Abstract
A porcine torovirus (PoTV) was identified and characterized; it is a novel member of the genus Torovirus (family Coronaviridae, order Nidovirales), closely related to but clearly distinct from the already recognized equine torovirus (ETV) and bovine torovirus (BoTV) representatives. Immunoelectron microscopy of feces from piglets revealed elongated, 120- by 55-nm particles which were recognized by a torovirus-specific antiserum. Amplification by reverse transcriptase (RT) PCR with primers designed to detect conserved regions (on the basis of the genomes of BoTV strain Breda and ETV strain Berne) resulted in the identification of the 489-bp nucleocapsid gene, encoding a 18.7-kDa protein. The sequence identity in this region between PoTV and both ETV and BoTV was only about 68%, whereas the latter two show 81% identity. Neutralizing antibodies directed against ETV were found in sera of adult and young pigs. In all 10 herds sampled, seropositive animals were present, and 81% of randomly selected adult sows possessed antibodies. A longitudinal study with RT PCR showed that piglets shed virus in the feces for 1 or more days, starting 4 to 14 days after weaning.
Toroviruses are spherical, oval, elongated, or kidney-shaped enveloped viruses with a single-stranded RNA genome of positive polarity; they belong to the second genus in the family Coronaviridae, in the recently established order Nidovirales (4; for reviews, see references 8 and 24). Toroviruses resemble coronaviruses in genome organization and replication strategy but differ in virion architecture, especially with regard to the nucleocapsid, which is a tubular structure responsible for the singular particle morphology (31).
The torovirus genome is nonsegmented, polyadenylated, and 25 to 30 kb in size. Its 5′ two thirds is occupied by two large overlapping open reading frames, ORF1a and ORF1b. These encode a polyprotein from which the viral polymerase is derived. Downstream of ORF1b are four smaller open reading frames which are expressed through a 3′-coterminal nested set of mRNAs. They code for the following structural proteins (from 5′ to 3′): (i) the 180,000-molecular-weight (180K) precursor of spike protein S, (ii) the 26K triple spanning integral membrane protein M, (iii) a 65K class I membrane protein (HE) exhibiting acetylesterase activity, and (iv) the 19K nucleocapsid protein N (5, 7, 15, 23, 25, 26).
Two torovirus species are recognized, bovine torovirus (BoTV, originally named Breda virus), evidenced in the feces of diarrheic calves (33), and equine torovirus (ETV, formerly Berne virus), isolated in cell culture from rectal swabs from a horse (31). There is serological evidence for the existence of toroviruses in other mammals (3, 32), including swine. During a serological survey in Switzerland, ETV-neutralizing antibodies were detected in the sera of 91 of 112 pigs tested (81%) (32). Furthermore, several authors have reported toroviruslike particles in the feces of swine (10, 20, 22, 35). It is of note, however, that in negatively stained preparations for electron microscopy (EM), torovirions are often pleiomorphic and may appear as spherical, oval, elongated, or kidney-shaped particles (31, 33) carrying either a single or a double fringe of surface projections (5, 31, 33). Without additional immunological confirmation, torovirions are difficult to distinguish from coronaviruses, other viral particles, and even nonviral fringed particles (1, 9, 33).
In this paper, we present formal evidence for the existence of a porcine torovirus (PoTV). By using a reverse transcriptase (RT) PCR targeted to the 3′-nontranslated region (NTR) of the genome, we detected torovirus RNA in the feces of piglets. Moreover, torovirions were identified in these samples by immunoelectron microscopy. Virus shedding, as monitored by RT PCR, started shortly after weaning and lasted for 1 or more days. Comparative sequence analysis of the N-protein gene indicated that PoTV is a novel torovirus closely related to but clearly distinct from BoTV and ETV.
MATERIALS AND METHODS
Cells, viruses, and antisera.
