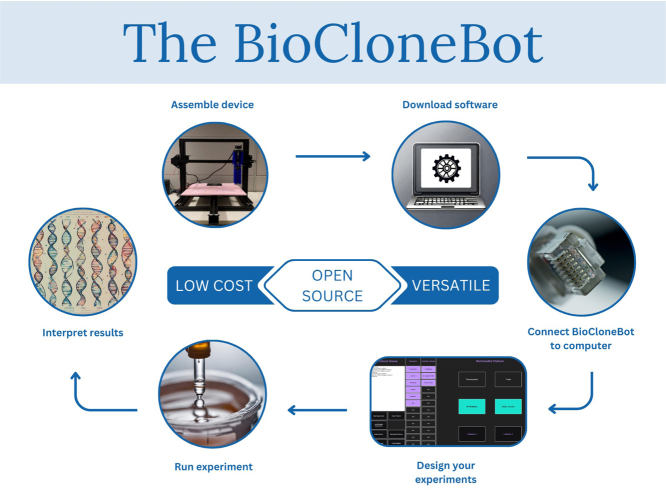
Abstract
Liquid handler systems can provide significant benefits to researchers by automating laboratory work, however, their unaffordable price provides a steep barrier to entry. Therefore, we provide the BioCloneBot, a versatile, low-cost, and open-source automated liquid handler. This system can be easily built with 3D-printed parts and readily available commercial components. The BioCloneBot is highly adaptive to user needs and facilitates various liquid handling tasks in research and diagnostics. Its user-friendly interface and programmable nature make it suitable for a wide range of applications, from small-scale experiments to larger laboratory setups. By utilizing BioCloneBot, researchers and scientists can streamline their liquid handling processes without the financial constraints posed by traditional systems. In this paper, we detail the design, construction, and validation of BioCloneBot, showcasing its precise control, accuracy, and repeatability in various liquid handling tasks. The open-source nature of the system encourages collaboration and customization, enabling researchers to contribute and adapt the technology to specific experimental requirements.
Keywords: Open-source, Liquid handler, Synthetic biology, DNA cloning, 3D printing
Graphical abstract
| Specifications table | |
| Hardware name | BioCloneBot (BCB) |
| Subject area |
Engineering and material science Biological sciences (e.g. microbiology and biochemistry) |
| Hardware type | Biological sample handling and preparation |
| Closest commercial analog | OpenTrons OT-2 |
| Open source license | GNU General Public License (GPL) 3.0 |
| Cost of hardware | $2000 CAD |
| Source file repository | https://doi.org/10.17605/OSF.IO/QDP7V |
1. Hardware in context
Synthetic biology, an emerging scientific discipline, focuses on the investigation of organisms through the manipulation of their genetic material [1]. This field holds significant potential for advancing various domains, including medicine [2], [3], agriculture [4], [5], and biofuel production [6], [7]. To expedite the transition from conceptualization to experimental outcomes, synthetic biologists are increasingly adopting automation technologies. This integration of automation provides numerous advantages, primarily enhanced throughput and a reduction in human-associated errors. However, the utilization of automation is often impeded by the high costs associated with commercial solutions. Consequently, do-it-yourself (DIY) biologists and engineers have taken on the task of developing affordable, open-source hardware to democratize automation. Currently, a multitude of open-source DIY solutions are available, encompassing syringe pumps [8], [9], [10], peristaltic pumps [11], plate-readers [12], microfluidic devices [13], [14], [15], [16], [17], [18] and a wide array of other innovations.
One of the pivotal components in laboratory automation is the automated liquid handler, which plays a crucial role in synthetic biology experiments by automating some or all of the numerous repetitive liquid handling procedures. These experiments encompass a wide range of tasks, including DNA extraction and the synthesis of customized DNA, which necessitate the precise movement of liquids between various types of labware, such as tubes, well plates, and reservoirs. Traditionally, scientists rely on manual pipettes to perform these liquid transfers involving DNA, enzymes, and reagents, at each stage of the experimental process. This is especially true when working with volumes of less than 1mL as can be seen in protocols such as Golden Gate cloning [19], Gibson assembly [20], and MIDAS [21]. While studying a single organism, a biologist could perform thousands of these manipulations. These steps often lead to human error and take significant time to perform. If error is introduced early on, it can propagate through the proceeding steps resulting in a skewed result, or sometimes a failed experiment. Depending on the complexity of the required procedures, repeating the entire process from initiation to completion may take multiple days. Automated liquid handlers alleviate these challenges by eliminating human error and streamlining the workflow for biologists through the automation of laborious manual manipulations.
The current lowest cost commercial liquid handling platforms, the OpenTrons-OT2, starts at $8,700 CAD and quickly rises to $12,000 CAD after adding a P20 and a P300 automated pipette [22]. These pipettes are two of the most common pipettes in a wet lab. Unlike the OpenTrons-OT2, most competing commercial liquid handlers begin at a comparable price points, but rapidly escalate into the low six-figure range. Consequently, numerous open-source liquid handling systems have been developed [23], [24], [25], [26], [27], [28], [29]. Most of the currently available open-source liquid handlers have one of two major flaws: they are limited to a single application/labware or they require a high level of training to use, sometimes involving code-based programming.
To overcome these limitations, we propose the BioCloneBot as an addition to existing open-source liquid handling solutions. The BioCloneBot represents a versatile, low-cost, and open-source automated liquid handler that addresses the aforementioned flaws. Notably, it incorporates an intuitive front-end interface for ease of use and can accommodate up to four different on-deck labware configurations, offering significant flexibility in experimental setups.
2. Hardware description
To realize the BioCloneBot, four major components were chosen: a micro-liter syringe pump to perform pipette actions (see Fig. 1), a 3-axis movement system and device frame for moving the syringe pump, a main board for controlling motors and related electronics, and a software front-end for designing and running experiments.
Fig. 1.

Custom Microliter Syringe Pump with P200 Tip Adapter.
2.1. Microliter syringe pump
In this study, we drew inspiration from various pump designs, namely the OpenTrons pump, EvoBot syringe modules and the re-usable DIY syringe pumps developed by Amarante et al. [9] and Gervasi, Cardol and Meyer [10]. Our aim was to create a syringe pump driven by a stepper motor, which offers several advantages such as high precision and ease of control. Notably, the use of stepper motors in 3D printing applications demonstrates their capability for precise movements required in a low-volume L pump.
The selected pump design comprises several important sub-components, including a Hamilton microliter syringe, a linear actuator featuring a 400-step bipolar stepper motor, a 2 mm pitch lead screw, a 5 mm to 6 mm shaft coupler, a lead screw nut, and 3D printed components. To enhance functionality, a limit switch, plus two springs that were incorporated for pipette tip removal, and auto-homing features. Considering the diverse range of volumes needed, the pump was designed to accommodate the Hamilton 700 series microliter syringes [30]. Specifically, we opted for the 50 L and 250 L syringes to cover volume ranges of 1.0–20.0 L and 10.0–200.0 L, respectively. Two custom tip adapters were securely attached to each syringe’s RN nut using a two-part epoxy, guaranteeing an airtight seal between the syringe and the tip adapter.
