Abstract
Label-free optical imaging employs natural and nondestructive approaches for the visualisation of biomedical samples for both biological assays and clinical diagnosis. Currently, this field revolves around multiple broad technology-oriented communities, each with a specific focus on a particular modality despite the existence of shared challenges and applications. As a result, biologists or clinical researchers who require label-free imaging are often not aware of the most appropriate modality to use. This manuscript presents a comprehensive review of and comparison among different label-free imaging modalities and discusses common challenges and applications. We expect this review to facilitate collaborative interactions between imaging communities, push the field forward and foster technological advancements, biophysical discoveries, as well as clinical detection, diagnosis, and monitoring of disease.
Editorial Summary
This Review covers a number of label free biomedical imaging techniques, their advantages over label-based methods and relevant applications.
1. Introduction
Biomedical optical imaging refers to multi-point measurement techniques that use light to capture images of biological samples in vitro or in vivo. Many biomedical samples, however, do not induce sufficient imaging contrast and important biological details might be missed in the resulting images. For example, some biological samples, such as cells in vitro, are optically semi-transparent and induce minimal light absorption, which is one of the primary sources of contrast in conventional optical imaging, so that only the cell edges can be scarcely seen in the resulting image. Exogenous labeling agents, such as fluorophores or specific stains, with chemical properties of binding to specific components in the biological samples, are widely used to induce the missing imaging contrast. Alternatively, it is possible to genetically modify the cells, organisms, or the resulting samples to express fluorescence or other useful optical properties. The former technique is typically used in both animal studies and in vitro diagnosis, when the sample is disposed shortly after imaging it, whereas the latter technique is typically used in animal studies only. Any form of exogenous labeling or genetic modifications is perturbing the natural biological processes, dynamics, and response of the cells or tissues, degrades the vitality of the sample, and might be fatal in longer-term longitudinal studies. The use of exogenous labeling often has confounding factors such as off-target binding, non-specific binding, or incomplete binding, making quantification and reproducibility a challenge. These are often some of the reasons why label-based imaging techniques are not recommended for use during in-vivo imaging for medical diagnosis and therapy, especially in human subjects. Furthermore, label-based imaging techniques are problematic to use even for in vitro imaging of live human cells when the sample is needed for further medical treatments, such as when imaging stem cells for personalized medicine or gamete cells during in vitro fertilization. In all these clinical settings, the introduction of label-free imaging is easier to perform since it does not require obtaining expensive and time-consuming approvals of the chemical marker substances as drugs.
Label-free optical imaging captures the intrinsic properties of the sample, such as the sample’s refractive index variations, autofluorescence, molecular vibrations, birefringence, scattering, or absorption properties, to reveal the needed imaging contrast. These techniques are nonperturbative to the measurement and the following analysis, as long as the optical energy needed is not so high as to perturb the structure, function, molecular composition, or physiology of the cells or tissues being measured. Based on the intrinsic source of imaging contrast mechanism in each label-free imaging technique, the optical setup needs to be adjusted. A few label-free imaging techniques, such as Zernike’s phase contrast of in-vitro cells, are rather simple to implement and use, and therefore are widely available to biologists and clinicians. Most label-free imaging techniques, however, require more complex and expensive optical setups. For example, if the source of contrast is Raman scattering, one would need to carefully design the illumination wavelength and intensity of the excitation source and use an imaging spectrograph to obtain the sample image. This higher degree of system complexity causes some label-free techniques to be intriguing and inaccessible to biologists and clinical researchers, leaving the field of label-free imaging to be centralized around several technology-oriented communities, each of which is leading one or a few label-free techniques, while missing important biomedical applications being pursued in other label-free imaging communities.
The current manuscript attempts to make label-free imaging accessible to cross-community investigators and users, by first discussing the working principles and special considerations in selecting the most suitable label-free imaging modality per application (Section 2), followed by discussing the future challenges of the field, as well as leading biomedical applications (Section 3).
2. Comparative analysis
The development and use of label-free imaging can be categorized to cases where labeling is not allowed and cases where labeling is allowed but label-free imaging provides better performance. The first type of cases might occur where the sample needs to be used for treatment after imaging it, for immunotherapy drug testing, and for regenerative medicine, where specific labeling agents do not exist, for toxicity measurements, for in-vivo applications, or for clinical applications where point-of-procedure or point-of-care detection, diagnosis, or guidance is needed, e.g., intraoperative diagnosis such as optical biopsy, and intra-vital microscopy. The second type of cases is where labeling is allowed, but still label-free imaging provides better measurements.
A comparison between various label-free imaging techniques is presented in Table 1. The key criterion that should be considered when choosing a label-free imaging technique per application is the intrinsic contrast mechanism and physics of the chosen technique, and its presence in the sample measured, where frequently label-free imaging can provide more quantitative measurements than label-based imaging. For example, a cell’s integral refractive index1–6 that can be measured via interference-based phase microscopy (PhM) (Figs. 1a–1d) is proportional to the cell dry mass surface density.4 This parameter cannot be measured via stain-based microscopy, in which the stain can indicate the labeled-organelle location but typically the grayscale value in each point on the image cannot be interpreted quantitatively. To obtain the three-dimensional (3D) image of an individual cell, with sectioning, interferometric computed tomography can be used,7,8 where quantitative PhM is performed from multiple angles to generate the cellular 3D refractive-index map with less than half a micron resolution (Figs. 1e and 1g).9 Polarization microscopy (PolM) is typically used to analyze optical anisotropy due to molecular order, where the average molecular orientation is not random. This may occur in biological samples containing extensive membranes, such as photoreceptors on the retina, or in biological samples containing filament arrays, such as collagen fibers10 and the cell mitotic spindles.11 Birefringence, anisotropy of refractive index, can be observed in sperm cells and oocytes12. Optical coherence tomography (OCT)13–15 uses light interference from a partially coherent light to obtain contrast from the tissue refractive index changes, with capability for optical sectioning and thus 3D imaging of multi-layer tissues ex vivo or in vivo, with a spatial resolution on the micron scale and a depth of penetration in most tissues on the 1–2 millimeter scale. Various clinical applications of OCT, including ophthalmic imaging15–17 coronary artery disease detection18,19, intraoperative optical biopsy in cancer detection,20–22 endoscopic evaluation of the gastrointestinal tract,23,24, skin imaging in dermatology25–27, and blood flow imaging in-vivo28,29 are demonstrated in Fig. 2. Furthermore, phase-variance OCT can sensitively detect movement, such as in dynamic cells30 or the retinal microvasculature.31,32 Harmonic generation microscopy (HGM) creates imaging contrast based on the high-order nonlinear susceptibility of the sample. Second-harmonic generation (SHG) occurs at non-centrosymmetric molecular structures or interfaces and most commonly from fibrillar structures, such as collagen or elastin in connective tissues, changing dramatically during carcinogenesis, or myosin and microtubules in muscle fibers and metastatic tumor cells.33–35 Third-harmonic generation (THG) occurs at interfaces across which there is a large change in refractive index, such as across a lipid-based biological membrane and the surrounding aqueous microenvironment.36,37 Auto-fluorescence microscopy (aFM) is based on measuring fluorescence from endogenous biomolecules, including nicotinamide adenine dinucleotide phosphate (NAD(P)H), flavin adenine dinucleotide (FAD), tryptophan, and others. Specifically, NAD(P)H and FAD images indicate the metabolic activity of cells and tissues. Optical excitation of these biomolecules can occur through single-photon interactions, but multiphoton interactions in two-photon autofluorescence (2PF), three-photon autofluorescence (3PF), or higher multi-photon autofluorescence, offer advantages of higher spatial resolution, higher SNR, deeper imaging penetration with near-infrared incident wavelengths, and optical sectioning capabilities.38 Simultaneous label-free autofluorescence multi-harmonic microscopy (SLAM)39 combines 2PF, 3PF, SHG, and THG all together (see Fig. 3). Multiple investigators have used the intensity values of this autofluorescence to calculate redox ratios, to characterize the metabolic properties of in vitro cells, living tissues in vivo, or fresh tissue specimens.40,41 Clearly such dynamic metabolic properties cannot be fully characterized in chemically processed, fixed, and stained sections, and the use of exogenous dyes or stains would affect the inherent metabolic processes being measured. Time correlated single-photon counting42 and, more recently, fast direct sampling techniques43 can capture the fluorescence lifetime, or the decay time of the fluorescence emission, in fluorescence lifetime imaging microscopy (FLIM). The decay profile offers another label-free contrast parameter that is sensitive to many of the micro-environmental conditions surrounding the autofluorescent molecules, such as chemical composition, pH, and any perturbation to these.44 Label-free hyperspectral imaging is able to capture the contiguous spectrum for each pixel in the image over a selected wavelength bandwidth, for detecting physiological changes by their absorption, reflectance, or scattering spectral signatures, with limited molecular specificity. Hyperspectral imaging can detect endogenous tissue chromophores such as hemoglobin, melanin, water, and collagen content.45 Specifically, in IR-absorption microscopy (IRAM), characteristic molecular vibrations of cell and tissue constituents are excited by absorption of radiation with wavelengths between 2.5 and 50 μm (4000–200 cm−1). The introduction of tunable quantum-cascade lasers has made IR excitation with a high photon density possible, partially compensating for the appearance of strong IR water absorption bands in the IR spectra of biomedical samples that mask other relevant bands.46–48 Photothermal IR microscopy is based on the non-radiative transformation of absorbed energy into heat, allowing sub-micrometer spatial resolution. Using mid-IR illumination, the absorbed heat leads to local expansion and refractive index change of the sample, which can be detected in the visible range and yield to better lateral resolution than classic IR spectroscopy.49,50
Table 1.
