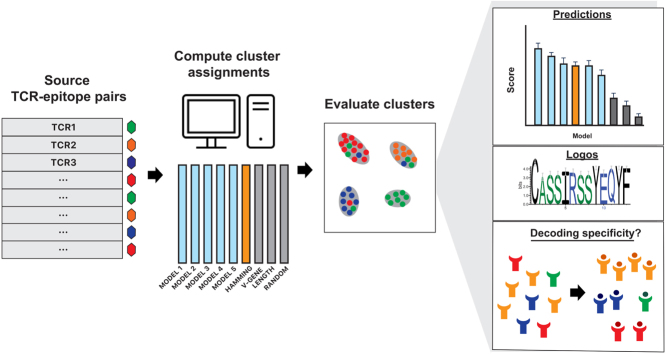
Abstract
The vast potential sequence diversity of TCRs and their ligands has presented an historic barrier to computational prediction of TCR epitope specificity, a holy grail of quantitative immunology. One common approach is to cluster sequences together, on the assumption that similar receptors bind similar epitopes. Here, we provide the first independent evaluation of widely used clustering algorithms for TCR specificity inference, observing some variability in predictive performance between models, and marked differences in scalability. Despite these differences, we find that different algorithms produce clusters with high degrees of similarity for receptors recognising the same epitope. Our analysis strengthens the case for use of clustering models to identify signals of common specificity from large repertoires, whilst highlighting scope for improvement of complex models over simple comparators.
Keywords: T cell antigen specificity, T cell receptor repertoire analysis, Clustering models, Deorphanizing TCRs
Graphical abstract
1. Introduction
T lymphocytes recognise peptide epitopes presented at the cell surface by Major Histocompatibility Complexes (MHC) in jawed vertebrates [1]. Recognition is mediated by diverse heterodimeric or TCR domains positioned on the cell surface. The chains of the more common TCR contain variable (V), joining (J) gene segments, constant (C) regions, and an additional diversity (D) segment in the polypeptide. Each cell expresses many copies of a single TCR, which bind to peptide-MHC (pMHC) via the complementarity determining regions (CDR) 1-3 of the TCR [2]. Productive TCR engagement triggers a context-dependent signalling cascade, which in turn promotes activation and differentiation of diverse immune effector cells [3].
The central role of the TCR in immune surveillance and response to disease has encouraged efforts to decode the rules of TCR-pMHC binding. Determination of specificity, or “receptor de-orphanisation”, can be achieved experimentally using sequencing and repertoire analysis, or with functional, multimer binding, or TCR screening methods, reviewed in [4], [5]. Indeed, the advent of high-throughput single cell methods has vastly increased the volume and functional annotation of cell repertoires. However, our understanding of epitope specificity remains limited to a small fraction of the universe of TCR-epitope pairs, and the ability to accurately predict the cognate epitope of any TCR in silico could vastly accelerate our understanding of fundamental and translational cell biology [6].
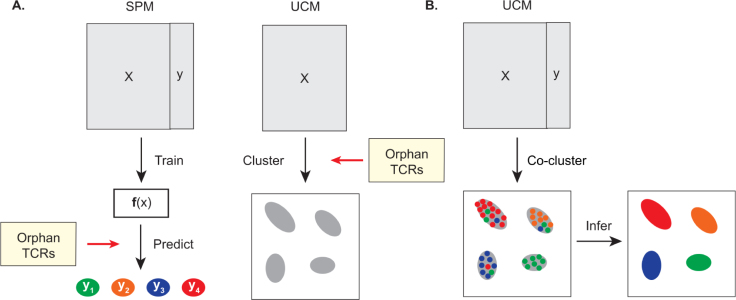
The availability of large repositories of TCR sequences and their known ligands has enabled the development of two major families of computational model for prediction of TCR antigen specificity: Supervised Predictive Models (SPMs) and Unsupervised Clustering Models (UCMs) (Fig. 1) [6]. These families are representative of two distinct approaches to machine learning. In supervised learning, predictive models are trained on a set of input instances having a known label (in this case, the cognate epitope for a given TCR). In unsupervised learning, models learn the underlying statistical features or patterns of a dataset to differentiate between input TCRs, applying techniques such as clustering or dimensionality reduction.
Fig. 1.
Supervised and unsupervised learning in cell epitope specificity inference. (A) SPMs (left) fit a predictive function f(x) to training data having an independent variable X (TCR sequences and other features) and dependent variable y (epitopes or pMHC complexes). This function may then be applied to predict the cognate epitopes of orphan TCRs. UCMs (right) generate a mapping from TCR sequences to a cluster allocation, such that each TCR is assigned to one or more clusters having common epitope specificity. (B) Application of UCMs to de-orphanise TCRs by co-clustering.