2.2. 3-Axis movement system & device frame
An Ender 3 Pro 3D printer was chosen to provide the basis of a mechanical frame and platform to hold labware. An Ender Xtender 400 kit was included to increase the size of the bed from 225 mm × 225 mm to 400 mm × 400 mm to allow the BioCloneBot to support multiple labware. The Ender 3 Pro and Ender Xtender 400 kit is $200-$300 cheaper than the next largest printer, the Creality Ender 5 Plus, which comes with a smaller bed dimension of 350 mm × 350 mm. This combo makes up the entire frame of the BioCloneBot and includes an already existing x, y, and z system with stepper motors and limit switches proven to work efficiently with highly precise movements required for 3D printing as shown in Fig. 2. With the larger bed size, the platform supports up to 6 labware. Given the maximum print width of 225 mm by the Ender 3 Pro, the platform is designed as 9 individual parts that connect with dove-tail fittings as seen in Fig. 3 below. Each labware slot is 128 mm × 86 mm to support the standard dimension of a well plate. To accommodate tube stands, a 3D-printed tube stand uploaded to Thingiverse by thehair [31] was used due to its matching length and width of a standard 96-well plate. thehair’s design allows for the use of up to 24 1.5 mL tubes and can be printed using the majority of 3D printers. A 20 L tip box was adapted from thehair’s design.
Fig. 2.
BioCloneBot frame and pump fully assembled.
Fig. 3.
3D model of platform where labware is placed and experiments are performed.
To control the movement of the syringe pump, the original stepper motors of the Ender are used. While the Ender 3 Pro mainboard could be utilized for controlling the motors, it lacks GPIO ports for expandability and is challenging to re-program. Therefore, an MKS Gen L V2.1 and four TMC2209 v2.0 stepper motor drivers were selected to control the x, y, z, and pump stepper motors. The TMC2209 supports microstepping with a minimum step size of 1/256, offering substantially higher precision compared to no microstepping. A microstepping factor of 1/8 was chosen to strike a balance between precision and speed.
2.3. Mainboard
While the Ender 3-Pro uses the open source 3D printer firmware, Marlin, the code base is large and has an even larger learning curve to understand and modify [32]. To reduce the complexity of the firmware code and ensure ease of modification for future improvements, the entire BioCloneBot firmware is implemented from scratch. An MKS Gen L v2.1 was chosen as the main board as it has matching headers for the stepper motors, motor drivers, and limit switches. In addition, it includes extra ports for additional hardware and features the same microprocessor as an Arduino Mega 2560 allowing for it to be programmed through standard Arduino C code. The firmware was designed to use a custom-made communication protocol. Each message is built as a string with the start character ‘#’ and the end character ‘%’. The first four characters of each message encode a 4-bit binary number. This allows for up to 16 different commands. The characters following the 4-bit encoding dictate the parameters of the command. A list of the commands is described in Table 1.
Table 1.
List of device commands.
| Command | 4-bits | Parameters | Description |
|---|---|---|---|
| Home | 0000 | None | Homes x, y, z, and pump stepper motors. |
| Move | 0001 | XYZ direction XXX.XX mm YYY.YY mm ZZZ.ZZ mm | Move pump in XYZ direction, XXX.XX mm, YYY.YY mm, and ZZZ.ZZ mm |
| Remove Tip | 0010 | None | Moves pump over trash bin, removes tip, and homes pump. |
| Aspirate | 0011 | XXX.XX L | Aspirates XXX.XX volume into pipette tip. |
| Dispense | 0100 | XXX.XX L | Dispenses XXX.XX volume into pipette tip. |
| Mix | 0101 | XYZ times XXX.XX volume | Mixes XXX.XX volume XYZ amount of times. |
| Enable Steppers | 1110 | None | Enables all stepper motor drivers. |
| Disable Steppers | 1111 | None | Disables all stepper motor drivers. |
As an example, a set of commands is described below.
-
1.
#0000%
-
2.
#0001100100.00000.00000.00%
-
3.
#0011100.00%
First, #0000% triggers the pump to move to the (0.00, 0.00, 0.00), identical to the origin of a standard Cartesian coordinate system. The start character # and end character % denote when a command starts and ends while being sent with serial-over-USB communication. Note that 0.00 on the -axis refers to the highest point instead of the lowest point. Once in the (0.00, 0.00, 0.00) position, the pump actuates until the limit switch is engaged, reverses directions until the switch is disengaged, and finally backs off a little further to create a trailing air gap. This set of actions homes the pump and removes backlash-induced error from the lead screw and lead screw nut.
Next, the command #0001100100.00000.00000.00% moves the pump to the position (100.00, 0.00, 0.00). The first four characters after the start character #, 0001, indicate the move command. The following three characters, 100, indicates the directions the pump will move in each axis. If the value is 0, the pump will move towards the limit switch. If the value is 1, it will move away from the limit switch. Thus, the 1 in 100 indicates to move toward the limit switch on the -axis, the following 0 indicates to move away from the limit switch on the -axis, and the final 0 indicates to move away from the limit switch on the -axis. The last portion can be broken up into three groups of 6 characters: XXX.XX, YYY.YY, and ZZZ.ZZ. 100.00 indicates that the pump should move 100.00 mm along the -axis. The remaining 000.00 and 000.00 indicate that the pump should move 000.00 mm along the -axis and -axis.
Finally, #0011100.00% tells the pump to aspirate 100.00 L. The first four characters, 0011, indicate the aspirate command, and the last 6 characters, 100.00, indicate that the pump should aspirate by 100.00 L. Thus ending the commands.
2.4. Front-end
Lastly, the BioCloneBot requires a front-end to allow the end user to design their own experiments. C# Sharp was chosen due to the easy-to-use graphical user interface design capabilities of Visual Studio. This limits the BioCloneBot to Windows devices which should be available in most lab settings. Within the front-end, the user must design experiments by first adding labware, and then initializing labware properties such as starting volumes, starting tip locations, etc. Once the labware has been decided, the user can generate an experiment by selecting operations. A user guide is available in Supplementary material UserGuide.docx.
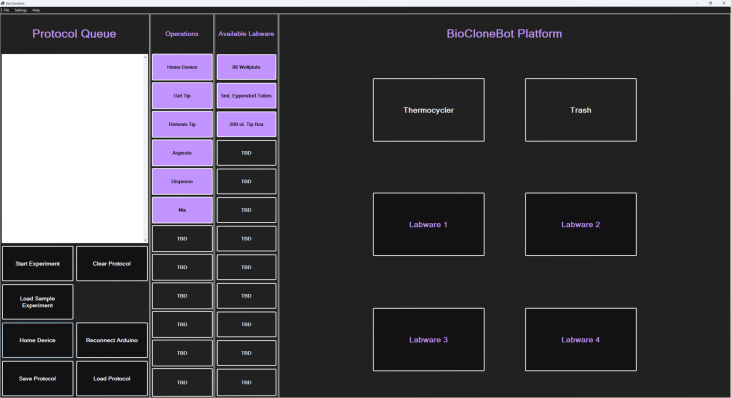
The front-end is designed to be simple and easy to use. Partially inspired by the user interfaces of the Biomek and the OpenTrons, the user interface is designed with the majority of the functionality built directly into the main window as seen in Fig. 4 below. Larger images of the left and right portions of the main window can be seen in Fig. 5 and Fig. 6 respectively.
Fig. 4.
BioCloneBot Front-End.
Fig. 5.
The left side of the front-end includes the Protocol Queue which displays the selected operations in order. Also included are the potential Operations and Available Labware. Underneath the Protocol Queue are buttons used to control the software such as Start Experiment, Clear Protocol, Load Sample Experiment, and more.
Fig. 6.
