Summary
Inducible expression systems can overcome the trade‐off between high‐level transgene expression and its pleiotropic effects on plant growth. In addition, they can facilitate the expression of biochemical pathways that produce toxic metabolites. Although a few inducible expression systems for the control of transgene expression in plastids have been developed, they all depend on chemical inducers and/or nuclear transgenes. Here we report a temperature‐inducible expression system for plastids that is based on the bacteriophage λ leftward and rightward promoters (pL/pR) and the temperature‐sensitive repressor cI857. We show that the expression of green fluorescent protein (GFP) in plastids can be efficiently repressed by cI857 under normal growth conditions, and becomes induced over time upon exposure to elevated temperatures in a light‐dependent process. We further demonstrate that by introducing into plastids an expression system based on the bacteriophage T7 RNA polymerase, the temperature‐dependent accumulation of GFP increased further and was ~24 times higher than expression driven by the pL/pR promoter alone, reaching ~0.48% of the total soluble protein. In conclusion, our heat‐inducible expression system provides a new tool for the external control of plastid (trans) gene expression that is cost‐effective and does not depend on chemical inducers.
Keywords: plastid transformation, pL/pR, cI857, heat‐inducible expression, T7 RNA polymerase, Nicotiana tabacum
Introduction
Due to several unique features of plastids (chloroplasts), including the high ploidy level of their genome and the absence of gene silencing, plastid engineering has become an attractive platform for the expression of recombinant proteins and the production of valuable metabolites (Bock, 2001; Cardi et al., 2010; Yang et al., 2022). To date, numerous recombinant proteins and valuable compounds have been produced by engineering plastid genomes (Bock, 2015). The expression level of transgenes in plastids can reach very high levels, with protein yields of up to 70% of the total soluble protein (TSP) of the plant (Oey et al., 2009). However, the expression of some transgenes and the extreme accumulation levels of some recombinant proteins have been shown to affect the normal function of plastids, leading to growth retardation, impaired photosynthesis or even loss of autotrophic growth of the transplastomic plants (Hennig et al., 2007; Lössl et al., 2003; Magee et al., 2004; Scotti and Cardi, 2014). To overcome this limitation, inducible expression systems for the external control of plastid transgene expression have been developed. They typically rely on chemical inducers and some of them require the expression of additional transgenes from the nuclear genome (Agrawal et al., 2022; Emadpour et al., 2015; Lössl et al., 2005; Muhlbauer and Koop, 2005; Rojas et al., 2019; Verhounig et al., 2010). Systems involving nuclear transgenes are based on the integration of a cis‐element (e.g. the T7 RNA polymerase promoter or a pentatricopeptide repeat (PPR)‐binding site) into the plastid genome to regulate transgene expression. The corresponding regulator (T7 RNA polymerase or PPR protein) is then expressed from the nuclear genome under the control of a chemically inducible promoter. Upon induction, the regulator protein is imported into plastids to trigger the expression of the plastid transgene by specific recognition of the upstream cis‐element. However, this approach is not only time‐consuming (in that it requires two successive transformation experiments), but also abrogates a key attraction of the transplastomic technology: the increased transgene containment that, due to maternal inheritance of the plastid DNA, greatly reduces the risk of undesired spread of transgenes via pollen (Rojas et al., 2019). Therefore, the development of plastid‐only inducible systems has been pursued, in which all required elements for control of transgene expression are encoded in the plastid. Successful approaches include a theophylline‐responsive riboswitch (Verhounig et al., 2010) and an IPTG‐inducible system based on the Lac repressor (Muhlbauer and Koop, 2005). While these systems provide inducible gene expression from the plastid genome without the requirement for additional (nuclear) transgenes, they all rely on the external application of chemical inducers to trigger the induction of transgenes in plastids. Moreover, some of the inducers used are expensive and highly toxic (e.g. IPTG).
To develop an easily applicable and scalable system for the external control of transgene expression in plastids, inducible systems that do not require the application of chemical inducers are highly desirable. In this study, we sought to develop a thermo‐regulated system to control gene expression in plastids.
The cI repressor is a key component controlling gene expression in bacteriophage λ. It acts as a genetic switch between lysogenic growth and lytic development by binding the leftward (pL) and rightward (pR) promoters. A specific amino acid substitution (Ala66 to Thr) in the cI protein release binding of the mutated repressor (referred to as variant cI857) to the pL/pR promoters at high temperature, while not affecting binding at low temperature. In this way, cI857 permits transcription by the RNA polymerase at elevated temperatures. Due to this property, the λpL/pR‐cI857 system was developed as a useful thermo‐regulated expression system for heterologous protein production in Escherichia coli, phages and viruses (Ahmad et al., 2018; Menart et al., 2003; Restrepo‐Pineda et al., 2022; Rosano and Ceccarelli, 2014; Valdez‐Cruz et al., 2010). Here, we have explored the possibility of engineering the λpL/pR‐cI857 expression system to function as thermo‐regulator of transgene expression in plastids.
Results
Generation of transplastomic tobacco lines with the phage λ pL/pR‐cI857 expression system
To establish a heat‐inducible expression system in plastids, we designed a negative feedback regulation system, in which the expression of the repressor protein cI857 and the reporter GFP were both driven by the phage pL/pR promoter. The two proteins were co‐expressed from a dicistronic transcription unit either without (pWB5) or with (pWB6) an intercistronic expression element (IEE) separating the two cistrons. The IEE mediates the processing of the polycistronic primary transcript into monocistronic mRNAs (Zhou et al., 2007). A construct harbouring only the gfp reporter gene driven by the pL/pR promoter served as control (pWB3; Figures 1a and S1a). The three constructs were introduced into tobacco plastids by biolistic bombardment (Svab et al., 1990). For each construct, several transformed plant lines were obtained and subjected to additional rounds of regeneration on spectinomycin‐containing medium. Two independently transformed plant lines for each construct (Nt‐pWB3#1, Nt‐pWB3#2, Nt‐pWB5#12, Nt‐pWB5#19, Nt‐pWB6#10 and Nt‐pWB6#11) were selected for in‐depth analysis, and integration of the transgenes into the plastid genome was verified by Southern blot analyses. The data revealed that all transformed lines showed the expected hybridization signals resulting from transgene integration into the plastid genome by homologous recombination (Nt‐pWB3: 7.68 kb; Nt‐pWB5: 8.56 kb; Nt‐pWB6: 8.61 kb), and complete absence of the hybridization signal of 4.72 kb that is diagnostic of the wild‐type plastid DNA (Figures 1b and S1a). Seed assays revealed that the progenies of all transplastomic lines showed no segregation on spectinomycin‐containing medium, thus ultimately demonstrating the homoplasmic state of the transplastomic lines (Figure S1b). When grown in soil under standard greenhouse conditions, all transplastomic plants grew normally and were indistinguishable from wild‐type plants (Figure S2).
