Abstract
The gypsy moth (Lymantria dispar) is nonpermissive for Autographa californica nucleopolyhedrovirus (AcNPV) infection. We previously isolated a gene, host range factor 1 (hrf-1), from L. dispar nucleopolyhedrovirus that promotes AcNPV replication in Ld652Y cells, a nonpermissive L. dispar cell line (S. M. Thiem, X. Du, M. E. Quentin, and M. M. Berner, J. Virol. 70:2221–2229, 1996). In the present study, we investigated the ability of hrf-1 to alter the larval host range of AcNPV. Bioassays using recombinant AcNPV bearing hrf-1 were conducted with insect larvae by use of oral infection. AcNPV bearing hrf-1 was infectious for neonate L. dispar larvae, with a 50% lethal concentration of 1.2 × 105 polyhedral inclusion bodies/ml of diet, which is similar to that of wild-type AcNPV for permissive hosts. AcNPV can kill neonate L. dispar larvae at high doses, but it does not kill third-instar larvae. However, electron microscopy studies of AcNPV-inoculated third-instar larvae revealed virus replication in the midgut cells. PCR analyses indicated that the virus was AcNPV. These results suggest that the block for AcNPV infection of L. dispar larvae is its inability to spread systematically from primary infection sites in the midgut epithelium and that this barrier is leaky in neonates. hrf-1 allows AcNPV to overcome this barrier. AcNPV recombinants bearing hrf-1 were also significantly more infectious for Helicoverpa zea, a resistant species, suggesting that the blocks for AcNPV infection of L. dispar and H. zea larvae may be similar.
Baculoviruses, nucleopolyhedroviruses (NPV) and granuloviruses (GV), are large double-stranded DNA viruses that infect invertebrates, primarily insects. Because they do not infect vertebrates or plants, baculoviruses are considered excellent prospects for development as highly specific insecticides and are registered for use on both agricultural and forest pests. One problem in developing baculovirus insecticides is that the host range of most baculoviruses is restricted to a single or a few related species. While limited host specificity is advantageous from a safety perspective, it is problematic economically since different baculoviruses will be required to control different pest insects. To modify the baculovirus host range to target specific pest species and to ensure that the host specificity of genetically engineered viruses for nontarget species is not altered, it is necessary to understand the genetic and molecular bases of host specificity.
Virus host range is a function of the ability of a virus to enter, replicate, and produce infectious progeny in a host organism. The successful completion of all these stages results in a productive infection. NPV produce two morphological forms that play different roles in the infection cycle. Occluded viruses (OV) are embedded in a crystalline protein matrix, called polyhedra or polyhedral inclusion bodies (PIBs), that stabilizes virions in the environment and mediates virus spread between insects. Budded viruses (BV) lack an external protein matrix, enter cells by absorptive endocytosis (33), and spread the virus within the insect host. Primary infection is initiated in midgut epithelial cells when PIBs are ingested by a susceptible insect. The occlusion matrix dissolves in the alkaline midgut lumen, releasing enveloped virions which enter midgut epithelial cells by membrane fusion (12, 16). Following virus replication in the midgut epithelial cells, BV spreads the virus to other tissues (9, 19).
The ability of viruses to efficiently spread from primary infection sites and to replicate in diverse tissues is essential to initiate a systemic infection. Electron microscopy studies coupled with titration of virus in insect hemolymph indicated that Autographa californica NPV (AcNPV) nucleocapsids budded through the midgut basal lamina into the hemocoel of Trichoplusia ni larvae within 0.5 h of ingestion (12). Studies employing immunocytochemistry in T. ni (19) or infection of Spodoptera exigua larvae with AcNPV carrying reporter genes under a different temporal control (9) showed that columnar and regenerative cells were the sites of primary infection and that secondary infections outside of the midgut were initiated only after virus replication in midgut cells. Studies on the permeability of the midgut basal lamina suggest that it may present a formidable barrier to virus passage (30). In T. ni larvae, AcNPV circumvents this barrier by spreading from the midgut via the tracheal system, which spans the basal lamina (7). Virus replication may also be constrained in a cell- and tissue-specific manner within the insect. Although reporter genes controlled by early baculovirus promoters were expressed in most tissues in AcNPV-infected S. exigua larvae, reporter gene activity from very late promoters was not observed in midgut goblet cells, salivary glands, or Malpighian tubules, indicating an aborted infection (21).
