Abstract
We investigated the relative importance of binding site occupancy and epitope specificity in antibody neutralization of human immunodeficiency virus (HIV) type 1 (HIV-1). The neutralization of a T-cell-line-adapted HIV-1 isolate (MN) was analyzed with a number of monovalent recombinant Fab fragments (Fabs) and monoclonal antibodies with a range of specificities covering all confirmed gp120-specific neutralization epitopes. Binding of Fabs to recombinant monomeric gp120 was determined by surface plasmon resonance, and binding of Fabs and whole antibodies to functional oligomeric gp120 was determined by indirect immunofluorescence and flow cytometry on HIV-infected cells. An excellent correlation between neutralization and oligomeric gp120 binding was observed, and a lack of correlation with monomeric gp120 binding was confirmed. A similar degree of correlation was observed between oligomeric gp120 binding and neutralization with a T-cell-line-adapted HIV-1 molecular clone (Hx10). The ratios of oligomer binding/neutralization titer fell, in general, within a relatively narrow range for antibodies to different neutralization epitopes. These results suggest that the occupancy of binding sites on HIV-1 virions is the major factor in determining neutralization, irrespective of epitope specificity. Models to account for these observations are proposed.
Antibody neutralization of viruses in vitro is an important phenomenon, since there is generally a good correlation between in vitro neutralization and in vivo antiviral efficacy (13, 33). The plausible mechanisms of neutralization of enveloped viruses have been debated from a number of standpoints. A series of studies have proposed the importance of the binding of a few antibody molecules to a virion to achieve neutralization (few-hit theory) (13, 14, 24). Elsewhere it has been argued that neutralization may result when the number of unoccupied sites on a virion falls below a critical minimum that is required for infectivity (occupancy model) (12, 20, 32). Another consideration is the importance of epitope specificity. In simple terms, does the binding of antibodies to distinct epitopes or different functional regions of a viral protein engender more or less neutralization, and thus can equal amounts of antibody bound to different epitopes on the virion produce different degrees of neutralization? A potential consequence of the influence of epitope specificity on neutralization is that different antibodies may inhibit viral infection of a target cell at different stages of the virus life cycle. In this respect, it has been argued that inhibition of attachment of virus to the target cell is a relatively rare mechanism of antibody neutralization and that processes following attachment, such as virus-cell membrane fusion, are more common targets (1, 13, 14, 22). Steric interference and physical constraints may also influence the neutralizing ability of an antibody; the size (Fab fragment versus immunoglobulin G [IgG] or IgM), orientation of attachment, and valency of attachment are all epitope-specific factors to be considered (13, 14). In the present study, we sought to investigate the importance of site occupancy and epitope specificity in the neutralization of human immunodeficiency virus (HIV) type 1 (HIV-1) by antibody.
Antibody neutralization of HIV-1 by antisera and monoclonal antibodies (MAbs) is well documented (reviewed recently in references 8, 27, 37, and 43). The neutralizing activity is directed overwhelmingly at the surface (gp120) envelope glycoprotein (8, 27, 37), although neutralization also can be mediated by transmembrane glycoprotein (gp41)-specific components (30, 31). The neutralizing antibody response to T-cell-line-adapted (TCLA) HIV-1 gp120 has been examined by the preparation and characterization of MAbs of diverse origin, allowing the identification of a number of neutralization epitopes on the envelope glycoproteins. The accessibility of such epitopes is considerably greater on TCLA strains than on primary isolates of HIV-1 (5, 16, 26, 27, 41). On TCLA viruses, neutralizing antibodies to gp120 have been described to react with the hypervariable loops V1/V2 and V3; a discontinuous epitope involving residues in the base of the V3 and V4 loops (2G12 epitope), the CD4 binding site (CD4bs), and the related C4 region; an epitope involving the CD4bs and residues in the V2 loop (b12 epitope); an epitope induced by the binding of CD4bs-specific antibodies; and an epitope partially induced by CD4 binding (reviewed in references 8 and 37). Only two gp120-specific neutralization epitopes have been well characterized as being present on a majority of primary isolates (b12 and 2G12 epitopes).
Primary isolates are clearly more relevant than TCLA strains of HIV-1 to human infection. However, the paucity of neutralizing antibodies to primary isolates, together with technical difficulties in measuring the binding of antibodies to functional primary isolate envelope glycoproteins, precluded their use in this study. As a result, we carried out analyses on TCLA viruses; the general principles established are, however, also likely to apply to primary isolates. The strategy adopted was to compare the binding of a number of antibodies to different gp120 epitopes presented in the form of functional oligomeric gp120 on infected cells with their capacity to neutralize the corresponding virus. A concentration of MAb yielding half-maximal binding (K50) and a neutralization titer of similar magnitude (ID50) would be consistent with antibody occupancy of virion binding sites playing a major role in HIV-1 neutralization. Similar K50/ID50 ratios for different epitopes would suggest that neutralization is broadly independent of the epitope recognized. Alternatively, a highly divergent ratio would imply a strong epitope-specific dependency.
Previous studies suggested a relationship between antibody affinity for oligomeric gp120 and neutralization (16, 39, 42). Here we carry out a quantitative and detailed analysis of the neutralization of two TCLA viruses by a range of monovalent recombinant Fab fragments (Fabs) and MAbs specific for all confirmed TCLA neutralization epitopes on gp120. We report that overall, neutralization correlates with the amount of antibody estimated to be bound to the virus. Although subtle epitope-specific effects may be present, there is no convincing evidence that the binding of antibody to any distinct neutralization epitope cluster yields a disproportionate effect on the loss of infectivity. These data are consistent with the occupancy model and demonstrate that epitope-specific and functional domain-specific effects on neutralization are, at most, subtle.
MATERIALS AND METHODS
Antibodies.
The recombinant Fabs b3, b6, b11, b12, b13, b14, DO8i, DA48, and 3B3 (specific for the CD4bs), DO142-10 and Loop 2 (specific for the V3 loop), and L17 (specific for the V2 loop) were obtained by screening phage display libraries for gp120 binding activity and HIV-1 neutralization as previously described (3, 7, 39, 44). Fabs b3, b6, b11, b12, b13, b14, DA48, and DO8i have been epitope mapped to residues which are considered to contribute to the CD4 binding region of gp120 or surrounding residues, and all have different fine specificities (6a, 39). Fab 3B3, from C. Barbas, was derived from Fab b12 by random mutagenesis in HCDR1 and HCDR3 and was selected for its increased affinity for soluble gp120 (4). IgG1 b12 and IgG1 Loop 2 were obtained by engineering the respective Fabs into IgG1 molecules (9). The anti-V3 loop Fabs Loop 2 and DO142-10 have been mapped and described elsewhere (33, 44). Fab L17 has been mapped to the V2 loop of gp120 (15). The human MAbs 19b (29) and F91 and 48d (47, 55) were from J. Robinson, Department of Pediatrics, University of Connecticut, Farmington; 2G12 (6, 50) was from H. Katinger, Institute of Applied Microbiology, Vienna, Austria; and 447-52D (11, 19) was from Cellular Products Inc., Buffalo, N.Y.
