Abstract
Fannia pusio, the chicken dung fly species, remains unexplored despite its forensic, sanitary, and veterinary importance in the Nearctic and Neotropical regions. In this study, we obtained the complete mitochondrial genome of Fannia pusio for the first time using next-generation sequencing. We compared it with previously published mitogenomes of the genus from the Palearctic region, and its phylogenetic position was studied based on the concatenated protein-coding genes (PCGs) dataset of Calyptratae flies. The circular mitochondrial genome of F. pusio is 16,176 bp in length, with a high A + T content (78.3%), whose gene synteny, codon usage analysis, and amino acid frequency are similar to previously reported Fannia mitogenomes. All PCGs underwent purifying selection except the nad2 gene. Interspecific K2P distances of PCGs of Fannia yielded an average of 12.4% (8.1%–21.1%). The Fannia genus is monophyletic and closely related to Muscidae based on molecular data. Further taxonomic sampling is required to deep into the phylogenetic relationships of the originally proposed species-groups and subgroups within the genus. These results provide a valuable dataset for studying the mitochondrial genome evolution and a resource for the taxonomy and systematics of Fannia.
Keywords: Illumina sequencing, Colombia, Neotropics, Phylogenomic
1. Introduction
The mitochondrial DNA (mtDNA) is inherited maternally and has a low recombination rate. It has evolutionary rates higher than the nuclear genome and a high copy number per cell, making it a valuable data source for species identification, evolutionary biology, and comparative genomics [1,2]. With the advent of next-generation sequencing (NGS) technologies, rapid data acquisition, lower cost, and time investment than Sanger sequencing have enabled researchers to increase the availability of insect mtDNA genomes [3]. This has led to comparative analyses based on all protein-coding genes (PCGs) rather than just relying on a single or few molecular markers [4].
Fannia Robineau-Desvoidy, 1830, is a genus of fly species within the Fanniidae family, encompassing approximately 350 species globally, with 113 documented in the Neotropics [5] and 11 groups and subgroups proposed based on adult morphology [6]. Previous phylogenetic analyses have relied mainly on morphological characters to support the monophyly of Fannia [7]. However, a phylogenetic approach based on all PCGs of the mitochondrial genomes and considering representative members of other Calyptratae has yet to be done. Species identification in this genus primarily relies on morphological characteristics of the male genitalia, while identifying females and immature stages is difficult or not always possible [8,9], and molecular reference data remain scarce [10,11].
Mitogenomes play an important role in providing data for evolutionary and phylogenetic analyses and revealing the genetic history of a population [12,13]. Despite the increase of published mitochondrial genomes, only few are available for most Calyptratae flies, particularly the Fannia genus [14]. Until now, the mitogenomes of Fannia scalaris Fabricius, 1794 (NC_053661) [15], Fannia canicularis (Linnaeus, 1761) (NC_068710) [16], and Fannia armata (Meigen, 1826) (MT628564) are available from specimens collected in the Palearctic region. However, mitogenomes of species from other biogeographical regions have not yet been published.
Fannia pusio (Wiedemann, 1830) has forensic, sanitary, and veterinary importance [[17], [18], [19]] and is widely recorded in the Nearctic and Neotropical regions [20]. Herein, we sequenced, assembled, and annotated the complete mitochondrial genome of Fannia pusio and performed comparative analyses with the previously published mitogenomes of the genus from specimens collected in the Palearctic region. In addition, we explored the phylogenetic position of Fannia pusio among other calyptrate flies based on all concatenated PCGs dataset.
