ABSTRACT
Whereas obligate human and animal bacterial pathogens may be able to depend upon the warmth and relative stability of their chosen replication niche, environmental bacteria such as Listeria monocytogenes that harbor the ability to replicate both within animal cells and in the outside environment must maintain the capability to manage life under a variety of disparate conditions. Bacterial life in the outside environment requires adaptation to wide ranges of temperature, available nutrients, and physical stresses such as changes in pH and osmolarity as well as desiccation. Following ingestion by a susceptible animal host, the bacterium must adapt to similar changes during transit through the gastrointestinal tract and overcome a variety of barriers associated with host innate immune responses. Rapid alteration of patterns of gene expression and protein synthesis represent one strategy for quickly adapting to a dynamic host landscape. Here, we provide an overview of the impressive variety of strategies employed by the soil-dwelling, foodborne, mammalian pathogen L. monocytogenes to straddle diverse environments and optimize bacterial fitness both inside and outside host cells.
A VARIETY OF MULTIFUNCTIONAL GENE PRODUCTS CONTRIBUTE TO LISTERIA MONOCYTOGENES ENTRY AND SURVIVAL WITHIN MAMMALIAN CELLS
The move from soil to cytosol requires the interplay of L. monocytogenes factors that promote survival in the gut, bacterial invasion, phagosomal escape, replication and movement within the cytosol, and spread to adjacent cells (Fig. 1). Bacterial gene products contributing to many key aspects of host infection continue to be identified, and new factors and novel functions often emerge (1, 2). Among the cast of well-known players are surface proteins that promote bacterial attachment to and invasion of nonprofessional phagocytic cells, such as the internalins InlA and InlB as well as Lap and InlP (which appears to be specific for placental invasion) (3–5). Following cell entry, L. monocytogenes escapes from host cell vacuoles via the secretion of the pore-forming cytolysin listeriolysin O (LLO) and two phospholipases, a phosphatidylinositol-specific phospholipase C (PI-PLC) encoded by plcA and a broad-range phospholipase C (PC-PLC) encoded by plcB (6). Entry into the cytosol requires metabolic adaptation as L. monocytogenes shifts from glycolysis to the oxidative pentose phosphate pathway and replicates by scavenging phosphorylated sugars, glycerol, lipoic acid, branched-chain amino acids, and peptides from conquered host cells (7). Bacteria spread to neighboring cells by usurping actin polymerization as a motile force, a process dependent upon expression of the bacterial surface protein ActA (8). The breaking and entering of L. monocytogenes into adjacent cells is further facilitated by InlC, which relieves cortical tension to allow the extension of membrane protrusions (9). Escape from the double membrane vacuoles formed as a result of L. monocytogenes cell-to-cell spread is once again dependent upon the activities of LLO, PC-PLC, and PI-PLC. Protein secretion of a variety of factors thus promotes bacterial survival and life within host cells, and central to the secretion of active protein products is the presence of the secretion chaperone PrsA2, located at the bacterial membrane-cell wall interface, where it contributes to the folding and activity of translocated polypeptide chains (10). L. monocytogenes is thereby able to maintain a complex and multifunctional protein arsenal to increase bacterial survival and replication within mammalian host cells.
FIGURE 1.
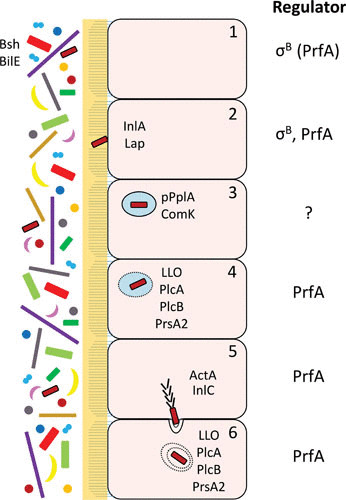
Schematic overview of L. monocytogenes colonization of the intestine by pathways controlled by σB and/or PrfA. The human microbiota is depicted to the left, lying beside the microvilli-containing epithelial cells (1). Survival of L. monocytogenes in the gut is mediated by different factors, among them Bsh and BilE (2). The bacterium adheres to cells using different adhesins (e.g., InlA and Lap) (3). Once inside the phagosome, the pPplA and ComK systems become activated, facilitating phagosomal escape (4). Different PrfA-regulated factors mediate lysis of the phagosome (5). Through recruitment of the Arp2/3 complex, ActA allows polymerization of actin at the pole of the bacterium, driving it through the cytoplasm. The entry of L. monocytogenes to adjacent cells requires InlC, which relieves cortical tension (6). The escape from the double membrane vacuole requires the action of LLO, PlcA, and PlcB. Regulators (σB or PrfA) involved in the different regulatory steps during intestinal passage of L. monocytogenes are shown at the right.
To better understand bacterial adaptation to the host environment, a number of studies have focused on the identification of bacterial genes expressed within tissue culture cells, within blood, or within infected animals. Microarray analyses of bacterial transcripts induced during L. monocytogenes infection of tissue culture cells revealed that approximately 20% of bacterial genes were differentially expressed, including genes with products that have established roles in bacterial virulence (11). Genes with increased expression in cytosolic bacteria included those with roles in general stress responses, cell division, modification of the cell wall, and the use of carbon sources such as glycerol and phosphorylated sugars. Transcriptional profiling of L. monocytogenes genes expressed during in vivo growth in mouse spleens also indicated that approximately 20% of bacterial genes were differentially expressed (12). Similar to the findings reported for bacteria grown within tissue culture cells, genes induced in vivo included those with defined roles in virulence, stress responses, cell wall metabolism, DNA metabolism, RNA/protein synthesis, and cell division. In contrast to tissue culture-based expression studies, transcripts from genes encoding enzymes involved in glycolysis were induced in vivo, while those involved in the nonoxidative phase of the pentose phosphate pathway had decreased levels of expression. These contrasting results may reflect differences observed between growth conditions within tissue culture cells versus growth in whole organs and animal tissues.
PrfA, THE MASTER REGULATOR OF L. MONOCYTOGENES INTRACELLULAR SURVIVAL
While life is never simple, a transcriptional activator known as PrfA does appear to suffice as the master virulence regulatory protein that enables L. monocytogenes to optimize life within a mammalian host while still managing a saprophytic existence in soil. PrfA is a 27-kDa transcriptional activator that is a member of the Crp/Fnr family of transcriptional regulators, and it regulates its target genes via the binding of a 14-bp palindromic DNA binding site, also known as the PrfA box, located in the –40 region of its target promoters (13–17). PrfA regulates the expression of a large number of gene products directly associated with bacterial virulence in mammals, and mutants lacking prfA are severely impaired for intracellular growth and are >100,000-fold less virulent in murine infection models (15). In addition to gene products required for host cell invasion, intracellular replication, and cell-to-cell spread, PrfA induces the expression of gene products required for surviving the journey through the gastrointestinal tract, including a bile salt hydrolase (encoded by bsh) and a bile exclusion system (encoded by bilE) (18, 19). Full PrfA activation occurs following bacterial entry into the cytosol of host cells and results in the expression of gene products that enable intracellular replication and bacterial spread to adjacent cells. A subset of genes directly regulated by PrfA is located on an L. monocytogenes pathogenicity island referred to as LIPI-1 (hly, plcA, prfA, mpl, actA, and plcB), while others (inlA, inlB, inlC, bsh, prsA2, and hpt) have distinct chromosomal locations. Comparison of the profiles of wild-type L. monocytogenes grown in brain heart infusion broth with those of a strain harboring a constitutively active PrfA protein suggests that the expression of at least 145 genes may be modulated by PrfA (20).
Given that PrfA is critical for enabling L. monocytogenes to mediate the balance between life in soil and life inside an infected host, the activity of PrfA is itself carefully regulated by a variety of mechanisms that include transcriptional, posttranscriptional, and posttranslational control. These critical regulatory circuits work together to tightly control prfA expression and PrfA activity as outlined below.
Transcriptional Regulation of prfA Expression
Transcriptional regulation of prfA expression occurs via three promoter elements (Fig. 2). Two promoters, prfAP1 and prfAP2, are located immediately upstream of the prfA translation initiation codon, while the third promoter lies immediately upstream of plcA and results in the generation of a plcA-prfA bicistronic transcript (14, 21, 22). The prfAP1 and prfAP2 promoters direct the synthesis of monocistronic transcripts of prfA that generate the initial levels of PrfA protein required to activate expression of hly and plcA, whose gene products are needed for efficient escape of L. monocytogenes from host cell phagosomes (22, 23). The plcA promoter, which is activated by PrfA, directs the synthesis of the plcA-prfA transcripts, resulting in the high levels of PrfA synthesis that are required to induce actA expression for efficient bacterial cell-to-cell spread. The prfAP1 promoter contains characteristics of a σA-dependent promoter, which is the vegetative sigma factor determining RNA polymerase specificity required for transcription in actively growing, unstressed bacterial cells. The prfAP2 promoter region contains sequences that resemble a PrfA binding box, a σA-dependent promoter, and the general stress response sigma factor σB-dependent promoters (23–26). σB directs RNA polymerase to the promoter regions of a large number of genes involved in adaptation to general environmental stresses, such as conditions of low pH, high osmolarity, oxidative stress, and carbon starvation (see below). A number of genes coregulated by PrfA and σB have been shown to contribute to pathogenesis of L. monocytogenes (e.g., bsh and inlAB), suggesting a cross-talk network between these two regulators and possibly other stress response regulators and alternative sigma factors (18, 27, 28). Interestingly, work by Guldimann et al. (29) suggests that activation of PrfA and σB occurs stochastically within individual bacterial cells under defined stress conditions, with evidence that PrfA activation appears population-wide, while σB activation is restricted to subpopulations.
FIGURE 2.
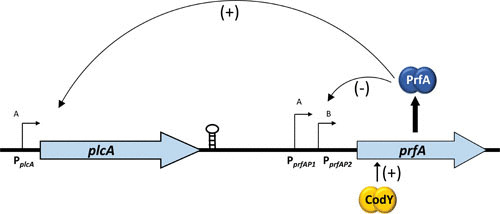
Transcriptional regulation of prfA expression. The prfA transcript is expressed from three different promoters, P1 (σA regulated), P2 (σA and σB regulated), and plcA (generating a plcA-prfA bicistronic messenger). The plcA promoter is positively regulated by active PrfA, creating a positive feedback loop of PrfA expression. CodY positively regulates expression of prfA by binding within its coding region. The stem-loop structure shows a putative transcriptional terminator located 3′ of the plcA gene.
In addition to stress-responsive promoter elements, Lobel et al. have recently identified CodY to activate transcription of the prfA gene (30). CodY is a regulatory protein that links prfA expression with the metabolic status of the bacterium. Intriguingly, CodY binds to a region located 15 bases downstream of the prfA start codon (30). CodY responds to the presence of branched chain amino acids and GTP, and L. monocytogenes appears to regulate CodY activity by controlling the internal isoleucine pool, a mechanism that has been postulated to have evolved to enable isoleucine to serve as a host signal and virulence effector (31, 32). L. monocytogenes thus appears to tie in multiple environmental signals that include both stress and metabolism in its quest to regulate virulence gene expression within host cells.
