Abstract
In addition to the genes involved in the structure of the viral particle, the bovine leukemia virus (BLV) genome contains a region called X which contains at least four genes. Among them, the tax and rex genes, respectively, are involved in transcriptional and posttranscriptional regulation of viral transcription. Two other genes, R3 and G4, were identified after cloning of the corresponding mRNAs from BLV-infected lymphocytes. Although the function of the two latter genes is still unknown, they appear to have important roles, since deletion of them restricts viral propagation in vivo. In order to assess the oncogenic potential of the R3 and G4 proteins, we first analyzed their ability to immortalize and/or transform primary rat embryo fibroblasts (Refs). In this assay, the G4 but not the R3 protein cooperated with the Ha-ras oncogene to induce tumors in nude mice. It thus appears that G4 exhibited oncogenic potential in vitro. To extend these observations in vivo, the pathology induced by recombinant viruses with mutations in G4 and in R3 and G4 was next evaluated with the sheep animal model. Viral propagation, as measured by semiquantitative PCR, appeared to be reduced when the R3 and G4 genes were deleted. These observations confirm and extend our previous data underlining the biological function of these genes. In addition, we present the results of a clinical survey that involves 39 sheep infected with six different BLV recombinants. Over a period of 40 months, 83% of the sheep infected with a wild-type virus developed leukemias and/or lymphosarcomas. In contrast, none out of 13 sheep infected with viruses with mutations in G4 or in R3 and G4 developed disease. We conclude that in addition to its oncogenic potential in vitro, G4 is required for pathogenesis in vivo. These observations should help us gain insight into the process of leukemogenesis induced by the related human T-cell leukemia virus type 1.
To date, the main problem in understanding leukemia concerns the low rate of cellular transformation. Considering the lifetime of an individual being, the rate of cellular renewal (mean lymphocyte values), and the number of cells that are prone to become transformed, it has been calculated that 1 cell out of 1012 will evolve into a leukemic clone (3, 8, 11). As a result, it is extremely difficult to analyze the mechanisms of cellular transformation in vivo. Therefore, in vitro models have been proposed to mimic the phenomena observed in vivo. In addition, the DNA recombinant techniques have permitted dissection of the molecular aspects of cell transformation. However, both cell culture and molecular data require to be confirmed in vivo to assess their biological relevance. In that respect, animal models are essential to analyze the leukemogenic process that evolves in humans. The goal of our research is to establish a link between in vitro and in vivo techniques to gain insight into the process of leukemogenesis. Our model system is the infection of sheep and cattle by bovine leukemia virus (BLV). The pathogenesis induced by BLV in cattle is similar to chronic lymphocytic leukemia in humans. Indeed, BLV induces a permanent increase in the number of circulating B cells, called persistent lymphocytosis, which can persist over extended periods of time. In about 5% of all of the infected cattle, a very aggressive expansion of a transformed clone evolves into massive tumors that finally kill the infected host. This kind of pathogenesis is also observed in individuals infected with the human T-cell leukemia virus type 1 (HTLV-1). After extended latency periods (up to 20 to 30 years), HTLV-infected people might develop a very aggressive disease called adult T-cell leukemia that is refractory to any type of medical care (19, 26). This phase of the disease is also observed in sheep infected by BLV, but in this model, the mean latency period is decreased to 30 months. In addition, the BLV and HTLV viruses belong to the Oncovirinae subfamily and share a common genomic organization (1, 16, 18). In fact, these viruses contain, in addition to the classical structural genes of all retroviruses, a region called X located between the envelope gene and the 3′ long terminal repeat. In the BLV genome, this region contains genes that encode at least four proteins: Tax, Rex, R3, and G4. The tax gene is a transcriptional activator of viral expression that has oncogenic potential in cell culture (6, 14, 20, 21). Indeed, primary cells (rat embryo fibroblasts) can be immortalized when they constitutively express the Tax protein. Furthermore, when another oncogene (Ha-ras) is coexpressed with tax, the cells containing these genes become fully transformed and induce tumors when injected into nude mice. Both functions of Tax, i.e., immortalization and transactivation, can be dissociated on the basis of specific mutations in the protein. For example, mutations in the zinc finger structure completely abrogate transactivation, whereas the transformation potential remains unaffected (22). On the contrary, substitution of the phosphorylation sites maintains transactivation but destroys immortalization of primary cells (25a). The second protein from the X region, Rex, is a posttranscriptional regulator of viral expression required for the synthesis of structural genes (7). Both tax and rex genes are essential for viral infectivity in vivo (23). The two other genes contained in the X region, R3 and G4, are considered to be accessory genes because deletion of them from the genome does not completely abrogate the infectious potential of recombinant proviruses (23). However, these genes, whose functions are still unknown, appear to have a biological function in vivo, since their absence drastically decreases the propagation of the virus within the infected host (5, 24). The goal of this report is to help gain insight into the role of R3 and G4 in the oncogenic potential of BLV.