2. Materials and methods
2.1. Sample collection and DNA extraction
One F. pusio male specimen was field collected in March 2011 in Pajarito, a locality from Medellin, Antioquia, Colombia (06°17′10.7″N, 75°36′43.7″W) at 1929 m above sea level and identified using morphological keys [[21], [22], [23]]. Specimen collection was done under collection permit 16455 issued by CORANTIOQUIA on May 18, 2011. The specimen was preserved in 95% ethanol and stored at −20 °C until DNA extraction. Then, the specimen was dried at room temperature and photographed using a digital camera OPTIKAM Pro 3 connected to a trinocular stereomicroscope (Nikon SMZ745T, Nikon Corp., Tokyo, Japan) as support for species identification. The abdomen was dissected for total genomic DNA using a GenElute™ Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich) following the manufacturer's protocol. The DNA was quantified using the Qubit dsDNA High Sensitivity assay (Thermo Fisher Scientific, Waltham, MA, USA) and stored at −20 °C until further processing. Species identification was confirmed by comparing COI sequences at the barcode of life data systems BOLD and GenBank databases [24,25].
2.2. Mitogenome sequencing
The F. pusio DNA library was sequenced using an Illumina NovaSeq 6000 (Illumina, Inc., California, US) with 2 × 150 bp pair-end reads, constructed using the TruSeq DNA Nano kit. Reads after trimming were required to have a minimum length of 50 bp. Adapters were removed, and quality was trimmed with Trimmomatic v. 039 [26]. Clean data were de novo assembled using SPAdes v. 3.15.4 with the mode read error correction and assembling with the following parameters: PHRED offset auto-detect, k = 55–77, repeat resolution enabled, mismatch careful mode OFF, mismatch corrector SKIPPED and coverage cutoff OFF [27]. A homology search was implemented to identify the contigs associated with the mitochondrial genome. All contigs were compared to the mitogenome of F. scalaris (NC_053661.1) using BLASTn v. 2.13.0 [28] with an e-value of 1e-15. To confirm that the mitogenome was completed, the ends of the identified contig were joined, and the inner region was split randomly, creating an alternative contig, which was used to map the reads with BWA v. 0.7.17 [29] and default parameters. The localization of the mapped reads was visualized in Tablet v. 1.21.02.08 [30].
The assembled mitogenome was annotated using the MITOS web server [31] based on invertebrate mitochondrial genetic code, and annotation curation was performed through comparison with closely related species within the graphical environment of the Geneious Prime software (Biomatters Ltd., v.11.0.15). In addition, PCGs were reconfirmed using NCBI ORFfinder [32], and tRNAs were rechecked using tRNAscan-SE v. 2.0 [33]. The circular map of the complete mitogenome was drawn with Geneious Prime software v.11.0.15 (Biomatters Ltd.).
2.3. Comparative analysis among mitogenomes of Fannia
Three species of Fannia with available mitogenomes to date (December 12, 2023) were downloaded for comparison with the new F. pusio mitogenome (Table 1). The eZmito pipeline was used to calculate and visualize strand, codon, and positional nucleotide biases with the following formulas: AT skew = [A−T]/[A + T], GC skew = [G–C]/[G + C] [34], also to calculate and visualize amino acid and codon usage, and Relative Synonymous Codon Usage (RSCU) across mitogenomes [35]. Genetic distances were calculated using the Kimura 2-parameter (K2P) model between each pair of the 13 core PCGs in MEGA v.11 [36]. Base composition and genome synteny analyses were performed on the Geneious Prime software v.11.0.15 (Biomatters Ltd.). We used DnaSP v. 6.12.03 [37] to calculate the nonsynonymous substitution rate (Ka) and the synonymous substitution rate (Ks), as well as the Ka/Ks ratio for all 13 core PCGs in the Fannia mitogenomes. Furthermore, we performed a sliding window analysis of whole mitogenomes using a window size of 200 bp and a step size of 20 bp.
Table 1.
Mitogenomes of the fly species sampled in this study.