Posttranscriptional Regulation
It has long been recognized that virulence factors as well as PrfA are mainly expressed at body temperature, 37°C, although the mRNA encoding the PrfA protein is present also at low temperatures (below 30°C) (21, 33). Computer prediction and chemical probing experiments showed that a 115-nucleotide (nt)-long 5′ untranslated RNA (UTR) present in front of the prfA encoding mRNA could form a long hairpin where the Shine-Dalgarno (SD) site was partially masked by an anti-SD site (34) (Fig. 3). In vitro translation of the wild-type prfA showed a thermoregulated expression, with PrfA mostly expressed at higher temperatures. Mutational analysis suggested that the actual thermosensing occurred in the middle part of the hairpin since mutations that destabilized the prfA-UTR structure abolished much of the thermoregulation (34). These data suggested a simple mechanism of thermosensing: at low temperatures, the SD site is closed due to an interaction with an anti-SD site, and binding of the ribosome is prevented. With increasing temperature, the region surrounding the prfA-SD site is destabilized, allowing binding of the ribosome and initiation of translation (34–36) (Fig. 3). However, a more complex mechanism of prfA translation initiation has been put forward, involving a ribosomal stand-by site inside the prfA coding RNA (37).
FIGURE 3.
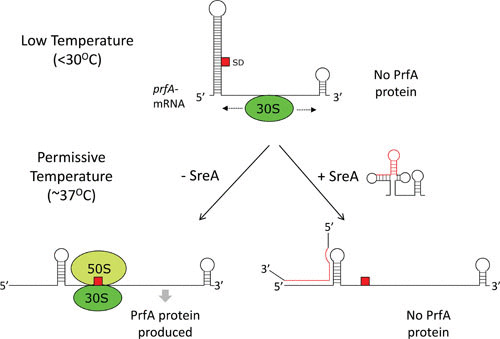
Posttranscriptional control of PrfA expression. At low temperatures (<30°C), the Shine-Dalgarno site (SD, red square) is sequestered in an RNA thermosensor, preventing binding of the 30S subunit (green sphere) of the ribosome. Possibly, the binding of the 30S subunit involves a ribosomal standby site, where it primarily binds to the coding region of the prfA mRNA, where it scans the transcript. At permissive temperatures (∼37°C), the RNA thermosensor dissociates, allowing access of the 30S subunit to the SD, where it can recruit the 50S subunit (light green sphere), and translation can initiate. Under conditions in which a terminated S-adenosylmethionine riboswitch is produced (+SreA), it can bind to the RNA thermosensor and by an unknown mechanism prevent PrfA production.
In addition, prfA translation is controlled by at least two prematurely terminated S-adenosyl methionine (SAM) responsive riboswitches (38). Riboswitches are RNA elements that are able to control downstream gene expression by directly binding specific metabolites with a high affinity. The metabolite-riboswitch interaction results in RNA restructuring, usually terminating transcription in Gram-positive bacteria (39, 40). When binding SAM, transcription is terminated and short (150 to 250 nt long) transcripts are generated. Two of the prematurely terminated SAM riboswitches in L. monocytogenes (SreA and SreB) can base-pair with the 5′ UTR of prfA and thus reduce PrfA expression by an unknown mechanism (38) (Fig. 3). Here, the function of the terminated riboswitches resembles the role played by several trans-acting small RNAs (41). Interestingly, SreA is unable to inhibit translation at low temperatures, possibly because its interaction site in the prfA thermosensor is in a closed conformation, preventing base-pairing with trans-acting RNA elements. Expression of PrfA can therefore be affected by SAM riboswitches only at temperatures encountered during infection. The entire SreA is not required for repression; instead, the central core part of the SreA is sufficient for efficient blocking of PrfA translation (S. Krajewski and J. Johansson, unpublished results). Intriguingly, SreA expression is positively controlled by PrfA, creating a negative feedback loop, where high levels of active PrfA turn off its own expression by increasing the level of the repressive element SreA (38). It therefore seems that the 5′ UTR of the prfA transcript can act as a sensor, integrating both the environmental temperature and the metabolic state of the bacterium through SAM sensing.
Posttranslational Regulation
The third critical mechanism for regulating PrfA activity occurs via posttranslational modification (Fig. 4). As stated above, PrfA protein is a member of the cyclic AMP receptor protein (Crp)-Fnr family of transcriptional regulators, of which there are approximately 400 members (42). Proteins in this family generally function as dimers and require the binding of small molecule cofactors (for example, cyclic AMP for Crp) or other forms of posttranslational modification (such as the binding of carbon monoxide by the heme moiety of CooA) for full activity. The PrfA cofactor has recently been identified as glutathione, and PrfA-glutathione cocrystals indicate that the cofactor binding site is located at the intraprotein tunnel site, located between the N- and C-terminal domains of the PrfA monomer (43–46). Working models have suggested that once inside the cytoplasm, both uptake of cytosolic glutathione and, most importantly, upregulation of the glutathione synthase, gshF, lead to increased glutathione concentrations in L. monocytogenes (43, 45). This is required since glutathione only binds PrfA with a modest affinity (0.5 mM), so the increased glutathione concentration thus signals to the bacterium its arrival into the cytosol. Recently, Krypotou et al. (47) proposed that Cys-containing peptides transported into L. monocytogenes via the Opp peptide transport systems provide for the generation of glutathione leading to PrfA activation. Interestingly, the authors further suggested that inhibitory peptides transported by this same system bind to PrfA and inhibit PrfA activation by inhibiting the binding of glutathione. The complexity of PrfA activation continues to increase.
FIGURE 4.
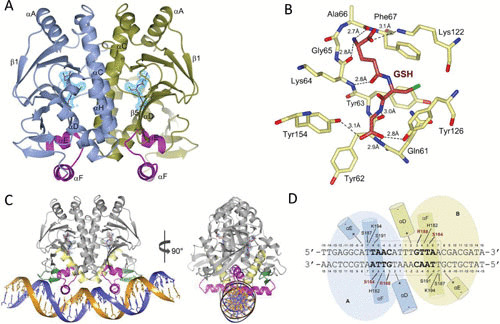
Structures of PrfA in complex with glutathione (GSH) and PrfA in complex with GSH and DNA. (A) Ribbon representation of the PrfA homodimer with GSH bound at the tunnel site of each monomer. The monomers A and B are colored blue and green, respectively. The helix-turn-helix motif is highlighted in magenta, and GSH has crimson carbon atoms with light blue electron density. (B) Local amino acid interactions and structural features of GSH-PrfA binding. Direct hydrogen bonds between amino acids and GSH are shown by dashed lines. (C) Ribbon representation of the DNA-bound homodimer in complex with GSH. The monomers A and B are colored gray, the helix-turn-helix motif is magenta, αD is yellow, and the winged β-hairpin is green. GSH is shown as sticks with the carbon atoms in crimson. The 30-bp palindromic operator is shown in dark blue and orange. (D) Schematic drawing of the PrfAWT-DNA interactions. Amino acids S184 and R188 make base-specific contacts and are highlighted in red. Other interactions are nonspecific between protein side chains and the DNA phosphate backbone. Figure adapted with permission from Hall et al. (44).
Years prior to the identification of glutathione as the PrfA cofactor, Ripio et al. identified an L. monocytogenes strain containing a single mutation within prfA coding sequences that resulted in the constitutive expression of PrfA-dependent virulence genes in broth culture (48). The substitution of a serine for a glycine at position 145 within PrfA was the first identification of a PrfA* mutation, so named because it appeared analogous to an A144T mutation identified within Crp that resulted in the constitutive expression of Crp-dependent gene products in the absence of cofactor (Crp* mutants) (49). Similar to Crp*, the PrfA G145S mutation was demonstrated by structural analyses to alter PrfA protein confirmation and increase the DNA binding affinity of PrfA for its target promoters via the repositioning of a helix-turn-helix DNA binding motif (50, 51).
There have now been a number of additional mutations identified that confer PrfA activation, albeit at differing levels of activation. Reported prfA* mutations include G145S, Y63C, S71C, E77K, A94T, L140F, Y154C, L148P, G155S, and P219S substitution mutants (52–61). These mutations in some cases map to very different regions of PrfA in comparison to the original G145S PrfA* mutation, and strains containing these different prfA* alleles exhibit levels of PrfA-dependent gene expression in broth culture that range from 4-fold to >200-fold greater than the levels of expression observed in wild-type bacteria (62, 63). With the exception of PrfA G145S, for which the structure has been solved using X-ray crystallography (51), the mechanisms by which the other prfA* mutations confer constitutive activation are not clear.
prfA* MUTATIONS REVEAL THE DIVERSITY OF GENE PRODUCTS REGULATED BY PrfA ACTIVATION
PrfA* mutants may be considered to phenotypically resemble L. monocytogenes under conditions of cytosolic activation (bound to glutathione), and thus the mutant strains have proven useful for the identification of novel virulence factors following transcriptomic and proteomic analyses. Milohanic et al. initially identified substantial overlap between genes whose expression was influenced by PrfA and stress-responsive genes regulated by the stress-responsive alternative sigma factor σB (20). However, studies by Ollinger et al. (64) using reverse transcriptase PCR reported that the transcript levels of some of PrfA-associated genes identified by Milohanic et al. (20) were not significantly affected by the presence or absence of PrfA. Discrepancies between these independent studies may reflect disparities between laboratory conditions, variations between strains used for examination (EGDe versus 10403S), or additional undefined complexities associated with PrfA-dependent gene expression.
Secreted proteins are often the first bacterial factors to interact with the host, and a comparison of secreted protein profiles derived from the culture supernatants of wild-type, ΔprfA, and prfA* mutants identified at least 17 proteins that were differentially secreted following PrfA activation (52). The majority of the genes encoding these proteins did not contain recognizable PrfA binding sites in their upstream promoter regions, suggesting that the synthesis and/or secretion of these proteins was indirectly influenced by PrfA activation. Proteins with increased abundance in the supernatants derived from prfA* cultures included a number of previously identified virulence factors as well as putative ABC transporters, cell wall-modifying enzymes, chitinases, antigenic lipoproteins, and chaperone proteins associated with protein secretion. Many of these PrfA-dependent secreted proteins also depended on the presence of the secretion chaperone PrsA2 for full activity (65). A significant number of secreted gene products that appear indirectly regulated by PrfA have been demonstrated to contribute to L. monocytogenes pathogenesis, and these gene products serve as further examples of the expansive influence of PrfA on L. monocytogenes life within the host.
