Abstract
Striatal-enriched protein tyrosine phosphatase (STEP) is a brain-specific enzyme that regulates the signaling molecules that control synaptic plasticity and neuronal function. Dysregulation of STEP is linked to the pathophysiology of Alzheimer’s disease and other neuropsychiatric disorders. Experimental results from neurological deficit disease models suggest that the modulation of STEP could be beneficial in a number of these disorders. This prompted our work to identify small-molecule modulators of STEP to provide the foundation of a drug discovery program. As a component of our testing funnel to identify small-molecule STEP inhibitors, we have developed a cellular target engagement assay that can identify compounds that interact with STEP46. We provide a comprehensive protocol to enable the use of this miniaturized assay, and we demonstrate its utility to benchmark the binding of newly discovered compounds.
Keywords: STEP, PTPN5, Cellular target engagement assay, CETSA, Cellular thermal shift, InCELL Pulse, Protein tyrosine phosphatase, Small molecule, Inhibitor, Neurodegenerative disorders, Alzheimer’s disease, Drug discovery, Protein–drug interaction
1. Introduction
Striatal-enriched protein tyrosine phosphatase (STEP), encoded by the PTPN5 gene, is a neuron-specific protein tyrosine phosphatase (PTP) that opposes the development of synaptic strengthening [1, 2]. High levels of active STEP contribute to the cognitive deficits in various neurodegenerative and neuropsychiatric disorders, including Alzheimer’s disease (AD) [3], Parkinson’s disease [4], schizophrenia [5], and fragile X syndrome [6]. Interestingly, STEP knockout (KO) mice show enhanced memory and learning abilities [7, 8]. Moreover, genetic reduction in STEP in mouse models of AD, schizophrenia, and fragile X syndrome reverses the cognitive and cellular deficits typically present in these models [5, 6, 9]. Those data suggest that small-molecule inhibitors of STEP could be beneficial for the treatment of AD and other neurological disorders. Indeed, a previously identified STEP inhibitor, TC-2153, was able to phenocopy the effects of STEP KO in a mouse model of AD [10]. While TC-2153 has served as a useful tool compound in multiple studies [11–14], its covalent and oxidative mechanism of action [10] and its potential to react with cellular thiols and modify DNA [15–18] have precluded this inhibitor from further preclinical advancement. Other reported STEP inhibitors suffer from poor selectivity for STEP and/or lack of efficacy under physiological conditions [19–22].
As part of our discovery platform to develop novel STEP inhibitors with improved properties, we have developed a cellular thermal shift assay (CETSA) protocol to assess the target engagement of candidate compounds in cells [23]. CETSA has found use as reliable means to validate and quantify small-molecule lead compound interactions in cells [24]. CETSA is based on the principle that the binding of a small molecule to a target protein can change its thermal stability. The original CETSA protocol uses heat pulses at varying temperatures and immunoblotting to quantify intact target protein [25]. However, several reporter-based systems that rely on the heterologous expression of targets have been developed and provide the potential for miniaturization and higher throughput [26, 27]. One such system is the InCELL Pulse™ platform (Eurofins DiscoverX). We previously utilized InCELL Pulse to study target engagement of small-molecule inhibitors with oncogenic forms of the SHP2 phosphatase [28, 29]. Based on this prior success, we have used InCELL Pulse to develop a cellular target engagement assay for STEP46, one of the two major splice variants of STEP.
InCELL Pulse is based on a β-galactosidase enzyme fragment complementation (EFC) assay (Fig. 1) [30]. The protein of interest is expressed as an N- or C-terminal fusion protein with an enhanced ProLabel® tag (ePL), a 42 amino acid fragment of β-galactosidase. After cell treatment with a candidate compound, application of a heat gradient, and cell lysis, a reporter enzyme acceptor (EA) is added, resulting in detectable β-galactosidase activity. Due to the applied temperature gradient, proteins will denature and aggregate as the temperature increases based on their thermal stability. The binding of a small molecule can stabilize (or destabilize) the target protein, and this change in thermal stability is quantified, as the ePL tag is only available for complementation when the target protein is intact and in solution. Melting curves are recorded for both candidate compound and vehicle treatment. A significant change in melting temperature (Tm) is indicative of cellular compound binding to the target protein. The CETSA protocol for STEP46 provided below utilizes advanced instrumentation for compound acoustic dispensing. However, the assay can be adapted to existing equipment in most laboratories. An example of assay performance for STEP46 is shown in Fig. 2. We have successfully used this protocol to confirm target engagement of novel STEP inhibitors developed in our laboratory (Fig. 3).