| Species | Family | GenBank accession number |
|---|---|---|
| Delia antiqua | Anthomyiidae | NC_028226 |
| Fucellia costalis | Anthomyiidae | NC_042770 |
| Hylemya nigrimana | Anthomyiidae | NC_063908 |
| Lispe assimilis | Anthomyiidae | NC_058292 |
| Pegoplata infirma | Anthomyiidae | NC_050312 |
| Aldrichina grahami | Calliphoridae | NC_026996 |
| Chrysomya albiceps | Calliphoridae | NC_019631 |
| Chrysomya rufifacies | Calliphoridae | NC_019634 |
| Lucilia cuprina | Calliphoridae | NC_019573 |
| Phormia regina | Calliphoridae | NC_026668 |
| Chlorops oryzae | Chloropidae | NC_059894 |
| Drosophila melanogaster | Drosophilidae | NC_024511 |
| Drosophila suzukii | Drosophilidae | NC_060762 |
| Drosophila pseudoobscura | Drosophilidae | NC_046603 |
| Drosophila curta | Drosophilidae | NC_060566 |
| Hirtodrosophila subflavohalterata | Drosophilidae | NC_070279 |
| Paraliodrosophila antennata | Drosophilidae | NC_070278 |
| Fannia canicularis | Fanniidae | NC_068710 |
| Fannia scalaris | Fanniidae | NC_053661 |
| Fannia armata | Fanniidae | MT628564 |
| Fannia pusio | Fanniidae | OQ692989a |
| Melophagus ovinus | Hippoboscidae | NC_037368 |
| Ornithomya biloba | Hippoboscidae | NC_061211 |
| Graphomya rufitibia | Muscidae | NC_038210 |
| Hydrotaea aenescens | Muscidae | NC_042952 |
| Hydrotaea chalcogaster | Muscidae | NC_041089 |
| Mesembrina meridiana | Muscidae | NC_063930 |
| Musca domestica | Muscidae | NC_024855 |
| Synthesiomyia nudiseta | Muscidae | NC_042953 |
| Azelia sp. | Muscidae | KP901269b |
| Nycteribia parvula | Nycteribiidae | NC_068095 |
| Phthiridium szechuanum | Nycteribiidae | NC_068222 |
| Cephalopina titillator | Oestridae | NC_046479 |
| Dermatobia hominis | Oestridae | NC_006378 |
| Gasterophilus intestinalis | Oestridae | NC_029834 |
| Gyrostigma rhinocerontis | Oestridae | NC_042379 |
| Hypoderma lineatum | Oestridae | NC_013932 |
| Oestrus ovis | Oestridae | NC_059851 |
| Rhinoestrus usbekistanicus | Oestridae | NC_045882 |
| Pollenia pediculata | Polleniidae | NC_053684 |
| Blaesoxipha lapidosa | Sarcophagidae | NC_063664 |
| Miltogramma oestracea | Sarcophagidae | NC_059872 |
| Oxysarcodexia thornax | Sarcophagidae | NC_041072 |
| Peckia collusor | Sarcophagidae | NC_041079 |
| Peckia resona | Sarcophagidae | NC_041077 |
| Ravinia pernix | Sarcophagidae | NC_026196 |
| Sarcophaga diminuta | Sarcophagidae | NC_053674 |
| Sarcophaga josephi | Sarcophagidae | NC_053666 |
| Sarcophaga pauciseta | Sarcophagidae | NC_053729 |
| Sarcophaga schuetzei | Sarcophagidae | NC_053681 |
| Sarcophaga tuberosa | Sarcophagidae | NC_047405 |
| Taxigramma karakulensis | Sarcophagidae | NC_069629 |
| Paradyschiria parvula | Streblidae | NC_044702 |
| Paratrichobius longicrus | Streblidae | NC_044652 |
| Clemelis pullata | Tachinidae | NC_039963 |
| Ectophasia rotundiventris | Tachinidae | NC_050938 |
| Exorista civilis | Tachinidae | NC_039824 |
| Lydina aenea | Tachinidae | NC_063609 |
| Peleteria iavana | Tachinidae | NC_063086 |
| Winthemia sumatrana | Tachinidae | NC_065138 |
Mitogenome obtained in this study.
The species was originally identified as Euryomma sp. but later assigned as Azelia sp., a member of the Muscidae family (Grzywacz et al., 2021b).
