Abstract
A non-negligible part of our DNA has been proven to be transcribed into non-protein coding RNA and its intricate involvement in several physiological processes has been highly evidenced. The significant biological role of non-coding RNAs (ncRNAs), including long non-coding RNAs (lncRNAs) has been variously reported. In the current review, the authors highlight the multifaceted role of myocardial infarction-associated transcript (MIAT), a well-known lncRNA, in hepatocellular carcinoma (HCC). Since its discovery, MIAT has been described as a regulator of carcinogenesis in several malignant tumors and its overexpression predicts poor prognosis in most of them. At the molecular level, MIAT is closely linked to the initiation of metastasis, invasion, cellular migration, and proliferation, as evidenced by several in-vitro and in-vivo models. Thus, MIAT is considered a possible theranostic agent and therapeutic target in several malignancies. In this review, the authors provide a comprehensive overview of the underlying molecular mechanisms of MIAT in terms of its downstream target genes, interaction with other classes of ncRNAs, and potential clinical implications as a diagnostic and/or prognostic biomarker in HCC.
Keywords: Long non-coding RNA (lncRNA), Myocardial infarction associated transcript (MIAT), Liver cancer, Metastasis, microRNA (miRNA), Theranostics
Graphical abstract
Abbreviations
- ABCG2
ATP Binding Cassette subfamily G member 2
- ACE
Angiotensin-converting enzyme gene
- ADGRL2
adhesion G protein-coupled receptor L2
- ANRIL
Antisense Non-coding RNA in the INK4 Locus
- ASO
Antisense Oligonucleotide
- ATG7
autophagy related 7
- ATM
Ataxia Telangiectasia Mutated
- BANCR
BRAF-Activated Non-Protein Coding RNA
- CASP1
Caspase 1
- CCAT1
Colon Cancer-Associated Transcript 1
- CCND1
Cyclin D1
- CD8
Cluster of Differentiation 8
- CDC16
Cell division cycle protein 16 homolog
- CDKN1A
Cyclin Dependent Kinase inhibitor 1A
- CDKN2B
Cyclin-Dependent Kinase 4 inhibitor B
- CEP170
Centrosomal Protein 170
- CK2
Checkpoint Kinase 2
- c-Met
Mesenchymal-Epithelial Transition factor
- CORO1C
Coronin-like actin-binding protein 1C
- CREBRF
CREB3 Regulatory Factor
- CTLA4
Cytotoxic T-Lymphocyte Antigen 4
- CTNNB1
catenin beta 1
- DAPK2
Death-associated protein kinase 2
- DDX5
DEAD box polypeptide 5
- Derlin-1
Degradation in Endoplasmic Reticulum protein 1
- DLG3
Disks large homolog 3
- DUSP7
Dual Specificity Phosphatase 7
- EGFR
Epidermal Growth Factor Receptor
- EMT
Epithelial-Mesenchymal Transition
- ENCORI
Encyclopedia of RNA Interactomes
- eNOS
Endothelial nitric oxide synthase
- EPHA2
Erythropoietin-Producing Hepatocellular receptor A2
- EZH2
Enhancer of Zest Homolog 2
- FASTKD5
FAST kinase domains 5
- FOXP3
Forkhead Box P3
- GAPDH
Glyceraldehyde-3-Phosphate Dehydrogenase
- GAS5
Growth arrest-specific 5
- GDI2
GDP Dissociation Inhibitor 2
- GENCODE
Human GENCODE database
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- GZMK
Granzyme K
- HAVCR2
hepatitis A virus cellular receptor 2
- HCC
Hepatocellular Carcinoma
- HDAC4
Histone Deacetylase 4
- HEIH
HCC-specific lncRNA Enhancer of Invasion and Migration
- HIF1A
Hypoxia Inducible Factor 1 Subunit Alpha
- HOTAIR
HOX Transcript Antisense RNA
- HOXA5
Homeobox A5
- HSC
hepatic stellate cell
- IER2
Immediate Early Response 2
- IL-17
Interleukin 17
- IPO7
Importin 7
- IST1
IST1 factor associated with ESCRT-III
- JAG1
Jagged canonical Notch ligand 1
- JAK2
Janus Kinase 2
- JNK
Jun N-terminal kinase
- KCND1
Potassium Voltage-Gated Channel Subfamily D Member 1
- KCNQ1
Potassium Voltage-Gated Channel Subfamily Q member 1
- KCNQ1OT1
KCNQ1 Overlapping Transcript 1
- LAG3
Lymphocyte-Activation Gene 3
- LASP1
LIM And SH3 Protein 1
- LDB1
LIM Domain-Binding protein 1
- LincRNA-p21
Long Intergenic Non-coding RNA-p21
- LINGO1
Leucine Rich Repeat And Ig Domain Containing 1
- LIPCAR
Long Intergenic Non-Protein Coding RNA for Cardiac Regeneration
- lncRNA
Long Non-coding RNA
- LncRNA-ATB
Long Non-coding RNA Activated by TGF-β
- LncRNA-LET
Long Non-coding RNA Low Expression in Tumor
- LONP2
Lon peptidase 2, peroxisomal
- LOXL2
Lysyl oxidase homolog 2
- LUCAT1
Lung Cancer-Associated Transcript 1
- MALAT1
Metastasis-Associated Lung Adenocarcinoma Transcript 1
- MEG3
Maternally expressed 3
- MEGF8
Multiple Epidermal Growth Factor-like Domains 8
- MEIS3
Meis Homeobox 3
- MIAT
Myocardial Infarction-Associated Transcript
- MIR34A
MicroRNA 34a
- miRNA
MicroRNA
- MLF2
Myeloid Leukemia Factor 2
- MMP14
Matrix metalloproteinase-14
- MTCH1
Mitochondrial carrier homolog 1
- MYO1B
Myosin IB
- NAD
Nicotinamide Adenine Dinucleotide
- NBR2
Neighbor Of BRCA1 LncRNA 2
- ncRNA
Non-coding RNA
- NEAT2
Nuclear Enriched Abundant Transcript 2
- NF-κB
Nuclear Factor kappa B
- NLM
National Library of Medicine
- NO
Nitric Oxide
- NONCODEV5
NONCODE database version 5
- NONO
Non-POU Domain Containing Octamer Binding
- NORAD
Non-Coding RNA Activated By DNA Damage
- Notch1
Neurogenic locus notch homolog protein 1
- PANDAR
Promoter of CDKN1A Antisense DNA Damage Activated RNA
- PCAT-1
Prostate cancer associated transcript-1
- PCDH1
Protocadherin 1
- PD-1
Programmed Cell Death 1
- PDCD1
Programmed cell death protein 1
- PD-L1
Programmed Death-Ligand 1 (CD274 molecule)
- PFN1
Profilin 1
- PGC-1a
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PI3K
Phosphatidylinositol 3-Kinase
- PLAGL2
Pleomorphic adenoma gene like-2
- PRODH
Proline Dehydrogenase 1
- PTENP1
Phosphatase and Tensin homolog Pseudogene 1
- PVT1
Plasmacytoma Variant Translocation 1
- RAN
Ras-related Nuclear protein
- Rfam
RNA Families database
- RIN1
Ras and Rab Interactor 1
- RPL13
Ribosomal Protein L13
- RPL23A
Ribosomal Protein L23a
- RPLP1
ribosomal protein lateral stalk subunit P1
- RPS3
Ribosomal Protein S3
- SART3
Spliceosome Associated factor 3
- SCHLAP1
SWI/SNF Complex Antagonist Associated With Prostate Cancer 1
- SF-1
Steroidogenic Factor 1
- SGK1
Serum/Glucocorticoid regulated Kinase 1
- siRNA
small interfering RNA
- SIRT1
Sirtuin 1
- SIX1
Sine oculis homeobox homolog 1
- SLC6A6
Solute Carrier Family 6 Member 6
- SNPs
Single Nucleotide Polymorphisms
- SOX4
SRY-Box Transcription Factor 4
- SP1
Specificity Protein 1
- SPHK2
Sphingosine Kinase 2
- STAT3
Signal Transducer and Activator of Transcription 3
- STAU1
Staufen-1
- TCF20
Transcription Factor 20
- TCGA
The Cancer Genome Atlas
- TGF-β2
Transforming Growth Factor beta 2
- TIM3
T-cell Immunoglobulin and Mucin-domain containing 3
- TME
Tumor Microenvironment
- TMEM147
Transmembrane Protein 147
- TP53
Tumor Protein p53
- TRAF6
TNF Receptor Associated Factor 6
- TTC37
Tetratricopeptide repeat domain 37
- TUBA1B
Tubulin Alpha 1B
- UBA2
Ubiquitin-Like Modifier-Activating Enzyme 2
- VEGF
Vascular endothelial growth factor
- VELUCT
Viability Enhancing in Lung cancer Transcript
- WNT9A
Wnt Family Member 9A
- XIST
X-linked X-inactive-specific transcript
- YAP1
Yes-associated protein 1
- YBX1
Y-box binding protein 1
- YWHAE
Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein Epsilon
- ZEB1
Zinc finger E-box binding homeobox 1
1. Introduction
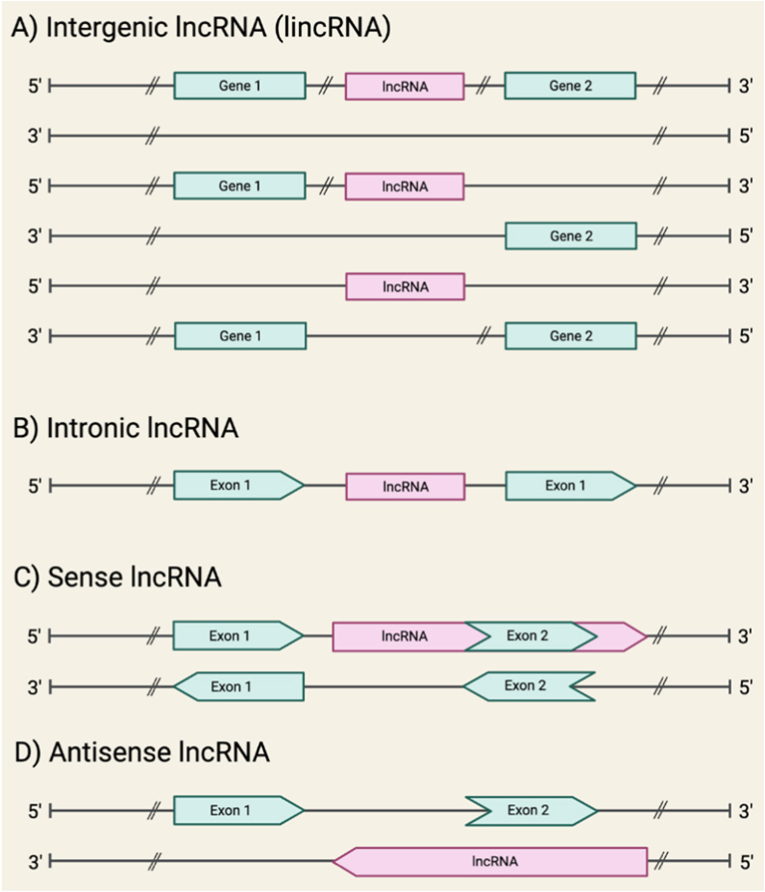
Following the completion of “The Human Genome Project” in 2003, the classification of the genome and its elements has been a challenge and the focus of all molecular scientists [1,2]. One of the broad classifications of the genome is categorizing it into protein-coding and non-protein-coding genes [3]. The protein-coding regions of the DNA were once thought to be the most fundamentally functional segment, and as a result, must make up the majority of the DNA [4,5]. Surprisingly, less than 3 % of the genome's transcribed region is translated into proteins [6,7]. Given this, non-coding RNAs (ncRNAs), or non-protein producing RNA, which were previously assumed to be the result of “junk DNA”, attracted attention as it became clear that they do serve a purpose [8,9]. According to databases such as Human GENCODE and NONCODEV5, the major class of ncRNAs are long non-coding RNAs (lncRNAs), with over 100,000 members. Yet to be determined is the precise number of functional lncRNAs [[10], [11], [12], [13], [14]]. Members of this class have transcripts larger than 200 nucleotides and are important regulators [15,16]. LncRNAs can also be divided into subclasses [17,18]. According to one of these classifications, the genomic location of ncRNAs can result in intergenic, intronic, sense, and antisense lncRNAs [9,19,20], as shown in Fig. 1.
