Abstract
Polypeptides of amino acids 1 to 241 (PVR241) and 1 to 330 (PVR330) of the human poliovirus receptor (hPVR) were produced in a baculovirus expression system. PVR241 contained extracellular domains 1 and 2 of hPVR, and PVR330 contained extracellular domains 1, 2, and 3. These peptides were purified by immunoaffinity column chromatography with an anti-hPVR monoclonal antibody (MAb). After the purification, PVR241 and PVR330 appeared to retain their native conformation as judged by reactivity with an anti-PVR MAb that recognized domain 1 of hPVR in a conformation-dependent manner. The virulent Mahoney strain of poliovirus type 1 was mixed with the purified PVRs in various concentrations. An average of at least 43 PVR330 molecules were able to bind to one virion particle under the conditions used. The equilibrium dissociation constant between the PVR330 molecule and the PVR binding site (canyon) on the virion was determined to be 4.50 ± (0.86) × 10−8 M at 4°C. Higher rates of conformational change of the virus (160S) to 135S and 80S particles were observed as the concentration of PVR330 was increased. In this in vitro system, the ratio of the amount of the 135S particle to that of the 80S particle seemed to be always constant. After the disappearance of the 160S particle, the amount of the 80S particle was not increased by further incubation at 37°C. These results suggested that the 80S particle was not derived from the 135S particle under the conditions used in this study.
Poliovirus, the causative agent of poliomyelitis, is a human enterovirus that belongs to the family Picornaviridae. The precise three-dimensional structure of the virion particle was elucidated by crystallographic studies (16). The nonenveloped capsid consists of 12 pentamers, each of which is composed of five protomers. A protomer is formed by three surface proteins, VP1, VP2, and VP3, and the internal protein VP4. A fivefold axis formed by the five capsid protein VP1s exists at the center of each pentamer. Surrounding each fivefold axis is a deep cleft, termed a canyon (35), that is proposed to attach to the poliovirus receptor (PVR) on the surface of permissive cells. These structural investigations suggest that each protomer carries a single attachment site for PVR, resulting in 60 PVR binding sites per virion. Indeed, experimental evidence involving soluble ICAM-1 and human rhinovirus, another member of the Picornaviridae, strongly suggests that 60 attachment sites for ICAM-1 per virion exist (17).
The genomic and complementary DNAs for human PVR (hPVR) have been isolated from HeLa cells (23, 31). hPVR is a member of the immunoglobulin (Ig) superfamily, with three linked extracellular Ig-like domains of V-C2-C2, followed by a membrane-spanning domain and a cytoplasmic domain. Analysis of the alternative splicing products of the hPVR gene has revealed that there are at least four mRNA isoforms. Two membrane-bound forms (hPVRα and hPVRδ) and two secreted forms (hPVRβ and hPVRγ) are potentially expressed in human cells (23). All of the isoforms have an identical nucleotide sequence encoding a signal peptide and the three Ig-like domains.
The extracellular domain of hPVR contains eight putative N-linked glycosylation sites. Some or all of the sites were suggested to be linked to sugar chains (3). Indeed, antibodies against hPVR detected membrane-associated molecules of 70 to 80 kDa (3, 23), although the molecular masses of hPVRα and hPVRδ calculated from the deduced amino acid sequences were approximately 45 and 43 kDa, respectively. Molecular genetic analysis has revealed that the poliovirus binding site resides in the N-terminal Ig-like domain (domain 1) (24, 32, 37) and that sugar moieties possibly attached to this domain are dispensable for the virus-receptor interaction leading to virus infection (25, 43). Furthermore, key amino acids that influence the binding of poliovirus type 1 have been identified (2, 4, 33).
It is well known that the susceptible cells convert the 160S infectious poliovirion to a 135S particle, sometimes called the A particle, which has lost the internal protein VP4 and is not an infectious particle (28). This conversion of the virion particle is considered to result in virus uncoating, that is, the formation of an 80S particle which has lost the RNA genome in addition to the VP4. A similar alteration of poliovirus particles is seen when susceptible cell extracts are mixed with poliovirus (9–11, 15). Conversion of poliovirus is also observed when the virus is mixed with nonsusceptible insect cells infected with a recombinant baculovirus carrying a cDNA encoding hPVR (20). The in vitro conversion product of 135S is indistinguishable from that observed in the early stage of productive infection in cultured cells (41). These observations suggest that the hPVR molecule itself triggers the uncoating process of poliovirus. To rule out the possibility that another molecule(s), in addition to PVR, contributes to the uncoating process of the virus, the effect of highly purified PVR on viral conformation should be tested. To date, only preliminary experiments involving a purified recombinant fusion protein, which carries the extracellular domain of hPVR and the Fc domain of human IgG, suggest that hPVR has an ability to convert the 160S infectious particle to 135S and 80S particles during incubation at 37°C (26).
The interaction of poliovirus with its cellular binding sites has been investigated by using a receptor-excess assay (6). Binding of poliovirus to HeLa cells fits a theoretical simple bimolecular noncooperative binding curve with an equilibrium dissociation constant (Kd) of 4.3 × 107 cells/ml at 4°C, or 2.1 × 10−10 M, assuming 3,000 virus binding sites per cell. This value, however, may not represent a real dissociation constant between hPVR and the attachment site (canyon), since there must be multiple PVR binding sites on a virion particle and since PVR may exist in small clusters on the cell surface (29). In this study, we produced extracellular portions of hPVRs by using a baculovirus expression system and purified them by an affinity purification method. The dissociation constant (Kd) for a recombinant PVR and a PVR binding site on the virion particle was calculated to be 4.50 × 10−8 ± 0.86 × 10−8 M at 4°C. The effect of the concentration of a recombinant PVR on the rate of the viral conformational change was investigated. Here we describe that PVR itself has an ability to convert 160S intact virions to 135S or 80S particles and that the 80S particle is not derived from the 135S particle under the reaction conditions used.
MATERIALS AND METHODS
Cells and viruses.
Suspension-cultured HeLa S3 cells were grown in RPMI 1640 medium supplemented with 5% newborn calf serum (NCS), and monolayer HeLa S3 cells were grown in Dulbecco modified Eagle’s medium supplemented with 5% NCS. These cells were used for preparation of poliovirus. African green monkey kidney cells were cultured in Dulbecco modified Eagle’s medium supplemented with 5% NCS and used for plaque assay (21). BmN insect cells were cultured in TC-100 medium supplemented with 10% fetal calf serum at 27°C and used for preparation of recombinant baculoviruses (Bombyx mori nuclear polyhedrosis viruses). Recombinant baculoviruses were amplified by passaging three times and used as inocula for preparation of recombinant PVRs. For preparation of recombinant PVRs, BmN cells were cultured in serum-free medium (EX-CELL 400; JRH Biosciences). The Mahoney strain of type 1 poliovirus, PV1(M), was produced in African green monkey kidney cells or HeLa cells transfected with an infectious RNA transcribed from the cDNA clone PV1(M)OM (39).