Fig. 1.
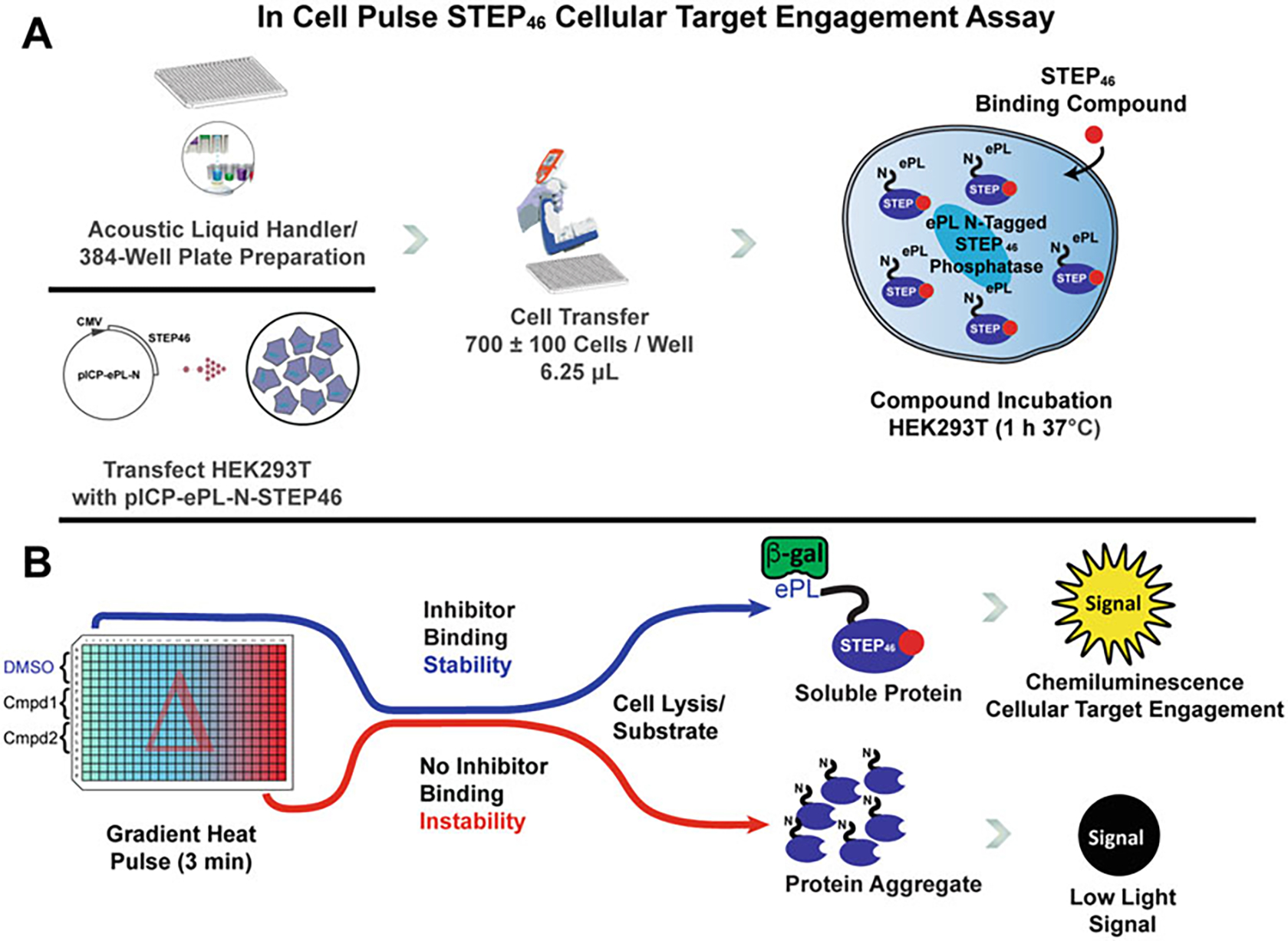
Principles and Workflow of the InCELL Pulse STEP46 Cellular Target Engagement Assay. The miniaturized STEP46 target engagement assay is a form of reporter-based cellular thermal shift assay that can be reliably integrated into a drug discovery campaign. (a) An assay plate is prepared using an Echo Liquid Handler (or similar) to spot compounds of interest in the wells to be probed. HEK293T cells are transiently transfected with a pICP-ePL-N-STEP46 plasmid that expresses STEP46 with an enhanced ProLabel (ePL, 42 amino acids) fusion tag. After 1 d, cells are detached, resuspended in fresh growth media, and are transferred to the assay plate. The cells are incubated with candidate compounds for 1 h. (b) For a thermal profile, the assay plate is subjected to a temperature gradient pulse for 3 min. Increasing temperatures