To this end, a first series of experiments were designed to test the transforming potential of the R3 and G4 genes in cell culture. Therefore, we used a general approach based on the transfection of primary cells which are normally programmed to die after a few rounds of division. This experimental protocol has already been used successfully to demonstrate the oncogenic potential of tax (15, 21). The detailed procedures of this strategy have been described in a previous paper (25).
Briefly, Fischer 344 rats at day 14 of gestation were anesthetized with chloroform, and the embryos were recovered after excision of the uterus. The embryonic cells were then dislodged in the presence of trypsin and cultivated at 37°C in a 5% CO2–95% air atmosphere in minimal essential medium (Gibco) supplemented with 10% fetal calf serum, 1 mM sodium pyruvate, 2 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Two days later, 2.5 million adherent cells (called rat embryo fibroblasts [Refs]) were transfected with the pSGR3 and pSGG4 effector plasmids carrying the R3 and G4 genes. These vectors contain the R3 and G4 genes from pRSR3 and pRSG4 (kindly provided by S. Alexandersen) inserted in the pSG5 expression plasmid (10). In parallel, Ref cells were transfected with an expression vector of tax as a positive control for cell transformation and with an empty plasmid (pSG5) to evaluate the background levels of the assay. All of these expression plasmids were cotransfected together with a vector that carries either the Ha-ras (pSV2neoEJ) or the myc (pSV2myc) oncogene. The goal of this cotransfection is to fully transform the Ref cells that are then capable of inducing tumors in nude mice. Indeed, the complete transformation of primary cells requires in general cooperation between at least two oncogenes to assess full malignancy (13). For example, tax alone does not induce tumors after transfection into Ref cells but necessitates the collaboration of Ha-ras. This cooperation between two oncogenes illustrates the multistep mechanism of carcinogenesis that includes a prior activation towards immortalization and a subsequent trigger by a second gene to fulfill complete transformation.
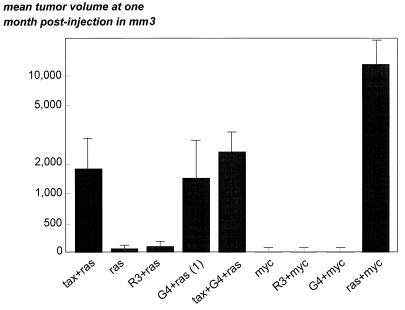
After cotransfection, the cells were cultivated for 2 days, harvested, and injected subcutaneously into the flanks of thymusless nude mice. A total of four mice in three independent experiments were injected for each reporter gene construct. The tumor volume was calculated by the ellipsoid formula: 4/3 Πab2, where a and b are, respectively, the length and the width of the tumor. The mean values among all of the tumor volumes indicated in cubic millimeters are illustrated on Fig. 1.
FIG. 1.
Oncogenic potential of G4 in cells. Adherent embryonic cells from Fischer 344 rats at day 14 of gestation were transfected with vectors containing tax (pSGTax), R3 (pSGR3), G4 (pSGG4), Ha-ras (pSV2neoEJ), and Myc (pSV2myc) as indicated. The amounts of DNA were kept constant with the pSG5 empty plasmid. After transfection, the cells were cultivated for 2 days, harvested, and injected subcutaneously into the flanks of thymusless nude mice (NMRI background). A total of four mice in three independent experiments were injected for each plasmid combination. The tumor volume was calculated by the ellipsoid formula 4/3 Πab2, where a and b are, respectively, the length and the width of the tumor. The mean values among all of the tumor volumes indicated in cubic millimeters are indicated. The cotransfection plasmids pSGG4 and pSV2neoEJ generated tumors in only one-third of the injected mice. Tumors appeared in one mouse out of three.