Figure 1.
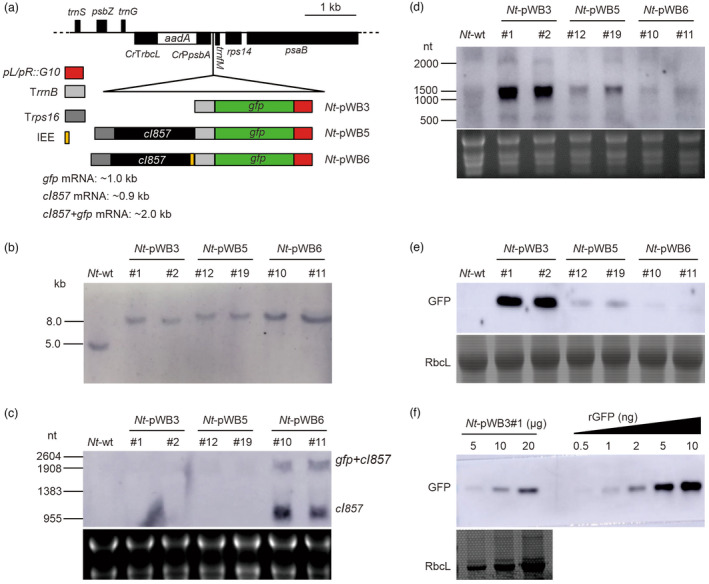
Generation and analysis of transplastomic tobacco plants containing the phage λ pL/pR‐cI857 expression system. (a) Physical maps of the transgene targeting region in the plastid genomes of the wild type (Nt‐wt) and the transplastomic Nt‐pWB3, Nt‐pWB5 and Nt‐pWB6 lines. The control of transgene expression is intended to be mediated by the binding of cI857 repressor to the pL/pR promoter. Genes above the lines are transcribed from left to right, genes below the line are transcribed in the opposite direction. (b) Southern blot analysis of transplastomic plants. Total DNA was digested with the restriction enzyme SpeI and hybridized to the DIG‐labelled psaB‐specific probe shown in Figure S1a. (c, d) Northern blot analysis of cI857 (c) and gfp (d) mRNA accumulation with gene‐specific RNA probes. The sizes of RNA marker bands are given on the left, and the dicistronic transcripts (gfp+cI857) and monocistronic transcripts (cI857) are indicated on the right (panel c). 30 μg and 5 μg of total RNA were loaded per lane in (c) and (d), respectively. The GelView‐stained gels prior to blotting are shown below each blot. (e) Western blot analysis of GFP accumulation in leaves with an anti‐GFP antibody. 100 μg of total protein were loaded in each lane. Coomassie staining of an identical gel is shown below the blot. (f) Determination of the GFP accumulation level in Nt‐pWB3#1 by semi‐quantitative analysis using a dilution series of recombinant GFP (rGFP; Vector Laboratories, USA). The amount of total protein loaded in each lane is indicated.
Analysis of transgene expression in transplastomic tobacco with the λ pL/pR‐cI857 system under normal condition
Having obtained the transplastomic tobacco plants expressing λpL/pR‐cI857, we next investigated transgene expression in transplastomic plants under normal growth conditions. Northern blot experiments were performed to examine cI857 and gfp transcript accumulation using hybridization probes specific for the coding regions of the transgenes. The results showed that cI857 transcripts were significantly more abundant in Nt‐pWB6 lines than in Nt‐pWB5 lines (Figure 1c), while they were not detectable in Nt‐pWB3 lines, as expected. The abundant presence of transcripts of ~1000 nt representing the monocistronic cI857 mRNA in Nt‐pWB6 lines, but not in the Nt‐pWB5 lines, is likely due to the presence of the IEE between the cI857 and gfp genes, which mediates the processing of the dicistronic primary transcript into monocistronic units and stabilizes the mRNA by PPR protein binding to its termini (Barkan, 2011; Legen et al., 2018; Zhou et al., 2007). Analysis of gfp transcript accumulation and GFP expression showed that in the absence of the repressor cI857 (Nt‐pWB3 lines), the expression of GFP reached much higher levels than in the lines with the repressor (Nt‐pWB5 and Nt‐pWB6 lines; Figure 1d,e). There was weak expression of GFP in the Nt‐pWB5 lines, while almost no GFP expression was detectable in the Nt‐pWB6 lines. This is likely due to more efficient repression of gfp transcription (and, consequently, GFP accumulation) in the Nt‐pWB6 lines (containing the IEE) than in the Nt‐pWB5 lines (lacking the IEE; Figures 1d,e and S3). These results indicate that the pL/pR promoter is active in tobacco plastids and can be controlled by the cI857 repressor, at least under normal growth conditions. However, GFP accumulation was relatively low, reaching only about 0.02% of TSP in the control transplastomic plants without the cI857 repressor (Nt‐pWB3 lines; Figure 1f). Although the −35 and −10 regions are largely conserved between the pL promoter and the PpsbA promoter, two T‐to‐G mutations in the first position of the −10 element and the extended −10 element may contribute to the low transcriptional activity of the heterologous pL/pR promoter from phage λ in plastids (Figure S1c).
