Abstract
We sequenced and phylogenetically analyzed the reverse transcriptase (RT) regions of the pol genes of 14 human immunodeficiency virus type 1 (HIV-1) isolates from Romanian patients, which were classified as subtype F on the basis of env gene structure. The RT sequences showed that the strains clustered phylogenetically and were equidistant from other HIV-1 subtypes as shown by the neighbor-joining and maximum-likelihood methods, allowing us to define HIV-1 subtype F according to the pol classification. The subtype F RT sequences differed from reported group M RT sequences by 10.94% (for nucleotides) and 7.6% (for amino acids). Phenotypic analysis of subtype F susceptibility to three classes of antiretroviral compounds showed an increase in the 50% inhibitory concentration of the tetrahydroimidazo[4,5,1-jk][1,4]-benzodiazepin-2-(1H)-one and -thione (TIBO) derivate R82913 for one strain which was naturally resistant to this compound. This first report of subtype F pol sequences confirms the perfect correlation between the phylogenetic positions determined by env and pol analyses and suggests that virus variability might influence the efficacy of antiretroviral treatments. This finding warrants a global evaluation of the phenotypic and genotypic susceptibility of HIV-1 subtypes to antiretroviral drugs.
Most human immunodeficiency virus type 1 (HIV-1) drug susceptibility studies have involved subtype B. Little information on the impact of viral diversity on natural susceptibility to antiretroviral drugs has been reported to date. However, HIV-1 group O viruses are naturally resistant to nonnucleoside reverse transcriptase (RT) inhibitors (8), as is HIV-2 (28). Subtypes are defined on the basis of the env (24, 25) or gag (19) gene. Most RT sequences reported to date belong to subtype B strains, which prevail in North America and western Europe (25), i.e., regions where antiretroviral drugs are developed and clinical trials are conducted. RT sequences of subtypes A to D are also available (25, 32, 34). The full sequences of Thai strains defined as subtype E according to env classification and defined as subtype A on the basis of the gag sequence (19) also corresponded to subtype A on the basis of the pol gene (5, 14). By contrast, full sequence analysis of a subtype G strain ruled out recombination events in the pol gene (6), in keeping with a previous report on the RT sequences of subtype G (17). To date, no information has been published on the pol genes of the other HIV-1 group M subtypes.
We present the first RT gene sequences and data on the phenotypic susceptibility to antiretroviral drugs of subtype F strains from Romanian patients. This is the dominant subtype in Romanian children and adults (1, 11) and is also a minor viral form in other countries, such as Brazil (23), Argentina (4), Cameroon (27), Russia (18), Taiwan (7), Martinique (10), Cyprus (16), France (33), Belgium (13), and The Netherlands (20). Recombinant F/B strains have also been reported (21, 30).
MATERIALS AND METHODS
Study population.
We studied 14 HIV-1 subtype F strains isolated from Romanian children (n = 9) and adults (n = 5). All but one of the infected children were nosocomially infected (by injections with nonsterile, reused needles and syringes); the remaining one was vertically infected. Clinical and epidemiological data are described elsewhere, together with virus isolation, env subtype determination, and strain codification (1). None of the patients had received antiretroviral therapy.
HIV-1 RT sequencing.
DNA was extracted with phenol-chloroform from cocultured peripheral blood mononuclear cells from (PBMC) infected patients, precipitated with ethanol, and quantified spectrophotometrically. The pol gene was then amplified in a nested PCR with outer primers RT-18 and RT out and inner primers RT-19 and RT-20 as previously described (26).
Each nested-PCR product (1,008 bp) was subjected to direct population sequencing with sense primer A20 (5′-ATTTTCCCATTAGTCCTATT-3′) and antisense primer NE120 (5′-ATGTCATTGACAGTCCAGCT-3′). Sequencing reactions were run with the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase (FS; Perkin-Elmer) on an automated sequencer (Applied Biosystems 373A).
Phylogenetic analysis.