The use of deep neural networks (DNNs) including large language models and convolutional neural networks has contributed significantly to recent improvements in UCM and SPM performance [7], [8], [9], [10], [11]. Despite these advances, no publicly available SPM is yet capable of accurately predicting the specificity of TCRs recognising “unseen” epitopes that were not encountered during model training [8], [12]. This is likely due at least in part to the limited volume of experimentally determined receptor-epitope pairs, which constitutes just a small fraction of the vast theoretical diversity of TCRs [6], [13].
Unlike SPMs, UCMs do not require receptor–ligand pairs as an input, but have been designed to group together TCRs on the basis of features thought to underly common antigen specificity [14], [15]. Such models are of particular use in an era when bulk and single-cell sequencing experiments can yield thousands of unique orphan TCRs per sample, applying them to group receptors sharing similar sequence, structure, or physico-chemical properties and so to shortlist TCRs of interest for later experimental de-orphanisation. Such approaches have been successfully applied to identify and characterise TCRs associated with mycobacterial and viral infection, cancer, and autoimmune disease [15], [16], [17], [18], [19], [20].
UCMs take as their input single or paired TCR CDR3 nucleotide or amino acid sequences, with or without V and J gene usage information, and return a mapping of sequences to unique clusters. This has historically been achieved using some form of distance measure, typically either direct sequence similarity and/ or the frequency enrichment of short sequence snippets (kmers) compared to a reference dataset. Recent approaches leverage DNNs to generate a compressed numeric representation of the input TCR as a precursor to clustering [11], [21], [22], [23].
Whilst this is not the purpose for which they were originally conceived, UCMs may be used to infer TCR epitope specificity from datasets having full or partial epitope labels, by assigning the most frequent epitope of a cluster as the predicted binder for all TCRs in that cluster (Fig. 1B). However, there has to date been no independent benchmarking study of UCMs as predictors of TCR specificity, despite their widespread use in the field. In the present work, we provide the first confirmation of the direct predictive capacity of UCMs by applying the models to sets of known TCR-epitope pairs, comparing performance using a balanced measure of precision and recall. We evaluate five commonly used UCMs, investigating the impact on performance of the number of TCR representatives provided per epitope, the receptor chain, the epitope studied, and the input dataset. We find that all five models outperform CDR3 length and random baselines, but that relative predictive performance is sensitive to each of these four factors. The five models studied are not consistently superior to of simple Hamming sequence distance and V-gene clustering models, highlighting significant room for improvement. Extending the analysis to qualitative comparison of the clusters formed, we observe that the five models also produce similar cluster motifs for common viral epitopes. Taken together, we conclude that whilst UCMs are of considerable value in de-orphanisation pipelines, further work is needed, and that in practice clustering algorithms, experimental methods, and predictive models should be combined to maximise the likelihood of identifying true epitope-specific clusters.
2. Methods
2.1. Datasets
A consolidated dataset of paired TCR amino acid sequences of human origin was developed using instances drawn from VDJdb [24] for initial model comparison, and TCR-epitope pairs from McPas-TCR [25] and from the MIRA dataset [26] were prepared as separate non-overlapping test sets (Table S1). Sequences derived from a 10X study of healthy donors, and CDR3 sequences containing non amino acid symbols, were removed from the input data. V and J gene codes were processed for consistency with IMGT reference sequences [27]. Duplicates were removed within and between datasets using CDR3-V-J bio-identities for both and chains, such that a given TCR was encoded in the format CDR3-TRAV-TRAJ-CDR3-TRBV-TRBJ. Only those TCRs having V or J genes included in the reference IMGT alleles of the tcrdist module of the CoNGA conda package (v.0.1.1) [28] were retained, to ensure that consistent numbers of sequences were provided to each model. Benchmarking experiments were performed on VDJdb data after chain selection and down-sampling as described in Results. Neither McPas-TCR nor MIRA datasets were downsampled, to give a representative view of model performance on imbalanced datasets more representative of real world datasets.
2.2. Models
A systematic review of the literature was conducted to identify studies presenting novel methods for prediction of antigen specificity from TCR sequences (Section S1). ClusTCR [29], GIANA [30], GLIPH2 [17], iSMART [20], and tcrdist3 [31] were shortlisted for analysis based on the availability of open-source python packages or executable files. ALICE [32] was excluded as more appropriately applied to the identification of expanded clones in individual patient repertoire data. To the five test models we added Hamming distance, V-gene, length and random baseline comparators. Implementation and background methodological detail is included for each of the selected algorithms in Section S2. All benchmarking experiments were run on a single remote Intel(R) Xeon(R) Gold 6126 CPU at 2.60 GHz to ensure fair comparison of algorithms with and without parallel processing capability.