Fig. 1.
The possible genomic locations of lncRNAs. Protein-coding genes and their exons are represented by green segments, while lncRNAs and their introns are represented by pink segments. (A) An intergenic lncRNA, transcribed from either DNA strand between two protein-coding genes. (B) An intronic lncRNA, transcribed from protein-coding gene introns. (C) A sense lncRNA, transcribed from the sense strand of protein-coding genes, overlapping with part (or entirely) of a protein-coding sequence and one or more introns. (D) An antisense lncRNA, transcribed from the antisense strand of protein-coding genes, overlapping with a part (or entirely) of a protein-coding sequence and 1 or more introns. (*) Sense and Antisense lncRNAs are subcategories of a larger class called genic lncRNAs.
2. Roles and functions of lncRNAs
Alternatively, lncRNAs can be classified according to their functional role [17]. As already indicated, lncRNAs have a significant regulatory function, modulating the activity of genes, RNAs, proteins, organelles, and nuclear condensates [21,22]. More specifically, lncRNA functions include one or more of the following: chromatin remodeling, histone modification, RNA-DNA-DNA triplex formation through direct pairing with the DNA, gene silencing, transcriptional regulation of genes through silencing or enhancing respective mRNA expression, nuclear condensate formation, post-transcriptional modifications exerted by direct protein-binding, miRNA sponging, and mRNA stabilization. Additionally, lncRNAs may affect mitochondrial function, intercellular exosomal transport, and cell regulation [12,20,23,24]. Table 1 lists functional roles played by lncRNAs.
Table 1.
Molecular and functional roles of lncRNAs.
| Molecular role | Functional role | References |
|---|---|---|
| Epigenetic regulation | Chromatin remodeling | [25] |
| Histone modification | [26] | |
| Gene silencing | [27] | |
| Triplex formation | [28] | |
| Transcriptional regulation | Promotion via promotor regions and/or transcription factor modulation | [29,30] |
| Suppression via promotor regions and/or transcription factor modulation | [31] | |
| Post-transcription regulation | Protein binding | [32] |
| mRNA stabilizing activity | [33] | |
| miRNA sponging (lncRNAs function as competing endogenous RNA) | [34,35] | |
| Nuclear scaffolding and condensates | – | [12] |
| Organelle regulation | Mitochondrial modulation of apoptosis, metabolism, and nucleus crosstalk | [36,37] |
| Exosomal release | [38] |
3. LncRNAs as potential theranostic agents (biomarkers and/or therapeutic agents)
The functional roles of lncRNAs unravel their crucial role in various aspects of cellular homeostasis [39]. Moreover, lncRNAs have the potential to serve as non-invasive prognostic markers, diagnostic markers, and/or potential therapeutic agents/targets in a variety of diseases, such as cancer, diabetes, and genetic diseases which perfectly classifies them as theranostic agents with great potential in several malignant and non-malignant contexts [12,14,40,41].
Specifically, several lncRNAs have been characterized as non-invasive biomarkers quantifiable in liquid biopsies [42]. LncRNAs show tissue-specific expression patterns, which aid in the traceability of different cancer types [43]. Additionally, they are abundant in plasma samples, enabling their easy detection. Although it is still unclear whether exosomal or free lncRNAs contribute more to the detectable fraction, exosomal lncRNAs are known to be stable in biological fluids, due to their resistance and stability against RNases [42,44,45].
Exosomal lncRNAs constitute promising candidates for therapeutic intervention in various diseases since they are increasingly recognized as key players in intercellular communication, controlling cellular functions, and affecting disease states. These characteristics also raise the possibility of employing non-oncogenic exosomal lncRNAs therapeutically for internalization and cell-specific effects after a disease mechanism has been thoroughly elucidated [42,45]. Other hypotheses have been put forth in which lncRNAs act as the therapeutic targets rather than the therapeutic agents [6,15].
LncRNAs that foster neovasculature, drug resistance, or cancer cell-to-cell communication could constitute promising targets [45]. Similarly, by identifying the binding domains of an oncogenic lncRNA that is responsible for triggering a certain signaling pathway, a complimentary chemical can be synthesized and delivered to block that lncRNA and subsequently inhibit its tumor-promoting action. Although such an approach has merit, it currently has the challenge of inadequate motif structural knowledge [12,[46], [47], [48]]. Additionally, lncRNAs were demonstrated to be the less toxic and more potent biological alternatives to proteins [12]. Jiang et al. highlighted how current technology can be used to pave the path for the usage of lncRNAs as therapeutic agents [12,49].
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) [14,15,18], HOX transcript antisense RNA (HOTAIR) [41,50], H19 imprinted maternally expressed transcript (H19) [16,22], colon cancer associated transcript 1 (CCAT1) [13,21], and hepatocellular carcinoma up-regulated EZH2-associated long non-coding RNA (HEIH) [7] are some of the well-known lncRNA members with the potential to serve as biomarkers or therapeutic agents/targets for several malignancies such as breast, liver, and brain tumors, while LIPCAR, CDKN2B antisense RNA 1 (ANRIL), and KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1) are well-studied lncRNAs in terms of non-malignant disorders such as cardiovascular diseases [42,51,52].
4. LncRNAs in oncology
Focusing on the role of lncRNAs in cancer, several studies have identified and correlated many lncRNAs to a diverse range of malignancies. LncRNAs are pivotal regulators in the complex landscape of oncology. Aberrant expression of lncRNAs is associated with various cancers (Table 2). In addition to the ones that were before listed, HEIH and sONE act as significant breast cancer regulators [7,53,54]. LncRNA activated by TGF-β (LNCRNA-ATB) and NPTN intronic transcript 1 (LNCRNA-LET) are associated with HCC. CCAT1, as its name suggests, is linked to colorectal cancer, while promoter of CDKN1A antisense DNA damage activated RNA (PANDAR) and colon cancer associated transcript 2 (CCAT2) are two examples of lncRNAs important for lung cancer progression and prognosis [20,55]. Lung cancer-related transcript 1 (LUCAT1), is involved in breast cancer, ovarian cancer, thyroid cancer, and renal cell carcinoma [56], as well as H19, plays a role in breast cancer, colorectal cancer, and lung cancer [20,34,51,55].
Table 2.
Potential oncogenic and tumor-suppressive lncRNAs in several malignant contexts.
| LncRNA | Cancer Type | Signaling pathway | Expression profile | Impact | Hallmark involved | Reference |
|---|---|---|---|---|---|---|
| H19 | Colorectal cancer | Sponging miR-141, thus activating the β-catenin pathway | Upregulated | Oncogenic | Cancer cell stemness and chemoresistance | [57] |
| Hepatocellular cancer | Exosomal release increases the expression of endothelial factors | Upregulated | Oncogenic | Angiogenesis | [58] | |
| HOTAIR | Non-small cell lung cancer | Suppression of matrix metalloproteinases and HOXA5 protein | Upregulated | Oncogenic | Cell invasion and metastasis | [59] |
| MIAT | Lung cancer | Promotor methylation of the MIR34A gene leads to decreased expression and subsequent activation of the PI3K/AKT signaling pathway | Upregulated | Oncogenic | Drug resistance | [60] |
| Glioma | Promotor of proliferation, migration, and metastasis of brain cancer cells | Upregulated | Oncogenic | Cancer progression | [61] | |
| PTENP1 | Bladder cancer | Exosomal release acts as a miR-17 decoy to regulate PTEN expression | Downregulated | Tumor-suppressive | Cancer progression | [62] |
| GAS5 | Non-small cell lung cancer | Suppression of miR-23a | Downregulated | Tumor-suppressive | Cancer tissue growth and apoptosis | [63,64] |
| MALAT1 (also known as NEAT2) | Non-small cell lung cancer | MALAT1 promoting activation through Specificity Protein 1 (SP1) | Upregulated | Oncogenic | Cancer cell growth and invasion | [65,66] |
