Abstract
Sequences of the human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) domain were determined by direct sequencing of HIV-1 RNA in successive plasma samples from eight seroconverting patients infected with virus bearing the T215Y/F amino acid substitution associated with zidovudine (ZDV) resistance. At baseline, additional mutations associated with ZDV resistance were detected. Three patients had the M41L amino acid change, which persisted. Two patients had both the D67N and the K70R amino acid substitutions; reversion to the wild type was seen at both positions in one of these patients and at codon 70 in the other one. Reversion to the wild type at codon 215 was observed in only one of eight patients. Unusual amino acids, such as aspartic acid (D) and cysteine (C), appeared at position 215 in four patients during follow-up. These variants isolated by coculturing were sensitive to ZDV. Overgrowth of these variants suggests that they have better fitness than the original T215Y variant. Intraindividual nucleoside substitutions over time were 10 times more frequent in codons associated with ZDV resistance (41, 67, 70, 215, and 219) than in other codons of the RT domain. The predominance of nonsynonymous substitutions observed over time suggests that most changes reflect adaptation of the RT function. The variance in sequence evolution observed among patients, in particular at codon 215, supports a role for chance in the evolution of the RT domain.
Human immunodeficiency virus (HIV) type 1 (HIV-1) with decreased in vitro sensitivity to zidovudine (ZDV) has been isolated from HIV-infected patients receiving prolonged ZDV therapy (17). Phenotypic resistance of HIV-1 to ZDV is associated with mutations affecting at least five codons of the reverse transcriptase (RT) domain of the pol gene (M41L, D67N, K70R, T215Y/F, and K219Q). Most viral isolates with reduced sensitivity to ZDV harbor the T215Y/F amino acid change (2, 14, 18). HIV-1 variants with M41L and/or T215Y/F were not detected before the introduction of ZDV, indicating reduced fitness of these variants compared to the wild type (9). In contrast, a low level of polymorphism has been described for codon 70 (22, 23, 32). Following removal of drug pressure, there is a slow rate of reversion of ZDV resistance mutations, suggesting that these mutations have only a modest impact on viral replication in the absence of drug (1, 3, 22, 27).
Because of the widespread use of ZDV in western countries, transmission of ZDV-resistant viruses has occurred in recent years in up to 10% of newly infected individuals (7, 12, 21, 31). Although the clinical impact of resistance has not been fully assessed, transmission of drug-resistant HIV-1 variants may impair the efficacy of antiviral treatment regimens (6, 13, 30).
The aim of this study was to analyze the evolution of the RT domain of the HIV-1 pol gene in sequential plasma samples from recently seroconverting patients infected with ZDV-resistant variants.
MATERIALS AND METHODS
Patients.
A total of 114 patients with documented HIV-1 seroconversion from 1988 to 1995 in Switzerland (31) and 36 patients with primary HIV-1 infection and enrolled in a multicenter controlled clinical trial of ZDV (16) were included in this study. Baseline samples were collected during the symptomatic phase of primary HIV-1 infection and before any antiviral treatment.
Selective PCR.
Blood was centrifuged twice for 5 min each time at 1,500 × g, divided into aliquots, and stored at −75°C within 2 h. Total RNA from 50 μl of plasma was extracted, reverse transcribed, and amplified by selective PCR (19, 30).
Sequence analysis.
Five microliters of the first PCR product was reamplified with 0.25 μg of primers NNA (5′ AAGCCAGGAATGGATGGCCCA) and E (biotinylated; 5′ CCATTTATCAGGATGGAGTTC) in a reaction mixture containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 3 mM MgCl2, 250 μM each deoxynucleoside triphosphate, and 2.5 U of AmpliTaq polymerase (Perkin-Elmer Cetus, Norwalk, Conn.). The reactions were carried out for 30 cycles of 20 s at 95°C, 30 s at 50°C, and 30 s at 72°C. The nucleotide sequence was determined by use of the AmpliTaq FS dyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) on an automatic sequencer (model 373; Applied Biosystems). Primer NNA was used to analyze the codon region from positions 30 to 130, and primer E was used to analyze the codon region from positions 130 to 228. Sequence alignment was performed with the CLUSTALW program. Phylogenetic analysis was performed by the maximum-likelihood method (PHYLIP version 3.5; University of Washington, Seattle).
Viral cultures.
