Abstract
RB1BUS6lacgpt, a Marek’s disease virus (MDV) mutant having a disrupted glycoprotein D (gD) homolog gene, established infection and induced tumors in chickens exposed to it by inoculation or by contact. Lymphoblastoid cell lines derived from RB1BUS6lacgpt-induced tumors harbored only the mutant virus. These results provide strong evidence that an intact gD homolog gene is not essential for oncogenicity or horizontal transmission of MDV.
Marek’s disease virus (MDV) is an avian herpesvirus principally known for causing T-cell lymphomas and/or nerve lesions in chickens (4, 27). In commercial poultry operations, Marek’s disease (MD) has been controlled since the early 1970s by vaccination with live, nononcogenic strains of MDV and/or a related virus, herpesvirus of turkeys. MDV was originally classified as a gammaherpesvirus based on its biological properties; however, its genome organization and content more closely resemble those of the alphaherpesviruses (3). Thus, even though MDV is somewhat difficult to manipulate experimentally, it has emerged as a most interesting herpesvirus, having molecular ties to the alphaherpesviruses but biological similarities to the gammaherpesviruses.
This report focuses on the MDV glycoprotein D (gD) homolog gene. The predicted MDV protein shows 43.8, 42.9, and 41% homology to the gD homologs of herpes simplex virus type 1 (HSV-1), pseudorabies virus (PRV), and equine herpesvirus type 1, respectively (31). It has been hypothesized that the MDV gD homolog is not essential for cell-to-cell spread of MDV but is important for the generation of cell-free infectious MDV from feather follicle epithelium. This hypothesis was based on the observations that (i) expression of an MDV gD homolog in cell culture where only cell-associated virus is produced has not been demonstrated; (ii) the gD homolog gene appears to be nonessential for cell culture propagation of MDV (11, 25); (iii) gp50, the gD homolog of PRV, is not required for cell-to-cell spread of PRV in a noncomplementing cell line or in mice (2, 20, 28); and (iv) varicella-zoster virus, also a strictly cell-associated virus, lacks a gD gene altogether (6). The hypothesis that MDV gD functions to produce cell-free virus from feather follicle epithelium predicts that the MDV gD gene would be important for horizontal transmission of MD during a natural infection but not for cell culture propagation of MDV or manifestation of disease in inoculated chickens.
Construction of the RB1BUS6lacgpt mutant.
MDV strain RB1B (32) at passage level 18 (RB1Bp18) was the parent strain for mutant construction. A mutagenesis cassette (Fig. 1A) containing the Escherichia coli lacZ and gpt genes was constructed by inserting a lacZ cassette (34) into pVEC12, a plasmid derived from pSV2gpt (21), which contains the E. coli gpt gene expressed from the simian virus 40 early promoter. The resulting plasmid, pMD181, contains the gpt and lacZ genes expressed in opposite orientations toward the bidirectional transcriptional terminator that was part of the original lacZ gene from pCH110. Plasmid pMD183 was made essentially as described for pMD175, the GAUS6lac transfer vector (25), except that the lacZ-gpt cassette from pMD181 was used instead of the lacZ cassette.
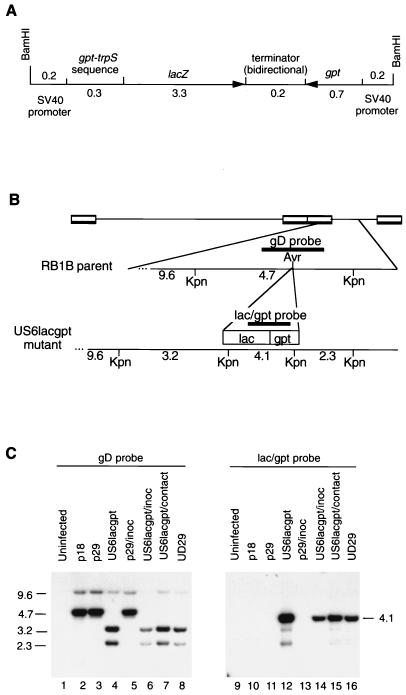
FIG. 1.