THE IMPACT OF CARBON SOURCES ON PrfA ACTIVATION AND THE STIMULATION OF PrfA-DEPENDENT VIRULENCE GENE EXPRESSION
The expression of virulence factors has long been known to respond to the presence of sugars in the medium (66–71). Sugars (e.g., cellobiose and glucose) taken up by the phosphoenolpyruvate):phosphotransferase system (PTS) repress PrfA activity, whereas non-PTS sugars (e.g., glycerol) modestly stimulate PrfA activity. Importantly, the PTS-mediated repression of PrfA activity is not mediated by the catabolite control protein A (CcpA), which controls expression of many genes in different Firmicutes in response to PTS sugars (72, 73). Instead, the phosphorylation state of the different PTS permeases appears to dictate the activation level of PrfA (74). In the presence of PTS sugars, the permeases are in an unphosphorylated state as they donate the phosphate group they received from the phosphoenolpyruvate system to the incoming sugar. In the absence of PTS sugars (and the presence of non-PTS sugars), the permeases are phosphorylated. It has been suggested that unphosphorylated permeases might bind and sequester PrfA directly, thereby blocking PrfA-mediated virulence gene expression (74). Glucose can be transported into Listeria by several different permeases. One of the mannose permeases, EIIABMan, was suggested to act as a PrfA-binding partner or, alternatively, to interact with a PrfA-activating molecule (75). In addition, the repressive effect of PTS sugars can be relieved by the addition of charcoal or amberlite to the medium during bacterial growth (76, 77). This suggests that a component released by L. monocytogenes could repress PrfA activity, either directly or indirectly. Precisely how the action of PTS-permeases or repressive extracellular molecules such as inhibitory peptides affect the level of glutathione (the PrfA cofactor) or PrfA binding of glutathione remains to be determined.
MAINTAINING THE BALANCE BETWEEN LIFE INSIDE AND OUTSIDE THE HOST: CONSTITUTIVE ACTIVATION OF PrfA IS NOT BENEFICIAL FOR L. MONOCYTOGENES SURVIVAL
At first glance, prfA* strains would appear to exhibit a number of advantages over wild-type bacteria. Strains containing prfA* are hyperinvasive, mediate more efficient phagosome escape, and initiate bacterial actin-based motility more rapidly. Activation of PrfA also shifts L. monocytogenes metabolism toward the preferred use of three carbon sugars and phosphorylated sugars, the principal carbon sources used by L. monocytogenes for growth within the cytosol (78). prfA* mutants are hypervirulent in mouse infection models and exhibit a competitive fitness advantage over wild-type strains during both oral and intravenous mixed infections in mice (78).
The fitness advantage observed for prfA* strains within the host does not, however, translate into bacterial fitness in the outside environment (78, 79). Constitutively activated prfA* mutants exhibit impaired flagellum-mediated swimming motility, a defect that would be expected to compromise bacterial fitness in environments where the bacteria must be able to detect and swim toward available nutrient sources and which would additionally impair biofilm formation. prfA* mutants also exhibit a pronounced fitness defect when grown in the presence of wild-type bacteria in mixed broth culture despite displaying no obvious growth defects in monoculture. Stress conditions such as high osmolarity or low pH exacerbate the competitive defects observed for prfA* strains in a manner that is independent of the stress-responsive sigma factor σB. Lastly, prfA* mutations appear to negatively impact the ability of L. monocytogenes to survive long periods of starvation. Interestingly, a PrfA* strain devoid of most PrfA-regulated genes did not display a fitness cost (80). The multiple mechanisms that thus exist to regulate PrfA activity appear to have evolved to carefully balance gene expression patterns so as to maintain bacterial fitness in the soil as well as in the cytosol.
BEYOND PrfA: OTHER MODES OF L. MONOCYTOGENES REGULATION OF GENE EXPRESSION
PrfA is a central factor in the regulation of L. monocytogenes virulence gene expression; however, numerous other regulatory factors and mechanisms exist that enable the bacterium to adapt its physiology in response to the diversity of environmental conditions encountered by this ubiquitous organism. A sample of some of these alternative regulatory modalities is presented below.
Posttranscriptional Regulation of Bacterial Gene Expression
Untranslated regions of mRNA (UTRs) have been demonstrated to regulate translation of the associated gene products for multiple genes. The 5′ UTRs located upstream of inlA, actA, and hly have been shown to control expression of their protein products (27, 81, 82). These 5′ UTRs appear, however, to support expression of InlA, ActA, and LLO, respectively, which is in contrast to the previously described prfA-UTR, which represses PrfA expression. It has been shown that partial deletions of the 5′ UTRs of inlA, actA, and hly decrease the protein production, but the exact mechanism(s) by which the 5′ UTRs act has not yet been revealed. Interestingly, it was shown that the coding region of hly could be important for LLO production (83). Whether this is due to alterations of secondary structures, binding of small regulatory RNAs (sRNAs), or some other factors remains to be investigated.
Regulation by sRNAs
Several hundred sRNAs have been identified in L. monocytogenes, ranging in size from less than 100 to more than 500 nt and with different expression patterns (84–88). However, the functions of only a subset of these sRNAs have been revealed, and an even smaller number have been shown to participate in virulence gene expression. Rli27 is an intracellularly induced sRNA able to interact with the 5′ UTR of lmo0514, encoding an LPXTG cell wall protein that is also upregulated during intracellular growth (89). Lmo0514 is required for survival of the bacteria in plasma and during infection of mice (90). The 5′ UTR of lmo0514 contains an inhibitory RNA structure that sequesters the SD region and blocks translation initiation. Binding of Rli27 to the 5′ UTR disrupts the inhibitory structure, thereby allowing intracellular expression of Lmo0514 (89).
Rli38 is a long (514 nt) sRNA that is highly induced when L. monocytogenes is exposed to blood (86). A Δrli38 mutant strain exhibited reduced bacterial loads in several organs compared to the wild-type strain. The target of Rli38 (RNA or protein) is not yet known, nor is its mechanism of action. Another sRNA induced when L. monocytogenes is exposed to blood is RliB (86). In contrast to Rli38, the absence of RliB increased bacterial loads in the liver compared to the wild-type strain. RliB is a substrate for polynucleotide phosphorylase, but it is also a clustered regularly interspaced short palindromic repeat (CRISPR) element, devoid of associated Cas proteins (91). However, presence of trans-encoded Cas proteins and polynucleotide phosphorylase allows RliB to exert DNA interference directed against matching protospacers. Why Listeria has a degenerated CRISPR system is not clear and deserves further study.
Possibly the most well-known sRNAs in L. monocytogenes are the sibling family of LhrC, consisting of seven members, all with varying degrees of conservation (86, 92–95). The LhrCs control expression of at least three virulence factors: lapB, encoding an adhesin, oppA, encoding an oligopeptide protein, and tcsA, encoding a CD4+ T cell-stimulating antigen. The different members of the LhrC family are able to bind to the 5′ UTRs of their targets. For lapB and oppA, the LhrCs directly interact with their SD region to inhibit translation, whereas the LhrCs destabilizes the tcsA transcript by binding a site distally from the SD region (93, 95, 96). Interestingly, two CU-rich binding sites in LhrC4 were identified to be important for oppA binding, giving the possibility of two target mRNAs being sequestered by one sRNA. Another exciting aspect of the LhrC family is that the expression of individual members is controlled by exposure to various environmental cues, such as blood and growth in the intestine and in macrophages through at least one bacterial two-component signaling system (93).
The functions of several sRNAs in bacteria are dependent on RNA chaperones. In Gram-negative bacteria, Hfq plays a vital role, stimulating interaction between short complementary sequences of the sRNA and the target mRNA (97). The role of Hfq appears to be more crucial if the level of complementarity between the RNAs is low. Although the phenotypes of a Δhfq strain are less pronounced in L. monocytogenes than a Δhfq strain in Gram-negative bacteria, Hfq has been shown to be associated with sRNAs and to contribute to L. monocytogenes virulence (84, 98). However, the interaction between the sRNA LhrA and the target mRNA lmo0850 is the only example so far described in L. monocytogenes when Hfq is needed for a functional regulatory outcome (99). Other RNA chaperones could in some cases replace Hfq.
The SpoVG protein of L. monocytogenes is an RNA binding protein that regulates lysozyme resistance, virulence, and swarming motility (100). The sRNA Rli31 was shown to interact with both the 5′ UTR of the spoVG transcript and the SpoVG protein (100). Surprisingly, Rli31 did not appear to affect the levels of either the spoVG transcript or the SpoVG protein, raising the possibility that Rli31 acts as a “sink” to inactivate SpoVG, in analogy with the CsrA/Rsm systems of posttranscriptional regulators in Gram-negative bacteria. Finally, RNA helicases have been shown to contribute to virulence gene expression in L. monocytogenes (101, 102). sRNAs are becoming increasingly recognized for their contributions to diverse aspects of L. monocytogenes physiology and virulence.
Regulation Induced in Response to Environmental Stress
During its life as a saprophyte and pathogen, L. monocytogenes encounters a diversity of stress-conditions, many of them potentially life-threatening. To survive disparate stresses, the bacterium makes use of a number of strategies. As mentioned previously, the expression of several proteins important for stress survival depends on the stress sigma factor σB, which competes with the vegetative sigma factor σA for binding to the RNA polymerase (103, 104). Activity of σB is controlled by a complex relay of protein interactions and phosphorylation events, ultimately freeing σB from the anti-sigma factor RsbW. At the top of this relay hierarchy lies a sensory organelle, the “stressosome,” which is a large (∼1.8 MDa) multiprotein complex, thus far primarily studied in Bacillus subtilis (105). The stressosome is able to detect and integrate environmental cues into a signal transduction pathway that eventually allows the liberation of σB and initiation of the expression of σB-regulated genes.
In L. monocytogenes, it has been suggested that there may be interplay between σB and PrfA activity, such that σB controls genes important for the initial steps of infection (i.e., during survival in the gut and adhesion to intestinal epithelial cells), whereas PrfA controls the expression of genes involved in subsequent steps (i.e., adhesion and intracellular growth as well as further dissemination in the host) (25, 26, 86, 106, 107). Interestingly, some genes, such as the genes encoding the two important invasins, InlA and InlB, are controlled by both σB and PrfA (27, 108). This dual regulation allows the expression of the bicistronic inlA-inlB message from two promoters, integrating several external cues and thereby permitting bacterial invasion of several host cell types. Both the σB- and PrfA-generated inlA-inlB transcripts contain unusually long 5′ UTRs (445 and 396 nt, respectively) possibly acting at the posttranscriptional level to sense environmental factors (27). Whether the stressosome is able to act as a central hub to integrate external signals to control virulence gene expression in concert with other regulators (e.g., PrfA) remains to be determined. Very little is known about the mechanisms underlying stressosome sensory perception and signal integration. It has been shown that a blue-light receptor can sense light and activate σB through the stressosome, but the exact mechanism is not known (109–111). A small peptide, Prli42, has been demonstrated to bind the stressosome, and its absence reduces σB-dependent gene expression while also affecting expression of virulence factors (112). Prli42 harbors a transmembrane domain and is associated with the membrane, potentially allowing the stressosome to sense stress exerted through membrane disturbance.