As demonstrated previously, the coexpression of both tax and Ha-ras into the Ref cells is able to induce tumors in nude mice (mean tumor volume at 1 month of 1,896 mm3) (21). In contrast, Ref cells transfected with the Ha-ras expression vector alone yielded only small nodules 60 mm3 in size. Similarly, coexpression of R3 and Ha-ras did not allow any growth of the transfected Ref cells. In contrast, an intermediate reaction was observed after cotransfection of G4 and Ha-ras: two-thirds of the mice were completely negative for tumor development (mean volume of 120 mm3), whereas the other animals displayed tumors 1,356 mm3 in size at 1 month postinjection. It thus appears that G4 exhibits a full oncogenic potential in only one-third of the injected mice. Coexpression of G4 together with tax and ras yielded tumors similar to those induced by tax and ras. Since G4 does not potentiate tax-induced tumor growth, it appears that both genes do not synergize in this transformation system. Finally, neither G4 nor R3 is able to cooperate with myc in transformation of Ref cells, indicating that these genes belong to different complementation groups. As a positive control, coexpression of myc and Ha-ras yielded huge tumors whose ellipsoid volume reached more than 10 cm3 at 1 month postinoculation. From these experiments, we conclude that G4, but not R3, exhibits an intermediate transforming potential in collaboration with the Ha-ras oncogene.
The goal of our project was to establish a link between experiments performed in cell culture and direct analyses in vivo. The BLV model system offers the main advantage of permitting the injection of recombinant proviruses into sheep and the analysis of their biological behavior in vivo (17, 23). We have previously described the construction of BLV recombinants which lack the R3 and G4 genes and have shown that these viruses are impaired in their ability to propagate efficiently in vivo (5, 23, 24). As a result, the proviral loads within the animals infected with the BLV mutants with deletions of the R3 and G4 genes are strongly decreased. To extend these observations, we analyzed the proviral loads in 39 sheep infected with six different BLV recombinants (Fig. 2). These mutants all derive from a wild-type provirus (strain 344), which was originally cloned from a BLV-induced sheep tumor. This proviral clone was shown to be infectious after direct inoculation into sheep, propagate efficiently within the infected animals, and induce tumors after latency periods similar to those observed after injection of blood samples contaminated with BLV. This molecular clone was used as a model template to obtain a series of mutants whose constructions were described in detail elsewhere (23) (Fig. 2). Three wild-type BLV variants were derived from provirus 344: (i) pBLV344/395, which is a hybrid between the 344 and 395 viruses harboring most of the 395 genome linked to the tax/rex region of the 344 strain; (ii) pBLVX3C, which contains the complete X3C open reading frame (ORF) corresponding to the 5′ end of the R3 second exon from the FLK/BLV strain (from fetal lamb kidney cells [18]); and (iii) pBLVIX, which contains a small deletion between two XbaI restriction sites at positions 6614 and 6732 (nucleotide positions according to reference 16). To evaluate viral propagation, these viruses were injected into a series of sheep (Table 1), and the proviral loads were measured by semiquantitative PCR (Fig. 3). Therefore, two oligonucleotides (5′-TGGAAAGAACTAACGCTGACGG-3′ at position 6450 [16] and 5′-CCCCAACCAACAACACTTGCTT-3′ at position 7060) were used to amplify viral sequences by 22 cycles of PCR as described in reference 5. As controls for semiquantification, serial dilutions of a blood lysate from sheep 210, which was infected with the 344 virus, were amplified in parallel under similar conditions (Fig. 3, sheep 210, 5× concentration, 1× lysate corresponding to 2 μl of blood, 5× and 25× dilutions). In contrast, no fragment (610 bp in size) was amplified when a lysate from an uninfected sheep (no. 120) was used. It should be mentioned here that a fragment of 490 bp instead of 610 bp was amplified in the lanes corresponding to mutants pBLV344/395 (sheep 250 and 251) and pBLVIX (sheep 247, 248, 259, 260, and 261), because these viruses contain a small deletion between two XbaI sites at positions 6614 and 6731. We should also recall that mutant pBLVDENV+DPOL was derived from the coinjection of two viruses with deletions in either the env or the pol gene, yielding the 344 clone after homologous recombination (23). It appeared that the pBLV344/395, pBLVX3C, and pBLVIX recombinants propagated at wild-type levels (pBLV344 and pBLVDENV+DPOL), as measured by the proviral loads within the infected animals (Fig. 3). These mutants may thus be considered as wild-type viruses in terms of infectivity and replication in vivo (23, 24) (Fig. 3). In contrast, the proviral loads in sheep infected with pBLVDX, pBLVRZ, and pBLVIG4 were strongly affected (Table 1 and Fig. 3). These mutants, which are illustrated in Fig. 2, harbor a mutation in the G4 gene (pBLVIG4) or deletions in both the R3 and G4 genes (pBLVDX and pBLVRZ) (see references 23 and 24 for their construction). The deletion of R3 and G4 that is responsible for the amplification of a smaller fragment of 230 bp also provides qualitative evidence for the presence of mutants pBLVDX and pBLVRZ. In two sheep (245 and 272), the proviral loads reached values which approached the wild-type levels after 40 months of infection (Fig. 3). This was not the case after a shorter latency period (24). It thus appears that the deletion of the R3 and G4 genes delays viral propagation but does not always restrict the absolute levels of proviral loads. To ensure that the injected proviruses were not revertants, the integrity of their genome was also evaluated by direct nucleotide sequence after PCR (data not shown).