(A) Diagram of the 4.9-kb mutagenesis cassette containing both the lacZ and gpt genes of E. coli. These genes are expressed from simian virus 40 (SV40) early promoter sequences and share a bidirectional transcription terminator. (B) Diagram of the relevant region of the MDV genome for the RB1B parent and the RB1BUS6lacgpt mutant. The positions of the gD-specific and lacZ-specific probes, as well as the KpnI sites, are shown. (C) Southern hybridization analysis of the RB1BUS6lacgpt mutant and corresponding parent strains. Approximately 10 μg of DNA from MDV-infected CEF was KpnI digested, and the resulting fragments were separated on a 0.6% agarose gel and transferred to nitrocellulose. The MDV gD-specific probe used for the Southern analysis was a 1-kb AvaI fragment purified from pMD100 (5) which traverses the AvrII insertion site. The cassette-specific probe was a 2.4-kb EcoRV fragment purified from pMD181 which spans the junction between the gpt and lacZ genes. Probe DNA fragments were gel purified and biotin labelled with the Gene Images random-primed DNA labelling kit (United States Biochemical Corp., Cleveland, Ohio). Lanes: 1 and 9, uninfected CEF DNA; 2 and 10, RB1Bp18-infected CEF DNA; 3 and 11, RB1Bp29-infected CEF DNA; 4 and 12, RB1BUS6lacgpt-infected CEF DNA; 5 and 13, DNA from RB1Bp29 reisolated from inoculated chickens onto CEF; 6 and 14, DNA from RB1BUS6lacgpt reisolated from inoculated chickens onto CEF; 7 and 15, DNA from RB1BUS6lacgpt reisolated from contact-exposed chickens onto CEF; 8 and 16, DNA from virus reactived from the MDCC-UD29 lymphoblastoid cell line, a cell line derived from an RB1BUS6lacgpt-induced tumor present in a contact-exposed chicken. Lanes 1 to 8 were probed with the gD-specific probe, and lanes 9 to 16 were probed with the lacZ-gpt cassette-specific probe. Sizes are indicated in kilobase pairs.
Cotransfection of 0.5 μg of pMD183 and 12 μg of RB1B DNA into secondary chicken embryo fibroblasts (CEF) was done as detailed previously for the construction of MDV mutants (5, 26), with some modifications, since we could select for the gpt gene (15, 22). The selective medium used consisted of M199 supplemented with 1% calf serum, 100 IU of penicillin G per ml, 100-μg/ml dihydrostreptomycin, 250-μg/ml xanthine, 13.6-μg/ml hypoxanthine, 8-μg/ml thymidine, 10-μg/ml glycine, 2-μg/ml aminopterin, and 1 to 10-μg/ml mycophenolic acid, depending on how efficient the selection appeared to be. The final mutant stock, RB1BUS6lacgpt, was obtained at passage level 29. A derivative of the original parent virus passed alongside the mutant during its construction was designated RB1Bp29.
Southern hybridization of KpnI-digested DNA from uninfected CEF or CEF infected with RB1Bp18, RB1Bp29, or RB1BUS6lacgpt indicated that the mutant stock was free of detectable contaminating parent virus (Fig. 1). In particular, a 4.7-kb KpnI fragment present in the parent viruses was absent in the mutant virus stock and this band was replaced by the expected 3.2-kb and 2.3-kb species (Fig. 1C, lane 4). The lacZ-specific probe detected an expected 4.1-kb band (Fig. 1C, lane 12). DNA from CEF infected with RB1BUS6lacgpt reisolated from chickens or reactivated from MDCC-UD29, a lymphoblastoid cell line established from an RB1BUS6lacgpt-induced tumor, had indistinguishable hybridization patterns (Fig. 1C, lanes 6 to 8 and 14 to 16). Additional Southern hybridizations using different restriction enzyme digestions and PCR analysis indicated that while no parental sequences were present in the mutant stock, a minor population of genomes had the mutagenesis cassette in the opposite orientation.
The AvrII insertion site, which was disrupted in the RB1BUS6lacgpt mutant, is positioned approximately one-third of the way into the MDV US6 coding sequence. It is possible that a truncated version of gD containing the first 154 amino acids but lacking the 249 carboxy-terminal amino acids could be produced. Based on the similarity of the coding sequence to that of HSV-1 gD, it is extremely unlikely that such a truncated gD protein could function. The predicted gD homologs of MDV and HSV-1 are strikingly similar with regard to the positioning of the six essential cysteine residues in HSV-1 gD. Two independent studies (7, 19) have demonstrated that deletions in the region containing these cysteine residues result in nonfunctional gD.
