Abstract
In the respiratory chain of the majority of aerobic organisms, the enzyme alternative oxidase (AOX) functions as the terminal oxidase and has important roles in maintaining metabolic and signaling homeostasis in mitochondria. AOX endows the respiratory system with flexibility in the coupling among the carbon metabolism pathway, electron transport chain (ETC) activity, and ATP turnover. AOX allows electrons to bypass the main cytochrome pathway to restrict the generation of reactive oxygen species (ROS). The inhibition of AOX leads to oxidative damage and contributes to the loss of adaptability and viability in some pathogenic organisms. Although AOXs have recently been identified in several organisms, crystal structures and major functions still need to be explored. Recent work on the trypanosome alternative oxidase has provided a crystal structure of an AOX protein, which contributes to the structure–activity relationship of the inhibitors of AOX. Here, we review the current knowledge on the development, structure, and properties of AOXs, as well as their roles and mechanisms in plants, animals, algae, protists, fungi, and bacteria, with a special emphasis on the development of AOX inhibitors, which will improve the understanding of respiratory regulation in many organisms and provide references for subsequent studies of AOX-targeted inhibitors.
1. Introduction
The classical mitochondrial respiratory system found in humans, which includes complexes I, III, and IV as its core proton-translocating elements; the auxiliary enzymes represented by complex II; and the small connecting molecules, including coenzyme Q (UQ) and cytochrome c (Cyt C),1 has been extensively investigated as to its structural and functional properties. The respiratory electron transport chain (ETC) is comprised of these complexes with ATP synthase, which plays a crucial role in facilitating the mitochondrial oxidative phosphorylation system. This system serves as the center of cellular metabolism and the primary mechanism for energy conversion in eukaryotic cells.2
In some organisms, the respiratory system may exhibit branching or modifications. For example, complex I can be substituted by the alternative NADH-quinone oxidoreductase (NDH-2), which can, in a variety of organisms, transfer an electron from NADH to quinone through FAD, without the need for proton pumping.3 In addition, this substitution occurs in the mitochondria of Saccharomyces cerevisiae, which lack the respiratory complex I but contain three rotenone-insensitive NADH dehydrogenases distributed on both the external (Nde1 and Nde2) and internal (Ndi1) surfaces of the inner mitochondrial membrane. The function of these enzymes involves the facilitation of electron transfer from NADH to ubiquinone while not causing the movement of protons across the membrane.4
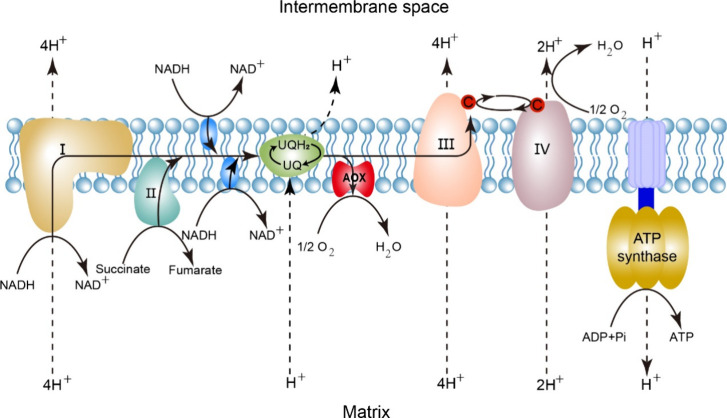
In the respiratory processes, the enzyme known as alternative oxidase (AOX) can be utilized by the respiratory system to diverge at the stage of ubiquinol oxidation. It is worth noting that in the majority of eukaryotes, cytochrome c oxidase serves as the final constituent of the mitochondrial respiratory chain.5 However, in this particular branch, the terminal complex is represented by AOX. The AOX facilitates the process of oxidizing ubiquinol while simultaneously reducing oxygen to form water. In AOX-catalyzed reactions, oxygen acquires protons from ubiquinol rather than from the mitochondrial matrix as found in the cytochrome c oxidase reaction.6 The catalytic process of AOX is a cyanide- and antimycin A-resistant process,7 which initiates with the binding of oxygen to one of the ferrous iron atoms, resulting in a four-electron reduction of oxygen to water, namely O2 + 2UQH2 → 2H2O + 2UQ.8,9 The entire respiratory chain containing AOX is illustrated in Figure 1.
Figure 1.
The conserved mitochondrial respiratory chain. The major mitochondrial respiratory pathway and the AOX pathway are shown.
Besides the typical respiratory cytochrome oxidase, mitochondria of various plants, algae, a majority of fungi, and certain protists that have been investigated thus far exhibit the presence of AOX. Beyond these organisms, the taxonomic distribution of AOX has been extended to some members of the animal kingdom.10 Over the past two decades, extensive sequencing of animal genomes has identified genes encoding unconventional respiratory system enzymes across several phyla. The genomes of Placozoa, Porifera, Cnidaria, Annelida, Echinodermata, Mollusca, Nematoda, Hemichordata, and Chordata have been identified to contain sequences that encode Ndh2 and AOX proteins.11 Genes encoding AOX proteins have also been identified in yeast, slime molds, free-living amoebae, eubacteria, and nematodes.12 Despite the occurrence of AOX in diverse organisms, the function of this oxidase may exhibit variability. In many organisms, AOX activity holds importance in the regulation of reactive oxygen species (ROS) and the preservation of metabolic homeostasis, which includes the management of carbon metabolism, cellular energy, and redox equilibrium. In thermogenic plants, the activity of AOX is also associated with the generation of heat.7 In addition, the trypanosome alternative oxidase (TAO) can function as a terminal oxidase in bloodstream forms (BSF) of trypanosomes to reoxidize NADH, which is accumulated during glycolysis because of the absence of functional oxidative phosphorylation.6 TAO, conserved among Trypanosoma brucei subspecies, is crucial for the respiration of BSF trypanosomes, and its potential as a target for antitrypanosomal drug has been well substantiated.13 Additionally, AOX is critical to the metabolism of opportunistic human fungal pathogens, including Candida albicans,14Candida auris, and Cryptococcus neoformans.15 Previous studies indicated that alternative respiration in C. albicans leads to decreased fluconazole susceptibility, thereby playing a significant role in the azole resistance and increased survival of C. albicans.16 Therefore, the investigation of AOX in the management of infectious ailments has emerged as a central area of interest in contemporary scholarly research.
Because AOX plays an important role in fundamental biological processes, it has attracted much attention. In this review, we describe aspects related to the structure, properties, and functions of AOX in different organisms, and emphasize AOX inhibitor studies directing design of AOX-targeted drugs.
2. Discovery of AOX
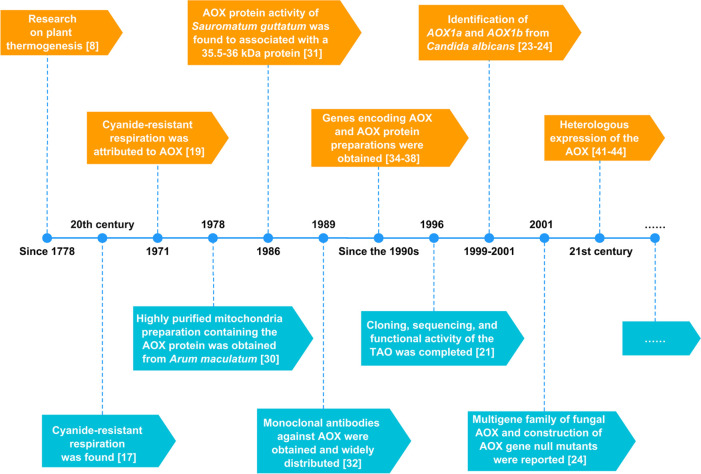
The timeline of discoveries of AOX is shown in Figure 2.
Figure 2.
Timeline of discoveries of AOX.
The exploration of AOX cannot be dissociated from the investigations on plant thermogenesis dating back to 1778, which was commenced with Araceae plants, and was represented by the largest thermogenic plant, Amorphophallus titanium.8 In the 20th century, studies suggested that some algal and fungal cells were able to maintain respiration in the presence of mitochondrial respiratory inhibitors, such as cyanide.17 Subsequently, the existence of cyanide-resistant respiration was also identified in both plants and some protists. Various hypotheses had been put forth in order to elucidate the properties and functions of alternate oxidase, which had been initially attributed to several candidate compounds, such as flavonoids with low affinity for oxygen, type A cytochrome, cytochrome B7, and others.17,18 In 1971, Bendall and Bonner disproved these hypotheses and attributed cyanide-resistant respiration to the function of AOX,19 which could act as the terminal oxidase by introducing a branch at the ubiquinone point of the ETC. At the time, this branch was thought to be a waste of energy because the AOX pathway would reduce the amount of ATP produced per oxygen consumption as the electron flux via this pathway allows for the circumvention of two of the three proton pump complexes within the ETC.20 In 1996, cloning, sequencing, and functional activity of the TAO was completed.21 Soon after, researchers established that the production of ROS, which is known for inducing oxidative harm in mitochondria and other constituents of the cell, would be suppressed, along with ATP production, through the action of AOX.22 In other species, Huh et al.23 reported the identification of AOX1a in C. albicans, which could be functionally expressed in S. cerevisiae, and two years later, AOX1b, a new member of the AOX gene family, was identified.24 A multigene family of fungal AOX, along with the construction of AOX gene null mutants, were reported in 2001.24 The application of molecular biology techniques has accelerated the research on AOX and has brought about many important discoveries and breakthroughs.
Recently, studies in plants have demonstrated the physiological role of AOX in developmental processes and response to environmental changes. AOX provides an extrachloroplastic means to optimize the status of chloroplast energy pools (ATP, NADPH) and to manage cellular carbohydrate pools in response to changing rates of carbon fixation and carbon demand for growth and maintenance. Transcriptional and posttranslational mechanisms play crucial roles in enabling AOX to effectively adapt to modifications in carbon and energy balance, which is especially noteworthy when facing abiotic stress conditions, like water deficit, high salinity, or temperature extremes.25 Similarly, in fungi, expression levels of AOX genes were upregulated in cells responding to oxidative or osmotic pressures,26 which suggests that AOX is involved in resistance to stressful conditions,27 response to oxidative stress,28 and in maintaining cellular homeostasis.29
The isolation and expression of AOX proteins was critical for the study of AOX. As early as 1978, a highly purified mitochondria preparation containing the AOX protein was successfully obtained from the metabolically active thermogenic tissues found in Arum maculatum plants.30 In 1986, the activity of the AOX protein in the thermogenic tissues of Sauromatum guttatum, was found to associated with a 35.5–36 kDa protein.31 By 1989, monoclonal antibodies against the proteins were obtained and widely distributed.32 The antibodies were effectively utilized to detect the presence of AOX in the mitochondria of the fungus Neurospora crassa.33 Following a span of two years, cDNA encoding a protein with a molecular weight of 38.9 kDa was obtained from a S. guttatum DNA library.34 Subsequently, genes encoding AOX were isolated from soybean35 and tobacco,36 while highly purified (from the alga Chlamydomonas reinhardtii(37)) and partially purified (the BSF of T. brucei(38)) AOX protein preparations have been obtained. In 2007 and 2009, the genes encoding AOX were isolated from methylotrophic yeast Pichia pastoris,39 the thermogenic tissue of Symplocarpus renifolius, and nonthermogenic Lysichiton camtschatcensis.40 In recent decades, numerous new technologies have emerged permitting researchers to acquire the AOX proteins, such as cloning and functional expression of the AOX1 of Aspergillus niger in Lactococcus lactis,41 expression of recombinant S. guttatum AOX in Escherichia coli membranes,42 heterologous expression of the AOX from Crassostrea gigas (Pacific oyster) and Moniliophthora perniciosa in the yeast S. cerevisiae,43,44 self-assembled proteoliposomes to functionally characterize the AOX from M. perniciosa,45 and identification of AOX encoding genes in Caulerpa cylindracea by de novo RNA sequencing (RNA-Seq) assembly analysis.46 The heterologous expression of AOX has made significant contributions to the research of function and mechanism of AOX.
3. Structure and Properties of AOX
3.1. Overview, Di-iron Center and Residues
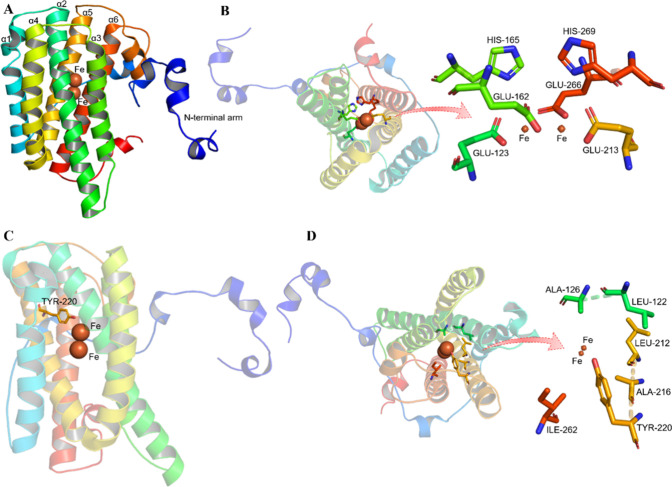
AOX localizes to mitochondria, thereby acquiring reducing equivalents at the substrate side of the antimycin A-sensitive element of the main respiratory chain. Structural models have indicated that AOX is an integral interfacial membrane protein containing a nonheme di-iron carboxylate active site, which interacts with one surface of the lipid bilayer. The asymmetric unit of the AOX consists of four monomers (Figure 3A), which come together to form homodimers. With the exception of the N-terminus (shown in Figure 3A), each monomer has a compact cylindrical shape (50 × 35 × 30 Å), and there are no obvious structural distinctions amid monomers in the asymmetric unit.47 It is believed that substrate binding sites are facilitated by a hydrophobic fissure extending from the membrane binding region to the di-iron center accompanied by various conserved protein folds, such as E-X-X-H motifs.48 It has been found that AOXs have an association with inner mitochondrial membrane.49 More recently, AOX has been identified as belonging to the family of di-iron carboxylate proteins by electron paramagnetic resonance, and the Fe(III)–Fe(III) oxide form with a single hydroxo-bridge was confirmed to be the structure of di-iron active site.50 In more recent time, analysis using electrochemistry coupled with Fourier transform infrared spectroscopy (FTIR) of the redox centers of a highly purified and stable recombinant AOX (rTAO) from T. brucei also confirmed the presence of a di-iron center.51 It should be noted that AOX is more hydrophobic than the insoluble di-iron carboxylate proteins.52
Figure 3.
Structure of TAO. (A) Monomer structure of TAO; α1-α6, six long α helices. (B) Structure of di-iron center in TAO. (C) Residue Tyr-220 in TAO. (D) Conserved hydrophobic residues in TAO. These figures were drawn by PyMol, and the AOX model used is TAO (PDB: 3vv9).
Beyond the hydroxo-bridge, Fe1 and Fe2 are bridged by two glutamate (Glu) and additionally coordinated in a bidentate fashion by two further Glu residues, which results in a five-coordinated di-iron center that possesses the geometry of warped pyramid (Figure 3B),47 and their conservations in different species are shown in the red solid box in Figure 4A. The coordination of the enzyme’s di-iron center necessitates the presence of a sequence of Glu and histidine (His) residues that are remarkably conserved among these proteins.53 Moreover, the activity of AOX requires a conserved tyrosine residue (TyrII/Tyr-275),50 while a conserved tryptophan (TrpI/Trp-206) residue is required for fixing AOX on the mitochondrial membrane.54 Mutations of the Tyr-253, Ser-256, His-261, and Arg-262 residues have suggested that the existence of hydroxyl, guanidino, imidazole, and aromatic groups, polar and charged residues, as well as the length of the amino-acid chain, are critical for the ubiquinone binding.52 There are also ubiquinone-binding entrance sites in complexes I–III.55 The binding of ubiquinone-10 to complex I and the conformational changes that occur when the ubiquinone site is occupied, are elucidated.56 For complex I, the stability of binding relies on the interactions between the ubiquinone head and finely balanced hydration of both ubiquinone tail and chamber residues.57 For AOX activity, related residues ThrI/Thr-179 and CysI/Cys-172 have been suggested to exert an impact on the catalytic cycle of the enzyme in regard to their mutual effects with oxygen. Thr-179 and Cys-172 may indirectly affect the catalysis of AOX by affecting the subtle secondary structure rearrangement and then iron-linked residues.54
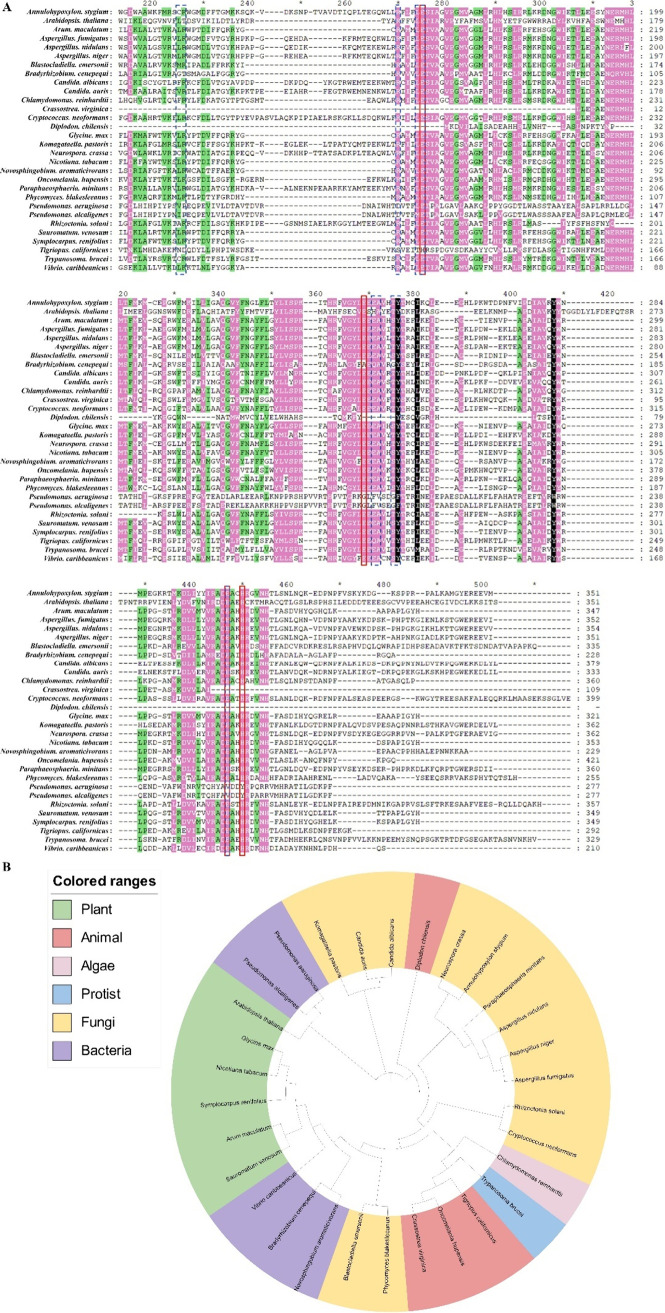
Figure 4.
