Abstract
The past decade has seen an increase in the prevalence of sequence type (ST) 45 methicillin-resistant Staphylococcus aureus (MRSA), yet the underlying drivers for its emergence and spread remain unclear. To better understand the worldwide dissemination of ST45 S. aureus, we performed phylogenetic analyses of Australian isolates, supplemented with a global population of ST45 S. aureus genomes. Our analyses revealed a distinct lineage of multidrug-resistant ST45 MRSA harbouring qacA, predominantly found in Australia and Singapore. Bayesian inference predicted that the acquisition of qacA occurred in the late 1990s. qacA was integrated into a structurally variable region of the chromosome containing Tn552 (carrying blaZ) and Tn4001 (carrying aac(6’)-aph(2”)) transposable elements. Using mutagenesis and in vitro assays, we provide phenotypic evidence that qacA confers tolerance to chlorhexidine. These findings collectively suggest both antimicrobial resistance and the carriage of qacA may play a role in the successful establishment of ST45 MRSA.
Subject terms: Antimicrobial resistance, Infectious-disease epidemiology, Bacterial genomics
A study underscores the emergence of a qacA-harbouring multidrug-resistant sequence type 45 methicillin-resistant Staphylococcus aureus lineage, highlighting the potential impact of biocide tolerance on its recent clonal spread in Australia and Asia.
Introduction
Infections caused by Staphylococcus aureus are among the most common bacterial infections worldwide1. These infections can lead to serious complications with increased morbidity, placing a substantial burden on healthcare systems. Colonisation by S. aureus is a known predisposing factor in the development of infections; as such, decolonisation using antimicrobials and/or biocides may be used to reduce healthcare-associated S. aureus infections2. Quaternary ammonium compounds (QACs), notably chlorhexidine, are routinely used as decolonising agents for hand hygiene and pre-surgical antisepsis3. However, it is possible that the widespread use of chlorhexidine exerts selective pressure on strains displaying tolerance to this biocide3,4.
Bacterial tolerance to chlorhexidine can be mediated by QAC multidrug efflux systems, with QacA (encoded by a plasmid-borne qacA gene) being the predominant type in S. aureus5. Of particular concern is the increasing prevalence of qacA carriage in methicillin-resistant S. aureus (MRSA)6. This is because MRSA is a primary target for infection prevention and control practices, and qacA carriage and possible development of co-resistance to mupirocin or/and other antimicrobials pose a potential risk to the success of these programmes6. In vitro studies have demonstrated an association between qacA carriage and chlorhexidine tolerance in S. aureus7,8. However, the clinical impact of low-level chlorhexidine tolerance conferred by qacA remains unclear, as in-use clinical concentrations are usually several orders of magnitude higher than the minimum concentrations required to inhibit bacterial growth, and chlorhexidine exposure may not always enrich for qacA-harbouring isolates as observed in clinical studies9,10. In addition, discrepancies between qacA carriage and phenotypic tolerance to chlorhexidine (minimum inhibitory concentration (MIC) < 4 mg/L) have been reported at the population level11,12. Understanding of qacA-mediated chlorhexidine tolerance is further complicated by the lack of a standardised method for biocide testing13. The presence of other drug efflux systems, e.g., those encoded by norA and mepA may partly mediate tolerance to chlorhexidine14. Hence, this knowledge gap highlights an urgent need for combined genomic and phenotypic analyses of biocide tolerance to help elucidate the contribution of specific genes, including qacA, to chlorhexidine tolerance in S. aureus, particularly MRSA.
While a recent study has adopted a combinatorial approach to uncovering the evolutionary benefit of qacA-mediated chlorhexidine tolerance in a dominant hospital-associated sequence type (ST) 239 MRSA lineage in Australia15, the molecular epidemiology of biocide tolerance in MRSA of other STs is largely unexplored. Currently in Australia, the rate of MRSA infections across the country remains stable, yet the clonal composition of MRSA is fluid and varies between states and territories over time16. Noticeably over the past decade, ST45 MRSA has become a major MRSA lineage associated with both hospital and community-onset infections16. Recent surveillance of S. aureus bacteraemia in Australia indicated that ST45 MRSA accounted for 10.1% of total MRSA infections, 67.3% of which were community onset17. Of particular note, the increased rate of multidrug resistance among community-associated MRSA (i.e., from 9.2% in 2013 to 13.7% in 2019) was primarily driven by ST4517. This clonal expansion was noted in one study from New South Wales (NSW), where the prevalence of ST45 MRSA increased from 0.4 to 14% between 2012 and 201718. An investigation into local outbreaks of ST45 MRSA in NSW revealed carriage of genetic determinants associated with resistance to aminoglycosides, macrolides, and tetracyclines19. Further, multidrug-resistant ST45 MRSA has been associated with hospital outbreaks outside Australia20,21, with a high carriage rate of qacA notified in the Singaporean healthcare system12. The acquisition of the above antimicrobial resistance (AMR) and biocide tolerance determinants may be an important contributor to the spread of ST45 MRSA in the healthcare setting. Nonetheless, to our knowledge, the contribution of qacA to the expansion of ST45 MRSA in Australia has not been explored. Given the worldwide dissemination of ST45 S. aureus22, this knowledge gap warrants the need to investigate the genomic correlation between qacA and the evolutionary dynamics of ST45 MRSA.
In this study, we conducted phylogenetic analyses using globally representative ST45 S. aureus, coupled with phenotypic characterisation of qacA-mediated biocide tolerance. Collectively, these data provide valuable information on the potential drivers for the widespread dissemination of ST45 MRSA.
Results
Complete genome sequence of a qacA-harbouring ST45 MRSA strain AUSMDU00020487
A representative qacA-harbouring ST45 MRSA, namely AUSMDU00020487, was selected as the reference strain for phylogenetic analyses and phenotypic characterisation in this study. This strain was obtained from a prospective state-wide surveillance study undertaken in Victoria, Australia in 201823. The genome of AUSMDU00020487 consisted of a single chromosome (GenBank accession number: CP138566, 2,895,690 bases, 32.8% GC content) and one plasmid (GenBank accession number: CP138567, 34,997 bases, 29.0% GC content). The chromosome harboured mecA located in a staphylococcal cassette chromosome mec (SCCmec) type V element; biocide tolerance genes (qacA, qacR); and resistance genes to penicillins (blaZ, encoding PC1 β-lactamase), tetracyclines (tetK), macrolides (ermC), and aminoglycosides (aac(6’)-aph(2”)). Additional resistance genes to macrolides (ermA) and tetracyclines (tetM) were found on the plasmid.
Divergent global evolution of ST45 S. aureus
Phylogenetic analyses utilised genomic data from 210 clonal complex (CC) 45 MRSA isolates recovered from Australian surveillance and epidemiological studies23–27 (Supplementary Note 1 & Supplementary Data 1). These sequences were supplemented with 1,503 ST45 S. aureus de novo genome assemblies from Staphopia28, and publicly available sequence data from 288 ST45 S. aureus genomes used by Effelsberg et al.22. In total, the final dataset contained 2,001 ST45 S. aureus genomes.
