Abstract
The great variability of protein sequences from human immunodeficiency virus (HIV) type 1 (HIV-1) isolates represents a major obstacle to the development of an effective vaccine against this virus. The surface protein (Env), which is the predominant target of neutralizing antibodies, is particularly variable. Here we examine the impact of variability among different HIV-1 subtypes (clades) on cytotoxic T-lymphocyte (CTL) activities, the other major component of the antiviral immune response. CTLs are produced not only against Env but also against other structural proteins, as well as some regulatory proteins. The genetic subtypes of HIV-1 were determined for Env and Gag from several patients infected either in France or in Africa. The cross-reactivities of the CTLs were tested with target cells expressing selected proteins from HIV-1 isolates of clade A or B or from HIV type 2 isolates. All African patients were infected with viruses belonging to clade A for Env and for Gag, except for one patient who was infected with a clade A Env-clade G Gag recombinant virus. All patients infected in France were infected with clade B viruses. The CTL responses obtained from all the African and all the French individuals tested showed frequent cross-reactions with proteins of the heterologous clade. Epitopes conserved between the viruses of clades A and B appeared especially frequent in Gag p24, Gag p18, integrase, and the central region of Nef. Cross-reactivity also existed among Gag epitopes of clades A, B, and G, as shown by the results for the patient infected with the clade A Env-clade G Gag recombinant virus. These results show that CTLs raised against viral antigens from different clades are able to cross-react, emphasizing the possibility of obtaining cross-immunizations for this part of the immune response in vaccinated individuals.
Genetic variability is one of the most remarkable hallmarks of human immunodeficiency virus (HIV), and it represents a major obstacle to the design of a vaccine against this virus. Due to this characteristic, neutralizing antibodies which are predominantly directed against the V3 loop of the envelope protein (gp120) react with only a small number of virus isolates (2, 27, 34). Other antibodies, especially those directed against conformational epitopes of the CD4 ligand of gp120 or transmembrane protein gp41, can neutralize a wider range of HIV type 1 (HIV-1) isolates (reviewed in reference 9). However, these antibodies are rarely, if ever, induced by vaccination. Cytotoxic T lymphocytes (CTLs) are thought to be another important component of the antiviral immune response. Indeed, the capacity of HIV-specific CTLs to efficiently limit viral replication is suggested by a large decrease in HIV load following the initial appearance of CTLs during primary infection (reviewed in reference 32) and by the temporal association between high CTL activity and stable viral load or CD4+ cell counts during asymptomatic stages (16, 28, 29). Furthermore, HIV-exposed but seronegative individuals, as well as uninfected children born to HIV-1-infected mothers, have exhibited anti-HIV CD8+ CTL reactivity as a unique sign of virus exposure (6, 31). Thus, it is generally accepted that vaccination must induce CTLs as well as neutralizing antibodies, so that infected cells can be killed before they produce any virus.
There are many target epitopes of CTLs, depending on donor HLA specificities; about 90 epitopes have been identified on the various structural and regulatory proteins of the virus (4, 13–15, 17, 18, 35, 36, 38, 41, 42; reviewed in reference 3). However, most experiments have involved lymphocytes from European or American donors infected with viruses of clade B. CTL activity has been reported for HIV type 2 (HIV-2)-infected patients (1, 12, 26, 31), but to our knowledge only one study has concerned African people infected with African HIV-1 isolates (31). We studied lymphoid cells from the blood of clade A virus-infected African patients and/or clade B virus-infected French patients. Both were tested against autologous target cells infected with recombinant viruses expressing various proteins from clade A or B viruses or from HIV-2. The large degree of cross-reactivities observed suggests that the variability of viral proteins will not be an obstacle in obtaining cross-reacting CTL in vaccinated individuals.
MATERIALS AND METHODS
Subjects.
Heparinized blood samples were collected from 16 consenting HIV-1-seropositive individuals, 7 in Bangui (Central African Republic) and 9 in France (1 was originally from Togo; patient W121). They were first diagnosed as HIV positive between 1989 and 1995. All had circulating anti-HIV-1 antibodies but not anti-HIV-2 antibodies. Peripheral blood mononuclear cells (PBMC) were isolated by density gradient centrifugation and frozen. HLA-A, -B, and -C types were determined serologically by the Laboratory of Immunology and Histocompatibility at Hospital Saint-Louis, Paris, France: B12 (HLA-A3/32, B41/−, C3/6); B15 (HLA-A3/31, B7/52, C6/−); B16 (HLA-A23/24, B13/47, C2/−); B18 (HLA-A2/31, B13/55, C2/6); B20 (HLA-A2/30, B7/13, C3/−); B22 (HLA-A2/30, B27/44, C2/−); B23 (HLA-A19.2/−, B44/57, C6/−); and W121 (HLA-A2/33, B50/70, C2/6).
Genetic subtyping of HIV-1 strains.
The genetic subtypes of HIV-1 were determined by a heteroduplex mobility assay (8). The C2V5 region of the env gene was amplified by nested PCR with the ED5 and ED12 primers for the first round and the ES8 primers for the second round. The gag gene was amplified by nested PCR with the G00 and G01 primers for the first round and the G60 and G25 primers for the nested PCR (33). The amplified fragment was then purified with a PCR product purification kit (33) and sequenced with a p24-specific internal primer by using dye terminator chemistry (Perkin-Elmer) on an automated DNA sequencer (Applied Biosystems 373A). DNA sequences were analyzed with the multiple sequence analysis program CLUSTALW (37). Reference strains for each subtype for the region analyzed were included in this study. Tree topology based on 634 nucleotides was inferred by the neighbor-joining method.