| Breast cancer | Exosomal | Upregulated | Oncogenic | Cell proliferation | [67] | |
| Esophageal squamous cell carcinoma | ATM-CHK2 dephosphorylation leading to unregulated G2/M cell cycle checkpoint | Upregulated | Oncogenic | Cancer cell proliferation, invasion, and metastasis | [68] | |
| CCAT1 | Non-small cell lung cancer | CCAT1/miR-130a-3p/SOX4 axis, boosting ABCG2-mediated drug efflux | Upregulated | Oncogenic | Cancer cell chemoresistance | [69] |
| MEG3 | Non-small cell lung cancer | Suppressive action on WNT/b-catenin pathway through TP53, b-catenin, and survivin | Downregulated | Tumor-suppressive | Cancer cell cycle regulation and chemoresistance | [70] |
| LC3 cleavage downregulation, suppressing intracellular components autophagy | Downregulated | Tumor-suppressive | Cancer cell chemoresistance | [71] | ||
| XIST | Non-small cell lung cancer | LC3 autophagy-factor cleavage promotion and overexpression of ATG7 through miR-17/autophagy axis | Upregulated | Oncogenic | Cancer cell chemoresistance | [72] |
| Pancreatic cancer | Compound action of a multitude of pathways, primarily by sponging miRNAs: EGFR/miR-133a, iASSP/miR-140/miR-124, YAP/miR-34a, ZEB1/miR-429, TGF-β2/miR-141–3p, and Notch1/miR-137 pathways | Upregulated | Oncogenic | Cancer cell growth, invasion, and migration | [73] | |
| NEAT1 | Triple-negative breast cancer | SOX2 mRNA downregulated expression | Upregulated | Oncogenic | Cancer cell stemness and chemoresistance | [74,75] |
| Hemangioma | Sponging of miR-33a-5p enhances NF-κB signaling thus increasing the expression of the HIF1A gene | Upregulated | Oncogenic | Cancer cell proliferation, invasion, and metastasis | [75,76] | |
| Acute myeloid leukemia | Negative feedback on miR-338-39, potentiating CREBRF | Downregulated | Tumor-suppressive | Cancer cell proliferation, invasion, and metastasis | [75,77] | |
| sONE | Triple-negative breast cancer | Suppression of MYC and enhancement of TP53 thus increasing the concentration of several downstream tumor suppressive mRNAs, including miR-34a, miR-15, miR-16, and let-7a. Additionally, NO production modulator via eNOS posttranscriptional regulation. | Downregulated | Tumor-suppressive | Cancer cell proliferation, invasion, and metastasis | [54,78] |
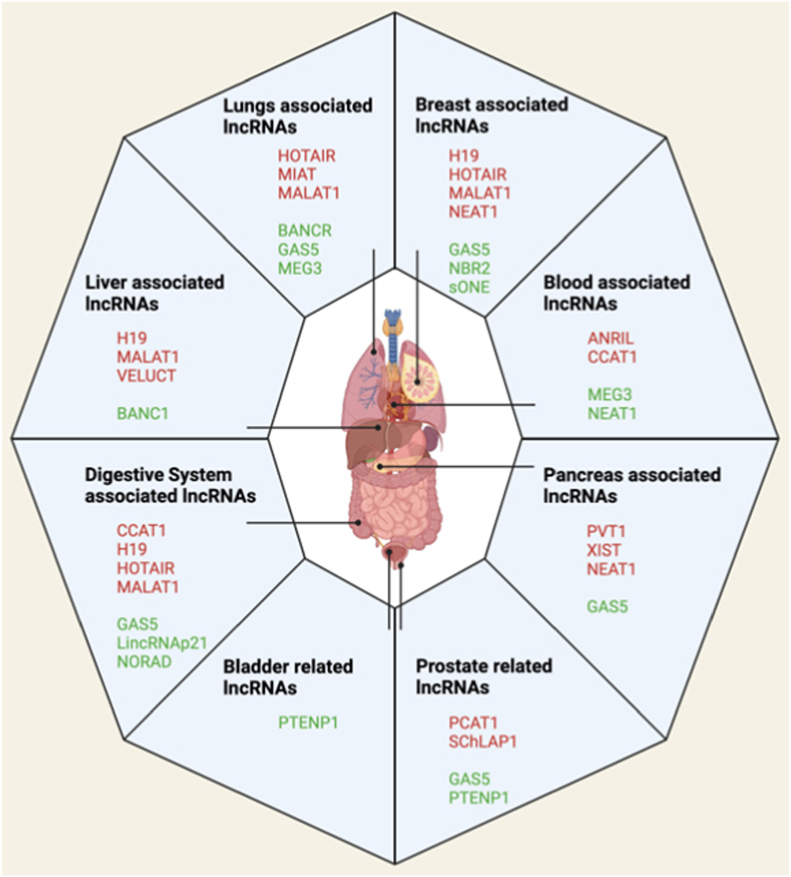
Remarkably, lncRNAs have the potential to be used as biomarkers and therapeutic agents if they are tumor-suppressive, or as therapeutic targets if they are oncogenic. Their potential to serve as therapeutic targets and diagnostic markers holds promise for improving early detection and treatment options in oncology. Fig. 2 represents a graphical presentation of the lncRNAs and their possible association with several solid malignancies. Each lncRNA has distinct effects mediated through different signaling pathways in each cancer context. This proposes a complexity in the mechanisms and interrelated functions of lncRNAs and their subsequent effect.
Fig. 2.
Graphical representation of human tumors and related lncRNAs. Red labels indicate oncogenic lncRNAs, while green labels indicate tumor-suppressive lncRNAs.
5. Methodology
In the current review, the authors aimed at exploring the role of MIAT in HCC. The authors screened the National Library of Medicine (PubMed). To search databases, the descriptors or keywords used were: “MIAT”, “myocardial infarction-associated transcript” “MIAT LncRNA”, “HCC”, “Hepaticellular carcinoma”, and “Oncology”, “LncRNAs” to cover as many articles as possible in the literature. Relevant publications with detailed information were included including research articles, review articles, and book chapters; These selected references were evaluated and summarized in order to fulfill the purpose of this review article.
6. Myocardial infarction associated transcript (MIAT)
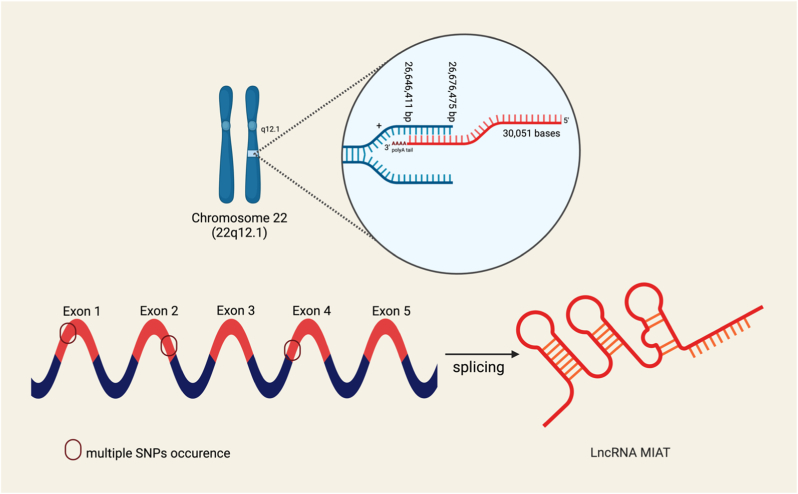
Myocardial infarction-associated transcript (MIAT) is a novel lncRNA that has recently been reported to have a fundamental role in several oncological contexts [13,41]. MIAT is located on chromosome 22q12.1 as shown in Fig. 3 [79]. MIAT gene length is around 30 Kb. MIAT is transcribed from the sense strand of the genome, producing a structure with 5 exons and several introns with multiple combinations of single nucleotide polymorphisms (SNPs), and a polyadenylated tail, as shown in Fig. 3. Post-transcriptional splicing gives rise to 4 different variants of spliced lncRNA [[80], [81], [82], [83]]. As indicated by its name, it is predicted that it has a role in cardiovascular diseases such as atherosclerosis, and coronary artery diseases. However, it has been discovered that not only does it influence these diseases, but it also plays a key role in several solid malignancies such as lung cancer, hepatocellular carcinoma, and breast cancer among countless others [20,60,84,85]. The biogenesis of MIAT-like all other lncRNAs is dependent on the cell type and stage, as previously reviewed [17]. Collectively, research studies focusing on MIAT shed light on its complex function and offer a possible path for therapeutic interventions, as discussed below. Different functional mechanisms of MIAT have been described, so far; moreover, this lncRNA has been described to decoy several microRNAs (miRNAs) and thus altering the downstream array of critical signaling pathways dictating cellular proliferation, apoptosis, and inflammation (Fig. 4).
Fig. 3.
Chromosomal locations and Single Nucleotide Polymorphisms (SNPs) in MIAT. MIAT is located on chromosome 22, band q12.1. A simplified view of MIAT is also illustrated with its exons with various splicing combinations and SNPs.
Fig. 4.
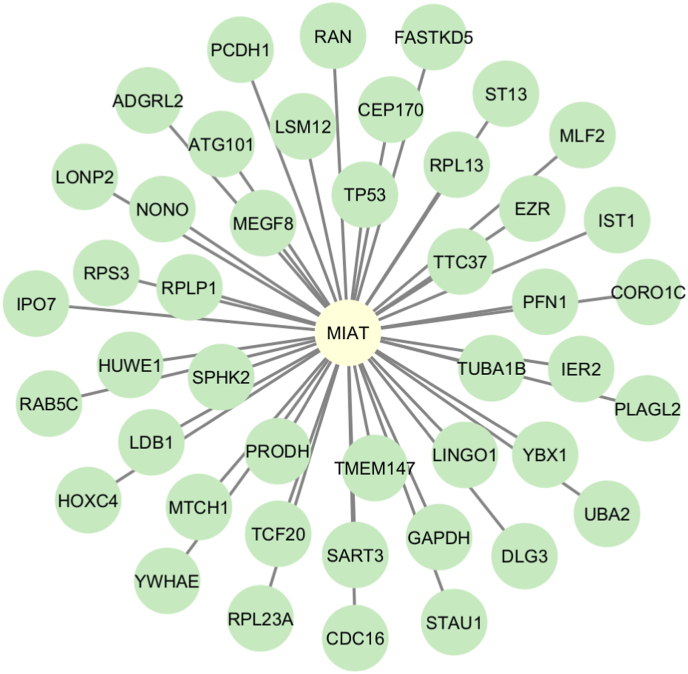
MIAT-mRNA interaction network. A network plot representing MIAT-mRNA regulatory network. Data was retrieved from the ENCODE database.
6.1. MIAT cellular localization and single nucleotide polymorphisms (SNPs)
The subcellular localization of MIAT is the nucleus, as demonstrated by a nuclear/cytosol fractionation assay that showed that it is mostly expressed in the nucleus of cardiomyocytes [86]. MIAT is known for its multiple SNPs that are strongly associated with increased susceptibility to myocardial infarction [87]. The fully elucidated sequence and SNP variations of lncRNA MIAT are present in multiple databases, including GeneCaRNA, Rfam, and the National Library of Medicine (NLM) gene dataset. For instance, in the Chinese Han population, the promoter polymorphisms in the MIAT gene Rs5752375 and Rs9608515 were found to be associated with acute myocardial infarction [83]. Furthermore, many different SNPs of MIAT have been shown to have different diverse effects as shown in Fig. 3. For instance, Rs1894720 MIAT polymorphism has been found to increase susceptibility to age-related loss of hearing by tuning the miR-29 b-3p/SIRT1/PGC-1α axis [88]. Another study demonstrated a notable correlation between Rs1894720 in MIAT and paranoid schizophrenia within the Chinese Han population [89].
6.2. MIAT-mRNA and MIAT-miRNA networks
Recent progress in high-throughput sequencing technologies has led to the identification of novel ncRNAs as well as an understanding the regulatory mechanisms for their co-expression including protein-coding genes and ncRNA molecules [90]. Databases are categorized into various classifications depending on their role in storing, displaying, and also analyzing data for ncRNAs.
RNAcentral is one of the most comprehensive databases for ncRNAs, where since its launch in 2014 about 10.2 million ncRNA sequences were identified, sequenced and recorded in it, thus providing invaluable information for subsequent recognition of ncRNAs once sequenced [91]. Also, RNAcentral integrates other ncRNA databases into one platform to allow accessible search for ncRNAs of interest.
NONCODE and LNCipedia are two of about 12 databases that integrate with RNAcentral, particularly to provide in-depth information regarding lncRNAs as well as tRNAs and rRNAs [92,93]. RNAcentral allows sequence similarity search, text search, and genome browsing of ncRNAs which aids in the identification of novel ncRNAs in different species as well as providing GO (Gene ontology) annotations for miRNAs and lncRNAs using RNAcentral identifiers.