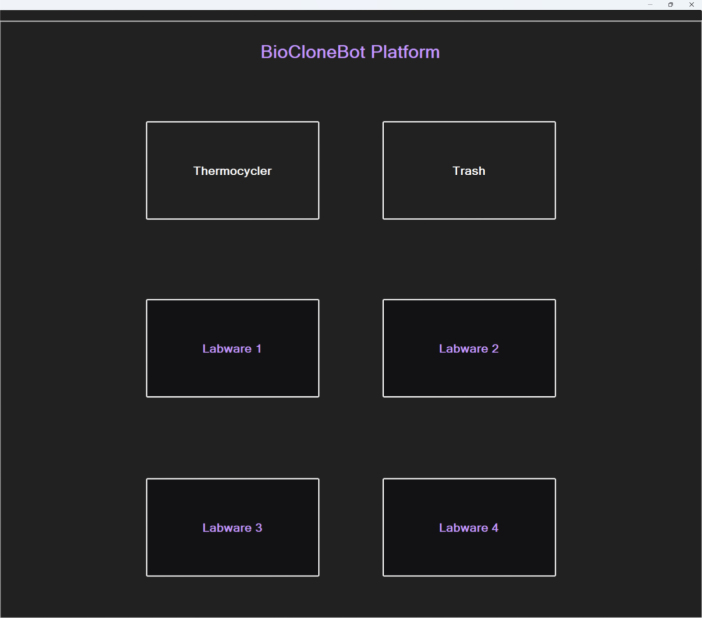
The right side of the front-end shows the platform for placing labware.
On the BioCloneBot Platform side of the front-end, any of the available labware can be added, removed, and modified. By default, a standard 96-well plate, a 5 mL Eppendorf tube stand consisting of a 4 × 6 configuration capable of holding 5 mL Eppendorf tubes, and a 200 L tip box are available. Once a labware is added, the properties can be modified by double clicking on the corresponding labware slot where the labware was added. The labware properties for each of the standard labware can be seen below in Fig. 7.
Fig. 7.
The properties window supports the selection of individual wells, tubes, or tips. The select all button allows for quick modification of reservoir volumes or tip availability. Modifications are only confirmed once the OK button is pressed.
The BioCloneBot:
-
•
Enables manufacture of an automated liquid handler for <$2000;
-
•
Provides a versatile and flexible platform to support up to 4 different labware;
-
•
Provides a microliter-scale liquid automation solution.
-
•
Provides an easy-to-use front-end interface.
3. Design files summary
| Design filename | File type | Open source license |
Location of the file |
|---|---|---|---|
| 10uLTipAdapter | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| 200uLTipAdapter | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| MotorHolder | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| SyringeCover | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| SyringeHolder | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| SyringePumpAssembly | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| SyringePumpBaseV3 | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| SyringePumpHolder | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| TipEjectorBottomEnd | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| TipEjectorShaft | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| [CIT Relay & CAD Switch] Limit-Switch | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| [Misumi]LeadScrew | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| [Misumi]LeadScrewNut | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| [Misumi]ShaftCoupler | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
| [MoonsSteppers]StepperMotor | CAD | GPL v3.0 | https://doi.org/10.17605/OSF.IO/QDP7V |
Each file not preceded by a [manufacturer] was designed using SolidWorks. The files from CIT Relay & CAD, Misumi, and MoonsSteppers are sourced from the manufacturers directly. STL files are available in NIH with the link .
4. Bill of materials summary
The bill of materials can be broken down into two separate tables. One table containing the required components for the assembly of the BioCloneBot, and one table containing the tools required to assemble the components together. 3D printers, filament, soldering irons, etc. are included in the tools table.
4.1. Components
The components listed below are the parts or inputs used to assemble the BioCloneBot. The Ender 3 Pro listed here will be used as the foundation of the BioCloneBot. It will be assembled and some parts will be removed or modified.
| Designator | Component | Number | Cost/unit (CAD) |
Total cost (CAD) |
Material type |
|---|---|---|---|---|---|
| 1 | Ender 3 Pro 3D Printer | 1 | $326.42 | $326.42 | Other |
| 2 | Ender Xtender 400 for Ender 3 Pro V1 | 1 | $247.58 | $247.58 | Other |
| 3 | MKS Gen L V2.1 | 1 | $37.48 | $37.48 | Other |
| 4 | TMC2209 V2.0 Stepper Motor Driver (5 pack) | 1 | $56.38 | $56.38 | Other |
| 5 | 400 Step/0.9 Degree 24 V/1.5 A NEMA17 Bipolar Stepper Motor | 1 | $57.15 | $57.15 | Other |
| 6 | Straight Lever Limit Switch | 1 | $8.84 | $8.84 | Other |
| 7 | Misumi MTSRA10-100-S30-Q6 Lead Screw | 1 | $31.79 | $31.79 | Metal |
| 8 | Misumi MRG-16-5-6 Shaft Coupler | 1 | $26.57 | $26.57 | Metal |
| 9 | Misumi MTSNR10 | 1 | $28.19 | $28.19 | Metal |
| 10 | Hamilton 50 L Microliter Syringe Switch | 1 | $110.19 | $110.19 | Other |
| 11 | Hamilton 250 L Microliter Syringe Switch | 1 | $145.12 | $145.12 | Other |
| 12 | JST XH Connector Kit | 1 | $34.47 | $34.47 | Other |
| 13 | 1–5/8” x 3/16” Compression Spring | 2 | $2.29 | $4.58 | Metal |
| 14 | Socket Head Cap | 1 | $36.99 | $36.99 | Metal |
The total cost of all components is $1,151.75 CAD.
4.2. Tools
The tools listed below are used to 3D print the parts or help manufacture some of the components from the prior table. The Ender 3 Pro listed here is used to print the 3D printed parts of the BioCloneBot. Alternative 3D printers can be used as long as their bed dimensions match those of the Ender 3 Pro (220 mm × 220 mm).
| Designator | Component | Number | Cost/unit (CAD) |
Total cost (CAD) |
Material type |
|---|---|---|---|---|---|
| 15 | Ender 3 Pro 3D Printer | 1 | $326.42 | $326.42 | Other |
| 16 | 1.75 mm Standard PLA Filament | 2KG | $49.95 | $49.95 | Plastic |
| 17 | Two Part Epoxy | 1 | $7.89 | $7.89 | Other |
| 18 | Digital Multimeter | 1 | $15.99 | $15.99 | Other |
| 19 | Soldering Iron Kit | 1 | $24.99 | $24.99 | Other |
The total cost of all the tools is $425.24 CAD
In total, the cost of the BioCloneBot components and assembly/manufacturing tools is $1,576.99.
5. Build instructions summary
The assembly guidelines can be segmented into four distinct phases: Disassembly of Stock Ender 3 Pro, Syringe Pump Assembly, Frame Assembly, and Electronic Connection. To initiate the disassembly of the frame, it is advised to meticulously adhere to the subsequent steps outlined below. A comprehensive assembly guide can be found in supplementary document BCB_Building_Instruction.pdf.
5.1. Disassembly of stock ender 3 pro
-
1.
Open the Ender 3 Pro box and remove all parts. (BI, Figure 1)
-
2.
Remove the four knobs and springs from the Ender 3 Pro Base. (BI, Figure 2)
-
3.
Carefully remove the Ender 3 Pro bed and set it aside. (BI, Figure 3)
-
4.
Check Y-Axis belt tension, then remove four screws from the Y-Axis Belt Tensioner. (BI, Figure 4)
-
5.
Unhook the Y-Axis belt from the Y-Axis Carriage and remove the tensioner. (BI, Figure 5)
-
6.
Remove the Y-Axis Carriage from the frame and set it aside. (BI, Figure 6)
-
7.
Unplug Y-Axis Limit Switch Connector and unscrew the Y-Axis Stepper Motor. (BI, Figure 7)
-
8.
Remove the Y-Axis Limit switch and screws, placing them aside. (BI, Figure 8)
-
9.
Remove the electronic housing, unscrewing screws from the top and bottom. (BI, Figure 9)
-
10.