Vaccinia viruses.
The viruses used to infect target cells were vaccinia viruses (Copenhagen strain) recombined with the complete env, gag, pol, or nef gene of HIV-1 (LAI strain) (resulting in viruses Env/LAI, Gag/LAI, Pol/LAI, and Nef/LAI). Recombinant viruses encoding various proteins, such as gp120, gp41, p24, p18, reverse transcriptase (RT), integrase, or protease, were also used. They were produced as previously described (20, 21). An initiation codon, a stop codon, and adequate restriction sites were introduced during the construction of the Nef-1 (codons 1 to 72), Nef-2 (codons 73 to 147), and Nef-3 (codons 145 to 206) regions of Nef by local mutagenesis immediately before and after the indicated positions. Other recombinant vaccinia viruses were also constructed to express genes from isolates of subtype A of HIV-1 or of HIV-2. Gag/CAR, Pol/CAR, and Nef/CAR vaccinia viruses expressed the corresponding genes from HIV-1 92CAR3253 (obtained from a Central African Republic patient). The corresponding DNA fragments were amplified by PCR from DNA extracted from human PBMC infected with this isolate (obtained from F. Barré-Sinoussi). Adequate restriction sites were introduced during the amplification procedure. Pol/CAR expressed the complete pol gene. The Gag protein produced by Gag/CAR lacked 36 amino acids at the C terminus and ended with PPAEI. The Nef protein produced by Nef/CAR lacked 3 amino acids at the C terminus and ended with MKPEF. Env/OUG vaccinia virus expressed the native envelope protein gene of HIV-1 92UG037 (from a Ugandan patient). Env/ROD, Gag/ROD, and Nef/ROD vaccinia viruses expressed the corresponding genes from HIV-2 (ROD strain) (obtained from a West African patient). These genes were excised from the genome of HIV-2ROD and subjected to local mutagenesis to introduce restriction sites before the ATG initiation codon and after the stop codon. They were then introduced into the genome of the vaccinia virus. The stop codon in the gp36 coding sequence was removed by local mutagenesis to restore the reading frame of the native envelope protein gene.
Peptides.
Peptides corresponding to epitopes previously identified in clade B viral sequences (23) were synthetized by Neosystem (Strasbourg, France): Gag 77-85 (SLYNTVATL) (39), Gag 263-272 (KRWIILGNK) (25), Nef 73-82 (QVPLRPMTYK) (19), and Nef 136-145 (PLTFGWCFKL) (15). They were supplied by the Agence Nationale de Recherche sur le SIDA. Lyophilized peptides were dissolved in water (2 mg/ml) and stored at −20°C.
Generation of anti-HIV cell lines.
Polyclonal anti-HIV cell lines were obtained by culturing PBMC (106/ml) with autologous phytohemagglutin-activated lymphocytes (2 × 105/ml) as described previously (20). The cells were incubated for 3 days in RPMI 1640 (GIBCO) supplemented with 2 mM l-glutamine, 10 mM HEPES buffer, and 10% fetal calf serum. They were then cultured at a concentration of 106/ml in medium supplemented with 10 U of human recombinant interleukin 2 (Boehringer) per ml. Cytolytic activity was tested after 14 to 21 days in culture.
Antipeptide cell lines were generated in some experiments by use of the same culture medium as that described above and by coculturing 107 PBMC (4 × 106/ml) with a similar number of autologous PBMC which had been treated with a pulse of 1 μg of peptide for 90 min and irradiated. Continuous cell lines were established by similar weekly stimulation as previously described (7).
CRT.
The target cells used in the chromium release test (CRT) were autologous lymphoblastoid cells obtained by transforming PBMC with Epstein-Barr virus (EBV-LCL). They were infected with recombinant vaccinia viruses by incubation with 5 PFU per cell for 18 h. Wild-type vaccinia virus (Vac/WT) was used as a control. EBV-LCL were labeled by incubation with 100 μCi of Na251CrO4 (Amersham) for 1 h and washed twice. EBV-LCL incubated with 1 μg of peptide for 90 min and then extensively washed were used as target cells in some experiments. The control consisted of target cells incubated with medium alone.
The CRT was performed with microculture plates by incubating various concentrations of effector cells and 5 × 103 target cells in RPMI 1640 supplemented with 10% fetal calf serum for 4 h. The supernatants were then harvested, and the chromium released was measured in a gamma counter. The spontaneous release was 10 to 25% of the total Cr incorporated. The specific chromium release was calculated as 100 × [(experimental − spontaneous release)/(total Cr incorporated − spontaneous release)]. HIV-specific activity was considered to be present when the specific chromium release was 10% greater than that of the control Vac/WT for two different effector/target cell ratios. Lytic units (LU) were calculated for 108 effector cells as 108/(5,000 × E/T30%), where E/T30% is the effector/target cell ratio that yields 30% specific lysis of 5,000 target cells. LU for Vac/WT were always less than 3.
Nucleotide sequence accession numbers.
The nucleotide sequences of the HIV-1 gag p24 region for patients B12, B15, B16, B18, B20, B22, B24, and W121 were deposited in the EMBL Nucleotide Sequence Database under accession no. Y16612 to Y16619, respectively.
RESULTS
Genetic subtyping of HIV-1 strains.
