Abstract
The chemokine receptors CCR5 and CXCR4, in combination with CD4, mediate cellular entry of macrophage-tropic (M-tropic) and T-cell-tropic strains of human immunodeficiency virus type 1 (HIV-1), respectively, while dualtropic viruses can use either receptor. We have constructed a panel of chimeric viruses and envelope glycoproteins in which various domains of the dualtropic HIV-1DH12 gp160 were introduced into the genetic background of an M-tropic HIV-1 isolate, HIV-1AD8. These constructs were employed in cell fusion and virus infectivity assays using peripheral blood mononuclear cells, MT4 T cells, primary monocyte-derived macrophages, or HOS-CD4 cell lines, expressing various chemokine receptors, to assess the contributions of different gp120 subdomains in coreceptor usage and cellular tropism. As expected, the dualtropic HIV-1DH12 gp120 utilized either CCR3, CCR5, or CXCR4, whereas HIV-1AD8 gp120 was able to use only CCR3 or CCR5. We found that either the V1/V2 or the V3 region of HIV-1DH12 gp120 individually conferred on HIV-1AD8 the ability to use CXCR4, while the combination of both the V1/V2 and V3 regions increased the efficiency of CXCR4 use. In addition, while the V4 or the V5 region of HIV-1DH12 gp120 failed to confer the capacity to utilize CXCR4 on HIV-1AD8, these regions were required in conjunction with regions V1 to V3 of HIV-1DH12 gp120 for efficient utilization of CXCR4. Comparison of virus infectivity analyses with various cell types and cell fusion assays revealed assay-dependent discrepancies and indicated that events occurring at the cell surface during infection are complex and cannot always be predicted by any one assay.
CD4-positive T lymphocytes and cells of the monocyte-macrophage lineage are the primary targets of human immunodeficiency virus type 1 (HIV-1) in vivo. Nearly all HIV-1 isolates are initially recovered by cocultivation with uninfected human peripheral blood mononuclear cells (PBMC), and all grow to varying extents in this cell type. Virus isolates have been divided into subgroups based on properties they exhibit during in vitro infectivity assays: (i) macrophage-tropic (M-tropic) isolates can infect cultures of primary human macrophages and PBMC but not continuous T-cell lines, (ii) T-cell-tropic (T-tropic) isolates can replicate in T-cell lines and PBMC, but not in monocyte-derived macrophage (MDM) cultures, and (iii) dualtropic isolates can infect both MDM and T-cell lines as well as PBMC. Several studies have reported that the G protein-coupled chemokine receptors CCR5 and CXCR4, in combination with CD4, mediate the entry of M-tropic and T-tropic strains of HIV-1, respectively (1, 12, 17–19, 22). Furthermore, chemokines that naturally bind to these receptors (i.e., SDF-1 for CXCR4 and RANTES, MIP-1α, or MIP-1β for CCR5) suppress HIV-1 replication at the level of virus entry (1, 3, 13, 17, 19, 30, 32). The chemokine receptors CCR2b, CCR3, and CCR8 have also been demonstrated to be used by some HIV-1 isolates (12, 18, 24, 33), although the significance of this usage is not clearly understood.
The cellular tropism of HIV-1 is determined largely at the level of virus entry and has been mapped primarily to the V3 variable region of the HIV-1 gp120 (6, 9, 16, 25, 31, 36, 40) although other portions of the envelope glycoprotein may also have some role (23, 26, 27, 35, 41). Consistent with such findings, (i) the ability of the M-tropic HIV-1JR-FL gp120 to block the binding of MIP-1β to CCR5 is prevented upon deletion of the V3 loop (39), (ii) antibodies against the V3 loop of HIV-1JR-FL gp120 prevent the protein from blocking MIP-1β binding to CCR5 (39), (iii) the utilization of CCR5 by M-tropic HIV-1 strains (ADA and YU2) has been mapped to the V3 region of gp120 (12), and (iv) the sensitivity of another M-tropic virus (HIV-1Bal) to β-chemokines (RANTES, MIP-1α, and MIP-1β) is conferred by the V3 region of gp120 (14). Dualtropic viruses (e.g., HIV-189.6) can infect PBMC and MDM as well as T-cell lines and are able to use either CCR5 or CXCR4 (15, 18, 37). However, relatively little is known about the determinants of dualtropic viral envelope proteins that enable them to use multiple chemokine receptors for cellular entry.
Viruses and vectors.
