Abstract
The cowpox virus (CPV) CrmA and the equivalent rabbitpox virus (RPV) SPI-2 proteins have anti-inflammatory and antiapoptosis activity by virtue of their ability to inhibit caspases, including the interleukin-1β-converting enzyme (ICE; caspase-1). Infection of LLC-PK1 pig kidney cells with a CPV CrmA mutant, but not with wild-type (wt) CPV, results in the induction of many of the morphological features of apoptosis (C. A. Ray and D. J. Pickup, Virology 217:384–391, 1996). In our study, LLC-PK1 cells infected with CPVΔcrmA, but not those infected with wt CPV, showed induction of poly(ADP-ribose) polymerase (PARP)- and lamin A-cleaving activities and processing of the CPP32 (caspase-3) precursor to a mature 18-kDa form. Surprisingly, infection of LLC-PK1 cells with either wt RPV (despite the presence of the SPI-2 protein) or RPVΔSPI-2 resulted in cleavage activity against PARP and lamin A and the appearance of the mature subunit of CPP32/caspase-3. The biotinylated specific peptide inhibitor Ac-Tyr-Val-Lys(biotinyl)-Asp-2,6-dimethylbenzoyloxymethylketone [AcYV(bio)KD-aomk] labeled active caspase subunits of 18, 19, and 21 kDa in extracts from LLC-PK1 cells infected with CPVΔcrmA, wt RPV, or RPVΔSPI-2 but not wt CPV. Mixed infection of LLC-PK1 cells with wt RPV and wt CPV gave no PARP-cleaving activity, and all PARP cleavage mediated by SPI-2 and CrmA mutants of RPV and CPV, respectively, could be eliminated by coinfection with wt CPV. These results suggest that the RPV SPI-2 and CPV CrmA proteins are not functionally equivalent and that CrmA, but not SPI-2 protein, can completely prevent apoptosis in LLC-PK1 cells under these conditions.
Orthopoxvirus infections are acute in nature and generally do not involve the establishment of a persistent or latent state (29). Therefore, recovery from a primary infection and the overall pathogenic profile of the infection are greatly influenced by nonspecific antiviral responses that operate early during infection. These responses include the development of an inflammatory response, the cytolytic action of natural killer cells, the antiviral activities of cytokines, and programmed cell death (apoptosis). The significance of nonspecific host responses to poxvirus infection is suggested by the identification of specific gene products, encoded by a number of poxviruses, that act to block cytokine maturation or binding of cytokines to receptors and interfere with inflammation, complement, and apoptosis (1, 2, 18, 19, 27, 33). Important and interesting examples of poxvirus proteins that modify the host response to infection are a number of members of the serpin superfamily of proteinase inhibitors. The poxvirus serpins serve to regulate both inflammation and programmed cell death (23, 32, 38, 48). To date, poxviruses are the only viruses known to encode functional serpins.
The cowpox virus (CPV) CrmA protein was the first poxvirus serpin identified (34) and has been the most thoroughly characterized. The 38-kDa CrmA protein is expressed early during infection and is found in the cytoplasm of the infected cell. Early studies showed that the deletion of the crmA gene from the Brighton Red strain of CPV resulted in a virus that produced white pocks upon infection of the chicken chorioallantoic membrane (CAM), instead of the red, hemorrhagic, noninflammatory pocks usually observed following infection with wild-type (wt) CPV (34). Subsequent studies showed that the production of white pocks on the CAM was the result of a massive infiltration of inflammatory cells into the site of infection, suggesting that the function of the CrmA protein is to block the production or activity of an important proinflammatory chemotactic factor (8, 11, 32). Indeed, the CrmA protein inhibits interleukin-1β (IL-1β)-converting enzyme (ICE) (37), an enzyme which cleaves pro-IL-1β to form active IL-1β (46), thereby mediating the inflammatory response. ICE is also known as caspase-1 and is the prototypic member of the caspase family (4) of cysteine proteinases, which cleave after aspartic acid residues.
During the last several years, apoptosis has become widely accepted as an important nonspecific host antiviral response. Premature death of an infected cell would have obvious deleterious consequences for the production of viral progeny, and therefore many viruses have evolved mechanisms to prevent apoptosis during infection (39). The significance of apoptosis as an antiviral defense against poxvirus infection is evidenced by the observation that a number of poxvirus genes encode proteins that can function to interfere with apoptosis in specific cell types. Among these genes are several that determine a host range phenotype, such that mutations in them render the virus unable to replicate in a given cell type. For example, the CPV CHOhr gene (15) and the vaccinia virus E3L gene (22) are required to prevent or delay cell death by apoptosis and allow fully productive infection in Chinese hamster ovary cells and primate fibroblasts, respectively. The SPI-1 gene of rabbitpox virus (RPV) has also been reported to encode a host range function, preventing the premature death of infected porcine kidney or human A549 epithelial cells during infection (3, 7). A recent study implicated two myxoma virus-encoded proteins, T2 and M11L, in the inhibition of lymphocyte apoptosis (25); lymphocytes infected with viruses bearing mutations in either the M11L or the T2 gene undergo apoptosis, resulting in a nonproductive infection.
Studies using cells transfected with a plasmid containing the crmA gene have shown that CrmA can block apoptosis induced by a variety of different stimuli, including growth factor deprivation (13) and signalling through the Fas or type 1 tumor necrosis factor (TNF) receptors (10, 24, 44), suggesting that in addition to its role in regulating inflammation, the CrmA protein blocks apoptosis mediated by proapoptotic caspase activity. Indeed, CrmA has been shown to inhibit caspase-1/ICE-mediated apoptosis (28), and more recent studies showed that CrmA is a particularly strong inhibitor of caspase-8 (MACH, FLICE, Mch5) (40, 42, 49), a proteinase that is recruited to the Fas and TNF receptor signalling complexes during delivery of the cell death signal (5, 31, 42). Caspase-8 is thought to be the first enzyme in a process by which activated caspases proteolytically activate other caspases in a cascade fashion, ultimately leading to cell death. CrmA also inhibits the proteolytic activity of granzyme B (35), a cytotoxic T-lymphocyte-encoded serine proteinase that has a substrate cleavage site specificity similar to that of the caspases (Asp residue at P1). Granzyme B itself is a key effector of cytotoxic T-lymphocyte-mediated cell death and may function through the activation of apoptotic proteinases such as caspase-3 (CPP32, apopain, Yama) (9, 36).
Although CrmA can block apoptosis when expressed in a number of heterologous systems, only one study has addressed the role that CrmA might play in regulating apoptosis induced during virus infection (38). Infection of the porcine cell line LLC-PK1 with CPV bearing a mutation in CrmA, but not with wt CPV, resulted in the induction of the morphological and nuclear changes associated with apoptosis (38), suggesting that the inhibition of proapoptotic proteolytic activity by CrmA may also be important in determining whether cells die by necrosis or apoptosis within the context of the virus infection. The observation that other cell types did not manifest signs of apoptotic cell death during infection with the CPV CrmA mutant suggests that the regulation of apoptosis may be multifactorial and cell line specific (38). For example, another CPV protein, the product of the CHOhr gene, facilitates virus growth on Chinese hamster ovary cells by preventing apoptosis (15), independent of the expression of the crmA gene.
To understand how poxviruses regulate apoptosis during virus infection, we must understand not only the biochemical functions of the individual gene products but also how the apoptotic program is induced in virus-infected cells. To this end, we have investigated in more detail the induction of apoptosis in LLC-PK1 cells infected with CPV and a CPV mutant deleted for the crmA gene (CPVΔcrmA). In addition, we also examined caspase activation by the related orthopoxvirus RPV. RPV, like the other orthopoxviruses, encodes a homolog of the CrmA protein, referred to as SPI-2 (3). Although studies have shown that purified RPV SPI-2 protein is able to block the cleavage of lamin A in a cell-free system (43) and can also function to regulate inflammation and pock color during infection of the chicken CAM (3), it is not known whether the RPV SPI-2 protein is functionally equivalent to the CPV CrmA protein in the inhibition of apoptosis during virus infection. In this regard, the SPI-2 protein contains the amino acids SV instead of CA at positions P6 and P5, respectively, within the serpin-reactive site loop as well as exhibiting a variety of amino acid differences from CrmA throughout the serpin backbone.
Here, we show that LLC-PK1 cells that are infected with CPVΔcrmA, but not those infected with wt CPV, contain at least five polypeptides corresponding to activated caspases, including a proteolytic activity responsible for the degradation of the nuclear proteins poly(ADP-ribose) polymerase (PARP) and lamin A. Furthermore, we discovered that unlike wt CPV, wt RPV induced infected LLC-PK1 cells to undergo apoptosis even in the presence of the CrmA homolog SPI-2. Mixed experiments suggested that the observed differences between CPV and RPV in controlling apoptosis are most likely representative of functional differences between the SPI-2 and CrmA proteins. Therefore, despite the apparent functional similarities between the CPV CrmA and RPV SPI-2 proteins in the regulation of inflammation during virus infection of the CAM (3), the regulation of apoptosis during infection by wt CPV clearly differs from that during infection with wt RPV.