FIG. 2.
Schematic representation of the recombinant BLV proviruses. Based on their ability to propagate efficiently in the animal model, the recombinant proviruses were separated into two groups. The mutants which behave similarly to the wild-type pBLV344 virus include pBLV344/395 (which is a hybrid between the 344 and 395 viruses harboring most of the 395 genome linked to the tax/rex region of the 344 strain), pBLVX3C (which contains the complete X3C ORF corresponding to the 5′ end of the R3 second exon from the FLK/BLV strain), and pBLVIX (which contains a small deletion between two XbaI restriction sites at positions 6614 and 6732). Three types of attenuated viruses were characterized: pBLVDX (which contains deletions of both the R3 and G4 genes), pBLVRZ (which is isogenic to pBLVDX but contains a ribozyme directed towards the tax gene sequences), and pBLVIG4 (in which a translational stop codon was inserted into the G4 gene). The precise description of these viruses was given elsewhere (23, 24). LTR, long terminal repeat.
TABLE 1.
Leukemia or lymphosarcoma latency period and cause of death for BLV-infected sheep in this studya
| Sheep no. | Provirus | Latency period (days):
|
Cause of death | |
|---|---|---|---|---|
| Before leukemia | Before death | |||
| 210 | pBLV344 | 1,227 | 1,231 | Lymphosarcoma |
| 214 | pBLV344 | 510 | Lymphosarcoma | |
| 224 | pBLV344 | 994 | 1,016 | Lymphosarcoma |
| 230 | pBLV344 | 420 | 448 | Lymphosarcoma |
| 234 | pBLV344 | 1,613 | Lymphosarcoma | |
| 235 | pBLV344 | |||
| 236 | pBLV344 | 867 | 1,440 | Lymphosarcoma |
| 243 | pBLV344 | 1,370 | Unrelated | |
| 244 | pBLV344 | 1,353 | Lymphosarcoma | |
| 252 | pBLV344 | 548 | 944 | Lymphosarcoma |
| 273 | pBLV344 | 306 | 691 | Lymphosarcoma |
| 274 | pBLV344 | 425 | 702 | Lymphosarcoma |
| 238 | pBLVDENV+DPOL | 1,425 | Lymphosarcoma | |
| 241 | pBLVDENV+DPOL | 1,028 | 1,208 | Lymphosarcoma |
| 250 | pBLV344/395 | 810 | 908 | Lymphosarcoma |
| 251 | pBLV344/395 | 747 | 802 | Lymphosarcoma |
| 253 | pBLVX3C | 1,249 | Unrelated | |
| 254 | pBLVX3C | 952 | 1,095 | Lymphosarcoma |
| 265 | pBLVX3C | |||
| 266 | pBLVX3C | 594 | 604 | Lymphosarcoma |
| 267 | pBLVX3C | 891 | Lymphosarcoma | |
| 247 | pBLVIX | |||
| 248 | pBLVIX | 994 | Unrelated | |
| 259 | pBLVIX | |||
| 260 | pBLVIX | 1,160 | Lymphosarcoma | |
| 261 | pBLVIX | 1,289 | Lymphosarcoma | |
| 245 | pBLVDX | |||
| 246 | pBLVDX | |||
| 262 | pBLVDX | |||
| 263 | pBLVDX | |||
| 264 | pBLVDX | |||
| 255 | pBLVRZ | |||
| 256 | pBLVRZ | 1,311 | Unrelated | |
| 257 | pBLVRZ | |||
| 237 | pBLVIG4 | |||
| 240 | pBLVIG4 | |||
| 249 | pBLVIG4 | 640 | Unrelated | |
| 271 | pBLVIG4 | 523 | Unrelated | |
| 272 | pBLVIG4 | |||
The leukemia threshold was 10,000 lymphocytes per mm3.
