Summary
Grain weight is an important determinant of grain yield. However, the underlying regulatory mechanisms for grain size remain to be fully elucidated. Here, we identify a rice mutant grain weight 9 (gw9), which exhibits larger and heavier grains due to excessive cell proliferation and expansion in spikelet hull. GW9 encodes a nucleus‐localized protein containing both C2H2 zinc finger (C2H2‐ZnF) and VRN2‐EMF2‐FIS2‐SUZ12 (VEFS) domains, serving as a negative regulator of grain size and weight. Interestingly, the non‐frameshift mutations in C2H2‐ZnF domain result in increased plant height and larger grain size, whereas frameshift mutations in both C2H2‐ZnF and VEFS domains lead to dwarf and malformed spikelet. These observations indicated the dual functions of GW9 in regulating grain size and floral organ identity through the C2H2‐ZnF and VEFS domains, respectively. Further investigation revealed the interaction between GW9 and the E3 ubiquitin ligase protein GW2, with GW9 being the target of ubiquitination by GW2. Genetic analyses suggest that GW9 and GW2 function in a coordinated pathway controlling grain size and weight. Our findings provide a novel insight into the functional role of GW9 in the regulation of grain size and weight, offering potential molecular strategies for improving rice yield.
Keywords: grain size and weight, floral organ identity, GW9, GW2, PRC2 complex, rice
Introduction
Grain weight is one of the three primary determinants of grain yield, which is tightly associated with grain size and filling (Xing and Zhang, 2010). A comprehensive exploration of the underlying molecular and genetic mechanisms of grain weight is beneficial to further improve crop yield. To date, numerous quantitative trait loci and genes related to grain size have been identified and characterized in rice, which are implicated in various pathways, including ubiquitin‐proteasome degradation, mitogen‐activated protein kinase (MAPK) signalling, G protein signalling, phytohormone signalling, and transcriptional regulation pathway (Li et al., 2018, 2019; Ren et al., 2023; Zuo and Li, 2014).
The ubiquitin‐proteasome pathway plays an important role in the precise regulation of grain size by affecting the stability, subcellular localization, and activity of the target proteins (Elsasser and Finley, 2005; Li et al., 2019). GW2, encoding a RING‐type E3 ubiquitin ligase, functions as a negative regulator of grain width and weight. Disruption of GW2 activity results in enhanced cell proliferation in spikelet hulls, resulting in wider and heavier grains (Song et al., 2007). The glutaredoxin protein WG1 has been identified as a substrate of GW2, which regulates grain width and weight by alleviating the inhibitory effect of WG1‐ASP1 on OsbZIP47 transcriptional activation activity (Hao et al., 2021). Additionally, GW2 targets the histone acetyltransferase protein GW6a, which acts as a positive regulator in controlling grain size and weight (Gao et al., 2021; Song et al., 2015). Another RING E3 ubiquitin ligase, CLG1, determines grain size by ubiquitinating the Gγ subunit GS3 and promoting its degradation via the endosome degradation pathway (Yang et al., 2021). Moreover, the HECT‐domain E3 ubiquitin ligase LARGE2 regulates grain number and width by modulating the stabilities of APO1 and APO2 (Huang et al., 2021). LG1 encodes ubiquitin‐specific protease 15 (OsUBP15), which has de‐ubiquitination activity and regulates grain length and width by promoting cell proliferation (Shi et al., 2019). The otubain‐like protease WTG1/OsOTUB1, also exhibiting deubiquitination activity, is involved in controlling grain size by restricting cell expansion (Huang et al., 2017). SMG3 and DGS1, functioning as an ERAD‐related E2‐E3 enzyme pair, positively regulate grain size and weight by ubiquitinating the BR receptor BRI1, thereby affecting its protein accumulation (Li et al., 2022).
Polycomb‐group (PcG) proteins are essential in plant development by forming multi‐protein complexes to repress target gene transcription (Bemer and Grossniklaus, 2012). There are two distinct complexes, PcG Repressive Complex 1 (PRC1) and PcG Repressive Complex 2 (PRC2) that are evolutionally conserved in eukaryotes (Forderer et al., 2016; Zhang et al., 2021a). In rice, PRC2 is composed of three core subunits: ESC homologues (OsFIE1 and OsFIE2), E(z)‐like homologues (OsCLF and OsiEZ1) and Su(z)12 homologues (OsEMF2a and OsEMF2b). These complexes play key roles in H3K27me3‐mediated epigenetic regulation, influencing critical developmental processes such as endosperm development, seed dormancy, plant height, flowering transition and floral organ development (Bieluszewski et al., 2021; Laugesen et al., 2019). Among them, OsFIE1, a maternally imprinted and confined to endosperm‐specific expression gene, is precisely regulated by DNA methylation (Zhang et al., 2012). The osfie1 mutants exhibit smaller grain size, delayed embryo development, and decreased seed set rate, while OsFIE1 overexpressing plants display short stature, reduced grain size and malformed floret (Cheng et al., 2020a; Huang et al., 2016; Zhang et al., 2012). OsFIE2, on the other hand, acts as an H3 methyltransferase and plays critical roles in reproductive growth, endosperm formation, grain yield and root development (Cheng et al., 2020b; Li et al., 2014; Liu et al., 2016). OsCLF/OsSDG711 and OsiEZ1/OsSDG718 have been identified as the enhancers of Zeste, which are primarily involved in regulating flowering time (Liu et al., 2014). Moreover, OsSDG711 has been reported to regulate panicle size via H3K27 methylation‐mediated suppression of key genes governing inflorescence meristem activity (Liu et al., 2015). OsEMF2a contributes to the developmental transitions post‐fertilization and endosperm cellularization, while OsEMF2b is predominantly involved in the regulation of flowering time and floral organ identity (Cheng et al., 2020b; Conrad et al., 2014; Deng et al., 2017; Luo et al., 2009; Tonosaki et al., 2021; Xie et al., 2015). In addition, RLB physically interacts with OsEMF2b and regulates lateral branching development through the mediation of H3K27me3 modification at the CKX4 promoter region (Wang et al., 2022). These reports provided substantial insights into the interpretation of PRC2 complex function, but the molecular mechanism underlying the PRC2 complex's involvement in regulating grain size remains to be further unveiled.
In this study, we isolated and characterized a rice mutant gw9, which exhibits increased plant height and grain weight. The gw9 phenotype is attributed a novel allelic mutation of OsEMF2b. Although previous research has demonstrated the involvement of GW9/OsEMF2b in the regulation of flowering time and floral organ identity, its function in modulating grain size and weight remains unexplored. Our results demonstrated that the C2H2‐ZnF and VEFS domain of GW9 play distinct roles in grain size and flower organ identity, respectively. Furthermore, we found that GW2 interacts physically with GW9 to regulate grain size via ubiquitination pathway.
Results
Phenotypic characterization of gw9 mutant
To gain deeper insights into the genetic networks controlling grain size, we identified and characterized a rice mutant gw9. Compared with wild‐type W7, the plant height and grain size of gw9 were markedly increased, but the tiller number was slightly reduced (Figure 1a–f). The average grain length, grain width, and 1000‐grain weight were increased by 22.81%, 12.33%, and 27.96%, respectively, but the ratio of grain length to width significantly decreased (Figure 1g–j). In addition, there were no significant differences observed in panicle length, the number of primary branches, or setting percentage, while grain number per panicle decreased due to a reduction in the number of secondary branches (Figure 1k–o). Furthermore, all internode lengths, diameters, and thicknesses of gw9 were significantly increased, with particular prominence in the third and fourth internodes (Figure S1). Moreover, dynamic observations of grain development indicated that both W7 and gw9 reached their maximum grain filling rates at 9 DAF (Day After Flowering), but with the rate in gw9 being significantly higher than that of W7 during the grain filling period (Figure S2).
Figure 1.
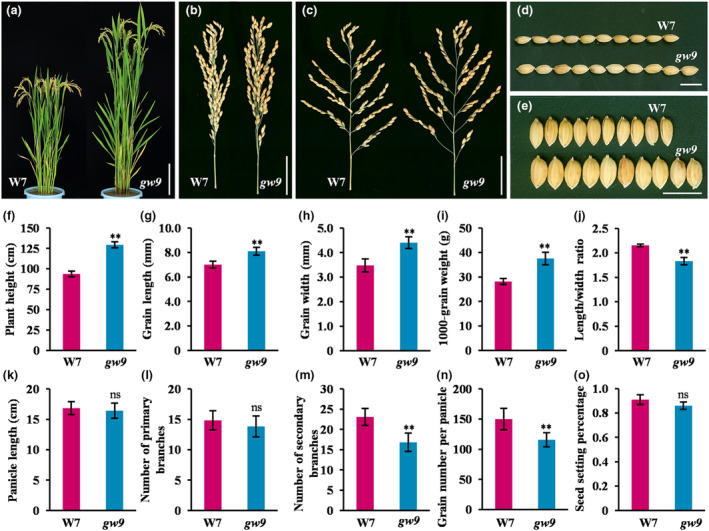
Phenotypic characterization of gw9 mutant. (a) Gross morphologies of W7 and gw9. Bars = 20 cm. (b, c) Panicle phenotypes of W7 and gw9. Bars = 5 cm. (d, e) Grain morphologies of W7 and gw9. Bars = 1 cm. (f–o) Comparison of agronomic traits between W7 and gw9 (n = 15). Error bars represent standard deviation (SD). **P < 0.01; ns, no significant difference (Student's t‐test).