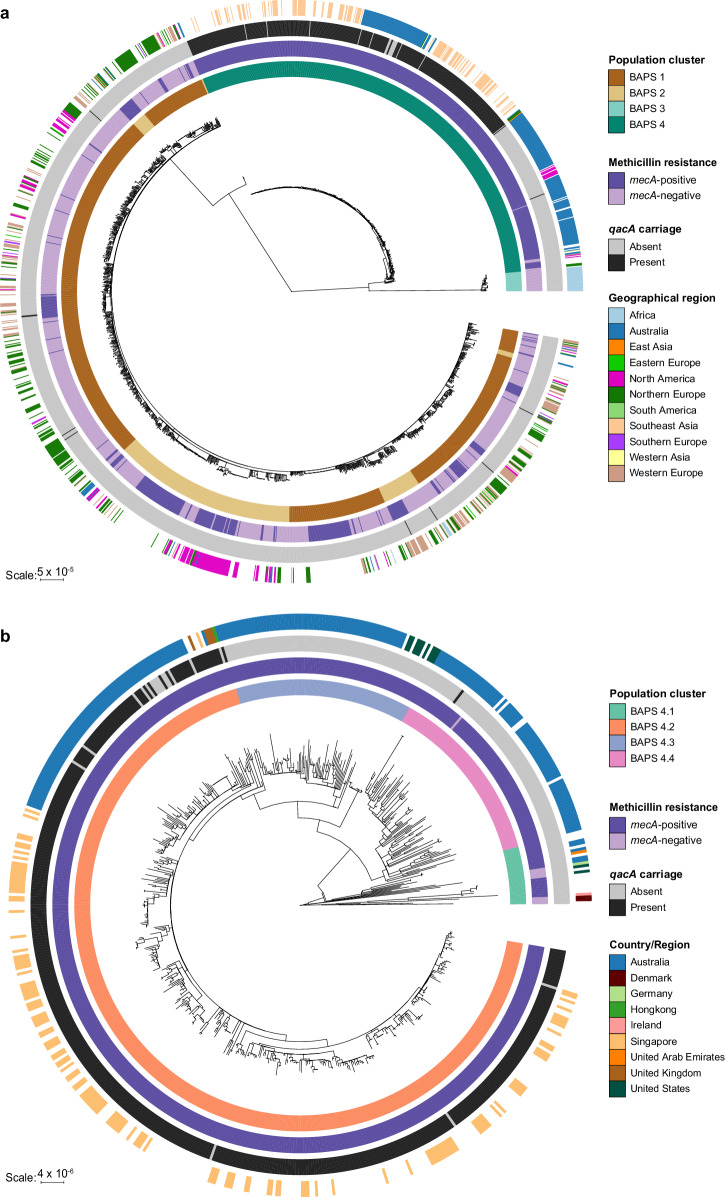
To investigate the evolution of qacA-harbouring ST45 MRSA, core genome single nucleotide polymorphism (SNP) analysis of the above global collection highlighted an early divergence of ST45 S. aureus into two distinct lineages (Fig. 1a). Bayesian analysis of population structure (BAPS) identified four major clades, with the BAPS 1 and 2 clades appearing interleaved (Fig. 1a). Some geographic patterns were observed. The largest lineages (BAPS 1 and 2) comprised isolates predominantly from Europe and North America. The smaller lineage (BAPS 3) consisted exclusively of isolates from Africa. The fourth lineage (BAPS 4) mainly contained isolates from Australia and Southeast Asia. Of the entire dataset, the vast majority of qacA-harbouring isolates (97.4%, 411/422) belonged to SCCmec type V (Supplementary Data 1). Notably, the isolates in the BAPS 4 clade were predominantly MRSA (98.7%, 608/616) and accounted for 97.6% (412/422) of qacA-harbouring S. aureus in the wider dataset, although this may represent sampling bias (as outlined in the discussion). Within BAPS 4, 99.8% (411/412) of qacA-harbouring isolates were found in the BAPS 4.2 subclade (Fig. 1b), consisting of isolates from Australia, Singapore, and the United Kingdom (UK). Of the qacA-positive isolates in this subset, ~1.0% (4/412) were from the UK, 32.0% (132/412) were from Singapore, 17.0% (70/412) were from Australia, and the remaining 206 isolates had an unknown geographic origin.
Fig. 1. Global phylogeny of ST45 S. aureus and qacA carriage.
a Maximum-likelihood tree was constructed using 37,549 core SNPs identified in the global collection of 2001 ST45 S. aureus. b Maximum-likelihood tree of 616 ST45 S. aureus in BAPS 4 was built using an alignment of 4716 core genome SNPs. From the innermost to the outermost ring, the heatmaps display (i) population clusters or sub-clusters determined by hierBAPS, (ii) whether an isolate was MRSA or MSSA based on mecA carriage, (iii) presence or absence of qacA carriage, (iv) geographical origin of the isolates. Isolates with unknown geographic information were denoted by the blank heatmaps on the outer ring.
Emergence of a distinct ST45 MRSA lineage is associated with qacA acquisition
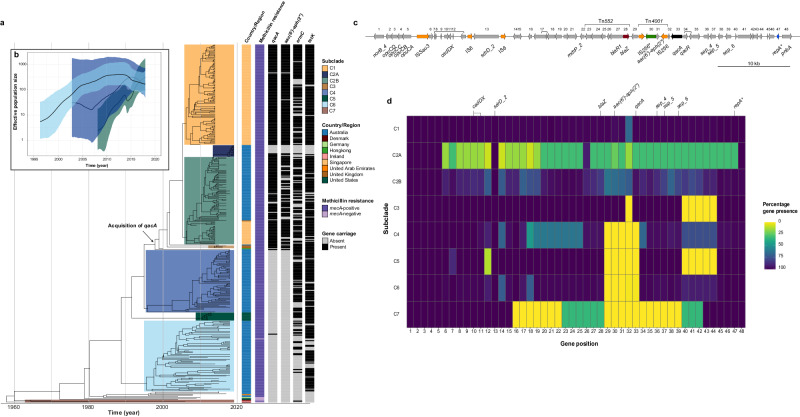
To gain insights into the temporal evolution of the BAPS 4 clade, 385 representative isolates with geographic information and year of collection details were selected for phylogenetic reconstruction. For population genetic and comparative genomic analyses, these isolates were divided into seven subclades, numbered C1 to C7, based on temporal phylogeny, geographic origin, and presence/absence of key antimicrobial resistance determinants including qacA (Fig. 2a). Noticeably, qacA was harboured by 99.1% (107/108), 95.8% (95/107), and 100% (3/3) of isolates in the C1 (Singaporean), C2 (Australian-Singaporean), and C3 (UK) subclades, respectively. The oldest qacA-harbouring isolate in the BAPS 4 clade was collected from Singapore in 2009, while our model predicted that the acquisition of qacA occurred in a MRSA background in the late 1990s (estimate: 1998, 95% highest posterior density [HPD] 1996–1999). qacA frequently co-occurred with aac(6’)-aph(2”) (85.5%, 185/219), ermC (71.7%, 157/219), and tetK (88.1%, 193/219) among isolates in the C1, C2, and C3 subclades. However, a lack of these AMR determinants including qacA was noted in the most recent Australian isolates (69.2%, 9/13) collected in 2019, resulting in the separation of C2A subclade for further investigation on gene loss. Of further note, SCCmec type differed between the qacA-negative (Type IV, 9/13) and qacA-positive isolates (Type V, 4/13) in the C2A subclade (Supplementary Data 1). Moreover, qacA-negative isolates formed two large C4 and C6 (Australian) subclades and a small C5 (American) subclade. The isolates in these qacA-negative subclades belonged to SCCmec type V (100%, 152/152) and lacked aac(6’)-aph(2”) (0%, 0/152), but were frequently associated with tetK (83.4%, 126/152) and less often carried ermC (49.3%, 75/152). The available metadata on the hospital and community origin of these isolates was limited (Supplementary Data 1). qacA was detected in 45.7% (16/35) of the community-associated isolates and 52% (13/25) of the hospital-associated isolates. Although 98.4% (379/385) of the isolates were MRSA, the most recent common ancestor for the BAPS 4 clade likely arose in the 1960s, with the oldest isolate found in the C7 (European) subclade being a methicillin-susceptible S. aureus (MSSA) of Danish origin, collected in 1970 (Fig. 2a).
