Abstract
“Image-based” computational fluid dynamics (CFD) simulations provide insights into each patient’s hemodynamic environment. However, current standard procedures for creating CFD models start with manual segmentation and are time-consuming, hindering the clinical translation of image-based CFD simulations. This feasibility study adopts deep-learning-based image segmentation (hereafter referred to as Artificial Intelligence (AI) segmentation) to replace manual segmentation to accelerate CFD model creation. Two published convolutional neural network-based AI methods (MIScnn and DeepMedic) were selected to perform CFD model extraction from three-dimensional (3D) rotational angiography data containing intracranial aneurysms.
In this study, aneurysm morphological and hemodynamic results using models generated by AI segmentation methods were compared with those obtained by two human users for the same data. Interclass coefficients (ICC), Bland-Altman plots, and Pearson’s correlation coefficients (PCC) were combined to assess how well AI-generated CFD models were performed. We found that almost perfect agreement was obtained between the human and AI results for all eleven morphological and five out of eight hemodynamic parameters, while a moderate agreement was obtained from the remaining three hemodynamic parameters. Given this level of agreement, using AI segmentation to create CFD models is feasible, given more developments.
Keywords: Convolution Neural Network, Image Segmentation, Computational Fluid Dynamics, Computational Hemodynamics, Aneurysm
1991 Mathematics Subject Classification: 22E46, 53C35, 57S20
1. Introduction
Intracranial aneurysms (IA) are pathologic, bulbous expansions of the arterial wall in the brain with a prevalence of 3–5%1,2. Although the risk of IA rupture is low (0.1% per year), mortality in patients with a ruptured IA is 45–50%, with survivors enduring significant neurological damage and physical or cognitive impairment3. Hemodynamic forces in the vasculature are known to alter vascular cell functionality and play an important role in destructive vascular remodeling such as IA formation, growth, and rupture4, motivating continued research in aneurismal hemodynamics. Recently, much research effort has been devoted to “image-based” computational hemodynamics5,6, which utilizes medical imaging data (computed tomography, digital subtraction angiography, magnetic resonance (MR) imaging, etc.) from individual patients to create “patient-specific” geometrical models and then compute hemodynamic parameters. As of now, “image-based” computational hemodynamics is gaining clinicians’ trust because it can produce more realistic gross hemodynamics compared with MR flow imaging7,8. In the last few years, computational hemodynamics has been utilized to predict the IA’s rupture risk. In a few recent studies9,10,11, when hemodynamic and morphological features were combined using machine learning, the prediction performance of IA’s rupture status was generally good (i.e.,the area under the curve [AUC] of receiving operating curve [ROC] ≈ 80%).
Recent progress has continued to motivate the translation of “image-based” computational hemodynamics into the clinical workflow. Recall that, in the framework of “image-based” computational hemodynamics5,6, “patient-specific” computational fluid dynamics (CFD) models are created through image segmentation, which is largely done manually and is a labor-intensive process (typically, > 1 hour). As clinicians do not have time in their schedules to manually build CFD models, manual model creation has been a roadblock preventing such a translation. As a result, developing an efficient software infrastructure to input patient-specific data and output clinically relevant (morphological and hemodynamic) parameters becomes a priority.
There have been some documented efforts in accelerated CFD model creation. Xiang et al. developed a clinical software platform12 for cerebral aneurysm flow simulation and visualization known as AViewa. In the AView software, CFD models are created in a branch-by-branch fashion. Each vascular branch grows from seed balloons supplied by a user, who first selects seed balloons using a published method13. Although the above-mentioned branch-by-branch method yielded accurate morphological parameters of IAs, CFD simulation results using AView were not compared with results through manual model creation. In another study, Seo et al. proposed a more automated method by constructing a level-set-based vessel “mask” for CFD model creation14. In their system, the lattice Boltzmann and boundary data immersion methods were combined to solve pressure and velocity fields. However, only calculated wall shear stresses (WSS) were qualitatively compared with prior studies.
It is also significant to note that the current protocol for “image-based” computational hemodynamics is computationally intensive and requires hours to solve. Hence, researchers are actively working on innovative ideas to reduce solution time. Recent studies, including CFDNet15 and DeepCFD16, have attempted to replace traditional CFD solvers with convolution neural network (CNN)-based solver models to accelerate the computing process. Although encouraging results have been obtained, there are many challenges remaining. As reported by Arzani et al.17, the current CNN-based solver models cannot readily handle complex boundary conditions and are only validated using simple benchmarks under the steady flow. Furthermore, machine-learning (ML)-based prediction of hemodynamic parameters is an active research field18,19,20. Yevtushenko et al. developed an ML-based centerline model to estimate pressure changes before and after coarctation of the aorta18. Fossan et al.19 trained fully-connected feed-forward neural networks (NN) to predict pressure losses in coronary arteries. They augmented the prediction accuracy with a reduced-order model. However, the performance of their ML-based model requires improvements since initial results are not entirely robust. Li et al. used a high-density point cloud structure to map CFD results onto the high-density 3D point cloud. Thus, after training, the predictions resolved complex fluid patterns with high resolution20. Interestingly, three applications all focused on pressure drop and/or fractional flow reserve (FFR). With the well-defined output metric, their preliminary results were promising. However, additional work must be carried out to investigate whether ML-based CFD predictors can accurately estimate time-resolved WSS and velocity in 3D when the vasculature is complex. In other words, the generalization of those ML-based CFD predictors needs to be further investigated.
In this study, our focus is to investigate whether CNN-based image segmentation can be used to accelerate the creation of CFD models. Results from the 2021 Cranial Aneurysm Detection and Analysis Challenge [CADA] indicate “recent deep learning methods delineate cerebral aneurysms with a DICE score around 0.9 and above”21. Given the excellent DICE scores, the deep-learning image segmentation model can probably be used for CFD model creation. However, can excellent DICE scores be directly translated into the notation that CNN-based image segmentation algorithms are ready to accelerate CFD model creation? Will we obtain similar hemodynamics results between the human- and CNN-created CFD models? To our knowledge, those questions have not been answered, and our study is the first of this kind. Moreover, CNN-based CFD model creation will be useful for ML-based CFD prediction in complex vasculature investigated by others18,19,20 as well.
To this end, leveraging the readily available CNN-based image segmentation methods, our preliminary goal is to investigate whether automated CFD model creation can be achieved simply by the adoption of a deep-learning-based image segmentation method (hereafter referred to as AI segmentation methods). Our secondary objective is to quantitatively compare hemodynamic parameters generated through manual segmentation and AI segmentation. Furthermore, to improve our study’s transparency and open science nature, we intentionally selected imaging data from a publicly available online database known as AneuriskWEBb. At the publication of this study, all data and relevant compute codes will be made available for public disseminationc, and thus, our study is reproducible elsewhere.
2. Materials and Methods
From the AneuriskWEB database, all 23 cases with imaging data (in Digital Imaging and Communications in Medicine [DICOM] data format) were collected for this study. The overall workflow of our study design is shown in Fig. 1. All aneurysms were saccular aneurysms: 15 located at the intracranial internal carotid artery (ICA), 6 located at the middle cerebral artery (MCA), and 2 located at the anterior cerebral artery (ACA). In the next few subsections, detailed procedures pertaining to manual segmentation, CNN-based segmentation (hereafter referred to as AI segmentation), CFD simulations, and data analytics are briefly presented.