MATERIALS AND METHODS
Cells and viruses.
LLC-PK1 cells and CV-1 cells were obtained from the American Type Culture Collection and grown in medium 199 or F11 (both from Gibco-BRL), respectively, containing 5% fetal bovine serum. HeLa S3 cells, kindly provided by J. B. Flanegan (University of Florida), were maintained in suspension in Joklik’s modified Eagle’s medium (Gibco-BRL) supplemented with 5% calf serum and 2% Fetal Clone II (HyClone). RPV (Utrecht strain) and vaccinia virus (strain WR) were obtained from the American Type Culture Collection, and CPV (Brighton Red strain) was obtained from David Pickup (Duke University). RPVΔSPI-2 and CPVΔcrmA (referred to elsewhere as CPVΔSPI-2) have been described previously (3). Virus stocks were routinely grown on CV-1 cells, which were also used for determination of virus titers.
DAPI staining of cells.
LLC-PK1 cells were grown in LabTek eight-well chamber slides to 50% confluence and infected with virus at a multiplicity of 10 in 100 μl of Gibco medium 199 without serum. Virus was adsorbed for 2 h at 37°C. After removal of the inoculum, the cells were washed with 300 μl of medium without serum. Cells were processed at 16 h postinfection, first being washed with 300 μl of phosphate-buffered saline (PBS) and then being fixed by incubation with 200 μl of PBS containing 3.5% paraformaldehyde for 20 min at room temperature and then with 200 μl of ice-cold methanol for 10 min. Cells were rinsed two times with 300 μl of PBS and then incubated with 100 μl of PBS containing 0.5 μg of DAPI (4′,6-diamidino-2-phenylindole) per ml for 30 min at room temperature. The cells were then rinsed three times with 300 μl of PBS, and the DNA was visualized by using a fluorescence microscope equipped with a DAPI filter. Fluorescent cells were photographed with Fuji 400 ASA film.
Preparation of infected-cell extracts.
Semiconfluent (80%) monolayers of LLC-PK1 cells in 100-mm-diameter dishes were either mock infected with medium alone or infected with RPV, RPVΔSPI-2, CPV, or CPVΔSPI-2. A multiplicity of infection (MOI) of 5 to 10 PFU/cell was used for the RPV derivatives, while an MOI of 25 to 50 PFU/cell was used for the CPV derivatives because CPV’s efficiency of plaque formation on LLC-PK1 cells is fivefold lower than that of RPV (data not shown). For single-virus infections, virus was adsorbed to cells for 60 to 90 min at 37°C, medium was added, and the infection was allowed to proceed for 14 h at 37°C. Where mixed infections were performed, the virus inocula were mixed prior to being added to the cells, using an MOI of 10 PFU/cell for RPV and of 50 PFU/cell for CPV, and virus adsorption was carried out at 4°C. Cells were harvested by scraping them into the medium with rubber policemen, pelleted, washed once with PBS (pH 7.4), pelleted again, washed once with cold extract preparation buffer [EPB; 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES, pH 7.0), 50 mM KCl, 5 mM EGTA, 2 mM MgCl2, 1 mM dithiothreitol (DTT), 20 μM cytochalasin B], pelleted, and resuspended in 100 to 200 μl of cold EPB containing the proteinase inhibitors phenylmethylsulfonyl fluoride (PMSF; 0.2 mM) and CLAP (containing chymostatin [20 μg/ml], leupeptin [5 μg/ml], antipain [20 μg/ml], and pepstatin A [5 μg/ml]). Cells were lysed by four cycles of freezing and thawing, and the lysates were subjected to centrifugation at 10,000 × g for 15 min. The supernatant cytoplasmic extract was aliquoted and either used immediately in assays or stored at −80°C. The protein concentration in the extract was estimated by the Bradford assay.
In vitro apoptosis assay.
Extracts were assayed for apoptotic activity by following the protocols described by Earnshaw and coworkers (20, 21). Nuclei were purified from rapidly growing HeLa S3 cells (maintained at 1 × 105 to 4 × 105/ml) as follows: cells were pelleted, washed in 20 volumes of PBS containing 1 mM MgCl2, repelleted, washed in 20 volumes of NB (10 mM PIPES [pH 7.4], 10 mM KCl, 2 mM MgCl2, 1 mM DTT, 10 μM cytochalasin B, PMSF, and CLAP), pelleted again, and then resuspended in 10 volumes of NB. The cells were allowed to swell on ice for 20 min and then were gently broken with a Dounce homogenizer. The homogenate was layered onto 30% sucrose in NB and centrifuged at 4°C and 800 × g to pellet the nuclei. The supernatant was aspirated, and the nuclei were washed once in cold NB and then quantitated in a hemacytometer. The nuclei were either used immediately or stored for 1 to 3 days in nuclear storage buffer (10 mM PIPES [pH 7.4], 80 mM KCl, 10 mM NaCl, 250 mM sucrose, 5 mM EGTA, 1 mM DTT, 0.5 mM spermidine, 0.2 mM spermine, PMSF, CLAP, 50% glycerol).
The presence of PARP-cleaving activity in the extracts was determined as described elsewhere (21); 25 to 50 μg of extract was brought up to 10 to 15 μl with EPB and mixed with 106 purified HeLa cell nuclei that had been washed and resuspended in 10 to 15 μl of mitotic dilution buffer (10 mM HEPES [pH 7.0], 40 mM α-glycerophosphate, 50 mM NaCl, 2 mM MgCl2, 5 mM EGTA, 1 mM DTT, 2 mM ATP, 10 mM creatine phosphate, 50 μg of creatine kinase per ml). In each experiment, protein in the extracts was quantitated by the Bradford assay, and equivalent concentrations of protein were added to each reaction. Reaction mixtures were incubated at 37°C for 15 to 60 min, and the reactions were stopped by the addition of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Proteins in the samples were resolved on SDS–10% polyacrylamide gels, transferred to nitrocellulose by the use of a semidry blotter (Fisher), and detected with a monoclonal antibody (C-2-10; Clontech) that recognizes human PARP; this was followed by addition of horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG) (Southern Biotechnology Associates, Inc.) and enhanced chemiluminescence (ECL) analysis (Amersham).
In vitro expression of human lamin A and cleavage assay.
A cDNA clone containing the complete coding sequence for human lamin A (kindly provided by B. Burke, University of Calgary, Calgary, Alberta, Canada) was subcloned into the EcoRI-XbaI sites of the plasmid pAlter-Ex1 (Promega Corporation), oriented such that lamin A could be expressed from the T7 promoter. 35S-labelled lamin A was synthesized by using the T7 Quick Coupled Transcription/Translation System (Promega Corporation) in accordance with the instructions provided by the manufacturer.
The presence of lamin A-cleaving activity in the extracts was determined by adding 2 μl of 35S-labelled lamin A from a 25-μl transcription-translation reaction mixture to 15 μg of extract from either mock- or virus-infected LLC-PK1 cells and then incubating at 37°C for 90 min. Proteins were resolved on SDS–12% polyacrylamide gels, the radioactive signal was enhanced with Amplify (Amersham), and proteins were visualized by autoradiography.
Expression and purification of His-tagged CrmA and SPI-2.
CrmA and SPI-2 were purified as His-tagged proteins, produced by standard methods by using the vaccinia virus-T7 expression system (12). Briefly, the PCR-amplified open reading frames for CrmA and SPI-2 were inserted into a derivative of the plasmid pTM1 (30), prepared by inserting the oligohistidine tag from the plasmid pET-16b. Clones were verified by sequencing, and vaccinia virus (strain WR) recombinants in which the PT7-His-tagged protein coding sequences were inserted into the viral thymidine kinase gene were generated. High-level expression of the His-tagged proteins was achieved by coinfection of HeLa S3 cells with the vaccinia virus His-CrmA or His–SPI-2 virus and the vaccinia virus T7 RNA polymerase-expressing recombinant vTF7-3 (12). His-tagged proteins were purified from extracts of infected cells by affinity chromatography on nickel columns by standard procedures (16).
Generation of an anti-CrmA–anti-SPI-2 monoclonal antibody and detection of SPI-2 in extracts.
The monoclonal antibody 2B12-4B1 was produced by immunizing female BALB/c mice with purified His-tagged RPV SPI-2 protein, prepared as described above. Splenocytes from immunized animals were fused with a murine myeloma cell line. Fused cells were selected and screened for clones producing culture supernatant that was immunopositive for purified CrmA and SPI-2 proteins by both enzyme-linked immunosorbent assay and immunoblot analyses. Clone 2B12-4B1 was selected on the basis of its strong reactivity with both the CrmA and SPI-2 proteins. The antibody was column purified prior to use.