LncTarD is a lncRNA-specific database that provides comprehensive information regarding lncRNA functions, regulatory mechanisms, as well as lncRNA-target insights following data retrieval from GEO (gene expression omnibus) database of NCBI as well as PubMed [94]. Another lncRNA database that provides lncRNA-target gene information is LncRNA2Target which includes curated data for both human and mice samples across different pathologies in order to display insightful results through combined analysis of stored lncRNA data in tabular format [95].
ENCORI (The Encyclopedia of RNA interactomes) is also a robust database that is particularly useful for understanding how RNA molecules integrate following results from high throughput sequencing data stored in the database. LncRNA-lncRNA, lncRNA-gene, miRNA-gene, and miRNA-ncRNA are useful integration techniques provided by ENCORI in order to understand how lncRNAs, miRNAs, and protein-coding genes interact together, particularly in cancer contexts [96].
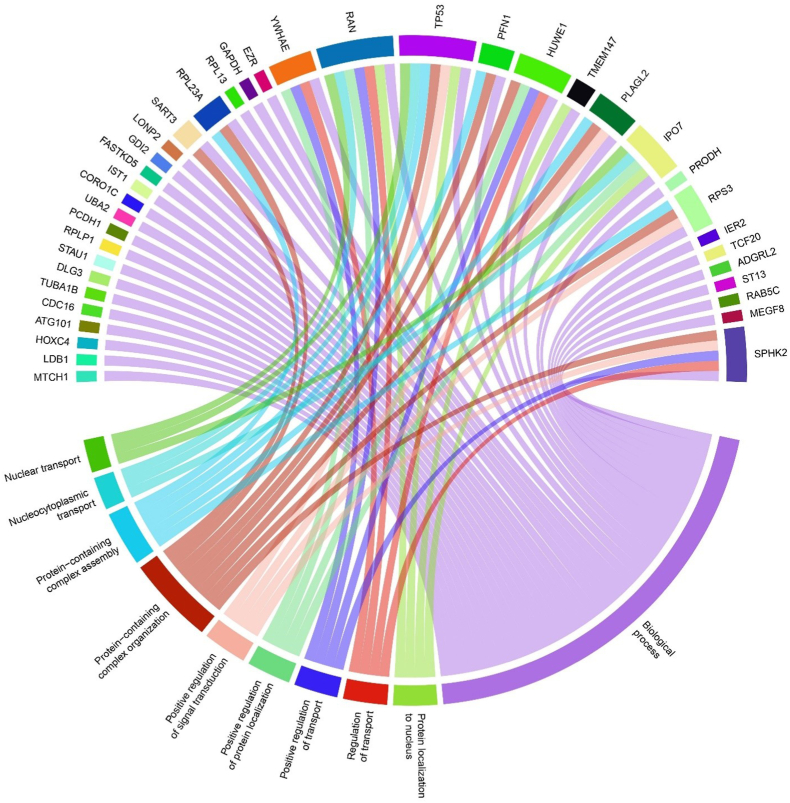
Following in-silico analysis of MIAT-mRNA and MIAT-miRNA interactions using ENCORI, LncTarD, and LncRNA2Target databases, results revealed a comprehensive list of protein-coding genes and miRNAs that directly interact with MIAT in different pathological conditions. MIAT-mRNA network was constructed using Cytoscape software (v.3.10) and revealed 45 interaction nodes with MIAT at different disease states as listed in Table 3, where tumor protein p53 (TP53), transmembrane protein 147 (TMEM147), importin 7 (IPO7), mitochondrial carrier homolog 1 (MTCH1), LIM domain-binding protein 1 (LDB1), ubiquitin-like modifier-activating enzyme 2 (UBA2), coronin‐like actin‐binding protein 1C (CORO1C), IST1 factor associated with ESCRT-III (IST1), and proline dehydrogenase 1 (PRODH) were highlighted in later annotation using gene ontology Fig. 4. Also, MIAT-miRNA interaction resulted in the detection of 9 miRNAs; miR-150, miR-145, miR-148 b, miR-206, miR-181 b, miR-149–5p, miR-214–3p, miR-641, and miR-181a-5p (Table 3); which could provide further understanding of MIAT role in HCC following further analysis. Moreover, gene ontology annotation of MIAT-mRNA interactions revealed several biological processes in which MIAT acts as a regulator of their action through suppressing the top 10 processes which MIAT-related protein-coding genes (n = 45) are directly regulating via GO scoring (Fig. 5), which explains the interconnection between all MIAT-related genes and subsequent GO functions following construction using R programming language (v. 4.0).
Table 3.
MIAT-mRNA and MIAT-miRNA selected interactionsa reported in non-coding RNA databases.
| mRNA or miRNA | Database | Reference |
|---|---|---|
| CORO1C | ENCORI |
[96] |
| IPO7 | ||
| IPO7 | ||
| IST1 | ||
| LDB1 | ||
| MTCH1 | ||
| PRODH | ||
| TMEM147 | ||
| TP53 | ||
|
UBA2 | ||
| miR-145 | LncTarD/LncRNA2Target | [94,95] |
| miR-148 b | ||
| miR-149–5p | ||
| miR-150 | ||
| miR-181a-5p | ||
| miR-181 b | ||
| miR-206 | ||
| miR-214–3p | ||
| miR-641 |
MIAT-mRNA and MIAT-miRNA interactions across multiple pathological conditions using ENCORI, LncTarD, and LncRNA2Target databases. Results were retrieved using search API tool identifier for MIAT-related interaction on the mentioned databases.
Fig. 5.
Chord diagram for MIAT gene-function network using GO and ENCORI databases; this diagram describes the top GO expressed biological functions for the 45 genes interacting with MIAT. Results showed that SPHK2, RPS3, IPO7, TP53, RAN, and YWHAE are highly expressed genes from MIAT-based network and are directly interacting with higher of functions compared to the other genes.
6.3. Role of MIAT in solid malignancies
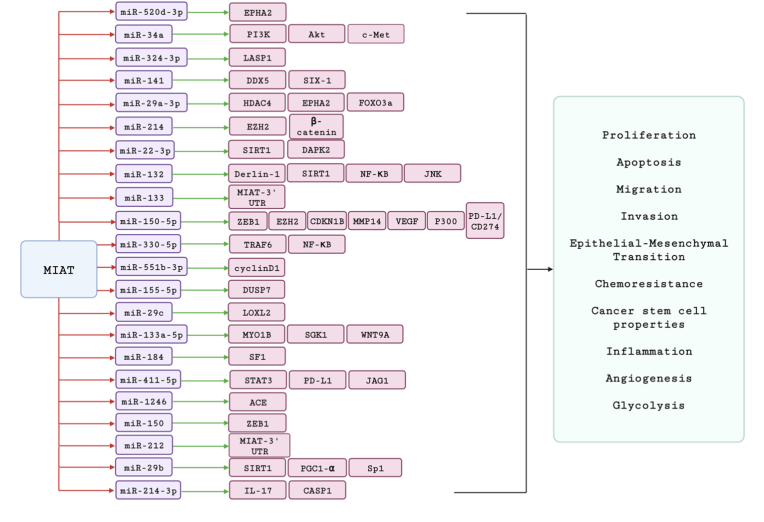
Recently, it has been experimentally reported that MIAT has distinct roles in the tumorigenesis and progression of distinct types of cancer. Briefly, MIAT has been shown to have a pro-tumorigenic activity against the following types of cancer including HCC, breast cancer, non-small cell lung cancer, colorectal cancer, pancreatic cancer, ovarian cancer, gastric cancer, cholangiocarcinoma, and renal cell carcinoma. Mainly, MIAT contributed to the previous types of cancers via increasing the proliferation, the invasion, and the migration. In addition to this, the sponging or interacting activity of MIAT with an array of miRNAs reveals how MIAT up-regulation could increase the cancerous activity of tumor cells via the downstream target genes/proteins of these miRNAs as shown in Fig. 6. We and others have extensively reported the direct involvement of miRNAs and lncRNAs in the hepatocarcinogenesis process [[97], [98], [99], [100], [101], [102], [103]]. The following section will discuss the role of MIAT as a puzzling lncRNA in HCC with its pro-tumorigenic or anti-tumorigenic nature, its mechanism of action in terms of the downstream target genes including their possible signaling pathways, crosstalk between different classes of ncRNAs and lastly, its clinical eligibility to be a predictive diagnostic and/or prognostic marker.
Fig. 6.
Validated MIAT lncRNA-miRNAs-mRNAs interaction network in carcinogenesis. Crosstalk between MIAT lncRNA, its microRNA prey and their respective targets, and the cancer hallmarks that are altered due to these interactions, in different malignant contexts. The potential of some microRNAs, such as miR-133 and miR-212, to target MIAT, is also highlighted. Red arrows signify downregulation effect mediated by lncRNA MIAT on the several microRNAs. Green arrows signify upregulation effect on the downstream targets, subsequent to the respective microRNA suppression.
7. Hepatocellular carcinoma (HCC)
HCC is considered one of the most lethal solid malignancies with high relapse rates and poor prognosis [97,98]. A myriad of factors could be listed under the etiology of the disease, as these factors have a profound impact on the initiation and progression of the disease. For instance, many research studies have revealed that unhealthy diet, alcohol, smoking, hepatitis C virus, and aflatoxin are the drivers for inflammation–causing conditions that eventually cause HCC. Although there are considerable scientific records for the causative reasons for the pathogenesis of HCC, there is still a gap that needs to be filled regarding how the non-coding part of our genome could have a non-negligible role in the aggravation of this disease.
7.1. Role of MIAT in chronic liver diseases
Chronic liver diseases (CLD), including cirrhosis, fibrosis, alcoholic liver disease, and chronic hepatitis, are important precursors of HCC. A recent study showed that elevated MIAT expression during liver fibrosis is linked to increased hepatic stellate cell (HSC) proliferation and collagen expression, while MIAT knockdown demonstrated a marked suppression of fibrosis progression and collagen accumulation in vivo. MIAT acts as a sponge for miR-3085–5p, showing a negative correlation with miR-3085–5p levels in cirrhotic patients and activated HSCs. The study underscores the role of MIAT in HSC activation through the miR-3085–5p/YAP1, where MIAT inhibition leads to reduced yes-associated protein 1 (YAP1) levels and subsequent suppression of the epithelial-to-mesenchymal transition (EMT) process [104]. While the detailed function of MIAT in alcoholic liver disease and chronic hepatitis has not yet been investigated, to the best of our knowledge, given its role in fibrosis and cirrhosis there is potential for a similar influential role in other forms of CLD. Going beyond merely acting as precursors to HCC, these observations imply that MIAT could contribute to the pathogenesis of HCC through inflammation, fibrosis, and other aspects inherent to CLD. Hence, understanding the role of MIAT in CLD could provide useful insights into the molecular mechanism of hepatocellular carcinogenesis.
