Abstract
Dendritic cells (DCs) develop in the bone marrow from haematopoietic progenitors that have numerous shared characteristics between mice and humans. Human counterparts of mouse DC progenitors have been identified by their shared transcriptional signatures and developmental potential. New findings continue to revise models of DC ontogeny but it is well accepted that DCs can be divided into two main functional groups. Classical DCs include type 1 and type 2 subsets, which can detect different pathogens, produce specific cytokines and present antigens to polarize mainly naive CD8+ or CD4+ T cells, respectively. By contrast, the function of plasmacytoid DCs is largely innate and restricted to the detection of viral infections and the production of type I interferon. Here, we discuss genetic models of mouse DC development and function that have aided in correlating ontogeny with function, as well as how these findings can be translated to human DCs and their progenitors.
Dendritic cells (DCs) are essential regulators of innate and adaptive immune responses. A diversity of functionally distinct DC subsets has been described, and emerging models have incorporated variability across tissues, species and genetically diverse individuals. Classical DCs (cDCs), also known as conventional DCs, have been defined by their discrete functions and transcriptional identities and include two major subsets, known as type 1 cDCs (cDC1s) and type 2 cDCs (cDC2s). The distinction between cDC1 and cDC2 subsets is supported by genetic models of DC development, which have been essential to define the specialized functions of cDC subsets1. For the most part, cDC1s present exogenous, cell-associated antigens to CD8+ T cells, thereby regulating cytotoxic T lymphocyte (CTL) responses to intracellular pathogens and cancer. By contrast, the cDC2 subset mainly presents soluble antigens to CD4+ T cells, thereby regulating immune responses to extracellular pathogens, parasites and allergens2. Plasmacytoid DCs (pDCs) are well established as a source of type I interferon during viral infections. However, the ontogeny and function of pDCs remain active areas of debate3. Genetic models of cellular functions are most developed for cDCs, and so this Review does not cover pDC function in great depth. Here, we review genetic models and in vivo descriptions that have led to the identification of human and mouse DCs and their progenitors, and the use of these models to advance our understanding of DC functions.
Transcriptional regulation of development
The specification of DC progenitors to discrete DC subsets occurs after the restriction of multipotent progenitors (MPPs) to the myeloid or lymphoid lineage4. Early work identified progenitors of the myeloid lineage that have DC, monocyte, macrophage and granulocyte potential. Recently, multiple DC-restricted progenitors have been identified that can give rise to the major DC subsets5–8. In this section, we review the current models of monocyte and DC development in mice (FIG. 1). These mouse models, in turn, provide a framework to highlight similarities and differences between mouse and human DC ontogeny9.
Fig. 1 |. Genetic models of mouse dendritic cell development and lineage restriction.
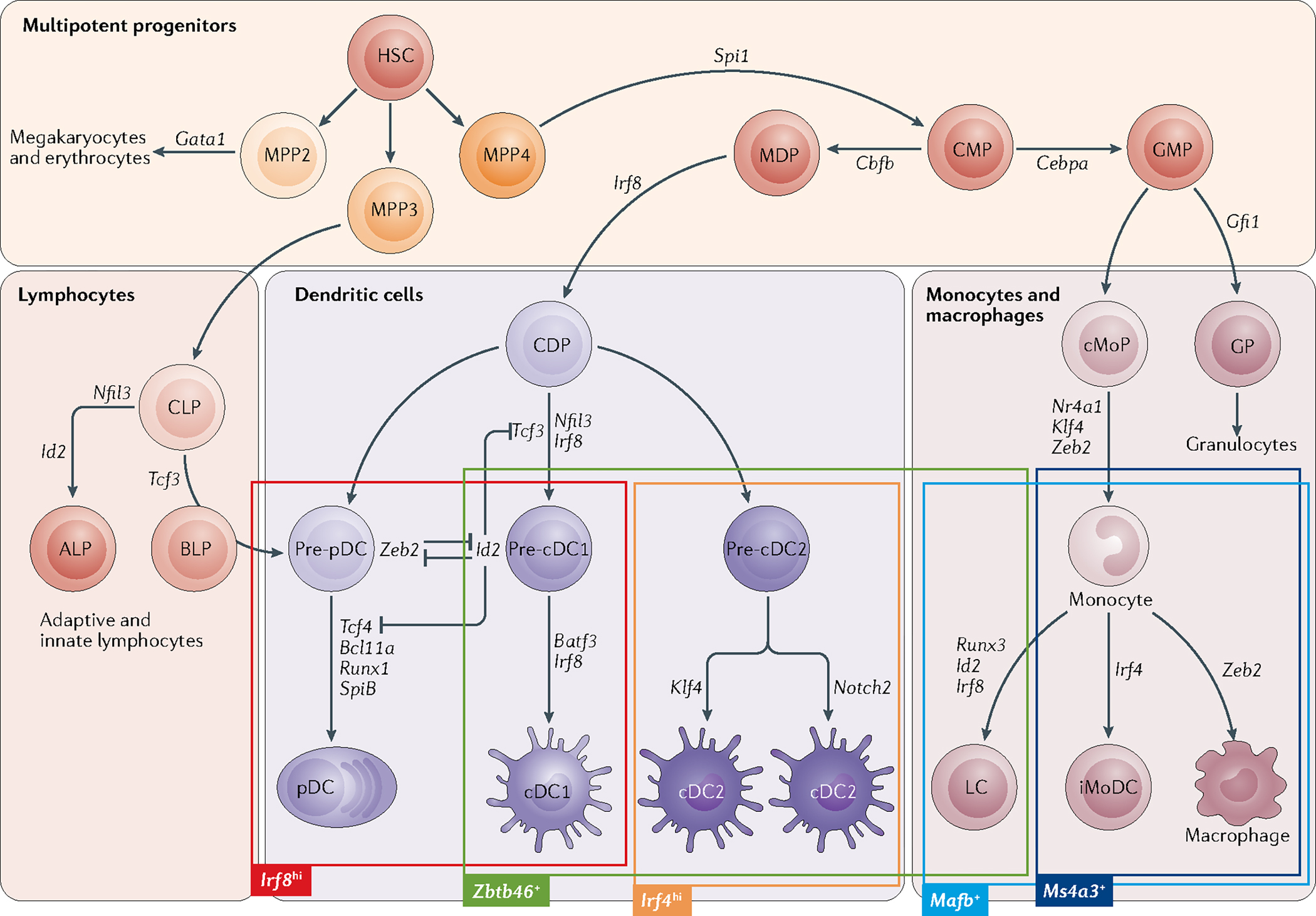
Hierarchical models of haematopoiesis are based on the developmental deficiencies that are observed in mice with mutations in transcription factor-encoding genes; these genes are shown adjacent to the associated stages of lineage restriction and specification. The figure highlights the development of mouse dendritic cell (DC), monocyte and macrophage subsets from shared progenitors. Haematopoietic stem cell (HSC)-derived multipotent progenitors (MPPs) undergo stages of differentiation to produce lineage-restricted progenitors of lymphocytes and myeloid cells — common lymphoid progenitors (CLPs) and common myeloid progenitors (CMPs). The CLP population can be separated into two subsets on the basis of Ly6D expression. The all-lymphoid progenitor (ALP) is Ly6D− and has B cell, T cell and innate lymphoid cell potential. The Tcf3-dependent B cell-biased lymphoid progenitor (BLP) is Ly6D+ and gives rise to B cells and plasmacytoid DCs (pDCs). High levels of expression of interferon regulatory factor 8 (Irf8) are associated with the specification of pDCs and type 1 classical DCs (cDC1s). Expression of Zbtb46 is a specific marker of cDC specification that is induced first in pre-cDC1s and precursor type 2 classical DCs (pre-cDC2s). cDC2 subsets are also characterized by high levels of Irf4 expression. Langerhans cells (LCs), monocytes and macrophages are all marked by Mafb-driven lineage tracing. Although LCs are not thought to be derived from the common DC precursor (CDP) population, given their embryonic origin, they do express Zbtb46 when they migrate to lymphoid organs from the skin. Lineage tracing based on Ms4a3 marks monocytes and monocyte-derived macrophages, as opposed to macrophages of embryonic origin. cMoP, common monocyte progenitor; GMP, granulocyte–macrophage progenitor; GP, granulocyte progenitor; iMoDC, immature monocyte-derived dendritic cell; MDP, monocyte–dendritic cell progenitor.
Lineage restriction of multipotent progenitors.
The primary function of haematopoietic stem cells (HSCs) in mice and humans is to generate all blood cell types. The capacity of HSCs to generate blood cells must be balanced by mechanisms to maintain stemness during an entire lifespan4,9,10. In the lifetime of a mouse, HSCs divide only a few times, giving rise to MPPs that are highly proliferative and populate the entire haematopoietic system11. Although HSCs can theoretically give rise to all haematopoietic lineages, methods of genetic barcoding and indexed cell sorting have uncovered patterns of bias towards one line-age or another, known as lineage imprinting or transcriptional priming12–14. So far, evidence for imprinting or priming of HSCs is mainly limited to correlations of transcriptional signatures with the developmental fate of individual clones. In the case of DC development, it has been suggested that specification occurs early in HSCs or MPPs that express high levels of interferon regulatory factor 8 (IRF8)10,15. However, although transcriptional signatures within early stem cells may correlate with granulocyte, monocyte or DC potential, they are not sufficient to recapitulate the chronologically ordered specification events that have been well established genetically across haematopoietic lineages16,17. Recently, single-cell analysis of clonal bias among sibling stem cells showed that they can have divergent developmental trajectories that are independent of transcriptional priming18.
Lymphoid and myeloid lineage divergence.
The common myeloid progenitor (CMP) and the common lymphoid progenitor (CLP) were initially described as cell populations that are committed to myeloid and lymphoid lineages, respectively19,20. Early models suggested that CMPs gave rise to granulocyte–monocyte progenitors (GMPs), which have lost megakaryocyte–erythroid progenitor (MEP) potential but can give rise to all myeloid lineages21. Deficiency in the transcription factor GATA1 results in embryonic lethality as a result of failed differentiation of MEPs, and PU.1 is expressed in progenitors that have excluded MEP potential22,23. Therefore, mutual repression between PU.1 and GATA1 was proposed as a mechanism to regulate the divergence of MEPs from CMPs. However, this remains an active area of debate because PU.1 is necessary for the development of both lymphoid and myeloid lineages, and so alone cannot enforce myeloid cell identity24. In addition, analysis of PU.1 and GATA1 protein expression did not identify a population that co-expresses these factors25,26. With the recent discovery that MEPs can be derived directly from a biased MPP population, mutual repression between PU.1 and GATA1 may not be necessary to explain the divergence of MEPs from a myeloid-restricted progenitor such as the CMP9,27.
Although the development of lymphoid lineages is beyond the scope of this Review, some discussion of the CLP is relevant to observations that DCs can also be derived from populations that are classically defined as lymphoid-restricted progenitors. CLPs induce expression of the transcription factors GATA3, TCF1 (also known as TCF7) and BCL-11B, and develop into T cells after encountering Notch ligands in the thymus. By contrast, an absence of Notch signalling in the bone marrow leads to the development of B cells, which requires the induction of TCF3 (also known as E2A), EBF1 and PAX5 (REF.17). pDCs have been reported to develop from a fraction of the CLP population that has excluded T cell potential but retained B cell potential (BOX 1).
Box 1 |. Lymphoid origin of plasmacytoid dendritic cells.
Early work on dendritic cell (DC) development suggested that classical DCs (cDCs) and plasmacytoid DCs (pDCs) could develop from both the common lymphoid progenitor (CLP) and the common myeloid progenitor30,230–232. Evidence reported over the past two decades, such as Rag1 expression, Il7ra expression and V(D)J recombination during pDC development, continues to support a lymphoid origin of pDCs216,230,233–235. As classically defined, both CLPs (Lineage−CD135+CD115−CD127+) and common DC precursors (CDPs; Lineage−CD135+CD117intCD115+CD127−) can give rise to pDCs20,41,232,234. This is supported by lineage tracing driven by expression of the lymphoid and myeloid markers, Il7ra and Csf1r, respectively216,219. pDCs are marked in both models in vivo.
