swabs reflecting community reporting, and hospitalization data. From February 2021 to August 2022 viral RNA levels in wastewater were closely correlated with the oscillating course of COVID-19 case and hospitalization numbers. However, from September 2022 cases remained low and hospitalization numbers dropped, whereas viral RNA levels in wastewater continued to oscillate. The low reported cases may reflect virulence reduction combined with abated inclination to report, and the divergence of virus levels in wastewater from reported cases may reflect COVID-19 shifting from pandemic to endemic. WBE, which also detects asymptomatic infections, can provide an early warning of impending cases, and offers crucial insights during pandemic waves and in the transition to the endemic phase.
1. INTRODUCTION
Traditionally, public health surveillance has relied on individual-level data, often acquired through clinical testing or surveys. However, these methods can suffer from limitations such as delayed reporting, incomplete coverage, and individual biases. In recent years, wastewater-based epidemiology (WBE) has emerged as a revolutionary tool, offering a unique glimpse into the collective health of a population through the analysis of municipal wastewater. WBE provides real-time, non-invasive, and often anonymous insights into the occurrence and prevalence of pathogens [[1], [2], [3], [4]], the consumption of illicit drugs and pharmaceuticals [[5], [6], [7]], and even the metabolic makeup of a population [7]. This field holds immense potential to transform public health surveillance, from informing targeted interventions to predicting and preventing outbreaks before they take hold [8].
SARS-CoV-2 is a positive-sense, single-stranded RNA virus that causes life-threatening coronavirus disease 2019 (COVID-19) in humans [[9], [10], [11], [12], [13]]. SARS-CoV-2 RNA is shed through the urine, feces, and sputum of individuals who have contracted COVID-19, and as a result, SARS-CoV-2 RNA can be detected in wastewater [[14], [15], [16], [17]]. Consequently, wastewater testing was implemented in cities around the world to determine SARS-CoV-2 prevalence in communities [[18], [19], [20]]. This led to the development of WBE to monitor COVID-19 transmission and to attempt pandemic mitigation through early detection of SARS-CoV-2 RNA in wastewater.
The COVID-19 pandemic has demonstrated the utility of WBE for monitoring outbreaks in small and medium-sized communities [21,22]. This study was borne out of a request from the city of Rochester, Minnesota, USA to monitor city-wide SARS-CoV-2 prevalence which could be used to inform pandemic mitigation policies. Rapid, cost-effective, and efficient methods were needed to test wastewater to provide precise data on changes in concentrations of the SARS-CoV-2 virus and to support public health decision-making.
Here we describe the strategies, tools, and workflow for processing wastewater with the goal of detecting SARS-CoV-2 RNA to monitor community wide COVID-19 prevalence and possibly, as an early indication of COVID-19 outbreaks in the community. Specifically, the development and performance characteristics are described for a quantitative reverse transcription droplet digital PCR (RT-ddPCR) test for the detection of SARS-CoV-2 in wastewater, which could be implemented and/or modified for other infectious disease targets in most community laboratories for monitoring and surveillance purposes. Our data provide a snapshot of the trend between SARS-CoV-2 viral RNA in wastewater and positive COVID-19 cases in a population of 121,395 in Rochester, Minnesota, from February 2021 to December 2022. We also compared the wastewater data to COVID-19 hospitalization data. In addition, we monitored the concentration of SARS-CoV-2 in wastewater along with new cases of COVID-19 in two smaller adjacent communities, Byron (population 6312) and Stewartville (population 6703).
Analysis of this data provided a clear indication of a positive correlation between the concentration of SARS-CoV-2 in wastewater and the new COVID-19 cases reported to the Olmsted County Public Health Services. However, this was only true for the period that required strict reporting guidelines and prior to the widespread availability of rapid home tests for COVID-19. The widespread availability of home-testing kits coincided with the waning of this correlation. This demonstrates the utility of wastewater surveillance to monitor community COVID-19 prevalence when there is no mandate to report positive cases. Interestingly, we noted a similar pattern with COVID-19 hospitalization data, where there was a loss of correlation with SARS-CoV-2 RNA levels in wastewater towards the latter half of 2022. In summary, we document a cohesive approach of multiple institutions and agencies cooperating to monitor pathogen components within wastewater which could be used as a basis for mitigation policies of emerging and re-emerging infections in the future.
2. Materials and methods
2.1. Wastewater sample collection
The influent wastewater at the Rochester Water Reclamation Plant (WRP) consists of approximately 48% residential, 47% commercial and 5% industrial facilities. At the Rochester WRP, influent wastewater was composited daily based on a flow weighting sampling regime. An average sampling rate of 188 mL/h was used to composite a total daily sample of 4.5 L. From Feb/2021 to May/2021 two times per week and from June/2021 to Dec/2022, four times per week, influent composite samples were clarified by centrifugation and 2 × 40 mL aliquots were stored at 4 °C [23] at WRP water chemistry lab prior to being delivered to Mayo Clinic, Rochester, MN. Byron and Stewartville samples were also flow-weighted samples that were collected by Rochester staff and centrifuged/stored at the WRP water chemistry lab.
Wastewater in the collection systems is subject to inflow meaning that outside sources of water (rainfall/snowmelt) can find its way into the sewer diluting the pollutants, including the SARS-CoV-2 RNA being carried to the WRP. To account for inflow, a dilution factor was applied based on an estimated baseline flow (flow known to be coming from human-derived wastewater). For this study the concentration of RNA was adjusted for inflow based on daily flow measurements at the Rochester plant. This inflow dilution factor averaged 1.09 for Rochester, which is extremely low indicating a dry period during the monitoring. This same exercise was not done with Byron and Stewartville as the baseline flow is not as well understood. Thus, while a dilution factor was not accounted for in Byron and Stewartville, it would be anticipated that it would be similar to Rochester due to the dry period, and thus dilution would have had little effect on the results.
2.2. Wastewater processing and total RNA extraction
The 40 mL wastewater sample was filtered through a 0.7 μm glass filter (Flipmate 50, Cat. No. SC0605, Environmental Express, Charleston, SC, USA) and subsequently concentrated with the Concentrating Pipette Select (InnovaPrep, Drexel, MO, USA) equipped with a 0.05 μm PS hollow fiber filter concentrating pipette tip (Cat. No. CC08020), according to the manufacturer's recommendations.
Approximately 200 μL of concentrated wastewater was recovered from this concentrating pipette and the RNA was extracted from this using a Qiagen Viral RNA Purification Kit (Qiagen, Hilden, Germany) according to the manufacturer's recommended protocol. The final elution volume of the RNA was 60 μL.
2.3. Quantification of SARS-CoV-2 by reverse transcription droplet digital PCR (RT-ddPCR)
The ddPCR assay designed to detect SARS-CoV-2 amplifies and quantifies 3 RNA targets using fluorescently labeled probes: N1 (SARS-CoV-2 nucleocapsid probe 1), N2 (SARS-CoV-2 nucleocapsid probe 2), and as a control, human RPP30 (human ribonuclease P/MRP subunit 30). The assay was performed according to the manufacturer's protocol, as instructed for the Bio-Rad SARS-CoV-2 ddPCR Test (Bio-Rad Laboratories, CA, USA) with slight modifications from the method used in previous reports [24,25]. Briefly, 12 μL of RNA eluate was added to 10 μL of ddPCR master mix containing 5.5 μL of 4x One Step-RT-ddPCR Supermix, 2.2 μL of reverse transcriptase, 1.1 μL of dithiothreitol, 1.1 μL of the 20 × 2019-nCoV CDC ddPCR triplex probe and 0.1 μL DNase/RNase free water. After the reaction was partitioned into droplets using the Bio-Rad AutoDG instrument, SARS-CoV-2 nucleocapsid (N1 and N2) target sequences, along with RPP30 target as a control, were amplified using Veriti thermocyclers (Thermo Fisher Scientific, Inc., Waltham, MA) using the following protocol: RT steps: 25 °C for 3 min and 50 °C for 60 min, followed by the PCR steps: 95 °C for 10 min, then 40 cycles of denaturation at 95 °C for 30 s and annealing/extension at 55 °C for 1 min, and a final enzyme deactivation at 98 °C for 10 min.