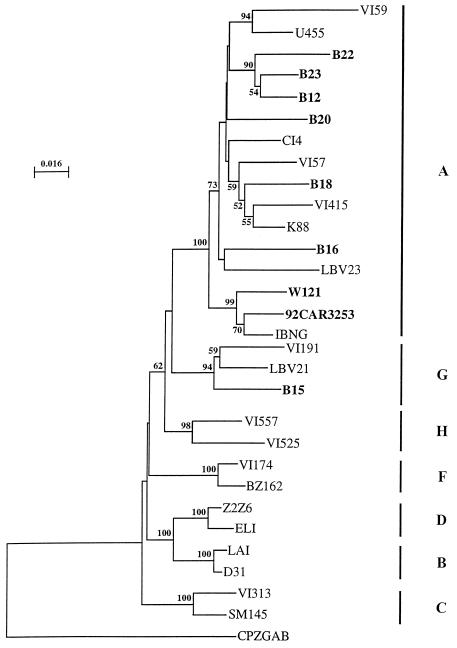
The samples from the Caucasian patients all formed fast-migrating heteroduplexes with the subtype B reference strain. The viruses originating from Bangui (B12, B15, B16, B18, B20, B22, and B23) all clearly formed fast-migrating heteroduplexes with the subtype A reference strain, as did the viral isolate from the patient from Togo (W121). Many different genetic subtypes have been found in the Central African Republic (24, 33a): A, E, D, C, H, G, and U (decreasing order of frequency); it is possible that some of the isolates studied were indeed recombinant genomes (11, 30). However, as no subtype B was found in this country, it is unlikely that any of the Bangui isolates were A-B recombinants. We sequenced part of the Gag region for Bangui isolates and for the virus from Togo. Seven of the eight viruses were identified as belonging to clade A (Fig. 1). One virus (B15) clustered with the subtype G isolates, showing that this virus was an A-G recombinant.
FIG. 1.
Phylogenetic analysis of gag nucleotide sequences. The phylogenetic tree was generated by the neighbor-joining method and drawn with Njplot. The numbers given at the branch points are the 50% threshold majority consensus values for 100 bootstrap replicates. The lengths of the horizontal branches are proportional to the relative evolutionary distances; vertical distances are for clarity only. The strains isolated from the African patients described in this study, as well as the strain used to produce the recombinant vaccinia viruses encoding the gag gene (92CAR3252), are shown in bold type.
Reactivity of CTLs stimulated with endogenous virus.
Lymphoid cells from the 16 patients were stimulated in vitro with autologous phytohemagglutinin-activated blast cells, so that the restimulating viral proteins were from endogenous viruses from the same patient. CTL reactivities were tested against a panel of structural proteins (Env, Gag, or Pol) from clade A or B viruses and against Nef (from clade A and B viruses), as Nef is the most frequently recognized regulatory protein (20). Reactivities with the Env, Gag, and Nef proteins of HIV-2ROD were also tested. The reagent for testing HIV-2 Pol was not available.
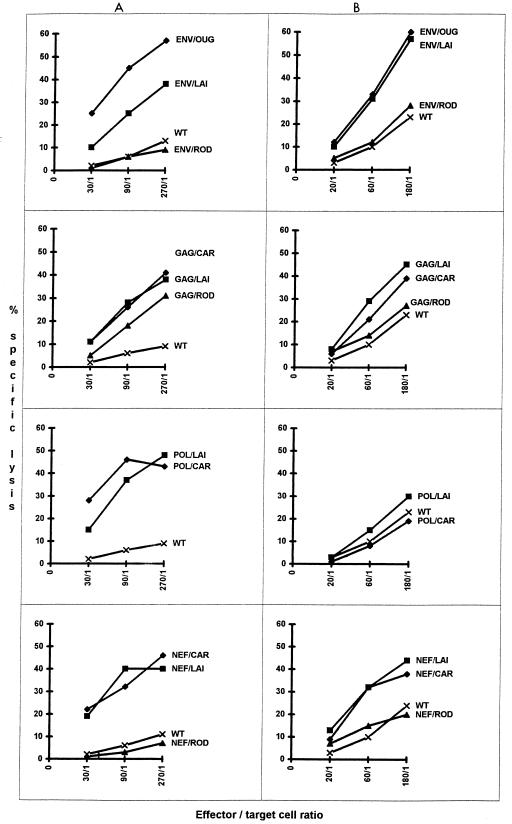
The results found with CTLs from clade A virus-infected patients are summarized in Table 1, and an example is shown in Fig. 2A. The CTLs from these eight patients clearly reacted with several clade A proteins; CTLs from three of them reacted with all four proteins tested, CTLs from three reacted with three proteins, and CTLs from two reacted with two proteins. Pol epitopes were recognized by CTLs from all donors, Gag and Nef epitopes were recognized by CTLs from most of them (seven of eight), but Env-reacting CTLs were found in only three of the eight donors. Figure 2A shows that B18-derived CTLs recognized equally well the Gag epitopes of CAR (clade A), LAI (clade B), and even ROD (HIV-2) viruses. Clear cross-reactivities were also found with Env, Pol, and Nef; in all cases, the levels of the responses were equivalent for CAR or LAI viruses. In contrast, cross-reactivities against ROD (HIV-2) were found only for Gag. CTLs from the other seven African patients also showed multiple cross-reactions with proteins of clade B viruses. The levels of their responses to Gag proteins of the two subtypes were similar. The reactivity with Pol was stronger with the homologous protein. Nevertheless, CTLs from all but one donor reacted with Pol/LAI. Finally, five of the seven donors had CTLs that recognized Nef/CAR and Nef/LAI in the same response range. In contrast, cross-reactivity with the Env protein was weaker; only one of the three donors (B18) whose CTLs reacted with Env/CAR had CTLs that also reacted with Env/LAI, but with weaker activity. CTLs from clade A-infected donors seldom cross-reacted with HIV-2 proteins. No cross-reactivity was found with Env or Nef, and reactivities against Gag/ROD were observed with CTLs from only three donors among the eight whose CTLs were capable of recognizing Gag/CAR.