7.2. Role of MIAT in HCC pathogenesis
As indicated in the literature, a unanimous consensus across multiple studies underscores the robust correlation between MIAT presence and the onset of HCC. A study focusing on lncRNAs associated with epithelial-mesenchymal transition (EMT) in HCC presented MIAT on top of the list of lncRNAs that were both differentially expressed in HCC and positively correlated with EMT in HCC. On the molecular level, it was reported that MIAT plays an oncogenic and metastatic role in HCC as upon MIAT knockdown in HCC cell lines, a significant reduction in the levels of Cadherin-1 (e-cadherin, an epithelial marker) and an elevation in the levels of Cadherin-2 (n-cadherin, a mesenchymal marker) was observed [105].
7.2.1. Crosstalk between MIAT and miR-214–3p
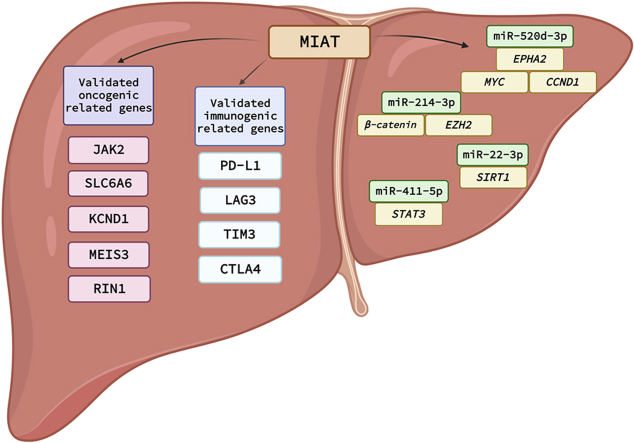
A study by Huang et al. discovered that HCC cell lines (HepG2, Huh-7, SK-HEP-1, and HLE) and patient tissue samples have higher levels of MIAT expression than adjacent normal tissues and normal hepatocyte cell line (L02); this could be considered as an indication that MIAT has a role in the pathogenesis of HCC. Mechanistically, it has been found that the H3/H4 epigenetic acetylation of the MIAT promoter in tumor tissues is the reason behind the elevated expression of MIAT in HCC tumor tissues and cell lines. Nonetheless, it has been experimentally validated that MIAT sponges the tumor-suppressor miRNA, miR-214–3p, in HCC cell lines and accordingly ranks MIAT as an oncogenic lncRNA in HCC [106]. In addition to this, MIAT knockdown in vivo resulted in the elevation of miR-214–3p levels, and subsequently, of catenin beta 1 (CTNNB1 or β-catenin) and enhancer of zest homolog 2 (EZH2) which are the downstream targets of miR-214–3p [107], as shown in Fig. 7.
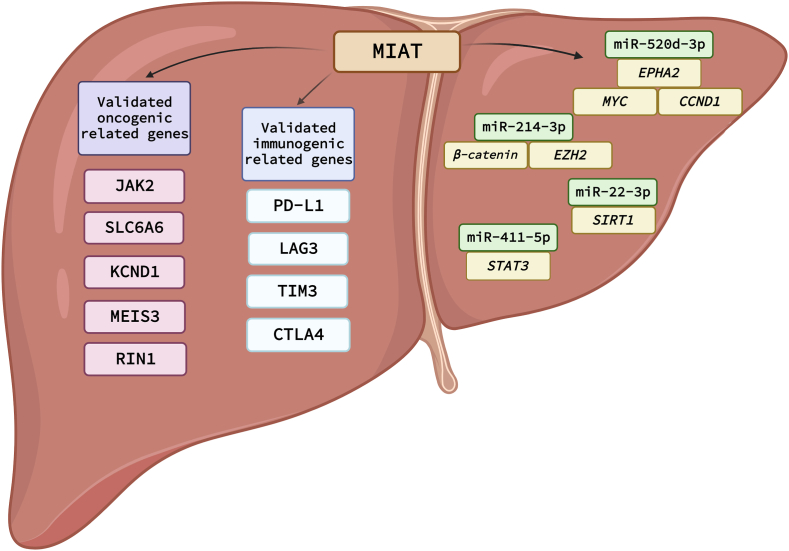
Fig. 7.
Graphical representation of signaling cascades and ncRNAs circuits drawn downstream MIAT in HCC. A summary for all reported oncogenic related genes, immunogenic related genes and ncRNA-miRNA circuits reported to be modulated by MIAT in HCC.
7.2.2. Crosstalk between MIAT and miR-520 d-3p
The role of MIAT as a miRNA sponge in the evolution of HCC was underlined in another study [108]. This work showed that miR-520 d-3p, which was previously proven to have an anti-tumorigenic function in HCC, is downregulated by MIAT. The authors found that MIAT expression affected the downstream target genes of miR-520 d-3p. Specifically, a positive association between MIAT and erythropoietin-producing hepatocellular receptor A2 (EPHA2), a miR-520 d-3p downstream target gene was recorded. It has been shown in this study and other studies that EPHA2 regulates MYC proto-oncogene (MYC) and cyclin D1 (CCND1), which are involved in cell cycle regulation and proliferation, and that inducing the expression of miR-520 d-3p in HCC cells results in a reduction in MYC and CCND1 expression by inhibition of EPHA2 [109,110], as shown in Fig. 7.
7.2.3. Crosstalk between MIAT and miR-22–3p
It was also shown that MIAT promotes the survival of HCC cells to survive against cellular senescence via regulating the miR-22–3p/SIRT1 axis (Fig. 7) [111]. Sirtuin 1 (SIRT1) protein is a NAD–dependent histone deacetylase that was reported for its inhibitory effect on cellular senescence via preventing apoptosis, maintaining cellular metabolism, and preventing cells from oxidative stress [112]. This work discovered that MIAT is a senescence-associated lncRNA in addition to being a differentially expressed lncRNA in HCC by analyzing The Cancer Genome Atlas (TCGA) liver cancer dataset. Experimental validation of whether cellular senescence is activated or not in the presence of MIAT was validated in human fibroblast 2BS cells and oncogene-induced 2BS cells. It was shown that MIAT was highly present in 2BS young cells whereas its expression decreased during the cellular senescence process of 2BS cells. The decrease of MIAT expression levels via its knockdown in 2BS cells led to an increase of senescence–associated beta-galactosidase activity, cell cycle arrest, and cell proliferation inhibition. Similar results were obtained, showing that the MIAT expression levels are lower in the HCC senescence models (HEPG2 and SMMC-7721) than in the normal cell lines [111].
7.3. Role of MIAT in HCC chemoresistance and immunotherapy
An important hallmark of cancer in general and HCC in particular is the immune escape [14,16,21,22,113,114]. Immune cells, infiltrating the tumor microenvironment (TME), have an enormous impact in stopping the tumor cells from proliferation, invasion, and dissemination [[115], [116], [117]]. Another considerable barrier in HCC therapeutic protocols is high incidence rates of resistance to the conventional anticancer agents that would consequently lead to more aggressive tumors and a rapid relapse in many cases [50,118]. MIAT is involved in the immune cellular response and associated non-cellular components at the TME. In a study performed by Peng et al. , a bioinformatics analysis was carried out using the TIMER database to explore the relationship between MIAT and immune cells and mediators in HCC. Analyzing the TIMER database showed a positive correlation between MIAT and immune cells: cytotoxic T lymphocytes, T helper cells, macrophages, dendritic cells, neutrophils, and B cells. This positive correlation was also found between MIAT, and the expression of immune checkpoint inhibitors programmed cell death 1 (PDCD1 or PD-1), CD274 molecule (PD-L1), cytotoxic T-lymphocyte associated protein 4 (CTLA4), lymphocyte activating 3 (LAG3), and hepatitis A virus cellular receptor 2 (HAVCR2, also known as TIM3). A deeper analysis was performed by using the single-cell sequencing techniques for the CD45+ immune cells in HCC. Results showed that MIAT contributes to tumor immunosuppression since MIAT expression was high in forkhead box P3 (FOXP3) and CD4 positive T cells and PD-1 and granzyme K (GZMK) and CD8 subunit alpha (CD8) positive T cells in tumors and blood, hepatic lymph nodes, and ascites. FOXP3+ CD4+ T cells and PDCD1+/GZMK + CD8+ T cells correspond to regulatory T cells and exhaustive T cells, respectively [119].
A significant aspect by which MIAT exerts its action is its cellular localization. With the assistance of lncLocator, the cellular location of MIAT was expected to be in the nucleus [120]. This was a sign that MIAT plays a role in gene expression regulation via interacting with transcription factors in the nucleus [120]. These transcription factors include Janus kinase 2 (JAK2), solute carrier family 6 member 6 (SLC6A6), potassium voltage-gated channel subfamily D member 1 (KCND1), Meis homeobox 3 (MEIS3), and Ras and Rab interactor 1 (RIN1). The correlation between MIAT and immune cells was further confirmed by exploring the correlation between the previously stated target genes and immune cells.
MIAT expression confers HCC resistance against sorafenib [121]. It was found that the resistance in sorafenib is associated with high expression of MIAT in HCC cells and this was also associated with the presence of PD-L1 [121]. Consistent with this, the mRNA and protein of PD-L1 were decreased upon MIAT knockdown in HepG2 and Huh7 cell lines, and the expression of both MIAT and PD-L1 were significantly elevated after treatment of HCC cells with sorafenib. Hence, the resistance of sorafenib in HCC could be attributed to the up-regulation of PD-L1 by MIAT and eventually lead to immune escape [121].
A similar issue of immune escape caused by MIAT in HCC was investigated in another study in terms of the potential regulatory network and mechanism of regulation of PD-L1 by MIAT in HCC cells. It was discovered that MIAT regulates miR-411–5p by functioning as a competitive endogenous RNA that binds to miR-411–5p and inhibits its actions [85]. Upon MIAT knockdown in HepG2 and Huh7 cells, a marked reduction in PD-L1 expression levels was witnessed. Signal transducer and activator of transcription 3 (STAT3), a transcription factor that controls PD-L1 by binding to its promoter, was also shown to be one of the putative targets of miR-411–5p. These results were supported by the transfection of a miR-411–5p oligonucleotide into HCC cells, which resulted in repression in PD-L1 and STAT3 on both mRNA and protein levels [121].
7.4. Potential clinical applications of MIAT
MIAT holds promise as a diagnostic biomarker for various cancers, including HCC. The reported upregulation of MIAT expression in tissue samples may serve as an early indicator of cancer, facilitating timely diagnosis [106]. Moreover, as the expression levels of MIAT have shown potential as prognostic indicators in other types of cancer [122], offering insights into clinical outcomes such as disease progression, metastasis, and overall survival in cancer patients, would be possible in HCC as well. Furthermore, exploring the association between MIAT expression and clinical features specific to HCC, such as tumor stage, grade, and vascular invasion, could enhance our understanding of the role of MIAT in HCC development and progression.
In the realm of cancer therapeutics, MIAT may present itself as a promising therapeutic target. MIAT promotes the growth and invasive abilities of HCC tumor cells [106]. Thus, inhibiting the expression of MIAT may be an effective way to treat HCC. This could potentially be achieved with targeted therapies like small interfering RNAs (siRNAs) or antisense oligonucleotides (ASOs) designed to bind to and cause degradation of specific lncRNAs.