Recently, single-cell RNA sequencing analysis revealed heterogeneity within populations of CLPs and CDPs that was used to identify clonogenic progenitors of pDCs, referred to as pre-pDCs5,7,236. One study found that bone marrow progenitors that represent a fraction of the CLP population (Lineage−CD127+CD135+CD117int/low Ly6D−SiglecH+) give rise exclusively to pDCs7. An independent group reported similar pDC potential within the Ly6D+ CLP population and could exclude pDC potential from the CDP population on the basis of CD81 expression5. The former group suggested that CDPs give rise to a distinct subset of Zbtb46–GFP-expressing pDCs in the bone marrow and spleen with functional characteristics of cDCs7. This conclusion is limited by the reliance on co-expression of Zbtb46 with canonical pDC markers that are known to be expressed by all DC progenitors at different stages8. Genetic models of pDC development, such as Bcl11a and Zeb2 deficiency, could help to resolve the ontogeny of pDCs in vivo55,56,79. Loss of pre-pDCs from both CLP and CDP populations in Zeb2-deficient mice, for example, could indicate that there is no genetic basis to model pre-pDCs as two ontogenetically unrelated populations. Alternatively, examination of Tcf3-deficient mice, which have a reduction in the Ly6D+ fraction of the CLP population, could support such a distinction237.
Restriction of granulocyte potential.
GMPs were originally proposed to give rise to monocyte–DC progenitors (MDPs), which lack granulocyte potential28–30. However, this model requires revision in the light of recent findings by multiple groups. Single cells within the GMP and MDP populations were shown to have surface marker heterogeneity that correlates with fate bias30–33. These studies suggest that monocytes and granulocytes can develop from intermediate progenitors in the bone marrow that do not retain DC potential. A restricted granulocyte progenitor and a common monocyte progenitor (cMoP) are now proposed to develop from a common GMP-like progenitor. A GMP lineage tracing model based on restricted expression of Ms4a3 identified granulocyte progenitors and cMoPs as GMP-derived populations31. However, a small percentage of monocytes in this study were untraced, which is consistent with the results of other groups that suggest monocytes can be derived from a progenitor not shared with granulocyte progenitors33. Lineage tracing driven by Mafb expression also supports a model whereby monocytes and macrophages are separate lineages from DCs and granulocytes34. Cbfb and Cebpa are required for monocyte and granulocyte development, respectively35,36. CBFβ has been shown to support the expression of IRF8, which is in turn associated with the exclusion of granulocyte potential35,37. Deletion of Irf8 results in unrestrained C/EBPα activity, a myeloproliferative disorder of expanded granulocyte progenitor and granulocyte populations, and defective development of Ly6C+ monocytes37–39.
Lineage tracing.
A method to identify cells that are expressing or have expressed a gene of interest at any point during their development, which enables the study of progenitor–progeny relationships.
Dendritic cell specification.
Initial work on the development of DCs from bone marrow progenitors identified a population referred to as the common dendritic cell precursor (CDP). Developing from MDPs, CDPs give rise to pDCs, cDC1s and cDC2s (REFs40,41). A mechanism to explain loss of monocyte and macrophage potential on transition from MDPs to CDPs has not so far been identified. Although CDPs are markedly reduced in Irf8−/− mice, pDCs and cDC2s can still develop despite significant surface marker, transcriptional and functional changes42. cDC1 and cDC2 subsets are functionally and transcriptionally more similar to each other than to pDCs43. Therefore, it was proposed that pDC progenitors diverge from CDPs before the specification of individual cDC subsets, which are derived from a putative precursor population known as pre-cDCs44–46. However, it has since been recognized that pre-cDCs are a heterogeneous population of cDC1-specified or cDC2-specified progenitors6,8,44. Pre-pDCs have also been identified recently and are thought to originate from CLPs as well as CDPs5,7 (BOX 1). After specification in the bone marrow, cDC-restricted progenitors may continue to undergo additional rounds of cell division in lymphoid organs and peripheral tissues, ultimately exiting the cell cycle on activation or homeostatic maturation46,47.
Mutual repression of cDC1 and pDC specification.
Recent advances have closely linked the genetic mechanisms of cDC1 and pDC specification. Development of cDC1s from bone marrow progenitors requires Irf8, Batf3, Id2 and Nfil3 (REFs48–50). Deficiencies in Batf3, Id2 or Nfil3 can be rescued by the ectopic expression of Irf8, which indicates that high levels of IRF8 expression are central to the genetic programme that controls cDC1 development51,52. Steady-state cDC1 development is impaired in Batf3−/− mice after specification occurs in the bone marrow as a result of insufficient maintenance of IRF8 expression levels6,53 (BOX 2; FIG. 2).
Box 2 |. Regulation of dendritic cell development by enhancer switching.
Enhancers surrounding transcription factor-encoding genes are dynamically regulated during haematopoiesis, which is necessary for lineage divergence and the maintenance of cell identity238. Lineage-defining transcription factors can enforce cellular identity through autoactivation, a mechanism by which a factor positively regulates its own expression. One example of a transcription factor that uses autoactivation during myeloid cell development is PU.1 (encoded by Spi1), which was shown to bind to an enhancer 14 kb upstream of Spi1 and increase its own expression by 100-fold239. Reminiscent of this model of PU.1 autoactivation, enhancers of interferon regulatory factor 8 (Irf8) were recently shown to mediate IRF8 autoactivation in cooperation with BATF3 (REF.6). Close examination of IRF8-binding sites in the Irf8 locus revealed three novel enhancers that are +41 kb, +32 kb and −50 kb from the transcription start site and are necessary to maintain high levels of Irf8 expression53. Germline deletion of the +41-kb enhancer resulted in reduced Irf8 expression in the monocyte–dendritic cell progenitor population and in plasmacytoid dendritic cells (pDCs), and blocked precursor type 1 classical dendritic cell (pre-cDC1) specification in the common dendritic cell precursor population. It was proposed that early activation of this enhancer depends on TCF3 (also known as E2A), which is consistent with TCF3 deficiency resulting in a failure of pDC and cDC1 development. The +32-kb enhancer is active in cDC1s and is bound by BATF3 but not TCF3. This suggested that a switch from IRF8–TCF3-mediated autoactivation at the +41-kb enhancer to IRF8–BATF3-mediated autoactivation at the +32-kb enhancer occurs on cDC1 specification. Pre-cDC1s develop normally in Irf8 +32−/− and Batf3−/− mice, but a failure to maintain high levels of IRF8 results in cDC1 deficiency in secondary lymphoid organs and peripheral tissues. Although deletion of the −50-kb enhancer had no effect on DC development, it resulted in reduced IRF8 levels and functional defects in monocytes. Studies of autoactivation by lineage-defining transcription factors have thus revealed novel mechanisms by which non-coding genetic elements may regulate haematopoiesis240.
Fig. 2 |. Stage-specific enhancer activation regulates Irf8-dependent specification of dendritic cell and monocyte progenitors.
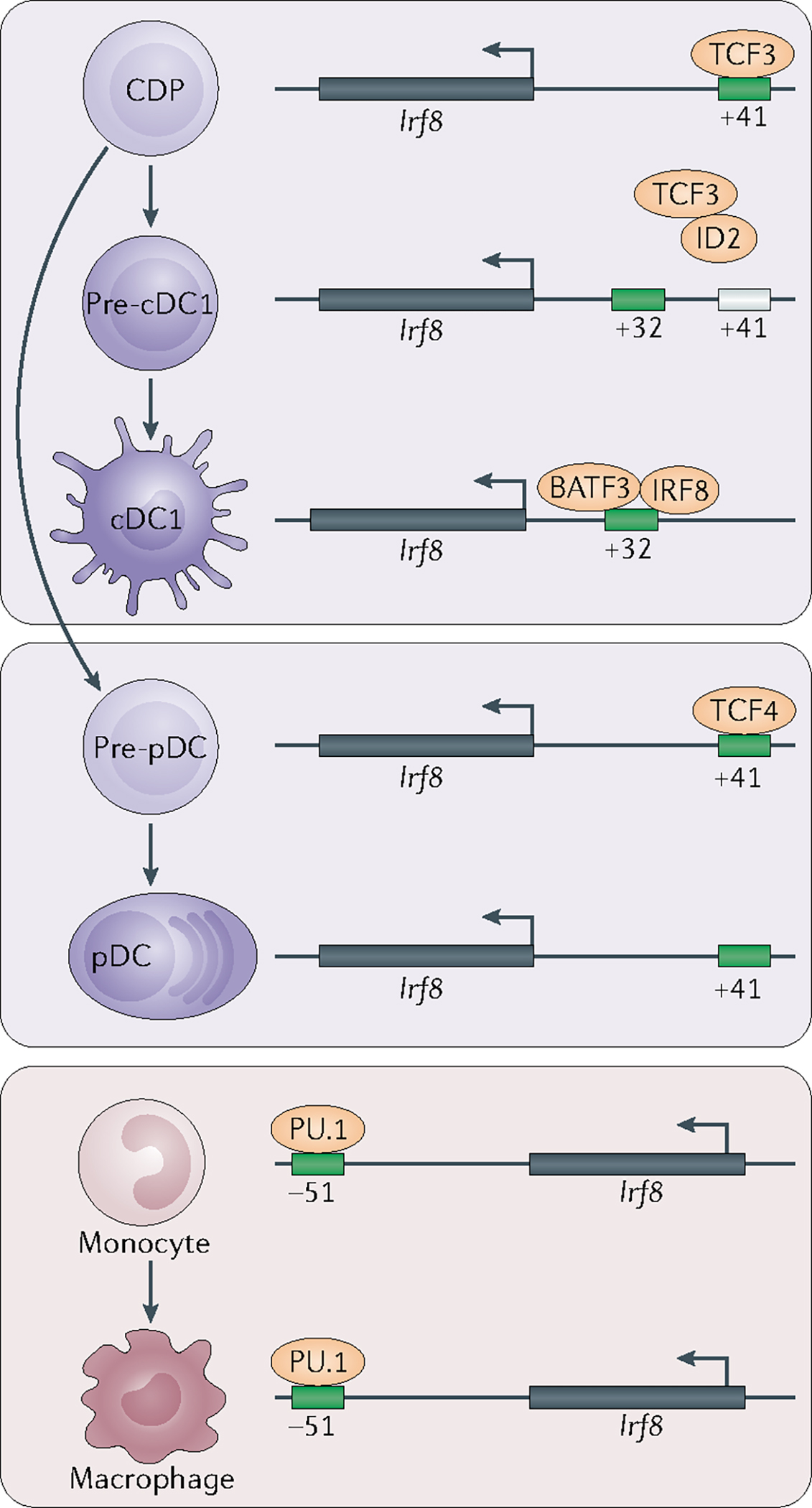
One mechanism for the development of type 1 classical dendritic cells (cDC1s) and plasmacytoid DCs (pDCs) from bone marrow progenitors is the strict regulation of interferon regulatory factor 8 (Irf8) expression by distal, evolutionarily conserved enhancer elements. E proteins — such as TCF3 (also known as E2A) and TCF4 (also known as E2–2) — support Irf8 expression through their actions at an E-box motif-containing enhancer located 41 kb upstream of the Irf8 transcription start site. An inactivating mutation at this locus impairs Irf8 expression in pDCs and blocks the specification of precursor cDC1s (pre-cDC1s) in the common DC precursor (CDP) population. On pre-cDC1 specification, Nfil3 is induced upstream of Id2. By blocking E protein activity, ID2 imposes a requirement for an alternative enhancer located 32kb upstream of Irf8 to promote Irf8 expression. In turn, Irf8 expression and cDC1 specification are supported by BATF3 and by IRF8-dependent autoactivation. An alternative enhancer located 51kb downstream of the Irf8 locus is required for Irf8 expression regulated by PU.1 in monocytes and macrophages.
Models of cDC1 development have been further expanded by a recent study that examined heterogeneity in the CDP population on the basis of in vivo expression of Zbtb46, Id2, Nfil3 and Zeb2 (REF.54). In the bone marrow, Zbtb46–GFP expression within the CDP population marks a CD11C−MHC class I−CD117int population of cDC1-specified progenitors, which give rise to a previously defined CD11C+MHC class IintCD117int pre-cDC1 population6,8. This early pre-cDC1 population expresses increased levels of Id2 and Nfil3 and decreased levels of Zeb2 when compared with bulk CDPs, and its development depends on Id2 and Nfil3. These results revealed the stage at which these factors function during cDC1 development. When cultured ex vivo, ZEB2low CDPs are biased towards cDC1s, which suggests that cDC1 specification involves the inhibition of Zeb2 (REF.54). Because ZEB2 regulates pDC development, it was hypothesized that early induction of Nfil3 functions to block pDC potential54–56. In support of this hypothesis, pre-cDC1s are restored in Nfil3−/−Zeb2−/− mice, which shows that the functions of NFIL3 are dispensable in the absence of ZEB2 (REF.54).