Carefully remove the bottom cover, unplug the fan, and remove the front screws. (BI, Figure 10)
-
11.
Remove the wiring bundle from the bottom frame notch. (BI, Figure 11)
-
12.
Locate and remove the extruder components, keeping the -axis roller aside. (BI, Figure 12)
-
13.
Open the electronics bay. (BI, Figure 13)
-
14.
Cut zip ties and remove tape from the wire bundle. (BI, Figure 14)
-
15.
Carefully unplug X, Y, Z, and E stepper motor wires from the mainboard, dealing with any glue. (BI, Figure 15)
-
16.
Cut the zip tie holding the wire bundle and carefully unplug X, Y, and Z limit switch connectors from the mainboard, dealing with glue. (BI, Figure 16)
-
17.
Remove the power cable from the original board. (BI, Figure 17)
-
18.
Set aside six wires for later use. (BI, Figure 18)
5.2. Syringe assembly
-
19.
Gather all parts for the syringe pump assembly. (BI, Figure 19)
-
20.
Attach the Syringe Pump Holder to the Syringe Pump Base using four M3x12 screws. (BI, Figure 20)
-
21.
Attach the Syringe Pump Stepper Motor to the Motor Holder using four M3x8 screws. (BI, Figure 21)
-
22.
Attach the Shaft Coupler to the Lead Screw using two set screws. Then attach the Shaft Coupler to the stepper motor shaft using the last two set screws, ensuring the Lead Screw is attached first for a proper fit. (BI, Figure 22)
-
23.
Attach the Lead Screw Nut to the Plunger Holder using two M2x12 screws, two M2 nuts, and two M2 washers. Ensure a tight fit, applying force if necessary. (BI, Figure 23)
-
24.
Assemble the Tip Ejector by screwing on two Tip Ejector Shafts to the Tip Ejector Top using two M2x8 screws. Slide the springs around the shafts, ensuring smooth movement. (BI, Figure 24)
-
25.
Insert the shafts into the Syringe Pump Base and compress the springs with your hand. Screw the Tip Ejector Bottom to the Tip Ejector Shafts using two M2x8 screws. (BI, Figure 25)
-
26.
Rotate the Plunger holder and Lead screw nut to the Lead Screw. (BI, Figure 26)
-
27.
Continue screwing in the Lead Screw until the Motor Holder is flush with the Syringe Pump Base. Screw the Motor Holder to the Syringe Pump base using four M3x8 screws. (BI, Figure 27)
-
28.
Put on the syringe cover and secure with four M2x8 screws. (BI, Figure 28)
-
29.
Install the limit switch as instructed. (BI, Figure 29)
5.3. Frame assembly
-
30.
Remove eight screws from left and right extrusions (four on each side). (BI, Figure 31)
-
31.
Install Vertical Extrusions to side extrusions using provided parts and screws. (BI, Figure 32)
-
32.
Use parts from Ender Xtender kit to attach Horizontal Extrusions to the frame using specific screws and T-nuts. (BI, Figure 33)
-
33.
Attach large Y-Axis Extrusion using T-nuts and align it properly. (BI, Figures 33, 34, 35, 36 and 37)
-
34.
Re-attach Y-Axis Carriage and feed Y-Axis Belt through the assembly. (BI, Figures 38 and 39)
-
35.
Re-attach Y-Axis Limit Switch and Stepper Motor Assembly. (BI, Figure 40)
-
36.
Attach Y-Axis Belt to Y-Axis Carriage and Tensioner, tightening appropriately. (BI, Figures 41-43)
-
37.
Prepare Heated Bed and Ender Xtender Platform by attaching screws, washers, and nuts. (BI, Figures 44-49)
-
38.
Attach the Heated Bed to the Ender Xtender Platform and secure the platform onto the Y-Axis Carriage using knobs. (BI, Figure 50)
-
39.
Gather X-Axis Stepper Motor Assembly, Z-Axis Roller, X-Axis Extrusion, and necessary screws. (BI, Figures 51 and 52)
-
40.
Attach Z-Axis Roller to the X-Axis Extrusion and slide the Syringe Pump onto the X-Axis Extrusion. (BI, Figures 53-55)
-
41.
Attach X-Axis Stepper Motor Assembly and align the rollers to Vertical Extrusions to secure X-Axis components. (BI, Figure 56)
-
42.
Attach Top Extrusion to the frame using provided screws. (BI, Figure 57)
-
43.
Modify the Z-Axis Limit Switch and attach it to the new Z-Axis Limit Switch Holder. (BI, Figure 58)
-
44.
Attach the Z-Axis Limit Switch to the Top Extrusion. (BI, Figure 58)
-
45.
Assemble the BioCloneBot platform components and place them over the platform. (BI, Figure 59)
5.4. Electronic connection
-
46.
Tune stepper motor drivers before connecting electronic parts, following specific current and set point values for X, Y, Z axis, and syringe pump motors. Be cautious to avoid motor damage during adjustments. (BI, Figure 60)
-
47.
Connect all electronic parts to the MKS GEN L V2.1 board as shown in a provided figure. (BI, Figure 61)
-
48.
Note that the original limit switch wires may not work and should be correctly configured following another provided figure. (BI, Figure 62)
-
49.
Confirm that the BioCloneBot frame and pump are now fully assembled. (BI, Figure 63)
6. Operation instructions
6.1. Safety concerns
While the overall usage of the BioCloneBot is safe, there are a few safety considerations to keep in mind during assembly and operation.
-
1.
When assembling the BioCloneBot, it is highly recommended to wear safety glasses and be careful to not get pinched when inserting and tightening screws.
-
2.
DO NOT PLUGIN THE POWER SUPPLY DURING ASSEMBLY - if the power supply is plugged in, there is a risk for electrical shock
-
3.
Do not interact with the platform while the BioCloneBot is performing an experiment as the stepper motors could crush fingers and hands
-
4.
When working with the mainboard ensure that power supply is turned off, unplugged from the wall, and unplugged from the mainboard
-
5.
Ensure the mainboard is not touching any metal portion of the BioCloneBot frame as there is a risk of shock or damaging the mainboard if accidentally electrically grounded or shorted.
-
6.
If an emergency stop is required, utilize the red switch on the power supply.
The GUI program, as seen in Fig. 8, features different lab experiment operations and labwares. Users have the ability to customize their laboratory procedures by leveraging a set of provided operations, including “Get Tip”, “Remove Tip”, “Aspirate”, “Dispense”, and “Mix”. Furthermore, users can tailor their experiments by either designing new labware or utilizing pre-existing standard labware options like the 96-well plate or the 200 L Tip Box. Additionally, users have the capability to create, save, and fine-tune their experimental setups for calibration and optimization of the device by clicking the “Setup” button on the top left. The following are detailed explanations for each operation.
Fig. 8.
BCB raphical User Interface for Lab Experiment.
-
•
Home Device: homes the x, y, z, and pump motors
-
•
Get Tip: gets a tip from a selected position on an available tip box
-
•
Remove Tip: unloads tip into the trash bin
-
•
Aspirate: aspirates a volume of L into the pipette tip
-
•
Dispense: dispenses a volume + 25 L to a destination reservoir
-
•
Mix: mixes a volume a specified number of times in a destination well
For in-depth and detailed operation instructions, kindly consult the following OSF link. https://doi.org/10.17605/OSF.IO/QDP7V
7. Validation and characterization
To validate and characterize the syringe pump, a procedure was developed for measuring the volume of each complete aspiration and dispensing. Each volume was found by using a Sartorius CP64 scale which is accurate down to 0.1 mg. The volumes were tested in two batches. The first batch consisted of volumes between 1 L and 20 L using the P20 tip adapter and the second batch consisted of volumes between 10 L and 200 L using the P200 tip adapter. An additional test using the P20 tip adapter was performed to measure the effectiveness of the pump when working with liquids of differing viscocities. Five different glyercol solutions were created by mixing glycerol into dH2O: 0%, 12.5%, 25%, 37.5%, and 50%. The percentage is defined by the percentage of mass of glycerol in the glycerol/dH2O mixture. The results of these experiments can be found in Table 2 and Table 4 respectively. The following equations are used to calculate accuracy (systematic error) and precision (random error):
Table 2.