GW9 regulates grain size by altering cell proliferation and cell expansion
Cell proliferation and expansion play vital roles in regulating spikelet hull growth. As expected, the spikelet hull of gw9 exhibited a significant increase in both length and width compared to those of W7 (Figure 2a). To clarify the cellular basis for the increase in grain size, we analysed the cell number and morphology of spikelet hulls by paraffin sections and scanning electron microscopy. The results showed that there are notable increases in the spikelet perimeters, cell layers and numbers, the average cell length as well as the width in outer and inner glumes of gw9 (Figure 2b–l), suggesting that the enlarged spikelet hull of gw9 was attributed to the increased cell number and size.
Figure 2.
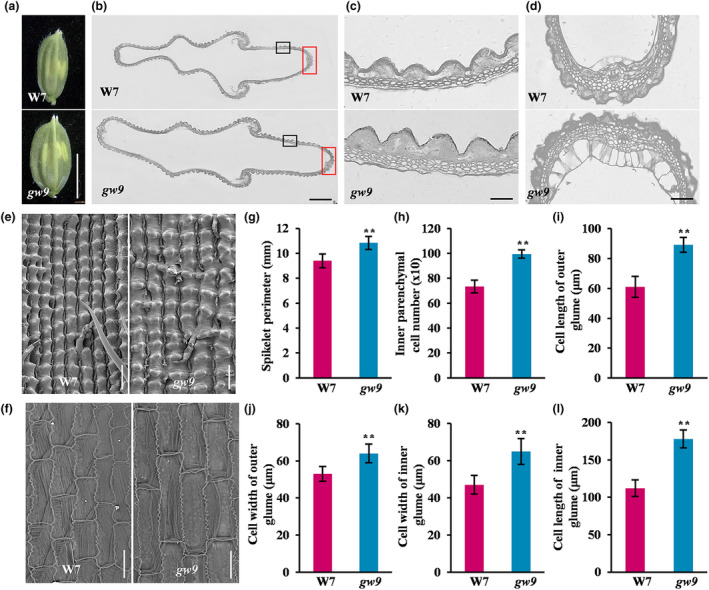
GW9 regulates rice grain size by controlling cell proliferation and cell expansion. (a) Young spikelet hulls of W7 and gw9. Bars = 5 mm. (b) Cross sections of spikelet hull. Bars = 500 μm. (c) Magnified views of the cross‐sections black boxed in (b). Bars = 50 μm. (d) Magnified views of the cross‐sections red boxed in (b). Bars = 50 μm. (e, f) Scanning electron micrographs of outer (e) and inner (f) surfaces of glums. Bars = 100 μm (e) and 50 μm (f), respectively. (g–l) Comparison analysis of W7 and gw9 for spikelet perimeter (g), inner parenchymal cell number (h), cell length and width in outer glume (i, j), cell width and length in inner glume (k, l). Error bars represent standard deviation (SD). **P < 0.01 (Student's t‐test).
Furthermore, we examined the expression levels of several genes known to regulate grain size by influencing cell proliferation and cell expansion. The results revealed that the expression of GL7, PGL1, and GLW7 were significantly enhanced, and GW8, FUWA, DST, and EP3 were dramatically reduced in gw9 (Figure S3a). In addition, we also detected the expression of several cell cycle‐related genes and observed significant upregulation in CAK1A, CDKB, CYCB2.1, CYCB2.2, MACM2, MACM3, and MACM4, along with remarkable decrease in CYCD2.1, CYCD4.1, CYCD4.2, CYCH1, and CYCT1.3 in gw9 (Figure S3b). These results implied that GW9 may modulate grain size by regulating the expression level of genes related to both cell expansion and cell proliferation.
Positional cloning of GW9 gene
Using a map‐based cloning strategy, we initially located GW9 on chromosome 9 between molecular markers P2 and P3 by employing 112 homozygous F2 plants. Then, we further narrowed down the region to a 50.2‐kb interval between the markers P8 and P9 by analysing 3469 F2 individuals (Figure 3a). Within this candidate region, four open reading frames (ORFs) were predicted based on the Rice Genome Annotation Project (RGAP) (Table S1), and only a single base substitution from G to A at site 7213 (from the start codon) was detected in the intron of LOC_Os09g13630 (Figure 3b,c). Intriguingly, this mutation was close to the 11th extron, which caused a 54‐bp alternative splicing deletion in the coding sequence (Figures 3d, S4a). Sequence analysis revealed that LOC_Os09g13630 encodes a polycomb group protein OsEMF2b, which contains a C2H2‐ZnF and VEFS domain. Further analysis showed the absence of 18 amino acids in the C2H2‐ZnF domain due to alternative splicing of the 11th exon in gw9 (Figure S4b).
Figure 3.
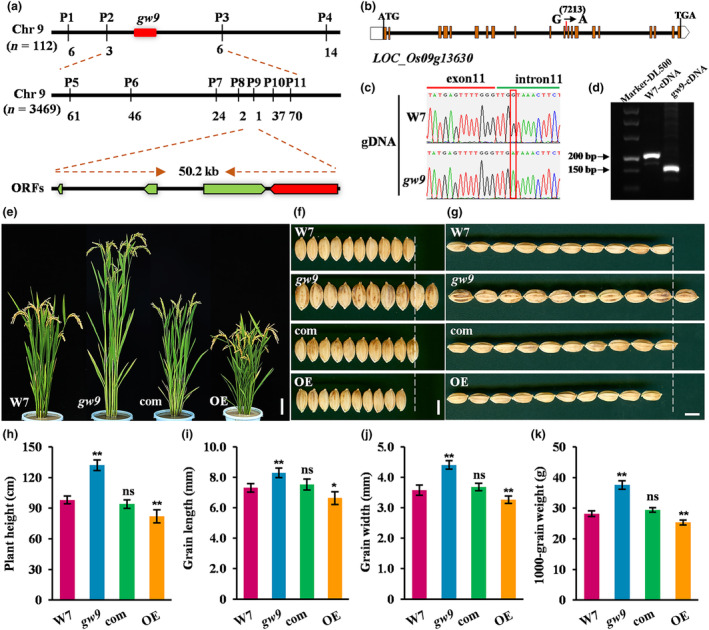
Map‐based cloning of GW9 and genetic complementation. (a) Gene mapping of GW9. The numerals indicate the number of recombinants. (b, c) Gene structure and mutation sites, the red box indicates the nucleotide substitution. (d) PCR analysis of the 54‐bp splicing deletion in cDNA. (e) Plant morphologies of W7, gw9, complementation lines (com) and overexpression lines (OE). Bar = 10 cm. (f, g) Grain morphologies of W7, gw9, com and OE. Bars = 5 mm. (h, i, j, k) Comparison analysis of plant height (h), grain length (i), grain width (j) and 1000‐grain weight (k). Error bars represent standard deviation (SD). **P < 0.01; *P < 0.05. ns, no significant difference.
GW9 negatively regulates grain size and plant height
To confirm the causal relationship between the phenotype of gw9 and the mutation of LOC_Os09g13630 gene, we performed a genetic complementation assay in gw9 background. A total of 36 positive complementary transgenic lines were obtained and all transgenic plants exhibited phenotypes similar to those of W7, including plant height, grain size, and grain weight (Figure 3e–k). To further elucidate the gene function of GW9, we introduced an overexpression vector of GW9 into W7, resulting in positive overexpression lines (OE) that displayed dwarfism and smaller grain size compared to W7 (Figure 3e–k). In addition, we performed expression level assay of GW9 in gw9 and different transgenic lines. The results showed that the expression level of GW9 was slightly decreased in gw9 and significantly increased in complementation and overexpression plants (Figure S5). Moreover, it has been reported that OsEMF2b is involved in regulating flowering time (Luo et al., 2009; Xie et al., 2015; Yang et al., 2013). We noticed that gw9 mutant flowered approximately 5.7 days earlier than W7, while the overexpression lines exhibited later flowering, occurring approximately 11.5 days (Figure S6). These results indicated that GW9/OsEMF2b plays a negative role in regulating grain size and plant height in rice, while positively controlling the heading date.
C2H2‐ZnF domain is required for grain size development
Different from all previously reported OsEMF2b mutations, the gw9 mutant here displayed increased plant height and grain size (Figure 1). In fact, all T‐DNA insertion and single‐base mutation in previous reports result in null alleles (Figure S7a,b) (Chen et al., 2017; Conrad et al., 2014; Deng et al., 2017; Wang et al., 2022; Xie et al., 2015; Zhong et al., 2018), but the 54‐bp fragment deletion within the C2H2‐ZnF domain of gw9 did not cause frameshift and had no effect on the integrity of the subsequent VEFS domain (Figure S7b). Consequently, we speculate that C2H2‐ZnF and VEFS domains of GW9 play distinct roles in the regulation of grain size and spikelet determinacy. To verify this hypothesis, we selected four target sites at different coding position within GW9 and obtained a number of novel alleles by using CRISPR/Cas9 technique (Figure S7c,d).