Protein analyses of AOX. (A) Comparison of protein sequences of AOX in different species mentioned in the text. The protein sequences are sourced from the National Center for Biotechnology Information (NCBI) website. White background, conserved percent is less than (<) 60%; green background, conserved percent is more than (≥) 60% and less than (<) 80%; pink background, conserved percent is more than (≥) 80% and less than (<) 100%; black background, conserved percent is (=) 100%; red solid box, amino acids of di-iron center in TAO (shown in Figure 3B); blue dotted box, amino acids of the conserved binding site of TAO (shown in Figure 7K). (B) The evolutionary tree of AOX in different species mentioned in the text.
The impact of CysI and CysII, both conserved Cys residues, on the activation of α-keto acids has been documented. The α-keto acids, particularly pyruvate, have the ability to enhance the function of AOX’s reducing dimers in plants, which is one of two interrelated features, and the other is thiol modulation. It has been proposed that ligands binding to these Cys residues can induce conformational alterations in the protein, which subsequently affects the di-iron active site.54 The highly conserved serine residue (Ser-256) also plays a key role in quinone binding of complex II.58 Gln-242 and Asn-247 were identified as the only significantly polar residues that are conserved in this hydrophobic region,59 and the most hydrophobic region of the protein is flanked by Trp/Tyr and Arg/Lys.52 Tyr-220, another tyrosine residue, with the addition of ubiquinol binding site, is extremely close to the di-iron center.60 Tyr-220 is extremely important for electron transport when the ubiquinol molecules (UQH2) are oxidized.9 In order to ascertain the function of highly conserved amino acid residues in AOX catalysis, site-directed mutants of Cys-172, Thr-179, Trp-206, Tyr-253, and Tyr-299 in AOX protein have been expressed in the yeast Schizosaccharomyces pombe. Evaluation of AOX activity in isolated mitochondria has showed that AOX activity was eliminated when Trp-206 was mutated to phenylalanine or tyrosine, while AOX activity was retained in either Tyr-253 or -299 mutants. Alanine replacement of Thr-179 or Cys-172 (a residue involved in AOX regulation by α-keto acids) contributes to a reduction in maximum AOX activity and a crucial increase in the affinity of the enzyme for oxygen (4- and 2-fold, respectively).54 Although the amino acid identity between plant and fungal AOX sequences is relatively low, there exists a significant level of conservation in the crucial residues, namely Gln-242, Asn-247, Tyr-253, His-261, and Arg-262.52
Despite the presence of numerous conserved amino acids in AOX (refer to ref (8) for information on the roles, locations, and effects of mutations in universally conserved AOX residues), variations still exist among different species and even among different AOXs within species. As a result, the activities of different AOXs exhibit inconsistency. To address this, certain scholars emphasized the importance of designing AOX inhibitors that are specific to particular species, which arise from the distinct functions of AOX in its natural host and xenotopic expression.61
3.2. Structure
Subsequent studies showed that the crystal structure of AOX contains four monomers per asymmetric unit, which link to form homodimers with the nonheme di-iron carboxylate active site buried within a four-helix bundle (α2, α3, α5, and α6 in Figure 3A). It is noteworthy that this active site is composed of merely four Glu residues in the absence of an oxidized inhibitor (AF2779OH).47 In contrast with various other di-iron proteins, including hemerythrin, Δ9-stearoyl-acyl carrier protein desaturase, methane monooxygenase hydroxylase, ribonucleotide reductase, and purple acid phosphatase, the TAO di-iron center exhibits a distinctive characteristic in its oxidized state, which is the absence of coordination by a His residue.9 However, there is a His-165 coordination with Fe1 in the ligand binding state. As previously mentioned, an exceedingly conserved Tyr-220 (Figure 3C) is located within 4 Å of the active site and plays an essential role in the facilitation of catalytic activity. In the dimer, because of their weak electron density, each observed monomer, which is missing approximately 30 residues at both the N- and C-terminal ends, comprises a long N-terminal arm, six long α helices, and four short helices.47
Each monomer has two hydrophobic cavities, and a large hydrophobic region is situated on one side of the dimer surface. It was proposed that the dimer binds to the mitochondrial inner membrane in an interfacial manner through hydrophobic regions as the opposite side of the dimer surface is relatively hydrophilic.47 CAVER protein analysis software predicts that another hydrophobic cavity may exist near the membrane surface, and this second cavity connects the di-iron active site with the membrane exterior. On this basis, the authors proposed that each hydrophobic cavity connects to a single ubiquinol near the active site with their quinol rings positioned at the lowermost part of each cavity.62 The active site, which is positioned within a hydrophobic region deep inside the molecule, consists of the di-iron center, along with four Glu residues (Glu-123, Glu-162, Glu-213, and Glu-266) and two His residues (His-165 and His-269), all of which display complete conservation.62 In addition, the conserved hydrophobic residues (Leu-122, Ala-126, Leu-212, Ala-216, Tyr-220, and Ile-262) are within 6 Å of the di-iron center (Figure 3D).8 The hydrogen bond network is significant for the stabilization of the AOX active site. The two monomers are connected by an amorphous biaxial approximately perpendicular to the bundle. In addition, the N-terminal arm of one monomer reaches the other monomer, which indicates that the arm is crucial for dimerization. In addition to the active site, the oxidation–reduction cycle of AOXs involves major conformational perturbations and carboxylate shifts.47
3.3. Properties
As with most redox proteins, infrared spectroscopy identified the largest IR bands in the amide I (peak/trough at 1658/1641 cm–1) and amide II (peak/trough at 1544/1554 cm–1) regions, with the most common alterations coming from the amide bond bands of the amide I (mainly C=O) and amide II (mainly N–H) polypeptide backbone.51 Alignment of the AOX crystal structure to ribonucleotide reductase (RNR), another protein of the di-iron carboxylate family, revealed a number of structural discrepancies around the central core.6 The O2 binding states of TAO were investigated by using quantum mechanics/molecular mechanics (QM/MM) theoretical level. The most stable O2 binding state is the end-on form, while the side-on type has higher energy. The O2 moiety unstably combines the Fe(II) atom in the O02 state rather than in the superoxo (O–2) or the peroxo (O2–2) state. The investigation also encompassed the coordination of other ligands, including water, Cl–, CN–, CO, N3–, and H2O2, and it was observed that H2O2 exhibits the highest affinity for the Fe(II) site.9
The sequencing analysis of AOX genes and proteins has also made some progress. Liu et al.28 showed that the DNA sequence of the Annulohypoxylon stygium AOX is 1320 nucleotides and involves three introns of 152, 54, and 58 nucleotides, respectively. The nucleotide composition of AsAOX is comprised of 1056 coding sequences, which in turn encode 351 amino acids. The molecular weight of AsAOX protein is 40.36 kDa, and the theoretical isoelectric point is 9.37 kDa. In-depth analysis of the conserved domains indicates that AsAOX protein is part of the ferritin-like superfamily, which facilitates dioxygen-dependent oxidation–hydroxylation reactions within di-iron centers.28 Besides, in an investigation of the sequence analysis pertaining to the 5′-flanking region of the AOX gene in A. niger, a possible heat shock element (HSE), as well as a stress response element (STRE), which could contribute to stress response, were identified.26
In regards to the catalytic activity of AOX across species, it has been demonstrated that the TAO displays the highest Vmax, followed by AOX in Arabidopsis thaliana and then AOX in S. guttatum.63 Another study has revealed that the activity of AOX in C. auris and C. albicans is comparable with TAO, albeit at a lower level.64 The Vmax of the wild-type TAO is 270 ± 28 μmol Q1H2 min–1 mg–1. The conserved residues in quinol binding within the active site of TAO is essential for catalytic activity, as the mutations within the hydrophobic cavity led to over 95% reduction in Vmax compared with the wild type.65 The inhibitory effects of several inhibitors on the wild-type AOX and some membrane-bound mutations were studied. As classic inhibitors of AOX, salicylic hydroxamic acid (SHAM) and octyl gallate had low efficacy, while quinol mimetic inhibitors, including ascochlorin, ascofuranone, colletochlorin B, and colletochlorin D, worked well. In contrast, the effect of inhibitor potency exhibited a noticeable difference on mutations. Arg-118, Glu-215, and Thr-219 mutations resulted in a complete loss of inhibition, whereas Arg-96 and Asp-100 resulted in a slight reduction.65 It should be noted that AOX activity is not only determined by its protein content, which is regulated by the concentration of oxygen,66 but also by the rate at which the NADH dehydrogenases can reduce UQ.67 Furthermore, the kinetic parameters (Km and Vmax) of the recombinant AOX of C. albicans and C. auris, which were expressed in a self-assembled proteoliposome system, were used to explore novel AOX inhibitors successfully, which will promote the development of new treatments for fungal infections.64 In summary, study of the crystal structure and the kinetic analysis of recombinant AOX will greatly facilitate the development of drugs targeting AOX.
4. Functions of AOX
4.1. Overview
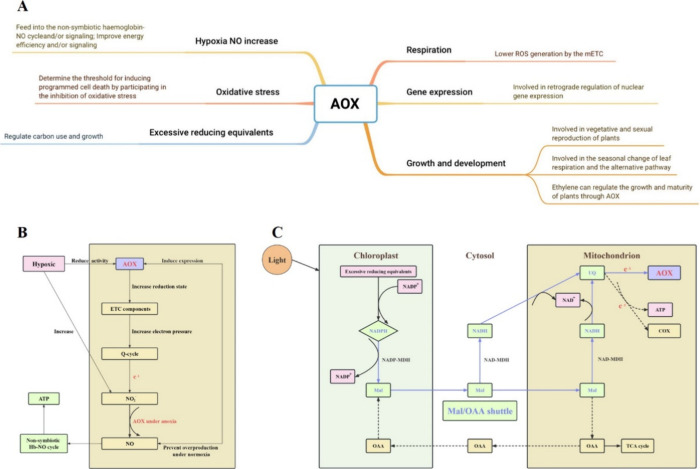
The alternative pathway (AP) can be regarded as a naturally evolved mechanism of rescue. When the ATP level is elevated or the cytochrome pathway (CP) is inhibited, the AP can ensure that electron flow, the glycolytic pathway, and the tricarboxylic acid (TCA) cycle run smoothly to maintain the general physiological activities.68 AOX contributes to the protection of the intracellular oxygen–water concentration, thereby forming an H2O molecule that allows four electrons to pass through AOX and ensure electron flow. Therefore, AOX suppresses the generation of ROS.10 A cycle of hypoxia or posthypoxia reoxidation, which produces ROS, often occurs in animals and plants. AOX participates in the antioxidant defense system when the ROS levels are increased and the concentrations of O2 together with its one-electron reductants in the mitochondria are decreased.69 It has been demonstrated that even minor alterations in electron transport rate or membrane potential can exert a significant influence on the production of ROS.25 Furthermore, AOX helps to remove the blockage between carbon metabolism, electron transport, and the ATP cycle, thus ensuring the integration of respiration with other metabolic processes.70 The existence of AOX gives the respiratory system inherent adaptability concerning the extent of coupling between carbon metabolism pathways, ETC activity, and ATP turnover71 that are the foundation of many biological functions. A contrast exists between the roles of AOX in maintaining metabolic and signaling homeostasis. Changes in the activity of AOX required to offer signal homeostasis may be tiny in comparison with changes in AOX activity that offer metabolic homeostasis.71
In microorganisms, the function of AOX is most often related to the relief of oxidative stress. A few microorganisms either do not express AOX normally or express it at very low levels. However, chemical inhibition of the classical respiratory chain by respiratory inhibitors, for instance, antimycin A (AMA) and potassium cyanide (KCN), can induce AOX expression.72
From an evolutionary point of view, a more noteworthy feature is the resistance of AOX to sulfide inhibition.10 Sulfides are able to block the CP by the inhibition of complex IV, which cuts off the use of oxygen as the terminal electron acceptor.73 During evolution of early metazoans, it is possible that the key function of AOX was to allow oxidative metabolism through sulfide-resistant respiration, which would allow more ATP to be produced under other inhibitory conditions.74
To systematically clarify the evolutionary relationship of AOX, the protein sequences of AOX in different species were compared (Figure 4A), and an evolutionary tree is shown on the basis of the organisms subsequently mentioned (Figure 4B).
4.2. AOX in Plants
AOX in plants is comprised of subfamilies AOX1 and AOX2. AOX1 and AOX2 exist in dicots, whereas only AOX1 is found in monocots.75 According to expression profiles and deep sequencing data, the AOX genes have been further divided into four subfamilies: AOX1a–c/1e, AOX1d, AOX2a–c, and AOX2d.75 It has been reported that AOX1 participates in responses to numerous biotic and abiotic pressures, as well as the various and complex signals of respiratory status.76 By contrast, AOX2 is thought to be developmentally and constitutively expressed. However, some emerging evidence has indicated that AOX2 can also respond to abiotic stresses, for instance, oxygen or copper deprivation, salinity, and osmotic stress.77,78
The main functions and mechanisms of AOX in plants are summarized in Figure 5A. The AP leads to heat transformation instead of ATP and reduces ROS production as electrons bypass complex III and IV.79 In addition, AOX plays a role in preventing the arrest of electron flow and facilitating the maintenance of intracellular oxygen–water concentration. Consequently, it enables the optimization of respiration both in normal and stressed conditions.80 AOX in plants is able to alleviate several types of stress, including oxidative stress, cold stress, heat stress, drought stress, and salt stress, with the reduction of ROS generation as a general mechanism.81 Stress conditions affect mitochondrial function, and this influences the rate of mitochondrial ETC-generated ROS.82 The role of the AP in sustaining the homeostasis of the redox state is of great importance, which may be attributed to the ability of AOX to reduce ROS leakage.71 In addition to its function in maintaining energy homeostasis in the cell, AOX can play a crucial role in stress responses by mitigating the excessive production of ROS in mitochondria.83 The mechanism is that excessive reducing equivalents in chloroplasts are shuttled to mitochondria in the form of NADPH via malate/oxaloacetate where they are effectively oxidized by the AP (Figure 5C).84 Then, the diminishment state of coenzyme Q, serving as the corresponding contributor to AOX reduction, decreases the generation of the superoxide anion radical and ultimately reduces the generation of hydrogen peroxide, the most stable ROS.18 AOX offers an extrachloroplastic means to enhance the status of chloroplast energy pools (ATP, NADPH), which is a mechanism to minimize metabolic imbalances by relaxing the tight coupling between NAD(P)H oxidation and ATP formation.25 AOX serves crucial functions in the plant metabolic adjustment to abiotic stresses through regulating the balance of carbon and nitrogen, carbon utilization efficiency, and the ratios of ATP/ADP and NAD(P)H/NAD(P)+.85 In addition to metabolic regulation, the expression and activities of AOX also change when the organism suffers from stress.85 For instance, in A. thaliana, the metabolic reactions of both roots and leaves to sublethal cadmium stress were influenced by the expression of AOX1a.86 α-Keto acids, such as pyruvate, can stimulate AOX activity.87 The defense elicitor flagellin (flg22) can also induce AOX to reduce superoxide, hydrogen peroxide, and peroxynitrite.88
Figure 5.
Main functions of AOX in plants. (A) Main functions of AOX in plants. (B) Interactions between AOX and NO production. (C) Stress responses regulated by AOX in plants. Mal, malate; OAA, oxaloacetate; NAD(P)-MDH, NAD(P)-malate dehydrogenase.
