Abstract
Breast milk serves as a vital source of essential nutrients for infants. However, human milk contamination via the transfer of environmental chemicals from maternal exposome is a significant concern for infant health. The milk to plasma concentration (M/P) ratio is a critical metric that quantifies the extent to which these chemicals transfer from maternal plasma into breast milk, impacting infant exposure. Machine learning-based predictive toxicology models can be valuable in predicting chemicals with a high propensity to transfer into human milk. To this end, we build such classification- and regression-based models by employing multiple machine learning algorithms and leveraging the largest curated data set, to date, of 375 chemicals with known milk-to-plasma concentration (M/P) ratios. Our support vector machine (SVM)-based classifier outperforms other models in terms of different performance metrics, when evaluated on both (internal) test data and an external test data set. Specifically, the SVM-based classifier on (internal) test data achieved a classification accuracy of 77.33%, a specificity of 84%, a sensitivity of 64%, and an F-score of 65.31%. When evaluated on an external test data set, our SVM-based classifier is found to be generalizable with a sensitivity of 77.78%. While we were able to build highly predictive classification models, our best regression models for predicting the M/P ratio of chemicals could achieve only moderate R2 values on the (internal) test data. As noted in the earlier literature, our study also highlights the challenges in developing accurate regression models for predicting the M/P ratio of xenobiotic chemicals. Overall, this study attests to the immense potential of predictive computational toxicology models in characterizing the myriad of chemicals in the human exposome.
Introduction
Breast milk is widely recognized as the optimal source of nutrition for infants and provides numerous benefits to both infants and the mother. Published studies, including studies by Rollins et al.,1 have shown that breastfeeding contributes toward a world that is healthier, better educated, more equitable, and more environmentally sustainable. Breast milk is also known to provide protection to the infant from health complications such as cardiovascular disease, sudden infant death syndrome, growth faltering, and inflammatory bowel disease.2 Notably, breastfeeding has been shown to be associated with reduced risk in mothers for premenopausal breast cancer, ovarian cancer, retained gestational weight gain, type 2 diabetes, myocardial infarction, and metabolic syndrome.3 Moreover, breast milk is an eco-friendly and cost-effective option compared to using infant formula.4
In spite of the benefits associated with breastfeeding, there is a legitimate concern about the potential exposure of infants to environmental chemicals (including drugs) through lactation.5,6 Pregnant women and lactating mothers are exposed to a wide range of environmental chemicals via food, medication, personal care products, and environmental pollutants.7−9 Exposure to such chemicals can affect the health of both mothers and the development of breastfed infants.10,11 Wild12 proposed the concept of “exposome” to describe the nongenetic factors that influence the health and disease of an individual, starting from the prenatal period. In essence, exposome captures the sum total of all environmental exposures that an individual experiences throughout their life and the associated health effects.12−15 Consequently, due to the potential impact on infant and maternal health, there is significant interest in characterizing environmental chemicals with high propensity to transfer from maternal plasma to human milk, i.e., potential human milk contaminants in the chemical exposome.5,6 In this direction, some of the authors of this study had previously built an online knowledgebase, ExHuMId,5 that compiles experimentally detected human milk contaminants from published studies analyzing breast milk samples from India.
Notably, experimentally measured milk-to-plasma concentration (M/P) ratio is used to identify the equilibrium concentration of a chemical in maternal plasma in comparison to breast milk, and this M/P ratio is used as an indicator for the propensity of a xenobiotic chemical to enter human milk.16−18 In terms of environmental chemicals in the human exposome, several studies have shown lipophilic substances to have a high potential to transfer into human milk from maternal plasma via passive diffusion.16−20 Furthermore, a systematic analysis based on available M/P ratios for human milk contaminants in ExHuMId revealed structural properties of chemicals that can influence their transfer into human milk.5 In a nutshell, previous analyses have led to the realization that the physicochemical properties of xenobiotic chemicals influence their potential to transfer from maternal plasma to human milk.5,16−20
Experimental measurement of a xenobiotic chemical’s propensity to enter human milk is both a difficult task and ethically impractical. Since the 1980s, there have been attempts to predict the M/P ratio for xenobiotic chemicals, and the initial studies were based on methods incorporating the physicochemical properties of the chemicals while ignoring clinical information or effects of active transport.21,22 Following the initial attempts, several studies employing machine learning algorithms and the quantitative structure–activity relationship (QSAR) principle have been proposed.23−29 Such predictive models have been built on the QSAR principle that the biological or chemical activity of a compound can be quantitatively related to its molecular structure and physicochemical properties. For instance, Yap and Chen developed a regression model based on a general regression neural network and reported an R2 value of 0.677 and mean squared error (MSE) value of 0.206 on test data.29 Katritzky et al. built a predictive model by dividing a data set of 100 chemicals into three subsets and reported an average R2 value of 0.763.27 Abraham et al. employed an artificial neural network to predict the logarithmically transformed M/P ratios of chemicals and reported a root-mean-square error (RMSE) of 0.109 on internal test data and 0.09 on external test data.23 Fatemi and Ghorbanzad’e developed a counter propagation artificial neural network-based classification model and the authors reported an accuracy of 100% on test data and 90% on external test data.25 Kar and Roy developed both classification and regression models using linear discriminant analysis (LDA) and multiple linear regression (MLR), respectively.26 For their regression model, the authors obtained an R2 value of 0.7 on train data, and for their classification model, the authors obtained an accuracy of 56.82%, sensitivity of 63.16%, and F-score of 55.81% on test data. Wanat et al. developed a random forest (RF)-based regression model and reported an R2 value of 0.29 on test data.28
Despite the availability of the above models, the prediction of transfer into milk from maternal plasma for actively transported drugs remains a challenging endeavor.30 The primary aim of this study is to build accurate machine learning models for predicting the high propensity of transfer of xenobiotic chemicals from maternal plasma to human breast milk by leveraging, to date, the largest curated data set of chemicals with experimentally determined M/P ratios. Notably, our approach upholds the three essential principles of data science: repeatability, reproducibility, and replicability. To accomplish this, we manually curated a data set of 375 chemicals along with their experimentally determined M/P ratios from the previously published scientific literature (Table S1). Thereafter, we computed 1875 molecular descriptors for each chemical in our data set, and the computed descriptors capture the structural and physicochemical features of the chemical data set. Subsequently, we utilized the computed descriptors for the chemicals as the features and known M/P ratios as the target variable along with multiple machine learning algorithms to build predictive models for the purpose. Our workflow (Figure 1) led to reliable classification models for predicting chemicals with a “high risk” of transfer from maternal plasma to human milk. To assess the applicability and generalizability of the built models, we validated our classification models by leveraging a large external test data set of 202 chemicals (Table S2), and this evaluation of an external test data set highlighted the robustness of the results obtained in this study. Additionally, we applied our best performing models on the approved drugs and experimental drugs to predict the propensity of a drug to transfer into human breast milk. Overall, this study also takes a step toward achieving FAIR31 compliance by publicly releasing our curated chemical data sets and computer codes through the associated GitHub repository (https://github.com/asamallab/M-by-P-ratio-Pred).
Figure 1.
Schematic diagram summarizing the workflow to build the classification- and regression-based machine learning models to predict xenobiotic chemicals with a high propensity to transfer from maternal plasma to human milk. This figure shows the key steps involved in data curation, feature generation, data preprocessing, feature selection, and the training and evaluation of classification- and regression-based machine learning models. The word clouds in this figure were generated using https://www.wordclouds.com.