Cell culture and virus infection.
H9 cells (from R. Gallo, National Institutes of Health, Bethesda, Md.) were cultured in growth medium (GM; RPMI medium with 10% fetal calf serum [FCS]) in 5% CO2. Infection of H9 cells with supernatant containing infectious virus of the nonclonal MN isolate of HIV-1 was done as follows. One million cells in 1 ml of medium were exposed to 104 50% tissue culture infective doses of virus-containing supernatant for 2 h at 37°C. After being washed, the cells were resuspended in GM and cultured for 8 to 10 days. At this time, 100% of the cells expressed large amounts of viral envelope glycoprotein, as determined by indirect immunofluorescence staining with anti-gp120 MAbs, but no CD4, as detected with MAbs to the first and fourth domains of CD4, as previously described for the Hx10 clone of HIV-1 (42). The majority of the viral gp120 observed at the infected cell surface was present on mature virus particles associated with the cell membrane, as determined by immunoelectron microscopic analysis (18a).
Analysis of antibody binding by flow cytometry.
H9 cells infected with MN virus as described above were washed twice in GM and resuspended at a concentration of 2 × 106 cells per ml. Fifty microliters of MAb previously diluted in phosphate-buffered saline (PBS)–1% FCS (wash buffer [WB]) was added to 50 μl of cell suspension in a U-bottom, 96-well microtiter plate, and the mixture was incubated with agitation at 37°C for 2 h. The cells were washed three times in WB and then fixed overnight at 4°C in WB containing 1% formaldehyde. After two further washes in WB, a 1:100 dilution of anti-human phycoerythrin was added (Immunotech Inc., Marseille, France). The cells were washed twice as before and then analyzed by flow cytometry by use of a FACScan (Becton Dickinson, San Jose, Calif.) with Consort 30 software.
Measurement of antibody affinity for gp120 by surface plasmon resonance.
Association and dissociation constants were deduced from the kinetic rate constants kon and koff, which were determined by surface plasmon resonance with a BIAcore 2000. Coupling of gp120 to the sensor chip and antibody binding to and elution from the immobilized antigen were achieved as described previously (39). HIV-1MN gp120 was obtained from Harvey Holmes (Medical Research Council AIDS Reagent Project, Potters Bar, United Kingdom).
HIV neutralization assay.
To analyze MAbs for neutralization activity, we used an assay based on the infection of HeLa cells expressing human CD4 and the HIV long terminal repeat fused to lacZ (10), which we have described previously (36). Infection of these cells with HIV-1 leads to the production of the viral Tat protein, which transactivates the transfected LTR and activates the lacZ gene; detergent lysis of the cells is followed by the addition of the soluble substrate and then by a spectrophotometric readout in an enzyme-linked immunosorbent assay format. The advantages of this assay system are that infection of the cells can be detected within 24 to 36 h (approximately one cycle of replication) and that a reduction in infectivity by pretreatment of virus with antibody can be quantitatively assessed rapidly. Briefly, 20 μl of a previously titrated suspension of HIV was preincubated with dilutions of MAbs in Dubecco’s minimal essential medium (DMEM)–5% FCS for 2 h at 37°C in a total volume of 40 μl before the addition of 10 μl of HeLa cells at a concentration of 5 × 106 cells/ml. After 1 h of incubation at 37°C with agitation, the cells were washed in PBS–10 mM EDTA and incubated with EDTA-trypsin (Gibco/BRL) for 15 min at 37°C. After being washed in DMEM–5% FCS, the cells were cultured for a further 24 to 36 h in flat-bottom 96-well microtiter plates. The medium was aspirated, and the cells were lysed in a solution of PBS containing 0.5% Nonidet P-40. An equal volume of a solution containing 16 mM chlorophenol red-β-d-galactopyranoside (Boehringer Mannheim, Meylan, France) was added; after 30 min of incubation with the substrate at 37°C, the optical density (OD) was read at 550 nm. The percentage of neutralization was calculated with the formula 100 − {[(t − c)/(m − c)] × 100}, where t represents the OD signal for the test sample in the presence of a neutralizing MAb, c represents the background signal in the absence of virus, and m represents the maximum signal obtained with virus but no inhibitor. Values obtained from the measurements of antibody binding and neutralization were evaluated for correlation by linear regression analysis of the log10 values for ID50, K50, Kd, kon, and koff. These and the Student t test and Mann-Whitney U tests were performed with the Statview program (Abacus, Berkeley, Calif.).
RESULTS
Binding affinity of Fabs for monomeric gp120 does not correlate with neutralizing ability.
There is conflicting evidence for the existence of a correlation between antibody affinity for recombinant monomeric gp120 and neutralization, in that some studies find a correlation (52), whereas others do not (16, 26, 39, 42). We wished to formally confirm or refute the notion of such a correlation by using a broad panel of neutralizing antibodies before proceeding to oligomeric gp120 binding and neutralization studies. We carried out a thorough analysis using Fabs and MAbs representing all confirmed TCLA gp120 neutralization epitopes and recombinant soluble gp120 derived from the MN strain of HIV-1. The approach chosen was surface plasmon resonance, as this allows a precise, real-time determination of the rates of association and dissociation. The Kd values obtained for the antibodies are shown in Table 1; they are similar to those previously described for strain LAI gp120 (39) and are within the range of 1.9 × 10−9 to 2.5 × 10−8 M, a maximum variation of about 10-fold. A comparison of these data with the ID50 values obtained for neutralization (see below) revealed no obvious relationship, in concordance with several earlier studies, and a statistical analysis showed no significant correlation when Fabs of all specificities were considered together or divided into two functional groups (see Table 3).
TABLE 1.