RNA expression in the RB1BUS6lacgpt mutant.
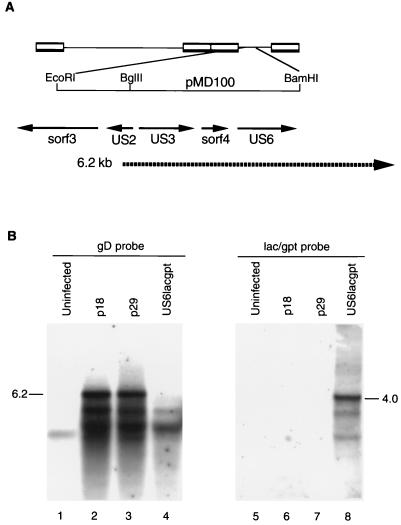
Northern hybridizations using RNA from uninfected CEF or CEF infected with RB1Bp18, RB1Bp29, or RB1BUS6lacgpt indicated that a 6.2-kb transcript present in RB1Bp18- or RB1Bp29-infected CEF was missing in cells infected with the mutant stock (Fig. 2, lane 4). This transcript has been reported previously to be present in CEF infected with an attenuated MDV, namely, GAatt85 (25), and it is believed to encode the US3 homolog or sorf4 gene product, both of which map 5′ to the gD homolog gene. The 4.0-kb lacZ transcript was present only in RNA from mutant-infected CEF (Fig. 2, lane 8).
FIG. 2.
Northern hybridization analysis of RB1BUS6lacgpt mutant and corresponding parent RNAs. Approximately 10 μg of total RNA from each sample was separated on a 1.2% agarose-formaldehyde gel and transferred to nitrocellulose. The gD-specific probe was a T7-derived riboprobe made from pMD168 by in vitro transcription with [32P]CTP. Plasmid pMD168 contains the 0.7-kb AvrII-BamHI fragment from pMD100 (5) and covers the 3′ half of the US6 coding region. The lacZ-specific probe was identical to that used for Southern hybridization. (A) Diagram of the relevant region of the MDV genome showing the positions of known open reading frames and the previously described 6.2-kb transcript that spans the gD coding region. (B) Northern blot analysis. Lanes: 1 and 5, uninfected CEF RNA; 2 and 6, RB1Bp18-infected CEF RNA; 3 and 7, RB1Bp29-infected CEF RNA; 4 and 8, RB1BUS6lacgpt-infected CEF RNA. Lanes 1 to 4 were probed with the gD-specific probe, and lanes 5 to 8 were probed with the lacZ-gpt cassette-specific probe. The positions of the 6.2-kb transcript and the 4.0-kb lacZ-specific transcript are indicated.
Reisolation of RB1BUS6lacgpt from infected chickens.
Two experiments were done to assess the phenotype of the RB1BUS6lacgpt mutant in inoculated and contact-exposed chickens. Briefly, 1-day-old specific-pathogen-free single-comb white Leghorn chickens (SPAFAS, Norwich, Conn.) were inoculated intra-abdominally with cell-associated RB1Bp18, RB1Bp29, or RB1BUS6lacgpt at the doses indicated in Table 1. At approximately 1, 2, and 3 weeks postinoculation (p.i.), spleen cells and peripheral blood lymphocytes (PBL) were isolated from three chickens per group as described previously (26). Two weeks after the initiation of the experiment, 1-day-old chicks were placed in the isolation units with the 14-day-old inoculates. These birds were exposed to MDV only by contact with the previously inoculated chickens, which at that time should have been shedding MDV. Over a 10-week period, chickens were observed daily for signs of MD, and at the end of the experiment, all remaining chickens were euthanized and representative tissues were processed for histological evaluation.
TABLE 1.