HIV-1 was isolated by standard procedures with peripheral blood mononuclear cells (PBMC) depleted of CD8 lymphocytes and cocultivated with phytohemagglutinin-stimulated PBMC from HIV-negative blood donors (11).
ZDV susceptibility assay.
ZDV susceptibility was determined in the PBMC assay, taking into account the replication kinetics of each strain, as previously described (4). Briefly, after conventional isolation of the HIV-1 strains from PBMC, the cell-free HIV-1-infected supernatants corresponding to the peak of RT activity were serially diluted (100 to 10−4) and incubated with fresh HIV-negative phytohemagglutinin-stimulated PBMC. After being washed, the cells were pipetted into 96-well plates containing increasing concentrations of ZDV (0, 0.01, 0.05, 0.25, 1.25, and 6.25 μM). The 50% tissue culture infective dose was assessed by measuring RT activity (26). On the day selected on the basis of replication kinetics criteria, the drug concentrations inhibiting 50 and 90% of the RT activity at a 50% tissue culture infective dose of 100 were calculated.
HIV-1 RNA quantitation.
The determination of HIV-1 RNA levels in plasma samples was performed with the Amplicor HIV Monitor according to the manufacturer’s instructions (Roche, Basel, Switzerland).
RESULTS
Patient characteristics.
HIV-1 RNA was successfully amplified from plasma samples from 136 (91%) of the 150 patients analyzed. A mutation at codon 215 was detected in baseline samples from 12 patients by selective PCR. Follow-up samples were collected from 8 of these 12 patients for at least 12 months and were retained for sequential evaluation. At baseline, the mean CD4 cell count was 517/mm3 (range, 210 to 754), and the mean HIV-1 RNA level was 4.68 log HIV-1 RNA copies/ml (range, 4.34 to 5.31). Four patients (A, C, D, and H) received antiviral treatment, started at the time of seroconversion, for 6 months. Patient B received ZDV for 6 months, starting 20 months after seroconversion.
Sequence analysis of the RT domain.
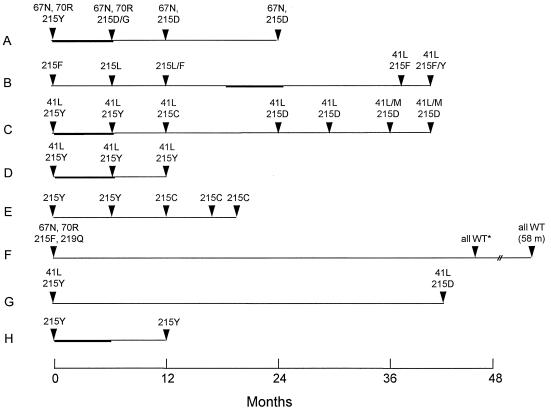
Amino acid substitutions associated with ZDV resistance over time are reported in Fig. 1. At baseline, the T215Y substitution was observed in six patients and the T215F substitution was observed in two patients. The M41L substitution was detected at baseline in three patients. The association of the D67N and K70R substitutions was detected in the baseline samples from two patients. During the follow-up (12 to 58 months), a reversion to the wild type at position 215 was observed in only one patient (F). This patient showed reversion to the wild type for the four initial mutations associated with ZDV resistance (D67N, K70R, T215F, and K219Q). One patient (A) showed reversion to the wild type at codon 67. The M41L substitution persisted in all patients during the follow-up (12 to 43 months).
FIG. 1.
Amino acid substitutions in the RT domain for the five codons associated with ZDV resistance. Bold lines indicate the period of antiretroviral treatment. All treated patients were on ZDV monotherapy treatment, except for patient C, who was on ZDV plus didanosine treatment. WT, wild type; m, months.
Unusual amino acids, such as aspartic acid (D), cysteine (C), and leucine (L), appeared at position 215 in five patients during follow-up (Fig. 1). Mutation Y215D (TAC→GAC) (changes are underlined) was observed in three patients (A, C, and G) and persisted for 18 months in two of them. At month 12, patient E showed substitution Y215C (TAC→TGC), which persisted up to month 21. Substitution Y215C was also transitorily observed in patient C, before the emergence of aspartic acid (D) at position 215, which persisted during the follow-up. In addition to Y215D and Y215C, F215L (TTC→CTC) was detected transitorily in patient B.
Genetic heterogeneity.