Fig. 2. Evolutionary history of qacA-harbouring ST45 MRSA.
a Maximum clade credibility tree of 385 ST45 S. aureus isolates. The heatmaps provide additional annotations regarding country/region of isolation, methicillin resistance determined by mecA carriage, and the presence or absence of additional AMR determinants, including qacA, aac(6’)-aph(2”), emrC, and tetK. The black arrow indicates the predicted time for qacA acquisition (median node age = 1998, 95% HPD = 1996 to 1999). These isolates were divided into seven subclades based on qacA carriage and geographic origin. b GMRF Bayesian skyride plot. EPS of the major qacA-positive C2B (light green) subclade was compared with those of the qacA-negative C4 (navy blue) and C6 (light blue) subclades. The solid black line indicates the median EPS, and the coloured boundary indicates the 95% HPD. c The chromosomal region surrounding qacA in AUSMDU00020487. Each CDS (grey arrow) in this region was assigned a positional number, and CDSs of the same ortholog groups are linked by black solid lines. ISs (orange), plasmid replication initiator gene repA (blue), and AMR genes, including blaZ (red), aac(6’)-aph(2”) (green), and qacA (black), are highlighted. Boundaries of Tn552 and TN4001 are depicted by the brackets. Fragmented genes are flagged with an asterisk. d Comparison of gene content surrounding qacA between the major subclades. Percentages of gene presence are displayed in the heatmap, with dark blue indicating the presence of the gene among all isolates within a given subclade, while yellow indicates the absence of the gene. A summary of gene functionality, dosage, and synteny can be accessed in Supplementary Data 3.
To determine the temporal change in the population structure and size of ST45 MRSA subclades in relation to qacA carriage in Australia, the effective population size (EPS) and effective reproduction number (Re) of major qacA-positive (C2B) and qacA-negative (i.e., C4 and C6) subclades were compared. Of note was the rapid increase in the median EPS of C2B between 2008 and 2018 (Fig. 2b), with the first peak observed in 2014 attributed to the Singaporean isolates. Extended analysis using the entire C2 subclade showed a population collapse in 2019 due to the inclusion of C2A subclade (Supplementary Fig. 1). In comparison, the median EPS of C4 appeared to have increased between 2007 and 2014, followed by a reduction from 2015 onwards. However, the broad 95% HPD interval indicates large uncertainty in estimating the EPS trajectory of this subclade prior to 2015. Similarly, the median EPS of the C6 subclade steadily increased until 2012, when it started to plateau and then declined between 2014 and 2018. The 95% HPD intervals of EPS of all three subclades overlapped between 2015 and 2018. Consistently, epidemic spikes in Re (95% HPD > 1, indicating exponential spread) were observed during the emergence of C2B (2011–2013) and C4 (2010–2012) as well as in recent years for C4 (2014–2017) and C6 (2013–2017) (Supplementary Fig. 2). In contrast, Re decreased or stabilised in C2B (95% HPD overlapping Re = 1, between 2013 and 2017). Collectively, these results demonstrate an overall increase in the population size of ST45 MRSA, regardless of qacA status over the past two decades, but dynamic changes in population growth were noted, including periods of likely exponential spread. However, there was no clear association between qacA acquisition and epidemic growth of qacA-harbouring isolates in the C2B subclade.
Chromosomal integration of qacA in ST45 MRSA
The presence of a chromosomal qacA (rather than plasmid-associated) in ST45 MRSA prompted further investigation into the genetic context of qacA. Gene conservation (Fig. 2c) and synteny (Supplementary Fig. 3) of each subclade were compared. The genomic configuration in all isolates belonging to subclades C4 – C7 showed early acquisition of multiple genetic elements, including mdrP (encoding a Na + /H+ antiporter), Tn552 carrying blaZ, and three enterotoxin encoding genes (sep) (Fig. 2c). These elements were likely brought into this region of the chromosome by the integration of a pF5-like plasmid, suggested by: (i) a fragmented repA gene, and (ii) a high level of similarity (>97%) to the genomic structure of a previously reported pF5 plasmid (GenBank accession number: AB765928) lacking Tn552 (Supplementary Fig. 4). The plasmid reported in S. aureus strain Fukuoka 5 has previously been associated with food poisoning-related illnesses29. Further, it was evident that changes to this region have occurred since the possible integration of a pF5-like plasmid, with the loss of a hypothetical protein CDS cluster upstream of repA in the C3 and C5 subclades (Fig. 2d, gene position 40–44).
Acquisition of qacA, possibly concurrent with Tn4001 containing aac(6’)-aph(2”), occurred in a MRSA background carrying blaZ as displayed in the C1, C2, and C3 subclades (Fig. 2d, gene position 29–33). Sequence alignment between AUSMDU00020487 and BPH2770 (GenBank accession number: GCA_027956625.1), a qacA-negative ST45 MRSA reference, revealed a 7981 bp cassette containing qacA and Tn4001 (Supplementary Fig. 5). This cassette shared 99% homology with the Tn4001-qacR spanning region of a pSK1 plasmid (GenBank accession number: NC_014369) with an inverted qacAR (Supplementary Fig. 4). However, the pSK1 plasmid did not contain the small putative membrane protein-encoding gene found between qacA and Tn4001, nor the hypothetical protein coding sequence between the truncated IS256 and blaZ (Supplementary Fig. 4). We did not identify evidence of other ISs or transposable elements flanking the putative cassette, and the mechanism for chromosomal integration remains unclear. Of note, this region was highly variable within the C2 subclade, in which the genes were less syntenic (Supplementary Fig. 3). Some isolates in the C2A subclade had lost the region (51,010 bp) spanning between ISSau3 and the fragmented repA, suggesting a potential ISSau3 transposition (Supplementary Fig. 6). These findings highlight the instability of the chromosomal region surrounding qacA, as the gain and loss of genes or mobile genetic elements appeared to have frequently occurred.
qacA in ST45 MRSA confers tolerance to quaternary ammonium compounds
To investigate the potential role of qacA in conferring tolerance to QACs in ST45 MRSA, a markerless deletion of qacA was made in the reference strain, AUSMDU00020487, using a previously described method30. Broth microdilution MIC assays showed that the deletion of qacA led to a twofold reduction in the chlorhexidine digluconate (CHG) MIC (from 2 to 1 mg/L) and a 16-fold reduction in acriflavine MIC (from 128 to 8 mg/L) compared to the wild-type strain (Table 1). The ratio of the minimum bactericidal concentration (MBC) to MIC was 2, as CHG and acriflavine MBCs were 2 mg/L and 16 mg/L for the ΔqacA strain, respectively; While CHG and acriflavine MBCs were 4 mg/L and 256 mg/L for the wild-type strain, respectively. Complementation of qacA restored the MICs and MBCs to the levels of the wild-type (Table 1). No secondary mutations confounding the observed phenotypic changes were detected in the ΔqacA and ΔqacA:qacA strains (Supplementary Data 2). The MIC and MBC results were consistent across three biological replicates.
Table 1.
