Summary
Acquired resistance to tyrosine kinase inhibitors (TKI), such as osimertinib used to treat EGFR-mutant lung adenocarcinomas, limits long-term efficacy and is frequently caused by non-genetic mechanisms. Here, we define the chromatin accessibility and gene regulatory signatures of osimertinib sensitive and resistant EGFR-mutant cell and patient-derived models and uncover a role for mammalian SWI/SNF chromatin remodeling complexes in TKI resistance. By profiling mSWI/SNF genome-wide localization, we identify both common and cancer cell line-specific gene targets underlying the resistant state. Importantly, genetic and pharmacologic disruption of the SMARCA4/SMARCA2 mSWI/SNF ATPases re-sensitizes a subset of resistant cell lines and an in vivo model to osimertinib via inhibition of mSWI/SNF-mediated regulation of cellular programs governing cell proliferation, epithelial-to-mesenchymal transition, epithelial cell differentiation, and NRF2 signaling. These data highlight the role of mSWI/SNF complexes in supporting TKI resistance and suggest potential utility of mSWI/SNF inhibitors in TKI-resistant lung cancers.
Introduction
Targeted therapies like tyrosine kinase inhibitors (TKIs) have brought precision medicine to the forefront of modern oncology. More than 70 different targeted therapies have been approved by the FDA (Food and Drug Administration) with utility across a broad range of cancers, including lung cancer1–5. Recent decreases in lung cancer mortality are partly attributed to the use of targeted therapies for subsets of oncogene-driven lung adenocarcinomas 6 such as Epidermal Growth Factor Receptor (EGFR)-mutant lung adenocarcinomas 7. Mutations in exons encoding the tyrosine kinase domain of EGFR account for ~15–50% of lung adenocarcinomas 8–10. Tumors harboring most EGFR mutations respond to TKIs which are approved for the first-line treatment of the disease and have improved patient outcomes11.
Targeted therapies face a major drawback: the emergence of acquired resistance which limits their long-term efficacy12. Although individual targeted therapies act on different targets, the mechanisms by which tumor cells become resistant are often shared. Mutations in the gene encoding the drug target (e.g. kinase) or alterations in genes encoding pathway members that bypass the target oncogene are amongst the most common mechanisms of resistance13. However, in many resistant tumors the mechanism of resistance cannot be identified, making their clinical management challenging. These issues are apparent in EGFR-mutant lung adenocarcinomas resistant to TKIs, including the third-generation TKI osimertinib 14, commonly used as first-line therapy for this disease 15. In contrast to tumors treated with earlier-generation TKIs, the proportion of tumors that develop on-target EGFR mutations at acquired resistance is lower with osimertinib 16,17. Emerging evidence reveals that tumors with off-target resistance mechanisms are not only more common, but they also have worse outcomes on TKI treatment 18,19. Therefore, understanding the cellular processes underpinning the emergence of off-target mechanisms of resistance is critical to offer alternative therapeutic approaches to patients.
Epigenetic processes can mediate resistance to targeted therapies and represent novel therapeutic targets, especially in tumors lacking clear genetic mechanisms of resistance20. For example, the histone lysine methyltransferase EZH2 plays a role in the neuroendocrine differentiation of prostate and lung cancer cells, leading to resistance to anti-androgens and TKIs, respectively 21,22. Furthermore, EZH2 inhibition overcomes resistance to anti-androgens in castration resistant prostate cancer 23. In EGFR-mutant lung cancer, inhibition of the histone demethylase KDM5A suppressed the growth of cells that persist upon TKI treatment 24,25 and global DNA methylation affecting key genes (e.g. HOXB9) has been linked to EGFR TKI resistance 26. Yet, overall, how epigenetic processes contribute to resistance is poorly understood.
Mammalian SWI/SNF (or BAF) complexes modulate chromatin architecture by altering DNA-nucleosome contacts and enabling chromatin accessibility 27–30. These complexes are 11–15-subunit entities pieced together from the products of 29 genes into three major forms: cBAF, PBAF, and ncBAF, each featuring unique subunits 27,31. Two ATPases, SMARCA4 and SMARCA2 hydrolyze ATP to power mSWI/SNF complexes. SMARCA4 is mutated in ~8% of non small cell lung cancer (NSCLC). Most of these are truncating/early frameshift mutations resulting in protein loss and rarely co-occur with EGFR mutations or ALK rearrangements, raising the possibility that mSWI/SNF complexes play a different role in these molecular subsets of lung cancer32,33. An emerging body of evidence implicates SMARCA4 in tumor maintenance and oncogenicity 34–40, which has prompted the development of small molecule inhibitors targeting the activity of mSWI/SNF complexes, that are currently being evaluated in clinical trials 41–43(NCT04879017, NCT04891757). For example, overexpression of SMARCA4 is associated with neuroendocrine prostate cancer35. Targeting mSWI/SNF ATPases has been shown to be an effective anti-tumor strategy for castration-resistant prostate tumors in mice that rely on SMARCA2/4 for activation of oncogenic gene expression programs44. Like prostate cancer, EGFR mutant lung adenocarcinomas can also undergo neuroendocrine differentiation following exposure to targeted agents. Altogether, these findings raise the intriguing possibility that SMARCA4 may play a tumor supportive role in EGFR-driven lung cancer.
Here, we discover that mSWI/SNF complexes, and specifically the ATPase activity supported by the SMARCA4/A2 subunits, play pivotal roles in mediating the osimertinib-resistance gene regulatory profile in EGFR-mutant lung adenocarcinoma. Genetic and pharmacologic inhibition of mSWI/SNF complexes results in improved osimertinib efficacy thus presenting these as therapeutic vulnerabilities in osimertinib-resistant lung cancers.
Results
Unique chromatin accessibility changes underpin osimertinib-resistant gene expression programs in EGFR-mutant lung cancer cells
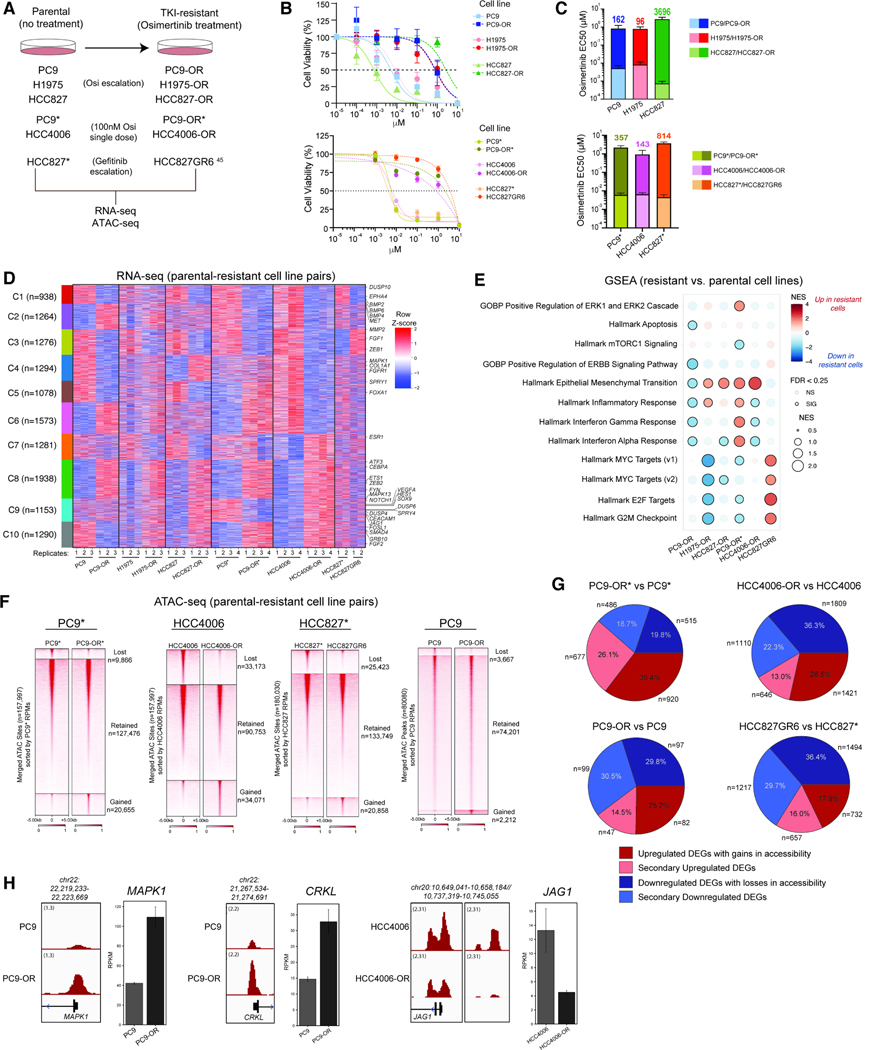
To investigate mechanisms of resistance to osimertinib, we generated five pairs of parental and osimertinib-resistant EGFR-mutant lung cancer cell lines (Figure 1A). We also studied the HCC827* cell line treated with increasing doses of gefitinib (HCC827* and HCC827GR6 cell line pair)45. Parental cell lines exhibited EC50s to osimertinib of 10nM or less while their resistant counterparts showed >90-fold higher EC50s (Figure 1B–C, Table S1). Exome sequencing studies performed on osimertinib-resistant cell lines did not reveal additional mutations in EGFR. Instead, we detected a previously-described loss of CIC in the H1975-OR cells (an R422* mutation) (Fig. S1A) and epithelial to mesenchymal transition (EMT) in the HCC827-OR and HCC4006-OR cells (Fig. S1B–C) 46,47. RAF1 amplification was detected in the PC9-OR cells (Fig. S1D), but neither knock-down of RAF1 nor trametinib (MEKi) treatment attenuated cell line proliferation in combination with osimertinib (Fig. S1E–I) 48. Similarly, PC9-OR* cells acquired a BRAF G469A TKI resistance mutation 49 which was not detected in PC9 parental cells, but PC9-OR* cells remained resistant to the combination of osimertinib and trametinib (Fig. S1J). Of note, HCC827GR6 cells are characterized by MET amplification45. Taken together, these data suggest that coding gene mutations do not play a key role in mediating TKI-resistance in these cell lines which rather may be due to non-genetic (i.e. epigenetic) mechanisms.
Figure 1. Chromatin accessibility and gene regulatory underpinnings of osimertinib resistance in EGFR-mutant lung cancer cell lines.
A. Schematic representation of the generation of the TKI- resistant cell lines. B. Osimertinib dose-response curves for the parental cell lines and their TKI-resistant counterparts (n=3 experimental replicates; mean ± SEM is shown). C. Bar graphs (mean ± SEM) of EC50 values for parental and TKI-resistant isogenic cell line pairs. The fold-change in osimertinib EC50 values between resistant and parental cells is indicated. D. Clustered heatmap performed on n=2–4 RNA-seq profiles (raw RPKM) from all parental and resistant cell line pairs. Examples of significant genes are labeled. RPKM signals were z-scored separately within each cell line pair and combined horizontally to highlight differences between parental and resistant states. E. GSEA for pathway enrichment using DEGs from parental and resistant cell line pairs. F. Heatmap representation of ATAC-seq peaks in PC9*, HCC4006, HCC827* and PC9 parental cell lines and their resistant counterparts sorted by RPKM values over all accessible genomic sites. G. Pie chart representation of proportion of DEGs (resistant cell line vs parental) near concordantly changed ATAC-seq peaks in PC9*, HCC4006, HCC827* and PC9 cell line pairs. H. ATAC-seq tracks over the MAPK1, CRKL and JAG1 loci in the PC9 and HCC4006 cell line pairs. Gene expression RPKM values are shown in bar graphs. Error bars represent the 95% confidence interval around the mean expression level for each cell line. See also Figure S1.
We performed RNA-seq on the cell line pairs to define the gene regulatory profiles of resistant cells. We identified differentially up- and down-regulated genes, many of which were shared between at least 2 cell lines (Figure 1D, S1K–L). Clusters 3 and 8 contained genes downregulated and upregulated, respectively, in each of the five osimertinib- but not gefitinib-resistant states, including genes such as MMP2, and FGF1 (downregulated) and ZEB2, ATF3, ETS-1 and FYN (upregulated). We identified many genes that were differentially regulated uniquely in one resistant cell line, underscoring the heterogeneity and complexity of the resistant state (Figure 1D, S1L). Pathways such as EMT and inflammatory response were upregulated whereas MYC targets, interferon alpha/gamma response signaling pathways were downregulated in more than one cell line (Figure 1E).
We next performed ATAC-seq across four cell line pairs, including PC9*, HCC4006, HCC827* and PC9 as they represent a diverse set of resistance-associated transcriptional programs (Figure 1F). We identified differentially-accessible chromatin regions that characterize resistant cells (Figure 1F). Integrating these data with RNA-seq data, we found that >50% of gene expression changes occurred in a concordant manner with DNA accessibility changes at or near gene promoters or enhancers (Figure 1G–H, S1M). Examination of these ‘primary’ gene targets revealed key gene sets/pathways that are hallmarks of the resistant states, including those pertaining to upregulation of EMT and RTK signaling pathways, and downregulation of epithelial cell differentiation and cell-cell adhesion (Figure S1N). Taken together, these findings establish that chromatin accessibility changes between resistant and parental states in EGFR-mutant NSCLC cancer cell lines underlie the gene regulatory programs that characterize TKI resistance.
Mammalian SWI/SNF complexes are upstream regulators of osimertinib resistance and target to a subset of accessible regions
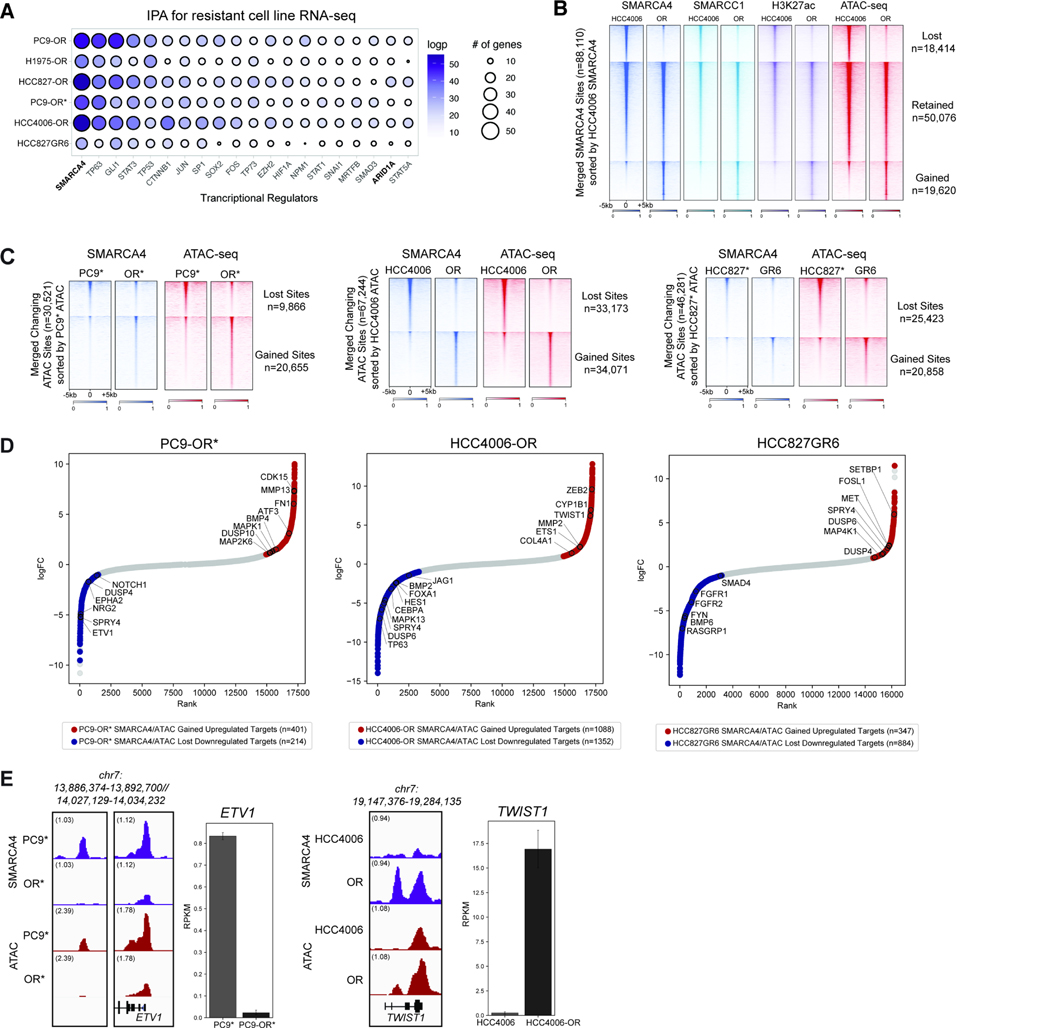
Given our findings that changes in chromatin accessibility underlie a subset of key gene regulatory features of the TKI-resistant state, we next sought to predict potential chromatin-associated regulators that govern these changes. Across all cell line pairs, Ingenuity Pathway Analysis (IPA) performed on differential gene expression profiles (RNA-seq) in the Osimertinib-resistant (OR) (or gefitinib-resistant) states revealed SMARCA4, the ATPase subunit of mSWI/SNF complexes, as the top hit (Figure 2A). Other top hits included TFs such as TP63, STAT3, and SOX2, several of which are tethered to chromatin by mSWI/SNF complexes 50,51, as well as ARID1A, the defining subunit of the canonical BAF (cBAF) mSWI/SNF subcomplex 27 (Figure 2A). Beta-catenin (CTNNB) was also within the top genes, and has been previously implicated in TKI resistance 52. We did not detect differences in SMARCA4 mRNA or protein levels between the parental and resistant states that would account for it emerging as a top hit (Figure S2A–B).
Figure 2. Mammalian SWI/SNF (BAF) complexes as critical regulators of resistance-associated gene loci.
A. Ingenuity Pathway Analysis (IPA) performed on differentially-regulated genes in parental versus resistant cell line pairs. Top 20 most significant transcriptional upstream regulators are shown; Circle size indicates the number of genes regulated. Circle color represents significance as measured by logpvalue <0.05. B. Heatmap for SMARCA4, SMARCC1, and H3K27ac occupancy (CUT&RUN) levels and ATAC-seq chromatin accessibility in HCC4006/HCC4006-OR cell lines across merged SMARCA4 sites. C. Heatmap displaying SMARCA4 occupancy levels and ATAC-seq chromatin accessibility in PC9*/PC9-OR*, HCC4006/HCC4006-OR and HCC827*/HCC827GR6 cell lines, across merged differential ATAC-seq sites. D. Hockey-stick plots representing the normalized rank and signals of RNA-seq in PC9-OR*, HCC4006-OR and HCC827GR6 cell lines. Representative SMARCA4/ATAC gained-associated genes that are upregulated are in red and representative BAF/ATAC lost-associated genes that are downregulated are in blue. E. SMARCA4 and ATAC-seq tracks at the ETV1 (in PC9*/OR*) and TWIST1 (in HCC4006/OR) loci. RNA-seq expression signal (RPKM) is shown for each; error bars represent the 95% confidence interval around the mean expression level. See also Figure S2.
