Abstract
Introduction
RET inhibitors with impressive overall response rates are now available for patients with NSCLC, yet the identification of RET fusions remains a difficult challenge. Most guidelines encourage the upfront use of next-generation sequencing (NGS), or alternatively, fluorescence in situ hybridization (FISH) or reverse transcriptase-polymerase chain reaction (RT-PCR) when NGS is not possible or available. Taken together, the suboptimal performance of single-analyte assays to detect RET fusions, although consistent with the notion of encouraging universal NGS, is currently widening some of the clinical practice gaps in the implementation of predictive biomarkers in patients with advanced NSCLC.
Methods
This situation prompted us to evaluate several RET assays in a large multicenter cohort of RET fusion–positive NSCLC (n = 38) to obtain real-world data. In addition to RNA-based NGS (the criterion standard method), all positive specimens underwent break-apart RET FISH with two different assays and were also tested by an RT-PCR assay.
Results
The most common RET partners were KIF5B (78.9%), followed by CCDC6 (15.8%). The two RET NGS-positive but FISH-negative samples contained a KIF5B(15)-RET(12) fusion. The three RET fusions not identified with RT-PCR were AKAP13(35)-RET(12), KIF5B(24)-RET(9) and KIF5B(24)-RET(11). All three false-negative RT-PCR cases were FISH-positive, exhibited a typical break-apart pattern, and contained a very high number of positive tumor cells with both FISH assays. Signet ring cells, psammoma bodies, and pleomorphic features were frequently observed (in 34.2%, 39.5%, and 39.5% of tumors, respectively).
Conclusions
In-depth knowledge of the advantages and disadvantages of the different RET testing methodologies could help clinical and molecular tumor boards implement and maintain sensible algorithms for the rapid and effective detection of RET fusions in patients with NSCLC. The likelihood of RET false-negative results with both FISH and RT-PCR reinforces the need for upfront NGS in patients with NSCLC.
Keywords: RET fusions, Next-generation sequencing, FISH, RT-PCR, Lung carcinoma
Introduction
The RET protooncogene is located on the long arm of chromosome 10 and encodes a transmembrane protein that consists of an extracellular ligand-binding domain, a transmembrane region, and an intracellular tyrosine kinase domain.1, 2, 3 RET activation occurs when the GDNF ligands bind to their receptors, causing homodimerization, autophosphorylation, and ultimately activation of downstream signaling pathways.4 Oncogenic activating fusions have been identified in a variety of malignant tumors, including papillary thyroid carcinomas and NSCLC.4,5 RET fusions are found in 1% to 2% of NSCLC, and there is a higher prevalence in never or light smokers, younger age, and adenocarcinoma (AC).4,5 In treatment-naive patients, RET fusions tend to be mutually exclusive with other major oncogenic drivers.4 The rearrangements typically involve the 3’ kinase domain of RET encoded by exons 12 to 18 to various 5’ heterologous upstream partner genes.4 In NSCLC, the most typically reported RET partners are KIF5B (∼70%), CCDC6 (∼20%), and NCOA4 (∼2%), and many other partners have been reported as isolated examples.6 Therefore, the molecular epidemiology of RET fusions is difficult to infer but the frequency of those uncommon RET partners with more overlap between the different series is usually around 1%: ERC1, TRIM24, TRIM27, TRIM33, DOCK1, KIF13A, and KIAA1468.7, 8, 9, 10, 11, 12, 13, 14, 15, 16 The development and approval of selective RET inhibitors in lung cancer, thyroid cancer or even in a tumor-agnostic strategy, with high efficacy, means that the relevance of accurately identifying RET fusions has never been greater.4,5,12,15, 16, 17, 18, 19, 20, 21, 22, 23, 24
The available diagnostic methodologies used to identify RET fusions include the increasingly popular next-generation sequencing (NGS) and single-gene approaches such as fluorescence in situ hybridization (FISH) and reverse transcriptase-polymerase chain reaction (RT-PCR).20,25 Accordingly, in clinical trials, there is vast heterogeneity in local testing methods, and between 18% to 42% of patients have been identified by either FISH or RT-PCR.12,15,16,19 Several professional organizations and academic groups have released recommendations on the standard methods to detect RET fusions in daily practice and clinical research.6,10,26,27 Most guidelines encourage the upfront use of NGS, or alternatively, FISH or RT-PCR when NGS is not possible or available.6,10,26 Although break-apart FISH has traditionally been the accepted standard test for the detection of fusions, RET FISH is especially difficult to interpret and may be susceptible to both false negatives and false positives.10 Moreover, the real-world performance of specific RT-PCR assays remains largely unknown.
Taken together, the suboptimal performance of single-analyte assays to detect RET fusions, although consistent with the notion of encouraging universal NGS, is currently widening some of the clinical practice gaps in the implementation of predictive biomarkers in advanced NSCLC.28 Therefore, we hypothesized that in-depth knowledge of the advantages and disadvantages of the different RET testing methodologies could help clinical and molecular tumor boards implement and maintain sensible algorithms for rapid and effective detection of predictive biomarkers (i.e., including RET) in patients with NSCLC. This situation prompted us to evaluate several RET assays (i.e., RNA-based NGS as criterion standard method, FISH, and RT-PCR) in a large multicenter cohort of RET-positive NSCLC to obtain real-world data.
Materials and Methods
Study Design and Tumor Samples
The flow diagram is depicted in Figure 1. There were 57 RET fusion–positive samples from patients with NSCLC that had been initially tested as part of routine clinical care in 16 different institutions, were used for this study (also known as RETING or RET and Individual gene assays & Next-Generation sequencing). To confirm the RET fusion–positive status, targeted RNA-based NGS analysis (the criterion standard method) was performed at the referral institution. Only cases with enough tissue available (i.e., a minimum of 20% tumor cell content) were included. In addition, 100 consecutive RET NGS-negative samples from NSCLC tested at the referral institution as part of routine clinical care were included as negative controls. The material available for all tumors was formalin-fixed and paraffin-embedded (FFPE). The specifics of formalin-fixation were unknown. All cases were reviewed by three pathologists (E.C., F.L.R., and J.L.R.C.). In addition to NGS, all positive specimens underwent break-apart RET FISH with two different assays using an automated scanning system and were also tested by an RNA PCR-based assay. In the negative cohort, only one RET FISH assay was investigated. The Institutional Ethics Committee at Fundacion de Investigation HM Hospitales and Hospital Universitario 12 de Octubre reviewed and approved this study. Each referring institution regulated the need for additional specific consent. Clinical data from the RET NGS-positive cohort were retrieved from the patient clinical records.
Figure 1.
Flowchart of samples in the RETING study. FISH, fluorescence in situ hybridization; NGS, next-generation sequencing; RT-PCR, reverse transcriptase–polymerase chain reaction.
NGS for RET Fusions
A targeted RNA-based NGS panel (Oncomine Comprehensive Assay v3 test [ThermoFisher Scientific, Waltham, MA]) was performed for all cases (positive and negative) on the Ion S5 sequencer with automated library preparation using the Ion Chef System, as described previously.29 For each FFPE tumor sample, freshly cut 5-μm–thick sections were collected on separate Eppendorf tubes for DNA and RNA extraction: three sections for surgical specimens and five sections for small biopsy specimens for each tube. The first and last sections were stained with hematoxylin-eosin and reviewed by two pathologists (E.C. and F.L-R.) to confirm that the percentage of tumor cells was greater than or equal to 20%. The DNA extraction was performed with the Cobas DNA Sample Preparation Kit (Roche Molecular Systems, Pleasanton, CA) following the manufactureŕs instructions. The RNA extraction was performed with the High Pure FFPET RNA Isolation Kit (Roche Molecular Systems) following the manufacturer’s instructions. The RNA was then purified and concentrated by using the GeneJET RNA cleanup and concentration micro kit (ThermoFisher Scientific). The protocol for the NGS analyses followed the manufactureŕs instructions, and a minimum of 500,000 mapped fusion panel reads was required for RET fusion analysis. The RET NGS result was used as the criterion standard method and the complete NGS report was only available for the RET NGS-positive cohort.