Individual reaction droplets were counted as negative or positive for one or more targets by the QX200 Droplet Reader and signal data were analyzed using QuantaSoft Analysis Pro software version 1.0.596 (Bio-Rad Laboratories, Hercules, CA, USA). For each run, two low positives [total 10 copies and 5 copies, Exact Diagnostics SARS-CoV-2 Standard (Exact Diagnostics LLC, Fort Worth, TX)] for SARS-CoV-2 and a no template control were included for quality control. QuantaSoft Analysis Pro software (version 1.0.596) was used for droplet cluster classification. The QX200 automated droplet reader counts every acceptable droplet and measures the fluorescence emissions from each droplet using 2 channels (FAM and HEX). Droplets of different color and intensity were displayed on 2-dimensional plots, allowing counting of negative droplets, as well as those positive for N1, N2, RPP30, or a combination of targets. For each fluorophore, the fraction of positive droplets, was fitted into a Poisson distribution equation, thereby providing absolute quantification of N1, N2, and RPP30 copies per well without a standard curve.
The number of droplets generated per reaction ranged from 9000 to 18299 droplets. Rejection criteria for excluding a result included an unusual spread of droplets or <9000 acceptable droplets measured per well.
2.4. Validation of wastewater processing and SARS-CoV-2 RNA quantification protocols
Rochester wastewater was sent to a commercial laboratory (PACE laboratory, Minneapolis, MN) for independent quantification of SARS-CoV-2 genome levels during the initial validation of our wastewater processing, RNA extraction and RT-ddPCR protocols. Furthermore, Rochester WRP participated in Center for Disease Control (CDC) phase 3 wastewater SARS-CoV-2 testing on a weekly basis from Mid-January to Mid-March 2021 by sending local wastewater samples to National Wastewater Surveillance System (NWSS) (https://www.cdc.gov/nwss/testing.html) [26].
2.5. SARS-CoV-2 variant analysis
Detection of SARS-CoV-2 variants in wastewater was performed at the University of Minnesota Genomics Center (Minneapolis, MN) which ran the targeted mutation ddPCR assays and the Metropolitan Council Environmental Services (Saint Paul, MN) interpreted the results. The assay was performed according to the manufacturer's protocol, as instructed for the Bio-Rad SARS-CoV-2 ddPCR Test (Cat. No. 1864021, Bio-Rad Laboratories, CA, USA).
2.6. COVID-19 cases and hospitalization data
Daily new COVID-19 cases and hospitalization data were collected from the Minnesota Electronic Disease Surveillance System (MEDSS) at the Minnesota Department of Health. Data in MEDSS was from reported positive PCR or antigen tests. Positive tests were considered confirmed (PCR+) or probable (antigen+). At-home tests are not reportable and are excluded from this analysis. New hospitalizations are the number of new admits for COVID-19 or with COVID-19 in Olmsted County residents on a given day. All patient records were deidentified and aggregated at the ZIP code level.
2.7. Statistical analysis
The data was analyzed and visualized using customized R and Python scripts from Python libraries including pandas [27], NumPy [28], and Matplotlib [29], as well as R packages such as dplyr and tidyverse [30]. To test the normality of the data, we used the Shapiro-Wilk normality test from the rstatix library [31], which assumes a null hypothesis for normally distributed data. However, our data rejected the null hypothesis (Supplementary Table 1), prompting us to employ Spearman's rank correlation coefficient, a non-parametric measure, to assess the strength and direction of the relationship between new COVID-19 cases, or hospitalizations and SARS-CoV-2 RNA in wastewater. Correlation analysis and plots were performed using R packages ggpubr [32] and ggplot2 [33], with statistical significance considered at a threshold of P < 0.05. The R package, changepoint [34] was used to detect a single divergence point in the ratio of SARS-CoV-2 concentration in wastewater and new COVID-19 cases or hospitalization. Our R and Python codes are available on GitHub (https://github.com/biruhalem/wastewater).
2.8. Attempts to determine lag period
To determine if we could use our data to predict COVID-19 case oscillations, based on SARS-CoV-2 RNA in wastewater, we applied three predictive models to the data before changes in COVID-19 case reporting guidelines (before 2022-04-25). There exist various mathematical and statistical predictive models for time series data [[35], [36], [37], [38]]. We worked with three predictive models— autoregressive integrated moving average (ARIMA),distributed lag (DL), and autoregressive distributed lag (ADL)—for lag analysis based on two primary considerations: 1) their common usage in analogous studies [14,[38], [39], [40], [41], [42], [43], [44]] and 2) their well-established frameworks, widespread application in time series analysis, and straightforward interpretability [45]. However, the models either failed to fit the data significantly or predicted lags that varied between models, indicating that our data set was not suitable for this type of predictive analysis. Several factors could affect the modeling results [37,46]. In our case wastewater samples were not collected daily or with equal spacing in consecutive samples, and interpolation of the missing data may affect the output. In addition, the weekend new cases of COVID-19 are significantly lower than the weekday cases which might contribute to the weekly significant cyclic lag patterns observed in the DL model.
3. Results
3.1. Multi-institution collaboration to monitor SARS-CoV-2 genome in wastewater
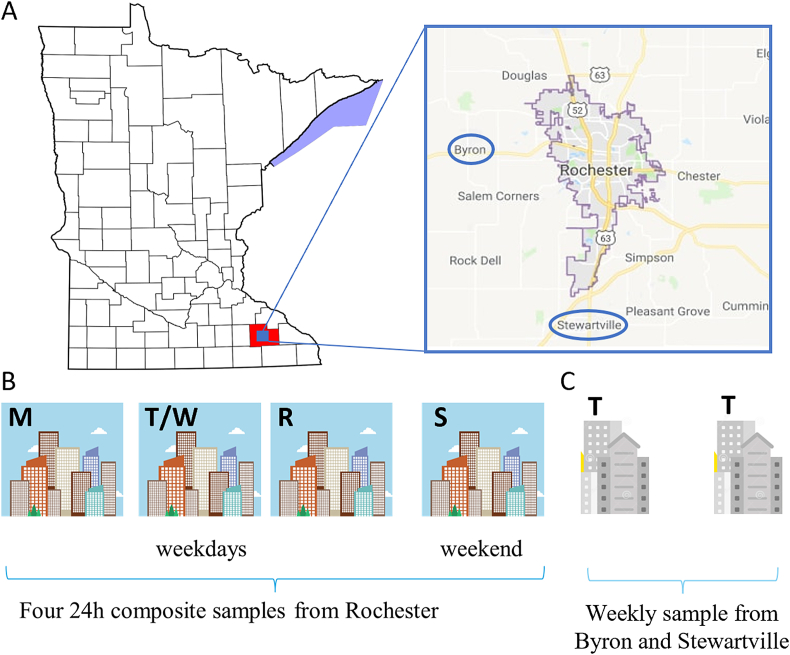
Wastewater samples were collected from the city of Rochester four times a week on the indicated days (Fig. 1A and B) and once a week from two smaller neighboring communities (Fig. 1A and C, Byron and Stewartville circled in blue). Several institutions collaborated to assist the city in this effort (Supplementary Fig. 1). The WRP that serves the city of Rochester (population 121,395) collected the wastewater. Researchers at the Mayo Clinic (Department of Molecular Medicine) processed it and extracted RNA with the help of undergraduate students from the University of Minnesota Rochester (UMR). SARS-CoV-2 detection by ddPCR was performed by the Advanced Diagnostics Laboratory (ADL) at Mayo Clinic and the data provided to the WRP for flow adjustment. The daily SARS-CoV-2 positive cases and COVID-19 hospitalization data were provided by the Olmsted County Department of Health.