Results and Discussion
Workflow for Building Classification and Regression Models
The aim of this study is to build machine learning models to predict xenobiotic chemicals with a high propensity to transfer from maternal plasma to human milk. First, we build classification-based models that can accurately categorize chemicals as either “high risk” or “low risk” of transfer from maternal plasma to human milk. Second, we build regression-based models that can predict the M/P concentration ratios for xenobiotic chemicals. Figure 1 presents a schematic workflow of the different steps undertaken in this study to build the classification- and regression-based machine learning models.
To build the machine learning models, we leveraged a curated data set of 375 chemicals with experimentally determined M/P ratios compiled from Vasios et al.18 and other published literature25,28,32,33 (Methods; Table S1). The descriptors for chemicals are generated as described in the Methods section. After descriptors are generated, the data are divided into train and test sets, with target variables designated for both classification and regression models (Methods). Figure S1 shows the distribution of M/P ratios for the complete data set of 375 chemicals, training set (300 chemicals), and internal test data set (75 chemicals). Prior to training, data normalization and feature selection are performed (Methods).
To build the classification models, we employed five different machine learning algorithms, namely, support vector machine (SVM), extreme gradient boosting (XGBoost), LDA, multilayer perceptron (MLP), and RF (Methods). Following the feature selection (Methods), to optimize our classification models, we performed hyperparameter tuning using GridSearchCV in Scikit-learn (Methods).34 For each of the five different classification algorithms, we selected the top 1, 2, 3, 4, and 5 ranked features and thereafter trained and evaluated the corresponding model. The best outcome for the five different classification algorithms is reported in Table 1. Notably, the five different classification algorithms were also evaluated using an external test data set of 202 chemicals that have been experimentally detected in human milk (Methods; Table S2; Table 1).
Table 1. Evaluation of the Best Classification Models Built Using Five Different Algorithms to Categorize Chemicals into “High Risk” or “Low Risk” Classes for Transfer from Maternal Plasma to Human Milka.
| algorithm | accuracy | sensitivity |
specificity | F-score | |
|---|---|---|---|---|---|
| (internal) test data (%) | (internal) test data (%) | external test data set (%) | (internal) test data (%) | (internal) test data (%) | |
| SVM | 77.33 | 64 | 77.78 | 84 | 65.31 |
| XGBoost | 78.67 | 48 | 50.79 | 94 | 60 |
| LDA | 69.33 | 24 | 71.96 | 92 | 34.29 |
| MLP | 74.67 | 60 | 77.22 | 82 | 61.22 |
| RF | 73.33 | 56 | 68.89 | 82 | 58.33 |
For each model, the classification accuracy is listed for the (internal) test data. The sensitivity is listed for both (internal) test data and the external test data set, respectively. The specificity and F-score are listed for the (internal) test data.
A regression algorithm is a statistical method that predicts the continuous values of the dependent variable based on the independent variables (features). To build the regression models to predict the M/P ratio of a chemical, we employed three different machine learning algorithms, namely, SVM, XGBoost, and RF (Methods). Upon completing feature selection, to optimize our regression models, we also performed hyperparameter tuning using GridSearchCV in Scikit-learn (Methods).34 For each of the three different regression algorithms, we selected the top 5, 10, 15, 20, 25, 30, 35, and 40 features and thereafter trained and evaluated the corresponding model. The best outcome for the three different regression algorithms is reported in Table 2.
Table 2. Evaluation of the Best Regression Models Built Using the Three Different Algorithms to Predict the M/P Ratio of the Chemicalsa.
| algorithm |
R2 |
MSE |
||
|---|---|---|---|---|
| train data | (internal) test data | train data | (internal) test data | |
| SVM | 0.6909 | 0.4460 | 0.0984 | 0.1978 |
| XGBoost | 0.9323 | 0.4785 | 0.02154 | 0.1862 |
| RF | 0.9277 | 0.4901 | 0.0230 | 0.1820 |
For each regression model, we report the R2 and the MSE value for both train data and (internal) test data.
Performance of the Classification Models
We next evaluated the performance of the five different classification algorithms, namely, SVM, XGBoost, LDA, MLP, and RF, which were used to build classification models in this study (Methods; Table 1).
In Table 1, we present the accuracy of the classification models on the (internal) test data of 75 chemicals. On train data, the models exhibited varying levels of accuracy. Specifically, SVM, XGBoost, LDA, MLP, and RF achieved an accuracy of 83.67, 96.33, 76, 91.33, and 94%, respectively, on train data. Moving on to (internal) test data (which is independent of the train data), the five models SVM, XGBoost, LDA, MLP, and RF achieved an accuracy of 77.33, 78.67, 69.33, 74.67, and 73.33%, respectively (Table 1). Although the accuracy values are relatively high for the five models on (internal) test data, it is important to consider other performance metrics such as sensitivity, specificity, and F-score.
Sensitivity measures a model’s ability to accurately predict “high risk” chemicals. On (internal) test data, SVM achieved a sensitivity of 64%, followed by MLP with 60%, RF with 56%, XGBoost with 48%, and LDA with 24% (Table 1). Similarly, specificity measures a model’s ability to accurately predict “low risk” chemicals. On (internal) test data, XGBoost achieved the highest specificity of 94%, followed by LDA with 92%, SVM with 84%, MLP with 82%, and RF with 82% (Table 1). F-score is a measure which combines precision and recall (sensitivity), and thus, provides an overall measure of a model’s performance. On (internal) test data, SVM achieved the highest F-score of 65.31%, followed by MLP with 61.22%, XGBoost with 60%, RF with 58.33%, and LDA with 34.29% (Table 1).
To summarize, in terms of accuracy, sensitivity, specificity, and F-score, SVM and MLP showed higher performance on (internal) test data. In contrast, LDA showed lower performance in terms of most evaluation metrics. In comparison, XGBoost and RF achieved reasonable performance but were slightly lower than SVM and MLP.
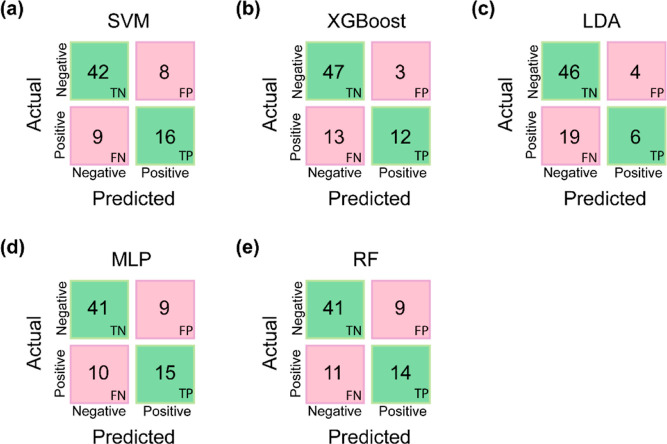
Figure 2 displays the confusion matrix for the five best models corresponding to the five different classification algorithms. The confusion matrix provides the true positive, true negative, false positive, and false negative predictions by the five different classification algorithms, namely, SVM, XGBoost, LDA, MLP, and RF on the (internal) test data of 75 chemicals. Thus, the confusion matrix provides a fine print of the performance of the classification models on (internal) test data which concurs with the results presented in Table 1.
Figure 2.
Confusion matrix depicting the performance of the best classification models corresponding to five classification algorithms, namely, (a) SVM, (b) XGBoost, (c) LDA, (d) MLP, and (e) RF on the (internal) test data of 75 chemicals. Note: TP, TN, FP, and FN stand for true positive, true negative, false positive, and false negative.