TABLE 1.
CTL activities in patients infected with subtype A HIV-1
| Target antigen | LUa for donor:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| B12 | B15 | B16 | B18 | B20 | B22 | B23 | W121 | |
| Subtype A HIV-1 | ||||||||
| Env/OUG | <3 | <3 | 190 | 277 | <3 | <3 | <3 | 233 |
| Gag/CAR | <3 | 33 | 85 | 65 | 99 | 95 | 97 | 25 |
| Pol/CAR | 92 | 47 | 583 | 62 | 61 | 52 | 101 | 75 |
| Nef/CAR | 73 | 24 | 285 | 116 | <3 | 63 | 98 | 30 |
| Subtype B HIV-1 | ||||||||
| Env/LAI | <3 | <3 | <3 | 80 | <3 | <3 | <3 | <3 |
| Gag/LAI | <3 | 27 | 65 | 58 | 64 | 42 | 61 | 130 |
| Pol/LAI | 79 | 32 | 143 | 28 | 36 | 21 | <3 | 17 |
| Nef/LAI | 47 | <3 | 234 | 93 | <3 | 33 | <3 | 23 |
| HIV-2 | ||||||||
| Env/ROD | <3 | <3 | <3 | <3 | <3 | <3 | <3 | <3 |
| Gag/ROD | <3 | 26 | <3 | 37 | 74 | <3 | <3 | <3 |
| Nef/ROD | <3 | <3 | <3 | <3 | <3 | <3 | <3 | <3 |
LU were calculated as described in Materials and Methods.
FIG. 2.
CTL activities in patients carrying viruses B18 (infected with clade A HIV-1) (A) and CO3M (infected with clade B HIV-1) (B). Effector cells were tested after in vitro restimulation with autologous blast cells. Target cells were infected with recombinant vaccinia viruses expressing the Env, Gag, Pol, or Nef protein of clade A and B isolates.
Similar experiments with lymphoid cells from clade B-infected European patients yielded similar results (Table 2). CTLs from all eight donors reacted with several proteins from clade B viruses, consistent with previous work (20). Figure 2B shows the strong cross-reactivities of CTLs from the donor carrying virus CO3M with Env, Gag, and Nef proteins of clades A and B. Altogether, the CTLs of most clade B-infected patients (seven of eight) reacted with Nef/CAR (clade A), and the response was in the same range as that for Nef/LAI. Cross-reactivities against Env/OUG (four of eight), Gag/CAR (five of eight), and Pol/CAR (six of eight) were also found. Finally, no CTLs from any of the clade B-infected patients were found to react with target cells expressing ROD (HIV-2) Env, Gag, or Nef.
TABLE 2.
CTL activities in patients infected with subtype B HIV-1
| Target antigen | LUa for donor:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| CO3M | T051 | BI02 | MIO7 | MI10 | MI21 | W39 | W44 | |
| Subtype B HIV-1 | ||||||||
| Env/LAI | 143 | <3 | 87 | 18 | 83 | 134 | 28 | 486 |
| Gag/LAI | 77 | 28 | 178 | 84 | 108 | 507 | 57 | 567 |
| Pol/LAI | <3 | 77 | 75 | 159 | 77 | 312 | 37 | 128 |
| Nef/LAI | 93 | 34 | 127 | 72 | 23 | 219 | 61 | <3 |
| Subtype A HIV-1 | ||||||||
| Env/OUG | 125 | <3 | <3 | <3 | 18 | <3 | 62 | 42 |
| Gag/CAR | 38 | <3 | <3 | 43 | 47 | <3 | 59 | 56 |
| Pol/CAR | <3 | 42 | 45 | 83 | 23 | <3 | 17 | 103 |
| Nef/CAR | 73 | 18 | 96 | 28 | 12 | 49 | 23 | <3 |
| HIV-2 | ||||||||
| Env/ROD | <3 | <3 | <3 | <3 | <3 | <3 | <3 | <3 |
| Gag/ROD | <3 | <3 | <3 | <3 | <3 | <3 | NT | <3 |
| Nef/ROD | <3 | <3 | <3 | <3 | <3 | <3 | <3 | <3 |
LU were calculated as described in Materials and Methods. NT, not tested.
Cross-reactivities of CTLs with various Env, Gag, Pol, and Nef subregions.
It is important for vaccine development to further determine the precise targets of cross-reacting CTLs. To do this, we used target cells infected with recombinant vaccinia viruses expressing Env gp120, Env gp41, Gag p24, Gag p18 (the recombinant for Gag p15 was not available), RT, integrase, or protease. Three recombinants expressing the N-terminal (Nef-1), central (Nef-2), or C-terminal (Nef-3) regions of Nef were also tested. All were from the clade B LAI isolate, the corresponding clade A reagents not being available. Previous experiments largely documented the capability of CTLs from patients infected with clade B viruses to react against these different proteins (reviewed in reference 40). This characteristic was tested with CTLs from African patients (Table 3). Reactivity against Env was rare, found in only one of the seven donors (B18), who produced CTLs specific for both gp120 and gp41 of the LAI isolate. On the contrary, broad cross-reactivities were detected with Pol and Gag, as p24 and p18 from the LAI isolate were recognized by CTLs from four of the six clade A-infected donors; RT was recognized by CTLs from two of the seven, protease was recognized by CTLs from only one of the seven, and remarkably enough integrase was recognized by CTLs from all seven of the African donors (at least four of them had a strong response). Finally, the reactivities detected against Nef in five of the seven patients always revealed cross-reacting epitopes in the central region of this protein.