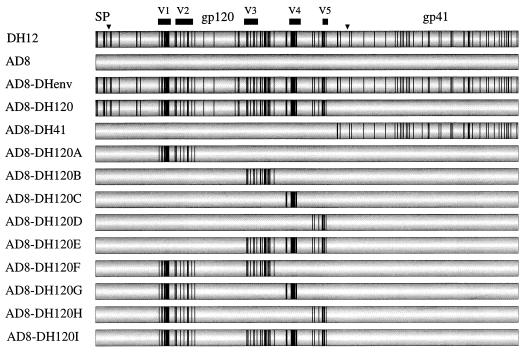
In a previous study (10), we generated a panel of viruses with chimeric envelope glycoproteins in which individual regions of the env gene of HIV-1DH12, a dualtropic virus strain, were transferred to HIV-1AD8, a full-length, infectious molecular clone of M-tropic HIV-1ADA, in order to identify envelope glycoprotein determinants conferring infectivity for chimpanzee PBMC (Fig. 1).
FIG. 1.
Structure of the chimeric viruses generated between HIV-1DH12 and HIV-1AD8. Shown is a schematic diagram of the HIV-1 envelope, with processing sites (inverted triangles) and cleavage products (signal peptide [SP], gp120, and gp41) indicated. Vertical lines, individual amino acid residues of HIV-1DH12 that are divergent from those of HIV-1AD8; solid bars, variable regions of gp120 (V1 to V5).
Recombinant vaccinia viruses and vectors used in this study were generated from these constructs as follows. The HIV-1DH125 (molecular clone no. 5 of HIV-1DH12) envelope gene was PCR amplified from pDH125 (34). The amplified DNA was digested with XmaI and SacII (introduced in the PCR primers) and inserted into the same sites in pNVV-3 (provided by M. Oldstone), resulting in pNVVDHenv*. Sequence analysis of pNVVDHenv* revealed two mutations introduced during PCR amplification. The mutations were removed by replacing KpnI (nucleotide 6340; numbering based on pAD8 [38])-MscI (in pNVV-3) with KpnI-Bpu102I (nucleotide 8853) subsequent to creation of a blunt end (for the Bpu1102I site) with Klenow fragment. The resulting plasmid was designated pNVVDHenv. Similarly, to generate pNVVADenv and pNVVDH120A, the KpnI-MscI fragment of pNVVDHenv* was replaced with the KpnI-Bpu1102I fragment from pAD8 and pAD8-DH120A (10), respectively. To generate plasmids pNVVDH120B to -I, the KpnI-AvrII (nucleotide 7990) fragment of pNVVADenv was replaced with the analogous fragment from pAD8-DH120B to -I (10). pNVVDHenv, pNVVADenv, and pNVVDH120A were used to generate recombinant vaccinia viruses (vvDHenv, vvADenv, and vv120Aenv, respectively) by methods described previously (11).
Coreceptor usage by HIV-1DH12–HIV-1AD8 chimeric envelope glycoproteins in cell fusion.
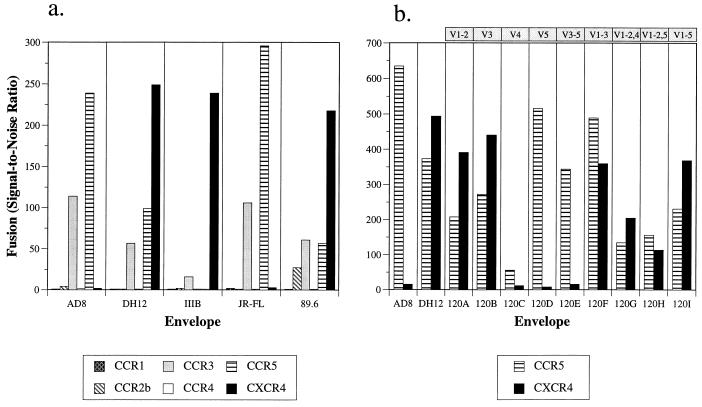
The availability of the panel of HIV-1DH12–HIV-1AD8 chimeric viruses provided an opportunity to assess the contributions of different gp120 subdomains from a dualtropic isolate to the use of different HIV-1 coreceptors. To do this, we first examined the abilities of the parental and chimeric envelope proteins to mediate fusion with quail QT6 cells expressing various chemokine receptors in conjunction with CD4. To quantitate cell fusion events, a luciferase-based gene reporter assay was used as previously described with some modification (18, 29). We found that HIV-1DH12 gp160 mediated fusion with cells expressing CXCR4, CCR5, and CCR3 (Fig. 2a). Unlike HIV-189.6, another dualtropic strain, HIV-1DH12 did not use CCR2b. In contrast, HIV-1AD8 gp160, like that of the M-tropic HIV-1JR-FL, utilized only CCR5 and CCR3. Thus, construction of chimeric envelope proteins provided an opportunity to determine which regions of HIV-1DH12 gp120, when introduced into HIV-1AD8, conferred use of CXCR4.