2.3. Metrics
Performance of each model was initially analysed using a broad panel of UCM-specific, entropy, and predictive measures. Clustering metrics included purity, consistency and retention, as described previously in [29], [33], where purity is the proportion of TCRs in a given cluster having the modal epitope; consistency is the proportion of epitope-specific TCRs that are assigned to a single cluster; and retention is the proportion of TCRs successfully assigned to a cluster. Entropy was calculated using a scikit learn implementation of adjusted mutual information (AMI) that accounts for class imbalance in computation of the mutual information of true and predicted labels [34]. To these were added the predictive metrics accuracy, precision, recall, and f1-score, as described in Eqs. (1a)–(1d). Each of these measures was weighted to adjust for per-epitope class imbalance, and computed for all input instances to account for differences in retention between models.
| (1a) |
| (1b) |
| (1c) |
| (1d) |
Eqs. (1a)–(1d): Predictive performance metrics, where TP True Positive, TN True Negative, FP False Positive, FN False Negative.
2.4. Synthetic TCRs
Synthetic TCR sequences were produced using OLGA [35] (see Study Limitations), which uses dynamic programming to generate TCR sequences (CDR3, V and J gene codes) with probabilities inferred from human T cell repertoires.
2.5. Statistics
Statistical significance was determined using a two-tailed Mann–Whitney test (consistent with [36]). Statistical analyses were performed with GraphPad Prism [37]. Plots were produced with GraphPad, Matplotlib, and Seaborn Python libraries [37], [38], [39].
2.6. CDR3 amino acid motifs
Sequence logos were produced from chain selections of dataset V1000 by retaining TCRs having the modal length from the largest cluster for each of the four epitopes present. Sequences were aligned with MUSCLEv5.1 [40], and logos were produced from the resulting multiple sequence alignments with WebLogo v3.7.12 [41].
3. Results
Five commonly used open-source UCMs were identified from the literature for which a python implementation was readily available: ClusTCR, GIANA, GLIPH2, iSMART, and tcrdist3 [17], [20], [29], [30], [31]. Four baseline models were added: A Hamming distance model that grouped together sequences having identical length and differing by not more than one amino acid, CDR3 length, V-gene clustering, and random baselines. A summary of the respective model methodologies is provided in Section S2.
Benchmarking analyses were performed on paired TCRs data drawn from VDJdb, a large, public, curated source of TCRs of known epitope specificity and which has been used as a reference for all of the five models selected [24]. However, past studies have presented comparisons using a single pre-processing strategy, for example by retaining only a subset of TCRs binding selected epitopes [17] or meeting a specific quality threshold [29]. To explore whether selective sub-setting of data might mask inherent differences between models, performance was analysed for data subsets generated by retaining epitopes having 10, 50, 100, 500, or 1000 cognate TCRs (datasets V10, V50, V100, V500, and V1000 respectively, Table S1 and Table S2). Instances were randomly down-sampled after pre-processing, such that each experimental run was performed on the same number of TCR sequences per epitope. All models were applied to or chain selections from the same set of paired TCRs. Sampling was repeated 25 times to account for variance between random TCR selections. We present performance on or chain selections independently (see Discussion and Study Limitations).
UCMs have historically been evaluated using cluster quality metrics such as purity, consistency, diversity, and retention (see Methods). By applying each model to sets of TCRs of known specificity, we were able to supplement these metrics with direct measures of predictive capacity following the schema depicted in Fig. 1B. Observing a positive correlation between performance according to cluster purity, consistency, adjusted mutual information, precision, recall, and F1-score (Fig. S2), we present results for F1-score alone, weighting scores to account for epitope-specific class imbalance and computing performance across all input TCRs to allow for differences in retention between models.