Purification of poliovirus.
HeLa S3 cells were infected with PV1(M) at a multiplicity of infection (MOI) of 10. The cells were collected at 7 h postinfection, and the virus was purified from cytoplasmic extracts of the infected cells by using DEAE-Sepharose CL-6B (Pharmacia Biotech) (18) followed by centrifugations on a sucrose density gradient and a CsCl density gradient as previously described (19). Purified virus was desalted by gel filtration on a PD-10 column (Pharmacia Biotech) equilibrated with phosphate-buffered saline (PBS) (10 mM phosphate buffer [pH 7.0], 137 mM NaCl, and 2.6 mM KCl). The concentration of poliovirions was determined by measuring absorbance at 260 nm, where 1.0 optical density unit is regarded as being equivalent to 9.4 × 1012 virions (36). The virus solutions were supplemented with bovine serum albumin (BSA) (Initial Fractionation by Heat Shock; Sigma) to give a final concentration of 1% and stored at −80°C. For preparation of [35S]methionine- and [35S]cysteine-labeled poliovirus, HeLa S3 cells in monolayer culture (2.0 × 107 cells) were infected with poliovirus at an MOI of 10. At 2 h postinfection, the medium was changed to methionine- and cysteine-free medium containing 4 mCi of [35S]methionine and [35S]cysteine (EXPRE35S35S Protein Labeling Mix, [35S]-; NEN). The labeled poliovirus was purified as described above.
Preparation and purification of recombinant PVRs.
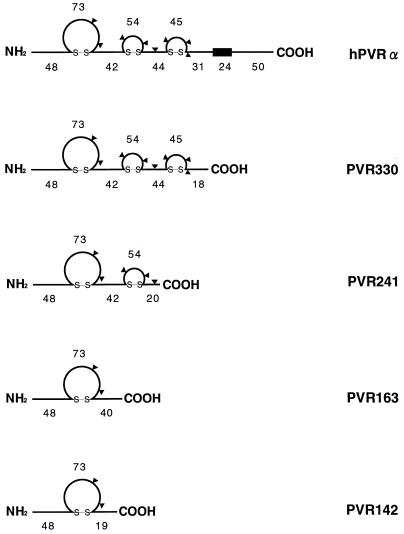
Recombinant baculoviruses were prepared by cotransfection of BmN cells with baculovirus DNA and a transfer vector (pBM.050) that carries a PVR cDNA encoding amino acids 1 to 142, 1 to 163, 1 to 241, or 1 to 330 of hPVR. Recombinant virus that did not produce the polyhedrin protein was chosen by limiting dilution of the virus (1). To produce hPVR extracellular domains in large numbers, BmN cells (107) were infected with recombinant baculoviruses at an MOI of 5, and the culture fluid collected at 60 to 66 h postinfection was subjected to centrifugation at 1,000 × g for 5 min at room temperature. The supernatant was filtered through a membrane filter (0.22-μm pore size; Millipore) and separated by immunoaffinity column chromatography (30) with the anti-PVR monoclonal antibody (MAb) p286, which recognizes domain 1 of hPVR in a conformation-dependent manner (42). Recombinant PVRs were eluted with 50 mM glycine-HCl (pH 3.0), neutralized immediately by the addition of 1/20 volume of 1 M Tris-HCl (pH 7.5), and concentrated with an Ultrafree CL concentrator (UFC4 LGC 25; Millipore). Recombinant PVRs carrying up to 142, 163, 241, and 330 amino acid residues from the N terminus of hPVR were designated PVR142, PVR163, PVR241, and PVR330, respectively (Fig. 1). For labeling recombinant PVRs, the medium was replaced at 24 h postinfection by methionine-free medium containing 4 mCi of [35S]methionine and [35S]cysteine (EXPRE35S35S Protein Labeling Mix, [35S]-), and the culture fluid was harvested at 60 to 66 h postinfection. The purification procedure for [35S]methionine-labeled recombinant PVRs was the same as that for unlabeled recombinant PVRs described above.
FIG. 1.
Schematic structures of recombinant PVRs expressed in a baculovirus system. The number of amino acids in each region is shown for hPVRα and recombinant PVRs. Possible N-linked glycosylation sites are indicated by triangles. The box represents a transmembrane domain.
The concentrations of purified recombinant PVRs in solution were determined by measuring the absorbance at 280 nm, taking their amino acid compositions into consideration (34), as described previously for soluble ICAM-1 (30). Western blot analysis was also employed for estimating amounts of PVRs (see below). Concentrations of [35S]methionine-labeled recombinant PVRs in solutions were determined by enzyme-linked immunosorbent assay (ELISA) (see below). In this expression system, 40 to 80 μg of purified PVRs was obtained from 5 × 107 BmN cells.
Gel filtration.
Purified PVRs were analyzed by gel filtration on Superdex 200 PC3.2/30 (Pharmacia Biotech) in PBS with a SMART System (Pharmacia Biotech). A low- and high-molecular-weight gel filtration calibration kit (Pharmacia Biotech) was used to estimate molecular weights according to the instructions of the manufacturer. Complexes of PVR330 and virion-related particles were analyzed on Sephacryl S-300 superfine.
Measurement of equilibrium dissociation constant.
Anti-PV1(M) MAb 7m012, which was able to bind to PV1(M) but not able to neutralize the virus, was purified by protein A-Sepharose column chromatography and adjusted to a concentration of 0.25 mg/ml in PBS. Each well of a microtitration plate (Immulon 2; Dynatech Laboratories) was treated with 50 μl of MAb 7m012 solution, kept at room temperature for 2 h, and washed with PBS twice. The wells were filled with 150 μl of 3% BSA in PBS, incubated at 4°C overnight, and washed three times with Tris-buffered saline (TBS) (20 mM Tris-HCl [pH 7.5], 137 mM NaCl) containing 0.05% Tween 20 (TBS-0.05T). The poliovirus suspension (100 μl/well) was added to the wells in a concentration range of 0.35 × 10−9 to 1.75 × 10−9 M and incubated at 4°C overnight. The wells were washed three times with TBS-0.05T and then treated with recombinant PVR (100 μl/well) in a concentration range of 2.29 × 10−8 to 21.2 × 10−8 M. After incubation at 4°C for 24 h, the solution (containing unbound recombinant PVR) was removed, and the radioactivity associated with the well (recombinant PVR bound to the well) was recovered in a 100-μl solution containing 0.2 M NaOH and 1% sodium dodecyl sulfate. The radioactivities of unbound PVR and PVR associated with the well were measured in a liquid scintillation counter. Approximately 2.5% of the total unbound-PVR radioactivity usually remained in the well after the removal of the solution. This value was used to revise the amount of PVR associated with the well. The Kd value was determined by plotting the data in a standard Scatchard plot.