The E protein transcription factor TCF4 (also known as E2–2) is also required for pDC development57,58. ID2 heterodimerizes with E proteins to block their activity by preventing binding to DNA59. This early observation is the basis for the hypothesis that ID2 functions during cDC1 specification to inhibit TCF4, thus blocking pDC development. Recently, it was shown that Id2 expression marks a subset of pre-cDC1s in the CDP population, further suggesting that it functions during cDC1 development to block E protein activity and pDC potential54. Therefore, it was hypothesized that Id2 would be dispensable for cDC1 development in a genetic background where pDC specification cannot occur. Accordingly, cDC1 development occurs normally in Id2−/−Zeb2−/− mice54. Although cDC2s also express Id2 and Zeb2, they do not require these genes for development. To shed light on the potential gene targets of ZEB2, whole transcriptome analysis of Zeb2-deficient cDC2s revealed increased expression of Id2, which suggests that ZEB2 might directly inhibit Id2 transcription54–56.
E protein.
A member of a family of transcription factors, including TCF3 (also known as E2A), TCF4 (also known as E2–2) and TCF12 (also known as HEB), that are essential for the development of several haematopoietic lineages and that bind conserved DNA motifs known as E-boxes.
Human progenitors of dendritic cells and monocytes.
The study of patients with combined immunodeficiencies has provided insight into the genes that regulate human haematopoiesis. For example, deficiency in GATA2 has been associated with a human syndrome known as DC, monocyte, B cell and natural killer cell lymphoid (DCML) deficiency60. It is not clear whether GATA2 functions in a shared progenitor of these lineages or whether it functions independently after the divergence of myeloid and lymphoid lineages. Humans with IKZF1 haploinsufficiency have a deficiency in pDC development that correlates with an increase in the size of cDC1 populations61. It has been suggested from a survey of chromatin immunoprecipitation followed by sequencing data that IKZF1 represses ID2, thereby promoting TCF4 activity and pDC specification62. Mutations in TCF4 are associated with a deficiency in pDC development in patients with Pitt–Hopkins syndrome57. Similar to the broad defects in the haematopoietic compartment that are associated with Irf8 deficiency in mice, several reports have linked human mutations in IRF8 to DC deficiencies, as well as defects in the development and function of granulocytes, B cells, T cells and natural killer cells63–65.
Several human counterparts of the mouse DC and monocyte progenitors have been identified on the basis of surface marker expression and developmental potential (TABLE 1). Although human DCs can develop from CMPs, early work suggested that human DCs can also be derived from CLPs66–69. This led to the conclusion that human DC ontogeny does not strictly conform to models of myeloid and lymphoid lineage divergence in mice69–71. A human CMP-derived granulocyte–monocyte–DC progenitor (GMDP) was recently reported to have combined potentials of mouse MDPs and GMPs72. Alternatively, heterogeneity in a GMDP-like population suggests that the DC potential of human GMPs is the result of contamination by a separate MDP population73. Human granulocyte progenitors, cMoPs and CDPs have also been identified but their ontogeny remains an active area of debate72,74–77.
Table 1 |.
Markers of human and mouse dendritic cell and monocyte progenitors
| Progenitor population | Human cell surface markers | Mouse cell surface markers |
|---|---|---|
| Pre-cDCl | CD135, CD117, CD116, CD123low/int, CD45RA, CADM1 (Lineage negativea) | CD135, CD115, CD117int, CD11C, CD226, MHC class IIlow/int (Lineage negativeb,c) |
| Pre-cDC2 | CD135, CD117, CD116, CD123low, CD45RA, CD172hi, CD1C (Lineage negativea) | CD135, CD117low, CD11C, Ly6C, ZBTB46 (Lineage negativeb,c) |
| Pre-cDC | CD135, CD117, CD116, CD123, CD45RA, CD172int, CD22 (Lineage negativea) | CD11C, CD135, CD172int (Lineage negativeb,c) |
| CDP | CD34, CD38, CD135, CD117, CD116, CD123hi, CD45RA (Lineage negativea) | CD135, CD115, CD117int (Lineage negativeb,d) |
| MDP | CD34, CD38, CD135, CD117, CD115, CD123 (Lineage negativea) | CD135, CD115, CD117hi (Lineage negativeb,d) |
| cMoP | CD34, CD38, CD45RA, CD135, CLEC12A, CD64 | CD135−, CD117, CD115, Ly6C (Lineage negativeb,d) |
| GMDP/GMP | CD34, CD38, CD135, CD117, CD123int (Lineage negativea) | CD117, CD34−, CD16/CD32low (Lineage negativeb,d) |
| CMP | CD34, CD38, CD135 (Lineage negativea) | CD117, CD34, CD16/CD32low (Lineage negativeb,d) |
| CLP | CD34, CD45RA, CD10 (Lineage negativea) | CD135, CD127, CD117int/low (Lineage negativeb,d) |
| Pre-pDC | ND | CD135, CD117int/low, CD127, Ly6D, SiglecH, CD81 (Lineage negativeb) |
cDC, classical dendritic cell; cDC1, type 1 classical dendritic cell; cDC2, type 2 classical dendritic cell; CDP, common dendritic cell precursor; CLP, common lymphoid progenitor; cMoP, common monocyte progenitor; CMP, common myeloid progenitor; GMDP, granulocyte–monocyte–dendritic cell progenitor; GMP, granulocyte–monocyte progenitor; MDP, monocyte–dendritic cell progenitor; ND, not determined; pDC, plasmacytoid dendritic cell.
Lineage negative for CD3, CD19, CD20, CD56, CD14, CD66B, CD303, CD335, CD10, CD123, CD1C.
Lineage negative for CD3, CD19, Ly6G, Ter-119, NK1.1, CD105, B220, MHC class II.
Lineage negative for SiglecH, CD127.
Lineage negative for CD5, CD11B, CD8a, CD4, CD127.
Outstanding questions in DC development.
Compared with cDC1s, the genetic mechanisms that regulate pDC and cDC2 specification are less well understood. The recent identification of clonogenic pDC progenitors, known as pre-pDCs, provides a new population with which CLPs and CDPs can be compared to identify the stages at which TCF4, ZEB2 and BCL-11A function to regulate pDC development5,7,55,56,78,79. Mechanisms of cDC2 specification are poorly understood and so are not discussed in this Review. Although pre-cDC2s that exclude cDC1 and pDC potential have been identified in the bone marrow, no genes necessary for pre-cDC2 development have so far been identified6,8. Notwithstanding, several genes that regulate cDC2 subset diversification in lymphoid organs and peripheral tissues have been identified and are described later in this Review.
Functional specialization of DC subsets
Human and mouse DCs have been organized into functional subsets based on various lines of evidence. In mice, genetic models of ontogeny have aided in the unification of functionally redundant subsets across tissues that are otherwise distinct on the basis of surface marker expression1 (TABLE 2). Similarly, these models have helped to separate phenotypically similar populations that have non-redundant functions. Advances along these lines have also been made for human DCs, in particular through the use of high-throughput single-cell analysis of transcriptomes and surface protein expression. However, independent groups have provided disparate definitions for seemingly overlapping human DC subsets, which necessitates a unification of the human DC subsets that have so far been described (TABLE 3). In this section, we review the diversity of human and mouse DC subsets, discuss the ontogenetic and functional similarities that are well established and highlight some unresolved differences (FIG. 3).
Table 2 |.
Useful models for the definition of dendritic cell ontogeny and function
| Gene | Expression by | Genetic mouse model | Refs | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| cDC1s | cDC2s | pDCs | DC progenitors | Reporter | Knockout | Floxed | Cre recombinase | Diphtheria toxin receptor | ||
| Markers | ||||||||||
| Itgax | + | + | + | + | No | No | No | Yes | Yes | 198–203 |
| CIec9a | + | − | + | + | Yes | Yes | No | Yes | Yes | 45,204 |
| Cd207 | + | + | − | − | No | No | No | Yes | Yes | 205–208 |
| SiglecH | − | − | + | + | No | No | No | Yes | Yes | 204,209,210 |
| Xcr1 | + | − | − | − | Yes | Yes | Yes | Yes | Yes | 99,211 |
| Karma/Gpr141b | + | − | − | − | No | No | No | No | Yes | 212 |
| Clec4a4 | − | − | − | − | No | No | No | No | Yes | 213 |
| Mgl2 | − | + | − | − | No | No | No | No | Yes | 134 |
| Clec4c | − | − | + | − | No | No | No | No | Yes | 152 |
| Cx3cr1 | − | + | − | + | Yes | Yes | No | No | Yes | 214,215 |
| IL7r | − | − | − | + | Yes | Yes | Yes | Yes | No | 216 |
| Csf2r | − | − | − | + | Yes | Yes | Yes | Yes | No | 217–219 |
| Transcription factors | ||||||||||
| Zbtb46 | + | + | − | + | Yes | Yes | No | Yes | Yes | 6,179,220 |
| Batf3 | + | + | − | + | No | Yes | No | No | No | 49 |
| Tcf3 | − | − | − | + | Yes | No | Yes | No | No | 221 |
| Tcf4 | − | − | + | + | No | No | Yes | No | No | 57 |
| Irf4 | − | + | − | − | Yes | Yes | Yes | No | No | 222 |
| Nfil3 | + | − | − | + | No | Yes | Yes | No | No | 50,223 |
| Id2 | + | + | − | + | No | No | Yes | No | No | 48,224,225 |
| Klf4 | − | + | − | − | No | No | Yes | No | No | 144,226 |
| Zeb2 | − | + | + | + | No | No | Yes | No | No | 55,56,227 |
| Irf8 | + | + | + | + | No | Yes | Yes | No | No | 228,229 |
| Irf8 + 32 kb | + | − | − | + | No | Yes | No | No | No | 53 |
| Irf8 + 41 kb | + | − | + | + | No | Yes | No | No | No | 53 |
cDC1, type 1 classical dendritic cell; cDC2, type 2 classical dendritic cell; DC dendritic cell; pDC, plasmacytoid dendritic cell.
Table 3 |.
Markers of human and mouse dendritic cell subsets
| DC subset | Human cell surface markers (Lineage negativea,b; Lineage positivec) | Mouse cell surface markers (Lineage positived) |
|---|---|---|
| cDC1 | XCR1, CD45, CADM1, CLEC9A, CD141 | XCR1, CD24 (Lineage negativeb,e) |
| Context-dependent cDC1 | CAMK2D, XCR1, CCR7, CD103, CD11C | CD103, CD207, CD326, CD8A, CLEC9A |
| cDC2 | CD45, CD1C, FceR1A, CD172A | CD172A (Lineage negativeb,e) |
| cDC2A | CD5 | ESAM, CD4, CD103, CD11B |
| cDC2B | CD14, CD163 | CD24, MGLL |
| Context-dependent cDC2 | CCR7, CD103, CD11B, CD11C, CD80, CD86 | CCR7, CD80, CD86, F4/80 |
| pDC | CD45RA, CD123, CD2 | B220, SiglecH, CD317, Ly6D, CCR9, Ly6C (Lineage negativee) |
Markers listed as ‘context-dependent’ can vary across tissues and physiological settings and so are separated from markers that generally have consistent patterns of expression for the given subsets. cDC1, type 1 classical dendritic cell; cDC2, type 2 classical dendritic cell; DC dendritic cell; pDC, plasmacytoid dendritic cell; XCR1, XC-chemokine receptor 1.
Lineage negative for CD3, CD19, CD20, CD56.
Lineage negative for B220, SiglecH, Bst2.
Lineage positive for HLA-DR, CD45.
Lineage positive for CD11C.
Lineage negative for CD3, CD19, Ly6G, Ter-119, NK1.1, CD64.
Fig. 3 |. Specialized functions of mouse classical dendritic cell subsets.