cause proteins to denature and form insoluble, inaccessible aggregates. Specific target engagement of a small molecule can stabilize STEP46 against aggregation. A mixture of enzyme acceptor (EA) complementation reagent and lysis buffer enables the quantification of soluble ePL-tagged STEP46 via the reporter enzyme chemiluminescence system. The signal for each well is recorded and the data are analyzed
Fig. 2.
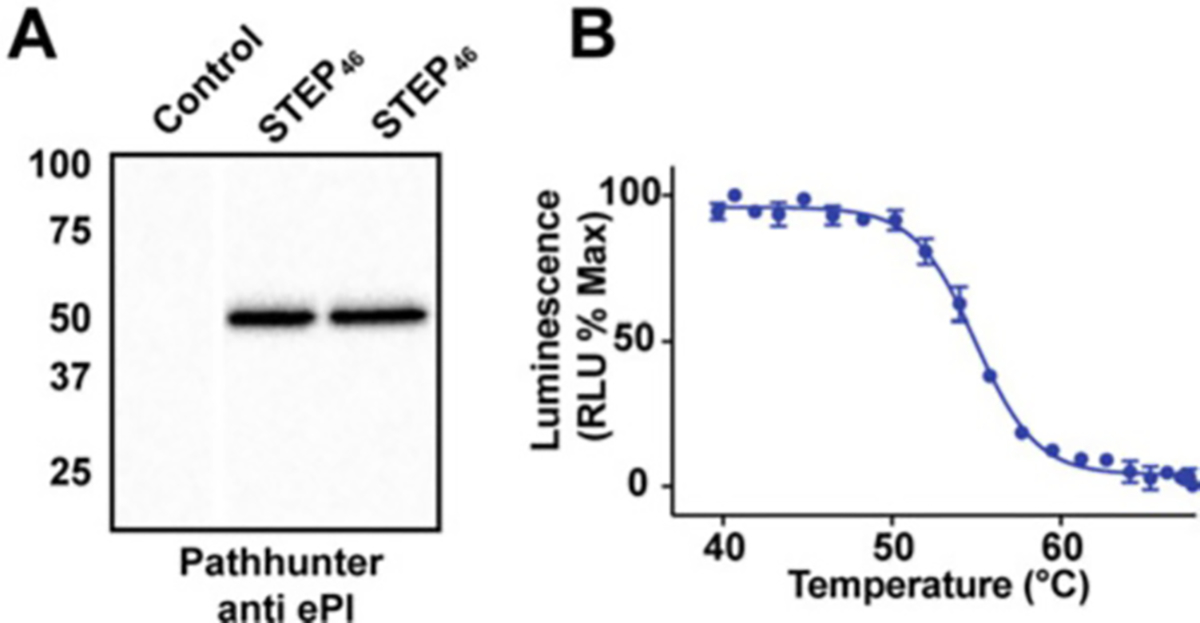
STEP46 Expression and Thermal Profile. (a) Transient transfection of HEK293T cells with pICP-ePL-N-STEP46. Western blot probing of two independently transfected HEK293T plate wells with an anti-EPL antibody (PathHunter) shows reliable expression of the STEP46 protein. (b) InCELL Pulse thermal profile for STEP46. The cellular melt curve for STEP46 exhibits the sigmoidal shape of a well-folded protein
Fig. 3.