In vitro susceptibility to quaternary ammonium compounds in AUSMDU00020487 and the derived mutants
| Strain | Genotype | CHG (mg/L) | Acriflavine (mg/L) | ||
|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | ||
| AUSMDU00020487_wild-type | Wild-type | 2 | 4 | 128 | 256 |
| AUSMDU00020487_qacA | ΔqacA | 1 | 2 | 8 | 16 |
| AUSMDU00020487_qacA:qacA | ΔqacA:qacA | 2 | 4 | 128 | 256 |
In vitro exposure to sub-MIC of chlorhexidine selects for qacA-harbouring ST45 MRSA
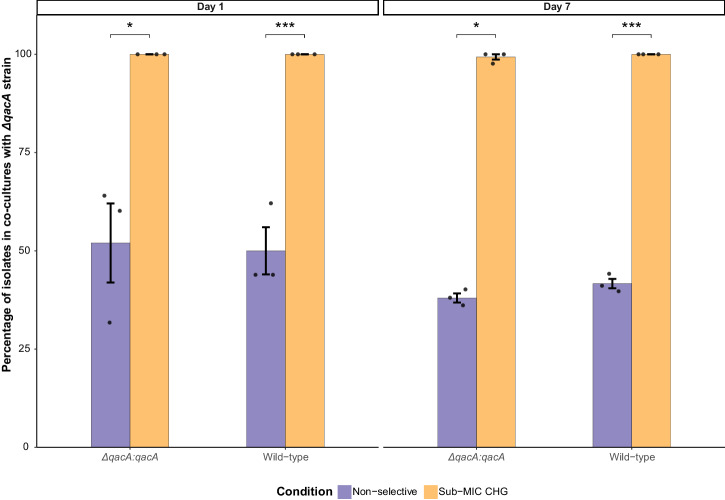
Competition assays were performed to determine the impact of qacA in conferring a competitive advantage in the presence of CHG, as previously described31. Co-cultures of the AUSMDU00020487 ΔqacA strain paired with either the wild-type or ΔqacA:qacA strain were established in a 1:1 ratio. These co-cultures were supplemented with or without CHG at a sub-MIC level (0.5 mg/L) for the ΔqacA strain. The results demonstrated that qacA-harbouring strains rapidly outcompeted the ΔqacA strain, with 100% of the isolates recovered from the co-cultures being wild-type or ΔqacA:qacA following 24 h exposure to CHG (day 1) (Fig. 3). The qacA-harbouring strains remained dominant at the conclusion of the assays on day 7, with 100% and >99% of the harvested isolates being wild-type and ΔqacA:qacA strains, respectively (Fig. 3). In contrast, no noticeable variation in the proportion of wild-type or ΔqacA:qacA strains relative to the mutant was observed on day 1 or day 7 in the absence of selective pressure. Additionally, the doubling time of the ΔqacA strain, ΔqacA:qacA and wild-type strains did not significantly differ (P > 0.05 determined by unpaired t test, Supplementary Fig. 7), indicating that the sub-MIC selection of qacA-harbouring strains was not associated with a faster bacterial growth rate.
Fig. 3. In vitro competition assays using co-cultures of AUSMDU00020487 wild-type paired with ΔqacA strain, and ΔqacA:qacA paired with ΔqacA strain.
These co-cultures were exposed to either non-selective (purple) or selective condition using 0.5 mg/L CHG (yellow) for 7 days. Three biological replicates denoted by the black dot points were performed for each condition tested. The mean percentages of wild-type or ΔqacA:qacA strains on day 1 and day 7 post-exposure are displayed, with the black error bars representing the standard error of the mean. Asterisks denote statistically significant differences as determined by paired t test (*P ≤ 0.05 and ***P ≤ 0.001).
Discussion
In this study, we investigated an emerging qacA-harbouring ST45 MRSA lineage using genomic and phenotypic analyses. In agreement with a recent study examining the emergence of ST45 S. aureus22, our analysis using a larger dataset revealed a clonally diverse global population structure with SCCmec Type V MRSA predominated among Australian isolates. Within the global dataset, we show that the presence of qacA was almost exclusively found in ST45 isolates from Australia and Singapore, with ST45 becoming a predominant MRSA lineage in these regions over the past decade16,32. Noticeably, a high prevalence (99%) of qacA has previously been reported in ST45 MRSA linked to universal chlorhexidine bathing implemented in Singaporean extended-care facilities12. However, the prevalence of qacA and its potential role in the evolutionary success of ST45 MRSA has not previously been investigated in Australia.
We observed a high carriage rate of qacA among ST45 MRSA isolates collected from the national staphylococcal bacteriemia surveillance programmes undertaken in 201526 and 201727. qacA-harbouring isolates in our study shared a similar AMR profile (aac(6’)-aph(2”), ermC, and tetK) with those responsible for local hospital outbreaks in the state of NSW between 2013 and 201719. Outside Australia, ST45 MRSA has been associated with healthcare settings in Singapore12 and Taiwan33, suggesting possible intercontinental dissemination of this lineage.
Phylodynamic analysis suggested that ST45 MRSA expanded over the past two decades, including qacA-harbouring sublineages. Population expansion was also observed in older lineages harbouring ermC and tetK, but lacking qacA. It is possible that the early introduction of these AMR determinants (i.e., ermC and tetK) into ST45 MRSA might have conferred an initial selective advantage for this lineage, with the subsequent acquisition of qacA and aac(6’)-aph(2”) in the late 1990s providing a further basis for selective expansion after 201318. The increased prevalence of ST45 MRSA coincided with the increase in chlorhexidine-based infection control and hand hygiene programmes designed predominantly to combat MRSA34. Dynamic changes in the Re occurred in both qacA-positive and qacA-negative sublineages, indicating that the sizes of these populations were fluctuating. While the acquisition of qacA may be an important contributor to the latest regional outbreaks caused by ST45 MRSA, further investigation using contemporary isolates is needed to evaluate the ongoing impact of these sublineages (both qacA-positive and qacA-negative) in contributing to the burden of S. aureus infections in Australia.
qacA was co-located with multiple AMR and virulence determinants in the chromosome of ST45 MRSA, similar to the chromosomal qacA integration in ST239 MRSA15. Comparative genomic analysis showed that early integration of a pF5-like plasmid (characterised by enterotoxin encoding genes) in historic ST45 isolates may have provided a ‘hotbed’ for subsequent acquisition of qacA and other mobile elements, including Tn552 and Tn4001. The gain of these mobile elements may also confer additional evolutionary benefits by co-selection of AMR (namely, blaZ and aac(6’)-aph(2”)). However, the inference drawn from this analysis was limited by the lack of isolates collected before 2009 (the year in which the first qacA-harbouring isolate was reported in our dataset), which may have impacted the accuracy of our prediction regarding the temporal emergence of qacA. Among the recent isolates collected in 2019, we observed a loss of regions surrounding qacA, possibly driven by ISSau3. This may provide opportunities for further spread of qacA and other co-located AMR and virulence determinants. Given chlorhexidine use has been linked to the enrichment of qacA carriage35–37, IS-mediated transposition could be reflective of demographic differences in infection control practices across geographic settings.
Our mutagenesis work provided experimental evidence that the presence of qacA was associated with increased tolerance to CHG, when the ST45 reference strain (MIC = 2 mg/L) was compared to its ΔqacA strain (MIC = 1 mg/L). However, the shift in MIC is below the cut-off (MIC ≥ 4 mg/L) used for determining chlorhexidine tolerance in some studies38,39. This underscores a known issue with using strict MIC cut-offs, in that subtle shifts in tolerance may be missed. The use of a higher cut-off might have led to the observation that qacA carriage in ST45 MRSA is not always associated with reduced susceptibility to chlorhexidine12. This inconsistency might be caused by the assay itself, which was originally intended for antibiotic susceptibility testing, and caution should be advised when used for biocide testing40. Of note, we observed that acriflavine may be a useful alternative for differentiating qacA-positive (MIC = 128 mg/L) from qacA-negative (MIC = 8 mg/L) isolates in vitro. Together, our findings and previous observations collectively emphasise the need for an improved and standardised methodology (validated across multiple laboratories) for determining biocide tolerance.
The demonstration of sub-MIC selection provides evidence that qacA may be an important determinant in enhancing the adaptation of ST45 MRSA to low levels of CHG exposure. While the in-use concentrations of chlorhexidine (e.g., 0.5, 2, and 4% (w/v)) are considerably higher than the sub-MIC tested41, it is worth noting that the selection of qacA-harbouring isolates has been reported in ST5, ST22, and ST239 MRSA backgrounds following chlorhexidine-based infection control measures42–44. Of concern is the co-selection of AMR and virulence genes as an unintentional consequence of chlorhexidine use. As such, this finding has implications for understanding the prolonged residual effect of chlorhexidine on enriching for qacA carriage as well as increasing the overall drug resistance in S. aureus.
This study has several strengths. First, the sample size for our global phylogenetic analysis was large and included representative isolates from various geographic regions. Second, the complete genome of a local reference strain was constructed to provide detailed insights into the emergence and divergence of major sublineages in Australia. Third, mutagenesis and in vitro assays were used to characterise qacA-mediated biocide tolerance in the reference strain.