Although AcNPV has a relatively broad host range compared to that of most baculoviruses (13), it does not infect the gypsy moth (Lymantria dispar). A few cell lines derived from L. dispar, such as the embryonic cell lines LdEG and LdEI, support AcNPV replication (23). Others, such as LdFB (23) and Ld652Y (11), derived from the L. dispar larval fat body and pupal ovary, respectively, are nonpermissive for AcNPV infection, suggesting that AcNPV replication in L. dispar may be restricted in a tissue-specific manner. In AcNPV-infected Ld652Y cells, a cytopathic effect is observed and no progeny viruses are produced (24). In these cells, global arrest of protein synthesis is observed by approximately 16 h postinfection (p.i.) (15), yet viral DNA is replicated and mRNA from early, late, and very late genes is transcribed (15, 26, 27). We previously identified a gene from L. dispar NPV (LdNPV), L. dispar host range factor 1 (hrf-1), that relieves the block for protein synthesis and promotes AcNPV replication in Ld652Y cells (31). Recombinant AcNPV bearing hrf-1 replicate normally in Ld652Y cells and produce high titers of BV and polyhedra in 100% of the cells (5). In this study, our objective was to determine if hrf-1 expanded the host range of AcNPV for larval insects by oral infection.
To determine if hrf-1 would enable AcNPV to infect L. dispar larvae by the normal oral route, bioassays were performed on L. dispar neonates by using LdNPV, AcNPV, or vAcLdPD, a recombinant virus bearing hrf-1 (5). LdNPV was propagated in L. dispar larvae hatched from eggs obtained from the USDA-APHIS Otis Methods Development Laboratory, Otis, Mass., and reared on a high-wheat-germ diet (28). AcNPV and vAcLdPD were propagated in T. ni larvae hatched from eggs obtained from the Forestry Canada Forest Pest Management Institute, Sault Ste. Marie, Ontario, Canada, and reared on an alfalfa and pinto bean diet (32). PIBs were isolated from infected larvae, suspended in sterile distilled water, and counted by using a Neubauer hemocytometer and phase-contrast microscopy. PIBs were incorporated into the diet (28) at concentrations ranging from 2 × 104 to 2 × 108 PIBs/ml of diet for AcNPV or vAcLdPD and 2 × 102 to 2 × 106 PIBs/ml of diet for LdNPV. L. dispar neonates (60 larvae/virus concentration) were allowed to feed on PIB-containing diet for 48 h, transferred to fresh diet cups, and observed daily for mortality. Experiments were terminated when all larvae infected with the highest dose of virus had died (15 days p.i. for AcNPV and vAcLdPD; 5 days p.i. for LdNPV). Data from two independent experiments were pooled. The concentrations of virus required to kill 50% of the larvae (LC50s) were determined by probit analysis (8) by using POLO-PC (LeOra Software, Berkeley, Calif.) (Table 1). Incorporation of highly concentrated AcNPV into the diet was required to observe L. dispar mortality. Because only 57% mortality (death of 34 of 60 larvae) was achieved with the highest dose (2 × 108 PIBs/ml), the LC50 for AcNPV-infected larvae could not be determined with greater than 90% confidence. The LC50 of vLdAcPD for L. dispar larvae was similar to that of AcNPV for the permissive hosts Spodoptera frugiperda (1.0 × 106 PIBs/ml of diet) and T. ni (1.8 × 105 PIBs/ml of diet) (22) and was significantly (1,800-fold) more infectious than AcNPV for L. dispar larvae (Table 1). However, vAcLdPD was significantly less infectious than LdNPV (Table 1) and killed L. dispar larvae much more slowly, requiring 15 days versus 5 days to kill all larvae at the highest dose of virus (data not shown). These results demonstrate that although hrf-1 expands the AcNPV host range to include L. dispar larvae, AcNPV bearing hrf-1 is less infectious and less virulent than LdNPV.
TABLE 1.
Dose response of L. dispar neonates infected per os with AcNPV, vAcLdPD, or LdNPV
| Virus | LC50 (PIB/ml)a | Fiducial limits (PIB/ml)b
|
|
|---|---|---|---|
| Upper | Lower | ||
| vAcLdPD | 1.2 × 105 a | 2.7 × 105 | 3.6 × 104 |
| AcNPV | 2.2 × 108 b | 9.2 × 108 | 1.4 × 108 |
| LdNPV | 1.1 × 104 c | 3.1 × 104 | 3.5 × 103 |
Significantly different LC50s are indicated by different letters.
Ninety percent confidence interval for AcNPV; 95% confidence interval for vAcLdPD and LdNPV.