To understand how SMARCA4 regulates gene expression, we profiled the occupancy of mSWI/SNF complexes genome-wide across the PC9*/OR*, HCC4006/OR, and HCC827*/GR6 cell lines using CUT&RUN (Figure 2B, S2C–D). We integrated these data with ATAC-seq profiles and identified a subset of sites that were gained or lost specifically in the resistant state (Figure 2C, S2E–F). Importantly, gained and lost sites were largely promoter-distal in nature, consistent with the positioning of cBAF complexes at distal enhancer sites 27, and were enriched over AP1-family motifs (Figure 2B–C, S2G–H). We then ranked differentially-expressed genes that exhibited differential mSWI/SNF binding as well as concordant changes in accessibility in the resistant state (Figure 2D, S2I–J). Genes corresponding to EMT (TWIST1, and ZEB2), cell migration (MMP13, BMP4, COL4A1) and RTK signaling were upregulated, while genes encoding members of epithelial cell differentiation and signaling (FGFR1/2, JAG1, NOTCH1) were downregulated in the resistant state. Expression of genes involved in MAPK signaling (MAPK1, DUSP6, SPRY4) were altered in a cell-line specific manner (Figure 2D, S2J). Concordant changes in mSWI/SNF occupancy, accessibility, and gene expression between the parental and resistant states are exemplified at the ETV1 and TWIST1 loci (Figure 2E). These findings point toward a key role for mSWI/SNF complex activity at resistance-associated gene loci across TKI-resistant EGFR-mutant cell lines.
Suppression of SMARCA4 re-sensitizes a subset of resistant tumor models to osimertinib
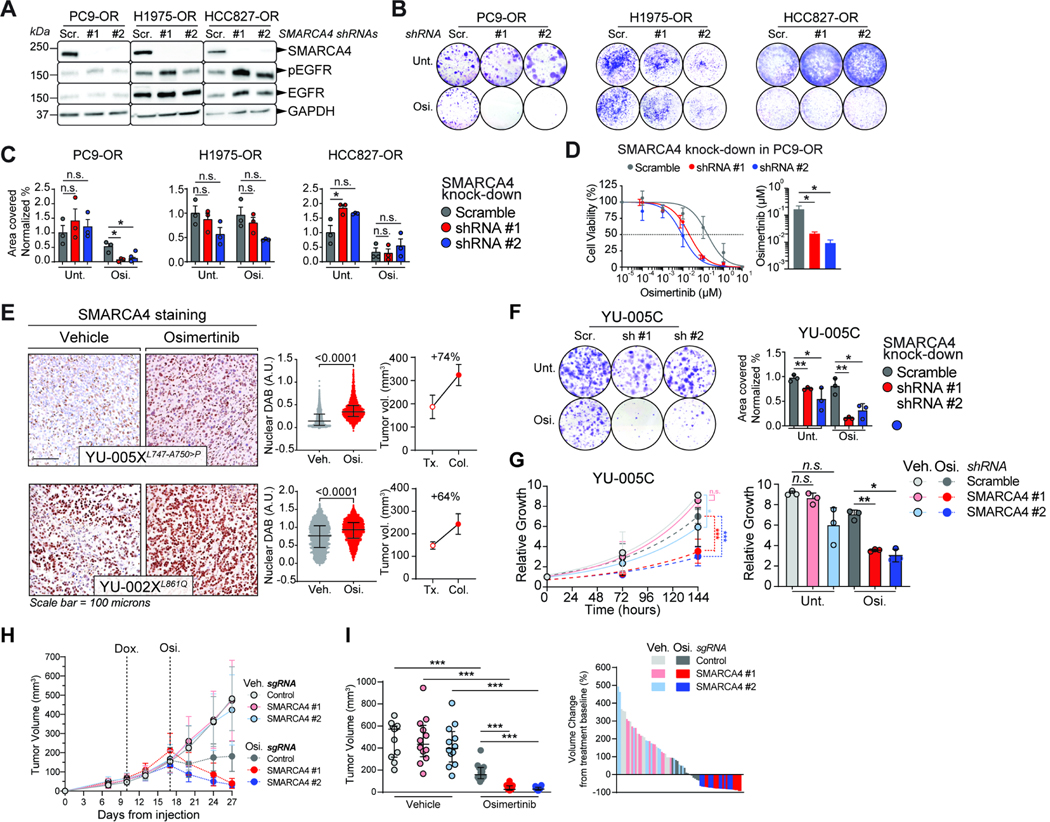
We next performed shRNA-mediated knock-down of SMARCA4 in PC9/PC9-OR, H1975/H1975-OR and HCC827/HCC827-OR parental and resistant cell line pairs (Figure 3A–D, S3A–B). EGFR phosphorylation was attenuated following osimertinib treatment in both parental and resistant cell lines, and SMARCA4 levels were held at relatively consistent levels across osimertinib treatments except for PC9 cells where the levels decreased following osimertinib treatment (Figure S3C–D). The most substantial impact of SMARCA4 knock-down was observed in PC9-OR cells upon osimertinib treatment as compared to H1975-OR and HCC827-OR cells (7 days post knockdown induction). Proliferation of PC9-OR cells was completely abrogated in the presence of osimertinib as compared to untreated cells (Figure 3B–C). Further, the osimertinib EC50 was decreased by ~8-fold upon SMARCA4 knockdown (Figure 3D). Knockdown of SMARCA4 in the osimertinib-sensitive counterparts of the three cell lines somewhat limited the growth of PC9 and H1975 cells, but not HCC827 cells (Figure S3E–F).
Figure 3. SMARCA4 loss sensitizes a subset of resistant tumors to osimertinib.
A. Western blots of OR cells transduced with SMARCA4-targeting shRNAs. Scramble (Scr.); SMARCA4 knockdown (#1 and #2). B-C. Representative colony formation assay in OR cells. (B). Quantification of the results for independent triplicates (C). Osimertinib doses: 750 nM (PC9-OR, HCC827-OR); 1500 nM (H1975-OR). D. Osimertinib dose-response curves for PC9-OR cells after 7 days of SMARCA4 knockdown. Significance was calculated using a paired t test and the Mean ± SEM is shown. *P<0.05. E. IHC staining for SMARCA4 on PDXs treated either with vehicle or osimertinib (left). Quantification of diaminobenzidine (DAB) intensity (middle) A.U., arbitrary units. Tumor volume change from the start of treatment (Tx.) to the day the tumor was collected (Col.) (right). F. Representative colony formation assay in YU-005C cells after one-week of SMARCA4 knock-down (left). Quantification of data from independent triplicates is shown (right). G. Proliferation curves of YU-005C cells one week after shRNA induction (left). Plot of the relative growth of the cells at the proliferation assay end-point. Data from three independent replicates are shown (right). H-I. Tumor volume of YU-005C cells injected subcutaneously in mice (left). ime-Dox., initiation of doxycycline to knock-out SMARCA4. Osi., start of osimertinib treatment. Tumor volume after two weeks of osimertinib treatment (middle). Waterfall plot after 2 weeks of osimertinib treatment (right). Significance was calculated using a Mann-Whitney test and the Median ± IQR for C, F-G. Significance was calculated using a paired t test and the Mean ± SEM is shown in I. ***P<0.001, **P<0.01, *P<0.05. See also Figure S3.
To extend these observations to additional EGFR-mutant tumors, we leveraged a collection of patient-derived xenografts (PDXs) of EGFR-mutant lung cancers with a range of sensitivities to osimertinib (Table S2). Examining the levels of SMARCA4 by immunohistochemistry (IHC) in these PDXs revealed that all tumors produced SMARCA4, albeit at variable levels (Figures 3E, S3G–H). We further performed functional studies using YU-005X since it exhibited primary resistance to osimertinib and a cell line derived from the tumor was available (YU-005C cells). Knock down of SMARCA4 significantly impaired the ability of YU-005C cells to proliferate and form colonies, which was accentuated by osimertinib treatment (Figure 3F–G). Finally, to confirm these findings in vivo, we knock-out SMARCA4 in established YU-005C-derived tumors (Figure 3H–I, S3J–M). Subcutaneous injection of YU-005C cells led to the formation of tumors that continued to grow upon doxycycline treatment and consequent SMARCA4 knock-out (Figure 3H). SMARCA4 knock-out and osimertinib treatment led to regression of established tumors (Figures 3H–I, S3M), while growth of control SMARCA4 wild-type tumors was slowed in the presence of osimertinib without regression (Figure 3H, YU-005C-derived tumors are somewhat more sensitive to the effects of osimertinib than YU-005X-derived tumors from Figure 3E). These results demonstrate that a subset of osimertinib-resistant tumors depend on SMARCA4 for proliferation in the presence of osimertinib.
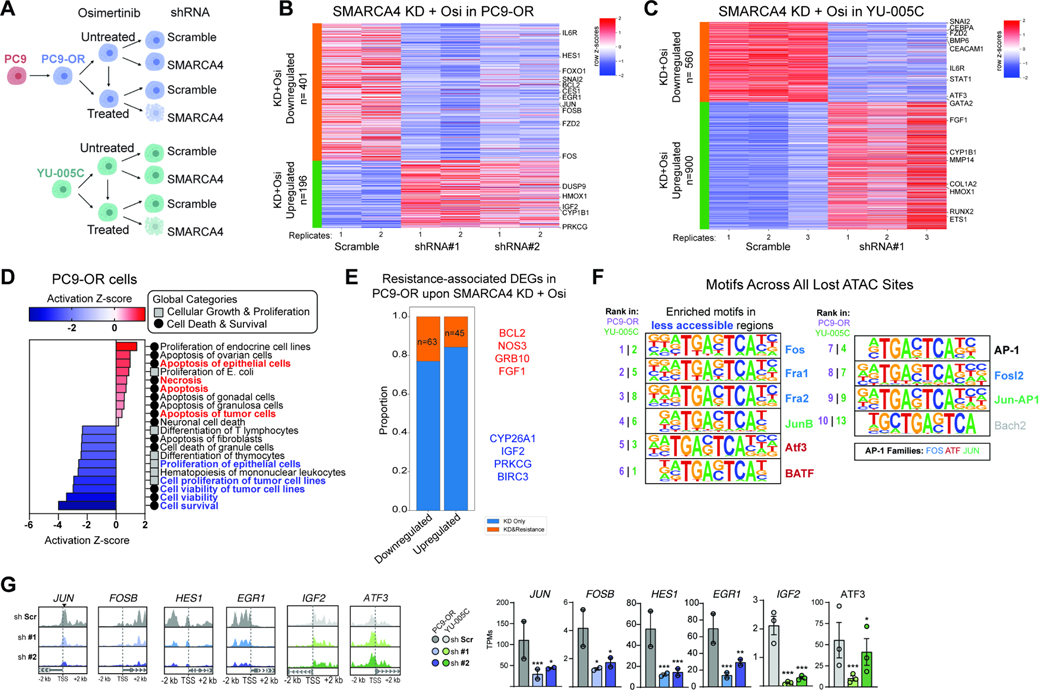
SMARCA4 loss alters the chromatin accessibility and expression of resistance- and proliferation-associated genes
To investigate how SMARCA4 sustains cancer cells in the presence of osimertinib, we next performed gene expression (RNA-seq) and chromatin accessibility (ATAC-seq) analyses on both PC9-OR and YU-005C cells treated with either scramble or SMARCA4 shRNA-mediated knockdown in the presence or absence of osimertinib (Figure 4A). We examined upregulated and downregulated genes in both cell contexts, identifying top genes whose expression was reversed upon SMARCA4 loss in the setting of osimertinib treatment as compared to the scramble condition since the differential cell viability phenotype was observed in these conditions (Figure 4B–C, S4A–B). Genes upregulated and downregulated upon SMARCA4 knockdown in PC9-OR and YU-005C cells were enriched in specific signaling pathways and biological processes related to cell proliferation and immune signaling (Figure 4D, S4C–D). Further, amongst the resistance-associated DEGs of PC9-OR cells (Figure S1K), we found that ~20% were deregulated upon SMARCA4 knockdown and osimertinib treatment (Figure 4E). Specific examples include genes such as BCL2 or CYP26a1, involved in cell survival and xenobiotic metabolism processes, respectively (Figure 4E, S4E). Integrating ATAC-seq studies, we identified key genes that were both reduced in accessibility and gene expression upon SMARCA4 suppression in PC9-OR and YU-005C cells (Figure 4F–G, S4F–G). A subset belongs to the AP-1 family of TFs (FOS and JUNB) as well as additional genes involved in cancer and proliferation such as HES1, EGR1, and JAG1 (Figure 4G, S4G). Taken together, these data highlight the impact of SMARCA4 loss on the maintenance of the resistant state and identify key genes impacted by its disruption.
Figure 4. SMARCA4 suppression in osimertinib-resistant cell lines regulates resistance-associated genes.
A. Experimental design for SMARCA4 knockdown (KD) in PC9-OR and YU-005C cells. Seven days after knockdown the cells were treated with osimertinib for 72 hours. B-C. Heatmaps reflecting row Z-score values for up- and- down- regulated genes in PC9-OR and YU-005C cells following SMARCA4 knockdown and osimertinib treatment. Top gene hits are indicated. D. Heatmap generated by IPA reflecting Z-scored enrichment of gene pathways affected in osimertinib treated PC9-OR cells upon SMARCA4 knockdown. E. Stacked bar graph depicting number of resistance-associated genes impacted by SMARCA4 knockdown + osi treatment in PC9-OR cells. Key up- and down-regulated genes are in red and blue, respectively. F. Motifs under differentially-accessible sites genome-wide in osimertinib treated PC9-OR (purple rank) and YU-005C (green rank) cells upon SMARCA4 KD. G. Representative ATAC-seq tracks and bar graphs showing altered gene expression for key resistance-associated genes. Mean ± SEM. Significance was calculated using DESeq2. ***P<0.001, *P<0.05. See also Figure S4.
Pharmacologic inhibition of mSWI/SNF ATPase activity reverses the osimertinib-resistant state in a subset of EGFR-mutant lung cancer cell lines
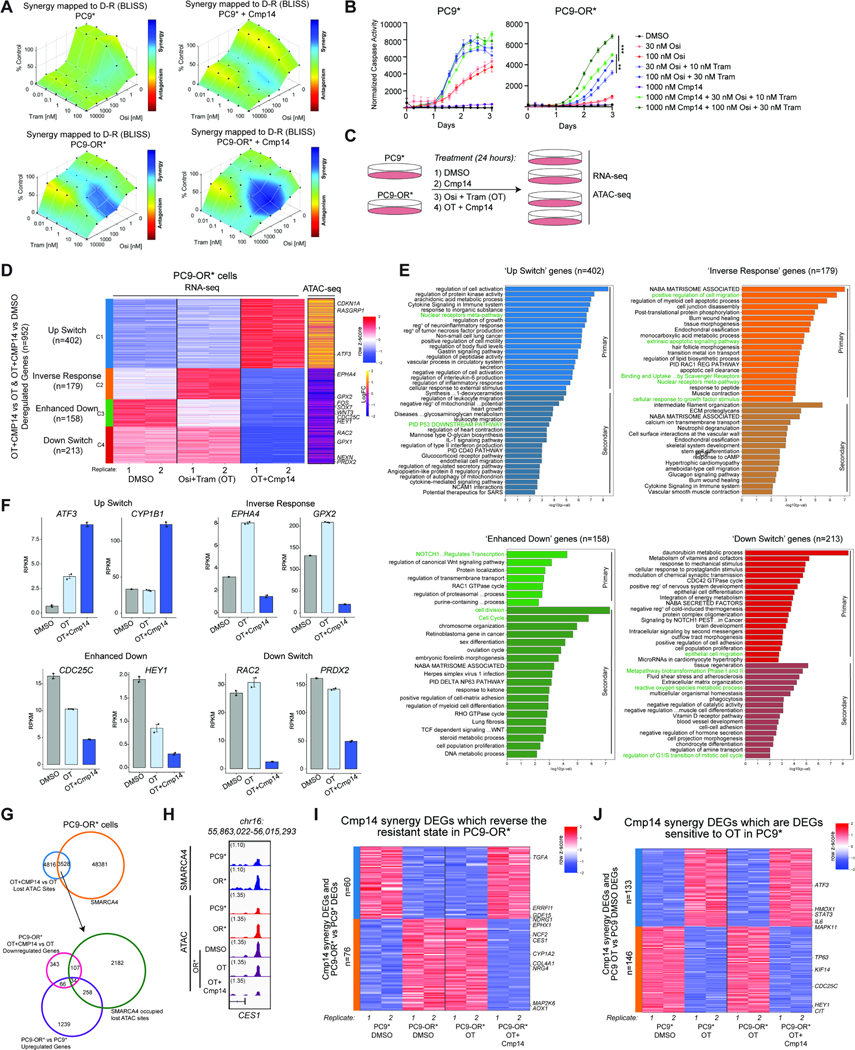
Recently, several small molecule inhibitors and degraders targeting the ATPase components of mSWI/SNF (BAF) complexes, SMARCA2/A4, have been developed and entered the clinic 41–43. We sought to investigate the impact of mSWI/SNF pharmacologic inhibition on the resistance associated chromatin and gene regulatory signature using a specific, allosteric SMARCA4/2 ATPase inhibitor, Compound14 (Cmp14) 41. To complement SMARCA4 knockdown experiments in selected cell line pairs, we used the PC9*, HCC4006*, and HCC827* cell line systems for these studies. Cell viability assays performed with Cmp14 alone across parental-resistant cell line pairs revealed modest effects (Figure S5A). To assess whether BAF inhibition could sensitize resistant cell lines to osimertinib treatment, we measured drug synergy using combenefit assays (Figure 5A). Notably, previous work from our group implicated ERK reactivation as a major determinant of EGFR TKI treatment failure, which could be circumvented by co-treatment with a MEK inhibitor 53,54. Since pERK was reactivated upon Cmp14 and osimertinib treatment in PC9-OR* cells (Figure S5B), PC9* and PC9-OR* cells were treated with both osimertinib and trametinib (OT) in the presence or absence of Cmp14 for combenefit assays (Figure 5A, S5C). Conversely, osimertinib alone was used for HCC827GR6 cells because the addition of Cmp14 was sufficient to dampen the pERK1/2 hyperactivation observed with osimertinib treatment alone (Figure S5D).
Figure 5. Pharmacologic targeting of mSWI/SNF complex ATPase activity reverses the TKI resistance program in a subset of EGFR-mutant cancer cell lines.