2.4. Phylogenetic analysis
Sixty complete or nearly complete mitochondrial genomes of representative species of the subsection Calyptratae (Diptera: Schizophora), including Fannia pusio with some Drosophilidae species and Chlorops oryzae (Diptera: Choloropidae) as outgroups (Table 1), were used for phylogenetic analyses. Entire mitogenome records were downloaded using NCBI Batch Entrez (https://www.ncbi.nlm.nih.gov/sites/batchentrez). Then, eZsplit and eZpipe were used to prepare the PCG files [35]. Each PCG was manually aligned, checked, and corrected in Geneious Prime software v.11.0.15 (Biomatters Ltd.) for quality control. A concatenated dataset from the PCGs was obtained to assess the phylogenetic relationships under probabilistic methods, Maximum Likelihood (ML) and Bayesian Inference (BI). We analyzed PCG matrices, including nucleotides in all three codon positions (PCG123) and the first and second codon positions of the protein codon genes (PCG12). Additional phylogenetic analyses were not feasible due to the unavailability of complete mitochondrial genomes for all the included taxa (i.e., rrnL, rrnS).
We used the best-fit model (GTR + G + I) determined with jModelTest2 v. 2.1.10 [38,39] for the ML analysis using IQ-TREE v.1.6.1 [40] with a combination of rapid hill-climbing and stochastic perturbation methods and 1000 bootstrap replicates.
BI analyses were performed using PhyloBayes MPI v. 1.9 [41] using the site-heterogeneous mixture model CAT + GTR, a model more suitable for larger multigene alignments to avoid phylogenetic biases [42]. In each analysis, two independent Markov Chain Monte Carlo (MCMC) chains were run after the removal of constant sites from the alignment and were stopped after the two runs had satisfactorily converged (maxdiff <0.1). A consensus tree was computed from the remaining trees combined from 2 runs after each run's initial 25% trees were discarded as burn-in.
All phylogenetic analyses were conducted on the CIPRES Science Gateway [43] in the High-Performance Computing Cluster at the University of Kentucky Analytics and Technologies. The resulting phylogenetic trees were visualized in iTOL v. 6.7.1 [44] and final editing was performed with Inkscape v.1.3.
3. Results and discussion
Overall, 7987207 reads were obtained during the sequencing. After the genome assembly, 603705 contigs were assembled with an N50 of 310 nt, an average coverage of 95.2X and a median coverage of 1.5X. The homology search identified only one contig with 88% identity to the F. scalaris genome and an e-value of 0. This contig exhibited a coverage of 225X and a length of 16443 nt. The mitogenome completeness analysis allowed us to detect an artefactual repetitive sequence produced during the assembly at both ends of the contig. After a trim, join, and read mapping strategy, the ends of the contigs were corrected.
3.1. Genome size
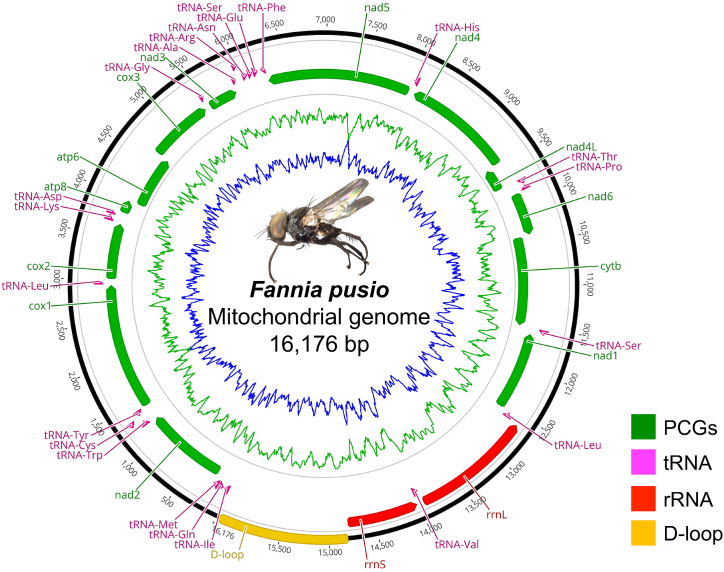
The circular mitochondrial genome of F. pusio is 16176 bp in length. It contains 13 PCGs, 22 tRNA genes, two rRNA genes, and an A + T-rich region (89.3%), also known as D-loop, with a length of 1365 bp (Fig. 1). The nucleotide composition of the whole mitogenome is A 40.1%, C 12.6%, G 9%, and T 38.3%, with a high AT content (78.3%), like other members in the genus: F. scalaris (77.9%), F. canicularis (79.3%), and F. armata (77.8%). Similar results have been found in other insect mitochondrial genomes [45]. In addition, the PCGs in the mitochondrial genome of Fannia pusio are typical of insect mitogenomes in terms of length and topology [46].