Equine dermis (EDERM) cells (American Type Culture Collection) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 100 IU of penicillin per ml, and 100 μg of streptomycin per ml. ETV strain Berne (P138/72) was propagated in EDERM cells as described previously (29). Tissue culture supernatant containing porcine epidemic diarrhea virus (PEDV) and porcine anti-PEDV serum (P-αPEDV) were obtained from Ghent University, Ghent, Belgium. Transmissible gastroenteritis virus (TGEV), strain Purdue, was propagated in PD5 (Philips Duphar) porcine kidney cells. Preparation of rabbit antiserum against BoTV (Ra-αBoTV) has been described elsewhere (14). An ascitic fluid sample (A40) from a cat that had succumbed to feline infectious peritonitis was used for the immunodetection of TGEV (19).
Collection of porcine serum and feces field samples.
Serum samples were obtained from four piglets at a commercial breeding and fattening farm in Belgium; the samples were obtained from the piglets at 2 to 11 weeks of age at 2-week intervals (13). In addition, serum samples were collected from 10 to 12 randomly selected adult sows at each of 10 commercial breeding farms in The Netherlands. At one of these, sequential serum samples were also collected from nine piglets in three litters (three samples per litter). The first samples were taken 1 to 3 days after birth (day 0), followed by bleedings at days 14, 21, 35, and 49. The piglets were weaned at day 21. Serum samples were also obtained from the sows after farrowing (day 0).
Fecal samples were collected from each piglet at days 14, 21, 23, 25, 27, 29, 31, 35, and 49.
Serum neutralization assay.
Serial twofold dilutions of heat-inactivated porcine sera were incubated with 100 50% infective doses of ETV for 1 h at 37°C. The mixtures were used to inoculate EDERM cell monolayers grown in 96-well microtiter plates. The cell cultures were examined for cytopathic effects (CPE) at 2 days after infection, and the neutralization titers were calculated by use of the Spearman-Kärber formula.
RT PCR and sequence analysis.
RNA was extracted from fecal samples and concentrated by a modification of the guanidinium isothiocyanate-silica protocol of Boom et al. (2), and an RT PCR was performed as described previously (11). Details of the primers used for RT PCR and for sequence analysis of the PCR products are given in Table 1. Torovirus RNA was detected by use of an RT PCR assay targeted to the NTR of the viral genome with oligonucleotides 293 and 294 as primers. For RT PCR amplification of the PoTV N-protein gene, oligonucleotides 294 and 253 and oligonucleotides 620 and 542 served as primers. The resulting products were cloned into vector pGEM-T (Promega Corp., Madison, Wis.) and sequenced with a T7 sequencing kit (Pharmacia Biotech) according to the manufacturer’s instructions. The nucleotide sequence was determined for both orientations with two or more independent clones. Nucleic acid and amino acid sequence data were analyzed with PC-DOS HIBIO DNASIS and PROSIS software from Pharmacia Biochemicals (Milwaukee, Wis.).
TABLE 1.
Oligonucleotide primers used for RT PCR and sequence analysis
| Primer | Sequence (5′ to 3′) | Positiona | Orientation | Derived from |
|---|---|---|---|---|
| 294 | CTTACATGGAGACACTCAACCA | 616 to 637 | Negative | ETV |
| 293 | AGCAACCTTGAGGTTGGGTCTGT | 502 to 524 | Positive | ETV |
| 253 | GAGAAAGAGCCAAGATGATT | −14 to 6 | Positive | ETV |
| 542 | GAGACACTATCTTTAG | −29 to −14 | Positive | ETV |
| 620 | CGCCAAACTCTGCAACTCAGGTGGA | 340 to 365 | Negative | PoTV |
| 593 | GTCAGAATAGATCACGCATT | 170 to 189 | Positive | PoTV |
Numerical position on the PoTV genome as determined from the start codon of the N protein gene.
EM and immunoelectron microscopy.