Neonates inoculated with vAcLdPD demonstrated typical symptoms of NPV infection such as larval melting and the presence of abundant polyhedra in infected cells and hemolymph. To confirm that mortality was due to vAcLdPD infection, DNA was isolated from polyhedra collected from vAcLdPD-infected larvae and compared with DNA isolated from AcNPV, LdNPV, and vAcLdPD by restriction analysis. DNAs isolated from larvae and vAcLdPD had identical PstI restriction patterns, including a diagnostic 3-kbp PstI fragment for vAcLdPD that is not present in AcNPV (data not shown). In contrast, dying larvae infected with AcNPV did not liquefy but had a shriveled appearance, and few polyhedra were observed upon microscopic examination of the dead larvae. Because of the atypical symptoms, we were concerned that the death observed for AcNPV-infected larvae may not have been due to AcNPV infection. One possibility is that feeding large doses of AcNPV may have induced a latent virus. A latent NPV in a laboratory colony of Mamestra brassicae was activated when larvae were challenged with other viruses (18).
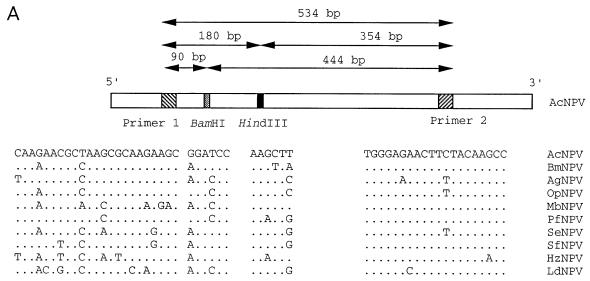
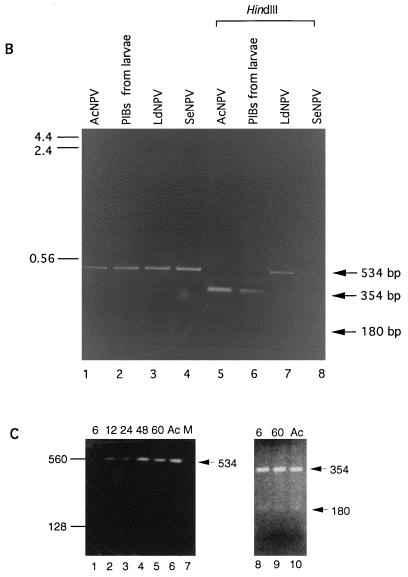
To address this possibility, we used a PCR approach. DNA was prepared from PIBs isolated from 30 pooled carcasses of AcNPV-infected L. dispar neonates by using standard protocols (29), and 1/30 of this pool was used as a template for PCR amplification. VentR(exo−) DNA polymerase (New England BioLab, Beverly, Mass.) was used with the reaction buffer provided by the manufacturer. Thirty cycles of amplification were performed by using the following cycling parameters: denaturing at 95°C for 30 s, annealing at 55°C for 1 min, and extending at 72°C for 45 s. Because the polyhedrin (polh) gene is highly conserved and its sequence is known for a large number of NPV, we designed primer pairs that would be able to amplify polh sequences from divergent NPV. The primer sequences were homologous to the AcNPV sequence and corresponded to conserved regions from known polh sequences (Fig. 1A). The primers were used successfully to amplify polh sequences from AcNPV, S. exigua NPV, and LdNPV, which is the most divergent of the known polh sequences (3, 37) (Fig. 1B). A 0.53-kbp DNA was amplified from AcNPV DNA (Fig. 1B, lane 1) and from DNA isolated from purified polyhedra from AcNPV-infected L. dispar larvae (Fig. 1B, lane 2). All amplified products could be digested with HindIII, resulting in two fragments with sizes of 354 and 180 bp (Fig. 1B, lane 5 and 6), and with BamHI, resulting in 90- and 444-bp fragments (data not shown) which are diagnostic for AcNPV (Fig. 1A). In contrast, the amplified products from LdNPV and S. exigua NPV DNA were not digested (Fig. 1B, lanes 7 and 8). To rule out the possibility of vAcLdPD cross-contamination, a primer pair specific to vAcLdPD (5) was used for PCR analysis of the DNA samples. No products were amplified from DNA isolated from AcNPV or from PIBs collected from infected larvae (data not shown).
FIG. 1.