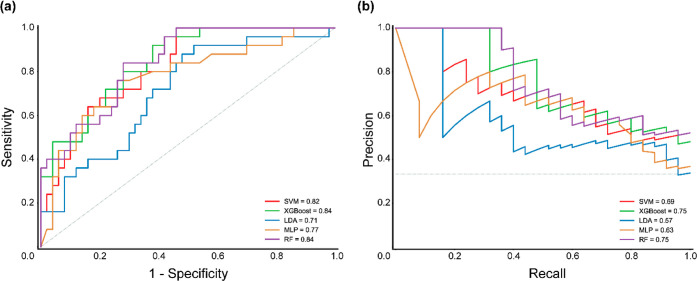
Furthermore, in addition to the evaluation metrics mentioned above, we also computed the area under the curve (AUC) values for the five different classification algorithms. Note that the AUC values provide a measure of a classifier’s ability to distinguish between the positive and negative classes in a data set. Specifically, for the best classification models obtained using SVM, XGBoost, LDA, MLP, and RF, the AUC values were 0.82, 0.84, 0.71, 0.77, and 0.84, respectively. These results indicate that XGBoost, RF, and SVM classifiers achieve high AUC values, and thus, are better at discriminating between “high risk” and “low risk” chemicals in test data. Figure 3a displays the receiver operating characteristic (ROC) curves for the best models built by using the five different classification algorithms.
Figure 3.
ROC curve and precision–recall curve for evaluating the best models built using the five different classification algorithms. (a) ROC curve plotted with sensitivity on the y-axis and 1-specificity on the x-axis. (b) Precision–recall curve plotted with precision on the y-axis and recall (sensitivity) on the x-axis.
Moreover, we employed the area under the precision–recall curve (AUPRC) as another evaluation metric to assess the performance of the five classification algorithms. Specifically, for the best classification models obtained using SVM, XGBoost, LDA, MLP, and RF, the AUPRC values were 0.69, 0.75, 0.57, 0.63, and 0.75, respectively. These results also indicate that XGBoost and RF classifiers followed by SVM achieve high AUPRC values, similar to the AUC value. Figure 3b displays the precision–recall curves for the best models built using the five different classification algorithms.
Subsequently, we evaluated the generalizability of our best classification models by leveraging an external test data set, comprising 202 chemicals, with a high risk of transfer from maternal plasma to human milk (Methods; Table S2). Note that we checked the domain of applicability of data points in the external test data set before evaluating our classification models (Methods). In Table 1, we present the sensitivity of the five different classification algorithms on an external test data set. By comparing the performance of the five different algorithms on an external data set, it is observed that SVM and MLP achieved the highest sensitivity values of 77.78 and 77.22%, respectively, followed by LDA with 71.96%, RF with 68.89%, and XGBoost with 50.79% (Table 1). In other words, these results indicate that SVM and MLP outperformed the remaining three algorithms in accurately classifying positive instances in the external test data set. Moreover, the high sensitivities achieved by SVM and MLP on the external test data set suggest that these two algorithms are better capable of capturing the relevant patterns and characteristics associated with positive instances (“high risk” chemicals), even though the train data is biased toward negative instances (“low risk” chemicals).
Overall, we find that SVM- and MLP-based classifiers have high predictive ability on both (internal) test data and external test data sets, and thus, the two models are robust and generalizable for the task of accurately predicting high-risk chemicals in real-world scenarios (Table 1). Furthermore, among the two models SVM and MLP, we believe that the SVM-based classifier is the more promising model for the classification of xenobiotic chemicals into “high risk” or “low risk” of transfer from maternal plasma to human milk.
We applied the SVM- and MLP-based classifier to predict the risk of approved and experimental drugs being transferred into human breast milk. For this purpose, the approved and experimental drugs were obtained from DrugBank35 and were processed similarly to the chemicals in the external test set. 722 and 622 approved drugs were classified as “high risk” to transfer into human breast milk by SVM and MLP, respectively. Similarly, 1112 and 951 experimental drugs were classified as “high risk” to transfer into human breast milk (Tables S3 and S4).
Importantly, the above-mentioned results also highlight the importance of assessing classifiers on external test data sets to ensure the generalizability of the built models.36,37 In particular, such an approach has practical implications while developing reliable machine learning models for drug safety assessment, environmental monitoring, and public health.36,37
Performance of the Regression Models
We next evaluated the performance of the three different regression algorithms, namely, SVM, XGBoost, and RF, which were used to build models for the prediction of the M/P ratio of xenobiotic chemicals (Methods; Table 2). For the best regression models obtained using SVM, XGBoost, and RF, the coefficient of determination (R2) values obtained on the train and (internal) test data were (0.6909, 0.4460), (0.9323, 0.4785), and (0.9277, 0.4901), respectively. The obtained R2 values not only suggest a moderate level of prediction performance but also indicate the possibility of overfitting, whereby the models may be fitting the train data more closely than the test data.
To the best of our knowledge, previous studies23,24,26−29 on building regression-based models for predicting the M/P ratio of xenobiotic chemicals have employed much smaller data sets in comparison to our data set of 375 chemicals with known M/P ratios, and moreover, in many of these published studies,23,24,26 the authors have not reported the R2 value on test data. Moreover, none of the previous studies on this topic have published the source code of the developed predictive models. Due to these reasons, it is difficult to compare the performance of our regression models with those reported in the previous studies.
In spite of our attempts to build a highly predictive regression model for the M/P ratio of xenobiotic chemicals, we were unable to improve the R2 values on the test data beyond those reported in Table 2 using different regression algorithms. In fact, other than SVM, XGBoost, and RF algorithms, we also tried another machine learning algorithm, MLP, to build regression models for prediction of the M/P ratio of xenobiotic chemicals. However, MLP yielded highly unsatisfactory regression-based models, and therefore, we decided not to report the results from the MLP-based regression model in this manuscript. We remark that the difficulty encountered by us in developing a highly predictive regression model for the M/P ratio of xenobiotic chemicals has also been faced by authors of previous studies; in particular, this issue is well documented in the published literature.23,27
While our regression models achieve moderate R2 values on (internal) test data, our classification models reported in the previous section achieve high accuracy on (internal) test data. Therefore, we decided to evaluate our best regression models for their ability to perform classification on (internal) test data. Note that if a regression model predicts the M/P ratio of a chemical to be ≥ log 2, then the chemical belongs to the “high risk” category, else the chemical belongs to the “low risk” category. We find that our best regression models for SVM, XGBoost, and RF achieve a classification based on the regression accuracy of 90.67, 95.33, and 95.33%, respectively, on train data, and 74.67, 72, and 73.33%, respectively, on (internal) test data (Table 3). We also evaluated the sensitivity, specificity, and F-score of the classification based on regression models (Table 3). The sensitivity, specificity, and F-score for SVM were 60, 82, and 61.22%, respectively, for XGBoost were 64, 76, and 60.38%, respectively, and for RF were 68, 76, and 62.96%, respectively (Table 3). Overall, the results from classification based on regression signify that the models perform reasonably well in categorizing the M/P ratio of xenobiotic chemicals into the two classes “high risk” and “low risk” on (internal) test data.