CrmA and SPI-2 were detected in extracts by immunoblotting by standard procedures. Proteins in extracts were resolved on SDS–12% polyacrylamide gels, transferred to nitrocellulose membranes by semidry blotting, and detected with antibody 2B12-4B1; this was followed by addition of HRP-conjugated goat anti-mouse IgG (Southern Biotechnology Associates, Inc.). Subsequently, ECL analysis (Amersham) was performed.
Detection of activated caspases in infected-cell extracts.
The processing of caspase-3 (CPP32) in infected-cell extracts was determined as follows. Extracts (15 to 30 μg each), prepared as described above, were resolved on SDS–15% polyacrylamide gels. Proteins were transferred to nitrocellulose membranes by semidry blotting, and caspase-3 (CPP32) was detected by addition of rabbit antiserum directed against the p17 subunit of human CPP32 (kindly provided by N. Thornberry, Merck Research Laboratories, Rahway, N.J.) followed by HRP-conjugated goat anti-rabbit IgG (Southern Biotechnology Associates, Inc.) and, subsequently, ECL analysis (Amersham).
Affinity labelling of activated caspases in extracts prepared as described above was conducted as follows. Extract (30 μg) from either mock- or virus-infected cells was incubated with 50 μM N-(acetyltyrosinylvalinyl-Nɛ-biotinyllysyl) aspartic acid [(2,6-dimethylbenzoyl)oxy] methyl ketone [YV(bio)KD-aomk] for 5 min at 37°C. Labelled proteins were resolved on SDS–16% polyacrylamide gels, transferred to nitrocellulose membranes, and detected by addition of streptavidin-HRP and subsequent performance of ECL analysis (Amersham).
ICE inhibition assay.
Purified His-tagged SPI-2 or CrmA (25 nM) was preincubated with 50 U (approximately 1 pmol) of purified recombinant human ICE (kindly provided by N. Thornberry, Merck Research Laboratories) for 5 min at room temperature in 1 ml of reaction buffer (100 mM HEPES [pH 7.5], 10% sucrose, 0.1% 3[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 10 mM DTT). The substrate Ac-Tyr-Val-Ala-Asp-7-amino-4-methylcoumarin (Ac-YVAD-AMC; Calbiochem, Inc.) was added to a concentration of 14 μM, and substrate cleavage was followed by fluorometry, with an excitation wavelength of 380 nm and an emission wavelength of 460 nm, using a Hoefer/Pharmacia DyNA Quant 200 fluorometer.
RESULTS
Morphological characteristics of LLC-PK1 cells infected with wt and CrmA/SPI-2 mutants of RPV and CPV.
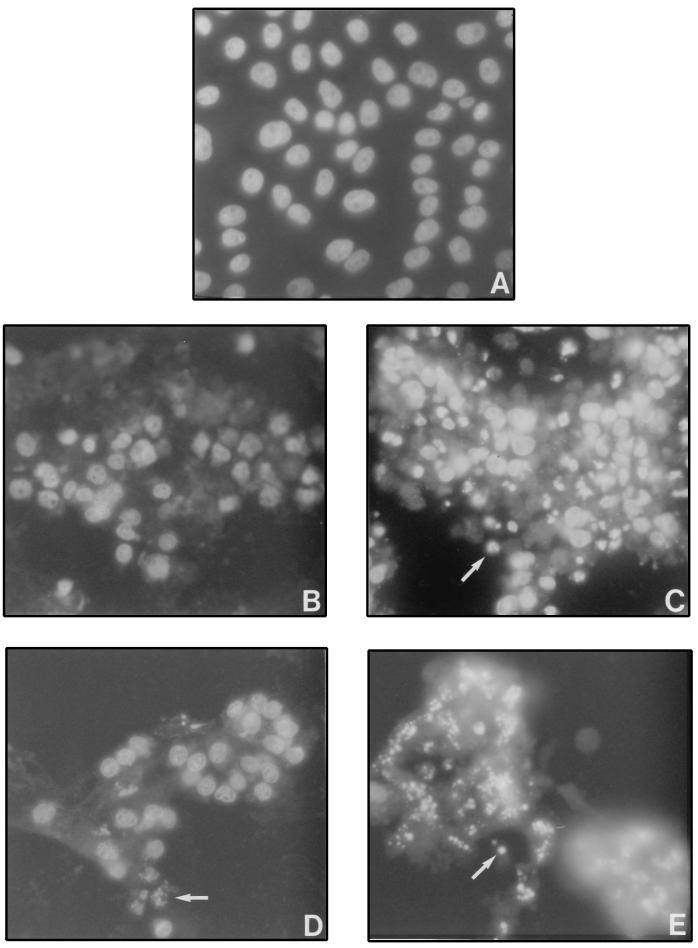
It has been shown that LLC-PK1 cells appear apoptotic when infected with CPVΔcrmA but not when infected with wt CPV (38). We confirmed this by staining the nuclei of infected cells with DAPI (Fig. 1B and C). RPVΔSPI-2 mutants behaved similarly to CPVΔcrmA, and by 16 h postinfection, many of the infected cells exhibited properties associated with apoptosis, such as densely staining apoptotic bodies (Fig. 1E). However, unlike cells infected with wt CPV, cells infected with wt RPV (Fig. 1D) (approximately 40% of the cells) also showed nuclear condensation and fragmentation by 16 h postinfection, consistent with apoptosis. Generally, we observed that RPV causes a more highly cytopathic infection than does CPV. There were more apoptotic nuclei in cells infected with RPVΔSPI-2 (∼60%) than in cells infected with wt RPV (∼40%) (compare Fig. 1E and D), and there was a larger proportion of apoptotic cells in RPVΔSPI-2 infections (∼60%) than in those caused by CPVΔcrmA (∼40%) (compare Fig. 1E and C). Apoptotic cells were only rarely observed in infections by wt CPV (less than 10%) (Fig. 1B). The finding that wt RPV induces apoptosis is surprising in view of the fact that RPV synthesizes a SPI-2 (CrmA) protein (7).
FIG. 1.
DAPI staining of CPV- or RPV-infected LLC-PK1 cells. Cells were mock infected (A) or infected with wt CPV (B), CPVΔcrmA (C), wt RPV (D), or RPVΔSPI-2 (E) and then stained with DAPI at 16 h postinfection as described in Materials and Methods. The arrows indicate examples of apoptotic nuclei.
PARP- and lamin A-cleaving activity in extracts prepared from infected LLC-PK1 cells.
We did studies to determine whether, in addition to the apoptosis-associated morphological changes observed in LLC-PK1 cells infected with CPVΔcrmA (38), the cells also manifest biochemical changes that normally occur during apoptosis, such as the activation of caspase activity leading to the cleavage of nuclear proteins, including PARP and lamins. Furthermore, we wanted to determine whether the RPV SPI-2 protein, which exhibits 93% amino acid identity to the CrmA protein, also functions to regulate apoptosis in LLC-PK1 cells infected with RPV. Using the in vitro apoptosis assay developed by Earnshaw and coworkers (20, 21), we prepared extracts from LLC-PK1 cells that had been either mock infected or infected with wt CPV, CPVΔcrmA, wt RPV, or RPVΔSPI-2 and tested them for the ability to cleave either PARP in purified HeLa cell nuclei or in vitro-transcribed- and -translated human lamin A.
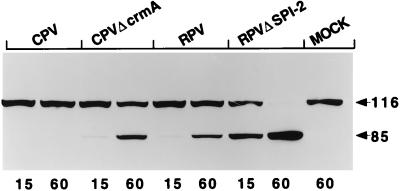
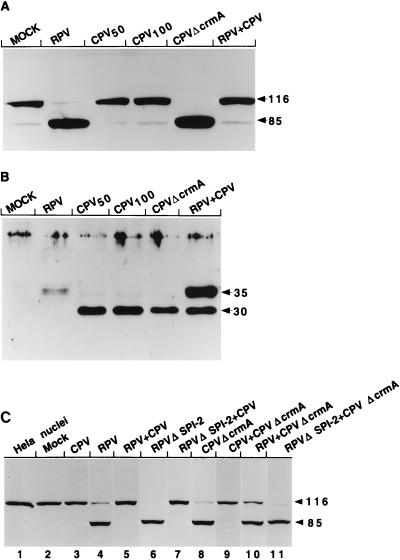
The data shown in Fig. 2 indicate that extracts prepared from LLC-PK1 cells infected with CPVΔcrmA, but not those from wt CPV-infected cells, contained activity that cleaved PARP from its native 116-kDa form to the signature 85-kDa fragment typically seen during apoptosis. This result suggests that the CPV CrmA protein blocks apoptosis in infected LLC-PK1 cells by either preventing the activation of PARP-cleaving activity or directly inhibiting the PARP-cleaving enzyme. Surprisingly, extracts prepared from wt RPV-infected LLC-PK1 cells also contained PARP-cleaving activity, unlike those prepared from wt CPV-infected cells (Fig. 2). In the case of RPV, the CrmA homolog SPI-2 alone is not sufficient to prevent PARP cleavage. However, extracts prepared from cells infected with RPVΔSPI-2 consistently resulted in more extensive cleavage of PARP than did extracts prepared from cells infected with wt RPV (Fig. 2), indicating that expression of the RPV SPI-2 protein has some role in reducing the extent of PARP cleavage. Infection of LLC-PK1 cells with RPVΔSPI-2 resulted in more PARP-cleaving activity than did infection with CPVΔcrmA (Fig. 2).