The influence of AOX on redox is not limited to ROS; the interactions between AOX and NO production (Figure 5B) are also important. As reported, NO was increased in transgenic tobacco plants lacking AOX, which indicated that AOX could prevent the overproduction of NO under normoxia.89 It was shown that AOX induction by NO had a protective effect against the tobacco mosaic virus and decreased the production of ROS.90 An additional investigation demonstrated the participation of AOX in the generation of NO produced by the ETC during hypoxic conditions. It was proposed that electron pressure within the Q-cycle of complex III can lead to the leakage of a single electron, which subsequently produces NO2– and ultimately NO (NO2– + 2H+ + e– → NO + H2O).91 The increase in NO, which is dependent on AOX, has the potential to enter the nonsymbiotic hemoglobin–NO (Hb-NO) cycle. Although it was previously believed that AOX caused energy wastage, as stated above, under hypoxic conditions, AOX indirectly improves energy efficiency by driving the Hb-NO cycle.88 Additionally, through a feedback process, NO treatment could also induce the AP in Arabidopsis seedlings92 and peach fruit.93 In previous studies, the role of AOX was believed to be associated with energetically wasteful processes, but under anoxic conditions, AOX indirectly improved the energy efficiency by reducing nitrite to NO and driving the Hb-NO cycle.88 On the basis of the important role of AOX under hypoxia, extensive work has been focused on the derivation of waterlog-resistant species, such as rice. It was suggested that AOX overexpression in crop plants could be a target for the development of flooding/waterlog resistant varieties.94
AOX is crucial for plant development, as well, as it participates in metabolic rearrangements in plants during embryogenesis and cell differentiation.95 The modulation of AOX can be influenced by light in two ways: first, through direct control by photoreceptors, and second, indirectly through photosynthetically dependent changes in metabolites that impact AOX transcriptional levels, protein abundance, and post-translational modifications.96 During greening of etiolated wheat seedlings, light increased the expression of AOX1a and subsequently sustained the chloroplast function.97 Similarly, AOX is also active in reaction to phosphate supply.98 Under a macronutrient stress, the AOX of tobacco cells has a regulatory influence on the efficiency of carbon use and growth.99 AOX is also crucial in controlling cell metabolism. In wild-grown Populus × canadensis, AOX is involved in the seasonal change of leaf respiration.100 AOX is a necessary condition for vegetative and sexual reproduction, and may determine the threshold for inducing programmed cell death by participating in the inhibition of oxidative stress.101,102 During germination in Vigna unguiculata, systemic coexpression of AOX1 and AOX2b was observed.103 In A. thaliana, overexpression of AOX1a has been observed to effectively prevent cell death induced by aluminum through attenuating ROS levels during the early stages of the stress reaction.104 In addition to regulating energy metabolism, AOX also participated the retrograde regulation of nuclear gene expression under stress and nonstress conditions. The functional interference of mitochondrial ETC alters the expression of many nuclear genes, including energy metabolism and protein synthesis genes in the mitochondria.105 As an endogenous plant hormone, ethylene can regulate the growth and maturity of plants through AOX. Ethylene changes the transcription of AOX1a in cadmium-exposed A. thaliana leaves, which plays a negative feedback role in ethylene biosynthesis and/or signaling.81 In the thermogenic flowering organs, mitochondria perform basic rearrangement of respiratory chain and make AOX the unique terminal high activity oxidase. The energy from substrate oxidation is converted into heat through this respiratory chain, which promotes evaporation of volatile substances that attract pollinators.106
4.3. AOX in Animals
AOX has been identified in a wide range of animal phyla, including tunicates, gastropods, bivalve molluscs,10 and copepods.107 However, it is absent in several species, including their sister groups, three highly advanced crown groups: vertebrates, insects, and cephalopod molluscs.108 A hypothesis is proposed to explain that the disappearance of AOX from most animals may be associated with the locomotive activity of the organism.109 Now, many studies have identified AOX-encoding genes in animal genomes and showed that these proteins could act as catalysts.43 AOX may inhibit the adverse consequences of the limited respiratory chain in animals.
In marine invertebrates, AOX plays a dominant role in tolerance to oxidative stress.110 In Drosophila, the introduction of AOX derived from Ciona intestinalis has the potential to rectify the defects of mitochondrial oxidative phosphorylation.111 In the fly, AOX may disturb sperm production through thermogenesis,112 while AOX in the molluscs Crassostrea virginica and Diplodon chilensis participates in tolerance to pressure.113 In Oncomelania hupensis, AOX can be significantly activated under niclosamide-induced stress, thus reducing oxidative stress in the snail.114
Widespread expression of AOX in a genetically tractable murine model confers robust resistance to inhibitors of the respiratory chain.115 Moreover, the expression of AOX allows bypass of the cytochrome C oxidase restriction and contributes to limiting mitochondrial ROS overproduction.116 The xenotropic expression of AOX from the tunicate Ciona intestinalis in murine mitochondria persists its freely diffusible ability.117 The effect of AOX on the physiological function of animals has also been studied. In mice, heterologously expressed AOX could attenuate inflammation.118 In Tigriopus californicus, the level of the AOX protein increased under cold and heat exposure relative to the normal feeding temperature, which suggests a potential role of AOX in transforming energy and heat.107 In Anadara broughtonii, AOX might contribute to tolerance to acute sulfide exposure.74
4.4. AOX in Algae
There are two pathways involved in regulating AOX1 transcription in C. reinhardtii; one is responsive to the metabolic shift caused by a transition from using ammonium as a nitrogen source to utilizing nitrate, while the other is responsive to stresses.76 The expression of AOX was upregulated in cadmium exposure119 and sulfur starvation120 in C. reinhardtii. AOX helps to protect the photosynthetic apparatus of C. reinhardtii from photodamage. When the electron carriers involved in the process of photosynthesis are in a state of high reduction, a chloroplast–mitochondria coupling reduces O2 through AOX-dependent respiration, especially using AOX1, thereby safely dissipating photosynthetically derived electrons.68 In addition, AOX is also involved in an increase of heat stress tolerance in C. reinhardtii, and the induction of AOX1 under heat stress depends on extracellular Ca2+ concentration.121
4.5. AOX in Protists
AOX is widespread in protists and mainly studied in T. brucei. T. brucei is the precipitating factor of human African trypanosomiasis, which affects 70 million underprivileged individuals mostly in sub-Saharan Africa and is spread by the biting of infected tsetse flies (Glossina spp.).122 Respiration of the BSF of T. brucei depends solely on TAO. In T. brucei, TAO is upregulated in the BSF and acts as the sole terminal oxidase under these conditions, thereby facilitating the reoxidation of cytosolic glycerol-3-phosphate.123 Indeed, throughout their life cycle, trypanosomes have demonstrated the ability to adjust their energy metabolism in response to the availability of nutrients within their surroundings. Thus, the procyclic form of T. brucei has a completely functional respiratory chain, and synthesizes ATP through mitochondrial oxidative phosphorylation. By contrast, glycolysis serves as the sole energy production for the bloodstream trypomastigotes of T. brucei, which exist in the mammalian host.124 Because of the essential role of AOX in BSF, it has become an important target against trypanosomiasis.
4.6. AOX in Fungi
It is clear that the role of AOX in mitigating or averting oxidative stress is of utmost importance. The decline in the reduction state of coenzyme Q, which is the AOX reduction equivalent donor, reduces the generation of superoxide anion radicals and hydrogen peroxide. The reduction of ROS is critical in fungal response to various kinds of stress. In Ganoderma lucidum, AOX affects the biosynthesis of ganoderic acid through regulation of intracellular ROS levels.125 In Aspergillus fumigatus, AOX confers resistance against oxidative stress,126 while in A. niger, the expression of AOX directly or indirectly responds to heat shock and oxidative and osmotic stresses, which potentially plays a role in carbohydrate metabolism and generation of citric acid.26 In Sclerotinia sclerotiorum, AOX is involved in regulating growth, development, and resistance to oxidative pressure.127 In Phycomyces blakesleeanus, AOX is involved in phosphate metabolism.128 In Rhizoctonia oryzae, the activity of alternative respiration is well correlated with fumaric acid fermentation.129 In Paracoccidioides brasiliensis, AOX contributes to maintaining cellular homeostasis, morphological transitions, oxidative stress, and mycelium-to-yeast differentiation.130 In Monascus ruber, AOX is partly responsible for the resistance of conidia to stress conditions.27
Previous studies have shown that AOX is associated with the virulence of several pathogenic fungi.131 AOX promotes sustained respiratory activity and is generally stimulated by stress conditions, for instance, AOX in C. albicans is linked to reduced fluconazole susceptibility.16 Heterologous expression of an AOX from M. perniciosa in S. cerevisiae provides antioxidant function.44 The role of mitochondrial AOX is crucial in the processes of growth and sporulation in the primitive fungus Blastocladiella emersonii.132 In Aspergillus nidulans, AOX regulates development, redirects cellular metabolism, maintains ATP and NADPH supplies, and sustains cellular ROS to affect aflatoxin biosynthesis.133 In A. stygium, a companion fungus of Tremella fuciformis, AOX participates in oxidative stress tolerance and melanin synthesis. In A. stygium, AOX participates in the biosynthesis of secondary metabolite.28
4.7. AOX in Bacteria
In the case of bacteria, AOX was first reported in Novosphingobium aromaticivorans in 2003. In N. aromaticivorans, AOX has a terminal oxidase activity, and the oxygen content and carbon source of the growth medium have a great influence on the expression of AOX.134 In Vibrio fischeri, the AOX gene was upregulated in response to nitric oxide and mediated the resistance to NO inhibition.135 AOX was also found in several nodules infected with rhizobia, such as Bradyrhizobium japonicum(136) and Rhizobium leguminosarum.137 Some studies have investigated the regulation of AOX activity in the symbiosis between Rhizobia and legumes.138
To summarize, the contribution of AOX is of paramount importance in multiple facets of the biological processes exhibited by diverse organisms.
5. Inhibitors of AOX
5.1. Overview and Classical Inhibitors of AOX
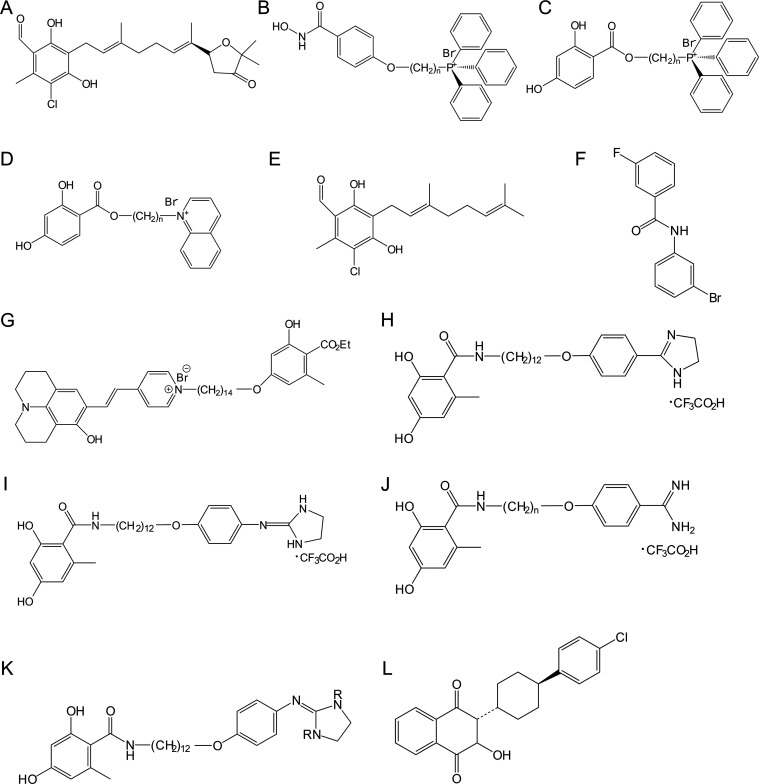
Because of the crucial function of AOX in many organisms, it is logical to develop drugs that block the activities of AOX. As early as 1971, an initial insight into AOX inhibitors was provided by Bendal and Bonner, who demonstrated that the activity of AOX is suppressed by chelators that combine variable valence cations, particularly Fe.19 Among them, thiocyanate, α,α′-dipyridyl, and 8-hydroxyquinoline were likely to be candidates that function as a terminal cyanide-resistant oxidase inhibitor. At present, SHAM is the most researched inhibitor and can suppress various cell reprogramming processes.139 SHAM was demonstrated as a specific inhibitor of TAO at 1 mM as early as 1976.140 Subsequently, studies reported the synergistic effect of SHAM with inhibitors of mitochondrial complex III (e.g., atovaquone)141 or complex IV (e.g., cyanide),142 as well as with glycerol.143 In addition, SHAM enabled the accumulation of glycero-3-phosphate in glycosomes and facilitated its conversion to glycerol.144 SHAM could also increase ROS levels within the mitochondria of procyclic trypanosomes by inhibiting TAO, thus leading to the oxidative damage of cellular proteins.145 In C. albicans, a synergistic effect of SHAM and fluconazole was also found.16 Additionally, the combination of SHAM and sodium nitroprusside could block the adaptability and viability of C. albicans.146 In P. brasiliensis, SHAM inhibited the transformation of mycelium to yeast, which is a key step in host colonization.147 In addition to trypanosomes and fungi, SHAM also has some effect on plants, as it could inhibit the growth of olive roots148,149 and hypoxic NO production.88 However, SHAM is not ideal in some ways because of its poor water solubility.150 Ascofuranone, an antibiotic purified from the fungus Ascochyta visiae, has been recognized as the most effective inhibitor of the AOX family enzymes discovered so far.151 Until the early 2000s, among the ascofuranone-derived compounds, ascofuranone (Figure 6A) was the sole TAO inhibitor that showed in vivo therapeutic activity in various murine trypanosomiasis models.152 In addition to the compounds previously mentioned, aluminum could also inhibit the AP in Jatropha curcas cells. As a consequence, the AOX expression was upregulated as a defense mechanism to maintain mitochondrial respiration.153
Figure 6.
Structures of representative AOX inhibitors: (A) ascofuranone, (B) SHAM-TPP conjugates, (C) 2,4-DHBA-TPP conjugates, (D) 2,4-DHBA-quinolinium conjugates, (E) ascofuranone derivative, (F) N-phenylbenzamide derivative, (G) one molecular rotor-based fluorescent inhibitor, (H) imidazoline, (I) 2-phenylimidazolin-3-ium, (J) benzamidine derivatives, (K) aminoimidazoline derivatives, and (L) atovaquone.
On the basis of these findings, researchers conducted further structure–activity relationship research on AOX inhibitors. These studies are mainly divided into two categories: one is based on using the crystal structure of AOX for design and synthesis, and the other is based on biological activity screening and structural modification. In addition, some scholars have combined the two to find and synthesize more effective inhibitors.