Fig. 1:
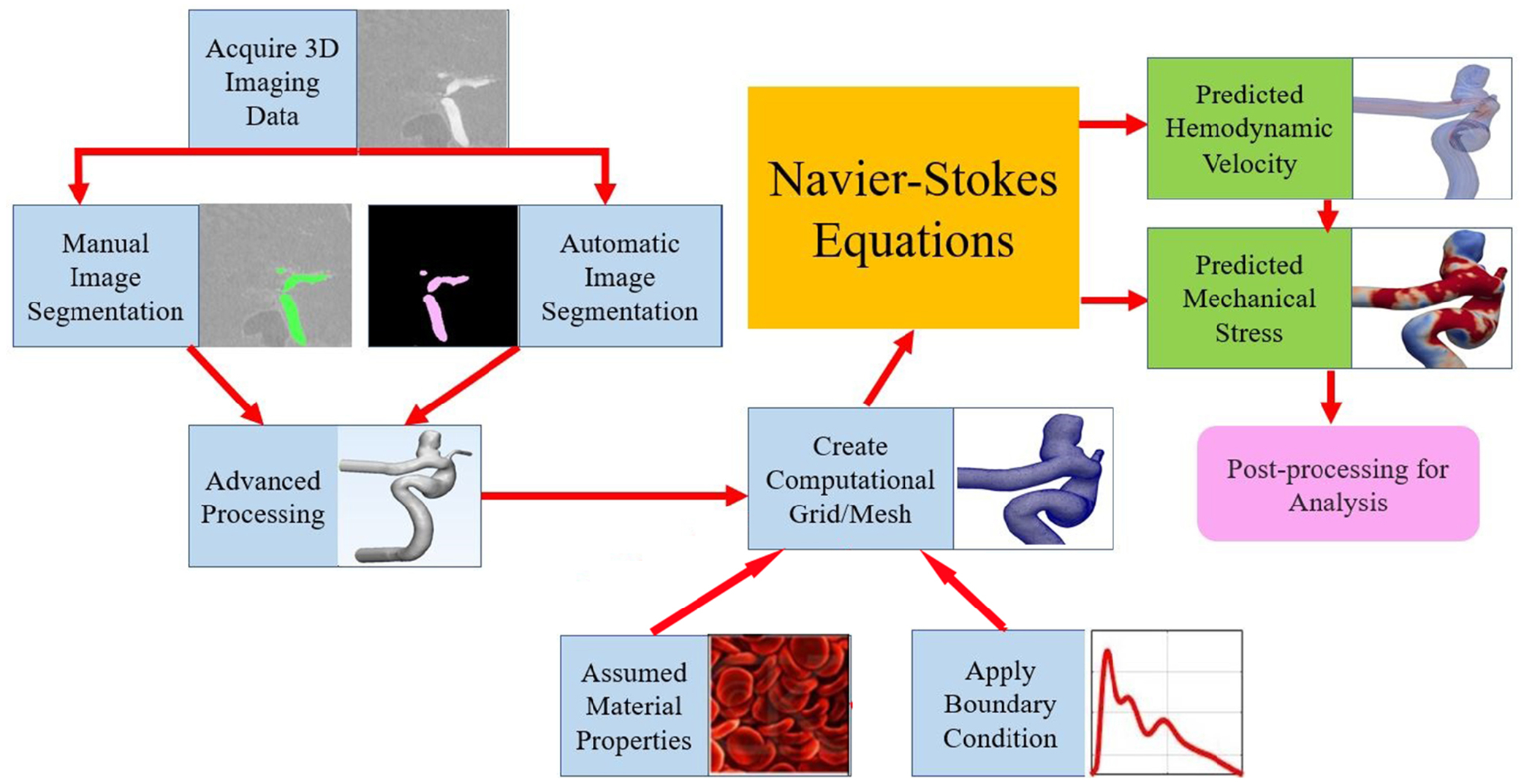
An overview of study design and typical computational hemodynamics workflow
2.1. Manual Segmentation
As represented in Figure 1, initially, three-dimensional rotational angiography (3DRA) files in DICOM format were loaded into Mimics Innovation Suite (version 22, Materialise Inc., Leuven, Belgium). In Mimics software, an intensity window was first used to identify the vasculature, followed by manual editing to isolate the region of interest (ROI). The ROI was transformed into a stereolithography (STL) geometric model for further processing by 3-Matic software (version 14, Materialise Inc., Leuven, Belgium). In 3-Matic software, a first-order Laplacian smoothing filter was locally applied on the surface. Surface smoothing reduced local irregularities on the reconstructed vessel surfaces due to noise while conserving the integrity of overall vessel structures. To minimize the influence of an inlet plug flow boundary condition, upstream vessel structures were retained22. Cylindrical flow extensions with a minimum size of 6 times the vessel diameter were added to all inlets and outlets using the open-source Vascular Modeling Toolkit (VMTK) package (version 1.4)23. Moreover, to manage simulation time, downstream vessels were removed. Two users (graduate students with 9 month’s experience in performing image-based CFD simulations) independently performed image segmentation and subsequent CFD simulations.
2.2. AI Segmentation
Two published AI segmentation packages (MIScnn24 and DeepMedic25) were selected to test automated image segmentation. Python implementations are available for both packages through Github. Brief descriptions of those two AI segmentation methods are provided below for the sake of completeness.
Among 23 sets of 3DRA data available to this study, 15 cases were selected for training, while the remaining 8 cases were used to test CFD model generation by the above-said two CNN models after training and by two users. Vessel labels used for training and evaluations of testing were generated by corroborating one human user’s manual segmentation results with the clinician-generated vessel geometries supplied by the AneuriskWEB project. Both training and testing datasets included a range of complexities: aneurysms with small secondary vessels, tortuous vasculature, narrow gap/spacing between an IA and its adjacent parent artery, and non-uniformly mixed contrast.
2.2.1. MIScnn
As shown in Fig. 2, MIScnn, a re-implementation of the well-known UNet model26, takes 3DRA images as input and outputs a binary mask defining the vasculature of interest. More specifically, MIScnn’s encoder consists of convolution and pooling operations to condense the input image to a set of best features. Then, the decoder in MIScnn (convolution, up-convolution, and softmax operations) transforms the above-mentioned features into the vasculature of interest. Of note, skip connections (i.e., dashed arrows) between the encoder and decoder allow spatial information lost during the encoding process to be recovered. More details can be found in the original publication24.
Fig. 2:
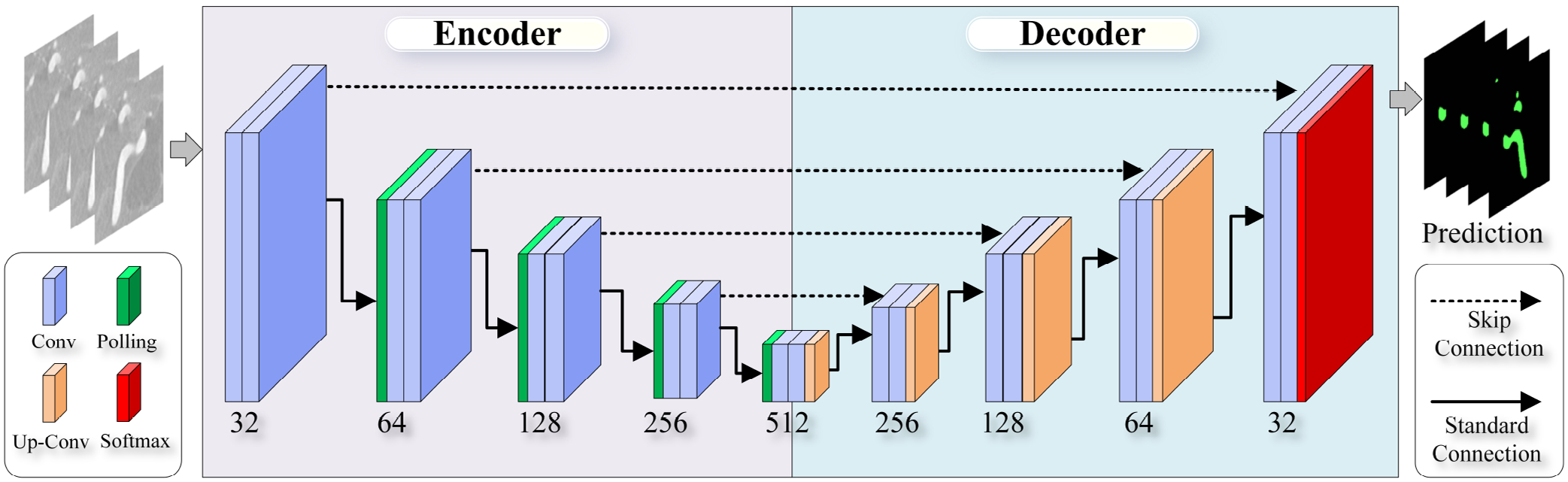
An illustrative diagram of MIScnn architecture. The model is a standard 3D U-Net without any batch normalization layer or dropout layer. The Tversky loss function was used for training.