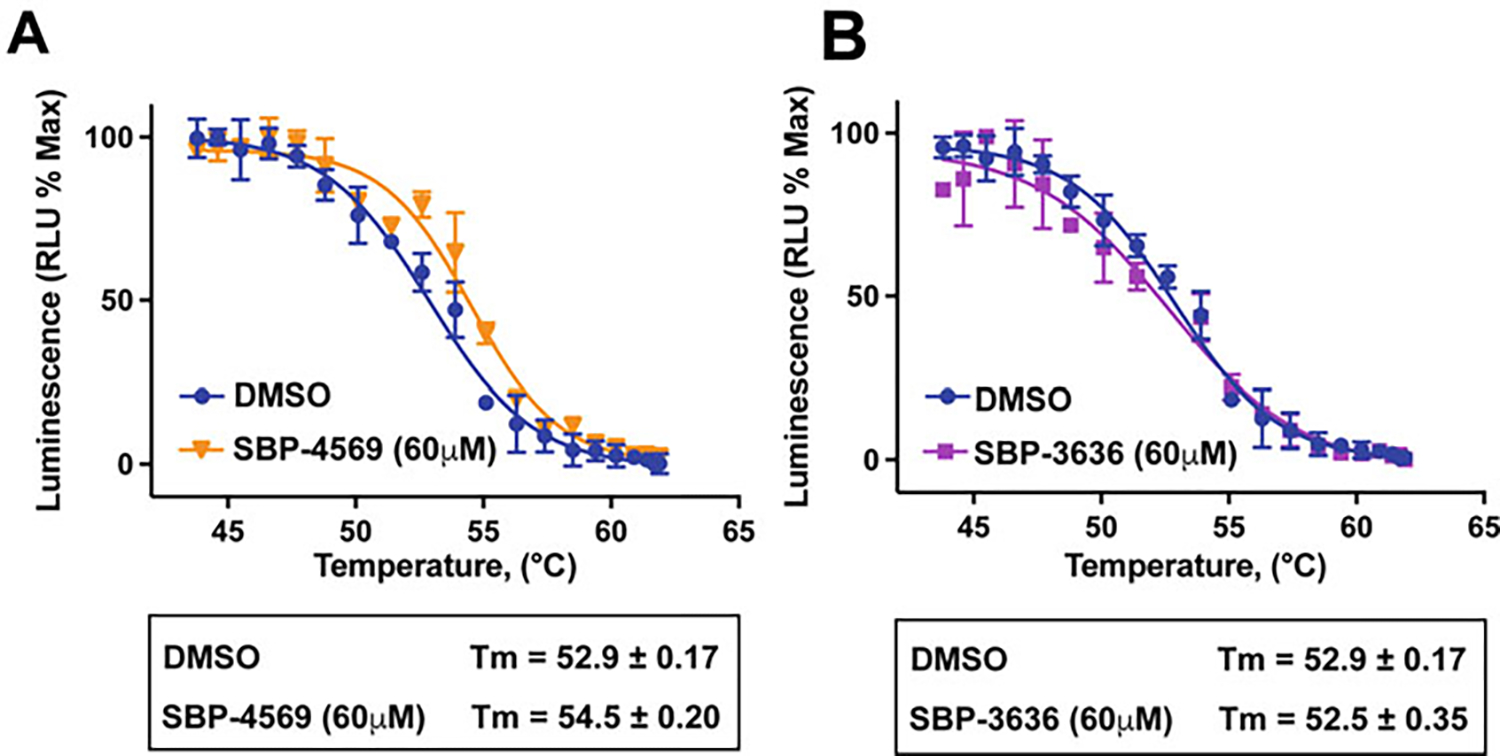
Small-Molecule Probesf of STEP46 can Stabilize the Enzyme (a) Cellular target engagement profiles of STEP46 with small-molecule inhibitor SBP-4569 (orange) and vehicle control (DMSO, blue). The stabilization of STEP46 by SBP-4569 is evident with adequate sampling of the transition temperature. (b) InCELL Pulse thermal profiles of STEP46 with small-molecule inhibitor SBP-3636 (magenta) and vehicle control (DMSO, blue). Although chemically similar to SBP-4569, SBP-3636 does not produce a shift of the STEP46 melting temperature, suggesting that this compound either does not enter cells or does not bind to STEP in cells
2. Materials
Measurement of cellular target engagement for compounds using the InCELL Pulse target engagement assay utilizes common commercial sources for the components and reagents. Storage conditions and specific handling measures are noted.