FIG. 3.
Proviral loads in the infected sheep. The proviral loads were estimated from blood lysates prepared at 40 months or just before the death of the animal (Table 1). Starting from an equivalent volume of 2 μl of blood, the viral sequences were amplified by 22 cycles of PCR with two oligonucleotides (5′- TGGAAAGAACTAACGCTGACGG-3′ at position 6450 [16] and 5′-CCCCAACCAACAACACTTGCTT-3′ at position 7060). The DNAs were then analyzed by Southern blot hybridization with a viral probe corresponding to the X3C sequences (positions 6528 to 6997 [16]). As controls for semiquantification, serial dilutions of a blood lysate from sheep 210, which was infected with the 344 virus, were amplified in parallel under similar conditions: 5× concentrated (conc.) (10 μl of lysate), 1× lysate (corresponding to 2 μl of blood), and 5× and 25× dilutions (dil.). A lysate from an uninfected sheep (no. 120) was used as a negative control. The sizes of the amplified fragments are indicated in base pairs.
We thus conclude that among the six recombinants which were derived from provirus 344, three of them, pBLV344/395, pBLVX3C, and pBLVIX, may be considered as wild-type viruses in terms of viral propagation in vivo. In contrast, the deletion of either R3 and G4 (in mutants pBLVDX and pBLVRZ) or G4 alone (in pBLVIG4) decreases the proviral loads. To evaluate the pathogenicity of these mutants, their ability to induce disease was monitored in the 39 infected sheep over a period of 40 months (Table 1). The wild-type 344 virus and three types of mutants (pBLV344/395, pBLVX3C, and pBLVIX) induced two kinds of pathologies characterized by a leukemia and/or a lymphosarcoma (Table 1). Leukemia, which may occur independently of the appearance of tumors (lymphosarcomas), is characterized by an increase in the absolute number of circulating blood lymphocytes. The mean latency period preceding the onset of leukemia ranged between 32 and 43 months for the variants, whereas a 33-month period was required for the wild-type 344 virus (Table 2). Despite some differences in their genome, the pBLV344, pBLV344/395, pBLVX3C, and pBLVIX proviruses thus exhibit wild-type behavior during the virus-induced pathogenesis in sheep. In contrast, the deletion of the R3 and G4 genes in the viruses pBLVDX, pBLVRZ, and pBLVIG4 strongly decreases their pathogenic potential (Table 1). Among 13 animals infected with these viruses, none of them exhibited lymphoproliferative syndromes or died from lymphosarcoma (Tables 1 and 2). We should mention here that three sheep (256, 249, and 271) died from unrelated causes linked to digestion disorders (enterotoxemia) or accidental causes. Among the wild-type virus-infected sheep, the same number of animals (sheep 243, 248, and 253) were also lost because of unrelated reasons. Thus the deletion of the G4 gene appears to be responsible not only for the decrease in the proviral loads but also for the attenuation of the pathogenic potential.
TABLE 2.
Development of leukemia or lymphosarcoma in sheep infected with BLV proviruses
| Provirus | No. of sheep with result/no. tested (%)
|
Latency period before death (mo) | |
|---|---|---|---|
| Leukemia | Lymphosarcoma | ||
| Wild type | |||
| pBLV344 | 7/11 | 10/11 | 32.7 |
| pBLVDENV+DPOL | 1/2 | 2/2 | 43.3 |
| pBLV344/395 | 2/2 | 2/2 | 28.1 |
| pBLVX3C | 2/4 | 3/4 | 28.4 |
| pBLVIX | 0/4 | 2/4 | 40.2 |
| Total | 12/23 (52) | 19/23 (83) | 33.4 |
| Attenuated | |||
| pBLVDX | 0/5 | 0/5 | |
| pBLVRZ | 0/2 | 0/2 | |
| pBLVIG4 | 0/3 | 0/3 | |
| Total | 0/10 | 0/10 | |
To summarize, among 23 animals infected with a wild-type virus, 19 of them (83%) developed a lymphosarcoma within the duration of the experiment (40 months). During the same period, none of the 13 sheep infected with the G4- or the R3- and G4-deleted proviruses developed either a leukemia or a lymphosarcoma. Altogether, these data underline the importance of the G4 gene in virus-induced pathogenesis in vivo and parallel its oncogenic potential in cell culture.