Results of syringe pump tests using the adjusted volumes derived from linear regressions 2 with n=10 for each tested volume. The error quickly decreases as the volume increases.
| Syringe/Pipette tip | Target volume (L) | Adjusted volume (L) | Accuracy (%) | Precision (%) |
|---|---|---|---|---|
| 50 L + P20 tip | 1.00 | 1.45 | 5.00% | 11.22% |
| 50 L + P20 tip | 2.00 | 2.52 | 12.50% | 7.03% |
| 50 L + P20 tip | 5.00 | 5.13 | 2.60% | 6.03% |
| 50 L + P20 tip | 10.00 | 11.13 | 0.80% | 5.17% |
| 50 L + P20 tip | 20.00 | 21.89 | 1.05% | 0.97% |
| 250 L + P200 tip | 10.00 | 11.12 | 2.60 % | 1.47 % |
| 250 L + P200 tip | 25.00 | 26.62 | 0.32 % | 0.39 % |
| 250 L + P200 tip | 50.00 | 52.47 | 0.74 % | 0.36 % |
| 250 L + P200 tip | 100.00 | 104.15 | 0.93 % | 4.80 % |
| 250 L + P200 tip | 200.00 | 207.52 | 0.95 % | 0.32 % |
Table 4.
Results of testing glycerol and dH2O mixtures at 0% glycerol, 12.5% glycerol, 25% glycerol, 37.5% glycerol, and 50% glycerol. Each volume was tested 10 times.
| Glycerol percentage | Target volume (L) | Adjusted volume (L) | Accuracy (%) | Precision (%) |
|---|---|---|---|---|
| 0% | 1.00 | 1.45 | 5.00% | 11.22% |
| 0% | 3.00 | 3.60 | 9.00% | 6.12% |
| 0% | 5.00 | 5.75 | 5.00% | 11.22% |
| 0% | 7.00 | 7.90 | 7.14% | 3.00% |
| 12.5% | 1.00 | 1.45 | 22.37% | 15.66% |
| 12.5% | 3.00 | 3.60 | 20.48% | 15.17% |
| 12.5% | 5.00 | 5.75 | 16.48% | 6.44% |
| 12.5% | 7.00 | 7.90 | 12.27% | 5.71% |
| 25% | 1.00 | 1.45 | 71.66% | 28.11% |
| 25% | 3.00 | 3.60 | 15.39% | 8.09% |
| 25% | 5.00 | 5.75 | 13.51% | 8.54% |
| 25% | 7.00 | 7.90 | 18.91% | 24.18% |
| 37.5% | 1.00 | 1.45 | 31.01% | 14.77% |
| 37.5% | 3.00 | 3.60 | 19.64% | 16.89% |
| 37.5% | 5.00 | 5.75 | 11.23% | 10.68% |
| 37.5% | 7.00 | 7.90 | 9.13% | 2.60% |
| 50% | 1.00 | 7.90 | 75.78% | 26.80% |
| 50% | 3.00 | 7.90 | 27.35% | 12.77% |
| 50% | 5.00 | 7.90 | 11.92% | 6.89% |
| 50% | 7.00 | 7.90 | 11.21% | 4.60% |
Accuracy (systematic error) can be found using
| (1) |
Before calculating precision, it is necessary to calculate the repeatability standard deviation,
| (2) |
With calculated, it is now possible to calculate the precision (random error)
| (3) |
7.1. Device characterization
Comparing the results of the BioCloneBot syringe pump in Table 2, Table 3, the BioCloneBot syringe pump produces comparable results to the OpenTrons-OT2 P20 Gen2 and P300 Gen2 pipette. In some cases, the BioCloneBot has better accuracy and precision than the OpenTrons as can be seen when comparing the results of the 250 L adapter with the P200 pipette tip to the OpenTrons P300 Gen2 pipettor. While the OpenTrons P20 Gen2 pipette out performs the BioCloneBot syringe pump with a 50 L syringe and P20 tip adaptor, the error is most likely induced by problem with the PLA material at such small dimensions. If alternative 3D printing materials or extra parts were added (such as a small O-ring) and the seal problem were resolved, the error could hypothetically decrease bringing the BioCloneBot closer to the OpenTrons-OT2 in performance.
Table 3.
OpenTrons-OT2 single-channel pipette specifications [33].
| Pipette | Volume | Accuracy D | Accuracy L | Precision CV | Precision L |
|---|---|---|---|---|---|
| P20 Gen2 | 1 | 15% | 0.15 L | 5% | 0.05 L |
| P20 Gen2 | 10 | 2% | 0.2 L | 1% | 0.1 L |
| P20 Gen2 | 20 | 1.5% | 0.3 L | 0.8% | 0.16 L |
| P300 Gen2 | 20 | 4% | 0.8 L | 2.5% | 0.5 L |
| P300 Gen2 | 150 | 1% | 1.5 L | 0.4% | 0.6 L |
| P300 Gen2 | 300 | 0.6% | 1.8 L | 0.3% | 0.9 L |
To validate the ability of the BioCloneBot to attach and remove tips, a simple test was performed. The setup for the test is as follows:
-
1.
Calibrate BioCloneBot for 20 L pipette tip box.
-
2.
Load tip box with 96 tips.
-
3.
In BioCloneBot software, add a Tip Box labware to labware slot 1.
-
4.
Add Home Device operation.
-
5.
Add Get Tip operation.
-
6.
Add Remove Tip operation.
-
7.
Repeat Get Tip and Remove Tip operations for half of the tip box.
-
8.
Add Home Device operation (to remove any hysteresis-induced error).
-
9.
Add Get Tip operation.
-
10.
Add Remove Tip operation.
-
11.
Repeat Get Tip and Remove Tip operations for remainder of the tip box.
-
12.
Run experiment.
After a duration of approximately 1.5 h of operation, the successful attachment and subsequent removal of all 96 pipette tips were observed. It should be noted that while this test does not guarantee a perfect seal for each tip, it effectively demonstrates the BioCloneBot’s reliability in attaching and detaching pipette tips. Furthermore, considering that the dimensions of the 96-tip box align with those of a standard 96-well plate, it can be inferred that the device possesses the necessary accuracy to effectively target a 96-well plate.
However, it is important to acknowledge that the speed of the BioCloneBot presents a drawback. In comparison, the OTTO liquid handler is capable of completing an entire 96-well plate qPCR reaction in less than an hour. Similarly, the OpenTrons system can perform a similar experiment involving the transfer of volumes between two 96-well plates, while changing tips between transfers, in just over 35 min.
By analyzing the footage of a performed experiment using VLC Media Player, rough velocities of the system were calculated. The -axis was found to have a velocity of 50.74 mm/s while traveling 202.95 mm, the -axis was found to have a velocity of 48.11 mm/s while traveling 250.55 mm, and the -axis was found to have a velocity of 8.23 mm/s while traveling 74.1 mm. These measurements were made by calculating the travel distance over a period of time according to the video. Frame-by-frame analysis was done to find exact seconds when the pump starts and stops moving for each axis.