A similar experiment was carried out to determine the number of PVR330 molecules bound per virion. In this case, however, the poliovirus suspension was added at a concentration of 0.18 × 10−9 M, and the PVR was at up to 4.3 × 10−7 M.
Sucrose density gradient centrifugation.
Poliovirions were incubated with recombinant PVR molecules in PBS containing 1% BSA in a total volume of 250 μl at 4°C overnight. After a temperature shift from 4 to 37°C, the mixtures were incubated for 0 to 60 min, applied to a 15 to 30% sucrose density gradient in PBS containing 0.1% BSA, and centrifuged at 39,000 rpm for 2 h at 4°C in a Beckman SW41 rotor. After fractionation, the radioactivity of each fraction (0.6 ml) was measured in a liquid scintillation counter. Native poliovirions (160S) and 80S particles prepared as reported by Lonberg-Holm et al. (28) and Kaplan et al. (20) were used as markers.
Western blot analysis.
Western blot analysis was performed by using a culture fluid of hybridoma cells which produced anti-PVR MAb 5D1 for quantification of purified PVRs. MAb 5D1 was prepared by Aoki et al. (2), and recognizes domain 1 of hPVR in a conformation-independent manner. The samples were subjected to 15% polyacrylamide gel electrophoresis in a Laemmli buffer system (27). The proteins in the gel were transferred to a polyvinylidene difluoride filter (Immobilon; Millipore) and blocked as described previously (39). The filters were incubated with MAb 5D1 solution (1:10 dilution) at room temperature for 1 h in TBS containing 0.5% Tween 20 (TBS-0.5T) and 2% nonfat dry milk. The filters were washed with TBS-0.5T three times for 5 min each, incubated with sheep anti-mouse IgG antibodies conjugated with horseradish peroxidase (1:1,000 dilution; Amersham Life Science) at room temperature for 1 h, and washed with TBS-0.5T five times for 5 min each. The filters were then treated with the ECL detection reagents (Amersham Life Science) for 1 min, and densities of visible bands were quantified by using a GS-525 Molecular Imager (Bio-Rad) and Molecular Analyst (Bio-Rad).
To investigate the components of poliovirus and its related particles, a similar Western blot analysis was performed with a mixture (1:1) of rabbit hyperimmune serum against PV1(M) virion and the capsid protein VP4 derived from the Sabin 1 strain of type 1 poliovirus.
ELISA.
ELISA was carried out as previously described (22, 40). Each well of a microtitration plate (Immulon 2; Dynatech Laboratories) was treated with 50 μl of purified anti-PVR MAb p286 (1:100 dilution of a 1-mg/ml solution) (42) at room temperature for 2 h. After washing with PBS, 150 μl of 3% BSA in PBS was added to each well and incubated at 4°C overnight. The wells were washed with TBS-0.05T three times and incubated with 100 μl of twofold serial dilutions of purified recombinant PVRs in PBS containing 1% BSA per well at 4°C overnight. The wells were washed with TBS-0.05T three times and incubated with rabbit hyperimmune serum against amino acid residues 32 to 46 of hPVR (1:1,000 dilution) at room temperature for 2 h. The wells were washed with TBS-0.05T three times, incubated with 100 μl of donkey anti-rabbit IgG antibodies conjugated with horseradish peroxidase (1:1,000 dilution; Amersham Life Science) per well at room temperature for 2 h, and washed five times. To each well was then added 100 μl of enzyme substrate solution (pH 5.0) containing 0.05 M citric acid, 0.1 M dibasic sodium phosphate, 0.04% ortho-phenylenediamine, and 0.006% H2O2. The enzyme reaction was performed in the dark at 37°C for 10 min and stopped by the addition of 50 μl of 2 M H2SO4 per well. The absorbance at 492 nm was measured for each well. Relative concentrations of recombinant PVR solutions were estimated by comparison with a standard curve that was obtained in a parallel experiment using a PVR preparation whose concentration had been determined by measuring absorbance at 280 nm as described above.
RESULTS
Purification of recombinant PVRs.
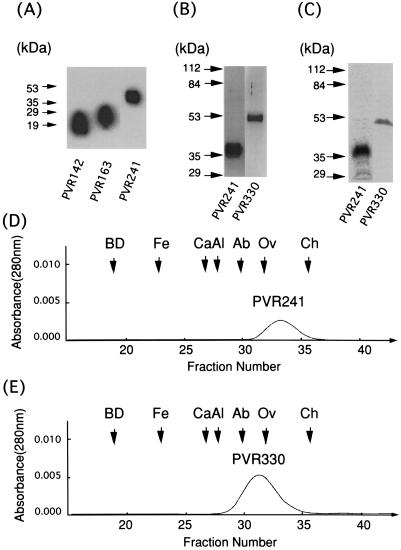
Recombinant PVRs, PVR142, PVR163, PVR241, and PVR330, were produced in BmN insect cells infected with the corresponding recombinant baculoviruses (Fig. 1) and purified by immunoaffinity column chromatography. Western blot analysis with MAb 5D1 was carried out on the preparation of PVR142, PVR163, or PVR241 after the purification process. As shown in Fig. 2A, only one protein was detected for each PVR preparation. Molecular masses calculated from the deduced amino acid sequences of these recombinant PVRs that lacked a putative signal peptide portion (27 amino acid residues) (5) were 12.8, 15.1, and 23.4 kDa, respectively; these values are smaller than those obtained from the data shown in Fig. 2A. These discrepancies in molecular mass may result from glycosylation occurring during the expression process. Similar observations were made for PVR330, whose calculated molecular mass was 33.1 kDa (data not shown; see Fig. 2B and C).
FIG. 2.
Purity of recombinant PVRs expressed by a baculovirus system. Recombinant PVRs were expressed in a baculovirus system and purified by immunoaffinity column chromatography as described in Materials and Methods. (A) Western blot analysis of purified PVR142, PVR163, and PVR241. (B) Silver staining of purified PVR241 and PVR330. (C) Autoradiography of [35S]methionine-labeled PVR241 and PVR330. (Positions of molecular mass markers are indicated on the left of panels A to C.) PVR241 (D) and PVR330 (E) were analyzed by a gel filtration. BD, Blue Dextran 2000; Fe, ferritin (440 kDa); Ca, catalase (232 kDa); Al, aldolase (158 kDa); Ab, albumin (67 kDa); Ov, ovalbumin (43 kDa), and Ch, chymotrypsinogen A (25 kDa).
Although PVR142 and PVR163 were observed in Western blot analysis with MAb 5D1 (Fig. 2A), these PVRs were not detected in an ELISA with MAb p286, which recognizes domain 1 of hPVR in a conformation-dependent manner, whereas PVR241 and PVR330 were detected in a similar assay (data not shown). This suggests that PVR142 and PVR163 had lost their native conformation during the affinity purification. These two recombinant PVRs were therefore not used for further studies.