5.2. Development of AOX Inhibitors
AOX, which is completely absent in primates, is conserved in T. brucei subspecies and is, thus, a potential curative target for trypanosomiasis. Since the reaction catalyzed by TAO in trypanosome BSF mitochondria is the only pathway for obtaining sufficient energy for trypanosome survival, inhibiting this pathway frequently results in the death of the parasite.154 The localization of TAO at the interface of the inner mitochondrial membrane has motivated the development of potent 4-hydroxybenzoate- and 4-alkoxybenzaldehyde-based inhibitors, for example, SHAM-TPP (triphenyl phosphonium) conjugates (Figure 6B), 2,4-DHBA-TPP conjugates (Figure 6C), and 2,4-DHBA-quinolinium conjugates (Figure 6D),47,155 which possess lipophilic cations as the mitochondrion-targeting moieties.13 Various sets of TAO inhibitors (Figure 6A,E) synthesized on the basis of ascofuranone, a natural isoprenoid antibiotic, have been reported.156 Purified recombinant AOX and isolated A. maculatum mitochondria show sensitivity to a range of AOX inhibitors, including SHAM, n-propyl gallate, ascofuranone, colletochlorin B, and ascochlorin. Ascochlorin, ascofuranone, and its derivative colletochlorin B are much more potent than the conventional range of AOX inhibitors, such as SHAM and octylgallate, and ascofuranone is more potent than both ascochlorin and colletochlorin B.157 Cationic and noncationic 4-hydroxybenzoate have inhibitory effects on TAO that lacks its N-terminal mitochondrial targeting signal (ΔMTS-TAO), and 4-alkoxybenzaldehyde derivatives inhibit the growth of T. brucei and Trypanosoma congolense.155 Further studies have investigated the SAR of 4-alkoxybenzoic acid.13 An important step in the biological activity of TAO inhibitors requires the compound accumulating in the mitochondria. It is expected that concentrating certain compounds in the T. brucei mitochondria will affect the mitochondrial membrane potential Ψm because of the accumulation of cations in the mitochondrial matrix and breakdown the mitochondrial function of maintaining the ion gradients.158 This works for various diamidines,159 choline-derived dications,160 phenanthridines,161 and bisphosphonium compounds.162 The latter compounds were shown to act on the mitochondrial FoF1 ATPase of T. brucei.163 It was proposed that ether-linked tails represent a potential class of more synthetically amenable TAO inhibitors.164 The activity against T. brucei in vitro was demonstrated to be promoted by the lipophilic cation attached to the 2,4-DHBA or SHAM scaffold. The 2,4-DHBA scaffold provided the most effective compounds, and the 14-methylene linker appeared to be ideal for TAO inhibition and trypanocidal action.165
Ferulenol is considered to be a competitive inhibitor of AOX, which has been applied to probe the binding site of ubiquinol.166 Crucially, inhibitors with the 2,4-dihydroxy-6-methyl scaffold are stable in serum liver fractions and active in the acutely infected STIB900 mice.155 It was possible to enhance the trypanocidal activity of TAO inhibitors using lipophilic cations, such as triphenylphosphonium or quinolinium salts, which were used as mitochondria-targeting moieties. The inhibitor and LC group must be separated by a long flexible chain, typically consisting of 14 methylene units, to allow optimal interaction with the TAO catalytic site.13 One or two more chlorine atoms in the TAO-binding pharmacophore were proposed to enhance TAO inhibition.164 However, subsequent studies have challenged this SAR and proposed that the addition of chlorine atoms correlated negatively with the activity against T. brucei.158
Some scholars have studied and compared the inhibitory effect of many compounds on AOX.13 The N-(3-bromophenyl)-3-fluorobenzamide (Figure 6F) was the best inhibitor against recombinant AOX from M. perniciosa. This compound interacted very well with AOX in the membrane mimetic system, as demonstrated by 1H-saturation transfer difference (STD)-NMR.167 The synthesis, photophysical characterization, and biological activity of fluorescent TAO inhibitor probes that combine a julolidine-based molecular rotor with a cationic pyridinium salt have been investigated for their potential to improve the targeting of mitochondria. Lipophilic cations were conjugated to inhibitors of TAO via C14 linkers to achieve optimal interaction with the active site of enzyme. Several fluorescent analogues were synthesized and characterized for their intracellular accumulation and distribution. Among them the most active compound for inhibiting TAO was (E)-1-{14-[4-(ethoxycarbonyl)-3-hydroxy-5-methylphenoxy]tetradecyl}-4-[2-(8-hydroxy-2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-9-yl)vinyl]pyridin-1-ium bromide (Figure 6G, IC50 = 1.6 nM), which showed the advantage of carboxylate probes.158 To further improve the activities of compounds with the 2,4-dihydroxy-6-methylbenzoic acid scaffold, the structure–activity relationship of imidazoline- and benzamidine-based TAO inhibitors was measured (Figure 6H–K). Compounds including 2-phenylimidazolin-3-ium (Figure 6I, EC50 = 1.72 μM), benzamidinium (n = 12 in Figure 6J, EC50 = 3.3 μM), and 2-(phenylamino)imidazolin-3-ium (R = H in Figure 6K, EC50 = 2.7 μM) had outstanding inhibitory effect on T. brucei.168 In addition to chemical synthesis, natural products are also important sources of AOX inhibitors. The anti-trypanosomal properties of Anogeissus leiocarpa extracts and their inhibitory effect on TAO have been identified. Only fraction four from EtOAc of A. leiocarpa, which contains carbohydrates, fatty acids, fatty acid esters, aromatic compounds, and organosulfide, significantly inhibited the activity of TAO (EC50 = 0.024 μg/μL). In the future, potential lead compounds that inhibit TAO may be isolated.169
The research of inhibitors of AOX in fungi is a hot topic nowadays. For instance, N-phenylbenzamide derivatives showed potent efficacy against the phytopathogen M. perniciosa.170 In fungal cells, the inactivation of complex I is one of the major sources of mitochondrial ROS accumulation and facilitates cell death.171 The utilization of fungal complex I inhibitors have the potential to induce fungistatic effects by restricting the production of ATP and fungicidal effects by increasing ROS levels.172 Honokiol, derived from M. officinalis, has been demonstrated to exert antifungal activities through inhibiting complex I.173S. cerevisiae expressing mitochondria-localized AOX could be used as an efficient platform for high-throughput screening of potential AOX inhibitors.44 It was reported that C. albicans was resistant to atovaquone (Figure 6L), which was not effective in inhibiting glucose-dependent growth. This study described the efficacy of atovaquone in suppressing respiration as comparable with antimycin A.174 In P. falciparum, when combined with AOX inhibitors, the efficacy of atovaquone was promoted significantly.141 On the basis of this observation, atovaquone failing to inhibit the glucose-dependent growth of C. albicans may be the result of alternative respiration induction. In a study of C. albicans responses to respiration inhibition caused by the NO donor, sodium nitroprusside (SNP), and the AOX inhibitor, SHAM, it was shown that cells exposed to inhibitors exhibited a more rapid transition to hyphal growth when inhibition was lifted.175 In an aoxΔmutant, no increase in hyphal switching was observed after pretreatment of SNP plus SHAM, which suggests that AOX plays an important role in the hyphal switching phenotype.175
It is worth noting that some of the inhibitors fall under the classification of pan-assay interference compounds (PAINS) or possess frequent attack segments that are capable of engaging in pleiotropic interactions with a significant number of proteins. This is a problem that necessitates further investigation in subsequent research endeavors.
5.3. Binding Sites of AOX Inhibitors
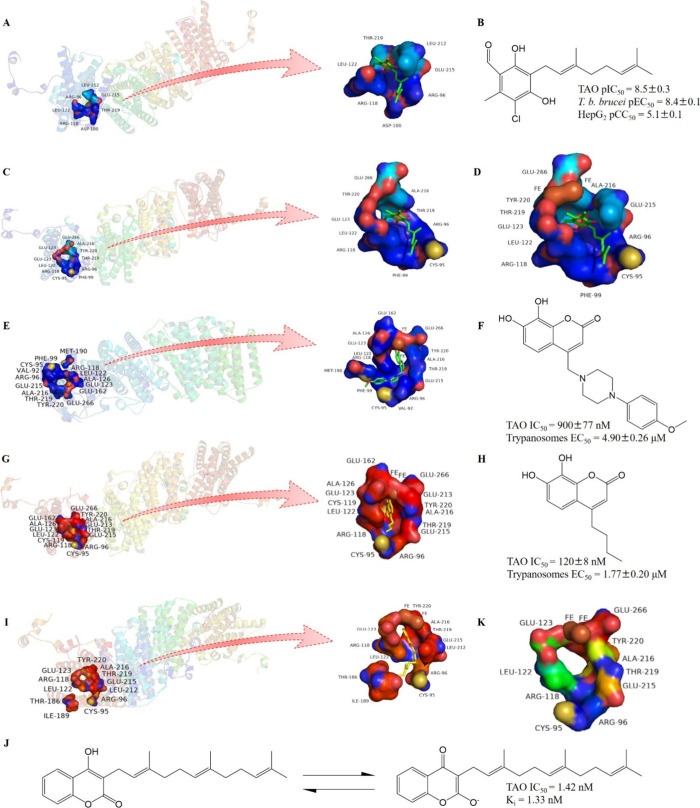
The key binding sites (Figure 7) of TAO with inhibitors have been also studied. In TAO, there is a hydrophobic cavity (Figure 7A) that could bind to inhibitor colletochlorin B (Figure 7B), quinol and quinone.65 Another site (Figure 7C) is also found to bind to colletochlorin B, as well as the inhibitor ascofuranone and derivatives. It has also been established that Leu-122, Arg-118, Thr-219, and Cys-95 are the residues that interact frequently with the ligands.62 There is also the binding site of AOX to the inhibitor 4-hydroxybenzoate and 4-alkoxybenzaldehyde derivatives in which Arg-96, Arg-118, Leu-122, Ala-216, and Thr-219 are considered important for forming hydrophobic interactions or hydrogen bonds.155 These two sites are adjacent, overlapping, and close to the di-iron center, which can be combined to obtain the important inhibitor binding site (Figure 7D). Located within a hydrophobic cavity extending from the di-iron center to the phospholipid bilayer, this site boasts the presence of two Leu residues positioned at the center of the channel. These Leu residues play a crucial role in directing both the substrate and AOX inhibitors toward the appropriate conformation for effective interaction with the di-iron center. Nevertheless, several studies have explored the impact of AOX antagonists on both wild-type and mutant forms of AOX to reveal that alterations in the bottleneck of the hydrophobic cavity can reduce their sensitivity to AOX inhibitors. Consequently, it has been proposed that future design of AOX inhibitors should develop structures that are less reliant on the orientation of the two-Leu residues.42 Other scholars have studied the binding sites of compound 17 (Figure 7E,F) and its derivative compound 17b (Figure 7G,H),175 as well as ferulenol (Figure 7I,J),60 with TAO. It can be observed that these four binding sites share some common amino acid residues, which are conserved in various species (Figure 4A, blue dotted box) and likely to be conserved binding sites for TAO inhibitors (Figure 7K). The binding mechanism of rotenone to respiratory complex I is contingent upon the flexibility of the ligand;176 it is also worthy to investigate the correlation between mechanism of this site and the flexibility of its ligands.
Figure 7.
The key binding sites of TAO with inhibitors. (A) The binding hydrophobic cavity of TAO with colletochlorin B. (B) Structure and activities of colletochlorin B. (C) The binding site of TAO with colletochlorin B. (D) The two binding sites mentioned in (G) and (H). (E) The binding site of TAO with compound 17. (F) Structure and activities of compound 17. (G) The binding site of TAO with compound 17b. (H) Structure and activities of compound 17b. (I) The binding site of TAO with ferulenol. (J) Structure and activities of ferulenol. (K) The conserved binding site of TAO. These figures of crystal structures were drawn with PyMol, and the AOX model used is TAO (A,C,D,K, PDB: 3W54; E, PDB: 5GN7; G, PDB: 5GN9; I, PDB: 5ZDP).
In Table 1, information about more representative inhibitors and derivatives has been listed.
Table 1. Inhibitors of AOX and Their Activities13,150,155,156,164,177−180.
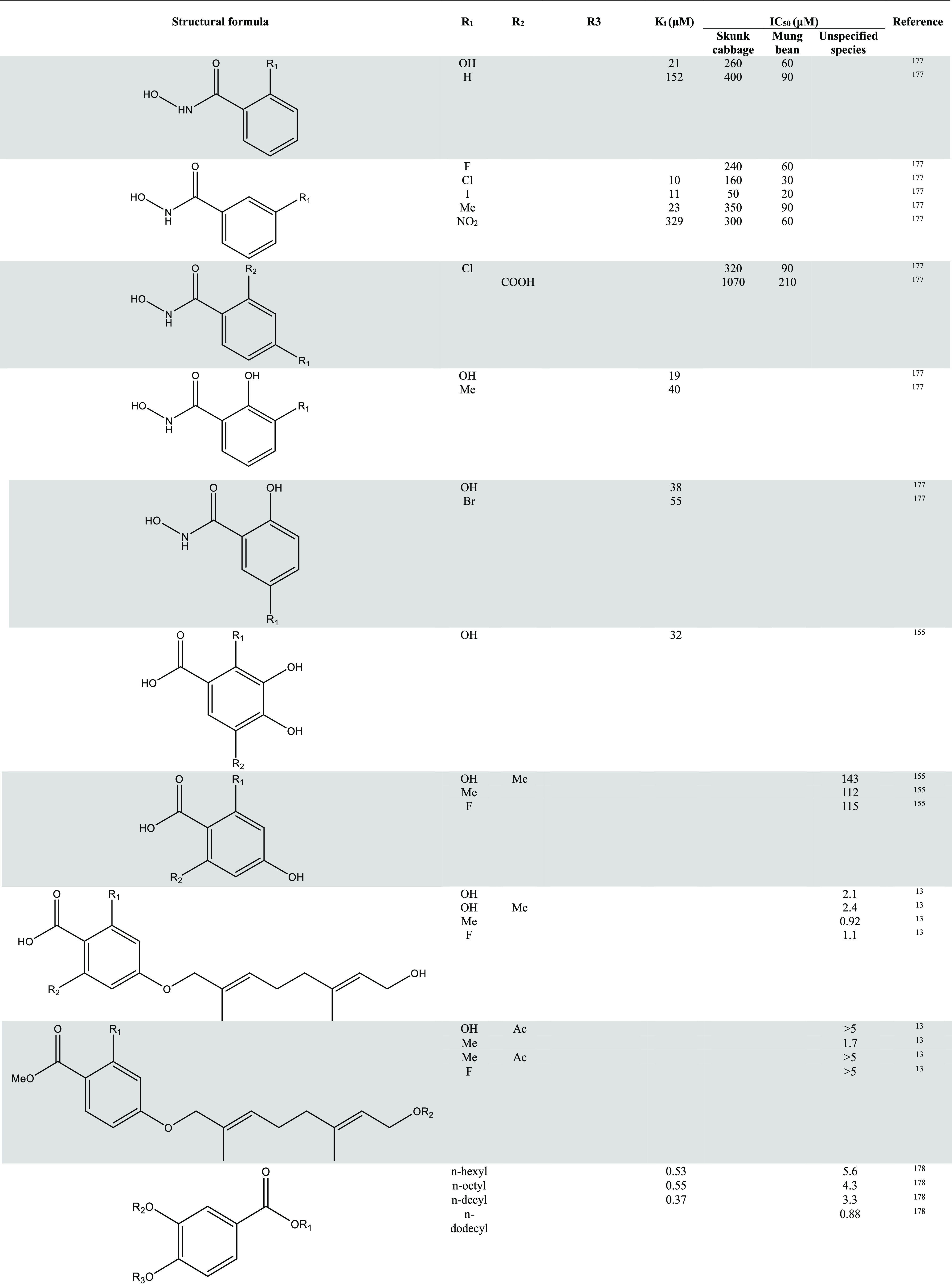
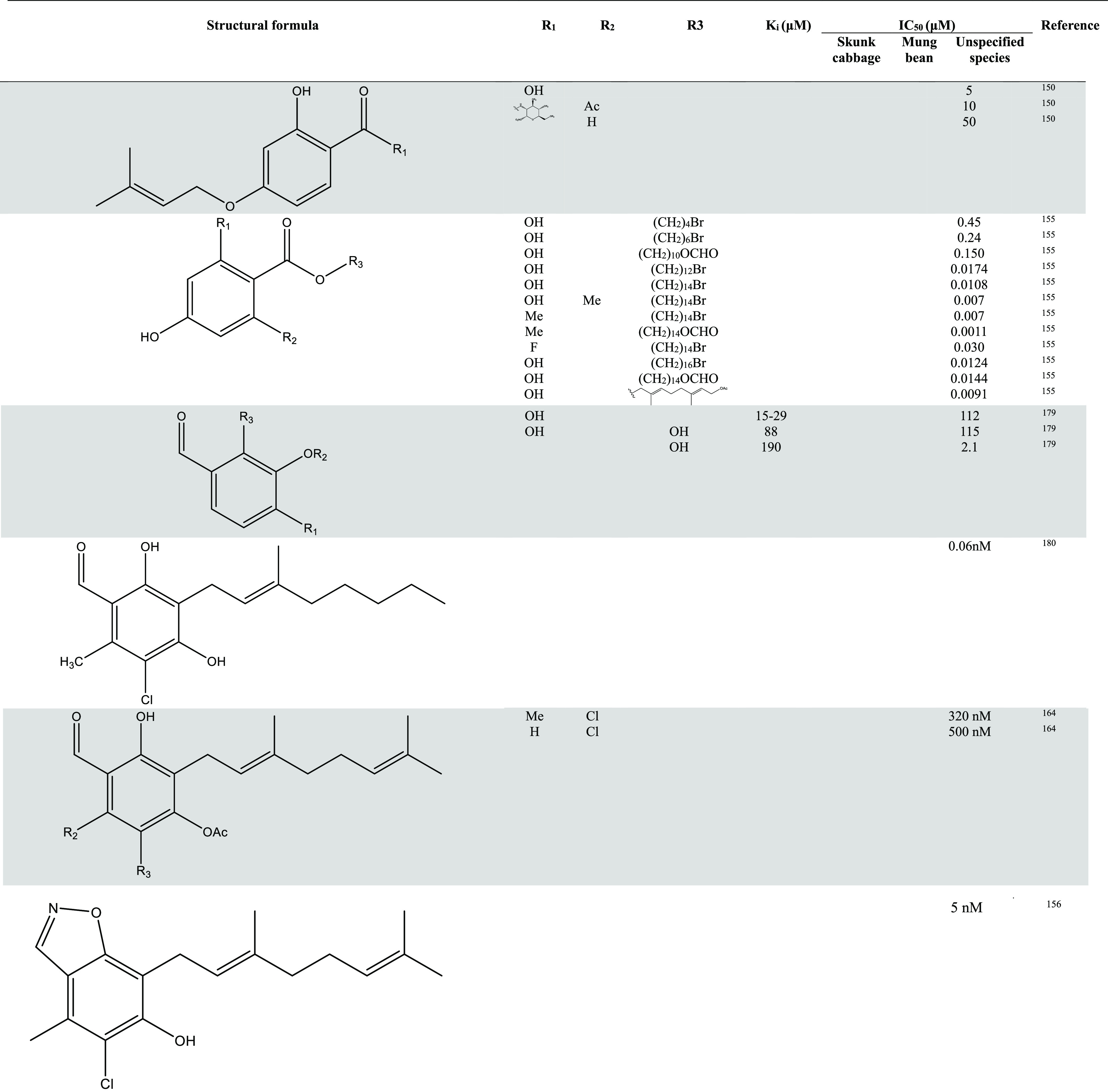
Despite the abundance of research on AOX inhibitors, the availability of compounds suitable for drug development or clinical trials is limited. This issue continues to be a central focus and presents a significant challenge for future investigations.