Immediately after collection, fecal samples were suspended in phosphate-buffered saline (approximately 1:2), and the suspension was centrifuged for 1 min at 12,000 × g. From the supernatant, 10-μl droplets were deposited on Parafilm, and a 300-mesh collodion- and carbon-coated copper grid was placed on top. Incubation was done for 5 min at room temperature. The preparations were negatively stained with 2% phosphotungstic acid (pH 6.9) for 1 min. For immunoelectron microscopy, the fecal suspension was spun through a 20% (wt/wt) sucrose solution onto a cushion of 50% (wt/wt) sucrose for 2 h at 150,000 × g and 4°C. The interphase was collected and used to prepare grids as described above. Preparations from TGEV and PEDV cell culture supernatants were used as controls. Blocking and immunostaining were performed as described previously (28); Ra-αBoTV and P-αPEDV sera were used at a 1:100 dilution in phosphate-buffered saline containing 0.5% bovine serum albumin (fraction V; Sigma), 1% acetylated bovine serum albumin (Aurion) or ascites A40 (diluted 1:200), and a protein A-colloidal gold (5 nm) conjugate (Aurion). The grids were examined with a Philips CM10 electron microscope at an accelerating voltage of 80 kV.
RESULTS
Serological evidence for torovirus infection in swine.
A serological survey was performed to study the occurrence of torovirus infections in swine. Sera collected from adult sows at 10 breeding farms in The Netherlands were tested for cross-reacting antibodies in an ETV neutralization assay. On each of the farms, seropositive animals were identified (Table 2). Of the 118 pigs tested, 96 (81.4%) had ETV-neutralizing antibody titers of ≥10; 13 pigs (11.9%) had titers of ≥160.
TABLE 2.
ETV-neutralizing antibody titers in adult pigs on 10 farms
| Farm | No. of animals with titers of:
|
No. of animals | ||
|---|---|---|---|---|
| <10 | ≥10 and <160 | ≥160 | ||
| 1 | 0 | 10 | 2 | 12 |
| 2 | 1 | 11 | 0 | 12 |
| 3 | 5 | 7 | 0 | 12 |
| 4 | 1 | 6 | 3 | 10 |
| 5 | 2 | 8 | 2 | 12 |
| 6 | 3 | 8 | 1 | 12 |
| 7 | 2 | 10 | 0 | 12 |
| 8 | 2 | 8 | 2 | 12 |
| 9 | 2 | 8 | 2 | 12 |
| 10 | 4 | 7 | 1 | 12 |
| Total | 22 | 83 | 13 | 118 |
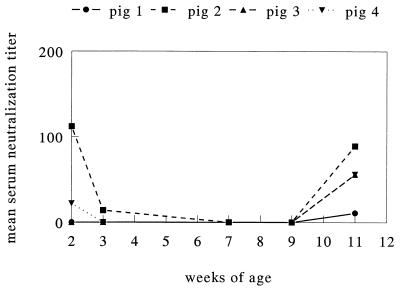
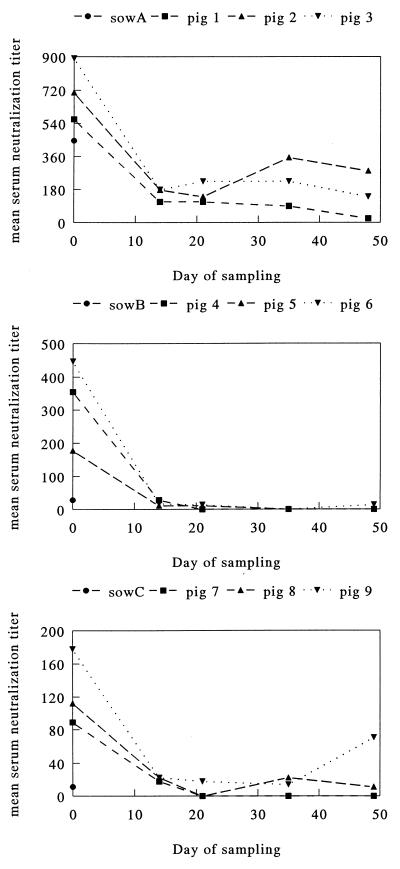
As we suspected infection to take place early in life, we tested sera obtained sequentially from four piglets at a breeding and fattening farm in Belgium. Two of the animals had ETV-neutralizing antibodies at 2 weeks of age. Titers then declined and, by 7 to 9 weeks, had dropped to values below 10; they then rose again, and at 11 weeks of age, three animals had ETV-neutralizing antibody titers between 10 and 90 (Fig. 1). In a more extensive longitudinal experiment on a breeding farm in The Netherlands, 9 piglets belonging to three litters (three animals per litter) were monitored until 7 weeks of age. Sera were obtained at regular intervals, and fecal samples also were collected. At the first sampling, at an age of 1 to 3 days, all piglets possessed ETV-neutralizing antibodies with titers ranging from 90 to 900 (Fig. 2). In each case, the titers in the piglets exceeded those in the sows. Again, titers declined rapidly in the following weeks; in a number of piglets, a moderate rise was noted at 2 to 4 weeks after weaning (Fig. 2).
