Abstract
The expression of poliovirus 2BC protein in yeast and mammalian cells leads to a number of metabolic and morphological alterations, such as growth inhibition, intracellular membrane proliferation, blockade of the exocytic pathway, and enhanced membrane permeability. Yeast cells that express poliovirus 2BC in an inducible manner were used to identify the regions of 2BC implicated in the modifications of these cellular functions. Several 2BC deletion mutants were generated to define the minimal portion of 2BC required to alter these activities. Additional deletion mutants that were obtained by random mutagenesis followed by selection in yeast cells provided new insights into the structure and mechanism of action of 2BC. The activity responsible for membrane proliferation is located in 2C, while the activities responsible for membrane permeabilization and inhibition of the exocytic pathway are located in 2B. Several regions of 2B and 2C required for the different functions of 2BC were identified. Thus, the integrity of the N termini of both 2B and 2C is necessary for 2BC-induced cytotoxicity. It is also possible to separate the different cellular alterations provoked by 2BC by the use of several 2BC variants. Deletion of amino acids 52 to 65 in 2B generates a 2BC deletion variant, 2bCΔAvrII, that still blocks yeast growth but is unable to enhance membrane permeability or to inhibit the exocytic pathway. On the other hand, 2Bc128*.32b and 2Bc128*.3c, which contain only 73 and 77 amino acids of 2B, interfere with yeast division and enhance membrane permeability but affect the exocytic pathway only weakly and do not induce membrane proliferation. Our findings indicate that Saccharomyces cerevisiae represents a useful model system to analyze the functions of poliovirus 2BC and show the feasibility of separating the activities assigned to this protein.
Poliovirus, a representative member of the Picornaviridae family, profoundly modifies cellular architecture and metabolism after infection. These alterations include enhanced membrane permeability (14, 17); inhibition of DNA, RNA, and protein syntheses (16, 44); blockade of membrane traffic (22); increased intracellular calcium levels (29); and modifications in phospholipase activities and lipid turnover (28, 30). There are also dramatic morphological alterations in chromatin structure and in the cytoskeleton (32). Moreover, the Golgi apparatus is not recognizable as such in poliovirus-infected cells (20, 42), and numerous membranous vesicles (50 to 400 nm) fill most of the cytoplasm at late times of infection (11, 21). All of these changes are referred to as the cytopathic effect (13, 15).
Membrane proliferation is essential for poliovirus genome replication, and viral RNA synthesis is associated with the newly generated vesicles (10, 28). For some time it was considered that these cytoplasmic vesicles originated by budding from the endoplasmic reticulum (ER) (9). However, more recent evidence suggests that the ER constitutes a significant but not exclusive source for the intracellular membranes induced by poliovirus infection (43). Thus, the cellular origin of these membranous vesicles, the exact mechanism used by poliovirus to induce them, and the mode by which the replicative complexes associate with these vesicles are not yet clear. It is clear, however, that protein 2BC and its proteolytic products, 2B and 2C, play an important role in these processes (2, 6, 11, 19, 22, 47). Understanding the functions of proteins 2B, 2C, and 2BC would help to elucidate the mechanisms of picornavirus genome replication and cytopathogenicity at the molecular level.
Processing of 2BC by 3Cpro gives rise to the mature products 2B and 2C, although a large fraction of 2BC remains uncleaved in picornavirus-infected cells. Genetic studies have suggested putative roles of 2B and 2C in RNA replication (4, 8, 31, 33, 36, 37, 45, 46). More recently, 2B, 2C, and 2BC were also implicated in the induction of diverse alterations in poliovirus-infected cells. Notably, proteins 2B and 2BC enhance membrane permeability and block the exocytic pathway when they are expressed individually in cultured cells and have been implicated in virus release (1, 22, 47, 48). The expression of proteins 2C and 2BC in mammalian cells with recombinant vaccinia viruses promotes membrane proliferation in the cytoplasm of transfected cells. Vesicle proliferation induced by 2BC is similar to that observed in poliovirus-infected cells, while 2C induces the formation of tubular membrane structures with a myelin-like arrangement (2, 20). In addition, the RNA replicative complexes contain a number of poliovirus proteins, including 2C (12). Three functions have been ascribed to 2C: ATPase and GTPase enzymatic activities (35, 38), interaction with viral RNA, and RNA binding by two regions of 2C (39) and interaction of 2C with membranes by means of an amphipathic helix at the N terminus (23). Based on these results, it was hypothesized that 2C or its precursor, 2BC, is responsible for poliovirus RNA binding to the induced cytoplasmic vesicles and participates in the spatial organization of the replicative complexes (39).
Inducible expression of poliovirus 2BC in yeast cells inhibits growth and provokes a number of morphological and metabolic modifications in these cells similar to those observed in mammalian cells (6). Thus, the synthesis of 2BC interferes with the exocytic pathway and promotes the formation of cytoplasmic vesicles in yeast cells. We now report the generation and characterization of a large collection of 2BC variants obtained by site-directed mutagenesis and random mutagenesis. Our results allow the dissection of the different activities assigned to the 2BC protein.
MATERIALS AND METHODS
Microbial strains.
Escherichia coli DH5 (41) was used to obtain all expression plasmids described. The Saccharomyces cerevisiae strain used was W303-1B (MATα ade2 his3, leu2 trp1 ura3).
General recombinant DNA protocols.
Construction of vectors was carried out by standard procedures (41). The yeast expression plasmid used was the yeast-E. coli shuttle vector pEMBLyex4 (18), a 2μm plasmid derivative. Poliovirus sequences were amplified by PCR from pT7XLD (a plasmid including the cDNA of poliovirus type 1, generously provided by E. Wimmer, Stony Brook, N.Y.).
The oligodeoxyribonucleotides used in this work were as follows: 5′2B.31, 5′-GCGGGATCCATGGTGACCAGTACCATCACTG, contains a BamHI restriction site and the poliovirus sequence from nucleotides (nt) 3923 to 3944; 5′2B.55, 5′-GCGGGATCCTCATGACTAGGAACTATGAAGACACC, contains a BamHI restriction site and the poliovirus sequence from nt 3995 to 4015; 5′2B.NheI, 5′-CAGTGGCTAGCAAAGAA(T/A)GCA, contains an NheI restriction site and the poliovirus sequence from nt 4067 to 4087; 5′2B.AvrII, 5′-ACAGTCCTAGGTACCC(G/T)GGCC, contains an AvrII restriction site and the poliovirus sequence from nt 4019 to 4039; 3′2B.HindIII, 5′-CCAACTGTAAGCTTGCTT, contains a HindIII restriction site and the poliovirus sequence from nt 4135 to 4118; 5′2CΔ40N, 5′-ATTCTAGAAGCTTGGGATAAGTTGGAA, contains a HindIII restriction site and the poliovirus sequence from nt 4239 to 4258; 3′2C-H2, 5′-GCG(A/T)TGCATAGATCTGTAAA, contains a BglII restriction site and the poliovirus sequence from nt 4164 to 4145; 3′2C.BglI, 5′-CTCCTTG(C/A)GCAGATCTATGAA, contains a BglII restriction site and the poliovirus sequence from nt 4225 to 4205; 3′2C.152, 5′-AGATTTAAGCTTACTACATTTCTAGTTGTCTAAGT, contains a HindIII restriction site, two stop codons, and the poliovirus sequence from nt 4288 to 4270; 3′2C.198, 5′-GTTTCTAAGCTTACTAGGCTTCCACTGCGTA, contains a HindIII restriction site, two stop codons, and the poliovirus sequence from nt 4417 to 4403; 3′2C.258, 5′-GGGCCCAAGCTTACTATGGATCCGGGGGTAGCGAGTAC, contains a HindIII restriction site, two stop codons, and the poliovirus sequence from nt 4606 to 4585; and 3′2C.B2, 5′-GGGCCCAAGCTTACTATTGAAACAAAGCCTCCATAC, contains a HindIII restriction site, two stop codons, and the poliovirus sequence from nt 5110 to 5091.
All oligonucleotides are shown in the 5′→3′ direction. The 5′ oligonucleotides contain poliovirus coding sequences, and the 3′ oligonucleotides contain complementary sequences (underlined). The mutated nucleotides are indicated with italic letters. DNA fragments obtained by PCR were sequenced by the dideoxy method (41).