Heat‐inducible expression of GFP in transplastomic plants mediated by the λ pL/pR‐cI857 system
Due to its much less leaky expression under normal growth conditions, we decided to use a transplastomic Nt‐pWB6 line (Nt‐pWB6#10) for heat induction experiments. To determine the optimal induction conditions, we exposed Nt‐pWB6#10 plants to different temperatures (33 °C, 36 °C, 39 °C and 42 °C) for 96 h under constant light. Northern and western blot analyses were then conducted to monitor GFP expression at different time points after the shift to elevated growth temperatures (Figure 2a–d). These experiments revealed that all temperatures tested induced the transcription of the gfp gene, and gfp mRNA accumulation increased with the time of the heat treatment. Similar transcript levels to that of the control (Nt‐pWB3#1) were reached after 72 h at 33 °C, after 60 h at 36 °C and after 24 h at 39 °C or 42 °C (Figure 2a–d). However, the GFP protein was almost undetectable at 33 °C (Figure 2a), and although it became detectable at 36 °C, its accumulation level was far below the GFP expression level in the control plant Nt‐pWB3#1 (Figure 2b). By contrast, the expression of GFP reached similarly high levels as in the control plants at 39 °C or 42 °C after 72 h (Figure 2c,d). Importantly, the expression of GFP remained unchanged in the control plant Nt‐pWB3#1 upon heat treatment at both the RNA and protein levels (Figure S4). Together, these data show that a temperature shift to at least 39 °C is required for the efficient induction of GFP expression by the λ pL/pR‐cI857 system.
Figure 2.
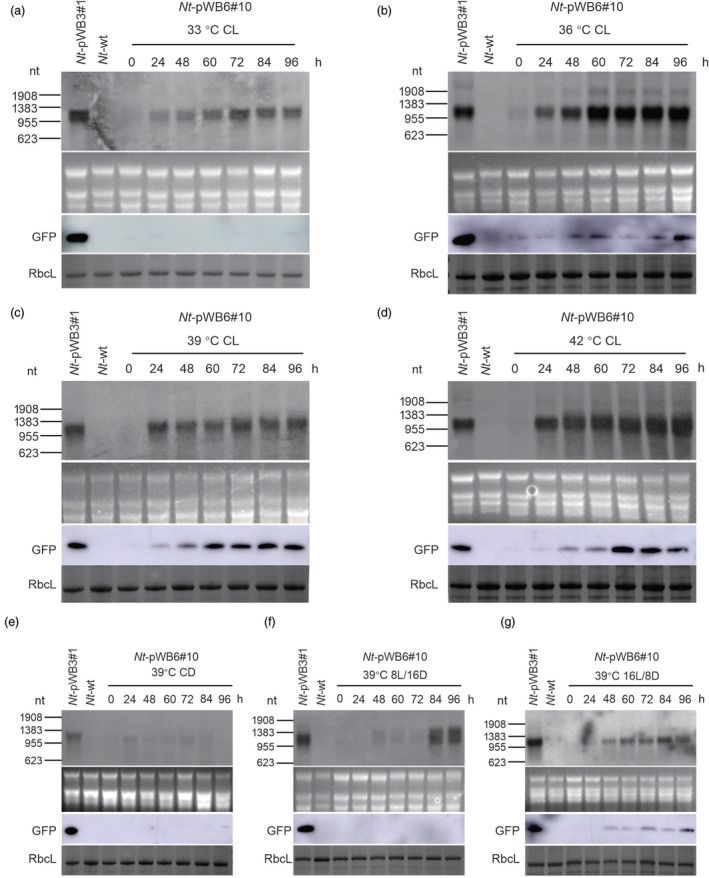
Heat‐inducible expression of the gfp transgene in transplastomic line Nt‐pWB6#10 upon shift to elevated temperatures and in dependence on the photoperiod. Northern blot and western blot analyses of gfp mRNA and protein accumulation in transplastomic line Nt‐pWB6#10 induced at 33 °C (a), 36 °C (b), 39 °C (c) and 42 °C (d) under constant light (CL) conditions is shown, and compared to gfp mRNA and protein accumulation in Nt‐pWB6#10 induced at 39 °C under constant darkness (CD; panel e), 8 h light and 16 h darkness (8L/16D; panel f), 16 h light and 8 h darkness (16L/8D; panel g). Samples of 5 μg total RNA and 10 μg total protein were loaded for northern blot and western blot analyses, respectively. Line Nt‐pWB3#1 grown under normal growth conditions served as control. The GelView‐stained RNA gels and the Coomassie‐stained protein gels prior to blotting are shown below each blot.
We next wanted to determine whether light is required for the induction of GFP expression by heat treatment. To this end, Nt‐pWB6#10 plants were treated at 39 °C under four different photoperiod conditions (constant darkness, 8 h light/16 h dark, 16 light/8 h dark and constant light). Under constant darkness, gfp transcripts and protein were not detectable (Figure 2e). Under 8 h light/16 h dark condition, the gfp transcripts became detectable after 48 h and signal intensity gradually increased with time. However, no GFP expression at the protein level was detectable during the whole 96 h induction process (Figure 2f). Under a 16 h light/8 h dark diurnal cycle, the presence of gfp transcripts was detected from time point 24 h on (Figure 2g). While GFP expression became detectable after 48 h, the protein accumulation level was still far below that in the control plant Nt‐pWB3#1 (Figure 2g). Under constant light, the gfp mRNA was efficiently induced at 24 h and remained stable at levels that were similarly high as those in the control line Nt‐pWB3#1 (Figure 2c). Taken together, these results demonstrate that light is required for efficient heat induction of the λ pL/pR‐cI857 expression system in plastids. We conclude that a temperature shift to at least 39 °C under constant light represents the optimal condition to efficiently induce the λ pL/pR‐cI857 expression system in plastids (Figure 2c).
Improved plastid transgene expression by introduction of an RNA amplification step into the heat‐inducible pL/pR‐cI857 system
Although we had successfully established the λ pL/pR‐cI857 expression system in plastids for the control of transgene expression by heat treatment, the GFP expression level in the fully induced state was rather low, probably due to the weak activity of the (heterologous) pL/pR promoter in plastids (Figure 1f). To increase the transgene expression levels in plastids, we attempted to introduce an RNA amplification step into our heat‐inducible system. In this design, the T7 RNA polymerase is under the control of the pL/pR promoter, with or without the cI857 repressor, while the gfp gene is driven by the T7 promoter (Figures 3a and S5a). Heat treatment of plants would result in the induction of T7 RNA polymerase expression at relatively low levels, which, however, should be sufficient to trigger strong transcriptional activation of the gfp transgene. We refer to this system as a heat induction coupled RNA amplification system or HICoRA.
Figure 3.