Incidence of tumors in chickens exposed to RB1BUS6lacgpt
| Virus and dose(s)a | Inoculates
|
Contacts
|
||
|---|---|---|---|---|
| No. of chickens | % with MDb | No. of chickens | % with MD | |
| Expt 1 | ||||
| None | 17 | 0 | 9 | 0 |
| RB1Bp18 | ||||
| 110 PFU | 23 | 78 | 8 | 88 |
| RB1Bp29 | ||||
| 80 PFU | 25 | 28 | 8 | 0 |
| 700 PFU | 18 | 72 | 8 | 0 |
| RB1BUS6lacgpt | ||||
| 60 PFU | 21 | 62 | 10 | 30 |
| 580 PFU | 19 | 42 | 10 | 60 |
| Expt 2 | ||||
| None | 9 | 0 | 5 | 0 |
| RB1Bp18 | ||||
| 120 PFU | 20 | 85 | 12 | 75 |
| 1,200 PFU | 18 | 100 | 13 | 77 |
| RB1Bp29 | ||||
| 60 PFU | 16 | 25 | 10 | 0 |
| 600 PFU | 19 | 68 | 9 | 0 |
| RB1BUS6lacgpt | ||||
| 50 PFU | 19 | 47 | 9 | 55 |
| 560 PFU | 20 | 45 | 10 | 60 |
For inoculates, the dose in PFU per chicken is indicated. For contacts, the indicated dose is that inoculated into cagemates.
Number of chickens with gross MD lesions/total × 100.
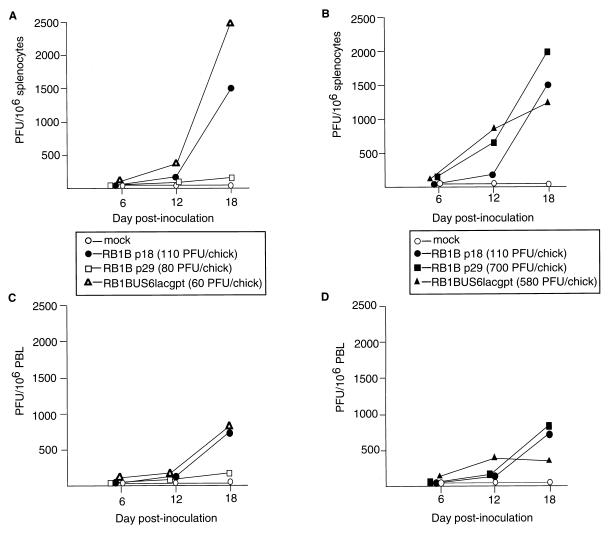
MDV was reisolated from both spleen cells and PBL of chickens inoculated with the RB1BUS6lacgpt mutant or with either of the control RB1B strains (Fig. 3). In most cases, virus reisolations were most efficient at 18 days p.i. of the three time points tested, the exception being PBL reisolations from chickens inoculated with a relatively high dose of the RB1BUS6lacgpt mutant. For chickens inoculated with the low doses (Fig. 3A and C), the RB1BUS6lacgpt mutant appeared to be more efficiently recovered than either parent virus at 18 days p.i., whereas for chickens inoculated with the high doses (Fig. 3B and D), the opposite occurred. These differences are probably of little importance, since cells were derived from only three chickens and one or more animals having high or low levels of spleen infection could alter the results obtained.
FIG. 3.
Virus reisolations from chickens inoculated with the RB1BUS6lacgpt and corresponding parent strains. Shown are results of virus reisolations from splenocytes (A and B) or PBL (C and D) of chickens inoculated with mock-infected CEF, RB1Bp18-infected CEF, RB1Bp29-infected CEF, or RB1BUS6lacgpt-infected CEF. Virus doses in PFU per chick are indicated in parentheses. Data from chickens inoculated with RB1Bp29 or RB1BUS6lacgpt at relatively low (A and C) or relatively high doses (B and D) are shown. Data are expressed as PFU obtained on the reisolation dishes per 106 cells.
Five weeks after contact-exposed chickens were placed in cages with inoculates, virus isolation attempts were made. MDV was isolated from splenocytes and PBL of chickens exposed by contact to either RB1BUS6lacgpt or RB1Bp18; however, no virus was detected in spleen cells or PBL of chickens exposed by contact to RB1Bp29 (data not shown and see below).
Regardless of whether the RB1BUS6lacgpt strain was reisolated from inoculates or contact-exposed chickens, all plaques on the reisolation dishes were Lac+. Southern hybridization and PCR on DNA prepared from reisolated viruses indicated that no detectable parent virus was present (Fig. 1C, lanes 6 and 7), the mutant retained the expected genomic structure after passage through the chicken, and the reisolated virus stocks continued to contain both genomic isomers with regard to the orientation of the lacZ-gpt cassette.