Gene knock‐out experiments revealed consistent results, with the frameshift mutants Cr‐2, Cr‐4, and Cr‐8 displaying similar phenotypes characterized by reduced plant stature, delayed flowering, malformed floral organs and sterility (Figures 4a–d, S8). Microscopic assay by paraffin section showed that the spikelet of three mutants displayed a series of abnormal morphologies, including the presence of extra glume‐like or mosaic organs, incurved and shrunken paleas, as well as deformities and extra carpels (Figures 4d,e, S8). In contrast, the non‐frameshift mutants Cr‐3, Cr‐5 and Cr‐6 within the C2H2‐ZnF domain exhibited increased plant height and larger grain size, which is similar to the phenotype of gw9 (Figure 4a,f–k). Notably, when the non‐frameshift mutation is close to the 5′ terminal, there were no significant phenotypes difference between Cr‐1 and W7 (Figure 4a–k). Therefore, we concluded that the VEFS domain is essential for maintaining floral organ identity, while the C2H2‐ZnF domain plays an important role in determining grain size and plant height.
Figure 4.
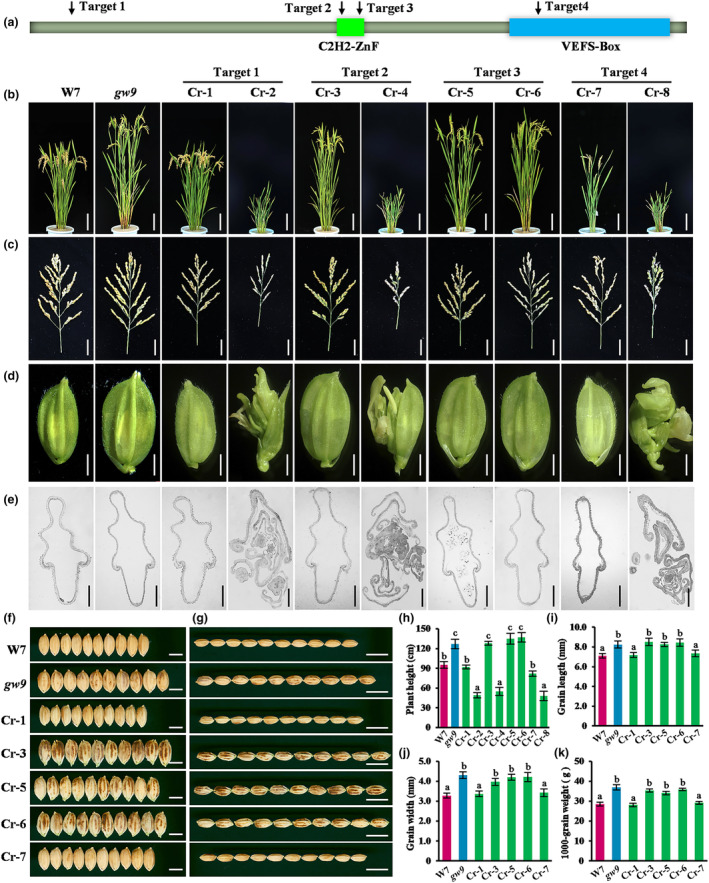
Gene knockouts of GW9. (a) Schematic diagram of GW9 protein structure. Green box indicates the C2H2‐ZnF domain, and blue box indicates the VEFS domain. Black arrows indicate the different target site of CRISPR/Cas9. (b) Plant morphologies of W7, gw9 and knockout plants. Cr‐1, Cr‐2, Cr‐3, Cr‐4, Cr‐5, Cr‐6, Cr‐7 and Cr‐8 represent the different homozygous knockout lines. Bars = 20 cm. (c, d) The panicle and spikelet morphologies of W7, gw9 and knockout plants. Bars = 5 cm (c) and 2 mm (d), respectively. (e) transverse section of W7, gw9 and knockout plants. Bars = 200 μm. (f, g) Grain morphologies of W7, gw9 and knockout plants. Bars = 0.5 cm (f) and 1 cm (g), respectively. (h–k) Comparison analysis of plant height (h), grain length (i), grain width (j) and 1000‐grain weight (k). Error bars represent standard deviation (SD). Statistical analyses were performed by Duncan's multiple range tests. The presence of the same lowercase letter denotes a non‐significant difference between means (P < 0.05).
Expression pattern and subcellular localization of GW9
To investigate the spatial expression pattern of GW9, we employed qRT‐PCR to analyse its expression levels in various organs, including the root, stem, leaf, leaf sheath, and panicle. The results showed that GW9 was generally expressed across all tested tissues, with higher expression level detected in leaf and young panicles (Figure 5a). Moreover, we transformed pGW9::GUS vector into W7 and performed histochemical staining, confirming GW9 activity in most of the examined organs (Figure 5b), consistent with qRT‐PCR result. To validate the subcellular localization of GW9 protein, we introduced the pActin::GW9‐GFP construct into gw9. The positive transgenic lines exhibited restored phenotypes, indicating the functionality of GW9‐GFP fusion protein (result not shown). We then examined the fluorescence of the GW9‐GFP signals in root tips by using fluorescence microscopy, and the results indicated that GW9 protein was located in the nucleus (Figure 5c). Moreover, the pUbi::GW9‐GFP vector was introduced into rice protoplasts and MADS29‐RFP was co‐expressed used as the nucleus marker. The co‐localization of GW9‐GFP and MADS29‐RFP signals confirmed that GW9 was localized in nucleus (Figure 5d).
Figure 5.
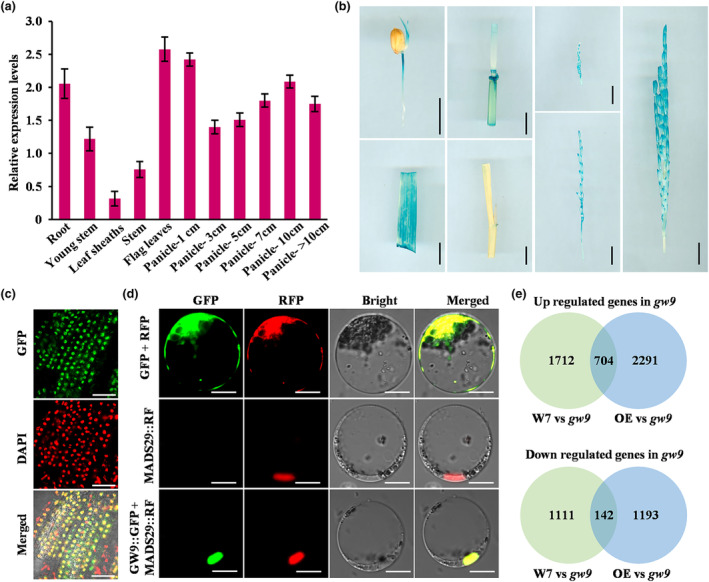
Subcellular localization and expression pattern of GW9. (a) Expression levels of GW9 in different organs. The number indicates the panicle length. OsActin was used as an internal control. (b) Tissue‐specific expression of the GUS gene driven by the GW9 promoter. Bars = 1 cm. (c) Nuclear localization of GW9 in GW9‐GFP transgenic plant roots. Bars = 50 μm. (d) Subcellular localization of GW9 in rice protoplasts. MADS29‐RFP was used as the nuclear marker. Bars = 10 μm. (e) Venn diagrams of differentially expressed genes. Error bars represent standard deviation (SD).
The transcriptome analyses of W7, gw9 and GW9‐OE were conducted by RNA‐seq, and the result showed that a total of 3669 differentially expressed genes (DEGs, false discovery rate <0.05 and induction fold >1) were identified between W7 and gw9, including 2416 up‐regulated and 1253 down‐regulated genes. Meanwhile, 4330 DEGs (2995 up‐regulated and 1335 down‐regulated) were identified between GW9‐OE and gw9 (Figure 5e, Table S3). Among these DEGs, numerous genes are related to the grain size and flowering time, such as DEP1, SPL7, SGL1, GW5, PGL1, CO3 and COL4. The heat map analysis further illustrated that the considerable alterations in their expression levels in W7, GW9‐OE and gw9 (Figure S9, Table S3). These results suggested that GW9 may regulate multiple growth and development processes by affecting the expression of these genes.
GW9 interacts with GW2
To elucidate the underlying mechanism of GW9 in grain size regulation, we conducted a yeast two‐hybrid assay (Y2H) screening to identify putative GW9‐interacting proteins. We identified a total of 15 proteins, among which only GW2 was found to be associated with grain size regulation (Figure S10). Previous research has shown that GW2 encodes a RING‐type protein with E3 ubiquitin ligase activity and is primarily located in the cytoplasm (Song et al., 2007). However, we found that GW9‐GFP fusion protein localized in nucleus, as shown above (Figure 5c,d). To verify whether GW2 and GW9 have overlapping subcellular localizations, pUbi::GW9‐GFP and pUbi::GW2‐RFP vectors were co‐expressed in rice protoplasts. The results demonstrated that GW2‐RFP fluorescence signal were detected in both the cytoplasm and nucleus, and there was an overlapping fluorescence pattern between GW9‐GFP and GW2‐RFP within the nucleus (Figure S11). Therefore, it can be concluded that GW9 has the potential to interact with GW2 in the nucleus.