Construction of plasmids encoding 2BC mutant proteins.
Plasmids pEMBL. 2B(−B), pEMBL.2C, pEMBL.2BC, pEMBL.2BcΔXbaI, pEMBL.2BcΔSphI, pEMBL.2BcΔGKS, pEMBL.2BcEcoRI, pEMBL.2BcSalI, pEMBL.2bC-S, pEMBL.2bC-D, and pEMBL.2bCΔ30N have been described elsewhere (6).
pEMBL.2bC-AD was constructed by the method of overlap extension. The first PCR (PCR1) was carried out with primers 5′2B.AvrII and 3′2C.B2, and PCR2 was carried out with primers 5′2B.30 and 3′2B.HindIII. Finally, PCR3 was carried out with primers 5′2B.30 and 3′2C.B2 with the overlap between the PCR1 and PCR2 products as a template. This PCR3 product was digested with SpeI and BamHI and ligated to pEMBL.2BC digested with the same enzymes. Clones with the AvrII and SmaI sites were selected. For pEMBL.2bC-ND, PCR1 was carried out with primers 5′2B.NheI and 3′2C.B2, and PCR2 was carried out with primers 5′2B.30 and 3′2B.HindIII. Finally, PCR3 was carried out with primers 5′2B.30 and 3′2C.B2 with the overlap between the PCR1 and PCR2 products as a template. This PCR3 product was digested with SpeI and BamHI and ligated to pEMBL.2BC digested with the same enzymes. Clones with the NheI and NsiI sites were selected.
For pEMBL.2Bc258, the PCR product obtained from pT7XLD with oligonucleotides 5′2B.31 and 3′2C.258 was digested with SpeI and HindIII and cloned in pEMBL.2BC digested with the same enzymes. For pEMBL.2Bc198, the PCR product obtained from pT7XLD with oligonucleotides 5′2B.31 and 3′2C.198 was digested with SpeI and HindIII and cloned in pEMBL.2BC digested with the same enzymes. For pEMBL.2Bc152, the PCR product obtained from pT7XLD with oligonucleotides 5′2B.31 and 3′2C.152 was digested with SpeI and HindIII and cloned in pEMBL.2BC digested with the same enzymes. For pEMBL. 2Bc128, the PCR product obtained from pT7XLD with oligonucleotides 5′2B.31 and 3′2C.BglII was digested with SpeI and BglII and cloned in pEMBL.2BC digested with SpeI and BamHI. For pEMBL.2Bc108, the PCR product obtained from pT7XLD with oligonucleotides 5′2B.31 and 3′2C.H2 was digested with SpeI and BglII and cloned in pEMBL.2BC digested with SpeI and BamHI. For pEMBL.2bcΔ127-257, the PCR product obtained from pT7XLD with oligonucleotides 5′2B.31 and 3′2C.BglII was digested with SpeI and BglII and cloned in pEMBL.2BC digested with SpeI and BamHI. For pEMBL.2bc(31-258) and pEMBL.2bc(31-152), the PCR products obtained from pT7XLD with oligonucleotides 5′2B.31 and 3′2C.258 or 3′2C.152 were digested with BamHI and HindIII and cloned in pEMBL.2BC digested with the same enzymes. For pEMBL.2bc(55-258) and pEMBL.2bc(55-152), the PCR products obtained from pT7XLD with oligonucleotides 5′2B.55 and 3′2C.258 or 3′2C.152 were digested with BamHI and HindIII and cloned in pEMBL.2BC digested with the same enzymes. For pEMBL.2bCΔNheI, the PCR product obtained from pT7XLD with oligonucleotides 5′2B.NheI and 3′2C.B2 was digested with NheI and BamHI and cloned in pEMBL.2BC digested with SpeI and BamHI. For pEMBL.2bCΔAvrII, the PCR product obtained from pT7XLD with oligonucleotides 5′2B.AvrII and 3′2C.B2 was digested with AvrII and BamHI and cloned in pEMBL.2BC digested with SpeI and BamHI. For pEMBL.2bcΔSphI(N/B), the PCR product obtained from pEMBL.2bcΔSphI (6) with oligonucleotides 5′2B.NheI and 3′2C.BglII was digested with NheI and BglII and cloned in pEMBL.2Bc258 digested with SpeI and BamHI.
For pEMBL.2BX, the PCR product obtained from pT7XLD with oligonucleotides 5′2B.31 and 3′2B.HindIII was digested with SpeI and HindIII and cloned in pEMBL.2BC digested with the same enzymes. This plasmid carries a protein with the 2B sequence followed by 61 amino acids of 2B-unrelated sequence and one stop codon. For pEMBL.2B-2CΔ40N, the PCR product obtained from pT7XLD with oligonucleotides 5′2CΔ40N and 3′2C.B2 was digested with HindIII and cloned in pEMBL.2BX digested with the same enzyme. For pEMBL.2b67, the PCR product obtained from pT7XLD with oligonucleotides 5′2B.31 and 3′2C.258 contained two fragments; the one with 711 bp corresponds to deletion mutant 2Bc258, and the other, with only 148 bp, corresponds to mutant 2b67. Both were cloned by digestion with SpeI and HindIII and ligation to pEMBL.2BC digested with the same enzymes. Oligonucleotide 3′2C.258 has two potential sites for hybridization to the poliovirus genome: its complementary sequence in the 2C gene and an alternative site in the 2B gene. The sequence of this mutant was predicted by PCR computer simulation programs (Amplify) and confirmed by sequencing of pEMBL.2b67. Thus, this deletion mutant contains 7 residues: PGSIVSL and one stop codon after amino acid 67 from 2B. For pEMBL.2bΔSpeI, pEMBL.2BC was digested with SpeI and XbaI and religated. This plasmid carries a protein with the initial 51 amino acids from 2B followed by the EMGN amino acid sequence.
Yeast growth, transformation, and induction.
Transformation of yeast by the lithium acetate procedure was done as previously described (40). Yeast growth and induction of the UASGAL-CYC promoter (18) was carried out as described previously (6). The different media used in this work (YNB.Glu, YNB.Gal, and YNB.LGal) have been defined previously (6).
Hydroxylamine mutagenesis and genetic assay.
Plasmid pEMBL.2Bc128 or pEMBL.2bcΔSphI(N/B) was randomly mutated with hydroxylamine as described previously (7, 40). The mutated DNA was used to transform S. cerevisiae W303-1B to obtain 100 to 200 colonies on each YNB.Glu plate. These colonies were replicated with YNB.Gal medium and incubated for 72 h. Colonies able to grow in YNB.Gal medium were considered potential 2BC variants. These clones were amplified, and their capacity to express poliovirus 2BC variant proteins was tested. Mutant plasmids were isolated from yeast cells by standard procedures (40). E. coli DH5 cells were transformed with these lysates. The location of the different mutations was determined by DNA sequencing by the dideoxy method (41).
Permeability changes tested with HB sensitivity.
The permeability changes in the plasma membrane were detected by testing the entry of the aminoglycoside antibiotic hygromycin B (HB) in yeast cells and the consequent inhibition of protein synthesis. The cultures were preincubated with or without 1 mM HB for 10 min before labeling of cells with 7.5 μCi of [35S]methionine per ml for 45 min. Finally, the samples were processed as described previously (51).
Other techniques.
Most of the other techniques used in this work have been described elsewhere (6). The isolation of antisera against poliovirus 2B and 2C was previously described (6). The antiserum against invertase was a generous gift from R. Schekman (University of California, Berkeley). The monoclonal antibodies against carboxypeptidase Y (CPY) were purchased from Molecular Probes, Inc. Cell labeling and immunoprecipitations were carried out as described previously (27). Yeast extracts for Western blot analysis were prepared as described previously (51). Immunoblot analyses were carried out as described elsewhere (6). The protocol for electron microscopy was described previously (6).
RESULTS
Site-directed mutagenesis of poliovirus 2BC protein.