2.2.2. DeepMedic
As illustrated in Fig. 3, also using 3DRA images as the input, the DeepMedic algorithm uses a different strategy to combine information at two spatial scales. Algorithmically, it has two parallel convolutional pathways: Converting 3DRA images at normal and lower resolutions into two sets of features, mainly through convolution operations. Those extracted features through the above-mentioned two parallel pathways are concentrated and input to a fully connected 3D Conditional Random Field (CRF) for regularization. Finally, a final 3D classification map is produced. More details can be found in the original publication25.
Fig. 3:
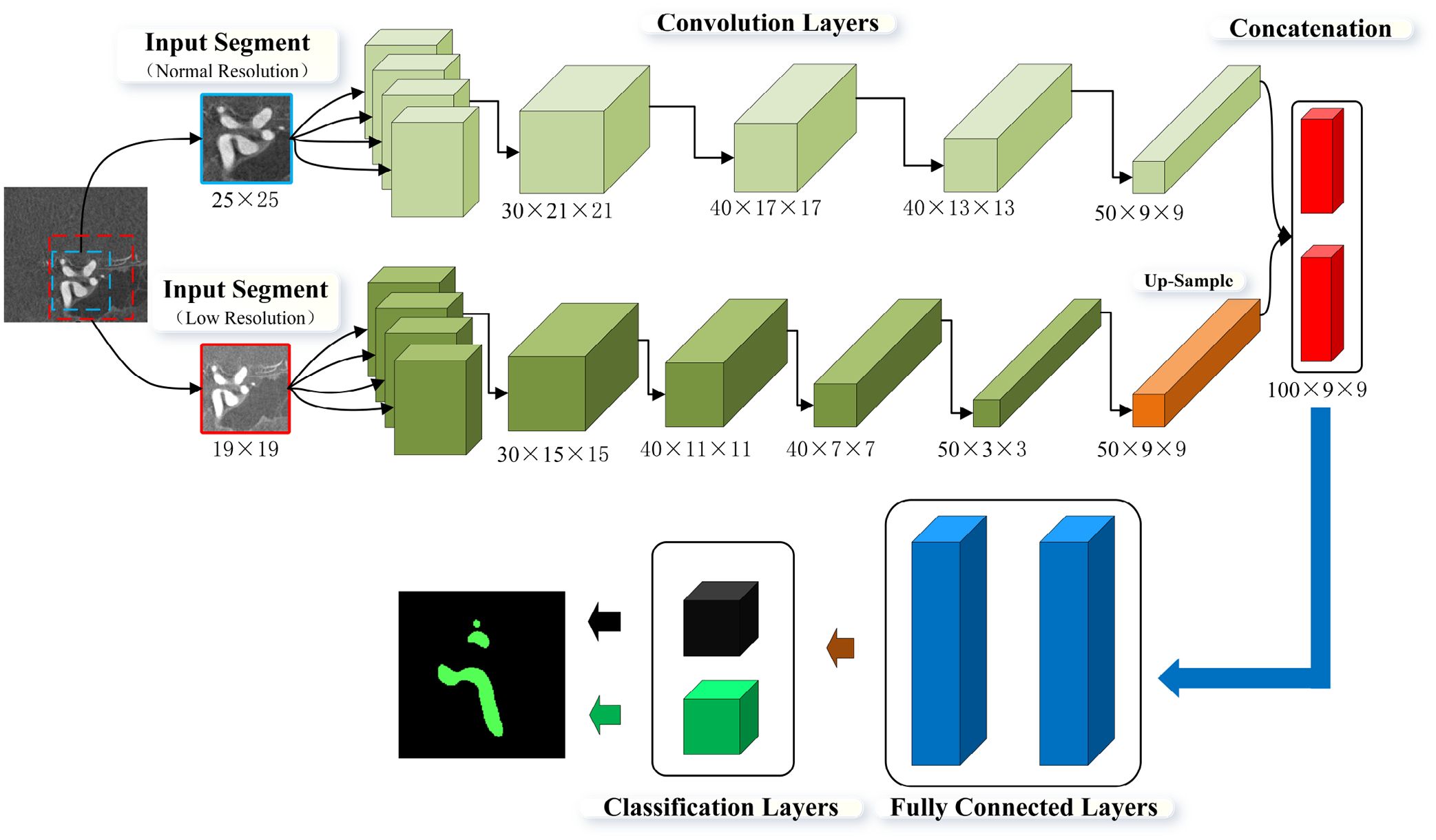
A schematic diagram showing DeepMedic’s model architecture. The model consists of two convolution pathways followed by two fully connected layers and one classification layer.
2.3. Mesh Generation
Once segmented vessel geometries became available through either manual or automated image segmentation (Fig. 1), a mesh-generator, TetGend(Version 1.4.2), was utilized to generate unstructured 3D tetrahedral meshes including five boundary layers. The mesh generation process was completed by a Python script originating from the VMTK (version 1.4). Approximately 1.5 million cells were generated per case in this process, with an average size of around 0.0022 mm2 and an average edge length of 0.12 mm. Furthermore, a mesh sensitivity test was applied to verify that the outcomes of our CFD simulations were not sensitive to the selected mesh size.
2.4. CFD Simulations
Once computer meshes became available, a commercial CFD solver (Version 21, Fluent, Ansys Inc., PA, USA) was used to solve Navier-stocks equations. Blood was assumed to be an incompressible, Newtonian fluid with a dynamic viscosity of 0.004 Pa · s and a mass density of 1050 kg/m327. Vessel walls were assumed to be rigid, with a no-slip boundary condition. A pulsatile flow (rate) waveform derived from averaged magnetic resonance flow measurements among healthy subjects’ internal carotid arteries (ICA)28 was first scaled to correspond to an average flow rate of 280 mL/min and then used as the inlet boundary condition. At the outlets, zero-pressure boundary conditions were prescribed during the CFD simulations. Four cardiac cycles were simulated at 2000 steps per period (0.0005 sec/timestep). By selecting 2000 time steps, Courant numbers were below 1 for the above-said waveform. Time-resolved hemodynamic data (20-time points) were saved for the last cardiac cycle for further processing. The systolic or peak Reynolds numbers were approximately between 350 to 550 for the ICAs, similar to those reported by others.
2.5. Data Analysis
Morphological indices and hemodynamics parameters are frequently used in predicting the IA rupture status9,10,11. Our ultimate goal is to develop an automated (software) platform that can be used to assess the risk of IA rupture, and thus, it is important to assess the degree of agreement between calculated morphological indices and hemodynamic parameters based on geometries extracted either using AI or manually. So, after the completion of either manual or AI segmentation, customized Python scripts derived from the open-source VMTK package were used to process data to obtain 11 geometric parameters: Aneurysm Volume in mm3, aneurysm height in mm, maximum aneurysm sac width in mm (Sac Max Width), size ratio between aneurysm height and parent vessel diameter (Size ratio height), size ratio between aneurysm width and parent vessel diameter (Size ratio width), ratio between multiplication of the aneurysm’s height and ostium’s circumference to four times of aneurysm ostium area (Aspect ratio*), (parent) Vessel diameter, minimal dimension of aneurysm ostium (Ostium_min), maximal dimension of aneurysm ostium (Ostium_max), Aneurysm surface area (Aneurysm Area) and Ostium Area.