Table 3. Evaluation of the Classification Based on Regression Models Built Using the Three Different Algorithms to Predict the M/P Ratio of the Chemicalsa.
| algorithm | accuracy | sensitivity |
specificity | F-score | |
|---|---|---|---|---|---|
| (internal) test data (%) | (internal) test data (%) | external test data set (%) | (internal) test data (%) | (internal) test data (%) | |
| SVM | 74.67 | 60 | 60.58 | 82 | 61.22 |
| XGBoost | 72 | 64 | 65.89 | 76 | 60.38 |
| RF | 73.33 | 68 | 75.97 | 76 | 62.96 |
For each model, the classification accuracy is listed for the (internal) test data. The sensitivity is listed for both (internal) test data and the external test data set. The specificity and F-score are listed for (internal) test data.
We also evaluated the classification based on regression on an external test data set. Prior to this evaluation, we applied a domain of applicability approach to ensure that the external test data set falls within the range of applicability for our models (Methods). On the external test data set, SVM, XGBoost, and RF achieved a sensitivity of 60.58, 65.89, and 75.97%, respectively (Table 3). These results underscore that the RF model exhibits the highest sensitivity on both (internal) test data and external test data sets, indicating its effectiveness in correctly detecting positive instances (“high risk” chemicals). In comparison, the XGBoost model displayed reasonable sensitivity, while the SVM model showed comparatively lower sensitivity (Table 3).
Overall, we find that the regression models achieve moderate R2 values, while the classification based on regression models displays reasonable performance on classification of xenobiotic chemicals into “high risk” and “low risk” categories. These results also highlight the complexity and challenges associated with accurate prediction of the M/P ratio of xenobiotic chemicals and emphasize the need for further research toward creation of improved experimental data sets in order to enhance model performance. Lastly, we remark that none of the three regression-based models for classifying the M/P ratio of xenobiotic chemicals, in terms of classification accuracy on the (internal) test data and external test data set, could outperform the SVM-based classification model, which was solely developed for the classification task (Table 1; Table 3).
We applied the classification based on RF regression to predict the risk of approved and experimental drugs to transfer into human breast milk. 709 approved drugs and 1082 experimental drugs were classified as “high risk” to transfer into human breast milk by classification based on RF regression (Tables S3 and S4).
Comparison with Earlier Models
In this subsection, we compare the performance of models built in this study with previously published models for the prediction of chemicals with a high propensity to transfer from maternal plasma to human milk. Earlier works employed both classification and regression to make such predictions.
Fatemi and Ghorbanzad’e built a counter propagation artificial neural network-based classification model using a train data set of 124 chemicals.25 When evaluated on a test data set of 20 chemicals, this model yielded 100% test classification accuracy (Table S5). While this may seem promising, the reported 100% test classification accuracy on a small test data set necessitates further investigation of the generalizability of this model. Kar and Roy built an LDA-based classification model using a train data set of 97 chemicals.26 When evaluated on a test data set of 88 chemicals, this model had a test classification accuracy of 56.82%, test sensitivity of 63.16%, and test F-score of 55.81% (Table S5). In the present study, we recognized the importance of a larger data set in improving classification performance. Therefore, we compiled and curated a larger data set of 375 chemicals with known M/P ratios, of which 300 chemicals are used as train data and 75 chemicals are used as (internal) test data. Notably, by evaluating our classification models on the (internal) test data of 75 chemicals (which are not part of the train data), we showed that our SVM-based classification model achieved a test classification accuracy of 77.33%, test sensitivity of 64%, and test F-score of 65.32%, which is a significant improvement over the model developed earlier by Kar and Roy (2013).26
Moving on to earlier publications on regression-based prediction of xenobiotic chemicals with a high propensity to transfer from maternal plasma to human milk, we find that there is limited reporting of regression R2 in such previous studies (Table S6). Agatonovic-Kustrin et al. developed a genetic neural network-based model to predict the degree of drug transfer into breast milk using a data set of 60 drugs and their experimentally derived M/P ratios, and the authors reported an R2 value >0.96 on train data and a root-mean-square (RMS) value of 0.425 on test data for their best model.24 Yap and Chen built their model using train data of 102 chemicals and test data of 20 chemicals, and they reported a test R2 of 0.677 and test MSE of 0.206 (Table S6).29 Katritzky et al. built a QSAR model by dividing a data set of 100 chemicals into three subsets.27 Three training data sets were prepared by considering the combinations of any two subsets, and thereafter, equations were obtained for each training data set. These equations were then used to predict the log(M/P) values for the corresponding test data sets. Katritzky et al. calculated the R2 obtained in the three test data sets and reported an average R2 of 0.763 (Table S6).27 Abraham et al. employed an artificial neural network to predict the logarithmically transformed M/P ratios of chemicals by using a data set of 179 drugs and environmental pollutants, of which 135 chemicals were in the train data, 22 chemicals were in the test data, and further 22 chemicals were in the external test data.23 For the best model, Abraham et al. reported an RMSE of 0.056 on train data, 0.109 on test data, and 0.09 on external test data.23 However, Abraham et al. did not report the R2 values for their model.23 Kar and Roy developed regression models using MLR and a data set consisting of 97 chemicals in the train data and 88 chemicals in the test data.26 For their regression model, the authors obtained an R2 value of 0.7 on train data. However, Kar and Roy did not report the R2 value for their regression model on test data.26 Wanat et al. built their model using train data of 58 chemicals and test data of 25 chemicals, and they reported a test R2 of 0.29 (Table S6).28 We remark that the above-mentioned previously published studies developed their regression models using relatively smaller data sets (in comparison to our models), and this may restrict the domain of applicability and limit the generalizability of the earlier published models. Additionally, the earlier studies also acknowledged the difficulty in achieving high test R2 values due to limited experimental data and the challenges associated with prediction of the M/P ratio for chemicals.23,27 Among the three regression-based models developed in this study, our RF-based model achieved a test R2 of 0.49 and an MSE of 0.1820 on the (internal) test data (Table 2). Thus, our results align with the limitations mentioned in earlier efforts, wherein previous studies also encountered challenges in achieving high R2 values on test data.
It is also worth noting that many previous studies on this subject do not adequately describe the methods (including technical information) used to build predictive models. This lack of transparency, along with limited reporting of train and test data sets and unavailability of codes for previous models, hindered direct comparison of our models with earlier works. To facilitate future research in this direction, our study provides detailed documentation of the technical aspects and methods employed to build the predictive models, and moreover, we have made the train data, test data, external test data set, and codes for the built models openly accessible via our GitHub repository: https://github.com/asamallab/M-by-P-ratio-Pred.
Overall, the present study highlights the significance of the chemical data set size in building machine learning models for both classification and regression tasks. Furthermore, our study shows the importance of an external test data set in evaluating the generalizability of built models. By utilizing a much larger train and test data of 375 chemicals plus an external test data set of 202 chemicals, we created classification- and regression-based models with better performance in terms of predicting the propensity of chemicals to transfer from maternal plasma into human milk.
Conclusions
In this study, we built and evaluated multiple machine learning models with the aim of classifying xenobiotic chemicals into “high risk” and “low risk” classes for potential transfer from maternal plasma to human milk based on M/P concentration ratios of chemicals. We find that our SVM-based classifier outperforms other classification models in terms of different evaluation metrics (Table 1; Figure 2). In particular, the SVM-based classifier on (internal) test data achieved a specificity of 84%, recall (sensitivity) of 64%, and F-score of 65.31%. Furthermore, from the confusion matrix, it can be seen that the SVM-based classifier achieves higher true positives and fewer false positives (Figure 2), and this suggests that the model is less likely to misclassify a “high risk” chemical as a “low risk” chemical. Importantly, based on evaluation of an external test data set, our SVM-based classification model is found to be generalizable, which further strengthens the validity of our approach. In brief, these results attest to the potential of our SVM-based classification model to serve as a valuable tool for predicting environmental chemicals whose exposure could pose a high risk to maternal and infant health. Notably, such computational toxicology models can serve as valuable alternatives to traditional methods for chemical risk assessment and have the potential to significantly accelerate the pace of characterizing the chemical exposome.