Nucleotide heterogeneity was compared for nucleotide sequences outside the five codons associated with ZDV resistance (41, 67, 70, 215, and 219), as well as for these five codons only (Table 1). The intraindividual differences for the entire sequence analyzed (594 nucleotides) did not exceed 1.7% of nucleotides, even when the samples were collected 43 or 46 months apart. Sequential RT sequences were absolutely identical only in patient D. The nucleotide heterogeneity of the five codons associated with ZDV resistance was much higher than that of the codons not associated with ZDV resistance. A total of 17 (4.7%) nucleotide substitutions were detected for these five codons over time. In contrast, only 59 (0.4%) nucleotide substitutions were observed for the other 193 codons analyzed (χ2 test, P < 0.0001). Nonsynonymous nucleotide changes were observed more often than synonymous nucleotide changes, and all substitutions observed in the five ZDV resistance-related codons were nonsynonymous (Table 1).
TABLE 1.
Nucleotide substitutions over time for the RT domain and codons associated with ZDV resistance
| Patient | Mo | No. of nucleotide substitutions
|
||
|---|---|---|---|---|
| Total RT (594 bp) | Truncated RTa (579 bp) (S/NS)b | Codons related to ZDV resistancec (15 bp) (S/NS)b | ||
| A | 0–6 | 6 | 2/1 | 0/3 |
| 6–12 | 7 | 3/4 | 0/0 | |
| 12–24 | 4 | 2/2 | 0/0 | |
| B | 0–6 | 2 | 0/1 | 0/1 |
| 6–12 | 5 | 0/4 | 0/1 | |
| 12–39 | 3 | 0/2 | 0/1 | |
| 39–42 | 1 | 0/0 | 0/1 | |
| C | 0–6 | 0 | 0/0 | 0/0 |
| 6–12 | 2 | 1/0 | 0/1 | |
| 12–24 | 5 | 0/4 | 0/1 | |
| 24–31 | 1 | 0/1 | 0/0 | |
| 31–36 | 3 | 0/3 | 0/0 | |
| 36–42 | 4 | 3/1 | 0/0 | |
| D | 0–6 | 0 | 0/0 | 0/0 |
| 6–9 | 0 | 0/0 | 0/0 | |
| 9–12 | 0 | 0/0 | 0/0 | |
| E | 0–9 | 0 | 0/0 | 0/0 |
| 9–12 | 4 | 1/2 | 0/1 | |
| 12–18 | 3 | 1/2 | 0/0 | |
| 18–21 | 3 | 1/2 | 0/0 | |
| F | 0–46 | 10 | 1/4 | 0/5 |
| 46–58 | 3 | 1/2 | 0/0 | |
| G | 0–43 | 7 | 2/3 | 0/2 |
| H | 0–12 | 3 | 1/2 | 0/0 |
Calculated for residues 121 to 657. The five codons associated with ZDV resistance were omitted.
S, synonymous substitution; NS, nonsynonymous substitution.
Calculated for residues of the five codons associated with ZDV resistance.
Distinct clusters of viral sequences corresponding to each patient over time were observed (data not shown), demonstrating the absence of PCR product contamination of viral sequences within this data set.
ZDV susceptibility assay.
ZDV susceptibilities were determined with viral stocks isolated from PBMC from patients C and E (Table 2). The baseline isolate from patient C, containing T215Y, was resistant to ZDV, whereas isolates obtained after 24 and 42 months (T215D) were susceptible to ZDV despite the persistence of the mutation M41L. PBMC were not available from patient E at baseline, but isolates obtained after 12 and 21 months (T215C) were susceptible to ZDV. For both patients, RT nucleotide sequences of isolates recovered from PBMC cocultures were identical to those determined by direct sequencing of PCR products derived from plasma HIV-1 RNA (data not shown).
TABLE 2.
ZDV susceptibility in two patients with T215D and T215C amino acid substitutionsa
| Patient | Time (mo) | RT genotype | ZDV IC50 (μM) | ZDV IC90 (μM) |
|---|---|---|---|---|
| C | 0 | 41L, 67D, 70K, 215Y, 219K | 0.18 | 2.19 |
| C | 24 | 41L, 67D, 70K, 215D, 219K | <0.01 | <0.02 |
| C | 42 | 41M/L, 67D, 70K, 215D, 219K | <0.01 | <0.01 |
| E | 0 | 41M, 67D, 70K, 215Y, 219K | NA | NA |
| E | 12 | 41M, 67D, 70K, 215C, 219K | <0.01 | 0.03 |
| E | 21 | 41M, 67D, 70K, 215C, 219K | <0.01 | <0.01 |
IC50 and IC90, concentrations inhibiting 50 and 90% of the RT activity at a 50% tissue culture infective dose of 100. NA, not available.