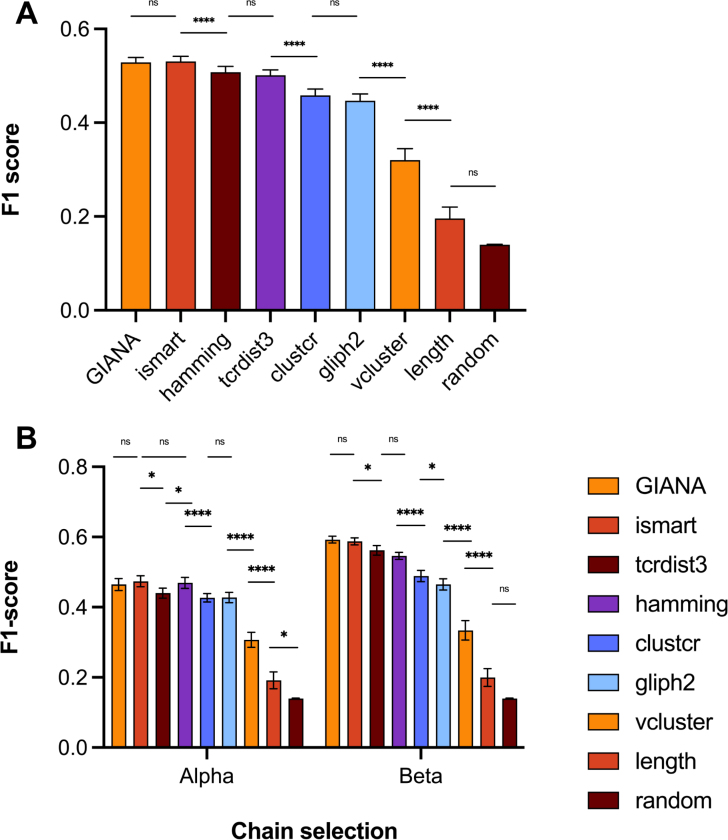
Differential model performance was first inspected at a global level for datasets V10 through V1000 combined, grouping over and chain selections and over all epitopes (Fig. 2A and Table S3). All study models outperformed length, V-gene, and random baselines (p0.0001). GIANA and iSMART performed equally well (p=0.67), surpassing both a Hamming distance and the other models studied (p0.0006). V-gene usage (vcluster) was better predictive of epitope specificity than CDR3 length, both of which outperformed a random baseline. However, significant variation in relative performance was observed for each model, sensitive to the choice of chain selection (Fig. 2B and Table S4), the number of representatives per epitope (Fig. S3A and Table S5), the epitope (Fig. S3B and Table S6), and the data source (Fig. S3C and Table S7), with no one model achieving superiority across all factors. The predictive capacity of all models was improved when provided with more TCR instances per epitope (Table S5), whereas performance was lower for than chain selections for all except V-gene, length and random baselines (Table. S4). Notably, clustering with V-gene identity alone achieved superior performance to the study models for AVFDRKSDAK and KLGGALQAK epitopes, and for McPas-TCR (Fig. S3) where performance was similar to a Hamming distance model.
Fig. 2.
Comparison of model performance across datasets. Weighted F1-scores shown for V10-V1000 grouped, (A) and chain selections combined; and (B) split by chain selection. Significance values: ****, p 0.0001; ***, p 0.001; **, p 0.01; *, p 0.05, n.s. not significant.
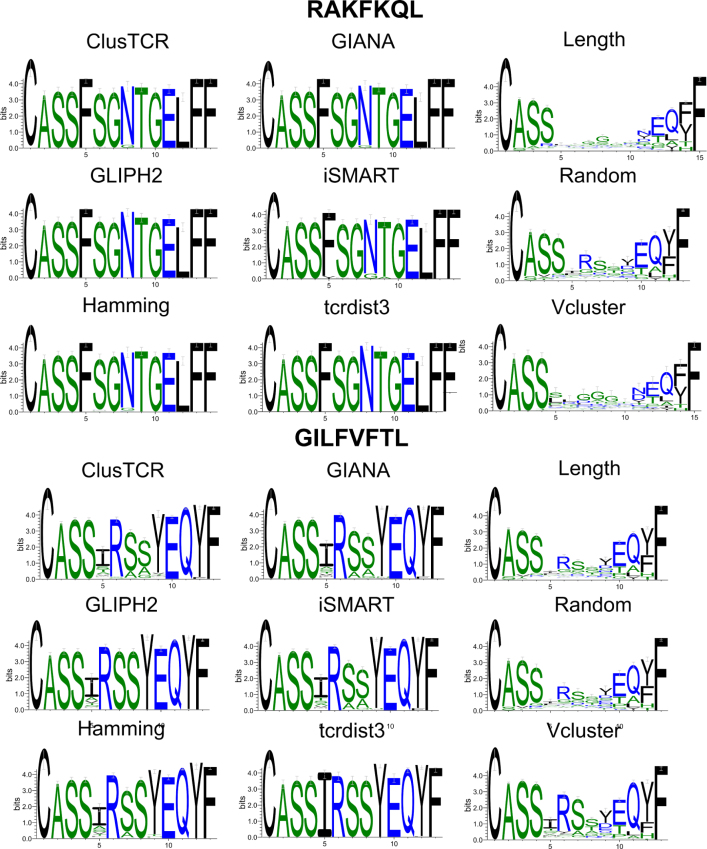
Whilst the size and epitope distribution of clusters varied between models (Fig. S4), CDR3 motifs for the largest clusters associated with each of these epitopes were strikingly similar for all except length, V-gene and random baselines, and were near-identical when applied to GILGFVFTL or RAKFKQLL (Fig. 3). Motifs were more varied for the other two epitopes in the V1000 dataset (KLGGALQAK and AVFDRKSDAK, Fig. S5).
Fig. 3.
CDR3 sequence logos for the largest clusters produced per epitope, dataset V1000. Logos were produced with WebLogo [41] for TCRs in the largest cluster produced for a given epitope per model following sequence alignment with MUSCLE [40].