A. Drug synergy plots in PC9* and PC9-OR* cells as assayed by Combenefit software. Bliss synergy scores were calculated for each drug combination, osimertinib (osi) and trametinib (Tram) in the absence or presence of Compound14 (Cmp14) after 72 hours. One representative experiment out of N=3 independent experiments is shown. B. Caspase-3/7 activity assays performed in PC9* and PC9-OR* cells across 3 days of drug treatment. A low and high concentration of osi and tram were used in these assays to highlight enhanced sensitization effects. Graphs represent fluorescent signals normalized to cellular confluency at each timepoint. One representative experiment out of N=5 independent experiments is shown. Data presented as Mean ± SEM with significance calculated at the last timepoint using an unpaired t test ***P<0.0005, **P<0.005. C. Experimental design for ATAC-seq and RNA-seq performed in PC9* and PC9-OR* cells following 24 hours of each treatment. D. RNA-seq clustered heatmap of Cmp14 synergy genes in PC9-OR* cells. Biological replicates are represented for DMSO, OT and OT+Cmp14 treatments. Expression signals were z-scored across the samples. Genes were k-means clustered (k=4) and clusters were reordered. The greatest coordinated ATAC-seq changes (in logFCs) between OT+CMP14 and OT in PC9-OR* for each gene are shown as a yellow/purple heatmap. Select genes are labeled. E. Metascape analysis of genes from each cluster of Cmp14 synergy genes separated by correlation to ATAC-seq signal. Primary analysis represents DEGs which have a closest associated change in ATAC peak while secondary analysis represents DEGs without an associated ATAC peak change. Cluster specific or common terms are highlighted. F. Bar graphs of key deregulated Cmp14 synergy genes from each RNA-seq cluster from (D) showing average RPKM values for each condition with one SEM for the error bars. G. Venn diagram representation of SMARCA4 occupied sites in PC9-OR* cells which overlap with lost ATAC sites at Cmp14 synergy DARs (upper). A subset of these sites overlap with upregulated DEGs which characterize the resistant state (PC9-OR* vs PC9*) and are subsequently downregulated by Cmp14 synergy treatment (lower). H. IGV tracks of SMARCA4 occupancy (Cut&Run) and accessibility (ATAC-seq) at the CES1 locus. I. RNA-seq heatmap of RPKM values of Cmp14 Synergy DEGs at resistant-state associated genes. Values are shown for PC9* and PC9-OR* cells under DMSO treatment as compared to PC9-OR* cells under OT and OT+Cmp14 treatments. J. RNA-seq heatmap of RPKM values of Cmp14 Synergy DEGs in PC9* cells under DMSO and OT treatments and in PC9-OR* cells under OT and OT+Cmp14 treatments. See also Figure S5.
Importantly, combenefit assays revealed substantial synergy between OT and Cmp14, specific to PC9-OR* cells (Figure 5A). We also observed striking synergy in HCC827GR6 cells, but not HCC4006-OR cells (Figure S5E). The addition of Cmp14 to OT augmented cellular apoptosis in PC9-OR* as compared to OT alone but was unsuccessful at eliciting further degrees of apoptosis in HCC827GR6 (Figure 5B and S5F–G). This difference in mechanism is likely attributable to the MET amplification in HCC827GR6 cells that can bypass apoptotic signaling45. These combination treatment experiments suggest that mSWI/SNF ATPase inhibitors can re-sensitize a subset of resistant cell lines to TKIs.
To define the transcriptional and chromatin accessibility changes underlying the observed drug synergy in PC9-OR* cells, we performed RNA-seq and ATAC-seq experiments (Figure 5C). Notably, protein levels for mSWI/SNF complex subunits, SMARCA4 and SMARCC1, were unaltered across conditions (Figure S5H). Clustering analyses performed on differentially regulated genes between DMSO, OT, and OT+Cmp14 (Cmp14 alone did not affect cell viability so was not included in this analysis) revealed four key sets of genes (Figure 5D, S5I–J). Cluster 1 (C1) included genes that switched from downregulated in either no treatment or OT conditions to strongly upregulated upon inclusion of Cmp14 (‘Up switch’), Cluster 2 (C2) contained genes that were activated upon OT treatment but strongly downregulated upon OT+Cmp14 (‘Inverse Response’), Cluster 3 (C3) included genes for which expression was mildly reduced by OT treatment (relative to control) but strongly reduced by OT+Cmp14 treatment (‘Enhanced down’), and Cluster 4 (C4) included genes that were on in both control and OT treatment conditions but strongly downregulated upon combination with Cmp14 (‘Down switch’) (Figure 5D). Integration of these data with ATAC-seq generated for each treatment condition, revealed that the chromatin accessibility of loci corresponding to genes in these four clusters correlated with gene expression changes (Figure 5D, S5K).
We next characterized the transcriptional responses across the four clusters (C1-C4) of Cmp14 synergy genes, highlighting both ‘primary’ targets (those with concordant chromatin accessibility (ATAC) changes), and ‘secondary’ targets (those lacking concordant changes in accessibility) (Figure 5E). Examples of genes that were downregulated by OT+Cmp14 treatment included those involved in cell cycle and apoptosis, cell migration and adhesion, metabolic processes, and nuclear receptor pathway factors, including NRF2 signaling and metabolism of toxins, exemplified by genes such as EPHA4, RAC2, CDC25C, and GPX2 (Figure 5D–F). Upregulated genes (C1) included genes involved in processes such as protein kinase activity, immune signaling, and nuclear receptors such as ATF3, and CYP1B1 (Figure 5E–F). Intriguingly, of the 8344 sites exhibiting reductions in accessibility upon OT+Cmp14 (but not OT alone), we found that over 40% contained a BAF complex peak in the PC9-OR* cells (Figure 5G, top). Further, a subset of these sites (green circle) mapped to genes that were selectively upregulated in the resistant state (purple circle) but downregulated only upon combined OT+Cmp14 treatment (magenta circle) (Figure 5G, bottom). This is exemplified at the CES1 locus at which we observed heightened BAF complex occupancy and accessibility in the OR* (resistant) setting that is substantially reduced upon OT+Cmp14 treatment (Figure 5H). Notably, CES1 is a critical NRF2-regulated enzyme which mediates xenobiotic metabolism 55 and is linked to chemotherapy resistance in hepatocellular carcinoma 56. Our results suggest that BAF inhibition may re-sensitize a subset of osimertinib resistant cells in part via antagonizing NRF2 signaling.
Finally, we aimed to identify whether genes that were uniquely up- or down-regulated in the resistant state (i.e. PC9-OR* versus PC9*) could be reversed selectively in the OT+Cmp14 combination treatment setting relative to OT only. We identified n=60 genes for which expression was downregulated in the resistant state, unaltered by OT alone, but reversed in expression (upregulated) upon OT+Cmp14 treatment (TGFA, GDF15, NDRG1) (Figure 5I). In parallel, we identified n=76 genes for which resistance-associated expression was reversed (downregulated) selectively in the OT+Cmp14 combination treatment COL4A1, MAP2K6, EPHX1) (Figure 5I, S5L–M). Additionally, we analyzed a collection of genes whose expression changed upon OT treatment in PC9* cells (up- or down-regulated genes) but did respond in a similar manner in PC9-OR* cells. Among these genes (involving cell proliferation and cytokine production pathways), we found that the combination of OT+Cmp14 in PC9-OR* cells was able to reverse their expression, mimicking the response observed in PC9* cells (Figure 5J, S5N). These transcriptional programs were broadly consistent with those observed upon SMARCA4 knockdown and osimertinib treatment (Figures 4C–D). Overall, these results demonstrate that the Cmp14 mSWI/SNF ATPase inhibitor can synergize with OT to re-sensitize resistant PC9-OR* cells to TKI treatment in part via rewiring chromatin accessibility to reverse a portion of the transcriptional programs underlying the drug resistant state.
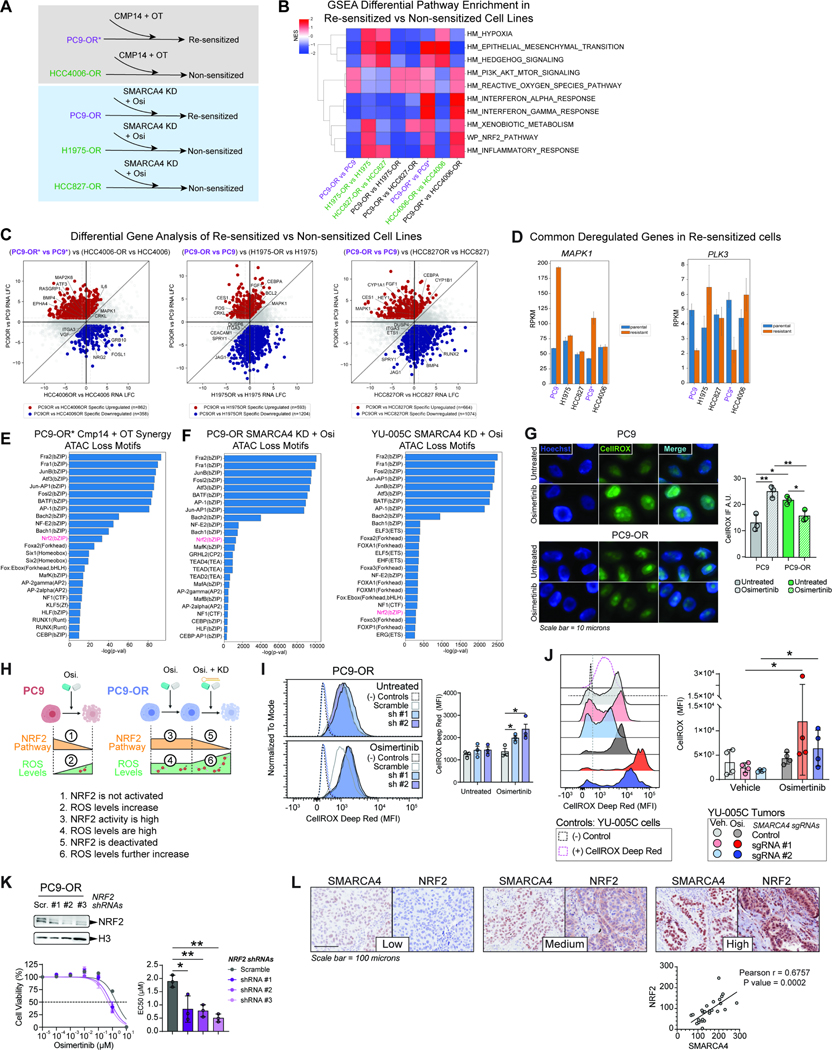
Attenuation of reactive oxygen species by SMARCA4 correlates with re-sensitization to osimertinib
We next sought to identify potential markers or hallmark signatures of EGFR-mutant cell lines re-sensitized to osimertinib either by SMARCA4/2 inhibition or SMARCA4 knockdown. We thus analyzed the differences in the resistant state DEGs between the cell lines that were re-sensitized to osimertinib and those that were not following SMARCA4 KD or pharmacologic inhibition. This revealed the differences between the PC9-OR (re-sensitized) cell line and the H1975-OR and HCC827-OR (non-sensitized) cell lines as well as between the PC9-OR* (re-sensitized) cell line and the HCC4006-OR (non-sensitized) cell lines (Figure 6A–B). GSEA pathway analysis revealed that the re-sensitized cell lines, PC9-OR and PC9-OR*, were both enriched for pathways such as PI3K signaling and Reactive Oxygen Species pathways relative to their non-sensitized cell lines (Figure 6B). Each re-sensitized cell line also exhibited specific positive or negative enrichment for Interferon α/γ signaling, suggesting cell line specific pathway functions (Figure 6B). In parallel, to identify gene targets which may potentiate the re-sensitization response to osimertinib or OT treatment in PC9-OR and PC9-OR* cells, we performed a differential gene analysis which enriches for DEGs specifically upregulated and downregulated in PC9-OR* and PC9-OR cells as compared to HCC4006-OR, H1975-OR or HCC827-OR DEGs (Figure 6C). While these cell line specific differential gene sets were enriched for their respective pathways (Figure S6A), we overlapped upregulated and downregulated differential genes to identify common regulators which may underpin or contribute to the re-sensitization of PC9-OR* and PC9-OR cells to osimertinib/OT upon SMARCA4 inhibition or KD (Figure S6B). Many terms are associated with MAPK signaling and detoxification (Figure S6C) and include upregulated genes MAPK1, CRKL, CES1, as well as downregulated genes PLK3 and ARNT2 (Figure 6D, S6D).
Figure 6. Resensitization of osimertinib-resistant cell lines reveals attenuation of reactive oxygen species by SMARCA4.
A. Schematic overview of cell lines which are responsive (resensitized; purple) and non-responsive (green) to osimertinib treatment upon inhibition or knock down of SMARCA4. B. GSEA pathway enrichment analysis of differentially expressed genes (DEGs) between each parental and osimertinib-resistant cell line pair as well as sensitized cell line DEGs vs. non sensitized cell line DEGs. C. Quadrant plots of differentially expressed genes specific to the resistant PC9-OR* state as compared to HCC4006-OR and specific to the resistant PC9-OR state as compared to H1975-OR and HCC827-OR. Upregulated genes (red), downregulated genes (blue). Gene examples are labelled. D. Bar graphs of key gene examples that are specifically upregulated and downregulated common to PC9-OR* and PC9-OR showing RPKM values across cell lines. E-F. Motif analysis of lost ATAC sites attributed to Cmp14+OT treatment in PC9-OR* cells (E) and attributed to SMARCA4 knock down in PC9-OR and YU-005C cells (F). G. Immunofluorescence images (IF) of cells stained using CellROX™ to quantify ROS in PC9 and PC9-OR cells in the presence and absence of 750 nM osimertinib. Hoechst staining was used to detect nuclear DNA (left). CellROX™ IF quantification of three independent replicates is shown (right). A.U., arbitrary units. H. Schematic model of ROS levels, NRF2 pathway activity, SMARCA4 regulation and osimertinib sensitivity. Upon osimertinib treatment NRF2 targets are downregulated, and ROS levels increase in PC9 cells (1 and 2). In treated PC9-OR cells NRF2 targets are activated, and ROS levels are high (3 and 4); upon SMARCA4 knockdown, NRF2 targets are downregulated and ROS levels further increase causing cellular toxicity. I-J. Flow cytometry using CellROX™ to measure ROS in PC9-OR cells (I) and in YU-005C tumors (J) in the presence and absence of osimertinib and with or without SMARCA4 knockdown. (−) Controls are from PC9-OR and YU-005C unstained cells respectively and (+) CellROX™ Deep Red control in (J) is from stained YU-005C cells. CellROX™ MFI was assessed in RFP+/shRNA-containing cells (I) and GFP+/sgRNA-containing cells (J). Representative MFI profile of CellROX™+ cells (I:, J: left). Quantification of three independent replicates (I: right) and four tumors (J: right). K. Western blot of PC9-OR cells transduced with three NRF2 shRNAs as indicated (upper). Osimertinib dose-response curves for PC9-OR cells after NRF2 knock-down (bottom left). Bar graph of EC50 values (bottom right). L. IHC staining for SMARCA4 and NRF2 in three representative cores of a TMA containing EGFR-mutant TKI-treated tumors (upper). Correlation plot of NRF2 and SMARCA4 H-Scores for all the tumors (lower). Significance was calculated using the Pearson r correlation test. Scr.: Scramble shRNA, sh #1: SMARCA4 shRNA #1; sh #2: SMARCA4 shRNA #2. Significance was calculated using a paired t test and the Mean ± SEM is shown in D, E and H. Significance was calculated using a Mann-Whitney test and the Median ± IQR is shown in F. **P<0.01, *P<0.05. See also Figure S6.
We next investigated the motifs underlying chromatin accessibility changes correlated with BAF-mediated gene expression changes upon SMARCA4 inhibition or knockout in PC9-OR* cells, PC9-OR cells and YU-005C cells (Figure 6E–F, S6E–F). At sites with reduced accessibility near Cmp14 synergy genes in PC9-OR* cells, we found motifs corresponding to AP-1 factors and NRF2 as putative candidates involved in the synergy response (Figure 6E) (gained sites would reflect indirect effects (Figure S6E)). Similarly, motif analysis at sites losing accessibility upon SMARCA4 KD and osimertinib treatment in PC9-OR and YU-005C cells also revealed NRF2 as a candidate factor (gained accessible sites represent indirect effects) (Figure 6F, S6F).
The NRF2 signaling pathway is responsible for scavenging reactive oxygen species (ROS) through the activation of antioxidant response elements under oxidative stress conditions 57. To validate the association between SMARCA4/2-mediated osimertinib resensitization, ROS and detoxification pathways, we investigated how osimertinib affects the levels of ROS in PC9-OR and YU-005C TKI-resistant cells. Under osimertinib treatment, the levels of ROS dramatically increased in PC9 cells, which is consistent with a global decrease in NRF2 signaling (Figures 6G–H, S6G). In contrast, the amount of baseline ROS was significantly higher in PC9-OR cells and osimertinib did not profoundly affect ROS levels, consistent with active NRF2 signaling (Figures 6G–H, S6G). SMARCA4 knock-down in osimertinib-treated PC9-OR cells further increased the levels of ROS while decreasing the activity of the NRF2 pathway, suggesting that SMARCA4 plays a key role in ROS neutralization (Figures 6H–I). Consistent with these observations, the levels of ROS in YU-005C subcutaneous tumors were highest in osimertinib-treated tumors upon SMARCA4 knock-out (Figure 6J, S6H–I). We next examined the functional role of NRF2 in maintaining the resistant phenotype. NRF2 knock-down reduced the sensitivity of PC9-OR cells to osimertinib by approximately half (Figure 6K), confirming that NRF2 is an important factor in maintaining resistance. Additionally, we tested whether the presence of ROS had a direct effect on how the cells respond to osimertinib. For this, we performed dose-response assays in the presence of a non-toxic concentration of the ROS scavenger N-acetyl-L-cysteine (NAC) 58. The incorporation of NAC significantly shifted the EC50 of osimertinib in PC9, PC9-OR and YU-005C cells to a similar extent (Figure S6J). However, the magnitude of the effect was small and very similar within the three cell lines independent of their response to osimertinib, suggesting that the accumulation of ROS is not directly involved in the resistance mechanism. Finally, we examined the levels of NRF2 and SMARCA4 in a tissue microarray of EGFR-mutant tumors (Table S3). The levels of SMARCA4 in these tumors were positively correlated with nuclear NRF2, further supporting that these two proteins may function together to regulate oxidative stress (Figure 6L). These results confirm a pivotal role for SMARCA4 chromatin remodeling activity in controlling the levels of osimertinib-induced oxidative stress in resistant cells, via NRF2 activation.