TABLE 3.
CTL activities against various proteins of HIV-1LAI
| Target antigen | LUa for donor:
|
||||||
|---|---|---|---|---|---|---|---|
| B12 | B15 | B16 | B18 | B20 | B22 | W121 | |
| Env/LAI | <3 | <3 | <3 | 80 | <3 | <3 | <3 |
| Gp120 | <3 | <3 | NT | 37 | NT | <3 | <3 |
| Gp41 | <3 | <3 | NT | 28 | NT | <3 | <3 |
| Gag/LAI | <3 | 27 | 65 | 58 | 64 | 42 | 130 |
| P24 | NT | 14 | <3 | 12 | <3 | 33 | 110 |
| P18 | NT | 19 | <3 | 35 | 51 | <3 | 68 |
| Pol/LAI | 79 | 32 | 143 | 28 | 36 | 52 | 17 |
| RT | 60 | <3 | <3 | 43 | <3 | <3 | <3 |
| Int | 43 | 41 | 179 | 56 | 18 | 37 | 26 |
| Pro | <3 | <3 | <3 | <3 | 12 | <3 | <3 |
| Nef/LAI | 47 | <3 | 234 | 93 | <3 | 63 | 23 |
| Nef-1 | <3 | <3 | NT | <3 | <3 | <3 | <3 |
| Nef-2 | 38 | <3 | 56 | 82 | <3 | 82 | 42 |
| Nef-3 | <3 | <3 | NT | <3 | <3 | <3 | <3 |
LU were calculated as described in Materials and Methods. NT, not tested.
Identification of conserved epitopes.
We investigated the presence in African patients of CTLs specific for peptides previously identified as epitopes in clade B viruses. This was feasible, since all of these donors had at least one class I molecule corresponding to a known epitope already studied in European or American patients. When PBMC were still available, antipeptide cell lines were produced, and CTL activity was tested against target cells sensitized with the corresponding peptide or infected with recombinant vaccinia virus. It is important to note that this experimental approach allows the detection of CTLs from PBMC of only infected (or vaccinated) individuals.
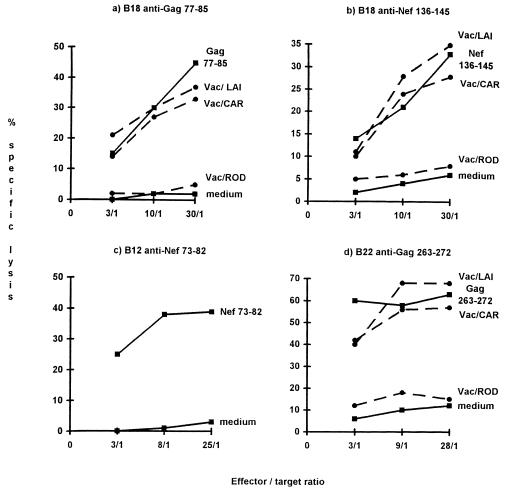
CTLs from patient B18 recognized epitopes Gag 77-85 (39) and Nef 136-145 (15), which are known to be HLA-A2 restricted (Fig. 3a and b). These CTLs also lysed target cells infected with recombinant vaccinia viruses carrying the corresponding genes from the LAI and CAR isolates but not the HIV-2ROD isolate. Similarly, epitope Nef 73-82, which is restricted to HLA-A3 (19), was recognized by CTLs from patient B12 (Fig. 3c). Finally, CTLs from patient B22 reacted with the well-known HLA-B27-restricted epitope Gag 263-272 (25) (Fig. 3d) but not with the HLA-A2-restricted epitope Nef 136-145.
FIG. 3.
Recognition of known epitopes by CTLs from African patients. CTLs were produced by in vitro stimulation with synthetic peptides. They were tested against target cells previously treated with the corresponding peptide or infected with recombinant vaccinia virus (Vac) (except for the B12 cell line, because effector cells were not available). The HLA typing was as follows: for B12, HLA-A3/32, B41/−, C3/6; for B22, HLA-A2/30, B27/44, C2/−; and for B18, HLA-A2/31, B13/55, C2/6. The target cells were heterologous EBV-LCL sharing HLA-A3 with B12 CTLs, HLA-B27 with B22 CTLs, or HLA-A2 with B18 CTLs.
DISCUSSION
Anti-HIV-1 CTL responses have been studied almost exclusively in Europe and North America with lymphoid cells from people infected with clade B viruses. They are directed against several proteins of the same virus and often against several epitopes of the same protein. This polymorphism of CTL responses is well documented for these viruses (reviewed in reference 40). Our results show that the same polymorphism exists for anti-clade A virus CTL responses (Table 1). CTLs from all eight patients tested responded to several structural proteins and also frequently to Nef, with at least three to five different CTL activities in the same donor (Table 3). It is interesting to note, however, that Env was recognized by the CTLs from only three of the eight patients, suggesting a low prevalence of broadly cross-reactive Env-specific CTLs among clade A virus-infected individuals, consistent with results reported for clade B viruses (5). Moreover, this study clearly demonstrates frequent cross-reactivities in the CTL responses obtained after infection with viruses belonging to two different clades. Several patients had almost identical CTL reactions to Gag and Nef proteins from clade A and B viruses, regardless of whether the original virus was of clade A or B. The homologous proteins sometimes elicited a stronger response, notably with Pol targets, but even in this case cross-reactivity was evident. Weaker cross-reactivities were detected against Env proteins. The CTLs of African patients were selective for homologous clade A Env. CTLs from only one donor showed a cross-reaction with Env/LAI. In the reverse situation, CTLs from four of the eight European donors cross-reacted with clade A Env. It is not surprising that Gag, Pol, and Nef were better targets for cross-reaction than Env, since the variability of Env is especially important and well known.