There are limitations to our sampling strategy. First, the missing geographic information for a large number of isolates included in the global phylogeny of ST45 S. aureus and the overrepresentation of Australian and Singaporean isolates sampled across limited time frames should be noted, limiting our capacity to fully understand the global emergence and dissemination of this lineage. Second, due to the lack of historic isolates, we were unable to fully elaborate on the sequential order of integration events leading to the chromosomal integration of qacA. Third, a population-level analysis of biocide tolerance, which could provide critical insights into the phenotypic contribution of qacA to biocide tolerance in different ST45 MRSA sublineages, was not assessed in this study. Fourth, we did not possess metadata regarding the community or hospital origin for the majority of isolates included in the phylogenetic analyses (Supplementary Data 1). This limitation limited our ability to estimate the burden of qacA-harbouring isolates and understand their adaption to settings with routine antiseptic use. Thus, future work should focus on (i) understanding the causative link between contemporary biocide usage and population-level phenotypic tolerance; (ii) evaluating the stability of the qacA-containing region in the chromosome as previously described45; (iii) determining the impact of clinically used concentrations of chlorhexidine in selecting for qacA carriage; and (iv) enhancing Bayesian phylogenetics on investigating origin and dissemination of the qacA-harbouring lineage/sublineages using additional historic and contemporary isolates with diverse geographic background. Regionally, the integration of epidemiological data and improved knowledge of regional differences in antiseptic use will also facilitate our understanding of whether AMR and chlorhexidine tolerance are key drivers in the selection of ST45 MRSA in Australia.
Despite decades of biocide use, adverse ecological impact associated with the widespread use of chlorhexidine has raised questions about the need to develop biocide stewardship. Here, we have described the emergence and clonal expansion of a qacA-harbouring ST45 MRSA lineage. Together with the phenotypic confirmation of its role in mediating chlorhexidine tolerance, our data suggest that qacA may have important implications for the recent clonal spread of ST45 MRSA in Australia and Asia.
Methods
Bacterial isolates and sequence data
S. aureus genomes used in this study were sourced from Staphopia28, European Nucleotide Archive (ENA) per the study conducted by Effelsberg et al.22, and surveillance programmes and national studies of S. aureus undertaken in Australia23–27. Further demographic information regarding the sources of isolates and selection criteria for sequence data are summarised in the Supplementary Information (Supplementary Table 1).
Construction of reference genome
A qacA-positive ST45 MRSA strain, AUSMDU00020487, was collected in Victoria, Australia, in 201823. To generate a complete reference genome, genomic DNA was sequenced on the Illumina NextSeq or iSeq (2 × 150 bp paired-end chemistry) and Oxford Nanopore GridION X5 (with FLO-MIN106D R9 flow cells) platforms. Demultiplexing and adaptor trimming of the Nanopore long-read data was performed using Porechop v0.2.4 (https://github.com/rrwick/Porechop). Trimmed data were filtered using Filtlong v0.2.1 (https://github.com/rrwick/Filtlong) by removing short reads (<1000 bases), and sampling to a target total bases value of 300 megabases, favouring the longest and highest quality reads. A hybrid assembly was then undertaken with Unicycler v0.4.846. The assembled genome underwent further error correction with the Illumina short-read data using Snippy v4.4 (https://github.com/tseemann/snippy) iteratively until no variants were called. The draft genome was annotated with Prokka v1.14.647 and analysed using mlst v2.19.0 (https://github.com/tseemann/mlst) to confirm that AUSMDU00020487 belonged to ST45 S. aureus. AMR genotyping analysis was performed using ABRicate v0.8.10 (https://github.com/tseemann/abricate) against the National Center for Biotechnology Information (NCBI) AMRFinderPlus database48. SCCmec typing was performed using staphopia-sccmec v1.0.028, and SCCmecFinder v.1.249 was used to distinguish between SCCmec Type V and VII.
Comparative genomic analysis
Illumina short reads or de novo assemblies of the global collection of ST45 S. aureus were mapped to the reference AUSMDU00020487 genome, using Snippy v4.4. Core SNPs identified were then used for constructing the maximum-likelihood phylogenetic tree with IQ-TREE v2.1.250, under a generalised time-reversible (GTR) model combined with the proportion of invariable sites, empirical base frequencies, a discrete gamma model with 4 rate categories (GTR + I + F + G4) and 1000 bootstrap replicates. BAPS clusters were identified using hierBAPS v1.0.151 with two levels and ten initial clusters on the core genome SNP alignment. All trees were midpoint rooted and visualised using R package ggtree v3.6.252.
Short reads were de novo assembled using SPAdes v3.1.253 and annotated by Prokka v1.14.6. MLST typing was performed on the genome assemblies with mlst v2.19.0. ABRicate v0.8.10 with the NCBI’s AMRFinderPlus database was then used for determining the AMR profile for each isolate. Isolates were categorised into MRSA or MSSA based on mecA carriage. Automated clustering of protein orthologs was performed using Roary v3.13.054 with a 95% amino acid homology. Gene content, synteny, and dosage were compared between subclades. Annotation for insertion sequences was checked by comparing the predicted IS CDS against the ISfinder database55.
Phylogenetic analysis
A core genome SNP alignment of 385 ST45 S. aureus was built, as described above. Recombination detection was performed using ClonalFrameML v1.12 (https://github.com/xavierdidelot/ClonalFrameML). Recombination sites, including 22 core SNPs, were masked using maskrc-svg v0.5 (https://github.com/kwongj/maskrc-svg). The strength of the temporal signal in the masked alignment was assessed using TempEst v1.5.356. To determine the optimal parameters for Bayesian phylogenetics, the masked alignment containing 3690 filtered core SNPs underwent further analysis using BEAST v1.10.457 under a GTR + G4 model of nucleotide substitution, with combinations of the strict and uncorrelated relaxed log-normal molecular clocks, and constant and exponential tree priors. The XML files generated by BEAUti were edited to reflect the number of invariant sites in the masked alignment. For each combination tested, marginal likelihood estimation was performed in triplicate using path sampling to calculate the Bayes factors. The best-fitting model was achieved by the uncorrelated relaxed log-normal clock with a coalescent exponential population tree prior. Using this parameter, ten replicates of 50 million Monte Carlo Markov Chain (MCMC) analyses were performed with sampling every 1000 steps and 20% burn-in removed. Replicates were combined using LogCombiner and re-sampled at a frequency that would achieve ~10,000 samples. Tracer v1.7.258 was used to check MCMC convergence and effective sample size value (greater than 200) to ensure adequate sampling of the posterior distribution. The maximum clade credibility tree with median heights was selected with TreeAnnotator.
Population genetic analysis
Subsets of the main core SNP alignment were created for several subclades of the above maximum clade credibility tree. These subclades were selected due to the number of available isolates, serial sampling dates, and their relevance to the emergence of ST45 in Australia. Following the same steps above for Bayesian phylogenetics, EPS was estimated using the uncorrelated relaxed log-normal clock coupled with a time-aware Gaussian Markov random field (GMRF) Bayesian skyride model59. The Bayesian skyride plots of median EPS were generated in Tracer v1.7.258 using the GMRF skyride reconstruction tool. Furthermore, serial birth–death skyline models in BEAST v2.660,61 were configured to infer changes in transmission dynamics at sublineage resolution. For all subclades, the model was run under a GTR + G4 model of nucleotide substitution using a strict clock model for simplicity, which was chosen due to the short timeframe of emergence (8–14 years) precluding assumptions of significant rate variation amongst clade members. Model priors, including a sliced effective reproductive number prior, were configured as described previously for S. aureus62. In brief, the Re prior was configured to a number of equally sized intervals over the maximum clade credibility tree; a suitable interval number was selected by running exploratory models for each lineage with 100 million iterations (dimensions = 5–10). A comparison of Re estimates under these configurations ensured the occurrence of stable posterior distributions and the absence of bi- or multimodal posteriors. Five equally sliced dimensions were then selected as a parsimonious configuration for the prior, allowing each slice to cover a range of 2–3 years depending on clade sampling times. Five replicates of 100 million MCMC steps with slight variation in the initial parameter values were run with sampling every 1000 steps and 20% burn-in removed. Tracer was used to check MCMC convergence and to ensure adequate sampling of the posterior distribution. Critter (https://github.com/esteinig/critter) was used to configure the models and to generate the plots depicting changes of average Re across slices and kernel density esimates of the underlying posterior distribution across slices (Supplementary Fig. 2).