Fig. 1.
Gene map of the complete mitochondrial genome of Fannia pusio. Arrows indicate the direction of gene transcription. Gene names are abbreviated as follows: nad for NADH dehydrogenase subunits 1–6 and 4l; cytb for cytochrome b; cox for cytochrome oxidase subunits 1–3; atp6 and atp8 for ATP synthase subunits; rrnL and rrnS for large and small rRNA subunits; tRNA genes are indicated; D-loop for control region. The GC content was plotted using a green sliding window and the AT content was blue.
3.2. Gene synteny analysis and tRNAs
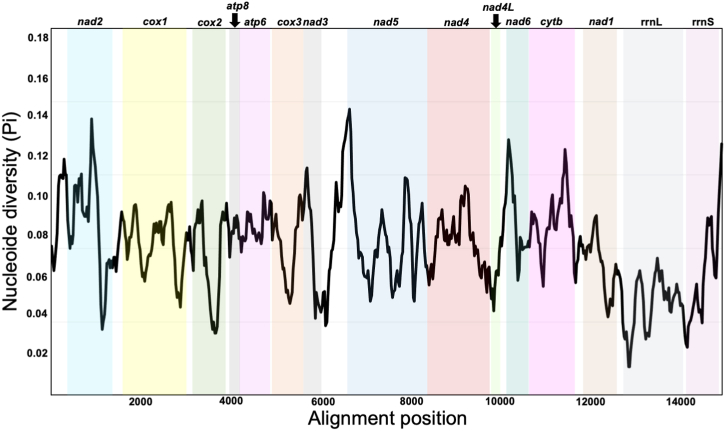
The four mitogenomes from Fannia encode an essential set of conserved genes, including three cytochrome c oxidase subunits (cox1, cox2, cox3), cytochrome b (cob), two ATP synthase subunits (atp6, atp8), seven subunits of NADH dehydrogenase (nad1, nad2, nad3, nad4, nad4L, nad5, nad6), and the small and large ribosomal RNA subunits (rrnS, rrnL). Comparison among members of the genus evidenced a highly conserved gene synteny among its mitogenomes [16,47] and other dipterans [48,49]. The highest nucleotide diversity values were detected for the nad2, nad5 and nad6 genes (Fig. 2). Previous studies in insect mitogenomes regarding to phylogenetic signal, highlight nad2 among the best genes for constructing a tree topology, while nad5 and nad6 are the best genes contributing to the branch lengths [50].
Fig. 2.
Nucleotide diversity (Pi) among the Fannia mitogenomes. The Pi values were calculated from a sliding window analysis of 200 bp in 25 bp steps, represented on the y-axis. The length values of the aligned sequence are represented on the x-axis. The limits of each PCG or rRNA are indicated by coloured bars.
In the new F. pusio mitogenome gene overlap exists between trnW/nad2 (2 bp), trnW/trnC (8 bp), trnY/cox1 (2 bp), cox1/trnL(4 bp), atp8/atp6 (7 bp), atp6/cox3 (1 bp), trnA/trnR (1 bp), nad4/nad4L (1 bp), nad6/cytb (2 bp), cob/trnS (2 bp) and trnV/rrnS (2 bp). Small non-coding regions are mostly 1–6 bp, with the longest between trnS2/nad1 (16 bp) and trnE/trnF (20 bp). Nine PCGs are transcribed on the majority strand (J-strand), whereas four are oriented on the minority strand (N-strand). On the other hand, it has 22 tRNA genes, ranging in size from 63 bp to 72 bp. J strand has 14 tRNAs, and the N-strand has 8 tRNAs. There is one tRNA for each amino acid except Leucine and Serine, which are encoded by two tRNAs each. Fannia species evidence a conserved arrangement pattern of tRNA genes, identical to other calyptrate flies [15,51].