(A) Sequence comparisons of various baculovirus polyhedrin gene sequences in the region used for PCR primers and restriction analysis. Abbreviations: Bm, Bombyx mori; Ag, Anticarsia gemmatalis; Op, Orgyia pseudotsugata; Mb, M. brassicae; Pf, Panolis flammea; Se, S. exigua; Sf, S. frugiperda; Hz, H. zea. Primer pairs F (5′-AGAACGCTAAGCGCAAGAAGCA-3′) and R (5′-GGCTTGTAGAAGTTCTCCCA-3′) are based on the AcNPV polyhedrin sequence and map to nucleotides 4603 to 4625 and 5136 to 5117 of the AcNPV genome, respectively (1). Locations of diagnostic HindIII and BamHI sites and sizes of predicted PCR product and restriction fragments are indicated. (For the polh sequence, see references 3 and 37). (B) PCR products amplified by using primer pairs specific to the AcNPV polyhedrin gene. AcNPV DNA (lane 1), DNA isolated from OV from moribund AcNPV-inoculated L. dispar larvae (lane 2), LdNPV DNA (lane 3), and S. exigua NPV DNA (lane 4) were amplified. The amplified products from lanes 1 to 4 were digested with HindIII (lanes 5 to 8, respectively). Predicted amplification and digestion products are indicated by arrows to the right of the panel. The size markers on the left are given in kilobase pairs. (C) PCR products amplified by using the AcNPV polh primer pair from DNA isolated from third-instar L. dispar midgut tissues or AcNPV DNA as the template. AcNPV-infected larvae at 6, 12, 24, 48, and 60 h p.i. (lanes 1 to 5, respectively), AcNPV DNA (lane 6), or mock-infected larvae (lane 7) were used. Amplified products from 6 and 60 h p.i. were digested with HindIII (lanes 8 and 9, respectively) and compared to the HindIII-digested amplification product from AcNPV DNA (lane 10). The predicted 354- and 180-bp restriction fragments are indicated with arrows. Size markers on the left are given in base pairs.
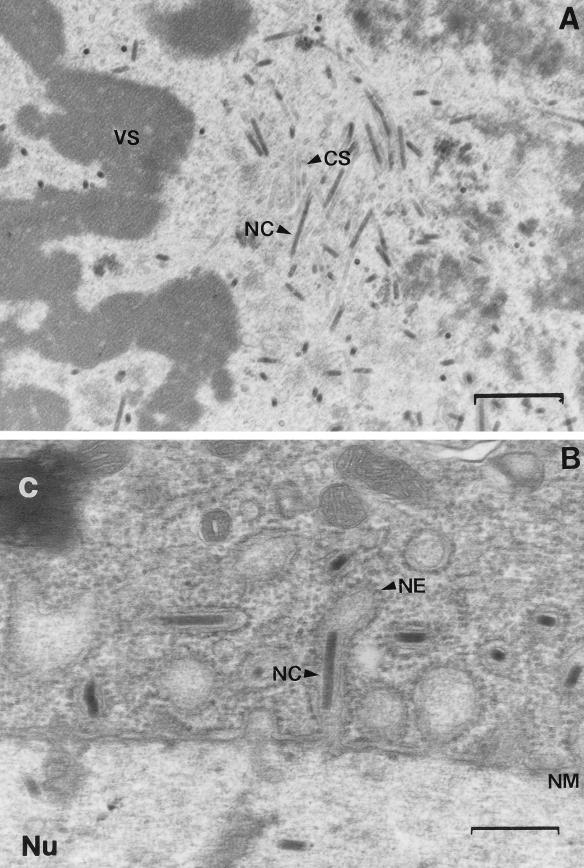
Second- and third-instar L. dispar larvae were susceptible to vAcLdPD but not to AcNPV infection. To determine if AcNPV entered or replicated in the midgut epithelium, midgut tissue from AcNPV-inoculated third-instar L. dispar was examined by transmission electron microscopy. Third-instar L. dispar larvae were fed AcNPV (1.15 × 106 PIBs/larva) on diet plugs and maintained on uncontaminated diet at 25°C until examination. At different times postinoculation, midguts were harvested, fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (0.1 M Na2HPO4 · 7H2O, 0.1 M KH2O4 [pH 7.2]), and prepared for microscopy. Tissues were postfixed in 1% osmium tetroxide (in 0.1 M phosphate buffer), dehydrated, and embedded in Poly/Bed 812 (51.13 g of Poly/Bed 812, 27.02 g of dodecenylsuccinic anhydride, 21.85 g of nadic methyl anhydride; Polysciences, Warrington, Pa.) containing 2% DMP30 (Polysciences). Ultrathin sections were prepared, mounted on copper grids, stained for 30 min with saturated aqueous uranyl acetate and 5 min with lead citrate, and then examined with a transmission electron microscope (model JEOL 100CX II TEMSCAN). Replicating virus was observed in the midgut epithelial cells of AcNPV-infected L. dispar. Virogenic stroma surrounded by nucleocapsids and empty capsid sheaths were observed in cell nuclei (Fig. 2A), and nucleocapsids were seen budding through the nuclear membrane into the cytoplasm (Fig. 2B).
FIG. 2.