To estimate the purities of PVR241 and PVR330, these recombinant PVRs were analyzed by polyacrylamide gel electrophoresis followed by silver staining (Fig. 2B) and autoradiography (Fig. 2C). Densitometric analyses of the stained gel and the autoradiography indicated that the purity of either PVR241 or PVR330 was more than 97% (data not shown). The data shows the high degree of purity of the 35S-labeled and unlabeled recombinant PVRs, PVR241 and PVR330, used in this study.
Gel filtration analyses were also employed for estimating the molecular masses of PVR241 and PVR330. As shown in Fig. 2D and E, the molecular masses of PVR241 and PVR330 were 36 and 63 kDa, respectively; those values were comparable to the values obtained by gel electrophoreses (Fig. 2B and C). The data also suggested that these molecules were present in monomer form in PBS.
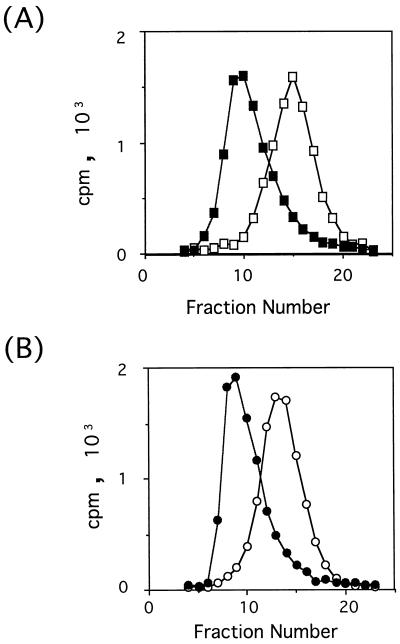
Reactivities of purified PVR241 and PVR330 with MAb p286 were examined by mixing 35S-labeled recombinant PVRs with the MAb followed by separation of the immune complexes by gel filtration column chromatography (Fig. 3). Peaks of those PVRs are shifted in the presence of MAb p286. The data indicate that almost all recombinant PVRs are recognized by MAb p286. Thus, the native conformation of domain 1 seems to have been retained in PVR241 and PVR330.
FIG. 3.
Recognition of purified PVR241 and PVR330 by MAb p286. 35S-labeled PVR241 (A) or PVR330 (B) was incubated with MAb p286 (closed symbols) or without MAb p286 (open symbols) in PBS containing 1% BSA at 4°C for 24 h. The final concentrations of PVRs and MAb p286 were 6.06 × 10−8 and 5.60 × 10−7 M, respectively. The mixtures were analyzed by chromatography on Sephacryl S-300 superfine (Pharmacia). The chromatography was carried out in PBS at 4°C. Radioactivities in aliquots of fractions were measured in a liquid scintillation counter.
PVR binding sites on virion particles.
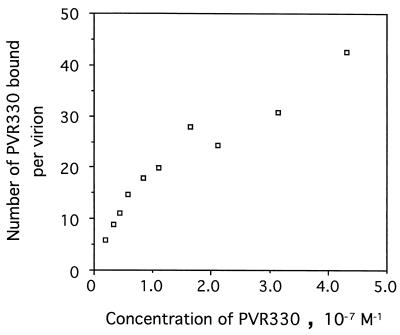
Structural studies of poliovirus have strongly suggested that 60 PVR binding sites are present on each virion particle. To prove this, PV1(M) was fixed to the well of a microtitration plate and reacted with various concentrations of PVR330 up to 4.3 × 10−7 M. The amount of PV1(M) bound per well was 1.33 × 10−14 mol. As shown in Fig. 4, the number of PVR330 molecules bound per virion increased with increasing concentrations of PVR330. The maximum average number of PVR330 molecules bound was 43 per virion in this experiment. Most probably, more PVR330 could bind per virion if a higher concentration of PVR330 was used; the data shown in Fig. 4 suggested that the virion surface was not yet saturated with PVR330 molecules. Thus, it is likely that a virion particle has the capacity to bind 60 PVR330 molecules. The binding affinity (see below) appeared not to be sufficiently high to demonstrate 60 PVR330 molecules bound per virion at the highest PVR330 concentration available for this study.
FIG. 4.
Number of PVR330 molecules bound per virion. Poliovirions (approximately 1.33 × 10−14 mol per well) bound to the well by means of MAb 7m012 were reacted with 35S-labeled PVR330 in various concentrations as described in Materials and Methods. Numbers of bound PVR330 molecules were calculated by using the specific activity of the labeled compound.
Measurement of Kd between poliovirus and recombinant PVR.
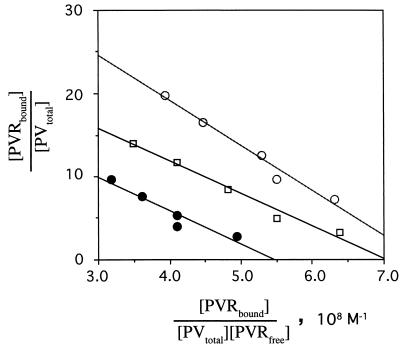
To estimate the affinity between a recombinant PVR330 molecule and a PVR binding site on PV1(M), the equilibrium dissociation constant (Kd) was measured at 4°C by two methods. The initial method involved separation of PVR bound to PV1(M) from unbound PVR on a gel filtration column. This method, however, gave a variety of Kd values of approximately 10−7 (data not shown), probably because of a low affinity between PVR and its binding site. PVR molecules were most likely released from virion particles during the experimental procedure. We therefore tried a second method that removed unbound PVR by washing. Washing the well after the binding reaction also appeared to result in release of PVR that had bound to PV1(M). Washing was omitted; the reaction supernatant was simply aspirated, and a correction factor for the lack of washing was accounted for in the calculation of binding as described in Materials and Methods. The affinity between the recombinant PVR and PV1(M) clearly seemed not to be very high.
Three different amounts of PV1(M) and various concentrations of PVR330 were used for binding reactions as described in Materials and Methods. As shown in Fig. 5, the Scatchard analysis indicates a fairly constant Kd value of 4.50 × 10−8 ± 0.86 × 10−8 M at 4°C under any of the conditions used here. When 2.60 × 10−14 mol of PV1(M) was bound to the well, the number of PVR330 molecules bound per PV1(M) particle (the point of intersection on the y axis) is about 40, which is close to that observed in Fig. 4. The number of PVR molecules bound per virion decreased as the number of PV1(M) particles bound to the well increased.
FIG. 5.
Kd value for recombinant PVR330 and its binding site on poliovirions. Poliovirions were fixed to the bottom of the well by means of MAb 7m012. Poliovirion concentrations of 1.31 × 10−13, 6.67 × 10−14, and 2.60 × 10−14 mol bound per well are indicated by closed circles, open squares, and open circles, respectively. Poliovirions in the well were incubated with various concentrations of PVR330 (67,000 cpm; adjusted to a final concentration of 2.29 × 10−8 to 21.2 × 10−8 M by the addition of unlabeled PVR) as described in Materials and Methods. Two independent experiments were performed.