DNA sequences were analyzed with the multiple-sequence editor Clustal W (35) and improved by visual inspection. The sequences were gapstripped and a pairwise matrix based on 591 sites was generated with the DNADIST program of the PHYLIP package, version 3.56 (12). Tree topology was inferred by the neighbor-joining method with the Kimura two-parameter distance matrix (PHYLIP) and a transition/transversion ratio of 2. Bootstrap analysis was performed with the SEQBOOT (100 resamplings), DNADIST, NEIGHBOR, and CONSENSE programs (PHYLIP package). Phylogenetic analysis was also performed by the maximum-likelihood method, using the DNAML program (12). The tree outliers were the HIV-1 group O sequences (MVP5180 and ANT70). In the tree construction we also included the RT sequences of six subtype B HIV-1 isolates (LAI, SF2, MN, OYI, JRFL, and JRCSF), three subtype D sequences (ELI, NDK, and Z2Z6), two subtype A sequences (UG037 and U455), the sequence of an A/G recombinant strain (IBNG) (14), and the sequence of a presumed A/D recombinant strain (MAL) from the Los Alamos National Laboratory database (25). The newly reported sequences for strains CM240 (5), 90CR402, and 93TH253 (14), recombinant A/E strains, were also used in the phylogenetic analyses.
Phenotypic susceptibility assay.
The phenotypic susceptibilities of the cellular HIV-1 subtype F isolates were analyzed in a PBMC assay, taking into account the replication kinetics of each strain (3). After conventional isolation of HIV from frozen PBMC, the cell-free HIV-1 subtype F supernatants corresponding to peak RT activity were serially diluted (100 to 10−4) and incubated with fresh normal phytohemagglutinin-stimulated PBMC. After being washed, the infected cells were placed in 96-well plates containing six serial dilutions of the antiretroviral drugs. Each dilution was tested in triplicate. On days 5, 7, and 10, the supernatants were collected and half the medium was replaced with fresh drug-containing medium. The 50% tissue culture infective dose was assessed by measuring RT activity in control drug-free supernatants collected on the same days. At the peak of RT activity we calculated the drug concentrations inhibiting 50 and 90% (IC50 and IC90, respectively) of the RT activity of 100 50% tissue culture infective doses. For zidovudine (ZDV), an IC50 cutoff of 0.05 μM has been defined to classify the virus isolates as ZDV sensitive or ZDV resistant, based on phenotypic analyses of several isolates from treated and untreated patients and on comparisons of these results to genotypic data. For the other antiretroviral drugs, it has not been possible to determine any cutoff value. In this study, phenotypic resistance to these compounds was defined as at least a fivefold increase in IC50s for the HIV-1 subtype F isolates compared to those for HIV-1 subtype B strains.
Antiretroviral agents.
We tested the nucleoside RT inhibitors ZDV (Wellcome, Dartford, United Kingdom) and lamivudine (3TC; Glaxo-Wellcome, Dartford, United Kingdom); the nonnucleoside RT inhibitors tetrahydroimidazo[4,5,1-jk][1,4]-benzodiazepin-2(1H)-one and -thione (TIBO) derivate R82913 (Janssen, Beerse, Belgium), delavirdine (DLV; Upjohn, Kalamazoo, Mich.), and nevirapine (NVP; Boehringer Ingelheim Pharmaceuticals, Ridgefield, Conn.); and the protease inhibitors saquinavir (SQV; Roche, Welwyn Garden City, United Kingdom) and ritonavir (RTV; Abbott, Abbott Park, Ill.). The purified drugs were kindly provided by the manufacturers.
Nucleotide sequence accession numbers.
The nucleotide and amino acid sequences of codons 33 to 235 of the RT genes and proteins from the 14 HIV-1 subtype F isolates have been submitted to GenBank (accession no. Y16138 to Y16151).
RESULTS
Genetic analysis.