Fig. 1.
(A) Water reclamation plant catchment area for city of Rochester and location of Byron/Stewartville (B) Testing schedule for Rochester (C) Testing schedule for Byron and Stewartville. M, Monday; T, Tuesday; W, Wednesday; R, Thursday; and S, Saturday.
3.2. Sensitivity of ddPCR assay
Droplet digital polymerase chain reaction (ddPCR) is an effective and sensitive tool for detection of pathogens in clinical and biological sources, particularly in the setting of PCR inhibitors, which may be present in wastewater [[47], [48], [49]]. Moreover, ddPCR can specifically measure single or multiple nucleotide targets in a single reaction [[50], [51], [52]].
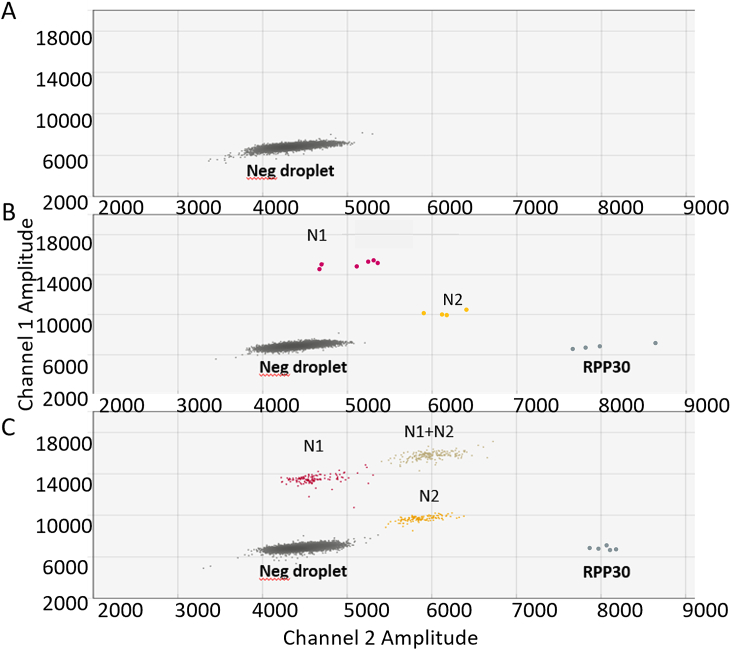
Fig. 2 shows representative plots of wastewater analysis by ddPCR. While only one droplet cluster comprised of negative droplets was present for the no-template negative control (Fig. 2A), target-positive droplet clusters were observed for SARS-CoV-2 positive specimens (Fig. 2B and C). These clusters included the human ribonuclease protein (RPP30)-control detecting RPP30-positive droplets. The wastewater sample shown in Fig. 2b had additional clusters for N1-positive and N2-positive droplets and the sample shown in Fig. 2C had an additional N1+N2 positive (double positive) cluster. Of note, human RPP30 positive droplets are typically present at a very low level in wastewater.
Fig. 2.
Representative plots of wastewater analysis by droplet digital PCR. (A) No template control. (B) Wastewater sample with low concentration of SARS-CoV-2 genomes. (C) Wastewater sample with a high concentration of SARS-CoV-2 genomes. N1, SARS-CoV-2 nucleocapsid probe 1 (red); N2, SARS-CoV-2 nucleocapsid probe 2 (gold); RPP30, probe for human ribonuclease P/MRP subunit p30 (dark blue). N1+N2; double positive droplets (tan). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Analytical sensitivity and the cut-off for final calls were set at > 4 N1 and N2 droplets. Based on the previously published experiments from our laboratories [24,25], 5 copies of viral RNA (equivalent to approximately 5 droplets of N1 and/or N2) can be reliably detected and is expected to be the approximate limit of detection. As there was a positive correlation between the SARS-CoV-2 genomes detected by N1 and N2 probes (Supplementary Fig. 2, R = 0.99, p < 2.2e−16), we used genome copies determined from each probe to calculate a mean genome copy number.
3.3. Validation and accuracy of the ddPCR assay
The accuracy of our assay was determined by comparing its results to those of an assay performed in a commercial laboratory (PACE laboratory, Minneapolis, MN). Near complete agreement (concentrations for N1 and N2, 16.6 & 25.9 copies/mL from PACE labs, respectively versus 15.9 & 29.6 copies/mL from our ddPCR assay) was observed although the method of extraction and assay procedures were different.
The results of our assay were also compared to results provided by the National Wastewater Surveillance System (NWSS) from our participation in the CDC phase 3 wastewater SARS-CoV-2 testing program. Although, the protocols for RNA extraction and viral RNA quantification (qPCR) utilized at NWSS were different, there is a similar trend to the SARS-CoV-2 RNA concentrations measured by our ddPCR analysis in Rochester for that period (Supplementary Fig. 3, compare panels A and B). It is noteworthy that the SARS-CoV-2 viral concentration showed a similar pattern whether normalized with pepper mild mottle virus (PMMoV, abundantly found in human fecal matter) concentration or not (Supplementary Fig. 3A, dashed red line and solid red line, respectively).