Fab binding and neutralization characteristics
| Specificity | Fab | Binding to gp120 monomera
|
Binding to gp120 oligomerb (K50) (nM) | Neutralizationc (ID50) (nM) | Ratiod (K50/ID50) | ||
|---|---|---|---|---|---|---|---|
| kon (M−1 s−1) | koff (s−1) | Kd (nM) | |||||
| CD4bs | b3 | 6.5 × 104 | 4.1 × 10−4 | 6.3 | 135 | 80.0 | 1.7 |
| b6 | NDe | ND | ND | 38.0 | 21.7 | 1.8 | |
| b11 | 5.2 × 104 | 2.2 × 10−4 | 4.2 | 18.7 | 40.0 | 0.5 | |
| b12 | 1.2 × 105 | 4.1 × 10−4 | 3.4 | 1.70 | 4.30 | 0.4 | |
| b13 | 4.6 × 104 | 4.6 × 10−4 | 10 | >200 | >200 | ND | |
| b14 | 5.1 × 104 | 3.4 × 10−4 | 6.7 | 60.0 | 166.7 | 0.4 | |
| DO8i | 1.5 × 104 | 5.2 × 10−5 | 3.5 | 46.7 | 18.3 | 2.5 | |
| DA48 | 1.3 × 104 | 2.1 × 10−4 | 16 | 118.0 | 61.7 | 1.9 | |
| 3B3 | 9.8 × 104 | 2.2 × 10−4 | 2.2 | 0.40 | 0.10 | 4.0 | |
| V3 loop | DO142-10 | 1.6 × 104 | 1.8 × 10−4 | 11 | 7.00 | 1.10 | 6.4 |
| Loop 2 | 1.2 × 104 | 2.3 × 10−5 | 1.9 | 13.0 | 3.70 | 3.5 | |
| V2 loop | L17 | 2.4 × 104 | 5.9 × 10−4 | 25 | 19.7 | 20.0 | 1.0 |
MN-derived soluble gp120 was attached to the solid phase, and real-time kinetic measurements of Fab binding were made by surface plasmon resonance.
Half-maximal binding of Fabs to oligomeric gp120 was determined by titration of Fabs on MN-infected cells and detection by indirect immunofluorescence and flow cytometric analysis.
Half-maximal neutralization was determined by preincubating serial dilutions of Fabs with a virus stock before infecting HeLa CD4+ LTR-LacZ cells.
The ratio of half-maximal Fab binding to half-maximal neutralization was determined for each Fab.
ND, not done.
TABLE 3.
Analysis of correlation between antibody binding and neutralization
| Comparison | ra | Approximate Pb |
|---|---|---|
| All Fabs | ||
| Monomer binding vs neutralization | 0.41 | 0.24 |
| Monomer kon vs neutralization | −0.15 | 0.68 |
| Monomer koff vs neutralization | 0.22 | 0.54 |
| Oligomer binding vs neutralization | 0.9 | 0.00020 |
| Monomer vs oligomer binding | 0.46 | 0.19 |
| CD4bs Fabs | ||
| Monomer binding vs neutralization | 0.71 | 0.074 |
| Oligomer binding vs neutralization | 0.92 | 0.0013 |
| Monomer vs oligomer binding | 0.76 | 0.046 |
| Specific MAbs | ||
| Oligomer binding vs neutralization (MN) | 0.98 | 0.00080 |
| Oligomer binding vs neutralization (Hx10) | 0.82 | 0.0020 |
| Oligomer binding vs neutralization (MN + Hx10) | 0.84 | 0.00010 |
| CD4bs oligomer binding vs neutralization (MN + Hx10) | 1.0 | 0.00010 |
| All MAbs (oligomer binding vs neutralization) | 0.77 | 0.00010 |
Calculated by simple linear regression of log10 ID50 plotted as a function of log10 kon, koff, Kd, or K50.
Calculated with F and t tests.
Binding of Fabs and MAbs to functional oligomeric gp120.
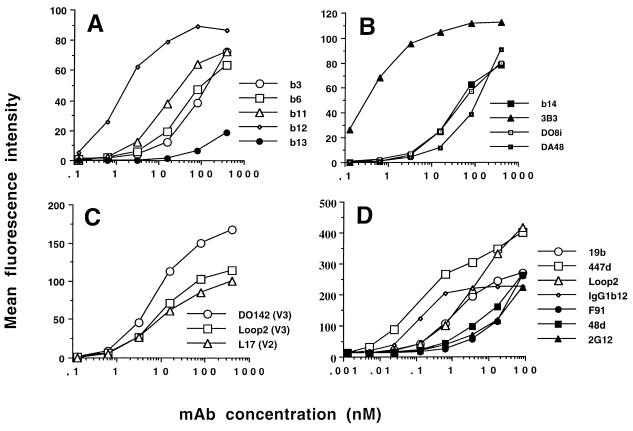
Quantitative ligand binding studies can be carried out by indirect fluorescence labeling of the surface of HIV-1-infected cells followed by flow cytometric analysis. This system was used to measure the binding of the panel of Fabs and MAbs to HIV-1MN-infected H9 cells. Figure 1 shows the binding curves obtained in one of five representative experiments. The antibodies are separated into three groups: CD4bs-specific Fabs, variable loop-specific (V2 and V3) Fabs, and IgG MAbs. Inspection of the binding curves for the CD4bs Fabs reveals large differences in their relative affinities. 3B3 stands out as the CD4bs-specific Fab requiring the lowest concentration to achieve 50% binding (0.4 nM), whereas b13 requires the highest (>200 nM), a difference of >500-fold between the two antibodies. Interestingly, the difference in Kd values for these two Fabs on recombinant monomeric gp120 was <5-fold. The rank order of affinity of Fab binding to recombinant monomeric gp120 (Loop 2 > 3B3 > b12 = DO8i > b11 > b3 > b14 > b13 > DO142-10 > DA48 > L17) was markedly different from that obtained for binding to the mature oligomeric form (3B3 > b12 > DO142-10 > Loop 2 > b11 > L17 > b6 > DO8i > b14 > DA48 > b3 > b13), implying that there is no general relationship between Fab binding to recombinant monomeric gp120 and that to functional oligomeric gp120. This suggestion was confirmed by the general lack of correlation in affinity measurements for the two forms of gp120 (see Table 3). There was, however, a weak correlation between binding to monomeric gp120 and that to oligomeric gp120 for the CD4bs-specific Fabs when they were considered as a separate group.
FIG. 1.
Fab (A, B, and C) and MAb (D) binding to HIV-1MN-infected H9 cells. Serial dilutions of Fabs and MAbs were incubated for 2 h at 37°C with MN-infected H9 cells before the cells were washed and fixed in 1% formaldehyde overnight. The cells were then stained with an anti-human or anti-mouse IgG-phycoerythrin conjugate and analyzed by flow cytometry. Each point represents 10,000 accumulated events, and the experiment shown is representative of five separate experiments.