2.1. Cell Culture
HEK293T cells (ATCC).
jetPRIME® reagent and buffer (Polyplus, Illkirch, France).
TrypLE™ Express (Gibco/Thermo Fisher).
6-well tissue culture (TC)-treated cell culture plates.
Growth media: Dulbecco’s modified Eagle’s medium (DMEM 1X + GlutaMAX™; Gibco/Thermo Fisher; 500 mL), fetal bovine serum (58 mL; 10%), 100X antimycotic–antibiotic (5.8 mL; 1X), 1 M HEPES (11.2 mL; 20 mM), and 100 mM sodium pyruvate (5.8 mL; 1 mM). Store at 4 °C.
2.2. Assay Components
InCELL Pulse Starter Kit (DiscoverX, Eurofins). Make 1 mL aliquots of the three assay components (EA reagent, lysis buffer, and substrate) and store at −20 °C.
ePL-tagged expression plasmid for STEP46: Prepare by PCR amplification of the STEP46 gene, digestion with restriction enzymes EcoRI and XbaI, and directional cloning into plasmid pICP-ePL-N. Propagate the pICP-ePL-N-STEP46 plasmid using E. coli strain DH5α and the GeneJET Plasmid Maxi Prep Kit (Thermo Fisher).
2.3. Instrumentation
Mastercycler X50h 384-Well Gradient-Capable Thermal Cycler (Eppendorf).
Echo® 555 Liquid Handler (Labcyte).
Tecan SPARK® Multimode Microplate Reader (Tecan).
E1-ClipTip™ Multichannel Pipette (Thermo Fisher).
384-Well Low Dead Volume (LDV) Echo-Qualified Plates Labcyte).
Armadillo High-Performance 384-Well White PCR Plates (Thermo Fisher).
Countess™ II FL Automated Cell Counter (Thermo Fisher).
3. Methods
3.1. Cell Culture and Transient Transfection with Target Engagement Plasmid
Revive HEK293T cells preserved in cryo-storage and maintain adherent cells in growth media in a low passage state at 37 °C, 5% CO2.
Split cells bi-weekly. Do not utilize cells that have been passaged greater than 25 times to ensure assay reproducibility.
Detach HEK293T cells from a 75-cm2 flask using TrypLE cell detachment solution (3 mL), dilute with growth media (12 mL), and collect the cells by centrifugation at 1400× g for 4 min. Resuspend cells in growth media (10 mL). Measure both the cell density and cell viability using Trypan blue and a countess cell counter. Plate 0.7 × 106 HEK293T cells per well in a 6-well TC-treated cell culture plate (2 mL per well). Incubate for 24 h at 37 °C, 5% CO2.
Dilute 2 μg DNA from a purified plasmid stock of pICP-ePL-N-STEP46 (500 ng/μL) into 200 μL of jetPRIME buffer. Vortex for 10 s, add 4 μL jetPRIME reagent, vortex, and incubate for 10 min at room temperature. Add the plasmid transfection mixture to one well of the 6-well cell culture plate containing 2 mL cells (see Note 1). Incubate at 37 °C, 5% CO2 for 24 h.