Fatty Acids and Regulation of Virulence
Many enterobacteria use fatty acids (FAs) to regulate virulence gene expression. For example, Vibrio cholerae, the causative agent of cholera, controls the expression of cholera toxin and toxin coregulated pilus using the transcriptional activator ToxT; unsaturated long-chain FAs were shown to directly bind to and repress activity of ToxT (113). Using the X-ray structure of palmitoleic acid bound to ToxT as a blueprint, highly effective ToxT inhibitors were synthesized that were able to inhibit intestinal colonization in mice (114). FAs have also been shown to affect virulence factor expression in L. monocytogenes and have been suggested to interfere with PrfA activity (115, 116). The addition of increasing amounts of non-branched-chain FAs (both saturated and nonsaturated) to bacterial cultures was found to reduce virulence factor expression (116). A similar effect was observed by decreasing the bacterial content of branched-chain FAs (115), possibly because this increases the amount of non-branched-chain FAs. Interestingly, a strain carrying a constitutively active PrfA protein (PrfAG155S) also exhibited reduced virulence factor expression in the presence of FA, suggesting that the repressive effect of FAs lies at a level subsequent to PrfA-activation (116). Additional work is required to decipher if FAs interact directly with PrfA or if the effect of the FAs is mediated through another pathway.
Regulation by Two-Component Sensing Systems
To sense and respond to environmental cues, many pathogenic bacteria use two-component systems (TCSs). Typically, the TCS consists of a histidine kinase (HK) that senses an external signal and transfers a phosphor group to activate a response regulator (RR) controlling the expression of genes important for the response of the particular stimulus (117). L. monocytogenes harbors 16 TCSs, one of which, DegU, is an orphan RR (118). Several of the Listeria TCSs have been implicated in virulence (119–122). One of them is part of the accessory gene regulator (Agr) system of L. monocytogenes, consisting of a quorum sensing/TCS system (119). In the Agr system, a small auto-inducing peptide (AIP, derivative of the agrD gene product) is processed and transported by AgrB to the outside of the bacterium. The AIP of L. monocytogenes was shown to be a cyclic pentapeptide (CFMFV) (123). Once outside, the AIP is recognized by the HK AgrC, which activates the RR AgrA. Exactly how the Agr system contributes to L. monocytogenes virulence is unclear, since the bacterium lacks RNAIII, an sRNA expressed divergently from the agr locus and which controls the expression of several virulence factors in Staphylococcus aureus (124). Another TCS implicated in L. monocytogenes virulence is LisrK (120). Loss of LisK (the HK) reduced bacterial growth in the spleens of mice. Interestingly, LisRK has been shown to be important for expression of the serine-protease HtrA and the sRNA LhrC, both of which contribute to L. monocytogenes pathogenesis (93, 125). The flagellin-associated TCSs CheY and CheA are important for listerial adhesion and invasion of cultured Caco-2 cells; however, bacteria lacking this system exhibited normal growth within the spleens of mice (121). The DegU RR appears to be of interest, because it lacks its associated HK, and the absence of DegU impairs L. monocytogenes growth in mice (126–128). DegU is also the epistatic regulator of motility by positively activating expression of the antirepressor GmaR, which in turn, inactivates the repressor of motility MogR (129). Having no accompanying kinase, DegU appears to be phosphorylated by intracellular acetyl phosphate (130). The VirRS TCS system was originally identified using a signature tagged mutagenesis screen seeking genes important for mouse infection (122). Surprisingly, VirR can be activated independently of the HK VirS, possibly through variations in bacterial intracellular acetyl phosphate concentrations. VirR controls the expression of several virulence-associated genes upregulated during infection (12). Recently, it was shown that the VirAB ABC transporter controls VirR function in response to the antimicrobial Nisin and/or host factors (131).
Regulation of Virulence via Peptide Pheromones
In addition to the peptide-responsive Agr system discussed above, an additional peptide pheromone system has been recently shown to contribute to L. monocytogenes virulence. A PrfA-inducible secreted peptide pheromone-encoding lipoprotein (PplA) was identified that shares significant homology with the Enterococcus faecalis Cad lipoprotein encoding the cAD1 peptide pheromone (132, 133). Similar to the cAD1 pheromone of E. faecalis, the pPplA peptide is processed from the released PplA lipoprotein N-terminal signal sequence (SS) peptide following secretion of the lipoprotein through the general secretory pathway. In contrast to E. faecalis, where the cAD1 pheromone stimulates a mating response between plasmid-containing and plasmid-free cells, the pPplA peptide pheromone has functionally evolved to enhance vacuolar escape of L. monocytogenes within nonprofessional phagocytic cells. Studies of mutants lacking both the PplA lipoprotein and its signal sequence-encoded peptide pheromone versus the lipoprotein alone have demonstrated that the pPplA peptide pheromone is a critical virulence factor that contributes to both bacterial aggregation in broth culture and survival in mouse models of infection. The PplA lipoprotein has no apparent role in vacuolar escape; however, its function as an extracellular electron transport protein that contributes to bacterial fitness within the gut has recently been described (134).
L. monocytogenes mutants lacking the pPplA peptide exhibit delays in escape from the vacuoles of nonprofessional phagocytic cells but escape with normal kinetics from the phagosomes of macrophage-like cell lines or in bone marrow-derived macrophages (132). Interestingly, loss of the pPplA pheromone did not impair LLO-dependent perforation of the vacuole, only bacterial escape into the cytosol. These data strongly suggest that complete vacuole escape requires an additional mechanism beyond initial pore formation mediated by the activity of the hemolysin LLO. The pPplA peptide was also found to contribute to maintenance of surface-associated and secreted proteins, and it is possible that some of these gene products may contribute to either the stabilization of LLO-induced membrane pores and/or the physical disruption of the vacuole membrane (Fig. 5).
FIGURE 5.
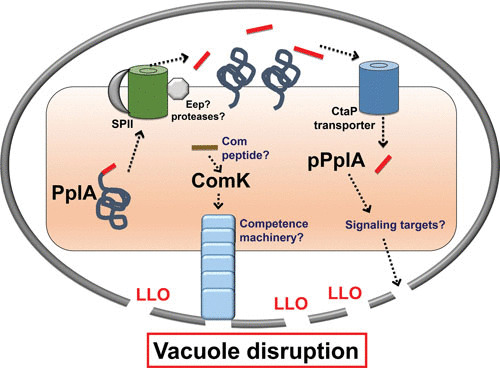
L. monocytogenes peptide signaling and expression of competence gene products via phage excision within the host vacuole. pplA encodes a lipoprotein (PplA) with a peptide pheromone (pPplA) located within the N terminal secretion signal peptide (shown in red). The signal sequence of prePplA is processed by signal peptidase II (SPII), and the released signal peptide is further cleaved by the protease Eep, releasing the pPplA pheromone, while the PplA protein becomes lipid modified and associated with the membrane. The confined space of the vacuole leads to import of the secreted pPplA pheromone, presumably stimulating a signaling cascade that results in the production of an unknown factor(s) that contributes to vacuole lysis. As part of a separate pathway, an unknown signal, potentially also involving a peptide pheromone, leads to the expression of ComK and the expression of a competence-associated pilus that may also aid in vacuole membrane disruption. Select L. monocytogenes comK genes harbor a lysogenic phage that must excise to enable ComK expression. The ComK pathway is required for L. monocytogenes vacuole escape from professional phagocytic cells; the pPplA system is required for bacterial escape from nonprofessional phagocytic cells.
Interestingly, all bacterial virulence defects associated with the loss of the pPplA peptide could be completely compensated for by the introduction of constitutively active PrfA*, suggesting a possible connection between the pPplA signaling pathway and PrfA activation (132). These studies suggest a model in which L. monocytogenes senses the confines of the host vacuoles as a result of the accumulation and import of the pPplA peptide, leading to PrfA activation and enhanced bacterial escape into the cytosol. The L. monocytogenes pPplA peptide thus appears to have evolved to enable the bacterium to sense the confined environment of the vacuole so as to induce the expression of gene products required for full membrane disruption and entry of the bacterium into the cytosol.
In addition to the pPplA peptide-signaling pathway, another potential peptide-based signaling system related to competence development has been implicated in enhancing escape of L. monocytogenes from host cell vacuoles, specifically, the phagosomes of macrophages (135). Competence is a brief physiological state during which the bacterial cells collectively become primed to transport extracellular DNA across the cell wall and bacterial membrane, at times resulting in integration of the newly acquired DNA into the bacterial genome (136). Several gene products, including those encoded by com genes, are required for peptide pheromone signaling and DNA uptake. Although it has not been demonstrated to exhibit natural competence, L. monocytogenes possesses a number of com genes that share homology with those involved in competence development in other bacteria; however, not all the genes required for competence are present (118). Most but not all of the regulatory components and late genes involved in assembly of the Com apparatus required for DNA transport are present in the Listeria genome. Because of these missing components, it is unclear if the competence system is functional in L. monocytogenes, and natural competence has not been demonstrated under laboratory conditions.
One competence regulator that is encoded by L. monocytogenes is ComK, a master transcriptional regulator that induces the expression of late com genes involved in assembly of the Com apparatus for DNA uptake, and it is this system which has been reported to be important for L. monocytogenes in vacuole escape of professional phagocytic cells (135). Interestingly, the comK gene in some strains of L. monocytogenes is inactivated by the presence of an A118-like prophage that integrates at a specific attachment site within the comK gene. However, Rabinovich et al. have found that the late competence genes regulated by ComK are highly expressed during intracellular growth as a result of prophage excision (135). In addition, mutants missing the specific components of the pseudopilus or the DNA translocation channel were impaired for vacuole escape in macrophages and were attenuated for bacterial virulence in mice, whereas the DNA binding components were dispensable for these processes. A strain cured of the prophage and containing an intact comK gene grew similarly to wild-type L. monocytogenes in macrophages, and many naturally occurring strains of L. monocytogenes lack this prophage and are capable of growth within macrophages. For strains that do contain prophages, data suggest that prophage integration/excision serves to regulate virulence gene expression and that the ComK system is somehow involved in sensing the presence of L. monocytogenes within host cell vacuoles in professional phagocytic cells. Thus, while no peptide pheromone has yet been identified to stimulate the induction of the L. monocytogenes com genes within the phagosome, it could be hypothesized that the bacterium has adapted components of the com peptide pheromone system for spatial sensing, similar to pPplA (Fig. 5).