Figure 1 shows the amino acid alignment of codons 33 to 235 of the RT genes of the different subtype F isolates. The subtype F consensus sequence differed from that of subtype B in 11 residues (V35T, T39A, E40D, D131E, I135L, S162Y, K173T, Q174K, T200A, Q207A, and R211K). Two of these mutations (A200 and K211) have been reported to occur in subtype B isolates. Although not yet reported for subtype B isolates, one residue (D40) of the subtype F consensus sequence has been described as occurring in consensus sequences of subtype A. The sequence T173-K174 has been described for the IBNG subtype A/G isolate (25). Residues A39, E131, L135, and Y162, which are frequently present in subtype F strains, have never been encountered in other HIV-1 group M subtypes. None of the sequences analyzed contained mutations previously linked to resistance to nucleoside or nonnucleoside RT inhibitors. The sequence of the RO-BCI23 isolate was more similar to that of the subtype B consensus sequence in the first part of the RT gene (subtype B structure in sequences encoding T39, E40, and I135) but was more variable than the other subtype F isolates in the second half of the RT gene (close to the binding pocket). Compared to the subtype B consensus sequence, the RO-BCI23 protein bore four mutations close to the active site of the RT protein (E169D, K173A, Q174K, and D177E).
FIG. 1.
Alignment of the deduced amino acid sequences of HIV-1 subtype F RT and the consensus (Cons) sequences of HIV-1 group M and group O strains. Strains RO-BCI17 to RO-BCI23 were isolated from HIV-1-infected adults. ∗, established following the analysis of the pol sequences originating from 11 Zimbabwean seroconverters (32). ∗∗, naturally resistant to TIBO derivate.
At position 60, all but one of the strains originating in nosocomially infected Romanian children (RO-BCI7, RO-BCI8, RO-BCI9, RO-BCI11, RO-BCI12, RO-BCI15, and RO-BCI16) bore a valine (as in the subtype B consensus sequence), whereas the strains isolated from adults (RO-BCI17, RO-BCI18, RO-BCI19, RO-BCI20, and RO-BCI23), a vertically infected child (RO-BCI13), and a nosocomially infected child from whom the strain was isolated in 1994 (RO-BCI1) bore an isoleucine. Conversely, at position 39, all but one (RO-BCI17) of the strains isolated from adults bore a threonine, like subtype B strains, whereas all those isolated from nosocomially infected children bore an alanine.
Phylogenetic analysis of HIV-1 RT sequences.
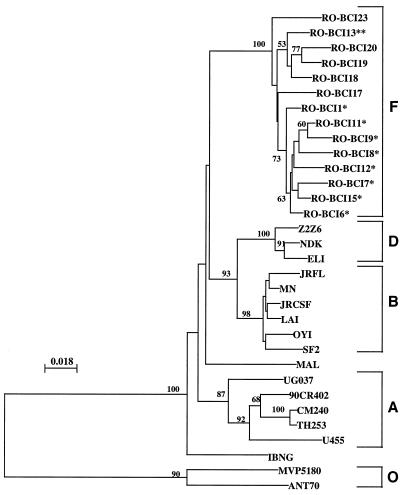
Phylogenetic analysis of the RT nucleotide sequences by the neighbor-joining and maximum-likelihood methods gave similar results. Phylogenetic trees were constructed by using the 14 Romanian HIV-1 RT sequences, representative RT sequences of subtype B strains, and sequences belonging to different subtypes obtained from the database (25). Figure 2 shows the phylogenetic tree constructed by neighbor joining. High bootstrap values were obtained at the relevant nodes, indicating that subtypes B, D, and F each form a consistent clade. Subtype A was composed of five different strains defined as A or E on the basis of the env classification, whereas recombinant strains IBNG and MAL were subtype outliers.
FIG. 2.
Phylogenetic analysis comparing the RT regions of HIV-1 pol genes from different strains. Tree topology was inferred by the neighbor-joining method. The tree was based on an alignment of nucleotides from which columns containing gaps have been deleted (597 nucleotides). The tree was rooted with HIV-1 group O sequences. The numbers given at the branch points are the 50% threshold majority consensus values for 100 bootstrap replicates. Vertical distances are given for clarity. The cluster of sequences from nosocomially infected children (∗) had already been observed by analyzing the env gene (1). Strain RO-BCI13 (∗∗) was isolated from a vertically infected child. Strains RO-BCI17, RO-BCI18, RO-BCI19, RO-BCI20, and RO-BCI23 were isolated from HIV-1-infected adults.