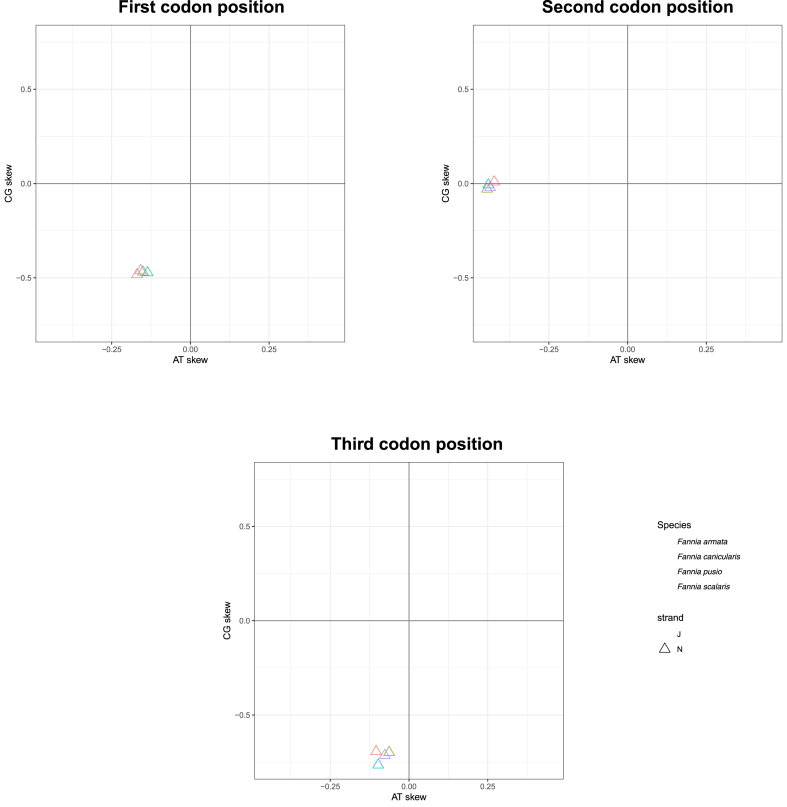
3.3. At and GC skew
The base composition of each codon position for the 13 PCGs shows they all have a high A + T percentage (70.4%–84%). Different codon positions of PCGs show various skew statistics (Fig. 3). The genes on the J-strand showed AT-skew at the first and second codon positions; in the third codon position, all species, except for F. pusio and F. armata, were AT-skewed. GC-skew was evidenced at the first codon position on the J-strand. The genes on the J-strand had a higher frequency of T (40.9%) and A (34.1%). All species were AT-skewed and GC-skewed on the N-strand for the three codon positions, except for F. armata at the second. The genes on the N-strand had a higher frequency of T (48.1%) and A (31.5%). Strand bias in nucleotide composition is concordant with most insect mitochondrial genomes [52].
Fig. 3.
Cytosine + Guanine (CG) vs. Adenine + Thymine (AT) skew by codon position with species colour-coded. Strand is indicated using shapes.