FIG. 2.
Use of chemokine receptors in a cell fusion assay. (a) The abilities of the HIV-1DH12 (vvDHenv), HIV-1AD8 (vvADenv), HIV-1IIIB (BH8 clone, vSC60 [5]), HIV-1JR-FL (vCB28 [5]), and HIV-189.6 (vBD3 [18]) envelope glycoproteins to mediate fusion with quail QT6 cells expressing CD4 and the indicated coreceptors were determined by a gene reporter fusion assay. The extent of fusion was expressed (as signal-to-noise ratio) relative to background levels (i.e., the spontaneous fusion seen with QT6 cells expressing CD4 and CXCR2). (b) Fusion of the chimeric envelope proteins (expressed by transfecting pNVVDH120A to -I) with cells expressing CCR5 or CXCR4 in conjunction with CD4 was determined. The results were averaged for three experiments, each of which gave identical patterns of coreceptor usage for each envelope protein. Absolute numbers varied between experiments due to differences in cell numbers and transfection efficiencies in independent experiments. It also should be noted that the time elapsing between the addition of the substrate and the determination of luciferase activity strongly influences the absolute light units obtained, though not the pattern of coreceptor use. The variable regions of HIV-1DH12 gp120 transferred into the chimeric envelope are indicated at the top.
We found that the chimeric envelopes containing either the V1/V2 or the V3 domain of HIV-1DH12 (AD8-DH120A and -120B, respectively) in an HIV-1AD8 envelope background acquired the ability to utilize CXCR4 in addition to CCR5 (Fig. 2b). In contrast, chimeric envelopes containing either the V4 or V5 region of HIV-1DH12 (AD8-DH120C and -120D, respectively), like their HIV-1AD8 parent, continued to use CCR5 only (albeit at a reduced level in the case of AD8-DH120C). Combining the V1/V2 and V3 regions of gp120 (AD8-DH120F) resulted in coreceptor usage similar to that observed with chimeric envelope glycoproteins containing either region individually. Other combinations of the different gp120 domains exhibited intermediate phenotypes. These results indicated that CXCR4 utilization in cell fusion assays by HIV-1DH12 is largely conferred by the V1/V2 or the V3 region of gp120.
Coreceptor usage by HIV-1DH12–HIV-1AD8 chimeric viruses during infections of HOS-CD4 cell lines.
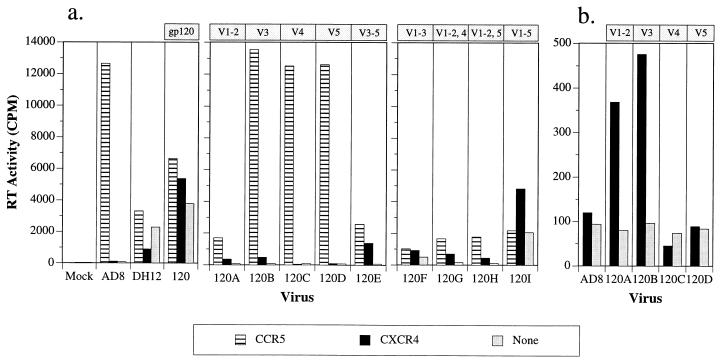
To extend our investigations of chemokine receptor utilization to cells undergoing spreading virus infections, individual HOS-CD4 cell lines expressing various chemokine receptors (8, 17) were infected with viruses containing the chimeric gp160 coding regions. Infectivity was monitored by measuring virion-associated reverse transcriptase (RT) activity released into the culture medium 4 days following infection. Only HOS-CD4 cells expressing CCR5 were susceptible to HIV-1AD8 infection (Fig. 3a and data not shown). In contrast, HIV-1DH12 and the chimeric virus encoding the entire HIV-1DH12 gp120 (AD8-DH120) established spreading infections in all of the HOS-CD4 cell lines, including the cell line expressing no cloned chemokine receptor. This result is undoubtedly due to the presence of low endogenous levels of CXCR4 on HOS cells, detectable by RT-PCR (28a) but not by fluorescence-activated cell sorter analyses (20) and is consistent with the observation that the envelope glycoproteins of some T-tropic HIV-1 isolates (HXB2 and SF2) also exhibit low levels of infectivity in HOS-CD4 cells (8, 17).
FIG. 3.