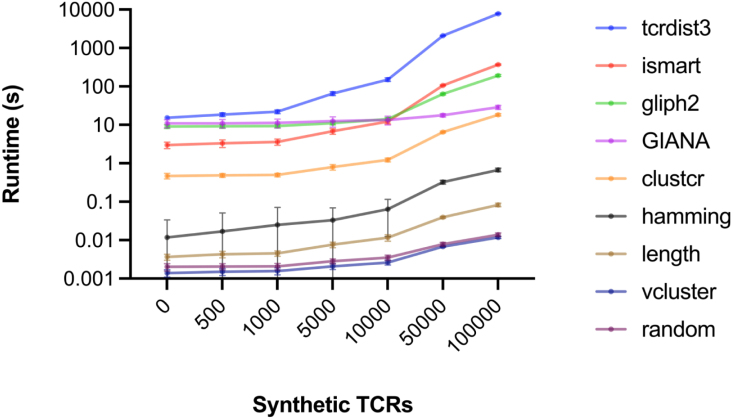
Finally, we compared UCM computational speeds by adding synthetic TCRs, produced with OLGA [35], to V1000 (Fig. 4). ClusTCR and GIANA were the most scalable of the five models tested, completing their cluster assignments within 10s for 100,000 TCRs, outpaced only by the baseline models. Time limitations prevented extension of the present analysis to comparative performance over parallel CPUs, and to the GPU-enabled versions of ClusTCR and GIANA. A batch-clustering implementation of tcrdist3 was used in order to avoid CPU overloading during distance calculation, which ran slowly compared to the other test models. Model predictions were negatively impacted by the addition of 100,000 synthetic TCR sequences, with F1-scores dropping to near zero for all but tcrdist3 (Fig. S6).
Fig. 4.
Investigating model scalability, comparing model runtimes as a function of the number of synthetic TCR sequences introduced with OLGA [35]. All experiments conducted on dataset V1000 ( chain selection).
4. Discussion
Despite the exponential growth of orphan TCR datasets, and the widespread use of UCMs in de-orphanisation pipelines, an independent comparison of the predictive capacity of clustering models has thus far been missing from the field. Here, we present an initial, modest attempt to address this need.
Our work suggests that clustering models can indeed be used to infer epitope specificity, by co-clustering TCR sequences having known and unknown cognate epitopes, and assigning the most common epitope as the predicted binder for a given TCR cluster. Whilst this analytical strategy has been deployed elsewhere [29], we report performance using metrics that can be applied equally to clustering and predictive models, over a wide variety of datasets and preprocessing strategies, which therefore give a more balanced view of true model performance. The comparative analytical framework and model datasets are made freely available at https://github.com/hudsondan/tcr-scapes.
We find that model ranking is sensitive to the number of TCRs provided per epitope, to the epitope studied, and to the dataset tested. This is most easily explained by the different model methodologies deployed (reviewed in Section S2). Whilst GIANA and iSMART perform well on VDJdb instances (Fig. 2), GLIPH2 provides superior predictions when applied to the large MIRA SARS-CoV-2 dataset, whilst Hamming distance and V-gene clustering baselines outperform the study models for McPas-TCR data (Fig. S3). The improved ability of V-gene identity clustering to group together TCRs recognising AVFDRKSDAK and KLGGALQAK compared with the study models (Fig. S3) may be reflective of the contribution of germline CDRs to recognition, although absolute performance was poor (under 10%). Only tcrdist3 was able to retain predictive performance in the presence of many synthetic TCRs, albeit not for all epitopes (Fig. S6). This is of particular relevance when applied to the task of extracting expanded antigen-specific clones from a mixed repertoire of effector and antigen-naive cells. Adding to the complexity, different models produced similar CDR3 motifs for TCRs recognising common viral epitopes, suggesting that one might reach a similar immunological conclusion irrespective of the model used.
With this in mind, should experimentalists deploy UCMs in the complex task of TCR de-orphanisation, and if so how? We observe that the five UCMs studied are able to group TCRs into antigen-specific clusters, even when provided with as few as ten representatives per epitope (Fig. S3). Whether these common specificity group are driven by recognition of unknown (unseen) or known (seen) antigens, will depend on the condition under investigation, the dataset in question, and the experimental methodology. We would nonetheless encourage their continued use, in combination with state of the art predictive models and experimental methods, as a means of improving confidence in putative assignment of epitope specificity. In the absence of a consistently superior model, one strategy could be to deploy two or more of the models studied to improve confidence in TCR cluster assignment in a manner robust to epitope-specific performance variations, for example combining a Hamming distance model with GIANA (a scalable follow-on to iSMART), ClusTCR (which performed equivalently to GLIPH2 for many datasets) or tcrdist3 (which appears to be more robust to noise). Choice of model will also depend on practical considerations including ease of use, and we note for example that at present on GLIPH2 is available as both a web tool and a command line executable file.