In an effort to support open science, we have made our complete workflow, train and test data sets, and computer codes for the built models publicly available via a GitHub repository. We believe that this will enable other researchers to reproduce our work without reinventing the wheel and, moreover, facilitate future efforts to leverage our results along with new information to build better predictive models.
While we were able to build highly predictive classification models, we encountered difficulties in developing regression models for predicting the M/P ratio of chemicals. Despite our best efforts, our best regression models for predicting the M/P ratio could achieve only moderate R2 values on (internal) test data (Table 2). This issue has been acknowledged by previous efforts in this direction and could be attributed to several factors including the lack of sufficient data to build the models and the influence of environmental factors on the outcome.23,27 Interestingly, while the best regression models achieved moderate R2 values for predicting the M/P ratio of chemicals, the same regression models performed well in classification based on regression (Table 2; Table 3). However, our best model for classification based on regression could not outperform the SVM model built solely for the classification task. Based on our results, future efforts in this direction may consider exploring different approaches to build separate models for the two tasks, regression, and classification.
Overall, our study attests to the potential of machine learning models in predicting xenobiotic chemicals with a high propensity to transfer from maternal plasma to human milk while also highlighting the challenges associated with developing accurate regression models for predicting the M/P ratio of such chemicals. While our classification models gave promising results, further investigation is needed to improve the accuracy of the regression models. Future efforts to build models with improved prediction power could explore the use of additional features such as genetics, food and lifestyle, physicochemical properties, environmental exposure, and toxicokinetic information or incorporate more sophisticated algorithms such as deep learning.
Methods
Chemical Data Set
In this study, we compiled from the published literature a curated data set of 375 chemicals (Table S1), consisting of xenobiotic chemicals (mainly, drugs), with known experimentally determined M/P concentration ratios. The majority (368) of the chemicals in our data set were compiled from Vasios et al.18 and information on few additional chemicals was obtained from other publications.25,28,32,33 For 4 chemicals in our data set, the M/P ratios were available from multiple publications, and in such cases, the mean of the reported M/P ratios for a chemical across publications was used. In a few instances, we did not include chemicals with a known M/P ratio in the published literature as the computation of the molecular descriptors (or features) failed for them. We have also annotated the 375 chemicals with ClassyFire-based chemical kingdom, superclass, class, and subclass (Table S1).38
For our curated data set of 375 chemicals with known M/P ratios, we obtained the two-dimensional (2D) structures from PubChem.39 We converted the 2D structures of these 375 chemicals to their corresponding three-dimensional (3D) structures as follows. Using RDKit, we first embedded a chemical in the 3D space by employing the ETKDG method.40 Thereafter, the 3D structure of a chemical was energy minimized using the MMFF94 force field in RDKit. Subsequently, we computed the molecular descriptors for the 375 chemicals in our data set using PaDEL version 2.21.41 A total of 1875 molecular descriptors were computed for each chemical using PaDEL, including topological, geometric, electrostatic, and several other types of descriptors. Note that the 2D structure of a chemical was used to compute one-dimensional (1D) and 2D descriptors, while the 3D structure was used to compute 3D descriptors in PaDEL. The computed descriptors were used as features to build machine learning models.
In this study, we build both classification and regression models to predict the propensity of a chemical to transfer from plasma to milk. For the classification models, chemicals with the M/P ratio ≥1 were designated as “high risk” while the remaining chemicals with the M/P ratio <1 were designated as “low risk”, following previously published studies.18,25,26 For the regression models, we treated the M/P ratio of chemicals in our data set as a continuous variable. A logarithmic transformation was used to reduce the skewness in the distribution of M/P ratios for chemicals in our data set. Note that a constant value of 1 was added to the M/P ratio of each chemical in our data set before logarithmic transformation in order to avoid undefined logarithmic values for chemicals with the M/P ratio equal to 0.
To build the classification and regression models, our data set of 375 chemicals (250 chemicals categorized as “low risk” and 125 chemicals categorized as “high risk”) with experimentally determined M/P ratios was randomly split into two sets with 80% of the data in the train set and the remaining 20% of the data in the test set (Table S1). Note that the initial data set of 375 chemicals exhibited an imbalanced distribution of chemicals, with a 2:1 ratio between “low risk” and “high risk” classes. During the process of splitting the data into the train and test set, we preserved the ratio between the “low risk” and “high risk” chemicals in both the train and test set. Moreover, we have also performed an analysis of the chemical structural diversity of the train and internal test data set based on the Soergel distance using MACCS key fingerprints.42 The intralibrary distance within the train and internal test set was found to be 0.70 and 0.67, respectively. The interlibrary distance between the train and internal test data set was found to be 0.69. This highlights that the train and internal test data sets were equally diverse among themselves and between them.
In addition to this train and test data set (together consisting of 375 chemicals with M/P ratios), we also compiled an external test data set of 202 chemicals from Karthikeyan et al.,5 Lehmann et al.,6 and Neveu et al.,43 which have been experimentally detected in human breast milk samples (Table S2). Since the 202 chemicals in the external test data set have been experimentally detected in human breast milk samples, they were categorized as “high risk” chemicals and thereafter used for the validation of classification models. However, the M/P ratio for these 202 chemicals in the external test data set has not been reported in the literature, and thus, the external test data set cannot be used to evaluate the regression models. For the 202 chemicals in the external test data set, we obtained the 2D structures from Karthikeyan et al.5 Thereafter, we followed the same procedure as described above for the 375 chemicals to generate the 3D structures and compute the 1875 molecular descriptors for the 202 chemicals in the external test data set.
Feature Selection
Each chemical in our data set has 1876 features including the dependent variable (M/P ratio). After randomly splitting the data set of 375 chemicals into the train set with 80% of the data and the test set with the remaining 20% of the data, we first removed the features with zero variance in the train set. Thereafter, the train and test sets were scaled using StandardScaler in Scikit-learn.34
While building classification models, we used BorutaPy, a wrapper built on the RF-based classifier, to implement feature selection.44,45 Notably, BorutaPy takes into account multivariable relationships and considers all features that are relevant to the dependent variable. The method involves taking a copy of the original features and shuffling them to create shadow features. These shadow features are concatenated with the train set to find the importance of individual features using the Z-score.45 If the importance of a feature is greater than the maximum importance of the shadow features, then such a feature is retained. Otherwise, the feature is considered unimportant and is dropped while building the classification models. While building regression models, we used the RF method within Scikit-learn to select the important features.46
Machine Learning Algorithms
In this study, we implemented several machine learning algorithms, namely, SVM, XGBoost, LDA, MLP, and RF, to build classification and regression models. The selection of these specific algorithms was informed by their well-established efficacy in the field of computational toxicology, particularly regarding their demonstrated use in predicting M/P ratios. In this subsection, we provide a concise overview of the algorithms employed here.