3.4. SARS-CoV-2 concentration in wastewater and COVID-19 cases in Rochester diverge after September 2022
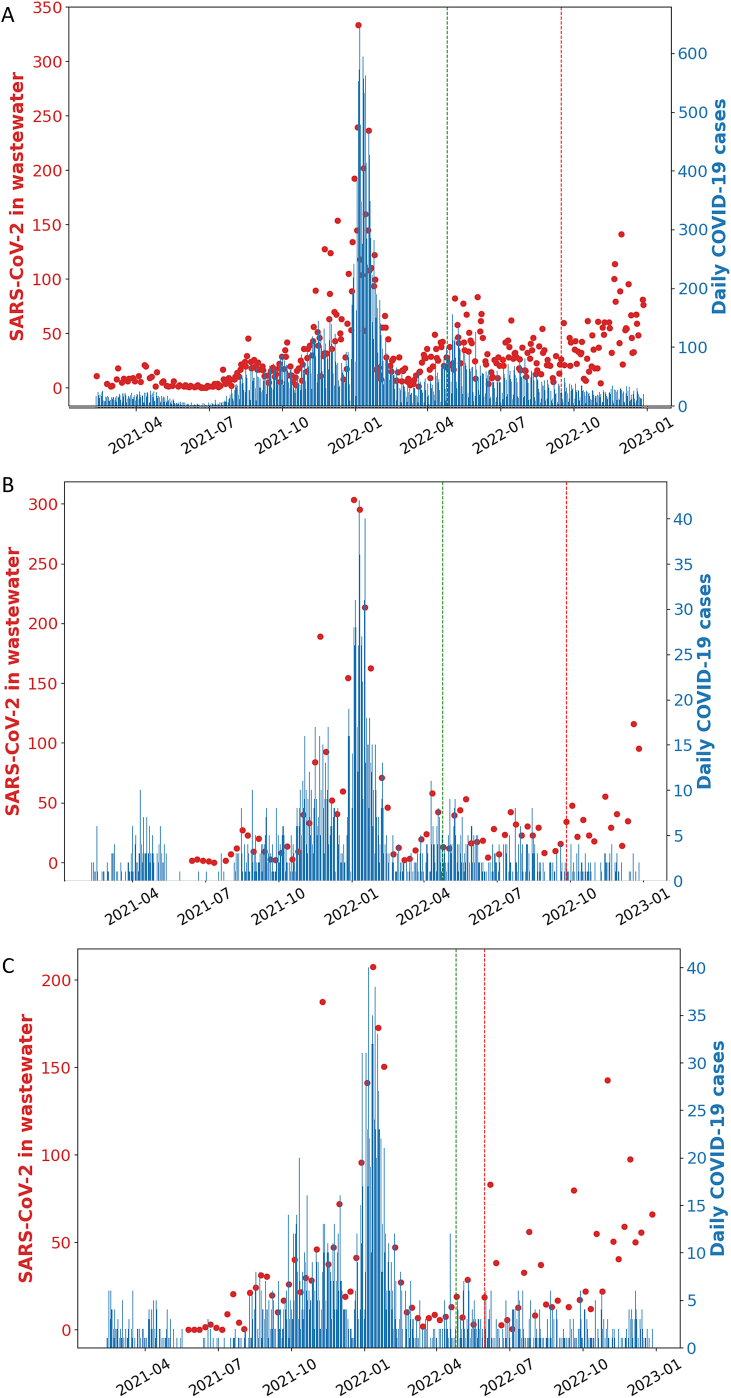
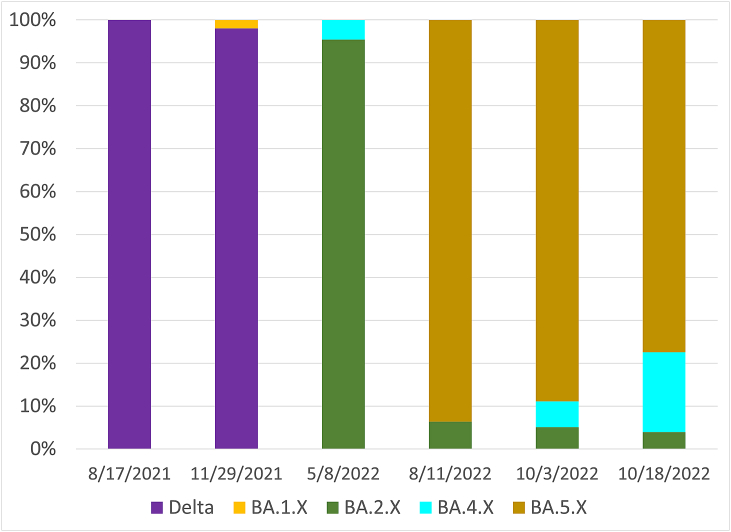
To assess whether the SARS-CoV-2 RNA concentrations correlated with the COVID-19 cases, the SARS-CoV-2 concentration in wastewater versus new COVID-19 cases was plotted daily over a period of 21 months (February 11, 2021–December 01, 2022), as shown in Fig. 3A (red dots). Daily new COVID-19 cases and SARS-CoV-2 concentrations in wastewater was low through the spring and mid-summer of 2021. Daily COVID-19 cases rose starting in August 2021 coincidentally with the circulation of the Delta variant (Fig. 4, purple columns). A wave of COVID-19 infections (Fig. 3A, blue dotted lines) with simultaneous rise in wastewater SARS-CoV-2 RNA levels (Fig. 3A, red dots) occurred from the mid-July until November 2021. From the beginning of December 2021 through March 2022 a steeper surge of COVID-19 cases was observed (Fig. 3A). From May 2022 onwards, SARS-CoV-2 variants of the Omicron lineage were detected in wastewater (Fig. 4, Omicron variants shown in different colors). Of note, from September 2022 through the end of the study period, there was no apparent correlation between virus levels estimated by wastewater and positive COVID-19 cases.
Fig. 3.
SARS-CoV-2 genome copies (red dots) and daily positive SARS-CoV-2 cases (blue lines) in (A) Rochester, (B) Byron, and (C) Stewartville. The dotted green line indicates when case reporting guidelines changed, and the dotted red line indicates the time point after which we observe a divergence between the reported positive cases and the SARS-CoV-2 genome copies. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
SARS-CoV-2 variant analysis at the indicated dates. The Delta variant and several Omicron sub-variants BA.1.X to BA.5.X were detected using specific ddPCR probes.
We also monitored the wastewater of two smaller communities adjacent to Rochester with a population of about 6500 each (Fig. 1, Byron, and Stewartville). We noted a very similar pattern of SARS-CoV-2 genome copies and daily positive cases (Fig. 3B and C). However, in Stewartville we observed an earlier divergence of the trends (Fig. 3C, May 2022).
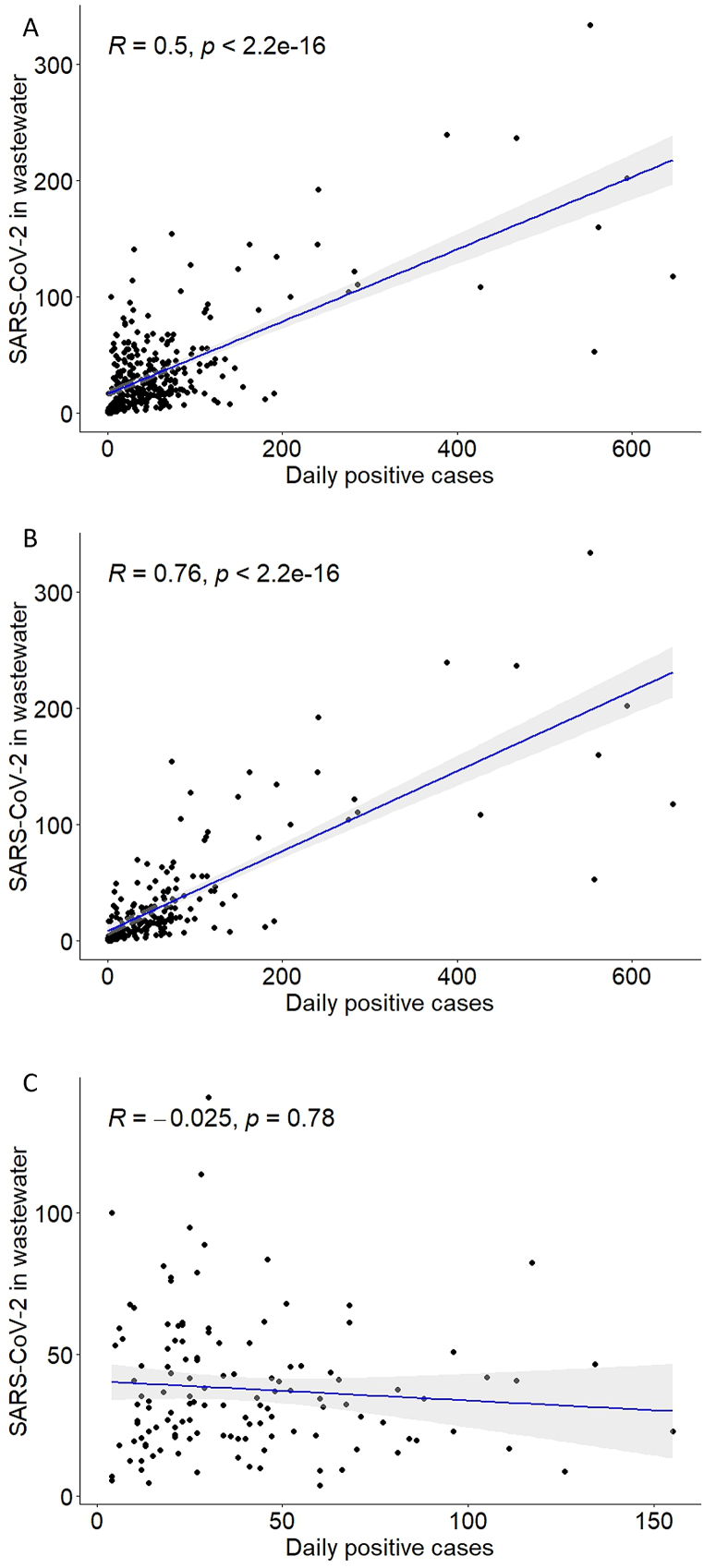
To assess whether the purported positive correlation between SARS-CoV-2 concentration in wastewater and new COVID-19 cases is statistically significant, we used the non-parametric Spearman's rank test. We observed a significant positive correlation between the concentration of SARS-CoV-2 in wastewater and daily new COVID-19 cases in Rochester (Fig. 5A), as well as in Byron and Stewartville (Supplementary Fig. 4A and Supplementary Fig. 5A). However, we noted a divergence in the trend. This divergence occurred after changes in case reporting guidelines (April 25, 2022), and the widespread availability of COVID-19 home-testing kits later in 2022 (Fig. 3, red dotted lines). Therefore, we re-analyzed the data in two groups: before and after April 25, 2022. We found a stronger positive correlation for the data before the changes in case reporting guidelines compared to the overall data in Rochester (Fig. 5B, R = 0.76, p < 2.2e-16), Byron (Supplementary Fig. 4B, R = 0.86, p < 1.3e-13), and Stewartville (Supplementary Fig. 5B, R = 0.88, p < 2.2e-16). Conversely, we did not observe a significant correlation between SARS-CoV-2 genome copies in wastewater and daily positive cases after the changes in case reporting guidelines (Fig. 5C, Rochester, R = −0.025, p = 0.78; Supplementary Fig. 4C, Byron, R = 0.081, p = 0.65; and Supplementary Fig. 5C, Stewartville, R = −0.084, p = 0.64).