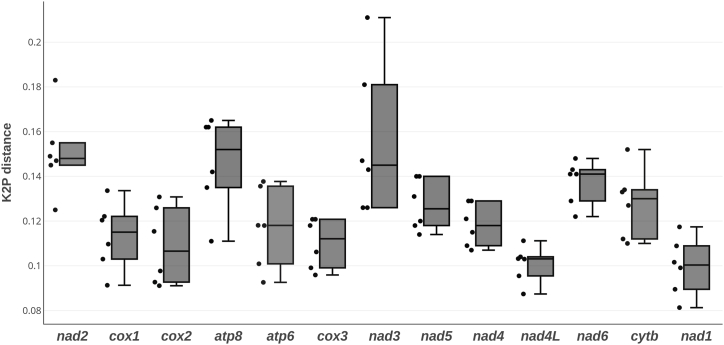
3.4. Interspecific genetic distances among all PCGs
Interspecific K2P distances yielded an average of 12.4%, with a minimum of 8.1% for the nad1 gene between F. canicularis and F. armata, and a maximum of 21.1% for the nad3 between F. armata and F. pusio (Fig. 4). The lowest variance was found for the nad5 gene, while the highest was detected in the nad3 gene. The interspecific K2P distance for the cox1 gene ranged from 9.1% to 13.4%, with an average of 11.3%. Interspecific K2P values for the PCGs are like those found in other flies [[53], [54], [55], [56]]. The success of the cox1 as a species diagnosis marker and the potential of the other ones requires further sampling to recognize the intraspecific diversity for each PCG [57,58]. Preliminary analyses have shown promising results with the barcode region of the cox1 gene for species identification of Fanniidae [11,59].
Fig. 4.
Interspecific pairwise Kimura 2-parameter (K2P) distances of 13 core protein-coding genes among five species of the Fannia genus. Box and whisker plot representing the median, upper and lower quartiles, and minimum and maximum values.
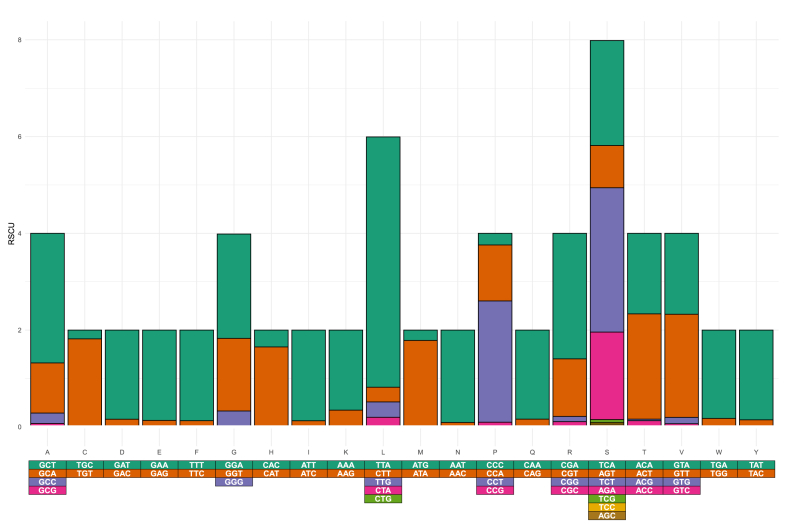
3.5. Codon usage analysis and amino acid frequency
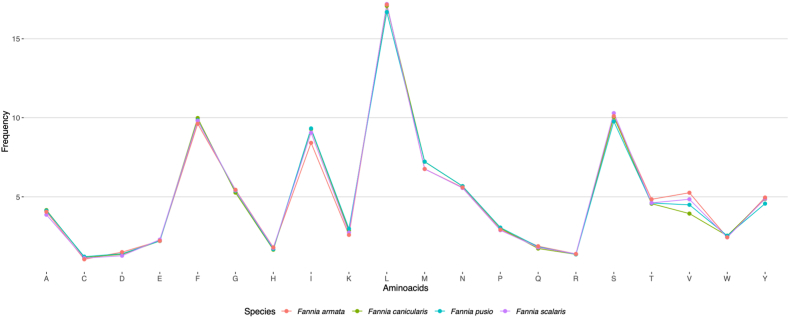
A codon usage bias was detected for F. pusio codons (52.1% of the RSCU values were higher than 1.0). The most frequently used codons for each amino acid are depicted in Fig. 5. The frequencies of amino acids encoded by the four Fannia mitogenomes are highly similar (Fig. 6). Most of the codons encode for non-polar amino acids like Leucine (L), Phenylalanine (F), and Isoleucine (I). In contrast, few codons encode for the polar-uncharged Cysteine (C), the basic Arginine (R), or the Aspartic acid (D), among others. The commonly encoded amino acids for the Fannia species mitogenomes are like those found in other insects [[60], [61], [62], [63]].
Fig. 5.