Type 1 classical dendritic cells (cDC1s) are specialized in the regulation of type I immune responses through the priming and activation of cytotoxic CD8+ T cells and CD4+ T helper 1 (TH1) cells. Relative to other DC subsets, cDC1s specifically express Toll-like receptor 3 (TLR3) and TLR11, which recognize double-stranded RNA (dsRNA) and the Toxoplasma gondii antigen profilin, respectively. cDC1s are an essential source of IL-12 and are necessary for resistance to intracellular viral, bacterial and parasitic infections. cDC1s are uniquely capable of acquiring antigens associated with host cells, through a process known as cross-presentation, which is essential for pathogen clearance and antitumour immune responses. Type 2 classical dendritic cells (cDC2s) regulate type II and type III immune responses and antibody responses to soluble antigens. cDC2s at barrier surfaces, such as in the lung, gut and skin, regulate type II immune responses to parasites, fungi and allergens and are required for the expansion of CD4+ T helper 2 (TH2) cell populations and activation of group 2 innate lymphoid cells (ILC2s). The regulation of such responses depends on cDC-intrinsic expression of interferon regulatory factor 4 (Irf4) and has been attributed to the Klf4-dependent cDC2 subset. Type III immune responses are regulated by a distinct subset of Notch 2-dependent cDCs, which are a necessary source of IL-23 during acute infection with Citrobacter rodentium. IL-23 is necessary to activate group 3 innate lymphoid cells (ILC3s) and to induce differentiation of CD4+ T helper 17 (TH17) cells. cDC2s have also been shown to regulate antibody responses though the induction of germinal centre responses to soluble antigens in lymphoid organs. Deficiency in Notch2-dependent cDCs results in a failure to induce CD4+ T follicular helper (TFH) cells and germinal centre B cells in the spleen, for example. TCR, T cell receptor.
Non-redundant functions of cDC1s in mice.
It has been appreciated for more than three decades that the DC lineage is specialized to prime antigen-specific T cells and that cDC1s are probably superior inducers of CD8+ CTL responses in vivo80,81. Initial work carried out ex vivo showed that cDC1s mediate CTL responses through a process known as cross-priming or cross-presentation, which involves the uptake of exogenous, cell-associated antigens, loading of processed peptides onto MHC class I molecules and their presentation to CD8+ T cells in lymphoid tissues82–84. The first in vivo demonstration that cDC1s mediate CTL responses was accomplished through the analysis of Batf3−/− mice. These mice have a deficiency in cDC1 development, which results in diminished CTL responses, susceptibility to viral infections and uncontrolled tumour growth49. This essential function of Batf3-dependent cDC1s in inducing CTLs has since been shown in a wide variety of model systems by numerous independent groups85–91.
With regard to the use of Batf3−/− mice as a genetic model to define the non-redundant functions of cDC1s in vivo, multiple groups have described a phenomenon in which cDC1 development can be restored, albeit partially, in some tissues of Batf3−/− mice. Infection with mycobacteria, IL-12 administration and irradiation can restore cDC1 development through compensation by other BATF factors, such as BATF and BATF2 (REFs51,92). Deletion of an enhancer bound by BATF3 blocks this compensation, suggesting that BATF and BATF2 function to support Irf8 expression at this site in the absence of BATF3 (REF.53) (BOX 2). By restoring cDC1 populations in Batf3−/− mice through transgenic overexpression of Irf8, it was recently shown that a restricted set of genes regulated by BATF3 is required for cDC1-intrinsic rejection of immunogenic tumours52,93. The genetic targets of BATF3 that regulate cDC1 identity and function remain an active area of investigation.
cDC1s can regulate innate immune responses through functions that are independent of cross-presentation to CTLs. cDC1s uniquely express Toll-like receptor 11 (TLR11), a pattern recognition receptor for the Toxoplasma gondii antigen profilin43. Early work identified IL-12 as a target of signalling through TLR11, and later analysis of Batf3−/− mice showed that cDC1s are a non-redundant source of IL-12 necessary to mediate resistance to acute infection with T. gondii94. It has been shown that IL-12 produced by cDC1s activates the production of interferon-γ (IFNγ) by natural killer cells, which in turn primes regulatory functions in developing monocytes95. TLR3 is another pattern recognition receptor that is selectively expressed by the cDC1 subset43. Stimulation of TLR3 with double-stranded RNA, including the model antigen poly(I:C), is sufficient to induce cDC1 maturation and promote antiviral and antitumour immune responses96,97.
Multiple models of conditional gene deletion have been developed to study the in vivo functions of cDC1s. Among these, the deletion of genes on the basis of Xcr1 expression is the most recently developed model and provides the best specificity for cDC1s (REFs98,99). XC-chemokine receptor 1 (XCR1) is expressed exclusively by the cDC1 subset in mice and humans86,100–102. These models have been used most recently to identify novel innate immune functions of cDC1s. For example, specific deletion of Vegfa in cDC1s results in decreased numbers of infiltrating neutrophils and reduced skin inflammation at sites infected with Propionibacterium acnes103. By contrast, CLEC9A (also known as DNGR1) was shown to inhibit the production of CXC-chemokine ligand 2 (CXCL2) by Batf3-dependent cDC1s, thus limiting inflammation associated with neutrophil recruitment104. These studies have highlighted previously unknown crosstalk between cDC1s and neutrophils, whereby the regulatory functions of cDC1s seem to be context dependent.
Checkpoint blockade.
A type of immunotherapy that inhibits immune signalling cascades that are normally engaged to prevent autoimmunity and uncontrolled inflammation but that can also prevent effective immune responses to cancer, for example.
Several cell surface receptors have been identified as being necessary for the regulation of adaptive immune responses by cDC1s. XCR1 and its ligand XCL1 are required for the appropriate localization of cDC1s and CD8+ T cells within lymphoid organs. Mice deficient in Xcr1 or Xcl1 have marked reductions in size and altered cellular localization of CD8+ and CD4+ T cell populations in the spleen, lymph nodes and intestines both at steady state and during inflammation99,102,105. Although its expression is not unique to the cDC1 subset, the receptor LY75 (also known as DEC-205) has been targeted to deliver antigens to cDC1s to improve T cell responses and can also function as a receptor to internalize CpG nucleotides for recognition by intravacuolar TLR9 (REFs106–108). CLEC9A has also been targeted for antibody-mediated delivery of model antigens to cDC1s, which can improve the efficacy of vaccines against model tumours and viral infections109–111. Similar strategies have been applied to target antigens to cDC1s through XCR1 (REFs112,113).
Mouse models of cDC1 function have highlighted the potency of this cell type in modulating cancer immunotherapy through the activation of tumour-specific CD8+ T cells. Therapeutic responses to checkpoint blockade in mice involving antibodies to TNFRSF9 (also known as CD137), PD1, PDL1 or CTLA4 require Batf3-dependent DCs88,114,115. Similarly, certain types of adoptive T cell therapy require cDC1s to induce tumour rejection89. Intratumoural injection of adjuvants can lead to a reduction in tumour mass and promote tumour rejection in a Batf3-dependent manner116–118. Similarly, administration of the growth factor FLT3L expands DC populations and promotes cDC1-mediated tumour rejection when combined with immunotherapy119–121. Intratumoural administration of XCL1 has also been shown to promote tumour rejection122. This is in line with observations in mice that natural killer cells recruit cDC1s to tumours through expression of XCL1 (REF.123).
Despite compelling evidence that cDC1s are the main cell type that mediates cross-presentation of cell-associated antigens in vivo, most studies of the molecular mechanisms of cross-presentation have been carried out using DCs derived in vitro from monocytes and bone marrow with granulocyte–macrophage colony-stimulating factor (GM–CSF) and IL-4 (known as GMDCs)124. Although these studies have been useful to advance our understanding of antigen processing and presentation, they do not necessarily recapitulate the cell-intrinsic functions of cDC1s in vivo (BOX 3). Therefore, we limit discussion here to a few recent studies that have identified genes that regulate cross-presentation by cDC1s in vivo. It was recently shown that an integral membrane protein, WDFY4, is specifically expressed by cDC1s and is required for the cross-presentation of cell-associated antigens to CD8+ T cells. cDC1s develop normally in Wdfy4−/− mice, but these mice have many of the same immunological defects as Batf3−/− mice125. It was hypothesized that WDFY4 regulates vesicular trafficking necessary for antigen processing, but the large size of the Wdfy4 gene has so far limited genetic, biochemical and structural analyses of its function. Another protein that is putatively involved in vesicular trafficking and that is required for cross-presentation by cDC1s in vivo is RAB43 (REF.126). Notably, GMDCs derived from Rab43−/− mice have no defect in cross-presentation, which further underscores the importance of distinguishing cross-presentation by cDC1s from that by GMDCs. Along these lines, the vesicular trafficking protein SEC22B was initially studied in GMDCs and shown to be required for cross-presentation through the regulation of phagosome maturation127. Conflicting results between groups have made it difficult to establish whether SEC22B is essential for cross-presentation in vivo, but preliminary results suggest that it may be necessary for antitumour immune responses128,129.
Box 3 |. Cross-presentation by BMDCs, GMDCs and MoDCs.
Significant effort has been made to identify the cell types and molecules that regulate cross-presentation. Whereas the focus of this Review is limited to the ontogeny and function of classical dendritic cells (cDCs) and plasmacytoid DCs, various cell types, such as monocytes and bone marrow-derived DCs (BMDCs), can be cultured in vitro with granulocyte–macrophage colony-stimulating factor (GM–CSF) alone or plus IL-4 to generate GMDCs with cross-presentation capacities241,242. Similarly, it has been suggested that monocytes can differentiate into DCs known as monocyte-derived DCs (MoDCs) that have the capacity to cross-present antigens in vivo243. However, when compared side by side with type 1 classical dendritic cells (cDC1s), GMDCs and MoDCs are phenotypically distinct and inferior at priming Tcells244. Unlike cDC1s, they do not require Batf3 for development but depend on alternative factors, such as Irf4, Ahr and Mafb in mice and humans126,245–247. The putative functions attributed to GMDCs, BMDCs and MoDCs have been recently reviewed by others and remain a matter of contentious debate248,249. Regardless of such controversy, monocytes remain a target antigen-presenting cell population for the development of novel immunotherapies250.
Type II immune responses.
On recognition of parasites or activation by allergens, cytokines such as IL-4, IL-5, IL-13 and IL-10 are produced, and naive CD4+ T cells are polarized to T helper 2 cells.
Type III immune responses.
On recognition of extracellular bacterial pathogens, cytokines such as IL-6, IL-17, IL-21, IL-22, IL-23 and transforming growth factor-β are produced, and naive T cells are polarized to T helper 17 cells.
Non-redundant functions of cDC2s in mice.
Multiple functional subsets of cDC2s have been identified, all of which are characterized by high relative levels of IRF4 expression130. cDC2s are an ontogenetically separate fraction of cDCs with subset-specific functions that are unable to compensate for cDC1 deficiencies. Here, we review various cDC2-intrinsic genetic deficiencies that have aided in the separation of cDC2s into discrete subsets. The broad functions of cDC2s have been revealed through the analysis of Irf4-deficient mice. A major caveat of this model is that it differentially affects cDC2 subsets in a tissue-specific manner131,132. Notwithstanding this caveat, extensive analysis of the immunological defects associated with Irf4 deficiency has highlighted the importance of cDC2s in the regulation of type II immune responses to allergens and parasites, type III immune responses to extra-cellular pathogens and the gut microbiota, and humoral immune responses to blood-borne antigens133–139.
More recently, analysis of cDC-intrinsic transcription factor deficiencies has enabled functions to be assigned to discrete cDC2 subsets. Notch2-dependent cDC2s are found in the spleen, lung and gut-associated lymphoid tissue140. This subset was also shown to be dependent on Ltbr and Rbpj141. Notch2-dependent cDC2s in the gut regulate type III immune responses to extracellular pathogens, such as Citrobacter rodentium140. cDC2-intrinsic expression of IL-23 activates group 3 innate lymphoid cells, which in turn confer resistance to infection through the production of IL-22 and activation of gut epithelial cells142. In the spleen, a Notch2-dependent cDC2 subset surveys the circulation for soluble antigens and is required for germinal centre formation and anti-body production. In the absence of this cDC2 subset, immunization with sheep red blood cells or heat-killed Listeria fails to induce the proliferation of T follicular helper cells or germinal centre B cells143. By contrast, the Klf4-dependent cDC2 subset migrates from barrier surfaces to draining lymph nodes, where these cells regulate type II responses to parasites and allergens144.
Antitumour immune responses and the effects of checkpoint blockade require intact CD8+ and CD4+ T cell responses114,145. Although a role for cDC1-mediated antigen presentation on MHC class II molecules and priming of CD4+ T cells has not been ruled out, non-redundant roles for cDC2s in priming CD4+ T cells are well established146. Consistent with these observations, tumour antigen-bearing cDC2s isolated from the tumour microenvironment can prime antigen-specific CD4+ T cells ex vivo. Ablation of CD11B+ cDC2s in tumour-draining lymph nodes of Cx3cr1LSL–DTRItgaxCre mice (in which diphtheria toxin treatment specifically depletes CX3CR1+CD11C+ cells, which include CD103−CD11B+ cDC2s) results in reduced priming and differentiation of naive CD4+ T cells147. To that end, various anticancer immunotherapies may require cDC2s for their efficacy, as reviewed recently by others148.