FIG. 2.
PARP-cleaving activity in infected LLC-PK1 cells. Extracts prepared from LLC-PK1 cells at 14 h postinfection were mixed with purified HeLa cell nuclei, and the mixtures were incubated for either 15 or 60 min (indicated by the numbers below the lanes) at 37°C. Proteins from the reaction were separated on SDS–10% polyacrylamide gels, and PARP was detected by immunoblotting with the anti-PARP monoclonal antibody C-2-10. The arrows indicate the intact 116-kDa molecule and the 85-kDa cleavage product. MOCK, mock-infected cell extract.
As shown in Fig. 3, extracts from LLC-PK1 cells infected with CPVΔcrmA, as well as those infected with wt RPV or RPVΔSPI-2, contained activity that cleaved human lamin A from an intact product, which migrates at approximately 80 kDa by SDS-PAGE, to 50- and 30-kDa fragments. This is consistent with the cleavage of lamin A at D230, mapped previously as the site at which lamin A becomes cleaved by caspase-6 (Mch2α) during apoptosis (41). Again no proteolytic activity was detected in extracts derived from wt CPV-infected cells. The presence of lamin A-cleaving activity in the extracts correlated exactly with the PARP-cleaving activity.
FIG. 3.
Human lamin A-cleaving activity in extracts from infected LLC-PK1 cells. Extracts prepared from LLC-PK1 cells at 14 h postinfection were mixed with 35S-labeled human lamin A, produced by in vitro transcription-translation, and incubated for 90 min at 37°C. The proteins were resolved by SDS–10% PAGE and visualized by autoradiography. The arrows indicate the two fragments resulting from lamin A cleavage. MOCK, mock-infected cell extract.
CrmA blocks proteolytic activity upstream of the PARP-cleaving activity in infected LLC-PK1 cells.
To determine whether the CPV CrmA protein present in extracts of infected LLC-PK1 cells is able to directly inhibit the activity of the PARP-cleaving proteinase in the extracts, the in vitro apoptosis assay was performed on mixtures of extracts derived from infected cells. We reasoned that if the CrmA present in extracts from cells infected with wt CPV is able to directly inhibit the enzyme that cleaves PARP in the assay, then PARP cleavage should be prevented when wt CPV extracts are incubated with extracts prepared from either CPVΔcrmA- or RPV-infected cells prior to addition to the purified HeLa cell nuclei.
The results of this experiment are shown in Fig. 4. Extracts or mixtures of extracts were either preincubated for 30 min at 37°C and then added to purified HeLa cell nuclei (Fig. 4, odd-numbered lanes) or added to HeLa cell nuclei directly with no preincubation (Fig. 4, even-numbered lanes) and then incubated a further 60 min at 37°C to allow PARP cleavage to occur. Preincubation of extracts prepared from mock-infected cells or cells infected with either RPV, CPV, or CPVΔcrmA did not affect PARP-cleaving activity present in the extracts (lanes 1 through 8). Furthermore, mixing extracts from CPV-infected cells with extracts from cells infected with either RPV (lanes 9 and 10) or CPVΔcrmA (lanes 13 and 14) did not inhibit PARP cleavage by either the RPV or the CPVΔcrmA extract, indicating that the CrmA present in the CPV extract was not able to inhibit the PARP-cleaving activity. Although a possible explanation for this result is that all of the CrmA present in the extract is already complexed with or cleaved by the enzyme(s) that it inhibits, this is not likely to be the case since uncleaved CrmA is present in the extracts (see Fig. 7 below) and no higher-molecular-weight complexes containing CrmA can be detected on native polyacrylamide gels (data not shown). Therefore, a more plausible explanation is that CrmA does not directly inhibit the PARP-cleaving enzyme in the extracts but rather blocks the activation of this activity by inhibiting a caspase that functions further upstream in the apoptotic proteolytic cascade.
FIG. 4.
Extracts from wt CPV-infected cells do not inhibit PARP-cleaving activity. Extracts prepared from infected LLC-PK1 cells at 14 h postinfection were mixed with purified HeLa cell nuclei, and the mixtures were incubated for 60 min at 37°C. Proteins from the reaction were separated by SDS–10% PAGE, and PARP was detected by immunoblotting with the monoclonal antibody C-2-10. In the odd-numbered lanes are extracts prepared from singly infected cells, mixed and preincubated at 37°C for 30 min before the nuclei were added. In the even-numbered lanes are the nuclear extracts that were mixed and added directly to the nuclei with no preincubation. The arrows indicate the intact 116-kDa molecule and the 85-kDa cleavage product. MOCK, mock-infected cell extract.
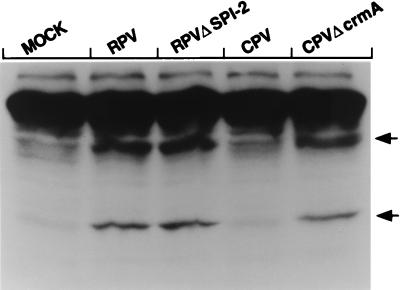
FIG. 7.
Expression of CrmA and SPI-2 in infected LLC-PK1 cells. LLC-PK1 cells were infected with either wt RPV or wt CPV at an MOI of 5 PFU/cell (RPV) or 25 PFU/cell (CPV). At the times indicated (in hours) above the lanes, extracts were prepared from the infected cells and an equal amount of protein from each was loaded onto an SDS–12% polyacrylamide gel. SPI-2 and CrmA were detected with the SPI-2- and CrmA-specific monoclonal antibody 2B12-4B1. The arrow indicates the position of the 38-kDa SPI-2/CrmA protein. In the RPV-infected cells, a lower-molecular-mass protein is also occasionally visualized at late times and may represent a minor cross-reacting protein or a further degradation product of SPI-2.
When an extract prepared from RPV-infected cells was preincubated with an extract from CPVΔcrmA-infected cells, we observed an increase in PARP-cleaving activity compared to that observed when the extracts were added directly to the HeLa cell nuclei without any preincubation (Fig. 3; compare lanes 11 and 12). This result suggests the possibility that the mixing of these extracts actually caused an increase in the amount of activation of the PARP-cleaving enzyme, possibly due to differences in the proteolytic activities that become activated in the cells. It is therefore possible that the induction of apoptotic proteolytic activity during infection of LLC-PK1 cells with RPV is somewhat different than that induced during infection with CPV.
Processing of CPP32 (caspase-3) in virus-infected LLC-PK1 cells.
Although CrmA and SPI-2 exhibit 93% overall amino acid identity and, within the reactive site loop, differ only at the P5 and P6 positions (3), it is formally possible that the two proteins possess differing inhibitory specificities with respect to caspases that become activated during virus-induced apoptosis. If this is the case, we might expect to see some difference in the spectra of caspases that become activated during the course of the infection with the different viruses. We therefore analyzed extracts from virus-infected LLC-PK1 cells by immunoblotting with a polyclonal antiserum that recognizes the p18 subunit of caspase-3 (CPP32), which is one of the proteinases capable of PARP cleavage during apoptosis.