6. Discussion
Extensive research has been conducted on the investigation of the categorization and functionality of AOX in plants. However, further examination is required to determine the specific growth conditions, tissues, and developmental stages affected by various AOX isoforms. Additionally, future endeavors should aim to differentiate the roles played by AOX in response to acute and chronic stresses, thereby elucidating situations where AOX is fundamentally advantageous or indispensable.25
Even if organisms do not naturally possess AOX, AOX can be introduced into the mitochondria through xenotopic expression to refold and catalytically participate when there is a blockage in the cytochrome component of the ETC. Consequently, AOX is regarded as a valuable tool for investigating disease etiologies.181 Over the past decade, AOX encoding genes from various sources have been expressed in a wide range of animals, including Drosophila, mice, and human cultured cells.182 In instances where AOX is expressed in disease models, it has been observed that there is a notable therapeutic benefit in certain cases, whereas in other cases it fails to produce any positive outcome and may even exacerbate the condition (refer to ref (181) for further insights on the utilization of AOX in diverse disease models). Under certain circumstances where there is involvement of respiratory complexes beyond those present in the cytochrome segment, it appears that AOX is unable to function effectively. This could potentially be due to a deficiency of complex I that is associated with these particular conditions.181 The biological functions of AOX have gradually been established. In the context of respiratory inhibition, expression of AOX in human cells hinders the accumulation of the β-amyloid (Aβ) peptide associated with Alzheimer’s disease. Furthermore, it mitigates the detrimental phenotypic effects caused by the pathogenic human Aβ variant expressed in Drosophila neurons.183 The underlying mechanism remains elusive and will serve as a topic for future investigations.108 The absence of AOX in human cells has led to its exploration as a potential gene therapy tool for addressing mitochondrial dysfunctions and disorders in humans, and a variety of studies have shown the feasibility of this idea. A potential pathway for rescuing electron flow is offered by the heterologous expression of AOX, which could potentially alleviate harmful complications linked to respiratory chain dysfunction in diseases, such as Parkinson’s.182 In addition, the application of AOX in healthcare and aquaculture is also worth exploring.107
AOXs are widespread among human parasites, such as T. brucei,124Cryptosporidium parvum,184Blastocystis hominis,185 and opportunistic human fungal pathogens of Candida spp.24 In anti-infective drug development, AOX is attracting increasing attention in therapeutic research directed at human and plant pathogens. Current treatments for human African trypanosomiasis are far from achieving success: only subspecies (T.b. gambiense and T.b. rhodesiense, respectively) could be treated by the first-stage inoculations using pentamidine and suramin, which necessitates the administration through intravenous (IV) or intramuscular (IM) routes.186 Likewise, the second stage is restricted to nifurtimox–eflornithine combination therapy (NECT) and melarsoprol, in which NECT has no effect on the treatment of T.b. rhodesiense infection.164 All current treatments are limited by high toxicity of the compounds. Each of these therapies necessitates clinicians for IV/IM injections,186 which poses a significant challenge because of the widespread distribution of the population across a considerable area of continental Africa.164 Because of the high incidences of drug resistance, the current drugs used to treat and manage trypanosomiasis are far from ideal. Drug resistance for melarsoprol, as well as pentamidine, has been increasingly observed both in vitro and in clinical isolates and is associated with mutated aquaglyceroporins, which are necessary for uptake of compounds by trypanosomes.187 The creation of trypanosome vaccines is frequently considered the most promising method, although it is a long way from clinical success because of the efficient immune evasion of trypanosoma.188 These challenges highlight a pressing requirement for novel antitrypanosomic drugs, and AOX shows promise as a viable target for the design of antitrypanosomic drugs.
Respiration is closely related to cell growth and proliferation, nutrient metabolism, secondary metabolites regulation, and stress responses. Chemobiology-based cell factory is a hit subject for the production of drug molecules, nutrients, and macromolecules. The production of drug molecules or nutrients can be specifically enhanced by modifying cellular metabolic pathways. The respiration and cell proliferation always influenced the productivity of cell factory. As the critical oxidase located in the cyanide-resistant respiration, the effect of AOX on cell synthesis of drug molecules is worth further exploration. In addition to regulating metabolism, AOX induction results in stress tolerance. Inhibition of AOX has been observed to enhance fluconazole inhibition of the C. albicans growth.24 With the increasing occurrence of fungal resistance to azoles in recent years, effective and specific drugs targeting AOX may be a promising direction to find synergistic agents with azoles. The development of fungal AOX inhibitors was complicated by the fact that the crystal structure of TAO is the only structure that has been determined.47 Meanwhile, the resilience and plasticity of fungal respiration and the conserved characteristics of eukaryotic respiration mechanisms present a barrier for drug development. Identification and characterization of respiratory proteins particular to fungi are required, as well as clarification of the functions of alternative respiratory pathways.172 Meanwhile, the research on the regulation of alternate respiration and AOX inhibitors is not limited to fungi. The comprehensive analysis of AOX from structure to function will greatly help us to understand the mechanisms of cellular adaptation to diverse environments.
Although numerous functions of AOX have been identified, the underlying mechanisms of many of these functions require further exploration. This knowledge gap hampers the development of inhibitors that target these pathways. Additionally, some inhibitors possess restricted potential, and certain studies lack depth, which prompts inquiries into how to enhance the development of exceptional inhibitors into effective drugs. Addressing these issues necessitates the sustained efforts of scholars and is essential for AOX to emerge as a significant target in the pharmaceutical industry. Furthermore, AOX has been extensively and comprehensively studied in the field of plants. However, its significance lies more in the safeguard and treatment of protists, fungi, bacteria, and associated ailments. The research on AOX inhibitors also predominantly centers around these organisms. Consequently, the exploration of AOX in connection to these organisms emerges as a crucial field for the future advancement of AOX research.
Acknowledgments
This work was supported by Shanghai International Science and Technology Cooperation Project (21430713000), the National Natural Science Foundation of China (No. 82173867), Shanghai Pujiang Program (No. 21PJD081), and Shanghai Natural Science Foundation (No. 23ZR1478800).
Author Contributions
▽ J.Li, S.Y., and Y.W. contributed equally to this work. J.Li, S.Y., Y.W.: original draft and figures preparation, review and editing (equal). R.W., Y.L., J.Liu, Z.Y., R.T.: original draft and figures editing (supporting). M.W.: review and editing (supporting). Q.L., L.Y.: writing—original draft and figures preparation, revising (equal).
The authors declare no competing financial interest.
References
- Lenaz G.; Genova M. L. Structure and organization of mitochondrial respiratory complexes: a new understanding of an old subject. Antioxid Redox Signal 2010, 12 (8), 961–1008. 10.1089/ars.2009.2704. [DOI] [PubMed] [Google Scholar]
- Vercellino I.; Sazanov L. A. The assembly, regulation and function of the mitochondrial respiratory chain. Nat. Rev. Mol. Cell Biol. 2022, 23 (2), 141–161. 10.1038/s41580-021-00415-0. [DOI] [PubMed] [Google Scholar]
- Iwata M.; Lee Y.; Yamashita T.; Yagi T.; Iwata S.; Cameron A. D.; Maher M. J. The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (38), 15247–15252. 10.1073/pnas.1210059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus-Ortega M. G.; Cárdenas-Monroy C. A.; Flores-Herrera O.; Mendoza-Hernández G.; Miranda M.; González-Pedrajo B.; Vázquez-Meza H.; Pardo J. P. New complexes containing the internal alternative NADH dehydrogenase (Ndi1) in mitochondria of Saccharomyces cerevisiae: Mitochondrial complexes containing the alternative NADH dehydrogenase. Yeast 2015, 32 (10), 629–641. 10.1002/yea.3086. [DOI] [PubMed] [Google Scholar]
- Wikström M.; Gennis R. B.; Rich P. R. Structures of the intermediates in the catalytic cycle of mitochondrial cytochrome c oxidase. Biochim Biophys Acta Bioenerg 2023, 1864 (2), 148933 10.1016/j.bbabio.2022.148933. [DOI] [PubMed] [Google Scholar]
- Young L.; May B.; Shiba T.; Harada S.; Inaoka D. K.; Kita K.; Moore A. L.. Structure and Mechanism of Action of the Alternative Quinol Oxidases. In Cytochrome Complexes: Evolution, Structures, Energy Transduction, and Signaling; Cramer W. A.; Kallas T., Eds.; Springer Netherlands: Dordrecht, Netherlands, 2016; pp 375–394. [Google Scholar]
- Ebiloma G. U.; Balogun E. O.; Cueto-Díaz E. J.; de Koning H. P.; Dardonville C. Alternative oxidase inhibitors: Mitochondrion-targeting as a strategy for new drugs against pathogenic parasites and fungi. Medicinal Research Reviews 2019, 39 (5), 1553–1602. 10.1002/med.21560. [DOI] [PubMed] [Google Scholar]
- Moore A. L.; Shiba T.; Young L.; Harada S.; Kita K.; Ito K. Unraveling the Heater: New Insights into the Structure of the Alternative Oxidase. Annual Review of Plant Biology 2013, 64 (1), 637–663. 10.1146/annurev-arplant-042811-105432. [DOI] [PubMed] [Google Scholar]
- Yamasaki S.; Shoji M.; Kayanuma M.; Sladek V.; Inaoka D. K.; Matsuo Y.; Shiba T.; Young L.; Moore A. L.; Kita K.; Shigeta Y. Weak O2 binding and strong H2O2 binding at the non-heme diiron center of trypanosome alternative oxidase. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2021, 1862 (4), 148356 10.1016/j.bbabio.2020.148356. [DOI] [PubMed] [Google Scholar]
- McDonald A. E.; Vanlerberghe G. C.; Staples J. F. Alternative oxidase in animals: unique characteristics and taxonomic distribution. J. Exp Biol. 2009, 212 (16), 2627–2634. 10.1242/jeb.032151. [DOI] [PubMed] [Google Scholar]
- McDonald A. E.; Gospodaryov D. V. Alternative NAD(P)H dehydrogenase and alternative oxidase: Proposed physiological roles in animals. Mitochondrion 2019, 45, 7–17. 10.1016/j.mito.2018.01.009. [DOI] [PubMed] [Google Scholar]
- McDonald A. E.; Vanlerberghe G. C. Origins, evolutionary history, and taxonomic distribution of alternative oxidase and plastoquinol terminal oxidase. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 2006, 1 (3), 357–364. 10.1016/j.cbd.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Meco-Navas A.; Ebiloma G. U.; Martin-Dominguez A.; Martinez-Benayas I.; Cueto-Diaz E. J.; Alhejely A. S.; Balogun E. O.; Saito M.; Matsui M.; Arai N.; Shiba T.; Harada S.; de Koning H. P.; Dardonville C. SAR of 4-Alkoxybenzoic Acid Inhibitors of the Trypanosome Alternative Oxidase. ACS Med. Chem. Lett. 2018, 9 (9), 923–928. 10.1021/acsmedchemlett.8b00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruy F.; Vercesi A. E.; Kowaltowski A. J. Inhibition of specific electron transport pathways leads to oxidative stress and decreased Candida albicans proliferation. Journal of Bioenergetics & Biomembranes 2006, 38 (2), 129–135. 10.1007/s10863-006-9012-7. [DOI] [PubMed] [Google Scholar]
- Trevijano-Contador N.; Rossi S. A.; Alves E.; Landin-Ferreiroa S.; Zaragoza O. Capsule Enlargement in Cryptococcus neoformans Is Dependent on Mitochondrial Activity. Front Microbiol 2017, 8, 1423. 10.3389/fmicb.2017.01423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L.; Li M.; Cao Y.; Gao P.; Cao Y.; Wang Y.; Jiang Y. The alternative oxidase of Candida albicans causes reduced fluconazole susceptibility. J. Antimicrob. Chemother. 2009, 64 (4), 764–773. 10.1093/jac/dkp273. [DOI] [PubMed] [Google Scholar]
- Lloyd D.; Edwards S. W.. Electron Transport Pathways Alternative to the Main Phosphorylating Respiratory Chain. In Functions of Alternative Terminal Oxidases; Degn H.; Lloyd D.; Hill G. C., Eds.; Pergamon, 1978; pp 1–10. [Google Scholar]
- Rogov A. G.; Sukhanova E. I.; Uralskaya L. A.; Aliverdieva D. A.; Zvyagilskaya R. A. Alternative oxidase: Distribution, induction, properties, structure, regulation, and functions. Biochemistry (Moscow) 2014, 79 (13), 1615–1634. 10.1134/S0006297914130112. [DOI] [PubMed] [Google Scholar]
- Bendall D. S.; Bonner W. D. Cyanide-insensitive Respiration in Plant Mitochondria. Plant Physiology 1971, 47 (2), 236–245. 10.1104/pp.47.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger H. G.; Guy R. D. Cytochrome and alternative pathway respiration in white spruce (Picea glauca) roots. Effects of growth and measurement temperature. Physiologia Plantarum 1991, 83 (4), 675–681. 10.1111/j.1399-3054.1991.tb02486.x. [DOI] [Google Scholar]
- Chaudhuri M.; Hill G. C. Cloning, sequencing, and functional activity of the Trypanosoma brucei brucei alternative oxidase. Mol. Biochem. Parasitol. 1996, 83 (1), 125–129. 10.1016/S0166-6851(96)02754-5. [DOI] [PubMed] [Google Scholar]
- Maxwell D. P.; Wang Y.; McIntosh L. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. U.S.A. 1999, 96 (14), 8271–8276. 10.1073/pnas.96.14.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K.; Kang S. O. Molecular cloning and functional expression of alternative oxidase from Candida albicans. J. Bacteriol. 1999, 181 (13), 4098–102. 10.1128/JB.181.13.4098-4102.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K.; Kang S. O. Characterization of the gene family encoding alternative oxidase from Candida albicans. Biochem. J. 2001, 356 (2), 595–604. 10.1042/bj3560595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe G. C.; Dahal K.; Alber N. A.; Chadee A. Photosynthesis, respiration and growth: A carbon and energy balancing act for alternative oxidase. Mitochondrion 2020, 52, 197–211. 10.1016/j.mito.2020.04.001. [DOI] [PubMed] [Google Scholar]
- Honda Y.; Hattori T.; Kirimura K. Visual expression analysis of the responses of the alternative oxidase gene (aox1) to heat shock, oxidative, and osmotic stresses in conidia of citric acid-producing Aspergillus niger. J. Biosci. Bioeng. 2012, 113 (3), 338–342. 10.1016/j.jbiosc.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Shao Y.; Li Q.; Zhou Y.; Chen F. Effects of an alternative oxidase gene on conidia viability under external stresses in Monascus ruber M7. Journal of Basic Microbiology 2017, 57 (5), 413–418. 10.1002/jobm.201600707. [DOI] [PubMed] [Google Scholar]
- Liu D.; Sun X.; Yan B.; Ma A. Alternative oxidase is involved in oxidative stress resistance and melanin synthesis in Annulohypoxylon stygium, a companion fungus of Tremella fuciformis. Antonie van Leeuwenhoek 2022, 115 (3), 365–374. 10.1007/s10482-021-01705-5. [DOI] [PubMed] [Google Scholar]
- Hernández Ruiz O.; Gonzalez A.; Almeida A. J.; Tamayo D.; Garcia A. M.; Restrepo A.; McEwen J. G. Alternative Oxidase Mediates Pathogen Resistance in Paracoccidioides brasiliensis Infection. PLoS Neglected Tropical Diseases 2011, 5 (10), e1353 10.1371/journal.pntd.0001353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq S.; Palmer J. M. Isolation of a cyanide-resistant duroquinol oxidase from Arum maculatum mitochondria. FEBS Lett. 1978, 95 (2), 217–20. 10.1016/0014-5793(78)80997-1. [DOI] [PubMed] [Google Scholar]