Geometries extracted using AI and by human users were used to create CFD models, and CFD simulations were completed following the protocols described in Section 2.4. Eight hemodynamic parameters were calculated for each CFD model: spatially-temporally averaged wall shear stress during systole (Systole_STAWSS), minimal wall shear stress during systole (Systole_WSSMin), maximal wall shear stress during systole (Systole_WSSMax), spatial mean oscillatory shear index (mean OSI), one standard deviation of the oscillatory shear index (Std_OSI), time-averaged low shear area (TA_LSA), one standard deviation of time-averaged low shear area (TA_LSA_Std) and temporally-averaged degree of overlap among vortex cores during systole (Systole_TADVO).
Both geometric and hemodynamic parameters were used in a prior study, and more details can be found elsewhere4. In this study, the segmentation processes and subsequent model creation were done by four independent agents, i.e., two human users, and two AI methods post-training. Consequently, quantitative analyses showing degrees of agreement between two different agents in terms of the above-mentioned geometric and hemodynamic parameters were conducted. Hereafter, such results regarding the degree of the agreement are referred to as user and AI results, respectively. A combination of Pearson’s correlation coefficient (PCC), interclass correlation coefficient (ICC)29, and Bland-Altman plot30 was used to assess the agreement in three categories: User1 versus User2, MIScnn versus DeepMedic, and an average of two users versus an average of two AI methods. As described above, there were two raters in each category, and we considered the rater effect, so a two-way mixed method seemed most appropriate. Thus, the ICC(3,2) formulation following the notations by Shrout and Fleiss was used31, where “3” refers to model 3 (two-way mixed method) and “2” refers to the fact that the mean of 2 raters was used in the calculation of ICC. Such a selection is consistent with suggestions in the literature32. Calculations of ICC (3,2) were completed using scriptse, that were implemented in MATLAB (Mathworks Inc., MA, USA).
3. Results
3.1. CFD Model Creation by AI Methods
The image segmentation results obtained from eight testing cases were quantitatively assessed, as summarized in Table 1. The averaged DICE scores showing the overlap between the segmented volumes and ground truth volumes were 0.88 and 0.89 for MIScnn and DeepMedic, respectively. Moreover, the average sensitivity values were above 0.90, while the average specificity values were nearly perfect. All quantitative assessments indicate that both AI segmentation methods performed reasonably well.
Table 1:
A summary of performance assessed using DICE (overlap between the segmented volume and ground truth), RVE (relative volume error), Sensitivity, Specificity, HD95 (maximum Hausdorff distance between segmented points and ground truth set in mm), and ASSD (average symmetric surface distance between the segmented points and ground truth set in mm).
| DICE | RVE | Sensitivity | Specificity | HD95 | ASSD | |
|---|---|---|---|---|---|---|
| MIScnn | 0.88±0.06 | 0.16±0.15 | 0.90±0.11 | 0.998±0.004 | 29.3±24.5 | 0.16±0.15 |
| DeepMedic | 0.89±0.04 | 0.13±0.11 | 0.90±0.09 | 0.998±0.002 | 15.9±10.8 | 0.13±0.11 |
In two cases (out of eight testing cases) segmented by MIScnn, we observed unwanted connections between adjacent vessel segments, and manual editing was subsequently used to remove those unneeded connections using the 3-Matics software (version 14, Materialise Inc., Leuven, Belgium). Then, mesh generation was performed to generate CFD models. When the DeepMedic package was used, four out of eight testing cases required the above-mentioned manual editing. Hence, the success rates for MIScnn and DeepMedic were 75% and 50%, respectively.
3.2. Consistency Assessments
A Pearson correlation coefficient was computed to assess the linear relationship between the outcomes of two independent agents. As summarized in Table 2, nearly perfect positive correlations among all three categories: User 1 versus User 2, MIScnn versus DeepMedic, and averaged user versus averaged AI, were obtained except for three hemodynamic parameters: Mean_OSI Std_OSI (moderate correlation) and Systole_TADVO (low positive correlation). The PCCs between the averaged user and averaged AI were the best among the above-said three categories.
Table 2:
A summary of estimated PCC values for geometrical and hemodynamic parameters in three categories: user versus user, MIScnn versus DeepMedic, and averaged user versus averaged AI. Geometrical and hemodynamic parameters were calculated following the workflow shown in Fig. 1 in 8 IA cases.
| User 1 .vs User2 | MIScnn .vs DeepMedic | user .vs AI | |
|---|---|---|---|
| Systole_STAWSS | 0.9463 | 0.9475 | 0.9900 |
| Systole_WSSMin | 0.9188 | 0.9113 | 0.9364 |
| Systole_WSSMax | 0.8992 | 0.9148 | 0.8886 |
| Mean_OSI | 0.7838 | 0.4868 | 0.7019 |
| Std_OSI | 0.5535 | 0.6015 | 0.5911 |
| TA_LSA | 0.9621 | 0.9391 | 0.9354 |
| TA-L_SA_Std | 0.9158 | 0.9320 | 0.9206 |
| Aneurysm Volume | 0.9994 | 0.9999 | 0.9999 |
| Aneurysm Height | 0.9982 | 0.9872 | 0.9882 |
| Sac_Max_Width | 0.9993 | 0.9984 | 0.9971 |
| Size_ratio_height | 0.8545 | 0.9917 | 0.9359 |
| Size_ratio_width | 0.9702 | 0.9930 | 0.9928 |
| AspectRatio* | 0.9527 | 0.9688 | 0.9738 |
| VesselDiameter | 0.8736 | 0.9548 | 0.9795 |
| Systole_TADVO | 0.3254 | 0.3076 | 0.9123 |
| Ostium Minimum | 0.9618 | 0.9760 | 0.9293 |
| Ostium Maximum | 0.9866 | 0.9622 | 0.9902 |
| Aneurysm Area | 0.9995 | 0.9998 | 0.9996 |
| Ostium Area | 0.9923 | 0.9628 | 0.9922 |
| Median PCC | 0.9572 | 0.9622 | 0.9738 |
| Mean PCC | 0.8890 | 0.8860 | 0.9292 |
Table 3 represents the degree of agreement between two different observers over 19 parameters in three different categories. Although the degree of agreement was moderate in three out of nineteen parameters, namely, Mean-OSI, Std-OSI and Systole-TADVO, the means of ICC values in all three columns were excellent.