SVM is a supervised machine learning algorithm used for classification and regression tasks. It maximizes predictive accuracy and addresses overfitting by finding an optimal hyperplane that maximizes the margin between training examples and class boundaries.47−49 SVM can handle both binary and multiclass classification tasks and can also handle nonlinear data by introducing a new dimension. The robustness and accuracy of such models can be improved through hyperparameter tuning. Additionally, there is a trade-off between correctly classified points and maximization of the margin. Support vector regressor (SVR) is a similar technique used for regression tasks and aims to find the hyperplane that maximizes the margin distance between data points by approximating the relationship between inputs and continuous targets. SVR is particularly effective for handling nonlinear and high-dimensional data.
XGBoost is a widely used machine learning algorithm for classification and regression tasks.50 It combines weak learners, known as base learners, to create a stronger learner by iteratively minimizing the overall error. XGBoost offers advantages such as high accuracy, fast execution speed, regularization techniques, and flexibility compared to other popular algorithms.51
LDA is a computationally efficient machine learning algorithm that is used for classification. LDA optimizes class separation by maximizing the variance between classes and minimizing the variance within classes through a linear discriminant function. It assumes Gaussian probability density functions with the same covariance for each class. LDA projects data onto a lower-dimensional space by maximizing the distances between the means of the classes while minimizing the within-class variance.52 LDA has advantages, such as handling high-dimensional data, generative modeling, and dimensionality reduction. However, the method has limitations such as assumptions of linearity and normal distribution of data, which may not hold in all cases.53
RF is a versatile machine learning algorithm used for classification and regression tasks. It consists of multiple tree predictors, where each tree’s outcome is influenced by a random vector sampled independently across all trees in the forest.46 In classification, the class predicted by the majority of trees determines the final result, while in regression, the final prediction is the average of predictions from all of the trees. RF excels in handling high-dimensional data sets and produces stable predictions.46,54 However, the computational cost of this algorithm increases when modeling large data sets.
Hyperparameter Tuning in Algorithms
Hyperparameter tuning is a technique used to exhaustively search for the best parameters among a given set of parameters for an algorithm to build predictive models. Hyperparameters of an estimator (algorithm) are a set of parameters that control the learning of the training data and determine the performance of the model on the test data. A notable challenge during model selection is to find the right combination of the hyperparameters for an estimator from the available hyperparameter space. In this study, we used GridSearchCV in Scikit-learn for hyperparameter tuning.34 In GridSearchCV, for an estimator and a set of hyperparameters, we performed hyperparameter tuning using repeated 10-fold cross-validation. Subsequent to hyperparameter tuning, the final model is trained based on the best set of parameters obtained from GridSearchCV. Table S7 lists the best sets of parameters obtained from GridSearchCV for the different estimators used in this study. Table S8 lists the final set of top-ranked features, which are used to build the classification and regression models in this study.
Domain of Applicability
In the Setubal workshop report,55 the applicability domain (AD) of a QSAR model is defined as “The AD of a (Q)SAR is the physico-chemical, structural, or biological space, knowledge or information on which the training set of the model has been developed, and for which it is applicable to make predictions for new compounds. The AD of a (Q)SAR should be described in terms of the most relevant parameters, i.e., usually those that are descriptors of the model. Ideally, the (Q)SAR should only be used to make predictions within that domain by interpolation and not extrapolation”. In other words, the prediction made by a QSAR model is reliable or acceptable only if the chemical in the test data lies in the AD of the model. Therefore, the domain of applicability can be used to better understand the scope of a predictive model.
In this study, we employed the standardized approach proposed by Roy et al. to find whether the chemicals in our test data fall within the AD of the built models for classification and regression.56
Acknowledgments
We thank N. Sukumar for discussion and Kishan Kumar for help with figures. Areejit Samal would like to acknowledge support from the Department of Atomic Energy (DAE), Government of India [Apex project to The Institute of Mathematical Sciences (IMSc), Chennai], and the Max Planck Society, Germany [Max Planck Partner Group in Mathematical Biology]. The funders have no role in the study design, data collection, data analysis, manuscript preparation, or decision to publish.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c09392.
Compiled data set of 375 xenobiotic chemicals with experimentally determined M/P concentration ratios from the published literature; compiled list of 202 chemicals in the external test data set which have been experimentally detected in human breast milk samples; list of approved drugs and their prediction of propensity to transfer from maternal plasma to milk; list of experimental drugs and their prediction of propensity to transfer from maternal plasma to milk; methodology and performance metrics for previously published classification models to predict chemicals with high propensity to transfer from maternal plasma into human milk; methodology and performance metrics for previously published regression models to predict the M/P ratio of chemicals; list of best sets of parameters obtained from GridSearchCV for different classification and regression algorithms used in this study to predict the propensity of chemicals to transfer from maternal plasma to milk; and list of top-ranked molecular descriptors (features) for the classification and regression algorithms used in this study for the prediction of the propensity of chemicals to transfer from maternal plasma to milk (XLSX)
Distribution of the M/P ratio of complete data set, train data, and internal test data (PDF)
Author Present Address
∥ Thorne HealthTech, New York, NY, United States
Author Contributions
Sudharsan Vijayaraghavan: Conceptualization, data compilation, data curation, formal analysis, software, visualization, and writing; Akshaya Lakshminarayanan: conceptualization, data compilation, data curation, formal analysis, software, and writing; Naman Bhargava: conceptualization, data compilation, data curation, formal analysis, software, and writing; Janani Ravichandran: data compilation, data curation, and writing; R.P. Vivek-Ananth: conceptualization, supervision, formal analysis, software, and writing; and Areejit Samal: conceptualization, supervision, formal analysis, and writing.
The authors declare no competing financial interest.