Infectivity of chimeric HIV-1 in HOS-CD4 cell lines expressing CCR5, CXCR4, or no chemokine coreceptor (HOS-CD4/pBABE-puro). Stocks of parental and chimeric HIV-1 were prepared as previously described (10). HOS cells were seeded (0.5 × 104 to 1 × 104 cells/well) in a 96-well plate 1 day prior to infection and were infected with equivalent amounts of viruses (normalized by virion-associated RT activity [42]). After an overnight incubation, the virus inoculum was removed and culture medium was added to a final volume of 250 μl. One hundred fifty microliters of culture medium was replaced on day 2 postinfection, and virus replication was monitored by measuring virion-associated RT activity on day 4 postinfection (a). The variable regions of HIV-1DH12 gp120 transferred into the chimeric envelope are indicated at the top. (b) Infectivities of HIV-1AD8 and the chimeric viruses AD8-DH120A to -D in HOS-CD4 cells expressing CXCR4 only. A smaller scale than that for panel a is shown here. The virion-associated RT activity in 0.8-μl aliquots of the culture supernatant is plotted.
As we observed in the quail cell fusion assay, the V3 but not the V4 or the V5 region of HIV-1DH12 gp120 conferred low but detectable CXCR4 utilization on HIV-1AD8 (Fig. 3b). Likewise, the chimera containing only the V1/V2 domain of HIV-1DH12 gp120 replicated above background levels in HOS-CD4 cells expressing CXCR4 (Fig. 3b) but lost significant infectivity for CCR5-expressing cells (Fig. 3a). While the V1/V2 or the V3 domain from HIV-1DH12 conferred on HIV-1AD8 the ability to replicate in HOS-CXCR4 cells, additional HIV-1DH12 domains were required for more efficient utilization of CXCR4. Thus, combining either the variable regions V3 to V5 (AD8-DH120E) or the variable regions V1 to V3 (AD8-DH120F) in the same chimeric gp120 resulted in a significant loss of infectivity for CCR5-expressing HOS-CD4 cells but an increased utilization of CXCR4 (Fig. 3a). Combining the V1/V2 region with the V3 (AD8-DH120F), but not the V4 (AD8-DH120G) or V5 (AD8-DH120H) region, resulted in a chimeric virus that began to resemble the parental HIV-1DH12, exhibiting the capacity to infect multiple HOS-CD4 cell lines, including the one expressing no cloned chemokine receptor. The chimeric virus AD8-DH120I, which contains all the variable regions of HIV-1DH12 gp120, produced higher levels of progeny virions than AD8-DH120F and also infected all of the HOS-CD4 cell lines. Taken together, these results strongly suggest that although variable regions V1/V2 and V3 together are sufficient to confer some of the properties of the parental HIV-1DH12 gp120 on HIV-1AD8, all of the HIV-1DH12 gp120 variable regions must be present (and presumably interact) for the acquisition of the complete functional phenotype.
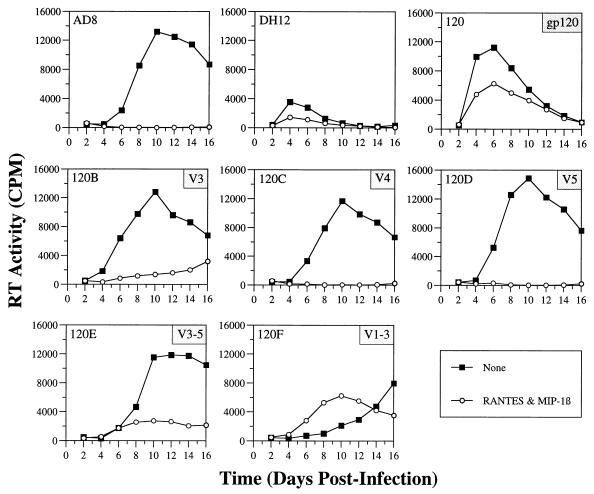
Effects of β-chemokines on the infectivity of HIV-1DH12–HIV-1AD8 chimeric viruses in human PBMC.
Although both cell fusion and HOS-CD4 cell infectivity analyses have been widely used to assess chemokine receptor usage by HIV-1 strains, overexpression of one or more of the various components associated with these assays (e.g., gp160, CD4, and/or the different chemokine receptors) could influence the results obtained. The ability of HIV-1 strains to use CCR3, for example, is strongly dependent upon cell surface expression levels (33). Therefore, to further assess coreceptor usage and cellular tropism of the chimeric viruses in a more physiologically relevant system, infectivity assays were carried out in three human cell types: PBMC, MDMs, and the MT4 T-cell line.