FISH for RET fusions
FISH was carried out on unstained 4-μm–thick FFPE tumor tissue sections from all cases. For all positive cases, we used two commercial break-apart RET FISH assays: Vysis RET FISH Break-Apart Probe RUO kit (Abbott Molecular, IL) and ZytoLight SPEC RET Dual Color BreakApart Probe (ZytoVision GMbH, Bremerhaven, Germany). In the negative cohort, we only investigated the Vysis RET FISH probe. The methodologies have been described in detail elsewhere.30,31 RET FISH assays were independently captured and scored with the automated BioView Duet scanning system (BioView, Rehovot, Israel) by a thoracic pathologist (E.C.) or molecular biologist (S.H.). A minimum of 50 tumor nuclei were counted. RET FISH-positive cases were defined as those with greater than or equal to 15% break-apart signals (separated by more than one signal diameter) or isolated 3’ signals in tumor cells.26,32,33 Using our own prespecified criteria, if the separation between the signals was greater than one but less than two signal diameters, the pattern was named “borderline positive break-apart.” RET FISH-negative samples were defined as those with fusion signals, isolated 5’ signals, or less than 15% of positive cells.26,32,33
RT- PCR Assay for RET Fusions
The AmoyDx Multigene Mutations Detection Kit (Amoy Diagnostics, Xiamen, People's Republic of China) was performed for all positive samples, according to the manufacturer's instructions. This RNA-based assay is designed to detect six different RET fusion variants (i.e., CCDC6[1]-RET[12], NCOA4[6]-RET[12], KIF5B[15]-RET[12], KIF5B[16]-RET[12], KIF5B[22]-RET[12] and KIF5B[23]-RET[12]) on a Cobas z 480 (user-defined function channel) instrument.
Results
The clinicopathologic characteristics of patients with RET fusions are presented in Table 1.
Table 1.
Clinicopathologic Features of Patients with RET Fusions
| Characteristic | Patients, n (%)a N = 38 |
|---|---|
| Tumor histology | |
| AC | 35 (92.1) |
| NSCLC-NOS | 3 (7.9) |
| Specimen type | |
| Surgical | 20 (52.6) |
| Small biopsy | 15 (39.5) |
| Cell block | 3 (7.9) |
| Age at diagnosis, yra | |
| Median (range) | 65 (39-89) |
| Distribution | |
| ≥18 to 64 yr | 17 (45.9) |
| ≥65 yr | 20 (54.1) |
| Sexa | |
| Female | 26 (70.3) |
| Male | 11 (29.7) |
| Smoking historya | |
| Never smoked | 26 (70.3) |
| Current / former smoker | 11 (29.7) |
| Stage at initial diagnosisa | |
| I | 6 (16.2) |
| II | 4 (10.8) |
| III | 6 (16.2) |
| IV | 21 (56.8) |
| Metastasis sites for stage IV diseasea | |
| Multiple organs | 16 (57.1) |
| Lung | 15 (53.6) |
| Bone | 13 (46.4) |
| Lymph node | 9 (32.1) |
| Liver | 7 (25) |
| Brain | 6 (21.4) |
| Pleura | 5 (17.9) |
| Others | 4 (14.3) |
| No. of previous lines before RET TKI therapya,b | |
| 0 | 7 (25) |
| 1 | 15 (53.6) |
| 2 | 3 (10.7) |
| ≥3 | 3 (10.7) |
| RET TKI therapya,b | |
| Pralsetinib | 10 (35.7) |
| Selpercatinib | 6 (21.4) |
| Others | 5 (17.9) |
| None | 7 (25) |
| Best overall response after RET TKI therapya,c | |
| Complete response | 3 (14.3) |
| Partial response | 10 (47.7) |
| Stable disease | 2 (9.5) |
| Progressive disease | 4 (19) |
| Not available | 2 (9.5) |
AC, adenocarcinoma; NSCLC-NOS, non-small cell lung carcinoma, not otherwise specified; TKI, tyrosine kinase inhibitor.
Clinical information was available for 37 patients.
Patients with stage IV disease (n=28).
Patients with stage IV disease treated with RET TKI therapy (n=21).
RET Fusions Assessed by NGS
Of the 57 RET fusion–positive lung carcinoma specimens, nine cases (9 of 57, 15.8%) were excluded for lack of sufficient tumor content (see above). Six samples (6 of 48, 12.5%) were negative for RET fusions and results could not be assessed in four cases (4 of 48, 8.3%) owing to insufficient sequencing coverage (Fig. 1). Therefore, the final size of the positive cohort was 38 tumors. There were 30 cases (30 of 38, 78.9%) that had a KIF5B-RET fusion (25 cases corresponding to the KIF5B[15]-RET[12] variant, two corresponding to the KIF5B[16]-RET[12] variant, and the remaining three cases corresponding to KIF5B[23]-RET[12], KIF5B[24]-RET[11] and KIF5B[24]-RET[9] variants, respectively), six cases (6 of 38, 15.8%) exhibited a CCDC6(1)-RET(12) fusion, one tumor (1 of 38, 2.6%) presented a NCOA4(6)-RET(12), and one sample (1 of 38, 2.6%) contained a AKAP13(35)-RET(12) fusion. Non-RET alterations were present in 44.7% (17 of 38) of RET-positive patients. The three more common co-occurring gene variants included TP53 (5 of 38, 13.2%), SETD2 (5 of 38, 13.2%), and CTNNB1 (2 of 38, 5.3%) mutations. Interestingly, isolated examples of copy number variations in genes MDM2 (1 of 38, 2.6%) and CDK6 (1 of 38, 2.6%) were also identified.
Because of the retrospective nature of the negative cohort, NGS had been successful in all 100 RET-negative tumors (Fig. 1).
RET Fusions Assessed by FISH
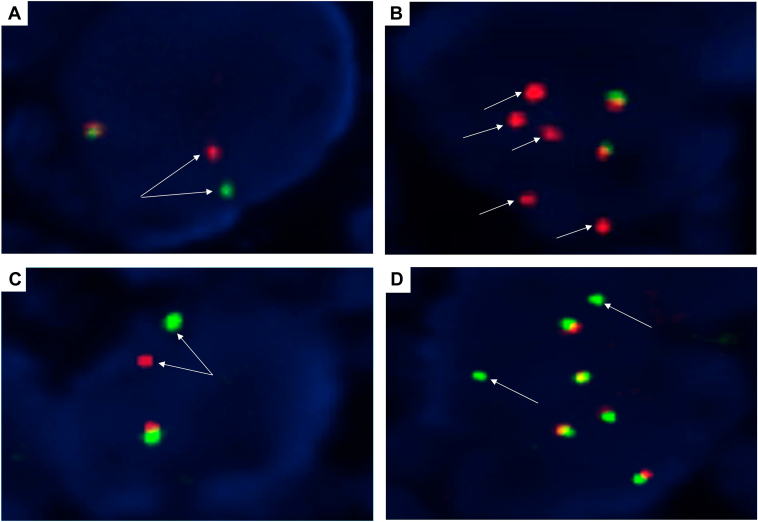
All 138 specimens (positive and negative) were successfully tested by FISH (Fig. 1). In agreement with the NGS results, 36 out of the 38 (94.7%) RET NGS-positive samples were RET FISH-positive by both probes. The overall results were very similar for both probes. The mean percentage of positive cells was 74.6% (median 77%, range 16%–100%) using the Vysis probe and 70.5% (median 74%, range 18%-96%) with the ZytoVision probe. The break-apart pattern was more frequently observed than the isolated 3’ signal pattern (30 of 36, 83.3% versus 6 of 36, 16.7%) (Fig. 2A–D). The number of cases with a borderline positive break-apart pattern (see definition above) was higher with one of the probes (13 of 36, 36.1% for ZytoVision versus 6 of 36, 16.7% for Vysis). Interestingly, this borderline pattern was identified in all fusion partners except AKAP13. The frequencies were higher for CCDC6 (3 of 6, 50% with ZytoVision and 1 of 6, 16.7% with Vysis) than for KIF5B (9 of 28, 32.1% with ZytoVision and 4 of 28, 14.3% with Vysis) (Fig. 3A). The two RET NGS-positive but FISH-negative samples contained a KIF5B(15)-RET(12) fusion (Fig. 3B). Both ACs exhibited psammoma bodies and were diagnosed in a surgical specimen. These two patients received a RET TKI and had partial responses. The FISH results for all cases from the negative cohort agreed with those obtained by NGS (Fig. 3C).