FIG. 1.
Serum neutralization titers against ETV in sera from four piglets 2 to 11 weeks old.
FIG. 2.
Serum neutralization titers against ETV in sera from three different sows (sows A, B, and C; one serum sample each at day 0) and nine piglets (three piglets per sow). On day 0, the piglets were 1 to 3 days old; on day 21, they were weaned. Note that the scales on the ordinate are different for the three graphs.
Torovirus detection in pig feces.
The serological findings indicated that torovirus infections are common in swine, with primary infection taking place shortly after weaning. To corroborate this observation and to obtain genetic evidence for the existence of a genuine PoTV, the fecal samples from the nine piglets were assayed with an RT PCR targeted to the NTR of the viral genome. The oligonucleotide primers had been designed according to conserved sequences in the genomes of BoTV (strain Breda) and ETV (strain Berne). For eight of the nine piglets, RT PCR yielded a product of the anticipated size of 135 bp. Sequence analysis confirmed that the product was torovirus specific, displaying 88.2 and 87.1% sequence identities with the NTRs of ETV and BoTV, respectively. Virus shedding, as monitored by RT PCR, started 4 to 14 days after weaning. Virus shedding was shown to last for 1 to 9 days in four of the eight PCR-positive piglets. Since no samples were collected between 14 and 28 days after weaning, the exact date of termination of shedding could not be determined for the other four animals (Table 3). During this period, the animals appeared healthy and passed normal stools.
TABLE 3.
RT PCR results for feces from nine piglets
| Pigleta | RT PCR result on the following day of samplingb:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 14 | 21 | 23 | 25 | 27 | 29 | 31 | 35 | 49 | |
| A1 | − | − | − | − | − | − | − | + | − | − |
| A2 | − | − | − | − | − | − | − | − | − | − |
| A3 | − | − | − | − | − | − | − | − | + | − |
| B4 | − | − | − | − | − | − | + | + | + | − |
| B5 | − | − | − | − | + | + | + | + | − | − |
| B6 | − | − | − | − | − | + | + | + | − | − |
| C7 | − | − | − | − | − | − | − | + | + | − |
| C8 | − | − | − | − | − | + | + | + | − | − |
| C9 | − | − | − | − | − | + | + | + | + | − |
A, piglet born to sow A; B, piglet born to sow B; C, piglet born to sow C.
Day 0, a few days after birth; day 21, day of weaning. +, positive PCR result; −, negative PCR result.