Poliovirus 2BC is a multifunctional protein that blocks cell growth and the exocytic pathway in yeast. In addition, 2BC induces the proliferation of intracellular cisternae that fill most of the cytoplasm. To define the 2BC regions involved in these activities, a variety of point and deletion mutations in 2BC were generated as described in Materials and Methods. The schematic representation and the nomenclature used for the 2BC mutants described in this work and in our previous work (6) are shown in Fig. 1. A summary of the results obtained with all of the 2BC mutants for yeast growth, inhibition of the exocytic pathway, and enhancement of membrane permeability is also shown in Fig. 1. The effects of several mutations in conserved regions of 2BC on cytotoxicity for yeast cells were reported previously (6). Now, the collection of 2BC mutants has been expanded with the aim of determining the minimal portion of 2BC able to block yeast growth, to permeabilize cells, and to inhibit glycoprotein processing.
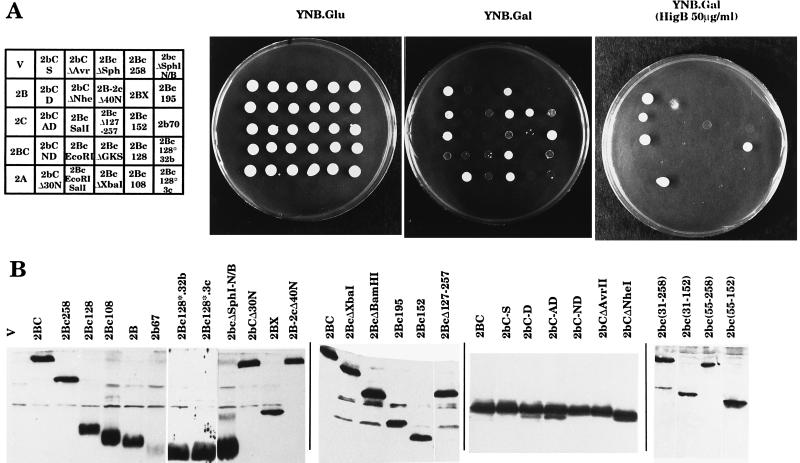
FIG. 1.
Schematic representation of 2BC mutants and summary of the effects of their expression in yeast cells. The amino acids of each protein are indicated. Black bars indicate sequences unrelated to 2BC. Growth agar (−HB/+HB) indicates growth on YNB.Gal plates supplied (+HB) or not supplied (−HB) with 50 μg of HB per ml; +++, growth rate similar to that of control cells; ++ growth at a rate lower than that of control cells; +, growth resulting in individual colony formation; −, no growth. The third value shown for 2B(1-97) corresponds to cells streaked for a second time on YNB.Gal plates supplemented with HB. Growth liquid indicates that exponentially growing yeast cultures were diluted in YNB.Lac medium, and galactose (2%) was added (initial A660, 0.150). Cells were harvested at 2-h intervals, and the A660 was measured to quantitate cell density: the optical density of the culture after 24 h of incubation increased more than 10-fold (+++), between 7- and 10-fold (++), between 5- and 7-fold (+), between 3- and 5-fold (−), or less than 3-fold (−−). pCPY arrest indicates that the ratio of precursor form of CPY (pCPY) to mature CPY in wild-type 2BC-expressing cells was calculated by densitometric analysis, and this value was taken as 100%. The relative proportion of pCPY was calculated for the rest of the mutants: +++, 100 to 70% wild-type 2BC activity; ++, 70 to 40% wild-type 2BC activity; +, 40 to 10% wild-type 2BC activity; −, no pCPY accumulation. HB sensitivity indicates that the ratio of protein synthesis in yeast cells expressing 2BC in the absence or in the presence of HB (1 mM) was calculated by densitometric analysis, and this value was taken as 100%. Therefore, 100% represents full permeabilization to HB. The relative inhibition of protein synthesis by HB was calculated for the rest of the mutants: +++, 100 to 70% wild-type 2BC activity; ++, 70 to 40% wild-type 2BC activity; +, 40 to 10% wild-type 2BC activity; −, no HB permeabilization.
The sequence of protein 2C is the most highly conserved among all picornaviruses. Several conserved domains in 2C can be identified in all the genera: a highly conserved central domain with a nucleoside triphosphate-binding motif flanked by two nonconserved regions at the N- and C-terminal ends (26). The N-terminal domain (with an amphipathic helix) has been implicated in membrane interaction (23). In contrast, the sequence of protein 2B is one of the least conserved among picornaviruses, particularly in comparisons of 2B from enteroviruses and aphthoviruses or hepatoviruses. In general, picornavirus 2B is a small (about 100 amino acids) hydrophobic protein, with a small conserved N-terminal region that may adopt an α-helical configuration, followed by an amphipathic helix (amino acids 34 to 53 in poliovirus), a potential transmembrane domain (amino acids 61 to 78 in poliovirus), and several positively charged amino acids (RKK in poliovirus). This general structure of 2B is apparent in most picornaviruses, although in members more distant from enteroviruses, such as foot-and-mouth disease virus, 2B contains 170 amino acids and a second transmembrane domain at the C terminus. Keeping in mind these conserved domains in 2B and 2C, a number of deletion and point mutations in poliovirus 2BC have been generated. The role of these conserved regions has been analyzed by use of several deletion mutations as well as point mutations in the nucleoside triphosphate-binding domain of 2C (2BcΔGKS, 2BcEcoRI, and 2BcSalI) and in the amphipathic helix (2bC-S and 2bC-D), transmembrane domain (2bC-AD and 2bCΔNhe), and positively charged region (2bC-ND) of 2B. The effects of these 2BC variants on several activities of yeast cells have been analyzed.
Effects of poliovirus 2BC and variants on yeast growth.
Poliovirus 2BC potently blocks yeast growth. The action of different 2BC variants on yeast growth both on solid and in liquid media was tested (Fig. 1 and 2). Two different types of solid media, supplemented or not supplemented with 50 μg of HB per ml, were used in order to test the effect of long-term exposure to this antibiotic.
FIG. 2.
Growth of yeast cells expressing different 2BC mutants. (A) Yeast cells expressing different 2BC variants were streaked on agar plates with the indicated composition and incubated for 3 days at 30°C. HigB, hygromycin B. (B) Extracts of yeast cells expressing the different 2BC variants indicated or bearing plasmid pEMBLyex4 (V) were obtained after 6 h of induction and assayed with antibodies against 2B or 2C proteins. Immunoblot analysis of these samples was performed as described in Materials and Methods.
The results obtained for growth in liquid medium and solid medium without HB indicate that deletion of 73 amino acids at the C terminus of 2C (2BcΔXbaI variant) produced a 2BC protein devoid of cytotoxity. However, longer deletions at the C terminus of 2C produced 2BC variants with restored growth-inhibitory effects. Thus, the mutant containing only 32 amino acids of 2C (2Bc128) was even more cytotoxic than wild-type 2BC. Even the mutant that contains only 11 amino acids of the N terminus of 2C (2Bc108) still retained some cytotoxicity. It is of interest to note that the N-terminal region of 2C (amino acids 108 to 132 of 2BC) contains an amphipathic helix involved in membrane interaction (23). The specificity of this region of 2C in mediating growth inhibition is shown by two additional observations: both the 2BX variant bearing the complete sequence of 2B followed by 63 amino acids unrelated to 2C and mutant 2B-2cΔ40N showed almost no cytotoxicity (Fig. 1 and 2). Other internal deletions and point mutations in 2C did not affect 2BC cytotoxicity, and only mutant 2BcΔGKS was devoid of cytotoxicity.
To analyze the sequences of 2B necessary for the inhibition of yeast growth, several 2BC variants with deleted 2B regions were obtained. Ablation of 30 amino acids at the N terminus of 2B (mutant 2bCΔ30N) abrogated the growth-inhibitory activity of 2BC, indicating that this region of 2B is crucial for 2BC cytotoxicity (Fig. 1 and 2). The involvement of regions at the C terminus of 2B in cytotoxicity was examined by generating variants 2bCΔNheI and 2bCΔAvrII, including deletions of amino acids 52 to 81 and 52 to 65 from 2B, respectively. These variants were still able to inhibit yeast growth (Fig. 1 and 2). This result suggests that the C-terminal region of 2B is not totally necessary for the cytotoxicity phenotype. This conclusion is reinforced by the results obtained with the double-deletion mutant 2bcΔSphI(N/B) which were also toxic for yeast cells. All the other deletion or point mutations in 2B had little or no effect on 2BC cytotoxicity. In summary, these results indicate that the N termini of both 2B and 2C are crucial for 2BC cytotoxicity (Fig. 1 and 2). Apart from the effects of 2BC and its variants on the ability of yeast cells to grow on solid medium, their action was also tested on plates supplemented with HB. Under these conditions, two processes overlapped: (i) the cytotoxicity of 2BC and its variants and (ii) permeabilization to the antibiotic HB and its consequent inhibition of yeast growth. These results on HB inhibition are discussed below.