Neutralizing IgG molecules to four distinct epitopes (CD4bs, V3 loop, CD4i, and 2G12) were analyzed for binding to virion-associated MN gp120, and binding curves for representatives of these epitope clusters are shown in Fig. 1D. Again, a broad spectrum of affinities was obtained, with the greatest difference being observed between IgG1 b12 (K50, 0.1 nM) and 2G12 (K50, 150 nM), a difference of 1,500-fold (Table 2). A comparison of the binding affinities for the two Fab fragments for which there were corresponding IgG molecules (b12 and Loop 2) revealed that conversion to a bivalent ligand increased b12 avidity by 17-fold and Loop 2 avidity by about 2-fold, within the expected range (33). The difference in the increases between the two ligands may represent the ability of one MAb (IgG1 b12) to bind bivalently, whereas IgG1 Loop 2 may bind solely with a single arm, resulting in a binding constant similar to that obtained for the Fab fragment.
TABLE 2.
IgG binding and neutralization characteristics
| Virus | Specificity | Antibody | K50 for oligomeric gp120 (nM)a | Neutralization (ID50) (nM)b | Ratio (K50/ID50)c |
|---|---|---|---|---|---|
| MN | CD4bs | IgG1 b12 | 0.10 | 0.010 | 10 |
| F91 | 65 | 68 | 1.0 | ||
| V3 loop | Loop 2 | 4.1 | 1.6 | 2.5 | |
| 19b | 3.0 | 2.5 | 1.2 | ||
| 447-52D | 0.60 | 0.050 | 12 | ||
| CD4i | 48d | 8.0 | 1.6 | 5.0 | |
| Other | 2G12 | 150 | >200 | <0.80 | |
| Hx10d | CD4bs | 15e | 20 | 4.8 | 4.2 |
| 21h | 20 | 6.0 | 3.3 | ||
| F91 | 8.0 | 2.4 | 3.3 | ||
| IgG1 b12 | 0.80 | 0.070 | 11 | ||
| C4 | G3-299 | >50e | 4.1 | >12 | |
| G3-519 | >50 | 98 | >0.50 | ||
| G3-536 | >50 | 270 | >0.18 | ||
| G3-508 | >50 | 190 | >0.27 | ||
| V3 | 110.5 | 0.050 | 0.20 | 0.25 | |
| 9284 | 0.12 | 0.93 | 0.13 | ||
| BAT123 | 0.080 | 0.13 | 0.62 | ||
| 110.I | 0.50 | 1.3 | 0.38 | ||
| 8/38c | 67 | 67 | 1.0 | ||
| V2 | G3-136 | >20 | 7.7 | >2.6 | |
| G3-4 | >70 | 44.4 | >1.6 | ||
| BAT085 | >70 | 133.4 | >0.52 | ||
| CD4i | 48d | 5.0 | 1.3 | 3.8 | |
| Other | 2G12 | 2.5 | 1.3 | 1.9 |
Half-maximal binding of MAbs to oligomeric gp120 was determined as described in Table 1, footnote b.
Half-maximal neutralization was determined as described in Table 1, footnote c.
The ratio of the concentration yielding 50% MAb binding to 50% neutralizing concentration was determined for each MAb.
Most of this data set is taken from a previous study (42). Data for MAbs F91, IgG1 b12, and 2G12 were obtained in the present study.
Saturation staining was not achieved under the experimental conditions used.
A data set obtained mostly in a previous study (42) with the HIV-1 molecular clone Hx10 was also included in Table 2 to allow a comparison of neutralization and MAb affinity for two different viruses. For many of the MAbs, saturation staining was not achieved, even at the highest concentration tested; increasing the incubation time did not lead to saturation (results not shown), suggesting a very low affinity. For these MAbs, we estimated the minimum 50% binding values; the true K50 values therefore will be higher. As with the MN-infected cells, MAb binding to Hx10-infected cells varied substantially: MAb 110.5 had the highest affinity (K50, 0.05 nM), whereas a number of MAbs to the C4 and V2 regions had a K50 of >50 nM, representing a difference of >1,000-fold.
Neutralization by Fabs and MAbs.
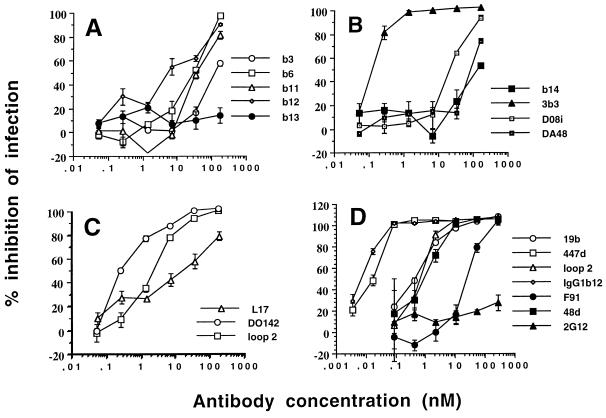
In order to evaluate the neutralization activity of the Fab and IgG molecules analyzed in this study, we used a previously described system based on the HIV-1 Tat-induced transactivation of the HIV-1 long terminal repeat fused to the lacZ reporter gene in human CD4+ HeLa cells. Neutralization curves for a representative experiment are shown for Fabs in Fig. 2A to C and for MAbs in Fig. 2D. ID50s for the Fab and IgG molecules showed variation over a range greater than 3 orders of magnitude (Tables 1 and 2), a range similar to that observed with Hx10 virus in a previous study (42).
FIG. 2.
Neutralization of MN virus produced in H9 cells by Fabs (A, B, and C) and MAbs (D). A virus-containing supernatant was incubated with serial dilutions of Fabs or MAbs for 2 h at 37°C before addition to HeLa CD4+ LTR-LacZ cells. After incubation for 2 h at 37°C, the cells were washed, trypsinized, and cultured for 24 h. The cells were then lysed, and the OD at 405 nm was measured after addition of the substrate. The OD values obtained were expressed as the percent inhibition of infection compared to that in the presence of virus and absence of inhibitory antibody (positive control) or absence of virus (negative control).
Analysis of the correlation between Fab and IgG binding and neutralization.
Visual inspection of the values suggested a relationship between neutralization and Fab binding to functional oligomeric gp120. In order to quantify the relationship, we analyzed the parameters by linear regression and calculated the strength and significance of the correlations. Table 3 summarizes the data obtained from scatter plots of neutralization versus antibody binding to oligomeric and monomeric gp120.