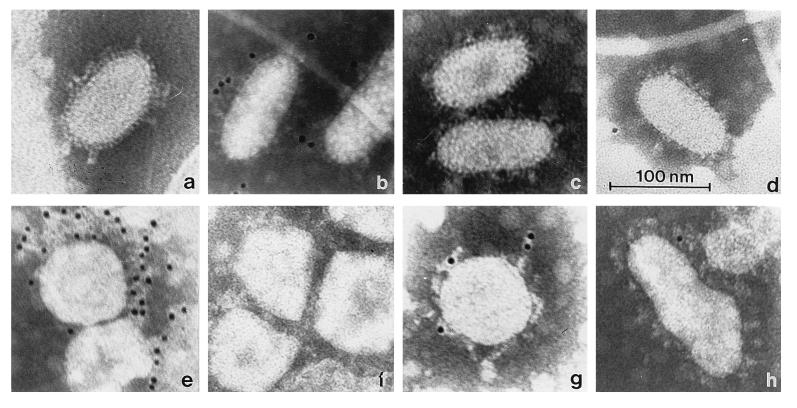
Direct evidence for torovirus shedding in the feces was obtained by EM. In fecal samples that had tested positive by RT PCR, torovirus-like particles were indeed observed. In fresh material they were mainly elongated, approximately 120 nm in length and approximately 55 nm in width, the surface projections excluded. Upon repeated freezing and thawing, the virions became more pleiomorphic, and round, kidney- and torus-shaped particles were found. Two sets of surface projections were discerned. The longer projections were drumstick or petal shaped, extending for approximately 19 nm. The short spikes measured about 6 nm (Fig. 3a).
FIG. 3.
Morphology of PoTV and immunogold labeling (controls include TGEV and PEDV) with different sera. (a) PoTV particles in a fresh fecal sample from a piglet 1 week after weaning. (b to h) Immunogold staining of PoTV with anti-BoTV serum and protein A–5-nm colloidal gold conjugate (b); PoTV with A40, specific for TGEV (c); PoTV with anti-PEDV serum (d); PEDV with anti-PEDV serum (e); PEDV with anti-BoTV serum (f); TGEV with A40, specific for TGEV (g); and TGEV with anti-BoTV serum (h). The bar applies to all of the micrographs.
Formal proof that the particles represented torovirions was obtained by immunoelectron microscopy. Ra-αBoTV specifically recognized the particles. This serum did not bind to virions of TGEV or PEDV, other members of the Coronaviridae family frequently found in porcine feces. Moreover, immunogold labeling of the particles did not occur when rabbit preimmune serum or antiserum against TGEV or PEDV was used (Fig. 3b to h).
Genetic analysis of PoTV.
To study the relationship of putative PoTV with BoTV and ETV, its N-protein gene was amplified by RT PCR. A schematic outline of the strategies for the RT PCR assay and sequence analysis is shown in Fig. 4. The N-protein gene of PoTV was found to be 489 bp in length and to encode an 18.7-kDa protein. The sequence variation in the N-protein gene between PoTVs sampled from two piglets was 0.2%. Comparison to the N-protein genes of BoTV and ETV (23) revealed sequence identities of 68.7 and 68.3%, respectively. In contrast, the N-protein genes of BoTV and ETV show 81.2% sequence identity. At the amino acid sequence level, the PoTV N protein showed 68.3 and 66.9% identities with the N proteins of BoTV and ETV, respectively. The BoTV and ETV N proteins are 82% identical (Table 4). An alignment is shown in Fig. 5.
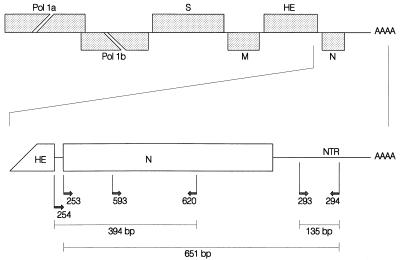
FIG. 4.
Genomic organization of toroviruses and a schematic outline of the strategies used for amplification and sequencing of the 3′ genomic region. The upper panel shows a schematic representation of the torovirus genome, with the genes represented by boxes. The genes for the polymerase (Pol 1a, Pol 1b), the spike protein (S), the membrane protein (M), the hemagglutinin-esterase (HE), and the nucleocapsid protein (N) are indicated. The lower panel shows a schematic outline of the RT PCR assay targeted to the NTR and the N-protein gene. The positions and orientations of the oligonucleotides on the PoTV genome are shown, as are the lengths of the products of the PCRs. Primer 593 was used only in the sequence analysis.