Considering the partial absence of C2H2‐ZnF domain in gw9 mutant, we constructed a series of GAL4‐BD vectors containing different N‐terminal and C‐terminal truncated fragments of GW9 to determine the regions responsible for GW9‐GW2 interaction (Figure 6a). The results showed that the full‐length fusion protein (GW9‐FL), the N‐terminal containing C2H2‐ZnF (GW9‐N1), the C‐terminal containing VEFS domain (GW9‐C1), and C‐terminal containing VEFS and C2H2‐ZnF domains (GW9‐C2) interacted with GW2, but the absence or partial absence of C2H2‐ZnF in N‐terminal domains (GW9‐N2 and GW9‐N3) did not (Figure 6a). We then conducted the BiFC and LCI assays to verify the interaction between GW9 and GW2, and the YFP and Luciferase activity signals were detected when GW9 and GW2 co‐expressed in Nicotiana benthamiana (N. benthamiana) leaves (Figure 6b,c). In addition, the interaction between GW9 and GW2 was further confirmed by GST pull‐down and Co‐IP assay in vitro and vivo, respectively (Figure 6d,e). Collectively, these results strongly supported the physical interaction between GW9 and GW2.
Figure 6.
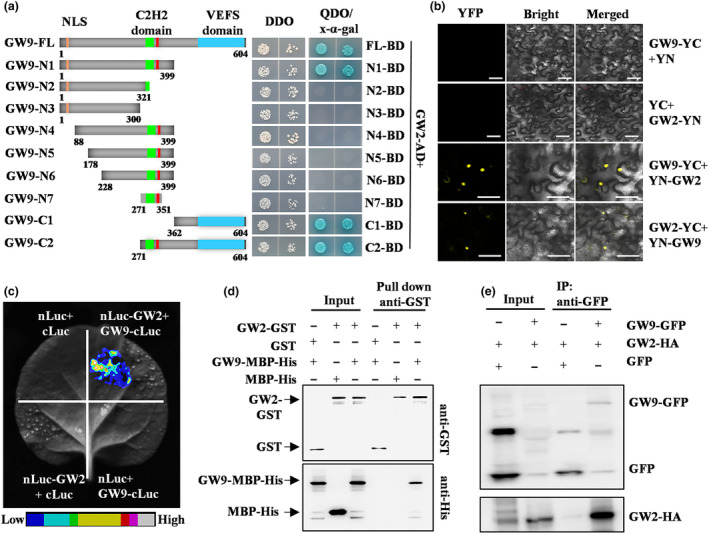
GW9 interacts with GW2 in vitro and in vivo. (a) Interactions of full‐length and truncated GW9 proteins with GW2 in yeast cells. At left, a schematic diagram of full‐length and truncated GW9 fusion proteins with pGBK‐T7 is shown; at right panel, the growth of transformed yeast on DDO (SD/‐Trp‐Leu) and QDO (SD/‐Trp‐Leu‐His‐Ade/x‐α‐gal) medium, respectively. α‐X‐gal staining shows the β‐galactosidase activity of the transformants. (b) BiFC assay for GW9 and GW2 interaction in N. benthamiana leaf cells. YN/GW9‐YC and YC/GW2‐YN are the negative controls. (c) LCI assay for GW9 and GW2 interaction in N. benthamiana leaf cells. nLuc/cLuc, nLuc‐GW2/cLuc, and nLuc/cLuc‐GW9 were used as the negative controls. Bars = 20 μm. (d) Pull‐down analysis of GW9‐GW2 interaction in vitro. (e) Co‐IP assay confirmation of GW9‐GW2 interaction in vivo. Anti‐GFP beads were used to immunoprecipitate GW2‐HA proteins.
GW9 acts in a coordinated pathway with GW2 to control grain size
Given the E3 ubiquitin ligase activity of GW2, we hypothesized that GW2 may promote the ubiquitination of GW9 protein. To assess this hypothesis, we conducted ubiquitination assay with GW9‐His and GW2‐GST fusion proteins in vitro. In the presence of E1, E2, and ubiquitin, smeared higher‐molecular‐mass bands could be detected by anti‐His, anti‐GST and anti‐Ubi antibodies, which indicated that GW9‐His protein can be ubiquitinated by GW2‐GST, and GW2‐GST also exhibited the ability of self‐ubiquitinated (Figure 7a). Moreover, we transiently co‐expressed the GW9‐GFP and GW2‐RFP‐HA in rice protoplasts to further confirm the ubiquitination of GW9. The proteins were immunoprecipitated with GFP‐Trap‐A beads and detected by immunoblot analysis using anti‐Ubi, anti‐GFP and anti‐HA antibodies. In the presence of both GW9‐GFP and GW2‐RFP‐HA, more dense ubiquitination bands were detected, which also confirmed that GW9 can be ubiquitinated by GW2 in vivo (Figure 7b). Taken together, our results demonstrated that GW9 can be ubiquitinated by GW2 in vivo and vitro.
Figure 7.
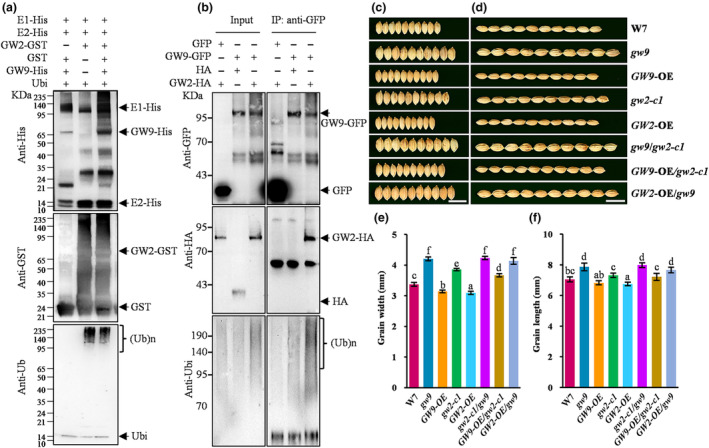
Genetic analysis of GW9 and GW2. (a) Ubiquitination analysis of GW9 by GW2 in vitro. GW2‐GST and GW9‐His fusion proteins were assayed for ubiquitination activity in the presence of E1‐His, E2‐His, and Ubiquitin. ‘+’ and ‘−’ indicate the presence and absence of the components in each reaction mixture, respectively. Ubiquitinated proteins were detected by immunoblotting with anti‐GST antibody, anti‐His antibody, and anti‐Ubi antibody. (b) Ubiquitination analysis of GW9 by GW2 in rice protoplast. GW9‐GFP and GW2‐RFP‐HA fusion proteins were co‐expressed in rice protoplast, and total protein immunoprecipitation with GFP‐Trap‐A. The immunoprecipitates were detected by immunoblotting with anti‐GFP, anti‐HA, anti‐Ubi. ‘+’ and ‘−’ indicate the presence and absence of the components in each reaction mixture, respectively. (c, d) Grain morphologies of W7, gw9, GW9‐OE, gw2‐c1, GW2‐OE, gw2‐c1/gw9, GW9‐OE/gw2‐c1, and GW2‐OE/gw9. Bars = 0.5 cm. (e, f) Statistical data of grain width and length. Error bars represent standard deviation (SD). Statistical analyses were performed by Duncan's multiple range tests (P < 0.05). The presence of the same lowercase letter denotes a non‐significant difference between means.
To analyse the genetic relationship, we generated two loss‐of‐function mutants, gw2‐c1 and gw2‐c2 in the W7 background using CRISPR/Cas9. Sequencing analysis revealed that gw2‐c1 and gw2‐c2 harboured 1‐bp deletion and 2‐bp insertion mutations, respectively, resulting in premature stop codons (Figure S12a). The qRT‐PCR analysis indicated that the expression levels of GW2 were significantly reduced in both mutant lines (Figure S12b). Consistent with the previous study results (Song et al., 2007), the grain length, grain width and 1000‐grain weight of gw2‐c1 and gw2‐c2 were significantly increased compared that of W7 (Figure S12c–g). We further generated the double mutations gw2‐c1/gw9 by crossing gw2‐c1 with gw9, and found the grain width and grain length were not statistically different from those of gw9 (Figure 7c–f). In addition, the overexpression of GW2 (GW2‐OE) in W7 resulted in a shorter and narrower grains, while GW2‐OE/gw9 plants displayed only slightly decreased grain size than gw9 (Figure 7c–f). These results suggested a cooperative regulation of grain size by GW9 and GW2, with GW9 acting downstream of GW2.
Discussion
GW9 is a negative regulator of grain size and weight
PRC2 complex exhibits chromatin‐associated methyltransferase activity, specifically mediating H3K27 methylation to repress the transcriptional expression of downstream genes (Bieluszewski et al., 2021; Godwin and Farrona, 2022; Laugesen et al., 2019). This activity is required for maintaining the gene expression patterns to ensure cell identity in organismal development (Laugesen et al., 2019; Yu et al., 2019). In plants, disruptions of core PRC2 subunits lead to the attenuation or elimination of H3K27me3 repression in target genes, resulting in a variety of developmental defects (Godwin and Farrona, 2022). As a component of PRC2 complex, EMF2b plays an important role in shoot and endosperm development, floral organ identity and flowering transition (Chen et al., 2017; Conrad et al., 2014; Deng et al., 2017; Xie et al., 2015). In this study, we identified a grain size mutant gw9, which exhibited significantly enlarged grain size and increased plant height (Figure 1a–e). Map‐based cloning revealed that GW9 encodes a polycomb group protein and is a new allele of OsEMF2b. Interestingly, the mutant phenotype of gw9 is different from all reported emf2 mutants in rice and Arabidopsis, indicating that GW9/OsEMF2b has a novel function in regulating grain size and plant height. Moreover, the GW9 knock‐out lines Cr‐3, Cr‐5 and Cr‐6 also exhibited large grains, while overexpression lines produced smaller grains (Figures 3e–g and 4), supporting the negative role of GW9 in regulating grain size and weight.