Table 3:
A summary of estimated ICC values for geometrical and hemodynamic parameters in three categories: user versus user, MIScnn versus DeepMedic, and averaged user versus averaged AI. Geometrical and hemodynamic parameters were calculated following the workflow shown in Fig. 1 in 8 IA cases.
| User 1 .vs User2 | MIScnn .vs DeepMedic | User .vs AI | |
|---|---|---|---|
| Systole_STAWSS | 0.9723 | 0.9725 | 0.9883 |
| Systole_WSSMin | 0.9499 | 0.9465 | 0.9090 |
| Systole_WSSMax | 0.9398 | 0.9047 | 0.9086 |
| Mean_OSI | 0.8761 | 0.6400 | 0.8055 |
| Std_OSI | 0.6978 | 0.7371 | 0.6925 |
| TA_LSA | 0.9785 | 0.9681 | 0.9655 |
| TA_LSA_Std | 0.9560 | 0.9643 | 0.9559 |
| Aneurysm Volume | 0.9996 | 0.9999 | 0.9995 |
| Aneurysm Height | 0.9997 | 0.9991 | 0.9975 |
| Sac_Max_Width | 0.9990 | 0.9991 | 0.9984 |
| Size_ratio_height | 0.9940 | 0.9935 | 0.9919 |
| Size_ratio_width | 0.9849 | 0.9964 | 0.9963 |
| AspectRatio* | 0.9714 | 0.9840 | 0.9866 |
| VesselDiameter | 0.9288 | 0.9768 | 0.9893 |
| Systole_TADVO | 0.4910 | 0.3791 | 0.9510 |
| Ostium Minimum | 0.9752 | 0.9691 | 0.9593 |
| Ostium Maximum | 0.9856 | 0.9753 | 0.9948 |
| Aneurysm Area | 0.9997 | 0.9998 | 0.9996 |
| Ostium Area | 0.9961 | 0.9772 | 0.9895 |
| Median ICC | 0.9752 | 0.9753 | 0.9883 |
| Mean ICC | 0.9313 | 0.9148 | 0.9513 |
Moreover, the Bland-Altman plot was used as an additional visual inspection to measure the level of agreement between outcomes of two different agents. In Fig. 4, four representative Bland-Altman plots for Aneurysm Volume, Ostium area, Systole-WSSmin, and Systole-TADVO show the degree of agreement between the outcomes of two raters using a 95% limit of agreement. In each of the four metrics, one out of eight cases was outside the 95% limit when human User 1 was compared with human User 2. In contrast, only the Ostium area metric in one case fell outside the 95% limit when MIScnn results were compared with those of DeepMedic. In general, AI-based estimations resulted in narrower 95% limits in Aneurysm Volume and Systole-WSSMin (see Fig. 4).
Fig. 4:
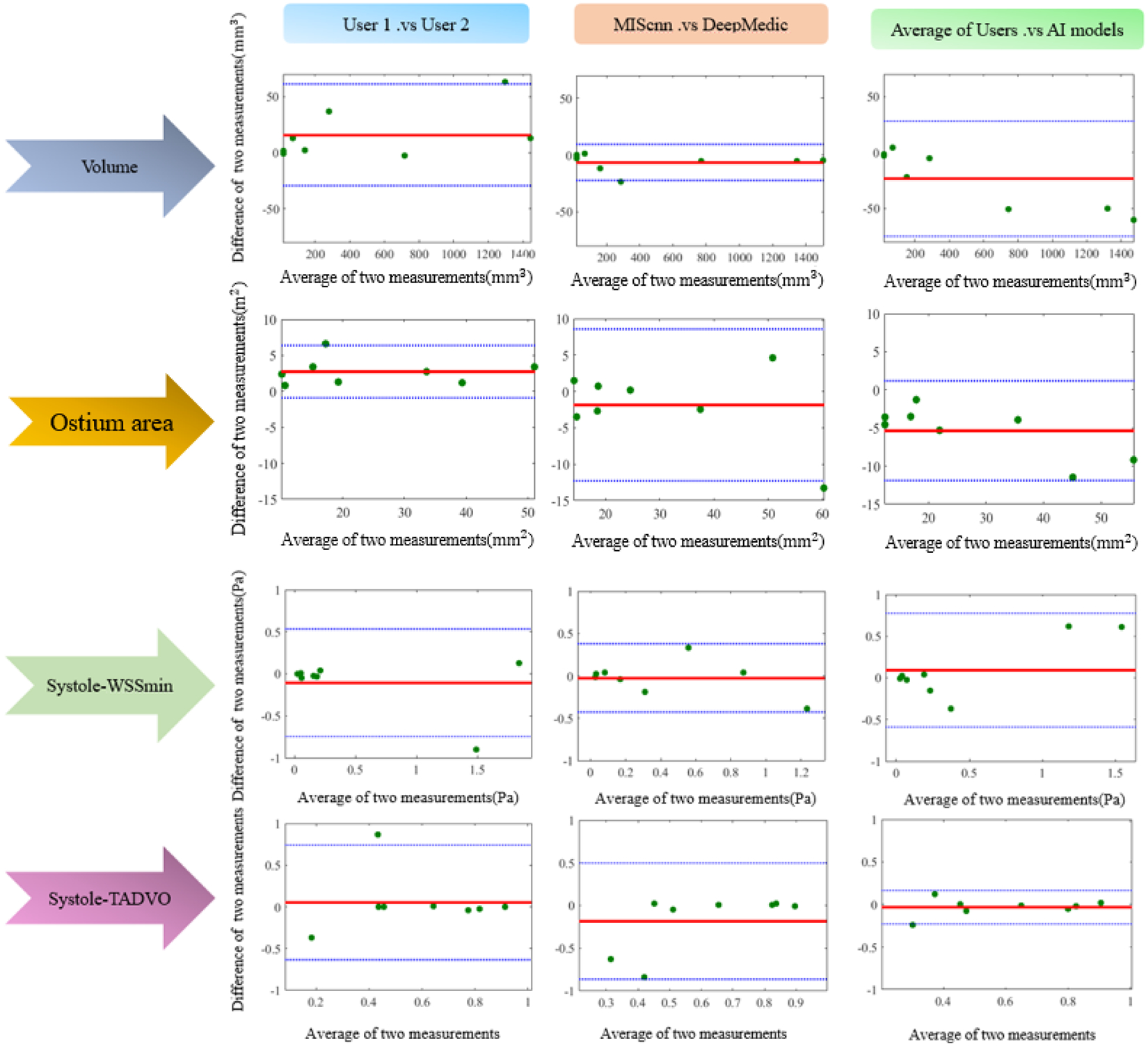
Bland-Altman plots for visualization of agreements between different raters’ outcomes in Aneurysm volume, Ostium area, Systole-WSSMin and Systole-TADVO. The blue dashed and red solid horizontal lines in each subplot represent the limits of agreement (95% confidence interval) and the biases, respectively.
It is worth noting that there were negative biases (roughly −20mm3 and −5mm2, respectively; see the top two right plots in Fig. 4) for both Aneurysm Volume and Ostium area between the averaged user and averaged AI segmentation, indicating AI segmentation underestimated the aneurysm size and ostium area.
4. Discussion
Our investigation started with image segmentation tasks by two human users and two AI segmentation methods. Based on quantitative results shown in Table 1, both MIScnn and DeepMedic essentially completed the segmentation process, as indicated by estimated average DICE scores (between 0.88 and 0.89). Furthermore, morphological parameters measured from IA geometries segmented by two AI methods had nearly perfect correlation coefficients and ICC values compared with two human users (see Tables 2 and 3). Our visual inspection confirmed that segmented vessel structures by two AI methods were almost the same as the ground truth, as shown in Supplemental Figure 1 in Supplemental Materials.
However, some minor (segmentation) errors that have not received sufficient attention in image segmentation literature caused significant problems for CFD model creation. Three examples are illustrated below. The first example is shown in Fig. 5-a, where MIScnn segmented vessel geometry appeared to have an overly smoothed aneurysm sac (see arrows). In Fig. 5-b, in the MIScnn segmented vessel geometry, the ophthalmic artery’s connection to the internal carotid artery was flattened out, likely due to the presence of image noise. Furthermore, the aneurysm apex was mistakenly connected to a downstream artery. Similarly, in Fig. 5-c, MIScnn segmented vessel geometry had two errors: (1) a missing ophthalmic artery and (2) an unwanted connection at the internal carotid artery (see arrows). Manual editing was required to fix unwanted connections prior to mesh generation; unwanted vessel connections resulted in a failure rate of 25% for MIScnn, whereas a flattened (Fig. 5-b) or missing (Fig. 5-b) small ophthalmic artery was not repaired for the subsequent CFD model generation.