Oncogenicity of the RB1BUS6lacgpt insertion mutant.
Tumors were evident in all groups inoculated with the various RB1B strains, including RB1BUS6lacgpt (Table 1). Chickens exposed to RB1BUS6lacgpt by contact also sustained tumors. This result clearly indicated that an intact gD gene is not required for horizontal transmission of virulent MDV. It was interesting that the RB1Bp29 equally passaged parent did not cause tumors in contact-exposed chickens. This result, coupled with the observation that this derivative could not be reisolated from spleens or PBL of contact-exposed chickens, led us to believe that it had sustained some attenuation during cell culture passage.
Establishment of lymphoblastoid cell lines from RB1BUS6lacgpt-induced lymphomas.
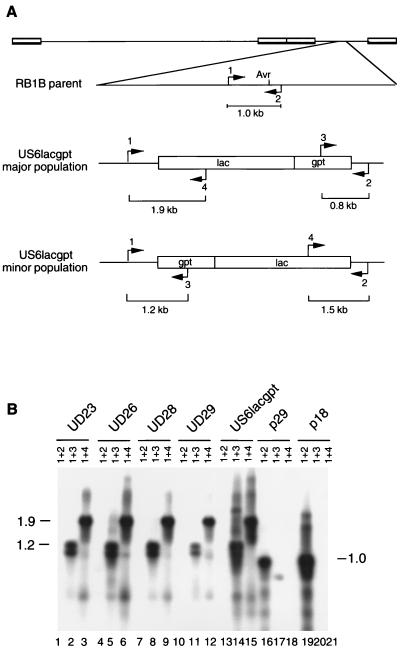
Lymphoblastoid cell lines were established from mutant-induced lymphomas as previously described (26). Eight lymphoblastoid cell lines were established from RB1BUS6lacgpt-induced tumors. The tumors from which these cell lines were derived were removed from five different chickens, namely, three inoculates and two contact-exposed chickens. PCR indicated that DNA present in all of the lymphoblastoid cell lines did not contain detectable parent virus, and data from four of these cell lines are shown in Fig. 4, lanes 1, 4, 7, and 10. Furthermore, these cell lines continued to contain both populations of the mutant with respect to the orientation of the lacZ-gpt cassette (Fig. 4, lanes 2, 3, 5, 6, 8, 9, 11, and 12). MDV that was spontaneously reactivated from these cell lines (26) was exclusively Lac+. PCR results obtained by using DNA purified from CEF infected with reactivated viruses were identical to those shown in Fig. 4.
FIG. 4.
PCR analysis of DNA from lymphoblastoid cell lines derived from RB1BUS6lacgpt-induced tumors. (A) Diagram of the structure of the RB1B parent and both isomers of the RB1BUS6lacgpt mutant with regard to the relevant region of the genome. The orientations of the mutagenesis cassette in the major and minor populations of the mutant are indicated. The positions of PCR primers, as well as the sizes of the predicted products, are shown. Primers 1 and 2, which flank the AvrII insertion site, should direct amplification of a 1.0-kb product from DNA purified from parent-infected CEF but no product from DNA purified from mutant-infected CEF under the PCR conditions used. (B) Southern blot of PCR products from analysis of lymphoblastoid cell line DNAs. PCR products were separated by agarose gel electrophoresis, transferred to nitrocellulose, and probed with the gD-specific probe described in the legend to Fig. 1. The primer pairs used are indicated above the lanes. Templates used are as follows: lanes 1 to 3, MDCC-UD23 DNA; lanes 4 to 6, MDCC-UD26 DNA; lanes 7 to 9, MDCC-UD28 DNA; lanes 10 to 12, MDCC-UD29 DNA; lanes 13 to 15, RB1BUS6lacgpt-infected CEF DNA; lanes 16 to 18, RB1Bp29-infected CEF DNA; lanes 19 to 21, RB1Bp18-infected CEF DNA. Sizes are indicated in kilobase pairs.