Supplementary Material
References
- Rollins N. C.; Bhandari N.; Hajeebhoy N.; Horton S.; Lutter C. K.; Martines J. C.; Piwoz E. G.; Richter L. M.; Victora C. G. Why Invest, and What It Will Take to Improve Breastfeeding Practices?. Lancet 2016, 387 (10017), 491–504. 10.1016/S0140-6736(15)01044-2. [DOI] [PubMed] [Google Scholar]
- Hauck F. R.; Thompson J. M. D.; Tanabe K. O.; Moon R. Y.; Vennemann M. M. Breastfeeding and Reduced Risk of Sudden Infant Death Syndrome: A Meta-Analysis. Pediatrics 2011, 128 (1), 103–110. 10.1542/peds.2010-3000. [DOI] [PubMed] [Google Scholar]
- Stuebe A. The Risks of Not Breastfeeding for Mothers and Infants. Rev. Obstet. Gynecol. 2009, 2 (4), 222–231. [PMC free article] [PubMed] [Google Scholar]
- Bartick M. C.; Schwarz E. B.; Green B. D.; Jegier B. J.; Reinhold A. G.; Colaizy T. T.; Bogen D. L.; Schaefer A. J.; Stuebe A. M. Suboptimal Breastfeeding in the United States: Maternal and Pediatric Health Outcomes and Costs. Matern. Child Nutr. 2017, 13 (1), e12366 10.1111/mcn.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan B. S.; Ravichandran J.; Aparna S. R.; Samal A. ExHuMId: A Curated Resource and Analysis of Exposome of Human Milk across India. Chemosphere 2021, 271, 129583. 10.1016/j.chemosphere.2021.129583. [DOI] [PubMed] [Google Scholar]
- Lehmann G. M.; LaKind J. S.; Davis M. H.; Hines E. P.; Marchitti S. A.; Alcala C.; Lorber M. Environmental Chemicals in Breast Milk and Formula: Exposure and Risk Assessment Implications. Environ. Health Perspect. 2018, 126 (9), 096001. 10.1289/EHP1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrigan P. J.; Sonawane B.; Mattison D.; McCally M.; Garg A. Chemical Contaminants in Breast Milk and Their Impacts on Children’s Health: An Overview. Environ. Health Perspect. 2002, 110 (6), A313. 10.1289/ehp.021100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead M. N. Contaminants in Human Milk: Weighing the Risks against the Benefits of Breastfeeding. Environ. Health Perspect. 2008, 116 (10), A427–A434. 10.1289/ehp.116-a426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane B. R. Chemical Contaminants in Human Milk: An Overview. Environ. Health Perspect. 1995, 103 (suppl 6), 197–205. 10.1289/ehp.95103s6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.-M.; Albrecht M.; Fromme H.; Schramm K.-W.; De Angelis M. Persistent Organic Pollutants in Human Breast Milk and Associations with Maternal Thyroid Hormone Homeostasis. Environ. Sci. Technol. 2020, 54 (2), 1111–1119. 10.1021/acs.est.9b06054. [DOI] [PubMed] [Google Scholar]
- Leibson T.; Lala P.; Ito S.. Drug and Chemical Contaminants in Breast Milk: Effects on Neurodevelopment of the Nursing Infant. Handbook of Developmental Neurotoxicology; Elsevier, 2018; pp 275–284. [Google Scholar]
- Wild C. P. Complementing the Genome with an “Exposome”: The Outstanding Challenge of Environmental Exposure Measurement in Molecular Epidemiology. Cancer Epidemiol., Biomarkers Prev. 2005, 14 (8), 1847–1850. 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Miller G. W.; Jones D. P. The Nature of Nurture: Refining the Definition of the Exposome. Toxicol. Sci. 2014, 137 (1), 1–2. 10.1093/toxsci/kft251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport S. M.; Smith M. T. Environment and Disease Risks. Science 2010, 330 (6003), 460–461. 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen R.; Schymanski E. L.; Barabási A. L.; Miller G. W. The Exposome and Health: Where Chemistry Meets Biology. Science 2020, 367 (6476), 392–396. 10.1126/science.aay3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agatonovic-Kustrin S.; Ling L. H.; Tham S. Y.; Alany R. G. Molecular Descriptors That Influence the Amount of Drugs Transfer into Human Breast Milk. J. Pharm. Biomed. Anal. 2002, 29 (1–2), 103–119. 10.1016/S0731-7085(02)00037-7. [DOI] [PubMed] [Google Scholar]
- Anadón A.; Martínez-Larrañaga M. R.; Ramos E.; Castellano V.. Transfer of Drugs and Xenobiotics through Milk. Reproductive and Developmental Toxicology; Elsevier, 2011; pp 57–71. [Google Scholar]
- Vasios G.; Kosmidi A.; Kalantzi O.-I.; Tsantili-Kakoulidou A.; Kavantzas N.; Theocharis S.; Giaginis C. Simple Physicochemical Properties Related with Lipophilicity, Polarity, Molecular Size and Ionization Status Exert Significant Impact on the Transfer of Drugs and Chemicals into Human Breast Milk. Expert Opin. Drug Metab. Toxicol. 2016, 12 (11), 1273–1278. 10.1080/17425255.2016.1230197. [DOI] [PubMed] [Google Scholar]
- Heinzow B. G. J.Endocrine Disruptors in Human Milk and the Health-Related Issues of Breastfeeding. Endocrine-Disrupting Chemicals in Food; Elsevier, 2009; pp 322–355. [Google Scholar]
- Zhao C.; Zhang H.; Zhang X.; Zhang R.; Luan F.; Liu M.; Hu Z.; Fan B. Prediction of Milk/Plasma Drug Concentration (M/P) Ratio Using Support Vector Machine (SVM) Method. Pharm. Res. 2006, 23 (1), 41–48. 10.1007/s11095-005-8716-4. [DOI] [PubMed] [Google Scholar]
- Begg E. J.; Atkinson H. C. Modelling of the Passage of Drugs into Milk. Pharmacol. Ther. 1993, 59 (3), 301–310. 10.1016/0163-7258(93)90072-L. [DOI] [PubMed] [Google Scholar]
- Wilson J. T.; Brown R. D.; Cherek D. R.; Dailey J. W.; Hilman B.; Jobe P. C.; Manno B. R.; Manno J. E.; Redetzki H. M.; Stewart J. J. Drug Excretion in Human Breast Milk1,2: Principles, Pharmacokinetics and Projected Consequences. Clin. Pharmacokinet. 1980, 5 (1), 1–66. 10.2165/00003088-198005010-00001. [DOI] [PubMed] [Google Scholar]
- Abraham M. H.; Gil-Lostes J.; Fatemi M. Prediction of Milk/Plasma Concentration Ratios of Drugs and Environmental Pollutants. Eur. J. Med. Chem. 2009, 44 (6), 2452–2458. 10.1016/j.ejmech.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Agatonovic-Kustrin S.; Tucker I. G.; Zecevic M.; Zivanovic L. J. Prediction of Drug Transfer into Human Milk from Theoretically Derived Descriptors. Anal. Chim. Acta 2000, 418 (2), 181–195. 10.1016/S0003-2670(00)00963-6. [DOI] [Google Scholar]
- Fatemi M. H.; Ghorbanzad’e M. Classification of Drugs According to Their Milk/Plasma Concentration Ratio. Eur. J. Med. Chem. 2010, 45 (11), 5051–5055. 10.1016/j.ejmech.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Kar S.; Roy K. Prediction of Milk/Plasma Concentration Ratios of Drugs and Environmental Pollutants Using In Silico Tools: Classification and Regression Based QSARs and Pharmacophore Mapping. Mol. Inform. 2013, 32 (8), 693–705. 10.1002/minf.201300018. [DOI] [PubMed] [Google Scholar]
- Katritzky A. R.; Dobchev D. A.; Hür E.; Fara D. C.; Karelson M. QSAR Treatment of Drugs Transfer into Human Breast Milk. Bioorg. Med. Chem. 2005, 13 (5), 1623–1632. 10.1016/j.bmc.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Wanat K.; Khakimov B.; Brzezińska E. Comparison of Statistical Methods for Predicting Penetration Capacity of Drugs into Human Breast Milk Using Physicochemical, Pharmacokinetic and Chromatographic Descriptors. SAR QSAR Environ. Res. 2020, 31 (6), 457–475. 10.1080/1062936X.2020.1772365. [DOI] [PubMed] [Google Scholar]