Additionally, given the deregulation of MIAT in HCC cell lines upon sorafenib treatment, MIAT expression levels could be leveraged to predict the response of cancer patients to specific treatments, guiding the development of personalized therapeutic strategies for improved outcomes. Monitoring changes in MIAT expression over time could serve as a valuable tool in tracking the progression of cancer. This longitudinal approach may provide insights into the evolving molecular landscape of tumors and help gauge treatment responses or the emergence of resistance [121].
Collectively, MIAT has immense potential in the clinical management of HCC, including as a non-invasive biomarker for early detection and prognosis, a therapeutic target, and a predictor of therapeutic response. However, further detailed studies and clinical trials are required to validate these applications of MIAT in HCC.
8. Conclusions and future perspectives
While the role of lncRNA MIAT is becoming increasingly apparent in HCC, there are some limitations in the current research. Firstly, most of the studies till now have utilized in vitro cell models, and human trials are lacking. Hence, the translation of these findings into clinical practice requires caution and remains a significant challenge.
Secondly, there is still a lot to be understood about the complex, multi-layered regulatory mechanisms of lncRNA MIAT in HCC. Much of the existing research provides evidence on a molecular level, but the comprehensive picture of physiological and pathological conditions remains incomplete. Another aspect that must be considered is that lncRNA MIAT might operate differently depending on the cellular context, which needs to be further investigated.
In terms of future directions, researchers need to focus on elucidating more detailed mechanisms by which MIAT regulates hepatocellular carcinoma progression. Also, more clinical studies are necessary to establish the potential of MIAT as a diagnostic marker or even a therapeutic target. Furthermore, considering the involvement of MIAT with other diseases like myocardial infarction and diabetic retinopathy, studying its systemic influence could provide profound insight and potentially higher clinical relevance.
In conclusion, while there is promising potential for the role of lncRNA MIAT in HCC research, it is important to acknowledge the limitations and challenges in the current state and to continue striving for a more detailed and comprehensive understanding.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Funding
Not applicable.
Declaration of competing interest
None.
CRediT authorship contribution statement
Rawan Amr Elmasri: Writing – original draft, Methodology, Investigation, Data curation. Alaa A. Rashwan: Writing – original draft, Methodology, Data curation. Sarah Hany Gaber: Visualization, Methodology, Investigation. Monica Mosaad Rostom: Writing – review & editing, Validation, Investigation, Formal analysis. Paraskevi Karousi: Data curation, Writing – original draft, Writing – review & editing. Montaser Bellah Yasser: Data curation, Formal analysis, Methodology, Software. Christos K. Kontos: Writing – review & editing, Validation, Supervision, Investigation, Formal analysis. Rana A. Youness: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization.
Acknowledgements
Not applicable.
References
- 1.Gibbs R.A. The human genome Project changed everything. Nat. Rev. Genet. 2020;21(10):575–576. doi: 10.1038/s41576-020-0275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Abaza T., El-Aziz M.K.A., Daniel K.A., Karousi P., Papatsirou M., Fahmy S.A., et al. Emerging role of circular RNAs in hepatocellular carcinoma immunotherapy. Int. J. Mol. Sci. 2023;24(22) doi: 10.3390/ijms242216484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawoud A., Ihab Zakaria Z., Hisham Rashwan H., Braoudaki M., Youness R.A. Circular RNAs: new layer of complexity evading breast cancer heterogeneity. Noncoding RNA Res. 2023;8(1):60–74. doi: 10.1016/j.ncrna.2022.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Daly S.M., Talaat R.M., Braoudaki M., Youness R.A., Cho W.C. Editorial: recent breakthroughs in the decoding of circulating nucleic acids and their applications to human diseases. Front. Mol. Biosci. 2023;10 doi: 10.3389/fmolb.2023.1203495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Youness R.A., Gad M.Z. Long non-coding RNAs: functional regulatory players in breast cancer. Noncoding RNA Res. 2019;4(1):36–44. doi: 10.1016/j.ncrna.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nafea H., Youness R.A., Abou-Aisha K., Gad M.Z. LncRNA HEIH/miR-939-5p interplay modulates triple-negative breast cancer progression through NOS2-induced nitric oxide production. J. Cell. Physiol. 2021;236(7):5362–5372. doi: 10.1002/jcp.30234. [DOI] [PubMed] [Google Scholar]
- 8.Hangauer M.J., Vaughn I.W., McManus M.T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9(6) doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson E.K., Covarrubias S., Carpenter S. The how and why of lncRNA function: an innate immune perspective. Biochim Biophys Acta Gene Regul Mech. 2020;1863(4) doi: 10.1016/j.bbagrm.2019.194419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uszczynska-Ratajczak B., Lagarde J., Frankish A., Guigo R., Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018;19(9):535–548. doi: 10.1038/s41576-018-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang S., Zhang L., Guo J., Niu Y., Wu Y., Li H., et al. NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018;46(D1):D308–D314. doi: 10.1093/nar/gkx1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Statello L., Guo C.J., Chen L.L., Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selem N.A., Youness R.A., Gad M.Z. What is beyond LncRNAs in breast cancer: a special focus on colon cancer-associated Transcript-1 (CCAT-1) Noncoding RNA Res. 2021;6(4):174–186. doi: 10.1016/j.ncrna.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramzy A., ElSafy S., Elshoky H.A., Soliman A., Youness R., Mansour S., et al. Drugless nanoparticles tune-up an array of intertwined pathways contributing to immune checkpoint signaling and metabolic reprogramming in triple-negative breast cancer. Biomed. Mater. 2022;18(1) doi: 10.1088/1748-605X/aca85d. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Latif M., Riad A., Soliman R.A., Elkhouly A.M., Nafae H., Gad M.Z., et al. MALAT-1/p53/miR-155/miR-146a ceRNA circuit tuned by methoxylated quercitin glycoside alters immunogenic and oncogenic profiles of breast cancer. Mol. Cell. Biochem. 2022;477(4):1281–1293. doi: 10.1007/s11010-022-04378-4. [DOI] [PubMed] [Google Scholar]
- 16.Abdallah R.M., Elkhouly A.M., Soliman R.A., El Mechawy N., El Sebaei A., Motaal A.A., et al. Hindering the synchronization between miR-486-5p and H19 lncRNA by hesperetin halts breast cancer aggressiveness through tuning ICAM-1. Anti Cancer Agents Med. Chem. 2022;22(3):586–595. doi: 10.2174/1871520621666210419093652. [DOI] [PubMed] [Google Scholar]
- 17.El-Aziz M.K.A., Dawoud A., Kiriacos C.J., Fahmy S.A., Hamdy N.M., Youness R.A. Decoding hepatocarcinogenesis from a noncoding RNAs perspective. J. Cell. Physiol. 2023;238(9):1982–2009. doi: 10.1002/jcp.31076. [DOI] [PubMed] [Google Scholar]
- 18.Mekky R.Y., Ragab M.F., Manie T., Attia A.A., Youness R.A. MALAT-1: immunomodulatory lncRNA hampering the innate and the adaptive immune arms in triple negative breast cancer. Transl. Oncol. 2023;31 doi: 10.1016/j.tranon.2023.101653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10(6):925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Zitello E., Guo R., Deng Y. The function of LncRNAs and their role in the prediction, diagnosis, and prognosis of lung cancer. Clin. Transl. Med. 2021;11(4):e367. doi: 10.1002/ctm2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selem N.A., Nafae H., Manie T., Youness R.A., Gad M.Z. Let-7a/cMyc/CCAT1/miR-17-5p circuit Re-sensitizes atezolizumab resistance in triple negative breast cancer through modulating PD-L1. Pathol. Res. Pract. 2023;248 doi: 10.1016/j.prp.2023.154579. [DOI] [PubMed] [Google Scholar]
- 22.Soliman A.H., Youness R.A., Sebak A.A., Handoussa H. Phytochemical-derived tumor-associated macrophage remodeling strategy using Phoenix dactylifera L. boosted photodynamic therapy in melanoma via H19/iNOS/PD-L1 axis. Photodiagnosis Photodyn. Ther. 2023;44 doi: 10.1016/j.pdpdt.2023.103792. [DOI] [PubMed] [Google Scholar]
- 23.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X., Wang W., Zhu W., Dong J., Cheng Y., Yin Z., et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 2019;20(22) doi: 10.3390/ijms20225573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Postepska-Igielska A., Giwojna A., Gasri-Plotnitsky L., Schmitt N., Dold A., Ginsberg D., et al. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell. 2015;60(4):626–636. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Bhan A., Mandal S.S. LncRNA hotair: a master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta. 2015;1856(1):151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Froberg J.E., Yang L., Lee J.T. Guided by RNAs: X-inactivation as a model for lncRNA function. J. Mol. Biol. 2013;425(19):3698–3706. doi: 10.1016/j.jmb.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Negishi M., Wongpalee S.P., Sarkar S., Park J., Lee K.Y., Shibata Y., et al. A new lncRNA, APTR, associates with and represses the CDKN1A/p21 promoter by recruiting polycomb proteins. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0095216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L., Lin C., Jin C., Yang J.C., Tanasa B., Li W., et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akerman I., Tu Z., Beucher A., Rolando D.M.Y., Sauty-Colace C., Benazra M., et al. Human pancreatic beta cell lncRNAs control cell-specific regulatory networks. Cell Metabol. 2017;25(2):400–411. doi: 10.1016/j.cmet.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin Q.F., Yang L., Zhang Y., Xiang J.F., Wu Y.W., Carmichael G.G., et al. Long noncoding RNAs with snoRNA ends. Mol. Cell. 2012;48(2):219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 33.Xu T.P., Liu X.X., Xia R., Yin L., Kong R., Chen W.M., et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34(45):5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 34.Zhou W., Ye X.L., Xu J., Cao M.G., Fang Z.Y., Li L.Y., et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci. Signal. 2017;10(483) doi: 10.1126/scisignal.aak9557. [DOI] [PubMed] [Google Scholar]