From the calculations, the maximum travel distance for each axis was calculated:
X-Axis: 400 mm/50.74 mm/s = 8 s
Y-Axis: 400 mm/48.11 mm/s = 8 s
Z-Axis: 135 mm/8.23 mm/s = 16 s
The accuracy and precision of the x, y, and z stepper motors were measured using a digital dial indicator probe. The manual control form was used to make n = 10 movements at 1.00, 5.00, 10.00, and 20.00 mm for each axis. The results can be seen in Table 5.
Table 5.
Results of x, y, and z stepper motor tests. Movements of n=10 at 1.00, 5.00, 10.00, and 20.00 mm were selected. All measurements were made using a digital dial indicator probe accurate to 0.001 mm.
| Axis | Test distance (mm) | Accuracy (%) | Precision (%) |
|---|---|---|---|
| X-Axis | 1.00 | 0.39% | 0.20% |
| 5.00 | 0.24% | 0.10% | |
| 10.00 | 0.31% | 0.04% | |
| 20.00 | 0.42% | 0.06% | |
| Y-Axis | 1.00 | 1.50% | 1.40% |
| 5.00 | 0.07% | 0.16% | |
| 10.00 | 0.23% | 0.05% | |
| 20.00 | 0.61% | 0.07% | |
| Z-Axis | 1.00 | 3.98% | 1.40% |
| 5.00 | 0.97% | 0.88% | |
| 10.00 | 0.50% | 0.18% | |
| 20.00 | 0.60% | 0.15% | |
7.2. Molecular biology application examples
After characterizing the functionality of the syringe pump, the overall functionality of the BioCloneBot was tested. Restriction Enzyme Digest and Ligation, Golden Gate Assembly, and Gibson Assembly were all run and compared to a manual manipulation via a pipette.
7.2.1. Restriction enzyme digest
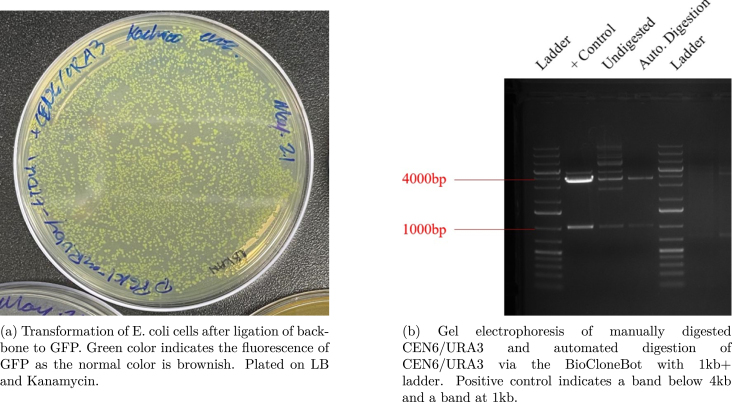
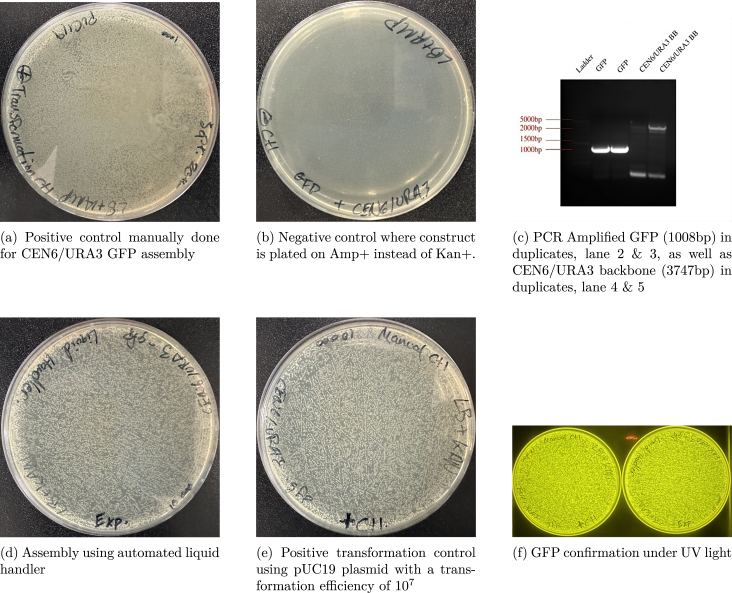
A restriction enzyme digest and ligation of a GFP-dropout insert was performed. Using a single plasmid containing GFP, a digestion was performed to remove the GFP flanking two BsaI cut sites. After digestion, the original GFP was religated back into the backbone. Using the tubestand labware, a total of three tubes were used. The pump picked up volumes ranging between 3 L to 11 L and placed them into a collection tube total 19 L. At the time of this test, only a 500 L syringe was available and as such, was used to perform the protocol. The P200 tip adapter was used with P200 pipette tips. In between each aspiration and dispensing step, the pipette tip was removed into the trash bin then replaced with a clean tip. Once each input was combined together, the resulting collection tube volume was moved to a PCR tube, then run through a thermocycler. The resulting DNA plasmid was transformed into E. coli cells and then plated. After plating, a mini-prep was run and the resulting DNA was run through a gel electrophoresis for final confirmation (see Fig. 9).
Fig. 9.
Restriction enzyme digest of CEN6/URA3 Kan+ plasmid with GFP dropout insert. Digestion with restriction enzyme BsaI results in a separation of the backbone at 3721 bp and the GFP insert at 1026 bp. The full plasmid length is 4747 bp.
7.2.2. Gibson assembly cloning
A Gibson Assembly was performed to insert a GFP insert into a linearized pCEN6/URA3 Kan+ backbone. The protocol was performed using three wells of a standard 96-well plate labware. The pump picked up volumes ranging between 1 L to 3 L and placed them into a collection well totaling 19.39 L. The 50 L syringe was used with the P20 tip adapter and P20 pipette tips. In between each aspiration and dispensing step, the pipette tip was removed into the trash bin then replaced with a clean tip. Once each input was combined together, the result was manually moved to a single PCR tube, then run through a thermocycler. The resulting DNA plasmid was transformed into E. coli cells and then plated. After plating, a mini-prep was run and the resulting DNA was run through a gel electrophoresis for initial confirmation (see Fig. 10). The final DNA was sent for sequencing and returned with 99.98% sequence match (the mismatches had no functional effect as they were outside the expression unit).
Fig. 10.
Gibson Assembly of CEN6/URA3 GFP dropout plasmid using automated liquid han- dler. Reaction was done under the Hifi Assembly protocol where each fragment, GFP and CEN6/URA3 Backbone were PCR amplified with specific 20 nucleotide overhangs.