Fig. 5.
Correlation between SARS-CoV-2 genome copies in wastewater and COVID-19 positive cases in Rochester: (A) over the entire period tested, (B) before the changes in COVID-19 case reporting guidelines, and (C) after the changes in COVID-19 case reporting guidelines.
3.5. SARS-CoV-2 concentration in wastewater and COVID-19 hospitalizations diverge from September 2022
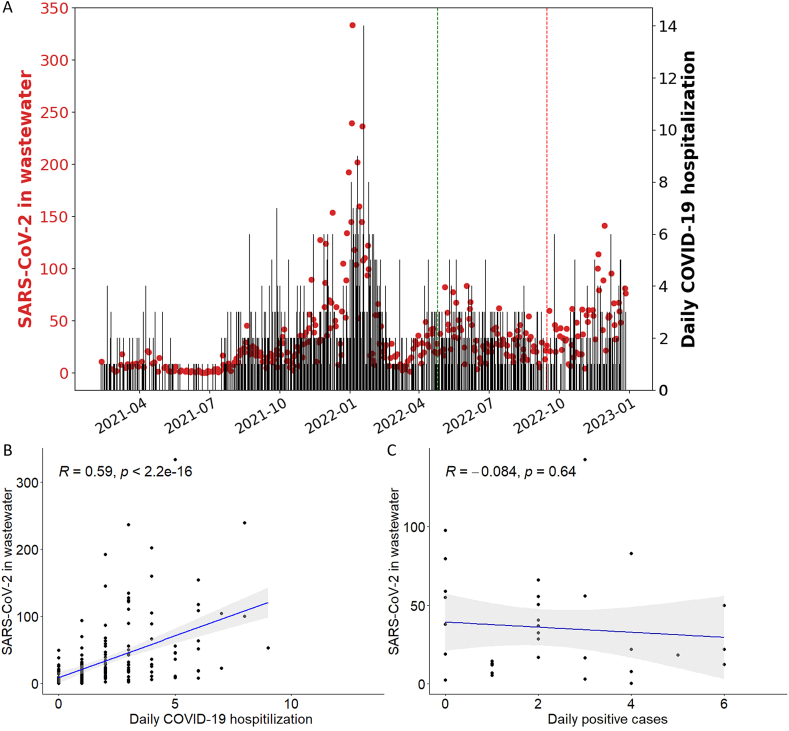
To assess the impact of SARS-CoV-2 infections on community health, we investigated the correlation between SARS-CoV-2 RNA levels in wastewater and COVID-19 hospitalization data. As with the daily new positive cases data, we noted that the hospitalization data correlated with SARS-CoV-2 RNA levels in wastewater only temporarily (Fig. 6A). Change-point statistical analysis was used to detect the divergence point in the ratio of wastewater SARS-CoV-2 RNA and COVID-19 hospitalizations. From September 2022, the COVID-19 hospitalizations and the SARS-CoV-2 genome copies in wastewater diverged (Fig. 6A, red line). There was a good correlation between SARS-CoV-2 genome copies in wastewater and daily COVID-19 hospitalization before September 2022 (Fig. 6B, R = 0.59, p < 2.2e-16) but not after (Fig. 6C, R = −0.084, p = 0.64).
Fig. 6.
(A) SARS-CoV-2 genome copies (red dots) and daily COVID-19 hospitalizations (black lines) in Rochester. The green line indicates when case reporting guidelines changed, and the red line indicates the time point after which we observe a divergence between the reported COVID-19 hospitalizations and the SARS-CoV-2 genome copies. (B) Correlation between SARS-CoV-2 genome copies in wastewater and daily COVID-19 hospitalization before September 2022, and (C) after September 2022. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
In Olmsted County, which encompasses Rochester, Byron and Stewartville, SARS-CoV-2 viral RNA concentrations in wastewater reflected the daily COVID-19 case trend through March 2022, specifically capturing two infection surges due to the different variants. The trend seen in our data is similar to US national wastewater and COVID-19 surveillance reports [53]. The temporal profile of SARS-CoV-2 concentration in wastewater mirrored the trends of COVID-19 new cases monitored through community infection. It is to be noted that the number of reported cases underestimates the infection number due to untested asymptomatic and symptomatic cases [54].
Starting in Spring 2022, due to changes in reporting behavior following the widespread availability of at-home testing kits, the reported cases further deviated from the actual total number of infections. This was borne out by the significant divergence between reported cases and SARS-CoV-2 RNA detected in the wastewater for Rochester, Byron and Stewartville observed in the last quarter of 2022. Similar conclusions were recently drawn from publicly available data from throughout the USA [55,56].
In addition to changes in COVID-19 case reporting guidelines, the reduced severity of Omicron variant infections may also have contributed to the divergence [[57], [58], [59]]. Consistently, the level of hospitalizations and intensive care unit (ICU) admissions in the study area were proportionally lower during the surge of the Omicron variant. Reduced Omicron variant infection severity may be due to a combination of increased immunization coverage [58] and virus attenuation [60,61]. Mild symptoms may reduce the test-seeking behavior in the community resulting in a low number of COVID-19 case reports. However, the virus is still being shed and detected in wastewater causing the divergence of COVID-19 cases and SARS-CoV-2 RNA levels in wastewater.
The divergence of the COVID-19 hospitalization data and wastewater SARS-CoV-2 RNA levels in the second half of 2022 is an indication for the transition of COVID-19 from pandemic to endemic [62,63]. Consistently, other recent wastewater surveillance data indicate seasonal patterns of SARS-CoV-2 similar to that of other endemic respiratory viruses like human parainfluenza, human rhinovirus, human metapneumovirus, and respiratory syncytial virus B [64]. This is further evidence suggesting the transition of SARS-CoV-2 from a pandemic to an endemic virus [65,66].
Due to the limiting factors of our dataset, it was not possible to determine the exact length of the anticipation afforded by the wastewater data. However, other studies have reported that wastewater data precede clinical case detection by 0–14 days [39,[67], [68], [69], [70], [71]]. This broad range suggests that the anticipation period is dynamic over time and space [46,70] and is affected by multiple factors including the strains in circulation and the vaccine coverage [71].