As previously reported, three of the intra-gp120 chimeric viruses (AD8-DH120A, -120G, and -120H) were unable to establish infections in human PBMC (10), although all three possessed envelope glycoproteins that utilized both CCR5 and CXCR4 in quail cell fusion and HOS-CD4 cell infectivity assays (Fig. 2 and 3). However, all three viruses used CCR5 much less efficiently than the parental virus, HIV-1AD8. All other chimeric viruses replicated in PBMC to varying degrees. To determine if infection of PBMC was mediated by CCR5, infections were performed in the presence of the β-chemokines RANTES and MIP-1β (Fig. 4). While infectivity of the M-tropic HIV-1AD8 was reduced to background levels by the two β-chemokines, HIV-1DH12 and AD8-DH120 were only partially sensitive, presumably due to their ability to utilize CXCR4. Infection of PBMC with the chimeric viruses revealed that AD8-DH120C and -120D (containing the V4 or the V5 region of HIV-1DH12 gp120, respectively) were completely sensitive to the β-chemokines, consistent with their inability to utilize CXCR4. In contrast, chimeric viruses AD8-DH120B and -120E, both of which contain the V3 region from HIV-1DH12 gp120 and could utilize CXCR4, were only partially sensitive. The chimeric virus AD8-DH120F, which carries both the V1/V2 and V3 regions of HIV-1DH12 gp120, was consistently more resistant to β-chemokines than AD8-DH120B and -120E. These results confirm that the chimeric viruses AD8-DH120C and -120D use CCR5 exclusively, that the HIV-1DH12 gp120 V3 region confers the capacity to utilize CXCR4, and that a cooperative interaction between the V1/V2 and V3 regions of gp120 very likely increases the affinity for CXCR4 compared to those of the chimeric gp120s containing either variable region alone.
FIG. 4.
Effects of β-chemokines on the infectivities of parental and chimeric viruses in human PBMC. Phytohemagglutinin-blasted PBMC (105 cells in 250 to 300 μl) were inoculated with equivalent amounts of virus (normalized by RT activity) in 96-well plates as previously described (10). One day postinfection, 200 μl of the inoculum was replaced with fresh medium. On the days indicated, 150 μl of the infection culture medium was harvested for RT assays and replaced with an equal volume of fresh medium. For infections with β-chemokines, PBMC were incubated with RANTES and MIP-1β (each at 200 ng/ml; R & D Systems) for 20 to 30 min at room temperature prior to the addition of virus. Subsequent to overnight adsorption, cells were cultured in medium containing β-chemokines at 100 ng/ml. Harvested samples were stored at −80°C and analyzed for virion-associated RT activity when samples had been collected at all the time points. Virion-associated RT activities in 0.8-μl aliquots of the culture supernatant are shown.
Infectivities of the HIV-1DH12–HIV-1AD8 chimeric viruses in MT4 T cells and human MDMs.
The tropic properties of the HIV-1DH12–HIV-1AD8 chimeric viruses were further examined by evaluating their infectivities in the MT4 human T-cell line. As expected, viruses bearing the entire DH12 gp160 or gp120 readily established spreading infections in MT4 cells, while HIV-1AD8 did not (data not shown). Surprisingly, when the replication competence of the intra-gp120 chimeric viruses for MT4 cells was evaluated, only AD8-DH120I, which contains all of the variable regions of HIV-1DH12 gp120, was infectious (data not shown), although many of the chimeric envelopes were able to utilize CXCR4 in fusion assays or in HOS-CD4 cell and PBMC infectivity assays.
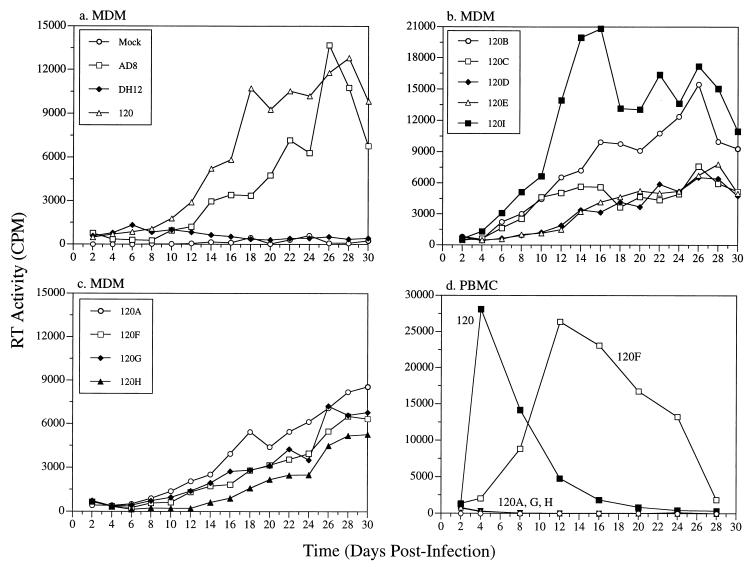
In contrast to the results obtained with MT4 cells, all the intra-gp120 chimeric viruses had demonstrable infectivity for MDMs (Fig. 5a to c), although their replication kinetics varied greatly. However, as noted earlier, three of these chimeric viruses, all of which carry the V1/V2 region of HIV-1DH12 gp120, were unable to infect PBMC (Fig. 5d). Combining the V3 (AD8-DH120F) region, but not the V4 or the V5 region (AD8-DH120G and -120H, respectively), with the V1/V2 region of HIV-1DH12 gp120 restored infectivity in PBMC. In MDMs, however, the chimeric viruses AD8-DH120A, -120G, and -120H replicated as well as AD8-DH120F (Fig. 5c). The ability of these three intra-gp120 chimeric viruses to replicate in MDMs but not in PBMC was not due to accumulation of mutations during passage in MDMs, because the progeny virus produced in MDMs was still unable to infect PBMC yet maintained replication competence for MDMs (data not shown).