Figure 2.
Representative examples of RET FISH-positive NSCLCs using the Vysis RET Probe (A,B) and the ZytoVision RET Probe (C,D). (A,C) A typical break-apart pattern is shown with one fused signal and one break-apart signal per nucleus (arrows). (B,D) An isolated 3’ signal pattern is depicted (red signals with the Vysis probe and green signals with the ZytoVision probe) (arrows). All four cases were scored using the BioView Duet scoring system and were RET NGS-positive. See text for details. Original magnification: x1000. FISH, fluorescence in situ hybridization; NGS, next-generation sequencing.
Figure 3.
Representative examples of RET FISH patterns: borderline break-apart positive (A), false-negative (B), and (C) typical negative fusion signal pattern (C). (A) A tumor with a CCDC6-RET fusion showing a borderline break-apart positive pattern. (B) A tumor with a KIF5B-RET fusion showing insufficient separation between the red and the green signals (i.e., FISH false-negative). (C) A typical example of a tumor without RET fusions exhibits two fused signals. All images correspond to the Vysis RET probe and were interpreted using the BioView Duet scoring system. The fusion status was confirmed by NGS. See text for details. Original magnification: x1000. FISH, fluorescence in situ hybridization.
RET Fusions Assessed by RT-PCR
Three RET NGS-positive cases were excluded for lack of tumor tissue after the previous analyses (Fig. 1). There were 32 out of the remaining 35 (32 of 35, 91.4%) NGS-positive samples that were RT-PCR–positive. The three RET fusions not identified with RT-PCR were AKAP13(35)-RET(12), KIF5B(24)-RET(9) and KIF5B(24)-RET(11). All three cases were AC that were diagnosed by surgical specimens (n = 2) or core-needle biopsy (n = 1). Both surgical specimens contained either signet ring cells or psammoma bodies. All three cases were FISH-positive, exhibited a typical break-apart pattern, and contained a very high number of positive tumor cells with both FISH assays (82%, 92%, and 90% for Vysis; 60%, 94%, and 96% for ZytoVision, respectively). Of note, two of these three patients received a RET TKI and had partial responses.
Histologic Characteristics
A total of 35 tumors (35 of 38, 92.1%) were AC and three (3 of 38, 7.9%) were NSCLC not otherwise specified. Of the AC, 16 (45.7%) were observed to have a predominant acinar pattern, 11 (31.4%) presented a solid architecture, five (14.3%) had a predominant lepidic pattern, two (5.7%) exhibited a papillary growth (KIF5B[15]-RET[12] and CCDC6[1]-RET[12]), and one (2.8%) had a predominant micropapillary pattern (KIF5B[15]-RET[12]). Signet ring cells, psammoma bodies, and pleomorphic features were frequently observed (in 13 of 38 [34.2%], 15 of 38 [39.5%], and 15 of 38 [39.5%] of tumors, respectively) (Fig. 4A–C). Interestingly, pleomorphism was only present with the KIF5B partner.
Figure 4.
Typical features of NSCLC with RET fusions. (A) signet ring cells, (B) psammoma bodies, and (C) pleomorphic nuclei (hematoxylin-eosin, original magnification X200 [A-C]).
Discussion
The information presented herein is very timely because a recent survey from more than 500,000 patients has identified that almost 50% of patients with advanced NSCLC were not candidates for targeted therapies because of biomarker testing issues.28 The clinical gaps can be summarized as follows: tissue (insufficient tissue or inaccurate estimation of tumor cell content), testing (appropriate assay was not ordered or results were inconclusive or false-negative), and time (turnaround time delays).28 Therefore, in some series the frequency of RET fusions falls below 1%11,34, 35, 36 and, unsurprisingly the percentage is within the expected range (i.e., 1%-2%) in fully genotyped cohorts.36 These results are consistent with mounting evidence of similar trends for other actionable fusions.37, 38, 39 Although broad molecular profiling is the recommended NSCLC testing option in most guidelines, NGS is not universally available or requested.6,26,39, 40, 41, 42 Until NGS is routinely performed in all patients with advanced NSCLC, a deep understanding of the concept of “molecular redundancy” is reassuring.43 This notion has been recommended and endorsed by all the major professional organizations in the field and can be summarized as follows: “Laboratories should ensure that test results that are unexpected, discordant, equivocal, or otherwise of low confidence are confirmed or resolved using an alternative method or sample.”43 Therefore, in this RETING study, we wanted to explore the performance of typically used single-gene RET assays as potential complementary tools to NGS in testing workflows for patients with advanced NSCLC.6,10,26
Reasoning that RNA sequencing is now becoming the accepted standard for fusion identification, because of its superior sensitivity,40,44 we decided to use as our standard criterion a large RNA-based NGS assay that required very little input RNA. The molecular landscape of RET fusions in our series is remarkably similar to previous reports (i.e., high frequency of co-occurring TP53, SETD2, and CTNNB1 mutations),7,8,10,45, 46, 47, 48 including the puzzling finding of MDM2 and CDK4/6 amplifications.8,46 Overall, the variety and individual frequencies of RET partners identified were like those described (Table 2).7, 8, 9, 10, 11,13,14,45, 46, 47,49, 50, 51, 52, 53, 54 The most common fusion partners are KIF5B, CCDC6 and NCOA4. Several conclusions can be drawn from our study. First, the performance of two typically used FISH probes was similar: two clear-cut false-negative results with both probes on the same samples. It is unfortunate that both suboptimal readings involved the most frequent RET fusion in patients with NSCLC (i.e., KIF5B[15]-RET[12] fusion) (Table 2). Despite the initial description that RET FISH false-negative results were restricted to the NCOA4 partner,10 isolated examples involving KIF5B fusions have been reported.33,46,47,55 Second, our absence of RET FISH false-positive results could be because of the use of an outstanding automated FISH scanning system and a large NGS panel as a FISH comparator. In agreement with other authors, we believe that the current false-positive rate of RET FISH could be overestimated for two main reasons: (1) the adoption of a low threshold of signal separation for positive break-apart signals or a low percentage of positive nuclei as the cutoff for positivity,56 and (2) the use of RT-PCR or small NGS panels as a standard criterion, which may miss some fusion partners.33,53,57 Moreover, similarly to other break-apart FISH probes,58 the presence of complex patterns in RET FISH assays (e.g., loss of signals) is clearly linked to false-positive results.10,32,33 Nevertheless, the literature on this topic should be interpreted with great caution because most series are small, and very different methods and criteria have been used (Table 3).10,32,33,46, 47, 48,53,59, 60, 61, 62, 63
Table 2.