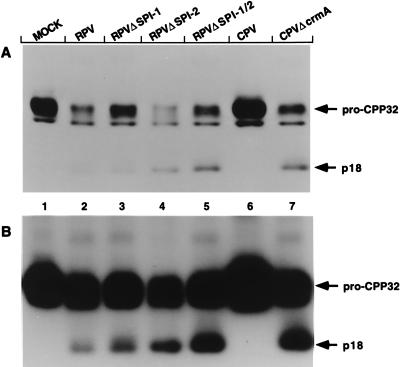
The results of immunoblotting for caspase-3 are shown in Fig. 5. The unprocessed form of the enzyme (pro-cpp32) is best seen in the blot subjected to a shorter exposure time (Fig. 5A) and is indicated by the uppermost arrow. The processed p18 subunit is most easily visualized in the blot subjected to a longer exposure period (Fig. 5B). Correlating with the presence of PARP-cleaving activity, we found that extracts prepared from cells infected with wt RPV, RPVΔSPI-2, or CPVΔcrmA all contained an immunopositive band migrating at approximately 18 kDa (Fig. 5B, lanes 2, 4, and 7), indicative of processing of the porcine homolog of caspase-3 zymogen to the activated form of the enzyme. There was no effect in either RPV or the RPVΔSPI-2 mutant upon further deletion of the related SPI-1 gene (Fig. 5, lanes 3 and 5) despite the fact that the SPI-1 gene has also been linked to control of apoptosis (7). There was no evidence of caspase-3 activation in either mock-infected cells or cells infected with wt CPV, even with the longer-exposure blot (Fig. 5B, lanes 1 and 6). Furthermore, we consistently noted that more processed caspase-3 was present in the extract from RPVΔSPI-2-infected cells than in that of cells infected with wt RPV (Fig. 5, lanes 2 and 4). This suggests that the SPI-2 protein may indeed have some function in the inhibition of the activation of caspase-3 (and hence PARP cleavage), but that this inhibition may be incomplete. Alternatively, it is possible that more than one upstream proteinase is involved in caspase-3 activation, and SPI-2 may not be able to inhibit both of them.
FIG. 5.
Processing of caspase-3 (CPP32) in extracts from infected LLC-PK1 cells. LLC-PK1 cells were either mock infected (MOCK) or infected with one of the following: RPV, RPV with a mutation in either the SPI-1 gene (a serpin with approximately 50% identity to SPI-2), the SPI-2 gene, or both SPI-1 and SPI-2; wild-type CPV; or the CPV CrmA mutant. Extracts prepared at 14 h postinfection were resolved on an SDS–15% polyacrylamide gel, and caspase-3 was detected by immunoblotting with a polyclonal rabbit antiserum directed against the p17 subunit of human caspase-3. (A) Blot with short exposure period; (B) longer exposure of the same blot shown in panel A. The arrows indicate the positions of the unprocessed proenzyme from porcine cells (pro-CPP32) and the cleaved subunit which runs at 18 kDa on SDS gels (p18).
Labelling of active caspases with a biotinylated tetrapeptide inhibitor.
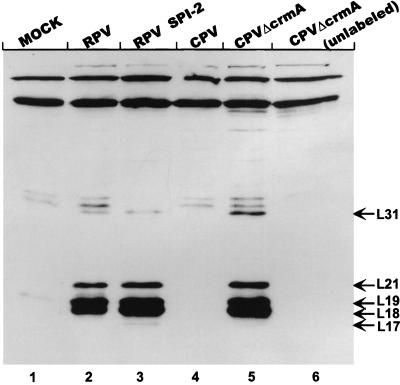
Extracts were analyzed for the presence and spectrum of other activated caspases by labelling them with the biotinylated synthetic peptide (acyloxy)methyl ketone inhibitor Ac-YV(bio)KD-aomk (47), which has been demonstrated to label at least five activated caspases in extracts from apoptotic cells (41). AcYV(bio)KD-aomk binds irreversibly to the larger subunits of a number of different caspases. We chose this approach in part because antisera to the wide variety of caspases now identified (4) are not readily available. The results of the peptide labelling are presented in Fig. 6. As expected, no activated caspases were detected in extracts prepared from mock-infected LLC-PK1 cells (lane 1). Furthermore, no caspases were labelled in extracts derived from cells infected with wt CPV (lane 4). In contrast, in extracts prepared from cells infected with RPV, RPVΔSPI-2, or CPVΔcrmA, three major activated caspases were detected (lanes 2, 3, and 5). In addition, minor bands appeared at 31 kDa (RPV, RPVΔSPI-2, and CPVΔcrmA) and 17 kDa (RPVΔSPI-2). Immunoblotting of the same samples led to the identification of the band labelled L18 as caspase-3 (CPP32) (data not shown). In agreement with the results of immunoblotting with anti-caspase-3 presented in Fig. 5, we noted that the band corresponding to L18 was more abundant in RPVΔSPI-2-derived extracts than in those corresponding to wt RPV. However, there was no overall difference in the patterns of major bands corresponding to the different caspases, suggesting that within the limits of the detection system used, RPV SPI-2 protein was not able to block the activation of caspases induced in infected cells. There remains the possibility that RPV activates a SPI-2/CrmA-insensitive proteinase that leads to caspase activation but is not detectable with available reagents.
FIG. 6.
Labelling of activated caspases in infected-cell extracts with Ac-YV(bio)KD-aomk. Lanes 1 to 5, extracts prepared from mock-infected (MOCK) or infected LLC-PK1 cells at 14 h postinfection, labelled with the biotinylated tetrapeptide inhibitor Ac-YV(bio)KD-aomk as described in Materials and Methods; lane 6, extract from CPVΔcrmA-infected cells incubated in the absence of the tetrapeptide. Proteins from the reactions were resolved on SDS–16% polyacrylamide gels. Labelled caspases were detected with streptavidin-HRP followed by ECL. Bands corresponding to labelled caspases were designated L followed by numbers indicating their approximate molecular masses (kilodaltons) and are indicated with arrows. This figure was prepared by using Adobe Photoshop 3.0 software for the Macintosh computer and an Epson GT-9500 scanner.
Expression of CPV CrmA and RPV SPI-2 in infected LLC-PK1 cells.
Because RPV encodes a homolog of the CPV CrmA protein, the finding that LLC-PK1 cells infected with wt RPV contain apoptotic proteolytic activity which does not become activated in the context of CPV infection in the presence of the CrmA protein was somewhat unexpected. A trivial explanation for this result could be that RPV simply does not express a significant amount of the SPI-2 protein or that the kinetics of SPI-2 expression differs from that of CPV CrmA. We therefore compared the expression of CrmA and SPI-2 in infected LLC-PK1 cells at various times postinfection. Equal amounts of protein from extracts prepared from LLC-PK1 cells infected for various amounts of time with either CPV or RPV were loaded onto an SDS-12% polyacrylamide gel, and CrmA/SPI-2 was detected by immunoblotting with the anti-crmA and anti-SPI-2 monoclonal antibody 2B12-4B1. This monoclonal antibody has a comparable avidity for each of these proteins. The results of this experiment are shown in Fig. 7. RPV was found to express levels of SPI-2 that are comparable to, if not slightly higher, than the amount of CrmA expressed during infection with CPV. RPV SPI-2 was detectable as early as 2.5 h postinfection (Fig. 7) and accumulated throughout the infection. A similar pattern of expression was observed for CPV CrmA, although there was somewhat less protein seen at the early time points. This pattern of expression is consistent with the observation that the SPI-2 and crmA genes are controlled by sequences conforming to early viral promoters and further suggests that the SPI-2 and CrmA proteins are stable within the cells during the course of the infection. From these data we can conclude that the differences in the activation of proteolytic activity seen in extracts prepared from CPV-infected LLC-PK1 cells and in those infected with RPV cannot be explained by differences in the expression of the CrmA and SPI-2 proteins.
Comparable inhibition of ICE by CrmA and SPI-2.
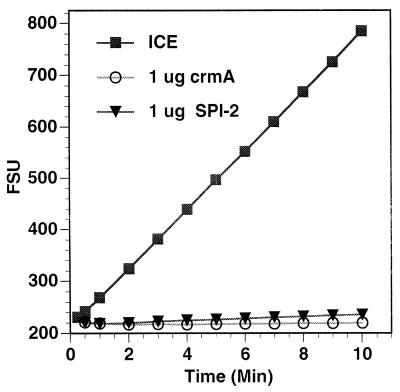
The first biochemical function ascribed to the CrmA protein was the inhibition of ICE (caspase-1) (37). Because the results presented above suggest that there may be functional differences between the CPV CrmA protein and the RPV SPI-2 protein, we tested the purified viral proteins directly for their ability to inhibit the activity of ICE against a fluorogenic peptide substrate in vitro. The data presented in Fig. 8 clearly indicate that there is no significant difference in the abilities of purified CrmA and SPI-2 to inhibit the activity of ICE in this assay. Thus, our results cannot be explained by assuming that the RPV protein is simply nonfunctional as a proteinase inhibitor. However, it remains possible that the slight differences in amino acid sequence in critical regions of the protein, such as the P5 and P6 residues in the reactive-site loop, define differing inhibitory specificities toward other members of the caspase family of proteinases, resulting in differing abilities to regulate apoptosis during virus infection. This possibility is currently under investigation.
FIG. 8.
Inhibition of ICE by both purified CrmA and purified SPI-2. His-tagged CrmA and SPI-2 proteins were purified as described in Materials and Methods, preincubated with purified human recombinant ICE for 5 min, and then incubated with a fluorogenic ICE substrate (Ac-YVAD-AMC). Fluorescence readings were taken at the indicated times and plotted as arbitrary fluorescence signal units (FSU).
CPV can block the activation of PARP-cleaving activity by RPV in LLC-PK1 cells coinfected with CPV and RPV.