The subtype F phylogenetic tree showed that the strains from nosocomially infected Romanian children formed a cluster, as was previously observed by analyzing the env sequence (1). The RT sequence of a strain from a vertically infected child (RO-BCI13) clustered with the adult sequences. The RT sequence of strain RO-BCI23 formed a separate branch within the subtype F tree.
The average intrasubtype F sequence divergence (597 nucleotides) was 3.56% (range, 1.54 to 6.36%), and the distance between the subtype F sequences and the sequences of other subtypes belonging to group M was 10.94% (range, 8.04 to 13.5%) (P < 0.001) (Table 1). The divergence among the subtype F amino acid sequences (197 residues) was 3.7% (range, 1.57 to 6.7%), whereas the mean distance between the subtype F RT amino acid sequences and other group M sequences was 7.6% (range, 6.00 to 11.59%).
TABLE 1.
Nucleotide and amino acid divergences between subtype F and other group M subtype and group O RT sequences
| Isolate group | % Divergence from subtype F (range)a
|
|
|---|---|---|
| Nucleotides | Amino acids | |
| Group M | 10.94 (8.04–14.84) | 7.6 (6.00–11.59) |
| Subtype B | 10.04 (8.04–13.15) | 7.48 (6.00–9.96) |
| Subtype A | 11.98 (9.35–14.84) | 7.63 (6.09–10.23) |
| Subtype D | 11.01 (9.55–11.96) | 8.36 (6.95–11.59) |
| Group O | 32.09 (29.9–33.52) | 19.09 (16.49–21.92) |
Average intrasubtype F divergence was 3.56% (range, 1.54 to 6.36%) for nucleotides and 3.7% (range, 1.57 to 6.7%) for amino acids.
Phenotypic susceptibility.
The IC50s of nucleoside analogs ZDV and 3TC were similar to those for wild-type subtype B field isolates. Although the IC50s were higher for the subtype F strains than for the subtype B strains, all the subtype F isolates were susceptible to protease inhibitors. All were also sensitive to the nonnucleoside RT inhibitors NVP and DLV. Susceptibility to the third nonnucleoside RT inhibitor, TIBO derivate R82913, was lower for two isolates (Table 2); one isolate (RO-BCI1) showed borderline susceptibility, with a moderate increase in IC50 and IC90 (0.30 and 1.1 μM, respectively), the second strain (RO-BCI23) showed a significant increase in both IC50 and IC90 (0.53 and 2.02 μM, respectively). This phenotype was not associated with any of the known mutations linked to TIBO resistance. However, the RO-BCI23 isolate showed the most variable RT sequence of the subtype F isolates and presented different residues close to the active site of the RT. We therefore assessed the phenotypic susceptibilities of all the available subtype F isolates to TIBO. The TIBO IC50 (mean ± standard deviation) for 12 subtype F isolates was 0.07 ± 0.06 μM (range, 0.01 to 0.2 μM), and the IC90 was 0.4 ± 0.27 μM (range, 0.01 to 0.88 μM), meaning that all 12 strains were susceptible.
TABLE 2.