PV1(M) was fixed to the bottom of the well by means of MAb 7m012. This fixation method may have reduced the numbers of virion binding sites available for PVR binding. In addition, virion particles may aggregate at higher concentrations. Indeed, the higher concentrations of PV1(M) used here resulted in clumping (data not shown), which may lower the number of free PVR binding sites per virion. In any case, all of the available binding sites on the virion appeared to have the same affinity for PVR.
Conformational change of poliovirus.
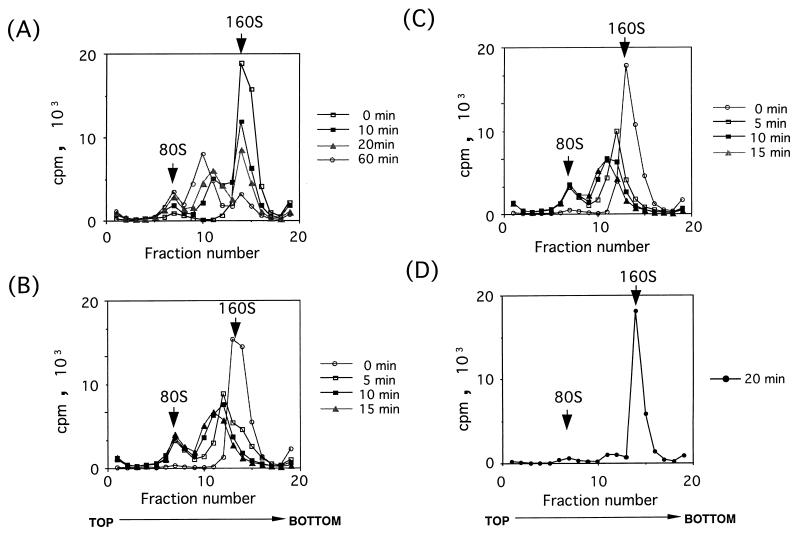
To examine whether intact poliovirions (160S) are converted to 135S and 80S particles by incubation with purified PVR330, PV1(M) was mixed with different concentrations of purified PVR330 at 4°C, followed by incubation at 37°C. As shown in Fig. 6A, 80S and 135S-like particles appeared and increased during the incubation at 37°C, while only slight increases of those particles were observed in the absence of PVR330 (Fig. 6D). Similar virus conversion was observed when the number of PVR330 molecules bound per virion was around 5 (data not shown). The data indicated that recombinant PVR330 itself is capable of virus conversion from the 160S particle to the smaller particles. The conversion rate was accelerated by using higher ratios of PVR molecules to PV1(M) (Fig. 6). Thus, a higher number of PVR330 molecules bound per virion seemed to result in a faster conformational change. Interestingly, the amount of 80S particle was constant during incubation at 37°C after the 160S particle had disappeared, and the ratio of 80S particle to 135S-like particle was also unchanged (Fig. 6C). Similar phenomena are shown in Fig. 6B. This data suggested that the 80S particle is not derived from the 135S peaks observed in the gradients.
FIG. 6.
Effect of concentration of PVR330 on rate of conformational change of poliovirions. [35S]methionine- and [35S]cysteine-labeled poliovirions were incubated with PVR330 at 37°C, and the conformational changes were analyzed at the indicated times by using sucrose density gradient centrifugation as described in Materials and Methods. Concentrations of 7.8 × 108 virions (1.8 × 105 cpm) per μl (A and D) and 3.9 × 108 virions (1.2 × 105 cpm) per μl (B and C) were mixed with 1.92 × 10−8 M (A), 3.37 × 10−8 M (B), 10.1 × 10−8 M (C), and no (D) PVR330. Fractions are numbered from the top to bottom of gradients. Positions of 80S and 160S particles are indicated by arrows.
Components in fractions of sucrose gradients.
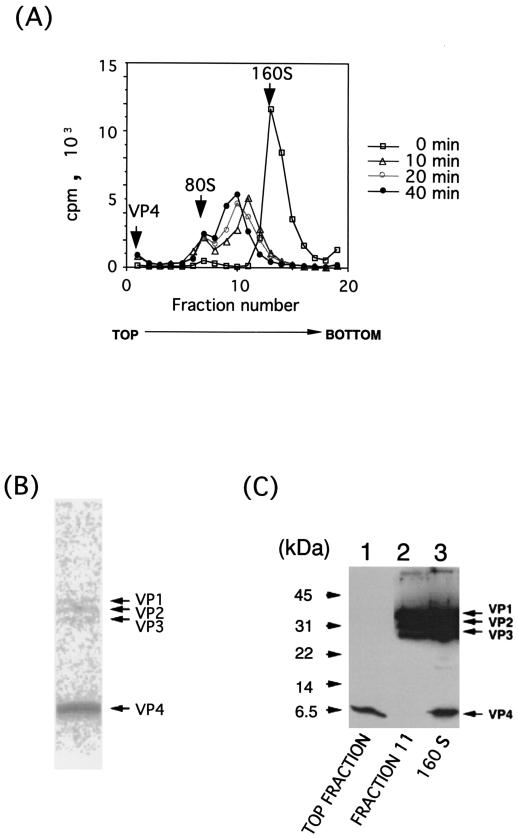
Ranging in size between the 160S and 80S particles, several forms of particles (135S-like particles) existed in sucrose gradient fractions (Fig. 6 and 7A). Because their S values appear not to be identical, the components of these particles may be different from those of the 135S particles so far observed. Particles from sucrose gradient fractions 9, 10, and 11, outlined in Fig. 7A, were disrupted and analyzed by Western blotting. The data for fraction 11 is shown in Fig. 7C, lane 2. The 135S-like particles appeared to have VP1, VP2, and VP3 but not VP4 when the pattern was compared with that of 160S particle (Fig. 7C, lane 3). Similar results were obtained for the other fractions. Thus, these particles seemed to be composed of the same materials as the 135S particle (41).
FIG. 7.
Materials in fractions of sucrose density gradients. (A) [35S]methionine- and [35S]cysteine-labeled poliovirus (1.4 × 105 cpm) at a concentration of 1.01 × 108 virions per ml was mixed with 4.13 × 10−8 M PVR, incubated at 37°C for indicated times, and analyzed under the same conditions described in the legend for Fig. 6. Positions of VP4, 80S particles, and 160S particles are indicated by arrows. (B) Autoradiography of top fractions (fractions 1, 2, and 3) after separation by polyacrylamide gel electrophoresis. In this fraction, radioactivities of VP1, VP2, VP3, and VP4 were 0.99, 1.77, 1.73, and 95.5% of the total radioactivity, respectively. (C) Materials in the top fraction (lane 1) and fraction 11 (lane 2) and 160S intact virion particles (lane 3) were analyzed by Western blot analysis with a mixture (1:1) of rabbit hyperimmune serum against poliovirus type 1 and its capsid protein VP4.