Finally, although recent structural [42], statistical [43], and predictive [9] analyses suggest that both polypeptide chains play an important role in epitope recognition, we observed consistently lower F1-scores for compared to chain selections (Table S4). One possible explanation is that the default model hyperparameters have been optimised for chain data, which make up the majority of published TCR-epitope pairs [6]. Alternatively, the chain may contribute more to determination of overall epitope specificity, as a product of its increased diversity relative to chains [13], as we will discuss further in Study Limitations. Modelling strategies that permit integration of and chain pairing with transcriptomic and phenotypic information, including graph network approaches such as CoNGA [28], may help efforts to decode the relative contribution of chain pairing to epitope specificity, which remains an open question in the field.
5. Study limitations
The significant scientific and economic potential of a generalisable solution to prediction of TCR epitope specificity has encouraged the development of a multitude of new SPMs and UCMs, summarised in [6]. However, the scope of the present study is limited to a handful of commonly used open-source models, on the basis of their widespread use and relative freedom from training data bias as compared to DNN-UCMs and SPMs. Nonetheless, an independent comparison of UCMs, DNN-UCMs and SPMs would be of great use to the community. We also limit our analysis to performance in an inference task, overlooking a host of useful features such as motif extraction, computation of probability of generation, and visualisation tools, which will likely influence user choice for a given immunological question.
There is also growing evidence that inclusion of both and chains improves predictive performance in SPMs and DNN-SPMs [9]. However, whilst ClusTCR, tcrdist3, and GLIPH2 may all theoretically be applied to paired chain data, cluster assignments are produced for a given CDR3 independent of the other chain for all but tcrdist3. An investigation of whether the integration of and chain information improves performance equally across models might reveal the relative merits of each, when applied to large orphan repertoires.
Finally, comparison of the generation probabilities of TCRs drawn from true repertoires and those produced with OLGA have concluded that the latter produces TCRs that are too distant from native repertoires to represent a true background [18]. It may be that the unique inclusion of a CDR2.5 loop in tcrdist3 distance calculations, or of a more permissive edit distance than other models, allows the model to differentiate between synthetic and input TCRs and so to cluster TCRs effectively with or without noise. Use of a mixed background [18] or of true orphan repertoires such as [44], might confirm this and so test the robustness of the models to other sources of noise.
CRediT authorship contribution statement
Dan Hudson: Software, Methodology, Formal analysis, Data curation. Alex Lubbock: Software. Mark Basham: Supervision. Hashem Koohy: Writing – review & editing, Writing – original draft, Supervision, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: D.H. provides consultancy services to companies active in T cell antigen discovery and vaccine development. The other authors declare no competing interests.
Acknowledgements
Our comparative framework borrowed from that developed by the Meysman group for ClusTCR (github.com/svalkiers/clusTCR), to whom the authors express their gratitude. Thanks are also due to Dr Ricardo A. Fernandes and Sam Farrar for critical review, and to Andreas Wilm for code suggestions. H.K. is supported by funding from the UK Medical Research Council grant number MC_UU_12010/3. D.H. receives administrative and financial support from the Biotechnology and Biological Sciences Research Council (BBSRC), UK (grant number BB_T008784_1) and from the Rosalind Franklin Institute. The computational aspects of this research were supported by the Wellcome Trust Core Award Grant Number 203141_Z_16_Z and the NIHR Oxford BRC .
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.immuno.2024.100033.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
Supplementary Material includes a supportive literature review, model methodology, supplementary figures and tables.
Data availability
The codes and data are available in our public github that will be publicly available upon publication.