The requirement of selected com gene products and the pPplA peptide for vacuole escape in different cell types makes it tempting to speculate that a competence-like pseudopilus may help stabilize the pores initially formed by LLO or that maybe the pseudopilus exerts physical pressure on the vacuole membrane to aid in membrane disruption and bacterial entry into the cytosol. It is possible that the pPplA and ComK systems are interconnected in some way and that fundamental differences in the vacuole membranes of professional phagocytic cells versus nonprofessional phagocytic cells necessitate the requirement for differentially regulated components. Additional characterization of the gene products regulated by pPplA and com gene products may clarify how these components function to enhance bacterial entry into the cytosol.
MAKING NEW FRIENDS: REGULATION AND THE EXAMINATION OF L. MONOCYTOGENES STRAINS BEYOND THE COMMONLY USED ISOLATES
Most studies of L. monocytogenes have been performed using EGDe, EGD, or 10403S strains, all belonging to lineage II. However, the most severe outbreaks of listeriosis are caused by L. monocytogenes strains belonging to a subset of lineage I (137, 138). These lineage I strains contain an additional pathogenicity island, LIP-III, which appears crucial for the higher virulence potential of lineage I strains. LIP-III encodes eight proteins involved in the synthesis, modification, and transport of listeriolysin S (LLS) (137). LLS was identified as a hemolysin and cytotoxin contributing to L. monocytogenes pathogenicity. This finding was challenged by a study suggesting that LLS was not important for infection or killing of eukaryotic cells (139). Instead, LLS was shown to act as a bacteriocin, potentiating survival of L. monocytogenes in the microbiota (140, 141). Bacteriocins are peptides capable of killing bacteria closely related to the bacteriocin-producing strain. It should, however, be noted that bacteriocins produced by another bacterium, Staphylococcus pseudintermedius, can act both as a bacteriocin and as a cytotoxin (142). An L. monocytogenes lineage I strain overexpressing LLS was shown to kill Lactococcus lactis and S. aureus as well as L. monocytogenes strains of lineage II in vitro (140). It was demonstrated that orally administered lineage I strains carrying LLS affected the microbiota in mice, whereas the microbiota remained undisturbed if infected with strains lacking LLS (140). Two of the bacterial genera that were significantly decreased in the gut in the presence of lineage I were Alloprevotella and Allobaculum. Interestingly, these two genera produced butyric acid, a fatty acid that has been shown to decrease L. monocytogenes virulence gene expression, possibly by interfering with PrfA activity (as mentioned earlier in this article). In contrast to LIP-I and LIP-II, expression of genes from LIP-III is not governed by PrfA, thereby allowing expression of LIP-III under conditions that repress PrfA activity (143). It can thus be speculated that LLS-producing L. monocytogenes kill bacteria that otherwise would impair PrfA-directed expression of factors needed for passage through the intestinal barrier. The production of LLS in lineage I strains may at least partially explain why this lineage is more often associated with outbreaks than linage II. The induced expression of LIP-III in lineage II strains might answer if other factors in lineage I strains also contribute to infection.
TARGETING REGULATION: THE SEARCH FOR NOVEL ANTIMICROBIALS EFFECTIVE AGAINST L. MONOCYTOGENES
Although resistance to traditional antibiotics is fortunately still limited, the increased occurrence of horizontal gene transfer makes the development of new antibacterial compounds targeting L. monocytogenes relevant. Stress survival mechanisms are clearly critical for L. monocytogenes existence, and a number of attempts have been undertaken to identify novel antibacterial and/or antivirulence chemical compounds targeting facets of L. monocytogenes stress resistance and/or virulence. Through a high-throughput screen, a substance (FPSS) was identified that could block the σB regulon and thereby also several genes encoding virulence factors (144). The exact mechanism of action of FPSS remains elusive (for example, direct interaction with σB or with another upstream component); however, the prospect of being able to inhibit the general stress response and virulence gene expression is exciting.
A different approach using analogs to metabolites that bind and control purine riboswitches led to the identification of 6-aminohydroxyl purine (6-N-HAP) as being able to exhibit both antibacterial and antivirulence properties (145). The target of 6-N-HAP has not yet been revealed, but addition of 6-N-HAP increased the bacterial mutation rate by almost 10,000-fold. Another screening approach identified a natural flavonoid (fisetin) as an efficient inhibitor of LLO activity (146, 147). Molecular modeling and interaction studies suggested that fisetin directly interacts with loops 2 and 3 of LLO, thereby preventing cholesterol binding and LLO oligomerization. Interestingly, fisetin prolonged the survival of mice infected with L. monocytogenes. Yet another screening-based approach was used to identify heterocyclic 2-pyridones as efficient inhibitors of L. monocytogenes infection and virulence factor expression; the target of the 2-pyridones was shown to be PrfA (148, 149). A 2-pyridone:PrfA cocrystal revealed that the compound bound in an intraprotein “tunnel” site, pulling the DNA-binding helix-turn-helix motif in an unfavorable position. The possibility of using tailor-made substances (such as fisetin and 2-pyridones) to inactivate specific targets (LLO and PrfA, respectively) is very appealing since it should only impair L. monocytogenes while leaving the remainder of the host microbiota untouched.
CONCLUDING REMARKS AND FUTURE OUTLOOK
L. monocytogenes is notable for its ability to survive and thrive both as an extraordinary pathogen of diverse animal hosts and as a saprophyte in the soil; to accomplish this bacterial gymnastic feat, L. monocytogenes utilizes an integrated myriad of regulatory systems (Fig. 6). The most centrally important virulence regulator identified to date is the transcriptional activator PrfA, which controls the expression of the majority of virulence factors in response to environmental cues such as redox status, temperature, energy levels, and potentially in combination with diffusion sensing of the limited space of the vacuole. PrfA is itself regulated by an impressive number of mechanisms that include transcriptional and posttranscriptional regulation as well as translational and posttranslational regulation, each combining to integrate environmental signals into an appropriate regulatory output of bacterial gene expression. It will be intriguing to study single bacterial cells to better understand how the PrfA regulatory network integrates into other sensory systems and how these systems collectively contribute to virulence. Despite being the subject of extensive study for the past 30 years, L. monocytogenes still appears to hold regulatory secrets that await discovery.
FIGURE 6.
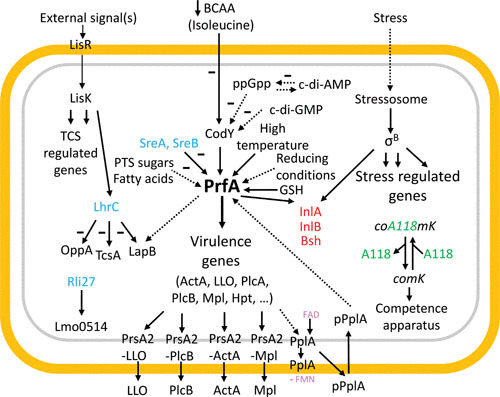
A summary of regulatory events underlying stress regulation and virulence factor expression in L. monocytogenes, especially highlighting the role of the PrfA system, but also TCS (here represented by LisRK), sRNAs (blue), and σB. Gene products regulated by both PrfA and σB are shown in red. Consult the text for further details.
Contributor Information
Jörgen Johansson, Department of Molecular Biology, Laboratory for Molecular Infection Medicine Sweden (MIMS) and Umeå Centre for Microbial Research (UCMR), Umeå University, 90187 Umeå, Sweden.
Nancy E. Freitag, Department of Microbiology and Immunology, University of Illinois at Chicago, Chicago IL
Vincent A. Fischetti, The Rockefeller University, New York, NY
Richard P. Novick, Skirball Institute for Molecular Medicine, NYU Medical Center, New York, NY
Joseph J. Ferretti, Department of Microbiology & Immunology, University of Oklahoma Health Science Center, Oklahoma City, OK
Daniel A. Portnoy, Department of Molecular and Cellular Microbiology, University of California, Berkeley, Berkeley, CA
Miriam Braunstein, Department of Microbiology and Immunology, University of North Carolina-Chapel Hill, Chapel Hill, NC.