TABLE 4.
Percentages of sequence identities
| Virus pair | % Identity in:
|
||
|---|---|---|---|
| N-protein gene
|
NTRa (nucleotides) | ||
| Nucleotide | Amino acid | ||
| PoTV-ETV | 68.3 | 66.9 | 88.2 |
| PoTV-BoTV | 68.7 | 68.3 | 87.1 |
| ETV-BoTV | 81.2 | 82.0 | 88.2 |
Between primers 293 and 294.
FIG. 5.
Amino acid sequence alignment of the N gene products of PoTV, BoTV, and ETV.
DISCUSSION
In this report, we describe the identification of a torovirus of swine and initial morphological and genetic data. The virus was detected in the feces of piglets by immunoelectron microscopy and RT PCR. In fresh fecal samples, PoTV particles appeared elongated, measuring 120 nm in length and 55 nm in width. Two types of surface projections were observed, the longer of which was petal shaped and 18 to 20 nm in length and most likely represented oligomers of the S protein (12). The shorter spikes were 6 nm long and presumably represented HE. Surface projections of this size are also seen in BoTV; they are absent in ETV (5), where the HE gene is truncated at its 5′ end (27). Preliminary observations from RT PCR amplification and sequence analysis indicate that, like BoTV, PoTV contains an intact HE gene (17).
Comparative sequence analysis of the torovirus N-protein genes showed that BoTV and ETV were closely related, with 81% sequence identity in this region. In contrast, PoTV showed only 68% sequence identity with the other two viruses for this region. The NTRs of PoTV, BoTV, and ETV were highly conserved, with sequence identities of about 88%. We conclude that PoTV is antigenically and genetically related to but clearly distinct from the bovine and equine representatives of the torovirus genus and should therefore be considered a new member.
In a heterotypic in vitro neutralization assay, >80% of the adult sows in The Netherlands were positive for torovirus antibody; similar observations had been reported for Switzerland (32). Torovirus infections are obviously as common and widespread in pigs as in cattle and horses (16, 31, 32, 34). Piglets are infected shortly after weaning, when protection by maternal antibodies and/or lactogenic immunity has waned. In this study, virus shedding in the feces, as monitored by RT PCR, lasted between 1 and 9 days, suggesting that PoTV predominantly causes acute enteric infections.
The high percentage of seropositive animals and the early occurrence of infection (shortly after weaning, when immunological protection has declined) are indicative of PoTV endemicity. The virus would persist in a herd because of the continuous presence of susceptible piglets and reinfection of partly immune animals. Also, chronically infected carriers may exist, as has been demonstrated for other members of the Nidovirales, such as coronaviruses (6, 11) and arteriviruses (for a review, see reference 21). A more sensitive nested RT PCR targeted to the conserved NTR may allow the identification of long-term shedders among the adult pig population. The physical stability of toroviruses in the environment is as yet unknown; ETV is surprisingly stable at a low pH but is less thermostable than TGEV (18, 30).
Piglets passed normal stools during PoTV shedding and did not show any sign of disease. As our longitudinal study included only a few animals and monitored only one herd, we cannot exclude the possibility that the virus causes disease at a low incidence, in other circumstances, or in combination with other agents. Future research will focus on the epidemiology and pathogenic potential of PoTV.
ACKNOWLEDGMENTS
We thank M. B. Pensaert and K. van Reeth, Laboratory of Virology, Faculty of Veterinary Medicine, University of Ghent, Ghent, Belgium, for providing the sequential sera from piglets, PEDV, and anti-PEDV serum. We also thank W. F. Voorhout for helping with the electron microscopy, A. A. P. M. Herrewegh for assistance with the RT PCR, and the Hargeerds Farm (Markelo, The Netherlands) for permission to take and help in taking samples on the premises.