Summary of key performance characteristics of major label-free biomedical imaging approaches
| Method | Spatial resolution | Imaging depth | Speed | Main source of contrast | Main applications | System complexity and cost | In vivo? | Clinically widely avaialble? |
|---|---|---|---|---|---|---|---|---|
| PhM | Sub μm | Tens of μm | **** | Refractive index | Cell structure | * | No | Yes |
| PolM | Sub μm | Tens of μm | **** | Birefringence | Cell and tissue structure (membranes and filament arrays, e.g., collagen fibers; cell spindle) | * | Yes (togther with other methods) | Yes |
| OCT | Several μm | Several mm | **** | Refractive index, speckle/phase variance, Doppler, strain and shear stress | Tissue structure; blood circulation; biomechanics | ** | Yes | Yes |
| HGM | Sub μm | Sub mm | *** | High-order nonlinear susceptibility | Cell and tissue structure SHG: Non-centrosymmetric structures and fibrillar structures, such as collagen or elastin in connective tissues, or myosin and microtubules in muscle fibers THG: Interfaces and optical heterogeneities, such as lipid-based biological membranes | **** | Yes | No |
| aFM | Sub μm | Sub mm | *** | Endogenous fluorochromes | Cell and tissue structure, NAD(P)H, FAD, keratin and elastin; redox; metabolic dynamics | *** | Yes | No |
| IRAM | Several μm | Sub mm | **** | Absorption | Endogenous tissue chromophores (hemoglobin, melanin, water and collagen) | ** | Yes | No |
| RM | Sub μm | Sub mm | * (linear) *** (nonlinear) |
Raman scattering of molecular vibrations | Selective macro-molecular vibrations of lipids, proteins, DNA, etc. | ** (linear) **** (nonlinear) |
Yes | No |
| PAT | Scalable: sub μm – tens of μm | Scalable: Sub mm – tens of mm | **** | Absorption | Cell and tissue structure; vasculature | **** | Yes | No |
PhM: Phase microscopy; PolM: Polarization microscopy; OCT: optical coherence tomography; HGM: Harmonic-generation microscopy; SHG: Second-harmonic generation; THG: Third-harmonic generation; aFM: Auto-fluorescence microscopy; IRAM: IR-absorption microscopy; RM: Raman microscopy; PAT: photoacoustic tomography. Rapid acquisition speed (****) refers to obtaining more than ten Mega-voxel/s in reasonable, clinical attainable resources.
Note: the table refers to the common version of each method.
Figure 1.
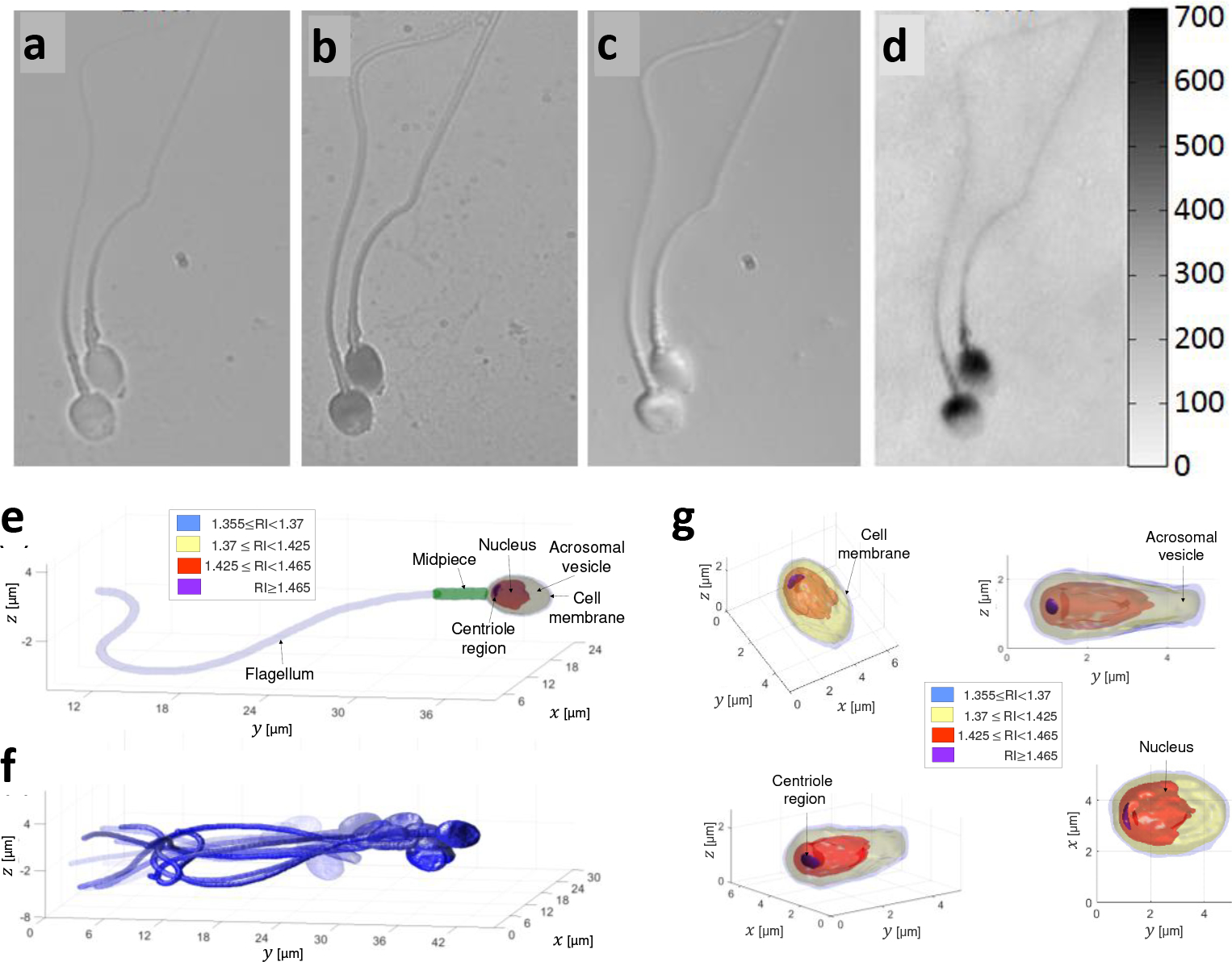
Comparison of imaging methods of human sperm cells. a, Label-free bright-field imaging, presenting low-contrast where the cell inner content cannot be seen. b, Label-based bright-field imaging (not allowed in human in-vitro fertilization). c, Label-free differential interference contrast (DIC) microscopy, a qualitative PhM method. d, Label-free quantitative PhM, where the colorbar on the right represents optical path delay values in nm.6 e-f, High-resolution label-free dynamic 3D imaging of a sperm cell swimming freely, acquired by interferometric computed tomography. Reproduced from Ref. 9. e, A single frame from the 3D motion, revealing the internal structure of the sperm cell. f, Overlay of 15 frames from the 3D motion. g, The sperm cell head 3D refractive-index profile from various perspectives. RI referred to refractive index.
Figure 2.