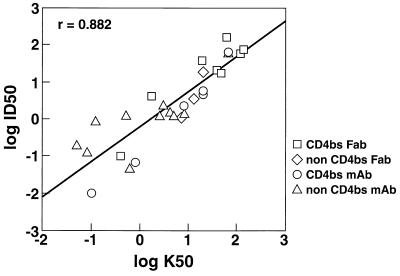
A strong (r = 0.9) and highly significant relationship was observed between log K50 for functional oligomeric gp120 and log ID50, but no significant correlation was found between log K50 for monomeric gp120 and log ID50 (r = 0.41). Likewise, no correlation was demonstrated between kon and koff rates for binding to monomeric gp120 and neutralization or between affinities for Fab binding to monomeric gp120 and oligomeric gp120. When CD4bs-specific Fabs were considered in isolation, there was an excellent correlation between oligomeric gp120 binding and neutralization (r = 0.92). There was a weak correlation between monomeric gp120 binding by CD4bs-specific Fabs and neutralization; this result may have arisen from the weak correlation between binding to monomeric gp120 and binding to oligomeric gp120 observed for this set of Fabs (Table 3). As observed for the Fabs, MAb binding to functional oligomeric gp120 from both the MN and Hx10 viruses correlated strongly with neutralization (Table 3). When the binding of CD4bs-specific MAbs to both Hx10 and MN was compared with neutralization, an excellent correlation was observed (r = 1.0). A less strong, but nevertheless highly significant, correlation was observed when the binding of all antibodies to both viruses was compared with neutralization (r = 0.88) (Fig. 3).
FIG. 3.
Neutralization as a function of MAb binding to oligomeric envelope glycoproteins. Log10 pooled ID50 and K50 values obtained from both viruses for all Fab and IgG molecules analyzed in this study were plotted on the y and x axes, respectively. Simple linear regression analysis showed the correlation to be strong (r = 0.882) and highly significant (P < 0.0001).
Importance of virion site occupancy and epitope specificity in antibody neutralization.
We estimated the antibody neutralization efficiency for a given epitope by calculating the K50/ID50 ratio for oligomeric gp120 (Tables 1 and 2). Significantly different ratios between antibodies against distinct epitopes would indicate that epitope-specific differences exist in neutralization efficiency. Neutralization of the virus would then occur at different levels of occupancy for different epitopes or epitope clusters. Despite the large ranges in K50 and ID50, most of the ratios in Tables 1 and 2 fell within a relatively narrow range, with a few exceptions. For the CD4bs-specific Fabs, most values were close to 1, as was that for the V2 loop-specific Fab. The V3 loop-specific Fabs had slightly higher values, suggesting a potentially greater neutralization efficiency. The K50/ID50 values for the IgG molecules for both MN and Hx10 viruses were more variable than those for the Fab molecules. For most the ratio was relatively close to 1, as for the Fabs. However, there were notable outliers, namely, IgG1 b12, 447-52D, and 48d for MN and IgG1 b12 and G3-299 for Hx10, where the values were closer to 10. It is unclear why a variation of such magnitude exists for these different antibodies, in particular because the values for K50 and ID50 used to calculate the ratios correlate with high significance (Table 3). It is unlikely that this variation represents differential antibody interference with different functional domains of gp120, since the antibodies that displayed the greatest divergence in neutralization efficiency came from diverse epitope clusters. Thus, IgG1 b12, 447-52D, and 48d represent three distinct functional regions of gp120: the CD4bs, the V3 loop, and the CD4i epitope. Other antibodies to these epitopes had values close to 1. That this variation is not due to epitope-specific effects is further demonstrated by the difference in the K50/ID50 ratios between Fab b12 (0.4) and Fab 3B3 (4.0). Fab 3B3 is an affinity-improved, closely related variant of Fab b12 and, by definition, binds to an almost identical epitope. The K50/ID50 values therefore would be expected to be very similar, and the observed difference is an indication of the range of data scatter.
In order to confirm that antibodies to one functional domain were not more potent neutralizers than those to other domains, we carried out a statistical analysis in which we compared the K50/ID50 values for CD4bs-specific antibodies with those for non-CD4bs-specific antibodies (results not shown). No significant difference was seen with either the unpaired t test or the Mann-Whitney U test between CD4bs-specific ligands (Fab or IgG molecules or both) and non-CD4bs-specific ligands (V2 or V3 loop, CD4i epitope, and 2G12 epitope). In summary, analysis of the individual data as well as statistical analysis provided no evidence for strong epitope-specific effects on the efficiency of neutralization.
DISCUSSION
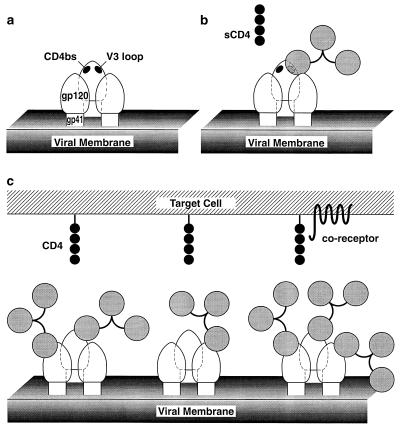
In this study, we show that antibody neutralization of two TCLA HIV-1 isolates, one a biological isolate and the other a molecular clone, is statistically highly correlated with binding affinity for mature oligomeric gp120 expressed on the surface of infected cells. In contrast, there is no significant correlation between neutralization by the panel of antibodies and binding to recombinant monomeric gp120. Overall, the most straightforward interpretation of our data is an occupancy model (Fig. 4) (see below), whereby neutralization is determined primarily by the fraction of antibody sites occupied on virions irrespective of epitope recognized.
FIG. 4.
Model for proposed interactions between the virion envelope glycoproteins and neutralizing IgG and CD4. These molecules are depicted roughly to scale based on their respective molecular weights and their known (IgG and soluble CD4 [sCD4]) or implied (gp120 and gp41) structures. (A) Envelope glycoprotein trimer on the viral surface. (B) Glycoprotein trimer with an IgG antibody molecule bound to the V3 loop on gp120. Soluble CD4 binding to the CD4bs is still possible. (C) Glycoprotein trimer with various numbers of antibody molecules associated; binding to membrane CD4 and/or coreceptor molecules is sterically inhibited.