Summary of Studies Addressing the Tissue Detection Rate of RET Fusions in Patients with NSCLCa
| Study | No. of Patients with Identified Upstream Partners | Frequencies of RET Partners Genes (%) |
Representation of RET Fusions not Identified by Single-Gene Assays in the Current Study (%b) |
|||||
|---|---|---|---|---|---|---|---|---|
| FISH False-Negative |
RT-PCR False-Negative |
|||||||
| KIF5B | CCDC6 | NCOA4 | KIF5B(15)-RET(12 ) | KIF5B(24)-RET(9) | KIF5B(24)-RET(11) | AKAP13(35)-RET(12) | ||
| Parimi et al.8 2023 | 523 | 66 | 18.2 | 2.9 | N/A | N/A | N/A | N/A |
| Wang et al.49 2022 | 262 | 48.5 | 16 | 2.3 | N/A | N/A | N/A | 0 |
| Feng et al.47 2022 | 167 | 68.2 | 16.8 | 1.2 | N/A | N/A | N/A | 0 |
| Aldea et al.7 2023 | 166 | 72 | 17 | 1.6 | N/A | N/A | N/A | 0 |
| Yang et al.10 2021 | 99 | 68.7 | 14 | 3 | 55.6 | 0 | 1 | 0 |
| Gautschi et al.50 2017 | 81 | 72 | 23 | 2 | N/A | N/A | N/A | 0 |
| Illini et al.14 2021 | 50 | 66 | 20 | 2 | N/A | N/A | N/A | N/A |
| Meng et al.51 2022 | 49 | 26.5 | 12.2 | 2.1 | N/A | N/A | N/A | 0 |
| Hess et al.11 2021 | 46 | 63 | 23.9 | 6.5 | N/A | N/A | N/A | 0 |
| Xiang et al.9 2022 | 41 | 68 | 12 | 0 | 56.1 | 0 | 0 | 2,4c |
| Tan et al.46 2020 | 40 | 62.5 | 30 | 0 | N/A | N/A | N/A | 0 |
| Passaro et al.54 2022 | 34 | 55.7 | 9.8 | 3.3 | N/A | N/A | N/A | N/A |
| Gao et al.45 2023 | 29 | 62 | 21 | 0 | 55.2 | 0 | 0 | 0 |
| Qiu et al.52 2020 | 23 | 60.9 | 26.1 | 4.3 | 26.1 | 0 | 0 | 0 |
| Jeon et al.13 2023 | 23 | 69.6 | 21.7 | 4.3 | N/A | N/A | N/A | N/A |
| Tsuta et al.53 2014 | 22 | 86.4 | 13.6 | 0 | 62,5d | 0d | 0d | 0d |
| Conde et al. 2024 | 38 | 78.9 | 15.8 | 2.6 | 65.8 | 2.6 | 2.6 | 2.6 |
NSCLC, non-small cell lung carcinoma.
Only studies with more than 20 RET-positive cases are included.
The denominator is the total number of RET fusions.
Corresponds to a AKAP13(35)-RET(11).
The specific breakpoint is only available for 16 of the 22 RET fusions.
Table 3.
Summary of Studies Addressing the use of FISH to Detect RET Fusions in Patients with NSCLCa
| Study | No. of Patients with RET Fusions | Genomic Confirmation | Probe | No. of Cells Evaluated | Cut-off for Positivity (%) | Range of Positive Signals (%) | Types of Positive Signals |
Inclusion of an Equivocal Category | ||
|---|---|---|---|---|---|---|---|---|---|---|
| BA Signals | Distance Between BA Signals (Signal Diameter) | Isolated Signals | ||||||||
| Yang et al.10 2021 | 48 | Yes | ZytoVision | 100 | ≥10 | N/A | Yes | ≥2 | Yes | No |
| Feng et al.47 2022 | 25 | Yes | Other | >100 | ≥15 | N/A | Yes | N/A | Yes | No |
| Baker et al.33 2022 | 23 | Yes | Vysis | 50 | ≥19 | 13-73 | Yes | >1 | Yes | Yes |
| Michels et al.61 2016 | 22 | Nob | ZytoVision | 100 | ≥20 and ≥15 | 21-100 | Yes | N/A | Yes | No |
| Tsuta et al.53 2014 | 22 | Yes | Other | 50 | ≥20 | 22-72 | Yes | >1 | Yes | No |
| Radonic et al.32 2021 | 18 | Yes | Several | 50 | ≥15 | N/A | Yes | >1 | Yes | Yes |
| Conde et al. 2024 | 38 | Yes | Vysis / ZytoVision | 50 | ≥15 | 16-100 / 18-96 | Yes | >1 | Yes | Yes |
BA, break-apart; NSCLC, non-small cell lung carcinoma.
Only series with genomic confirmation of more than 15 cases and reproducible FISH protocols are included.
Despite of the lack of genomic confirmation this series is included due to the high quality of the FISH data.
When using RT-PCR it is important to understand the concept of “diagnostic sensitivity,” which relates to the comprehensiveness of the assay, or the percentage of all RET fusions described for the gene detectable by the given assay.6,64 Users of these assays should be constantly aware that “pseudo false-negatives” (i.e., because fusion partners are not included in the design of an assay) are unavoidable. Accordingly, three RET fusions were missed by the RT-PCR kit, emphasizing the need to always consider NGS testing in patients with driver-negative NSCLC.6 A review of the literature in light of our findings suggests that the presence of an AKAP13 partner is a rare event. Unfortunately, the lack of detail regarding the specific KIF5B breakpoints in some large series prevents drawing definitive conclusions regarding the molecular epidemiology of KIF5B(24)-RET(9) and KIF5B(24)-RET(11) fusions (Table 2). According to Mizukami et al.,65 the frequency of the KIF5B(24)-RET(11) fusion across several cohorts comprising 60 patients is 2%, which is similar to our experience (2.6%). Nevertheless, the occasional presence of this fusion in two very small series (13 and 14 patients with a frequency of around 7%) remains worrisome and highlights the difficulty in calculating the risk of false-negative results when using RT-PCR for RET testing.59,62 Single-analyte assays are still very popular across the globe for cost reasons or because exclusionary testing is implemented in high EGFR mutation prevalence regions.6 In exclusionary testing, several biomarkers are tested first, followed by NGS in driver-negative patients. Despite contradictory reports on the cost-effectiveness of this strategy,66, 67, 68 recently released expert consensus or recommendations from the Asia-Pacific region support the use of upfront NGS in patients with NSCLC.27,69
Although RET immunohistochemistry to detect RET fusions is not currently recommended because of its wide range of sensitivity (50%–100%) and specificity (30%–90%),6,10,26,27 several comments might be helpful for the future implementation and development of RET antibodies: (1) evidence on the topic is still inconclusive because of the small sample size of many reports and the insufficient representation of non-KIF5B partners26; (2) only antibodies directed to the C-terminal portion of RET should be used to identify the chimeric protein26; and (3) the clone EPR2871 is probably the most frequently used and well characterized, with an interesting association between the fusion partner and the expression of the protein.10 Some authors have reported higher H-scores for KIF5B fusions, which resulted in perfect sensitivity for the detection of KIF5B-RET fusions.10
The histologic characteristics of our NSCLC with RET fusions is concordant with the literature. A careful review of published studies identifies that most cases are AC (range: 82%–100%, mean: 92.6%, median: 94%).7, 8, 9, 10,45,47,50, 51, 52, 53,62,63 That RET fusion–positive AC can contain signet ring cells (range: 27-36%, mean: 30.7%, median: 30%) and psammoma bodies are well known,62,70 but the predictive value of these features is not fully recognized in clinical practice. Of note, four of the five false-negative FISH/RT-PCR samples contained either signet ring cells or psammoma bodies. Accordingly, pathologists should always report them and persevere in the search for actionable fusions in those circumstances, as they can also be found in NSCLC with ALK or ROS1 fusions.30,31,70 Another interesting and underrecognized feature is the presence of papillary or micropapillary patterns in RET fusion–positive lung AC: almost 9% of the AC in the present series exhibited either one and reported rates to range from 9% to 36% (median: 20%, media: 22%).48,62,63 In agreement with other authors, both KIF5B and non-KIF5B partners were involved in papillary formation.62,63,71 Finally, it is important to emphasize that RET fusions have been reported in other lung carcinoma subtypes, including squamous cell carcinomas,8,18,50,52,72,73 adenosquamous carcinomas,8,47,52,63,73 sarcomatoid carcinomas,51 pleomorphic carcinomas,10 and neuroendocrine carcinomas.7, 8, 9, 10,18,52,72,74 Interestingly, neuroendocrine differentiation can also be found in pancancer studies of RET fusion–positive solid tumors, highlighting the need to also use histologic classification as a way to increase the likelihood of finding an actionable fusion in tumor-agnostic approaches, as counterintuitive as it might seem.21,22,29,49
In conclusion, the potential for false-negative results with single-analyte assays reinforces the need for upfront NGS in patients with NSCLC. A consideration of the clinical problem of NSCLC highlights the need to be aware of how the methods that we use perform in the real-world setting.