Diversity and function of mouse and human pDCs.
The non-redundant functions of mouse pDCs have been difficult to uncover owing to a lack of in vivo infection models that require pDCs for survival. Notwithstanding, descriptive studies of pDC activities during immune responses in mice have shown that they are poised for the recognition of viruses and production of type I interferon. The rapid response of pDCs to viral infection is enabled by their expression of pattern recognition receptors, such as TLR7 and TLR9, which recognize single-stranded RNA and CpG dinucleotides, respectively149,150. However, these receptors are also expressed by other immune cell lineages, such as monocytes, B cells and cDCs, which may provide a redundant source of innate recognition during viral infection43. It is unclear whether a redundant source of type I interferon exists in humans, as patients with Pitt–Hopkins syndrome have a deficiency in pDC development that correlates with impaired IFNα production57.
Several models have been developed to specifically ablate pDCs (TABLE 2), which show that pDCs have a role in the control of acute and chronic viral infections. Conditional deletion of the transcription factor Tcf4 in ItgaxCre mice results in steady-state loss of pDCs in vivo57. Mice lacking pDCs in this model fail to control acute viral hepatitis or infection with chronic lymphocytic choriomeningitis virus151. A widely used model of pDC ablation is based on pDC-specific expression of the human gene encoding CLEC4C (also known as BDCA2 and CD303). Although this gene is not conserved in mice, transcriptional activation by its promoter drives pDC-intrinsic expression of diphtheria toxin receptor (DTR) when inserted as a transgene152. Several studies using this model of pDC deficiency have shown that pDCs contribute to the control of viral infection by regulating the population expansion of natural killer cells and antigen-specific CTLs and provide a local source of type I interferon that can induce cDC1 maturation105,152,153. CLEC4C-based or Tcf4-based models have provided strong evidence to validate long-standing claims that pDCs contribute to autoimmunity, most notably in models of systemic lupus erythematosus154–157. Unlike cDCs, which exit the cell cycle in peripheral tissues and secondary lymphoid organs, pDCs undergo terminal differentiation before exiting the bone marrow, which depends on the transcription factor RUNX2 (REFs41,158,159). The maintenance of pDC function as measured by type I interferon production is, in turn, regulated by IRF8 in terminally differentiated cells42.
The role of cross-presentation by pDCs and translation of mouse models to human pDC function remain active and unresolved areas of debate160. In mice and humans, both cDC1s and pDCs have been shown to cross-present antigens161–164. However, in vivo immune responses in mice that require cross-presentation for survival do not depend on an intact pDC population165. Furthermore, when human and mouse cDC1s are compared side by side with pDCs, cDC1s are far superior in terms of their ability to cross-present cell-associated antigens to CTLs100,166. Reports that have shown the ability of pDCs to cross-present have used soluble peptides of varying lengths as model antigens164,167. It is known that the molecular mechanisms for the cross-presentation of soluble antigens are distinct from those for the cross-presentation of cell-associated antigens. The latter requires direct transfer of antigens from infected, apoptotic or necrotic cells to the antigen-presenting cell, followed by processing and presentation162. Despite the controversies regarding non-redundant functions of pDCs, several groups are working to harness the ability of pDCs to produce type I interferon and internalize exogenous antigens for novel vaccines and immunotherapies168–171.
Most recently, the discovery of heterogeneity in the classically defined human pDC population (HLA-DR+CD45RA+CD123+) has called into question whether the functions that have been attributed to human pDCs could be the result of contamination with a functionally distinct DC subset with cDC-like characteristics. Two independent groups identified a DC population, referred to here as AS-DC, that was AXL+SIGLEC6+CD123+CD11Clow (REFs172,173). Another group identified a similar CD33+CX3CR1+ population that was shown to contaminate classically defined pDCs174. This population had phenotypical similarities to a previously described human pre-DC progenitor, leading the last group to conclude that AS-DCs are actually DC progenitors with a phenotype similar to pre-cDCs76,119. When this population was excluded, functions that were formerly attributed to pDCs, such as priming of naive T cells and IL-12p40 production, could not be recapitulated164,175. Subsequently, a putative mouse homologue of the AS-DC was reported and named a transitional DC, although with limited genetic evidence to support its ontogeny176. Regardless of AS-DC ontogeny, the identification of phenotypical and functional heterogeneity within human pDC populations revives the question of whether they are a non-redundant subset of antigen-presenting cells.
Diversity and function of human cDC1s.
Only one functional subset of cDC1s has so far been identified in mice and humans. Markers used to identify human cDC1s in vivo and in vitro include XCR1, CADM1, thrombomodulin (also known as CD141), CLEC9A and amino-peptidase N (also known as CD13). The expression of lineage-defining transcription factors is conserved between mouse and human cDC1s, including IRF8, BATF3, ID2 and ZBTB46 (REFs174,177–180). The requirement for IRF8 in cDC1 development and survival is evolutionarily conserved between mice and humans, as supported by the observation that rare mutations in the human IRF8 locus result in cDC1 deficiency64. As discussed for mice, the effects of IRF8 deficiency in humans extend beyond the cDC1 lineage, resulting in multilineage defects and immunodeficiency63. The ability to generate cDC1-like cells from human fibroblasts using a protocol that involves the overexpression of SPI1, IRF8 and BATF3 lends further support to the assertion that cDC1 development and ontogeny are conserved between mice and humans181.
The major in vivo functions attributed to cDC1s in mice are consistent with the descriptions of human cDC1 activities so far reported, although there is some divergence in cytokine expression profiles. For example, several groups have observed that human cDC1s produce less IL-12 than cDC2s, which is the opposite to mouse models in which cDC1s have been shown to be the obligate source of IL-12 for certain infections94,166,182,183. However, there is no effect on the production of IL-12 by leukocytes from patients with SPPL2A mutations, who lack cDC2s (REF.184). This result suggests that IL-12 can be produced outside the cDC2 compartment. However, these patients have a defect in T helper 1 cell-mediated responses to mycobacteria, highlighting the possibility that cDC2s can regulate type I immune responses, a function that is restricted to cDC1s in mice184. The superior ability of cDC1s to cross-present cell-associated antigens seems to be conserved between mice and humans86,100,182,185,186. In both mice and humans, CD8+ T cells and natural killer cells produce XCL1, which supports a role for XCR1+ cDC1s in CTL responses187. This is consistent with observations in humans that the size of cDC1 populations positively correlates with CTL-dependent antitumour and antiviral immune responses123,185,188–190.
Type I immune responses.
On recognition of viral or intracellular bacterial pathogens, cytokines such as IL-2, IL-12 and interferon-γ are produced, and naive CD8+ and CD4+ T cells are polarized to cytotoxic T lymphocytes and T helper 1 cells, respectively.
Diversity and function of human cDC2s.
Defining cDC2 subset diversity is complicated by the highly variable expression of surface markers across tissues in mice and humans. Heterogeneity in surface marker expression within human cDC2s has been dissected using high-dimensional flow cytometry and single-cell RNA sequencing by various groups. Subsets of human cDC2s can be identified using CD1A, CD1C, CD1D, CD172A, dipeptidyl peptidase 4 (also known as CD26), CLEC4C, FcεRIα, HLA-DQ and CD163 (REFs130,172,174,191,192). Multiple studies have documented overlapping surface phenotypes between cDC2s and monocytes, particularly for CD115, CD1C, CD14, CD64, CCR2 and CX3CR1 expression193. Exclusive expression of C5AR1 (also known as CD88) and FcαR (also known as CD89) by human monocytes was recently shown to allow for their separation from human cDC2s, which express FcεRIα uniformly191.
Although CD14 has been used as a lineage marker for monocytes, cDC2s were recently shown to include CD14+ and CD14− subsets in some tissues173,191. Two subsets exist within the CD14− fraction of cDC2s on the basis of CD5 expression, referred to here as cDC2A (CD5+) and cDC2B (CD5−). Comparison of their transcriptomes indicates that cDC2A and cDC2B subsets correspond to populations that have been identified independently and named DC2 and DC3, respectively173. However, the extent to which these populations phenotypically and functionally overlap remains to be examined, and these studies do not provide sufficient evidence to determine the genetic basis for functional and phenotypical heterogeneity in human cDC2 subsets. It should be noted that the identity of a putative DC population, named DC4, has since been revised to a subset of CD16+ monocytes191,194. Functionally, cDC2A and cDC2B subsets were equally capable of inducing type I responses ex vivo, as shown by IFNγ production and the proliferation of naive CD4+ T cells173. cDC2As were poor inducers of type II and type III responses. Under inflammatory conditions, an additional CD14+CD163+ population clustered with CD5− cDC2Bs was identified and correlated with increased production of IL-4 and IL-17A by T helper 2 cells and T helper 17 cells, respectively191. These results are consistent with previous studies in mice and humans showing that cDC2s regulate type II and type III immune responses195. Although a specific cDC2 subset was not identified, human cDC2s can polarize naive CD4+ T cells to a T follicular helper cell phenotype by direct priming and production of polarizing cytokines, such as transforming growth factor-β196.
Recent work that compared the transcriptomes of cDC2s suggests that at least two subsets are ontogenetically conserved between mice and humans197. Reporter analysis of Tbx21 (which encodes the transcription factor T-bet) and Rorc (which encodes nuclear receptor RORγ) expression revealed two cDC2 subsets that were phenotypically and functionally distinct with respect to their induction of polarizing cytokine production by naive CD4+ T cells197. Notably, markers associated with Notch2-dependent and Klf4-dependent cDC2s in mice, such as ESAM and MGLL (also known as MGL2), are also expressed by the human cDC2A and cDC2B subsets, respectively140,141,144. NOTCH2 and KLF4 expression were recently reported to correlate with two subsets of IRF4-expressing human cDC2s, which were CD1C+CD14− (NOTCH2lowKLF4hi) and CD1C−CD14+ (NOTCH2hiKLF4low)191. However, more detailed analyses are required to establish functional differences between human cDC2s on the basis of NOTCH2 and KLF4 expression.
Conclusions
Advances in our understanding of DC development have been essential to the elucidation of specialized DC functions. There is significant overlap between humans and mice in terms of DC development, but caution must be used in interpreting results as the molecular mechanisms that regulate DC physiology may not be evolutionarily conserved. Along these lines, the discovery of human mutations in genes that regulate DC development and function has provided novel links to blood disorders, immunodeficiencies and autoimmunity62. Mouse models of DC development have established cDC1s as potent regulators of type I immune responses through interactions with the innate and adaptive immune systems. Therefore, the functions of cDC1s are central to the development of immunotherapies that modulate CTL responses to prevent and treat viral infections and cancer148. In mice and humans, cDC2s can regulate humoral, type I, type II and type III immune responses through antigen presentation and polarizing cytokine production. There are several differences between mice and humans in terms of the behaviour of cDC2s, particularly with respect to type I immune responses. Recent advances in the description of human cDC2 diversity should provide novel insights into the non-redundant functions of human cDC2s. Although the non-redundant functions of human and mouse pDCs and GMDCs remain controversial, substantial efforts have been made to harness their functions ex vivo. Continued research in the field of DC development and function should advance models of the underlying cellular and molecular mechanisms that are used by DCs to maintain homeostasis and regulate immune responses to diverse pathogens and cancers.
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Immunology thanks S. Naik, C. Reis e Sousa and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
References
- 1.Guilliams M et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol. 14, 571–578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vu Manh TP, Bertho N, Hosmalin A, Schwartz-Cornil I & Dalod M Investigating evolutionary conservation of dendritic cell subset identity and functions. Front. Immunol. 6, 260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reizis B Plasmacytoid dendritic cells: development, regulation, and function. Immunity 50, 37–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laurenti E & Gottgens B From haematopoietic stem cells to complex differentiation landscapes. Nature 553, 418–426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dress RJ et al. Plasmacytoid dendritic cells develop from Ly6D+ lymphoid progenitors distinct from the myeloid lineage. Nat. Immunol. 20, 852–864 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Grajales-Reyes GE et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α+ conventional DC clonogenic progenitor. Nat. Immunol. 16, 708–717 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodrigues PF et al. Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat. Immunol. 19, 711–722 (2018). Together with Dress et al. (2019), this study defines mouse pre-pDCs as a progenitor population within the lymphoid compartment on the basis of IL-7Rα and Ly6D expression.