Fig. 5:
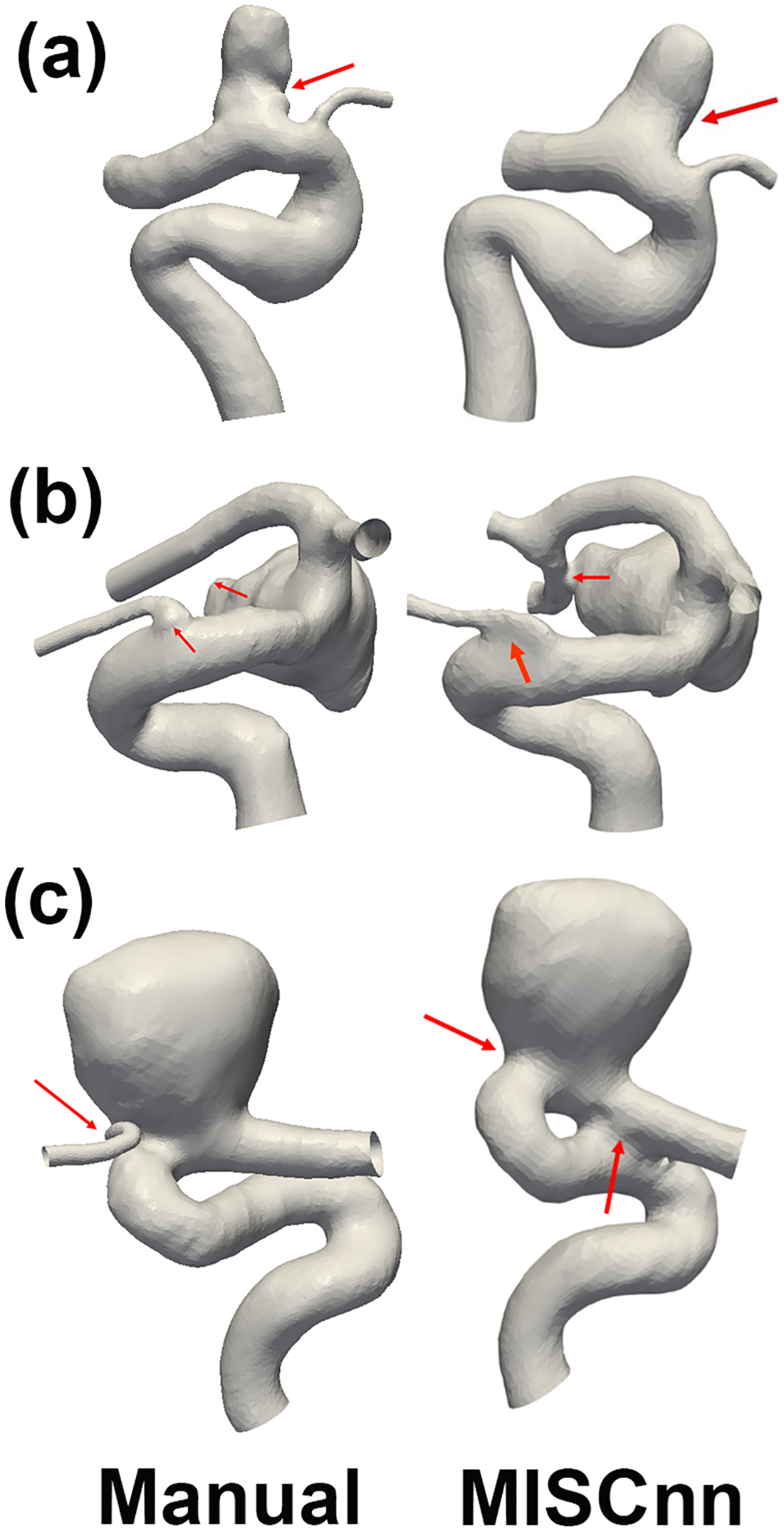
Comparison segmentation results between MIScnn method (right column) and a human user (left column). The human user results have been corroborated with the clinicians’ results provided by AueuriskWEB and, therefore, can be considered as ground truth. Arrows in the right column point to problematic areas visible from MIScnn results, while arrows in the left column guide readers to view the correct segmentation results.
Using AI-based image segmentation is in part, motivated by the goal of streamlining CFD model creation in the clinical workflow. Our preliminary data suggest that a good fraction of cases (e.g., 25% for MISCnn) require manual editing. In our initial experience, a trained engineer can typically complete such manual editing within 5–10 minutes because the scope of the manual editing is limited, as shown in Fig. 5. However, the required manual editing still challenges clinicians in using computational hemodynamics.
Our data suggested that some discrepancies in the created CFD models (e.g., missing or imperfectly connected to a small artery) did not significantly influence hemodynamic parameters. As shown in Tables 2 and 3, there were almost perfect positive correlations and ICC values for five hemodynamic parameters, except for Mean_OSI, Std_OSI, and Systole_TADVO. It is interesting to note that the consistency for Mean_OSI, Std_OSI, and Systole_TADVO was low among the pair of human users as well. Inconsistency in Mean_OSI and Std_OSI is largely due to the fact that the oscillatory shear index (OSI) is a sensitive metric. OSI often falls into a narrow range (e.g., between 0.006–0.025), and small changes in the surface smoothness of a segmented aneurysm may cause relatively substantial changes in OSI values. Similarly, Systole-TADVO is also a sensitive metric related to tracking aneurysmal flow vortex cores over the systole phase because flow vortices are estimated using velocity derivatives33. For instance, slight modifications of aneurysm ostium size (i.e., critical flow pathway entering aneurysm dome) could lead to significant changes in Systole-TADVO. As shown in Fig. 4, on average, two AI segmentation methods overestimated the ostium area by 5mm2, equivalent to 20%. Further improving AI segmentation methods to avoid such a critical overestimation is an important future research topic.
It is interesting to note that, based on performance metrics that are widely used to assess image segmentation methods (see Table 1), DeepMedic slightly outperformed MIScnn. But our experience with CFD model creation suggested otherwise. We found that the MIScnn model performed better in (1) avoiding unwanted vessel connections and (2) reducing the missing small but important arteries. As shown in Fig. 2, MIScnn is a re-implementation of the U-Net model. Thus, in MIScnn, extracted features from five resolution levels were effectively combined to generate a final vessel segmentation mask. In contrast, DeepMedic (see Fig. 3) first combined features from two resolution levels and then used a fully-connected layer to obtain a finalized vessel mask. In theory, the feature fusion at the five different resolutions, instead of at two levels in DeepMedic, allowed the MIScnn model to detect small vessels and avoid unwanted vessel-to-vessel or vessel-to-aneurysm connections.
To further improve the performance of CNN-based image segmentation for CFD model creation involving cerebral vasculature, we probably need to investigate all three aspects of the CNN-based image segmentation: (1) pre-processing, (2) post-processing, and (3) feature extraction and fusion. Many pre- and post-processing techniques surveyed in Salvi et al.’s review paper34 can be used to improve the outcomes of image segmentation. Particularly, using conditional random field (3DCRF) and 3D connected component optimization (3DCCO) to improve vessel connectivity while removing artifacts has been very popular. Referring to feature extraction and fusion, it is well recognized that convolution operations are local, and thus, CNN cannot learn global and long-distance semantic information interaction well. Thus, Attention and Transformer mechanisms have been popular. Particularly, the combination of a global Transformer with local low-level details through convolutions may be an effective way to improve the segmentation of complex cerebral vasculature, including IAs. Our ongoing work is designing innovative CNN-based image segmentation models to improve cerebral vessel segmentation.