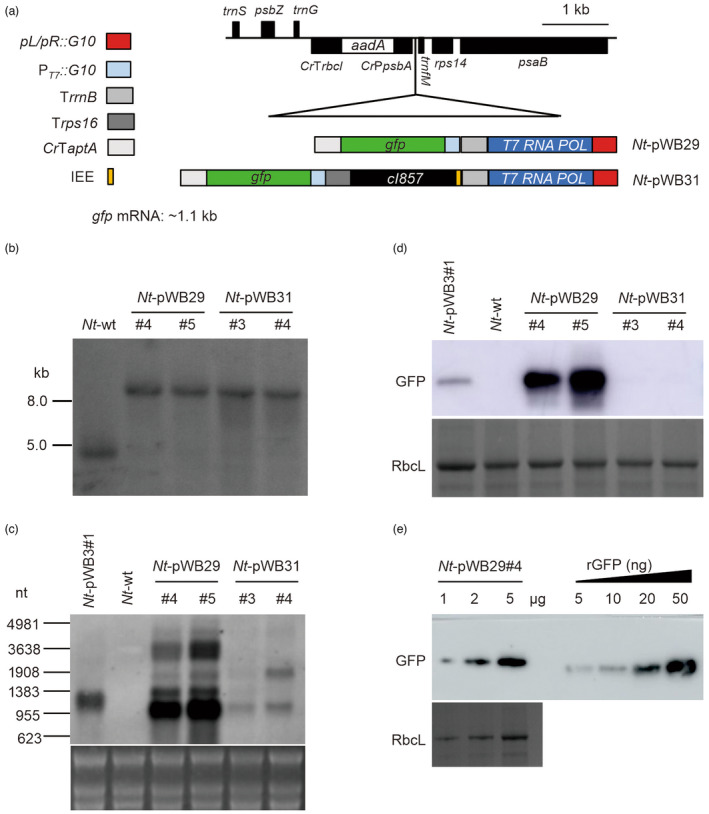
Generation and analysis of transplastomic tobacco plants expressing the HICoRA system under normal growth conditions. (a) Physical maps of the region in the plastid genomes of the transplastomic Nt‐pWB29 and Nt‐pWB31 lines. POL: polymerase. (b) Southern blot analysis of transplastomic plants. Total DNA was digested with the restriction enzyme SphI and hybridized to the DIG‐labelled psaB‐specific probe shown in Figure S3a. (c) Northern blot analysis of gfp mRNA accumulation detected with a gene‐specific RNA probe. The sizes of RNA marker bands are given at the left. 5 μg of total RNA were loaded in each lane. The GelView‐stained gel prior to blotting is shown below the blot. (d) Western blot analysis of GFP protein accumulation in leaves using an anti‐GFP antibody. 10 μg of total protein were loaded in each lane. Coomassie staining of an identical gel is shown below the blot. (e) Determination of the GFP accumulation level in Nt‐pWB29#4 by semi‐quantitative analysis using a dilution series of recombinant GFP (rGFP). The amount of total protein loaded in each lane is indicated.
We next introduced the HICoRA system into plastids with our recently developed protocol for plastid transformation with fragmented DNA (Ren et al., 2022). The 5′ end of fragment WB28 has 539 bp homologous sequence to the 3′ ends of fragments WB29 and WB31, respectively (Figure S5a). Mixtures of WB28 and WB29 or WB28 and WB31 were used for biolistic transformation. For each transformation, several transplastomic lines were obtained and subjected to additional rounds of regeneration to isolate homoplasmic lines. Two independent transplastomic lines from each transformation (Nt‐pWB29#4 and Nt‐pWB29#5, Nt‐pWB31#3 and Nt‐pWB31#4) were selected for further molecular analysis. The homoplasmic state of transplastomic lines was confirmed by Southern blot analyses, and evidenced by the exclusive presence of the hybridization signals diagnostic of the transformed plastid genomes (Nt‐pWB29: 9.62 kb; Nt‐pWB31: 10.54 kb), and absence of the hybridization signal (4.72kb fragment) specific to the wild types (Figure 3b). Moreover, all seedlings germinated from seeds of transplastomic lines were shown to be resistant to spectinomycin, further indicating that the transplastomic plants are homoplasmic (Figure S5b). When grown in soil under standard greenhouse conditions, transplastomic plants showed no phenotypic difference to wild‐type plants (Figure S5c).
Under normal growth conditions, the results of northern and western blot analyses demonstrated that the hybridization signals of the transplastomic Nt‐pWB29 lines (HICoRA system, without the cI857 repressor) were much stronger than those of Nt‐pWB3#1 (λ pL/pR‐cI857 system, without the cI857 repressor; Figure 3c,d). The GFP accumulation level of an Nt‐pWB29 line was quantified and found to correspond to approximately 1% TSP, which is ~50 folds higher than that of the Nt‐pWB3 line (Figures 1f and 3e). These findings indicate that the introduction of the T7 expression system into plastids can significantly elevate transgene expression levels by RNA amplification. Notably, GFP expression in the Nt‐pWB31 lines (HICoRA system, with the cI857 repressor) under normal growth conditions was significantly suppressed by the cI857 repressor, and almost no GFP accumulation was detectable (Figure 3c,d).
Heat‐inducible expression of GFP in transplastomic plants with the HICoRA system
We next tested whether the HICoRA system can be induced by heat treatment. Using our optimized induction conditions, transplastomic Nt‐pWB31#4 plants were treated at 39 °C and constant light for 96 h (4 days). In these experiments, the GFP expression was found to be only moderately induced (Figure 4a,b). However, when the induction time was extended to 20 days, the gfp transcripts in the Nt‐pWB31#4 plants were induced from day 4 on and remained at high levels, albeit lower than those of control plants (Nt‐pWB29#4; Figure 4c). GFP accumulation in the Nt‐pWB31#4 plants gradually increased and reached a steady level (ca. 0.48% of TSP) at day 12 (Figure 4e). Interestingly, after the heat‐treated plants had been transferred back to room temperature, GFP accumulation levels in the Nt‐pWB6#10 and the Nt‐pWB31#4 plants remained high for 24 h, but then slightly decreased (Figure S6).
Figure 4.