Viral capsid protein VP4, which was missing in the conformationally altered particles, must exist at the top fraction of the gradient. To confirm this, the radioactive materials of the top fraction were analyzed by polyacrylamide gel electrophoresis followed by autoradiography (Fig. 7B). The data showed that most of the radioactivity was due to VP4. Densitometric analysis demonstrated that more than 95% of the radioactivity was VP4, and VP4 in the top fraction was also detected by Western blot analysis (Fig. 7C, lane 1).
PVR binding to virion-related particles.
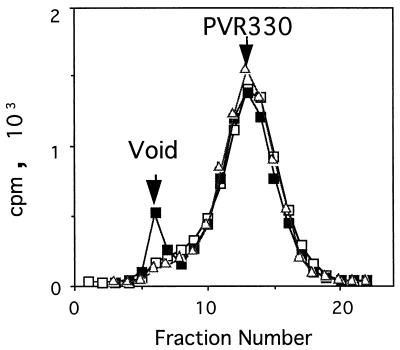
To determine whether PVR330 stays bound to the virion-related particles after the conversion from the 160S particle, the binding of PVR330 to 135S and 80S particles was examined (Fig. 8). 35S-labeled PVR330 was mixed with unlabeled 160S particles under the conditions used for the experiments described in Fig. 6C, incubated at 37°C for 20 min, and further incubated at 4°C for 20 min. At the end of the first incubation, all of the 160S particles should have been converted to 135S and 80S particles. After the reaction, the mixture was separated by gel filtration with Sephacryl S-300 superfine (Pharmacia Biotech) (Fig. 8). As a control, 35S-labeled PVR330 was incubated with unlabeled 160S particles and treated similarly. As shown in Fig. 8, a peak at the void volume was observed only when PVR330 was incubated with the 160S particle. The data indicated that PVR330 binds only to the 160S particle, suggesting that PVR330 is released from the virion-related particles after the conversion of the 160S particle.
FIG. 8.
Gel filtration of mixtures of PVR330 and virion-related particles. PVR330 was incubated at 4°C with 160S intact poliovirions (▪) or their conversion products (135S) (□) and analyzed by gel filtration. ▵, PVR330 alone.
Possible reduction of PVR activity during the purification procedure.
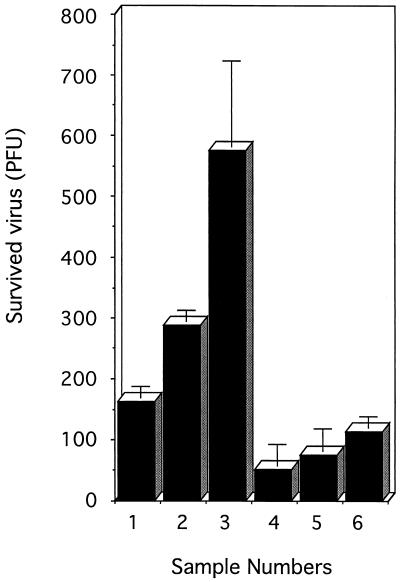
Domain 1 of purified PVR330 appeared to be intact as judged by its reactivity with MAb p286 (Fig. 3). However, virus conversion activity may have been reduced during elution at pH 3.0 from the immunoaffinity column. To exclude this possibility, the pH of PVR330 culture fluid from baculovirus-infected cells was raised to 3.0, and the fluid was examined for its virus-neutralizing activity (Fig. 9). The culture fluid was incubated with 1.5 × 106 PFU of PV1(M) at 37°C for 1 h (Fig. 9, bar 1). Residual activity of the virus was less than 200 PFU. These remaining infectious particles might have been PVR-resistant poliovirus mutants (7). The number of residual infectious particles appeared to increase when 0.9 and 0.8 volumes of the culture fluid were used in similar experiments (Fig. 9, bars 2 and 3). The data strongly suggested that the virus-neutralizing activity of the culture fluid was marginal; the conditions used for the experiment shown in Fig. 9, bar 1, are useful to examine the possible reduction of PVR330 virus-neutralizing activity during the purification procedure. The culture fluids treated at pH 3.0 for 1, 3, or 5 min at 4°C were adjusted to neutral pH, dialyzed against PBS, and examined for their virus-neutralizing activities (Fig. 9, bars 4, 5, and 6). Fewer than 200 PFU remained after incubation with the treated fluids. The data indicated that reduction of the virus conversion activity by the acid treatment was negligible. Similar results were obtained for the culture fluid containing PVR241. The exposure time for recombinant PVRs under acidic conditions was less than 5 min and should not have affected the virus conversion activities of PVR241 and PVR330.
FIG. 9.
Neutralizing activity of culture fluids containing PVR330. Culture fluid (134 μl [bar 1], 121 μl [bar 2], or 107 μl [bar 3]) containing PVR330 was mixed with 1.5 × 106 PFU of PV1(M) in PBS containing 1% BSA and incubated at 4°C for 3 h and then at 37°C for 1 h. Plaque assay was performed as described in Materials and Methods. Similar experiments with 134 μl of the culture fluid were carried out after the treatment of the fluid at pH 3.0 in a glycine-HCl buffer at 4°C for 1 (bar 4), 3 (bar 5), and 5 (bar 6) min, neutralization, and dialysis against PBS. Experiments were performed in triplicate, and error bars indicate standard deviations.
DISCUSSION
Reduction of poliovirus infectivity mediated by two N-terminal domains of hPVR has been reported (44). In this study, we have demonstrated that the purified extracellular region of hPVR itself has an activity that changes the conformation of the intact poliovirion (160S) to 135S-like and 80S particles. Previous studies (24, 32, 37, 38) have shown that domains 2 and 3 of hPVR are dispensable for establishment of binding and infection of poliovirus. PVR142 and PVR163, which carried only domain 1 of hPVR, however, failed to retain their intact conformation during the purification procedure used in this study. Purified PVR142 and PVR163 did not show significant virus-neutralizing activity (data not shown), whereas purified PVR241 (data not shown) and PVR330 (Fig. 9) did. Thus, the virus conversion activity appeared to be mainly due to the intact conformation of hPVR domain 1. Our data indicated that domain 2 is necessary for maintenance of domain 1 conformation, at least during the purification process.
Based on the results of structural studies (16), as many as 60 PVR binding sites reside on the poliovirus particle, although 43 PVR molecules was the observed maximum average number that bound per virion (Fig. 3). A higher ratio of PVR molecules to virion particles resulted in a faster conformational change of the virions. Acceleration of the virion conversion rate by increasing the concentration of soluble receptor has been reported for human rhinovirus (14, 17). Furthermore, our data strongly suggested that, after the conversion, PVR is released from the virion-related particles. According to our preliminary experiments, the released PVR330 still appeared to have virion conversion activity. It is therefore possible that the virion conversion activity of PVR330 is catalytic.