Construction of ΔqacA strain by allelic exchange
To make a markerless deletion of qacA in AUSMDU00020487, a 1.5-kb gene cassette containing jointed flanking regions upstream and downstream of qacA was amplified by spliced overlap extension PCR. The cassette was cloned into the pIMAY-Z shuttle vector30 using seamless ligation cloning extract63. The cloned plasmid was electroporated into Escherichia coli IM30B to obtain methylation profiles compatible with ST45 S. aureus. Successful E. coli transformants were selected following overnight incubation at 37 °C on Luria agar (LA) containing 10 mg/L of chloramphenicol and 50 mg/L of X-gal (5-bromo-4-chloro-3-indo-lyl-D-galactopyranoside; Melford). Extracted plasmid from E. coli transformants was then introduced into AUSMDU00020487 by electroporation. AUSMDU00020487 transformants (indicated by blue colonies) were passaged in brain heart infusion (BHI) broth at 30 °C (i.e., non-permissive temperature for plasmid replication) to promote chromosomal integration of pIMAY-Z. To perform allelic exchange, dilutions of the broth culture were spread onto BHI agar supplemented with 100 mg/L of X-gal, followed by overnight incubation at 37 °C. White colonies were cross-patched onto BHI agar containing 10 mg/L of chloramphenicol and 100 mg/L of X-gal; and BHI agar containing 100 mg/L of X-gal only. Following overnight incubation at 37 °C, white colonies displaying susceptibility to chloramphenicol (indicating loss of pIMAY-Z plasmid) were confirmed as mutants by PCR and whole genome sequencing. For qacA complementation, a substitution at nucleotide 720 (c.720 A > G) resulting in a silent mutation at codon 240 (p. Pro240Pro) was introduced to the ΔqacA:qacA. Secondary mutations in the ΔqacA and ΔqacA:qacA strains were screened using Snippy v4.4 (Supplementary Data 2). All primers used in the experiments are listed in Table 2.
Table 2.
Primers used in this study
| Primer | Sequence (5’ – > 3’) | Description |
|---|---|---|
| qacA_Fp | CCTCACTAAAGGGAACAAAAGCTGGGTACCCTTTTAATTCTAGCGTGCCTAC | Flanking primers used for amplification of gene cassettes for qacA deletion and complementation |
| qacA_Rp | CGACTCACTATAGGGCGAATTGGAGCTCCGTAATTTAGAAATAATATTTATTGGTATTTCAAG | |
| qacA_SOE_Fp | CTAATCTACAATATCTAAAAATATATGTTTAGTATTAAGTTCCCTCCAATCCTTATAG | Construction of gene cassette for qacA deletion by Splicing by Overlap Extension PCR |
| qacA_SOE_Rp | CTATAAGGATTGGAGGGAACTTAATACTAAACATATATTTTTAGATATTGTAGATTAG | |
| qacA_comp_Fp | CTATTACAACCCACGGAATAATATCTGCTAGTC | Construction of gene cassette for qacA complementation by Splicing by Overlap Extension PCR |
| qacA_comp_Rp | GACTAGCAGATATTATTCCGTGGGTTGTAATAG |
Antimicrobial susceptibility testing and growth assay
Broth microdilution MIC and MBC assays were performed in accordance with the Clinical & Laboratory Standards Institute (CLSI) guidelines64 using 20% CHG solution (C9394; Sigma-Aldrich) or acriflavine (A8126; Sigma-Aldrich). Briefly, a twofold serial dilution of an antimicrobial was performed in 100 μL of Cation-adjusted Mueller Hinton (CAMH) broth in a 96-well microplate. A fresh bacterial culture was used to prepare a 0.5 MacFarland suspension in CAMH broth, and 100 μL of the suspension was dispensed into the wells containing the antimicrobial. The microplate was incubated at 37 °C overnight before MIC determination. MIC was defined as the lowest antimicrobial concentration inhibiting observable bacterial growth. To perform MBC testing without neutralisation of the antimicrobial, the contents in each well of the microplate was mixed thoroughly. 10 μL of the resuspension was spot-plated onto CAMH agar, which was incubated for 48 h at 37 °C. MBC was defined as the lowest antimicrobial concentration required to prevent bacterial growth on agar. Three biological replicates were performed for both MIC and MBC testing.
Growth assays were performed as previously described65. Briefly, an overnight bacterial broth culture was diluted in fresh BHI broth to an optical density of 0.05 at 600 nm (OD600). Subsequently, 200 μL of the bacterial suspension was distributed into a 96-well microplate. The microplate was incubated in a microplate reader (CLARIOstar, BMG LABTECH) at 37 °C for 16 h with agitation at 200 rpm, and the OD600 was measured at 15-min intervals. Six biological replicates were performed for each strain tested. The bacterial growth rates denoted as doubling times were determined using the R package Growthcurver v0.3.166.
In vitro competition assay
To determine the competitive advantage conferred by qacA carriage following exposure to CHG, an in vitro assay was performed according to a previously established protocol31. This assay tested the pairing of (i) AUSMDU00020487 wild-type and ΔqacA strain, and (ii) ΔqacA:qacA and ΔqacA strain. These strains were grown in BHI broth overnight prior to the assay. In brief, an overnight bacterial culture was first diluted with fresh BHI broth to an OD600 of 0.10. A bacterial suspension was prepared by mixing the competing strains in a 1:1 ratio. The mixed suspension was then diluted 1:100 in 10 mL (i) non-selective BHI broth or (ii) BHI broth supplemented with 0.5 mg/L of CHG (i.e., 0.5 × MIC for the ΔqacA strain). The co-cultures were incubated at 37 °C with shaking at 200 rpm for 7 days. Following 24 h of incubation (day 1), a 300 µl sample was withdrawn and tenfold serially diluted in phosphate-buffered saline. In total, 100 µl of appropriate dilutions were spread onto BHI agar and incubated at 37 °C overnight. Following the incubation, 50 single colonies were randomly selected and cross-patched onto cation-adjusted Mueller Hinton agar supplemented with or without 64 mg/L of acriflavine to determine the ratio of the two competing strains on day 1. The same process was repeated on day 7 post-exposure. Three biological replicates were conducted for each pair/condition tested. For strain validation and analysis of secondary mutations, two output isolates collected at the conclusion of the experiment under the selective condition were selected for Illumina short-read sequencing (Supplementary Data 2). The results were visualised using R package ggplot2 v3.4.167.
Statistics and reproducibility
Unpaired and paired two-sided t tests (two-tailed) were performed using R package rstatix68 to compare bacterial growth rate and in vitro competition, respectively. A P value lower than 0.05 was determined to be statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors would like to thank the staff in the Microbiological Diagnostic Unit Public Health Laboratory, Victoria, Australia and in the PathWest Laboratory Medicine, Western Australia, Australia. This work is funded by a National Health and Medical Research Council (NHMRC) Investigator Grant (APP1174555).
Author contributions
D.A.W., B.P.H., G.P.C., S. Pasricha, S.L.B., and Y.N. contributed to the conception and design of the work. G.W.C., D.A.D., and S. Pang provided the genomic data from the Australian Staphylococcal Sepsis Outcome Program (ASSOP). P.N.A.H. and B.M.F. provided the genomic data from the Queensland surveillance of MRSA program. B.P.H. and S.L.B. provided genomic data from the Australian and New Zealand Cooperative on Outcome in Staphylococcal Sepsis (ANZCOSS) study and the Vancomycin Efficacy in Staphylococcal Sepsis in Australasia (VANESSA) study. B.P.H. and N.L.S. provided the genomic data and reference isolates from the ‘Controlling Superbugs’ study. G.P.C., I.R.M. and Y.N. designed the experimental procedures and provided materials. Y.N. performed the genomic and phylogenetic analyses with input from E.S. and guidance from S.L.B. Y.N. performed experiments with input from G.L.P. and guidance from G.P.C. and I.R.M. D.A.W., G.P.C., S. Pasricha, S.L.B., G.L.P., E.S., G.T. and Y.N. contributed to the interpretation of the results. Y.N. conducted the statistical analyses, produced the figures and tables, and drafted the manuscript. All authors revised and approved the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Wendy Mok and Tobias Goris. A peer review file is available.