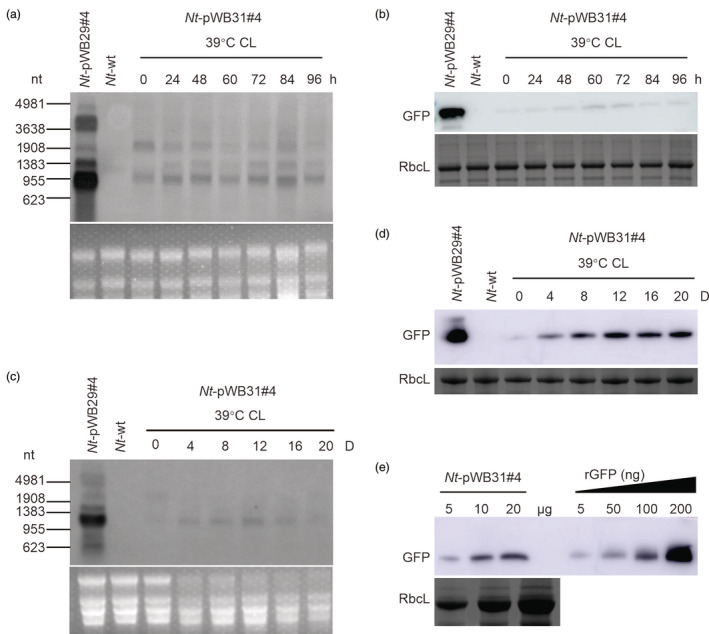
Heat‐inducible transgene expression in transplastomic line Nt‐pWB31#4 under optimized conditions. Shown are northern blot and western blot analyses of gfp mRNA (a, c) and protein accumulation (b, d) in transplastomic Nt‐pWB6#10 plants induced at 39 °C under constant light (CL). 5 μg total RNA and 10 μg total protein were loaded per lane for northern blot (a, c) and western blot analyses (b, d), respectively. (e) Determination of the GFP accumulation level in Nt‐pWB31#4 plants after 12 day of heat induction at 39 °C under constant light (CL) by semi‐quantitative analysis using a dilution series of rGFP. The amount of total protein loaded in each lane is indicated. Line Nt‐pWB29#4 grown under normal growth conditions served as control. The GelView‐stained RNA gels and Coomassie‐stained protein gels prior to blotting are shown below each blot. h: hours; D: days.
Discussion
In this work, we have adopted the bacteriophage λ pL/pR‐cI857 system as a temperature‐regulated system to control gene expression in plastids without the need to use chemical inducers. Our data show that the pL/pR promoter can be faithfully recognized by the RNA polymerase in plastids, but its activity was found to be low (Figure 1e,f). This resulted in relatively low GFP levels, even though strong translation signals (gene10 leader from bacteriophage T7) (Oey et al., 2009) had been introduced into the 5’ UTR of the gfp reporter. In plastids, the first two positions of the −10 box and the TGn motif upstream of the −10 box (referred to as extended −10 element) are crucial for promoter activity, as demonstrated for the psbA promoter (Toyoshima et al., 2005). Thus, it seems possible that the T‐to‐G mutations at the first positions of these two elements (Figure S1c) result in the relatively weak activity of the pL/pR promoter.
Our data revealed that the induction of transgene expression was dependent on light, with the most efficient and stable induction obtained under continuous illumination (Figure 2). We also found that the −35 and −10 regions of the pL/pR promoter were highly similar to those of the PpsbA promoter (Figure S1c), a light‐regulated plastid‐encoded promoter (Leivar et al., 2009) driven by the eubacterial‐type plastid‐encoded RNA polymerase (Tozawa et al., 1998). The high sequence similarity of the core sequences of the pL/pR and PpsbA promoters may in part explain the requirement for light to achieve full activation of transcription from the pL/pR promoter.
Introduction of the HICoRA system into plastids has increased GFP expression in comparison to the pL/pR‐cI857 system (Figure 4d), but the GFP accumulation level is lower than with the HICoRA system in the absence of the cI857 repressor (Nt‐pWB29; ~1% TSP) (Figure 3e). Previous studies have shown that the subunit stoichiometry of multimeric protein complexes in plastids is controlled at the translational level and additionally by protein degradation to mediate adaptation to long‐term heat exposure (Lukoszek et al., 2016). Under long‐term heat treatment, the translation machinery is slowed down in plastids (Zoschke and Bock, 2018). Consistent with this heat sensitivity of translation, we observed that, over the time course of induction, the translation of the gfp mRNA lagged behind the heat induction of gfp transcription (Figures 2 and 4).
Upon optimal induction conditions, the GFP accumulation reached 0.48% of the TSP (Figure 4d), a moderate expression level compared with constitutively expressed GFP in plastids (Wu et al., 2017). A possible reason could be that the gene expression machinery of plastids is negatively affected by the high‐temperature treatment. Although the transgene expression levels attainable with the HICoRA system are not very high, the system shows relatively tight repression and only low‐level leakiness of transgene expression. Therefore, this system offers potential advantages when the expression of toxic products or metabolites is attempted (Agrawal et al., 2022). Moreover, since the levels of GFP accumulation are highly correlated with the induction conditions (light, time and temperature), the HICoRA system may provide a useful tool for quantitative tuning of gene expression in synthetic biology.
In summary, we report here a novel inducible expression system for plastids that is based on temperature. It offers the benefits of not requiring nuclear transgenes and being independent of chemical inducers. We have demonstrated that, with the HICoRA system, the induction of transgene expression can be finely tuned. Since temperature changes can be easily applied upon greenhouse cultivation of plants, the HICoRA system should be easily scalable, while avoiding the use of toxic and/or expensive chemical inducers. Thus, the heat‐inducible expression system developed in this study is expected to expand the range of applications of the transplastomic technology.
Materials and methods
Plant material and growth conditions
Aseptic tobacco (Nicotiana tabacum cv. Petit Havana) plants were grown on Murashige & Skoog (MS) medium containing 3% sucrose and 0.54% micro agar (Duchefa, Netherlands) (Murashige and Skoog, 1962). Bombarded tobacco leaves were selected on the RMOP medium containing spectinomycin (500 mg/L) (Svab and Maliga, 1993). Regenerated shoots were rooted on MS medium supplemented with NAA (0.1 mg/L), 3% sucrose, 0.54% micro agar and spectinomycin (500 mg/L). Homoplasmic transplastomic tobacco plants were transferred to soil and grown in controlled environmental conditions under a diurnal cycle of 16 h light (16L) at 25 °C and 8 h darkness (8D) at 22 °C.