The nature of so-called cell-free MDV produced in feather follicle epithelium needs to be briefly discussed. We noted the presence of both mutant isomers in DNA purified directly from lymphoblastoid cells established from contact-exposed chickens, as well as in virus reactivated from the cells. These results indicate that even in a contact-exposed chicken, target cells for transformation are infected with more than a single viral particle. One explanation for this is that the infectious unit released from feather follicles contains more than a single viral particle. In the contact-exposed chicken, this dust particle, or infectious unit, may be phagocytosed or otherwise taken into cells as infection proceeds. If this is so, then the cell-free MDV released from feather follicles differs significantly from cell-free herpesvirus particles in general. Our results could also be explained if the lacZ-gpt mutagenesis cassette is continually isomerizing within the MDV genome. If this isomerization occurs, however, it does so to a relatively constant equilibrium between the amounts of the two isomers present, since the relative proportions of the two isomers did not change among the mutant stocks and reisolated viruses. In addition, this isomerization does not result in detectable loss or alteration of the mutagenesis cassette.
It is indeed unusual for a herpesvirus gD gene, if present, to be nonessential or without apparent function. Homologs of HSV-1 gD (16, 38) have been reported for HSV-2 (17, 37), PRV (29), bovine herpesvirus type 1 (36), and equine herpesvirus type 1 (1, 35, 40). In all cases, the glycoprotein is essential for productive infection (8, 10, 28, 30, 40). Much effort has been placed on elucidating the function of gD. HSV-1 gD is not required for virus binding to heparin sulfate but is necessary for fusion and penetration into cells (9, 12–14, 24). The structure-function relationships of domains of gD-1 are complex, with different structural domains being important in different functional assays (23). Recent work indicates that gD binds directly to herpesvirus entry mediator (HVEM), a member of the tumor necrosis factor/nerve growth factor receptor family, to mediate HSV entry into cells (18, 39). In the case of MDV, other glycoproteins may have evolved to compensate for lack of gD expression. Such appears to be the case for gD-negative derivatives of PRV (2, 20, 28) and bovine herpesvirus type 1 (33).
In summary, the RB1BUS6lacgpt mutant was able to efficiently establish infection in chicken spleen cells and PBL, induce tumors in inoculated chickens, and spread horizontally, causing tumors in contact-exposed animals. In addition, lymphoblastoid cell lines established from tumors present in contact-exposed chickens harbored only the mutant virus. These results provide compelling evidence that an intact gD gene is not required for MDV oncogenicity or horizontal transmission.
Acknowledgments
We thank Paul Sondermeijer for the pVEC12 plasmid.
This work was supported by grants from the U.S. Department of Agriculture (92-37204-8039), the American Cancer Society (VM-44), the Delaware Research Partnership, and Intervet International, Boxmeer, The Netherlands.
Footnotes
Paper no. 1627 in the Journal Series of the Delaware Agricultural Experiment Station.
REFERENCES
- 1.Audonnet J-C, Winslow J, Allen G, Paoletti E. Equine herpesvirus type 1 unique short fragment encodes glycoproteins with homology to herpes simplex virus type 1 gD, gI and gE. J Gen Virol. 1991;71:2969–2978. doi: 10.1099/0022-1317-71-12-2969. [DOI] [PubMed] [Google Scholar]
- 2.Babic N, Mettenleiter T C, Flamand A, Ugolini G. Role of essential glycoproteins gII and gp50 in transneuronal transfer of pseudorabies virus from the hypoglossal nerve of mice. J Virol. 1993;67:4421–4426. doi: 10.1128/jvi.67.7.4421-4426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckmaster A E, Scott S D, Sanderson M J, Boursnell M E G, Ross N L J, Binns M M. Gene sequence and mapping data from Marek’s disease virus and herpesvirus of turkeys: implication for herpesvirus classification. J Gen Virol. 1988;69:2033–2042. doi: 10.1099/0022-1317-69-8-2033. [DOI] [PubMed] [Google Scholar]