- 35.Paci P., Colombo T., Farina L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst. Biol. 2014;8:83. doi: 10.1186/1752-0509-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y., Sun L., Wang R.R., Hu J.F., Cui J. The effects of mitochondria-associated long noncoding RNAs in cancer mitochondria: new players in an old arena. Crit. Rev. Oncol. Hematol. 2018;131:76–82. doi: 10.1016/j.critrevonc.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Leucci E., Vendramin R., Spinazzi M., Laurette P., Fiers M., Wouters J., et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531(7595):518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 38.Fatima F., Nawaz M. Vesiculated long non-coding RNAs: offshore packages deciphering trans-regulation between cells, cancer progression and resistance to therapies. Non-Coding RNA. 2017;3(1):10. doi: 10.3390/ncrna3010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nafea H., Youness R.A., Dawoud A., Khater N., Manie T., Abdel-Kader R., et al. Dual targeting of H(2)S synthesizing enzymes; cystathionine beta-synthase and cystathionine gamma-lyase by miR-939-5p effectively curbs triple negative breast cancer. Heliyon. 2023;9(10) doi: 10.1016/j.heliyon.2023.e21063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youness R.A., Mohamed A.H., Efthimiadou E.K., Mekky R.Y., Braoudaki M., Fahmy S.A. A snapshot of photoresponsive liposomes in cancer chemotherapy and immunotherapy: opportunities and challenges. ACS Omega. 2023;8(47):44424–44436. doi: 10.1021/acsomega.3c04134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fahmy S.A., Dawoud A., Zeinelabdeen Y.A., Kiriacos C.J., Daniel K.A., Eltahtawy O., et al. Molecular engines, therapeutic targets, and challenges in pediatric brain tumors: a special emphasis on hydrogen sulfide and RNA-based nano-delivery. Cancers. 2022;14(21) doi: 10.3390/cancers14215244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szilagyi M., Pos O., Marton E., Buglyo G., Soltesz B., Keseru J., et al. Circulating cell-free nucleic acids: main characteristics and clinical application. Int. J. Mol. Sci. 2020;21(18) doi: 10.3390/ijms21186827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramon Y.C.S., Segura M.F., Hummer S. Interplay between ncRNAs and cellular communication: a proposal for understanding cell-specific signaling pathways. Front. Genet. 2019;10:281. doi: 10.3389/fgene.2019.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Y., Wang Y., Wei M., Han X., Xu T., Cui M. Advances in the study of exosomal lncRNAs in tumors and the selection of research methods. Biomed. Pharmacother. 2020;123 doi: 10.1016/j.biopha.2019.109716. [DOI] [PubMed] [Google Scholar]
- 45.Dragomir M., Chen B., Calin G.A. Exosomal lncRNAs as new players in cell-to-cell communication. Transl. Cancer Res. 2017:S243–S252. doi: 10.21037/tcr.2017.10.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hawkes E.J., Hennelly S.P., Novikova I.V., Irwin J.A., Dean C., Sanbonmatsu K.Y. COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep. 2016;16(12):3087–3096. doi: 10.1016/j.celrep.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Somarowthu S., Legiewicz M., Chillon I., Marcia M., Liu F., Pyle A.M. HOTAIR forms an intricate and modular secondary structure. Mol. Cell. 2015;58(2):353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D.N., Thiel B.C., Mrozowich T., Hennelly S.P., Hofacker I.L., Patel T.R., et al. Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution. Nat. Commun. 2020;11(1):148. doi: 10.1038/s41467-019-13942-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang M.C., Ni J.J., Cui W.Y., Wang B.Y., Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019;9(7):1354–1366. [PMC free article] [PubMed] [Google Scholar]
- 50.ZeinElAbdeen Y.A., AbdAlSeed A., Youness R.A. Decoding insulin-like growth factor signaling pathway from a non-coding RNAs perspective: a step towards precision oncology in breast cancer. J. Mammary Gland Biol. Neoplasia. 2022;27(1):79–99. doi: 10.1007/s10911-022-09511-z. [DOI] [PubMed] [Google Scholar]
- 51.Mo Y.-Y., Zou D., Koirala P. Long non-coding RNAs as key regulators of cancer metastasis. J.Cancer Metastasis Treat. 2015;0(0):1–10. [Google Scholar]
- 52.Boon R.A., Jae N., Holdt L., Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J. Am. Coll. Cardiol. 2016;67(10):1214–1226. doi: 10.1016/j.jacc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 53.Youness R.A., Assal R.A., Abdel Motaal A., Gad M.Z. A novel role of sONE/NOS3/NO signaling cascade in mediating hydrogen sulphide bilateral effects on triple negative breast cancer progression. Nitric Oxide. 2018;80:12–23. doi: 10.1016/j.niox.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 54.Youness R.A., Hafez H.M., Khallaf E., Assal R.A., Abdel Motaal A., Gad M.Z. The long noncoding RNA sONE represses triple-negative breast cancer aggressiveness through inducing the expression of miR-34a, miR-15a, miR-16, and let-7a. J. Cell. Physiol. 2019;234(11):20286–20297. doi: 10.1002/jcp.28629. [DOI] [PubMed] [Google Scholar]
- 55.Wang M., Zhou L., Yu F., Zhang Y., Li P., Wang K. The functional roles of exosomal long non-coding RNAs in cancer. Cell. Mol. Life Sci. 2019;76(11):2059–2076. doi: 10.1007/s00018-019-03018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xing C., Sun S.G., Yue Z.Q., Bai F. Role of lncRNA LUCAT1 in cancer. Biomed. Pharmacother. 2021;134 doi: 10.1016/j.biopha.2020.111158. [DOI] [PubMed] [Google Scholar]
- 57.Ren J., Ding L., Zhang D., Shi G., Xu Q., Shen S., et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8(14):3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conigliaro A., Costa V., Lo Dico A., Saieva L., Buccheri S., Dieli F., et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer. 2015;14(1):155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X.H., Liu Z.L., Sun M., Liu J., Wang Z.X., De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu Y., Li C., Luo Y., Li L., Liu J., Gui R. Silencing of long non-coding RNA MIAT sensitizes lung cancer cells to gefitinib by epigenetically regulating miR-34a. Front. Pharmacol. 2018;9:82. doi: 10.3389/fphar.2018.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amirmahani F., Vallian S., Asadi M.H. The LncRNA MIAT is identified as a regulator of stemness-associated transcript in glioma. Mol. Biol. Rep. 2023;50(1):517–530. doi: 10.1007/s11033-022-07962-5. [DOI] [PubMed] [Google Scholar]
- 62.Zheng R., Du M., Wang X., Xu W., Liang J., Wang W., et al. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol. Cancer. 2018;17(1):143. doi: 10.1186/s12943-018-0880-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamel L.M., Atef D.M., Mackawy A.M.H., Shalaby S.M., Abdelraheim N. Circulating long non-coding RNA GAS5 and SOX2OT as potential biomarkers for diagnosis and prognosis of non-small cell lung cancer. Biotechnol. Appl. Biochem. 2019;66(4):634–642. doi: 10.1002/bab.1764. [DOI] [PubMed] [Google Scholar]
- 64.Mei Y., Si J., Wang Y., Huang Z., Zhu H., Feng S., et al. Long noncoding RNA GAS5 suppresses tumorigenesis by inhibiting miR-23a expression in non-small cell lung cancer. Oncol. Res. 2017;25(6):1027–1037. doi: 10.3727/096504016X14822800040451. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Li S., Wang Q., Qiang Q., Shan H., Shi M., Chen B., et al. Sp1-mediated transcriptional regulation of MALAT1 plays a critical role in tumor. J. Cancer Res. Clin. Oncol. 2015;141(11):1909–1920. doi: 10.1007/s00432-015-1951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang R., Xia Y., Wang Z., Zheng J., Chen Y., Li X., et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017;490(2):406–414. doi: 10.1016/j.bbrc.2017.06.055. [DOI] [PubMed] [Google Scholar]
- 67.Zhang P., Zhou H., Lu K., Lu Y., Wang Y., Feng T. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. OncoTargets Ther. 2018;11:291–299. doi: 10.2147/OTT.S155134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu L., Wu Y., Tan D., Meng H., Wang K., Bai Y., et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2015;34(1):7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu B., Zhang H., Wang Z., Zhang F., Wei H., Li L. LncRNA CCAT1/miR-130a-3p axis increases cisplatin resistance in non-small-cell lung cancer cell line by targeting SOX4. Cancer Biol. Ther. 2017;18(12):974–983. doi: 10.1080/15384047.2017.1385679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xia Y., He Z., Liu B., Wang P., Chen Y. Downregulation of Meg3 enhances cisplatin resistance of lung cancer cells through activation of the WNT/beta-catenin signaling pathway. Mol. Med. Rep. 2015;12(3):4530–4537. doi: 10.3892/mmr.2015.3897. [DOI] [PubMed] [Google Scholar]
- 71.Xia H., Qu X.L., Liu L.Y., Qian D.H., Jing H.Y. LncRNA MEG3 promotes the sensitivity of vincristine by inhibiting autophagy in lung cancer chemotherapy. Eur. Rev. Med. Pharmacol. Sci. 2018;22(4):1020–1027. doi: 10.26355/eurrev_201802_14384. [DOI] [PubMed] [Google Scholar]
- 72.Sun W., Zu Y., Fu X., Deng Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol. Rep. 2017;38(6):3347–3354. doi: 10.3892/or.2017.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang J., Qi M., Fei X., Wang X., Wang K. Long non-coding RNA XIST: a novel oncogene in multiple cancers. Mol. Med. 2021;27(1):159. doi: 10.1186/s10020-021-00421-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shin V.Y., Chen J., Cheuk I.W., Siu M.T., Ho C.W., Wang X., et al. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019;10(4):270. doi: 10.1038/s41419-019-1513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li K., Yao T., Zhang Y., Li W., Wang Z. NEAT1 as a competing endogenous RNA in tumorigenesis of various cancers: role, mechanism and therapeutic potential. Int. J. Biol. Sci. 2021;17(13):3428–3440. doi: 10.7150/ijbs.62728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu L., Shu H., Xing L., Lv M.X., Li L., Xie Y.C., et al. Silencing long non-coding RNA NEAT1 suppresses the tumorigenesis of infantile hemangioma by competitively binding miR-33a-5p to stimulate HIF1alpha/NF-kappaB pathway. Mol. Med. Rep. 2020;22(4):3358–3366. doi: 10.3892/mmr.2020.11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feng S., Liu N., Chen X., Liu Y., An J. Long non-coding RNA NEAT1/miR-338-3p axis impedes the progression of acute myeloid leukemia via regulating CREBRF. Cancer Cell Int. 2020;20:112. doi: 10.1186/s12935-020-01182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Youness R., Assal R., Hafez M., Motaal A.A., Gad M. PO-347 sONE, a novel tumour suppressor lncRNA, with diminished expression level in young triple negative breast cancer (TNBC) patients with lymphnode metastasis and large tumour size. ESMO Open. 2018;3:A364–A365. [Google Scholar]
- 79.Ghafouri-Fard S., Azimi T., Taheri M. Myocardial infarction associated transcript (MIAT): review of its impact in the tumorigenesis. Biomed. Pharmacother. 2021;133 doi: 10.1016/j.biopha.2020.111040. [DOI] [PubMed] [Google Scholar]