- 8. Schlitzer A et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 16, 718–728 (2015). Together with Grajales-Reyes et al. (2015), this study defines mouse pre-cDC1s and pre-cDC2s as bone marrow progenitors found within the previously defined, but heterogeneous, pre-cDC population.
- 9.Notta F et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 351, aab2116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee J et al. Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat. Immunol. 18, 877–888 (2017). This work suggests that DC specification occurs as early as HSCs through an unknown mechanism of lineage priming that correlates with IRF8 expression levels.
- 11.Bernitz JM, Kim HS, MacArthur B, Sieburg H & Moore K Hematopoietic stem cells count and remember self-renewal divisions. Cell 167, 1296–1309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naik SH et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature 496, 229–232 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Naik SH, Schumacher TN & Perie L Cellular barcoding: a technical appraisal. Exp. Hematol. 42, 598–608 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Velten L et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 19, 271–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurotaki D et al. Epigenetic control of early dendritic cell lineage specification by the transcription factor IRF8 in mice. Blood 133, 1803–1813 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goode DK et al. Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev. Cell 36, 572–587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothenberg EV Transcriptional control of early T and B cell developmental choices. Annu. Rev. Immunol. 32, 283–321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinreb C, Rodriguez-Fraticelli A, Camargo FD & Klein AM Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science 367, eaaw3381 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akashi K, Traver D, Miyamoto T & Weissman IL A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193–197 (2000). This work provides the first description of the mouse CMP as the precursor of all myeloid populations.
- 20. Kondo M, Weissman IL & Akashi K Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91, 661–672 (1997). This work provides the first description of the mouse CLP as the precursor of all lymphoid populations.
- 21.Iwasaki H & Akashi K Myeloid lineage commitment from the hematopoietic stem cell. Immunity 26, 726–740 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Iwasaki H et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106, 1590–1600 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klemsz MJ, McKercher SR, Celada A, Van Beveren C & Maki RA The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell 61, 113–124 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Busslinger M Transcriptional control of early B cell development. Annu. Rev. Immunol. 22, 55–79 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Hoppe PS et al. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature 535, 299–302 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Kueh HY, Champhekar A, Nutt SL, Elowitz MB & Rothenberg EV Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science 341, 670–673 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Fraticelli AE et al. Clonal analysis of lineage fate in native haematopoiesis. Nature 553, 212–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fogg DK et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 311, 83–87 (2006). This work defines the mouse MDP as a bone marrow progenitor with monocyte, macrophage and DC potential.
- 29. Hettinger J et al. Origin of monocytes and macrophages in a committed progenitor. Nat. Immunol. 14, 821–830 (2013). This work defines the mouse cMoP as a bone marrow-resident clonogenic progenitor of mouse monocytes.
- 30.Sathe P et al. Lymphoid tissue and plasmacytoid dendritic cells and macrophages do not share a common macrophage–dendritic cell-restricted progenitor. Immunity 41, 104–115 (2014). [DOI] [PubMed] [Google Scholar]
- 31. Liu Z et al. Fate mapping via Ms4a3-expression history traces monocyte-derived cells. Cell 178, 1509–1525 (2019). This work describes populations that are derived from the GMP in mice on the basis of lineage tracing driven by Ms4a3 expression.
- 32.Olsson A et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698–702 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanez A et al. Granulocyte–monocyte progenitors and monocyte–dendritic cell progenitors independently produce functionally distinct monocytes. Immunity 47, 890–902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X et al. Mafb lineage tracing to distinguish macrophages from other immune lineages reveals dual identity of Langerhans cells. J. Exp. Med. 213, 2553–2565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satpathy AT et al. Runx1 and Cbfβ regulate the development of Flt3+ dendritic cell progenitors and restrict myeloproliferative disorder. Blood 123, 2968–2977 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang P et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBPα. Immunity 21, 853–863 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Becker AM et al. IRF-8 extinguishes neutrophil production and promotes dendritic cell lineage commitment in both myeloid and lymphoid mouse progenitors. Blood 119, 2003–2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurotaki D et al. IRF8 inhibits C/EBPα activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nat. Commun. 5, 4978 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Kurotaki D et al. Transcription factor IRF8 governs enhancer landscape dynamics in mononuclear phagocyte progenitors. Cell Rep. 22, 2628–2641 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Naik SH et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8, 1217–1226 (2007). [DOI] [PubMed] [Google Scholar]
- 41. Onai N et al. Identification of clonogenic common Flt3+ M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8, 1207–1216 (2007). This work describes the CDP as a population giving rise to pDCs, cDC1s and cDC2s in vivo.
- 42. Sichien D et al. IRF8 transcription factor controls survival and function of terminally differentiated conventional and plasmacytoid dendritic cells, respectively. Immunity 45, 626–640 (2016). This work defines the role of IRF8 in the function and development of terminally differentiated populations of DCs and shows that pDC development is not dependent on IRF8, as was suggested by earlier studies.
- 43.Miller JC et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13, 888–899 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naik SH et al. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat. Immunol. 7, 663–671 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Schraml BU et al. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell 154, 843–858 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Liu K et al. In vivo analysis of dendritic cell development and homeostasis. Science 324, 392–397 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cabeza-Cabrerizo M et al. Tissue clonality of dendritic cell subsets and emergency DCpoiesis revealed by multicolor fate mapping of DC progenitors. Sci. Immunol. 4, eaaw1941 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hacker C et al. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat. Immunol. 4, 380–386 (2003). [DOI] [PubMed] [Google Scholar]
- 49. Hildner K et al. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008). This work reports the first genetic model in mice that specifically ablates cDC1 development in vivo.
- 50.Kashiwada M, Pham NL, Pewe LL, Harty JT & Rothman PB NFIL3/E4BP4 is a key transcription factor for CD8α+ dendritic cell development. Blood 117, 6193–6197 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seillet C et al. CD8α+ DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3. Blood 121, 1574–1583 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Theisen DJ et al. Batf3-dependent genes control tumor rejection induced by dendritic cells independently of cross-presentation. Cancer Immunol. Res. 7, 29–39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Durai V et al. Cryptic activation of an Irf8 enhancer governs cDC1 fate specification. Nat. Immunol. 20, 1161–1173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bagadia P et al. An Nfil3–Zeb2–Id2 pathway imposes Irf8 enhancer switching during cDC1 development. Nat. Immunol. 20, 1174–1185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott CL et al. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J. Exp. Med. 213, 897–911 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu X et al. Transcription factor Zeb2 regulates commitment to plasmacytoid dendritic cell and monocyte fate. Proc. Natl Acad. Sci. USA 113, 14775–14780 (2016). Together with Scott et al. (2016), this work demonstrates a role for ZEB2 in mouse pDC development.
- 57. Cisse B et al. Transcription factor E2–2 is an essential and specific regulator of plasmacytoid dendritic cell development 1. Cell 135, 37–48 (2008). This work reports the dependence of pDC development on the transcription factor TCF4 in mice and humans.
- 58.Grajkowska LT et al. Isoform-specific expression and feedback regulation of E protein TCF4 control dendritic cell lineage specification. Immunity 46, 65–77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun XH, Copeland NG, Jenkins NA & Baltimore D Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix–loop–helix proteins. Mol. Cell Biol. 11, 5603–5611 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dickinson RE et al. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood 118, 2656–2658 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cytlak U et al. Ikaros family zinc finger 1 regulates dendritic cell development and function in humans. Nat. Commun. 9, 1239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bigley V, Cytlak U & Collin M Human dendritic cell immunodeficiencies. Semin. Cell Dev. Biol. 86, 50–61 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Bigley V et al. Biallelic interferon regulatory factor 8 mutation: a complex immunodeficiency syndrome with dendritic cell deficiency, monocytopenia, and immune dysregulation. J. Allergy Clin. Immunol. 141, 2234–2248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hambleton S et al. IRF8 mutations and human dendritic-cell immunodeficiency. N. Engl. J. Med. 365, 127–138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salem S et al. Functional characterization of the human dendritic cell immunodeficiency associated with the IRF8(K108E) mutation. Blood 124, 1894–1904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doulatov S et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 11, 585–593 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Chicha L, Jarrossay D & Manz MG Clonal type I interferon-producing and dendritic cell precursors are contained in both human lymphoid and myeloid progenitor populations. J. Exp. Med. 200, 1519–1524 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galy A, Travis M, Cen D & Chen B Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity 3, 459–473 (1995). [DOI] [PubMed] [Google Scholar]
- 69.Ishikawa F et al. The developmental program of human dendritic cells is operated independently of conventional myeloid and lymphoid pathways. Blood 110, 3591–3660 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geissmann F et al. Development of monocytes, macrophages, and dendritic cells. Science 327, 656–661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Helft J et al. Dendritic cell lineage potential in human early hematopoietic progenitors. Cell Rep. 20, 529–537 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee J et al. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J. Exp. Med. 212, 385–399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kawamura S et al. Identification of a human clonogenic progenitor with strict monocyte differentiation potential: a counterpart of mouse cMoPs. Immunity 46, 835–848 (2017). This work identifies a human counterpart to the mouse cMoP.
- 74.Buenrostro JD et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 173, 1535–1548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu YP et al. Identification of an early unipotent neutrophil progenitor with pro-tumoral activity in mouse and human bone marrow. Cell Rep. 24, 2329–2341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Breton G et al. Circulating precursors of human CD1c+ and CD141+ dendritic cells. J. Exp. Med. 212, 401–413 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee J et al. Clonal analysis of human dendritic cell progenitor using a stromal cell culture. J. Immunol. Methods 425, 21–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ghosh HS, Cisse B, Bunin A, Lewis KL & Reizis B Continuous expression of the transcription factor E2–2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity 33, 905–916 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu X et al. Bcl11a controls Flt3 expression in early hematopoietic progenitors and is required for pDC development in vivo. PLoS ONE 8, e64800 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Inaba K, Young JW & Steinman RM Direct activation of CD8+ cytotoxic T lymphocytes by dendritic cells. J. Exp. Med. 166, 182–194 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kronin V et al. A subclass of dendritic cells regulates the response of naive CD8 T cells by limiting their IL-2 production. J. Immunol. 157, 3819–3827 (1996). [PubMed] [Google Scholar]
- 82.Bevan MJ Cross-priming for a secondary cytotoxic response to minor H antigens with H-2 congenic cells which do not cross-react in the cytotoxic assay. J. Exp. Med. 143, 1283–1288 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.den Haan JM, Lehar SM & Bevan MJ CD8+ but not CD8− dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 192, 1685–1696 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pooley JL, Heath WR & Shortman K Cutting edge: intravenous soluble antigen is presented to CD4 T cells by CD8− dendritic cells, but cross-presented to CD8 T cells by CD8+ dendritic cells. J. Immunol. 166, 5327–5330 (2001). [DOI] [PubMed] [Google Scholar]
- 85.Atif SM et al. Cutting edge: roles for Batf3-dependent APCs in the rejection of minor histocompatibility antigen-mismatched grafts. J. Immunol. 195, 46–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bachem A et al. Expression of XCR1 characterizes the Batf3-dependent lineage of dendritic cells capable of antigen cross-presentation. Front. Immunol. 3, 214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martinez-Lopez M, Iborra S, Conde-Garrosa R & Sancho D Batf3-dependent CD103+ dendritic cells are major producers of IL-12 that drive local TH1 immunity against Leishmania major infection in mice. Eur. J. Immunol. 45, 119–129 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanchez-Paulete AR et al. Cancer immunotherapy with immunomodulatory anti-CD137 and anti-PD-1 monoclonal antibodies requires BATF3-dependent dendritic cells. Cancer Discov. 6, 71–79 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spranger S, Dai D, Horton B & Gajewski TF Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell 31, 711–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Becker M et al. Ontogenic, phenotypic, and functional characterization of XCR1+ dendritic cells leads to a consistent classification of intestinal dendritic cells based on the expression of XCR1 and SIRPα. Front. Immunol. 5, 326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roberts EW et al. Critical role for CD103+/CD141+ dendritic cells bearing CCR7 for tumour antigen trafficking and priming of T cell immunity in melanoma. Cancer Cell 30, 324–336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tussiwand R et al. Compensatory dendritic cell development mediated by BATF–IRF interactions. Nature 490, 502–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schonheit J et al. PU.1 level-directed chromatin structure remodeling at the Irf8 gene drives dendritic cell commitment. Cell Rep. 3, 1617–1628 (2013). [DOI] [PubMed] [Google Scholar]
- 94. Mashayekhi M et al. CD8a+ dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity 35, 249–259 (2011). This work uses the model of Batf3 deficiency to confirm early reports of cDC1s as a source of IL-12 during innate immune responses in mice.