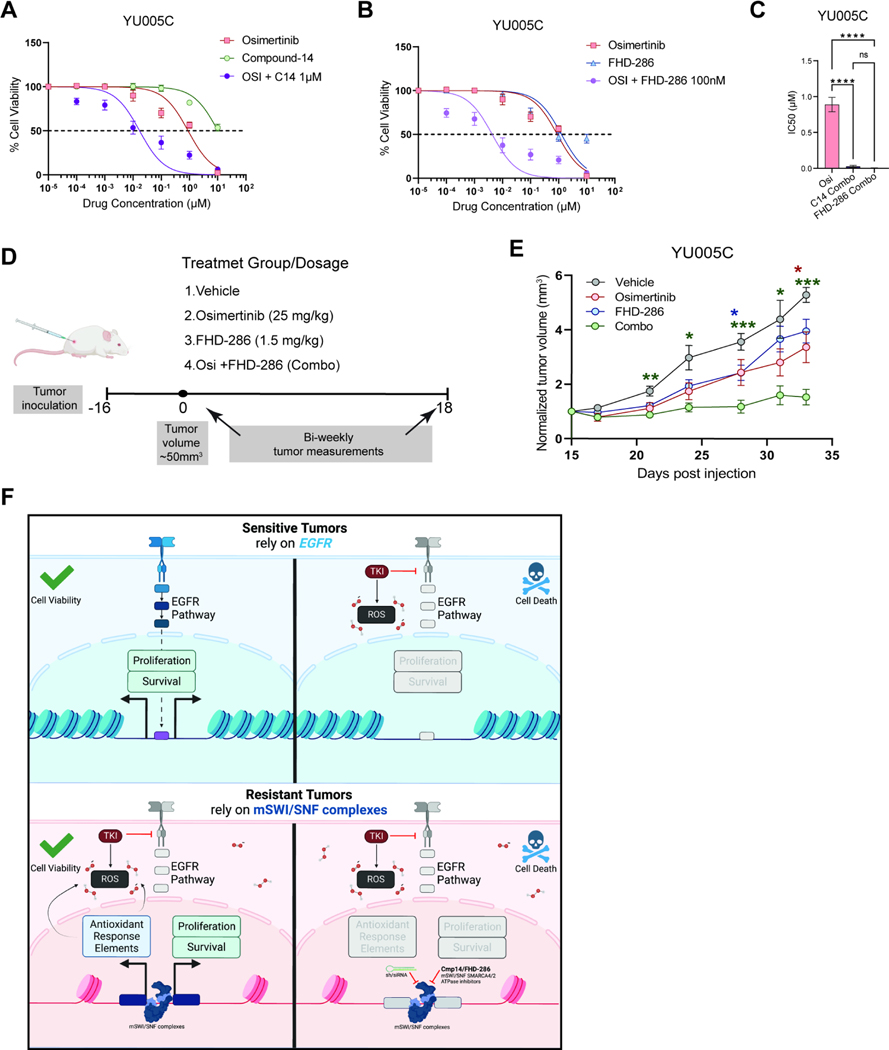
Pharmacological inhibition of mSWI/SNF ATPase activity attenuates tumor growth in an osimertinib-resistant PDX mouse model
We next explored the utility of pharmacological inhibition of mSWI/SNF ATPase activity in the SMARCA4-dependent YU-005 patient-derived model. YU005C cells were resistant to the treatment of Compound 14 (Cmp14) or FHD-286 (a clinical-stage SMARCA4/SMARCA2 inhibitor alone (Figure S7A, S7B). However, both Cmp14 and FHD-286 sensitized YU005C cells to osimertinib (Figures 7A–C) consistent with our findings in isogenic cell lines (Figures 5 and S5). We, therefore, investigated the potential of combining SMARCA4/2 inhibition with osimertinib in vivo using FHD-286 (Figure 7D). Tumor bearing mice were treated with vehicle, osimertinib, FHD-286 or the combination of osimertinib and FHD-286 for 18 days. While each drug alone modestly slowed tumor growth, the osimertinib+FHD-286 combination treatment significantly suppressed tumor growth as compared to baseline (Figure 7E and S7C).
Figure 7. Pharmacological inhibition of mSWI/SNF ATPase activity sensitizes a patient-derived tumor to osimertinib.
A. Osimertinib, compound-14, and combination (titrated osimertinib + 1μM of Compound-14) dose-response curves for YU-005C cells. B. Osimertinib, FHD-286, and combination (titrated osimertinib + 100nM of FHD-286) dose-response curves for YU-005C cells. (A,B) N=4. The mean ± standard deviation is shown. C. Bar graph of IC50 values for YU-005C cells treated with osimertinib (alone), or in combination with compound-14 or FHD-286. The mean ± SEM is shown. Significance was calculated using the one-way repeated measures ANOVA test and Tukey’s multiple comparisons test. ***P<0.001, **P<0.01, *P<0.05. D. Experimental design. YU-005C cells were injected subcutaneously in mice that were treated with either vehicle, osimertinib, FHD-286 or the combination of both. E. Normalized tumor volume of YU-005C cells treated with either vehicle, osimertinib, FHD-286 or the combination. Individual tumor volumes reflect the change in volume from treatment baseline. Tumor volume mean and ± standard error of the mean is shown. Significance was calculated using the two-way repeated measures ANOVA test and Dunnett’s multiple comparisons test, with a single pooled variance. ***P<0.001, **P<0.01, *P<0.05. G. Schematic representation of the mechanistic model by which SMARCA4 promotes osimertinib resistance. Sensitive tumors rely on EGFR signaling pathway. Osimertinib blocks EGFR and generates ROS killing the cells. Resistant tumors rely on SMARCA4 to keep proliferating and neutralizing the accumulated ROS. Blocking SMARCA4 activity generates stress killing the cells. Created with Biorender.com. See also Figure S7.
Discussion
Non-genetic mechanisms of resistance are likely responsible for TKI resistance in a large fraction of human tumors, yet detailed mechanistic understanding is lacking. We uncover that mSWI/SNF complexes are retargeted genome-wide in TKI-resistant lung cancer cell lines, promoting extensive chromatin accessibility changes which underlie the transcriptional programs that characterize the resistant state. mSWI/SNF inhibition allowed for reversal of the resistant state and resensitization to osimertinib, which promoted cell death in a subset of Osimertinib-resistant models. These data suggest that some osimertinib-resistant tumors evolve during treatment to require SMARCA4 for survival in the presence of osimertinib. These findings demonstrate that SMARCA4 can have a pro-growth role in lung cancer cells, in addition to its well-known role as a tumor suppressor32,59–61. Recent studies have revealed tumor-promoting roles for SMARCA4 in cancers 38,35,40,62,34,39,63,64,37, 36 which, in some cases, is dependent on the differentiation state of the tumors 39. It is well established that mSWI/SNF complexes are involved in several differentiation processes during normal development and cancer 28,30,65, including during the process of EMT 66–68. Our results in the EMT-like HCC827-OR cells and HCC4006-OR cells indicate that disruption of mSWI/SNF activity does not affect their proliferation or resistance phenotype. However, it is possible that BAF complexes participate in EMT, perhaps explaining the role for SMARCA4 in regulating the transcriptome of these cells. SMARCA4 knock-down upon osimertinib treatment does not have a significant effect on the proliferation of H1975-OR cells either, which suggests that mSWI/SNF activity is not the only factor supporting the resistant state (i.e. CIC loss). In these cells, it is plausible that SMARCA4 contributes to the drug-tolerant persister states which allow the cells to survive in the presence of TKI 69 until a genetic alteration or another mechanism leads to overt resistance.
Our studies suggest that mSWI/SNF complexes target to and act over many genomic sites in the TKI-resistant state. Previous studies have suggested that such retargeting can be a result of many mechanisms including interactions with the histone landscape 70 or binding to transcription factors which target complexes to specific motifs genome wide 62,63,71. It is possible that kinases alter post-translational modifications on either the chromatin landscape or on BAF complex subunits themselves which in turn affect interactions with other factors such as TFs. This agrees with our findings that the AP-1 family of TFs are amongst the most enriched motifs at sites for which BAF occupancy is changed in the resistant state. Since the AP-1 family of TFs govern many biological processes deemed hallmarks of cancer, it is not surprising. The interplay between these TFs and other TF effectors of downstream signaling cascades affected by TKIs or BAF inhibition (immune, PI3K, cytokine, MAPK etc), or of bromodomains on mSWI/SNF subunits, may be fine-tuning events that modulate therapeutic sensitivity.
We found that chromatin accessibility of genes with NRF2 binding motifs are regulated by mSWI/SNF complexes. Our data suggest that mSWI/SNF increases accessibility at these loci, which facilitates expression of antioxidant response genes and allows cells to withstand osimertinib-induced oxidative stress and survive (Figure 7F). Indeed, we recently reported that Keap1 loss is protective for EGFR-mutant mouse tumors treated with osimertinib 72. Finally, activation of antioxidant programs to counter osimertinib resistance including in drug-tolerant persister cells has been observed 58,73. We show that the oxidative stress caused by osimertinib is partially attenuated by SMARCA4-mediated chromatin remodeling and transcriptional regulation. It has been reported that SMARCA4 can physically interact with NRF2 to regulate oxidative stress 74 consistent with concordance in the levels of the two proteins observed in EGFR-mutant tumors. Given that SMARCA4 knock-down does not modify NRF2 levels, it is likely that NRF2 and SMARCA4 both translocate to the nucleus and cooperate on chromatin to activate expression of antioxidant genes. In contrast, in KRAS mutant LUAD cell lines and a squamous cell carcinoma cell line, loss of mSWI/SNF chromatin remodeling was found to cause increased NRF2 activity 75. While this further substantiates a link between the two pathways, it also highlights how the role of SMARCA4 may be different in different biological contexts. These results underscore how epigenetics can play a role in the regulation of oxidative stress in osimertinib resistant tumors.
Our finding that EGFR-mutant tumors can rely on SMARCA4 for their survival is consistent with the observation that mutations in SMARCA4 (mostly loss-of-function) and EGFR are mutually exclusive (LUAD/TCGA, PanCancer Atlas, P<0.030; 76 NSCLC/GENIE Cohort v9.1-public P<0.001; 77; cbioportal.org) (Figure S7D) 78,79. Although occasional cases of SMARCA4 mutations have been reported in EGFR-mutant tumors 80, these events are rare suggesting that SMARCA4 function is important for EGFR-mutant tumors. While several studies of osimertinib-resistant tumors have not uncovered SMARCA4 mutations 16,32,81,82, a recent study found SMARCA4 mutations in tumors with sub-optimal responses to osimertinib indicating that these can occur perhaps in tumors that lose dependence on EGFR for survival19. Interestingly, a large-scale analysis of SMARCA4 mutations in thousands of solid tumors showed that they are not only mutually exclusive with EGFR alterations in lung adenocarcinomas, but also with other common oncogenic driver alterations in this disease83, suggesting potentially more wide-spanning impact of our findings.
Limitations of the Study
While we identify commonly altered genes in specific NSCLC cell lines exhibiting resensitization upon mSWI/SNF ATPase inhibition and osimertinib treatment, the identification of reliable ‘biomarkers’ that may faithfully predict sensitivity to mSWI/SNF ATPase inhibitors is limited. Another limitation is that the cell lines used have not undergone whole-genome sequencing. As such, it is unclear whether mutations in the non-coding genome act in cis on selected genes to generate or support the resistant state or to contribute to the features dictating synergy between mSWI/SNF inhibitors and osimertinib.
Star Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Katerina Politi (katerina.politi@yale.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All sequencing raw and processed data have been deposited in the Gene Expression Omnibus (GEO) database and are publicly available upon publication under the series GSE202857: Whole Exome Sequencing data (GSE202863), RNA-sequencing data (GSE202859), ATAC-sequencing data (GSE202857). All raw and processed Cut and Run, ATAC and RNA sequencing data for PC9*/OR*, HCC4006/OR and HCC827*/GR6 lines is publicly available upon publication under the series GSE227999. Accession numbers are listed in the Key Resource Table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Brg1 (D1Q7F) | CST | Cat#49360; AB_2728743 |
| Recombinant Anti-BRG1 | Abcam | Cat#ab108318; AB_10889900 |
| Phospho-EGF Receptor (Tyr1068) (D7A5) | CST | Cat#3777; AB_2096270 |
| EGF Receptor | CST | Cat#2232; AB_901981 |
| GAPDH (14C10) | CST | Cat#2118; AB_561053 |
| E-Cadherin (24E10) (AF594 Conjugate) | CST | Cat#7687; AB_2797633 |
| Vimentin (D21H3) (AF488 Conjugate) | CST | Cat#9854; AB_10829352 |
| c-Raf (D4B3J) | CST | Cat#53745; AB_2799444 |
| MEK1/2 (L38C12) | CST | Cat#4694; AB_10695868 |
| Phospho-MEK1/2 (Ser217/221) (41G9) | CST | Cat#9154; AB_2138017 |
| p44/42 MAPK (Erk1/2) (137F5) | CST | Cat#4695; AB_390779 |
| P-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) | CST | Cat#4370; AB_2315112 |
| Lamin A/C | CST | Cat#2032; AB_2136278 |
| CRISPR-Cas9 Antibody (7A9–3A3) | Novus Biologicals | Cat#NBP2–36440 |
| Anti-Nrf2 | Abcam | Cat#ab137550; AB_2687540 |
| Anti-mouse IgG, HRP-linked Antibody | CST | Cat#7076; AB_330924 |
| Anti-rabbit IgG, HRP-linked Antibody | CST | Cat#7074; AB_2099233 |
| Rabbit monoclonal anti-EGF Receptor (D38B1) XP® | Cell Signaling Technology | Cat# 4267, RRID:AB_2246311 |
| Rabbit monoclonal anti-phospho-Akt (Ser473) (D9E) XP® | Cell Signaling Technology | Cat# 4060, RRID:AB_2315049 |
| Rabbit polyclonal anti-Akt | Cell Signaling Technology | Cat# 9272, RRID:AB_329827 |
| Rabbit monoclonal anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (197G2) | Cell Signaling Technology | Cat# 4377, RRID:AB_331775 |
| Rabbit polyclonal p44/42 MAPK (Erk1/2) | Cell Signaling Technology | Cat# 9102, RRID:AB_330744 |
| Rabbit monoclonal HSP90 (C45G5) | Cell Signaling Technology | Cat# 4877, RRID:AB_2233307 |
| Rabbit monoclonal Vinculin (E1E9V) XP® | Cell Signaling Technology | Cat# 7074, RRID:AB_2099233 |
| Goat polyclonal anti-rabbit HRP-linked IgG | Cell Signaling Technology | Cat# 49360, RRID:AB_2728743 |
| Rabbit monoclonal Axl (C89E7) | Cell Signaling Technology | Cat# 8661, RRID:AB_11217435 |
| Rabbit monoclonal E-cadherin (24E10) | Cell Signaling Technology | Cat# 3195, RRID:AB_2291471 |
| Rabbit polyclonal Vimentin (R28) | Cell Signaling Technology | Cat# 3932, RRID:AB_2288553 |
| Rabbit monoclonal Brg1 (D1Q7F) | Cell Signaling Technology | Cat# 365062, RRID:AB_1084786 |
| Mouse monoclonal anti-GAPDH (G-9) | Santa Cruz | Cat# 11956, RRID:AB_2797776 |
| Rabbit monoclonal SMARCC1/BAF155 (D7FS8) | Cell Signaling Technology | Cat# 8173, RRID:AB_10949503 |
| CUTANA Rabbit IgG CUT&RUN Negative Control Antibody | Epicypher | Cat# 130042, RRID:AB_2923178 |
| Rabbit monoclonal Acetyl-Histone H3 (Lys27) (D5E4) XP® | Cell Signaling Technology | Cat# 2729, RRID:AB_1031062 |
| Bacterial and virus strains | ||
| MAX Efficiency® DH5α Competent Cells | Thermo Fisher | Cat#18258012 |
| Biological samples | ||
| EGFR-mutant tumors tissue micro array | Toki MI. et al, 2018 (PMID: 30267840) | YTMA-356 |
| Patient Derived Xenografts (PDXs) | This Paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Osimertinib | Selleckchem | Cat#S7297 |
| Trametinib | Selleckchem | Cat#S2673 |
| Osimertinib | MedChemExpress | Cat# HY-15772A |
| Osimertinib | AstraZeneca | AZD9291 |
| Compound14 | Jun Qi’s Laboratory | N/A |
| FHD-286 | Jun Qi’s Laboratory | N/A |
| High Sensitivity D100 ScreenTape | Agilent | Cat# 5067–5584 |
| High Sensitivity D100 Reagents | Agilent | Cat# 5067–5585 |
| BioMag Plus Concanavalin A beads | PolySciences | Cat# 8605 |
| AMPure XP Beads | Beckman Coulter | Cat# A6388 |
| Spermidine | Sigma-Aldrich | Cat# S2626 |
| pMD2.G | Addgene | Addgene_12259 |
| psPAX2 | Addgene | Addgene_12260 |
| pInducer10-mir-RUP-PheS | Meerbrey KL et al, 2011 (PMID: 21307310) | Addgene_44011 |
| TLCV2 | Addgene | Addgene_87360 |
| pLKO.1 puro | Addgene | Addgene_8453 |
| NRF2-sh1 | Sigma-Aldrich | TRCN0000007556 |
| NRF2-sh2 | Sigma-Aldrich | TRCN0000007557 |
| NRF2-sh2 | Sigma-Aldrich | TRCN0000007558 |
| Critical commercial assays | ||
| RPMI 1640 Medium | Thermo Fisher | Cat#A1049101 |
| Fetal Bovine Serum | Thermo Fisher | Cat#16140–071 |
| Penicillin-Streptomycin | Thermo Fisher | Cat#15140122 |
| 0.25% Trypsin with EDTA | Thermo Fisher | Cat#25200056 |
| TaqMan CNV Assay EGFR | Thermo Fisher | Cat#Hs02088787_cn |
| TaqMan CNV Assay EGFR | Thermo Fisher | Cat#Hs02190396_cn |
| TaqMan CNV Assay RAF1 | Thermo Fisher | Cat#Hs02614899_cn |
| TaqMan CNV Assay RAF1 | Thermo Fisher | Cat#Hs04252880_cn |
| Power SYBR Green Master mix | Thermo Fisher | Cat#4367659 |
| Protease and Phosphatase inhibitor cocktail | Thermo Fisher | Cat#78440 |
| SuperSignal West Pico PLUS | Thermo Fisher | Cat#34579 |
| SuperSigna West Femto Maximum Sensitivity Substrat | Thermo Fisher | Cat# 34096 |
| Triton X-100 | Sigma Aldrich | Cat#X-100 |
| Hoechst dye | Thermo Fisher | Cat#62249 |
| Tumor Dissociation Kit, human | Miltenyi Biotec | Cat#130–095-92 |
| Matrigel | Corning | Cat#356237 |
| Doxycycline diet | Envigo | Cat#TD.00426 |
| ProLong® Gold Antifade Mountant | Thermo Fisher | Cat#P10144 |
| CellROX® Deep Red Reagent | Thermo Fisher | Cat#C10422 |
| CellROX® Green Reagent | Thermo Fisher | Cat#C10444 |
| DNeasy Blood & Tissue kit | Qiagen | Cat#69504 |
| SuperScript III reverse transcriptase | Thermo Fisher | Cat#18080093 |
| RNeasy extraction kit | Thermo Fisher | Cat#74004 |
| MycoAlertTM Mycoplasma Detection Kit | Lonza | Cat#LT07–118 |
| CellTiter-Blue® Cell Viability Assay | Promega | Cat#G8081 |
| GenePrint 10 System | Promega | Cat#B9510 |
| Lipofectamine® RNAiMAX Transfection Reagent | Thermo Fisher | Cat#13778150 |
| TransIT®−293 Transfection Reagent | Mirus | Cat#MIR2704 |
| Antigen Unmasking Solution, Citrate-Based | Vector Laboratories | Cat#H-3300–250 |
| Antigen Unmasking Solution, Tris-Based | Vector Laboratories | Cat#H-3301–250 |
| VECTASTAIN® ABC-HRP Kit, Peroxidase (Mouse IgM ) | Vector Laboratories | Cat#PK-4010 |
| VECTASTAIN® ABC-HRP Kit, Peroxidase (Rabbit IgG) | Vector Laboratories | Cat#PK-4001 |
| Vector® NovaRED® Substrate Kit, Peroxidase (HRP) | Vector Laboratories | Cat#SK-4800 |
| CellTiter-Glo® Luminescent Cell Viability Assay | Promega | Cat# G7573 |
| CellEvent® Caspase-3/7 Green ReadyProbes® Reagent | Invitrogen | Cat# R37111 |
| CUTANA® ChIC/CUT&RUN Kit | Epicypher | Cat# 141048 |
| CUTANA® CUT&RUN Library Prep Ki | Epicypher | Cat# 141001 |
| RNAeasy Mini Kit | Qiagen | |
| MinElute PCR Purification kit | Qiagen | Cat# 28006 |
| NEBNext Ultra II Directional RNA Library Prep Kit for Illumina | New England Biolabs | Cat# E7760L |
| Tagment DNA Enzyme and Buffer Large Kit | Illumina | Cat# 20034197 |
| NextSeq 500/550 High Output Kit v2.5 (75 cylcles) | Illumina | Cat# 20024906 |
| Deposited data | ||
| All sequencing data SuperSeries | GEO Database | GSE202864 |
| Whole Exome Sequencing | GEO Database | GSE202863 |
| RNA-sequencing | GEO Database | GSE202859 |
| ATAC-sequencing | GEO Database | GSE202857 |
| Cut&Run, ATAC-seq, RNA-seq | GEO Database | GSE227999 |
| Experimental models: Cell lines | ||
| PC9 | ATCC / This paper | CVCL_B260; N/A |
| PC9-OR | This paper | N/A |
| H1975 | ATCC | CVCL_1511 |
| H1975-OR | This paper | N/A |
| HCC827 | ATCC | CVCL_2063 |
| HCC827-OR | This paper | N/A |
| YU-005C | This paper | N/A |
| HEK293T | ATCC | CVCL_0063 |
| PC-9*; human EGFR-mutant NSCLC, male | Dr. Kazuto Nishio (Kindai University, Osaka, Japan) | RRID:CVCL_B26 |
| PC-9_OR*; human EGFR-mutant NSCLC, male | This paper | N/A |
| HCC4006; human EGFR-mutant NSCLC, male | ATCC | CRL-2871; RRID:CVCL_1269 |
| HCC4006_OR; human EGFR-mutant NSCLC, male | This paper | N/A |
| HCC827; human EGFR-mutant NSCLC, female | Dr. Adi Gazdar (UT Southwestern, Dallas, TX) | RRID:CVCL_2063 |
| HCC827GR6; human EGFR-mutant NSCLC, female | Engelman et al. 2007 Science | N/A |
| Experimental models: Organisms/strains | ||
| NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice | Jackson Labs | IMSR_JAX:005557 |
| Oligonucleotides | ||
| See Table S4 - Oligonucleotides | N/A | N/A |
| Software and algorithms | ||
| QuPath | Bankhead P. et al 2017 (PMID: 29203879) | SCR_018257 |
| ImageJ | Schneider et al., 2012 | SCR_003070 |
| ApE | https://jorgensen.biology.utah.edu/wayned/ape/ | SCR_014266 |
| Graphpad 9.1.1 | PRISM | SCR_000306 |
| FlowJo v10 | BD | SCR_008520 |
| Ingenuity Pathway Analysis | Qiagen | SCR_008653 |
| RStudio (1.3.1093) | N/A | SCR_000432 |
| Kent Tools | UCSC | https://github.com/ucscGenomeBrowser/kent |
| IGV (2.8.2) | Robinson JT et al. 2012 (PMID: 21221095) | SCR_011793 |
| HOMER (4.11) | Heinz S. et al. 2010 (PMID: 20513432) | SCR_010881 |
| Primer3 | https://primer3.ut.ee/ | SCR_003139 |
| Combenefit | Di Veroli et al. 2016 Bioinformatics | N/A |
| Graphpad Prism 9 | https://www.graphpad.com/scientific-software/prism/ | N/A |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| STAR | Dobin etal., 2013 | https://github.com/alexdobin/STAR |
| MACS2 | Zhang etal., 2008 | https://github.com/taoliu/MACS |
| ngsplot | Shen et al., 2014 | https://github.com/shenlab-sinai/ngsplot |
| EdgeR | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| GSEA | Subramanian et al., 2005 | http://software.broadinstitute.org/gsea/index.jsp |
| Bedtools | Quinlan and Hall, 2010 | http://bedtools.readthedocs.io/en/latest/ |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Picard | Broad Institute | http://broadinstitute.github.io/picard |
| Trimmomatic | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| Gene Ontology | Gene Ontology Consortium, 2015 | http://geneontology.org/ |
| BWA-MEM | Li, 2013 | bio-bwa.sourceforge.net/ |
| Matplotlib | Hunter, 2007 | https://matplotlib.org/stable/ |
| Seaborn | Michael Waskom | https://seaborn.pydata.org/ |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animal Models
All in vivo experiments were performed in NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Labs, IMSR_JAX:005557). Tumors to generate patient-derived xenografts (PDXs) were digested according to the manufacturer instructions (Miltenyi Biotec, Cat#130–095-92) prior to subcutaneous injection. Cells were counted, re-suspended in PBS and mixed 1:1 with matrigel (Corning, Cat#356237) and injected in the right flanks of the mice. Tumor volume was measured using calipers and calculated with the formula [(Length × Width × Width)/2]. For the CRISPR/Cas9 knock-out of SMARCA4 in vivo experiment, cells were transduced with lentiviral particles as described above, selected with 1 μg/mL puromycin in culture for one week, and 5×106 cells were injected per mouse. After monitoring tumor growth for 10 days, the diet was changed to doxycycline to activate Cas9 (Envigo, Cat#TD.00426). Treatment was initiated after 7 days on a doxycycline diet. The mice were treated daily with either vehicle (5% DMSO + 40% PEG300 + 5%Tween 80 + 50% MilliQ-H2O) or osimertinib (25 mg/kg, Selleckchem, Cat#S7297) by oral gavage. Tumor volume was measured twice a week and the mice were euthanized after 2 weeks of treatment. Six mice per group were used in two independent replicates (n=12) from which: 4 tumors were used to obtain paraffin-embedded tissue, 4 were used for flow cytometry CellROX™ (Thermo Fisher, Cat#C10422) experiments and 4 were flash frozen to archive at −80 °C.