- 80.Ribatti D., Tamma R., Annese T. Epithelial-mesenchymal transition in cancer: a historical overview. Transl. Oncol. 2020;13(6) doi: 10.1016/j.tranon.2020.100773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun C., Huang L., Li Z., Leng K., Xu Y., Jiang X., et al. Long non-coding RNA MIAT in development and disease: a new player in an old game. J. Biomed. Sci. 2018;25(1):23. doi: 10.1186/s12929-018-0427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ishii N., Ozaki K., Sato H., Mizuno H., Susumu S., Takahashi A., et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006;51(12):1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 83.Ma R., He X., Zhu X., Pang S., Yan B. Promoter polymorphisms in the lncRNA-MIAT gene associated with acute myocardial infarction in Chinese Han population: a case-control study. Biosci. Rep. 2020;40(2) doi: 10.1042/BSR20191203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guo X., Wang Y., Zheng D., Cheng X., Sun Y. LncRNA-MIAT promotes neural cell autophagy and apoptosis in ischemic stroke by up-regulating REDD1. Brain Res. 2021;1763 doi: 10.1016/j.brainres.2021.147436. [DOI] [PubMed] [Google Scholar]
- 85.Zhang X., Pan B., Qiu J., Ke X., Shen S., Wang X., et al. lncRNA MIAT targets miR-411-5p/STAT3/PD-L1 axis mediating hepatocellular carcinoma immune response. Int. J. Exp. Pathol. 2022;103(3):102–111. doi: 10.1111/iep.12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou J. LncRNA MIAT promotes hypoxia-induced H9C2 cell pyroptosis via binding to SF1 to inhibit CGRP transcription. Exp. Physiol. 2022;107(1):58–67. doi: 10.1113/EP089833. [DOI] [PubMed] [Google Scholar]
- 87.Yang L., Deng J., Ma W., Qiao A., Xu S., Yu Y., et al. Ablation of lncRNA Miat attenuates pathological hypertrophy and heart failure. Theranostics. 2021;11(16):7995–8007. doi: 10.7150/thno.50990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hao S., Wang L., Zhao K., Zhu X., Ye F. Rs1894720 polymorphism in MIAT increased susceptibility to age-related hearing loss by modulating the activation of miR-29b/SIRT1/PGC-1alpha signaling. J. Cell. Biochem. 2019;120(4):4975–4986. doi: 10.1002/jcb.27773. [DOI] [PubMed] [Google Scholar]
- 89.Rao S.Q., Hu H.L., Ye N., Shen Y., Xu Q. Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr. Res. 2015;166(1–3):125–130. doi: 10.1016/j.schres.2015.04.032. [DOI] [PubMed] [Google Scholar]
- 90.Huang Z., Zhou J.K., Peng Y., He W., Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer. 2020;19(1):77. doi: 10.1186/s12943-020-01188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.The R.C., Petrov A.I., Kay S.J.E., Kalvari I., Howe K.L., Gray K.A., et al. RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 2017;45(D1):D128–D134. doi: 10.1093/nar/gkw1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiyuan L., Dechao B., Liang S., Yang W., Shuangsang F., Hui L., et al. Using the NONCODE database resource. Curr. Protoc. Bioinformatics. 2017;58 doi: 10.1002/cpbi.25. 12 16 11-12 16 19. [DOI] [PubMed] [Google Scholar]
- 93.Volders P.J., Helsens K., Wang X., Menten B., Martens L., Gevaert K., et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41(Database issue):D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao H., Yin X., Xu H., Liu K., Liu W., Wang L., et al. LncTarD 2.0: an updated comprehensive database for experimentally-supported functional lncRNA-target regulations in human diseases. Nucleic Acids Res. 2023;51(D1):D199–D207. doi: 10.1093/nar/gkac984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cheng L., Wang P., Tian R., Wang S., Guo Q., Luo M., et al. LncRNA2Target v2.0: a comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019;47(D1):D140–D144. doi: 10.1093/nar/gky1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rincon-Riveros A., Morales D., Rodriguez J.A., Villegas V.E., Lopez-Kleine L. Bioinformatic tools for the analysis and prediction of ncRNA interactions. Int. J. Mol. Sci. 2021;22(21) doi: 10.3390/ijms222111397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Youness R.A., El-Tayebi H.M., Assal R.A., Hosny K., Esmat G., Abdelaziz A.I. MicroRNA-486-5p enhances hepatocellular carcinoma tumor suppression through repression of IGF-1R and its downstream mTOR, STAT3 and c-Myc. Oncol. Lett. 2016;12(4):2567–2573. doi: 10.3892/ol.2016.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Youness R.A., Rahmoon M.A., Assal R.A., Gomaa A.I., Hamza M.T., Waked I., et al. Contradicting interplay between insulin-like growth factor-1 and miR-486-5p in primary NK cells and hepatoma cell lines with a contemporary inhibitory impact on HCC tumor progression. Growth Factors. 2016;34(3–4):128–140. doi: 10.1080/08977194.2016.1200571. [DOI] [PubMed] [Google Scholar]
- 99.Shaalan Y.M., Handoussa H., Youness R.A., Assal R.A., El-Khatib A.H., Linscheid M.W., et al. Destabilizing the interplay between miR-1275 and IGF2BPs by Tamarix articulata and quercetin in hepatocellular carcinoma. Nat. Prod. Res. 2018;32(18):2217–2220. doi: 10.1080/14786419.2017.1366478. [DOI] [PubMed] [Google Scholar]
- 100.Youssef S.S., Abbas E., Youness R.A., Elemeery M.N., Nasr A.S., Seif S. PNPLA3 and IL 28B signature for predicting susceptibility to chronic hepatitis C infection and fibrosis progression. Arch. Physiol. Biochem. 2022;128(2):483–489. doi: 10.1080/13813455.2019.1694039. [DOI] [PubMed] [Google Scholar]
- 101.ElKhouly A.M., Youness R.A., Gad M.Z. MicroRNA-486-5p and microRNA-486-3p: multifaceted pleiotropic mediators in oncological and non-oncological conditions. Noncoding RNA Res. 2020;5(1):11–21. doi: 10.1016/j.ncrna.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Youssef S.S., Youness R.A., Abbas E.A.E., Osman N.M., A E.L., El-Kassas M. miR-516a-3P, a potential circulating biomarker in hepatocellular carcinoma, correlated with rs738409 polymorphism in PNPLA3. Méd.. 2022;19(6):483–493. doi: 10.2217/pme-2022-0005. [DOI] [PubMed] [Google Scholar]
- 103.Ahmed Youness R., Amr Assal R., Mohamed Ezzat S., Zakaria Gad M., Abdel Motaal A. A methoxylated quercetin glycoside harnesses HCC tumor progression in a TP53/miR-15/miR-16 dependent manner. Nat. Prod. Res. 2020;34(10):1475–1480. doi: 10.1080/14786419.2018.1509326. [DOI] [PubMed] [Google Scholar]
- 104.Zhan Y., Tao Q., Meng Q., Zhang R., Lin L., Li X., et al. LncRNA-MIAT activates hepatic stellate cells via regulating Hippo pathway and epithelial-to-mesenchymal transition. Commun. Biol. 2023;6(1):285. doi: 10.1038/s42003-023-04670-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang Z., Wang S., Liu W. EMT-related long non-coding RNA in hepatocellular carcinoma: a study with TCGA database. Biochem. Biophys. Res. Commun. 2018;503(3):1530–1536. doi: 10.1016/j.bbrc.2018.07.075. [DOI] [PubMed] [Google Scholar]
- 106.Huang X., Gao Y., Qin J., Lu S. lncRNA MIAT promotes proliferation and invasion of HCC cells via sponging miR-214. Am. J. Physiol. Gastrointest. Liver Physiol. 2018;314(5):G559–G565. doi: 10.1152/ajpgi.00242.2017. [DOI] [PubMed] [Google Scholar]
- 107.Xia H., Ooi L.L., Hui K.M. MiR-214 targets beta-catenin pathway to suppress invasion, stem-like traits and recurrence of human hepatocellular carcinoma. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0044206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiang Y., Huang Y., Sun H., Pan Y., Wu M., Zhang J. Deregulation of miR-520d-3p promotes hepatocellular carcinoma development via lncRNA MIAT regulation and EPHA2 signaling activation. Biomed. Pharmacother. 2019;109:1630–1639. doi: 10.1016/j.biopha.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 109.Huang C., Chen Z., He Y., He Z., Ban Z., Zhu Y., et al. EphA2 promotes tumorigenicity of cervical cancer by up-regulating CDK6. J. Cell Mol. Med. 2021;25(6):2967–2975. doi: 10.1111/jcmm.16337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li H., Sun Q., Han B., Yu X., Hu B., Hu S. MiR-26b inhibits hepatocellular carcinoma cell proliferation, migration, and invasion by targeting EphA2. Int. J. Clin. Exp. Pathol. 2015;8(5):4782–4790. [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao L., Hu K., Cao J., Wang P., Li J., Zeng K., et al. lncRNA miat functions as a ceRNA to upregulate sirt1 by sponging miR-22-3p in HCC cellular senescence. Aging (Albany NY) 2019;11(17):7098–7122. doi: 10.18632/aging.102240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rahman S., Islam R. Mammalian Sirt1: insights on its biological functions. Cell Commun. Signal. 2011;9(1):11. doi: 10.1186/1478-811X-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abdel-Latif M., Youness R.A. Why natural killer cells in triple negative breast cancer? World J. Clin. Oncol. 2020;11(7):464–476. doi: 10.5306/wjco.v11.i7.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rahmoon M.A., Youness R.A., Gomaa A.I., Hamza M.T., Waked I., El Tayebi H.M., et al. MiR-615-5p depresses natural killer cells cytotoxicity through repressing IGF-1R in hepatocellular carcinoma patients. Growth Factors. 2017;35(2–3):76–87. doi: 10.1080/08977194.2017.1354859. [DOI] [PubMed] [Google Scholar]
- 115.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 116.Elemam N.M., Talaat I.M., Assal R.A., Youness R.A. Editorial: understanding the crosstalk between immune cells and the tumor microenvironment in cancer and its implications for immunotherapy. Front. Med. 2023;10 doi: 10.3389/fmed.2023.1202581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Elemam N.M., Youness R.A., Hussein A., Shihab I., Yakout N.M., Elwany Y.N., et al. Expression of GPR68, an acid-sensing orphan G protein-coupled receptor, in breast cancer. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.847543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Housman G., Byler S., Heerboth S., Lapinska K., Longacre M., Snyder N., et al. Drug resistance in cancer: an overview. Cancers. 2014;6(3):1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sakuishi K., Ngiow S.F., Sullivan J.M., Teng M.W., Kuchroo V.K., Smyth M.J., et al. TIM3(+)FOXP3(+) regulatory T cells are tissue-specific promoters of T-cell dysfunction in cancer. OncoImmunology. 2013;2(4) doi: 10.4161/onci.23849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cao Z., Pan X., Yang Y., Huang Y., Shen H.B. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34(13):2185–2194. doi: 10.1093/bioinformatics/bty085. [DOI] [PubMed] [Google Scholar]
- 121.Peng L., Chen Y., Ou Q., Wang X., Tang N. LncRNA MIAT correlates with immune infiltrates and drug reactions in hepatocellular carcinoma. Int. Immunopharm.. 2020;89(Pt A) doi: 10.1016/j.intimp.2020.107071. [DOI] [PubMed] [Google Scholar]
- 122.Ye T., Feng J., Cui M., Yang J., Wan X., Xie D., et al. LncRNA MIAT services as a noninvasive biomarker for diagnosis and correlated with immune infiltrates in breast cancer. Int. J. Womens Health. 2021;13:991–1004. doi: 10.2147/IJWH.S312714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.