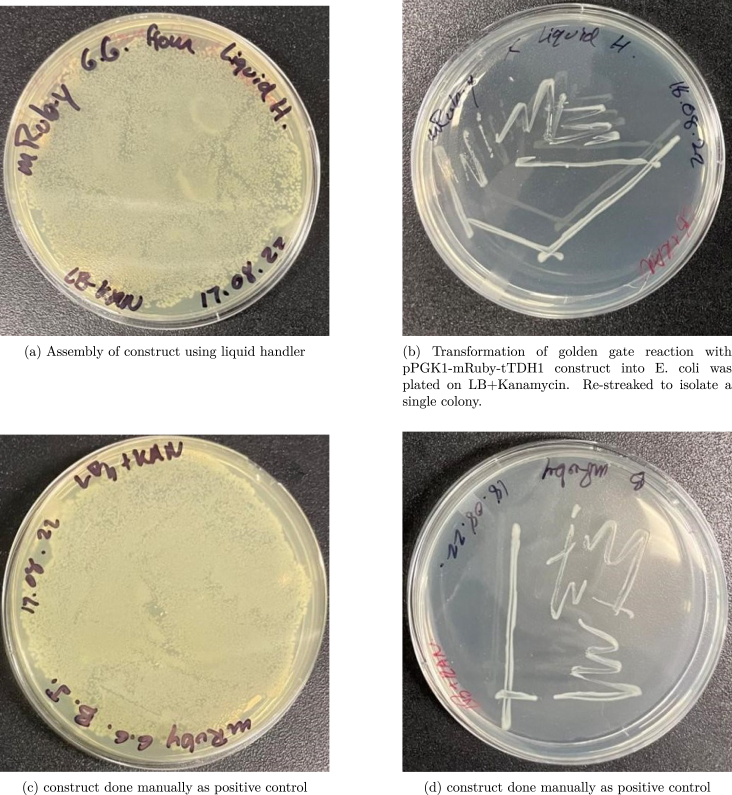
7.2.3. Golden gate cloning
A Golden Gate Cloning was performed to insert a promotor, mRuby gene, and terminator into a Kan+ plasmid for E. coli. Using the tubestand labware, the assembly consisted of four DNA parts. The 250 L syringe was used inside of the syringe pump. The pump aspirated then dispensed volumes between 3 L and 4 L into a final collection tube resulting in a total of 19 L. The 19 L tube was run through a thermocycler. The resulting DNA plasmid was transformed into E. coli cells then plated (see Fig. 11). After plating, a mini-prep was run and the resulting DNA was run through a gel electrophoresis for an initial confirmation (see Fig. 12). The final DNA was sent for sequencing and returned with 100% sequence match.
Fig. 11.
Golden Gate Cloning assembly of mRuby construct using the BioCloneBot. Diges- tion and ligation using the NEB Golden Gate assembly kit and plasmid: pYTK011 (pPGK1), pYTK034 (mRuby), pYTK056 (tTDH1) and CEN6/URA3-GFP dropout backbone.
Fig. 12.
Miniprep of isolated colonies from automated and manual assembly. The full length of assembly is 5733 bp. Digestion with NotI results in bands at 3614bp and 1763 bp.
8. Discussion and future work
Despite the initial high cost associated with acquiring commercial automation equipment, it is worth noting that viable alternatives in the form of low-cost open-source devices are within reach. For less than $2,000 CAD, an audacious scientist can purchase all the necessary components and tools to assemble the BioCloneBot. Once assembled, it is fully capable of performing a wide variety of liquid handling steps involving a combination of well plates, tubestands, and tip boxes. The device stands out not only for its affordability and adaptability but also for its potential to match, and at times surpass, the precision offered by competing commercial counterparts. Consequently, even research laboratories with constrained budgets can gain access to fully capable automation tools, thereby augmenting their experimental throughput and accelerating turnaround times. This enhanced performance imparts an accelerated pace to scientific advancements, characterized by heightened accuracy that often supersedes human execution capabilities.
Furthermore, the advent of open-source devices heralds a novel era of experiential learning for scientists and engineers alike. During the assembly and calibration process of the BioCloneBot, one can learn about topics including 3D printing, programming, and hardware assembly. Proficiency in these domains holds increasing significance within laboratory environments. Leveraging 3D printing technology can lead to cost-effective replication of proprietary labware, customized adaptations for various equipment, and even the restoration of components employed in commercial hardware. With a little bit a of programming exposure, biologists can start to delve into the world of bioinformatics allowing for data processing with higher throughput, the development of custom electronics, and the automation of time-consuming development processes such as DNA cloning [34]. With the acquisition of these skills through open-source hardware and software, laboratories can be empowered to engineer economical, in-house solutions tailored to their unique needs.
There are a few aspects of the BCB that merit further refinement. The imperfections in the 20 L tip adapter compromises the seal of pipette tips, leading to volume discrepancies. This concern could be alleviated by exploring alternative 3D printing materials or incorporating a small rubber o-ring. Addressing another issue, the use of dH2O in the Hamilton syringes, while practical during development, presents challenges due to its tendency to dry out over time. Employing a non-contaminating lubricant could mitigate this issue.
Although the software was conceived for user-friendliness, updating the current front-end with a guided calibration process integrated into the software interface would drastically enhance usability. This feature would streamline the calibration process, enabling end-users to configure the BioCloneBot without necessitating code modifications. Additionally, while the current labware adaptability extends to different dimensions, modifying labware specifications currently entails altering the source code. Enhancing the front-end to allow for the addition of new labware and the modification of existing labware properties would also improve usability.
It is possible to extend the functionality of the BCB by allowing for the inclusion of additional hardware such as a custom-made thermocycler. The current board does not allow for these expansions, which could be resolved by creating a custom-made PCB with both a higher performing processor as well as the hardware capable of driving thermocycler grade Peltier modules. Finally, the stepper motor function could be improved by implementing timing register based PWM control to drive the stepper motors. By using a simple bit-banging implementation, the current 3-axis system maintains high-accuracy over a long period of time at the cost of speed. With the PWM control, proper acceleration can be implemented allowing for substantially higher speed with high accuracy during a long experiment.
Lastly, improving stepper motor functionality can be achieved by implementing timing register based PWM control to govern stepper motor operation. Moving beyond the current bit-banging implementation, this upgrade would preserve precision during lengthy experiments while allowing for substantially increased speed through effective acceleration control.
9. Conclusion
In conclusion, we developed a highly accurate, low-cost liquid handler versatile enough to support up to four tip boxes, tube stands, and/or 96-well plates. The included syringe pump features accuracy that rivals commercial equivalents at volumes ranging from 1 L to 200 L. The front-end software is easy-to-use and supports the major liquid handling operations: getting a tip, removing a tip, aspirating a volume, dispensing a volume, and mixing a volume a specified number of times. With the creation of low-cost, versatile, and highly accurate automated liquid handlers such as the BioCloneBot, cost conscious labs can begin adopting cutting edge automation technology to improve the throughput of their workflows.
Funding
This work was supported by Concordia University GCS (Gina Cody School of Engineering and Computer Science), FRS (Faculty’s Research Support), and NSERC (CREATE) in Concordia Synthetic Biology Applications (SynBioApps). Hamilton, the company, provided us, free of charge, with some of the glass syringes used in the making of the BCB.
CRediT authorship contribution statement
Ke’Koa CDH Wells: Conceptualization, Methodology, Software, Validation. Nawwaf Kharma: Conceptualization, Methodology, Validation, Writing – review & editing. Brandon B. Jaunky: Validation. Kaiyu Nie: Software, Writing – review & editing. Gabriel Aguiar-Tawil: Validation. Daniel Berry: Conceptualization.
Declaration of Generative AI and AI-assisted technologies in the writing process
During the preparation of this work the authors used ChatGPT in order to improve clarity, flow, and proofread the manuscript and Canva to generate some of the images used in the visual abstract. After using these tools/services, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Biography

Ke’Koa Wells is a lead software developer for the Canada Excellence Research Chair in Smart, Sustainable, and Resilient Communities which is the engineering branch of the Next-Generations Cities Institute at Concordia University in Montréal, Québec. His work focuses on the creation software tools and services to aid researchers, public officials, and community stakeholders in the decision making process for the development of communities. He recently graduated from Concordia University with a Masters of Applied Sciences in Electrical and Computer Engineering as a SynBioApps fellow. His primary interests include software development, robotics, and Internet of Things devices.