Construction of vectors or fragmented DNA for plastid transformation
The pLpR fragment was PCR amplified with primers pLpR(SalI)‐F1/pLpR‐R1/pLpR(NcoI)‐R2 (Table S1) by overlap extension using plasmid pBV220 as template (Zhang et al., 1990), and introducing the 5′ untranslated region (UTR) of bacteriophage T7 gene 10 (Ye et al., 2001). The PCR products were digested with SalI/NcoI and cloned into the similarly cut vector pYY12 (Wu et al., 2017), generating vector pWB3 (accession number: OR584051). The cI857 gene was PCR amplified with primer pair cI857(ApaI)‐F1/cI857‐R1 (Table S1) using pBV220 as template (Table S1), adding the Shine‐Dalgarno (SD) sequence from the tobacco rbcL gene. The terminator of the tobacco rps16 gene (Trps16) was amplified using vector pZF75 as template (Zhou et al., 2007) with primer pair Trps16‐F1/Trps16(KpnI)‐R1 (Table S1). The cI857 and Trps16 fragments were then fused by overlap‐extension PCR with primer pair cI857(ApaI)‐F1/Trps16(KpnI)‐R1 (Table S1). The obtained PCR products were digested with ApaI/KpnI and inserted into the similarly cut vector pWB3, resulting in vector pWB5 (accession number: OR584052). The DNA sequence of a plastid intercistronic expression element (IEE) (Zhou et al., 2007) was fused to the 5′ end of cI857 gene by overlap‐extension PCR with primers IEE‐F1/IEE(ApaI)‐F2/Trps16(KpnI)‐R1 (Table S1) using pWB5 as template. The PCR products were digested with ApaI/KpnI and cloned into the similarly cut vector pWB3, generating vector pWB6 (accession number: OR584053).
To additionally introduce the T7 expression elements while omitting further cloning steps, we employed a protocol for plastid transformation with fragmented DNA (Ren et al., 2022). A DNA fragment containing a partial pL sequence and a partial T7 RNA polymerase sequence was obtained by PCR amplification with primer pair T7RH‐F/T7‐pL‐R using plasmid pME16 (Emadpour et al., 2015) as template (Table S1). Another DNA fragment containing flanking sequences from the tobacco chloroplast genome and the pLpR promoter was PCR amplified with primer pair pLpR‐R1/psaB‐R (Table S1) using pWB3 as template. Finally, DNA fragment WB28 (accession number: OR621291) was amplified with primer pair T7RH‐F/psaB‐R (Table S1) using the obtained two DNA fragments as templates by overlap‐extension PCR (Figure S3a).
To obtain fragments WB29 (accession number: OR621292) and WB30 (accession numbers: OR621293), the green fluorescent protein (gfp) sequence was cloned as NcoI/XbaI fragment from pWB3 into the similarly cut pME16, resulting in plasmid pME16‐gfp. PCR amplification was performed using primer pair T7P‐F/T7T‐R (Table S1) using pME16‐gfp as template to obtain the intermediate fragment T7gfp, which was subsequently digested with KpnI and cloned into vectors pWB3 and pWB6, generating vectors pWB3‐T7gfp and pWB6‐T7gfp, respectively. A DNA fragment (P1) containing flanking sequences from the tobacco chloroplast genome, the selectable marker gene aadA and the gfp expression cassette was then PCR amplified with primer pair Trrn‐T7‐R/psbZ‐F (Table S1) using pWB3‐T7gfp as a template. Another DNA fragment (P2) covering part of the T7 RNA polymerase gene and a partial TrrnB sequence was obtained by PCR amplification with primer pair T7LH‐R/T7‐Trrn‐F (Table S1) using pME16 as template. Finally, DNA fragment WB29 (Figure S3a) was amplified with primer pair psbZ‐F/T7LH‐R using the obtained two DNA fragments as templates by overlap‐extension PCR (Table S1). Similarly, a fragment (P3) containing the cI857 gene in addition to fragment P1 was amplified using primer pair Trrn‐T7‐R/psbZ‐F (Table S1) and pWB6‐T7gfp as a template. Finally, DNA fragment WB31 (Figure S3a) was amplified with primer pair psbZ‐F/T7LH‐R (Table S1) using fragments P1 and P3 as templates by overlap‐extension PCR. The sequences of all plasmid vectors and DNA fragments were verified through sequencing.
Plastid transformation and selection of transplastomic tobacco lines
Tobacco plastid transformation experiments were carried out as previously described (Svab and Maliga, 1993). Plasmid DNA for plastid transformation was prepared using the Nucleobond Xtra Plasmid Midi Kit (Macherey‐Nagel, Germany). Young leaves from sterile tobacco plants were bombarded with DNA‐coated gold particles (0.6 μm) using a PDS‐1000/He Biolistic Particle Delivery System (Bio‐Rad, USA). The bombarded leaf samples were cut into 5 × 5 mm squares and transferred onto RMOP medium containing spectinomycin (500 mg/L). For each construct, several independent transplastomic lines were obtained and subjected to one or two additional rounds of regeneration on the same medium to select for homoplasmy.
For tobacco plastid transformation with fragmented DNA, previously described protocols were followed (Ren et al., 2022). The DNA fragments were obtained by PCR amplification using the PrimeSTAR® Max DNA Polymerase (TaKaRa, Japan). The PCR products were purified by the E.Z.N.A. Gel Extraction Kit (Omega, USA), and the concentration of each fragment was measured using the Nano Photometer (IMPLEN, Germany). DNA fragment WB28 (3334 bp, 8.24 μg) was mixed with WB29 (4745 bp, 11.73 μg) and WB31 (5676 bp, 14.03 μg), respectively, in a total volume of 20 μL (in Ultrapure water; for one bombardment), adjusting the molar concentration of each DNA fragment to 4 pmol. The mass of 1 pmol of each DNA fragments was calculated using the software SnapGene 3.2.1 (https://www.snapgene.com/).
Isolation of DNA and Southern blot analysis
Total DNA was extracted from leaves of wild‐type and selected spectinomycin‐resistant plants using a cetyltrimethylammonium bromide‐based method (Doyle and Doyle, 1990). DNA samples (5 μg) were digested with the restriction enzymes SpeI or SphI, separated by electrophoresis in 1% agarose gels and transferred onto a positively charged nylon membrane (GE Healthcare, USA) by capillary action using the semi‐dry transfer method. A 550‐bp fragment of the psaB gene was amplified with primer pair psaB‐Fp/psaB‐Rp (Table S1) and used as a hybridization probe to verify plastid transformation (Wu et al., 2017). Labelling of the probe and hybridization was performed with the DIG‐High Prime DNA Labeling and Detection Starter Kit I following the manufacturer's instructions (Roche, Switzerland). The hybridization signals were analysed using the BCIP/NBT reagent in the same kit.