In a recent study (51), we found that antibodies to all the neutralization epitopes on gp120, including the CD4bs, V2 and V3 loop, and 2G12 epitopes, neutralize HIV-1MN and Hx10 at least in part by blocking attachment of virus to cells. Indeed, the only antibody that did not interfere with HIV-cell attachment was the anti-gp41 MAb 2F5, which interacts with an epitope close to the transmembrane domain of the molecule. Taken together, the data of Ugolini et al. (51) and the present study suggest two plausible mechanisms for HIV-1 neutralization. The first invokes coating of the viral surface, which obstructs the close approach of virus and target cell membranes, as the principal mechanism. Individual epitopes play a minor role in this model because of the size of the antibody molecule relative to the proximity of the neutralization epitopes on gp120 to the CD4 binding region (Fig. 4). In this model, the high degree of glycosylation (about 50%) of gp120 reduces antibody accessibility to the protein surface to a relatively low level. The available protein surface is further reduced on the virion surface by the heterodimeric interaction of gp120 with gp41 and the homotrimerization of gp41 and gp120. Neutralization epitopes, such as the CD4bs, V2 and V3 loop, and 2G12 epitopes, are known to be proximal and are probably located within a confined region termed the “neutralizing face” (28). However, there is sufficient separation to ensure that the binding of IgG to an epitope such as the V3 loop does not inhibit the binding of either soluble CD4 or IgG to oligomeric gp120 or vice versa (Fig. 4b). For attachment of virus to the target cell to occur, it is presumed that multiple contacts in a localized area must be established. Unlike the binding of soluble CD4, this process may be readily inhibited by antibodies to epitopes other than the CD4bs, since the binding of an array of CD4 molecules anchored to the membrane has far more stringent geometric constraints than does the binding of individual soluble CD4 molecules (Fig. 4c). An antibody bound to gp41 would probably project less from the surface of the virion than an antibody bound to gp120, potentially explaining the reason why MAb 2F5 is unable to interfere with HIV-cell attachment. Fabs would provide a smaller steric interference with HIV-cell binding, but since two Fab molecules would be expected to bind in place of one IgG molecule, the overall effect on virus-cell binding would probably be similar. The K50/ID50 ratios for MAbs and Fabs are 3.6 ± 1.8 and 2.2 ± 3.9, respectively (t test, P = 0.25; Mann-Whitney U test, P = 0.53), indicating that there is no significant difference in the neutralization efficiencies of these two sets of ligands. As discussed above, minor variations in the ability of individual antibodies to interfere with virus-cell binding may come from the orientation of the antibody molecule with respect to the gp120 oligomer or from cross-linking of epitopes by bivalent binding to two gp120 molecules. Thus, in this model, the important factor in neutralization is the fraction of virion binding sites occupied; epitope-specific and functional domain-specific effects are relegated to a secondary role.
The second, related mechanism is based on the idea that two sites on gp120 are thought to interact with the target cell: the CD4bs and a site for interaction with a chemokine receptor. In this model, efficient virus-cell binding can be achieved only by the interaction of HIV-1 gp120 with both CD4 and the appropriate coreceptor. Thus, neutralizing antibodies that do not bind to the CD4bs bind to a site overlapping the chemokine receptor binding site and thereby also block viral attachment. Evidence to support such a model comes from recent studies in which neutralizing MAbs to regions of gp120 other than the CD4bs interfered with a gp120-CCR5 interaction (49, 54). Similar results have been obtained in our laboratory: neutralizing MAbs prevented the interaction of TCLA soluble gp120 from strains IIIB and MN with CXCR4 (24a). Further experiments designed to establish the correct model are under way.
Despite the strong correlation between antibody affinity for oligomeric gp120 and neutralization found here, there were clear differences in the neutralization efficiencies of a small number of antibodies; however, these do not change the major conclusion of this paper. It seems unlikely that this finding represents the result of ligand binding to more or less functionally sensitive domains on gp120, since these antibodies belong to different epitope clusters. Possible explanations for the outliers are experimental error in K50/ID50 ratios for antibodies with low ID50 values or secondary effects due to antibody-induced shedding (36), epitope cross-linking (this may explain why the K50/ID50 values for the Fab molecules were within a narrower range than those for the IgG molecules), and/or minor orientation differences for antibodies binding to a given epitope.
Our study confirms unequivocally previous observations which showed that antibody binding to functional oligomeric gp41-gp120 complexes correlates with neutralization (16, 39, 42). However, there are a few reports that indicate that some antibodies may bind relatively well to virion-associated oligomeric gp120 without neutralizing the virus. An anti-V2 MAb (62c) bound to Hx10-infected H9 cells but was unable to neutralize this virus even at relatively high concentrations (45). Likewise, Stamatatos and colleagues (46) found that the binding of another anti-V2 MAb (G3.4) to primary isolate virion gp120 did not correlate with neutralizing activity. In both cases, however, the apparent affinity constant could not be determined accurately. In the first study, binding saturation of MAb 62c was not reached (45). In the second study, virion gp120-antibody complexes were measured in an enzyme-linked immunosorbent assay only after oligomeric gp120 was disrupted into monomeric gp120 by detergent (46). The severe limitations of such a format for performing affinity measurements have been eloquently discussed by Fouts et al. for a very similar assay (16).
Synergy in neutralization has been described between MAbs recognizing the CD4bs and the V2 (53) or V3 (21, 23, 25, 38, 48, 53) loop. This effect, however, is rather weak. Moreover, the idea that the binding of a MAb to one functional domain of gp120 increases the affinity of another MAb for another domain and thereby causes synergistic neutralization is fully consistent with the occupancy model.
Passive transfer studies with an animal model showed that a potent neutralizing antibody can protect against challenge with primary isolates of HIV-1 and implied that a vaccine that induced such an antibody could be successful in preventing transmission (18, 35). The finding presented in the present study, that antibody affinity is of primary importance in HIV neutralization, may have practical consequences for vaccination. It suggests that antibodies to all conserved and well-exposed epitopes on the mature envelope may be equally effective in virus neutralization and that it may therefore be unnecessary to target multiple epitopes. We suggest that vaccine design efforts should focus on increasing the immunogenicity of the native oligomeric (mature) envelope for presentation to the immune system irrespective of the epitope involved. This suggestion is based on the assumption that neutralization is an indicator for protection. In the case of HIV-1, we and others have indeed shown an excellent correlation among antibody affinity, neutralization, and protection against HIV-1 challenge of hu-PBL-SCID mice (17, 18, 34, 40). An antibody against the V3 loop, for example, protected the mice against a TCLA virus which was neutralized, but this antibody was ineffective against primary viruses which were not (17). We further found a good association between protective doses of a potent neutralizing antibody and the neutralization sensitivities of both TCLA and primary viruses (34, 35). Bachmann et al. recently suggested a breakdown in the correlation between protection against vesicular stomatitis virus and in vitro neutralization for a subset of high-affinity antibodies against the vesicular stomatitis virus surface glycoprotein (2). While many studies have shown a good correlation between in vitro neutralization and protection, this observation is not without precedence, and such disparities have also been described for other viruses (13, 33). However, for HIV-1 the correlation appears to hold, and eliciting neutralizing antibodies should be a major goal of HIV-1 vaccine development.
ACKNOWLEDGMENTS
We thank C. F. Barbas III for the kind gift of Fab 3B3. We are grateful to J. Robinson and H. Katinger for contributing antibodies to this study.