It must be emphasized that the classical methods used to test CTLs allow demonstration only of responses directed to conserved epitopes, as patients are stimulated with epitopes of their own viruses, which are variable, while the tests are carried out with target cells infected with a single recombinant vaccinia virus. The target cells for clade B viruses generally express viral proteins from HIV-1LAI, so only epitopes conserved between LAI and the infecting virus can be revealed in the reaction. Similarly, the recently produced panel of recombinants expressing different clade A proteins was produced with a single clade A virus, so only epitopes conserved among clade A virus-infected donors can be detected. The same epitopes are probably responsible for most of the cross-reactivity between the two clades, as few differences were detected in cross-reactions involving Gag, Pol, or Nef. The target epitopes map to Gag p18 and p24 and the central region of Nef, as has already been shown for clade B viruses (3). Our results suggest a particular importance for integrase epitopes with constant cross-reactivities between clade A and B viruses (Table 3). Reactions directed against integrase are poorly documented. However, we previously identified this enzyme as a good CTL target for clade B viruses (21). Cross-reactivities with HIV-2 are only occasional. We found that anti-clade A virus CTLs sometimes cross-reacted with HIV-2 Gag protein, as previously reported (12, 26, 31), whereas they did not cross-react with other proteins. On the other hand, CTLs from patients infected with clade B viruses did not cross-react with HIV-2 proteins.
It is very probable that only some of the epitopes recognized on homologous proteins are responsible for cross-reactivity. However, our results suggest that obstacles to vaccination because of the induction of broadly cross-reactive neutralizing antibodies seem not to affect CTL responses so extensively. This idea is not surprising because the target proteins of CTLs are less variable than the V3 loop and because the African patients tested shared with northern populations at least one HLA specificity capable of presenting previously identified epitopes, although we did not select for this. Further studies are required to investigate cross-reactivities with viruses of other clades, including C, D, E, and G, and even viruses of type 0. It is likely that strong cross-reactivities will be found, as one of the donors bore a virus (B15) belonging to clade G (for the Gag proteins) but reacting with clade A and B Gag proteins, including p24 and p18. Viruses of clade E also carry a clade A gag gene (11), so cross-reactions probably will be identified at least against Gag.
Results obtained with synthetic epitopes showed that African patients have CTL precursors which react with previously identified epitopes in Gag and Nef proteins of clade B isolates. Similar experiments performed with European patients allowed us to discover such cross-reactivities at the epitope level (data not shown). For example, CTLs specific for the Nef 84-92 epitope (7) were able to recognize target cells infected with Vac/CAR. This result is not surprising, since the peptide sequence is conserved between LAI and CAR isolates. A similar finding was obtained for the RT 325-333 epitope (41), suggesting that the mutation Ser (LAI isolate) → Ala (CAR isolate) at position 8 did not affect the epitope. Similarly, mutations observed in CAR isolates at the level of the Nef 73-82, Gag 77-85, and Gag 263-272 epitopes did not induce escape of recognition by CTLs from African patients (Fig. 3). These results are consistent with previous reports of conserved epitopes shown with CTLs from patients infected with clade B isolates (reviewed in reference 22) or from uninfected vaccinated volunteers (10). They show that cross-reactivity can be due to conservation of epitope sequences as well as to cross-recognition of epitopes which differ in amino acid sequences. However, further studies are necessary to identify the precise epitopes involved in cross-reactions. This identification may be of value in developing a vaccination system based on peptides or lipopeptides, although the level of CTL response required for possible in vivo protection is not known.
ACKNOWLEDGMENTS
This work was supported by grants from the Agence Nationale de Recherche sur le Sida (ANRS) and Ensemble Contre le SIDA, Sidaction, Paris, France. Deniz Durali was supported by a fellowship from ANRS.
We thank Jean-Gérard Guillet for support; Françoise Barré-Sinoussi and Marie-Paule Kiény for providing the HIV-1 92CAR3253 isolate and the recombinant vaccinia viruses; and Jean-Christophe Deschemin, Julienne Ipero, Josiane Leal, and Karine Dott for excellent technical assistance. We also acknowledge the generous participation of the patients involved in these studies. The English text was edited by Julie Knight.