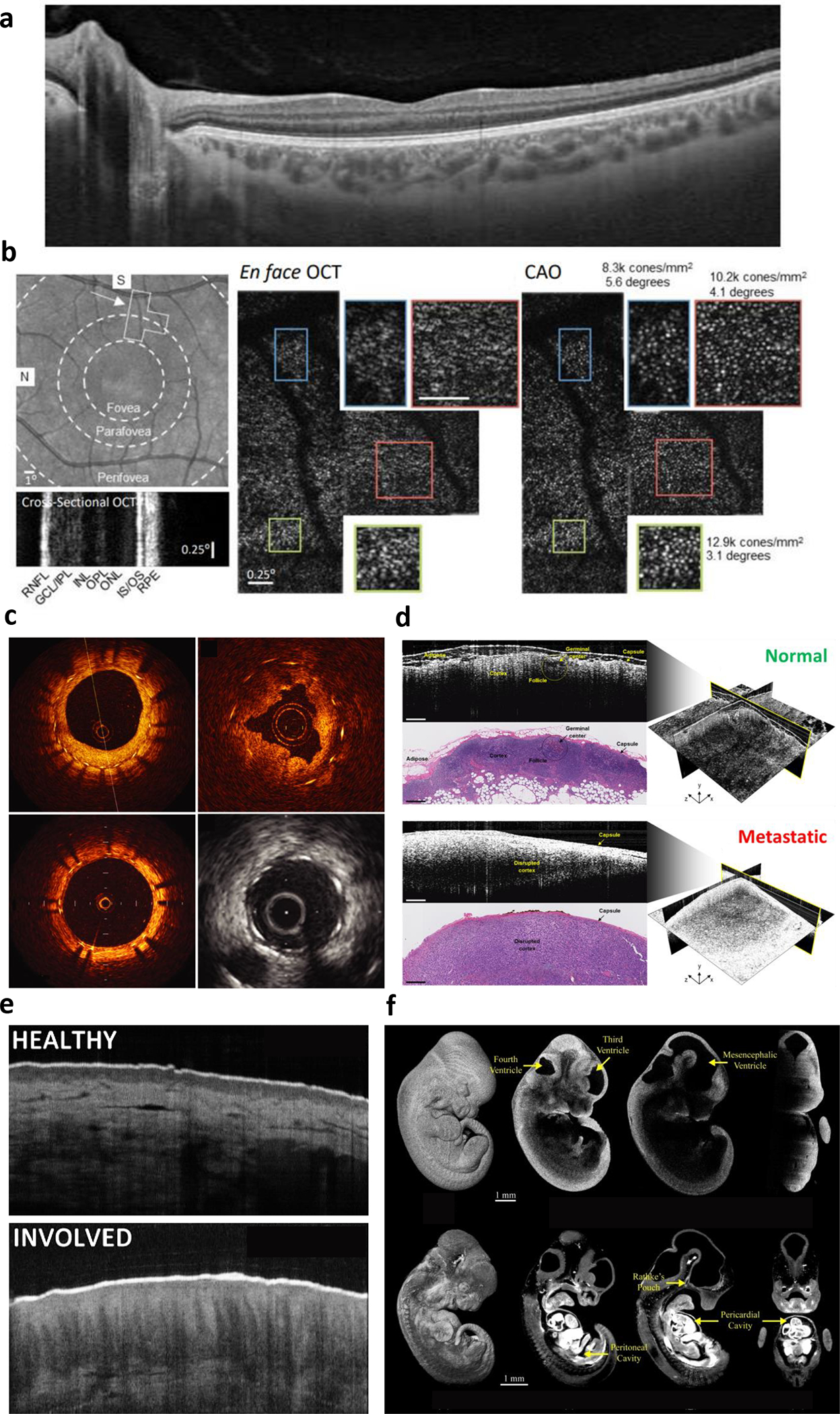
Label-free optical coherence tomography (OCT) applications. a, Ophthalmic OCT of the human retina (cross-section) delineating individual retina layers. b, Computational adaptive optics to correct optical aberrations in the human eye to enable en face OCT of individual photoreceptors in the mosaic. Reproduced from Ref. 17. c, Fiber-optic catheter-based radial OCT of the human coronary artery to assess stent apposition, compared to intravascular ultrasound (greyscale image). Reproduced from Ref. 19. d, Intraoperative OCT for surgical oncology guidance. 3D images of human lymph nodes reveal increased scattering following metastatic involvement. Reproduced from Ref. 21. e, OCT of human skin, revealing architectural differences from atopic dermatitis. Adapted from Ref. 27. f, 3D OCT of mouse embryonic development, with real-time functional assessment of cardiac dynamics. Reproduced from Ref. 29.
Figure 3.
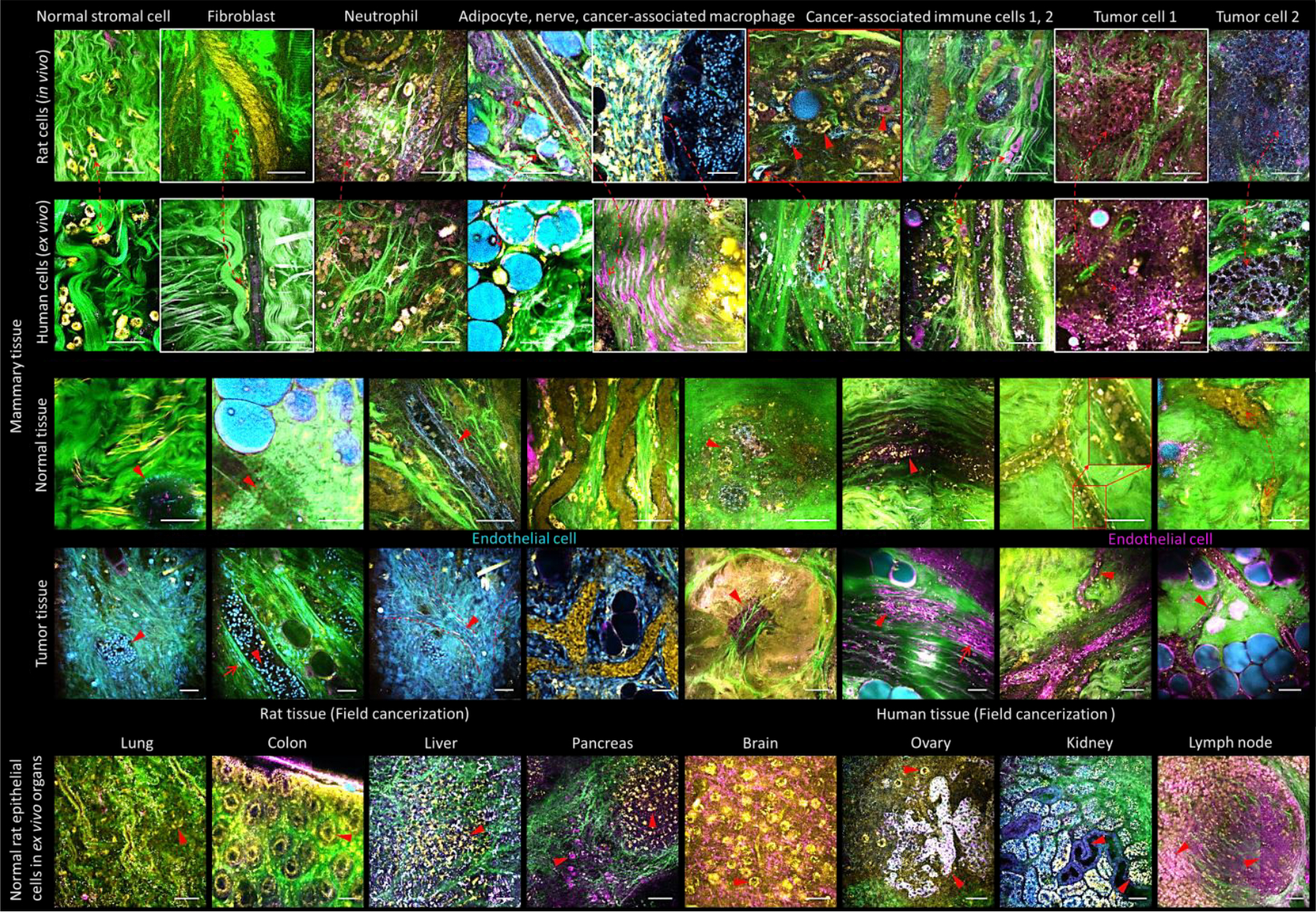
Image atlas obtained by simultaneous label-free autofluorescence multiharmonic (SLAM) microscopy of various normal and cancer tissues from fresh, unstained human and rat biopsy specimens, in vivo and ex vivo, showing various endogenous contrast channels. Yellow: 2PF of FAD, Blue: 3PF of NAD(P)H, Green: SHG of fibrous structures (collagen), Magenta: THG of lipid-aqueous interfaces. Other colors appear based on the relative color mixing between the overlaid channels. Scale bars represent 25 μm.
Raman microscopy (RM) is based on the detection of inelastic Raman scattering via molecular vibrations that are specific for chemical bonds inside the molecule, i.e., lipids, carbohydrates, pigments, DNA, RNA, proteins, etc.51–54 Linear (spontaneous) Raman spectroscopy is characterized by intrinsically small scattering cross-sections, which makes it challenging to acquire hyperspectral Raman images of large tissue areas. This disadvantage can be overcome by using nonlinear coherent Raman scattering, including coherent anti-Stokes Raman scattering (CARS)55 (see Fig. 4, as combined with SLAM56,57) and stimulated Raman scattering (SRS).58,59 These techniques enhance the intrinsically weak Raman signal and avoid the appearance of a large autofluorescence background, but have reduced molecular selectivity because they can image only one or a few characteristic Raman bands. Photoacoustic tomography (PAT)60 provides 3D imaging based on the photoacoustic effect. When light is absorbed by molecules and converted to heat, an acoustic wave is generated due to thermoelastic expansion. The acoustic wave is detected with negligible scattering to form a high-resolution tomographic image. Consequently, PAT combines optical contrast of molecular absorption and ultrasonic resolution despite optical diffusion, providing multiscale high-resolution structural and functional imaging61–63 in-vivo at depths beyond the optical diffusion limit (~1 mm in the skin). In focused scanning PAT,64 the acoustic focusing of an ultrasonic transducer or the optical focusing of an objective lens provides lateral resolution, whereas the acoustic time of flight offers axial resolution. In photoacoustic computed tomography, usually hundreds to thousands of unfocused ultrasonic transducers receive photoacoustic waves in parallel. An inverse reconstruction algorithm65 is used to reconstruct a tomographic image. Whereas the first mode costs less, the second mode provides greater speed and more uniform spatial resolution. PAT is capable of in-vivo imaging66–70 as well as in-vitro imaging71 at multiple length scales ranging from subcellular organelles to human organs or small-animal organisms72 with the same contrast origin (see Fig. 5).
Figure 4.