A possible explanation for the above-presented results is that the apoptotic signal induced by RPV, but not that induced by CPV, activates a CrmA/SPI-2-insensitive activity that can go on to activate the PARP- and lamin A-cleaving enzymes in the infected cells. Alternatively, it is also possible either that the RPV SPI-2 protein is not functionally equivalent to the CrmA protein or that CPV encodes another factor which in addition to CrmA is necessary to block the apoptotic cascade in these cells. To investigate these possibilities, we prepared extracts from LLC-PK1 cells coinfected with both CPV and RPV and assayed for PARP-cleaving activity by the in vitro apoptosis assay. Here, we reasoned that if CPV coinfection cannot block the activation of PARP-cleaving activity, then it is likely that RPV activates a CrmA/SPI-2-insensitive apoptotic pathway in infected LLC-PK1 cells. However, if the activation of PARP-cleaving activity is blocked, then SPI-2 is not functionally equivalent to CrmA and/or another CPV-encoded factor in addition to CrmA/SPI-2 is required to prevent apoptosis.
The data presented in Fig. 9A clearly show that extracts prepared from cells coinfected with CPV and RPV were unable to cleave PARP in this assay. To exclude the possibility that CPV infection somehow prevented coinfection with RPV, the same extracts were assayed by immunoblotting for the expression of the orthopoxvirus chemokine-binding protein, which is expressed as an early protein by both CPV and RPV (14). Because the CPV and RPV versions of the chemokine-binding protein migrate differently on SDS-polyacrylamide gels (30 and 35 kDa, respectively [31a]), this protein could be used as a marker to determine whether both viruses are expressing early proteins within the same cells. Figure 9B clearly shows that both the CPV and RPV homologs of the chemokine-binding protein are expressed in the coinfected cells. Together, the results suggest that it is not likely that RPV activates a CrmA/SPI-2-insensitive pathway in the infected cells. Therefore, the SPI-2 protein either has an inhibitory specificity different from that of the CrmA protein or requires a CPV-encoded cofactor in order to block apoptosis in infected LLC-PK1 cells.
FIG. 9.
PARP-cleaving activity in LLC-PK1 cells coinfected with RPV and CPV derivatives. Extracts were prepared from cells that had been either mock infected (MOCK), singly infected with RPV (MOI = 10 [A and B] or 15 [C]), CPV (MOI = 50 [CPV50] [A and B], 100 [CPV100] [A and B], or 15 [C]), CPVΔcrmA (MOI = 15 [A and B] or 5 [C]), or RPVΔSPI-2 (MOI = 5 [C]), or dually infected with two of the following: CPV (MOI = 15), RPV (MOI = 15), CPVΔcrmA (MOI = 5), and RPVΔSPI-2 (MOI = 5). All samples were harvested at 14 h postinfection. (A) Extracts were assayed for PARP-cleaving activity, using purified HeLa cell nuclei and immunoblotting with the anti-PARP monoclonal antibody. The arrows indicate the intact 116-kDa molecule and the 85-kDa cleavage fragment. (B) Extracts were separated on an SDS–12% polyacrylamide gel and immunoblotted with a polyclonal rabbit antiserum specific for the orthopoxvirus chemokine-binding protein. The CPV version of this protein migrates at approximately 30 kDa, while the RPV homolog migrates at 35 kDa, as indicated by the arrows. (C) PARP immunoblot of HeLa cell nuclei that either were untreated (lane 1) or had been incubated with extracts from mock-infected (lane 2) or infected (lanes 3 to 11) cells. The viruses used to generate the various extracts are indicated above the lanes.
The questions of whether SPI-2 and CrmA are equivalent and whether CPV encodes a second factor, in addition to CrmA, to prevent apoptosis were approached genetically through a series of mixed infections with wt and SPI-2/CrmA mutants of RPV and CPV (Fig. 9C). PARP cleavage induced by either RPVΔSPI-2 or CPVΔcrmA could be completely reversed by coinfection with wt CPV (lanes 7 and 9, respectively). However, the apoptosis induced by CPVΔCrmA infections could not be eliminated by coinfection with wt RPV (lane 10); i.e., the properties of the extracts most closely resemble those of cells singly infected with wt RPV. Control mixed infections with RPVΔSPI-2 and CPVΔCrmA mutants (lane 11) yielded high levels of PARP cleavage, as expected. These results are most easily explained by functional differences between SPI-2 and CrmA and the fact that SPI-2, unlike CrmA, is incapable of completely blocking apoptosis within the context of a virus infection.
DISCUSSION
The CPV CrmA protein is an important natural regulator of apoptosis. CrmA is capable of inhibiting apoptosis induced by a variety of signals in transfected mammalian cells (see, for example, references 6, 10, 13, 24, 28, and 44), as well as during CPV infection of LLC-PK1 cells (38). Nevertheless, the mechanism by which CPV induces apoptosis during infection and the specific cellular proteins with which CrmA interacts to prevent apoptosis have yet to be elucidated. Furthermore, the role that the CrmA-mediated inhibition of apoptosis plays during the course of viral infection in vivo is not clear. Whereas disruptions in the CPV crmA gene as well as in the highly similar RPV SPI-2 gene have been shown to result in an increased inflammatory response in infected chicken CAMs (3), it is not known whether any of the infected cells undergo apoptosis. Studies of BALB/c mice inoculated intranasally with either CPV, vaccinia virus, or RPV CrmA/SPI-2 mutants are equivocal as to whether deletion of the gene leads to attenuation (17, 45).
To gain a more complete understanding of the regulation of apoptosis during viral infection, we undertook a more detailed investigation of the infection of LLC-PK1 cells with wt CPV and a CPV crmA mutant (CPVΔcrmA). In addition, we also examined the phenotype of cells infected with the related orthopoxvirus RPV and an RPV SPI-2 gene (the homolog of the crmA gene) mutant (RPVΔSPI-2).
In agreement with earlier results (38), we found that LLC-PK1 cells infected with CPVΔcrmA, but not those infected with wt CPV, showed morphological features of apoptosis (Fig. 1). We showed here that extracts prepared from the infected cells contained activated caspase activity that resulted in the proteolytic cleavage of the nuclear enzyme PARP in vitro (Fig. 2), as well as in vitro-transcribed and -translated human lamin A (Fig. 3). Extract mixing experiments (Fig. 4) suggested that CrmA blocked PARP cleavage indirectly by inhibiting an activating proteinase further upstream in the apoptotic proteolytic cascade. In experiments using a peptide inhibitor (Fig. 6), we found that at least five caspases, including the porcine homolog of caspase-3 (CPP32), became activated during CPVΔcrmA infection. During wt CPV infection, the CrmA protein was sufficient to block the activation of all of these caspases, suggesting that the cellular target for CrmA is a proapoptotic proteinase that lies at or near the apex of the presumed proteolytic cascade. Indeed, CrmA has been shown to be an effective inhibitor of caspase-8 (FLICE) (40, 42, 49), which is believed to act very early in the apoptotic proteinase cascade following engagement of the Fas or TNF receptors (5, 31). Our results are consistent with the hypothesis that the cellular target for the CrmA protein is a key caspase such as caspase-8 or a closely related enzyme that regulates the activation of the apoptotic program.
We were surprised to find that unlike LLC-PK1 cells infected with wt CPV or CPVΔcrmA, both wt RPV- and RPVΔSPI-2-infected cells underwent apoptosis, as judged by nuclear morphology and several biochemical criteria. Infection with RPVΔSPI-2 consistently resulted in an increased amount of processed caspase-3/CPP32 compared with wt RPV infection (Fig. 5), concomitant with an increased activation of PARP-cleaving activity (Fig. 2). It is important to note that RPV and RPVΔSPI-2 viruses both produce plaques on LLC-PK1 cells. Examination of the expression of both the CrmA and SPI-2 proteins during LLC-PK1 cell infection with either CPV or RPV (Fig. 7) indicated that the proteins were expressed to similar levels and with similar kinetics. In addition, when cells were coinfected with equivalent amounts of CPV and RPV, we were unable to detect proteolytic activity against PARP (Fig. 9A), indicating that the RPV-induced apoptosis of infected LLC-PK1 cells can be inhibited by CPV.