REFERENCES
- 1.Beards G M, Brown D W G, Green J, Flewett T H. Preliminary characterization of torovirus-like particles of humans: comparison with Berne virus of horses and Breda virus of calves. J Med Virol. 1986;20:67–78. doi: 10.1002/jmv.1890200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, Van Der Noordaa J. A rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown D W G, Beards G M, Flewett T H. Detection of Breda virus antigen and antibody in humans and animals by enzyme immunoassay. J Clin Microbiol. 1987;25:637–640. doi: 10.1128/jcm.25.4.637-640.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 5.Cornelissen L A H M, Wierda C M H, Van Der Meer F J, Herrewegh A A P M, Horzinek M C, Egberink H F, De Groot R J. Hemagglutinin-esterase, a novel structural protein of torovirus. J Virol. 1997;71:5277–5286. doi: 10.1128/jvi.71.7.5277-5286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crouch C F, Bielefeldt Ohrmann H, Watts T C, Babiuk L A. Chronic shedding of bovine enteric coronavirus antigen-antibody complexes by clinically normal cows. J Gen Virol. 1985;66:1489–1500. doi: 10.1099/0022-1317-66-7-1489. [DOI] [PubMed] [Google Scholar]
- 7.Den Boon J A, Snijder E J, Krijnse Locker J, Horzinek M C, Rottier P J M. Another triple-spanning envelope protein among intracellularly budding RNA viruses: the toroviral E protein. Virology. 1991;182:655–663. doi: 10.1016/0042-6822(91)90606-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Vries A A F, Horzinek M C, Rottier P J M, De Groot R J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doane F W, Anderson N. Electron microscopy in diagnostic virology. A practical guide and atlas. Cambridge, England: Cambridge University Press; 1987. [Google Scholar]
- 10.Durham P J K, Hassard L E, Norman G R, Yemen R L. Viruses and virus-like particles detected during examination of feces from calves and piglets with diarrhea. Can Vet J. 1989;30:876–881. [PMC free article] [PubMed] [Google Scholar]
- 11.Herrewegh A A P M, De Groot R J, Cepica A, Egberink H F, Horzinek M C, Rottier P J M. Detection of feline coronavirus RNA in feces, tissues, and body fluids of naturally infected cats by reverse transcriptase PCR. J Clin Microbiol. 1995;33:684–689. doi: 10.1128/jcm.33.3.684-689.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horzinek M C, Ederveen J, Kaeffer B, De Boer D, Weiss M. The peplomers of Berne virus. J Gen Virol. 1986;67:2475–2483. doi: 10.1099/0022-1317-67-11-2475. [DOI] [PubMed] [Google Scholar]
- 13.Houben A, van Reeth K, Pensaert M B. Pattern of infection with the porcine reproductive and respiratory syndrome virus on swine farms in Belgium. Zentralbl Veterinaermed Reihe B. 1995;42:209–215. doi: 10.1111/j.1439-0450.1995.tb00704.x. [DOI] [PubMed] [Google Scholar]
- 14.Koopmans M, Cremers H, Woode G, Horzinek M C. Breda virus (Toroviridae) infection and systematic antibody response in sentinel calves. Am J Vet Res. 1990;51:1443–1448. [PubMed] [Google Scholar]
- 15.Koopmans M, Ederveen J, Woode G N, Horzinek M C. Surface proteins of Breda virus. Am J Vet Res. 1986;47:1896–1900. [PubMed] [Google Scholar]
- 16.Koopmans M, Van Den Boom U, Woode G, Horzinek M C. Seroepidemiology of Breda virus in cattle using ELISA. Vet Microbiol. 1989;19:233–243. doi: 10.1016/0378-1135(89)90069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroneman, A. Unpublished data.