The Kd value for poliovirus binding sites on HeLa cells and poliovirions has been calculated to be 2.1 × 10−10 M at 4°C (6), assuming 3,000 binding sites on HeLa cells. In this study, the Kd between recombinant PVR and the PVR binding site on poliovirus was determined to be 4.50 × 10−8 ± 0.86 × 10−8 M at 4°C. Thus, the value obtained here is much higher than that previously reported. Possibly more than two PVR molecules per virion mediate poliovirus binding to HeLa cells. Several PVRs in a cluster may contribute to binding of a virion to the cell, because PVRs are reported to be present in small clusters on the cell surface (29). It should be noted that oligomerization of purified PVR was not observed in this study (Fig. 2D and E). It is possible that purified recombinant PVR has a reduced virus binding activity compared with the native PVR present on the cell surface, although virus conversion activity appeared not to be lowered during the purification process (Fig. 9). Other domains, such as a transmembrane domain and/or a cytoplasmic domain, might be required in part for the virus binding activity of hPVR. In addition, the activity may be influenced by glycosylation that differs in mammalian cells and insect cells.
In vitro experiments involving purified recombinant PVR and virion particles demonstrated that the 80S particle was not formed from the 135S particle after the 160S particle disappeared. This indicated that the 80S particle is formed directly from 160S intact virions. Although the 135S particle has long been thought not to be infectious, Curry et al. (8) have recently described that poliovirus 135S particles can infect Chinese hamster ovary cells and murine L cells. Neither of those cell types are susceptible to infection by native poliovirus, because they lack a PVR. The 135S infectivity may be associated with the N terminus of VP1, which is exposed on the surface of the 135S particle and has been shown to mediate interactions of the particle with lipid membranes (13). These results suggested that the poliovirus 135S particle is a potential intermediate in the cell entry pathway. There may be a pathway in virus conversion from the 135S particle to the 80S particle in these cells. If this is the case, a cellular factor(s) other than PVR may be required for the conversion. The experimental system described here might be a powerful tool to search for such a possible factor(s).
Recent studies involving cold-adapted poliovirus mutants (12) have demonstrated that the mutants do not convert to 135S particles at 25°C and that the particle-to-PFU ratio of poliovirus does not change at 25°C in the absence of the conformational alteration. Furthermore, mutation sites that influenced the phenotype were identified in the RNA encoding the noncapsid protein 2C. The results do not support a model that a transition to the 135S particle is obligatory for poliovirus entry into cells. The relationship between virus conformational change and establishment of infection is not yet clear. Poliovirus infection may be established by any of several pathways, such as PVR-mediated fusion resulting in release of the viral genome into the cell cytoplasm, PVR-mediated endocytosis followed by uncoating in endosomes, and PVR-independent penetration and uncoating. Indeed, our preliminary results suggested that the PVR-independent pathway serves to establish poliovirus infection (data not shown). All these pathways may exist together in cultured cells susceptible to poliovirus. Investigations of individual pathways will give insights into the molecular mechanisms that drive the early events of poliovirus infection.
ACKNOWLEDGMENTS
We are grateful to S. Kuge, K. Shiroki, and H. Toyoda for helpful suggestions and discussions. We thank Y. Sasaki and K. Iwasaki for expert technical assistance and E. Suzuki and M. Watanabe for help in preparation of the manuscript.
This work was supported in part by a grant-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan and the Ministry of Health and Welfare of Japan and by the Science and Technology Agency of Japan.
REFERENCES
- 1.Aoki J, Koike S, Asou H, Ise I, Suwa H, Tanaka T, Miyasaka M, Nomoto A. Mouse homolog of poliovirus receptor-related gene 2 product, mPRR2, mediates homophilic cell aggregation. Exp Cell Res. 1997;235:374–384. doi: 10.1006/excr.1997.3685. [DOI] [PubMed] [Google Scholar]
- 2.Aoki J, Koike S, Ise I, Sato-Yoshida Y, Nomoto A. Amino acid residues on human poliovirus receptor involved in interaction with poliovirus. J Biol Chem. 1994;269:8431–8438. [PubMed] [Google Scholar]
- 3.Bernhardt G, Bibb J A, Bradley J, Wimmer E. Molecular characterization of the cellular receptor for poliovirus. Virology. 1994;199:105–113. doi: 10.1006/viro.1994.1102. [DOI] [PubMed] [Google Scholar]
- 4.Bernhardt G, Harber J, Zibert A, deCrombrugghe M, Wimmer E. The poliovirus receptor: identification of domains and amino acid residues critical for virus binding. Virology. 1994;203:344–356. doi: 10.1006/viro.1994.1493. [DOI] [PubMed] [Google Scholar]
- 5.Bibb J A, Bernhardt G, Wimmer E. Cleavage site of the poliovirus receptor signal sequence. J Gen Virol. 1994;75:1875–1881. doi: 10.1099/0022-1317-75-8-1875. [DOI] [PubMed] [Google Scholar]
- 6.Bibb J A, Witherell G, Bernhardt G, Wimmer E. Interaction of poliovirus with its cell surface binding site. Virology. 1994;201:107–115. doi: 10.1006/viro.1994.1270. [DOI] [PubMed] [Google Scholar]
- 7.Colston E, Racaniello V R. Soluble receptor-resistant poliovirus mutants identify surface and internal capsid residues that control interaction with the cell receptor. EMBO J. 1994;13:5855–5862. doi: 10.1002/j.1460-2075.1994.tb06930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curry S, Chow M, Hogle J M. The poliovirus 135S particle is infectious. J Virol. 1996;70:7125–7131. doi: 10.1128/jvi.70.10.7125-7131.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Sena J, Mandel B. Studies on the in vitro uncoating of poliovirus. I. Characterization of the modifying factor and the modifying reaction. Virology. 1976;70:470–483. doi: 10.1016/0042-6822(76)90288-9. [DOI] [PubMed] [Google Scholar]
- 10.De Sena J, Mandel B. Studies on the in vitro uncoating of poliovirus. II. Characteristics of the membrane-modified particle. Virology. 1977;78:554–566. doi: 10.1016/0042-6822(77)90130-1. [DOI] [PubMed] [Google Scholar]
- 11.De Sena J, Torian B. Studies on the in vitro uncoating of poliovirus. III. Roles of membrane-modifying and -stabilizing factors in the generation of subviral particles. Virology. 1980;104:149–163. doi: 10.1016/0042-6822(80)90373-6. [DOI] [PubMed] [Google Scholar]