CRediT Authorship Contribution Statement
Esther Conde: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Resources; Supervision; Visualization; Roles/Writing - original draft; Writing - review & editing.
Susana Hernandez: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Resources; Supervision; Visualization; Roles/Writing - original draft; Writing - review & editing.
Jose Luis Rodriguez Carrillo: Formal analysis; Investigation; Methodology; Visualization; Review & editing.
Rebeca Martinez: Review & editing.
Marta Alonso: Review & editing.
Daniel Curto: Clinical data compilation, review & editing.
Beatriz Jimenez: Resources; Review & editing.
Alejandra Caminoa: Resources; Review & editing.
Amparo Benito: Resources; Review & editing.
Pilar Garrido: Resources; Review & editing.
Sergi Clave: Resources; Review & editing.
Edurne Arriola: Resources; Review & editing.
Isabel Esteban-Rodriguez: Resources; Review & editing.
Javier De Castro: Resources; Review & editing.
Irene Sansano: Resources; Review & editing.
Enriqueta Felip: Resources; Review & editing.
Federico Rojo: Resources; Review & editing.
Manuel Dómine: Resources; Review & editing.
Ihab Abdulkader: Resources; Review & editing.
Jorge Garcia-Gonzalez: Resources; Review & editing.
Cristina Teixido: Resources; Review & editing.
Noemi Reguart: Resources; Review & editing.
Desamparados Compañ: Resources; Review & editing.
Amelia Insa: Resources; Review & editing.
Nuria Mancheño: Resources; Review & editing.
Sarai Palanca: Resources; Review & editing.
Oscar Juan-Vidal: Resources; Review & editing.
Nuria Baixeras: Resources; Review & editing.
Ernest Nadal: Resources; Review & editing.
Maria Cebollero: Resources; Review & editing.
Antonio Calles: Resources; Review & editing.
Paloma Martin: Resources; Review & editing.
Clara Salas: Resources; Review & editing.
Mariano Provencio: Resources; Review & editing.
Ignacio Aranda: Resources; Writing - review & editing.
Bartomeu Massuti: Resources; Review & editing.
Laura Lopez-Vilaro: Resources; Review & editing.
Margarita Majem: Resources; Writing - review & editing.
Luis Paz-Ares: Resources; Review & editing.
Fernando Lopez-Rios: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Resources; Supervision; Visualization; Roles/Writing - original draft; Writing - review & editing.
Disclosure
Dr. Conde has received research funding from Eli Lilly, AstraZeneca, and ThermoFisher Scientific; and honoraria from Pfizer, Roche, AstraZeneca, Janssen, and Eli Lilly. Dr. Hernandez has received research funding from Eli Lilly, AstraZeneca, and ThermoFisher Scientific, and honoraria from Pfizer, Roche, AstraZeneca, ThermoFisher Scientific, and Eli Lilly. Mr. Alonso has received research funding from AstraZeneca, and honoraria from Pfizer, Roche, and AstraZeneca. Dr. Jimenez has received honoraria from Roche. Dr. Garrido has received research grants from Amgen, AstraZeneca, Blueprint, Bristol-Myers Squibb, Boehringer Ingelheim, Daiichi-Sankyo, GlaxoSmithKline, Janssen, IO Biotech, Eli Lilly, Merck Sharp & Dohme, Roche, Takeda; and honoraria from AbbVie, Amgen, AstraZeneca, Bayer, Bristol-Myers Squibb, Boehringer Ingelheim, Daiichi-Sankyo, GlaxoSmithKline, Janssen, Eli Lilly, Merck Sharp & Dohme, Novartis, Pfizer, Roche, Sanofi, Takeda, Medscape, and Touch Medical. Dr. Clave has received honoraria from AstraZeneca, Pfizer, Roche, Eli Lilly, and Takeda. Dr. Arriola has received honoraria from AstraZeneca, Boehringer Ingelheim, Pfizer, Roche/Genentech, Eli Lilly and Company, Novartis, Takeda, Merck Sharp & Dohme, Bayer, and Bristol Myers Squibb. Dr. Esteban-Rodriguez has received honoraria from AstraZeneca, Pfizer, and Merck Sharp & Dohme. Dr. De Castro has received honoraria from AstraZeneca, Bristol Myers Squibb, Hoffmann- La Roche, Merck Sharp and Dohme, Boehringer-Ingelheim, Janssen, Eli Lilly, Sanofi, Takeda, Pfizer, Glaxo, and Gilead. Dr. Sansano has received honoraria from F. Hoffmann La Roche AG, Merck Sharp & Dohme, Pfizer, Takeda, AstraZeneca, and Boehringer Ingelheim. Dr. Felip has received honoraria from AbbVie, Amgen, AstraZeneca, Bayer, Beigene, Boehringer Ingelheim, Bristol Myers Squibb, Daiichi-Sankyo, Eli Lilly, F. Hoffmann – La Roche, Gilead, Glaxo Smith Kline, Genentech, Janssen, Medical Trends, Medscape, Merck Serono, Merck Sharp & Dohme, Novartis, Peptomyc, Peervoice, Pfizer, Regeneron, Sanofi, Takeda, Turning Point, and Touch Oncology. Dr. Rojo has received research funding from Roche, AstraZeneca, Menarini, Novartis, Merck, Merck Sharp & Dohme, Bristol-Myers Squibb, Pfizer, GlaxoSmithKline, Palex, Amgen, Agilent, and Janssen, and honoraria from Roche, AstraZeneca, Menarini, Novartis, Merck, Merck Sharp & Dohme, Bristol-Myers Squibb, Pfizer, GlaxoSmithKline, Palex, Amgen, Agilent, Janssen. Dr. Dómine has received honoraria from AstraZeneca, Boehringer Ingelheim, Pfizer, Roche/Genentech, Takeda, Merck Sharp & Dohme, and Bristol Myers Squibb. Dr. Abdulkader has received honoraria from AstraZeneca, Eli Lilly, Pfizer, Roche, Merck Sharp & Dohme, Bristol Myers Squibb, Takeda, and Agilent Technologies S.A. Dr. Garcia-Gonzalez has received honoraria from Amgen, AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Merck Sharp & Dohme, Novartis, Roche, Sanofi, Pierre Fabre, Eli Lilly, Pfizer, and Takeda. Dr. Teixido has received honoraria from Novartis, AstraZeneca, Roche, Merck Sharp Dohme, Pfizer, Janssen, Eli Lilly, and, Bristol Myers Squibb. Dr. Reguart has received honoraria from Amgen, AstraZeneca, Bayer, Bristol-Myers Squibb, Boehringer, Guardant, Janssen, Merck Sharp & Dohme, Novartis, Pfizer, Roche, Sanofi, and Takeda. Dr. Insa has received honoraria from Roche, Bristol Myers Squibb, Sanofi, Pfizer, Boehringer Ingelheim, AstraZeneca, Takeda, Bayer, Merck Sharp & Dohme, and Eli Lilly. Dr. Mancheño has received honoraria from Roche, AstraZeneca, and Pfizer. Dr. Palanca has received honoraria from Roche Pharma, Pfizer, Amgen, AstraZeneca, Takeda, Eli Lilly, and Janssen. Dr. Juan-Vidal has received honoraria from Boehringer Ingelheim, Bristol Myers Squibb, Merck Sharp & Dohme, Roche/Genetech, AstraZeneca, Pfizer, Eli Lilly, and Takeda. Dr. Baixeras has received honoraria from AstraZeneca and Eli Lilly. Dr. Nadal has received research funding from Roche, Pfizer, Bristol-Myers Squibb and Merck Serono, and honoraria from Roche, Bristol Myers Squibb, Merck Sharp Dohme, Merck Serono, Sanofi, Pfizer, Eli Lilly, Janssen, Amgen, Daiichi-Sankyo, Boehringer Ingelheim, AstraZeneca, Takeda, Sanofi, Pierre Fabre, Qiagen, Janssen, and Bayer. Dr. Calles has received research funding from Merck Sharp & Dome, and honoraria from AstraZeneca, Boehringer Ingelheim, Pfizer, Roche/Genentech, Eli Lilly and Company, Novartis, Takeda, Merck Sharp & Dohme, and Bristol Myers Squibb. Dr. Martin has received honoraria from Daiichi-Sankyo and Pfizer. Dr. Salas has received honoraria from Boehringer Ingelheim, Pfizer, and Merck Sharp & Dohme. Dr. Provencio has received honoraria