- Elthon T. E.; McIntosh L. Characterization and Solubilization of the Alternative Oxidase of Sauromatum guttatum Mitochondria. Plant Physiol 1986, 82 (1), 1–6. 10.1104/pp.82.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elthon T. E.; Nickels R. L.; McIntosh L. Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol 1989, 89 (4), 1311–7. 10.1104/pp.89.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz A. M.; Sabourin J. R.; Bertrand H.; Nickels R.; McIntosh L. Immunological identification of the alternative oxidase of Neurospora crassa mitochondria. Mol. Cell. Biol. 1989, 9 (3), 1362–1364. 10.1128/mcb.9.3.1362-1364.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D M; McIntosh L Isolation and characterization of a cDNA clone encoding an alternative oxidase protein of Sauromatum guttatum (Schott). Proc. Natl. Acad. Sci. U. S. A. 1991, 88 (6), 2122–2126. 10.1073/pnas.88.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan J.; McIntosh L.; Day D. A. Sequencing of a soybean alternative oxidase cDNA clone. Plant Physiol 1993, 103 (4), 1481. 10.1104/pp.103.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe G. C.; McIntosh L. Mitochondrial electron transport regulation of nuclear gene expression. Studies with the alternative oxidase gene of tobacco. Plant Physiol 1994, 105 (3), 867–74. 10.1104/pp.105.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M.; Gardestrom P.; Samuelsson G. Isolation, Purification, and Characterization of Mitochondria from Chlamydomonas reinhardtii. Plant Physiol 1995, 107 (2), 479–483. 10.1104/pp.107.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri M.; Ajayi W.; Temple S.; Hill G. C. Identification and partial purification of a stage-specific 33 kDa mitochondrial protein as the alternative oxidase of the Trypanosoma brucei brucei bloodstream trypomastigotes. J. Eukaryot Microbiol 1995, 42 (5), 467–72. 10.1111/j.1550-7408.1995.tb05892.x. [DOI] [PubMed] [Google Scholar]
- Kern A.; Hartner F. S.; Freigassner M.; Spielhofer J.; Rumpf C.; Leitner L.; Frohlich K. U.; Glieder A. Pichia pastoris ″just in time″ alternative respiration. Microbiology (Reading) 2007, 153 (4), 1250–1260. 10.1099/mic.0.2006/001404-0. [DOI] [PubMed] [Google Scholar]
- Ito-Inaba Y.; Hida Y.; Inaba T. What is critical for plant thermogenesis? Differences in mitochondrial activity and protein expression between thermogenic and non-thermogenic skunk cabbages. Planta 2009, 231 (1), 121–130. 10.1007/s00425-009-1034-z. [DOI] [PubMed] [Google Scholar]
- Papagianni M.; Avramidis N. Cloning and functional expression of the mitochondrial alternative oxidase gene (aox1) of Aspergillus niger in Lactococcus lactis and its induction by oxidizing conditions. Enzyme Microb Technol. 2012, 50 (1), 17–21. 10.1016/j.enzmictec.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Young L.; May B.; Pendlebury-Watt A.; Shearman J.; Elliott C.; Albury M. S.; Shiba T.; Inaoka D. K.; Harada S.; Kita K.; Moore A. L. Probing the ubiquinol-binding site of recombinant Sauromatum guttatum alternative oxidase expressed in E. coli membranes through site-directed mutagenesis. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2014, 1837 (7), 1219–1225. 10.1016/j.bbabio.2014.01.027. [DOI] [PubMed] [Google Scholar]
- Robertson A.; Schaltz K.; Neimanis K.; Staples J. F.; McDonald A. E. Heterologous expression of the Crassostrea gigas (Pacific oyster) alternative oxidase in the yeast Saccharomyces cerevisiae. J. Bioenerg Biomembr 2016, 48 (5), 509–520. 10.1007/s10863-016-9685-5. [DOI] [PubMed] [Google Scholar]
- Moretti-Almeida G.; Thomazella D. P. T.; Pereira G. A. G.; Monteiro G. Heterologous expression of an alternative oxidase from Moniliophthora perniciosa in Saccharomyces cerevisiae: Antioxidant function and in vivo platform for the study of new drugs against witches’ broom disease. Fungal Genet Biol. 2019, 126, 50–55. 10.1016/j.fgb.2019.02.006. [DOI] [PubMed] [Google Scholar]
- Barsottini M. R. O.; Copsey A.; Xu F.; Pereira G. A. G.; Moore A. L. Self-assembled proteolipossomes to functionally characterize the alternative oxidase from Moniliophthora perniciosa. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2018, 1859, e65–e66. 10.1016/j.bbabio.2018.09.196. [DOI] [Google Scholar]
- Unlu E. S.; Unuvar O. C.; Aydin M. Identification of alternative oxidase encoding genes in Caulerpa cylindracea by de novo RNA-Seq assembly analysis. Mar Genomics 2019, 46, 41–48. 10.1016/j.margen.2019.03.004. [DOI] [PubMed] [Google Scholar]
- Shiba T.; Kido Y.; Sakamoto K.; Inaoka D. K.; Tsuge C.; Tatsumi R.; Takahashi G.; Balogun E. O.; Nara T.; Aoki T.; Honma T.; Tanaka A.; Inoue M.; Matsuoka S.; Saimoto H.; Moore A. L.; Harada S.; Kita K. Structure of the trypanosome cyanide-insensitive alternative oxidase. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (12), 4580–5. 10.1073/pnas.1218386110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold D. A.; Andersson M. E.; Nordlund P. New insight into the structure and function of the alternative oxidase. Biochim. Biophys. Acta 2000, 1460 (2–3), 241–54. 10.1016/S0005-2728(00)00149-3. [DOI] [PubMed] [Google Scholar]
- Nihei C.; Fukai Y.; Kita K. Trypanosome alternative oxidase as a target of chemotherapy. Biochimica Et Biophysica Acta 2002, 1587 (2–3), 234–239. 10.1016/S0925-4439(02)00086-8. [DOI] [PubMed] [Google Scholar]
- Moore A. L.; Carre J. E.; Affourtit C.; Albury M. S.; Crichton P. G.; Kita K.; Heathcote P. Compelling EPR evidence that the alternative oxidase is a diiron carboxylate protein. Biochim. Biophys. Acta 2008, 1777 (4), 327–30. 10.1016/j.bbabio.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Marechal A.; Kido Y.; Kita K.; Moore A. L.; Rich P. R. Three redox states of Trypanosoma brucei alternative oxidase identified by infrared spectroscopy and electrochemistry. J. Biol. Chem. 2009, 284 (46), 31827–33. 10.1074/jbc.M109.059980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albury M. S.; Elliott C.; Moore A. L. Ubiquinol-binding site in the alternative oxidase: mutagenesis reveals features important for substrate binding and inhibition. Biochim. Biophys. Acta 2010, 1797 (12), 1933–9. 10.1016/j.bbabio.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Siedow J. N.; Umbach A. L.; Moore A. L. The active site of the cyanide-resistant oxidase from plant mitochondria contains a binuclear iron center. FEBS Lett. 1995, 362 (1), 10–4. 10.1016/0014-5793(95)00196-G. [DOI] [PubMed] [Google Scholar]
- Crichton P. G.; Albury M. S.; Affourtit C.; Moore A. L. Mutagenesis of the Sauromatum guttatum alternative oxidase reveals features important for oxygen binding and catalysis. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2010, 1797 (6–7), 732–737. 10.1016/j.bbabio.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Teixeira M. H.; Arantes G. M. Effects of lipid composition on membrane distribution and permeability of natural quinones. RSC Adv. 2019, 9 (29), 16892–16899. 10.1039/C9RA01681C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I.; Wright J. J.; Bridges H. R.; Ivanov B. S.; Biner O.; Pereira C. S.; Arantes G. M.; Hirst J. Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy. Nat. Commun. 2022, 13 (1), 2758. 10.1038/s41467-022-30506-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoias Teixeira M.; Menegon Arantes G. Balanced internal hydration discriminates substrate binding to respiratory complex I. Biochim Biophys Acta Bioenerg 2019, 1860 (7), 541–548. 10.1016/j.bbabio.2019.05.004. [DOI] [PubMed] [Google Scholar]
- Horsefield R.; Yankovskaya V.; Sexton G.; Whittingham W.; Shiomi K.; Omura S.; Byrne B.; Cecchini G.; Iwata S. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J. Biol. Chem. 2006, 281 (11), 7309–16. 10.1074/jbc.M508173200. [DOI] [PubMed] [Google Scholar]
- Kido Y.; Sakamoto K.; Nakamura K.; Harada M.; Suzuki T.; Yabu Y.; Saimoto H.; Yamakura F.; Ohmori D.; Moore A.; Harada S.; Kita K. Purification and kinetic characterization of recombinant alternative oxidase from Trypanosoma brucei brucei. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2010, 1797 (4), 443–450. 10.1016/j.bbabio.2009.12.021. [DOI] [PubMed] [Google Scholar]
- Shiba T.; Inaoka D. K.; Takahashi G.; Tsuge C.; Kido Y.; Young L.; Ueda S.; Balogun E. O.; Nara T.; Honma T.; Tanaka A.; Inoue M.; Saimoto H.; Harada S.; Moore A. L.; Kita K. Insights into the ubiquinol/dioxygen binding and proton relay pathways of the alternative oxidase. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2019, 1860 (5), 375–382. 10.1016/j.bbabio.2019.03.008. [DOI] [PubMed] [Google Scholar]
- Szibor M.; Schenkl C.; Barsottini M. R. O.; Young L.; Moore A. L. Targeting the alternative oxidase (AOX) for human health and food security, a pharmaceutical and agrochemical target or a rescue mechanism?. Biochemical Journal 2022, 479 (12), 1337–1359. 10.1042/BCJ20180192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell-Hidalgo A.; Young L.; Moore A. L.; Ghafourian T. QSAR and molecular docking for the search of AOX inhibitors: a rational drug discovery approach. Journal of Computer-Aided Molecular Design 2021, 35 (2), 245–260. 10.1007/s10822-020-00360-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F.; Copsey A. C.; Barsottini M. R. O.; Moore A. L. Comparison of the kinetic parameters of alternative oxidases purified from Trypanosoma brucei, Sauromatum guttatum and Arabidopsis thaliana. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2018, 1859, e69 10.1016/j.bbabio.2018.09.206. [DOI] [Google Scholar]
- Copsey A. C.; Barsottini M. R. O.; May B.; Xu F.; Albury M. S.; Young L.; Moore A. L. Kinetic characterisation and inhibitor sensitivity of Candida albicans and Candida auris recombinant AOX expressed in a self-assembled proteoliposome system. Sci. Rep. 2021, 11 (1), 14748. 10.1038/s41598-021-94320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L.; Rosell-Hidalgo A.; Inaoka D. K.; Xu F.; Albury M.; May B.; Kita K.; Moore A. L. Kinetic and structural characterisation of the ubiquinol-binding site and oxygen reduction by the trypanosomal alternative oxidase. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2020, 1861 (10), 148247 10.1016/j.bbabio.2020.148247. [DOI] [PubMed] [Google Scholar]
- Szal B.; Jolivet Y.; Hasenfratz-Sauder M. P.; Dizengremel P.; Rychter A. M. Oxygen concentration regulates alternative oxidase expression in barley roots during hypoxia and post-hypoxia. Physiologia Plantarum 2003, 119 (4), 494–502. 10.1046/j.1399-3054.2003.00161.x. [DOI] [Google Scholar]
- Wagner A. M.; Krab K.; Wagner M. J.; Moore A. L. Regulation of thermogenesis in flowering Araceae: The role of the alternative oxidase. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2008, 1777 (7–8), 993–1000. 10.1016/j.bbabio.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Kaye Y.; Huang W.; Clowez S.; Saroussi S.; Idoine A.; Sanz-Luque E.; Grossman A. R. The mitochondrial alternative oxidase from Chlamydomonas reinhardtii enables survival in high light. J. Biol. Chem. 2019, 294 (4), 1380–1395. 10.1074/jbc.RA118.004667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyda-Ploszczyca A.; Koziel A.; Antos-Krzeminska N.; Jarmuszkiewicz W. Impact of oxidative stress on Acanthamoeba castellanii mitochondrial bioenergetics depends on cell growth stage. Journal of Bioenergetics 2011, 43 (3), 217–225. 10.1007/s10863-011-9351-x. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe G. C.; Cvetkovska M.; Wang J. Is the maintenance of homeostatic mitochondrial signaling during stress a physiological role for alternative oxidase?. Physiol Plant 2009, 137 (4), 392–406. 10.1111/j.1399-3054.2009.01254.x. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe G. C. Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 2013, 14 (4), 6805–47. 10.3390/ijms14046805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais P.; Piazzon C.; Lamas J.; Mallo N.; Leiro J. M. Effect of Resveratrol on Oxygen Consumption by Philasterides dicentrarchi, a Scuticociliate Parasite of Turbot. Protist 2013, 164 (2), 206–217. 10.1016/j.protis.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Nicholls P.; Marshall D. C.; Cooper C. E.; Wilson M. T. Sulfide inhibition of and metabolism by cytochrome c oxidase. Biochem. Soc. Trans. 2013, 41 (5), 1312–6. 10.1042/BST20130070. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Zhang X. The critical roles of mitochondrial alternative chains in juvenile ark shells (Anadara broughtonii) exposed to acute hypoxia with or without sulfide. Aquatic Toxicology 2021, 241, 105996 10.1016/j.aquatox.2021.105996. [DOI] [PubMed] [Google Scholar]
- Costa J. H.; McDonald A. E.; Arnholdt-Schmitt B.; Fernandes de Melo D. A classification scheme for alternative oxidases reveals the taxonomic distribution and evolutionary history of the enzyme in angiosperms. Mitochondrion 2014, 19 (Pt B), 172–183. 10.1016/j.mito.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Molen T. A.; Rosso D.; Piercy S.; Maxwell D. P. Characterization of the alternative oxidase of Chlamydomonas reinhardtii in response to oxidative stress and a shift in nitrogen source. Physiologia Plantarum 2006, 127 (1), 74–86. 10.1111/j.1399-3054.2006.00643.x. [DOI] [Google Scholar]
- Ostroukhova M.; Zalutskaya Z.; Ermilova E. New insights into AOX2 transcriptional regulation in Chlamydomonas reinhardtii. European Journal of Protistology 2017, 58, 1–8. 10.1016/j.ejop.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Cavalcanti J. H. F.; Oliveira G. M.; Saraiva K. D. d. C.; Torquato J. P. P.; Maia I. G.; Fernandes de Melo D.; Costa J. H. Identification of duplicated and stress-inducible Aox2b gene co-expressed with Aox1 in species of the Medicago genus reveals a regulation linked to gene rearrangement in leguminous genomes. Journal of Plant Physiology 2013, 170 (18), 1609–1619. 10.1016/j.jplph.2013.06.012. [DOI] [PubMed] [Google Scholar]
- He Q.; Wang X.; He L.; Yang L.; Wang S.; Bi Y. Alternative respiration pathway is involved in the response of highland barley to salt stress. Plant Cell Reports 2019, 38 (3), 295–309. 10.1007/s00299-018-2366-6. [DOI] [PubMed] [Google Scholar]
- Chien L. F.; Wu Y. C.; Chen H. P. Mitochondrial energy metabolism in young bamboo rhizomes from Bambusa oldhamii and Phyllostachys edulis during shooting stage. Plant Physiology and Biochemistry 2011, 49 (4), 449–457. 10.1016/j.plaphy.2011.01.024. [DOI] [PubMed] [Google Scholar]
- Saha B.; Borovskii G.; Panda S. K. Alternative oxidase and plant stress tolerance. Plant Signaling & Behavior 2016, 11 (12), e1256530 10.1080/15592324.2016.1256530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.; Van Aken O.; Schwarzländer M.; Belt K.; Millar A. H. Roles of Mitochondrial Reactive Oxygen Species in Cellular Signaling and Stress Response in Plants. Plant Physiol. 2016, 171 (3), 1551–1559. 10.1104/pp.16.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R.; Day D. A.; Fricke W.; Watt M.; Arsova B.; Barkla B. J.; Bose J.; Byrt C. S.; Chen Z. H.; Foster K. J.; et al. Energy costs of salt tolerance in crop plants. New Phytologist 2020, 225 (3), 1072–1090. 10.1111/nph.15864. [DOI] [PubMed] [Google Scholar]
- Zhang L. T.; Zhang Z. S.; Gao H. Y.; Meng X. L.; Yang C.; Liu J. G.; Meng Q. W. The mitochondrial alternative oxidase pathway protects the photosynthetic apparatus against photodamage in Rumex K-1 leaves. BMC Plant Biol. 2012, 12, 40. 10.1186/1471-2229-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan P. M.; Soole K. L.; Umbach A. L., Alternative Mitochondrial Electron Transport Proteins in Higher Plants. In Plant Mitochondria: From Genome to Function, Vol. 17; Day D. A.; Millar A. H.; Whelan J., Eds.; Springer Netherlands: Dordrecht, Netherlands, 2004; pp 163–230. [Google Scholar]