Oncogenes can be classified into two main groups on the basis of their ability to cooperate to fully transform primary rat embryo fibroblasts (13). A first series of oncogenes that includes tax, myc, adenovirus E1A, and the polyomavirus large-T antigen gene (large-T) are able to increase the rate of immortalization of the Ref cells. These genes are able to complement another class of oncogenes (like ras and polyomavirus middle-T) implicated in morphological alteration and anchorage-independent cell growth. The collaboration between immortalizing and transforming oncogenes contributes to the complete tumorigenesis of primary cells that are then capable of inducing tumors in nude mice. We have shown here that G4 belongs to the immortalizing class of oncogenes (i.e., complements a transforming oncogene), since it is able to cooperate with Ha-ras but not with myc in cell transformation. However, G4 appears to be a mild oncogene, since only one-third of the mice developed a tumor, whereas the assay was completely negative in the other animals. The tumor development in these mice does not appear to be linked to the genotype, age, or sex of the animals. Such an intermediate tumorigenicity has been described in the literature for Ref cells transfected with polyomavirus middle-T and myc (13). In the HTLV-1 system (2, 12), the homologous counterparts of R3 and G4 have also been shown to exhibit oncogenic potential. Indeed, the p12I protein is able to potentiate cellular transformation by bovine papillomavirus E5 in C127 mouse cells (9).
The cell culture experiments corroborate the in vivo observations with the recombinant proviruses that are deficient for G4 expression. However, the fact that G4 is able to immortalize primary cells does not imply that it has oncogenic potential in vivo. Indeed, since the deletion of G4 also has a drastic effect on the proviral loads, it appears that this gene is also required for viral propagation. Whether or not the immortalizing and replicating functions of G4 are linked is currently unknown. It is indeed possible that a decrease in the number of infected cells by a defect in viral propagation is sufficient to destroy the oncogenic potential of the recombinant provirus. In this case, the limiting factor during leukemogenesis is the number of cells which are prone to become transformed, and the occurrence of a leukemia will be delayed eventually beyond the normal lifetime of the animal. Alternatively, the oncogenic potential of G4 revealed in vitro could be required for viral persistence within the infected cell and allow further propagation in the animal in vivo. It should be mentioned here that reduced viral propagation in vivo does not always correlate with a defect in oncogenicity in vitro. Indeed, a mutant virus with a deletion in R3 but not in G4 is also attenuated in terms of proviral loads in vivo (our unpublished data), but R3 is inactive in the Ref transformation assay (this report).
Keeping in mind some of its properties, we can speculate about possible functions of G4. Several pieces of evidence indicate that G4 could play a role in transcription. Indeed, the G4 gene harbors a domain homologous to the myb gene (1) and contains arginine-rich motifs (RHRLPRRALQALRDPLPDNDK) which could interact with nucleic acids. In this report, we have shown that G4 belongs to the class of immortalizing oncogenes which includes transcriptional transactivators like myc and adenovirus E1A. It is therefore possible that G4 transactivates the expression of specific cellular genes involved in cell proliferation which remain to be identified. The key role of G4 in vivo is underlined by its specific expression during the leukemogenic phase of BLV-induced pathogenesis in cattle (1). This G4 gene is also required for efficient viral propagation in vivo (see references 5 and 24 and this report). Since the pBLVIG4-infected lymphocytes also exhibit reduced apoptotic levels in ex vivo short-term cultures (5), the decrease in the proviral loads is not due to a lack of protection against programmed cell death. In fact, the number of infected cells, as measured by expression of the p24 major capsid protein, can in some cases reach high levels (about 10% in sheep 272 infected with pBLVIG4 and 21% in sheep 245 infected with pBLVDX) (5). These values are confirmed by semiquantitative PCR of viral sequences (see reference 5 and this report). However, the periods of time required to reach similar proviral loads are quite different in wild-type and G4-deleted viruses. It thus appears that the deletion of G4 does not restrict the absolute numbers of infected cells but decreases the efficiency of viral propagation in term of kinetics. It is therefore possible that the G4-deleted mutants still exhibit oncogenic potential and that the infected sheep might develop tumors much later.