FIG. 5.
Replication of chimeric viruses in primary MDMs and PBMC. MDMs were prepared by differentiating freshly elutriated monocytes on Sterilin plates (Bibby Sterilin, Stone, Staffordshire, United Kingdom) over a 14-day period in macrophage medium (Dulbecco modified Eagle medium [DMEM], 10% pooled normal human serum, 1 mM sodium pyruvate, 2 mM glutamine, 25 U of penicillin/ml, and 25 μg of streptomycin/ml) as previously described (21). Differentiated MDMs were frozen in DMEM containing 20% human serum and 7.5% dimethyl sulfoxide. Cells were thawed and seeded 1 day prior to infection in 96-well plates (Nunc) (105 cells in 200 μl per well). MDMs (a through c) or PBMC (d) were infected with the parental virus or one of the chimeric viruses. Infection, sample collection, and analysis of progeny virus production were carried out as described for Fig. 4.
The unusual property of the three chimeric viruses (AD8-DH120A, -120G, and -120H) to infect MDMs but not PBMC (or MT4 cells), coupled with their capacity to utilize CCR5 and CXCR4 in both fusion and HOS-CD4 infectivity assays, prompted us to examine whether PBMC and MT4 cells were intrinsically unable to interact with the chimeric envelope glycoproteins associated with these viruses. Human PBMC and MT4 cells were therefore infected with recombinant vaccinia viruses expressing the gp160 of HIV-1DH12, HIV-1AD8, or AD8-DH120A. Infection of PBMC with any of these three vaccinia recombinants induced syncytium formation (data not shown). In contrast, only vvDHenv and vv120Aenv induced syncytia in MT4 cells (data not shown). These results suggest that AD8-DH120A gp160, when overexpressed by using a recombinant vaccinia virus vector, is able to interact with coreceptors on PBMC and MT4 cells and to induce cell fusion.
The utilization of distinct chemokine receptors by HIV-1 strains largely accounts for the mechanisms underlying viral tropism at the level of virus entry (1, 12, 17–19, 22). Thus, M-tropic virus strains require CCR5 in conjunction with CD4 for cellular entry, while T-tropic virus strains, which typically emerge in individuals several years after infection, utilize the chemokine receptor CXCR4. Dualtropic viruses, which may represent evolutionary intermediates in the transition from M to T tropism, can use either CCR5 or CXCR4 as a coreceptor (18, 37). The V3 region of gp120 plays an important role in governing whether CCR5 or CXCR4 is used (12, 14, 39). However, non-V3 regions in gp120 have also been implicated as playing a role in cellular tropism (23, 26, 27, 35, 41), and little is known about the regions in gp120 that enable dualtropic viruses to use both CCR5 and CXCR4 for cellular entry. We found that the HIV-1DH12 envelope protein, unlike that of the M-tropic strain HIV-1AD8, was able to efficiently utilize either CCR5 or CXCR4. By using chimeric HIV-1DH12–HIV-1AD8 envelope glycoproteins, we identified gp120 determinants that contribute to the dualtropic phenotype. Furthermore, by using both cell fusion and virus infection assays, we observed a number of assay-dependent discrepancies which suggest that events occurring at the cell surface during the establishment of HIV-1 infections are complex and cannot necessarily be predicted by any one assay.
The ability of HIV-1DH12 to efficiently utilize both CXCR4 and CCR5 as coreceptors in both cell fusion and virus infection assays indicates that HIV-1DH12 gp120 contains determinants that enable it to utilize two coreceptors that differ by approximately 79% in amino acid sequence in their extracellular regions. We found that either the V1/V2 or the V3 region of HIV-1DH12 gp120, when placed in the background of the M-tropic strain HIV-1AD8, resulted in chimeric envelope proteins that could utilize either CXCR4 or CCR5 for cell fusion and virus infection (Fig. 2 and 3). One interpretation of these results is that both the V1/V2 and V3 regions of HIV-1DH12 gp120 may interact physically with CXCR4 during the induction of cell fusion. Alternatively, it is possible that the presence of V1/V2 or V3 significantly alters the conformation of the resulting chimeric gp120, thereby permitting interaction with CXCR4. By contrast, the V4 and V5 domains of HIV-1DH12 did not, individually, confer on HIV-1AD8 the ability to use CXCR4.