In Arabidopisis, the emf2 mutant showed a 54% decrease in H3K27 trimethylated genes (Kim et al., 2012). To clarify whether gw9 mutation causes H3K27me3 status changes for downstream target genes, ChIP‐qPCR assay was performed to analysis some relate genes expression levels in young panicles of W7, gw9, and GW9‐OE plants. It has been reported that MADS1, MADS6, MADS34, and MADS58 regulating floral organ identity and grain size development by H3K27me3 methylation (Conrad et al., 2014; Mao et al., 2022; Zheng et al., 2015). The results revealed that the methylation levels of MADS1, MADS6, MADS34, and MADS58 genes were significantly lower in the gw9 mutant than in W7 plant, while the methylation levels were significantly higher in GW9‐OE plants than in W7 plant (Figure S13a–d). This suggests that GW9 as a member of the PRC2 complex could regulate the downstream target genes expression by influencing their H3K27me3 status.
Previous studies had reported that PRC2 complex directly regulates cell cycle and controls proliferation (Adhikari and Davie, 2020; Grossniklaus and Paro, 2014; Simonini et al., 2021; Sun et al., 2014). We also found that the enlarged spikelet hulls of gw9 resulted from increased cell number and cell size (Figure 2). Correspondingly, the expression levels of several genes control grain size by modulating cell proliferation and expansion, such as GL7, PGL1 and GLW7 were significantly increased in gw9 (Figure S3a). In addition, the expression levels of several cell cycle‐related genes also significantly changed in gw9 (Figure S3b). So, we believe that GW9 plays a negatively role in controlling grain size by restricting cell proliferation and expansion.
C2H2‐ZnF domain plays an important role in the control of grain size
The GW9 contains C2H2‐ZnF and VEFS domains, and the splicing deletion of exon 11 of coding sequence eventually disrupts the C2H2‐ZnF domain in gw9 (Figures 3b–d, S7). C2H2‐type zinc finger protein (C2H2‐ZFP) is one of the most important Zinc finger proteins, which activates or inhibits downstream gene transcription to regulate plant growth and development or respond to environmental singles (Huang et al., 2022; Lyu and Cao, 2018). However, previous studies mainly focused on abiotic stress responses, disease resistance, and plant growth, but few in grain or seed size development (Huang et al., 2022; Lyu and Cao, 2018). In rice, disruption of C2H2‐ZFP genes, such as Du13 and LRG1, result in small and short‐wide grains, respectively (Cai et al., 2022; Xu et al., 2020). Similarly, in Arabidopsis, the ectopic expression of C2H2‐ZFP gene ENY produces big seeds (Feurtado et al., 2011). To confirm the role of C2H2‐ZnF domain in grain size development, we designed different knock‐out targets within the GW9 coding sequence and the results indicated that only deletion of C2H2‐ZnF domain led to a significant increase in grain size, while disruption of the VEFS domain caused floral organ defects and indeterminacy (Figures 4a–g, S8). Consistent with this result, the H3K27me3 were markedly decreased in disruption of VEFS domain lines, and slightly decreased in the C2H2‐ZnF domain deletions lines (Figure S13e). ChIP‐qPCR analysis of young panicles from W7, gw9 and GW9‐OE also revealed that the methylation levels of MADS1, MADS6, MADS34 and MADS58 were declined in gw9 and significantly improved in GW9‐OE (Figure S13a–d). Therefore, we conclude that the C2H2‐ZnF domain of GW9 plays an important role in controlling grain size, and VEFS domain is essential for maintaining floral organ identity. These findings indicate the potential for future directional genome editing of GW9 gene to enhance grain size and ultimately improve grain yield.
GW9 is involved in GW2 ubiquitination pathway
By screening rice yeast library, we found an interaction between GW9 and the GW2 protein (Figures 6, S10). Previous studies have shown that GW2 encodes a RING‐type E3 ubiquitin ligase, and is a negative regulator responsible for grain width and weight (Song et al., 2007). It is well known that the E3‐ubiquitin ligase plays a key role in recognizing target proteins and catalysing its ligation to ubiquitin (Dikic and Robertson, 2012). Therefore, GW2 should be able to interact with target genes and treat it as substrate to promote its degradation. It has been confirmed that WG1 and GW6a are the targets of GW2 and involved in ubiquitination pathway to control grain size (Gao et al., 2021; Hao et al., 2021). In addition, OsEXPLA1, a cell wall‐loosening protein that promotes cell growth, is also a substrate for ubiquitination by GW2 (Choi et al., 2018). Although OsEXPLA1 has not been reported to be directly involved in regulating grain size in rice, the heterologous overexpression of cell expansion gene BdEXPA27 and IbEXP1 significantly increased seed size in Arabidopsis (Bae et al., 2014; Chen et al., 2020). In this study, GW9 physically interacts with GW2, and GW2 can promote the ubiquitination of GW9 (Figures 6 and 7a,b), indicating that GW9 is also a ubiquitin target protein of GW2. We further constructed double mutant and found that there was no significant difference between gw2/gw9 and gw9 in grain length and width. Moreover, GW2 overexpression in gw9 background failed to rescue gw9 grain size, but GW9 overexpression partially rescued the gw2 phenotype (Figure 7b–e). These results supported that GW9 acts in a common pathway with GW2 and functions downstream of GW2. To be sure, GW9 is not the only substrate for GW2 in regulating grain size. Therefore, we speculate that may be other targets in the GW2 ubiquitination pathway, such as WG1, GW6a, EXPLA1, which may influence GW9‐OE to fully restore GW2. However, it was confusing that GW2 has been reported to promote the degradation of target proteins, while both GW2 and GW9 are play as a negative regulators of grain size in our study. In fact, it is unlikely that GW9 is a substrate of GW2 for protein degradation, but more possible that GW2 may activate or stabilize GW9 by ubiquitination. It is also possible that there may have another unknown E3 ubiquitin ligase that synergistically increases protein ubiquitination to regulate grain size. Based on these results, we propose a working model for the functional role of GW9 in regulating grain size and weight. GW6 and WG1, as known substrates of GW2, can be ubiquitinated by GW2 to regulate grain size and weight. Meanwhile, GW9 was also ubiquitinated by GW2 and further regulated the downstream target genes expression by mediating H3K27me3 modification, thereby promoting cell proliferation and expansion in spikelet hull, ultimately leading to increased grain size and weight (Figure 8).
Figure 8.
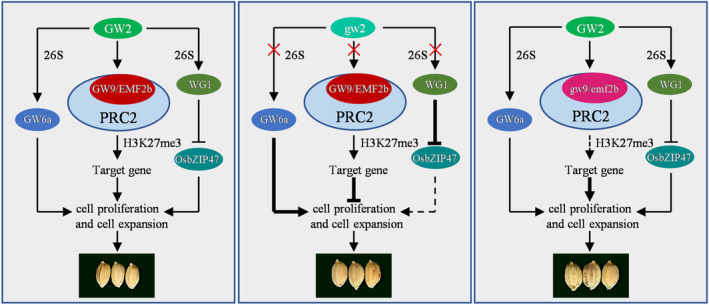
Proposed model of GW9 regulating grain size and weight in rice. The ubiquitination of GW9, GW6a, and WG1 are regulated by GW2. GW6a and WG1 positively regulates grain size by promoting cell proliferation and expansion. Meanwhile, GW9 is a component of the PRC2 complex, which negatively regulates grain size by inhibiting the expression of downstream target genes through H3K27me3 modification. In the presence of GW2, the mutation of GW9 removes the repressive effect of downstream target genes, improved the expression level of cell proliferation and expansion related genes, and thus increasing grain size and weight.
Our findings enrich the genetic regulatory network of GW2‐mediated ubiquitin pathway controlling grain size and provide a potential target for future molecular breeding of high yield in rice.
Materials and methods
Plant materials and growth conditions
The gw9 mutant was identified from the EMS‐treated mutant population of the Japonica cultivar Wuyungeng7 (W7). In this study, W7 was used as the wild type in all analyses, and the mutant was backcrossed with W7 to produce gw9 mutant to eliminate interference. All plants were cultivated in Hangzhou (Zhejiang province, China) and Lingshui (Hainan province, China) under natural growth conditions.
Map‐based cloning of GW9
To map the mutated gene, gw9 was crossed with Taichung Native 1 (TN1), an indica rice cultivar, and F1 plants were self‐pollinated to generate an F2 mapping population. New In/Del molecular markers were developed to fine‐map GW9. The gene was finally mapped within a 50.2‐kb region on chromosome 9, and all open reading frames (ORFs) were amplified and sequenced. The candidate genes within the 50.2‐kb region are listed in Table S1.