- Keunen E.; Florez-Sarasa I.; Obata T.; Jozefczak M.; Remans T.; Vangronsveld J.; Fernie A. R.; Cuypers A. Metabolic responses of Arabidopsis thaliana roots and leaves to sublethal cadmium exposure are differentially influenced by ALTERNATIVE OXIDASE1a. Environmental and Experimental Botany 2016, 124, 64–78. 10.1016/j.envexpbot.2015.11.015. [DOI] [Google Scholar]
- Carre J. E.; Affourtit C.; Moore A. L. Interaction of purified alternative oxidase from thermogenic Arum maculatum with pyruvate. FEBS Lett. 2011, 585 (2), 397–401. 10.1016/j.febslet.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Vishwakarma A.; Kumari A.; Mur L. A. J.; Gupta K. J. A discrete role for alternative oxidase under hypoxia to increase nitric oxide and drive energy production. Free Radic Biol. Med. 2018, 122, 40–51. 10.1016/j.freeradbiomed.2018.03.045. [DOI] [PubMed] [Google Scholar]
- Cvetkovska M.; Vanlerberghe G. C. Alternative oxidase modulates leaf mitochondrial concentrations of superoxide and nitric oxide. New Phytologist 2012, 195 (1), 32. 10.1111/j.1469-8137.2012.04166.x. [DOI] [PubMed] [Google Scholar]
- Cvetkovska M.; Dahal K.; Alber N. A.; Jin C.; Cheung M.; Vanlerberghe G. C. Knockdown of mitochondrial alternative oxidase induces the ‘stress state’ of signaling molecule pools in Nicotiana tabacum, with implications for stomatal function. New Phytologist 2014, 203 (2), 449–461. 10.1111/nph.12773. [DOI] [PubMed] [Google Scholar]
- Alber N. A.; Sivanesan H.; Vanlerberghe G. C. The occurrence and control of nitric oxide generation by the plant mitochondrial electron transport chain. Plant Cell Environ 2017, 40 (7), 1074–1085. 10.1111/pce.12884. [DOI] [PubMed] [Google Scholar]
- Royo B.; Moran J. F.; Ratcliffe R. G.; Gupta K. J. Nitric oxide induces the alternative oxidase pathway in Arabidopsis seedlings deprived of inorganic phosphate. J. Exp Bot 2015, 66 (20), 6273–80. 10.1093/jxb/erv338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.; Zhao Y.; Li A.; Qi S.; Lin Q.; Duan Y. Postharvest nitric oxide treatment induced the alternative oxidase pathway to enhance antioxidant capacity and chilling tolerance in peach fruit. Plant Physiol Biochem 2021, 167, 113–122. 10.1016/j.plaphy.2021.07.036. [DOI] [PubMed] [Google Scholar]
- Singh S. K.; Chien C. T.; Chang I. F. The Arabidopsis glutamate receptor-like gene GLR3.6 controls root development by repressing the Kip-related protein gene KRP4. J. Exp Bot 2016, 67 (6), 1853–69. 10.1093/jxb/erv576. [DOI] [PubMed] [Google Scholar]
- Zulet A.; Gil-Monreal M.; Zabalza A.; van Dongen J. T.; Royuela M. Fermentation and alternative oxidase contribute to the action of amino acid biosynthesis-inhibiting herbicides. J. Plant Physiol 2015, 175, 102–12. 10.1016/j.jplph.2014.12.004. [DOI] [PubMed] [Google Scholar]
- Igamberdiev A. U.; Eprintsev A. T.; Fedorin D. N.; Popov V. N. Phytochrome-mediated regulation of plant respiration and photorespiration. Plant Cell Environ 2014, 37 (2), 290–9. 10.1111/pce.12155. [DOI] [PubMed] [Google Scholar]
- Garmash E. V.; Velegzhaninov I. O.; Grabelnych O. I.; Borovik O. A.; Silina E. V.; Voinikov V. K.; Golovko T. K. Expression profiles of genes for mitochondrial respiratory energy-dissipating systems and antioxidant enzymes in wheat leaves during de-etiolation. J. Plant Physiol 2017, 215, 110–121. 10.1016/j.jplph.2017.05.023. [DOI] [PubMed] [Google Scholar]
- Gonzàlez-Meler M. A.; Giles L.; Thomas R. B.; Siedow J. N. Metabolic regulation of leaf respiration and alternative pathway activity in response to phosphate supply. Plant, Cell & Environment 2001, 24 (2), 205–215. 10.1111/j.1365-3040.2001.00674.x. [DOI] [Google Scholar]
- Sieger S. M.; Kristensen B. K.; Robson C. A.; Amirsadeghi S.; Eng E. W.; Abdel-Mesih A.; M?ller I. M.; Vanlerberghe G. C. The role of alternative oxidase in modulating carbon use efficiency and growth during macronutrient stress in tobacco cells. J. Exp Bot 2005, 56 (416), 1499–1515. 10.1093/jxb/eri146. [DOI] [PubMed] [Google Scholar]
- Searle S. Y.; Turnbull M. H. Seasonal variation of leaf respiration and the alternative pathway in field-grown Populus × canadensis. Physiologia Plantarum 2011, 141 (4), 332–342. 10.1111/j.1399-3054.2010.01442.x. [DOI] [PubMed] [Google Scholar]
- Van Aken O.; Giraud E.; Clifton R.; Whelan J. Alternative oxidase: a target and regulator of stress responses. Physiol Plant 2009, 137 (4), 354–61. 10.1111/j.1399-3054.2009.01240.x. [DOI] [PubMed] [Google Scholar]
- Chai T.-T.; Simmonds D.; Day D. A.; Colmer T. D.; Finnegan P. M. A GmAOX2b antisense gene compromises vegetative growth and seed production in soybean. Planta 2012, 236 (1), 199–207. 10.1007/s00425-012-1601-6. [DOI] [PubMed] [Google Scholar]
- Costa J. H.; Mota E. F.; Cambursano M. V.; Lauxmann M. A.; de Oliveira L. M.; Silva Lima M. d. G.; Orellano E. G.; Fernandes de Melo D. Stress-induced co-expression of two alternative oxidase (VuAox1 and 2b) genes in Vigna unguiculata. J. Plant Physiol 2010, 167 (7), 561–570. 10.1016/j.jplph.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Liu J.; Li Z.; Wang Y.; Xing D. Overexpression of ALTERNATIVE OXIDASE1a alleviates mitochondria-dependent programmed cell death induced by aluminium phytotoxicity in Arabidopsis. J. Exp Bot 2014, 65 (15), 4465–78. 10.1093/jxb/eru222. [DOI] [PubMed] [Google Scholar]
- Welchen E.; García L.; Mansilla N.; Gonzalez D. H. Coordination of plant mitochondrial biogenesis: keeping pace with cellular requirements. Front Plant 2014, 4 (2), 551. 10.3389/fpls.2013.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K.; Ogata T.; Kakizaki Y.; Elliott C.; Albury M. S.; Moore A. L. Identification of a gene for pyruvate-insensitive mitochondrial alternative oxidase expressed in the thermogenic appendices in Arum maculatum. Plant Physiol 2011, 157 (4), 1721–32. 10.1104/pp.111.186932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tward C. E.; Singh J.; Cygelfarb W.; McDonald A. E. Identification of the alternative oxidase gene and its expression in the copepod Tigriopus californicus. Comp Biochem Physiol B Biochem Mol. Biol. 2019, 228, 41–50. 10.1016/j.cbpb.2018.11.003. [DOI] [PubMed] [Google Scholar]
- Jacobs H. T.; Ballard J. W. O. What physiological role(s) does the alternative oxidase perform in animals?. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2022, 1863, 148556 10.1016/j.bbabio.2022.148556. [DOI] [PubMed] [Google Scholar]
- Matus-Ortega M. G.; Salmerón-Santiago K. G.; Flores-Herrera O.; Guerra-Sánchez G.; Martínez F.; Rendón J. L.; Pardo J. P. The alternative NADH dehydrogenase is present in mitochondria of some animal taxa. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 2011, 6 (3), 256–263. 10.1016/j.cbd.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Abele E.; Philip E.; Gonzalez P. M.; Puntarulo S. Marine invertebrate mitochondria and oxidative stress. Front Biosci 2007, 12, 933–46. 10.2741/2115. [DOI] [PubMed] [Google Scholar]
- Fernandez-Ayala D. J.; Sanz A.; Vartiainen S.; Kemppainen K. K.; Babusiak M.; Mustalahti E.; Costa R.; Tuomela T.; Zeviani M.; Chung J.; O’Dell K. M.; Rustin P.; Jacobs H. T. Expression of the Ciona intestinalis alternative oxidase (AOX) in Drosophila complements defects in mitochondrial oxidative phosphorylation. Cell Metab 2009, 9 (5), 449–60. 10.1016/j.cmet.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Saari S.; Andjelkovic A.; Garcia G. S.; Jacobs H. T.; Oliveira M. T. Expression of Ciona intestinalis AOX causes male reproductive defects in Drosophila melanogaster. BMC Dev Biol. 2017, 17 (1), 9. 10.1186/s12861-017-0151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusseppone M. S.; Rocchetta I.; Sabatini S. E.; Luquet C. M.; Rios de Molina M. D. C.; Held C.; Abele D. Inducing the Alternative Oxidase Forms Part of the Molecular Strategy of Anoxic Survival in Freshwater Bivalves. Front Physiol 2018, 9, 100. 10.3389/fphys.2018.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N.; Li S.-Z.; Zhang Y.-W.-Q.; Habib M. R.; Xiong T.; Xu S.; Dong H.; Zhao Q.-P. The identification of alternative oxidase in intermediate host snails of Schistosoma and its potential role in protecting Oncomelania hupensis against niclosamide-induced stress. Parasites & Vectors 2022, 15 (1), 97. 10.1186/s13071-022-05227-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szibor M.; Dhandapani P. K.; Dufour E.; Holmström K. M.; Zhuang Y.; Salwig I.; Wittig I.; Heidler J.; Gizatullina Z.; Gainutdinov T.; German Mouse Clinic Consortium; Fuchs H.; Gailus-Durner V.; de Angelis M. H.; Nandania J.; Velagapudi V.; Wietelmann A.; Rustin P.; Gellerich F. N.; Jacobs H. T.; Braun T. Broad AOX expression in a genetically tractable mouse model does not disturb normal physiology. Disease Models & Mechanisms 2016, 10 (2), 163–171. 10.1242/dmm.027839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khoury R.; Dufour E.; Rak M.; Ramanantsoa N.; Grandchamp N.; Csaba Z.; Duvillié B.; Bénit P.; Gallego J.; Gressens P.; et al. Alternative Oxidase Expression in the Mouse Enables Bypassing Cytochrome c Oxidase Blockade and Limits Mitochondrial ROS Overproduction. PLoS Genetics 2013, 9 (1), e1003182. 10.1371/journal.pgen.1003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szibor M.; Gainutdinov T.; Fernandez-Vizarra E.; Dufour E.; Gizatullina Z.; Debska-Vielhaber G.; Heidler J.; Wittig I.; Viscomi C.; Gellerich F.; Moore A. L. Bioenergetic consequences from xenotopic expression of a tunicate AOX in mouse mitochondria: Switch from RET and ROS to FET. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2020, 1861 (2), 148137 10.1016/j.bbabio.2019.148137. [DOI] [PubMed] [Google Scholar]
- Mills E. L.; Kelly B.; Logan A.; Costa A. S. H.; Varma M.; Bryant C. E.; Tourlomousis P.; Dabritz J. H. M.; Gottlieb E.; Latorre I.; Corr S. C.; McManus G.; Ryan D.; Jacobs H. T.; Szibor M.; Xavier R. J.; Braun T.; Frezza C.; Murphy M. P.; O’Neill L. A. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 2016, 167 (2), 457–470e13. 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalutskaya Z.; Ostroukhova M.; Ermilova E. The Chlamydomonas alternative oxidase 1 is regulated by cadmium stress: New insights into control of expression. Environmental and Experimental Botany 2016, 130, 133–140. 10.1016/j.envexpbot.2016.05.015. [DOI] [Google Scholar]
- Zalutskaya Z.; Filina V.; Ostroukhova M.; Ermilova E. Regulation of alternative oxidase 1 in Chlamydomonas reinhardtii during sulfur starvation. Eur. J. Protistol 2018, 63, 26–33. 10.1016/j.ejop.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Zalutskaya Z.; Lapina T.; Ermilova E. The Chlamydomonas reinhardtii alternative oxidase 1 is regulated by heat stress. Plant Physiology and Biochemistry 2015, 97, 229–234. 10.1016/j.plaphy.2015.10.014. [DOI] [PubMed] [Google Scholar]
- Bobba V.; Li Y.; Afrin M.; Dano R.; Zhang W.; Li B.; Su B. Synthesis and biological evaluation of imidamide analogs as selective anti-trypanosomal agents. Bioorg. Med. Chem. 2022, 61, 116740 10.1016/j.bmc.2022.116740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri M.; Ott R. D.; Hill G. C. Trypanosome alternative oxidase: from molecule to function. Trends in Parasitology 2006, 22 (10), 484–491. 10.1016/j.pt.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Verner Z.; Basu S.; Benz C.; Dixit S.; Dobakova E.; Faktorova D.; Hashimi H.; Horakova E.; Huang Z.; Paris Z.; Pena-Diaz P.; Ridlon L.; Tyc J.; Wildridge D.; Zikova A.; Lukes J. Malleable mitochondrion of Trypanosoma brucei. Int. Rev. Cell Mol. Biol. 2015, 315, 73–151. 10.1016/bs.ircmb.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Shi D. K.; Zhu J.; Sun Z. H.; Zhang G.; Liu R.; Zhang T. J.; Wang S. L.; Ren A.; Zhao M. W. Alternative oxidase impacts ganoderic acid biosynthesis by regulating intracellular ROS levels in Ganoderma lucidum. Microbiology (Reading) 2017, 163 (10), 1466–1476. 10.1099/mic.0.000527. [DOI] [PubMed] [Google Scholar]
- Magnani T.; Soriani F. M.; Martins V. P.; Nascimento A. M.; Tudella V. G.; Curti C.; Uyemura S. A. Cloning and functional expression of the mitochondrial alternative oxidase of Aspergillus fumigatus and its induction by oxidative stress. FEMS Microbiol Lett. 2007, 271 (2), 230–8. 10.1111/j.1574-6968.2007.00716.x. [DOI] [PubMed] [Google Scholar]
- Xu T.; Yao F.; Liang W. S.; Li Y. H.; Li D. R.; Wang H.; Wang Z. Y. Involvement of alternative oxidase in the regulation of growth, development, and resistance to oxidative stress of Sclerotinia sclerotiorum. J. Microbiol 2012, 50 (4), 594–602. 10.1007/s12275-012-2015-7. [DOI] [PubMed] [Google Scholar]
- Stanic M.; Zakrzewska J.; Hadzibrahimovic M.; Zizic M.; Markovic Z.; Vucinic Z.; Zivic M. Oxygen regulation of alternative respiration in fungus Phycomyces blakesleeanus: connection with phosphate metabolism. Res. Microbiol 2013, 164 (7), 770–8. 10.1016/j.resmic.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Gu S.; Xu Q.; Huang H.; Li S. Alternative respiration and fumaric acid production of Rhizopus oryzae. Appl. Microbiol. Biotechnol. 2014, 98 (11), 5145–52. 10.1007/s00253-014-5615-9. [DOI] [PubMed] [Google Scholar]
- Hernandez O.; Araque P.; Tamayo D.; Restrepo A.; Herrera S.; McEwen J. G.; Pelaez C.; Almeida A. J. Alternative oxidase plays an important role in Paracoccidioides brasiliensis cellular homeostasis and morphological transition. Med. Mycol 2015, 53 (3), 205–14. 10.1093/mmy/myu091. [DOI] [PubMed] [Google Scholar]
- Lin Z.; Wu J.; Jamieson P. A.; Zhang C. Alternative Oxidase Is Involved in the Pathogenicity, Development, and Oxygen Stress Response of Botrytis cinerea. Phytopathology 2019, 109 (10), 1679–1688. 10.1094/PHYTO-01-19-0012-R. [DOI] [PubMed] [Google Scholar]
- Luévano-Martínez L. A.; Caldeira da Silva C. C.; Nicastro G. G.; Schumacher R. I.; Kowaltowski A. J.; Gomes S. L. Mitochondrial alternative oxidase is determinant for growth and sporulation in the early diverging fungus Blastocladiella emersonii. Fungal Biology 2019, 123 (1), 59–65. 10.1016/j.funbio.2018.11.005. [DOI] [PubMed] [Google Scholar]
- Tian F.; Lee S. Y.; Woo S. Y.; Chun H. S. Alternative Oxidase: A Potential Target for Controlling Aflatoxin Contamination and Propagation of Aspergillus flavus. Front Microbiol 2020, 11, 419. 10.3389/fmicb.2020.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark P.; Nordlund P. A prokaryotic alternative oxidase present in the bacterium Novosphingobium aromaticivorans. FEBS Lett. 2003, 552 (2–3), 189–192. 10.1016/S0014-5793(03)00920-7. [DOI] [PubMed] [Google Scholar]
- Dunn A. K.; Karr E. A.; Wang Y.; Batton A. R.; Ruby E. G.; Stabb E. V. The alternative oxidase (AOX) gene in Vibrio fischeri is controlled by NsrR and upregulated in response to nitric oxide. Mol. Microbiol. 2010, 77 (1), 44–55. 10.1111/j.1365-2958.2010.07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. H.; Finnegan P. M.; Whelan J.; Drevon J. J.; Day D. A. Expression and kinetics of the mitochondrial alternative oxidase in nitrogen-fixing nodules of soybean roots. Plant Cell & Environment 1997, 20 (10), 1273–1282. 10.1046/j.1365-3040.1997.d01-25.x. [DOI] [Google Scholar]
- Matamoros M. A.; Fernandez-Garcia N.; Wienkoop S.; Loscos J.; Saiz A.; Becana M. Mitochondria are an early target of oxidative modifications in senescing legume nodules. New Phytol 2013, 197 (3), 873–885. 10.1111/nph.12049. [DOI] [PubMed] [Google Scholar]
- Ortiz J.; Sanhueza C.; Romero-Munar A.; Hidalgo-Castellanos J.; Castro C.; Bascunan-Godoy L.; Coba de la Pena T.; Lopez-Gomez M.; Florez-Sarasa I.; Del-Saz N. F. In Vivo Metabolic Regulation of Alternative Oxidase under Nutrient Deficiency-Interaction with Arbuscular Mycorrhizal Fungi and Rhizobium Bacteria. International Journal of Molecular Sciences 2020, 21 (12), 4201. 10.3390/ijms21124201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederico A. M.; Campos M. D.; Cardoso H. G.; Imani J.; Arnholdt-Schmitt B. Alternative oxidase involvement in Daucus carota somatic embryogenesis. Physiol Plant 2009, 137 (4), 498–508. 10.1111/j.1399-3054.2009.01278.x. [DOI] [PubMed] [Google Scholar]