IC50s and IC90s of nucleoside and nonnucleoside RT inhibitors and protease inhibitors of HIV-1 subtype F isolates, reference HIV-1 isolates, and HIV-2
| Isolate | ZDV
|
3TC
|
DLV
|
NVP
|
TIBO
|
SQV
|
RTV
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IC50 (μM) | IC90 (μM) | IC50 (μM) | IC90 (μM) | IC50 (μM) | IC90 (μM) | IC50 (μM) | IC90 (μM) | IC50 (μM) | IC90 (μM) | IC50 (nM) | IC90 (nM) | IC50 (nM) | IC90 (nM) | |
| HIV-1 subtype Ba | <0.03 | <0.03 | 0.05 ± 0.03 | 0.23 ± 0.1 | 0.02 ± 0.01 | 0.10 ± 0.07 | 0.04 ± 0.01 | 0.22 ± 0.01 | 0.09 ± 0.05 | 0.44 ± 0.12 | 11 ± 2 | 19 ± 5 | 55 ± 26 | 150 ± 57 |
| HIV-2 ROD | <0.03 | 0.20 | 0.02 | 0.04 | >62.5 | >62.5 | >20 | >20 | >6.25 | >6.25 | 10 | 19 | 80 | 433 |
| HIV-1 group Ob | <0.03 | 0.04 ± 0.03 | 0.03 ± 0.02 | 0.16 ± 0.13 | 15.5 ± 13 | 31 ± 29 | <0.02–>20 | <0.02–>20 | 5.4 ± 2.1 | 5.6 ± 1.7 | 15 ± 4 | 26 ± 6 | 108 ± 79 | 453 ± 379 |
| HIV-1 subtype F | ||||||||||||||
| RO-BCI1 | <0.03 | 0.05 | 0.08 | 0.21 | <0.10 | <0.10 | 0.03 | 0.11 | 0.3 | 1.10 | 21 | 35 | 65 | 239 |
| RO-BCI16 | <0.03 | 0.01 | 0.04 | 0.16 | <0.10 | 0.16 | 0.02 | 0.12 | 0.17 | 0.27 | 21 | 33 | 108 | 309 |
| RO-BCI18 | <0.03 | 0.02 | 0.04 | 0.16 | <0.10 | <0.10 | 0.03 | 0.13 | 0.16 | 0.57 | 36 | 48 | 165 | 480 |
| RO-BCI23 | <0.03 | 0.03 | 0.03 | 0.10 | <0.10 | <0.10 | <0.002 | 0.005 | 0.53 | 2.02 | 14 | 26 | 109 | 295 |
IC50s and IC90s were measured for isolates from untreated patients (n = 25).
Performed on 11 HIV-1 group O isolates as previously described (9).
DISCUSSION
Studies of HIV’s genetic diversity have shown different rates of variability for the different viral genes, the most conserved structural gene being pol (29, 31). Analysis of the pol gene was not considered relevant for genotyping (36), but several recent reports have shown that the numerous selection pressures on the pol gene, generally reflected by synonymous substitutions, make it suitable for phylogenetic studies (32, 34). The different pol subtypes described so far correspond to env or gag subtypes (18, 25, 32, 34).
We analyzed the RT coding regions of 14 isolates originating in different parts of Romania and characterized as subtype F on the basis of the env sequence (1). Based on a pol nucleotide and amino acid sequence comparison, these strains clustered phylogenetically and were equidistant from other HIV-1 subtypes. This first report of subtype F pol sequences confirms the perfect correlation between the phylogenetic positions determined by env and gag analysis. The similar results obtained by the neighbor-joining and maximum-likelihood methods support the reliability of the phylogenetic tree of HIV-1 RT sequences. Furthermore, phylogenetic analysis of the pol gene of Romanian HIV-1 isolates revealed a cluster similar to that obtained by analyzing the env gene sequences. The sequences of isolates from nosocomially infected children formed a separate branch within the Romanian sequence cluster with a high bootstrap value (i.e., 73), which supports our hypothesis of a unique introduction of HIV in horizontally infected children from Romania, probably resulting from the use of an infected blood product (1).
None of the patients in this study had ever received antiretroviral therapy. To investigate the impact of viral variability on susceptibility to antiretroviral drugs, we analyzed the phenotypic susceptibilities of four subtype F isolates to different classes of antiretroviral drugs. All but two of the strains had drug susceptibilities similar to those of wild-type subtype B field isolates in the same assay (Table 2). One isolate (RO-BCI1) showed borderline susceptibility, i.e., there was a moderate increase in both the IC50 and IC90 of a nonnucleoside RT inhibitor, TIBO, whereas another isolate (RO-BCI23) showed significantly diminished susceptibility to TIBO. Although this compound is not used to treat HIV infection, the phenotypic susceptibility analysis showed that diversity within group M might affect drug susceptibility. Nonnucleoside RT inhibitors are a promising class of antiretroviral compounds for combined therapy because of their low toxicity, facility of use, and reasonable cost. However, the use of these agents might be hindered by the existence of naturally resistant variants. HIV-1 group O isolates are naturally resistant to nonnucleoside RT inhibitors, as is HIV-2 (9). Because point mutations can lead to marked reductions in drug susceptibility (22), intra- or intersubtype pol diversity might be reflected by particular drug susceptibility profiles. HIV-1 subtype F is only a minor form of HIV-1 but is widely distributed (4, 7, 10, 13, 16, 18, 20, 21, 23, 27, 33), with major circulation in Romania (1, 11, 15). Phylogenetic studies of isolated strains from different countries have revealed that separate epidemiological events contributed to the worldwide distribution of this subtype (1, 2, 16, 18). Our study revealed a natural reduction in susceptibility to a nonnucleoside RT inhibitor in HIV-1 subtype F strains and reinforces the need for global screening for HIV-1 group M isolates by sequencing and phenotyping susceptibility analysis.