Electron micrographs of AcNPV-infected third-instar L. dispar larval midgut cells at 48 h p.i. (A) Nucleocapsids assembling around virogenic stroma in cell nucleus. Magnification, ×19,000. Bar, 0.76 μm. (B) A nucleocapsid budding through the nuclear membrane. Magnification, ×36,000. Bar, 0.4 μm. Abbreviations: C, cytoplasm; CS, capsid sheath; NE, nuclear envelope; NC, nucleocapsid; NM, nuclear membrane; Nu, nucleus; VS, virogenic stroma.
To verify the presence of AcNPV, we employed PCR analysis with the polh primers previously described. DNA was prepared (25) from midgut tissues collected at 6, 12, 24, 48, and 60 h p.i. from AcNPV-inoculated third-instar L. dispar larvae. To minimize AcNPV contamination from the gut lumen, peritrophic membranes containing gut contents were carefully removed and the tissues were washed extensively in phosphate-buffered saline prior to DNA isolation. DNA equivalent to that of 0.5 midgut was used as a template for PCR amplification with the cycling parameters previously described. One-tenth of the first PCR-amplified products were subjected to secondary PCR amplification. A 534-bp amplification product was obtained by using DNA isolated from AcNPV-infected midgut tissues (Fig. 1C, lanes 1 to 5) or AcNPV DNA (Fig. 1C, lane 6). No products were amplified from DNA isolated from mock-infected larval midgut tissue (Fig. 1C, lane 7). Fragments with sizes of 354 and 180 bp were observed following HindIII digestion of PCR products from AcNPV-infected midguts, confirming the presence of AcNPV (Fig. 1C, lanes 8 and 9). AcNPV DNA was detected in L. dispar midguts from 6 to 60 h p.i. DNA amplification at 6 h p.i. could be from AcNPV that had entered midgut cells or from PIBs in the inoculum. Although our PCR results are not quantitative, the persistence of AcNPV genomic DNA in midguts from 6 to 60 h p.i. along with electron micrographs showing replicating virus indicated that L. dispar midgut tissues were permissive for AcNPV replication. These data do not support the hypothesis that a latent virus was activated, although they cannot rule out the presence of a baculovirus with a more divergent or missing polh sequence. A possible explanation for neonate susceptibility at high AcNPV dosage is that midgut epithelial cells are infected to such a great extent in neonates that the larvae cannot feed effectively and die from starvation. Older larvae which are larger and have more extensive energy reserves are likely to have relatively fewer infected cells or be better able to survive until infected midgut cells can be replaced with healthy cells. The shriveled appearance, lack of larval melting, and low number of PIBs observed for neonate L. dispar larvae that died following infection with high doses of AcNPV are consistent with this hypothesis.
The resistance of older L. dispar instars to AcNPV infection could be explained as developmental resistance, a term used to describe a well-known but poorly understood phenomenon whereby insect larvae become increasingly resistant to baculovirus infection as they proceed through development. With older larvae, we observed virus replication in the midgut epithelium but saw no evidence of systemic spread, suggesting that a physical or physiological barrier prevented virus infection from spreading beyond the midgut. In support of this hypothesis, gene activity was observed when L. dispar larvae were injected with AcNPV bearing reporter genes but not when the virus was administered orally as PIBs (17). The midgut basal lamina may be a more effective barrier in older instars than in neonates and thus may have been occasionally breached when the neonates were exposed to high doses of AcNPV. If a physiological mechanism is responsible, neonates may not be fully competent. Alternatively, virus spread might be prevented simply by the inability of AcNPV to replicate efficiently in some tissues. Our previous studies with cell culture demonstrated that AcNPV replication is blocked in Ld652Y cells as a result of global protein synthesis arrest, which is precluded by hrf-1 (5, 6, 31). For example, if AcNPV traversed the midgut basal lamina via the tracheal system in L. dispar and protein synthesis was arrested in tracheal cells, systemic spread would be curtailed.