Another limitation of this study was that our ddPCR technique did not differentiate between circulating strains. This was mitigated by sending select samples to the University of Minnesota Genomics Center (Minneapolis, MN) which allowed quantification of the levels of different variants within these samples. As a future perspective, daily wastewater testing coupled with variant analysis may overcome the limitations of our current dataset and allow for reliable forecasting of positive cases.
Based on our experience and that of others, wastewater surveillance has been particularly helpful in detecting community spread of SARS-CoV-2 and other viral pathogens such as polio [72] and monkeypox [73] outbreaks not always represented in local clinical testing data [56]. This report presents a cohesive approach of multiple institutions and agencies to monitor pathogen components within wastewater. This template may be used to predict and control outbreaks of emerging and re-emerging infections in the future, allowing hospitals and communities to plan for upswings in cases that might require additional resources [74,75].
Wastewater-based epidemiology for SARS-CoV-2 detection has rapidly transcended its niche status to become a valuable tool in the public health arsenal. Demonstrating sensitivity to even asymptomatic infections and capable of early warning before clinical cases rise, WBE offers crucial insights during pandemic waves and in the transition to the endemic phase. However, challenges remain in accurately quantifying case numbers and distinguishing variants. WBE's potential for cost-effective surveillance and targeted interventions positions it as a promising complement to traditional methods, shaping the future of infectious disease monitoring.
Data availability statement
The data associated with this study has been deposited into a publicly available repository which is linked here: https://github.com/biruhalem/wastewater.
CRediT authorship contribution statement
Ramanath Majumdar: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation. Biruhalem Taye: Writing – review & editing, Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Data curation. Corey Bjornberg: Writing – review & editing, Methodology, Investigation, Data curation, Conceptualization. Matthew Giljork: Writing – review & editing, Methodology, Formal analysis, Data curation. Danielle Lynch: Writing – review & editing, Investigation. Fadumasahra Farah: Writing – review & editing, Investigation. Intisar Abdullah: Writing – review & editing, Investigation. Kristin Osiecki: Writing – review & editing, Writing – original draft, Validation, Methodology, Data curation. Iris Yousaf: Writing – review & editing, Investigation. Aaron Luckstein: Writing – review & editing, Supervision, Data curation. Wendy Turri: Writing – review & editing, Methodology, Conceptualization. Priya Sampathkumar: Writing – review & editing, Methodology, Conceptualization. Ann M. Moyer: Writing – review & editing, Writing – original draft, Validation, Supervision, Methodology, Formal analysis. Benjamin R. Kipp: Writing – review & editing, Supervision, Methodology, Funding acquisition. Roberto Cattaneo: Writing – review & editing, Writing – original draft, Supervision, Project administration, Methodology, Funding acquisition, Conceptualization. Caroline R. Sussman: Writing – review & editing, Project administration, Funding acquisition, Conceptualization. Chanakha K. Navaratnarajah: Writing – review & editing, Writing – original draft, Visualization, Supervision, Methodology, Investigation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge Carol Wilusz (Colorado State University) for help with setting up the wastewater analysis pipeline, Sagar Chowdhury (Olmstead County Public Health) for providing the Concentrating Pipette Select instrument for wastewater processing, Matthew Binnicker and Dragana Milosevic (Department of Laboratory Medicine and Pathology, Mayo Clinic) for initial help with SARS-CoV-2 RNA quantification, the University of Minnesota Genomics Center (Minneapolis, MN) for running the targeted mutation ddPCR assays, and Metropolitan Council Environmental Services (Saint Paul, MN) for providing interpretation of those results. The Mayo Clinic COVID-19 task force provided funding for the project.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27974.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Ansari S.A., Farrah S.R., Chaudhry G.R. Presence of human immunodeficiency virus nucleic acids in wastewater and their detection by polymerase chain reaction. Appl. Environ. Microbiol. 1992;58(12):3984–3990. doi: 10.1128/aem.58.12.3984-3990.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deshpande J.M., Shetty S.J., Siddiqui Z.A. Environmental surveillance system to track wild poliovirus transmission. Appl. Environ. Microbiol. 2003;69(5):2919–2927. doi: 10.1128/AEM.69.5.2919-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oghuan J., Chavarria C., Vanderwal S.R., Gitter A., Ojaruega A.A., Monserrat C., Bauer C.X., Brown E.L., Cregeen S.J., Deegan J., et al. Wastewater surveillance suggests unreported Mpox cases in a low-prevalence area. medRxiv preprint. 2023 doi: 10.1101/2023.05.28.23290658. [DOI] [Google Scholar]
- 4.Sherchan S.P., Solomon T., Idris O., Nwaubani D., Thakali O. Wastewater surveillance of Mpox virus in Baltimore. Sci. Total Environ. 2023;891 doi: 10.1016/j.scitotenv.2023.164414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas K.V., Bijlsma L., Castiglioni S., Covaci A., Emke E., Grabic R., Hernandez F., Karolak S., Kasprzyk-Hordern B., Lindberg R.H., et al. Comparing illicit drug use in 19 European cities through sewage analysis. Sci. Total Environ. 2012;432:432–439. doi: 10.1016/j.scitotenv.2012.06.069. [DOI] [PubMed] [Google Scholar]
- 6.Escola Casas M., Schroter N.S., Zammit I., Castano-Trias M., Rodriguez-Mozaz S., Gago-Ferrero P., Corominas L. Showcasing the potential of wastewater-based epidemiology to track pharmaceuticals consumption in cities: comparison against prescription data collected at fine spatial resolution. Environ. Int. 2021;150 doi: 10.1016/j.envint.2021.106404. [DOI] [PubMed] [Google Scholar]
- 7.O'Brien J.W., Choi P.M., Li J., Thai P.K., Jiang G., Tscharke B.J., Mueller J.F., Thomas K.V. Evaluating the stability of three oxidative stress biomarkers under sewer conditions and potential impact for use in wastewater-based epidemiology. Water Res. 2019;166 doi: 10.1016/j.watres.2019.115068. [DOI] [PubMed] [Google Scholar]
- 8.Faraway J., Boxall-Clasby J., Feil E.J., Gibbon M.J., Hatfield O., Kasprzyk-Hordern B., Smith T. Challenges in realising the potential of wastewater-based epidemiology to quantitatively monitor and predict the spread of disease. J. Water Health. 2022;20(7):1038–1050. doi: 10.2166/wh.2022.020. [DOI] [PubMed] [Google Scholar]
- 9.Lowery S.A., Sariol A., Perlman S. Innate immune and inflammatory responses to SARS-CoV-2: implications for COVID-19. Cell Host Microbe. 2021;29(7):1052–1062. doi: 10.1016/j.chom.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badraoui R., Alrashedi M.M., El-May M.V., Bardakci F. Acute respiratory distress syndrome: a life threatening associated complication of SARS-CoV-2 infection inducing COVID-19. J. Biomol. Struct. Dyn. 2021;39(17):6842–6851. doi: 10.1080/07391102.2020.1803139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19(3):141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 16.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran H.N., Le G.T., Nguyen D.T., Juang R.S., Rinklebe J., Bhatnagar A., Lima E.C., Iqbal H.M.N., Sarmah A.K., Chao H.P. SARS-CoV-2 coronavirus in water and wastewater: a critical review about presence and concern. Environ. Res. 2021;193 doi: 10.1016/j.envres.2020.110265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orive G., Lertxundi U., Barcelo D. Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Sci. Total Environ. 2020;732 doi: 10.1016/j.scitotenv.2020.139298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill K., Zamyadi A., Deere D., Vanrolleghem P.A., Crosbie N.D. SARS-CoV-2 known and unknowns, implications for the water sector and wastewater-based epidemiology to support national responses worldwide: early review of global experiences with the COVID-19 pandemic. Water Quality Research Journal. 2021;56(2):57–67. doi: 10.2166/wqrj.2020.100. [DOI] [Google Scholar]