from AstraZeneca, Boehringer Ingelheim, Pfizer, Roche/Genentech, Takeda, Merck Sharp & Dohme, and Bristol Myers Squibb. Dr. Massuti has received research funding from Bristol Myers Squibb, and honoraria from Bristol Myers Squibb, Roche, Janssen, Merck Sharp & Dohme, and AstraZeneca. Dr. Majem has received research funding from Amgen Inc., AstraZeneca, Bristol Myers Squibb, and Roche, and honoraria from AstraZeneca, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Kyowa Kyrin, Merck Sharp & Dohme, Novartis, Pierre Fabre, Roche, Sanofi, and Takeda. Dr. Paz-Ares has received research funding from Merck Sharp & Dohme, AstraZeneca, Pfizer, and Bristol-Myers Squibb, and honoraria from Eli Lilly, Merck Sharp & Dohme, Roche, Pharmamar, Merck, AstraZeneca, Novartis, Servier, Amgen, Pfizer, Sanofi, Bayer, Bristol-Myers Squibb, Mirati, GlaxoSmithKline, Janssen, Takeda, and Mirati. Dr. Lopez-Rios has received research funding from Eli Lilly, AstraZeneca, Roche, Pfizer, and ThermoFisher Scientific, and honoraria from Abbvie, Astellas, AstraZeneca, Bayer, Bristol-Myers Squibb, Daiichi-Sankyo, Janssen, Eli Lilly, Merck Sharp & Dohme, Merck, Pfizer, Roche, Sanofi, Takeda, and Thermo Fisher. The remaining authors declare no conflict of interest.
Acknowledgments
This study was mainly funded by Eli Lilly through an agreement with Fundacion de Investigacion HM Hospitales and Hospital Universitario 12 de Octubre. The authors thank the Fundacion Mutua Madrileña [AP18051-2022], Instituto de Salud Carlos III (ISCIII) Fondos FEDER and Plan Estatal I+D+I 2008–2011 [PI11-02866] and 2013–2016 [PI14-01176, PI17-01001], Instituto de Salud Carlos III (ISCIII) through the project PI22-01700 and co-funded by the European Union and Comunidad de Madrid iLUNG Program [B2017/BMD-3884, P2022/BMD-7437]. L. P-A is supported by Instituto de Salud Carlos III (ISCIII) project INGENIO [PMP21/00107, PMPTA22/00167] and the Next-Generation EU funds, EC (TOPMESO Transcan) and Fundación Cris (PI20/00870). Dr. Lopez-Rios is supported by the Tom Crean expedition.
Footnotes
Drs. Conde and Hernandez contributed equally to this work.
Cite this article as: Conde E, Hernandez S, Carrillo JLR, et al. RET fusion testing in patients with NSCLC: the RETING study. JTO Clin Res Rep. 2024;5:100653.
References
- 1.Ju Y.S., Lee W.C., Shin J.Y., et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;22:436–445. doi: 10.1101/gr.133645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipson D., Capelletti M., Yelensky R., et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohno T., Ichikawa H., Totoki Y., et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Addeo A., Miranda-Morales E., den Hollander P., et al. RET aberrant cancers and RET inhibitor therapies: current state-of-the-art and future perspectives. Pharmacol Ther. 2023;242 doi: 10.1016/j.pharmthera.2023.108344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin J.J., Gainor J.F. Selective targeting of RET fusions in lung cancer. J Clin Oncol. 2023;41:410–412. doi: 10.1200/JCO.22.01644. [DOI] [PubMed] [Google Scholar]
- 6.International Association for the Study of Lung Cancer IASLC atlas of molecular testing for targeted therapy in lung cancer. https://www.iaslc.org/iaslc-atlas-molecular-testing-targeted-therapy-lung-cancer
- 7.Aldea M., Marinello A., Duruisseaux M., et al. RET-MAP: an international multicenter study on Clinicobiologic features and treatment response in patients with lung cancer harboring a RET fusion. J Thorac Oncol. 2023;18:576–586. doi: 10.1016/j.jtho.2022.12.018. [DOI] [PubMed] [Google Scholar]
- 8.Parimi V., Tolba K., Danziger N., et al. Genomic landscape of 891 RET fusions detected across diverse solid tumor types. NPJ Precis Oncol. 2023;7:10. doi: 10.1038/s41698-023-00347-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang C., Guo L., Zhao R., et al. Identification and validation of noncanonical RET fusions in non–small-cell lung cancer through DNA and RNA sequencing. J Mol Diagn. 2022;24:374–385. doi: 10.1016/j.jmoldx.2021.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Yang S.R., Aypar U., Rosen E.Y., et al. A performance comparison of commonly used assays to detect RET fusions. Clin Cancer Res. 2021;27:1316–1328. doi: 10.1158/1078-0432.CCR-20-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hess L.M., Han Y., Zhu Y.E., Bhandari N.R., Sireci A. Characteristics and outcomes of patients with RET-fusion positive non-small lung cancer in real-world practice in the United States. BMC Cancer. 2021;21:28. doi: 10.1186/s12885-020-07714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou C., Solomon B., Loong H.H., et al. First-line selpercatinib or chemotherapy and pembrolizumab in RET fusion–positive NSCLC. N Engl J Med. 2023;389:1839–1850. doi: 10.1056/NEJMoa2309457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeon Y., Jung H.A., Park S., et al. Expanded access program pralsetinib in advanced non–small cell lung cancer with rearranged during transfection (RET) gene rearrangement. Cancer Res Treat. 2023;55:1144–1151. doi: 10.4143/crt.2023.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Illini O., Hochmair M.J., Fabikan H., et al. Selpercatinib in RET fusion-positive non-small-cell lung cancer (SIREN): a retrospective analysis of patients treated through an access program. Ther Adv Med Oncol. 2021;13 doi: 10.1177/17588359211019675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drilon A., Oxnard G.R., Tan D.S.W., et al. Efficacy of selpercatinib in RET fusion–positive non–small-cell lung cancer. N Engl J Med. 2020;383:813–824. doi: 10.1056/NEJMoa2005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gainor J.F., Curigliano G., Kim D.W., et al. Pralsetinib for RET fusion-positive non-small-cell lung cancer (ARROW): a multi-cohort, open-label, phase 1/2 study. Lancet Oncol. 2021;22:959–969. doi: 10.1016/S1470-2045(21)00247-3. [DOI] [PubMed] [Google Scholar]
- 17.Selpercatinib shifts treatment paradigm for MTC and NSCLC. Cancer Discov. 2023;13 doi: 10.1158/2159-8290.CD-ND2023-0011. [DOI] [PubMed] [Google Scholar]
- 18.Drilon A., Subbiah V., Gautschi O., et al. Selpercatinib in patients with RET fusion–positive non–small-cell lung cancer: updated safety and efficacy from the registrational LIBRETTO-001 phase I/II trial. J Clin Oncol. 2023;41:385–394. doi: 10.1200/JCO.22.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griesinger F., Curigliano G., Thomas M., et al. Safety and efficacy of pralsetinib in RET fusion–positive non-small-cell lung cancer including as first-line therapy: update from the ARROW trial. Ann Oncol. 2022;33:1168–1178. doi: 10.1016/j.annonc.2022.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Novello S., Califano R., Reinmuth N., Tamma A., Puri T. RET fusion-positive non-small cell lung cancer: the evolving treatment landscape. Oncologist. 2023;28:402–413. doi: 10.1093/oncolo/oyac264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subbiah V., Wolf J., Konda B., et al. Tumour-agnostic efficacy and safety of selpercatinib in patients with RET fusion-positive solid tumours other than lung or thyroid tumours (LIBRETTO-001): a phase 1/2, open-label, basket trial. Lancet Oncol. 2022;23:1261–1273. doi: 10.1016/S1470-2045(22)00541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subbiah V., Cassier P.A., Siena S., et al. Pan-cancer efficacy of pralsetinib in patients with RET fusion–positive solid tumors from the phase 1/2 ARROW trial. Nat Med. 2022;28:1640–1645. doi: 10.1038/s41591-022-01931-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Aiello A., Halmos B. Tissue-agnostic RET inhibition: can you trust your target? Lancet Oncol. 2022;23:1235–1237. doi: 10.1016/S1470-2045(22)00556-3. [DOI] [PubMed] [Google Scholar]