Julian I. Rood, Infection and Immunity Program, Monash Biomedicine Discovery Institute, Monash University, Melbourne, Australia
REFERENCES
- 1.Radoshevich L, Cossart P. 2018. Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol 16:32–46 10.1038/nrmicro.2017.126. [PubMed] [DOI] [PubMed] [Google Scholar]
- 2.Pizarro-Cerdá J, Cossart P. 2018. Listeria monocytogenes: cell biology of invasion and intracellular growth. Microbiol Spectr 6:GPP3-0013-2018 10.1128/microbiolspec.GPP3-0013-2018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drolia R, Bhunia AK. 2019. Crossing the intestinal barrier via Listeria adhesion protein and internalin A. Trends Microbiol 27:408–425 10.1016/j.tim.2018.12.007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 4.Lowe DE, Robbins JR, Bakardjiev AI. 2018. Animal and human tissue models of vertical Listeria monocytogenes transmission and implications for other pregnancy-associated infections. Infect Immun 86:e00801-17 10.1128/IAI.00801-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamond NM, Freitag NE. 2018. Vertical transmission of Listeria monocytogenes: Probing the balance between protection from pathogens and fetal tolerance. Pathogens 7:E52 10.3390/pathogens7020052. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnupf P, Portnoy DA. 2007. Listeriolysin O: a phagosome-specific lysin. Microbes Infect 9:1176–1187 10.1016/j.micinf.2007.05.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 7.Grubmüller S, Schauer K, Goebel W, Fuchs TM, Eisenreich W. 2014. Analysis of carbon substrates used by Listeria monocytogenes during growth in J774A.1 macrophages suggests a bipartite intracellular metabolism. Front Cell Infect Microbiol 4:156. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillich H, Puri M, Chakraborty T. 2017. ActA of Listeria monocytogenes and its manifold activities as an important listerial virulence factor. Curr Top Microbiol Immunol 399:113–132 10.1007/82_2016_30. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Rajabian T, Gavicherla B, Heisig M, Müller-Altrock S, Goebel W, Gray-Owen SD, Ireton K. 2009. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat Cell Biol 11:1212–1218 10.1038/ncb1964. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cahoon LA, Freitag NE. 2014. Listeria monocytogenes virulence factor secretion: don’t leave the cell without a chaperone. Front Cell Infect Microbiol 4:13 10.3389/fcimb.2014.00013. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun 74:1323–1338 10.1128/IAI.74.2.1323-1338.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camejo A, Buchrieser C, Couvé E, Carvalho F, Reis O, Ferreira P, Sousa S, Cossart P, Cabanes D. 2009. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog 5:e1000449 10.1371/journal.ppat.1000449. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leimeister-Wächter M, Haffner C, Domann E, Goebel W, Chakraborty T. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc Natl Acad Sci USA 87:8336–8340 10.1073/pnas.87.21.8336. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mengaud J, Dramsi S, Gouin E, Vazquez-Boland JA, Milon G, Cossart P. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol Microbiol 5:2273–2283 10.1111/j.1365-2958.1991.tb02158.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 15.Chakraborty T, Leimeister-Wächter M, Domann E, Hartl M, Goebel W, Nichterlein T, Notermans S. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J Bacteriol 174:568–574 10.1128/jb.174.2.568-574.1992. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freitag NE, Youngman P, Portnoy DA. 1992. Transcriptional activation of the Listeria monocytogenes hemolysin gene in Bacillus subtilis. J Bacteriol 174:1293–1298 10.1128/jb.174.4.1293-1298.1992. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Böckmann R, Dickneite C, Middendorf B, Goebel W, Sokolovic Z. 1996. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol Microbiol 22:643–653 10.1046/j.1365-2958.1996.d01-1722.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 18.Dussurget O, Cabanes D, Dehoux P, Lecuit M, Buchrieser C, Glaser P, Cossart P, European Listeria Genome Consortium. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol 45:1095–1106 10.1046/j.1365-2958.2002.03080.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 19.Sleator RD, Wemekamp-Kamphuis HH, Gahan CG, Abee T, Hill C. 2005. A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol Microbiol 55:1183–1195 10.1111/j.1365-2958.2004.04454.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 20.Milohanic E, Glaser P, Coppée JY, Frangeul L, Vega Y, Vázquez-Boland JA, Kunst F, Cossart P, Buchrieser C. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol 47:1613–1625 10.1046/j.1365-2958.2003.03413.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Leimeister-Wächter M, Domann E, Chakraborty T. 1992. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J Bacteriol 174:947–952 10.1128/jb.174.3.947-952.1992. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freitag NE, Rong L, Portnoy DA. 1993. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect Immun 61:2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freitag NE, Portnoy DA. 1994. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol Microbiol 12:845–853 10.1111/j.1365-2958.1994.tb01070.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 24.Schwab U, Bowen B, Nadon C, Wiedmann M, Boor KJ. 2005. The Listeria monocytogenes prfAP2 promoter is regulated by sigma B in a growth phase dependent manner. FEMS Microbiol Lett 245:329–336 10.1016/j.femsle.2005.03.025. [PubMed] [DOI] [PubMed] [Google Scholar]
- 25.Nadon CA, Bowen BM, Wiedmann M, Boor KJ. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect Immun 70:3948–3952 10.1128/IAI.70.7.3948-3952.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauch M, Luo Q, Müller-Altrock S, Goebel W. 2005. SigB-dependent in vitro transcription of prfA and some newly identified genes of Listeria monocytogenes whose expression is affected by PrfA in vivo. J Bacteriol 187:800–804 10.1128/JB.187.2.800-804.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stritzker J, Schoen C, Goebel W. 2005. Enhanced synthesis of internalin A in aro mutants of Listeria monocytogenes indicates posttranscriptional control of the inlAB mRNA. J Bacteriol 187:2836–2845 10.1128/JB.187.8.2836-2845.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. 2003. Listeria monocytogenes sigma B regulates stress response and virulence functions. J Bacteriol 185:5722–5734 10.1128/JB.185.19.5722-5734.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guldimann C, Guariglia-Oropeza V, Harrand S, Kent D, Boor KJ, Wiedmann M. 2017. Stochastic and differential activation of σB and PrfA in Listeria monocytogenes at the single cell level under different environmental stress conditions. Front Microbiol 8:348 10.3389/fmicb.2017.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobel L, Sigal N, Borovok I, Belitsky BR, Sonenshein AL, Herskovits AA. 2015. The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene prfA. Mol Microbiol 95:624–644 10.1111/mmi.12890. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brenner M, Lobel L, Borovok I, Sigal N, Herskovits AA. 2018. Controlled branched-chain amino acids auxotrophy in Listeria monocytogenes allows isoleucine to serve as a host signal and virulence effector. PLoS Genet 14:e1007283 10.1371/journal.pgen.1007283. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lobel L, Herskovits AA. 2016. Systems level analyses reveal multiple regulatory activities of CodY controlling metabolism, motility and virulence in Listeria monocytogenes. PLoS Genet 12:e1005870 10.1371/journal.pgen.1005870. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renzoni A, Klarsfeld A, Dramsi S, Cossart P. 1997. Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infect Immun 65:1515–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551–561 10.1016/S0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 35.Neupert J, Karcher D, Bock R. 2008. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res 36:e124 10.1093/nar/gkn545. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loh E, Kugelberg E, Tracy A, Zhang Q, Gollan B, Ewles H, Chalmers R, Pelicic V, Tang CM. 2013. Temperature triggers immune evasion by Neisseria meningitidis. Nature 502:237–240 10.1038/nature12616. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loh E, Memarpour F, Vaitkevicius K, Kallipolitis BH, Johansson J, Sondén B. 2012. An unstructured 5′-coding region of the prfA mRNA is required for efficient translation. Nucleic Acids Res 40:1818–1827 10.1093/nar/gkr850. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. 2009. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139:770–779 10.1016/j.cell.2009.08.046. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Lotz TS, Suess B. 2018. Small-molecule-binding riboswitches. Microbiol Spectr 6:RWR-0025-2018 10.1128/microbiolspec.RWR-0025-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ignatov D, Johansson J. 2017. RNA-mediated signal perception in pathogenic bacteria. Wiley Interdiscip Rev RNA 8:e1429 10.1002/wrna.1429. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Thorsing M, Dos Santos PT, Kallipolitis BH. 2018. Small RNAs in major foodborne pathogens: from novel regulatory activities to future applications. Curr Opin Biotechnol 49:120–128 10.1016/j.copbio.2017.08.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 42.Kolb A, Busby S, Buc H, Garges S, Adhya S. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem 62:749–795 10.1146/annurev.bi.62.070193.003533. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Reniere ML, Whiteley AT, Hamilton KL, John SM, Lauer P, Brennan RG, Portnoy DA. 2015. Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517:170–173 10.1038/nature14029. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall M, Grundström C, Begum A, Lindberg MJ, Sauer UH, Almqvist F, Johansson J, Sauer-Eriksson AE. 2016. Structural basis for glutathione-mediated activation of the virulence regulatory protein PrfA in Listeria. Proc Natl Acad Sci USA 113:14733–14738 10.1073/pnas.1614028114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reniere ML, Whiteley AT, Portnoy DA. 2016. An in vivo selection identifies Listeria monocytogenes genes required to sense the intracellular environment and activate virulence factor expression. PLoS Pathog 12:e1005741 10.1371/journal.ppat.1005741. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Feng H, Zhu Y, Gao P. 2017. Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes. Protein Cell 8:308–312 10.1007/s13238-017-0390-x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krypotou E, Scortti M, Grundström C, Oelker M, Luisi BF, Sauer-Eriksson AE, Vázquez-Boland J. 2019. Control of bacterial virulence through the peptide signature of the habitat. Cell Rep 26:1815–1827.e5 10.1016/j.celrep.2019.01.073. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ripio MT, Domínguez-Bernal G, Lara M, Suárez M, Vazquez-Boland JA. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J Bacteriol 179:1533–1540 10.1128/jb.179.5.1533-1540.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garges S, Adhya S. 1988. Cyclic AMP-induced conformational change of cyclic AMP receptor protein (CRP): intragenic suppressors of cyclic AMP-independent CRP mutations. J Bacteriol 170:1417–1422 10.1128/jb.170.4.1417-1422.1988. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vega Y, Dickneite C, Ripio MT, Böckmann R, González-Zorn B, Novella S, Domínguez-Bernal G, Goebel W, Vázquez-Boland JA. 1998. Functional similarities between the Listeria monocytogenes virulence regulator PrfA and cyclic AMP receptor protein: the PrfA* (Gly145Ser) mutation increases binding affinity for target DNA. J Bacteriol 180:6655–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eiting M, Hagelüken G, Schubert WD, Heinz DW. 2005. The mutation G145S in PrfA, a key virulence regulator of Listeria monocytogenes, increases DNA-binding affinity by stabilizing the HTH motif. Mol Microbiol 56:433–446 10.1111/j.1365-2958.2005.04561.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 52.Port GC, Freitag NE. 2007. Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect Immun 75:5886–5897 10.1128/IAI.00845-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mueller KJ, Freitag NE. 2005. Pleiotropic enhancement of bacterial pathogenesis resulting from the constitutive activation of the Listeria monocytogenes regulatory factor PrfA. Infect Immun 73:1917–1926 10.1128/IAI.73.4.1917-1926.