References
- 1.Davis M.M., Bjorkman P.J. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334(6181):395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 2.Bosselut R. T cell antigen recognition: Evolution-driven affinities. Proc Natl Acad Sci USA. 2019;116(44):21969–21971. doi: 10.1073/pnas.1916129116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sckisel G.D., Bouchlaka M.N., Monjazeb A.M., Crittenden M., Curti B.D., Wilkins D.E., et al. Out-of-sequence signal 3 paralyzes primary CD4(+) T-cell-dependent immunity. Immunity. 2015;43(2):240–250. doi: 10.1016/j.immuni.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joglekar A.V., Li G. T cell antigen discovery. Nature Methods. 2021;18(8):873–880. doi: 10.1038/s41592-020-0867-z. [DOI] [PubMed] [Google Scholar]
- 5.Valkiers S., de Vrij N., Gielis S., Verbandt S., Ogunjimi B., Laukens K., et al. Recent advances in T-cell receptor repertoire analysis: Bridging the gap with multimodal single-cell RNA sequencing. Immunoinformatics. 2022;5 [Google Scholar]
- 6.Hudson D., Fernandes R.A., Basham M., Ogg G., Koohy H. Can we predict T cell specificity with digital biology and machine learning? Nat Rev Immunol. 2023 doi: 10.1038/s41577-023-00835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber A., Born J., Rodriguez Martínez M. TITAN: T cell receptor specificity prediction with bimodal attention networks. Bioinformatics. 2021;37(S1):I237–I244. doi: 10.1093/bioinformatics/btab294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moris P., Postovskaya A., Gielis S., De Neuter N., Bittremieux W., Ogunjimi B., et al. Current challenges for unseen-epitope TCR interaction prediction and a new perspective derived from image classification. Brief Bioinform. 2021;22(4):1–12. doi: 10.1093/bib/bbaa318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montemurro A., Schuster V., Povlsen H.R., Bentzen A.K., Jurtz V., Chronister W.D., et al. NetTCR-2.0 enables accurate prediction of TCR-peptide binding by using paired alpha and beta sequence data. Nat Commun Bio. 2021;4(1) doi: 10.1038/s42003-021-02610-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu K., Yost K.E., Daniel B., Belk J.A., Xia Y., Egawa T., et al. 2021. TCR-BERT: Learning the grammar of T-cell receptors for flexible antigen-xbinding analyses. Preprint at https://www.biorxiv.org/content/10.1101/2021.11.18.469186v1. [Google Scholar]
- 11.Zhang W., Hawkins P.G., He J., Gupta N.T., Liu J., Choonoo G., et al. A framework for highly multiplexed dextramer mapping and prediction of T cell receptor sequences to antigen specificity. Sci Adv. 2021;7(20):eabf5835. doi: 10.1126/sciadv.abf5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meysman P., Barton J., Bravi B., Cohen-Lavi L., Karnaukhov V., Lilleskov E., et al. Benchmarking solutions to the T-cell receptor epitope prediction problem: IMMREP22 workshop report. ImmunoInformatics. 2023;9 [Google Scholar]
- 13.Arstila T.P., Casrouge A., Baron V., Even J., Kanellopoulos J., Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 14.Dash P., Fiore-Gartland A.J., Hertz T., Wang G.C., Sharma S., Souquette A., et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547(7661):89–93. doi: 10.1038/nature22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glanville J., Huang H., Nau A., Hatton O., Wagar L.E., Rubelt F., et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547(7661):94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi F., Isobe N., Glanville J., Matsushita T., Maimaitijiang G., Fukumoto S., et al. A new clustering method identifies multiple sclerosis-specific T-cell receptors. Ann Clin Transl Neurol. 2021;8(1):163–176. doi: 10.1002/acn3.51264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H., Wang C., Rubelt F., Scriba T.J., Davis M.M. Analyzing the mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening. Nat Biotechnol. 2020;38:1194–1202. doi: 10.1038/s41587-020-0505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pogorelyy M.V., Rosati E., Minervina A.A., Mettelman R.C., Scheffold A., Franke A., et al. Resolving SARS-CoV-2 CD4+ T cell specificity via reverse epitope discovery. Cell Rep Med. 2022;3(8) doi: 10.1016/j.xcrm.2022.100697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Duan F., Zhu Z., Yu M., Jia X., Dai H., et al. Analysis of TCR repertoire by high-throughput sequencing indicates the feature of T cell immune response after SARS-CoV-2 infection. Cells. 2021;11(1):68. doi: 10.3390/cells11010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H., Liu L., Zhang J., Chen J., Ye J., Shukla S., et al. Investigation of antigen-specific T-cell receptor clusters in human cancers. Clin Canc Res. 2020;26(6):1359–1371. doi: 10.1158/1078-0432.CCR-19-3249. [DOI] [PubMed] [Google Scholar]
- 21.Sidhom J.-W., Larman H.B., Pardoll D.M., Baras A.S. DeepTCR is a deep learning framework for revealing sequence concepts within T-cell repertoires. Nature Commun. 2021;12(1) doi: 10.1038/s41467-021-21879-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Springer I., Tickotsky N., Louzoun Y.a. Contribution of T cell receptor alpha and beta CDR3, MHC typing, V and J genes to peptide binding prediction. Front Immunol. 2021;12:1436. doi: 10.3389/fimmu.2021.664514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drost F., An Y., Dratva L.M., Lindeboom R.G., Haniffa M., Teichmann S.A., et al. 2021. Integrating T-cell receptor and transcriptome for large-scale single-cell immune profiling analysis. Preprint at https://www.biorxiv.org/content/10.1101/2021.06.24.449733v2. [Google Scholar]