While the V3 domain is clearly a major determinant of viral tropism, non-V3 regions of gp120 have also been implicated. Our results confirmed that non-V3 domains of gp120 can affect both tropism and coreceptor use. Specifically, the V1/V2 region of HIV-1DH12 conferred on HIV-1AD8 gp120 the ability to use CXCR4, although not as efficiently as the V3 domain. However, the insertion of both the V1/V2 and the V3 region of HIV-1DH12 resulted in a chimeric virus (AD8-DH120F) with augmented infectivity for CXCR4-expressing HOS-CD4 cells, suggesting a functional interaction between the V1/V2 and V3 regions of gp120, consistent with earlier reports (2, 7, 10, 23, 26, 35, 41). However, while the V1/V2 region from T-tropic gp120 can render M-tropic envelope proteins competent to mediate syncytium formation with T-cell lines (2, 23), reciprocal chimeras fail to confer M tropism on T-tropic envelope proteins (12). Thus, the V1/V2 domain from T-tropic and dualtropic viruses may confer the ability to use CXCR4 on M-tropic viruses, but the V1/V2 domain from M-tropic viruses may not be sufficient to impart the ability to use CCR5 on T-tropic envelope proteins. Finally, usage of CXCR4 by the intra-gp120 chimeric viruses was further enhanced by also including the V4 and V5 regions (AD8-DH120I). These results suggest that although the V1/V2 or the V3 region of gp120 by itself may interact with CXCR4, optimal utilization of the CXCR4 coreceptor likely requires all five variable regions of the dualtropic HIV-1DH12 gp120. This notion is consistent with the result showing that AD8-DH120I was the only intra-gp120 chimeric virus able to infect MT4 cells.
Our use of fusion and infection assays with several cell types revealed assay-dependent differences that not only could influence conclusions about envelope determinants of coreceptor usage, but also hint at some of the complexities at the cell surface required for a productive virus infection. We found excellent agreement between different assays in analysis of the parental viruses and AD8-DH120I, which contains all of the HIV-1DH12 variable regions in an HIV-1AD8 background. Assay-dependent differences were observed, however, with some of the other chimeric envelope proteins. For the most part, the coreceptor usage exhibited by the chimeric envelope glycoproteins in the fusion assay was mirrored by the infectivities of the corresponding viruses in HOS-CD4 cells. As in fusion assays, the V1/V2 or V3, but not the V4 or V5, region of HIV-1DH12 conferred CXCR4 usage (Fig. 3). Differences were sometimes observed in the relative utilization of CCR5 and CXCR4. For example, AD8-DH120A used CXCR4 well in the cell fusion assay but only marginally during infection of HOS cells. By contrast, AD8-DH120C used CCR5 very well during infection of HOS cells, yet relatively poorly for cell fusion. A more significant difference between the assays was observed with AD8-DH120E (containing the V3 through V5 regions of HIV-1DH12), which used only CCR5 in cell fusion assays but used both CCR5 and CXCR4 during infection of HOS cells. Some of these inconsistencies could merely reflect inherent differences in the two experimental systems. Since what was measured in the HOS infection assay was complete virus replication cycles rather than just entry, how the chimeric envelope proteins are processed, transported to the plasma membrane, and/or incorporated onto virus particles could affect the results.
The experiments showing that many of the chimeric envelope proteins and corresponding HIVs utilized CXCR4 and/or CCR5 to induce cell fusion or to infect HOS-CD4 cells yet were unable to infect PBMC and/or MT4 cells might, at first glance, seem contradictory. However, these divergent results could be, in part, due to differences in the surface concentrations of CD4, the coreceptors, and/or gp120 as well as to the innate property of a particular envelope protein (e.g., its affinity for a coreceptor). The infectivity of HIV-1 has been shown to be highly dependent on cell surface concentrations of CCR5 and CD4, as well as on the affinity between gp120 and CD4 (28, 43). Differences in CD4 and/or coreceptor molecule concentrations may also explain why some mutations in gp120 allowed replication of the virus in some T-cell lines but not in others (4). Additional studies quantitating various components involved in the fusion reaction in various cell types might allow better understanding of HIV-1 tropism.
Acknowledgments
We thank Alicia J. Buckler-White for sequence analysis and oligonucleotide synthesis, George Englund for preparing MDMs, and Michael Oldstone for providing pNVV-3. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: HOS-CD4 cell lines expressing CCR1, CCR2b, CCR3, CCR4, CCR5, and HOS-CD4/pBABE-puro from Nathaniel Landau. We thank Ron Willey, Riri Shibata, and Eric Freed for critical reviews of the manuscript.