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong KK, Freitag NE. 2004. A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products. J Bacteriol 186:6265–6276 10.1128/JB.186.18.6265-6276.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miner MD, Port GC, Bouwer HG, Chang JC, Freitag NE. 2008. A novel prfA mutation that promotes Listeria monocytogenes cytosol entry but reduces bacterial spread and cytotoxicity. Microb Pathog 45:273–281 10.1016/j.micpath.2008.06.006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miner MD, Port GC, Freitag NE. 2008. Functional impact of mutational activation on the Listeria monocytogenes central virulence regulator PrfA. Microbiology 154:3579–3589 10.1099/mic.0.2008/021063-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xayarath B, Smart JI, Mueller KJ, Freitag NE. 2011. A novel C-terminal mutation resulting in constitutive activation of the Listeria monocytogenes central virulence regulatory factor PrfA. Microbiology 157:3138–3149 10.1099/mic.0.049957-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xayarath B, Volz KW, Smart JI, Freitag NE. 2011. Probing the role of protein surface charge in the activation of PrfA, the central regulator of Listeria monocytogenes pathogenesis. PLoS One 6:e23502 10.1371/journal.pone.0023502. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vega Y, Rauch M, Banfield MJ, Ermolaeva S, Scortti M, Goebel W, Vázquez-Boland JA. 2004. New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulator PrfA. Mol Microbiol 52:1553–1565 10.1111/j.1365-2958.2004.04052.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.Deshayes C, Bielecka MK, Cain RJ, Scortti M, de las Heras A, Pietras Z, Luisi BF, Núñez Miguel R, Vázquez-Boland JA. 2012. Allosteric mutants show that PrfA activation is dispensable for vacuole escape but required for efficient spread and Listeria survival in vivo. Mol Microbiol 85:461–477 10.1111/j.1365-2958.2012.08121.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Velge P, Herler M, Johansson J, Roche SM, Témoin S, Fedorov AA, Gracieux P, Almo SC, Goebel W, Cossart P. 2007. A naturally occurring mutation K220T in the pleiotropic activator PrfA of Listeria monocytogenes results in a loss of virulence due to decreasing DNA-binding affinity. Microbiology 153:995–1005 10.1099/mic.0.2006/002238-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Shetron-Rama LM, Marquis H, Bouwer HG, Freitag NE. 2002. Intracellular induction of Listeria monocytogenes actA expression. Infect Immun 70:1087–1096 10.1128/IAI.70.3.1087-1096.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shetron-Rama LM, Mueller K, Bravo JM, Bouwer HG, Way SS, Freitag NE. 2003. Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol Microbiol 48:1537–1551 10.1046/j.1365-2958.2003.03534.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Ollinger J, Wiedmann M, Boor KJ. 2008. SigmaB- and PrfA-dependent transcription of genes previously classified as putative constituents of the Listeria monocytogenes PrfA regulon. Foodborne Pathog Dis 5:281–293 10.1089/fpd.2008.0079. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alonzo F III, Freitag NE. 2010. Listeria monocytogenes PrsA2 is required for virulence factor secretion and bacterial viability within the host cell cytosol. Infect Immun 78:4944–4957 10.1128/IAI.00532-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Behari J, Youngman P. 1998. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect Immun 66:3635–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Datta AR, Kothary MH. 1993. Effects of glucose, growth temperature, and pH on listeriolysin O production in Listeria monocytogenes. Appl Environ Microbiol 59:3495–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brehm K, Ripio MT, Kreft J, Vázquez-Boland JA. 1999. The bvr locus of Listeria monocytogenes mediates virulence gene repression by beta-glucosides. J Bacteriol 181:5024–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Milenbachs AA, Brown DP, Moors M, Youngman P. 1997. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol Microbiol 23:1075–1085 10.1046/j.1365-2958.1997.2711634.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Park SF, Kroll RG. 1993. Expression of listeriolysin and phosphatidylinositol-specific phospholipase C is repressed by the plant-derived molecule cellobiose in Listeria monocytogenes. Mol Microbiol 8:653–661 10.1111/j.1365-2958.1993.tb01609.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 71.Park SF. 1994. The repression of listeriolysin O expression in Listeria monocytogenes by the phenolic beta-d-glucoside, arbutin. Lett Appl Microbiol 19:258–260 10.1111/j.1472-765X.1994.tb00958.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 72.Herro R, Poncet S, Cossart P, Buchrieser C, Gouin E, Glaser P, Deutscher J. 2005. How seryl-phosphorylated HPr inhibits PrfA, a transcription activator of Listeria monocytogenes virulence genes. J Mol Microbiol Biotechnol 9:224–234 10.1159/000089650. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Deutscher J, Herro R, Bourand A, Mijakovic I, Poncet S. 2005. P-Ser-HPr: a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochim Biophys Acta 1754:118–125 10.1016/j.bbapap.2005.07.029. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Joseph B, Mertins S, Stoll R, Schär J, Umesha KR, Luo Q, Müller-Altrock S, Goebel W. 2008. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J Bacteriol 190:5412–5430 10.1128/JB.00259-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aké FM, Joyet P, Deutscher J, Milohanic E. 2011. Mutational analysis of glucose transport regulation and glucose-mediated virulence gene repression in Listeria monocytogenes. Mol Microbiol 81:274–293 10.1111/j.1365-2958.2011.07692.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Ripio MT, Domínguez-Bernal G, Suárez M, Brehm K, Berche P, Vázquez-Boland JA. 1996. Transcriptional activation of virulence genes in wild-type strains of Listeria monocytogenes in response to a change in the extracellular medium composition. Res Microbiol 147:371–384 10.1016/0923-2508(96)84712-7. [DOI] [PubMed] [Google Scholar]
- 77.Ermolaeva S, Novella S, Vega Y, Ripio MT, Scortti M, Vázquez-Boland JA. 2004. Negative control of Listeria monocytogenes virulence genes by a diffusible autorepressor. Mol Microbiol 52:601–611 10.1111/j.1365-2958.2004.04003.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 78.Bruno JC, Jr, Freitag NE. 2010. Constitutive activation of PrfA tilts the balance of Listeria monocytogenes fitness towards life within the host versus environmental survival. PLoS One 5:e15138 10.1371/journal.pone.0015138. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bruno JC Jr, Freitag NE. 2011. Listeria monocytogenes adapts to long-term stationary phase survival without compromising bacterial virulence. FEMS Microbiol Lett 323:171–179 10.1111/j.1574-6968.2011.02373.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vasanthakrishnan RB, de Las Heras A, Scortti M, Deshayes C, Colegrave N, Vázquez-Boland JA. 2015. PrfA regulation offsets the cost of Listeria virulence outside the host. Environ Microbiol 17:4566–4579 10.1111/1462-2920.12980. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wong KK, Bouwer HG, Freitag NE. 2004. Evidence implicating the 5′ untranslated region of Listeria monocytogenes actA in the regulation of bacterial actin-based motility. Cell Microbiol 6:155–166 10.1046/j.1462-5822.2003.00348.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Shen A, Higgins DE. 2005. The 5′ untranslated region-mediated enhancement of intracellular listeriolysin O production is required for Listeria monocytogenes pathogenicity. Mol Microbiol 57:1460–1473 10.1111/j.1365-2958.2005.04780.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Schnupf P, Hofmann J, Norseen J, Glomski IJ, Schwartzstein H, Decatur AL. 2006. Regulated translation of listeriolysin O controls virulence of Listeria monocytogenes. Mol Microbiol 61:999–1012 10.1111/j.1365-2958.2006.05286.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 84.Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Søgaard-Andersen L, Kallipolitis BH. 2006. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 12:1383–1396 10.1261/rna.49706. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. 2007. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res 35:962–974 10.1093/nar/gkl1096. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Régnault B, Coppée JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956 10.1038/nature08080. [PubMed] [DOI] [PubMed] [Google Scholar]
- 87.Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T. 2011. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res 39:4235–4248 10.1093/nar/gkr033. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Bécavin C, Archambaud C, Cossart P, Sorek R. 2012. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol 8:583 10.1038/msb.2012.11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quereda JJ, Ortega AD, Pucciarelli MG, García-Del Portillo F. 2014. The Listeria small RNA Rli27 regulates a cell wall protein inside eukaryotic cells by targeting a long 5′-UTR variant. PLoS Genet 10:e1004765 10.1371/journal.pgen.1004765. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Quereda JJ, García-Del Portillo F, Pucciarelli MG. 2016. Listeria monocytogenes remodels the cell surface in the blood-stage. Environ Microbiol Rep 8:641–648 10.1111/1758-2229.12416. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Sesto N, Touchon M, Andrade JM, Kondo J, Rocha EP, Arraiano CM, Archambaud C, Westhof É, Romby P, Cossart P. 2014. A PNPase dependent CRISPR system in Listeria. PLoS Genet 10:e1004065 10.1371/journal.pgen.1004065. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mollerup MS, Ross JA, Helfer AC, Meistrup K, Romby P, Kallipolitis BH. 2016. Two novel members of the LhrC family of small RNAs in Listeria monocytogenes with overlapping regulatory functions but distinctive expression profiles. RNA Biol 13:895–915 10.1080/15476286.2016.1208332. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sievers S, Sternkopf Lillebæk EM, Jacobsen K, Lund A, Mollerup MS, Nielsen PK, Kallipolitis BH. 2014. A multicopy sRNA of Listeria monocytogenes regulates expression of the virulence adhesin LapB. Nucleic Acids Res 42:9383–9398 10.1093/nar/gku630. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dos Santos PT, Menendez-Gil P, Sabharwal D, Christensen JH, Brunhede MZ, Lillebæk EMS, Kallipolitis BH. 2018. The small regulatory RNAs LhrC1-5 contribute to the response of Listeria monocytogenes to heme toxicity. Front Microbiol 9:599 10.3389/fmicb.2018.00599. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sievers S, Lund A, Menendez-Gil P, Nielsen A, Storm Mollerup M, Lambert Nielsen S, Buch Larsson P, Borch-Jensen J, Johansson J, Kallipolitis BH. 2015. The multicopy sRNA LhrC controls expression of the oligopeptide-binding protein OppA in Listeria monocytogenes. RNA Biol 12:985–997 10.1080/15476286.2015.1071011. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ross JA, Thorsing M, Lillebæk EMS, Teixeira Dos Santos P, Kallipolitis BH. 2019. The LhrC sRNAs control expression of T cell-stimulating antigen TcsA in Listeria monocytogenes by decreasing tcsA mRNA stability. RNA Biol 16:270–281 10.1080/15476286.2019.1572423. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Santiago-Frangos A, Woodson SA. 2018. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip Rev RNA 9:e1475 10.1002/wrna.1475. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Christiansen JK, Larsen MH, Ingmer H, Søgaard-Andersen L, Kallipolitis BH. 2004. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J Bacteriol 186:3355–3362 10.1128/JB.186.11.3355-3362.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Valentin-Hansen P, Kallipolitis BH. 2010. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res 38:907–919 10.1093/nar/gkp1081. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Burke TP, Portnoy DA. 2016. SpoVG is a conserved RNA-binding protein that regulates Listeria monocytogenes lysozyme resistance, virulence, and swarming motility. MBio 7:e00240 10.1128/mBio.00240-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bäreclev C, Vaitkevicius K, Netterling S, Johansson J. 2014. DExD-box RNA-helicases in Listeria monocytogenes are important for growth, ribosomal maturation, rRNA processing and virulence factor expression. RNA Biol 11:1457–1466 10.1080/15476286.2014.996099. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Netterling S, Bäreclev C, Vaitkevicius K, Johansson J. 2015. RNA helicase important for Listeria monocytogenes hemolytic activity and virulence factor expression. Infect Immun 84:67–76 10.1128/IAI.00849-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wiedmann M, Arvik TJ, Hurley RJ, Boor KJ. 1998. General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol 180:3650–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.O’Byrne CP, Karatzas KA. 2008. The role of sigma B (σB) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Adv Appl Microbiol 65:115–140 10.1016/S0065-2164(08)00605-9. [DOI] [PubMed] [Google Scholar]