This study was supported by the Centre National de la Recherche Scientifique, the Institute National de la Santé et la Recherche Médicale, the Agence Nationale de Recherches sur le SIDA, the Fondation pour la Recherche Médicale (SIDACTION), the European Shared-Cost Action “Antibody-medicated enhancement and neutralization of lentivirus infections: role in immune pathogenesis and vaccine development,” and NIH grants AI33292 (to D.R.B.) and AI40377 (to P.W.H.I.P.). P.W.H.I.P. acknowledges a scholarship award from the Pediatric AIDS Foundation (77290-20-PF). P.J.K. was supported by an MRC fellowship.
REFERENCES
- 1.Armstrong S J, McInerney T L, McLain L, Wahren B, Hinkula J, Levi M, Dimmock N J. Two neutralizing anti-V3 monoclonal antibodies act by affecting different functions of human immunodeficiency virus type 1. J Gen Virol. 1996;77:2931–2941. doi: 10.1099/0022-1317-77-12-2931. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann M F, Kalinke U, Althage A, Freer G, Burkhart C, Roost H-P, Aguet M, Hengartner H, Zinkernagel R M. The role of antibody concentration and avidity in antiviral protection. Science. 1997;276:2024–2027. doi: 10.1126/science.276.5321.2024. [DOI] [PubMed] [Google Scholar]
- 3.Barbas C F, Collet T A, Amberg W, Roben P, Binley J M, Hoekstra D, Cababa D, Jones T M, Williamson R A, Pilkington G R, Haigwood N L, Cabezas E, Satterthwait A C, Sanz I, Burton D R. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230:812–823. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- 4.Barbas C F, III, Hu D, Dunlop N, Sawyer L, Cababa D, Hendry R M, Nara P L, Burton D R. In vitro evolution of a neutralizing human antibody to human immunodeficiency virus type 1 to enhance affinity and broaden strain cross-reactivity. Proc Natl Acad Sci USA. 1994;91:3809–3813. doi: 10.1073/pnas.91.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bou-Habib D C, Roderiquez G, Oravecz T, Berman P W, Lusso P, Norcross M A. Cryptic nature of envelope V3 region epitopes protects primary human immunodeficiency virus type 1 from antibody neutralization. J Virol. 1994;68:6006–6013. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauder A, Katinger H. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 6a.Burton, D. Unpublished results.
- 7.Burton D R, Barbas C F, Persson M A, Koenig S, Chanock R M, Lerner R A. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton, D. R., and D. C. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl. A):S87–S98. [PubMed]
- 9.Burton D R, Pyati J, Koduri R, Sharp S J, Thornton G B, Parren P W H I, Sawyer L S W, Hendry R M, Dunlop N, Nara P L, Lamacchia M, Garratty E, Stiehm E R, Bryson Y J, Cao Y, Moore J P, Ho D D, Barbas C F. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 10.Clavel F, Charneau P. Fusion from without directed by human immunodeficiency virus particles. J Virol. 1994;68:1179–1185. doi: 10.1128/jvi.68.2.1179-1185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conley A J, Gorny M K, Kessler II J A, Boots L J, Ossorio-Castro M, Koenig S, Lineberger D W, Emini E A, Williams C, Zolla-Pazner S. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody 447-52D. J Virol. 1994;68:6994–7000. doi: 10.1128/jvi.68.11.6994-7000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Della-Porte A J, Westaway E G. A multi-hit model for the neutralization of animal viruses. J Gen Virol. 1977;38:1–19. doi: 10.1099/0022-1317-38-1-1. [DOI] [PubMed] [Google Scholar]
- 13.Dimmock N J. Neutralization of animal viruses. Curr Top Microbiol Immunol. 1993;183:1–149. doi: 10.1007/978-3-642-77849-0. [DOI] [PubMed] [Google Scholar]
- 14.Dimmock N J. Update on the neutralization of animal viruses. Rev Med Virol. 1995;5:165–179. [Google Scholar]
- 15.Ditzel H J, Parren P W H I, Binley J M, Sodroski J, Moore J P, Barbas C F, Burton D R. Mapping the protein surface of human immunodeficiency virus type 1 gp120 using human monoclonal antibodies from phage display libraries. J Mol Biol. 1997;267:684–695. doi: 10.1006/jmbi.1997.0912. [DOI] [PubMed] [Google Scholar]
- 16.Fouts T R, Binley J M, Trkola A, Robinson J E, Moore J P. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gauduin M C, Safrit J T, Weir R, Fung M S, Koup R A. Pre- and post-exposure protection against human immunodeficiency virus type 1 infection mediated by a monoclonal antibody. J Infect Dis. 1995;171:1203–1209. doi: 10.1093/infdis/171.5.1203. [DOI] [PubMed] [Google Scholar]
- 18.Gauduin M-C, Parren P W H I, Weir R, Barbas C F, Burton D R, Koup R A. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nat Med. 1997;3:1389–1393. doi: 10.1038/nm1297-1389. [DOI] [PubMed] [Google Scholar]
- 18a.Gelderblom, H., and Q. Sattentau. Unpublished results.
- 19.Gorny M K, Conley A J, Karwowska S, Buchbinder A, Xu J-Y, Emini E A, Koenig S, Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klasse P J, Moore J P. Quantitative model of antibody- and soluble CD4-mediated neutralization of primary isolates and T-cell-line-adapted strains of human immunodeficiency virus type 1. J Virol. 1996;70:3668–3677. doi: 10.1128/jvi.70.6.3668-3677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laal S, Burda S, Gorny M K, Karwowska S, Buchbinder A, Zolla-Pazner S. Synergistic neutralization of human immunodeficiency virus type 1 by combinations of human monoclonal antibodies. J Virol. 1994;68:4001–4008. doi: 10.1128/jvi.68.6.4001-4008.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McInerney T L, McLain L, Armstrong S J, Dimmock N J. A human IgG1 (b12) specific for the CD4 binding site of HIV-1 neutralizes by inhibiting the virus fusion entry process, but b12 Fab neutralizes by inhibiting a post-fusion event. Virology. 1997;233:313–326. doi: 10.1006/viro.1997.8547. [DOI] [PubMed] [Google Scholar]
- 23.McKeating J A, Cordell J, Dean C J, Balfe P. Synergistic interaction between ligands binding to the CD4 binding site and V3 domain of human immunodeficiency virus type 1 gp120. Virology. 1992;191:732–742. doi: 10.1016/0042-6822(92)90249-o. [DOI] [PubMed] [Google Scholar]
- 24.McLain L, Dimmock N J. Single- and multi-hit kinetics of immunoglobulin G neutralization of human immunodeficiency virus type 1 by monoclonal antibodies. J Gen Virol. 1994;75:1457–1460. doi: 10.1099/0022-1317-75-6-1457. [DOI] [PubMed] [Google Scholar]
- 24a.Mondor, I., and Q. J. Sattentau. Unpublished results.