- 24.Castinetti F., Borson-Chazot F. Thirty years of progress thanks to the RET oncogene. N Engl J Med. 2023;389:1916–1918. doi: 10.1056/NEJMe2311331. [DOI] [PubMed] [Google Scholar]
- 25.Herbst R.S., Aisner D.L., Sonett J.R., Turk A.T., Weintraub J.L., Lindeman N.I. Practical considerations relating to routine clinical biomarker testing for non–small cell lung cancer: focus on testing for RET fusions. Front Med. 2021;7 doi: 10.3389/fmed.2020.562480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belli C., Penault-Llorca F., Ladanyi M., et al. ESMO recommendations on the standard methods to detect RET fusions and mutations in daily practice and clinical research. Ann Oncol. 2021;32:337–350. doi: 10.1016/j.annonc.2020.11.021. [DOI] [PubMed] [Google Scholar]
- 27.Pu X., Xu C., Wang Q., et al. Expert consensus on the diagnosis and treatment of RET gene fusion non-small cell lung cancer in China. Thorac Cancer. 2023;14:3166–3177. doi: 10.1111/1759-7714.15105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadik H., Pritchard D., Keeling D.M., et al. Impact of clinical practice gaps on the implementation of personalized medicine in advanced non–small-cell lung cancer. JCO Precis Oncol. 2022;6 doi: 10.1200/PO.22.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez S., Conde E., Molero A., et al. Efficient identification of patients with NTRK fusions using a supervised tumor-agnostic approach. Arch Pathol Lab Med. 2024;148:318–326. doi: 10.5858/arpa.2022-0443-OA. [DOI] [PubMed] [Google Scholar]
- 30.Conde E., Suárez-Gauthier A., Benito A., et al. Accurate identification of ALK positive lung carcinoma patients: novel FDA-cleared automated fluorescence in situ hybridization scanning system and ultrasensitive immunohistochemistry. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conde E., Hernandez S., Martinez R., et al. Assessment of a new ROS1 immunohistochemistry clone (SP384) for the identification of ROS1 rearrangements in patients with non–small cell lung carcinoma: the ROSING study. J Thorac Oncol. 2019;14:2120–2132. doi: 10.1016/j.jtho.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Radonic T., Geurts-Giele W.R.R., Samsom K.G., et al. RET fluorescence in situ hybridization analysis is a sensitive but highly unspecific screening method for RET fusions in lung cancer. J Thorac Oncol. 2021;16:798–806. doi: 10.1016/j.jtho.2021.01.1619. [DOI] [PubMed] [Google Scholar]
- 33.Baker J.A., Sireci A.N., Marella N., et al. Analytical accuracy of RET fusion detection by break-apart fluorescence in situ hybridization. Arch Pathol Lab Med. 2022;146:351–359. doi: 10.5858/arpa.2020-0376-OA. [DOI] [PubMed] [Google Scholar]
- 34.Griesinger F., Eberhardt W., Nusch A., et al. Biomarker testing in non-small cell lung cancer in routine care: analysis of the first 3,717 patients in the German prospective, observational, nation-wide CRISP Registry (AIO-TRK-0315) Lung Cancer. 2021;152:174–184. doi: 10.1016/j.lungcan.2020.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Kuang S., Fung A.S., Perdrizet K.A., et al. Upfront next generation sequencing in non-small cell lung cancer. Curr Oncol. 2022;29:4428–4437. doi: 10.3390/curroncol29070352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mack P.C., Klein M.I., Ayers K.L., et al. Targeted next-generation sequencing reveals exceptionally high rates of molecular driver mutations in never-smokers with lung adenocarcinoma. Oncologist. 2022;27:476–486. doi: 10.1093/oncolo/oyac035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin H.M., Wu Y., Yin Y., et al. Real-world ALK testing trends in patients with advanced non–small-cell lung cancer in the United States. Clin Lung Cancer. 2023;24:e39–e49. doi: 10.1016/j.cllc.2022.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Lee D.H., Tsao M.S., Kambartel K.O., et al. Molecular testing and treatment patterns for patients with advanced non-small cell lung cancer: PIvOTAL observational study. PLoS One. 2018;13 doi: 10.1371/journal.pone.0202865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sireci A.N., Krein P.M., Hess L.M., et al. Real-world biomarker testing patterns in patients with metastatic non-squamous non-small cell lung cancer (NSCLC) in a US community-based oncology practice setting. Clin Lung Cancer. 2023;24:429–436. doi: 10.1016/j.cllc.2023.03.002. [DOI] [PubMed] [Google Scholar]
- 40.National Comprehensive Cancer Network NCCN Guidelines. Version 3.2023 Non-Small Cell Lung Cancer. https://www.nccn.org/home/
- 41.Normanno N., Apostolidis K., Wolf A., et al. Access and quality of biomarker testing for precision oncology in Europe. Eur J Cancer. 2022;176:70–77. doi: 10.1016/j.ejca.2022.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Hess L.M., Krein P.M., Haldane D., Han Y., Sireci A.N. Biomarker testing for patients with advanced/metastatic nonsquamous NSCLC in the United States of America, 2015 to 2021. JTO Clin Res Rep. 2022;3 doi: 10.1016/j.jtocrr.2022.100336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalemkerian G.P., Narula N., Kennedy E.B., et al. Molecular testing guideline for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the college of American pathologists/ international association for the. J Clin Oncol. 2018;36:911–919. doi: 10.1200/JCO.2017.76.7293. [DOI] [PubMed] [Google Scholar]
- 44.Benayed R., Offin M., Mullaney K., et al. High yield of RNA sequencing for targetable kinase fusions in lung ACs with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res. 2019;25:4712–4722. doi: 10.1158/1078-0432.CCR-19-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao Q.Y., Xiao F.M., Lin X.C., et al. Pathological characteristics and tumour immune microenvironment of lung malignancies with RET rearrangement. Cancer Treat Res Commun. 2023;35 doi: 10.1016/j.ctarc.2023.100707. [DOI] [PubMed] [Google Scholar]
- 46.Tan A.C., Seet A.O.L., Lai G.G.Y., et al. Molecular characterization and clinical outcomes in RET-rearranged NSCLC. J Thorac Oncol. 2020;15:1928–1934. doi: 10.1016/j.jtho.2020.08.011. [DOI] [PubMed] [Google Scholar]