Taken together, the results presented above suggest at least two possible hypotheses. The first possibility is that the RPV SPI-2 protein is not functionally equivalent to the CPV CrmA protein. However, it is clear that the purified SPI-2 protein was able to inhibit the proteolytic activity of caspase-1 (ICE) in a fashion nearly identical to that of the purified CrmA protein in an in vitro assay (Fig. 8). While SPI-2 is a fully functional inhibitor of caspase-1, we cannot exclude the possibility that SPI-2 has an inhibitory specificity different from that of CrmA with respect to other members of the caspase family. Indeed, SPI-2 does not appear to be as effective as CrmA at inhibiting the activity of caspase-8 (42). SPI-2 and CrmA exhibit 93% amino acid identity, including an aspartic acid in the P1 position of the predicted reactive-site loop, the region of the serpin molecule that is critical for determining the inhibitory specificity. Nevertheless, the two proteins do differ at positions P6 and P5 (CA in CPV; SV in RPV) as well as at a number of sites in the serpin backbone. Although these represent fairly conservative substitutions, these differences may contribute to the inhibitory specificity of the proteins. Indeed, a recent study of a number of caspases indicated that at least one caspase relies on substrate and inhibitor residues as far away from the cleavage site as P5 (26), suggesting that there may be more specificity among the natural substrates and inhibitors of the caspases than has been suggested by studies relying on the use of tetrapeptide substrates and inhibitors (26).
A second hypothesis predicts that CPV encodes an additional antiapoptotic protein absent from RPV and that CrmA/SPI-2 is necessary, but not sufficient, for inhibition of apoptosis during LLC-PK1 cell infection. This factor or protein would then be required in addition to CrmA for inhibition of apoptosis during infection. In this model, RPV would be unable to block apoptosis during infection despite producing a functional homolog of the CrmA protein. If the RPV-infected cells were coinfected with CPV, the cells would be prevented from undergoing apoptosis since this factor would be supplied by CPV.
Distinguishing between these two possibilities can be accomplished by swapping the two genes between the viruses. If the SPI-2 protein indeed has a somewhat different inhibitory specificity than the CrmA protein, then we would expect that LLC-PK1 cells infected with CPV expressing the RPV SPI-2 protein in place of the CrmA protein would undergo apoptosis while cells infected with RPV expressing CrmA would not. However, if CrmA-mediated inhibition of apoptosis during infection requires an additional CPV-encoded factor, then inhibition of apoptosis will only be observed in the context of a CPV infection.
A second approach to distinguish between these two models is to examine cells infected with different combinations of wt and SPI-2/CrmA mutant viruses (Fig. 9). All apoptosis induced by RPV and RPVΔSPI-2 can be alleviated by coinfection with wt CPV (Fig. 9A and C). However, coinfection of CPVΔcrmA and wt RPV generates an infection with induction of apoptosis at levels reminiscent of that generated by infection with wt RPV alone. This result suggests that there is not a second gene unique to CPV which together with CrmA serves to block apoptosis. Rather, the most likely explanation for our results is that the SPI-2 and CrmA proteins are not functionally equivalent and that the SPI-2 protein of RPV is not as effective as the CrmA protein in controlling apoptosis.
If we consider these results in light of the distinctive pathobiologies observed for the two viruses during infection in animal models (chicken CAMs and BALB/c mice), some interesting correlations can be drawn. First, both wt CPV and wt RPV produce red pocks on chicken CAM, but those of wt RPV are not as red as those of CPV and frequently appear pink. Both the CPVΔcrmA and RPVΔSPI-2 viruses produce white pocks (3). Thus, both the CrmA and SPI-2 proteins function to regulate inflammation during infection, a function that is most likely mediated by the regulation of the proteolytic activation of IL-1β by inhibition of caspase-1 (ICE). Our results showing comparable inhibition of caspase-1 activity in the in vitro assay (Fig. 8) are in complete agreement with such a scheme. Second, although the mutation of CrmA/SPI-2 in both CPV and RPV resulted in altered pulmonary pathology in intranasally infected BALB/c mice, infection with CPVΔcrmA resulted in reduced inflammation while infection with RPVΔSPI-2 resulted in increased inflammation (45). Furthermore, during infection with the wt viruses, CPV infection resulted in an overall greater influx of inflammatory cells than did infection with wt RPV (45). If we reexamine these phenomena in light of the observations presented here, it is conceivable that in the context of a wt CPV infection, the infected pulmonary epithelial cells die by necrosis and thereby elicit a higher-level inflammatory response (in essence negating the anti-inflammatory effects of the crmA gene) than would be expected if the cells died by apoptosis during infection with the CrmA mutant. Conversely, in the case of wt RPV infection, if the infected cells die by apoptosis, one would expect to observe a lesser degree of inflammation. In this context, SPI-2 would function only in the regulation of the activation of IL-1β, and therefore, when SPI-2 is inactivated in the context of RPV infection, an increase in inflammation is observed relative to that produced by the wt virus. Clearly, understanding the distinctive pathologies of infection with these viruses requires a more comprehensive understanding of the interactions between the viruses and the host cells. Elucidation of the functions of viral proteins such as CrmA and SPI-2 in poxvirus infection will facilitate a better understanding of the roles that both inflammation and apoptosis play during acute viral pathogenesis.
ACKNOWLEDGMENTS
We thank Mike Duke for technical assistance. A.T. also acknowledges the support and encouragement of M. Sasada of Kyoto University.
This work was supported by grant AI-15722 from the National Institutes of Health. A.T. is a Research Fellow for the Japan Foundation for Aging and Health.
REFERENCES
- 1.Alcami A, Smith G L. Cytokine receptors encoded by poxviruses: a lesson in cytokine biology. Immunol Today. 1995;16:474–478. doi: 10.1016/0167-5699(95)80030-1. [DOI] [PubMed] [Google Scholar]
- 2.Alcami A, Smith G L. Soluble interferon-gamma receptors encoded by poxviruses. Comp Immunol Microbiol Infect Dis. 1996;19:305–317. doi: 10.1016/0147-9571(96)00013-6. [DOI] [PubMed] [Google Scholar]
- 3.Ali A N, Turner P C, Brooks M A, Moyer R W. The SPI-1 gene of rabbitpox virus determines host range and is required for hemorrhagic pock formation. Virology. 1994;202:306–314. doi: 10.1006/viro.1994.1347. [DOI] [PubMed] [Google Scholar]
- 4.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Human ICE/CED-3 protease nomenclature. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 5.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 6.Boudreau N, Sympson C J, Werb Z, Bissell M J. Suppression of ICE and apoptosis in mammary epithelial cells by extracellular matrix. Science. 1995;267:891–893. doi: 10.1126/science.7531366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks M A, Ali A N, Turner P C, Moyer R W. A rabbitpox virus serpin gene controls host range by inhibiting apoptosis in restrictive cells. J Virol. 1995;69:7688–7698. doi: 10.1128/jvi.69.12.7688-7698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chua T P, Smith C E, Reith R W, Williamson J D. Inflammatory responses and the generation of chemoattractant activity in cowpox virus-infected tissues. Immunology. 1990;69:202–208. [PMC free article] [PubMed] [Google Scholar]
- 9.Darmon A J, Nicholson D W, Bleackley R C. Activation of the apoptotic protease CPP32 by cytotoxic T-cell derived granzyme B. Nature. 1996;377:446–448. doi: 10.1038/377446a0. [DOI] [PubMed] [Google Scholar]
- 10.Enari M, Hug H, Nagata S. Involvement of an ICE-like protease in Fas-mediated apoptosis. Nature. 1995;375:78–81. doi: 10.1038/375078a0. [DOI] [PubMed] [Google Scholar]
- 11.Fredrickson T N, Sechler J M, Palumbo G J, Albert J, Khairallah L H, Buller R M. Acute inflammatory response to cowpox virus infection of the chorioallantoic membrane of the chick embryo. Virology. 1992;187:693–704. doi: 10.1016/0042-6822(92)90472-2. [DOI] [PubMed] [Google Scholar]
- 12.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagliardini V, Fernandez P A, Lee R K, Drexler H C, Rotello R J, Fishman M C, Yuan J. Prevention of vertebrate neuronal death by the crmA gene. Science. 1994;263:826–828. doi: 10.1126/science.8303301. [DOI] [PubMed] [Google Scholar]