Phenotypical characterization
Plant height, panicle length, grain number per panicle, number of primary and secondary panicle branches were measured at the maturity stage. Fully filled grains were used to measured grain length, grain width, and 1000‐grain weight by using a ScanMaker i800 (MICROTEK, China). The endosperm fresh weight and dry weight at 3, 6, 9, 12, 15, 18, 21, 24, 27, and 30 days after fertilization (DAF) were measured to investigate the grain filling rate, and the grain filling rate was calculated by using ‘Richards equation’.
Histological analysis
Spikelet hulls were fixed in 50% FAA (50% ethanol, 5% glacial acetic acid, and 5% formaldehyde) overnight and then stored at 4 °C. The fixed samples were dehydrated in a gradient of ethanol solutions, cleared in a graded xylene‐ethanol solution, and finally embedded in Paraplast Plus (Sigma). The samples were sliced into 8‐μm‐thick sections with a microtome (HM340E), and observed by a light microscopy (90I, Nikon) (Hu et al., 2022). For glume cell observation, young spikelet hulls were fixed in 2.5% glutaraldehyde and observed by scanning electron microscopy (HITACHI, S‐3000N) (Ren et al., 2022). Cell size and cell number were measured by Image J software (version 1.53c).
Plasmid construction and plant transformation
For the complementation test, the full‐length genomic sequence of GW9, containing a 2.2‐kb upstream region, coding sequence, and 1.1‐kp of downstream region, was amplified from W7 and cloned into the pCAMBIA1300 to generate the vector pGW9::GW9. The entire cDNA of GW9 was amplified and inserted into pActin::GFP to generate the pActin::GW9‐GFP plasmid for overexpression analysis and was inserted into Ubiquitin::GFP to generate the vector for protein subcellular localization. A 2.2‐kb DNA fragment containing the promoter of GW9 was cloned into the pCAMBIA1300‐GUS to generate the vector pGW9::GUS for GUS staining analysis. Gene editing constructs via CRISPR/Cas9 system was performed as previously described (Xie et al., 2017). All constructs were confirmed by sequencing and then transformed into rice callus or protoplasts. The PCR primers used are listed in Table S2.
GUS staining and subcellular localization of GW9
The tissues from the transgenic plants carrying the pGW9::GUS were collected at different developmental stages and stained in GUS staining buffer overnight at 37 °C. Then, samples were then bleached in 75% ethanol and photographed (Zhang et al., 2021b). For the subcellular localization of the GW9 protein, the pUbi::GW9‐GFP vectors were introduced into rice protoplasts using PEG‐mediated transformation methods. MADS29‐RFP was used as the nuclear marker (Yin and Xue, 2012). In addition, the roots of 3‐day‐old seedlings of pUbi::GW9‐GFP transgenic plants were used to determine the subcellular localization of GW9 protein. For nuclear staining, the nuclear dye DAPI (1 μg/mL) was applied to the root for 10 min before observation. The GFP fluorescence and RFP fluorescence were observed at the excitation wavelength of 488 nm and 561 nm, respectively, while DAPI fluorescence was observed at the excitation wavelength of 460 nm. Fluorescence signals were observed using a confocal microscope (Carl Zeiss, LSM700).
Yeast two‐hybrid (Y2H) assay
For yeast two‐hybrid assay, the full‐length coding sequences of GW9 and different truncated forms were cloned into PGBKT7 vector, while full‐length cDNA of GW2 was cloned into PGADT7 vector. The prey and bait vectors were co‐transformed in yeast strain Y2H‐gold and the yeast cells were cultured on selection medium DDO (SD/‐Trp‐Leu) and QDO (SD/‐Trp‐Leu‐His‐Ade/x‐α‐gal) at 30 °C. PGBKT7‐53 and PGADT7‐T were used as positive control, and PGBKT7‐empty and PGADT7‐empty were used as negative control. The primers used are given in Table S2.
Bimolecular fluorescence complementary (BiFC) assay
Full‐length coding sequences of GW9 and GW2 were cloned into 1300‐YC and 1300‐YN plasmid, respectively. The vectors were co‐infiltrated into leaves of N. benthamiana. Fluorescence signals were detected using confocal microscope (Carl Zeiss, LSM700). The primers used are listed in Table S2.
Split luciferase complementation (LCI) assay
Full‐length coding sequences of GW9 and GW2 were cloned into 1300‐cLUC and 1300‐nLUC to generate the cLUC‐GW9 and GW2‐nLUC fusion vectors, respectively. Different vector combinations were then transformed into N. benthamiana leaves. After 2 days of incubation, luciferin was smeared on the leaves and the activated luciferase reconstitution signals were imaged using a plant imaging system (PI Lumazone 1300B). The primers used are listed in Table S2.
Co‐IP assays
Full‐length coding sequences of GW9 and GW2 were cloned into pUbi‐GFP and pUbi‐RFP‐HA to produce the fusion protein Ubi::GW9‐GFP and Ubi::GW2‐RFP‐HA, respectively. Different combinations of plasmids were transiently expressed into rice protoplast. Total proteins were extracted with extraction buffer (50 mM Tris–HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA [pH 8.0], 1% NP40, 5% glycerol, protease inhibitor cocktail, and 1 mM PMSF) and incubated with GFP‐trap beads (ChromoTek, gtma‐20) for 1–2 h at 4 °C. Beads were washed five times with protein extraction buffer and then proteins were boiling for 10 min after adding SDS‐loading buffer (5×). Protein samples were separated by 10% SDS‐PAGE gel and detected by immunoblotting with anti‐GFP (Abmart, M20004M, 1:2000) and anti‐HA (Abmart, M20003M, 1:2000) antibodies, respectively. The primers used are listed in Table S2.
Protein purification and ubiquitination assay
The full‐length coding sequences of GW9 and GW2 were cloned into the PET28a and pGEX‐4T‐1 to generate GW9‐His and GW2‐GST vector, respectively. These constructs were transformed into E. coli ArcticExpress (DE3) pRARE2 competent cell, and the recombinant proteins were induced with 0.2 mM IPTG at 18 °C for 16 h. Fusion proteins were purified using the His Resin (TransGen, DP101) or GST Resin (Sangon, C600327‐0001) according to the manufacturer's protocols. The ubiquitination assay in vitro was performed using a Ubiquitinylation Kit (Enzo Life Sciences, BMLUW9920‐0001) according to the manufacturer's instructions and the UbcH5b in the kit was used as E2 enzyme in this study. Polyubiquitinated proteins were detected by immunoblotting with anti‐His (Abmart, M30111M, 1:5000), anti‐GST (Abmart, M20007M, 1:5000), and anti‐ubiquitin (Abcam, ab7254, 1:2000) antibodies, respectively. The primers used are listed in Table S2.
GST pull‐down assays
GST or GST‐GW2 coupled GST beads (Sigma, G0924) were incubated with MBP‐His‐GW9 for 2 h at 4 °C, and then washed thoroughly, boiled in SDS‐loading buffer (1×). Protein samples were separated by 10% SDS‐PAGE gel and detected by immunoblotting with anti‐GST (Abmart, M20007M, 1:5000) and anti‐His antibodies (Abmart, M30111M, 1:5000), respectively.
RNA extraction and qRT‐PCR analysis
Total RNA was extracted from various tissues using the AxyPrep™ total RNA Miniprep Kit (Axygen) according to the manufacturer's instructions. The first‐strand cDNA was synthesized based on the ReverTra Ace kit (Toyobo) and the cDNA was diluted as a template for qRT‐PCR. qRT‐PCR was performed with the Bio‐Rad CFX96 real‐time PCR detection system using the SYBR Green PCR Master Mix kit (Applied Biosystems). The rice gene Actin (LOC_Os03g50885) was used an as the internal control. Three replicates were performed for all experiments and Student's t‐test was used to analyse the significance of differences. The primers used are listed in Table S2.
RNA‐seq and data analysis
Total RNA samples were isolated from young panicle of W7, gw9 and GW9‐OE plants by using RNAiso Plus (Takara, 9108) according to the manufacturer's instructions. Different expression genes were identified by edgeR with FDR <0.05 and |log2(Fold change)| > 1 using normalized expression values among all samples. The DEGs are listed in Table S3.
ChIP‐qPCR assay
For ChIP‐qPCR, young panicles from W7, gw9 and GW9‐OE plants were harvested and cross‐linked in 1% (v/v) formaldehyde for 15 min at 4 °C, and subsequently added glycine to 125 mM to stop the reaction. The chromatin was extracted, purified, and sheared into 200–500 bp fragments by sonication, prepared sample were precleared with Dynabeads™ protein A/G (Invitrogen, 10001D and 10003D) for 1 h before incubation with anti‐H3K27me3 (abcam, ab6002) antibodies with new Dynabeads™ protein A/G overnight at 4 °C. The protein‐DNA complexes eluted from the beads were collected and reverse crosslinked, and then, the purified DNA fragments were analysed with qPCR.