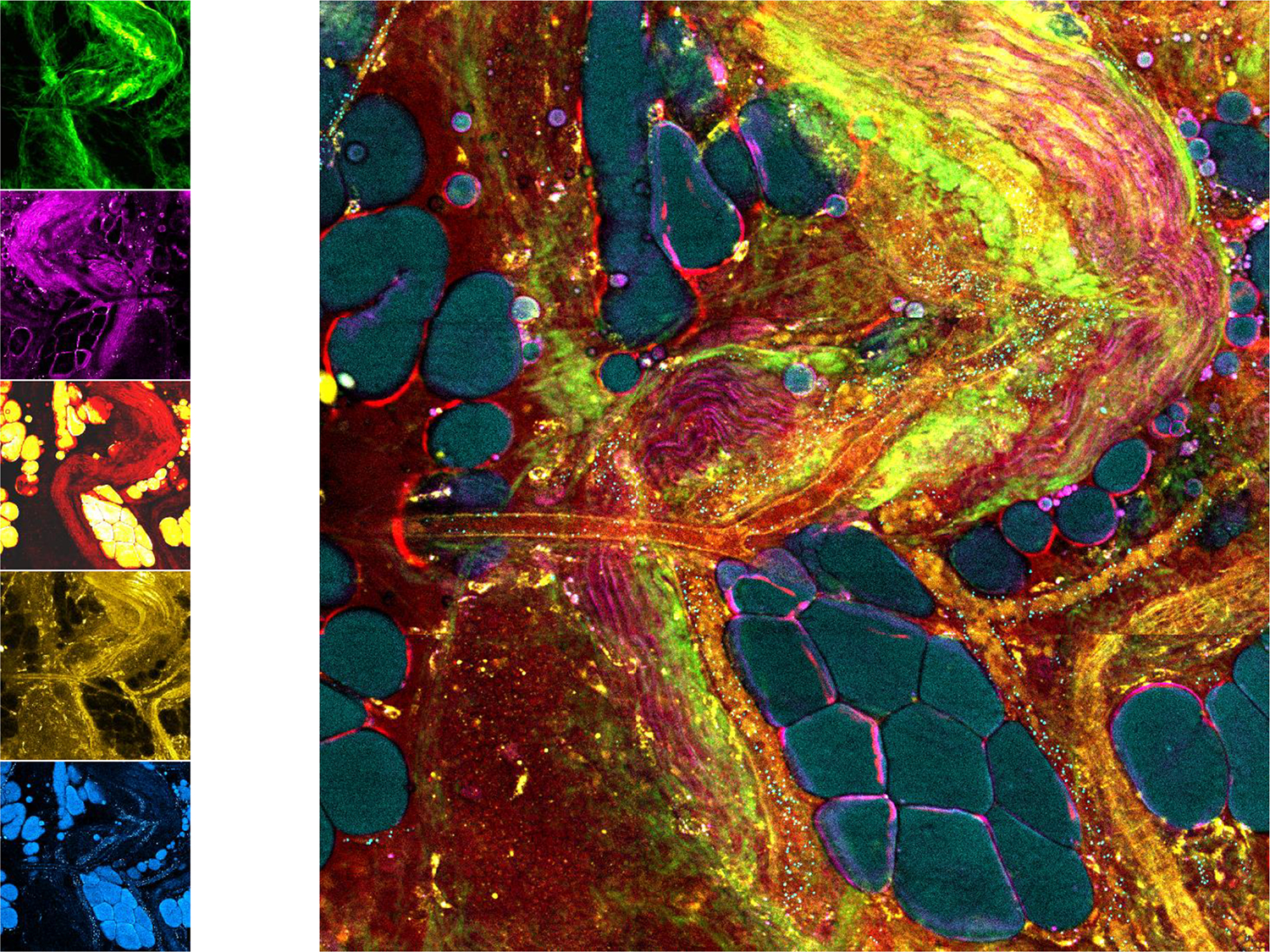
Single-shot label-free multimodal nonlinear imaging (SHG, THG, CARS, 2PF, 3PF). Using a single fiber-laser pumped photonic crystal fiber source to generate supercontinuum illumination, along with parallel multi-channel photomultiplier tube detectors, multiple nonlinear processes can be excited in tissue simultaneously and detected to generate a spatially and temporally co-registered label-free image of tissue microstructure, molecular composition, function, and metabolism. Adapted from Ref. 56. Multimodal images, radar plots, and spectra for normal and tumor tissues can be found in Ref. 57.
Figure 5.
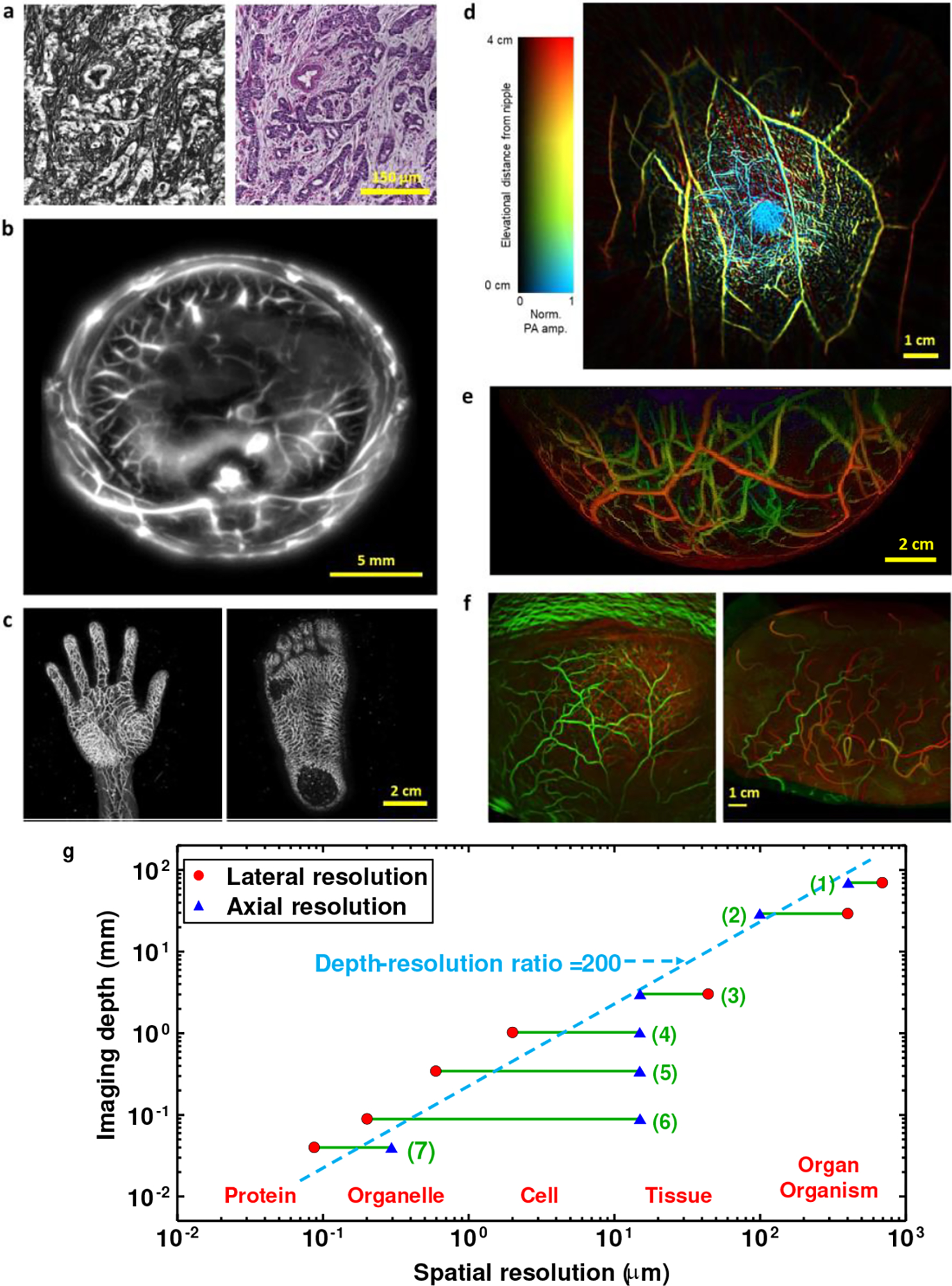
Multiscale label-free photoacoustic imaging of molecular absorption. a, Photoacoustic pathology. Left: photoacoustic microscopic image of a breast cancer specimen without staining. Right: conventional histologic image of the same breast cancer specimen with hematoxylin and eosin staining. Adapted from Ref. 71. b, In-vivo whole-body photoacoustic image of a rodent acquired in full-ring detection geometry. Adapted from Ref. 72. c, In-vivo 3D photoacoustic images of human extremities. Adapted from Ref. 67. d, Single-breath-hold photoacoustic image of the breast acquired in full-ring detection geometry. Adapted from Ref. 68. e, Photoacoustic image of the breast acquired in hemispherical detection geometry. Adapted from Ref. 69. f, Functional photoacoustic image of the human brain acquired in hemispherical detection geometry (left) versus functional MRI image (right). Adapted from Ref. 70. g, Graph illustrating the scalability in photoacoustic imaging. Imaging ranges indicated on the graph: (1) Low-frequency photoacoustic tomography; (2) Photoacoustic macroscopy; (3) Acoustic-resolution photoacoustic microscopy; (4) Optical-resolution photoacoustic microscopy; (5) Submicron photoacoustic microscopy; (6) Sub-wavelength photoacoustic microscopy; (7) Super-resolution photoacoustic microscopy.