In contrast to the R3 and G4 genes, the intermediate sequences located at the 5′ end of the X region are not essential for the leukemogenic potential of BLV. An mRNA containing this region has been identified in cells transfected with a BLV molecular clone (4); this mRNA contains an ORF consisting of the Tax AUG linked in frame with the X3C ORF and is predicted to produce a 94-amino-acid protein. The 344 proviral clone and some natural BLV variants (16) contain a premature stop codon that would truncate this putative protein near the carboxy terminus, yielding a 51-amino-acid polypeptide. These X3C sequences have been deleted in the pBLVIX recombinant analyzed in this report. Two out of four sheep infected with this provirus developed lymphosarcoma after a period of 40 months, which is similar to the wild-type latency. It should be mentioned here that the animals infected with the pBLVIX provirus yielded higher levels of transient viral expression around the seroconversion period (our unpublished results). We cannot therefore exclude that this intermediate region of the genome does encode a functional gene required for expression and/or replication in vivo. If so, it appears that the 51-amino-acid truncated protein is sufficient to induce this phenotype, since proviruses pBLV344 (having the stop codon) and pBLVX3C (harboring the complete X3C ORF) both induce lymphoproliferation in the circulating bloodstream (7 out of 11 and 2 out of 4 sheep, respectively). We should, however, be cautious about these data because of the lack of statistical relevance due to the low number of infected animals.
Anyway, the pathology induced by the pBLVIX provirus demonstrates that some regions in the BLV genome might be deleted without affecting its leukemogenic potential. In contrast, the deletion of the G4 gene sequences completely abrogates the ability of the BLV virus to induce both leukemia and lymphosarcoma in all infected animals. Altogether, these data underline the importance of the G4 gene in virus-induced pathogenesis in vivo.
Acknowledgments
We thank the Association belge contre le Cancer, the Bekales Foundation, the Caisse générale d’Epargne et de Retraite, the FNRS, and the Service de Programmation pour la Politique scientifique (SSTC P4/30) for financial support.
We are grateful to C. Dillen, F. de Foresta, R. Martin, P. Ridremont, and G. Vandendaele for excellent technical help, and we thank V. Ciminale for helpful discussions.
REFERENCES
- 1.Alexandersen S, Carpenter S, Christensen J, Storgaard T, Viuff B, Wannemuehler Y, Belousov J, Roth J A. Identification of alternatively spliced mRNAs encoding potential new regulatory proteins in cattle infected with bovine leukemia virus. J Virol. 1993;67:39–52. doi: 10.1128/jvi.67.1.39-52.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berneman Z, Gartenhaus R, Reitz M, Blattner A, Manns A, Hanchard B, Ikehara O, Gallo R, Klotman M. Expression of alternatively spliced human T-lymphotropic virus type I pX mRNA in infected cell lines and in primary uncultured cells from patients with adult T-cell leukemia/lymphoma and healthy carriers. Proc Natl Acad Sci USA. 1992;89:3005–3009. doi: 10.1073/pnas.89.7.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burny A, Cleuter Y, Kettmann R, Mammerickx M, Marbaix G, Portetelle D, Van den Broeke A, Willems L, Thomas R. Bovine leukemia: facts and hypothesis derived from the study of an infectious cancer. Cancer Surv. 1987;6:139–159. [PubMed] [Google Scholar]
- 4.Ciminale V, Pavlakis G N, Derse D, Cunningham C P, Felber B K. Complex splicing in the human T-cell leukemia virus (HTLV) family of retroviruses: novel mRNAs and proteins produced by HTLV type I. J Virol. 1992;66:1737–1745. doi: 10.1128/jvi.66.3.1737-1745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dequiedt F, Hanon E, Kerkhofs P, Pastoret P-P, Portetelle D, Burny A, Kettmann R, Willems L. Both wild-type and strongly attenuated bovine leukemia viruses protect peripheral blood mononuclear cells from apoptosis. J Virol. 1997;71:630–639. doi: 10.1128/jvi.71.1.630-639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derse D. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J Virol. 1987;61:2462–2471. doi: 10.1128/jvi.61.8.2462-2471.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derse D. trans-Acting regulation of bovine leukemia virus mRNA processing. J Virol. 1988;62:1115–1119. doi: 10.1128/jvi.62.4.1115-1119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrer J. Bovine lymphosarcoma. Adv Vet Sci Comp Med. 1980;24:1–68. [PubMed] [Google Scholar]