ACKNOWLEDGMENTS
This work was supported by Agence Nationale de Recherches sur le SIDA (ANRS), France (grant 96009). C.A. is an ANRS postdoctoral fellow. The samples were obtained with the support of the Romanian Association against AIDS.
REFERENCES
- 1.Apetrei C, Loussert-Ajaka I, Collin G, Letourneur F, Duca M, Saragosti S, Simon F, Brun-Vézinet F. HIV type 1 subtype F sequences in Romanian children and adults. AIDS Res Hum Retroviruses. 1997;13:363–365. doi: 10.1089/aid.1997.13.363. [DOI] [PubMed] [Google Scholar]
- 2.Bandea C I, Ramos A, Pienazek D, Pascu F R, Tanuri A, Schochetman G, Rayfield M A. Epidemiologic and evolutionary relationships between Romanian and Brazilian HIV-1 subtype F strains. Emerg Infect Dis. 1995;1:91–93. doi: 10.3201/eid0103.950305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brun Vézinet F, Ingrand D, Deforges L, Gochi K, Ferchal F, Schmitt M, Jung M, Masquelier B, Aubert J, Buffet-Janvresse C, Fleury H. HIV-1 sensitivity to zidovudine: a consensus culture technique validated by genotypic analysis of the reverse-transcriptase. J Virol Methods. 1991;37:177–188. doi: 10.1016/0166-0934(92)90045-f. [DOI] [PubMed] [Google Scholar]
- 4.Campodonico M, Janssens W, Heyndrickx L, Fransen K, Leonaers A, Fay F F, Taborda M, van der Groen G, Fay O H. HIV type 1 subtypes in Argentina and genetic heterogeneity of the V3 region. AIDS Res Hum Retroviruses. 1996;12:79–81. doi: 10.1089/aid.1996.12.79. [DOI] [PubMed] [Google Scholar]
- 5.Carr J K, Salminen M O, Koch C, Gotte D, Artenstein A W, Hegerich P A, St. Louis D, Burke D S, McCutchan F E. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol. 1996;70:5935–5943. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr J K, Salminen M O, Lenikki P, Johansson B, Burke D S, McCutchan F E. First full length sequence prototypes of HIV-1 clades C, E and G: two of three are mosaic genomes, abstr. TuA 100. XIth International Conference on AIDS. 1996. [Google Scholar]
- 7.Chang K S S, Lin C I, Chen J H, Shih C H, Lin H C, Twu S C, Salminen M O. HIV type 1 env gene diversity detected in Taiwan. AIDS Res Hum Retroviruses. 1997;13:201–204. doi: 10.1089/aid.1997.13.201. [DOI] [PubMed] [Google Scholar]
- 8.Descamps D, Collin G, Loussert-Ajaka I, Saragosti S, Simon F, Brun-Vézinet F. HIV-1 group O sensitivity to antiretroviral drugs. AIDS. 1995;8:977–978. [PubMed] [Google Scholar]
- 9.Descamps D, Collin G, Letourneur F, Apetrei C, Damond F, Loussert-Ajaka I, Simon F, Saragosti S, Brun-Vézinet F. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J Virol. 1997;71:8893–8898. doi: 10.1128/jvi.71.11.8893-8898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desgranges C, Fillon S, Audoly G, Neisson-Vernant C, Bera O, Ouka M, Cesaire R, Buzelay L, Barin F. Presence of HIV-1 subtypes B and F and HTLV-I in HIV/HTLV coinfected individuals of Martinique. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:468–470. doi: 10.1097/00042560-199612150-00015. [DOI] [PubMed] [Google Scholar]