Detection of GW9 ubiquitination in vivo
Different combinations of Ubi::GW9‐GFP and Ubi::GW2‐RFP‐HA vectors were transiently co‐expressed in rice protoplasts. The expressed proteins were extracted with NB1 protein extraction buffer (50 mM Tris‐MES [pH 8], 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA [pH 8.0], 5 mM DTT, protease inhibitor cocktail, and 1 mM PMSF) and immunoprecipitated with GFP‐trap beads (ChromoTek, gtma‐20) with 50 μM MG132 for 1–2 h at 4 °C with gently shaking. Then, beads were washed three times with NB1 buffer and boiling for 10 min after adding SDS‐loading buffer (5×). Protein samples were loaded on 10% SDS‐PAGE gel and immunoblotted with anti‐Ubi (Abcam, ab7254, 1:5000), anti‐GFP (Abmart, M20004M, 1:2000) and anti‐HA (Abmart, M20003M, 1:2000) antibodies, respectively.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
JH and QQ designed and supervised research; PH and YW wrote the manuscript; PH and YW performed experiments; PH, YT, YW, HW, JW, KW and BC contributed to the phenotype and data analysis; YF, LZ, GZ, ZG, DR, DZ, LS, GD, QZ, QL, and GX provided technical assistances. PH, YW and YF contributed equally to this work. All authors have read and agreed to the published version of the manuscript.
Supporting information
Figure S1 Culm structure analysis of W7 and gw9 mutant.
Figure S2 Grain filling analysis of W7 and gw9 mutant.
Figure S3 Transcript levels of grain size‐related and cell cycle‐related genes in W7 and gw9 mutant.
Figure S4 Coding sequence and protein structure analysis.
Figure S5 Transcription level analysis.
Figure S6 GW9 regulates the heading date.
Figure S7 Schematic representation and gene edition of GW9.
Figure S8 The spikelet phenotype and pollen fertility of mutants.
Figure S9 Heat map analysis of W7, gw9 and OE.
Figure S10 GW9 interacts with GW2 in yeast cells.
Figure S11 GW2 and GW9 are co‐located in the nucleus of rice protoplasts.
Figure S12 Gene edition of GW2.
Figure S13 GW9 mediated histone H3K27me3 modification.
Table S1 List of genes in the 50.2‐kb target region.
Table S2 Primers used in this study.
Table S3 DEGs from RNA‐seq compared between with W7, gw9 and OE.
Acknowledgements
This study was supported by the National Key Research and Development Program of China (2022YFF1002904), the Natural Science Foundation of China (32272109), the Hainan Yazhou Bay Seed Laboratory (B21HJ0215), and the Nanfan special project (ZDXM2315) of CAAS, and the academician workstation of Qianqian. We thank Wangshu Mou for her help in language Polish.
Contributor Information
Dawei Xue, Email: dwxue@hznu.edu.cn.
Qian Qian, Email: qianiqn188@hotmail.com.
Jiang Hu, Email: hujiang588@163.com.
Data availability statement
The original contributions presented in the study are included in the article and Supplementary materials, further inquiries can be directed to the corresponding author.
References
- Adhikari, A. and Davie, J.K. (2020) The PRC2 complex directly regulates the cell cycle and controls proliferation in skeletal muscle. Cell Cycle, 19, 2373–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, J.M. , Kwak, M.S. , Noh, S.A. , Oh, M.J. , Kim, Y.S. and Shin, J.S. (2014) Overexpression of sweetpotato expansin cDNA (IbEXP1) increases seed yield in Arabidopsis. Transgenic Res. 23, 657–667. [DOI] [PubMed] [Google Scholar]
- Bemer, M. and Grossniklaus, U. (2012) Dynamic regulation of polycomb group activity during plant development. Curr. Opin. Plant Biol. 15, 523–529. [DOI] [PubMed] [Google Scholar]
- Bieluszewski, T. , Xiao, J. , Yang, Y. and Wagner, D. (2021) PRC2 activity, recruitment, and silencing: a comparative perspective. Trends Plant Sci. 26, 1186–1198. [DOI] [PubMed] [Google Scholar]
- Cai, Y. , Zhang, W. , Fu, Y. , Shan, Z. , Xu, J. , Wang, P. , Kong, F. et al. (2022) Du13 encodes a C2H2 zinc‐finger protein that regulates Wx b pre‐mRNA splicing and microRNA biogenesis in rice endosperm. Plant Biotechnol. J. 20, 1387–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M. , Xie, S. , Ouyang, Y. and Yao, J. (2017) Rice PcG gene OsEMF2b controls seed dormancy and seedling growth by regulating the expression of OsVP1 . Plant Sci. 260, 80–89. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Luo, Y. , Wang, G. , Feng, C. and Li, H. (2020) Genome‐wide identification of expansin genes in brachypodium distachyon and functional characterization of BdEXPA27 . Plant Sci. 296, 110490. [DOI] [PubMed] [Google Scholar]
- Cheng, X. , Pan, M. , Er, Z. , Zhou, Y. , Niu, B. and Chen, C. (2020a) Functional divergence of two duplicated fertilization independent endosperm genes in rice with respect to seed development. Plant J. 104, 124–137. [DOI] [PubMed] [Google Scholar]
- Cheng, X. , Pan, M. , Er, Z. , Zhou, Y. , Niu, B. and Chen, C. (2020b) The maternally expressed polycomb group gene OsEMF2a is essential for the endosperm cellularization and imprinting in rice. Plant Commun. 2, 100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, B.S. , Kim, Y.J. , Markkandan, K. , Koo, Y.J. , Song, J.T. and Seo, H.S. (2018) GW2 functions as an E3 ubiquitin ligase for rice expansin‐like 1. Int. J. Mol. Sci. 19, 1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, L.J. , Khanday, I. , Johnson, C. , Guiderdoni, E. , An, G. , Vijayraghavan, U. and Sundaresan, V. (2014) The polycomb group gene EMF2B is essential for maintenance of floral meristem determinacy in rice. Plant J. 80, 883–894. [DOI] [PubMed] [Google Scholar]
- Deng, L. , Zhang, S. , Wang, G. , Fan, S. , Li, M. , Chen, W. , Tu, B. et al. (2017) Down‐regulation of OsEMF2b caused semi‐sterility due to anther and pollen development defects in rice. Front. Plant Sci. 8, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic, I. and Robertson, M. (2012) Ubiquitin ligases and beyond. BMC Biol. 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser, S. and Finley, D. (2005) Delivery of ubiquitinated substrates to protein‐unfolding machines. Nat. Cell Biol. 7, 742–749. [DOI] [PubMed] [Google Scholar]
- Feurtado, J.A. , Huang, D. , Wicki‐Stordeur, L. , Hemstock, L.E. , Potentier, M.S. , Tsang, E.W. and Cutler, A.J. (2011) The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell, 23, 1772–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forderer, A. , Zhou, Y. and Turck, F. (2016) The age of multiplexity: recruitment and interactions of polycomb complexes in plants. Curr. Opin. Plant Biol. 29, 169–178. [DOI] [PubMed] [Google Scholar]
- Gao, Q. , Zhang, N. , Wang, W. , Shen, S. , Bai, C. and Song, X. (2021) The ubiquitin‐interacting motif‐type ubiquitin receptor HDR3 interacts with and stabilizes the histone acetyltransferase GW6a to control the grain size in rice. Plant Cell, 33, 3331–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin, J. and Farrona, S. (2022) The importance of networking: plant polycomb repressive complex 2 and its interactors. Epigenomes, 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U. and Paro, R. (2014) Transcriptional silencing by polycomb‐group proteins. Cold Spring Harb. Perspect. Biol. 6, a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, J. , Wang, D. , Wu, Y. , Huang, K. , Duan, P. , Li, N. , Xu, R. et al. (2021) The GW2‐WG1‐OsbZIP47 pathway controls grain size and weight in rice. Mol. Plant, 14, 1266–1280. [DOI] [PubMed] [Google Scholar]
- Hu, P. , Tan, Y. , Wen, Y. , Fang, Y. , Wang, Y. , Wu, H. , Wang, J. et al. (2022) LMPA regulates lesion mimic leaf and panicle development through ros‐induced pcd in rice. Front. Plant Sci. 13, 875038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K. , Wang, D. , Duan, P. , Zhang, B. , Xu, R. , Li, N. and Li, Y. (2017) WIDE AND THICK GRAIN 1, which encodes an otubain‐like protease with deubiquitination activity, influences grain size and shape in rice. Plant J. 91, 849–860. [DOI] [PubMed] [Google Scholar]
- Huang, L. , Hua, K. , Xu, R. , Zeng, D. , Wang, R. , Dong, G. , Zhang, G. et al. (2021) The LARGE2‐APO1/APO2 regulatory module controls panicle size and grain number in rice. Plant Cell, 33, 1212–1228. [DOI] [PubMed] [Google Scholar]
- Huang, X. , Lu, Z. , Wang, X. , Ouyang, Y. , Chen, W. , Xie, K. , Wang, D. et al. (2016) Imprinted gene OsFIE1 modulates rice seed development by influencing nutrient metabolism and modifying genome H3K27me3. Plant J. 87, 305–317. [DOI] [PubMed] [Google Scholar]