- 21.Rainey A.L., Loeb J.C., Robinson S.E., Lednicky J.A., McPherson J., Colson S., Allen M., Coker E.S., Sabo-Attwood T., Maurelli A.T., et al. Wastewater surveillance for SARS-CoV-2 in a small coastal community: effects of tourism on viral presence and variant identification among low prevalence populations. Environ. Res. 2022;208 doi: 10.1016/j.envres.2021.112496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lancaster E., Byrd K., Ai Y., Lee J. Socioeconomic status correlations with confirmed COVID-19 cases and SARS-CoV-2 wastewater concentrations in small-medium sized communities. Environ. Res. 2022;215(Pt 2) doi: 10.1016/j.envres.2022.114290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerin-Rechdaoui S., Bize A., Levesque-Ninio C., Janvier A., Lacroix C., Le Brizoual F., Barbier J., Amsaleg C.R., Azimi S., Rocher V. Fate of SARS-CoV-2 coronavirus in wastewater treatment sludge during storage and thermophilic anaerobic digestion. Environ. Res. 2022;214(Pt 4) doi: 10.1016/j.envres.2022.114057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milosevic D., Moyer A.M., Majumdar R., Kipp B.R., Yao J.D. A reverse-transcription droplet digital PCR assay to detect and quantify SARS-CoV-2 RNA in upper respiratory tract specimens. J. Clin. Virol. 2022;153 doi: 10.1016/j.jcv.2022.105216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majumdar R., Vrana J.A., Koepplin J.W., Milosevic D., Roden A.C., Garcia J.J. Kipp BR, Moyer AM: SARS-CoV-2 RNA detection in Formalin-Fixed Paraffin-Embedded (FFPE) tissue by droplet digital PCR (ddPCR) Clin. Chim. Acta. 2022;532:181–187. doi: 10.1016/j.cca.2022.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CDC: wastewater surveillance testing methods. National Wastewater Surveillance System (NWSS) 2023 https://www.cdc.gov/nwss/testing.html [Google Scholar]
- 27.McKinney W. In Proceedings of the 9th Python in Science Conference. 2010. Data structures for statistical computing in Python; pp. 51–56. [Google Scholar]
- 28.Harris C.R., Millman K.J., van der Walt S.J., Gommers R., Virtanen P., Cournapeau D., Wieser E., Taylor J., Berg S., Smith N.J., et al. Array programming with NumPy. Nature. 2020;585(7825):357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9(3):90–95. [Google Scholar]
- 30.Wickham H., Averick M., Bryan J., Chang W., McGowan L., François R., Grolemund G., Hayes A., Henry L., Hester J., et al. Welcome to the tidyverse. J. Open Source Softw. 2019;4(43) doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- 31.Kassambara A. 2023. Rstatix: Pipe-Friendly Framework for Basic Statistical Tests. [Google Scholar]
- 32.Kassambara A. Based Publication Ready Plots; 2023. Ggpubr: 'ggplot2. [Google Scholar]
- 33.Wickham H. Springer-Verlag; New York: 2009. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 34.Killick R. Eckley IA: changepoint: an R package for changepoint analysis. J. Stat. Software. 2014;58(3):1–19. doi: 10.18637/jss.v058.i03. [DOI] [Google Scholar]
- 35.Ahmad S., Ullah A., Al-Mdallal Q.M., Khan H., Shah K., Khan A. Fractional order mathematical modeling of COVID-19 transmission. Chaos, Solit. Fractals. 2020;139 doi: 10.1016/j.chaos.2020.110256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan A., Alshehri H.M., Abdeljawad T., Al-Mdallal Q.M., Khan H. Stability analysis of fractional nabla difference COVID-19 model. Results Phys. 2021;22 doi: 10.1016/j.rinp.2021.103888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauch W., Schenk H., Insam H., Markt R., Kreuzinger N. Data modelling recipes for SARS-CoV-2 wastewater-based epidemiology. Environ. Res. 2022;214(Pt 1) doi: 10.1016/j.envres.2022.113809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torabi F., Li G., Mole C., Nicholson G., Rowlingson B., Smith C.R., Jersakova R., Diggle P.J., Blangiardo M. Wastewater-based surveillance models for COVID-19: a focused review on spatio-temporal models. Heliyon. 2023;9(11) doi: 10.1016/j.heliyon.2023.e21734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schoen M.E., Wolfe M.K., Li L., Duong D., White B.J., Hughes B., Boehm A.B. SARS-CoV-2 RNA wastewater settled solids surveillance frequency and impact on predicted COVID-19 incidence using a distributed lag model. ACS ES T Water. 2022;2(11):2167–2174. doi: 10.1021/acsestwater.2c00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanchan S., Ogden E., Kesheri M., Skinner A., Miliken E., Lyman D., Armstrong J., Sciglitano L., Hampikian G. COVID-19 hospitalizations and deaths predicted by SARS-CoV-2 levels in Boise, Idaho wastewater. Sci. Total Environ. 2024;907 doi: 10.1016/j.scitotenv.2023.167742. [DOI] [PubMed] [Google Scholar]
- 41.Zhao L., Zou Y., Li Y., Miyani B., Spooner M., Gentry Z., Jacobi S., David R.E., Withington S., McFarlane S., et al. Five-week warning of COVID-19 peaks prior to the Omicron surge in Detroit, Michigan using wastewater surveillance. Sci. Total Environ. 2022;844 doi: 10.1016/j.scitotenv.2022.157040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karthikeyan S., Ronquillo N., Belda-Ferre P., Alvarado D., Javidi T., Longhurst C.A., Knight R. High-throughput wastewater SARS-CoV-2 detection enables forecasting of community infection dynamics in San Diego county. mSystems. 2021;6(2) doi: 10.1128/mSystems.00045-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ansari N., Kabir F., Khan W., Khalid F., Malik A.A., Warren J.L., Mehmood U., Kazi A.M., Yildirim I., Tanner W., et al. Environmental surveillance for COVID-19 using SARS-CoV-2 RNA concentration in wastewater - a study in District East, Karachi, Pakistan. Lancet Reg Health Southeast Asia. 2024;20 doi: 10.1016/j.lansea.2023.100299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galani A., Aalizadeh R., Kostakis M., Markou A., Alygizakis N., Lytras T., Adamopoulos P.G., Peccia J., Thompson D.C., Kontou A., et al. SARS-CoV-2 wastewater surveillance data can predict hospitalizations and ICU admissions. Sci. Total Environ. 2022;804 doi: 10.1016/j.scitotenv.2021.150151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demirhan H., dLagM An R package for distributed lag models and ARDL bounds testing. PLoS One. 2020;15(2) doi: 10.1371/journal.pone.0228812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schill R., Nelson K.L., Harris-Lovett S., Kantor R.S. The dynamic relationship between COVID-19 cases and SARS-CoV-2 wastewater concentrations across time and space: considerations for model training data sets. Sci. Total Environ. 2023;871 doi: 10.1016/j.scitotenv.2023.162069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H., Bai R., Zhao Z., Tao L., Ma M., Ji Z., Jian M., Ding Z., Dai X., Bao F., et al. Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci. Rep. 2018;38(6) doi: 10.1042/BSR20181170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dingle T.C., Sedlak R.H., Cook L., Jerome K.R. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin. Chem. 2013;59(11):1670–1672. doi: 10.1373/clinchem.2013.211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amoah I.D., Abunama T., Awolusi O.O., Pillay L., Pillay K., Kumari S., Bux F. Effect of selected wastewater characteristics on estimation of SARS-CoV-2 viral load in wastewater. Environ. Res. 2022;203 doi: 10.1016/j.envres.2021.111877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinheiro L., Emslie K.R. Basic concepts and validation of digital PCR measurements. Methods Mol. Biol. 2018;1768:11–24. doi: 10.1007/978-1-4939-7778-9_2. [DOI] [PubMed] [Google Scholar]