Confirm expression levels of STEP46 using immunoblotting and PathHunter® anti-PK/PL antibodies (DiscoverX, Eurofins) (Fig. 2a).
3.2. Cell Detachment and Assay Plate Preparation
Aspirate growth media and add 0.3 mL TrypLE cell detachment reagent to the adherent surface-bound cells. Incubate cells at 23 °C for 2 min. Add 1 mL of growth media to cells. Gently dislodge cells by pipet (3x) and transfer cells to a 15-mL Falcon tube. Centrifuge at 1400× g for 4 min. Aspirate media and replace with ~3 mL growth media. Measure the concentration and viability of cells with a cell counter. Dilute cells to 0.125 × 106 cell/mL and use cells within 2 h (see Note 2).
Spot compounds in quadruplicate into a 384-well Twin.tec 384 real-time PCR plate using an Echo Liquid Handler or equivalent (e.g., 20 nL of a 20 mM compound stock solution; see Note 3). Add cells to a sterile single-channel trough and add 6.25 μL cells to each assay well using a multichannel pipette. Centrifuge plate at 42× g for 30 s, apply a lid seal, and incubate the assay plate at 37 °C, 5% CO2 for 1 h.
Prepare the InCELL Pulse Master Mix according to the manufacturer’s protocol (EA-10; 3 mL Master Mix for one 384-well plate; volume fractions: substrate buffer (0.67), EA reagent (0.17), and lysis buffer (0.17)).
3.3. Thermal Pulse and Assay Quantification
Remove the assay plate from the incubator and apply a 3 min heat pulse using a gradient-capable thermal cycler with a desired temperature gradient (e.g., horizontal gradient of 42–62 °C across 24 wells). Employ a 15 s countdown to enable stable temperatures to be established when the plate is placed on the grid. (see Note 4). Add a recovery step of 3 min at 20 °C after the heat pulse has been applied.
Add to each assay well 6.25 μL of the InCELL Pulse Master Mix. Centrifuge plate at 42× g for 30 s and incubate at ambient temperature for 30–60 min.
Measure chemiluminescence with the use of a Tecan Spark Multimode microplate reader or equivalent instrument capable of reading chemiluminescence (integration time, 1000 ms; settle time, 0 s).
Analyze chemiluminescence data using GraphPad Prism or an equivalent program. Calculated curve fits are as follows: normalize chemiluminescent values with the maximum and minimum value defined as 100% or 0%, respectively. Apply a Boltzmann sigmoidal nonlinear least squared fit of the normalized chemiluminescence and calculate an EC50 value, which corresponds to the Tm value (see Note 5) (Fig. 2b).
4. Notes
Transfect from a single source of concentrated plasmid for uniform assay results. One well of transfected cells (0.7 × 106 cells) will be enough for two 384-well CETSA plates.
Cell viability (>90% survival) is a critical parameter that assures that meaningful data about cellular penetrance and target engagement are obtained. The one-hour incubation usually does not result in significant cell death for most compounds. Increasing the scale of the assay could be useful in adapting it to a high-throughput campaign. However, high cell viability should not be compromised.
If compound stock solutions are in DMSO, the amount of stock solution added should be chosen so that the final DMSO concentration is ≤0.5%.
As a depletion assay, meaningful data from a thermal profile would optimally be obtained by sampling more data points at the melt temperature. For STEP46, we set the temperature gradient from 38 to 68 °C or 42 to 62 °C, with the narrow temperature span best able to assess the effect of target engagement.
A sigmoidal melt curve with a narrow transition temperature is observed for STEP46. This is indicative of a well-folded protein in a cellular environment. Melt curves that exhibit broad, indiscrete transitions could indicate that the cellular protein is not correctly folded under the chosen expression conditions. Interpretation of melt curve shape due to compound binding has been described [31]. Optimization of the transfection protocol can be performed to modulate the expression level and can influence the observed temperature profile.