- 4.Calnek B W, Witter R L. Marek’s disease. In: Calnek B W, editor. Diseases of poultry. Ames: Iowa State University Press; 1997. pp. 369–413. [Google Scholar]
- 5.Cantello J L, Anderson A S, Francesconi A, Morgan R W. Isolation of a Marek’s disease virus (MDV) recombinant containing the lacZ gene of Escherichia coli stably inserted within the MDV US2 gene. J Virol. 1991;65:1584–1588. doi: 10.1128/jvi.65.3.1584-1588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davison A J, McGeoch D J. Evolutionary comparisons of the S segments in the genomes of herpes simplex virus type 1 and varicella-zoster virus. J Gen Virol. 1986;67:597–611. doi: 10.1099/0022-1317-67-4-597. [DOI] [PubMed] [Google Scholar]
- 7.Feenstra V, Hodaie M, Johnson D C. Deletions in herpes simplex virus glycoprotein D define nonessential and essential domains. J Virol. 1990;64:2096–2102. doi: 10.1128/jvi.64.5.2096-2102.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fehler F, Hermann J M, Saalmuller A, Mettenleiter T C, Keil G M. Glycoprotein IV of bovine herpesvirus 1-expressing cell line complements and rescues a conditionally lethal viral mutant. J Virol. 1992;66:831–839. doi: 10.1128/jvi.66.2.831-839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller A O, Spear P G. Anti-glycoprotein D antibodies that permit adsorption but block infection by herpes simplex virus 1 prevent virion-cell fusion at the cell surface. Proc Natl Acad Sci USA. 1987;84:5454–5458. doi: 10.1073/pnas.84.15.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Highlander S L, Sutherland S L, Gage P J, Johnson D C, Levine M, Glorioso J C. Neutralizing monoclonal antibodies specific for herpes simplex virus glycoprotein D inhibit virus penetration. J Virol. 1987;61:3356–3364. doi: 10.1128/jvi.61.11.3356-3364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isfort R J, Qian Z, Jones D, Silva R F, Witter R, Kung H J. Integration of multiple chicken retroviruses into multiple chicken herpesviruses: herpesviral gD as a common target of integration. Virology. 1994;203:125–133. doi: 10.1006/viro.1994.1462. [DOI] [PubMed] [Google Scholar]
- 12.Johnson D C, Burke R L, Gregory T. Soluble forms of herpes simplex virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson D C, Ligas M W. Herpes simplex viruses lacking glycoprotein D are unable to inhibit virus penetration: quantitative evidence for virus-specific cell surface receptors. J Virol. 1988;62:4605–4612. doi: 10.1128/jvi.62.12.4605-4612.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall D R, Reilly J D, Liu X, Silva R F. Selection of Marek’s disease virus recombinants expressing the Escherichia coli gpt gene. Virology. 1993;195:638–648. doi: 10.1006/viro.1993.1415. [DOI] [PubMed] [Google Scholar]
- 16.McGeoch D J, Doland A, Donald S, Rixon F J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1985;181:1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- 17.McGeoch D J, Moss H W M, McNab D, Frame M C. DNA sequence and genetic content of the HindIII 1 region in the short unique component of the herpes simplex virus type 2 genome: identification of the gene encoding glycoprotein G, and evolutionary comparisons. J Gen Virol. 1987;68:19–38. doi: 10.1099/0022-1317-68-1-19. [DOI] [PubMed] [Google Scholar]
- 18.Montgomery R I, Warner M S, Lum B J, Spear P J. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/HBF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 19.Muggeridge M I, Wilcox W C, Cohen G H, Eisenberg R J. Identification of a site on herpes simplex virus type 1 glycoprotein D that is essential for infectivity. J Virol. 1990;64:3617–3626. doi: 10.1128/jvi.64.8.3617-3626.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulder W, Pol J, Kimman T, Kok G, Priem J, Peeters B. Glycoprotein D-negative pseudorabies virus can spread transneuronally via direct neuron-to-neuron transmission in its natural host, the pig, but not after additional inactivation of gE or gI. J Virol. 1996;70:2191–2200. doi: 10.1128/jvi.70.4.2191-2200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulligan R C, Berg P. Expression of a bacterial gene in mammalian cells. Science. 1980;209:1422–1427. doi: 10.1126/science.6251549. [DOI] [PubMed] [Google Scholar]