To determine if hrf-1 increased AcNPV infectivity for other species, bioassays were conducted on two resistant species, Helicoverpa zea and Plutella xylostella, and one susceptible species, S. exigua. AcNPV, vAcLdPD, and vAcLdPS, an AcNPV recombinant containing hrf-1 and fusolin (2, 5, 31), were assayed on second-instar larvae. Larvae were allowed to feed on insect diet that was surface contaminated with different amounts of PIBs. PIBs were serially diluted in 0.01% sodium dodecyl sulfate, and virus suspensions (0.4 ml) were applied to a 16-cm2 area of Stoneville artificial diet (20) (Southland Products Incorporated, Lake Village, Ark.). Doses ranging from 105 to 107 PIBs/ml were used for bioassays of H. zea, and doses of 102 to 104 PIBs/ml were used for S. exigua and P. xylostella. Larvae were allowed to feed on the contaminated diet for the duration of the test. Data from three replicates, each containing 32 larvae per treatment, were collected, pooled, and analyzed by probit analysis (8) (Table 2). The infectivities of vAcLdPD and vAcLdPS were significantly greater than that of AcNPV for H. zea, 5.8- and 10-fold, respectively. No significant differences in infectivity were found for P. xylostella or S. exigua (Table 2). These data indicate that the ability of hrf-1 to increase AcNPV infectivity for different species is limited. The ability of hrf-1 to increase AcNPV infectivity for H. zea suggests that the mechanisms that preclude efficient AcNPV replication in L. dispar and H. zea may be similar. It is possible that H. zea NPV carries a hrf-1 homolog or a gene with a similar function. However, we have not been able to identify hrf-1 homologs in H. zea by Southern hybridization (data not shown). The increase in AcNPV infectivity for H. zea afforded by hrf-1 was small compared to that for L. dispar, given that the reductions in LC50 were 5.8- and 1,800-fold, respectively, compared to that of wild-type AcNPV. It is likely that there are additional mechanisms that limit AcNPV infection in H. zea that were not overcome by hrf-1. A recent study suggested that resistance to AcNPV infection in H. zea was due to an insect immune response in which infected cells were encapsulated by hemocytes (34).
TABLE 2.
Dose response of P. xylostella, S. exigua, and H. zea second-instar larvae infected per os with AcNPV, vAcLdPD, or vAcLdPS
| Species | Virus | LC50 (PIB/ml of diet)a | 95% Fiducial limits (PIB/ml of diet)
|
|
|---|---|---|---|---|
| Upper | Lower | |||
| H. zea | AcNPV | 3.17 × 106 a | 7.92 × 106 | 1.73 × 106 |
| vAcLdPD | 5.49 × 105 b | 1.04 × 106 | 3.07 × 105 | |
| vAcLdPS | 3.29 × 105 b | 6.05 × 105 | 1.72 × 105 | |
| S. exigua | AcNPV | 5.46 × 102 a | 7.27 × 102 | 4.08 × 102 |
| vAcLdPD | 4.51 × 102 a | 5.77 × 102 | 3.45 × 102 | |
| vAcLdPS | 1.01 × 103 a | 1.51 × 103 | 0.70 × 103 | |
| P. xylostella | AcNPV | 5.67 × 105 a | 3.09 × 106 | 1.09 × 105 |
| vAcLdPD | 7.16 × 105 a | 1.09 × 106 | 4.87 × 105 | |
| vAcLdPS | 1.41 × 106 a | 2.62 × 106 | 0.88 × 106 | |
Significantly different LC50s within each species are indicated by different letters.
hrf-1 alone and expressed at low levels from its own promoter was sufficient to increase AcNPV infectivity for H. zea. No significant differences in infectivity were observed between vAcLdPD and vAcLdPS (Table 2). vAcLdPS differs from vAcLdPD in that hrf-1 transcription is controlled by a strong synthetic late promoter and it bears a second LdNPV gene, fusolin or gp37 (2, 5, 31), a gene with unknown function that is found in baculoviruses (2, 14, 35) and entomopoxviruses (4, 10, 36). These results demonstrated that neither overexpression of hrf-1 nor the presence of fusolin contributes to the increased infectivity of AcNPV for H. zea larvae.
To further investigate the progression of AcNPV infection and study the function of hrf-1 in larvae, more direct and sophisticated approaches are needed. To study pathogenesis, we plan to use viruses bearing reporter genes to monitor the infection course of wild-type AcNPV carrying a reporter gene and AcNPV carrying both hrf-1 and a reporter gene (7, 9, 34). Studies are currently in progress to determine how hrf-1 counteracts protein synthesis arrest in Ld652Y cells, which should provide clues to understanding its function in vivo. Although hrf-1 enabled AcNPV to infect L. dispar larvae, recombinants were less virulent than LdNPV, suggesting that AcNPV requires additional virulence factors to effectively infect L. dispar larvae. To employ hrf-1 for generating NPV with improved insecticidal properties, it will be important to identify these factors.
Acknowledgments
C.-J. Chen and M. E. Quentin contributed equally to this work.
We thank Bob McCron at Forest Pest Management Institute, Forestry Canada, for providing T. ni eggs and Gary Bernon at Otis Methods Development Laboratory, USDA-APHIS, for providing L. dispar eggs.
This work was supported by USDA NRI grant 93-37302-9573 and Public Health Service grant GM48608 from the National Institute of General Medical Sciences to S.M.T.
REFERENCES
- 1.Ayres M D, Howard S C, Kuzio J, Lopezferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 2.Cassar, S. S., and S. M. Thiem. Unpublished data.