- 95.Askenase MH et al. Bone-marrow-resident NK cells prime monocytes for regulatory function during infection. Immunity 42, 1130–1142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Diebold SS et al. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature 424, 324–328 (2003). [DOI] [PubMed] [Google Scholar]
- 97.Schulz O et al. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature 433, 887–892 (2005). [DOI] [PubMed] [Google Scholar]
- 98.Mattiuz R et al. Novel Cre-expressing mouse strains permitting to selectively track and edit type 1 conventional dendritic cells facilitate disentangling their complexity in vivo. Front. Immunol. 9, 2805 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ohta T et al. Crucial roles of XCR1-expressing dendritic cells and the XCR1–XCL1 chemokine axis in intestinal immune homeostasis. Sci. Rep. 6, 23505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bachem A et al. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 207, 1273–1281 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Crozat K et al. Cutting edge: expression of XCR1 defines mouse lymphoid-tissue resident and migratory dendritic cells of the CD8α+ type. J. Immunol. 187, 4411–4415 (2011). [DOI] [PubMed] [Google Scholar]
- 102.Dorner BG et al. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 31, 823–833 (2009). [DOI] [PubMed] [Google Scholar]
- 103.Janela B et al. A subset of type I conventional dendritic cells controls cutaneous bacterial infections through VEGFα-mediated recruitment of neutrophils. Immunity 50, 1069–1083 (2019). [DOI] [PubMed] [Google Scholar]
- 104.del Fresno C et al. DNGR-1 in dendritic cells limits tissue damage by dampening neutrophil recruitment. Science 362, 351–356 (2018). [DOI] [PubMed] [Google Scholar]
- 105.Brewitz A et al. CD8+ T cells orchestrate pDC–XCR1+ dendritic cell spatial and functional cooperativity to optimize priming. Immunity 46, 205–219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bonifaz LC et al. In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J. Exp. Med. 199, 815–824 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lahoud MH et al. DEC-205 is a cell surface receptor for CpG oligonucleotides. Proc. Natl Acad. Sci. USA 109, 16270–16275 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bozzacco L et al. DEC-205 receptor on dendritic cells mediates presentation of HIV gag protein to CD8+ T cells in a spectrum of human MHC I haplotypes. Proc. Natl Acad. Sci. USA 104, 1289–1294 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Caminschi I et al. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood 112, 3264–3273 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li J et al. Antibodies targeting Clec9A promote strong humoral immunity without adjuvant in mice and non-human primates. Eur. J. Immunol. 45, 854–864 (2015). [DOI] [PubMed] [Google Scholar]
- 111.Tullett KM et al. Targeting CLEC9A delivers antigen to human CD141+ DC for CD4+ and CD8+ T cell recognition. JCI Insight 1, e87102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fossum E et al. Vaccine molecules targeting Xcr1 on cross-presenting DCs induce protective CD8+ T-cell responses against influenza virus. Eur. J. Immunol. 45, 624–635 (2015). [DOI] [PubMed] [Google Scholar]
- 113.Hartung E et al. Induction of potent CD8 T cell cytotoxicity by specific targeting of antigen to cross-presenting dendritic cells in vivo via murine or human XCR1. J. Immunol. 194, 1069–1079 (2015). [DOI] [PubMed] [Google Scholar]
- 114. Gubin MM et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 (2014). This work shows that cDC1s are required for therapeutic effects of checkpoint blockade and antigen-specific tumour rejection.
- 115.Garris CS et al. Successful anti-PD-1 cancer immunotherapy requires T cell-dendritic cell crosstalk involving the cytokines IFN-γ and IL-12. Immunity 49, 1148–1161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Aznar MA et al. Immunotherapeutic effects of intratumoral nanoplexed poly I:C. J. Immunother. Cancer 7, 116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wculek SK et al. Effective cancer immunotherapy by natural mouse conventional type-1 dendritic cells bearing dead tumor antigen. J. Immunother. Cancer 7, 100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang H et al. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc. Natl Acad. Sci. USA 114, 1637–1642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Breton G, Lee J, Liu K & Nussenzweig MC Defining human dendritic cell progenitors by multiparametric flow cytometry. Nat. Protoc. 10, 1407–1422 (2015). This work describes a protocol to isolate human DC progenitors from human bone marrow and cord blood samples.
- 120.Guermonprez P et al. Inflammatory Flt3l is essential to mobilize dendritic cells and for T cell responses during Plasmodium infection. Nat. Med. 19, 730–738 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Salmon H et al. Expansion and activation of CD103+ dendritic cell progenitors at the tumor site enhances tumor responses to therapeutic PD-L1 and BRAF inhibition. Immunity 44, 924–938 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sanchez-Paulete AR et al. Intratumoral immunotherapy with XCL1 and sFlt3L encoded in recombinant Semliki Forest virus-derived vectors fosters dendritic cell-mediated T-cell cross-priming. Cancer Res. 78, 6643–6654 (2018). [DOI] [PubMed] [Google Scholar]
- 123.Bottcher JP et al. NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 172, 1022–1037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Theisen D & Murphy K The role of cDC1s in vivo: CD8 T cell priming through cross-presentation. F1000Res 6, 98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Theisen DJ et al. WDFY4 is required for cross-presentation in response to viral and tumor antigens. Science 362, 694–699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kretzer NM et al. RAB43 facilitates cross-presentation of cell-associated antigens by CD8α+ dendritic cells. J. Exp. Med. 213, 2871–2883 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cebrian I et al. Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell 147, 1355–1368 (2011). [DOI] [PubMed] [Google Scholar]
- 128.Alloatti A et al. Critical role for Sec22b-dependent antigen cross-presentation in antitumor immunity. J. Exp. Med. 214, 2231–2241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu SJ et al. A critical analysis of the role of SNARE protein SEC22B in antigen cross-presentation. Cell Rep. 19, 2645–2656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Guilliams M et al. Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity 45, 669–684 (2016). This work provides a comprehensive analysis and description of all known human and mouse DC subsets so far defined.
- 131.De Silva NS, Simonetti G, Heise N & Klein U The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol. Rev. 247, 73–92 (2012). [DOI] [PubMed] [Google Scholar]
- 132.Huber M & Lohoff M IRF4 at the crossroads of effector T-cell fate decision. Eur. J. Immunol. 44, 1886–1895 (2014). [DOI] [PubMed] [Google Scholar]
- 133.Calabro S et al. Bridging channel dendritic cells induce immunity to transfused red blood cells. J. Exp. Med. 213, 887–896 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kumamoto Y et al. CD301b+ dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity 39, 733–743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Linehan JL et al. Generation of TH17 cells in response to intranasal infection requires TGF-β1 from dendritic cells and IL-6 from CD301b+ dendritic cells. Proc. Natl Acad. Sci. USA 112, 12782–12787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Persson EK et al. IRF4 transcription-factor-dependent CD103+CD11b+ dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 38, 958–969 (2013). [DOI] [PubMed] [Google Scholar]
- 137.Plantinga M et al. Conventional and monocyte-derived CD11b+ dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity 38, 322–335 (2013). [DOI] [PubMed] [Google Scholar]
- 138.Reboldi A et al. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science 352, aaf4822 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schlitzer A et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity 38, 970–983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Satpathy AT et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14, 937–948 (2013). This work provides a genetic model to establish a role for cDC2 subsets in type III immune responses.
- 141.Lewis KL et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 35, 780–791 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.von Moltke J, Ji M, Liang HE & Locksley RM Tuft-cell-derived IL-25 regulates an intestinal ILC2–epithelial response circuit. Nature 529, 221–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Briseno CG et al. Notch2-dependent DC2s mediate splenic germinal center responses. Proc. Natl Acad. Sci. USA 115, 10726–10731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Tussiwand R et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 42, 916–928 (2015). This work provides a genetic model to establish a role for cDC2 subsets in type II immune responses.
- 145.Alspach E et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 574, 696–671 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bennett SR, Carbone FR, Karamalis F, Miller JF & Heath WR Induction of a CD8+ cytotoxic T lymphocyte response by cross-priming requires cognate CD4+ T cell help. J. Exp. Med. 186, 65–70 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Binnewies M et al. Unleashing type-2 dendritic cells to drive protective antitumor CD4+ T cell immunity. Cell 177, 556–571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wculek SK et al. Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 20, 7–24 (2020). [DOI] [PubMed] [Google Scholar]
- 149.Diebold SS, Kaisho T, Hemmi H, Akira S & Sousa CRE Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531 (2004). [DOI] [PubMed] [Google Scholar]
- 150.Sasai M, Linehan MM & Iwasaki A Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science 329, 1530–1534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cervantes-Barragan L et al. Plasmacytoid dendritic cells control T-cell response to chronic viral infection. Proc. Natl Acad. Sci. USA 109, 3012–3017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Swiecki M, Gilfillan S, Vermi W, Wang Y & Colonna M Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8+ T cell accrual. Immunity 33, 955–966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Swiecki M, Wang Y, Gilfillan S & Colonna M Plasmacytoid dendritic cells contribute to systemic but not local antiviral responses to HSV infections. PLoS Pathog. 9, e1003728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ah K et al. Plasmacytoid dendritic cells promote systemic sclerosis with a key role for TLR8. Sci. Transl. Med. 10, eaam8458 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Davison LM & Jorgensen TN Sialic acid-binding immunoglobulin-type lectin H-positive plasmacytoid dendritic cells drive spontaneous lupus-like disease development in B6.Nba2 mice. Arthritis Rheumatol. 67, 1012–1022 (2015). [DOI] [PubMed] [Google Scholar]
- 156.Rowland SL et al. Early, transient depletion of plasmacytoid dendritic cells ameliorates autoimmunity in a lupus model. J. Exp. Med. 211, 1977–1991 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sisirak V et al. Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J. Exp. Med. 211, 1969–1976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chopin M et al. RUNX2 mediates plasmacytoid dendritic cell egress from the bone marrow and controls viral immunity. Cell Rep. 15, 866–878 (2016). [DOI] [PubMed] [Google Scholar]
- 159.Sawai CM et al. Transcription factor Runx2 controls the development and migration of plasmacytoid dendritic cells. J. Exp. Med. 210, 2151–2159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Villadangos JA & Young L Antigen-presentation properties of plasmacytoid dendritic cells. Immunity 29, 352–361 (2008). [DOI] [PubMed] [Google Scholar]
- 161.Di Pucchio T et al. Direct proteasome-independent cross-presentation of viral antigen by plasmacytoid dendritic cells on major histocompatibility complex class I. Nat. Immunol. 9, 551–557 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Joffre OP, Segura E, Savina A & Amigorena S Cross-presentation by dendritic cells. Nat. Rev. Immunol. 12, 557–569 (2012). [DOI] [PubMed] [Google Scholar]
- 163.Mouries J et al. Plasmacytoid dendritic cells efficiently cross-prime naive T cells in vivo after TLR activation. Blood 112, 3713–3722 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Segura E, Durand M & Amigorena S Similar antigen cross-presentation capacity and phagocytic functions in all freshly isolated human lymphoid organ-resident dendritic cells. J. Exp. Med. 210, 1035–1047 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.GeurtsvanKessel CH et al. Clearance of influenza virus from the lung depends on migratory langerin+CD11b− but not plasmacytoid dendritic cells. J. Exp. Med. 205, 1621–1634 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mittag D et al. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J. Immunol. 186, 6207–6217 (2011). [DOI] [PubMed] [Google Scholar]
- 167.Aspord C, Leloup C, Reche S & Plumas J pDCs efficiently process synthetic long peptides to induce functional virus- and tumour-specific T-cell responses. Eur. J. Immunol. 44, 2880–2892 (2014). [DOI] [PubMed] [Google Scholar]
- 168.Hoeffel G et al. Antigen crosspresentation by human plasmacytoid dendritic cells. Immunity 27, 481–492 (2007). [DOI] [PubMed] [Google Scholar]
- 169.Kranz LM et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016). [DOI] [PubMed] [Google Scholar]