- 105.Marles-Wright J, Lewis RJ. 2010. The stressosome: molecular architecture of a signalling hub. Biochem Soc Trans 38:928–933 10.1042/BST0380928. [PubMed] [DOI] [PubMed] [Google Scholar]
- 106.Kazmierczak MJ, Wiedmann M, Boor KJ. 2006. Contributions of Listeria monocytogenes sigmaB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology 152:1827–1838 10.1099/mic.0.28758-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.Guariglia-Oropeza V, Orsi RH, Yu H, Boor KJ, Wiedmann M, Guldimann C. 2014. Regulatory network features in Listeria monocytogenes-changing the way we talk. Front Cell Infect Microbiol 4:14 10.3389/fcimb.2014.00014. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim H, Marquis H, Boor KJ. 2005. SigmaB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 151:3215–3222 10.1099/mic.0.28070-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ondrusch N, Kreft J. 2011. Blue and red light modulates SigB-dependent gene transcription, swimming motility and invasiveness in Listeria monocytogenes. PLoS One 6:e16151 10.1371/journal.pone.0016151. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tiensuu T, Andersson C, Rydén P, Johansson J. 2013. Cycles of light and dark co-ordinate reversible colony differentiation in Listeria monocytogenes. Mol Microbiol 87:909–924 10.1111/mmi.12140. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.O’Donoghue B, NicAogáin K, Bennett C, Conneely A, Tiensuu T, Johansson J, O’Byrne C. 2016. Blue-light inhibition of Listeria monocytogenes growth is mediated by reactive oxygen species and is influenced by σB and the blue-light sensor Lmo0799. Appl Environ Microbiol 82:4017–4027 10.1128/AEM.00685-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Impens F, Rolhion N, Radoshevich L, Bécavin C, Duval M, Mellin J, García Del Portillo F, Pucciarelli MG, Williams AH, Cossart P. 2017. N-terminomics identifies Prli42 as a membrane miniprotein conserved in Firmicutes and critical for stressosome activation in Listeria monocytogenes. Nat Microbiol 2:17005 10.1038/nmicrobiol.2017.5. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lowden MJ, Skorupski K, Pellegrini M, Chiorazzo MG, Taylor RK, Kull FJ. 2010. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc Natl Acad Sci USA 107:2860–2865 10.1073/pnas.0915021107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Woodbrey AK, Onyango EO, Kovacikova G, Kull FJ, Gribble GW. 2018. A modified ToxT inhibitor reduces Vibrio cholerae virulence in vivo. Biochemistry 57:5609–5615 10.1021/acs.biochem.8b00667. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sun Y, Wilkinson BJ, Standiford TJ, Akinbi HT, O’Riordan MX. 2012. Fatty acids regulate stress resistance and virulence factor production for Listeria monocytogenes. J Bacteriol 194:5274–5284 10.1128/JB.00045-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sternkopf Lillebæk EM, Lambert Nielsen S, Scheel Thomasen R, Færgeman NJ, Kallipolitis BH. 2017. Antimicrobial medium- and long-chain free fatty acids prevent PrfA-dependent activation of virulence genes in Listeria monocytogenes. Res Microbiol 168:547–557 10.1016/j.resmic.2017.03.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 117.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215 10.1146/annurev.biochem.69.1.183. [PubMed] [DOI] [PubMed] [Google Scholar]
- 118.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couvé E, de Daruvar A, Dehoux P, Domann E, Domínguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, García-del Portillo F, Garrido P, Gautier L, Goebel W, Gómez-López N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Pérez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, et al. 2001. Comparative genomics of Listeria species. Science 294:849–852. [DOI] [PubMed] [Google Scholar]
- 119.Autret N, Raynaud C, Dubail I, Berche P, Charbit A. 2003. Identification of the agr locus of Listeria monocytogenes: role in bacterial virulence. Infect Immun 71:4463–4471 10.1128/IAI.71.8.4463-4471.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cotter PD, Emerson N, Gahan CG, Hill C. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J Bacteriol 181:6840–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dons L, Eriksson E, Jin Y, Rottenberg ME, Kristensson K, Larsen CN, Bresciani J, Olsen JE. 2004. Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence. Infect Immun 72:3237–3244 10.1128/IAI.72.6.3237-3244.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mandin P, Fsihi H, Dussurget O, Vergassola M, Milohanic E, Toledo-Arana A, Lasa I, Johansson J, Cossart P. 2005. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol Microbiol 57:1367–1380 10.1111/j.1365-2958.2005.04776.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 123.Zetzmann M, Sánchez-Kopper A, Waidmann MS, Blombach B, Riedel CU. 2016. Identification of the agr peptide of Listeria monocytogenes. Front Microbiol 7:989 10.3389/fmicb.2016.00989. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Le KY, Otto M. 2015. Quorum-sensing regulation in staphylococci: an overview. Front Microbiol 6:1174 10.3389/fmicb.2015.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stack HM, Sleator RD, Bowers M, Hill C, Gahan CG. 2005. Role for HtrA in stress induction and virulence potential in Listeria monocytogenes. Appl Environ Microbiol 71:4241–4247 10.1128/AEM.71.8.4241-4247.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Knudsen GM, Olsen JE, Dons L. 2004. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol Lett 240:171–179 10.1016/j.femsle.2004.09.039. [PubMed] [DOI] [PubMed] [Google Scholar]
- 127.Williams T, Joseph B, Beier D, Goebel W, Kuhn M. 2005. Response regulator DegU of Listeria monocytogenes regulates the expression of flagella-specific genes. FEMS Microbiol Lett 252:287–298 10.1016/j.femsle.2005.09.011. [PubMed] [DOI] [PubMed] [Google Scholar]
- 128.Gueriri I, Cyncynatus C, Dubrac S, Arana AT, Dussurget O, Msadek T. 2008. The DegU orphan response regulator of Listeria monocytogenes autorepresses its own synthesis and is required for bacterial motility, virulence and biofilm formation. Microbiology 154:2251–2264 10.1099/mic.0.2008/017590-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 129.Kamp HD, Higgins DE. 2009. Transcriptional and post-transcriptional regulation of the GmaR antirepressor governs temperature-dependent control of flagellar motility in Listeria monocytogenes. Mol Microbiol 74:421–435 10.1111/j.1365-2958.2009.06874.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gueriri I, Bay S, Dubrac S, Cyncynatus C, Msadek T. 2008. The Pta-AckA pathway controlling acetyl phosphate levels and the phosphorylation state of the DegU orphan response regulator both play a role in regulating Listeria monocytogenes motility and chemotaxis. Mol Microbiol 70:1342–1357 10.1111/j.1365-2958.2008.06496.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 131.Grubaugh D, Regeimbal JM, Ghosh P, Zhou Y, Lauer P, Dubensky TW Jr, Higgins DE. 2018. The VirAB ABC transporter is required for VirR regulation of Listeria monocytogenes virulence and resistance to nisin. Infect Immun 86:e00901-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xayarath B, Alonzo F III, Freitag NE. 2015. Identification of a peptide-pheromone that enhances Listeria monocytogenes escape from host cell vacuoles. PLoS Pathog 11:e1004707 10.1371/journal.ppat.1004707. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Clewell DB, Brown BL. 1980. Sex pheromone cAD1 in Streptococcus faecalis: induction of a function related to plasmid transfer. J Bacteriol 143:1063–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Light SH, Su L, Rivera-Lugo R, Cornejo JA, Louie A, Iavarone AT, Ajo-Franklin CM, Portnoy DA. 2018. A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature 562:140–144 10.1038/s41586-018-0498-z. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits AA. 2012. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 150:792–802 10.1016/j.cell.2012.06.036. [PubMed] [DOI] [PubMed] [Google Scholar]
- 136.Sun D. 2018. Pull in and push out: mechanisms of horizontal gene transfer in bacteria. Front Microbiol 9:2154 10.3389/fmicb.2018.02154. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cotter PD, Draper LA, Lawton EM, Daly KM, Groeger DS, Casey PG, Ross RP, Hill C. 2008. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog 4:e1000144 10.1371/journal.ppat.1000144. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Maury MM, Tsai YH, Charlier C, Touchon M, Chenal-Francisque V, Leclercq A, Criscuolo A, Gaultier C, Roussel S, Brisabois A, Disson O, Rocha EPC, Brisse S, Lecuit M. 2016. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet 48:308–313 10.1038/ng.3501. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Quereda JJ, Nahori MA, Meza-Torres J, Sachse M, Titos-Jiménez P, Gomez-Laguna J, Dussurget O, Cossart P, Pizarro-Cerdá J. 2017. Listeriolysin S is a streptolysin S-like virulence factor that targets exclusively prokaryotic cells in vivo. MBio 8:e00259-17 10.1128/mBio.00259-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Quereda JJ, Dussurget O, Nahori MA, Ghozlane A, Volant S, Dillies MA, Regnault B, Kennedy S, Mondot S, Villoing B, Cossart P, Pizarro-Cerda J. 2016. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc Natl Acad Sci USA 113:5706–5711 10.1073/pnas.1523899113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Quereda JJ, Meza-Torres J, Cossart P, Pizarro-Cerdá J. 2017. Listeriolysin S: a bacteriocin from epidemic Listeria monocytogenes strains that targets the gut microbiota. Gut Microbes 8:384–391 10.1080/19490976.2017.1290759. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wladyka B, Piejko M, Bzowska M, Pieta P, Krzysik M, Mazurek Ł, Guevara-Lora I, Bukowski M, Sabat AJ, Friedrich AW, Bonar E, Międzobrodzki J, Dubin A, Mak P. 2015. A peptide factor secreted by Staphylococcus pseudintermedius exhibits properties of both bacteriocins and virulence factors. Sci Rep 5:14569 10.1038/srep14569. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Quereda JJ, Andersson C, Cossart P, Johansson J, Pizarro-Cerdá J. 2018. Role in virulence of phospholipases, listeriolysin O and listeriolysin S from epidemic Listeria monocytogenes using the chicken embryo infection model. Vet Res (Faisalabad) 49:13 10.1186/s13567-017-0496-4. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Palmer ME, Chaturongakul S, Wiedmann M, Boor KJ. 2011. The Listeria monocytogenes σB regulon and its virulence-associated functions are inhibited by a small molecule. MBio 2:e00241-11 10.1128/mBio.00241-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Krajewski SS, Isoz I, Johansson J. 2017. Antibacterial and antivirulence effect of 6-N-hydroxylaminopurine in Listeria monocytogenes. Nucleic Acids Res 45:1914–1924. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang J, Qiu J, Tan W, Zhang Y, Wang H, Zhou X, Liu S, Feng H, Li W, Niu X, Deng X. 2015. Fisetin inhibits Listeria monocytogenes virulence by interfering with the oligomerization of listeriolysin O. J Infect Dis 211:1376–1387 10.1093/infdis/jiu520. [PubMed] [DOI] [PubMed] [Google Scholar]
- 147.Wang J, Zhou X, Liu S, Li G, Zhang B, Deng X, Niu X. 2015. Novel inhibitor discovery and the conformational analysis of inhibitors of listeriolysin O via protein-ligand modeling. Sci Rep 5:8864 10.1038/srep08864. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Good JA, Andersson C, Hansen S, Wall J, Krishnan KS, Begum A, Grundström C, Niemiec MS, Vaitkevicius K, Chorell E, Wittung-Stafshede P, Sauer UH, Sauer-Eriksson AE, Almqvist F, Johansson J. 2016. Attenuating Listeria monocytogenes virulence by targeting the regulatory protein PrfA. Cell Chem Biol 23:404–414 10.1016/j.chembiol.2016.02.013. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kulén M, Lindgren M, Hansen S, Cairns AG, Grundström C, Begum A, van der Lingen I, Brännström K, Hall M, Sauer UH, Johansson J, Sauer-Eriksson AE, Almqvist F. 2018. Structure-based design of inhibitors targeting PrfA, the master virulence regulator of Listeria monocytogenes. J Med Chem 61:4165–4175 10.1021/acs.jmedchem.8b00289. [PubMed] [DOI] [PubMed] [Google Scholar]