- Yap C. W.; Chen Y. Z. Quantitative Structure-Pharmacokinetic Relationships for Drug Distribution Properties by Using General Regression Neural Network. J. Pharm. Sci. 2005, 94 (1), 153–168. 10.1002/jps.20232. [DOI] [PubMed] [Google Scholar]
- Anderson P. O.; Momper J. D. Clinical Lactation Studies and the Role of Pharmacokinetic Modeling and Simulation in Predicting Drug Exposures in Breastfed Infants. J. Pharmacokinet. Pharmacodyn. 2020, 47 (4), 295–304. 10.1007/s10928-020-09676-2. [DOI] [PubMed] [Google Scholar]
- Wilkinson M. D.; Dumontier M.; Aalbersberg Ij. J.; Appleton G.; Axton M.; Baak A.; Blomberg N.; Boiten J.-W.; da Silva Santos L. B.; Bourne P. E.; Bouwman J.; Brookes A. J.; Clark T.; Crosas M.; Dillo I.; Dumon O.; Edmunds S.; Evelo C. T.; Finkers R.; Gonzalez-Beltran A.; Gray A. J. G.; Groth P.; Goble C.; Grethe J. S.; Heringa J.; ’t Hoen P. A. C.; Hooft R.; Kuhn T.; Kok R.; Kok J.; Lusher S. J.; Martone M. E.; Mons A.; Packer A. L.; Persson B.; Rocca-Serra P.; Roos M.; van Schaik R.; Sansone S.-A.; Schultes E.; Sengstag T.; Slater T.; Strawn G.; Swertz M. A.; Thompson M.; van der Lei J.; van Mulligen E.; Velterop J.; Waagmeester A.; Wittenburg P.; Wolstencroft K.; Zhao J.; Mons B. The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 2016, 3 (1), 160018. 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N.; Ito K.; Ikebuchi Y.; Toyoda Y.; Takada T.; Hisaka A.; Oka A.; Suzuki H. Prediction of Drug Transfer into Milk Considering Breast Cancer Resistance Protein (BCRP)-Mediated Transport. Pharm. Res. 2015, 32, 2527. 10.1007/s11095-015-1641-2. [DOI] [PubMed] [Google Scholar]
- Larsen L. A.; Ito S.; Koren G. Prediction of Milk/Plasma Concentration Ratio of Drugs. Ann. Pharmacother. 2003, 37 (9), 1299–1306. 10.1345/aph.1C379. [DOI] [PubMed] [Google Scholar]
- Pedregosa F.; Varoquaux G.; Gramfort A.; Michel V.; Thirion B.; Grisel O.; Blondel M.; Prettenhofer P.; Weiss R.; Dubourg V.; Vanderplas J.; Passos A.; Cournapeau D.; Brucher M.; Perrot M.; Duchesnay E. ´. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12 (85), 2825–2830. 10.5555/1953048.2078195. [DOI] [Google Scholar]
- Wishart D. S.; Feunang Y. D.; Guo A. C.; Lo E. J.; Marcu A.; Grant J. R.; Sajed T.; Johnson D.; Li C.; Sayeeda Z.; Assempour N.; Iynkkaran I.; Liu Y.; Maciejewski A.; Gale N.; Wilson A.; Chin L.; Cummings R.; Le D.; Pon A.; Knox C.; Wilson M. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46 (D1), D1074–D1082. 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker S. E.; Moll H. A.; Steyerberg E. W.; Donders A. R. T.; Derksen-Lubsen G.; Grobbee D. E.; Moons K. G. M. External Validation Is Necessary in Prediction Research:. J. Clin. Epidemiol. 2003, 56 (9), 826–832. 10.1016/S0895-4356(03)00207-5. [DOI] [PubMed] [Google Scholar]
- Cabitza F.; Campagner A.; Soares F.; García de Guadiana-Romualdo L.; Challa F.; Sulejmani A.; Seghezzi M.; Carobene A. The Importance of Being External. Methodological Insights for the External Validation of Machine Learning Models in Medicine. Comput. Methods Progr. Biomed. 2021, 208, 106288. 10.1016/j.cmpb.2021.106288. [DOI] [PubMed] [Google Scholar]
- Djoumbou Feunang Y.; Eisner R.; Knox C.; Chepelev L.; Hastings J.; Owen G.; Fahy E.; Steinbeck C.; Subramanian S.; Bolton E.; Greiner R.; Wishart D. S. ClassyFire: Automated Chemical Classification with a Comprehensive, Computable Taxonomy. J. Cheminf. 2016, 8 (1), 61. 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Chen J.; Cheng T.; Gindulyte A.; He J.; He S.; Li Q.; Shoemaker B. A.; Thiessen P. A.; Yu B.; Zaslavsky L.; Zhang J.; Bolton E. E. PubChem 2023 Update. Nucleic Acids Res. 2023, 51 (D1), D1373–D1380. 10.1093/nar/gkac956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum G.RDKit: Open-Source Cheminformatics, 2022.
- Yap C. W. PaDEL-Descriptor: An Open Source Software to Calculate Molecular Descriptors and Fingerprints. J. Comput. Chem. 2011, 32 (7), 1466–1474. 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- Vivek-Ananth R. P.; Sahoo A. K.; Baskaran S. P.; Samal A. Scaffold and Structural Diversity of the Secondary Metabolite Space of Medicinal Fungi. ACS Omega 2023, 8 (3), 3102–3113. 10.1021/acsomega.2c06428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu V.; Moussy A.; Rouaix H.; Wedekind R.; Pon A.; Knox C.; Wishart D. S.; Scalbert A. Exposome-Explorer: A Manually-Curated Database on Biomarkers of Exposure to Dietary and Environmental Factors. Nucleic Acids Res. 2017, 45 (D1), D979–D984. 10.1093/nar/gkw980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel H.Boruta_py, 2022. https://github.com/scikit-learn-contrib/boruta_py.
- Kursa M. B.; Rudnicki W. R. Feature Selection with the Boruta Package. J. Stat. Software 2010, 36 (11), 1. 10.18637/jss.v036.i11. [DOI] [Google Scholar]
- Breiman L. Random Forests. Mach. Learn. 2001, 45 (1), 5–32. 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Boser B. E.; Guyon I. M.; Vapnik V. N.. A Training Algorithm for Optimal Margin Classifiers. Proceedings of the fifth annual workshop on Computational learning theory; ACM: Pittsburgh Pennsylvania USA, 1992; pp 144–152.
- Cortes C.; Vapnik V. Support-Vector Networks. Mach. Learn. 1995, 20 (3), 273–297. 10.1007/BF00994018. [DOI] [Google Scholar]
- Drucker H.; Burges C. J. C.; Kaufman L.; Smola A.; Vapnik V.. Support Vector Regression Machines. Advances in Neural Information Processing Systems; MIT Press, 1996; Vol. 9. [Google Scholar]
- Friedman J. H. Greedy Function Approximation: A Gradient Boosting Machine. Annu. Stat. 2001, 29 (5), 1189. 10.1214/aos/1013203451. [DOI] [Google Scholar]
- Chen T.; Guestrin C.. XGBoost: A Scalable Tree Boosting System. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; ACM: San Francisco California USA, 2016; pp 785–794.
- Fisher R. A. The Use of Multiple Measurements in Taxonomic Problems. Ann. Eugen. 1936, 7 (2), 179–188. 10.1111/j.1469-1809.1936.tb02137.x. [DOI] [Google Scholar]
- Tharwat A.; Gaber T.; Ibrahim A.; Hassanien A. E. Linear Discriminant Analysis: A Detailed Tutorial. AI Commun. 2017, 30 (2), 169–190. 10.3233/AIC-170729. [DOI] [Google Scholar]
- Liaw A.; Wiener M. Classification and Regression by randomForest. R News 2002, 2 (3), 18–22. [Google Scholar]
- Jaworska J.; Nikolova-Jeliazkova N.; Aldenberg T. QSAR Applicability Domain Estimation by Projection of the Training Set in Descriptor Space: A Review. Altern. Lab. Anim. 2005, 33 (5), 445–459. 10.1177/026119290503300508. [DOI] [PubMed] [Google Scholar]
- Roy K.; Kar S.; Ambure P. On a Simple Approach for Determining Applicability Domain of QSAR Models. Chemom. Intell. Lab. Syst. 2015, 145, 22–29. 10.1016/j.chemolab.2015.04.013. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.