- 3.Cowan P, Bulach D, Goodge K, Robertson A, Tribe D E. Nucleotide sequence of the polyhedrin gene region of Helicoverpa zea single nucleocapsid nuclear polyhedrosis virus: placement of the virus in lepidopteran nuclear polyhedrosis virus group II. J Gen Virol. 1994;75:3211–3218. doi: 10.1099/0022-1317-75-11-3211. [DOI] [PubMed] [Google Scholar]
- 4.Dall D, Sriskantha A, Vera A, Lai-Fook J, Symonds T. A gene encoding a highly expressed spindle body protein of Heliothis armigera entomopoxvirus. J Gen Virol. 1993;74:1811–1818. doi: 10.1099/0022-1317-74-9-1811. [DOI] [PubMed] [Google Scholar]
- 5.Du X, Thiem S M. Characterization of host range factor 1 (hrf-1) expression in Lymantria dispar M nucleopolyhedrovirus- and recombinant Autographa californica M nucleopolyhedrovirus-infected IPLB-Ld652Y cells. Virology. 1997;227:420–430. doi: 10.1006/viro.1996.8356. [DOI] [PubMed] [Google Scholar]
- 6.Du X, Thiem S M. Responses of insect cells to baculovirus infection: protein synthesis shutdown and apoptosis. J Virol. 1997;71:7866–7872. doi: 10.1128/jvi.71.10.7866-7872.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelhard E K, Kammorgan L N W, Washburn J O, Volkman L E. The insect tracheal system—a conduit for the systemic spread of Autographa californica M nuclear polyhedrosis virus. Proc Natl Acad Sci USA. 1994;91:3224–3227. doi: 10.1073/pnas.91.8.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finney D J. Probit analysis, a statistical treatment of the sigmoid response curve. 2nd ed. Cambridge, United Kingdom: Cambridge University Press; 1952. [Google Scholar]
- 9.Flipsen J T M, Martens J W M, van Oers M M, Vlak J M, van Lent J W M. Passage of Autographa californica nuclear polyhedrosis virus through the midgut epithelium of Spodoptera exigua larvae. Virology. 1995;208:328–335. doi: 10.1006/viro.1995.1156. [DOI] [PubMed] [Google Scholar]
- 10.Gauthier L, Cousserans F, Veyrunes J C, Bergoin M. The Melolontha melolontha entomopoxvirus (MmEPV) fusolin is related to the fusolins of lepidopteran EPVs and to the 37K baculovirus glycoprotein. Virology. 1995;208:427–436. doi: 10.1006/viro.1995.1173. [DOI] [PubMed] [Google Scholar]
- 11.Goodwin R H, Tompkins G J, McCawley P. Gypsy moth cell lines divergent in viral susceptibility. In Vitro. 1978;14:485–494. doi: 10.1007/BF02616088. [DOI] [PubMed] [Google Scholar]
- 12.Granados R R, Lawler K A. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology. 1981;108:297–308. doi: 10.1016/0042-6822(81)90438-4. [DOI] [PubMed] [Google Scholar]
- 13.Gröner A. Specificity and safety of baculoviruses. In: Granados R R, Federici B A, editors. The biology of baculoviruses. 1. Biological properties and molecular biology. Boca Raton, Fla: CRC Press, Inc.; 1986. pp. 177–202. [Google Scholar]
- 14.Gross C H, Wolgamot G M, Russell R L Q, Pearson M N, Rohrmann G F. A 37-kilodalton glycoprotein from a baculovirus of Orgyia pseudotsugata is localized to cytoplasmic inclusion bodies. J Virol. 1993;67:469–475. doi: 10.1128/jvi.67.1.469-475.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzo D, Rathburn H, Guthrie K, Dougherty E. Viral and host cellular transcription in Autographa californica nuclear polyhedrosis virus-infected gypsy moth cell lines. J Virol. 1992;66:2966–2972. doi: 10.1128/jvi.66.5.2966-2972.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton H M, Burand J P. Saturable attachment sites for polyhedron-derived baculovirus on insect cells and evidence for entry via direct membrane fusion. J Virol. 1993;67:1860–1868. doi: 10.1128/jvi.67.4.1860-1868.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X-P, Davis T R, Hughes P, Wood A. Potential replication of recombinant baculoviruses in non-target insect species: reporter gene products as indicators of infection. J Invertebr Pathol. 1997;69:234–245. [Google Scholar]
- 18.Hughes D S, Possee R D, King L A. Activation and detection of a latent baculovirus resembling Mamestra brassicae nuclear polyhedrosis virus in M. brassicae insects. Virology. 1993;194:608–615. doi: 10.1006/viro.1993.1300. [DOI] [PubMed] [Google Scholar]