- 18.Laude H. Thermal inactivation studies of a coronavirus, transmissible gastroenteritis virus. J Gen Virol. 1981;56:235–240. doi: 10.1099/0022-1317-56-2-235. [DOI] [PubMed] [Google Scholar]
- 19.Osterhaus A D M E, Horzinek M C, Reynolds D J. Seroepidemiology of feline infectious peritonitis virus infections using transmissible gastroenteritis virus as antigen. Zentralbl Veterinaermed Reihe B. 1977;24:835–841. doi: 10.1111/j.1439-0450.1977.tb00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penrith M L, Gerdes G H. Breda virus-like particles in pigs in South Africa. J S Afr Vet Assoc. 1992;63:102. [PubMed] [Google Scholar]
- 21.Plagemann P G W, Moennig V. Lactate dehydrogenase-elevating virus, equine arteritis virus, and simian hemorrhagic fever virus: a new group of positive-strand RNA viruses. Adv Virus Res. 1992;41:99–192. doi: 10.1016/S0065-3527(08)60036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott A C, Chaplin M J, Stack M J, Lund L J. Porcine torovirus. Vet Rec. 1987;120:583. doi: 10.1136/vr.120.24.583-a. [DOI] [PubMed] [Google Scholar]
- 23.Snijder E J, Den Boon J A, Spaan W J M, Verjans G M G M, Horzinek M C. Identification and primary structure of the gene encoding the Berne virus nucleocapsid protein. J Gen Virol. 1989;70:3363–3370. doi: 10.1099/0022-1317-70-12-3363. [DOI] [PubMed] [Google Scholar]
- 24.Snijder E J, Horzinek M C. The molecular biology of toroviruses. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 219–238. [Google Scholar]
- 25.Snijder E J, Horzinek M C, Spaan W J M. A 3′-coterminal nested set of independently transcribed mRNAs is generated during Berne virus replication. J Virol. 1990;64:331–338. doi: 10.1128/jvi.64.1.331-338.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snijder E J, Den Boon J A, Spaan W J M, Weiss M, Horzinek M C. Primary structure and post-translational processing of the Berne virus peplomer protein. Virology. 1990;178:355–363. doi: 10.1016/0042-6822(90)90332-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snijder E J, Den Boon J A, Horzinek M C, Spaan W J M. Comparison of the genome organization of toro- and coronaviruses: evidence for two non-homologous RNA recombination events during Berne virus evolution. Virology. 1991;180:448–452. doi: 10.1016/0042-6822(91)90056-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vennema H, Godeke G J, Rossen J W, Voorhout W F, Horzinek M C, Opstelten D J, Rottier P J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss M, Horzinek M C. Morphogenesis of Berne virus (proposed family Toroviridae) J Gen Virol. 1986;67:1305–1314. doi: 10.1099/0022-1317-67-7-1305. [DOI] [PubMed] [Google Scholar]
- 30.Weiss M, Horzinek M C. Resistance of Berne virus to physical and chemical treatment. Vet Microbiol. 1986;11:41–49. doi: 10.1016/0378-1135(86)90005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss M, Steck F, Horzinek M C. Purification and partial characterization of a new enveloped RNA virus (Berne virus) J Gen Virol. 1983;64:1849–1858. doi: 10.1099/0022-1317-64-9-1849. [DOI] [PubMed] [Google Scholar]
- 32.Weiss M, Steck F, Kaderli R. Antibodies to Berne virus in horses and other animals. Vet Microbiol. 1984;9:523–531. doi: 10.1016/0378-1135(84)90014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woode G N, Reed D E, Runnels P L, Herrig M A, Hill H T. Studies with an unclassified virus isolated from diarrheic calves. Vet Microbiol. 1982;7:221–240. doi: 10.1016/0378-1135(82)90036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woode G N, Saif L J, Quesada M, Winand N J, Pohlenz J F, Kelso Gourley N. Comparative studies on three isolates of Breda virus of calves. Am J Vet Res. 1985;46:1003–1010. [PubMed] [Google Scholar]
- 35.Woode G N. Breda and Breda-like viruses: diagnosis, pathology and epidemiology. In: Bock G, Whelan J, editors. Novel diarrhea viruses. Ciba Foundation Symposium 128. Chichester, England: John Wiley & Sons; 1987. pp. 175–191. [DOI] [PubMed] [Google Scholar]