DISCUSSION
This investigation was performed to assess the genetic evolution of the RT domain of the HIV-1 pol gene in recently seroconverting patients infected with ZDV-resistant variants. The generation of viral genetic variants occurs principally through random mutational and recombinational events. However, the rate of emergence of particular variants is determined by factors such as the strength of selective pressures, the complexity of the preexisting genetic pool, and the rate of viral turnover (5, 10, 28). In our seroconverting patients, high levels of viremia were observed over time, thus providing the basis for the emergence of mutations.
During a median follow-up of 33 months, changes in amino acids were predominantly detected in codons associated with ZDV resistance and, in particular, in codon 215. In five patients, the changes at codon 215 resulted in amino acids (aspartic acid, cysteine, and leucine) that did not belong to the natural HIV-1 polymorphism. These amino acid substitutions were linked to a single nucleotide change, whereas return to the wild-type codon would have required two nucleotide changes. The T215C and T215D variants were reported recently for a very small number of patients, most likely in the background of T215Y (8, 24, 29). These variants were found to be sensitive to ZDV and were not seen in patients on ZDV therapy. The persistence of T215Y was observed in only two patients; these patients had only 12 months of follow-up and were on ZDV during the first 6 months following seroconversion, at the time of maximal viral turnover. Our data suggest that most patients would present a switch from T215Y to other amino acids within 1 year in the absence of drug pressure. Interestingly, the lower frequency of reversion to the wild type observed for the other codons associated with ZDV resistance is suggestive of a lower impact of these codons on RT fitness, as shown recently for the M41L mutation (8). In contrast, variants with T215D were shown to display a 10 to 25% higher relative fitness than the initial T215Y variants (8). The ability of T215D or T215C viruses to overgrow the original T215Y variant in vivo suggests a selective advantage of these variants. Moreover, the growth potential of the T215D and T215C variants was demonstrated by their selective isolation from PBMC bulk cultures. The transient expression of T215C before the emergence of T215D in patient C suggests better fitness of the T215D variant.
As for patients receiving antiretroviral therapy (15, 25), more nonsynonymous rather than synonymous changes were observed in our patients with primary HIV-1 infection, most of the time in the absence of drug pressure. In addition, nucleotide substitutions were 10 times more frequent in codons associated with ZDV resistance than in other codons, and all nucleotide substitutions affecting ZDV resistance-associated codons were nonsynonymous. This result suggests that selective pressure, most probably on RT function, is the driving force for RT sequence evolution in vivo. At the same time, the high variance in sequence evolution observed among patients and the rapid emergence of various unusual amino acids at codon 215 suggest that the evolutionary pathway of viral populations is modulated by chance events (20) and that HIV-1 is able to take new evolutionary pathways starting from viruses that do not naturally occur.
Finally, the frequency of transmission of drug-resistant mutants should be considered in the design of antiretroviral regimens and raises the issue of systematic screening for mutations associated with resistance in drug-naive patients before the initiation of therapy.
ACKNOWLEDGMENTS
This work was supported by the National AIDS Research Program, by the Swiss Federal Office of Public Health (cohort study part A), and the Swiss National AIDS Research Program (grant 3239.041951.94).
We thank W. Caveng, C. Gaille, and E. Ramirez for excellent technical help and S. Emler, G. Schockmel, and S. Saragosti for helpful discussions. This study took advantage of the infrastructure of the Swiss HIV Cohort Study, whose members are M. Battegay, P. Bürgisser, R. Doorly, M. Egger, P. Erb, W. Fierz, M. Flepp, P. Francioli, P. Grob, U. Grüninger, B. Hirschel, B. Ledergerber, R. Lüthy, R. Malinverni, L. Matter, M. Opravil, F. Paccaud, L. Perrin, W. Pichler, M. Rickenbach, O. Rutschmann, P. Vernazza, and J. von Overbeck.