- 11.Dumitrescu O, Kalish M L, Kilks S C, Bandea C I, Levy J A. Characterization of human immunodeficiency virus type 1 isolates from children in Romania: identification of a new envelope subtype. J Infect Dis. 1994;169:281–288. doi: 10.1093/infdis/169.2.281. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein J. PHYLIP (phylogeny interference package), version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 13.Fransen K, Buvé A, Nkengasong J N, Laga M, van der Groen G. Longstanding presence in Belgians of multiple non-B HIV-1 subtypes. Lancet. 1996;347:1403. doi: 10.1016/s0140-6736(96)91042-9. [DOI] [PubMed] [Google Scholar]
- 14.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, Fultz P N, Girard M, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm-Hansen C, Grothues D, Rusad S, Rosok B, Pascu F R, Asjö B. Characterization of HIV type 1 from Romanian children: lack of correlation between V3 loop amino acid sequence and syncytium formation in MT-2 cells. AIDS Res Hum Retroviruses. 1995;11:597–603. doi: 10.1089/aid.1995.11.597. [DOI] [PubMed] [Google Scholar]
- 16.Kostrikis L G, Bagdades E, Cao Y, Zhang L, Dimitriou D, Ho D D. Genetic analysis of human immunodeficiency virus type 1 strains from patients in Cyprus: identification of a new subtype designated subtype I. J Virol. 1995;69:6122–6130. doi: 10.1128/jvi.69.10.6122-6130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leitner T, Escanilla D, Marquina S, Wahlberg J, Broström C, Hansson H B, Uhlén M, Albert J. Biological and molecular characterization of subtype D, G and A/D recombinant HIV-1 transmission in Sweden. Virology. 1995;209:136–146. doi: 10.1006/viro.1995.1237. [DOI] [PubMed] [Google Scholar]
- 18.Leitner T, Korovina G, Marquina S, Smolskaya T, Albert J. Molecular epidemiology and MT-2 tropism of Russian HIV type 1 variants. AIDS Res Hum Retroviruses. 1996;12:1595–1603. doi: 10.1089/aid.1996.12.1595. [DOI] [PubMed] [Google Scholar]
- 19.Louwagie J, McCutchan F E, Peeters M, Brennan T P, Sanders-Buell E, Eddy G A, van der Groen G, Fransen K, Gershy-Damet G D, Deleys R, Burke D S. Phylogenetic analysis of gag genes from 70 international HIV-1 isolates provides evidence for multiple genotypes. AIDS. 1993;7:769–780. doi: 10.1097/00002030-199306000-00003. [DOI] [PubMed] [Google Scholar]
- 20.Lukhasov V V, Kuiken C L, Boer K, Goudsmit J. HIV type 1 subtype in The Netherlands circulating among women originating from AIDS-endemic regions. AIDS Res Hum Retroviruses. 1996;12:951–953. doi: 10.1089/aid.1996.12.951. [DOI] [PubMed] [Google Scholar]
- 21.Marquina S, Leitner T, Rabinovich R D, Benetucci J, Libonatti O, Albert J. Coexistence of subtypes B, F and a B/F env recombinant of HIV type 1 in Buenos Aires, Argentina. AIDS Res Hum Retroviruses. 1996;12:1651–1654. doi: 10.1089/aid.1996.12.1651. [DOI] [PubMed] [Google Scholar]
- 22.Mellors J W, Schinazi R F, Larder B A. Mutations in retroviral genes associated with drug resistance. In: Myers G, Foley B, Mellors J W, Korber B, Jeang K T, Wain-Hobson S, editors. Human retroviruses and AIDS 1996. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. pp. III-206–III-241. [Google Scholar]
- 23.Morgado M G, Sabino E C, Sapaer E G, Bongertz V, Brigido L, Guimaraes M D C, Castilho E A, Galvao-Castro B, Mullins J I, Hendry R M, Mayer A. V3 region polymorphism in HIV-1 from Brazil: prevalence of subtype B strains divergent from North American/European prototype and detection of subtype F. AIDS Res Hum Retroviruses. 1994;10:569–576. doi: 10.1089/aid.1994.10.569. [DOI] [PubMed] [Google Scholar]