- 19.Keddie B A, Aponte G W, Volkman L E. The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science. 1989;243:1728–1730. doi: 10.1126/science.2648574. [DOI] [PubMed] [Google Scholar]
- 20.King E G, Hartley G G. Heliothis virescens. In: Singh P, Moore R F, editors. Handbook of insect rearing. II. Amsterdam, The Netherlands: Elsevier Science Publishers; 1985. pp. 323–328. [Google Scholar]
- 21.Knebel-Mörsdorf D, Flipsen J T M, Ronsarati J, Kleefsman A W F, Vlak J M. Baculovirus infection of Spodoptera exigua larvae: lacZ expression driven by promoters of early gene pe38 and me53 in larvae tissue. J Gen Virol. 1996;77:815–824. doi: 10.1099/0022-1317-77-5-815. [DOI] [PubMed] [Google Scholar]
- 22.Lu A, Miller L K. Species-specific effects of the hcf-1 gene on baculovirus virulence. J Virol. 1996;70:5123–5130. doi: 10.1128/jvi.70.8.5123-5130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynn D E, Dougherty E M, McClintock J T, Loeb M. Development of cell lines from various tissues of Lepidoptera. In: Kuroda Y, Kurstak E, Maramorsch K, editors. Invertebrate and fish tissue culture. New York, N.Y: Springer-Verlag; 1988. pp. 39–242. [Google Scholar]
- 24.McClintock J T, Dougherty E M, Weiner R M. Semipermissive replication of a nuclear polyhedrosis virus of Autographa californica in a gypsy moth cell line. J Virol. 1986;57:197–204. doi: 10.1128/jvi.57.1.197-204.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meinkoth J, Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984;138:267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- 26.Morris T D, Miller L K. Promoter influence on baculovirus-mediated gene expression in permissive and nonpermissive insect cell lines. J Virol. 1992;66:7397–7405. doi: 10.1128/jvi.66.12.7397-7405.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris T D, Miller L K. Characterization of productive and non-productive AcMNPV infection in selected insect cell lines. Virology. 1993;197:339–348. doi: 10.1006/viro.1993.1595. [DOI] [PubMed] [Google Scholar]
- 28.ODell T M, Butt C A, Bridgeforth A W. Lymantria dispar. In: Singh P, Moore R F, editors. Handbook of insect rearing. II. Amsterdam, The Netherlands: Elsevier Science Publishers; 1985. pp. 355–367. [Google Scholar]
- 29.O’Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. W. H. New York, N.Y: Freeman & Co.; 1992. [Google Scholar]
- 30.Reddy J T, Locke M. The size limited penetration of gold particles through insect basal laminae. J Insect Physiol. 1990;36:387–407. [Google Scholar]
- 31.Thiem S M, Du X, Quentin M E, Berner M M. Identification of a baculovirus gene that promotes Autographa californica nuclear polyhedrosis virus replication in a nonpermissive insect cell line. J Virol. 1996;70:2221–2229. doi: 10.1128/jvi.70.4.2221-2229.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Treat T L, Halfhill J E. Rearing alfalfa loopers and celery loopers on an artificial diet. J Econ Entomol. 1973;66:569–570. [Google Scholar]
- 33.Volkman L E, Goldsmith P A. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: inhibition of entry by adsorptive endocytosis. Virology. 1985;143:185–195. doi: 10.1016/0042-6822(85)90107-2. [DOI] [PubMed] [Google Scholar]
- 34.Washburn J, Kirkpatrick B A, Volkman L E. Insect protection against viruses. Nature (London) 1996;383:767. [Google Scholar]
- 35.Wu J, Miller L K. Sequence, transcription and translation of a late gene of the Autographa californica nuclear polyhedrosis virus encoding a 34.8K polypeptide. J Gen Virol. 1989;70:2449–2459. doi: 10.1099/0022-1317-70-9-2449. [DOI] [PubMed] [Google Scholar]
- 36.Yuen L, Dionne J, Arif B, Richardson C. Identification and sequencing of the spheroidin gene of Choristoneura biennis entomopoxvirus. Virology. 1990;175:427–433. doi: 10.1016/0042-6822(90)90427-s. [DOI] [PubMed] [Google Scholar]
- 37.Zanotto P M D, Sampaio M J A, Johnson D W, Rocha T L, Maruniak J E. The Anticarsia gemmatalis nuclear polyhedrosis virus polyhedron gene region: sequence analysis, gene product and structural comparisons. J Gen Virol. 1992;73:1049–1056. doi: 10.1099/0022-1317-73-5-1049. [DOI] [PubMed] [Google Scholar]