From the Bland-Altman plots shown in Fig. 4, the 95% limits between two AI methods were actually better than those between two human users for Aneurysm Volume, Systole-WSSMin. Referring to the Systole-TADVO metric, the 95% limits were essentially the same between two categories: User1 versus User2 and MIScnn versus DeepMedic. Also, biases in all three above-mentioned parameters were nearly zero, and no outlier was found between the two AI segmentation methods. This observation is encouraging but still needs to be verified in a large study and should not be over-interpreted. Nevertheless, our preliminary results suggest that AI-segmentation has the potential to be used for CFD model creation. More technological developments must be devoted to resolving the two significant geometric errors mentioned above.
In this feasibility study, one limitation is that we selected only two AI segmentation methods, MIScnn and DeepMedic, which are two classic but different CNN-based image segmentation configurations. More specifically, as shown in Figs. 2 and 3, MIScnn contains only convolution layers, while DeepMedic concatenates a multi-resolution CNN model with fully-connected layers. Of note, many CNN-based vessel segmentation methods were designed for 2D vessel extraction (e.g., FrangiNet35) and were not considered in this study. In this sense, our results are encouraging because the DICE scores achieved were around 0.9 and close to the state-of-the-art, as reported by the Medical Image Computing and Computer Assisted Intervention (MICCAI Society) challenge21. If newer models and deep-learning techniques36,37 are adopted, fine-tuned CNN-based image segmentation methods can further improve their performance. We also attempted to use traditional 3D image segmentation methods. After an extensive search followed by trials, only two semi-automatic 3D image segmentation methods, a level-set-based method23 and a graph-cuts-based method38, can be used to obtain reasonable geometries for CFD model creation. However, both methods require human input. For instance, the Level-Sets model requires the selection of two specified thresholds as the initial level sets and requires a user to place two seeds on the imaging data. In contrast, the Graph-Cuts method requires a user to specify the foreground and background seeds on the imaging data. Since we evaluate automatic image segmentation for CFD model creation, the level-set and graph-cuts methods were not included for comparison in this study.
The second limitation of this study is that only a subset of summary morphological and hemodynamic parameters was selected for comparison. This is largely due to the fact that those parameters are extensively used in conjunction with machine learning methods for predicting the risk of aneurysm rupture in the literature. Over 90 morphological and hemodynamic parameters have been reported, as summarized in a recent survey paper39. Hence, it isn’t easy to include all of them while keeping our presentation concise. Furthermore, more detailed investigations comparing spatially-distributed hemodynamic parameters are ongoing. Given the feasibility nature of this study, our study design is a good first step.
We also recognize that only 23 data sets were used and considere this another limitation. Although the results are encouraging, more testing and development are needed. To demonstrate the repeatability of this study, upon the acceptance of our work, all source codes used in this study will be uploaded to Githubf along with the 23 sets of imaging data.
5. Conclusion
Our study demonstrated the feasibility of using AI segmentation methods for the creation of CFD models. No significant discrepancies were found between human-created and AI-segmented CFD models in terms of morphological and hemodynamic parameters for a vast majority of parameters investigated. Our observations also identified that further developments in AI segmentation methods need to resolve (1) unwanted vessel connections and (2) missing small arteries. The unmet needs motivate our ongoing work.
Supplementary Material
Acknowledgments
The project is funded by a research grant from US National Institutes of Health/National Institute of Biomedical Imaging and Bioengineering (R01-EB029570A1). Mr. Mostafa Rezaeitaleshmahalleh receives a research fellowship from the Health Research Institute at Michigan Technological University and Dr. N. Mu is supported from a Post-doctoral Fellowship Award from the American Heart Association (AHA-23Post1022454). We also would like to thank Dr. Min Wang from the University of Texas at San Antonio for his advice on statistical analysis.
Footnotes
Conflicts of Interest
There are no conflicts to declare.
Ethical Compliance
Research experiments conducted in this article with human subject data were approved by the Ethical Committee and responsible authorities (Institutional Review Board) of our research organization(s) following all guidelines, regulations, legal, and ethical standards as required for human subjects. Since the open database was used, no consent was required.
Upon acceptance, data and codes will be available in https://github.com/jjiang-mtu/
References
- 1.Revilla-Pacheco F, Escalante-Seyffert MC, Herrada-Pineda T, Manrique-Guzman S, Perez-Zuniga I, Rangel-Suarez S, Rubalcava-Ortega J, Loyo-Varela M, Prevalence of incidental clinoid segment saccular aneurysms, World neurosurgery 115:e244–e251, 2018. [DOI] [PubMed] [Google Scholar]
- 2.Hackenberg KA, Hänggi D, Etminan N, Unruptured intracranial aneurysms: contemporary data and management, Stroke 49(9):2268–2275, 2018. [DOI] [PubMed] [Google Scholar]
- 3.Villablanca JP, Duckwiler GR, Jahan R, Tateshima S, Martin NA, Frazee J, Gonzalez NR, Sayre J, Vinuela FV, Natural history of asymptomatic unruptured cerebral aneurysms evaluated at ct angiography: growth and rupture incidence and correlation with epidemiologic risk factors, Radiology 269(1):258–265, 2013. [DOI] [PubMed] [Google Scholar]
- 4.Sunderland K, Jiang J, Zhao F, Disturbed flow’s impact on cellular changes indicative of vascular aneurysm initiation, expansion, and rupture: A pathological and methodological review, Journal of Cellular Physiology n/a(n/a):1–23, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jou LD, Quick CM, Young WL, Lawton MT, Higashida R, Martin A, Saloner D, Computational approach to quantifying hemodynamic forces in giant cerebral aneurysms, American Journal of Neuroradiology 24(9):1804–1810, 2003. [PMC free article] [PubMed] [Google Scholar]
- 6.Steinman DA, Milner JS, Norley CJ, Lownie SP, Holdsworth DW, Image-based computational simulation of flow dynamics in a giant intracranial aneurysm, American Journal of Neuroradiology 24(4):559–566, 2003. [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang J, Johnson K, Valen-Sendstad K, Mardal KA, Wieben O, Strother C, Flow characteristics in a canine aneurysm model: A comparison of 4d accelerated phase-contrast mr measurements and computational fluid dynamics simulations, Medical Physics 38(11):6300–6312, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain K, Jiang J, Strother C, Mardal KA, Transitional hemodynamics in intracranial aneurysms - comparative velocity investigations with high resolution lattice boltzmann simulations, normal resolution ansys simulations, and mr imaging, Medical Physics 43(11):6186–6198, 2016. [DOI] [PubMed] [Google Scholar]
- 9.Detmer FJ, Luckehe D, Mut F, Slawski M, Hirsch S, Bijlenga P, von Voigt G, Cebral JR, Comparison of statistical learning approaches for cerebral aneurysm rupture assessment, International Journal of Computer Assisted Radiology and Surgery 15:141–150, 2020. [DOI] [PubMed] [Google Scholar]
- 10.Tanioka S, Ishida F, Yamamoto A, Shimizu S, Sakaida H, Toyoda M, Kashiwagi N, Suzuki H, Machine learning classification of cerebral aneurysm rupture status with morphologic variables and hemodynamic parameters, Radiology: Artificial Intelligence 2(1):e190077, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sunderland K, Wang M, Pandey A, Gemmete J, Huang Q, Goudge A, Jiang J, Quantitative analysis of flow vortices: differentiation of unruptured and ruptured medium-sized middle cerebral artery aneurysms, Acta Neurochirurgica 163(8):2339–2349, 2021. [DOI] [PubMed] [Google Scholar]
- 12.Xiang J, Antiga L, Varble N, Snyder KV, Levy EI, Siddiqui AH, Meng H, Aview: An image-based clinical computational tool for intracranial aneurysm flow visualization and clinical management, Ann Biomed Eng 44:1085–1096, 2016. [DOI] [PubMed] [Google Scholar]
- 13.Antiga L, Ene-Iordache B, Caverni L, Paolo Cornalba G, Remuzzi A, Geometric reconstruction for computational mesh generation of arterial bifurcations from ct angiography, Computerized Medical Imaging and Graphics 26(4):227–235, 2002. [DOI] [PubMed] [Google Scholar]
- 14.Seo JH, Eslami P, Caplan J, Tamargo RJ, Mittal R, A highly automated computational method for modeling of intracranial aneurysm hemodynamics, Frontiers in Physiology 9, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obiols-Sales O, Vishnu A, Malaya N, Chandramowliswharan A, Cfdnet: A deep learning-based accelerator for fluid simulations, Proceedings of the 34th ACM International Conference on SupercomputingICS ‘20, ICS ‘20, Association for Computing Machinery, New York, NY, USA, 2020. ISBN 9781450379830. doi: 10.1145/3392717.3392772, URL 10.1145/3392717.3392772. [DOI] [Google Scholar]
- 16.Ribeiro MD, Rehman A, Ahmed S, Dengel A, DeepCFD: Efficient Steady-State Laminar Flow Approximation with Deep Convolutional Neural Networks, 2020.