- 22.Mulligan R C, Berg P. Selection for animal cells that express the Escherichia coli gene coding for xanthine-guanine phosphoribosyltransferase. Proc Natl Acad Sci USA. 1981;78:2072–2076. doi: 10.1073/pnas.78.4.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noble A G, Lee G T-Y, Sprague R, Parish M L, Spear P G. Anti-gD monoclonal antibodies inhibit cell fusion induced by herpes simplex virus type 1. Virology. 1983;129:218–224. doi: 10.1016/0042-6822(83)90409-9. [DOI] [PubMed] [Google Scholar]
- 25.Parcells M S, Anderson A S, Morgan R W. Characterization of a Marek’s disease virus mutant containing a lacZ insertion in the US6 (gD) homologue gene. Virus Genes. 1994;9:5–13. doi: 10.1007/BF01703430. [DOI] [PubMed] [Google Scholar]
- 26.Parcells M S, Anderson A S, Morgan R W. Retention of oncogenicity by a Marek’s disease virus mutant lacking six unique short region genes. J Virol. 1995;69:7888–7898. doi: 10.1128/jvi.69.12.7888-7898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Payne L N. Marek’s disease. Scientific basis and control. Boston, Mass: Martinus Nijhoff; 1985. [Google Scholar]
- 28.Peeters B, Pol J, Gielkens A, Moormann R. Envelope glycoprotein gp50 of pseudorabies virus is essential for virus entry but is not required for viral spread in mice. J Virol. 1993;67:170–177. doi: 10.1128/jvi.67.1.170-177.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrovskis E A, Timmins J G, Armentrout M A, Marchioli C C, Yancey R J, Post L E. DNA sequence of the gene for pseudorabies virus gp50, a glycoprotein without N-linked glycosylation. J Virol. 1986;59:216–223. doi: 10.1128/jvi.59.2.216-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rauh I, Mettenleiter T C. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J Virol. 1991;65:5348–5356. doi: 10.1128/jvi.65.10.5348-5356.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross L J N, Binns M M. Properties and evolutionary relationships of the Marek’s disease virus homologues of protein kinase, glycoprotein D and glycoprotein I of herpes simplex virus. J Gen Virol. 1991;72:939–947. doi: 10.1099/0022-1317-72-4-939. [DOI] [PubMed] [Google Scholar]
- 32.Schat K A, Calnek B W, Fabricant J. Characterization of two highly oncogenic strains of Marek’s disease virus. Avian Pathol. 1982;11:593–605. doi: 10.1080/03079458208436134. [DOI] [PubMed] [Google Scholar]
- 33.Schroder C, Linde G, Fehler F, Keil G M. From essential to beneficial: glycoprotein D loses importance for replication of bovine herpesvirus 1 in cell culture. J Virol. 1997;71:25–33. doi: 10.1128/jvi.71.1.25-33.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sondermeijer P J A, Claessens J A J, Jenniskens P E, Mockett A P A, Thijssen R A J, Willemse M J, Morgan R W. Avian herpesvirus as a live viral vector for the expression of heterologous antigens. Vaccine. 1993;11:349–358. doi: 10.1016/0264-410x(93)90198-7. [DOI] [PubMed] [Google Scholar]
- 35.Telford E A R, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 36.Van Drunen Littel-Van den Hurk S, Van den Hurk J V, Babiuk L A. Topographical analysis of bovine herpesvirus type-1 glycoproteins: use of monoclonal antibodies to identify and characterize functional epitopes. Virology. 1985;144:216–227. doi: 10.1016/0042-6822(85)90319-8. [DOI] [PubMed] [Google Scholar]
- 37.Watson R J. DNA sequence of the herpes simplex virus type 2 glycoprotein D gene. Gene. 1983;26:307–312. doi: 10.1016/0378-1119(83)90203-2. [DOI] [PubMed] [Google Scholar]
- 38.Watson R J, Weis J H, Salstrom J S, Enquist L W. Herpes simplex virus type 1 glycoprotein D gene: nucleotide sequence and expression in E. coli. Science. 1982;218:381–384. doi: 10.1126/science.6289440. [DOI] [PubMed] [Google Scholar]
- 39.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulike A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittaker G R, Taylor L A, Elton D M, Giles L E, Bonass W A, Halliburton I W, Killington R A, Meredith D M. Glycoprotein 60 of equine herpesvirus type 1 is a homologue of herpes simplex virus glycoprotein D and plays a major role in penetration of cells. J Gen Virol. 1992;73:801–809. doi: 10.1099/0022-1317-73-4-801. [DOI] [PubMed] [Google Scholar]