The expression of all of the 2BC variants was analyzed by Western blotting of cell extracts obtained after 6 h of growth in YNB.LGal medium with anti-2B- or anti-2C-specific antisera (Fig. 2B). All constructs expressed the recombinant protein at similar levels, although variants 2b67 and 2bΔSpeI probably were not efficiently recognized by the anti-2B antibodies used. This result was not unexpected, since 2b67 and 2bΔSpeI could be immunoprecipitated in mammalian cells but showed weak or undetectable signals in Western blot analysis (1). The expression of other 2BC variants not shown in Fig. 2B has already been reported (6).
Enhanced membrane permeability induced by poliovirus 2BC in yeast cells.
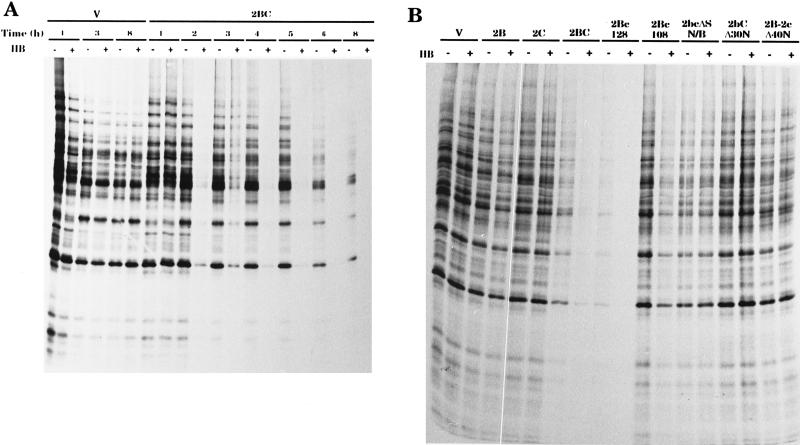
Poliovirus proteins 2B and 2BC enhance membrane permeability in mammalian cells, as demonstrated by the entry of HB, a nonpermeating inhibitor of translation. First, we analyzed membrane permeabilization induced by poliovirus proteins 2B, 2C, and 2BC using the HB test with yeast cells. 2BC strongly promoted the entry of HB into yeast cells soon after its induction (Fig. 3A). In contrast to mammalian cells, yeast cells were not permeabilized by 2B (Fig. 3B). This result is in agreement with the finding that 2BC enhances membrane permeability more strongly than 2B in mammalian cells. Other poliovirus proteins, such as 2C (Fig. 3B), 2A, which is strongly cytotoxic for yeast cells (7), 3A, and 3AB, had no effect on the entry of HB into yeast cells (results not shown for 2A, 3A, and 3AB).
FIG. 3.
Action of 2BC variants on HB permeabilization. (A) 2BC induces permeability changes. Protein synthesis in yeast cells bearing plasmids pEMBLyex4 (V) and pEMBL.2BC (2BC) in the presence (+) or absence (−) of HB (1 mM) was assayed. The samples were obtained as described in Materials and Methods at the postinduction times indicated in hours. (B) Effects of 2BC variants on membrane permeability. Protein synthesis in yeast cells expressing the 2BC variants indicated or bearing plasmid pEMBLyex4 (V) in the presence (+) or absence (−) of HB (1 mM) was assayed. The samples were obtained as described in Materials and Methods at 5 h postinduction. This experiment was performed with all of the mutants obtained, and the results of the densitometric analysis of the corresponding sodium dodecyl sulfate-polyacrylamide gel electrophoresis autoradiograms are included in Fig. 1.
The action of several 2BC variants on the inhibition of translation by HB in yeast cells was also assayed. To this end, two types of assays were used: growth in solid medium supplemented with 50 μg of HB per ml (Fig. 2A) or direct measurement of protein synthesis during 1 h of incubation in liquid medium containing [35S]methionine and 500 μg of HB per ml (Fig. 3B and summary in Fig. 1). The results obtained with these assays were slightly different, particularly for some variants, such as 2BX, 2B-2cΔ40N and, less drastically, 2B, which did not permeabilize cells at short times, while continued exposure rendered the cells susceptible to HB. This effect was enhanced when cells were replicated for a second time on YNB.Gal plates supplemented with HB. Thus, cells expressing 2B did not grow under these conditions, while control or 2C-expressing cells did (results not shown).
The requirement for an intact N terminus in both 2B and 2C for membrane permeabilization is documented by the lack of inhibition of protein synthesis with 2bCΔ30N and 2B-2cΔ40N (Fig. 3B). On the other hand, most of the 2C sequence is not required for permeabilization of the yeast membrane, since inhibition of [35S]methionine uptake with 2Bc128 was as strong as that with wild-type 2BC. The remaining point or deletion variants of 2B, which partially inhibited yeast growth, did not permeabilize cells to HB. As an exception, the 2bC-ND variant was as active as 2BC in promoting the entry of HB into cells. In summary, the integrity of both the amphipathic helix and the transmembrane domain is essential for membrane permeabilization, while the positively charged residues located just after the transmembrane domain are not required for this activity. It should be noted that 2BC was inhibitory for yeast translation and that this effect was more pronounced with 2Bc128. The molecular basis for the interference of 2BC or 2Bc128 with yeast protein synthesis remains to be investigated; it could be an indirect consequence of cell death.
Effects of 2BC variants on the exocytic pathway.
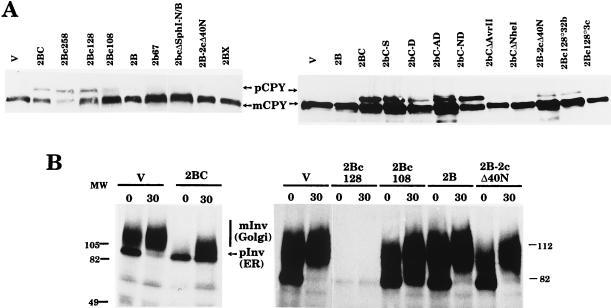
The action of 2BC and its variants on the exocytic pathway in yeast cells was assayed by analyzing the processing of CPY and invertase (Fig. 4). CPY is a vacuolar enzyme that is synthesized as a proenzyme in the ER, where it is core glycosylated (67 kDa). CPY is further glycosylated in the Golgi apparatus and is cleaved to generate the mature form of the enzyme (61 kDa) within the vacuole (5, 27). Yeast invertase is also synthesized as a precursor that is extensively processed in the ER (79 to 81 kDa) and in the Golgi apparatus (different forms of 100 to 140 kDa) to produce the mature enzyme, which is secreted into the medium (24, 25). In general, there was a good correlation between the enhancement of membrane permeability and the ability to block the exocytic pathway for most of the 2BC variants tested. The majority of the 2BC variants obtained affected CPY processing to different degrees (Fig. 4A). The expression of mutant 2Bc128 resulted in the accumulation of the CPY precursor form to levels similar to those obtained with wild-type 2BC. However, the synthesis of 2Bc108 led to only small amounts of the precursor form of CPY, while mutants 2B-2cΔ40N and 2BX had no effect on CPY processing. No accumulation of the CPY precursor form was seen with 2bcΔSphI(N/B) or with the 2B deletion variants, while the 2B point variants resulted in CPY precursor form accumulation to different degrees (Fig. 4A and summary in Fig. 1).
FIG. 4.
Action of 2BC variants on the exocytic pathway. (A) Extracts of yeast cells expressing the different 2BC variants indicated or bearing plasmid pEMBLyex4 (V) were obtained after 6 h of induction and assayed with antibodies against CPY. mCPY, mature CPY; pCPY, precursor form of CPY. (B) Yeast cells at 4 h postinduction in YNB.LGal medium were labeled for 10 with [35S]methionine and chased for 0 or 30 min. Invertase immunoprecipitation of control cells (bearing pEMBLyex4 [V]) or cells expressing the different 2BC variants indicated was performed as described in Materials and Methods. This pulse-chase experiment was carried out three times for mutant 2Bc128. In all cases, the amount of invertase precipitated was much smaller than that obtained with any other 2BC variant. mInv, mature invertase; pInv, precursor forms of invertase; MW, molecular weight (in thousands).