All in vivo experiments were performed in female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Labs, #005557). YU-005C cells were counted, re-suspended in PBS and mixed 1:3 with matrigel (Corning, #356237) and 5×106 cells were injected in the right flank of each mouse. Tumor volume was measured using a caliper and calculated with the formula [(Length × Width × Width)/2]. Treatment was initiated 16-days after injection when the tumors reached an average size of ~50 mm3. The mice were treated daily with either vehicle (5% DMSO + 40% PEG300 + 5%Tween 80 + 50% MilliQ-H2O), osimertinib (25 mg/kg, Selleckchem, # S7297), FHD-286 (1.5 mg/kg, Jun Qi laboratory), or combination (osimertinib and FHD-286) by oral gavage (n=7 mice per group). Tumor volume was measured twice a week and the mice were euthanized 18-days posttreatment initiation.
Cell Lines
Isogenic osimertinib-sensitive and resistant cell lines were generated independently by both the Politi and Kadoch/Jänne labs and analyses of the lines were integrated as described in the manuscript. PC9, H1975, HCC827 and YU-005C cells were maintained in RPMI 1640 Medium (Thermo Fisher, Cat#A1049101) supplemented with 10% Fetal Bovine Serum (Thermo Fisher, Cat#16140–071) and 1% Penicillin-Streptomycin (Thermo Fisher, Cat#15140122). These cell lines were authenticated at the Yale DNA Analysis Facility using the GenePrint 10 System (Promega. Cat#B9510). Cells were passaged using 0.25% Trypsin with EDTA (Thermo Fisher, Cat#25200056) when necessary and routinely tested for mycoplasma (Lonza, Cat#LT07–118). To generate osimertinib resistant PC9, H1975 and HCC827 cells (Politi lab), sensitive parental cells were sequentially treated with increasing concentrations of osimertinib. The starting concentration was 25 nM and the drug-containing complete media was changed every 2–3 days. When the surviving cell population exhibited signs of proliferation and reached confluence in 10 cm plates, the cells were transferred to a new plate and the drug concentration was subsequently increased by 25 nM, 50 nM and 100 nM. The process was repeated until EC50 of the cells increased by ~100-fold compared to the sensitive cells. The final concentrations were 1 μM for PC9-OR, 1 μM for HCC827-OR and 2 μM for H1975-OR cells. All OR cells were maintained in the respective final concentration of the drug to ensure the survival of the resistant population.
PC9*, PC9-OR*, HCC4006, HCC4006-OR, HCC827*, and HCC827GR6 were maintained in RMPI-1640 (Gibco) supplemented with 10% fetal bovine serum (GeminiBio) and 1% penicillin-streptomycin (Gibco). Cell line identity was confirmed for PC-9*, HCC4006, and HCC827* by DNA fingerprinting. PC9-OR* and HCC4006-OR cell lines were generated by continuously culturing the respective parental cell lines in 100 nM osimertinib (MedChemExpress) for at least 2 months before being characterized (Kadoch/Jänne labs). Unless stated otherwise, these OR lines were maintained in 100 nM osimertinib (MedChemExpress) but the drug was removed for 1 week before use in drug experiments. HCC827GR6 was previously established in the Jӓnne laboratory 45. Cells were routinely tested and confirmed to be mycoplasma negative (ATCC Cat#30–1012K).
METHOD DETAILS
Cell Growth, Viability & Drug Synergy Assays
For PC9, H1975 and HCC827 cell line pairs and YU-005C, dose-response curves and cell viability assays were performed in 96-well plates in three technical replicates. For dose-response curves, cells were plated at 20–40% confluency and treated for 72 hours with the drugs suspended in DMSO. The media was replaced after the treatment and viability was measured by a fluorescence-based viability assay (Promega, Cat#G8081). Conditions without drug and 0.1% Triton X-100 were used as 100% and 0% viability controls, respectively. IC50 values and dose-response models were calculated using the following formula in GraphPad Prism software: normalized viability=100/(1+ [inhibitor]/IC50). For proliferation assays, the viability was measured using the same system at the indicated times. Colony formation assays were performed in 6-well plates in which 1000–3000 cells/well were plated and treated with the specified conditions for 10–14 days. Cells were washed with PBS, fixed with 10% NBF and stained with Crystal Violet solution (5 mg/mL crystal violet powder, 20% methanol in water). Copy number assays were performed following the manufacturer instructions. The following TaqMan assays from Thermo Fisher were used: EGFR (Cat#Hs02088787_cn & Cat#Hs02190396_cn) and RAF1 (Cat#Hs02614899_cn & Cat#Hs04252880_cn). 3–4 biological replicates were performed for in vitro functional experiments as indicated in each specific figure. Quantitative RT-PCRs were performed using the Power SYBR Green Master mix (Thermo Fisher, Cat#4367659) and custom designed primers (https://primer3.ut.ee/, RRID:SCR_003139) in a Viia 7 Real-Time PCR System (Thermo Fisher, RRID:SCR_019582).
For PC9*, HCC4006 and HCC827* cell line pairs, 500 cells per well were seeded into 384-well plates and 2000 cells per well were seeded into 96-well plates. Cells were drugged the following morning in triplicate. Cell viability was assessed after 72 hours with CellTiter-Glo® Luminescent Cell Viability Assay (Promega) according to manufacturer’s instructions. Plates were read using a POLARstar Omega microplate reader (BMG Labtech). Drug synergy was assessed with Combenefit software as previously described 84. Cells were drugged in a 6-by-6 drug concentration matrix in triplicate and viability was assessed by CellTiter-Glo® after 72 hours. Bliss synergy scores were calculated for each drug combination and were mapped relative to cell proliferation using Combenefit v2.021.
Caspase Assays
Apoptosis was measured in real-time using CellEvent™ Caspase-3/7 Green ReadyProbes™ Reagent (Invitrogen) as previously described 85. Briefly, 2000 cells per well were seeded in 96-well plates. The following morning, media was replaced with CellEvent™ dye and drug containing media. Plates were housed in a BioSpa 8 automated incubator (Agilent) and were scanned at regular intervals using a Cytation5 cell imaging multimode reader (Agilent) or were housed and scanned with an Incucyte S3 (Sartorius). Refer to figure legends for further details on specific experiments. Fluorescent signal was normalized to cellular confluence at each timepoint.
DNA sequencing
Genomic DNA for whole-exome sequencing (WES) or Sanger sequencing of the sensitive and the osimertinib resistant PC9, H1975 and HCC827 cell line pairs was extracted from cells in culture using the DNeasy Blood & Tissue kit (Qiagen, Cat#69504). cDNA used for Sanger sequencing was obtained from RNA using SuperScript III reverse transcriptase (Thermo Fisher, Cat#18080093) following the manufacturer instructions. Sanger sequencing was performed in the Yale Keck Biotechnology Resource Laboratory following their guidelines (https://medicine.yale.edu/keck/). ABI sequencing trace files were visualized using the software ApE (https://jorgensen.biology.utah.edu/wayned/ape/). WES library preparation and Illumina sequencing was performed at the Yale Center for Genome Analysis (YCGA, https://medicine.yale.edu/keck/ycga/). One ug of genomic DNA was sheared to a mean fragment length of about 140 base pairs using focused acoustic energy (Covaris E210). Exome sequencing was performed by exome capture using the IDT xGen capture probe panel with an additional “spike-in” of ~2,500 regions, totaling ~620 kb, of RefGene coding regions that were not included or were poorly covered by the IDT panel. Captured fragments were sequenced using 101 bp paired-end sequencing reads in an Illumina NovaSeq 6000 with an S4 flowcell according to Illumina protocols. Sequencing reads were aligned to human genome build 38 (GRCh38/hg38) using the BWA-MEM, aggregated into a BAM file, and further processed to produce somatic variants with GATK v3.4 and MuTect, following the GATK Best Practices workflow 86. Identified variants were further filtered based on their presence in repositories of common variations (1000 Genomes, NHLBI exome variant server, and 2,577 noncancer exomes sequenced at Yale). CNV analysis was based on the read depth ratio differences between tumor and normal using custom scripts.
Genomic DNA for PC9*, PC9-OR*, HCC4006, and HCC4006-OR was extracted using the DNeasy Blood & Tissue kit (Qiagen, Cat#69504) and was submitted for WES at the Broad Institute of MIT and Harvard using the “Express Somatic Human WES v6” workflow. Captured fragments were sequenced on a NovaSeq 6000 with an S4 flow cell according to standard Illumina protocols using 151 bp paired-end sequencing reads and achieved a median target coverage of ~200X. Sequencing reads were aligned to human genome build 37 (GRCh37/hg19), aggregated into a BAM file, and further processed to identify somatic variants with GATK v4.0.4.0, MuTect2, and Oncotator v1.9.8.0. A panel of normal tissue samples were used to filter out germline variants, identify sequencing artifacts, and to serve as a reference for CNV analysis. For follow up studies, RNA was extracted from PC9*, PC9* treated with 100 nM osi for 4 days, and PC9-OR* cells using the RNeasy Plus Mini Kit (Qiagen, Cat#74136) and was reverse transcribed to cDNA using the QuantiTect Reverse Transcription kit (Qiagen, Cat#205313). PCR amplicons encompassing the BRAF G469A site were generated using the Platinum SuperFi II PCR mix (Invitrogen, Cat #12368250), and were submitted for Sanger sequencing at Genewiz according to standard protocols.
siRNA Design and Expression Vectors
All the siRNAs were designed using i-Score Designer and siRNA Scales 87,88 and obtained from Sigma-Aldrich. For siRNA-mediated inhibition, cells were plated on unsupplemented RPMI media and transfected with Lipofectamine and 30 nM of siRNA following the manufacturer instructions (Thermo Fisher, Cat#13778150). The SMARCA4 shRNAs were custom designed (http://katahdin.mssm.edu/siRNA/RNAi.cgi?type=shRNA, 89 and cloned into the pINDUCER10 lentiviral construct following the standard protocol 90. For NRF2 shRNA inhibition, we used the pLKO.1-puro vector (Addgene_8453; Sigma Millipore) and the empty vector as control. The SMARCA4 sgRNA sequences were previously described 91 and cloned into the TLCV2 (Addgene_87360) lentiviral construct following the standard protocol 92. All the sequences are in Table S4. Lentiviral particles were produced in 293T cells using the pMD2.G (Addgene_12259) and psPAX2 (Addgene_12260) vector system and a DNA transfection reagent optimized for 293T cells following the vendor instructions (Mirus, Cat#MIR2704). Doxycycline inducible expression was achieved by adding 1 μg/mL (shRNAs/PC9-OR, sgRNAs/YU-005C) or 2 μg/mL (shRNAs/YU-005C) DMSO-dissolved doxycycline into the culture media and replacing the media every 2–3 days. Knock-down of SMARCA4 with shRNAs was performed for a week prior to treatment in all experiments as show in Figure S3A unless stated otherwise in the figure legends.
Western Blotting
Whole cell and nuclear lysates were generated using RIPA lysis buffer (50 mmol/L Tris, pH 8.0, 150 mmol/L NaCl, 5 mmol/L MgCl2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) and no salt EB0 buffer (50mM Tris, 0.1% NP-40, 1mM EDTA, 1mM MgCl2) followed by high salt EB300 buffer (50mM Tris, 1% NP-40, 1mM EDTA, 1mM MgCl2, 300mM NaCl) supplemented with protease and phosphatase inhibitor cocktail (Thermo Fisher, Cat#78440), respectively. Equal amounts of total protein were separated by SDS-PAGE and blots were probed as indicated (all antibodies are found in KRT). Signals were detected using either SuperSignal West Pico PLUS (Thermo Fisher, Cat#34579) or Femto chemiluminescent substrates (Thermo Fisher, Cat# 34096) or imaged on LI-COR Odyssey CLx.
IHC and IF
Subcutaneous tumors were collected, fixed in 4% paraformaldehyde overnight at room temperature (RT), and rehydrated in 70% ethanol. Paraffin-embedding and sectioning was performed by the Yale Pathology Tissue Services (YPTS). Four-micrometer sections were used for hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) or immunofluorescence (IF) staining using standard protocols. The YTMA-356 preparation has been previously described 93 and purchased from YPTS. Antigen retrieval was performed by steam heating of slides at 95 °C for 30 minutes using either citrate-based pH 6.0 (Vector, Cat#H-3300–250) or Tris-based pH 9.0 (Vector, Cat#H-3301–250) solutions. Biotinylated molecule detection (Vector Cat#PK-4001, Cat#PK-4010) and peroxidase reactions (Vector, Cat#SK-4800) were performed using the manufacturer protocols. For IF staining, permeabilization was achieved by adding 0.25% Triton X-100 (Sigma Aldrich, Cat#X-100) in PBS for 45 minutes at RT and blocking was performed with 3% BSA + 0.05% Tween 20 in PBS for 1 hour. Nuclear staining was performed by adding Hoechst dye (2 μg/mL, Thermo Fisher, Cat#62249) using the manufacturer’s instructions prior to mounting the slides.
The following antibodies were used for IHC: SMARCA4 (Cat#ab108318; AB_10889900), NRF2 (Cat#ab137550; AB_2687540) and ɣH2A.X (Cat#2577; AB_2118010, CST). For IF staining of cultured cells, cells were plated in 8-well chambers previously coated with collagen and standard protocols were applied for the stainings. The following antibodies were used for IF staining: CDH1-AF594 (Cat#7687; AB_2797633), VIM-AF488 (Cat#9854; AB_10829352) and ɣH2A.X-AF647 (Cat#9720; AB_10692910) from Cell Signaling Technology. ImageJ was used to quantify the bands of the immunoblots and QuPath 94 to quantify IHC and IF staining. All antibodies were used at the dilutions suggested by the manufacturer. Primary and secondary antibody incubations were performed overnight at 4 °C and for 1 hour at RT, respectively.