REFERENCES
- 1.Albert J, Wahlberg J, Lundeberg J, Cox S, Sadstrom E, Wahren B, Uhlen M. Persistence of azidothymidine-resistant human immunodeficiency virus type 1 RNA genotypes in posttreatment sera. J Virol. 1992;66:5627–5630. doi: 10.1128/jvi.66.9.5627-5630.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher C A, O’Sullivan E, Mulder J W, Ramautarsing C, Kellam P, Darby G, Lange J M, Goudsmit J, Larder B A. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J Infect Dis. 1992;165:105–110. doi: 10.1093/infdis/165.1.105. [DOI] [PubMed] [Google Scholar]
- 3.Boucher C A, van Leeuven R, Kellam P, Schipper P, Tijnagel J, Lange J M, Larder B A. Effects of discontinuation of zidovudine treatment on zidovudine sensitivity of human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1525–1530. doi: 10.1128/aac.37.7.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brun-Vezinet F, Ingrand D, Desforges L, Gochi K, Ferchal F, Schmitt M P, Jung M, Masquelier B, Aubert J, Buffet-Janvresse C, Fleury H. HIV-1 sensitivity to zidovudine: a consensus culture technique validated by genotypic analysis of the reverse transcriptase. J Virol Methods. 1992;37:177–188. doi: 10.1016/0166-0934(92)90045-f. [DOI] [PubMed] [Google Scholar]
- 5.Coffin J M. HIV population dynamics in vivo: implication for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 6.D’Aquila R T, Johnson V A, Welles S L, Japour A J, Kuritzkes D R, DeGruttola V, Reichelderfer P S, Coombs R W, Crumpacker C S, Kahn J O, Richman D D. Zidovudine resistance and HIV-1 disease progression during antiretroviral therapy. Ann Intern Med. 1995;122:401–408. doi: 10.7326/0003-4819-122-6-199503150-00001. [DOI] [PubMed] [Google Scholar]
- 7.De Ronde A, Schuurman R, Goudsmit J, van den Hoek A, Boucher C A. First case of new infection with zidovudine-resistant HIV-1 among prospectively studied intravenous drug users and homosexual men in Amsterdam, The Netherlands. AIDS. 1996;10:231–232. doi: 10.1097/00002030-199602000-00017. [DOI] [PubMed] [Google Scholar]
- 8.Goudsmit J, de Ronde A, de Rooij E, Boer R. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J Virol. 1997;71:4479–4484. doi: 10.1128/jvi.71.6.4479-4484.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrigan, P. R., S. Bloor, and B. A. Larder. 1996. Quantitation of viral fitness and AZT selection pressure of AZT resistant HIV-1 variants. Antivir. Ther. 1(Suppl. 1):55.
- 10.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virion and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 11.Hollinger F B, editor. ACTG virology manual for HIV laboratories, 2nd ed. U.S. Public Health Service publication no. NIH-94-3828. Bethesda, Md: Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health; 1993. [Google Scholar]
- 12.Imrie A, Carr A, Duncombe C, Finlayson R, Vizzard J, Law M, Kaldor J, Penny R, Cooper D A the Sydney Primary HIV Infection Study Group. Primary infection with zidovudine-resistant human immunodeficiency virus type 1 does not adversely affect outcome at 1 year. J Infect Dis. 1996;174:195–198. doi: 10.1093/infdis/174.1.195. [DOI] [PubMed] [Google Scholar]
- 13.Japour A J, Welles S, D’Aquila R T, Johnson V A, Richman D D, Coombs R W, Reichelderfer P S, Kahn J O, Crumpacker C S, Kuritzkes D R. Prevalence and clinical significance of zidovudine resistance mutations in human immunodeficiency virus isolated from patients after long-term zidovudine treatment. J Infect Dis. 1995;171:1172–1179. doi: 10.1093/infdis/171.5.1172. [DOI] [PubMed] [Google Scholar]
- 14.Kellam P, Boucher C A, Larder B A. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high level resistance to zidovudine. Proc Natl Acad Sci USA. 1992;89:1934–1938. doi: 10.1073/pnas.89.5.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keulen W, Boucher C A, Berkhout B. Nucleotide substitution patterns can predict the requirements for drug-resistance of HIV-1 proteins. Antiviral Res. 1996;31:45–57. doi: 10.1016/0166-3542(96)00944-8. [DOI] [PubMed] [Google Scholar]