- 9.Franchini G, Mulloy J C, Koralnik I J, Lo Monico A, Sparkowski J J, Andresson T, Goldstein D J, Schlegel R. The human T-cell leukemia/lymphotropic virus type I p12I protein cooperates with the E5 oncoprotein of bovine papillomavirus in cell transformation and binds the 16-kilodalton subunit of the vacuolar H+ ATPase. J Virol. 1993;67:7701–7704. doi: 10.1128/jvi.67.12.7701-7704.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green S, Isseman I, Sheer E. A versatile in vivo and in vitro expression vector for protein engineering. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kettmann R, Burny A, Callebaut I, Droogmans L, Mammerickx M, Willems L, Portetelle D. Bovine leukemia virus. In: Levy J A, editor. The Retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 39–81. [Google Scholar]
- 12.Koralnik I, Gessain A, Klotman M, Lo Monico A, Berneman Z, Franchini G. Protein isoforms encoded by the pX region of human T-cell leukemia/lymphotropic virus type I. Proc Natl Acad Sci USA. 1992;89:8813–8817. doi: 10.1073/pnas.89.18.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Land H, Parada L, Weinberg R. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 14.Powers M A, Radke K. Activation of bovine leukemia virus transcription in lymphocytes from infected sheep: rapid transition through early to late gene expression. J Virol. 1992;66:4769–4777. doi: 10.1128/jvi.66.8.4769-4777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pozzatti R, Vogel J, Jay G. The human T-lymphotropic virus type I tax gene can cooperate with the ras oncogene to induce neoplastic transformation of cells. Mol Cell Biol. 1990;10:413–417. doi: 10.1128/mcb.10.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice N, Stephens R, Gilden R. Sequence analysis of the bovine leukemia virus genome. In: Burny A, Mammerickx M, editors. Enzootic bovine leukosis and bovine leukemia virus. The Hague, The Netherlands: Nijhoff; 1987. pp. 115–144. [Google Scholar]
- 17.Rovnak J, Boyd A L, Casey J W, Gonda M A, Jensen W A, Cockerell G L. Pathogenicity of molecularly cloned bovine leukemia virus. J Virol. 1993;67:7096–7105. doi: 10.1128/jvi.67.12.7096-7105.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sagata N, Yasunaga T, Tsuzuku-Kawamura J, Ohishi K, Ogawa Y, Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci USA. 1985;82:677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waldmann T A. The promiscuous IL-2/IL-15 receptor: a target for immunotherapy of HTLV-I-associated disorders. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;13:S179–S185. doi: 10.1097/00042560-199600001-00027. [DOI] [PubMed] [Google Scholar]
- 20.Willems L, Gegonne A, Chen G, Kettmann R, Ghysdael J. The bovine leukemia virus p34 is a transactivator protein. EMBO J. 1987;6:3385–3389. doi: 10.1002/j.1460-2075.1987.tb02661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willems L, Heremans H, Chen G, Portetelle D, Billiau A, Burny A, Kettmann R. Cooperation between bovine leukemia virus transactivator protein and Ha-ras oncogene product in cellular transformation. EMBO J. 1990;9:1577–1581. doi: 10.1002/j.1460-2075.1990.tb08277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willems L, Grimonpont C, Heremans H, Rebeyrotte N, Chen G, Portetelle D, Burny A, Kettmann R. Mutations in the bovine leukemia tax protein can abrogate the LTR-directed transactivating activity without concomitant loss of transforming potential. Proc Natl Acad Sci USA. 1992;89:3957–3961. doi: 10.1073/pnas.89.9.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willems L, Kettmann R, Dequiedt F, Portetelle D, Vonèche V, Cornil I, Kerkhofs P, Burny A, Mammerickx M. In vivo infection of sheep by bovine leukemia virus mutants. J Virol. 1993;67:4078–4085. doi: 10.1128/jvi.67.7.4078-4085.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willems L, Kerkhofs P, Dequiedt F, Portetelle D, Mammerickx M, Burny A, Kettmann R. Attenuation of bovine leukemia virus by deletion of R3 and G4 open reading frames. Proc Natl Acad Sci USA. 1994;91:11532–11536. doi: 10.1073/pnas.91.24.11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willems L, Heremans H, Burny A, Kettmann R. Rat embryo fibroblasts immortalization by bovine leukemia virus Tax protein. Methods Cell Sci. 1995;17:137–140. [Google Scholar]
- 25a.Willems, L., et al. Phosphorylation of bovine leukemia virus Tax protein is required for in vitro transformation but not for transactivation. Oncogene, in press. [DOI] [PubMed]
- 26.Yoshida M. Molecular biology of HTLV-I: recent progress. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;13:S63–S68. doi: 10.1097/00042560-199600001-00012. [DOI] [PubMed] [Google Scholar]