Drug Treatments for Genomics
For Figures 3,4, and 6 and associated supplemental figures cells were treated with 750nM osimertinib (Selleckchem) unless stated otherwise. For Figure 5 and S5, PC9* and PC9-OR* cells were treated with DMSO, 1uM Compound 14 (Cmp14), 100nM Osimertinib (Osi) (MedChemExpress) and 100nM Osimertinib + 30nM Trametinib (OT) for 24hrs before harvesting for ATAC-seq and RNA-seq (see Methods below). The PC9-OR* cells were washed to remove continuous osimertinib growth media for 7 days prior to experimental setup and then rechallenged with osimertinib (MedChemExpress) or OT as appropriate (see Figure 5 and S5). This work represents efforts from several different labs and as such different sources of osimertinib were used based on protocols in effect in the labs when the experiments were conducted.
CellROX™ oxidative stress detection
CellROX™ Green (Thermo Fisher, Cat#C10444) and Deep Red (Thermo Fisher, Cat#C10422) were used for IF imaging and flow cytometry quantification respectively. For IF imaging, cells were plated on 8-well collagen-coated plates and treated as specified for 72 hours. The cells were washed with PBS, incubated with 5 μM CellROX™ Green in culture media at concentration for 30 minutes at 37 °C, washed with PBS again and incubated with Hoechst dye (2 ug/mL) for 15 minutes in the dark at RT. The slides were mounted with ProLong™ Gold Antifade Mountant (Thermo Fisher, #P10144) and imaged in an inverted fluorescence microscope (Zeiss). The quantification of the IF signal was performed using QuPath 94. For in vitro flow cytometry experiments, cells were treated as described in this section in 6-well plates. Cells were trypsinized, collected, resuspended in FACS buffer (2% FBS in PBS) and kept on ice. For in vivo flow cytometry experiments, subcutaneous tumors were extracted, weighed, and digested as previously described. After 3 washes with PBS, tumor cells were suspended in complete RPMI culture media containing 5 μM CellROX™ DeepRed and incubated for 30 minutes at 37 °C. Cells were washed with PBS, resuspended in FACS buffer and kept on ice. All experiments were performed in a BD LSRII flow cytometer at the Yale Flow Cytometry Facility and analyzed using FlowJo version 10. Quantification of the MFI for YU-005C in vivo tumors was performed after gating GFP+ (Cas9/sgRNA+) and APC+ (CellROX™ DeepRed+) cells.
CUT&RUN
CUT&RUN was performed following EpiCypher’s protocol for their CUTANA ChIC/CUT&RUN kit with slight modifications to day 1. Prior to wash steps, cells were lysed using a nuclear extraction buffer 20mM HEPES–KOH, pH 7.9, 10mM KCl, 0.1% Triton X-100 and 20% glycerol, supplemented with fresh 0.5mM spermidine, and cOmplete Mini, EDTA-free protease inhibitor (Sigma). Next, cells were pelleted at 500g for 3 mins. Cell pellets were then washed using protocol’s permeabilization buffer and subsequently incubated with activated Concanavalin A beads at room temperature for 15 mins. Next, beads mixed in Antibody Buffer were incubated with appropriate primary antibodies overnight (antibodies in key resource table). Library amplification was carried out using Epicypher’s CUT&RUN library kit using 12 cycles of amplification. Quality of CUT&RUN libraries were assessed by TapeStation (Agilent). Libraries were sequenced on NextSeq 500 (Illumina) using 37 bp paired-end sequencing.
ATAC-seq
The Omni-ATAC protocol 95 was used with slight modifications as follows. 100,000 cells per condition were used and washed with PBS before lysis. Cell pellets were lysed in 50μl of cold ATAC-seq resuspension buffer containing 10mM Tris–HCl pH 7.4, 10mM NaCl and 3mM MgCl2, supplemented with fresh NP-40 (final 0.1% v/v), Tween-20 (final 0.1% v/v) and digitonin (final 0.01% v/v) for 3–5min on ice. Next cells were washed with 1ml of resuspension buffer supplemented with Tween-20 (final 0.1% v/v) and pelleted at 500g for 10min at 4 °C. Cell pellets were resuspended in 50μl of transposition reaction mix containing 25μl of 2× TD buffer, 2.5μl of transposase, 16.5μl of 1× PBS, 0.5μl of 1% digitonin (final 0.01% v/v), 0.5μl of 10% Tween-20 (final 0.1% v/v) and 5μl of nuclease-free water. The transposition reaction was incubated at 37°C for 30 minutes with constant shaking on a thermomixer. The Qiagen MinElute Reaction Cleanup Kit was used for DNA purification. A standard ATAC-seq amplification protocol with 5–7 cycles of amplification was used to amplify the tagmented library96. ATAC-seq libraries were sequenced on NovaSeq 6000 (Illumina) using 37-bp paired-end sequencing or performed by YCGA following their standard protocols (https://medicine.yale.edu/keck/ycga/). ATAC-seq experiments were performed in biological duplicates for all cell lines and three replicates for YU-005C cells. Note: ATAC-seq studies in PC9-OR (Figure 1) were performed on cells with washout of osimertinib; washout was not applied to all other cell lines.
RNA-seq
One million cells were harvested and washed with cold PBS to remove trypsin and stored in RLT buffer (Qiagen) until further processing. RNA was purified using the Qiagen RNeasy kit (Qiagen). PC9*/OR*, HCC4006/OR and HCC827*/GR6 RNA was further processed using the Illumina NEBNext Ultra II Directional RNA Library Prep Kit and quality control was assessed by TapeStation (Agilent) and libraries were quantified by Qubit Fluorometer. These RNA–seq libraries were sequenced on the Illumina NextSeq 500 with 75-bp single-end sequencing. Quality control and library preparation for RNA from PC9/OR, H1975/OR and HCC827/OR cell lines was performed at the Yale Center for Genome Analysis (YCGA) following their standard protocols. Replicates for specific RNA-seq experiments are as indicated in figure legends.
QUANTIFICATION AND STATISTICAL ANALYSIS
Enrichment and Statistical Analysis
The Upstream Regulator & Cellular Functions enrichment analysis were generated using Ingenuity Pathway Analysis (https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis). Student t-tests for paired and unpaired samples, depending on the experimental setting, were performed for in vitro experiments in which we performed 3–4 biological replicates. Shapiro-Wilk and Kolmogorov-Smirnov normality tests were performed for datasets with an n≥10. Datasets following a normal distribution were analyzed with a parametric Student t-test and whilst datasets that did not meet normality were analyzed with the non-parametric Mann-Whitney test. Statistical P values for RNA-seq and ATAC-seq data were obtained from the DESeq2 analyses. The Coefficient of Drug Interaction (CDI) was calculated as follows: CDI = AB/(AxB); where AB is % combinatorial effect of osimertinib and SMARCA4 knock-down, A is the % effect of osimertinib alone and B is the % effect of SMARCA4-knock down alone. Other specific tests, different sample sizes as well as depicted values and error bars are indicated in figure legends. All tests were performed in GraphPad Prism 9.1.1.
Data processing for RNA, ATAC and CUT & RUN
RNA-seq reads were mapped to the hg19 human genome assembly using STAR v2.3.1 97 with default parameters. Alignment files in BAM format were generated using samtools v0.1.19 98. ATAC-seq paired-end reads were processed as follows: reads were trimmed to 30bp using Trimmomatic v0.35 99, mapped to the hg19 human genome assembly using Bowtie2 v2.1.0 (−X2000), and filtered for duplicates using Picard MarkDuplicates (Broad Institute, 2019). Cut&Run reads were processed using the Cut&RunTools pipeline with default parameters 100.
RNA-seq gene counts for cell-line data were generated with using STAR (-quantMode GeneCounts, last column of GeneReadsOut.tab) against the hg19 refFlat annotation. The raw counts were prefiltered to exclude genes with less than one read per sample on average. These prefiltered raw count matrices were converted to RPKMS using a hg19 refFlat annotation with the median isoform length for gene length and the total gene counts per sample (in millions) for the per-million scaling factor.
Raw counts for Cut&Run, and ATAC samples across a subset of sites were generated using bedtools intersect with default parameters on coverage bed files. Paired-end reads for Cut&Run and ATAC samples were converted to the appropriate paired-end bed file using samtools view (-h) and a custom perl script to filter the SAM entries. Raw counts were converted to RPKMs using site widths and total million mapped reads from samtools idxstats as the scaling factor.
Peaks were called using MACS2 v2.1.0 (-q 0.001) 101 with the narrow peak caller for all marks in this study and broad peak caller for ATAC. Duplicate reads were excluded using samtools rmdup and used for downstream analyses. ATAC and Cut&Run tracks were generated using deepTools bamCoverage (--normalizeUsing CPM -bs 50 --smoothLength 600 --ignoreDuplicates --samFlagInclude 64) and RNA tracks were generated using deepTools bamCoverage (--normalizeUsingRPKM).
CUT & RUN, ATAC-seq and RNA-seq Data Analysis
SMARCA4, SMARCC1, H3K27ac, IgG and ATAC peaks were merged across conditions for each antibody using the default bedtools merge call. Upon peak calling and visualization of IgG negative control samples, IgG signal was negligible and no further processing to account of IgG was needed. Venn diagrams of peak overlaps were generated using the ChIPpeakAnno package in R with the findOverlapsOfPeaks and makeVennDiagram functions with default parameters. Unless otherwise noted, gained and lost ATAC or SMARCA4 Cut&Run sites, and upregulated or downregulated genes across the indicated conditions were determined using edgeR (glmQLFit, log2FC=1, FDR=0.05). Perceived batch effects were modeled as covariates in the analysis if necessary. Volcano plots for changes in expression and accessibility across the indicated conditions were visualized as scatter plots in -log10(adj.p) vs logFC using matplotlib. Interaction terms were modelled to assess significant differences in accessibility and expression between the parental and resistance state between the indicated cell lines for the quadrant analyses plots. Quadrant analyses were visualized as scatter plots of the logFCs between the parental and resistant state for the indicated cell lines.
Changes in SMARCA4 occupancy across the indicated conditions were determined using logFCs of the log RPKM values (logFC=1). These changes were visualized as a scatterplot of the log RPKM signal in each condition and colored by gains or losses in BAF occupancy using matplotlib. FASTA sequences across these sets of sites were generated using site centers with flanking windows of 200bp (total window size of 400bp). Enriched motifs across these sets of sites were determined using HOMER findMotifsGenome.pl 102 against genome-background (-size 400).
HOMER motif known results were visualized as barplots using matplotlib. Unless otherwise noted, barplots and Venn diagrams were visualized using matplotlib, and heatmaps were created using seaborn clustermap.
The Metascape web server was used to assess the enrichment of different pathways and gene sets using an input gene list. These gene lists were determined by up and downregulated genes across the indicated conditions, and the overlap of these deregulated genes with coordinated or anti-coordinated changes in accessibility and BAF localization (using genes assigned to changing peaks by nearest distance to TSS) as indicated in the figures. The significance of enriched pathways and gene sets were visualized as horizontal bar charts using the description column of the metascape_results.xlsx files. To elaborate, sites of interest were annotated to their nearest protein-coding gene against the hg19 refFlat gene annotation and distance to transcription start site (TSS) were calculated with a custom perl script. Gene intersections with gene subsets were visualized as volcano plots of logFC vs -log(adj.p). Gene intersections were input into downstream gene enrichment analysis such as Metascape with default parameters. Gene enrichment results from Metascape were visualized as barplots of -log(p-val) by each enriched term, process, or pathway as horizontal bar charts using matplotlib. RPKMs of different genes were visualized as bar plots across the indicated conditions using matplotlib or ggplot in R. Different sets of sites were assigned to their nearest protein-coding gene using distances to TSS. These distances were visualized as stacked bar charts using ggplot in R, highlighting the proportion of promoter, promoter proximal, and distal enhancer regions.
PCA plots were generated using the quantile-normalized log RPKM signal of the expression and accessibility profiles as input and visualized as scatterplots using seaborn scatterplot.
Heatmaps and metaplots were generated for each antibody over subsets of sites using ngs.plot.r with either 10kb windows centered on each site (-G hg19 -FL 150 -L 5000 -R bed). Heatmaps and metaplots were visualized on the RPM-scale using matplotlib.
Unless otherwise noted, gene heatmaps were visualized as z-scored RPKMs across the samples using a blue-white-red heatmap in the clustermap function of seaborn. For gene heatmaps with rectangular black boxes within it, the RPKMs were z-scored across the samples within each box separately and horizontally concatenated together to accentuate the different between the indicated conditions and to help visualize the most biologically meaningful differences between the samples. Whenever indicated, k-means clustering was used cluster the genes with similar expression profiles using the scikit-learn k-means clustering utility, and clusters were reordered and colored to present the data more meaningfully.
The number of common and specific deregulated genes between the parental and resistant state across all six cell lines were visualized as a bar chart using matplotlib (Figure S1L). The number and proportion of deregulated genes that have coordinated changes in accessibility were visualized as pie charts using matplotlib.
LogFCs estimated by EdgeR across the indicated conditions (such as parental vs resistant in each cell line) were used to rank genes for input into GSEA in pre-ranked mode (-norm meandiv -nperm 20000 -scoring_scheme weighted -set_ max 500 -set_min 15) against a variety of MSigDB databases, including Hallmark 103, BioCarta, GO Biological Processes, and Wikipathways 104 databases. GSEA results were visualized as bubble charts of normalized enrichment scores (NES) and FDR values using ggplot in R or as blue-white-red clustered heatmaps of NES values using seaborn clustermap. Differences in logFCs between the parental and resistant state between cell lines were also used as input into GSEA in pre-rank mode using the same parameters and the logFC difference as the ranking metric.
Changes in the expression profiles characterizing resistance for each cell line were also visualized as an ordered scatter plot (e.g. hockey stick plot) of the logFC between the parental and resistant state and the rank of the logFC for each cell line and colored by coordinated changes in BAF localization and accessibility.
Gene overlaps between the indicated conditions were visualized as venn diagrams using matplotlib.
In Figure 5D, the expression profiles of a subset of deregulated genes as defined in the text were z-scored across the indicated samples (RPKM z-score values) and k-means clustered using scikit-learn (k=4). The largest coordinated change in accessibility for the ATAC peaks assigned to these genes (by nearest distance to TSS analysis) were plotted as a blue-purple-yellow heatmap using seaborn heatmap. Genes with no matched ATAC peak were assigned an ATAC logFC of 0.
Supplementary Material
EC50 values, fold changes, and calculations are shown for each cell line pair used in this study.
Table S2, Related to Figure 3 and S3. Properties of patient-derived xenografts (PDX) of EGFR-mutant lung cancers.
Table S3, Related to Figure 3 and S3. Clinicopathological information of patients from the YTMA-356.
Table S4. Related to STAR Methods. Oligonucleotides used throughout the study.
Highlights.
Osimertinib resistance is associated with extensive chromatin accessibility changes
mSWI/SNF complex targeting and activity govern the resistance signature
mSWI/SNF sustains a proliferative and ROS-attenuating profile in resistant cells
Genetic and pharmacological inhibition of SMARCA4/2 reverses osimertinib resistance
In this study, de Miguel et al. find that mammalian SWI/SNF chromatin remodeling complexes promote osimertinib resistance in EGFR-mutant lung cancer model systems via changes in chromatin accessibility which underlie resistance-associated gene expression and cancer cell proliferation. Genetic and pharmacological inhibition of mSWI/SNF complexes leads to osimertinib resensitization in a subset of osimertinib-resistant cell and in vivo tumor models.
Acknowledgements
We thank members of the Politi, Kadoch and Jänne labs for their critical feedback. This work was supported by P50CA196530–06 and U01CA235747 (to K.P. and D.X.N) and a Yale Cancer Center (YCC) pilot grant (to K.P., Q.Y. and A.Z.X.). F.d.M was supported by T32 CA193200–01A1. C.G. was supported by the Helen Gurley Brown Fellowship (DFCI). F.E was supported by the Fundación Ramón Areces Life and Matter Sciences Postdoctoral Fellowship. C.K. was supported by the Howard Hughes Medical Institute (HHMI). The YCC Shared Resources used were in part supported by P30 CA016359. The authors also thank the Molecular Biology Core Facility (MBCF) at the DFCI (especially Zach Herbert and Maura Sullivan) and the Yale Center for Genome Analysis (especially Sameet Mehta) as well as Drs. Monte Winslow and Arnaud Augert for feedback.
C.K. is the Scientific Founder, Scientific Advisor to the Board of Directors, Scientific Advisory Board member, shareholder, and consultant for Foghorn Therapeutics, Inc. (Cambridge, MA), serves on the Scientific Advisory Boards of Nereid Therapeutics, Nested Therapeutics, Accent Therapeutics, and Fibrogen, Inc. and is a consultant for Cell Signaling Technologies and Google Ventures. C.K. is also a member of the Molecular Cell and Cell Chemical Biology Editorial Boards. D.L. Rimm reports grants and personal fees from Amgen, Astra Zeneca, Cepheid, Konica – Minolta, Lilly, NextCure and personal fees from Cell Signaling Technology, Danaher, Fluidigm, GSK, Merck, Monopteros, NanoString, Odonate, Paige.AI, Regeneron, Roche, Sanofi, Ventana, Verily. K. Politi reports grants from the NCI/NIH; grants and personal fees from AstraZeneca; grants from Kolltan, Roche/Genentech, Boehringer Ingelheim, D2G Oncology and Symphogen; and personal fees from Janssen, Dynamo Therapeutics, Halda, Maverick Therapeutics, and Tocagen; and a patent for EGFRT790M mutation testing issued, licensed, and with royalties paid from Molecular Diagnostics/Memorial Sloan Kettering Cancer Center. P.A.J. is an equity owner in Gatekeeper Pharmaceuticals; consults for AstraZeneca, Boehringer Ingelheim, Pfizer, Roche/Genentech, Chugai Pharmaceuticals, Eli Lilly Pharmaceuticals, Araxes Pharmaceuticals, SFJ Pharmaceuticals, Voronoi, Daiichi Sankyo, Biocartis, Novartis, Sanofi, Takeda Oncology, Mirati Therapeutics, Transcenta, Silicon Therapeutics, Syndax, Nuvalent, Bayer, Esai, Allorion Therapeutics, Accutar Biotech, and Abbvie; receives research support from AstraZeneca, Daiichi Sankyo, PUMA, Eli Lilly, Boehringer Ingelheim, Revolution Medicines, and Takeda Oncology and is a co-inventor and receives postmarketing royalties on a DFCI owned patent on EGFR mutations licensed to LabCorp. Q.Y. reports grants and personal fees from AstraZeneca, and is a Scientific Advisory Board member of AccuraGen Inc. D.X.N received research funding from AstraZeneca.
Footnotes
Declaration of Interests
The other authors declare no competing interests.