- 47.Feng J., Li Y., Wei B., et al. Clinicopathologic characteristics and diagnostic methods of RET rearrangement in Chinese non-small cell lung cancer patients. Transl Lung Cancer Res. 2022;11:617–631. doi: 10.21037/tlcr-22-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song Z., Yu X., Zhang Y. Clinicopathologic characteristics, genetic variability and therapeutic options of RET rearrangements patients in lung adenocarcinoma. Lung Cancer. 2016;101:16–21. doi: 10.1016/j.lungcan.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Wang T., Wei L., Lu Q., et al. Landscape of potentially targetable receptor tyrosine kinase fusions in diverse cancers by DNA-based profiling. NPJ Precis Oncol. 2022;6:84. doi: 10.1038/s41698-022-00325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gautschi O., Milia J., Filleron T., et al. Targeting RET in patients with RET -Rearranged lung cancers: results from the global, multicenter RET registry. J Clin Oncol. 2017;35:1403–1410. doi: 10.1200/JCO.2016.70.9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng Y., Yang Y., Fang Y., et al. The treatment status of patients in NSCLC with RET fusion under the prelude of selective RET-TKI application in China: a multicenter retrospective research. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.864367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu Z., Ye B., Wang K., et al. Unique genetic characteristics and clinical prognosis of female patients with lung cancer harboring RET fusion gene. Sci Rep. 2020;10 doi: 10.1038/s41598-020-66883-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsuta K., Kohno T., Yoshida A., et al. RET-rearranged non-small-cell lung carcinoma: a clinicopathological and molecular analysis. Br J Cancer. 2014;110:1571–1578. doi: 10.1038/bjc.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Passaro A., Russo G.L., Passiglia F., et al. Pralsetinib in RET fusion-positive non-small-cell lung cancer: a real-world data (RWD) analysis from the Italian expanded access program (EAP) Lung Cancer. 2022;174:118–124. doi: 10.1016/j.lungcan.2022.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Ambrosini-Spaltro A., Farnedi A., Calistri D., et al. The role of next-generation sequencing in detecting gene fusions with known and unknown partners: a single-center experience with methodologies’ integration. Hum Pathol. 2022;123:20–30. doi: 10.1016/j.humpath.2022.02.005. [DOI] [PubMed] [Google Scholar]
- 56.Uguen A., Each R.E.T. Break-apart fluorescence in situ hybridization probe requires proper interpretation criteria. J Thorac Oncol. 2021;16:e55. doi: 10.1016/j.jtho.2021.03.021. [DOI] [PubMed] [Google Scholar]
- 57.Pecciarini L., Brunetto E., Grassini G., et al. Gene fusion detection in NSCLC routine clinical practice: targeted-NGS or FISH? Cells. 2023;12:1135. doi: 10.3390/cells12081135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hernandez S., Conde E., Alonso M., et al. A narrative review of methods for the identification of ALK fusions in patients with non-small cell lung carcinoma. Transl Lung Cancer Res. 2023;12:1549–1562. doi: 10.21037/tlcr-22-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeuchi K., Soda M., Togashi Y., et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 60.Pan Y., Zhang Y., Li Y., et al. ALK, ROS1 and RET fusions in 1139 lung ACs: a comprehensive study of common and fusion pattern-specific clinicopathologic, histologic and cytologic features. Lung Cancer. 2014;84:121–126. doi: 10.1016/j.lungcan.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Michels S., Scheel A.H., Scheffler M., et al. Clinicopathological characteristics of RET rearranged lung cancer in European patients. J Thorac Oncol. 2016;11:122–127. doi: 10.1016/j.jtho.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 62.Lee S.E., Lee B., Hong M., et al. Comprehensive analysis of RET and ROS1 rearrangement in lung adenocarcinoma. Mod Pathol. 2015;28:468–479. doi: 10.1038/modpathol.2014.107. [DOI] [PubMed] [Google Scholar]
- 63.Wang R., Hu H., Pan Y., et al. RET fusions define a unique molecular and clinicopathologic subtype of non–small-cell lung cancer. J Clin Oncol. 2012;30:4352–4359. doi: 10.1200/JCO.2012.44.1477. [DOI] [PubMed] [Google Scholar]
- 64.Pennell N.A., Arcila M.E., Gandara D.R., West H. Biomarker testing for patients with advanced non–small cell lung cancer: real-world issues and tough choices. Am Soc Clin Oncol Educ Book. 2019;39:531–542. doi: 10.1200/EDBK_237863. [DOI] [PubMed] [Google Scholar]
- 65.Mizukami T., Shiraishi K., Shimada Y., et al. Molecular mechanisms underlying oncogenic RET fusion in lung AC. J Thorac Oncol. 2014;9:622–630. doi: 10.1097/JTO.0000000000000135. [DOI] [PubMed] [Google Scholar]
- 66.Tan A.C., Lai G.G.Y., Tan G.S., et al. Utility of incorporating next-generation sequencing (NGS) in an Asian non-small cell lung cancer (NSCLC) population: incremental yield of actionable alterations and cost-effectiveness analysis. Lung Cancer. 2020;139:207–215. doi: 10.1016/j.lungcan.2019.11.022. [DOI] [PubMed] [Google Scholar]
- 67.Yang S.C., Yeh Y.C., Chen Y.L., Chiu C.H. Economic analysis of exclusionary EGFR test versus up-front NGS for lung adenocarcinoma in high EGFR mutation prevalence areas. J Natl Compr Canc Netw. 2022;20:774–782.e4. doi: 10.6004/jnccn.2021.7120. [DOI] [PubMed] [Google Scholar]
- 68.Loong H.H., Wong C.K.H., Chan C.P.K., et al. Clinical and economic impact of upfront next-generation sequencing for metastatic NSCLC in East Asia. JTO Clin Res Rep. 2022;3 doi: 10.1016/j.jtocrr.2022.100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Loong H.H., Shimizu T., Prawira A., et al. Recommendations for the use of next-generation sequencing in patients with metastatic cancer in the Asia-Pacific region: a report from the APODDC working group. ESMO Open. 2023;8 doi: 10.1016/j.esmoop.2023.101586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mukhopadhyay S., Pennell N.A., Ali S.M., Ross J.S., Ma P.C., Velcheti V. RET-rearranged lung adenocarcinomas with lymphangitic spread, psammoma bodies, and clinical responses to cabozantinib. J Thorac Oncol. 2014;9:1714–1719. doi: 10.1097/JTO.0000000000000323. [DOI] [PubMed] [Google Scholar]
- 71.Suehara Y., Arcila M., Wang L., et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18:6599–6608. doi: 10.1158/1078-0432.CCR-12-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cai W., Su C., Li X., et al. KIF5B-RET fusions in Chinese patients with non-small cell lung cancer. Cancer. 2013;119:1486–1494. doi: 10.1002/cncr.27940. [DOI] [PubMed] [Google Scholar]
- 73.Zhang K., Chen H., Wang Y., et al. Clinical characteristics and molecular patterns of RET-rearranged lung cancer in Chinese patients. Oncol Res. 2019;27:575–582. doi: 10.3727/096504018X15344979253618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Digumarthy S.R., Mendoza D.P., Lin J.J., et al. Imaging features and patterns of metastasis in non-small cell lung cancer with RET rearrangements. Cancers (Basel) 2020;12:693. doi: 10.3390/cancers12030693. [DOI] [PMC free article] [PubMed] [Google Scholar]