Acknowledgements
Research reported in this publication was supported by the National Institutes of Health under awards numbers R01AG065387 and R21AG067155 (to L. T.) and by the NCI Cancer Center Support Grant P30CA030199. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Lombroso P, Murdoch G, Lerner M (1991) Molecular characterization of a protein-tyrosine-phosphatase enriched in striatum. Proc Natl Acad Sci U S A 88:7242–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lombroso PJ, Ogren M, Kurup P et al. (2016) Molecular underpinnings of neurodegenerative disorders: striatal-enriched protein tyrosine phosphatase signaling and synaptic plasticity. F1000Res 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu J, Kurup P, Nairn AC et al. (2012) Striatal-enriched protein tyrosine phosphatase in Alzheimer’s disease. Adv Pharmacol 64:303–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurup PK, Xu J, Videira RA et al. (2015) STEP61 is a substrate of the E3 ligase parkin and is upregulated in Parkinson’s disease. Proc Natl Acad Sci U S A 112:1202–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carty N, Xu J, Kurup P et al. (2012) The tyrosine phosphatase STEP: implications in schizophrenia and the molecular mechanism underlying antipsychotic medications. Transl Psychiatry 2:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goebel-Goody S, Wilson-Wallis E, Royston S et al. (2012) Genetic manipulation of STEP reverses behavioral abnormalities in a fragile X syndrome mouse model. Genes Brain Behav 11:586–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venkitaramani D, Paul S, Zhang Y et al. (2009) Knockout of striatal enriched protein tyrosine phosphatase in mice results in increased ERK1/2 phosphorylation. Synapse 63:69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkitaramani D, Moura P, Picciotto M et al. (2011) Striatal-enriched protein tyrosine phosphatase (STEP) knockout mice have enhanced hippocampal memory. Eur J Neurosci 33:2288–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Kurup P, Xu J et al. (2010) Genetic reduction of striatal-enriched tyrosine phosphatase (STEP) reverses cognitive and cellular deficits in an Alzheimer’s disease mouse model. Proc Natl Acad Sci U S A 107:19014–19019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J, Chatterjee M, Baguley TD et al. (2014) Inhibitor of the tyrosine phosphatase STEP reverses cognitive deficits in a mouse model of Alzheimer’s disease. PLoS Biol 12:e1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulikova EA, Khotskin NV, Illarionova NB et al. (2018) Inhibitor of striatal-enriched protein tyrosine phosphatase, 8-(Trifluoromethyl)-1,2,3,4,5-Benzopentathiepin-6-amine hydrochloride (TC-2153), produces antidepressant-like effect and decreases functional activity and protein level of 5-HT2A receptor in the brain. Neuroscience 394:220–231 [DOI] [PubMed] [Google Scholar]
- 12.Siemsen BM, Lombroso PJ, McGinty JF (2018) Intra-prelimbic cortical inhibition of striatal-enriched tyrosine phosphatase suppresses cocaine seeking in rats. Addict Biol 23:219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee M, Kwon J, Benedict J et al. (2021) STEP inhibition prevents Abeta-mediated damage in dendritic complexity and spine density in Alzheimer’s disease. Exp Brain Res 239:881–890 [DOI] [PubMed] [Google Scholar]
- 14.Lee ZF, Huang TH, Chen SP et al. (2021) Altered nociception in Alzheimer disease is associated with striatal-enriched protein tyrosine phosphatase signaling. Pain 162:1669–1680 [DOI] [PubMed] [Google Scholar]
- 15.Chatterji T, Gates KS (1998) DNA cleavage by 7-methylbenzopentathiepin: a simple analog of the antitumor antibiotic varacin. Bioorg Med Chem Lett 8:535–538 [DOI] [PubMed] [Google Scholar]