The results obtained for invertase processing (Fig. 4B) agreed well with those obtained for CPY processing. Only cells that expressed 2Bc128 accumulated the ER form of invertase. The small amounts of invertase immunoprecipitate detected in 2Bc128-expressing yeast cells might have been a consequence of the general inhibition of protein synthesis that occurs in these cells. In addition, these cells showed an altered ER, as observed by electron microscopy (see below); therefore, glycoprotein synthesis could have been particularly affected.
Modification of the ultrastructure of yeast cells by 2BC variants.
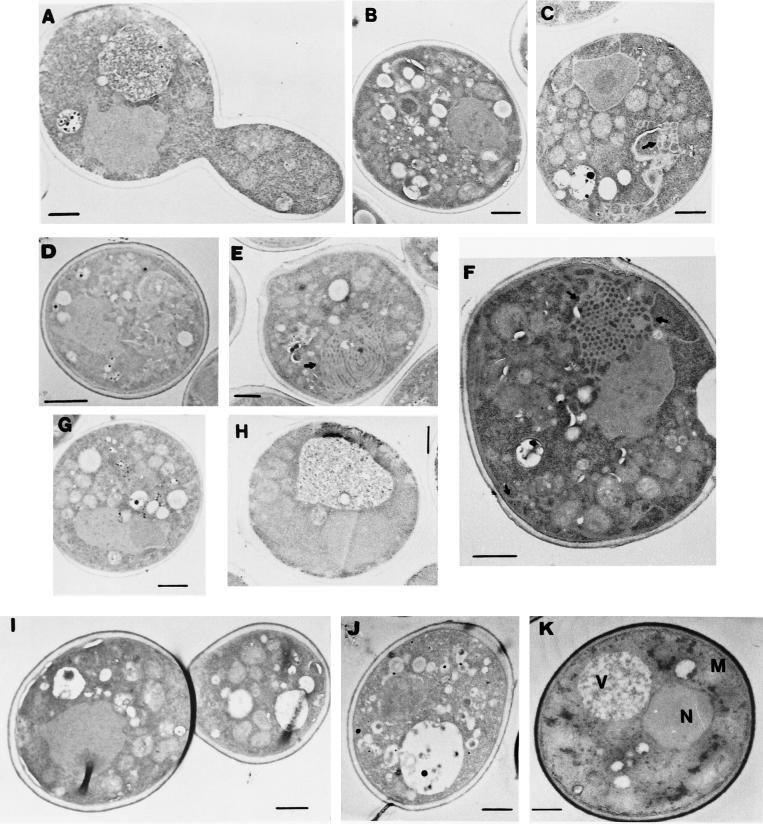
The ultrastructure of yeast cells that expressed some of the more interesting 2BC mutants was analyzed by electron microscopy (Fig. 5). The wild-type form of poliovirus 2BC induces the proliferation of a large number of cytoplasmic vesicles, together with the disruption of the yeast vacuole (Fig. 5B). Of the mutants tested, only 2bC-D (Fig. 5D) was able to reproduce the ultrastructural modifications found with 2BC, although it did so at a lower rate. Mutants 2Bc258 (Fig. 5C) and 2Bc128 (Fig. 5E and F), which showed the same behavior as wild-type 2BC in all of the previous assays, induced ultrastructural changes in yeast cells clearly different from those induced by 2BC. Protein 2Bc258 (Fig. 5C) induced the generation of vesicles in the cytoplasm that were larger (±500 nm) than those observed in 2BC-expressing cells (Fig. 5B). A similar phenotype was observed with 2Bc.EcoRI (results not shown). The expression of protein 2Bc128 (Fig. 5E and F) induced membranous structures that probably corresponded to ER swelling. Most yeast cells that expressed 2Bc128 had a cytoplasm occupied by honeycomb-like structures, where ER stacks were piled. This phenomenon has been described for the overexpression of several residential ER proteins, such as the HMG coenzyme A reductase (50) and cytochrome P-450 (49). On the contrary, cells expressing mutants 2Bc108 (Fig. 5G) and 2B-2cΔ40N (Fig. 5I) showed a phenotype more similar to that of control yeast cells. However, cells that synthesized 2Bc108 or 2B-2cΔ40N did not contain the large vacuole typical of control cells (Fig. 5K). Finally, cells expressing 2bCΔ30N (Fig. 5A) and 2B (Fig. 5H) looked like control yeast cells that did not express any poliovirus gene (Fig. 5K). Thus, the region of the 2C protein next to 2B dictates the type of membrane generated in yeast cells, while the N terminus of 2B is essential to induce structural changes.
FIG. 5.
Ultrastructure of yeast cells expressing different 2BC variants. Thin-section electron microscopy was carried out for yeast cells expressing 2bCΔ30N (A), 2BC (B), 2Bc258 (C), 2bC-D (D), 2Bc128 (E and F), 2Bc108 (G), 2B (H), 2B-2cΔ40N (I), or 2Bc128*3c (J) or transformed with plasmid pEMBLyex4 (K). Cells were chemically fixed at 20 h postinduction and were processed for electron microscopy as described in Materials and Methods. N, nucleus; V, vacuole; M, mitochondria. Black arrows indicate ER swelling. Bars, 500 nm.
Random mutagenesis of 2BC variants.
In principle, the inhibition of yeast cell growth induced by 2BC can be used as a genetic assay to obtain 2BC variants deficient in cytotoxicity. This approach has been successfully used in the case of poliovirus 2Apro (7), and it would be particularly interesting to obtain thermosensitive variants of poliovirus proteins that inhibit yeast cell growth.
The isolation of random mutants of 2BC has two main handicaps compared to that of 2Apro. (i) The 2BC gene is three times larger than the 2Apro gene, making identification of the potential 2BC variants isolated more cumbersome. For this reason, we decided not to work with the entire 2BC gene, using instead some of the shortest deletion mutants that were cytotoxic. (ii) A high frequency of cells resistant to 2BC cytotoxicity appear spontaneously. In principle, we cannot differentiate cells resistant to 2BC from cells bearing authentic mutations in the 2BC gene. The frequency of resistant mutants obtained can be reduced by use of basic pH-buffered plates and, to a lesser degree, by use of plates supplemented with HB. Thus, depending on the degree of stringency required, the kind of plate used in each assay varies.
Taking into account these considerations, we carried out three experiments to obtain mutants of 2BC. The results obtained are summarized in Table 1. The shortest 2BC cytotoxic mutant, 2bcΔSphI(N/B), was used in the first experiment. Because it has an attenuated cytotoxicity phenotype, the most restrictive conditions, i.e., plates buffered at pH 7.5, were used. For subsequent analyses, mutant 2Bc128 was used. This 2BC mutant is still easy to sequence, was even more toxic, and caused inhibition of the protein secretory pathway and increased membrane permeability comparable to those caused by wild-type 2BC.
TABLE 1.
Mutations identified in the genetic analysis of 2BC variants
| Expt | Mutant | Mutated nucleotide | Amino acid change | Toxicity at second transformationa |
|---|---|---|---|---|
| Ab | 1a | C→T | Q19Stop | − |
| 2a | C→T | Q20Stop | − | |
| 3a | —c | − | ||
| 4a | C→T | Q20Stop | − | |
| 5a | G→A | −ATG0d | − | |
| 8a | C→T | Q19Stop | − | |
| 9a | —c | − | ||
| 14a | C→T | Q20Stop | − | |
| 18a | C→T | Q20Stop | − | |
| 20a | C→T | Q20Stop | − | |
| Be | 2b | C→T | Q20Stop | − |
| 3b | G→A | Q97Stop | − | |
| 7b | C→T | Q20Stop | − | |
| 26b | C→T | E38Stop | − | |
| 30b | G→A | Q97Stop | − | |
| 32bf | Δ5bpi | 2b73+6 | ++ | |
| 34bg | − | |||
| Ch | 3cf | G→A | W78Stop | +++ |
| 4cg | − | |||
| 17cg | − | |||
| 18cg | − |
Yeast cells were streaked on YNB.Gal plates. +++, growth rate similar to that of control cells; ++, growth at a rate lower than that of control cells; −, no growth.