REFERENCES
- 1.Ariyoshi K, Cham F, Berry N, Jaffar S, Sabally S, Corrah T, Whittle H. HIV-2-specific cytotoxic T-lymphocyte activity is inversely related to proviral load. AIDS. 1995;9:555–559. doi: 10.1097/00002030-199506000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Bolognesi D. HIV antibodies and vaccine design. AIDS. 1989;3:S111–S119. doi: 10.1097/00002030-198901001-00016. [DOI] [PubMed] [Google Scholar]
- 3.Brander C, Walker B D. The HLA-class I-restricted CTL response in HIV-1 infection; identification of optimal epitopes. HIV Molecular Immunology Database. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. . IV-1 to IV-8. [Google Scholar]
- 4.Buseyne F, Stevanovic S, Rammensee H-G, Rivière Y. Characterization of an HIV-1 p24gag epitope recognized by a CD8+ cytotoxic T-cell clone. Immunol Lett. 1997;55:145–149. doi: 10.1016/s0165-2478(97)02696-5. [DOI] [PubMed] [Google Scholar]
- 5.Carmichael A, Jin X, Sissons P. Analysis of the human env cytotoxic T-lymphocyte (CTL) response in natural immunodeficiency virus type 1 infection: low prevalence of broadly cross-reactive env-specific CTL. J Virol. 1996;70:8468–8476. doi: 10.1128/jvi.70.12.8468-8476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheynier R, Langlade-Demoyen P, Marescot M R, Blanche S, Blondin G, Wain-Hobson S, Griscelli C, Vilmer E, Plata F. Cytotoxic T lymphocyte responses in the peripheral blood of children born to HIV-1-infected mothers. Eur J Immunol. 1993;22:2211–2217. doi: 10.1002/eji.1830220905. [DOI] [PubMed] [Google Scholar]
- 7.Culmann-Penciolelli B, Lamhamedi-Cherradi S, Couillin I, Guegan N, Levy J-P, Guillet J-G, Gomard E. Identification of multirestricted immunodominant regions recognized by cytolytic T lymphocytes in the human immunodeficiency virus type 1 Nef protein. J Virol. 1994;68:7336–7343. doi: 10.1128/jvi.68.11.7336-7343.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delwart E W, Shpaet E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1992;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 9.Dimmock N J. Neutralization of animal viruses. Curr Top Microbiol Immunol. 1993;183:1–149. doi: 10.1007/978-3-642-77849-0. [DOI] [PubMed] [Google Scholar]
- 10.Ferrari G, Humphrey W, McElrath M J, Excler J-L, Duliège A-M, Clements M L, Corey L C, Bolognesi D P, Weinhold K J. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc Natl Acad Sci USA. 1997;94:1396–1401. doi: 10.1073/pnas.94.4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, Fultz P N, Girard M, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotch F, McAdam S N, Allsopp C E L, Gallimore A, Elvin J, Kiény M-P, Andrew A V S, McMichael A J, Whittle H C. Cytotoxic T cells in HIV-2 seropositive Gambians. Identification of a virus-specific MHC-restricted peptide epitope. J Immunol. 1993;151:3361–3369. [PubMed] [Google Scholar]
- 13.Goulder P, Conlon C, McIntyre K, McMichael A. Identification of a novel human leukocyte antigen A26-restricted epitope in a conserved region of Gag. AIDS. 1996;10:1442–1443. doi: 10.1097/00002030-199610000-00025. [DOI] [PubMed] [Google Scholar]
- 14.Goulder P J R, Edwards A, Phillips R E, McMichael A J. Identification of a novel HLA-B3501-restricted cytotoxic T lymphocyte epitope using overlapping peptides. AIDS. 1997;11:930–931. [PubMed] [Google Scholar]
- 15.Haas G, Plikat U, Debré P, Lucchiari M, Katlama C, Dudoit Y, Bonduelle O, Bauer M, Ihlenfeldt H-G, Jung G, Maier B, Meyermans A, Autran B. Dynamics of viral variants in HIV-1 Nef and specific cytotoxic T lymphocytes in vivo. J Immunol. 1996;157:4212–4221. [PubMed] [Google Scholar]
- 16.Harrer T, Harrer E, Kalams S A, Elbeik T, Staprans S I, Feinberg M B, Cao Y, Ho D H, Yilma T, Caliendo A M, Johnson R P, Buchbinder S P, Walker B D. Strong cytotoxic T cell and weak neutralizing antibody responses in a subset of persons with stable nonprogressing HIV type 1 infection. AIDS Res Hum Retroviruses. 1996;12:585–592. doi: 10.1089/aid.1996.12.585. [DOI] [PubMed] [Google Scholar]
- 17.Harrer T, Harrer E, Kalams S A, Barbosa P, Trocha A, Johson R P, Elbeik T, Feinberg M B, Buchbinder S P, Walker B D. Cytotoxic T lymphocytes in asymptomatic long-term nonprogressing HIV-1 infection: breadth and specificity of the response and relation to in vivo viral quasispecies in a person with prolonged infection and low viral load. J Immunol. 1996;156:2616–2623. [PubMed] [Google Scholar]
- 18.Klenerman P, Luzzi G, McIntyre K, Phillips R, McMichael A. Identification of a novel HLA-A25-restricted epitope in a conserved region of p24 gag (positions 71–80) AIDS. 1996;10:348–349. doi: 10.1097/00002030-199603000-00023. [DOI] [PubMed] [Google Scholar]
- 19.Koenig S, Fuerst T R, Wood L V, Woods R M, Suzich J A, Jones G, de la Crux V, Davey R, Venkatesan S, Moss B, Biddison W, Fauci A. Mapping the fine specificity of a cytolytic T cell response to HIV-1 nef protein. J Immunol. 1990;145:127–135. [PubMed] [Google Scholar]
- 20.Lamhamedi-Cherradi S, Culmann-Penciolelli B, Guy B, Kiény M-P, Dreyfus F, et al. Qualitative and quantitative analysis of human cytotoxic T lymphocyte responses to HIV-1 proteins. AIDS. 1992;6:1249–1258. doi: 10.1097/00002030-199211000-00002. [DOI] [PubMed] [Google Scholar]
- 21.Lamhamedi-Cherradi S, Culmann-Penciolelli B, Guy B, Ly T H, Goujard C, Guillet J-G, Gomard E. Different patterns of HIV-1 specific cytotoxic T-lymphocyte activity after primary infection. AIDS. 1995;9:421–426. [PubMed] [Google Scholar]