- 14.Graham K A, Lalani A S, Macen J L, Ness T L, Barry M, Liu L Y, Lucas A, Clark-Lewis I, Moyer R W, McFadden G. The T1/35kDa family of poxvirus secreted proteins bind chemokines and modulate leukocyte influx into virus infected tissues. Virology. 1997;229:12–24. doi: 10.1006/viro.1996.8423. [DOI] [PubMed] [Google Scholar]
- 15.Ink B S, Gilbert C S, Evan G I. Delay of vaccinia virus-induced apoptosis in nonpermissive Chinese hamster ovary cells by the cowpox virus CHOhr and adenovirus E1B 19K genes. J Virol. 1995;69:661–668. doi: 10.1128/jvi.69.2.661-668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janknecht R, de Martynoff G, Lou J, Hipskind R A, Nordheim A, Stunnenberg H G. Rapid and efficient purification of native histidine-tagged protein expressed by recombinant vaccinia virus. Proc Natl Acad Sci USA. 1991;88:8972–8976. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kettle S, Blake N W, Law K M, Smith G L. Vaccinia virus serpins B13R (SPI-2) and B22R (SPI-1) encode Mr 38.5 and 40K, intracellular polypeptides that do not affect virus virulence in a murine intranasal model. Virology. 1995;206:136–147. doi: 10.1016/s0042-6822(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 18.Kotwal G J, Isaacs S N, McKenzie R, Frank M M, Moss B. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science. 1990;250:827–830. doi: 10.1126/science.2237434. [DOI] [PubMed] [Google Scholar]
- 19.Kotwal G J, Moss B. Vaccinia virus encodes a secretory polypeptide structurally related to complement control proteins. Nature. 1988;335:176–178. doi: 10.1038/335176a0. [DOI] [PubMed] [Google Scholar]
- 20.Lazebnik Y A, Cole S, Cooke C A, Nelson W G, Earnshaw W C. Nuclear events of apoptosis in vitro in cell-free mitotic extracts: a model system for analysis of the active phase of apoptosis. J Cell Biol. 1993;123:7–22. doi: 10.1083/jcb.123.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazebnik Y A, Kaufmann S H, Desnoyers S, Poirier G G, Earnshaw W C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 22.Lee S B, Esteban M. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology. 1994;199:491–496. doi: 10.1006/viro.1994.1151. [DOI] [PubMed] [Google Scholar]
- 23.Lomas D A, Evans D L, Upton C, McFadden G, Carrell R W. Inhibition of plasmin, urokinase, tissue plasminogen activator, and C1S by a myxoma virus serine proteinase inhibitor. J Biol Chem. 1993;268:516–521. [PubMed] [Google Scholar]
- 24.Los M, Van de Craen M, Penning L C, Schenk H, Westendorp M, Baeuerle P A, Droge W, Krammer P H, Fiers W, Schulze-Osthoff K. Requirement of an ICE/CED-3 protease for Fas/APO-1-mediated apoptosis. Nature. 1995;375:81–83. doi: 10.1038/375081a0. [DOI] [PubMed] [Google Scholar]
- 25.Macen J L, Graham K A, Lee S F, Schreiber M, Boshkov L K, McFadden G. Expression of the myxoma virus tumor necrosis factor receptor homologue and M11L genes is required to prevent virus-induced apoptosis in infected rabbit T lymphocytes. Virology. 1996;218:232–237. doi: 10.1006/viro.1996.0183. [DOI] [PubMed] [Google Scholar]
- 26.Margolin N, Raybuck S A, Wilson K P, Chen W, Fox T, Gu Y, Livingston D J. Substrate and inhibitor specificity of interleukin-1 beta-converting enzyme and related caspases. J Biol Chem. 1997;272:7223–7228. doi: 10.1074/jbc.272.11.7223. [DOI] [PubMed] [Google Scholar]
- 27.McFadden G. DNA viruses that affect cytokine networks. In: Aggarwal B B, Puri R K, editors. Human cytokines: their role in health and disease. Cambridge, Mass: Blackwell Scientific; 1994. pp. 404–422. [Google Scholar]
- 28.Miura M, Zhu H, Rotello R, Hartwieg E A, Yuan J. Induction of apoptosis in fibroblasts by IL-1β converting enzyme, a mammalian homolog of the C. elegans cell death gene CED-3. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 29.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2637–2672. [Google Scholar]
- 30.Moss B, Elroy Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 31.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 31a.Ness, T. L., and R. W. Moyer. Unpublished data.
- 32.Palumbo G J, Pickup D J, Fredrickson T N, McIntyre L J, Buller R M. Inhibition of an inflammatory response is mediated by a 38-kDa protein of cowpox virus. Virology. 1989;172:262–273. doi: 10.1016/0042-6822(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 33.Pickup D J. Poxviral modifiers of cytokine responses to infection. Infect Agents Dis. 1994;3:116–127. [PubMed] [Google Scholar]
- 34.Pickup D J, Ink B S, Hu W, Ray C A, Joklik W K. Hemorrhage in lesions caused by cowpox virus is induced by a viral protein that is related to plasma protein inhibitors of serine proteases. Proc Natl Acad Sci USA. 1986;83:7698–7702. doi: 10.1073/pnas.83.20.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quan L T, Caputo A, Bleackley R C, Pickup D J, Salvesen G S. Granzyme B is inhibited by the cowpox virus serpin cytokine response modifier A. J Biol Chem. 1995;270:10377–10379. doi: 10.1074/jbc.270.18.10377. [DOI] [PubMed] [Google Scholar]
- 36.Quan L T, Tewari M, O’Rourke K, Dixit V, Snipas S J, Poirier G G, Ray C, Pickup D J, Salvesen G S. Proteolytic activation of the cell death protease Yama/CPP32 by granzyme B. Proc Natl Acad Sci USA. 1996;93:1972–1976. doi: 10.1073/pnas.93.5.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ray C A, Black R A, Kronheim S R, Greenstreet T A, Sleath P R, Salvesen G S, Pickup D J. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 38.Ray C A, Pickup D J. The mode of death of pig kidney cells infected with cowpox virus is governed by the expression of the crmA gene. Virology. 1996;217:384–391. doi: 10.1006/viro.1996.0128. [DOI] [PubMed] [Google Scholar]
- 39.Razvi E S, Welsh R M. Apoptosis in viral infections. Adv Virus Res. 1995;45:1–60. doi: 10.1016/s0065-3527(08)60057-3. [DOI] [PubMed] [Google Scholar]
- 40.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Litwack G, Alnemri E S. Molecular ordering of the Fas-apoptotic pathway: the Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases. Proc Natl Acad Sci USA. 1996;93:14486–14491. doi: 10.1073/pnas.93.25.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi A, Alnemri E S, Lazebnik Y A, Fernandes-Alnemri T, Litwack G, Moir R D, Goldman R D, Poirier G G, Kaufmann S H, Earnshaw W C. Cleavage of lamin A by Mch2α but not CPP32: multiple interleukin 1β converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc Natl Acad Sci USA. 1996;93:8395–8400. doi: 10.1073/pnas.93.16.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi A, Hirata H, Yonehara S, Imai Y, Lee K K, Moyer R W, Turner P C, Mesner P W, Okazaki T, Sawai H, Kishi S, Yamamoto K, Okuma M, Sasada M. Affinity labeling displays the stepwise activation of ICE-related proteases by Fas, staurosporine, and CrmA-sensitive caspase-8. Oncogene. 1997;14:2741–2752. doi: 10.1038/sj.onc.1201131. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi A, Musy P Y, Martins L M, Poirier G G, Moyer R W, Earnshaw W C. CrmA/SPI-2 inhibition of an endogenous ICE-related protease responsible for lamin A cleavage and apoptotic nuclear fragmentation. J Biol Chem. 1996;271:32487–32490. doi: 10.1074/jbc.271.51.32487. [DOI] [PubMed] [Google Scholar]
- 44.Tewari M, Dixit V M. Fas- and tumor necrosis factor-induced apoptosis is inhibited by the poxvirus crmA gene product. J Biol Chem. 1995;270:3255–3260. doi: 10.1074/jbc.270.7.3255. [DOI] [PubMed] [Google Scholar]
- 45.Thompson J P, Turner P C, Ali A N, Crenshaw B C, Moyer R W. The effects of serpin gene mutations on the distinctive pathobiology of cowpox and rabbitpox virus following intranasal inoculation of Balb/c mice. Virology. 1993;197:328–338. doi: 10.1006/viro.1993.1594. [DOI] [PubMed] [Google Scholar]
- 46.Thornberry N A, Bull H G, Calaycay J R, Chapman K T, Howard A D, Kostura M J, Miller D K, Molineaux S M, Weidner J R, Aunins J, Elliston K O, Ayala J M, Casano F J, Chin J, Ding G J F, Egger L A, Gaffney E P, Limjuco G, Palyha O C, Raju S M, Rolando A M, Salley J P, Yamin T T, Lee T D, Shiveley J E, MacCross M, Mumford R A, Schmidt J A, Tocci M J. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 47.Thornberry N A, Peterson E P, Zhao J J, Howard A D, Griffin P R, Chapman K T. Inactivation of interleukin-1 beta converting enzyme by peptide (acyloxy)methyl ketones. Biochemistry. 1994;33:3934–3940. doi: 10.1021/bi00179a020. [DOI] [PubMed] [Google Scholar]
- 48.Turner P C, Musy P Y, Moyer R W. Poxvirus serpins. In: McFadden G, editor. Viroceptors, virokines and related immune modulators encoded by DNA viruses. R. G. Galveston, Tex: Landes; 1995. pp. 67–88. [Google Scholar]
- 49.Zhou Q, Snipas S, Orth K, Muzio M, Dixit V M, Salvesen G S. Target protease specificity of the viral serpin CrmA—analysis of five caspases. J Biol Chem. 1997;272:7797–7800. doi: 10.1074/jbc.272.12.7797. [DOI] [PubMed] [Google Scholar]