- 170.Lui G et al. Plasmacytoid dendritic cells capture and cross-present viral antigens from influenza-virus exposed cells. PLoS ONE 4, e7111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Perez CR & De Palma M Engineering dendritic cell vaccines to improve cancer immunotherapy. Nat. Commun. 10, 5408 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Alcantara-Hernandez M et al. High-dimensional phenotypic mapping of human dendritic cells reveals interindividual variation and tissue specialization. Immunity 47, 1037–1050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Villani AC et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, 283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.See P et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science 356, 1044 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Krug A et al. Toll-like receptor expression reveals CpG DNA as a unique microbial stimulus for plasmacytoid dendritic cells which synergizes with CD40 ligand to induce high amounts of IL-12. Eur. J. Immunol. 31, 3026–3037 (2001). [DOI] [PubMed] [Google Scholar]
- 176.Leylek R et al. Integrated cross-species analysis identifies a conserved transitional dendritic cell population. Cell Rep. 29, 3736–3750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Breton G et al. Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs. J. Exp. Med. 213, 2861–2870 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Carpentier S et al. Comparative genomics analysis of mononuclear phagocyte subsets confirms homology between lymphoid tissue-resident and dermal XCR1+ DCs in mouse and human and distinguishes them from Langerhans cells. J. Immunol. Methods 432, 35–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Meredith MM et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J. Exp. Med. 209, 1153–1165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Robbins SH et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 9, R17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rosa FF et al. Direct reprogramming of fibroblasts into antigen-presenting dendritic cells. Sci. Immunol. 3, eaau4292 (2018). [DOI] [PubMed] [Google Scholar]
- 182.Haniffa M et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity 37, 60–73 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nizzoli G et al. Human CD1c+ dendritic cells secrete high levels of IL-12 and potently prime cytotoxic T-cell responses. Blood 122, 932–942 (2013). [DOI] [PubMed] [Google Scholar]
- 184.Kong XF et al. Disruption of an antimycobacterial circuit between dendritic and helper T cells in human SPPL2a deficiency. Nat. Immunol. 19, 973–985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jongbloed SL et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 207, 1247–1260 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Crozat K et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8α+ dendritic cells. J. Exp. Med. 207, 1283–1292 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kroczek RA & Henn V The role of XCR1 and its ligand XCL1 in antigen cross-presentation by murine and human dendritic cells. Front. Immunol. 3, 14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Meyer MA et al. Breast and pancreatic cancer interrupt IRF8-dependent dendritic cell development to overcome immune surveillance. Nat. Commun. 9, 1250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zilionis R et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 50, 1317–1334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Silvin A et al. Constitutive resistance to viral infection in human CD141+ dendritic cells. Sci. Immunol 2, eaai8071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dutertre CA et al. Single-cell analysis of human mononuclear phagocytes reveals subset-defining markers and identifies circulating inflammatory dendritic cells. Immunity 51, 573–589 (2019). [DOI] [PubMed] [Google Scholar]
- 192.Granot T et al. Dendritic cells display subset and tissue-specific maturation dynamics over human life. Immunity 46, 504–515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Richter L et al. Transcriptional profiling reveals monocyte-related macrophages phenotypically resembling DC in human intestine. Mucosal Immunol. 11, 1512–1523 (2018). [DOI] [PubMed] [Google Scholar]
- 194.Calzetti F et al. Human dendritic cell subset 4 (DC4) correlates to a subset of CD14dim/–CD16++ monocytes. J. Allergy Clin. Immunol. 141, 2276–2279 (2018). [DOI] [PubMed] [Google Scholar]
- 195.Collin M & Bigley V Human dendritic cell subsets: an update. Immunology 154, 3–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Durand M et al. Human lymphoid organ cDC2 and macrophages play complementary roles in T follicular helper responses. J. Exp. Med. 216, 1561–1581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Brown CC et al. Transcriptional basis of mouse and human dendritic cell heterogeneity. Cell 179, 846–863 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Caton ML, Smith-Raska MR & Reizis B Notch–RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J. Exp. Med. 204, 1653–1664 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Griffin MD et al. Dendritic cell modulation by 1α,25 dihydroxyvitamin D3 and its analogs: a vitamin D receptor-dependent pathway that promotes a persistent state of immaturity in vitro and in vivo. Proc. Natl Acad. Sci. USA 98, 6800–6805 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Griffin MD, Dong X & Kumar R Vitamin D receptor-mediated suppression of RelB in antigen presenting cells: a paradigm for ligand-augmented negative transcriptional regulation. Arch. Biochem. Biophys. 460, 218–226 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hochweller K, Striegler J, Hammerling GJ & Garbi N A novel CD11c.DTR transgenic mouse for depletion of dendritic cells reveals their requirement for homeostatic proliferation of natural killer cells. Eur. J. Immunol. 38, 2776–2783 (2008). [DOI] [PubMed] [Google Scholar]
- 202. Jung S et al. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity 17, 211–220 (2002). This work provides a widely used model of Cre-mediated gene deletion driven by CD11C (also known as Itgax) expression.
- 203.Stranges PB et al. Elimination of antigen-presenting cells and autoreactive T cells by Fas contributes to prevention of autoimmunity. Immunity 26, 629–641 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Piva L et al. Cutting edge: Clec9A+ dendritic cells mediate the development of experimental cerebral malaria. J. Immunol. 189, 1128–1132 (2012). [DOI] [PubMed] [Google Scholar]
- 205.Bennett CL et al. Inducible ablation of mouse Langerhans cells diminishes but fails to abrogate contact hypersensitivity. J. Cell Biol. 169, 569–576 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kaplan DH, Jenison MC, Saeland S, Shlomchik WD & Shlomchik MJ Epidermal Langerhans cell-deficient mice develop enhanced contact hypersensitivity. Immunity 23, 611–620 (2005). [DOI] [PubMed] [Google Scholar]
- 207.Kissenpfennig A et al. Dynamics and function of Langerhans cells in vivo: dermal dendritic cells colonize lymph node areas distinct from slower migrating Langerhans cells. Immunity 22, 643–654 (2005). [DOI] [PubMed] [Google Scholar]
- 208.Zahner SP et al. Conditional deletion of TGF-βR1 using Langerin-Cre mice results in Langerhans cell deficiency and reduced contact hypersensitivity. J. Immunol. 187, 5069–5076 (2011). [DOI] [PubMed] [Google Scholar]
- 209.Puttur F et al. Absence of Siglec-H in MCMV infection elevates interferon α production but does not enhance viral clearance. PLoS Pathog. 9, e1003648 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Takagi H et al. Plasmacytoid dendritic cells are crucial for the initiation of inflammation and T cell immunity in vivo. Immunity. 35, 958–971 (2011). [DOI] [PubMed] [Google Scholar]
- 211.Yamazaki C et al. Critical roles of a dendritic cell subset expressing a chemokine receptor, XCR1. J. Immunol. 190, 6071–6082 (2013). [DOI] [PubMed] [Google Scholar]
- 212.Alexandre YO et al. XCR1+ dendritic cells promote memory CD8+ T cell recall upon secondary infections with Listeria monocytogenes or certain viruses. J. Exp. Med. 213, 75–92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Muzaki AR et al. Intestinal CD103+CD11b− dendritic cells restrain colitis via IFN-γ-induced anti-inflammatory response in epithelial cells. Mucosal Immunol. 9, 336–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Diehl GE et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature 494, 116–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Jung S et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell Biol. 20, 4106–4114 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Schlenner SM et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity 32, 426–436 (2010). [DOI] [PubMed] [Google Scholar]
- 217.Dai XM et al. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99, 111–120 (2002). [DOI] [PubMed] [Google Scholar]
- 218.Dai XM, Zong XH, Sylvestre V & Stanley ER Incomplete restoration of colony-stimulating factor 1 (CSF-1) function in CSF-1-deficient Csf1op/Csf1op mice by transgenic expression of cell surface CSF-1. Blood 103, 1114–1123 (2004). [DOI] [PubMed] [Google Scholar]
- 219.Loschko J et al. Inducible targeting of cDCs and their subsets in vivo. J. Immunol. Methods 434, 32–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Loschko J et al. Absence of MHC class II on cDCs results in microbial-dependent intestinal inflammation. J. Exp. Med. 213, 517–534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kwon K et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28, 751–762 (2008). [DOI] [PubMed] [Google Scholar]
- 222.Klein U et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7, 773–782 (2006). [DOI] [PubMed] [Google Scholar]
- 223.Kamizono S et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 206, 2977–2986 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jackson JT et al. Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages. EMBO J. 30, 2690–2704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rawlins EL, Clark CP, Xue Y & Hogan BL The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 136, 3741–3745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Katz JP et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129, 2619–2628 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Higashi Y et al. Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis 32, 82–84 (2002). [DOI] [PubMed] [Google Scholar]
- 228.Aliberti J et al. Essential role for ICSBP in the in vivo development of murine CD8α+ dendritic cells. Blood 101, 305–310 (2003). [DOI] [PubMed] [Google Scholar]
- 229. Schiavoni G et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8α+ dendritic cells. J. Exp. Med. 196, 1415–1425 (2002). This work is an early report on the dependence of cDC1 development on IRF8, and the since-corrected observation that pDCs also require IRF8 for development.
- 230.Manz MG, Traver D, Miyamoto T, Weissman IL & Akashi K Dendritic cell potentials of early lymphoid and myeloid progenitors. Blood 97, 3333–3341 (2001). [DOI] [PubMed] [Google Scholar]
- 231.Sathe P, Vremec D, Wu L, Corcoran L & Shortman K Convergent differentiation: myeloid and lymphoid pathways to murine plasmacytoid dendritic cells. Blood 121, 11–19 (2013). [DOI] [PubMed] [Google Scholar]
- 232.D’Amico A & Wu L The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt31. J. Exp. Med. 198, 293–303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wu L et al. Development of thymic and splenic dendritic cell populations from different hemopoietic precursors. Blood 98, 3376–3382 (2001). [DOI] [PubMed] [Google Scholar]
- 234.Onai N et al. A clonogenic progenitor with prominent plasmacytoid dendritic cell developmental potential. Immunity 38, 943–957 (2013). [DOI] [PubMed] [Google Scholar]
- 235.Shigematsu H et al. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity 21, 43–53 (2004). [DOI] [PubMed] [Google Scholar]
- 236.Herman JS, Sagar & Grun, D. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat. Methods 15, 379–386 (2018). [DOI] [PubMed] [Google Scholar]
- 237.Inlay MA et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 23, 2376–2381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Huang J et al. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev. Cell 36, 9–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Okuno Y et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol. Cell Biol. 25, 2832–2845 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Steidl U et al. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J. Clin. Invest. 117, 2611–2620 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Caux C, Dezutter-Dambuyant C, Schmitt D & Banchereau J GM-CSF and TNF-α cooperate in the generation of dendritic Langerhans cells. Nature 360, 258–261 (1992). [DOI] [PubMed] [Google Scholar]
- 242.Inaba K et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 176, 1693–1702 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Jakubzick CV, Randolph GJ & Henson PM Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 17, 349–362 (2017). [DOI] [PubMed] [Google Scholar]
- 244.Tamoutounour S et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 39, 925–938 (2013). [DOI] [PubMed] [Google Scholar]
- 245. Helft J et al. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c+MHCII+ macrophages and dendritic cells. Immunity 42, 1197–1211 (2015). This work presents a contemporary analysis that reveals heterogeneity within GMDC populations.
- 246. Goudot C et al. Aryl hydrocarbon receptor controls monocyte differentiation into dendritic cells versus macrophages. Immunity 47, 582–596 (2017). This work reveals a genetic requirement for Ahr in GMDC development.
- 247. Briseno CG et al. Distinct transcriptional programs control cross-priming in classical and monocyte-derived dendritic cells. Cell Rep. 15, 2462–2474 (2016). This work reveals a genetic requirement for Irf4 but not Batf3 in GMDC development.
- 248.Lutz MB, Inaba K, Schuler G & Romani N Still alive and kicking: in-vitro-generated GM-CSF dendritic cells! Immunity 44, 1–2 (2016). [DOI] [PubMed] [Google Scholar]
- 249.Lutz MB, Strobl H, Schuler G & Romani N GM-CSF monocyte-derived cells and Langerhans cells as part of the dendritic cell family. Front. Immunol. 8, 1388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Huang MN et al. Antigen-loaded monocyte administration induces potent therapeutic antitumor T cell responses. J. Clin. Invest. 130, 774–788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]