References
- 1.Synthetic Biology . 2022. Genome.gov. https://www.genome.gov/about-genomics/policy-issues/Synthetic-Biology. (Accessed 03 July 2023) [Google Scholar]
- 2.Xie M., Haellman V., Fussenegger M. Synthetic biology — application-oriented cell engineering. Curr. Opin. Biotechnol. 2016;40:139–148. doi: 10.1016/j.copbio.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Yan X., Liu X., Zhao C., Chen G.-Q. Applications of synthetic biology in medical and pharmaceutical fields. Sig. Transduct. Target Ther. 2023;8(1):199. doi: 10.1038/s41392-023-01440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyagi A., Kumar A., Aparna S.V., Mallappa R.H., Grover S., Batish V.K. Synthetic biology: Applications in the food sector. Crit. Rev. Food Sci. Nutr. 2016;56(11):1777–1789. doi: 10.1080/10408398.2013.782534. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhary P., et al. Application of synthetic consortia for improvement of soil fertility. Pollut. Remediat. Agric. Product. Rev. Agron. 2023;13(3):643. doi: 10.3390/agronomy13030643. [DOI] [Google Scholar]
- 6.Pfleger B.F., Takors R. Recent progress in the synthesis of advanced biofuel and bioproducts. Curr. Opin. Biotechnol. 2023;80 doi: 10.1016/j.copbio.2023.102913. [DOI] [PubMed] [Google Scholar]
- 7.Kajla S., Kumari R., Das A., Patel V.K. Current advancements in biofuel production from microalgae: Harvesting and conversion technologies. Chem. Eng. Technol. 2023 doi: 10.1002/ceat.202300010. ceat.202300010. [DOI] [Google Scholar]
- 8.Wijnen B., Hunt E.J., Anzalone G.C., Pearce J.M. Open-source syringe pump library. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amarante L.M., Newport J., Mitchell M., Wilson J., Laubach M. An open source syringe pump controller for fluid delivery of multiple volumes. eNeuro. 2019;6(5) doi: 10.1523/ENEURO.0240-19.2019. ENEURO.0240-19.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gervasi A., Cardol P., Meyer P.E. Open-hardware wireless controller and 3D-printed pumps for efficient liquid manipulation. HardwareX. 2021;9 doi: 10.1016/j.ohx.2021.e00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behrens M.R., et al. Open-source, 3D-printed peristaltic pumps for small volume point-of-care liquid handling. Sci. Rep. 2020;10(1):1543. doi: 10.1038/s41598-020-58246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szymula K.P., Magaraci M.S., Patterson M., Clark A., Mannickarottu S.G., Chow B.Y. An open-source plate reader. Biochemistry. 2019;58(6):468–473. doi: 10.1021/acs.biochem.8b00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shih S.C.C., et al. A versatile microfluidic device for automating synthetic biology. ACS Synth. Biol. 2015;4(10):1151–1164. doi: 10.1021/acssynbio.5b00062. [DOI] [PubMed] [Google Scholar]
- 14.Gach P.C., Iwai K., Kim P.W., Hillson N.J., Singh A.K. Droplet microfluidics for synthetic biology. Lab Chip. 2017;17(20):3388–3400. doi: 10.1039/C7LC00576H. [DOI] [PubMed] [Google Scholar]
- 15.Schuster B., et al. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nature Commun. 2020;11(1):5271. doi: 10.1038/s41467-020-19058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goyal G., Elsbree N., Fero M., Hillson N.J., Linshiz G. Repurposing a microfluidic formulation device for automated DNA construction. PLoS One. 2020;15(11) doi: 10.1371/journal.pone.0242157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dettinger P., et al. An automated microfluidic system for efficient capture of rare cells and rapid flow-free stimulation. Lab Chip. 2020;20(22):4246–4254. doi: 10.1039/D0LC00687D. [DOI] [PubMed] [Google Scholar]
- 18.Diep J., et al. Microfluidic chip-based single-cell cloning to accelerate biologic production timelines. Biotechnol. Progress. 2021 doi: 10.1002/btpr.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engler C., Kandzia R., Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3(11) doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson D.G. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Res. 2009;37(20):6984–6990. doi: 10.1093/nar/gkp687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Dolleweerd C.J., et al. MIDAS: A modular DNA assembly system for synthetic biology. ACS Synth. Biol. 2018;7(4):1018–1029. doi: 10.1021/acssynbio.7b00363. [DOI] [PubMed] [Google Scholar]
- 22.2023. OT-2 Robot - Opentrons. https://shop.opentrons.com/ot-2-robot/_gl=1*op8qmp*_ga*MTMxNDY5MDcxOC4xNjg2MTk3Mjk5*_ga_66HK7MC5D7*MTY4ODk1NTY5NC4yLjEuMTY4ODk1NTczNi4xOC4wLjA.*_ga_GNSMNLW4RY*MTY4ODk1NTY5NC4yLjEuMTY4ODk1NTczNi4xOC4wLjA. (Accessed 09 July 2023) [Google Scholar]
- 23.Zhang C., Wijnen B., Pearce J.M. Open-source 3-D platform for low-cost scientific instrument ecosystem. SLAS Technol. 2016;21(4):517–525. doi: 10.1177/2211068215624406. [DOI] [PubMed] [Google Scholar]
- 24.Faa A., Nejatimoharrami F., Stoy K., Theodosiou P., Taylor B., Ieropoulos I. Proceedings of the Artificial Life Conference 2016. MIT Press; Cancun, Mexico: 2016. EvoBot: An open-source, modular liquid handling robot for nurturing microbial fuel cells; pp. 626–633. [DOI] [Google Scholar]
- 25.Nejatimoharrami F., Faina A., Stoy K. New capabilities of EvoBot: A modular, open-source liquid-handling robot. SLAS Technol. 2017;22(5):500–506. doi: 10.1177/2472630316689285. [DOI] [PubMed] [Google Scholar]
- 26.2020. OTTO: The Open-Source Automatic Liquid Handler. [Online Video]. Available: https://www.youtube.com/watch?v=35Zv_xLzYRs. (Accessed 29 November 2022) [Google Scholar]
- 27.Faiña A., Nejati B., Stoy K. EvoBot: An open-source, modular, liquid handling robot for scientific experiments. Appl. Sci. 2020;10(3):814. doi: 10.3390/app10030814. [DOI] [Google Scholar]
- 28.Storch M., Haines M.C., Baldwin G.S. DNA-BOT: a low-cost, automated DNA assembly platform for synthetic biology. Synth. Biol. 2020;5(1) doi: 10.1093/synbio/ysaa010. ysaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barthels F., Barthels U., Schwickert M., Schirmeister T. FINDUS: An open-source 3D printable liquid-handling workstation for laboratory automation in life sciences. SLAS Technol. 2020;25(2):190–199. doi: 10.1177/2472630319877374. [DOI] [PubMed] [Google Scholar]
- 30.2022. Hamilton syringes - 700 series Hamilton company. https://www.hamiltoncompany.com/laboratory-products/syringes/general-syringes/microliter-syringes/700-series. (Accessed 29 November 2022) [Google Scholar]
- 31.Thingiverse.com . 2023. OpenTrons tube rack by thehair. https://www.thingiverse.com/thing:3405002. (Accessed 07 August 2023) [Google Scholar]
- 32.jbrazio . 2022. What is Marlin? Marlin Firmware. https://marlinfw.org/docs/basics/introduction.html. (Accessed 19 October 2022) [Google Scholar]
- 33.2022. OT-2 Pipettes by Opentrons New Pipettes for Lab Automation. https://opentrons.com/products/pipettes/#tech-specs. (Accessed 03 October 2022) [Google Scholar]
- 34.Hillson N.J., Rosengarten R.D., Keasling J.D. j5 DNA assembly design automation software. ACS Synth. Biol. 2012;1(1):14–21. doi: 10.1021/sb2000116. [DOI] [PubMed] [Google Scholar]