- 17.Arzani A, Wang JX, Sacks MS, Shadden SC, Machine learning for cardiovascular biomechanics modeling: Challenges and beyond, Annals of Biomedical Engineering pp. 1–13, 2022. [DOI] [PubMed] [Google Scholar]
- 18.Yevtushenko P, Goubergrits L, Gundelwein L, Setio A, Ramm H, Lamecker H, Heimann T, Meyer A, Kuehne T, Schafstedde M, Deep learning based centerline-aggregated aortic hemodynamics: An efficient alternative to numerical modeling of hemodynamics, IEEE Journal of Biomedical and Health Informatics 26(4):1815–1825, 2021. [DOI] [PubMed] [Google Scholar]
- 19.Fossan FE, Müller LO, Sturdy J, Bråten AT, Jørgensen A, Wiseth R, Hellevik LR, Machine learning augmented reduced-order models for ffr-prediction, Computer Methods in Applied Mechanics and Engineering 384:113892, 2021. [Google Scholar]
- 20.Li G, Wang H, Zhang M, Tupin S, Qiao A, Liu Y, Ohta M, Anzai H, Prediction of 3d cardiovascular hemodynamics before and after coronary artery bypass surgery via deep learning, Communications biology 4(1):1–12, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivantsits M, Goubergrits L, Kuhnigk JM, Huellebrand M, Bruening J, Kossen T, Pfahringer B, Schaller J, Spuler A, Kuehne T, Jia Y, Li X, Shit S, Menze B, Su Z, Ma J, Nie Z, Jain K, Liu Y, Lin Y, Hennemuth A, Detection and analysis of cerebral aneurysms based on x-ray rotational angiography - the cada 2020 challenge, Medical Image Analysis 77:102333, 2022. [DOI] [PubMed] [Google Scholar]
- 22.Poethke J, Spuler A, Petz C, Hege HC, Goubergrits L, Affeld K, Kertzscher U, Cerebral aneurysm hemodynamics and a length of parent vessel, World Congress on Medical Physics and Biomedical Engineering, September 7–12, 2009, Munich, Germany, Springer, pp. 1608–1611, 2009. [Google Scholar]
- 23.Piccinelli M, Veneziani A, Steinman DA, Remuzzi A, Antiga L, A framework for geometric analysis of vascular structures: application to cerebral aneurysms, IEEE transactions on medical imaging 28(8):1141–1155, 2009. [DOI] [PubMed] [Google Scholar]
- 24.Müller D, Kramer F, Miscnn: a framework for medical image segmentation with convolutional neural networks and deep learning, BMC Medical Imaging 21(1):1–11, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B, Efficient multi-scale 3d cnn with fully connected crf for accurate brain lesion segmentation, Medical image analysis 36:61–78, 2017. [DOI] [PubMed] [Google Scholar]
- 26.Ronneberger O, Fischer P, Brox T, U-net: Convolutional networks for biomedical image segmentation, Navab N, Hornegger J, Wells WM, Frangi AF (eds.), Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, Springer International Publishing, Cham, pp. 234–241, 2015. [Google Scholar]
- 27.Sunderland K, Jiang J, Multivariate analysis of hemodynamic parameters on intracranial aneurysm initiation of the internal carotid artery, Medical engineering & physics 74:129–136, 2019. [DOI] [PubMed] [Google Scholar]
- 28.Gwilliam MN, Hoggard N, Capener D, Singh P, Marzo A, Verma PK, Wilkinson ID, Mr derived volumetric flow rate waveforms at locations within the common carotid, internal carotid, and basilar arteries, Journal of Cerebral Blood Flow & Metabolism 29(12):1975–1982, 2009. [DOI] [PubMed] [Google Scholar]
- 29.McGraw KO, Wong SP, Forming inferences about some intraclass correlation coefficients., Psychological methods 1(1):30, 1996. [Google Scholar]
- 30.Bland JM, Altman D, Statistical methods for assessing agreement between two methods of clinical measurement, The lancet 327(8476):307–310, 1986. [PubMed] [Google Scholar]
- 31.Koo TK, Li MY, A guideline of selecting and reporting intraclass correlation coefficients for reliability research, Journal of chiropractic medicine 15(2):155–163, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ranganathan P, Pramesh C, Aggarwal R, Common pitfalls in statistical analysis: Measures of agreement, Perspectives in clinical research 8(4):187, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sunderland K, Huang Q, Strother C, Jiang J, Two Closely Spaced Aneurysms of the Supraclinoid Internal Carotid Artery: How Does One Influence the Other?, Journal of Biomechanical Engineering 141(11), 2019. [DOI] [PubMed] [Google Scholar]
- 34.Salvi M, Acharya UR, Molinari F, Meiburger KM, The impact of pre- and post-image processing techniques on deep learning frameworks: A comprehensive review for digital pathology image analysis, Computers in Biology and Medicine 128:104129, 2021. doi: 10.1016/j.compbiomed.2020.104129, URL https://www.sciencedirect.com/science/article/pii/S0010482520304601. [DOI] [PubMed] [Google Scholar]
- 35.Fu W, Breininger K, Schaffert R, Ravikumar N, Würfl T, Fujimoto J, Moult E, Maier A, Frangi-net, Maier A, Deserno TM, Handels H, Maier-Hein KH, Palm C, Tolxdorff T (eds.), Bildverarbeitung für die Medizin 2018, Springer Berlin Heidelberg, Berlin, Heidelberg, pp. 341–346, 2018. [Google Scholar]
- 36.Mu N, Lyu Z, Rezaeitaleshmahalleh M, Tang J, Jiang J, An attention residual u-net with differential preprocessing and geometric postprocessing: Learning how to segment vasculature including intracranial aneurysms, Medical Image Analysis 84:102697, 2023. doi: 10.1016/j.media.2022.102697, URL https://www.sciencedirect.com/science/article/pii/S1361841522003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mu N, Lyu Z, Rezaeitaleshmahalleh M, Zhang X, Rasmussen T, McBane R, Jiang J, Automatic segmentation of abdominal aortic aneurysms from ct angiography using a context-aware cascaded u-net, Computers in Biology and Medicine p. 106569, 2023. doi: 10.1016/j.compbiomed.2023.106569, URL https://www.sciencedirect.com/science/article/pii/S0010482523000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jirk M, Lukes V, Svobodova M, Zelezny M, Image segmentation in medical imaging via graph-cuts, 2014.
- 39.Liang L, Steinman DA, Brina O, Chnafa C, Cancelliere NM, Pereira VM, Towards the clinical utility of cfd for assessment of intracranial aneurysm rupture – a systematic review and novel parameter-ranking tool, Journal of NeuroInterventional Surgery 11(2):153–158, 2019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