Data availability
Sequence data for the isolates retrieved from publications and public databases can be accessed in accordance to Supplementary Note 1 and Supplementary Data 1. Additional genomic data generated in this study have been deposited under the BioProject PRJNA984755 in the NCBI database. The complete genome assembly of the ST45 S. aureus reference AUSMDU00020487 strain (GenBank accession numbers: CP138566-7) has been uploaded under the BioProject PRJNA565795. Numerical source data for competition assays, growth assays, and Re analysis are available on Figshare (10.26188/25201073).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sarah L. Baines, Deborah A. Williamson.
Contributor Information
Yi Nong, Email: yi.nong@unimelb.edu.au.
Deborah A. Williamson, Email: deborah.williamson@unimelb.edu.au
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06012-z.
References
- 1.Esposito S, et al. Diagnosis and management of skin and soft-tissue infections (Ssti). A literature review and consensus statement: an update. J. Chemother. 2017;29:197–214. doi: 10.1080/1120009X.2017.1311398. [DOI] [PubMed] [Google Scholar]
- 2.Ryu S, et al. Colonization and infection of the skin by S. aureus: immune system evasion and the response to cationic antimicrobial peptides. Int. J. Mol. Sci. 2014;15:8753–8772. doi: 10.3390/ijms15058753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaBreck PT, et al. Systematic analysis of efflux pump-mediated antiseptic resistance in Staphylococcus aureus suggests a need for greater antiseptic stewardship. mSphere. 2020;5:e00959-19. doi: 10.1128/mSphere.00959-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardy K, et al. Increased usage of antiseptics is associated with reduced susceptibility in clinical isolates of Staphylococcus aureus. mBio. 2018;9:e00894-18. doi: 10.1128/mBio.00894-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson DA, et al. Current and emerging topical antibacterials and antiseptics: agents, action, and resistance patterns. Clin. Microbiol. Rev. 2017;30:827–860. doi: 10.1128/CMR.00112-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madden GR, Sifri CD. Antimicrobial resistance to agents used for staphylococcus aureus decolonization: is there a reason for concern? Curr. Infect. Dis. Rep. 2018;20:26. doi: 10.1007/s11908-018-0630-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baraldi MM, et al. Risks and benefits of using chlorhexidine gluconate in handwashing: a systematic literature review. Am. J. Infect. Control. 2019;47:704–714. doi: 10.1016/j.ajic.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Babiker A, et al. Assessing the potential for unintended microbial consequences of routine chlorhexidine bathing for prevention of healthcare-associated infections. Clin. Infect. Dis. 2020;72:891–898. doi: 10.1093/cid/ciaa1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlett CD, et al. Prevalence of chlorhexidine-resistant methicillin-resistant Staphylococcus aureus following prolonged exposure. Antimicrob. Agents Chemother. 2014;58:4404–4410. doi: 10.1128/AAC.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritz SA, et al. Mupirocin and chlorhexidine resistance in Staphylococcus aureus in patients with community-onset skin and soft tissue infections. Antimicrob. Agents Chemother. 2013;57:559–568. doi: 10.1128/AAC.01633-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JT, et al. Longitudinal analysis of chlorhexidine susceptibilities of nosocomial methicillin-resistant Staphylococcus aureus isolates at a teaching hospital in Taiwan. J. Antimicrob. Chemother. 2008;62:514–517. doi: 10.1093/jac/dkn208. [DOI] [PubMed] [Google Scholar]
- 12.Htun HL, et al. Chlorhexidine and octenidine use, carriage of qac genes, and reduced antiseptic susceptibility in methicillin-resistant Staphylococcus aureus isolates from a healthcare network. Clin. Microbiol. Infect. 2019;25:1154.e1–1154.e7. doi: 10.1016/j.cmi.2018.12.036. [DOI] [PubMed] [Google Scholar]
- 13.Roedel A, et al. Genetic but no phenotypic associations between biocide tolerance and antibiotic resistance in Escherichia coli from German Broiler Fattening Farms. Microorganisms. 2021;9:651. doi: 10.3390/microorganisms9030651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassanzadeh S, et al. Epidemiology of efflux pumps genes mediating resistance among Staphylococcus aureus; a systematic review. Microb. Pathog. 2020;139:103850. doi: 10.1016/j.micpath.2019.103850. [DOI] [PubMed] [Google Scholar]
- 15.Baines, S. L. et al. Remodeling of Psk1 family plasmids and enhanced chlorhexidine tolerance in a dominant hospital lineage of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents and Chemother.63, 10–1128 (2019). [DOI] [PMC free article] [PubMed]
- 16.Coombs, G. et al. MRSA in Australia: MRSA Bacteraemia – 2013 to 2018 (Australian Commission on Safety and Quality in Health Care, Sydney, 2020). https://www.safetyandquality.gov.au/sites/default/files/2020-09/methicillinresistant_staphylococcus_aureus_in_australia_mrsa_bacteraemia_2013_to_2018.pdf.
- 17.Coombs, G. W. et al. Australian group on antimicrobial resistance (Agar) Australian Staphylococcus aureus sepsis outcome programme (Assop) annual report 2019. Communicable Diseases Intelligence2018. 44, 18 (2020). [DOI] [PubMed]
- 18.Dotel R, et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolates in New South Wales, Australia, 2012–2017. Infect. Dis. Health. 2019;24:134–140. doi: 10.1016/j.idh.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Beukers AG, et al. A multicentre outbreak of St45 Mrsa containing deletions in the spa gene in New South Wales, Australia. J. Antimicrob. Chemother. 2020;75:1112–1116. doi: 10.1093/jac/dkz560. [DOI] [PubMed] [Google Scholar]
- 20.Htun HL, et al. Methicillin-resistant Staphylococcus aureus colonisation: epidemiological and molecular characteristics in an acute-care tertiary hospital in Singapore. Epidemiol. Infect. 2018;146:1785–1792. doi: 10.1017/S0950268818001966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, Y.-C. et al. Detection, spread and phylogeny of meticillin-resistant Staphylococcus aureus sequence type 45 in Taiwan. Microb. Genomics7, 000555 (2021). [DOI] [PMC free article] [PubMed]
- 22.Effelsberg, N. et al. Global epidemiology and evolutionary history of Staphylococcus aureus St45. J. Clin. Microbiol.59, 10–1128 (2020). [DOI] [PMC free article] [PubMed]
- 23.Sherry, N. et al. Genomic interrogation of the burden and transmission of multidrug-resistant pathogens within and across hospital networks. bioRxiv10.1101/764787 (2019).