After understanding whether a certain intrinsic property exists in the sample, which will lead to the selection of the label-free imaging method as reviewed above, other key parameters should be considered. As shown in Table 1, these include the target imaging depth, spatial resolution, and acquisition time. Selection of these parameters requires knowledge in the physics of the optical system and the contrast mechanism. As a rule of thumb, there is a tradeoff between obtaining a high resolution and a large imaging depth. For example, PAT offers a variable tradeoff between imaging depth and spatial resolution due to the ultrasonic physics. The ultrasonic attenuation coefficient is approximately proportional to the ultrasonic frequency while the ultrasonic spatial resolution in length is inversely proportional to the ultrasonic frequency. Within the reach of diffuse light in biological tissues, the ratio of the imaging depth to the spatial resolution is approximately constant on the order of 200, resulting in the dashed line shown in Fig. 5g. As another example, the lateral resolution limit in time-domain OCT is proportional to the wavelength of the light source divided by the numerical aperture of the imaging system, whereas the depth of focus (imaging depth range) is proportional to the wavelength of the light source divided by the square of the numerical aperture of the imaging system; thus, earning a smaller resolution limit comes at a cost of significantly decreasing the depth of focus. The axial resolution in time-domain OCT, on the other hand, is proportional to the square of the central wavelength of the light source divided by its wavelength bandwidth. Thus, increasing the central wavelength will damage both the lateral and axial resolutions, but will increase the imaging depth range.
Exogenous fluorescence labeling agents tend to photobleach under high excitation powers, resulting in a variable and often unpredictable time-dependent loss of imaging contrast. When rapid imaging is needed, the number of fluorescence photons might be too small to be detected, resulting in low single-to-noise ratio. Thus, label-free imaging can be beneficial for imaging very rapid dynamic phenomena where fluorescence imaging fails. Depending on the speed of the dynamics, one might prefer label-free imaging techniques that do not require sample scanning, such as PhM, PolM, FLIM or optical parametrically gated microscopy.73
Of course, the system construction and use complexity as well as its overall cost are additional considerations. Nonlinear label-free nonlinear imaging techniques, such as HGM, RM, etc., typically require the use of ultrashort (femtosecond or picosecond) optical pulses with sufficient peak power to induce these nonlinear effects in biological samples, which are generally weak and require more complex imaging systems with ultrafast lasers. Recent development of turn-key high-intensity ultrashort lasers in label-free imaging resulted in significant improvements in penetration depth, optical resolution, and acquisition speed. For example, simultaneous absorption of two or three photons leads to high localization of the autofluorescent light (2PF and 3PF, respectively) or the high harmonic generation signal, since such nonlinear absorption processes can only take place in an extremely small volume. Multi-photon imaging using near-IR femtosecond lasers is also characterized by high penetration depths. However, nonlinear imaging approaches might require higher illumination intensity. Attention must be given to maximizing the optical power to generate a larger signal, while minimizing it to avoid damage to the cells and tissues due to photothermal or photomechanical effects. The interplay of several factors should be considered in the optical power optimization. These include the pulse energy, which relates to the laser repetition rate per a given optical power; the pixel dwell time, which relates to the scanning speed; the illumination wavelengths, where the IR regime close to the visible is preferable than the visible regime due to lower photon absorption; and the illumination pattern, which relates to the laser irradiance per sample space and the exposure times per each sample position. This tradeoff can be managed by utilizing increasingly sensitive optical detectors and optical signal amplification to detect the weak harmonic generation signals for imaging.73
3. Future challenges
Label-free imaging utilizes endogenous intrinsic signals rather than specific exogenous markers, while displaying both morphology and chemical composition, and presenting no confounding factors associated with targeting, no biochemical perturbations, and no potential toxicity. Therefore, it allows rapid clinical translation, without drug approvals, as well as imaging of fresh tissues, even in-vivo. In addition, it allows recording new forms of contrast and extracting new features, which cannot be obtained when using exogenous markers, and thus it can provide high dimensionality for AI analysis. Specifically, label-free imaging can provide structural, functional, and metabolic imaging, especially with highly multiplexed and multimodal imaging that is based on multiple contrast mechanisms and physics. Despite these great advantages, the main disadvantage of label-free imaging is limited sources of endogenous contrast. For some of the methods, this might result in lower molecular specificity and weak signals with less clear origin. For these methods, optical energy deposition may occur when using higher incident energy/power while attempting to compensate for the weaker signals. Furthermore, for exogenous labeling and in most cases, the same hardware system can be used to image various biological molecules. The burden of system complexity is then shifted to chemistry. In contrast, typically for each label-free imaging method, a specific hardware system must be tuned to image various molecules, prompting the development of multi-modal imaging systems. Accordingly, the challenges in the field of label-free imaging and possible future solutions are discussed below.
Some internal contrast mechanisms used in label-free imaging are not specific to certain cell organelles, receptors on cell membranes, or biomolecules. For these techniques, obtaining label-free specificity is a great challenge. For example, the refractive index of the cell nucleus might be close or even lower than that of the cell cytoplasm.74 Therefore, it is difficult to determine the nucleus boundaries via refractive-index-based label-free imaging techniques; whereas this is a relatively simple task when exogenous labeling agents are used. Recent AI-based approaches have enabled virtual tissue and cell staining,75–78 also referred to as computational staining, or virtual histology. In this case, a deep neural network is trained on label-free and label-based images, so that after training, the network can take a label-free image of the same type used for training and make it look as if the sample has been chemically labeled. These techniques have been shown to be useful for virtual histopathology of tissue slices and individual cells in vitro, avoiding standardization problems that might occur when using chemical staining, as well as providing virtually stained images when chemical cell staining is not allowed, such as during in-vitro fertilization.76 These AI-based methods work as long as the label-free images can sufficiently define, in a collective manner, the basis for training the deep network, even if in the single-image level, virtual staining cannot be done in a first sight. AI can also help in identifying the source of label-free signal, or even discern between several sources of signals when multimodal label-free imaging is implemented. Specifically, automated interpretation of label-free hyperspectral datasets with AI instead a naked eye opens new possibilities in the derivation of secondary data and conclusions from the primary information.79 We believe that in the near future, with increasing computational processing power and fast acquisition and imaging techniques, these AI-based techniques will be more widely used to obtain in-vivo virtual staining as well.80,81 Furthermore, intensive cross-fertilization between photonics and AI is expected to enable new technological concepts in the field of label-free imaging, allowing new hardware-based AI-integrated systems.
Obtaining label-free nanoscopy, meaning imaging nano-scale objects without exogenous chemical labeling, is another great challenge yet to be addressed. Far-field optical microscopy is typically restricted by the diffraction of light to approximately 200–500 nm. So far, the main advances have been achieved by super-resolved fluorescence microscopy, such as stimulated emission depletion (STED) and localization microscopies (PALM/STORM),82 which resulted in the 2014 Nobel Prize in Chemistry. These techniques enabled label-based nanoscopy of biological cells utilizing specific nanoscale fluorescence emitters. On the other hand, overcoming the far-field diffraction limit without cell labeling is extremely challenging due to the low number of photons originating from unlabeled nanoscale objects, especially when performing rapid 3D imaging. Preliminary label-free super-resolution results have been demonstrated for quantitative phase imaging83 and Raman spectroscopy,84,85 as well as by using photoacoustic86 and photothermal87 effects. Another potential approach to obtain label-free nanoscopy is label-free localization of nanoparticles. Specifically, interferometric scattering microscopy (iSCAT)88 allows label-free localization of nanoparticles as small as 5 nm, including viruses and proteins, via recording their scattering signal. Mass photometry has been used together with iSCAT to record specific protein assembly and disassembly.89,90 As the size of the nanoparticles detected decreases, the signal to noise ratio decreases exponentially, requiring extremely sensitive detectors to allow full-field imaging. Finding stochastic or switchable mechanisms of the scatterers, similarly to fluorophores in PALM and STOM, which can be activated naturally during label-free imaging, might allow transforming label-free localization techniques into label-free nanoscopy techniques. Of course, future approaches also might utilize a combination of experimental label-free optical techniques with computational optics or with AI-based methods.
Another challenge is in-vivo label-free imaging. Being able to ignore a patient’s natural dynamics, such as respiratory or cardiac motion, is a challenge of any in-vivo imaging technique. While some label-free imaging techniques, such as OCT and PAT, are regularly used for in-vivo imaging, other techniques, such as PhM, are more challenging to implement in vivo mainly due to the low-photon generation and collection, and the scattering properties of tissues, which yield noisy images that are hard to interpret, as well as the fact that some of these techniques are typically implemented in transmission mode rather than reflection mode. Fiber-based label-free imaging via various catheters, endoscopes, and needle probes has made techniques with limited imaging penetration depths of several millimeters, such as OCT, relevant for deep in-vivo imaging, utilizing portable imaging systems in clinical environments such as the operating room.91,92 To avoid ex-vivo examination of fixed and stained tissue samples by a pathologist, in-vivo endoscopy can be performed. Specifically, new approaches are needed for a reliable intraoperative tissue diagnosis when time-consuming procedures cannot be used, with clear preference to label-free imaging approaches. As another example, clinically usable Raman fiber probes in combination with field-deployable compact Raman microscopes and endoscopes have been used for intraoperative detection of tumors.54,93 Still, future innovations are needed in the beam-delivery devices, such as hand-held probes, fiber-optic based catheters and endoscopes, and needle-based probes, which will permit high-resolution label-free nonlinear optical imaging at deeper sites within the human body. The implementation of such a nonlinear multimodal imaging approach for in-vivo tissue screening requires new endo-spectroscopic probe concepts, which is a major technological challenge.94 On-chip implementations might also reduce the complexity of the optical systems and make label-free imaging techniques more attainable for direct clinical in-vivo or in-vitro uses. For example, photonic integrated waveguide gratings were recently used for on-chip OCT,95 and nanophotonic waveguides were used to excite and collect signals in close vicinity of a waveguide for on-chip RM.96 Efficient on-chip implementations typically require advanced fabrication techniques, which are expected to be further improved in the future. Specifically, recent advances in optical metasurfaces97,98 might bring to new efficient on-chip implementations of various label-free imaging techniques.