- Huang, Y. , Du, L. , Wang, M. , Ren, M. , Yu, S. and Yang, Q. (2022) Multifaceted roles of zinc finger proteins in regulating various agronomic traits in rice. Front. Plant Sci. 13, 974396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.Y. , Lee, J. , Eshed‐Williams, L. , Zilberman, D. and Sung, Z.R. (2012) EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet. 8, e1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen, A. , Hojfeldt, J.W. and Helin, K. (2019) Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell, 74, 8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Zhang, B. , Duan, P. , Yan, L. , Yu, H. , Zhang, L. , Li, N. et al. (2022) An ERAD‐related E2‐E3 enzyme pair controls grain size and weight through the brassinosteroid signaling pathway in rice. Plant Cell, 35, 1076–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. , Xu, R. , Duan, P. and Li, Y. (2018) Control of grain size in rice. Plant Reprod. 31, 237–251. [DOI] [PubMed] [Google Scholar]
- Li, N. , Xu, R. and Li, Y. (2019) Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 70, 435–463. [DOI] [PubMed] [Google Scholar]
- Li, S. , Zhou, B. , Peng, X. , Kuang, Q. , Huang, X. , Yao, J. , Du, B. et al. (2014) OsFIE2 plays an essential role in the regulation of rice vegetative and reproductive development. New Phytol. 201, 66–79. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Wei, X. , Sheng, Z. , Jiao, G. , Tang, S. , Luo, J. and Hu, P. (2016) Polycomb protein OsFIE2 affects plant height and grain yield in rice. PLoS One, 11, e0164748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Zhou, C. , Zhao, Y. , Zhou, S. , Wang, W. and Zhou, D. (2014) The rice enhancer of zeste [E(z)] genes SDG711 and SDG718 are respectively involved in long day and short day signaling to mediate the accurate photoperiod control of flowering time. Front. Plant Sci. 5, 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Zhou, S. , Wang, W. , Ye, Y. , Zhao, Y. , Xu, Q. , Zhou, C. et al. (2015) Regulation of histone methylation and reprogramming of gene expression in the rice inflorescence meristem. Plant Cell, 27, 1428–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M. , Platten, D. , Chaudhury, A. , Peacock, W.J. and Dennis, E.S. (2009) Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol. Plant, 2, 711–723. [DOI] [PubMed] [Google Scholar]
- Lyu, T. and Cao, J. (2018) Cys2/His2 Zinc‐Finger proteins in transcriptional regulation of flower development. Int. J. Mol. Sci. 19, 2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, D. , Tao, S. , Li, X. , Gao, D. , Tang, M. , Liu, C. , Wu, D. et al. (2022) The Harbinger transposon‐derived gene PANDA epigenetically coordinates panicle number and grain size in rice. Plant Biotechnol. J. 20, 1154–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, D. , Ding, C. and Qian, Q. (2023) Molecular bases of rice grain size and quality for optimized productivity. Sci. Bull. 68, 314–350. [DOI] [PubMed] [Google Scholar]
- Ren, D. , Xie, W. , Xu, Q. , Hu, J. , Zhu, L. , Zhang, G. , Zeng, D. et al. (2022) LSL1 controls cell death and grain production by stabilizing chloroplast in rice. Sci. China Life Sci. 65, 2148–2161. [DOI] [PubMed] [Google Scholar]
- Shi, C. , Ren, Y. , Liu, L. , Wang, F. , Zhang, H. , Tian, P. , Pan, T. et al. (2019) Ubiquitin specific protease 15 has an important role in regulating grain width and size in rice. Plant Physiol. 180, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini, S. , Bemer, M. , Bencivenga, S. , Gagliardini, V. , Pires, N.D. , Desvoyes, B. , van der Graaff, E. et al. (2021) The polycomb group protein MEDEA controls cell proliferation and embryonic patterning in Arabidopsis. Dev. Cell, 56, 1945–1960 e1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X. , Huang, W. , Shi, M. , Zhu, M.Z. and Lin, H. (2007) A QTL for rice grain width and weight encodes a previously unknown RING‐type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. [DOI] [PubMed] [Google Scholar]
- Song, X. , Kuroha, T. , Ayano, M. , Furuta, T. , Nagai, K. , Komeda, N. , Segami, S. et al. (2015) Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. USA, 112, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, B. , Looi, L.S. , Guo, S. , He, Z. , Gan, E. , Huang, J. , Xu, Y. et al. (2014) Timing mechanism dependent on cell division is invoked by polycomb eviction in plant stem cells. Science, 343, 1248559. [DOI] [PubMed] [Google Scholar]
- Tonosaki, K. , Ono, A. , Kunisada, M. , Nishino, M. , Nagata, H. , Sakamoto, S. , Kijima, S.T. et al. (2021) Mutation of the imprinted gene OsEMF2a induces autonomous endosperm development and delayed cellularization in rice. Plant Cell, 33, 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Tong, X. , Tang, L. , Wang, Y. , Zhao, J. , Li, Z. , Liu, X. et al. (2022) RLB (RICE LATERAL BRANCH) recruits PRC2‐mediated H3K27 tri‐methylation on OsCKX4 to regulate lateral branching. Plant Physiol. 188, 460–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, S. , Chen, M. , Pei, R. , Ouyang, Y.D. and Yao, J. (2015) OsEMF2b acts as a regulator offlowering transition and floral organ identity by mediating H3K27me3 deposition at OsLFL1 and OsMADS4 in rice. Plant Mol. Biol. Rep. 33, 121–132. [Google Scholar]
- Xie, X. , Ma, X. , Zhu, Q. , Zeng, D. , Li, G. and Liu, Y. (2017) CRISPR‐GE: a convenient software toolkit for CRISPR‐based genome editing. Mol. Plant, 10, 1246–1249. [DOI] [PubMed] [Google Scholar]
- Xing, Y. and Zhang, Q. (2010) Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. [DOI] [PubMed] [Google Scholar]
- Xu, Q. , Yu, H. , Xia, S. , Cui, Y. , Yu, X. , Liu, H. , Zeng, D. et al. (2020) The C2H2 zinc‐finger protein LACKING RUDIMENTARY GLUME 1 regulates spikelet development in rice. Sci. Bull. 65, 753–764. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Lee, S. , Hang, R. , Kim, S.R. , Lee, Y.S. , Cao, X. , Amasino, R. et al. (2013) OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J. 73, 566–578. [DOI] [PubMed] [Google Scholar]
- Yang, W. , Wu, K. , Wang, B. , Liu, H. , Guo, S. , Guo, X. , Luo, W. et al. (2021) The RING E3 ligase CLG1 targets GS3 for degradation via the endosome pathway to determine grain size in rice. Mol. Plant, 14, 1699–1713. [DOI] [PubMed] [Google Scholar]
- Yin, L. and Xue, H. (2012) The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development. Plant Cell, 24, 1049–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.R. , Lee, C.H. , Oksuz, O. , Stafford, J.M. and Reinberg, D. (2019) PRC2 is high maintenance. Genes Dev. 33, 903–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Cheng, Z. , Qin, R. , Qiu, Y. , Wang, J. , Cui, X. , Gu, L. et al. (2012) Identification and characterization of an epi‐allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell, 24, 4407–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P. , Zhu, C. , Geng, Y. , Wang, Y. , Yang, Y. , Liu, Q. , Guo, W. et al. (2021a) Rice and Arabidopsis homologs of yeast CHROMOSOME TRANSMISSION FIDELITY PROTEIN 4 commonly interact with polycomb complexes but exert divergent regulatory functions. Plant Cell, 33, 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Yang, C. , Lin, H. , Wang, J. and Xue, H. (2021b) Rice SPL12 coevolved with GW5 to determine grain shape. Sci. Bull. 66, 2353–2357. [DOI] [PubMed] [Google Scholar]
- Zheng, M. , Wang, Y. , Wang, Y. , Wang, C. , Ren, Y. , Lv, J. , Peng, C. et al. (2015) DEFORMED FLORAL ORGAN1 (DFO1) regulates floral organ identity by epigenetically repressing the expression of OsMADS58 in rice (Oryza sativa). New Phytol. 206, 1476–1490. [DOI] [PubMed] [Google Scholar]
- Zhong, J. , Peng, Z. , Peng, Q. , Cai, Q. , Peng, W. , Chen, M. and Yao, J. (2018) Regulation of plant height in rice by the polycomb group genes OsEMF2b, OsFIE2 and OsCLF . Plant Sci. 267, 157–167. [DOI] [PubMed] [Google Scholar]
- Zuo, J. and Li, J. (2014) Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48, 99–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Culm structure analysis of W7 and gw9 mutant.
Figure S2 Grain filling analysis of W7 and gw9 mutant.
Figure S3 Transcript levels of grain size‐related and cell cycle‐related genes in W7 and gw9 mutant.
Figure S4 Coding sequence and protein structure analysis.
Figure S5 Transcription level analysis.
Figure S6 GW9 regulates the heading date.
Figure S7 Schematic representation and gene edition of GW9.
Figure S8 The spikelet phenotype and pollen fertility of mutants.
Figure S9 Heat map analysis of W7, gw9 and OE.
Figure S10 GW9 interacts with GW2 in yeast cells.
Figure S11 GW2 and GW9 are co‐located in the nucleus of rice protoplasts.
Figure S12 Gene edition of GW2.
Figure S13 GW9 mediated histone H3K27me3 modification.
Table S1 List of genes in the 50.2‐kb target region.
Table S2 Primers used in this study.
Table S3 DEGs from RNA‐seq compared between with W7, gw9 and OE.
Data Availability Statement
The original contributions presented in the study are included in the article and Supplementary materials, further inquiries can be directed to the corresponding author.