- 24.Turnidge JD, et al. Staphylococcus aureus bacteraemia: a major cause of mortality in Australia and New Zealand. Med. J. Aust. 2009;191:368–373. doi: 10.5694/j.1326-5377.2009.tb02841.x. [DOI] [PubMed] [Google Scholar]
- 25.Holmes NE, et al. Morbidity from in-hospital complications is greater than treatment failure in patients with Staphylococcus aureus bacteraemia. BMC Infect. Dis. 2018;18:107. doi: 10.1186/s12879-018-3011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coombs, G. W. et al. Australian Group on Antimicrobial Resistance (Agar) Australian Staphylococcus aureus Sepsis Outcome Programme (Assop) Annual Report 2015. Communicable Diseases Intelligence42, S2209-6051(18)00016-7 (2018). [PubMed]
- 27.Coombs, G. W. et al. Australian Group on Antimicrobial Resistance (Agar) Australian Staphylococcus aureus Sepsis Outcome Programme (Assop) Annual Report 2017. Communicable Diseases Intelligence43, 10.33321/cdi.2019.43.43 (2019). [DOI] [PubMed]
- 28.Petit RA, 3rd, Read TD. Staphylococcus aureus viewed from the perspective of 40,000+ genomes. PeerJ. 2018;6:e5261. doi: 10.7717/peerj.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ono HK, et al. Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect. Immun. 2008;76:4999–5005. doi: 10.1128/IAI.00045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monk, I. R. et al. Complete bypass of restriction systems for major Staphylococcus aureus lineages. mBio6, 10–1128 (2015). [DOI] [PMC free article] [PubMed]
- 31.Nong, Y. et al. Clinical relevance of topical antibiotic use in co-selecting for multidrug-resistant Staphylococcus aureus: insights from in vitro and ex vivo models. Antimicrob. Agents Chemother.65, 10–1128 (2021). [DOI] [PMC free article] [PubMed]
- 32.Hon PY, et al. Changing molecular epidemiology and high rates of mupirocin resistance among meticillin-resistant Staphylococcus aureus in Singaporean hospitals. J. Glob. Antimicrob. Resist. 2014;2:53–55. doi: 10.1016/j.jgar.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Liu C-Y, et al. Predominance of methicillin-resistant Staphylococcus aureus in the residents and environments of long-term care facilities in Taiwan. J. Microbiol. Immunol. Infect. 2019;52:62–74. doi: 10.1016/j.jmii.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Weinstein RA, et al. Chlorhexidine: expanding the armamentarium for infection control and prevention. Clin. Infect. Dis. 2008;46:274–281. doi: 10.1086/524736. [DOI] [PubMed] [Google Scholar]
- 35.Miyazaki NHT, et al. The presence of Qaca/B gene in Brazilian methicillin-resistant Staphylococcus aureus. Mem.órias do Inst. Oswaldo Cruz. 2007;102:539–540. doi: 10.1590/S0074-02762007000400018. [DOI] [PubMed] [Google Scholar]
- 36.Hasanvand A, et al. Antiseptic resistance in methicillin sensitive and methicillin resistant Staphylococcus aureus isolates from some major hospitals, Iran. Recent Pat. Anti-Infect. Drug Discov. 2015;10:105–112. doi: 10.2174/1574891X10666150623093259. [DOI] [PubMed] [Google Scholar]
- 37.Mayer S, et al. Distribution of the antiseptic resistance genes Qaca, Qacb and Qacc in 497 methicillin-resistant and -susceptible European isolates of Staphylococcus aureus. J. Antimicrob.Chemother. 2001;47:896–897. doi: 10.1093/jac/47.6.896. [DOI] [PubMed] [Google Scholar]
- 38.Horner C, et al. Reduced susceptibility to chlorhexidine in Staphylococci: is it increasing and does it matter? J. Antimicrob. Chemother. 2012;67:2547–2559. doi: 10.1093/jac/dks284. [DOI] [PubMed] [Google Scholar]
- 39.Conceição T, et al. Prevalence of biocide resistance genes and chlorhexidine and mupirocin non-susceptibility in Portuguese hospitals during a 31-year period (1985–2016) J. Glob. Antimicrob. Resist. 2021;24:169–174. doi: 10.1016/j.jgar.2020.12.010. [DOI] [PubMed] [Google Scholar]
- 40.Cieplik F, et al. Resistance toward chlorhexidine in oral bacteria—is there cause for concern? Front. Microbiol. 2019;10:587. doi: 10.3389/fmicb.2019.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim K-S, Kam PCA. Chlorhexidine-pharmacology and clinical applications. Anaesth. Intensive Care. 2008;36:502–512. doi: 10.1177/0310057X0803600404. [DOI] [PubMed] [Google Scholar]
- 42.Otter JA, et al. Selection for Qaca carriage in Cc22, but Not Cc30, methicillin-resistant Staphylococcus aureus bloodstream infection isolates during a successful institutional infection control programme. J. Antimicrob. Chemother. 2013;68:992–999. doi: 10.1093/jac/dks500. [DOI] [PubMed] [Google Scholar]
- 43.Hong, S. I. et al. Clinical and molecular characteristics of Qaca- and Qacb-positive methicillin-resistant Staphylococcus aureus causing bloodstream infections. Antimicrob. Agents Chemother.63, e02157 (2019). [DOI] [PMC free article] [PubMed]
- 44.Ho J, Branley J. Prevalence of antiseptic resistance genes Qaca/B and specific sequence types of methicillin-resistant Staphylococcus aureus in the era of hand hygiene. J. Antimicrob. Chemother. 2012;67:1549–1550. doi: 10.1093/jac/dks035. [DOI] [PubMed] [Google Scholar]
- 45.Carter GP, et al. Topical antibiotic use coselects for the carriage of mobile genetic elements conferring resistance to unrelated antimicrobials in Staphylococcus aureus. Antimicrob. Agents Chemother. 2018;62:AAC.02000–17. doi: 10.1128/AAC.02000-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wick RR, et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Computat. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 48.Feldgarden M, et al. Validating the Amrfinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 2019;63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaya, H., et al. Sccmecfinder, a web-based tool for typing of staphylococcal cassette chromosome Mec in Staphylococcus aureus using whole-genome sequence data. mSphere10.1128/msphere.00612-17 (2018). [DOI] [PMC free article] [PubMed]
- 50.Nguyen L-T, et al. Iq-Tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tonkin-Hill G, et al. Rhierbaps: an R implementation of the population clustering algorithm hierbaps. Wellcome Open Res. 2018;3:93–93. doi: 10.12688/wellcomeopenres.14694.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu G, et al. Ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017;8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 53.Bankevich A, et al. Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Page AJ, et al. Roary: rapid large-scale prokaryote Pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siguier P, et al. Isfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rambaut A, et al. Exploring the temporal structure of heterochronous sequences using tempest (formerly Path-O-Gen) Virus Evol. 2016;2:vew007–vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drummond AJ, Rambaut A. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rambaut A, et al. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minin VN, et al. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 2008;25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bouckaert R, et al. Beast 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stadler T, et al. Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV) Proc. Natl. Acad. Sci. USA. 2013;110:228–233. doi: 10.1073/pnas.1207965110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steinig, E. et al. Phylodynamic inference of bacterial outbreak parameters using nanopore sequencing. Mol. Biol. Evol.39, msac040 (2022). [DOI] [PMC free article] [PubMed]
- 63.Zhang Y, et al. Seamless ligation cloning extract (slice) cloning method. Methods Mol. Biol. 2014;1116:235–244. doi: 10.1007/978-1-62703-764-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.CLSI. Performance standards for antimicrobial susceptibility testing. In M07-A10: Methods for Dilution Antimicrobial Susceptibility (ed. CLSI) 27–41 (Clinical & Laboratory Standards Institute, 2015).
- 65.Guérillot R, et al. Convergent evolution driven by rifampin exacerbates the global burden of drug-resistant Staphylococcus aureus. mSphere. 2018;3:e00550-17. doi: 10.1128/mSphere.00550-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sprouffske K, Wagner A. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinforma. 2016;17:172. doi: 10.1186/s12859-016-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Villanueva, R. A. M. & Chen, Z. J. Ggplot2: Elegant Graphics for Data Analysis (Taylor & Francis, 2019).
- 68.Kassambara, A. Comparing Groups: Numerical Variables, Vol. 192 (Datanovia, 2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Sequence data for the isolates retrieved from publications and public databases can be accessed in accordance to Supplementary Note 1 and Supplementary Data 1. Additional genomic data generated in this study have been deposited under the BioProject PRJNA984755 in the NCBI database. The complete genome assembly of the ST45 S. aureus reference AUSMDU00020487 strain (GenBank accession numbers: CP138566-7) has been uploaded under the BioProject PRJNA565795. Numerical source data for competition assays, growth assays, and Re analysis are available on Figshare (10.26188/25201073).