Since each label-free imaging technique might be based on a different overall contrast mechanism, as well as provide different quantitative values, multimodal imaging methods are beneficial. One can think of two end-member strategies for this combination of different label-free imaging methods. On one end, imaging modalities with similar acquisition speed and resolution can be combined, all of which are efficiently excited by the same laser source and can be detected in parallel.99 Specifically, SLAM microscopy38 offers precise spatiotemporal correlation of 2PF and 3PF, SHG, THG, and even CARS through single-shot excitation of ultrafast pulses from a supercontinuum source, followed by fast parallel detection for each of these modalities, as demonstrated in Figs. 3 and 4, presenting convincing potential for label-free cancer identification, even in vivo. A future focus in this field is investigating the dynamic living tumor-tissue microenvironment without perturbative dyes or stains, as well as being able to do this in real-time at point-of-care sites. For example, the potential for label-free cancer diagnosis via label-free imaging of extracellular vesicles in situ with single-vesicle spatiotemporal resolution has been demonstrated in tissue, serum, and urine.56,100 In contrast, all other current methods require extraction of tissue and isolation of vesicles, while losing the spatiotemporal context of these vesicles and their signatures of cancer. The fact that there is no drug (exogenous marker) involved enables rapid translation to clinical studies, trials, and eventually use. More examples for applications of interest for label-free multimodal imaging include characterizing the metabolic dynamics in neurons and astrocytes, and label-free detection of neural activity and connectivity in neuronal cultures via rapid FLIM, OCT, and other multimodal techniques,101,102 label-free detection and characterization of amyloid-beta plaques in brain tissue slices via 3PF and THG,103 and tumor-boundary label-free detection via combined SHG, 3PF, and CARS during surgery. For the latter application a major step forward would be the implementation of spectroscopic-guided ablation combined with an endoscope104 in a seek-and-treat manner, allowing real-time monitoring of the ablated tissue features. The co-registered multimodal image datasets are ideally suited for high-dimensional AI analysis and correlations between the various contrast mechanisms and the underlying physics.105 On the other end, imaging modalities of different imaging speed and tissue penetration can be synergistically combined, so that a fast but chemically less specific method provides an overview on the tissue volume, while a slower, molecule-specific second method is used to classify tissues detected by the faster modality in suspicious areas. One such approach would be to combine OCT or FLIM with RM.21,22,106
The development of these label-free imaging modalities also offers the advantage of longitudinal imaging, such as fast imaging capturing dynamics over time periods of seconds, or longer time-lapse imaging over periods of minutes to hours. Without the concerns of dye photobleaching, potential toxicity, and perturbative changes to the biological processes under investigation, these label-free imaging modalities can be used to explore various time-dependent cellular activities and biological functions, such as rapid sperm dynamics9 and neural activity,101 cell-death processes,102 and intercellular communications via extracellular vesicles and organelle trafficking.56,104 Future opportunities remain for using these unique dynamic nonperturbing features of label-free biomedical imaging for discovering new biological principles as well as new biomarkers indicative of disease.
Finally, the measurability of new characteristics of the light field, beyond the intensity distribution, opens new conceptual and technological possibilities for biomedical research. New light sources generating noise-free quantum states might yield future label-free imaging methods that rely on the correlations between photons and detector systems.108,109
To conclude, when compared to label-based imaging, label-free imaging wins in studying live biological processes in cells and tissues, where exogenous markers perturb the biology studied, especially in longitudinal studies or monitoring. In the near future, label-free imaging will clearly win for in-vivo applications in general and for intraoperative diagnosis in particular, where using exogenous markers is both time consuming and requires regulations and safety approvals that may take years to obtain. We thus expect that in spite of the challenges in the field, yet to be addressed, label-free imaging will become more and more attractive and popular for both biological assays and clinical applications.
Acknowledgments:
Horizon2020 ERC grant (678316) (PI: NTS). NIH Center for Label-free Imaging and Multiscale Biophotonics (CLIMB) at the University of Illinois Urbana-Champaign (http://climb.beckman.illinois.edu) P41 EB031772 and NIH grants R01 CA241618 and R01 CA213149 (PI: SAB). NIH grants R01 NS102213, U01 EB029823 (BRAIN Initiative), R35 CA220436 (Outstanding Investigator Award), and R01 EB028277 (PI: LW).
Biographies
Authors’ Short Resumes:
Natan T. Shaked is a Professor and the Chair of the Department of Biomedical Engineering at Tel Aviv University, Israel. He directs a large experimental group dealing with label-free biomedical imaging with focus on phase imaging. He is the author >100 refereed journal papers and >170 conference papers, as well as a book on biomedical phase microscopy and nanoscopy. He is the Founder and Chair of the SPIE Label-Free Imaging and Sensing (LBIS) annual conference in SPIE Photonics West, San Francisco, CA, USA (founded in 2019). He is a Fellow in the Optica Society (previously OSA) and a Fellow in the SPIE.
Stephen A. Boppart is a Professor and Grainger Distinguished Chair in Engineering at the University of Illinois Urbana-Champaign with appointment in the Departments of Electrical and Computer Engineering, Bioengineering, and Medicine. His Biophotonics Imaging Laboratory at the Beckman Institute for Advanced Science and Technology is focused on developing novel optical biomedical diagnostic and imaging technologies and translating these into clinical applications. He has published over 450 invited and contributed publications, delivered over 1000 invited and contributed presentations, has over 50 patents and co-founded 4 start-ups to disseminate his technologies, and has mentored over 200 young researchers. He is a Fellow of AAAS, IEEE, Optica, SPIE, AIMBE, BMES, and IAMBE, and has been elected into the U.S. National Academy of Inventors. He played an active role in the visioning and growth of the new engineering-based Carle Illinois College of Medicine, and some of his recognitions include the international Hans Sigrist Prize in the field of Diagnostic Laser Medicine, the IEEE Technical Achievement Award, and the SPIE Biophotonics Technology Innovator Award.
Lihong V. Wang is Bren Professor of Medical and Electrical Engineering and Department Chair of Medical Engineering at Caltech. He has published 580 journal articles (h-index = 152, citations = 100,000) and delivered 580 keynote/plenary/invited talks. He published the first functional photoacoustic CT, 3D photoacoustic microscopy, and compressed ultrafast photography (world’s fastest camera). He served as Editor-in-Chief of the Journal of Biomedical Optics. He received the Goodman Book Award, NIH Outstanding Investigator and Director’s Pioneer Awards, OSA Mees Medal, IEEE Technical Achievement and Biomedical Engineering Awards, SPIE Chance Award, IPPA Senior Prize, OSA Feld Biophotonics Award, and an honorary doctorate from Lund University, Sweden. He was inducted into the National Academy of Engineering.
Jürgen Popp is the Chair for Physical Chemistry at the Friedrich-Schiller University Jena. He is also the Scientific Director of the Leibniz Institute of Photonic Technology, Jena. His core research focus is biophotonics and optical health technology. In particular, his expertise is the development and application of innovative linear and non-linear spectroscopy approaches (with focus on Raman spectroscopy) for multi-contrast and multi-parameter imaging for pathology, oncology, and infection/sepsis. He has published more than 950 papers and has been named as an inventor on 15 patents. Prof. Popp is the recipient of the 2016 Pittsburgh Spectroscopy Award. In 2016, he was elected to the American Institute for Medical and Biological Engineering College of Fellows. In 2018, he won the third prize of the Berthold Leibinger Innovationspreis and received the Kaiser-Friedrich-Forschungspreis. In 2020, he became an Optica (previously OSA) Senior Fellow. In 2021, he became a Fellow of the Royal Society of Chemistry.
Footnotes
Competing Interests
The authors declare no competing interests.
Peer review information
Nature Photonics thanks Paul Campagnola, Ping Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Contributor Information
Natan T. Shaked, Department of Biomedical Engineering, Faculty of Engineering, Tel Aviv University, Tel Aviv, Israel
Stephen A. Boppart, Beckman Institute for Advanced Science and Technology, Department of Electrical and Computer Engineering,; Department of Bioengineering, University of Illinois Urbana-Champaign, Urbana, IL, USA
Lihong V. Wang, Caltech Optical Imaging Laboratory, Andrew and Peggy Cherng Department of Medical Engineering, Department of Electrical Engineering, California Institute of Technology, Pasadena, CA, USA
Jürgen Popp, Leibniz Institute of Photonic Technology, Member of Leibniz Health Technologies, Member of the Leibniz Centre for Photonics in Infection Research, Jena, Germany; Institute of Physical Chemistry and Abbe Center of Photonics, Friedrich Schiller University Jena, Jena, Germany.