Random mutagenesis of pEMBL.2bcΔSphI(N/B) and selection with YNB. Gal (pH 7.5) plates. Of the 12,300 clones analyzed, 21 (0.17%) grew on YNB. Gal replica plates. Plasmid DNA could not be recovered from 11 of these 21; the plasmid present in the 10 remaining clones was sequenced.
Recombinant plasmids with an abnormal restriction pattern.
−ATG0, Mutant which has lost the initiation codon ATG (→GTG). Translation began at the next ATG, which was located by chance at the same reading frame. This mutant encodes a 2Bc128 variant without the first 30 amino acids; the reading frame starts at Met31.
Random mutagenesis of pEMBL.2Bc.128 and selection with YNB.Gal plates. Of the 4,700 clones analyzed, 54 (1.15%) grew on YNB.Gal replica plates. After discarding of false-positives and clones from which plasmid DNA could not be recovered, the six remaining clones were sequenced. In addition, we also sequenced a false-positive which showed the expression of a protein with a mobility slower than that of 2B in Western blot analysis.
False mutants that were cytotoxic at the second transformation. They were sequenced to identify the protein with a mobility slower than that of the 2B protein.
Proteins with no substitutions in the 2Bc.128 sequence but present at very low levels or not at all.
Random mutagenesis of pEMBL.2Bc.128 and selection with YNB.Gal plates supplied with 50 μg of HB per ml. Of the 2,000 clones analyzed, 18 (0.9%) grew on YNB.Gal replica plates. After discarding of false-positives and clones from which plasmid DNA could not be recovered, the three remaining clones were sequenced. In addition, we also sequenced a false-positive which showed the expression of a protein with a mobility slower than that of 2B in Western blot analysis.
Δ5bp, frameshift mutant encoding a truncated 2B protein with six residues (FTMAVA) and one stop codon after amino acid 73 of protein 2B.
To obtain noncytotoxic variants of both 2bcΔSphI(N/B) and 2Bc128, the same procedure was followed. (i) Yeast cells were transformed by an in vitro-mutated plasmid and growth on YNB.Glu plates. (ii) Replica plating on galactose plates was done in order to induce the synthesis of 2BC variants. (iii) The clones thus obtained were regrown on YNB.Gal plates to ensure that they were noncytotoxic. Some of them did not grow on the second plates and were classified as false mutants. (iv) Plasmid DNA and protein were extracted from every clone that showed the mutant phenotype. (v) Protein samples were blotted against an anti-2B antiserum to test the induction of the protein. (vi) E. coli cells were transformed with the DNA samples from the clones that exhibited an inducible 2BC variant. Sometimes no transformant colonies were obtained, suggesting that the plasmid in those yeast clones could not be analyzed. In other instances, the restriction pattern of the plasmid was abnormal as a result of the reorganization of plasmid DNA. (vii) The plasmids amplified in E. coli were used to retransform yeast cells. (viii) The ability of the new yeast clones to grow on galactose plates was assayed. This ability was lost for some of the clones; therefore, these clones, which showed a noncytotoxicity phenotype in the first yeast transformation but were cytotoxic in the second one (T2 in Table 1), really corresponded to yeast cells which spontaneously acquired resistance to 2BC cytotoxicity. (ix) The 2BC variant genes were sequenced from the clones in which the mutant phenotype (no cytotoxicity for yeast cells) was conserved in the second transformation.
The procedure described above yielded a number of poliovirus 2Apro variants readily (7). However, we did not obtain any point mutations that inactivated 2BC, even though more than 12,000 clones were analyzed (Table 1). This negative result could have been a consequence of the multiple mechanisms of 2BC cytotoxicity. Although one amino acid alteration inactivated one of the 2BC cytotoxic activities, the other toxic functions of the protein were still present in other regions of 2BC. Therefore, only multiple mutations would inactivate the cytotoxicity of 2BC.
When buffered plates were used, only those clones from which a plasmid was not recovered on bacteria or that expressed very short polypeptides were able to grow (Table 1, experiment A), while when less restrictive conditions for growth were used, more than 75% of the clones were false mutants (Table 1, experiments B and C). Two clones that expressed a deletion mutant with 96 amino acid residues were obtained from 2Bc128 (mutants 3b and 30b in Table 1). Thus, the protein encoded by these mutants was 2B, except for the absence of the last amino acid residue, the Gln at the C terminus. Cells that expressed this protein (2B lacking one amino acid) showed the same phenotype as 2B-expressing cells (results not shown). Two clones that expressed proteins shorter than 2B, as evidenced by Western blot analysis, were also obtained from 2Bc.1.28 mutagenesis (mutants 32b and 3c in Table 1). However, these clones were false mutants of the second transformation, since they inhibited yeast growth on galactose plates. These results indicate that deletion mutants shorter than 2B showed cytotoxic activity absent in 2B. We decided to analyze in detail the phenotype of yeast cells expressing these 2B deletion proteins.
Analysis of 2B deletion mutants.
The mutants obtained by random mutagenesis of pEMBL.2Bc128, known as 32b and 3c, showed a phenotype that was, in principle, unexpected. Although 2B alone has almost no effect on yeast growth, short deletions at the C terminus give rise to truncated 2B proteins that are cytotoxic (2Bc128*.32b and 2Bc128*.3c). Note that longer deletions of 10 or 26 additional residues generate nontoxic 2B variants (2b67 and 2bΔSpeI). The sequences of these mutants are shown in Fig. 6.
FIG. 6.
Sequences of 2B deletion mutants. Amino acid sequences from position 51 to the C terminus of the indicated mutants (Mut) are shown. Shaded sequences correspond to the wild-type 2B (2Bwt) sequence.
Characterization of the 2B deletion mutants included analyses of growth on solid and in liquid media, inhibition of the exocytic pathway, ultrastructure observed by electron microscopy, and effects on membrane permeability. The results of these experiments are shown in Fig. 1, 2, 4, and 5 in order to facilitate comparison with the results for the site-directed mutants. Three major effects were found after the induction of 32b and 3c expression: (i) yeast growth inhibition (Fig. 1 and 2A), with a more potent inhibitory effect of 3c than of 32b in liquid medium; (ii) enhanced membrane permeability, as tested with HB (Fig. 1); and (iii) low-level accumulation of the CPY precursor form by both mutants (Fig. 4). However, the 2B deletion mutants did not induce the membrane proliferation that was observed with 2BC or 2Bc128 (Fig. 5J).
These results led to the conclusion that 2B sequences are sufficient to inhibit yeast growth and to enhance membrane permeability in a manner similar to that observed with 2BC. These short 2B variants also exhibit a slight inhibition of the exocytic pathway. However, the structural changes observed with wild-type 2BC or 2Bc128 are not reproduced by the 2B deletion mutants. The fact that these 2B variants are cytotoxic suggests that not only the presence of a certain sequence but also the particular folding of that sequence is important to manifest the toxicity phenotype.
DISCUSSION
Activities of 2B, 2C, and 2BC proteins in yeast and human cells.
Our knowledge of the mode of action of picornavirus proteins 2B, 2C, and 2BC has benefited from the individual expression of the corresponding genes in both mammalian and yeast cells. These studies have provided evidence that 2B and 2BC block the exocytic pathway and interfere with the correct processing of glycoproteins (6, 22). In addition, 2B and 2BC enhance membrane permeability to low-molecular-weight compounds, such as uridine or HB, and to ions, such as calcium, in mammalian cells (1, 3, 22, 47, 48). These effects are more pronounced with 2BC than with 2B in the case of poliovirus (1, 3). Besides, poliovirus 2B is only slightly toxic for yeast, and only when its expression is prolonged for several generations is there an inhibition of yeast growth. This inhibition is clearly observed when HB is continuously present in the growth medium of 2B-expressing yeast cells. Therefore, yeast cells seem to be more resistant to the effects of 2B than mammalian cells, and only when a more potent protein such as 2BC is expressed are the cytotoxic and permeabilizing activities of the protein clearly observed. In addition, two noncytotoxic 2BC deletion mutants randomly isolated in yeast cells encoded proteins almost identical to 2B (variants 3b and 30b in Table 1).