- 12.Dove A W, Racaniello V R. Cold-adapted poliovirus mutants bypass a postentry replication block. J Virol. 1997;71:4728–4735. doi: 10.1128/jvi.71.6.4728-4735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fricks C E, Hogle J M. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol. 1990;64:1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greve J M, Forte C P, Marlor C W, Meyer A M, Hoover-Litty H, Wunderlich D, McClelland A. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J Virol. 1991;65:6015–6023. doi: 10.1128/jvi.65.11.6015-6023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttman N, Baltimore D. A plasma membrane component able to bind and alter virions of poliovirus type 1: studies on cell-free alteration using a simplified assay. Virology. 1977;82:25–36. doi: 10.1016/0042-6822(77)90029-0. [DOI] [PubMed] [Google Scholar]
- 16.Hogle J M, Chow M, Filman D J. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 17.Hoover-Litty H, Greve J M. Formation of rhinovirus-soluble ICAM-1 complexes and conformational change in the virion. J Virol. 1993;67:390–397. doi: 10.1128/jvi.67.1.390-397.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horodniceanu F, Panon G, Le Fur R, Barme M. Purification of poliovirus obtained from human diploid cells by means of DEAE-Sepharose. Dev Biol Stand. 1979;42:75–80. [PubMed] [Google Scholar]
- 19.Kajigaya S, Arakawa H, Kuge S, Koi T, Imura N, Nomoto A. Isolation and characterization of defective-interfering particles of poliovirus Sabin 1 strain. Virology. 1985;142:307–316. doi: 10.1016/0042-6822(85)90339-3. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan G, Freistadt M S, Racaniello V R. Neutralization of poliovirus by cell receptors expressed in insect cells. J Virol. 1990;64:4697–4702. doi: 10.1128/jvi.64.10.4697-4702.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawamura N, Kohara M, Abe S, Komatsu T, Tago K, Arita M, Nomoto A. Determinants in the 5′ noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J Virol. 1989;63:1302–1309. doi: 10.1128/jvi.63.3.1302-1309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohara M, Omata T, Kameda A, Semler B L, Itoh H, Wimmer E, Nomoto A. In vitro phenotypic markers of a poliovirus recombinant constructed from infectious cDNA clones of the neurovirulent Mahoney strain and the attenuated Sabin 1 strain. J Virol. 1985;53:786–792. doi: 10.1128/jvi.53.3.786-792.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koike S, Horie H, Ise I, Okitsu A, Yoshida M, Iizuka N, Takeuchi K, Takegami T, Nomoto A. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 1990;9:3217–3224. doi: 10.1002/j.1460-2075.1990.tb07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koike S, Ise I, Nomoto A. Functional domains of the poliovirus receptor. Proc Natl Acad Sci USA. 1991;88:4104–4108. doi: 10.1073/pnas.88.10.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koike S, Ise I, Sato Y, Yonekawa H, Gotoh O, Nomoto A. A second gene for the African green monkey poliovirus receptor that has no putative N-glycosylation site in the functional N-terminal immunoglobulin-like domain. J Virol. 1992;66:7059–7066. doi: 10.1128/jvi.66.12.7059-7066.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koike S, Ise I, Sato Y, Mitsui K, Horie H, Umeyama H, Nomoto A. Early events of poliovirus infection. Semin Virol. 1992;3:109–115. [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lonberg-Holm K, Gosser L B, Kauer J C. Early alteration of poliovirus in infected cells and its specific inhibition. J Gen Virol. 1975;27:329–342. doi: 10.1099/0022-1317-27-3-329. [DOI] [PubMed] [Google Scholar]
- 29.Mannweiler K, Nobis P, Hohenberg H, Bohn W. Immunoelectron microscopy on the topographical distribution of the poliovirus receptor. J Gen Virol. 1990;71:2737–2740. doi: 10.1099/0022-1317-71-11-2737. [DOI] [PubMed] [Google Scholar]
- 30.Martin S, Martin A, Staunton D E, Springer T A. Functional studies of truncated soluble intercellular adhesion molecule 1 expressed in Escherichia coli. Antimicrob Agents Chemother. 1993;37:1278–1285. doi: 10.1128/aac.37.6.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendelsohn C L, Wimmer E, Racaniello V R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 32.Morrison M E, Racaniello V R. Molecular cloning and expression of a murine homolog of the human poliovirus receptor gene. J Virol. 1992;66:2807–2813. doi: 10.1128/jvi.66.5.2807-2813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrison M E, He Y-J, Wien M W, Hogle J M, Racaniello V R. Homolog-scanning mutagenesis reveals poliovirus receptor residues important for virus binding and replication. J Virol. 1994;68:2578–2588. doi: 10.1128/jvi.68.4.2578-2588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pace C N, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossmann M G, Arnold E, Erickson J W, Frankenberger E A, Griffith J P, Hecht H-J, Johnson J E, Kamer G, Luo M, Mosser A G, Rueckert R R, Sherry B, Vriend G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 36.Rueckert R R. On the structure and morphogenesis of picornaviruses. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. Vol. 6. New York, N.Y: Plenum Press; 1976. pp. 131–213. [Google Scholar]
- 37.Selinka H-C, Zibert A, Wimmer E. Poliovirus can enter and infect mammalian cells by way of an intercellular adhesion molecule 1 pathway. Proc Natl Acad Sci USA. 1991;88:3598–3602. doi: 10.1073/pnas.88.9.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selinka H-C, Zibert A, Wimmer E. A chimeric poliovirus/CD4 receptor confers susceptibility to poliovirus on mouse cells. J Virol. 1992;66:2523–2526. doi: 10.1128/jvi.66.4.2523-2526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shiroki K, Ishii T, Aoki T, Kobashi M, Ohka S, Nomoto A. A new cis-acting element for RNA replication within the 5′ noncoding region of poliovirus type 1 RNA. J Virol. 1995;69:6825–6832. doi: 10.1128/jvi.69.11.6825-6832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Marel P, Hazendonk T G, Henneke M A C, van Wezel A L. Induction of neutralizing antibodies by poliovirus capsid polypeptides VP1, VP2, and VP3. Vaccine. 1983;1:17–22. doi: 10.1016/0264-410x(83)90007-5. [DOI] [PubMed] [Google Scholar]
- 41.Yafal A G, Kaplan G, Racaniello V R, Hogle J M. Characterization of poliovirus conformational alteration mediated by soluble cell receptors. Virology. 1993;197:501–505. doi: 10.1006/viro.1993.1621. [DOI] [PubMed] [Google Scholar]
- 42.Yang W-X, Terasaki T, Shiroki K, Ohta S, Aoki J, Tanabe S, Nomura T, Terada E, Sugiyama Y, Nomoto A. Efficient delivery of circulating poliovirus to the central nervous system independently of poliovirus receptor. Virology. 1997;229:421–428. doi: 10.1006/viro.1997.8450. [DOI] [PubMed] [Google Scholar]
- 43.Zibert A, Wimmer E. N glycosylation of the virus binding domain is not essential for function of the human poliovirus receptor. J Virol. 1992;66:7368–7373. doi: 10.1128/jvi.66.12.7368-7373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zibert A, Selinka H-C, Elroy-Stein O, Wimmer E. The soluble form of two N-terminal domains of the poliovirus receptor is sufficient for blocking viral infection. Virus Res. 1992;25:51–61. doi: 10.1016/0168-1702(92)90099-u. [DOI] [PubMed] [Google Scholar]