References
- 1.Zernike F How I discovered phase contrast. Science 121, 3141, 345–349 (1955). [DOI] [PubMed] [Google Scholar]
- 2.Lang W Nomarski differential interference-contrast microscopy. ZEISS Information 70, 114–120 (1968). [Google Scholar]
- 3.Marquet P et al. Digital holographic microscopy: a noninvasive contrast imaging technique allowing quantitative visualization of living cells with subwavelength axial accuracy. Opt. Lett. 30, 468–470 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Girshovitz P & Shaked NT Generalized cell morphological parameters based on interferometric phase microscopy and their application to cell life cycle characterization. Biomed. Opt. Express 3, 1757–1773 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park YK, Depeursinge C & Popescu G Quantitative phase imaging in biomedicine. Nature Photonics 12, 578–589 (2018). [Google Scholar]
- 6.Haifler M, Girshovitz P, Band G, Dardikman G, Madjar I & Shaked NT Interferometric phase microscopy for label-free morphological evaluation of sperm cells. Fertility and Sterility 104, 43–47, 2015. [DOI] [PubMed] [Google Scholar]
- 7.Choi W et al. Tomographic phase microscopy. Nature Methods 717–719 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Jin D, Zhou R, Yaqoob Z & So PTC Tomographic phase microscopy: principles and applications in bioimaging. J Opt Soc Am B. 34, B64–B77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dardikman-Yoffe G, Mirsky SK, Barnea I & Shaked NT, High-resolution 4-D acquisition of freely swimming human sperm cells without staining. Science Advances 6, eaay7619 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oldenbourg R, Polarized light microscopy: principles and practice. In Imaging: A Laboratory Manual (ed. Yuste). CSHL Press, Cold Spring Harbor, NY, USA, 2011. [DOI] [PubMed] [Google Scholar]
- 11.Oldenbourg R Polarized light microscopy of spindles. in Methods in Cell Biology 61, 175–208 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Koike-Tani M, Tani T, Mehta SB, Verma A & Oldenbourg R, Polarized light microscopy in reproductive and developmental biology. Molecular Reproduction and Development 82, 548–562 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drexler W & Fujimoto JG Optical Coherence Tomography: Technology and Applications. (Springer International Publishing; 2008). [Google Scholar]
- 14.Leitgeb R, Hitzenberger CK & Fercher AF Performance of Fourier domain vs. time domain optical coherence tomography. Opt. Express 11, 889–894 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Hillmann D et al. Aberration-free volumetric high-speed imaging of in vivo retina. Sci. Rep. 6, 35209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duker JS, Waheed NK & Goldman D Handbook of Retinal OCT: Optical Coherence Tomography E-Book (Elsevier Health Sciences, Oxford, UK; 2013). [Google Scholar]
- 17.Shemonski ND, Adie SG, South FA, Liu Y-Z, Carney PS & Boppart SA Computational high-resolution optical imaging of the living human retina. Nature Photonics 9, 440–443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tearney GJ et al. Three-dimensional coronary artery microscopy by intracoronary optical frequency domain imaging. JACC Cardiovascular Imaging 1, 752–761 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raffel OC, Akasaka T & Jang I-K, Cardiac optical coherence tomography. Heart 94, 1200–1210 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Tearney GJ et al. In vivo endoscopic optical biopsy with optical coherence tomography. Science 276, 2037–2039 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Nolan RM et al. Intraoperative optical coherence tomography for assessing human lymph nodes for metastatic cancer. BMC Cancer 16, 144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erickson-Bhatt SJ, et al. Real-time imaging of the resection bed using a handheld probe to reduce incidence of microscopic positive margins in cancer surgery. Cancer Research 75. 3706–3712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poneros JM & Nishioka NS Diagnosis of Barrett’s esophagus using optical coherence tomography. Gastrointest Endosc Clin N Am 13, 309–323 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Dong J, et al. Feasibility and safety of tethered capsule endomicroscopy in patients with Barrett’s esophagus in a multi-center study. Clin Gastroenterol Hepatol 20, 756–765 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sattler E, Kästle R & Welzel J Optical coherence tomography in dermatology. J Biomed Opt.18, 061224 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Gambichler T, Moussa G, Sand M, Sand D, Altmeyer P & Hoffmann K Applications of optical coherence tomography in dermatology. J Dermatological Science 40, 85–94 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Byers RA et al. Sub-clinical assessment of atopic dermatitis severity using angiographic optical coherence tomography. Biomedical Optics Express 9, 2001–2017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larina IV et al. Live imaging of blood flow in mammalian embryos using Doppler swept-source optical coherence tomography. J Biomed Opt 13, 060506 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Singh M, et al. Applicability, usability, and limitations of murine embryonic imaging with optical coherence tomography and optical projection tomography. Biomedical Optics Express 7, 2295–2310 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park S et al. Quantitative evaluation of the dynamic activity of HeLa cells in different viability states using dynamic full-field optical coherence microscopy. Biomed Opt Express 12, 6431–6441 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mecê P, Scholler J, Groux K & Boccara C High-resolution in-vivo human retinal imaging using full-field OCT with optical stabilization of axial motion. Biomed Opt Express 11, 492–504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ralston TS, Marks DL, Carney PS & Boppart SA Interferometric synthetic aperture microscopy. Nature Physics 3, 129–134 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohler W, Millard AC & Campagnola PJ Second harmonic generation imaging of endogenous structural proteins. Methods 29, 97–109 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Conklin MW et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J Pathol 178, 1221–1232 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinn KP et al. Optical metrics of the extracellular matrix predict compositional and mechanical changes after myocardial infarction. Sci Rep 6, 35823 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu S-W, Tai S-P, Ho C-L, Lin C-H & Sun C-K High-resolution simultaneous three-photon fluorescence and third-harmonic-generation microscopy. Microsc Res Tech 66, 193–197 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Tsai M-R, Chen S-Y, Shieh D-B, Lou P-J & Sun C-K In vivo optical virtual biopsy of human oral mucosa with harmonic generation microscopy. Biomed Opt Express 2, 2317–2328 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh AJ et al. Classification of T-cell activation via autofluorescence lifetime imaging. Nat Biomed Eng 5, 77–88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You S et al. Intravital imaging by simultaneous label-free autofluorescence-multiharmonic microscopy. Nature Communications 9, 2125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skala MC et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci U S A 104, 19494–19499 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Meng J, Quinn KP & Georgakoudi I Tissue imaging and quantification relying on endogenous contrast. Adv Exp Med Biol 3233, 257–288 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Becker W, Bergmann A & Biskup C Multispectral fluorescence lifetime imaging by TCSPC. Microsc Res Tech 70, 403–409 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Sorrells JE et al. Computational photon counting using multi-threshold peak detection for fast fluorescence lifetime imaging microscopy. ACS Photonics 9, 2748–2755 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bower AJ, Marjanovic M, Zhao Y, Li J, Chaney EJ, Boppart SA Label-free in vivo cellular-level detection and imaging of apoptosis. J Biophotonics 10, 143–150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q, He X, Wang Y, Liu H, Xu D & Guo F Review of spectral imaging technology in biomedical engineering: achievements and challenges. J Biomedical Optics 18, 100901 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Kole MR, Reddy RK, Schulmerich MV, Gelber MK, & Bhargava R Discrete frequency infrared microspectroscopy and imaging with a tunable quantum cascade laser. Analytical Chemistry 84, 10366–10372 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pilling MJ, Henderson A & Gardner P Quantum cascade laser spectral histopathology: Breast cancer diagnostics using high throughput chemical imaging. Analytical Chemistry 89, 7348–7355 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Kuepper C, Kallenbach-Thieltges A, Juette H, Tannapfel A, Großerueschkamp F & Gerwert K Quantum cascade laser-based infrared microscopy for label-free and automated cancer classification in tissue sections. Scientific Reports 8, 7717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang D, Li C, Zhang C, Slipchenko MN, Eakins G & Cheng J-X Depth-resolved mid-infrared photothermal imaging of living cells and organisms with submicrometer spatial resolution. Science Advances 2, e1600521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nedosekin DA, Galanzha EI, Dervishi E, Biris AS & Zharov VP Super-resolution nonlinear photothermal microscopy. Small 10, 135–142 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Brauchle E & Schenke-Layland K Raman spectroscopy in biomedicine – non-invasive in vitro analysis of cells and extracellular matrix components in tissues, Biotechnol J 8, 288–297 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krafft C, et al. , Label-free molecular imaging of biological cells and tissues by linear and nonlinear Raman spectroscopic approaches. Angew Chem Int Ed 56, 4392–4431 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Lee KS et al. Raman microspectroscopy for microbiology. Nat Rev Methods Primers 1, 80 (2021). [Google Scholar]
- 54.Matanfack GA, Rüger J, Stiebing C, Schmitt M & Popp J Imaging the invisible—Bioorthogonal Raman probes for imaging of cells and tissues. J Biophotonics 13, e202000129 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Zumbusch A, Holtom GR & Xie XS Three-dimensional vibrational imaging by coherent anti-Stokes Raman scattering. Phys Rev Lett 82, 4142–4145 (1999). [Google Scholar]