- 22.McMichael A J, Walker B D. Cytotoxic T lymphocyte epitopes: implications for HIV vaccines. AIDS. 1994;8:S155–S173. [Google Scholar]
- 23.Meyers G, Korber B, Hahn B, et al., editors. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. [Google Scholar]
- 24.Murphy E, Korber B, Geaoges-Courbot M C, You B, Pinter A, Cook D, Kiény M-P, Georges A, Mathiot C, Barré-Sinoussi F, et al. Diversity of V3 region sequences of human immunodeficiency virus type 1 from the Central African Republic. AIDS Res Hum Retroviruses. 1993;10:997–1006. doi: 10.1089/aid.1993.9.997. [DOI] [PubMed] [Google Scholar]
- 25.Nixon D F, Townsend A R M, Elvin J G, Rizza C R, Gallwey J, McMichael A J. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature. 1988;336:484–487. doi: 10.1038/336484a0. [DOI] [PubMed] [Google Scholar]
- 26.Nixon D F, Huet S, Rothbard J, Kiény M-P, Delchambre M, Thiriart C, Rizza C R, Gotch F M, McMichael A J. An HIV-1 and HIV-2 cross-reactive cytotoxic T-cell epitope. AIDS. 1990;4:841–845. doi: 10.1097/00002030-199009000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Palker T J, Clarck M L, Langlois A J, Matthews T J, Weinhold K J, Randall R, Bolognesi D P, Haynes B F. Type-specific neutralization of HIV with antibodies to env-encoded synthetic peptides. Proc Natl Acad Sci USA. 1988;85:1932–1936. doi: 10.1073/pnas.85.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinaldo C, Huang X-L, Fan Z, Ding M, Beltz L, Logar A, Panicali D, Mazzara G, Liebmann J, Cottrill M, Gupta P. High levels of anti-human immunodeficiency virus type 1 (HIV-1) memory cytotoxic T-lymphocyte activity and low viral load are associated with lack of disease in HIV-1-infected long-term nonprogressors. J Virol. 1995;69:5838–5842. doi: 10.1128/jvi.69.9.5838-5842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivière Y, McChesnay M B, Porrot F, et al. Gag-specific cytotoxic responses to HIV type 1 are associated with a decreased risk of progression to AIDS-related complex or AIDS. AIDS Res Hum Retroviruses. 1995;8:903–907. doi: 10.1089/aid.1995.11.903. [DOI] [PubMed] [Google Scholar]
- 30.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature (London) 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 31.Rowland-Jones S, Sutton J, Ariyoshi K, Dong T, Gotch F, McAdam S, Whitby D, Sabally S, Gallimore A, et al. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 32.Safrit J T, Koup R A. The immunology of primary HIV infections: which immune responses control HIV replication? Curr Opin Immunol. 1995;7:456–461. doi: 10.1016/0952-7915(95)80088-3. [DOI] [PubMed] [Google Scholar]
- 33.Sanders-Buell E, Salminen M O, McCutchan F E. Sequencing primers for HIV-1, human retroviruses and AIDS. III. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. pp. 15–21. [Google Scholar]
- 33a.Saragosti, S. Unpublished data.
- 34.Scott C F, Silver S, Profy A T, Putney S D, Langlois A, Weinhold K, Robinson J E. Human monoclonal antibody that recognizes the V3 region of human immunodeficiency virus gp120 and neutralizes the human T-lymphotropic virus type III-MN strain. Proc Natl Acad Sci USA. 1990;87:8597–8603. doi: 10.1073/pnas.87.21.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiga H, Shiado T, Tomiyama H, Takamiya Y, Oka S, Kimura S, Yamaguchi Y, Gojoubori T, Rammensse H-G, Miwa K, Takiguchi M. Identification of multiple HIV-1 cytotoxic T-cell epitopes presented by human leukocyte antigen B35 molecules. AIDS. 1996;10:1075–1083. [PubMed] [Google Scholar]
- 36.Sipsas N V, Kalams S A, Trocha A, He S, Blattner W A, Walker B D. Identification of type-specific T lymphocyte responses to homologous viral proteins in laboratory workers accidentally infected with HIV-1. J Clin Invest. 1997;99:752–762. doi: 10.1172/JCI119221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson J D, Higgind D G, Gibson T J. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gag penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomiyama H, Miwa K, Shiga H, Moore Y I, Oka S, Iwamoto A, Kaneko Y, Takiguchi M. Evidence of presentation of multiple HIV-1 cytotoxic T lymphocyte epitopes by HLA-B3501 molecules that are associated with the accelerated progression of AIDS. J Immunol. 1997;158:5026–5034. [PubMed] [Google Scholar]
- 39.Tsomides T J, Aldovini A, Johnson R P, Walker B D, Young R A, Eisen H N. Naturally processed viral peptides recognized by cytotoxic T lymphocytes on cells chronically infected by human immunodeficiency virus type 1. J Exp Med. 1994;180:1283–1293. doi: 10.1084/jem.180.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venet A, Walker B D. Cytotoxic T-cell epitopes in HIV/SIV infection. AIDS. 1993;7:S117–S126. [PubMed] [Google Scholar]
- 41.Wilson C C, Kalams S A, Wilkes B M, Ruhl D J, Gao F, Hahn B H, Hanson I C, Luzuriaga K, Wolinsky S, Koup R, Buchbinder S P, Johnson R P, Walker B D. Overlapping epitopes in human immunodeficiency virus type 1 gp120 presented by HLA-A, -B, and -C molecules: effects of viral mutation on cytotoxic T-lymphocyte recognition. J Virol. 1997;71:1256–1264. doi: 10.1128/jvi.71.2.1256-1264.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang O O, Kalams S A, Rosenzweig M, Trocha A, Jones N, Koziel M, Walker B D, Johnson R P. Efficient lysis of human immunodeficiency virus type 1-infected cells by cytotoxic T lymphocytes. J Virol. 1996;70:5799–5806. doi: 10.1128/jvi.70.9.5799-5806.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]