- 25.Montefiori D C, Graham B S, Zhou J, Schwartz D H, Cavacini L A, Posner M R NIH-NIAID AIDS Vaccine Clinical Trials Network. V3-specific neutralizing antibodies in sera from HIV-1 gp160-immunized volunteers block virus fusion and act synergistically with human monoclonal antibody to the conformation-dependent CD4 binding site of gp120. J Clin Invest. 1993;92:840–847. doi: 10.1172/JCI116658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore J P, Cao Y, Qing L, Sattentau Q J, Pyati J, Koduri R, Robinson J, Barbas C F, Burton D R, Ho D D. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J Virol. 1995;69:101–109. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117–S136. [PubMed]
- 28.Moore J P, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore J P, Trkola A, Korber B, Boots L J, Kessler II J A, McCutchan F E, Mascola J, Ho D D, Robinson J, Conley A J. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J Virol. 1995;69:122–130. doi: 10.1128/jvi.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muster T, Guinea R, Trkola A, Purtscher M, Klima A, Steindl F, Palese P. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J Virol. 1994;68:4031–4034. doi: 10.1128/jvi.68.6.4031-4034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Rüker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nara P L, Garrity R R, Goudsmit J. Neutralization of HIV-1: a paradox of humoral proportions. FASEB J. 1991;5:2437–2455. doi: 10.1096/fasebj.5.10.1712328. [DOI] [PubMed] [Google Scholar]
- 33.Parren P W H I, Burton D R. Antibodies against HIV-1 from phage display libraries: mapping of an immune response and progress towards anti-viral immunotherapy. Chem Immunol. 1997;65:18–56. doi: 10.1159/000319346. [DOI] [PubMed] [Google Scholar]
- 34.Parren P W H I, Ditzel H J, Gulizia R J, Binley J M, Barbas C F, Burton D R, Mosier D E. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS. 1995;9:F1–F6. doi: 10.1097/00002030-199506000-00001. [DOI] [PubMed] [Google Scholar]
- 35.Parren P W H I, Gauduin M-C, Koup R A, Poignard P, Sattentau Q J, Fisicaro P, Burton D R. Relevance of the antibody response against human immunodeficiency virus type 1 envelope to vaccine design. Immunol Lett. 1997;58:125–132. doi: 10.1016/s0165-2478(97)00109-0. [DOI] [PubMed] [Google Scholar]
- 36.Poignard P, Fouts T, Naniche D, Moore J P, Sattentau Q J. Neutralizing antibodies to human immunodeficiency virus type-1 gp120 induce envelope glycoprotein subunit dissociation. J Exp Med. 1996;183:473–484. doi: 10.1084/jem.183.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poignard P, Klasse P-J, Sattentau Q J. Antibody neutralization of HIV-1. Immunol Today. 1996;17:239–246. doi: 10.1016/0167-5699(96)10007-4. [DOI] [PubMed] [Google Scholar]
- 38.Potts B J, Field K G, Wu Y, Posner M, Cavacini L, White-Scharf M. Synergistic inhibition of HIV-1 by CD4 binding domain reagents and V3-directed monoclonal antibodies. Virology. 1993;197:415–419. doi: 10.1006/viro.1993.1604. [DOI] [PubMed] [Google Scholar]
- 39.Roben P, Moore J P, Thali M, Sodroski J, Barbas C F, Burton D R. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Safrit J T, Fung M S C, Andrews C A, Braun D G, Sun W N C, Chang T W, Koup R A. Hu-PBL-SCID mice can be protected from HIV-1 infection by passive transfer of monoclonal antibody to the principal neutralizing determinant of envelope gp120. AIDS. 1993;7:15–21. doi: 10.1097/00002030-199301000-00002. [DOI] [PubMed] [Google Scholar]
- 41.Sattentau Q J. HIV-1 neutralization: antibody-gp120 interactions. In: Girard M, Dodet B, editors. Retroviruses of human AIDS and related animal diseases. Lyon, France: Fondation Marcel Merieux; 1994. pp. 135–139. [Google Scholar]
- 42.Sattentau Q J, Moore J P. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J Exp Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sattentau Q J. Neutralization of HIV-1 by antibody. Curr Opin Immunol. 1996;8:540–545. doi: 10.1016/s0952-7915(96)80044-6. [DOI] [PubMed] [Google Scholar]
- 44.Seligman S J, Binley J M, Gorny M K, Burton D R, Zolla-Pazner S, Sokolowski K A. Characterization by serial deletion competition ELISAs of HIV-1 V3 loop epitopes recognized by monoclonal antibodies. Mol Immunol. 1996;33:737–745. doi: 10.1016/0161-5890(96)00044-2. [DOI] [PubMed] [Google Scholar]
- 45.Shotton C, Arnold C, Sattentau Q, Sodroski J, McKeating J A. Identification and characterization of monoclonal antibodies specific for polymorphic antigenic determinants within the V2 region of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1995;69:222–230. doi: 10.1128/jvi.69.1.222-230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatatos L, Zolla-Pazner S, Gorny M K, Cheng-Mayer C. Binding of antibodies to virion-associated gp120 molecules of primary-like human immunodeficiency virus type 1 (HIV-1) isolates: effect on HIV-1 infection of macrophages and peripheral blood mononuclear cells. Virology. 1997;229:360–369. doi: 10.1006/viro.1997.8443. [DOI] [PubMed] [Google Scholar]
- 47.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tilley S A, Honnen W J, Racho M E, Chou T C, Pinter A. Synergistic neutralization of HIV-1 by human monoclonal antibodies against the V3 loop and the CD4 binding site of gp120. AIDS Res Hum Retroviruses. 1992;8:461–467. doi: 10.1089/aid.1992.8.461. [DOI] [PubMed] [Google Scholar]
- 49.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 50.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ugolini S, Mondor I, Parren P W H I, Burton D R, Tilley S A, Klasse P J, Sattentau Q J. Inhibition of attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J Exp Med. 1997;186:1287–1298. doi: 10.1084/jem.186.8.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.VanCott T C, Bethke F R, Polonis V R, Gorny M K, Zolla-Pazner S, Refield R R, Birx D L. Dissociation rate of antibody-gp120 binding interactions is predictive of V3-mediated neutralization of HIV-1. J Immunol. 1994;153:449–459. [PubMed] [Google Scholar]
- 53.Vijh-Warrier S, Pinter A, Honnen W J, Tilley S A. Synergistic neutralization of human immunodeficiency virus type 1 by a chimpanzee monoclonal antibody against the V2 domain of gp120 in combination with monoclonal antibodies against the V3 loop and the CD4-binding site. J Virol. 1996;70:4466–4473. doi: 10.1128/jvi.70.7.4466-4473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu L, Gerard N, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cordoso A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 55.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]