Inclusion and Diversity Statement
We support inclusive, diverse and equitable conduct of research. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bedard PL, Hyman DM, Davids MS, and Siu LL (2020). Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet 395, 1078–1088. 10.1016/S0140-6736(20)30164-1. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P, Cross D, and Janne PA (2021). Kinase drug discovery 20 years after imatinib: progress and future directions. Nat Rev Drug Discov 20, 551–569. 10.1038/s41573-021-00195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim G, McKee AE, Ning YM, Hazarika M, Theoret M, Johnson JR, Xu QC, Tang S, Sridhara R, Jiang X, et al. (2014). FDA approval summary: vemurafenib for treatment of unresectable or metastatic melanoma with the BRAFV600E mutation. Clin Cancer Res 20, 4994–5000. 10.1158/1078-0432.CCR-14-0776. [DOI] [PubMed] [Google Scholar]
- 4.Tan AC, and Tan DSW (2022). Targeted Therapies for Lung Cancer Patients With Oncogenic Driver Molecular Alterations. J Clin Oncol 40, 611–625. 10.1200/JCO.21.01626. [DOI] [PubMed] [Google Scholar]
- 5.Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, et al. (2017). Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med 377, 523–533. 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 6.Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA, Mariotto AB, Lowy DR, and Feuer EJ (2020). The Effect of Advances in Lung-Cancer Treatment on Population Mortality. N Engl J Med 383, 640–649. 10.1056/NEJMoa1916623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JR., Cohen M., Sridhara R., Chen YF., Williams GM., Duan J., Gobburu J., Booth B., Benson K., Leighton J., et al. (2005). Approval summary for erlotinib for treatment of patients with locally advanced or metastatic non-small cell lung cancer after failure of at least one prior chemotherapy regimen. Clin Cancer Res 11, 6414–6421. 10.1158/1078-0432.CCR-05-0790. [DOI] [PubMed] [Google Scholar]
- 8.Carrot-Zhang J, Soca-Chafre G, Patterson N, Thorner AR, Nag A, Watson J, Genovese G, Rodriguez J, Gelbard MK, Corrales-Rodriguez L, et al. (2021). Genetic Ancestry Contributes to Somatic Mutations in Lung Cancers from Admixed Latin American Populations. Cancer Discov 11, 591–598. 10.1158/2159-8290.CD-20-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arrieta O, Cardona AF, Martin C, Mas-Lopez L, Corrales-Rodriguez L, Bramuglia G, Castillo-Fernandez O, Meyerson M, Amieva-Rivera E, Campos-Parra AD, et al. (2015). Updated Frequency of EGFR and KRAS Mutations in NonSmall-Cell Lung Cancer in Latin America: The Latin-American Consortium for the Investigation of Lung Cancer (CLICaP). J Thorac Oncol 10, 838–843. 10.1097/JTO.0000000000000481. [DOI] [PubMed] [Google Scholar]
- 10.Midha A, Dearden S, and McCormack R (2015). EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res 5, 2892–2911. [PMC free article] [PubMed] [Google Scholar]
- 11.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura F, Nogami N, Kurata T, et al. (2018). Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 378, 113–125. 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 12.Wood KC (2015). Mapping the Pathways of Resistance to Targeted Therapies. Cancer Res 75, 4247–4251. 10.1158/0008-5472.CAN-15-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabnis AJ, and Bivona TG (2019). Principles of Resistance to Targeted Cancer Therapy: Lessons from Basic and Translational Cancer Biology. Trends Mol Med 25, 185–197. 10.1016/j.molmed.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MR, Ward RA, Mellor MJ, et al. (2014). AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 4, 1046–1061. 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Noia V, D’Aveni A, D’Argento E, Rossi S, Ghirardelli P, Bortolotti L, Vavassori V, Bria E, and Ceresoli GL (2021). Treating disease progression with osimertinib in EGFR-mutated non-small-cell lung cancer: novel targeted agents and combination strategies. ESMO Open 6, 100280. 10.1016/j.esmoop.2021.100280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Passaro A, Jänne PA, Mok T, and Peters S (2021). Overcoming therapy resistance in EGFR-mutant lung cancer. Nature Cancer 2, 377–391. 10.1038/s43018-021-00195-8. [DOI] [PubMed] [Google Scholar]
- 17.Schoenfeld AJ, Chan JM, Kubota D, Sato H, Rizvi H, Daneshbod Y, Chang JC, Paik PK, Offin M, Arcila ME, et al. (2020). Tumor Analyses Reveal Squamous Transformation and Off-Target Alterations As Early Resistance Mechanisms to First-line Osimertinib in EGFR-Mutant Lung Cancer. Clin Cancer Res 26, 2654–2663. 10.1158/1078-0432.CCR-19-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oxnard GR, Hu Y, Mileham KF, Husain H, Costa DB, Tracy P, Feeney N, Sholl LM, Dahlberg SE, Redig AJ, et al. (2018). Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 4, 1527–1534. 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chmielecki J, Gray JE, Cheng Y, Ohe Y, Imamura F, Cho BC, Lin MC, Majem M, Shah R, Rukazenkov Y, et al. (2023). Candidate mechanisms of acquired resistance to first-line osimertinib in EGFR-mutated advanced non-small cell lung cancer. Nat Commun 14, 1070. 10.1038/s41467-023-35961-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammad HP, Barbash O, and Creasy CL (2019). Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med 25, 403–418. 10.1038/s41591-019-0376-8. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y., Zheng D., Zhou T., Song H., Hulsurkar M., Su N., Liu Y., Wang Z., Shao L., Ittmann M., et al. (2018). Androgen deprivation promotes neuroendocrine differentiation and angiogenesis through CREB-EZH2-TSP1 pathway in prostate cancers. Nat Commun 9, 4080. 10.1038/s41467-018-06177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murai F, Koinuma D, Shinozaki-Ushiku A, Fukayama M, Miyaozono K, and Ehata S (2015). EZH2 promotes progression of small cell lung cancer by suppressing the TGF-beta-Smad-ASCL1 pathway. Cell Discov 1, 15026. 10.1038/celldisc.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai Y, Zhang Z, Cheng L, Wang R, Chen X, Kong Y, Feng F, Ahmad N, Li L, and Liu X (2019). Inhibition of enhancer of zeste homolog 2 (EZH2) overcomes enzalutamide resistance in castration-resistant prostate cancer. J Biol Chem 294, 9911–9923. 10.1074/jbc.RA119.008152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. (2010). A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80. 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitsui E, Yoshida S, Shinoda Y, Matsumori Y, Tsujii H, Tsuchida M, Wada S, Hasegawa M, Ito A, Mino K, et al. (2019). Identification of ryuvidine as a KDM5A inhibitor. Sci Rep 9, 9952. 10.1038/s41598-019-46346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su SF, Liu CH, Cheng CL, Ho CC, Yang TY, Chen KC, Hsu KH, Tseng JS, Chen HW, Chang GC, et al. (2021). Genome-Wide Epigenetic Landscape of Lung Adenocarcinoma Links HOXB9 DNA Methylation to Intrinsic EGFR-TKI Resistance and Heterogeneous Responses. JCO Precis Oncol 5. 10.1200/PO.20.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mashtalir N, D’Avino AR, Michel BC, Luo J, Pan J, Otto JE, Zullow HJ, McKenzie ZM, Kubiak RL, St Pierre R, et al. (2018). Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 175, 1272–1288 e1220. 10.1016/j.cell.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittal P, and Roberts CWM (2020). The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat Rev Clin Oncol 17, 435–448. 10.1038/s41571-020-0357-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Centore RC, Sandoval GJ, Soares LMM, Kadoch C, and Chan HM (2020). Mammalian SWI/SNF Chromatin Remodeling Complexes: Emerging Mechanisms and Therapeutic Strategies. Trends Genet 36, 936–950. 10.1016/j.tig.2020.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Kadoch C, and Crabtree GR (2015). Mammalian SWI:SNF chromatin remodeling complexes and cancer- Mechanistic insights gained from human genomics. Sci. Adv. 1(5). 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michel BC, D’Avino AR, Cassel SH, Mashtalir N, McKenzie ZM, McBride MJ, Valencia AM, Zhou Q, Bocker M, Soares LMM, et al. (2018). A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat Cell Biol 20, 1410–1420. 10.1038/s41556-018-0221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoenfeld AJ, Bandlamudi C, Lavery JA, Montecalvo J, Namakydoust A, Rizvi H, Egger J, Concepcion CP, Paul S, Arcila ME, et al. (2020). The Genomic Landscape of SMARCA4 Alterations and Associations with Outcomes in Patients with Lung Cancer. Clin Cancer Res 26, 5701–5708. 10.1158/1078-0432.CCR-20-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernando TM, Piskol R, Bainer R, Sokol ES, Trabucco SE, Zhang Q, Trinh H, Maund S, Kschonsak M, Chaudhuri S, et al. (2020). Functional characterization of SMARCA4 variants identified by targeted exome-sequencing of 131,668 cancer patients. Nature Communications 11, 5551. 10.1038/s41467-020-19402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buscarlet M, Krasteva V, Ho L, Simon C, Hebert J, Wilhelm B, Crabtree GR, Sauvageau G, Thibault P, and Lessard JA (2014). Essential role of BRG, the ATPase subunit of BAF chromatin remodeling complexes, in leukemia maintenance. Blood 123, 1720–1728. 10.1182/blood-2013-02-483495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cyrta J., Augspach A., De Filippo MR., Prandi D., Thienger P., Benelli M., Cooley V., Bareja R., Wilkes D., Chae SS., et al. (2020). Role of specialized composition of SWI/SNF complexes in prostate cancer lineage plasticity. Nat Commun 11, 5549. 10.1038/s41467-020-19328-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerrero-Martinez JA, and Reyes JC (2018). High expression of SMARCA4 or SMARCA2 is frequently associated with an opposite prognosis in cancer. Sci Rep 8, 2043. 10.1038/s41598-018-20217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jubierre L, Soriano A, Planells-Ferrer L, Paris-Coderch L, Tenbaum SP, Romero OA, Moubarak RS, Almazan-Moga A, Molist C, Roma J, et al. (2016). BRG1/SMARCA4 is essential for neuroblastoma cell viability through modulation of cell death and survival pathways. Oncogene 35, 5179–5190. 10.1038/onc.2016.50. [DOI] [PubMed] [Google Scholar]
- 38.Kim SY, Shen Q, Son K, Kim HS, Yang HD, Na MJ, Shin E, Yu S, Kang K, You JS, et al. (2021). SMARCA4 oncogenic potential via IRAK1 enhancer to activate Gankyrin and AKR1B10 in liver cancer. Oncogene. 10.1038/s41388-021-01875-6. [DOI] [PubMed] [Google Scholar]
- 39.Roy N, Malik S, Villanueva KE, Urano A, Lu X, Von Figura G, Seeley ES, Dawson DW, Collisson EA, and Hebrok M (2015). Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev 29, 658–671. 10.1101/gad.256628.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun A, Tawfik O, Gayed B, Thrasher JB, Hoestje S, Li C, and Li B (2007). Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate 67, 203–213. 10.1002/pros.20521. [DOI] [PubMed] [Google Scholar]
- 41.Papillon JPN, Nakajima K, Adair CD, Hempel J, Jouk AO, Karki RG, Mathieu S, Mobitz H, Ntaganda R, Smith T, et al. (2018). Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers. J Med Chem 61, 10155–10172. 10.1021/acs.jmedchem.8b01318. [DOI] [PubMed] [Google Scholar]
- 42.Farnaby W, Koegl M, Roy MJ, Whitworth C, Diers E, Trainor N, Zollman D, Steurer S, Karolyi-Oezguer J, Riedmueller C, et al. (2019). BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat Chem Biol 15, 672–680. 10.1038/s41589-019-0294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kofink C, Trainor N, Mair B, Wohrle S, Wurm M, Mischerikow N, Roy MJ, Bader G, Greb P, Garavel G, et al. (2022). A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo. Nat Commun 13, 5969. 10.1038/s41467-022-33430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao L, Parolia A, Qiao Y, Bawa P, Eyunni S, Mannan R, Carson SE, Chang Y, Wang X, Zhang Y, et al. (2022). Targeting SWI/SNF ATPases in enhancer-addicted prostate cancer. Nature 601, 434–439. 10.1038/s41586-021-04246-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. (2007). MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043. 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 46.Inoue Y, Nikolic A, Farnsworth D, Shi R, Johnson FD, Liu A, Ladanyi M, Somwar R, Gallo M, and Lockwood WW (2021). Extracellular signal-regulated kinase mediates chromatin rewiring and lineage transformation in lung cancer. Elife 10. 10.7554/eLife.66524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okimoto RA, Breitenbuecher F, Olivas VR, Wu W, Gini B, Hofree M, Asthana S, Hrustanovic G, Flanagan J, Tulpule A, et al. (2017). Inactivation of Capicua drives cancer metastasis. Nat Genet 49, 87–96. 10.1038/ng.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCormick F (2018). c-Raf in KRas Mutant Cancers: A Moving Target. Cancer Cell 33, 158–159. 10.1016/j.ccell.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohashi K., Sequist LV., Arcila ME., Moran T., Chmielecki J., Lin YL., Pan Y., Wang L., de Stanchina E., Shien K., et al. (2012). Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A 109, E2127–2133. 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, and Crabtree GR (2011). esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nature Cell Biology 13, 903–913. 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saladi SV, Ross K, Karaayvaz M, Tata PR, Mou H, Rajagopal J, Ramaswamy S, and Ellisen LW (2017). ACTL6A Is Co-Amplified with p63 in Squamous Cell Carcinoma to Drive YAP Activation, Regenerative Proliferation, and Poor Prognosis. Cancer Cell 31, 35–49. 10.1016/j.ccell.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arasada RR, Shilo K, Yamada T, Zhang J, Yano S, Ghanem R, Wang W, Takeuchi S, Fukuda K, Katakami N, et al. (2018). Notch3-dependent beta-catenin signaling mediates EGFR TKI drug persistence in EGFR mutant NSCLC. Nat Commun 9, 3198. 10.1038/s41467-018-05626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ercan D, Xu C, Yanagita M, Monast CS, Pratilas CA, Montero J, Butaney M, Shimamura T, Sholl L, Ivanova EV, et al. (2012). Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer Discov 2, 934–947. 10.1158/2159-8290.Cd-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tricker EM, Xu C, Uddin S, Capelletti M, Ercan D, Ogino A, Pratilas CA, Rosen N, Gray NS, Wong KK, and Janne PA (2015). Combined EGFR/MEK Inhibition Prevents the Emergence of Resistance in EGFR-Mutant Lung Cancer. Cancer Discov 5, 960–971. 10.1158/2159-8290.CD-15-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu W, Hellerbrand C, Köhler UA, Bugnon P, Kan YW, Werner S, and Beyer TA (2008). The Nrf2 transcription factor protects from toxin-induced liver injury and fibrosis. Lab Invest 88, 1068–1078. 10.1038/labinvest.2008.75. [DOI] [PubMed] [Google Scholar]
- 56.Li G, Li X, Mahmud I, Ysaguirre J, Fekry B, Wang S, Wei B, Eckel-Mahan KL, Lorenzi PL, Lehner R, and Sun K (2023). Interfering with lipid metabolism through targeting CES1 sensitizes hepatocellular carcinoma for chemotherapy. JCI Insight 8. 10.1172/jci.insight.163624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen T, Nioi P, and Pickett CB (2009). The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 284, 13291–13295. 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marcar L, Bardhan K, Gheorghiu L, Dinkelborg P, Pfaffle H, Liu Q, Wang M, Piotrowska Z, Sequist LV, Borgmann K, et al. (2019). Acquired Resistance of EGFR-Mutated Lung Cancer to Tyrosine Kinase Inhibitor Treatment Promotes PARP Inhibitor Sensitivity. Cell Rep 27, 3422–3432 e3424. 10.1016/j.celrep.2019.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, and Sanchez-Cespedes M (2008). Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat 29, 617–622. 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 60.Orvis T., Hepperla A., Walter V., Song S., Simon J., Parker J., Wilkerson MD., Desai N., Major MB., Hayes DN., et al. (2014). BRG1/SMARCA4 inactivation promotes non-small cell lung cancer aggressiveness by altering chromatin organization. Cancer Res 74, 6486–6498. 10.1158/0008-5472.CAN-14-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez-Nieto S, and Sanchez-Cespedes M (2009). BRG1 and LKB1: tales of two tumor suppressor genes on chromosome 19p and lung cancer. Carcinogenesis 30, 547–554. 10.1093/carcin/bgp035. [DOI] [PubMed] [Google Scholar]
- 62.Sandoval GJ, Pulice JL, Pakula H, Schenone M, Takeda DY, Pop M, Boulay G, Williamson KE, McBride MJ, Pan J, et al. (2018). Binding of TMPRSS2-ERG to BAF Chromatin Remodeling Complexes Mediates Prostate Oncogenesis. Mol Cell 71, 554–566 e557. 10.1016/j.molcel.2018.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, Awad ME, Rengarajan S, Volorio A, McBride MJ, et al. (2017). Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 171, 163–178 e119. 10.1016/j.cell.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McBride MJ, Pulice JL, Beird HC, Ingram DR, D’Avino AR, Shern JF, Charville GW, Hornick JL, Nakayama RT, Garcia-Rivera EM, et al. (2018). The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell 33, 1128–1141 e1127. 10.1016/j.ccell.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ronan JL, Wu W, and Crabtree GR (2013). From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet 14, 347–359. 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang T, Peng B, Luo T, Tian D, Zhao Z, Fu Z, and Li Q (2022). ZEB1 recruits BRG1 to regulate airway remodelling epithelial-to-mesenchymal transition in asthma. Exp Physiol 107, 515–526. 10.1113/EP090212. [DOI] [PubMed] [Google Scholar]
- 67.Tomihara H, Carbone F, Perelli L, Huang JK, Soeung M, Rose JL, Robinson FS, Lissanu Deribe Y, Feng N, Takeda M, et al. (2021). Loss of ARID1A Promotes Epithelial-Mesenchymal Transition and Sensitizes Pancreatic Tumors to Proteotoxic Stress. Cancer Res 81, 332–343. 10.1158/0008-5472.CAN-19-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanchez-Tillo E, Lazaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, Engel P, and Postigo A (2010). ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 29, 3490–3500. 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 69.Mikubo M, Inoue Y, Liu G, and Tsao MS (2021). Mechanism of Drug Tolerant Persister Cancer Cells: The Landscape and Clinical Implication for Therapy. J Thorac Oncol 16, 1798–1809. 10.1016/j.jtho.2021.07.017. [DOI] [PubMed] [Google Scholar]
- 70.Mashtalir N, Dao HT, Sankar A, Liu H, Corin AJ, Bagert JD, Ge EJ, D’Avino AR, Filipovski M, Michel BC, et al. (2021). Chromatin landscape signals differentially dictate the activities of mSWI/SNF family complexes. Science 373, 306–315. 10.1126/science.abf8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vierbuchen T, Ling E, Cowley CJ, Couch CH, Wang X, Harmin DA, Roberts CWM, and Greenberg ME (2017). AP-1 Transcription Factors and the BAF Complex Mediate Signal-Dependent Enhancer Selection. Mol Cell 68, 1067–1082 e1012. 10.1016/j.molcel.2017.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Foggetti G, Li C, Cai H, Hellyer JA, Lin WY, Ayeni D, Hastings K, Choi J, Wurtz A, Andrejka L, et al. (2021). Genetic Determinants of EGFR-Driven Lung Cancer Growth and Therapeutic Response In Vivo. Cancer Discov. 10.1158/2159-8290.CD-20-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oren Y, Tsabar M, Cuoco MS, Amir-Zilberstein L, Cabanos HF, Hutter JC, Hu B, Thakore PI, Tabaka M, Fulco CP, et al. (2021). Cycling cancer persister cells arise from lineages with distinct programs. Nature 596, 576–582. 10.1038/s41586-021-03796-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang J, Ohta T, Maruyama A, Hosoya T, Nishikawa K, Maher JM, Shibahara S, Itoh K, and Yamamoto M (2006). BRG1 interacts with Nrf2 to selectively mediate HO-1 induction in response to oxidative stress. Mol Cell Biol 26, 7942–7952. 10.1128/MCB.00700-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song S, Nguyen V, Schrank T, Mulvaney K, Walter V, Wei D, Orvis T, Desai N, Zhang J, Hayes DN, et al. (2020). Loss of SWI/SNF Chromatin Remodeling Alters NRF2 Signaling in Non-Small Cell Lung Carcinoma. Mol Cancer Res 18, 1777–1788. 10.1158/1541-7786.MCR-20-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, Paczkowska M, Reynolds S, Wyczalkowski MA, Oak N, et al. (2018). Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 173, 355–370 e314. 10.1016/j.cell.2018.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Consortium, A.P.G. (2017). AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 7, 818–831. 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, Zhang H, Marquez VE, Hammerman PS, Wong KK, and Kim CF (2015). EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature 520, 239–242. 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oike T, Ogiwara H, Tominaga Y, Ito K, Ando O, Tsuta K, Mizukami T, Shimada Y, Isomura H, Komachi M, et al. (2013). A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res 73, 5508–5518. 10.1158/0008-5472.CAN-12-4593. [DOI] [PubMed] [Google Scholar]
- 80.Kunimasa K, Hirotsu Y, Miyashita Y, Goto T, Amemiya K, Mochizuki H, Samamoto I, Ohki T, Oyama T, Honma K, et al. (2020). Multiregional sequence revealed SMARCA4 R1192C mutant clones acquired EGFR C797S mutation in the metastatic site of an EGFR-mutated NSCLC patient. Lung Cancer 148, 28–32. 10.1016/j.lungcan.2020.07.035. [DOI] [PubMed] [Google Scholar]
- 81.Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, and Tiseo M (2019). Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 121, 725–737. 10.1038/s41416-019-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Piotrowska Z, Isozaki H, Lennerz JK, Gainor JF, Lennes IT, Zhu VW, Marcoux N, Banwait MK, Digumarthy SR, Su W, et al. (2018). Landscape of Acquired Resistance to Osimertinib in EGFR-Mutant NSCLC and Clinical Validation of Combined EGFR and RET Inhibition with Osimertinib and BLU-667 for Acquired RET Fusion. Cancer Discov 8, 1529–1539. 10.1158/2159-8290.CD-18-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fernando TM, Piskol R, Bainer R, Sokol ES, Trabucco SE, Zhang Q, Trinh H, Maund S, Kschonsak M, Chaudhuri S, et al. (2020). Functional characterization of SMARCA4 variants identified by targeted exome-sequencing of 131,668 cancer patients. Nat Commun 11, 5551. 10.1038/s41467-020-19402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, and Jodrell DI (2016). Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 32, 2866–2868. 10.1093/bioinformatics/btw230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kurppa KJ, Liu Y, To C, Zhang T, Fan M, Vajdi A, Knelson EH, Xie Y, Lim K, Cejas P, et al. (2020). Treatment-Induced Tumor Dormancy through YAP-Mediated Transcriptional Reprogramming of the Apoptotic Pathway. Cancer Cell 37, 104–122.e112. 10.1016/j.ccell.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, and Getz G (2013). Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31, 213–219. 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ichihara M, Murakumo Y, Masuda A, Matsuura T, Asai N, Jijiwa M, Ishida M, Shinmi J, Yatsuya H, Qiao S, et al. (2007). Thermodynamic instability of siRNA duplex is a prerequisite for dependable prediction of siRNA activities. Nucleic Acids Res 35, e123. 10.1093/nar/gkm699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matveeva O., Nechipurenko Y., Rossi L., Moore B., Saetrom P., Ogurtsov AY., Atkins JF., and Shabalina SA. (2007). Comparison of approaches for rational siRNA design leading to a new efficient and transparent method. Nucleic Acids Res 35, e63. 10.1093/nar/gkm088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cleary MA, Kilian K, Wang Y, Bradshaw J, Cavet G, Ge W, Kulkarni A, Paddison PJ, Chang K, Sheth N, et al. (2004). Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat Methods 1, 241–248. 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 90.Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, et al. (2011). The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A 108, 3665–3670. 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL, Strine MS, Zhang SM, Graziano VR, Schmitz CO, et al. (2021). Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell 184, 76–91 e13. 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784. 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Toki MI, Mani N, Smithy JW, Liu Y, Altan M, Wasserman B, Tuktamyshov R, Schalper K, Syrigos KN, and Rimm DL (2018). Immune Marker Profiling and Programmed Death Ligand 1 Expression Across NSCLC Mutations. J Thorac Oncol 13, 1884–1896. 10.1016/j.jtho.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, et al. (2017). QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878. 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14, 959–962. 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–29. 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhu Q, Liu N, Orkin SH, and Yuan GC (2019). CUT&RUNTools: a flexible pipeline for CUT&RUN processing and footprint analysis. Genome Biol 20, 192. 10.1186/s13059-019-1802-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425. 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Slenter DN., Kutmon M., Hanspers K., Riutta A., Windsor J., Nunes N., Mélius J., Cirillo E., Coort SL., Digles D., et al. (2018). WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res 46, D661–d667. 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EC50 values, fold changes, and calculations are shown for each cell line pair used in this study.