- 24.Bagaev D.V., Vroomans R.M., Samir J., Stervbo U., Rius C., Dolton G., et al. VDJdb in 2019: Database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res. 2020;48(D1):D1057–D1062. doi: 10.1093/nar/gkz874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tickotsky N., Sagiv T., Prilusky J., Shifrut E., Friedman N.a. McPAS-TCR: A manually curated catalogue of pathology-associated T cell receptor sequences. Bioinformatics. 2017;33(18):2924–2929. doi: 10.1093/bioinformatics/btx286. [DOI] [PubMed] [Google Scholar]
- 26.Nolan S., Vignali M., Kaplan I.M., Biotechnologies A., Svejnoha E., Craft T., et al. 2020. A large-scale database of T-cell receptor beta (TCR) sequences and binding associations from natural and synthetic exposure to SARS-CoV-2. [Google Scholar]
- 27.Lefranc M.P., Giudicelli V., Duroux P., Jabado-Michaloud J., Folch G., Aouinti S., et al. IMGT, the international ImMunoGeneTics information system 25 years on. Nucleic Acids Res. 2015;43(D1):D413–D422. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schattgen S.A., Guion K., Crawford J.C., Souquette A., Barrio A.M., Stubbington M.J.T., et al. Integrating T cell receptor sequences and transcriptional profiles by clonotype neighbor graph analysis (CoNGA) Nat Biotechnol. 2022;40(1):54–63. doi: 10.1038/s41587-021-00989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valkiers S., Van Houcke M., Laukens K., Meysman P. ClusTCR: A Python interface for rapid clustering of large sets of CDR3 sequences with unknown antigen specificity. Bioinformatics. 2021;37(24):4865–4867. doi: 10.1093/bioinformatics/btab446. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H., Zhan X., Li B. GIANA allows computationally-efficient TCR clustering and multi-disease repertoire classification by isometric transformation. Nature Commun. 2021;12(4699):1–11. doi: 10.1038/s41467-021-25006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayer-Blackwell K., Schattgen S., Cohen-Lavi L., Crawford J.C., Souquette A., Gaevert J.A., et al. TCR meta-clonotypes for biomarker discovery with tcrdist3 enabled identification of public, HLA-restricted clusters of SARS-CoV-2 TCRs. eLife. 2021;10 doi: 10.7554/eLife.68605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pogorelyy M.V., Minervina A.A., Shugay M., Chudakov D.M., Lebedev Y.B., Mora T., et al. Detecting T cell receptors involved in immune responses from single repertoire snapshots. PLoS Biol. 2019;17(6) doi: 10.1371/journal.pbio.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meysman P., De Neuter N., Gielis S., Bui Thi D., Ogunjimi B., Laukens K. On the viability of unsupervised T-cell receptor sequence clustering for epitope preference. Bioinformatics. 2018;35(9):1461–1468. doi: 10.1093/bioinformatics/bty821. [DOI] [PubMed] [Google Scholar]
- 34.Pedregosa F., Michel V., Grisel O., Blondel M., Prettenhofer P., Weiss R., et al. Scikit-learn: Machine learning in Python. JMLR. 2011;12:2825–2830. [Google Scholar]
- 35.Sethna Z., Elhanati Y., Callan C.G., Walczak A.M., Mora T. OLGA: Fast computation of generation probabilities of B-and T-cell receptor amino acid sequences and motifs. Bioinformatics. 2019;35(17):2974–2981. doi: 10.1093/bioinformatics/btz035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postovskaya A., Vujkovic A., deBlock T., van Petersen L., van Frankenhuijsen M., Brosius I., et al. Leveraging T-cell receptor – epitope recognition models to disentangle unique and cross-reactive T-cell response to SARS-CoV-2 during COVID-19 progression/resolution. Front Immunol. 2023;14 doi: 10.3389/fimmu.2023.1130876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.GraphPad v 10.0.3 for Mac, GraphPad Software, Boston, Massachusetts USA,www.graphpad.com.
- 38.Hunter J.D. Matplotlib: A 2D graphics environment. Comput Sci Eng. 2007;9(3):90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 39.Waskom M.L. Seaborn: Statistical data visualization. J Open Source Softw. 2021;6(60):3021. doi: 10.21105/joss.03021. [DOI] [Google Scholar]
- 40.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raybould M.I.J., Nissley D.A., Kumar S., Deane C.M. Computationally profiling peptide:MHC recognition by T-cell receptors and T-cell receptor-mimetic antibodies. Front Immunol. 2023;13 doi: 10.3389/fimmu.2022.1080596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayer A., Callan C.G. Measures of epitope binding degeneracy from T cell receptor repertoires. Proc Natl Acad Sci USA. 2023;120(4) doi: 10.1073/pnas.2213264120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emerson R.O., Dewitt W.S., Vignali M., Gravley J., Hu J.K., Osborne E.J., et al. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet. 2017;49(5):659–665. doi: 10.1038/ng.3822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material includes a supportive literature review, model methodology, supplementary figures and tables.
Data Availability Statement
The codes and data are available in our public github that will be publicly available upon publication.