Isolation of RNA and northern blot analysis
Total RNA was extracted using the RNAiso plus Reagent (TaKaRa, Japan) according to the manufacturer's protocol. RNA samples were denatured and separated by electrophoresis in formaldehyde‐containing 1% agarose gels, and then transferred from the gel to a positively charged nylon membrane (GE Healthcare, USA). PCR products generated by amplification with gene‐specific primers were used as templates for the synthesis of RNA probes (Table S1). The probes were labelled with DIG using the DIG Northern Starter Kit following the manufacturer's instructions (Roche, Switzerland). The hybridization signals were analysed by chemiluminescence immunoassays using an Amersham Imager 600 (GE Healthcare, USA) or by using the BCIP/NBT Alkaline Phosphatase Colour Development Kit (Solarbio, China).
Protein isolation and western blot analysis
Leaf samples (∼100 mg fresh materials) of tobacco plants were ground in liquid nitrogen. Total protein was isolated using a phenol‐based method (Cahoon et al., 1992). The protein concentration was determined with the Easy II Protein Quantitative Kit (TransGen Biotech, China). Protein samples were separated by electrophoresis in 10% SDS‐containing polyacrylamide gels (PAGE Gel Fast Preparation Kit, Epizyme Biotech, China). Proteins were visualized by Coomassie blue staining or transferred onto polyvinylidene difluoride (PVDF) membranes (GE Healthcare, USA). Protein detection was performed with a monoclonal mouse antibody raised against GFP (ABclonal, UK). All results were confirmed by three biological replicates.
Heat treatment and induction conditions
Seeds were germinated on MS medium, seedlings were transferred to soil after 2 weeks and grown under standard greenhouse conditions. For heat treatment, 7‐week‐old tobacco plants were transferred to an incubator (QHX‐250BSH‐III, CIMO Medical Instrument, China) and treated at different temperatures (25 °C, 33 °C, 36 °C, 39 °C and 42 °C) under different photoperiods (constant darkness, 16 h light/8 h dark, 8 h light/16 h dark, or constant light). The relative humidity was kept constant at 70%. Leaves number from 3 to 5 (counted from the top) were sampled at the indicated time points for further analysis.
Author contributions
J.Z. designed research; W.X. and J.Z. performed research; W.X., S.L., R.B. and J.Z. analysed data; J.Z. and R.B. wrote the paper with input from other authors.
Competing interest statement
The authors declare no conflict of interest.
Supporting information
Figure S1 Generation of transplastomic tobacco plants harbouring the phage λ pL/pR‐cI857 expression system.
Figure S2 Phenotypes of soil‐grown transplastomic tobacco plants in greenhouse.
Figure S3 Northern blot analysis of gfp mRNA accumulation detected with a gfp‐specific RNA probe.
Figure S4 Northern blot analysis of gfp mRNA accumulation and western blot analysis of GFP accumulation in transplastomic tobacco line Nt‐pWB3#1 upon heat induction.
Figure S5 Generation and characterization of transplastomic tobacco plants containing the HICoRA system.
Figure S6 Western blot analysis of GFP accumulation in transplastomic tobacco lines Nt‐pWB6#10 and Nt‐pWB31#4 when shifted back to normal temperature after heat induction.
Table S1 List of oligonucleotides used in this study.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32271912) and Key Program in AGIS under Grant No. AGIS‐ZDXM‐202304.
Data availability statement
The data that supports the findings of this study are available in the supplementary material of this article.
References
- Agrawal, S. , Karcher, D. , Ruf, S. , Erban, A. , Hertle, A.P. , Kopka, J. and Bock, R. (2022) Riboswitch‐mediated inducible expression of an astaxanthin biosynthetic operon in plastids. Plant Physiol. 188, 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, I. , Nawaz, N. , Darwesh, N.M. , Ur Rahman, S. , Mustafa, M.Z. , Khan, S.B. and Patching, S.G. (2018) Overcoming challenges for amplified expression of recombinant proteins using Escherichia coli . Protein Expr. Purif. 144, 12–18. [DOI] [PubMed] [Google Scholar]
- Barkan, A. (2011) Expression of plastid genes: organelle‐specific elaborations on a prokaryotic scaffold. Plant Physiol. 155, 1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock, R. (2001) Transgenic plastids in basic research and plant biotechnology. J. Mol. Biol. 312, 425–438. [DOI] [PubMed] [Google Scholar]
- Bock, R. (2015) Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 66, 211–241. [DOI] [PubMed] [Google Scholar]
- Cahoon, E.B. , Shanklin, J. and Ohlrogge, J.B. (1992) Expression of a coriander desaturase results in petroselinic acid production in transgenic tobacco. Proc. Natl. Acad. Sci. USA 89, 11184–11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardi, T. , Lenzi, P. and Maliga, P. (2010) Chloroplasts as expression platforms for plant‐produced vaccines. Expert Rev. Vaccines 9, 893–911. [DOI] [PubMed] [Google Scholar]
- Doyle, J.J. and Doyle, J.L. (1990) Isolation of plant DNA from fresh tissue. Focus 12, 13–15. [Google Scholar]
- Emadpour, M. , Karcher, D. and Bock, R. (2015) Boosting riboswitch efficiency by RNA amplification. Nucleic Acids Res. 43, e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig, A. , Bonfig, K. , Roitsch, T. and Warzecha, H. (2007) Expression of the recombinant bacterial outer surface protein A in tobacco chloroplasts leads to thylakoid localization and loss of photosynthesis. FEBS J. 274, 5749–5758. [DOI] [PubMed] [Google Scholar]
- Legen, J. , Ruf, S. , Kroop, X. , Wang, G. , Barkan, A. , Bock, R. and Schmitz‐Linneweber, C. (2018) Stabilization and translation of synthetic operon‐derived mRNAs in chloroplasts by sequences representing PPR protein‐binding sites. Plant J. 94, 8–21. [DOI] [PubMed] [Google Scholar]
- Leivar, P. , Tepperman, J.M. , Monte, E. , Calderon, R.H. , Liu, T.L. and Quail, P.H. (2009) Definition of early transcriptional circuitry involved in light‐induced reversal of PIF‐imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21, 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lössl, A. , Eibl, C. , Harloff, H.J. , Jung, C. and Koop, H.U. (2003) Polyester synthesis in transplastomic tobacco (Nicotiana tabacum L.): significant contents of polyhydroxybutyrate are associated with growth reduction. Plant Cell Rep. 21, 891–899. [DOI] [PubMed] [Google Scholar]