The expression of 2C and 2BC in mammalian cells with the vaccinia virus VT7 system promotes the formation of different types of cytoplasmic membranous structures (2, 20). 2BC induces the proliferation of small vesicles similar to those observed in poliovirus-infected cells, while myelin-like sheaths and large vesicles are observed in 2C-expressing cells (2, 20). 2BC expression in yeast cells mimics all the effects observed in mammalian cells. On the other hand, deletion mutant 2Bc128 induces in yeast some structures generated by 2C in mammalian cells, while 2C expression in S. cerevisiae does not produce any particular phenotype. It is possible that a particular vaccinia virus gene complements or redirects 2C function to alter intracellular membrane formation in human cells. Therefore, additional studies on the effects of 2C in mammalian cells in the absence of any other viral infection are necessary.
Structure-activity relationships of 2BC.
The ease of manipulation of S. cerevisiae makes this microorganism a useful model system for understanding the mode of action of 2BC and facilitates genetic studies of this protein. The analysis of the effects of the different 2BC variants described in this work leads to a number of conclusions concerning the regions of 2BC involved in the different activities tested.
(i) Deletion of 30 residues at the N terminus of 2B [variants 2bCΔ30N and 2bc(31-258)] abolishes all the functions typical of 2BC. Most likely this region of 2B is involved in a process essential for 2BC function, such as folding, oligomerization, insertion in the membrane, and so forth. In addition, point mutations in this region of 2B cause defects in poliovirus RNA replication (8, 31).
(ii) Mutations targeted to the amphipathic helix of 2B, spanning residues 34 to 53 (e.g., 2bC-S or 2bC-D), as well as mutations in the second hydrophobic domain (residues 61 to 78) (e.g., 2bC-AD), can disrupt the ability of 2BC to modify membrane permeability and to inhibit protein secretion. However, deletion of the positively charged residues (amino acids 82 to 84) present just after the transmembrane domain of 2B (2bC-ND) is not important for these activities. These results are in agreement with the proposal that both the amphipathic helix and the hydrophobic domain of coxsackievirus 2B participate in these functions, traversing the lipid bilayer and leading to the formation of an aqueous pore contributed by both domains (48).
(iii) Progressive deletions at the C terminus of 2C have little or no effect on cytotoxicity, HB permeabilization, blockade of the exocytic pathway, or membrane proliferation but dictate the type of membrane generated. Only when the complete sequence of 2C is present in 2BC does the protein generate the typical vesicles found in poliovirus-infected cells. The phenotype found for 2BcΔXbaI or 2BcΔGKS (note that the point mutation in this domain did not impair the cytotoxic effects of 2BC) could be explained by considering that some internal regions of 2C are necessary for the correct folding of other domains involved in cytotoxicity. On the other hand, the presence of the N-terminal amphipathic helix of 2C (23) following the 2B sequence (2Bc128 variant) makes this protein even more cytotoxic than wild-type 2BC. Moreover, deletion of the initial 38 residues of 2C, containing this amphipathic helix, results in a 2BC variant (2B-2cΔ40N) devoid of all the activities tested. Therefore, this 2C region seems to be crucial to manifest 2BC cytotoxic activities.
(iv) The 3c and 32b variants obtained by random mutagenesis indicate that 2B sequences suffice to enhance membrane permeability and to interfere weakly with the exocytic pathway. These activities are hidden in the complete 2B protein or in the 2B-2cΔ40N variant. These findings could be rationalized if the C terminus of 2B had inhibitory activity over the cytotoxic functions located in the other two thirds of 2B. The interaction of the C-terminal region of 2B with the N-terminal region of 2C makes the protein active and therefore toxic, as has been observed with wild-type 2BC and all the other cytotoxic variants.
(v) Although there are 2BC variants, such as 2bC-AD and 2bC-D, that are defective in blocking the exocytic pathway and enhancing membrane permeability, both activities can be separated. Thus, mutants 3c and 32b enhance membrane permeability but exert only marginal inhibition of protein trafficking. Mutant 2bC-S shows the opposite behavior. The results obtained with the coxsackievirus 2B mutants (48) are similar. An explanation of these results is that the different 2BC variants could be preferentially targeted to different membranes (ER, Golgi apparatus, or plasma membrane) and, once there, the activity of each variant is dictated by its particular structure.
Functioning of 2B, 2C, and 2BC in the poliovirus replicative cycle.
The 2BC precursor is very abundant in cells infected by poliovirus, producing the mature products 2B and 2C after 3Cpro cleavage. It now seems clear that 2BC has activities that are not present or are clearly diminished in the mature products 2B and 2C. This assertion is particularly clear when yeast cells are assayed. It seems logical to propose that the initial generation of 2BC targets this protein to the ER, redirecting membrane trafficking toward the formation of small vesicles. The formation of these membranous vesicles is a requisite for viral RNA synthesis. Therefore, polioviruses mutated in 2B or 2C are deficient in replicating viral RNA. Coxsackievirus 2B has been described as a viroporin, because of its ability to enhance membrane permeability, perhaps by pore formation, thus facilitating the release of progeny virions (47, 48). In addition, coxsackievirus 2B (47) and poliovirus 2BC (3) enhance intracellular calcium levels, a process that could facilitate the assembly of new virions. In conclusion, the release of 2B from the 2BC precursor may locate this protein at the plasma membrane to act as a viroporin. The generation of mature 2C could lead to the transport of mature viral RNA bound to the protein to special locations where the virion assembly process takes place. In fact, some polioviruses mutated in 2C show defects in the assembly of new virions (34).
ACKNOWLEDGMENTS
The expert technical assistance of M. A. Sanz and M. T. Rejas is acknowledged. We thank F. Were and J. A. Lewis for critical reading of the manuscript and R. Adabe for his collaboration and stimulating discussions.
A.B. holds a CSIC postdoctoral fellowship. We acknowledge financial support provided by DGICYT project PB94-0148 and an institutional grant to CBM from Fundación Ramón Areces.