- Opperdoes F. R.; Borst P.; Fonck K. The potential use of inhibitors of glycerol-3-phosphate oxidase for chemotherapy of African trypanosomiasis. Febs Letters 1976, 62 (2), 169–172. 10.1016/0014-5793(76)80045-2. [DOI] [PubMed] [Google Scholar]
- Murphy A. D.; Lang-Unnasch N. Alternative Oxidase Inhibitors Potentiate the Activity of Atovaquone against Plasmodium falciparum. Antimicrob. Agents Chemother. 1999, 43 (3), 651–654. 10.1128/AAC.43.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. W.; Lloyd D. Properties of mitochondria isolated from cyanide-sensitive and cyanide-stimulated cultures of Acanthamoeba castellanii. Biochemical Journal 1978, 174 (1), 203–211. 10.1042/bj1740203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson A. B.; Brohn F. H. Trypanosomiasis: an approach to chemotherapy by the inhibition of carbohydrate catabolism. Science (New York, N.Y.) 1976, 194 (4261), 204–206. 10.1126/science.986688. [DOI] [PubMed] [Google Scholar]
- Baker J. Progress in human African Trypanosomiasis, sleeping sickness. Transactions of the Royal Society of Tropical Medicine and Hygiene 2000, 94 (1), 82–82. 10.1016/S0035-9203(00)90449-8. [DOI] [Google Scholar]
- Fang J.; Beattie D. S. Alternative oxidase present in procyclic Trypanosoma brucei may act to lower the mitochondrial production of superoxide. Archives of Biochemistry & Biophysics 2003, 414 (2), 294–302. 10.1016/S0003-9861(03)00196-6. [DOI] [PubMed] [Google Scholar]
- Duvenage L.; Walker L. A.; Bojarczuk A.; Johnston S. A.; MacCallum D. M.; Munro C. A.; Gourlay C. W. Inhibition of Classical and Alternative Modes of Respiration in Candida albicans Leads to Cell Wall Remodeling and Increased Macrophage Recognition. mBio 2019, 10 (1), e02535-18. 10.1128/mBio.02535-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins V. P.; Dinamarco T. M.; Soriani F. M.; Tudella V. G.; Oliveira S. C.; Goldman G. H.; Curti C.; Uyemura S. A. Involvement of an alternative oxidase in oxidative stress and mycelium-to-yeast differentiation in Paracoccidioides brasiliensis. Eukaryotic Cell 2011, 10 (2), 237–248. 10.1128/EC.00194-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Macedo E.; Sircar D.; Cardoso H. G.; Peixe A.; Arnholdt-Schmitt B. Involvement of alternative oxidase (AOX) in adventitious rooting of Olea europaea L. microshoots is linked to adaptive phenylpropanoid and lignin metabolism. Plant Cell Reports 2012, 31 (9), 1581–1590. 10.1007/s00299-012-1272-6. [DOI] [PubMed] [Google Scholar]
- Porfirio S.; Calado M. L.; Noceda C.; Cabrita M. J.; da Silva M.; Azadi P.; Peixe A. Tracking biochemical changes during adventitious root formation in olive (Olea europaea L.). Scientia horticulturae 2016, 204, 41–53. 10.1016/j.scienta.2016.03.029. [DOI] [Google Scholar]
- Ott R.; Chibale K.; Anderson S.; Chipeleme A.; Chaudhuri M.; Guerrah A.; Colowick N.; Hill G. C. Novel inhibitors of the trypanosome alternative oxidase inhibit Trypanosoma brucei brucei growth and respiration. Acta Tropica 2006, 100 (3), 172–184. 10.1016/j.actatropica.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Kumar S. R.; Sathishkumar R. Gene technology applied for AOX functionality studies. Alternative respiratory pathways in higher plants 2015, 287–297. 10.1002/9781118789971.ch18. [DOI] [Google Scholar]
- Yabu Y.; Yoshida A.; Suzuki T.; Nihei C. I.; Kawai K.; Minagawa N.; Hosokawa T.; Nagai K.; Kita K.; Ohta N. The efficacy of ascofuranone in a consecutive treatment on Trypanosoma brucei brucei in mice. Parasitology International 2003, 52 (2), 155–164. 10.1016/S1383-5769(03)00012-6. [DOI] [PubMed] [Google Scholar]
- Vicentini T. M.; Cavalheiro A. H.; Dechandt C. R. P.; Alberici L. C.; Vargas-Rechia C. G. Aluminum directly inhibits alternative oxidase pathway and changes metabolic and redox parameters on Jatropha curcas cell culture. Plant Physiol Biochem 2019, 136, 92–97. 10.1016/j.plaphy.2019.01.012. [DOI] [PubMed] [Google Scholar]
- Menzies S. K.; Tulloch L. B.; Florence G. J.; Smith T. K. The trypanosome alternative oxidase: a potential drug target?. Parasitology 2018, 145 (2), 175–183. 10.1017/S0031182016002109. [DOI] [PubMed] [Google Scholar]
- Ebiloma G. U.; Ayuga T. D.; Balogun E. O.; Gil L. A.; Donachie A.; Kaiser M.; Herraiz T.; Inaoka D. K.; Shiba T.; Harada S.; Kita K.; de Koning H. P.; Dardonville C. Inhibition of trypanosome alternative oxidase without its N-terminal mitochondrial targeting signal (ΔMTS-TAO) by cationic and non-cationic 4-hydroxybenzoate and 4-alkoxybenzaldehyde derivatives active against T. brucei and T. congolense. Eur. J. Med. Chem. 2018, 150, 385–402. 10.1016/j.ejmech.2018.02.075. [DOI] [PubMed] [Google Scholar]
- West R. A.; Cunningham T.; Pennicott L. E.; Rao S. P. S.; Ward S. E. Toward More Drug Like Inhibitors of Trypanosome Alternative Oxidase. ACS Infect Dis 2018, 4 (4), 592–604. 10.1021/acsinfecdis.7b00218. [DOI] [PubMed] [Google Scholar]
- Elliott C.; Young L.; May B.; Shearman J.; Albury M. S.; Kido Y.; Kita K.; Moore A. L. Purification and characterisation of recombinant DNA encoding the alternative oxidase from Sauromatum guttatum. Mitochondrion 2014, 19 (Pt B), 261–268. 10.1016/j.mito.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Cueto-Diaz E. J.; Ebiloma G. U.; Alfayez I. A.; Ungogo M. A.; Lemgruber L.; Gonzalez-Garcia M. C.; Giron M. D.; Salto R.; Fueyo-Gonzalez F. J.; Shiba T.; Gonzalez-Vera J. A.; Ruedas Rama M. J.; Orte A.; de Koning H. P.; Dardonville C. Synthesis, biological, and photophysical studies of molecular rotor-based fluorescent inhibitors of the trypanosome alternative oxidase. Eur. J. Med. Chem. 2021, 220, 113470 10.1016/j.ejmech.2021.113470. [DOI] [PubMed] [Google Scholar]
- Lanteri C. A.; Tidwell R. R.; Meshnick S. R. The Mitochondrion Is a Site of Trypanocidal Action of the Aromatic Diamidine DB75 in Bloodstream Forms of Trypanosoma brucei. Antimicrob. Agents Chemother. 2008, 52 (3), 875–882. 10.1128/AAC.00642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim H. M.; Al-Salabi M. I.; El Sabbagh N.; Quashie N. B.; Alkhaldi A. A.; Escale R.; Smith T. K.; Vial H. J.; de Koning H. P. Symmetrical choline-derived dications display strong anti-kinetoplastid activity. J. Antimicrob. Chemother. 2011, 66 (1), 111–25. 10.1093/jac/dkq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eze A. A.; Gould M. K.; Munday J. C.; Tagoe D. N.; Stelmanis V.; Schnaufer A.; De Koning H. P. Reduced Mitochondrial Membrane Potential Is a Late Adaptation of Trypanosoma brucei brucei to Isometamidium Preceded by Mutations in the gamma Subunit of the F1Fo-ATPase. PLoS Negl Trop Dis 2016, 10 (8), e0004791 10.1371/journal.pntd.0004791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardonville C.; Alkhaldi A. A. M.; De Koning H. P. SAR Studies of Diphenyl Cationic Trypanocides: Superior Activity of Phosphonium over Ammonium Salts. ACS Med. Chem. Lett. 2015, 6 (2), 151–155. 10.1021/ml500408d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhaldi A. A. M.; Martinek J.; Panicucci B.; Dardonville C.; Zikova A.; de Koning H. P. Trypanocidal action of bisphosphonium salts through a mitochondrial target in bloodstream form Trypanosoma brucei. Int. J. Parasitol Drugs Drug Resist 2016, 6 (1), 23–34. 10.1016/j.ijpddr.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R. A.; O’Doherty O. G.; Askwith T.; Atack J.; Beswick P.; Laverick J.; Paradowski M.; Pennicott L. E.; Rao S. P. S.; Williams G.; Ward S. E. African trypanosomiasis: Synthesis & SAR enabling novel drug discovery of ubiquinol mimics for trypanosome alternative oxidase. Eur. J. Med. Chem. 2017, 141, 676–689. 10.1016/j.ejmech.2017.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fueyo Gonzalez F. J.; Ebiloma G. U.; Izquierdo Garcia C.; Bruggeman V.; Sanchez Villamanan J. M.; Donachie A.; Balogun E. O.; Inaoka D. K.; Shiba T.; Harada S.; Kita K.; de Koning H. P.; Dardonville C. Conjugates of 2,4-Dihydroxybenzoate and Salicylhydroxamate and Lipocations Display Potent Antiparasite Effects by Efficiently Targeting the Trypanosoma brucei and Trypanosoma congolense Mitochondrion. J. Med. Chem. 2017, 60 (4), 1509–1522. 10.1021/acs.jmedchem.6b01740. [DOI] [PubMed] [Google Scholar]
- Inaoka D. K.; Shiba T.; Chiaki T.; Kido Y.; Balogun E. O.; Young L.; Moore A.; Saimoto H.; Harada S.; Kita K. Targeting the alternative oxidase for antitrypanosomal drug development. Biochimica et Biophysica Acta (BBA) - Bioenergetics 2018, 1859, e25–e26. 10.1016/j.bbabio.2018.09.080. [DOI] [Google Scholar]
- Costa P. C. S.; Barsottini M. R. O.; Vieira M. L. L.; Pires B. A.; Evangelista J. S.; Zeri A. C. M.; Nascimento A. F. Z.; Silva J. S.; Carazzolle M. F.; Pereira G. A. G.; Sforça M. L.; Miranda P. C. M. L.; Rocco S. A. N-Phenylbenzamide derivatives as alternative oxidase inhibitors: Synthesis, molecular properties, 1H-STD NMR, and QSAR. J. Mol. Struct. 2020, 1208, 127903 10.1016/j.molstruc.2020.127903. [DOI] [Google Scholar]
- Cisneros D.; Cueto-Díaz E. J.; Medina-Gil T.; Chevillard R.; Bernal-Fraile T.; López-Sastre R.; Aldfer M. M.; Ungogo M. A.; Elati H. A. A.; Arai N.; Otani M.; Matsushiro S.; Kojima C.; Ebiloma G. U.; Shiba T.; de Koning H. P.; Dardonville C. Imidazoline- and Benzamidine-Based Trypanosome Alternative Oxidase Inhibitors: Synthesis and Structure–Activity Relationship Studies. ACS Med. Chem. Lett. 2022, 13 (2), 312–318. 10.1021/acsmedchemlett.1c00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauheed A. M.; Mamman M.; Ahmed A.; Suleiman M. M.; Balogun E. O. Antitrypanosomal properties of Anogeissus leiocarpa extracts and their inhibitory effect on trypanosome alternative oxidase. Phytomedicine Plus 2022, 2 (2), 100223 10.1016/j.phyplu.2022.100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsottini M. R.; Pires B. A.; Vieira M. L.; Pereira J. G.; Costa P. C.; Sanitá J.; Coradini A.; Mello F.; Marschalk C.; Silva E. M.; Paschoal D.; Figueira A.; Rodrigues F. H.; Cordeiro A. T.; Miranda P. C.; Oliveira P. S.; Sforça M. L.; Carazzolle M. F.; Rocco S. A.; Pereira G. A. Synthesis and testing of novel alternative oxidase (AOX) inhibitors with antifungal activity against Moniliophthora perniciosa (Stahel), the causal agent of witches’ broom disease of cocoa, and other phytopathogens. Pest Management Science 2019, 75 (5), 1295–1303. 10.1002/ps.5243. [DOI] [PubMed] [Google Scholar]
- Li D.; Chen H.; Florentino A.; Alex D.; Sikorski P.; Fonzi W. A.; Calderone R. Enzymatic Dysfunction of Mitochondrial Complex I of the Candida albicans goa1Mutant Is Associated with Increased Reactive Oxidants and Cell Death. Eukaryotic Cell 2011, 10 (5), 672–682. 10.1128/EC.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvenage L.; Munro C. A.; Gourlay C. W. The potential of respiration inhibition as a new approach to combat human fungal pathogens. Current Genetics 2019, 65 (6), 1347–1353. 10.1007/s00294-019-01001-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.; Liao K.; Wang D. Honokiol induces superoxide production by targeting mitochondrial respiratory chain complex I in Candida albicans. PLoS One 2017, 12 (8), e0184003 10.1371/journal.pone.0184003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa N.; Uehara M.; Seki S.; Nitta A.; Kogawara K. Effects of combined addition of atovaquone and lithium on the in vitro cell growth of the pathogenic yeast Candida albicans. Yakugaku Zasshi 2010, 130 (2), 247–251. 10.1248/yakushi.130.247. [DOI] [PubMed] [Google Scholar]
- Balogun E. O.; Inaoka D. K.; Shiba T.; Tsuge C.; May B.; Sato T.; Kido Y.; Nara T.; Aoki T.; Honma T.; Tanaka A.; Inoue M.; Matsuoka S.; Michels P. A. M.; Watanabe Y.-I.; Moore A. L.; Harada S.; Kita K. Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase of Trypanosoma brucei brucei. FASEB journal: official publication of the Federation of American Societies for Experimental Biology 2019, 33 (11), 13002–13013. 10.1096/fj.201901342R. [DOI] [PubMed] [Google Scholar]
- Pereira C. S.; Teixeira M. H.; Russell D. A.; Hirst J.; Arantes G. M. Mechanism of rotenone binding to respiratory complex I depends on ligand flexibility. Sci. Rep. 2023, 13 (1), 6738. 10.1038/s41598-023-33333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonbaum G. R.; Bonner W. D.; Storey B. T.; Bahr J. T. Specific inhibition of the cyanide-insensitive respiratory pathway in plant mitochondria by hydroxamic acids. Plant Physiology 1971, 47 (1), 124–128. 10.1104/pp.47.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady R. W.; Bienen E. J.; Clarkson A. B. Esters of 3,4-dihydroxybenzoic acid, highly effective inhibitors of the sn-glycerol-3-phosphate oxidase of Trypanosoma brucei brucei. Mol. Biochem. Parasitol. 1986, 21 (1), 55–63. 10.1016/0166-6851(86)90079-4. [DOI] [PubMed] [Google Scholar]
- Minagawa N.; Yabu Y.; Kita K.; Nagai K.; Ohta N.; Meguro K.; Sakajo S.; Yoshimoto A. An antibiotic, ascofuranone, specifically inhibits respiration and in vitro growth of long slender bloodstream forms of Trypanosoma brucei brucei. Mol. Biochem. Parasitol. 1996, 81 (2), 127. 10.1016/0166-6851(96)02665-5. [DOI] [PubMed] [Google Scholar]
- Saimoto H.; Kido Y.; Haga Y.; Sakamoto K.; Kita K. Pharmacophore identification of ascofuranone, potent inhibitor of cyanide-insensitive alternative oxidase of Trypanosoma brucei. Journal of Biochemistry 2013, 153 (3), 267–273. 10.1093/jb/mvs135. [DOI] [PubMed] [Google Scholar]
- Viscomi C.; Moore A. L.; Zeviani M.; Szibor M. Xenotopic expression of alternative oxidase (AOX) to study mechanisms of mitochondrial disease. Biochim Biophys Acta Bioenerg 2023, 1864 (2), 148947 10.1016/j.bbabio.2022.148947. [DOI] [PubMed] [Google Scholar]
- El-Khoury R.; Kemppainen K. K.; Dufour E.; Szibor M.; Jacobs H. T.; Rustin P. Engineering the alternative oxidase gene to better understand and counteract mitochondrial defects: state of the art and perspectives. Br. J. Pharmacol. 2014, 171 (8), 2243–2249. 10.1111/bph.12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khoury R.; Kaulio E.; Lassila K. A.; Crowther D. C.; Jacobs H. T.; Rustin P. Expression of the alternative oxidase mitigates beta-amyloid production and toxicity in model systems. Free Radic Biol. Med. 2016, 96, 57–66. 10.1016/j.freeradbiomed.2016.04.006. [DOI] [PubMed] [Google Scholar]
- Roberts C. W.; Roberts F.; Henriquez F. L.; Akiyoshi D.; Samuel B. U.; Richards T. A.; Milhous W.; Kyle D.; McIntosh L.; Hill G. C.; Chaudhuri M.; Tzipori S.; McLeod R. Evidence for mitochondrial-derived alternative oxidase in the apicomplexan parasite Cryptosporidium parvum: a potential anti-microbial agent target. Int. J. Parasitol 2004, 34 (3), 297–308. 10.1016/j.ijpara.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Tsaousis A. D.; Hamblin K. A.; Elliott C. R.; Young L.; Rosell-Hidalgo A.; Gourlay C. W.; Moore A. L.; van der Giezen M. The Human Gut Colonizer Blastocystis Respires Using Complex II and Alternative Oxidase to Buffer Transient Oxygen Fluctuations in the Gut. Front Cell Infect Microbiol 2018, 8, 371. 10.3389/fcimb.2018.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stich A.; Ponte-Sucre A.; Holzgrabe U. Do we need new drugs against human African trypanosomiasis?. Lancet Infect Dis 2013, 13 (9), 733–4. 10.1016/S1473-3099(13)70191-9. [DOI] [PubMed] [Google Scholar]
- Alsford S.; Eckert S.; Baker N.; Glover L.; Sanchez-Flores A.; Leung K. F.; Turner D. J.; Field M. C.; Berriman M.; Horn D. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature 2012, 482 (7384), 232–6. 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black S. J.; Mansfield J. M. Prospects for vaccination against pathogenic African trypanosomes. Parasite Immunol 2016, 38 (12), 735–743. 10.1111/pim.12387. [DOI] [PubMed] [Google Scholar]