- Lössl, A. , Bohmert, K. , Harloff, H. , Eibl, C. , Muhlbauer, S. and Koop, H.U. (2005) Inducible trans‐activation of plastid transgenes: expression of the R. eutropha phb operon in transplastomic tobacco. Plant Cell Physiol. 46, 1462–1471. [DOI] [PubMed] [Google Scholar]
- Lukoszek, R. , Feist, P. and Ignatova, Z. (2016) Insights into the adaptive response of Arabidopsis thaliana to prolonged thermal stress by ribosomal profiling and RNA‐Seq. BMC Plant Biol. 16, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee, A. , Coyne, S. , Murphy, D. , Horvath, E. , Medgyesy, P. and Kavanagh, T. (2004) T7 RNA polymerase‐directed expression of an antibody fragment transgene in plastids causes a semi‐lethal pale‐green seedling phenotype. Transgenic Res. 13, 325–337. [DOI] [PubMed] [Google Scholar]
- Menart, V. , Jevsevar, S. , Vilar, M. , Trobis, A. and Pavko, A. (2003) Constitutive versus thermoinducible expression of heterologous proteins in Escherichia coli based on strong PR,PL promoters from phage lambda. Biotechnol. Bioeng. 83, 181–190. [DOI] [PubMed] [Google Scholar]
- Muhlbauer, S.K. and Koop, H.U. (2005) External control of transgene expression in tobacco plastids using the bacterial lac repressor. Plant J. 43, 941–946. [DOI] [PubMed] [Google Scholar]
- Murashige, T. and Skoog, F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue culture. Physiol. Plantarum 15, 473–497. [Google Scholar]
- Oey, M. , Lohse, M. , Kreikemeyer, B. and Bock, R. (2009) Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J. 57, 436–445. [DOI] [PubMed] [Google Scholar]
- Ren, K. , Xu, W.B. , Ren, B.L. , Fu, J.Q. , Jiang, C.M. and Zhang, J. (2022) A simple technology for plastid transformation with fragmented DNA. J. Exp. Bot. 73, 6078–6088. [DOI] [PubMed] [Google Scholar]
- Restrepo‐Pineda, S. , Sanchez‐Puig, N. , Perez, N.O. , Garcia‐Hernandez, E. , Valdez‐Cruz, N.A. and Trujillo‐Roldan, M.A. (2022) The pre‐induction temperature affects recombinant HuGM‐CSF aggregation in thermoinducible Escherichia coli . Appl. Microbiol. Biotechnol. 106, 2883–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas, M. , Yu, Q. , Williams‐Carrier, R. , Maliga, P. and Barkan, A. (2019) Engineered PPR proteins as inducible switches to activate the expression of chloroplast transgenes. Nat. Plants 5, 505–511. [DOI] [PubMed] [Google Scholar]
- Rosano, G.L. and Ceccarelli, E.A. (2014) Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol. 5, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti, N. and Cardi, T. (2014) Transgene‐induced pleiotropic effects in transplastomic plants. Biotechnol. Lett. 36, 229–239. [DOI] [PubMed] [Google Scholar]
- Svab, Z. and Maliga, P. (1993) High‐frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 90, 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab, Z. , Hajdukiewicz, P. and Maliga, P. (1990) Stable transformation of plastids in higher plants. Proc. Natl. Acad. Sci. USA 87, 8526–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima, Y. , Onda, Y. , Shiina, T. and Nakahira, Y. (2005) Plastid transcription in higher plants. Crit. Rev. Plant Sci. 24, 59–81. [Google Scholar]
- Tozawa, Y. , Tanaka, K. , Takahashi, H. and Wakasa, K. (1998) Nuclear encoding of a plastid sigma factor in rice and its tissue‐ and light‐dependent expression. Nucleic Acids Res. 26, 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez‐Cruz, N.A. , Caspeta, L. , Perez, N.O. , Ramirez, O.T. and Trujillo‐Roldan, M.A. (2010) Production of recombinant proteins in E. coli by the heat inducible expression system based on the phage lambda pL and/or pR promoters. Microb. Cell Fact. 9, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhounig, A. , Karcher, D. and Bock, R. (2010) Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc. Natl. Acad. Sci. USA 107, 6204–6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , You, L. , Li, S. , Ma, M. , Wu, M. , Ma, L. , Bock, R. et al. (2017) In vivo assembly in Escherichia coli of transformation vectors for plastid genome engineering. Front. Plant Sci. 8, 1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Deng, Y. and Li, S. (2022) Advances in plastid transformation for metabolic engineering in higher plants. aBIOTECH 3, 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, G.‐N. , Hajdukiewicz, P.T.J. , Broyles, D. , Rodriguez, D. , Xu, C.W. , Nehra, N. and Staub, J.M. (2001) Plastid‐expressed 5‐enolpyruvylshikimate‐3‐phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J. 25, 261–270 . [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Yao, L. and Hou, Y. (1990) Construction and application of a high level expression vector containing PRPL promoter. Chin. J. Virol. 2, 111–116. [Google Scholar]
- Zhou, F. , Karcher, D. and Bock, R. (2007) Identification of a plastid intercistronic expression element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J. 52, 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke, R. and Bock, R. (2018) Chloroplast translation: structural and functional organization, operational control and regulation. Plant Cell 30, 745–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Generation of transplastomic tobacco plants harbouring the phage λ pL/pR‐cI857 expression system.
Figure S2 Phenotypes of soil‐grown transplastomic tobacco plants in greenhouse.
Figure S3 Northern blot analysis of gfp mRNA accumulation detected with a gfp‐specific RNA probe.
Figure S4 Northern blot analysis of gfp mRNA accumulation and western blot analysis of GFP accumulation in transplastomic tobacco line Nt‐pWB3#1 upon heat induction.
Figure S5 Generation and characterization of transplastomic tobacco plants containing the HICoRA system.
Figure S6 Western blot analysis of GFP accumulation in transplastomic tobacco lines Nt‐pWB6#10 and Nt‐pWB31#4 when shifted back to normal temperature after heat induction.
Table S1 List of oligonucleotides used in this study.
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