Table S2, Related to Figure 3 and S3. Properties of patient-derived xenografts (PDX) of EGFR-mutant lung cancers.
Table S3, Related to Figure 3 and S3. Clinicopathological information of patients from the YTMA-356.
Table S4. Related to STAR Methods. Oligonucleotides used throughout the study.
Data Availability Statement
All sequencing raw and processed data have been deposited in the Gene Expression Omnibus (GEO) database and are publicly available upon publication under the series GSE202857: Whole Exome Sequencing data (GSE202863), RNA-sequencing data (GSE202859), ATAC-sequencing data (GSE202857). All raw and processed Cut and Run, ATAC and RNA sequencing data for PC9*/OR*, HCC4006/OR and HCC827*/GR6 lines is publicly available upon publication under the series GSE227999. Accession numbers are listed in the Key Resource Table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Brg1 (D1Q7F) | CST | Cat#49360; AB_2728743 |
| Recombinant Anti-BRG1 | Abcam | Cat#ab108318; AB_10889900 |
| Phospho-EGF Receptor (Tyr1068) (D7A5) | CST | Cat#3777; AB_2096270 |
| EGF Receptor | CST | Cat#2232; AB_901981 |
| GAPDH (14C10) | CST | Cat#2118; AB_561053 |
| E-Cadherin (24E10) (AF594 Conjugate) | CST | Cat#7687; AB_2797633 |
| Vimentin (D21H3) (AF488 Conjugate) | CST | Cat#9854; AB_10829352 |
| c-Raf (D4B3J) | CST | Cat#53745; AB_2799444 |
| MEK1/2 (L38C12) | CST | Cat#4694; AB_10695868 |
| Phospho-MEK1/2 (Ser217/221) (41G9) | CST | Cat#9154; AB_2138017 |
| p44/42 MAPK (Erk1/2) (137F5) | CST | Cat#4695; AB_390779 |
| P-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) | CST | Cat#4370; AB_2315112 |
| Lamin A/C | CST | Cat#2032; AB_2136278 |
| CRISPR-Cas9 Antibody (7A9–3A3) | Novus Biologicals | Cat#NBP2–36440 |
| Anti-Nrf2 | Abcam | Cat#ab137550; AB_2687540 |
| Anti-mouse IgG, HRP-linked Antibody | CST | Cat#7076; AB_330924 |
| Anti-rabbit IgG, HRP-linked Antibody | CST | Cat#7074; AB_2099233 |
| Rabbit monoclonal anti-EGF Receptor (D38B1) XP® | Cell Signaling Technology | Cat# 4267, RRID:AB_2246311 |
| Rabbit monoclonal anti-phospho-Akt (Ser473) (D9E) XP® | Cell Signaling Technology | Cat# 4060, RRID:AB_2315049 |
| Rabbit polyclonal anti-Akt | Cell Signaling Technology | Cat# 9272, RRID:AB_329827 |
| Rabbit monoclonal anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (197G2) | Cell Signaling Technology | Cat# 4377, RRID:AB_331775 |
| Rabbit polyclonal p44/42 MAPK (Erk1/2) | Cell Signaling Technology | Cat# 9102, RRID:AB_330744 |
| Rabbit monoclonal HSP90 (C45G5) | Cell Signaling Technology | Cat# 4877, RRID:AB_2233307 |
| Rabbit monoclonal Vinculin (E1E9V) XP® | Cell Signaling Technology | Cat# 7074, RRID:AB_2099233 |
| Goat polyclonal anti-rabbit HRP-linked IgG | Cell Signaling Technology | Cat# 49360, RRID:AB_2728743 |
| Rabbit monoclonal Axl (C89E7) | Cell Signaling Technology | Cat# 8661, RRID:AB_11217435 |
| Rabbit monoclonal E-cadherin (24E10) | Cell Signaling Technology | Cat# 3195, RRID:AB_2291471 |
| Rabbit polyclonal Vimentin (R28) | Cell Signaling Technology | Cat# 3932, RRID:AB_2288553 |
| Rabbit monoclonal Brg1 (D1Q7F) | Cell Signaling Technology | Cat# 365062, RRID:AB_1084786 |
| Mouse monoclonal anti-GAPDH (G-9) | Santa Cruz | Cat# 11956, RRID:AB_2797776 |
| Rabbit monoclonal SMARCC1/BAF155 (D7FS8) | Cell Signaling Technology | Cat# 8173, RRID:AB_10949503 |
| CUTANA Rabbit IgG CUT&RUN Negative Control Antibody | Epicypher | Cat# 130042, RRID:AB_2923178 |
| Rabbit monoclonal Acetyl-Histone H3 (Lys27) (D5E4) XP® | Cell Signaling Technology | Cat# 2729, RRID:AB_1031062 |
| Bacterial and virus strains | ||
| MAX Efficiency® DH5α Competent Cells | Thermo Fisher | Cat#18258012 |
| Biological samples | ||
| EGFR-mutant tumors tissue micro array | Toki MI. et al, 2018 (PMID: 30267840) | YTMA-356 |
| Patient Derived Xenografts (PDXs) | This Paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Osimertinib | Selleckchem | Cat#S7297 |
| Trametinib | Selleckchem | Cat#S2673 |
| Osimertinib | MedChemExpress | Cat# HY-15772A |
| Osimertinib | AstraZeneca | AZD9291 |
| Compound14 | Jun Qi’s Laboratory | N/A |
| FHD-286 | Jun Qi’s Laboratory | N/A |
| High Sensitivity D100 ScreenTape | Agilent | Cat# 5067–5584 |
| High Sensitivity D100 Reagents | Agilent | Cat# 5067–5585 |
| BioMag Plus Concanavalin A beads | PolySciences | Cat# 8605 |
| AMPure XP Beads | Beckman Coulter | Cat# A6388 |
| Spermidine | Sigma-Aldrich | Cat# S2626 |
| pMD2.G | Addgene | Addgene_12259 |
| psPAX2 | Addgene | Addgene_12260 |
| pInducer10-mir-RUP-PheS | Meerbrey KL et al, 2011 (PMID: 21307310) | Addgene_44011 |
| TLCV2 | Addgene | Addgene_87360 |
| pLKO.1 puro | Addgene | Addgene_8453 |
| NRF2-sh1 | Sigma-Aldrich | TRCN0000007556 |
| NRF2-sh2 | Sigma-Aldrich | TRCN0000007557 |
| NRF2-sh2 | Sigma-Aldrich | TRCN0000007558 |
| Critical commercial assays | ||
| RPMI 1640 Medium | Thermo Fisher | Cat#A1049101 |
| Fetal Bovine Serum | Thermo Fisher | Cat#16140–071 |
| Penicillin-Streptomycin | Thermo Fisher | Cat#15140122 |
| 0.25% Trypsin with EDTA | Thermo Fisher | Cat#25200056 |
| TaqMan CNV Assay EGFR | Thermo Fisher | Cat#Hs02088787_cn |
| TaqMan CNV Assay EGFR | Thermo Fisher | Cat#Hs02190396_cn |
| TaqMan CNV Assay RAF1 | Thermo Fisher | Cat#Hs02614899_cn |
| TaqMan CNV Assay RAF1 | Thermo Fisher | Cat#Hs04252880_cn |
| Power SYBR Green Master mix | Thermo Fisher | Cat#4367659 |
| Protease and Phosphatase inhibitor cocktail | Thermo Fisher | Cat#78440 |
| SuperSignal West Pico PLUS | Thermo Fisher | Cat#34579 |
| SuperSigna West Femto Maximum Sensitivity Substrat | Thermo Fisher | Cat# 34096 |
| Triton X-100 | Sigma Aldrich | Cat#X-100 |
| Hoechst dye | Thermo Fisher | Cat#62249 |
| Tumor Dissociation Kit, human | Miltenyi Biotec | Cat#130–095-92 |
| Matrigel | Corning | Cat#356237 |
| Doxycycline diet | Envigo | Cat#TD.00426 |
| ProLong® Gold Antifade Mountant | Thermo Fisher | Cat#P10144 |
| CellROX® Deep Red Reagent | Thermo Fisher | Cat#C10422 |
| CellROX® Green Reagent | Thermo Fisher | Cat#C10444 |
| DNeasy Blood & Tissue kit | Qiagen | Cat#69504 |
| SuperScript III reverse transcriptase | Thermo Fisher | Cat#18080093 |
| RNeasy extraction kit | Thermo Fisher | Cat#74004 |
| MycoAlertTM Mycoplasma Detection Kit | Lonza | Cat#LT07–118 |
| CellTiter-Blue® Cell Viability Assay | Promega | Cat#G8081 |
| GenePrint 10 System | Promega | Cat#B9510 |
| Lipofectamine® RNAiMAX Transfection Reagent | Thermo Fisher | Cat#13778150 |
| TransIT®−293 Transfection Reagent | Mirus | Cat#MIR2704 |
| Antigen Unmasking Solution, Citrate-Based | Vector Laboratories | Cat#H-3300–250 |
| Antigen Unmasking Solution, Tris-Based | Vector Laboratories | Cat#H-3301–250 |
| VECTASTAIN® ABC-HRP Kit, Peroxidase (Mouse IgM ) | Vector Laboratories | Cat#PK-4010 |
| VECTASTAIN® ABC-HRP Kit, Peroxidase (Rabbit IgG) | Vector Laboratories | Cat#PK-4001 |
| Vector® NovaRED® Substrate Kit, Peroxidase (HRP) | Vector Laboratories | Cat#SK-4800 |
| CellTiter-Glo® Luminescent Cell Viability Assay | Promega | Cat# G7573 |
| CellEvent® Caspase-3/7 Green ReadyProbes® Reagent | Invitrogen | Cat# R37111 |
| CUTANA® ChIC/CUT&RUN Kit | Epicypher | Cat# 141048 |
| CUTANA® CUT&RUN Library Prep Ki | Epicypher | Cat# 141001 |
| RNAeasy Mini Kit | Qiagen | |
| MinElute PCR Purification kit | Qiagen | Cat# 28006 |
| NEBNext Ultra II Directional RNA Library Prep Kit for Illumina | New England Biolabs | Cat# E7760L |
| Tagment DNA Enzyme and Buffer Large Kit | Illumina | Cat# 20034197 |
| NextSeq 500/550 High Output Kit v2.5 (75 cylcles) | Illumina | Cat# 20024906 |
| Deposited data | ||
| All sequencing data SuperSeries | GEO Database | GSE202864 |
| Whole Exome Sequencing | GEO Database | GSE202863 |
| RNA-sequencing | GEO Database | GSE202859 |
| ATAC-sequencing | GEO Database | GSE202857 |
| Cut&Run, ATAC-seq, RNA-seq | GEO Database | GSE227999 |
| Experimental models: Cell lines | ||
| PC9 | ATCC / This paper | CVCL_B260; N/A |
| PC9-OR | This paper | N/A |
| H1975 | ATCC | CVCL_1511 |
| H1975-OR | This paper | N/A |
| HCC827 | ATCC | CVCL_2063 |
| HCC827-OR | This paper | N/A |
| YU-005C | This paper | N/A |
| HEK293T | ATCC | CVCL_0063 |
| PC-9*; human EGFR-mutant NSCLC, male | Dr. Kazuto Nishio (Kindai University, Osaka, Japan) | RRID:CVCL_B26 |
| PC-9_OR*; human EGFR-mutant NSCLC, male | This paper | N/A |
| HCC4006; human EGFR-mutant NSCLC, male | ATCC | CRL-2871; RRID:CVCL_1269 |
| HCC4006_OR; human EGFR-mutant NSCLC, male | This paper | N/A |
| HCC827; human EGFR-mutant NSCLC, female | Dr. Adi Gazdar (UT Southwestern, Dallas, TX) | RRID:CVCL_2063 |
| HCC827GR6; human EGFR-mutant NSCLC, female | Engelman et al. 2007 Science | N/A |
| Experimental models: Organisms/strains | ||
| NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice | Jackson Labs | IMSR_JAX:005557 |
| Oligonucleotides | ||
| See Table S4 - Oligonucleotides | N/A | N/A |
| Software and algorithms | ||
| QuPath | Bankhead P. et al 2017 (PMID: 29203879) | SCR_018257 |
| ImageJ | Schneider et al., 2012 | SCR_003070 |
| ApE | https://jorgensen.biology.utah.edu/wayned/ape/ | SCR_014266 |
| Graphpad 9.1.1 | PRISM | SCR_000306 |
| FlowJo v10 | BD | SCR_008520 |
| Ingenuity Pathway Analysis | Qiagen | SCR_008653 |
| RStudio (1.3.1093) | N/A | SCR_000432 |
| Kent Tools | UCSC | https://github.com/ucscGenomeBrowser/kent |
| IGV (2.8.2) | Robinson JT et al. 2012 (PMID: 21221095) | SCR_011793 |
| HOMER (4.11) | Heinz S. et al. 2010 (PMID: 20513432) | SCR_010881 |
| Primer3 | https://primer3.ut.ee/ | SCR_003139 |
| Combenefit | Di Veroli et al. 2016 Bioinformatics | N/A |
| Graphpad Prism 9 | https://www.graphpad.com/scientific-software/prism/ | N/A |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| STAR | Dobin etal., 2013 | https://github.com/alexdobin/STAR |
| MACS2 | Zhang etal., 2008 | https://github.com/taoliu/MACS |
| ngsplot | Shen et al., 2014 | https://github.com/shenlab-sinai/ngsplot |
| EdgeR | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| GSEA | Subramanian et al., 2005 | http://software.broadinstitute.org/gsea/index.jsp |
| Bedtools | Quinlan and Hall, 2010 | http://bedtools.readthedocs.io/en/latest/ |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Picard | Broad Institute | http://broadinstitute.github.io/picard |
| Trimmomatic | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| Gene Ontology | Gene Ontology Consortium, 2015 | http://geneontology.org/ |
| BWA-MEM | Li, 2013 | bio-bwa.sourceforge.net/ |
| Matplotlib | Hunter, 2007 | https://matplotlib.org/stable/ |
| Seaborn | Michael Waskom | https://seaborn.pydata.org/ |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.