Portions of this work were supported by National Institutes of Health grants AI-35383, AI-40880, and AI-38225 to R.W.D. J.F.B. is a predoctoral fellow of the Howard Hughes Medical Institute.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Andeweg A C, Leeflang P, Osterhaus A D M E, Bosch M L. Both the V2 and V3 regions of the human immunodeficiency virus type 1 surface glycoprotein functionally interact with other envelope regions in syncytium formation. J Virol. 1993;67:3232–3239. doi: 10.1128/jvi.67.6.3232-3239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 4.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broder C C, Berger E A. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cann A J, Churcher M J, Boyd M, O’Brien W, Zhao J Q, Zack J, Chen I S. The region of the envelope gene of human immunodeficiency virus type 1 responsible for determination of cell tropism. J Virol. 1992;66:305–309. doi: 10.1128/jvi.66.1.305-309.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrillo A, Ratner L. Cooperative effects of the human immunodeficiency virus type 1 envelope variable loops V1 and V3 in mediating infectivity for T cells. J Virol. 1996;70:1310–1316. doi: 10.1128/jvi.70.2.1310-1316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng-Mayer C, Liu R, Landau N R, Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chesebro B, Nishio J, Perryman S, Cann A, O’Brien W, Chen I S Y, Wehrly K. Identification of human immunodeficiency virus envelope gene sequences influencing viral entry into CD4-positive HeLa cells, T-leukemia cells, and macrophages. J Virol. 1991;65:5782–5789. doi: 10.1128/jvi.65.11.5782-5789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho M W, Shibata R, Martin M A. Infection of chimpanzee peripheral blood mononuclear cells by human immunodeficiency virus type 1 requires cooperative interaction between multiple variable regions of gp120. J Virol. 1996;70:7318–7321. doi: 10.1128/jvi.70.10.7318-7321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho M W, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 13.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 α, and MIP-1 β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 14.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 15.Collman R, Balliet J W, Gregory S A, Friedman H, Kolson D L, Nathanson N, Srinivasan A. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Jong J J, Goudsmit J, Keulen W, Klaver B, Krone W, Tersmette M, de Ronde A. Human immunodeficiency virus type 1 clones chimeric for the envelope V3 domain differ in syncytium formation and replication capacity. J Virol. 1992;66:757–765. doi: 10.1128/jvi.66.2.757-765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 18.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 19.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 20.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 21.Englund G, Theodore T S, Freed E O, Engelman A, Martin M A. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J Virol. 1995;69:3216–3219. doi: 10.1128/jvi.69.5.3216-3219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 23.Groenink M, Fouchier R A, Broersen S, Baker C H, Koot M, van’t Wout A B, Huisman H G, Miedema F, Tersmette M, Schuitemaker H. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 24.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 25.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 26.Koito A, Harrowe G, Levy J A, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koito A, Stamatatos L, Cheng-Mayer C. Small amino acid sequence changes within the V2 domain can affect the function of a T-cell line-tropic human immunodeficiency virus type 1 envelope gp120. Virology. 1995;206:878–884. doi: 10.1006/viro.1995.1010. [DOI] [PubMed] [Google Scholar]
- 28.Kozak S L, Platt E J, Madani N, Ferro F, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Landau, N. Personal communication.
- 29.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 31.O’Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 32.Oravecz T, Pall M, Norcross M A. Beta-chemokine inhibition of monocytotropic HIV-1 infection. Interference with a postbinding fusion step. J Immunol. 1996;157:1329–1332. [PubMed] [Google Scholar]
- 33.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibata R, Hoggan M D, Broscius C, Englund G, Theodore T S, Buckler-White A, Arthur L O, Israel Z, Schultz A, Lane H C, Martin M A. Isolation and characterization of a syncytium-inducing, macrophage/T-cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J Virol. 1995;69:4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 36.Shioda T, Levy J A, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theodore T S, Englund G, Buckler-White A, Buckler C E, Martin M A, Peden K W C. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res Hum Retroviruses. 1996;12:191–194. doi: 10.1089/aid.1996.12.191. [DOI] [PubMed] [Google Scholar]
- 39.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 40.Westervelt P, Gendelman H E, Ratner L. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc Natl Acad Sci USA. 1991;88:3097–3101. doi: 10.1073/pnas.88.8.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westervelt P, Trowbridge D B, Epstein L G, Blumberg B M, Li Y, Hahn B H, Shaw G M, Price R W, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willey R L, Smith D H, Lasky L A, Theodore T S, Earl P L, Moss B, Capon D J, Martin M A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]