- 16.Kinloch-de Loes S, Hirschel B, Hoen B, Cooper D A, Tindall B, Carr A, Saurat J H, Clumeck N, Lazzarin A, Mathiesen L, Raffi F, Antunes F, von Overbeck J, Luthy R, Glauser M, Hawkins D, Baumberger C, Yerly S, Perneger T V, Perrin L. A controlled trial of zidovudine in primary human immunodeficiency virus infection. N Engl J Med. 1995;333:408–413. doi: 10.1056/NEJM199508173330702. [DOI] [PubMed] [Google Scholar]
- 17.Larder B A, Darby G, Richman D D. HIV-1 with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 18.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 19.Larder B A, Kellam P, Kemp S D. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS. 1991;5:137–144. doi: 10.1097/00002030-199102000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Leigh Brown A J, Richman D D. HIV-1: gambling on the evolution of drug resistance? Nat Med. 1997;3:268–271. doi: 10.1038/nm0397-268. [DOI] [PubMed] [Google Scholar]
- 21.Mayers D L, Yerly S, Perrin L, Imrie A, Cooper D A, Karney W W, Brown A E, Rakik A, Harris R, Gambel J, Weislow O S, Lennox J L, Burke D S. Program and abstracts of the 34th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1994. Prevalence of AZT-resistant HIV-1 in persons seroconverting in Switzerland, Australia, and the United States between 1988 and 1994. [Google Scholar]
- 22.Mohri H, Singh M K, Ching W T, Ho D D. Quantitation of zidovudine-resistant human immunodeficiency virus type 1 in the blood of treated and untreated patients. Proc Natl Acad Sci USA. 1993;90:25–29. doi: 10.1073/pnas.90.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Najera I, Holguin A, Quinones-Mateu M E, Munoz-Fernandez M A, Najera R, Lopez-Galindez C, Domingo E. Pol gene quasispecies of human immunodeficiency virus: mutations associated with drug resistance in virus from patients undergoing no drug therapy. J Virol. 1995;69:23–31. doi: 10.1128/jvi.69.1.23-31.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quigg M, Rebus S, France A J, McMenamin J, Darby G, Leigh Brown A J. Mutations associated with zidovudine resistance in HIV-1 among recent seroconvertors. AIDS. 1997;11:835–836. [PubMed] [Google Scholar]
- 25.Quinones-Mateu M E, Holguin A, Dopazo J, Najera I, Domingo E. Point mutation frequencies in the pol gene of human immunodeficiency virus type 1 are two- to threefold lower than those of env. AIDS Res Hum Retroviruses. 1996;12:1117–1128. doi: 10.1089/aid.1996.12.1117. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz O, Henin Y, Marechal V, Montagnier L. A rapid and simple colorimetric test for the study of anti-HIV agents. AIDS Res Hum Retroviruses. 1988;4:441–448. doi: 10.1089/aid.1988.4.441. [DOI] [PubMed] [Google Scholar]
- 27.Smith M S, Koerber K L, Pagano J S. Long-term persistence of zidovudine resistance mutations in plasma isolates of human immunodeficiency virus type 1 of dideoxyinosine-treated patients removed from zidovudine therapy. J Infect Dis. 1994;169:184–188. doi: 10.1093/infdis/169.1.184. [DOI] [PubMed] [Google Scholar]
- 28.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in HIV-1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 29.Wong J K, Ignacio C C, Torriani F, Havlir D, Fitch N J S, Richman D D. In vivo compartmentalization of human immunodeficiency virus: evidence from examination of pol sequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yerly S, Denereaz N, Mermillod B, Hirschel B, Perrin L. Predictive value of codon 215 reverse transcriptase mutation on the efficacy of didanosine in HIV-infected, zidovudine-experienced patients. Antiviral Ther. 1996;1:167–171. [PubMed] [Google Scholar]
- 31.Yerly S, Rakik A, Kinloch-de Loes S, Erb P, Vernazza P, Hirschel B, Perrin L. Prévalence de la transmission de virus résistant à la zidovudine en Suisse. Schweiz Med Wochenschr. 1996;126:1845–1848. [PubMed] [Google Scholar]
- 32.Zhang L Q, Simmonds P, Ludlam C A, Leigh Brown A J. Detection, quantification and sequencing of HIV-1 from the plasma of seropositive individuals and from factor VIII concentrates. AIDS. 1991;5:675–681. doi: 10.1097/00002030-199106000-00006. [DOI] [PubMed] [Google Scholar]