- 56.Tu H et al. , Concurrence of extracellular vesicle enrichment and metabolic switch visualized label-free in the tumor microenvironment. Science Advances 3, e1600675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, Tu H, You S, Chaney EJ, Marjanovic M & Boppart SA Label-free molecular profiling for identification of biomarkers in carcinogenesis using multimodal multiphoton imaging. Quant. Imaging Med. Surg, 9:742–756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Freudiger CW et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 322, 1857–1861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng J-X, Min W, Ozeki Y & Polli D Stimulated Raman Scattering Microscopy: Techniques and Applications (ScienceDirect, 2022). [Google Scholar]
- 60.Wang LV & Hu S, Photoacoustic tomography: in vivo imaging from organelles to organs. Science 335, 1458–1462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang XD, Pang YJ, Ku G, Xie XY, Stoica G & Wang LV Noninvasive laser-induced photoacoustic tomography for structural and functional in vivo imaging of the brain. Nat Biotechnol 21, 803–806 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Siphanto RI et al. Serial noninvasive photoacoustic imaging of neovascularization in tumor angiogenesis. Optics Express 13, 89–95 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Laufer J, Delpy D, Elwell C & Beard P Quantitative spatially resolved measurement of tissue chromophore concentrations using photoacoustic spectroscopy: application to the measurement of blood oxygenation and haemoglobin concentration. Phys Med Biol 52, 141–168 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Zhang HF, Maslov K, Stoica G & Wang LV Functional photoacoustic microscopy for high-resolution and noninvasive in vivo imaging. Nat Biotechnol 24, 848–851 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Xu MH & Wang LV Universal back-projection algorithm for photoacoustic computed tomography. Physical Review E 71, 016706 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Wang XD, Pang YJ, Ku G, Xie XY, Stoica G & Wang LV Noninvasive laser-induced photoacoustic tomography for structural and functional in vivo imaging of the brain. Nat Biotechnol 21, 803–806 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Nagae K et al. Real-time 3D photoacoustic visualization system with a wide field of view for imaging human limbs. F1000Research 7, 1813 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin L, et al. Single-breath-hold photoacoustic computed tomography of the breast. Nature Communications 9, 2352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.PDantuma M, Lucka F, Kruitwagen S, Javaherian A, Alink L, van Meerdervoort R, et al. Fully three-dimensional sound speed-corrected multi-wavelength photoacoustic breast tomography. arXiv preprint arXiv:230806754 2023. [Google Scholar]
- 70.Na S et al. Massively parallel functional photoacoustic computed tomography of the human brain. Nature Biomedical Engineering 6, 584–592 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong TT et al. Fast label-free multilayered histology-like imaging of human breast cancer by photoacoustic microscopy. Science Advances, 3, e1602168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L et al. Single-impulse panoramic photoacoustic computed tomography of small-animal whole-body dynamics at high spatiotemporal resolution. Nature Biomedical Engineering 1, 0071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun Y, Tu H, You S, Zhang C, Liu Y-Z & Boppart SA Detection of weak near-infrared optical imaging signals under ambient light by optical parametric amplification. Optics Letters 44, 4391–4394 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schürmann M, Scholze J, Müller P, Guck J & Chan CJ Cell nuclei have lower refractive index and mass density than cytoplasm. J Biophotonics 9, 1068–1076 (2016). [DOI] [PubMed] [Google Scholar]
- 75.Rivenson Y et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nature Biomedical Engineering 3, 466–477 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Nygate YN et al. Holographic virtual staining of individual biological cells. Proc Natl Acad Sci USA 117, 9223–9231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kandel ME et al. Phase imaging with computational specificity (PICS) for measuring dry mass changes in sub-cellular compartments. Nature Communications 11, 6256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.You S, Chaney EJ, Tu H, Sinha S & Boppart SA Label-free deep profiling of the tumor microenvironment. Cancer Research 81, 2534–2544 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krafft C & Popp J Opportunities of optical and spectral technologies in intraoperative histopathology. Optica 10, 214–231 (2023). [Google Scholar]
- 80.Pradhan P et al. Computational tissue staining of non-linear multimodal imaging using supervised and unsupervised deep learning. Biomedical Optics Express 12, 2280–2298 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.You S, et al. Real-time intraoperative diagnosis by deep neural network driven multiphoton virtual histology. Precision Oncology 3, 33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hell SW et al. The 2015 super-resolution microscopy roadmap. Journal of Physics D: Applied Physics 48, 443001 (2015). [Google Scholar]
- 83.Cotte Y et al. Marker-free phase nanoscopy. Nature Photonics 7, 113–117 (2013). [Google Scholar]
- 84.Bi Y et al. Near-resonance enhanced label-free stimulated Raman scattering microscopy with spatial resolution near 130 nm. Light: Science & Applications 7, 81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gong L, Zheng W, Ma Y & Huang Z Higher-order coherent anti-Stokes Raman scattering microscopy realizes label-free super-resolution vibrational imaging. Nature Photonics 14, 115–122 (2020). [Google Scholar]
- 86.Danielli A et al. Label-free photoacoustic nanoscopy, J Biomed Opt 19, 086006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fu P et al. Super-resolution imaging of non-fluorescent molecules by photothermal relaxation localization microscopy. Nature Photonics 17, 330–337 (2023). [Google Scholar]
- 88.Lindfors K, Kalkbrenner T, Stoller P & Sandoghdar V Detection and spectroscopy of gold nanoparticles using supercontinuum white light confocal microscopy. Phys. Rev. Lett. 93, 037401 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Foley EDB, Kushwah MS, Young G & Kukura P Mass photometry enables label-free tracking and mass measurement of single proteins on lipid bilayers. Nature Methods 18, 1247–1252 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heermann T, Steiert F, Ramm B, Hundt N & Schwille P Mass-sensitive particle tracking to elucidate the membrane-associated MinDE reaction cycle. Nature Methods 18, 1239–1246 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sun Y et al. Intraoperative visualization of the tumor microenvironment and quantification of extracellular vesicles by label-free nonlinear imaging. Science Advances, 4, eaau5603 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Monroy GM, Won J, Spillman DR, Dsouza R & Boppart SA Clinical translation of handheld optical coherence tomography: practical considerations and recent advances. J Biomedical Optics 22, 121715 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jermyn M et al. Intraoperative brain cancer detection with Raman spectroscopy in humans. Science Translational Medicine 7, 274ra19 (2015). [DOI] [PubMed] [Google Scholar]
- 94.Pshenay-Severin E el al., Multimodal nonlinear endomicroscopic imaging probe using a double-core double-clad fiber and focus-combining micro-optical concept. Light: Science & Applications 10, 207 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rank EA et al. Toward optical coherence tomography on a chip: in vivo three-dimensional human retinal imaging using photonic integrated circuit-based arrayed waveguide gratings. Light: Science & Applications 10, 6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wuytens PC, Skirtach AG & Baets R On-chip surface-enhanced Raman spectroscopy using nanosphere-lithography patterned antennas on silicon nitride waveguides. Opt Express 25, 12926–12934 (2017). [DOI] [PubMed] [Google Scholar]
- 97.Yu N & Capasso F Flat optics with designer metasurfaces. Nature Materials 13, 139–150 (2014). [DOI] [PubMed] [Google Scholar]
- 98.Neshev D & Aharonovich I Optical metasurfaces: new generation building blocks for multi-functional optics. Light: Science & Applications 7, 58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meyer T et al. A compact microscope setup for multimodal nonlinear imaging in clinics and its application to disease diagnostics. Analyst 138, 4048–4057 (2013). [DOI] [PubMed] [Google Scholar]
- 100.You S et al. Label-free visualization and characterization of extracellular vesicles in breast cancer. Proc Natl Acad Sci USA, 116, 24012–24018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Iyer RR. et al. Ultra-parallel label-free optophysiology of neural activity. iScience 25, 104307 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bower AJ, Li J, Chaney EJ, Marjanovic M, Spillman DR Jr & Boppart SA. High-speed imaging of transient metabolic dynamics using two-photon fluorescence lifetime imaging microscopy. Optica 5, 1290–1296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tehrani KF, Park J, Renteria C & Boppart SA Label-free identification of Alzheimer’s disease plaques using multiple co-registered nonlinear optical biomarkers. SPIE Photonics West BIOS, San Francisco, CA, January 28 – February 2, (2023). [Google Scholar]
- 104.Lai C et al. Design and test of a rigid endomicroscopic system for multimodal imaging and femtosecond laser ablation. Journal of Biomedical Optics (2023). Submitted [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chernavskaia O et al. Beyond endoscopic assessment in inflammatory bowel disease: real-time histology of disease activity by non-linear multimodal imaging. Scientific Reports 6, 29239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fitzgerald S et al. Multimodal Raman spectroscopy and optical coherence tomography for biomedical analysis. Journal of Biophotonics, e202200231 (2023). In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Barreto Lemos G, Borish V, Cole GD, Ramelow S, Lapkiewicz R & Zeilinger A Quantum imaging with undetected photons. Nature 512, 409–412 (2014). [DOI] [PubMed] [Google Scholar]
- 108.Kalashnikov DA, Paterova AV, Kulik SP & Krivitsky LA Infrared spectroscopy with visible light. Nature Photonics 10, 98–101 (2016). [Google Scholar]