- 24.Myers G. HIV: between past and future. AIDS Res Hum Retroviruses. 1994;10:1317–1324. doi: 10.1089/aid.1994.10.1317. [DOI] [PubMed] [Google Scholar]
- 25.Myers G, Foley B, Mellors J W, Korber B, Jeang K T, Wain-Hobson S, editors. Human retroviruses and AIDS 1996. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. [Google Scholar]
- 26.Nihjhuis M, Boucher C A B, Schuurman R. Sensitive procedure for the amplification of HIV-1 RNA using combined reverse-transcription and amplification reaction. BioTechniques. 1995;19:178–180. [PubMed] [Google Scholar]
- 27.Nkengasong J N, Janssens W, Heyndrickx L, Fransen K, Ndumbe P M, Motte J, Leonaers A, Ngolle M, Ayuk J, Piot P, van der Groen G. Genotypic subtypes of HIV-1 in Cameroon. AIDS. 1994;8:1405–1412. doi: 10.1097/00002030-199410000-00006. [DOI] [PubMed] [Google Scholar]
- 28.Pauwels R, Andries K, Desmyter J, Schols D, Kukla M, Breslin H, Raeymaeckers A, Van Gelder J, Woestenborghs R, Heykants J, Schellekens K, Janssen M, De Clercq E, Janssen P. Potent and selective inhibition of HIV-1 replication in vitro by a novel series of TIBO derivatives. Nature. 1990;343:470–474. doi: 10.1038/343470a0. [DOI] [PubMed] [Google Scholar]
- 29.Quinones-Mateu M E, Holguin A, Dopazo J, Najera I, Domingo E. Point mutation frequencies in the pol gene of human immunodeficiency virus type 1 are two- to threefold lower than those of env. AIDS Res Hum Retroviruses. 1996;12:1117–1128. doi: 10.1089/aid.1996.12.1117. [DOI] [PubMed] [Google Scholar]
- 30.Sabino E C, Shpaer E G, Morgado M G, Korber B T M, Diaz R S, Bongertz V, Cavalcante S, Galvão-Castro B, Mullins J I, Mayer A. Identification of human immunodeficiency virus type 1 envelope genes recombinant between subtypes B and F in two epidemiologically linked individuals from Brazil. J Virol. 1994;68:6340–6346. doi: 10.1128/jvi.68.10.6340-6346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seibert S A, Howell C Y, Hughes M K, Hughes A L. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1) Mol Biol Evol. 1995;12:803–813. doi: 10.1093/oxfordjournals.molbev.a040257. [DOI] [PubMed] [Google Scholar]
- 32.Shafer R W, Eisen J A, Merigan T C, Katzenstein D A. Sequence and drug susceptibility of subtype C reverse transcriptase from human immunodeficiency virus type 1 seroconverters in Zimbabwe. J Virol. 1997;71:5441–5448. doi: 10.1128/jvi.71.7.5441-5448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon F, Loussert-Ajaka I, Damond F, Saragosti S, Barin F, Brun-Vézinet F. HIV type 1 diversity in Northern Paris, France. AIDS Res Hum Retroviruses. 1996;12:1427–1433. doi: 10.1089/aid.1996.12.1427. [DOI] [PubMed] [Google Scholar]
- 34.Soto-Ramirez L E, Tripathy S, Renjifo B, Essex M. HIV pol sequences from India fit distinct subtype pattern. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:299–307. doi: 10.1097/00042560-199612010-00001. [DOI] [PubMed] [Google Scholar]
- 35.Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.WHO Network for HIV Isolation and Characterization. HIV type 1 variation in World Health Organization-sponsored vaccine evaluation sites: genetic screening, sequence analysis, and preliminary biological characterization of selected viral strains. AIDS Res Hum Retroviruses. 1994;10:1327–1343. doi: 10.1089/aid.1994.10.1327. [DOI] [PubMed] [Google Scholar]