- 16.Lee AH, Chan AS, Li T (2002) Acid-accelerated DNA-cleaving activities of antitumor antibiotic varacin. Chem Commun:2112–2113 [DOI] [PubMed] [Google Scholar]
- 17.Lee AH, Chen J, Liu D et al. (2002) Acid-promoted DNA-cleaving activities and total synthesis of varacin C. J Am Chem Soc 124:13972–13973 [DOI] [PubMed] [Google Scholar]
- 18.Greer A (2001) On the origin of cytotoxicity of the natural product varacin. A novel example of a pentathiepin reaction that provides evidence for a triatomic sulfur intermediate. J Am Chem Soc 123:10379–10386 [DOI] [PubMed] [Google Scholar]
- 19.National Center for Biotechnology Information. PubChem BioAssay Database; AID=588619. https://pubchem.ncbi.nlm.nih.gov/bioassay/588619. Accessed 3 Feb 2019 [Google Scholar]
- 20.Masaki Suzuki [JP], Kazumi Kondo [JP], Muneaki Kurimura [JP], Reddy Valluru Krishna [In], Akira Takahahi [JP], Takeshi Kuroda [JP], Haruka Takahashi [JP], Tae Fukushima [JP], Shin Miyamura [JP], Indranath Ghosh [US], Abhishek Dogra [US], Geraldine Harriman [US], Amy Elder [US], Satoshi Shimiza [JP], Kevin J Hodgetts [US], Jason S Newcom [US]. Quinazolines as therapeutic compounds and related methods of use. (2014) US2014315886 (A1) [Google Scholar]
- 21.Masaki Suzuki [JP], Kazumi Kondo [JP], Muneaki Kurimura [JP], Reddy Valluru Krishna [In], Akira Takahahi [JP], Takeshi Kuroda [JP], Haruka Takahashi [JP], Tae Fukushima [JP], Shin Miyamura [JP], Indranath Ghosh [US], Abhishek Dogra [US], Geraldine Harriman [US], Amy Elder [US], Satoshi Shimiza [JP], Kevin J Hodgetts [US], Jason S Newcom [US]. Therapeutic compounds and related methods of use. (2015) US2015307477 (A1) [Google Scholar]
- 22.Witten MR, Wissler L, Snow M et al. (2017) X-ray characterization and structure-based optimization of striatal-enriched protein tyrosine phosphatase inhibitors. J Med Chem 60:9299–9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molina DM, Jafari R, Ignatushchenko M et al. (2013) Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341:84–87 [DOI] [PubMed] [Google Scholar]
- 24.Prabhu N, Dai L, Nordlund P (2020) CETSA in integrated proteomics studies of cellular processes. Curr Opin Chem Biol 54:54–62 [DOI] [PubMed] [Google Scholar]
- 25.Martinez Molina D, Jafari R, Ignatushchenko M et al. (2013) Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341:84–87 [DOI] [PubMed] [Google Scholar]
- 26.Dart ML, Machleidt T, Jost E et al. (2018) Homogeneous assay for target engagement utilizing bioluminescent thermal shift. ACS Med Chem Lett 9:546–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez NJ, Asawa RR, Cyr MG et al. (2018) A widely-applicable high-throughput cellular thermal shift assay (CETSA) using split Nano luciferase. Sci Rep 8:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romero C, Lambert LJ, Sheffler DJ et al. (2020) A cellular target engagement assay for the characterization of SHP2 (PTPN11) phosphatase inhibitors. J Biol Chem 295:2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambert LJ, Romero C, Sheffler DJ et al. (2020) Assessing cellular target engagement by SHP2 (PTPN11) phosphatase inhibitors. J Vis Exp [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNulty DE, Bonnette WG, Qi H et al. (2018) A high-throughput dose-response cellular thermal shift assay for rapid screening of drug target engagement in living cells, exemplified using SMYD3 and IDO1. SLAS Discov 23:34–46 [DOI] [PubMed] [Google Scholar]
- 31.Henderson MJ, Holbert MA, Simeonov A et al. (2020) High-throughput cellular thermal shift assays in research and drug discovery. SLAS DISCOVERY: Advancing the Science of Drug Discovery 25:137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]