- 51.Pinheiro L.B., Coleman V.A., Hindson C.M., Herrmann J., Hindson B.J., Bhat S., Emslie K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012;84(2):1003–1011. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quan P.L., Sauzade M., Brouzes E. dPCR: a technology review. Sensors. 2018;18(4) doi: 10.3390/s18041271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.CDC . 2023. Trends in United States COVID-19 Hospitalizations, Deaths, Emergency Visits, and Test Positivity by Geographic Area.https://covid.cdc.gov/covid-data-tracker/#trends_weeklyhospitaladmissions_select_00 [Google Scholar]
- 54.Wu S.L., Mertens A.N., Crider Y.S., Nguyen A., Pokpongkiat N.N., Djajadi S., Seth A., Hsiang M.S., Colford J.M., Jr., Reingold A., et al. Substantial underestimation of SARS-CoV-2 infection in the United States. Nat. Commun. 2020;11(1):4507. doi: 10.1038/s41467-020-18272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boehm A.B., Wolfe M.K., White B., Hughes B., Duong D. Divergence of wastewater SARS-CoV-2 and reported laboratory-confirmed COVID-19 incident case data coincident with wide-spread availability of at-home COVID-19 antigen tests. PeerJ. 2023;11 doi: 10.7717/peerj.15631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varkila M.R.J., Montez-Rath M.E., Salomon J.A., Yu X., Block G.A., Owens D.K., Chertow G.M., Parsonnet J., Anand S. Use of wastewater metrics to track COVID-19 in the US. JAMA Netw. Open. 2023;6(7) doi: 10.1001/jamanetworkopen.2023.25591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Markov P.V., Ghafari M., Beer M., Lythgoe K., Simmonds P., Stilianakis N.I., Katzourakis A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023;21(6):361–379. doi: 10.1038/s41579-023-00878-2. [DOI] [PubMed] [Google Scholar]
- 58.Puhach O., Meyer B., Eckerle I. SARS-CoV-2 viral load and shedding kinetics. Nat. Rev. Microbiol. 2023;21(3):147–161. doi: 10.1038/s41579-022-00822-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prasek S.M., Pepper I.L., Innes G.K., Slinski S., Betancourt W.Q., Foster A.R., Yaglom H.D., Porter W.T., Engelthaler D.M., Schmitz B.W. Variant-specific SARS-CoV-2 shedding rates in wastewater. Sci. Total Environ. 2023;857(Pt 1) doi: 10.1016/j.scitotenv.2022.159165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Doremalen N., Singh M., Saturday T.A., Yinda C.K., Perez-Perez L., Bohler W.F., Weishampel Z.A., Lewis M., Schulz J.E., Williamson B.N., et al. SARS-CoV-2 Omicron BA.1 and BA.2 are attenuated in rhesus macaques as compared to Delta. Sci. Adv. 2022;8(46) doi: 10.1126/sciadv.ade1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki R., Yamasoba D., Kimura I., Wang L., Kishimoto M., Ito J., Morioka Y., Nao N., Nasser H., Uriu K., et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022;603(7902):700–705. doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Antia R., Halloran M.E. Transition to endemicity: understanding COVID-19. Immunity. 2021;54(10):2172–2176. doi: 10.1016/j.immuni.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Are E.B., Song Y., Stockdale J.E., Tupper P., Colijn C. COVID-19 endgame: from pandemic to endemic? Vaccination, reopening and evolution in low- and high-vaccinated populations. J. Theor. Biol. 2023;559 doi: 10.1016/j.jtbi.2022.111368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boehm A.B., Hughes B., Duong D., Chan-Herur V., Buchman A., Wolfe M.K., White B.J. Wastewater concentrations of human influenza, metapneumovirus, parainfluenza, respiratory syncytial virus, rhinovirus, and seasonal coronavirus nucleic-acids during the COVID-19 pandemic: a surveillance study. Lancet Microbe. 2023;4(5):e340–e348. doi: 10.1016/S2666-5247(22)00386-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoogeveen M.J., Hoogeveen E.K. Comparable seasonal pattern for COVID-19 and flu-like illnesses. One Health. 2021;13 doi: 10.1016/j.onehlt.2021.100277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wiemken T.L., Khan F., Puzniak L., Yang W., Simmering J., Polgreen P., Nguyen J.L., Jodar L., McLaughlin J.M. Seasonal trends in COVID-19 cases, hospitalizations, and mortality in the United States and Europe. Sci. Rep. 2023;13(1):3886. doi: 10.1038/s41598-023-31057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larsen D.A., Wigginton K.R. Tracking COVID-19 with wastewater. Nat. Biotechnol. 2020;38(10):1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hutchison J.M., Li Z., Chang C.N., Hiripitiyage Y., Wittman M., Sturm B.S.M. Improving correlation of wastewater SARS-CoV-2 gene copy numbers with COVID-19 public health cases using readily available biomarkers. FEMS Microbes. 2022;3 doi: 10.1093/femsmc/xtac010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swift C.L., Isanovic M., Correa Velez K.E., Norman R.S. SARS-CoV-2 concentration in wastewater consistently predicts trends in COVID-19 case counts by at least two days across multiple WWTP scales. Environ Adv. 2023;11 doi: 10.1016/j.envadv.2023.100347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keshaviah A., Huff I., Hu X.C., Guidry V., Christensen A., Berkowitz S., Reckling S., Noble R.T., Clerkin T., Blackwood D., et al. Separating signal from noise in wastewater data: an algorithm to identify community-level COVID-19 surges in real time. Proc. Natl. Acad. Sci. U. S. A. 2023;120(31) doi: 10.1073/pnas.2216021120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olesen S.W., Imakaev M., Duvallet C. Making waves: defining the lead time of wastewater-based epidemiology for COVID-19. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ryerson A.B., Lang D., Alazawi M.A., Neyra M., Hill D.T., St George K., Fuschino M., Lutterloh E., Backenson B., Rulli S., et al. Wastewater testing and detection of poliovirus type 2 genetically linked to virus isolated from a paralytic polio case - New York, March 9-october 11, 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71(44):1418–1424. doi: 10.15585/mmwr.mm7144e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolfe M.K., Yu A.T., Duong D., Rane M.S., Hughes B., Chan-Herur V., Donnelly M., Chai S., White B.J., Vugia D.J., et al. Use of wastewater for Mpox outbreak surveillance in California. N. Engl. J. Med. 2023;388(6):570–572. doi: 10.1056/NEJMc2213882. [DOI] [PubMed] [Google Scholar]
- 74.Kirby A.E., Walters M.S., Jennings W.C., Fugitt R., LaCross N., Mattioli M., Marsh Z.A., Roberts V.A., Mercante J.W., Yoder J., et al. Using wastewater surveillance data to support the COVID-19 response - United States, 2020-2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70(36):1242–1244. doi: 10.15585/mmwr.mm7036a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kilaru P., Hill D., Anderson K., Collins M.B., Green H., Kmush B.L., Larsen D.A. Wastewater surveillance for infectious disease: a systematic review. Am. J. Epidemiol. 2023;192(2):305–322. doi: 10.1093/aje/kwac175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data associated with this study has been deposited into a publicly available repository which is linked here: https://github.com/biruhalem/wastewater.