REFERENCES
- 1.Aldabe R, Barco A, Carrasco L. Membrane permeabilization by poliovirus proteins 2B and 2BC. J Biol Chem. 1996;271:23134–23137. doi: 10.1074/jbc.271.38.23134. [DOI] [PubMed] [Google Scholar]
- 2.Aldabe R, Carrasco L. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem Biophys Res Commun. 1995;206:64–76. doi: 10.1006/bbrc.1995.1010. [DOI] [PubMed] [Google Scholar]
- 3.Aldabe R, Irurzun A, Carrasco L. Poliovirus protein 2BC increases cytosolic free calcium concentrations. J Virol. 1997;71:6214–6217. doi: 10.1128/jvi.71.8.6214-6217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baltera R F J, Tershak D R. Guanidine-resistant mutants of poliovirus have distinct mutations in peptide 2C. J Virol. 1989;63:4441–4444. doi: 10.1128/jvi.63.10.4441-4444.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banta L M, Robinson J S, Klionsky D J, Emr S D. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988;107:1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barco A, Carrasco L. A human virus protein, poliovirus protein 2BC, induces membrane proliferation and blocks the exocytic pathway in the yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3349–3364. doi: 10.1002/j.1460-2075.1995.tb07341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barco A, Ventoso I, Carrasco L. The yeast Saccharomyces cerevisiae as a genetic system for obtaining variants of poliovirus protease 2A*. J Biol Chem. 1997;272:12683–12691. doi: 10.1074/jbc.272.19.12683. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein H D, Sarnow P, Baltimore D. Genetic complementation among poliovirus mutants derived from an infectious cDNA clone. J Virol. 1986;60:1040–1049. doi: 10.1128/jvi.60.3.1040-1049.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bienz K, Egger D, Pasamontes L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology. 1987;160:220–226. doi: 10.1016/0042-6822(87)90063-8. [DOI] [PubMed] [Google Scholar]
- 10.Bienz K, Egger D, Rasser Y, Bossart W. Kinetics and location of poliovirus macromolecular synthesis in correlation to virus-induced cytopathology. Virology. 1980;100:390–399. doi: 10.1016/0042-6822(80)90530-9. [DOI] [PubMed] [Google Scholar]
- 11.Bienz K, Egger D, Rasser Y, Bossart W. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology. 1983;131:39–48. doi: 10.1016/0042-6822(83)90531-7. [DOI] [PubMed] [Google Scholar]
- 12.Bienz K, Egger D, Troxler M, Pasamontes L. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J Virol. 1990;64:1156–1163. doi: 10.1128/jvi.64.3.1156-1163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caliguiri L A, Tamm I. Membranous structures associated with translation and transcription of poliovirus RNA. Science. 1969;166:885–886. doi: 10.1126/science.166.3907.885. [DOI] [PubMed] [Google Scholar]
- 14.Carrasco L. Membrane leakiness after viral infection and a new approach to the development of antiviral agents. Nature. 1978;272:694–699. doi: 10.1038/272694a0. [DOI] [PubMed] [Google Scholar]
- 15.Carrasco L. Modification of membrane permeability by animal viruses. Adv Virus Res. 1995;45:61–112. doi: 10.1016/S0065-3527(08)60058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrasco L, Castrillo J L. The regulation of translation in picornavirus-infected cells. In: Carrasco L, editor. Mechanisms of viral toxicity in animal cells. Boca Raton, Fla: CRC Press, Inc.; 1987. pp. 115–146. [Google Scholar]
- 17.Carrasco L, Otero M J, Castrillo J L. Modification of membrane permeability by animal viruses. Pharmacol Ther. 1989;40:171–212. doi: 10.1016/0163-7258(89)90096-x. [DOI] [PubMed] [Google Scholar]
- 18.Cesareni G, Murray J A H. Plasmid vectors carrying the replication origin of filamentous single-stranded phages. Genet Eng. 1987;9:135–153. [Google Scholar]
- 19.Cho M W, Richards O C, Dmitrieva T M, Agol V, Ehrenfeld E. RNA duplex unwinding activity of poliovirus RNA-dependent RNA polymerase 3Dpol. J Virol. 1993;67:3010–3018. doi: 10.1128/jvi.67.6.3010-3018.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho M W, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 21.Dales S, Eggers H J, Tamm I, Palade G E. Electron microscopic study of the formation of poliovirus. Virology. 1965;26:379–389. doi: 10.1016/0042-6822(65)90001-2. [DOI] [PubMed] [Google Scholar]
- 22.Doedens J R, Kirkegaard K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995;14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Echeverri A C, Dasgupta A. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology. 1995;208:540–553. doi: 10.1006/viro.1995.1185. [DOI] [PubMed] [Google Scholar]
- 24.Esmon B, Novick P, Schekman R. Compartmentalized assembly of oligosaccharides on exported glycoproteins in yeast. Cell. 1981;25:451–460. doi: 10.1016/0092-8674(81)90063-5. [DOI] [PubMed] [Google Scholar]
- 25.Franzusoff A, Schekman R. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorbalenya A E, Koonin E V, Wolf Y I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 27.Graham T R, Scott P A, Emr S D. Brefeldin A reversibly blocks early but not late protein transport steps in the yeast secretory pathway. EMBO J. 1993;12:869–877. doi: 10.1002/j.1460-2075.1993.tb05727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guinea R, Carrasco L. Phospholipid biosynthesis and poliovirus genome replication, two coupled phenomena. EMBO J. 1990;9:2011–2016. doi: 10.1002/j.1460-2075.1990.tb08329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irurzun A, Arroyo J, Alvarez A, Carrasco L. Enhanced intracellular calcium concentration during poliovirus infection. J Virol. 1995;69:5142–5146. doi: 10.1128/jvi.69.8.5142-5146.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irurzun A, Perez L, Carrasco L. Enhancement of phospholipase C activity during poliovirus infection. J Gen Virol. 1993;74:1063–1071. doi: 10.1099/0022-1317-74-6-1063. [DOI] [PubMed] [Google Scholar]
- 31.Johnson K, Sarnow P. Three poliovirus 2B mutants exhibit noncomplementable defects in viral RNA amplification and display dosage-dependent dominance over wild-type poliovirus. J Virol. 1991;65:4341–4349. doi: 10.1128/jvi.65.8.4341-4349.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenk R, Penman S. The cytoskeletal framework and poliovirus metabolism. Cell. 1979;16:289–301. doi: 10.1016/0092-8674(79)90006-0. [DOI] [PubMed] [Google Scholar]
- 33.Li J P, Baltimore D. Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J Virol. 1988;62:4016–4021. doi: 10.1128/jvi.62.11.4016-4021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J P, Baltimore D. An intragenic revertant of a poliovirus 2C mutant has an uncoating defect. J Virol. 1990;64:1102–1107. doi: 10.1128/jvi.64.3.1102-1107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirzayan C, Wimmer E. Biochemical studies on poliovirus polypeptide 2C: evidence for ATPase activity. Virology. 1994;199:176–187. doi: 10.1006/viro.1994.1110. [DOI] [PubMed] [Google Scholar]
- 36.Pincus S E, Diamond D C, Emini E A, Wimmer E. Guanidine-selected mutants of poliovirus: mapping of point mutations to polypeptide 2C. J Virol. 1986;57:638–646. doi: 10.1128/jvi.57.2.638-646.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pincus S E, Wimmer E. Production of guanidine-resistant and -dependent poliovirus mutants from cloned cDNA: mutations in polypeptide 2C are directly responsible for altered guanidine sensitivity. J Virol. 1986;60:793–796. doi: 10.1128/jvi.60.2.793-796.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodríguez P L, Carrasco L. Poliovirus protein 2C has ATPase and GTPase activities. J Biol Chem. 1993;268:8105–8110. [PubMed] [Google Scholar]
- 39.Rodríguez P L, Carrasco L. Poliovirus protein 2C contains two regions involved in RNA binding activity. J Biol Chem. 1995;270:10105–10112. doi: 10.1074/jbc.270.17.10105. [DOI] [PubMed] [Google Scholar]
- 40.Rose M D, Winston F, Hieter P. Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Sandoval I V, Carrasco L. Poliovirus infection and expression of the poliovirus protein 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro-090179. J Virol. 1997;71:4679–4693. doi: 10.1128/jvi.71.6.4679-4693.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlegel A, Giddings T H, Ladinsky M S, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonenberg N. Poliovirus translation. Curr Top Microbiol Immunol. 1990;161:23–47. doi: 10.1007/978-3-642-75602-3_2. [DOI] [PubMed] [Google Scholar]
- 45.van Kuppeveld F J M, Galama J M D, Zoll J, Van den Hurk P J J C, Melchers W J G. Coxsackie B3 virus protein 2B contains a cationic amphipathic helix that is required for viral RNA replication. J Virol. 1996;70:3876–3886. doi: 10.1128/jvi.70.6.3876-3886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Kuppeveld F J M, Galama J M D, Zoll J, van den Hurk J J C, Melchers W J G. Genetic analysis of a hydrophobic domain of coxsackie B3 virus protein 2B: a moderate degree of hydrophobicity is required for a cis-acting function in viral RNA synthesis. J Virol. 1995;69:7782–7790. doi: 10.1128/jvi.69.12.7782-7790.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Kuppeveld F J M, Hoenderop J G J, Smeets R L L, Willems P H G M, Kijkman H B P M, Galama J M D, Melchers W J G. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 1997;16:3519–3532. doi: 10.1093/emboj/16.12.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Kuppeveld F J M, Melchers W J G, Kirkegaard K, Doedens J R. Structure-function analysis of coxsackie B3 virus protein 2B. J Virol. 1997;227:111–118. doi: 10.1006/viro.1996.8320. [DOI] [PubMed] [Google Scholar]
- 49.Wiedmann B, Silver P, Schunck W-H, Wiedmann M. Overexpression of the ER-membrane protein P-450 CYP52A3 mimics sec mutant characteristics in Saccharomyces cerevisiae. Biochim Biophys Acta. 1993;1153:267–276. doi: 10.1016/0005-2736(93)90415-v. [DOI] [PubMed] [Google Scholar]
- 50.Wright R, Basson M, D’Ari L, Rine J. Increased amounts of HMG-CoA reductase induce “Karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J Cell Biol. 1988;107:101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yaffe M P, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]