Relative Synonymous Codon Usage (RSCU) of Fannia pusio. Codons are color-coded.
Fig. 6.
Complete amino acid frequencies by mitogenome. Genomes are color-coded.
3.6. Evolutionary rates of PCGs
The mean nucleotide frequencies of the PCGs are A = 33.3%, T/U = 43.1%, C = 11.4%, and G = 10.9%, respectively. Among the 13 mitochondrial PCGs, the ratios of Ka/Ks were less than 1 for almost all, except for nad2, indicating a strong purifying selection on most PCGs [64]. A higher Ka/Ks ratio in nad2, compared to the other PCGs, has been reported in other mitochondrial genomes [[65], [66], [67]], likely related to its small distance from the origin of replication (D-Loop) that exposes it to a higher mutation rate than other PCGs [68], its function [69] or a positive selection in the nad2 gene [64]. It is an exception, considering that PCGs belonging to complex I region (nad genes) have shown a weaker purification pressure compared to PCGs from other mitochondrial complex regions [70].
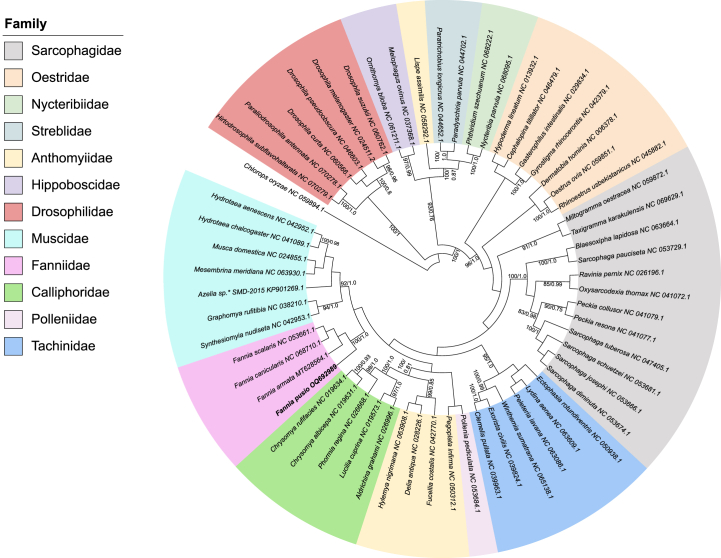
3.7. Phylogenetic analysis
Both the single gene and concatenated analyses, based on ML and BI approaches, supported the monophyly of the Fannia and the position of F. pusio as a member of the genus (Fig. 7). Interestingly, including the third codon position (i.e., PCG123 matrix) allowed a better tree resolution than just the PCG12 matrix. Despite third codon positions usually being highly saturated and recommended for exclusion [71,72], our dataset did not reach saturation at this position, adding information for phylogenetic reconstruction. The monophyletic status of Fannia based on PCGs of the mitochondrial genome is congruent with previous data based on adult external morphology, female and male terminalia [7], as well as molecular data [73]. Though some authors proposed species groups and subgroups within Fannia based on morphological data and a cladistic analysis [7], with the current available molecular data is not possible to test this phylogenetic hypothesis. Thus, we highlight the need for further taxonomic sampling within the Fannia genus. Based on morphological data, Fanniidae was initially considered a subfamily of Muscidae during the 1960s (i.e., Fanniinae) [74,75], but later, most specialists recognized it as a separate family within Muscoidea [7,21,[76], [77], [78], [79]]. Herein, the Fanniidae family was closer to the Muscidae, Calliphoridae and Anthomyiidae families, as previously indicated based on a molecular phylogeny of the Calyptratae using mitochondrial and nuclear markers [80] and the musculature of the male terminalia [81].
Fig. 7.
Phylogenetic analysis of Fannia pusio among other members of the Calyptratae (Diptera: Schizophora) based on 13 protein-coding genes. The branches show ML bootstrap support (80–100%) and Bayesian posterior probabilities (0.8–1.0).
4. Conclusions
The mitochondrial genome of Fannia pusio has been sequenced for the first time, revealing a size of 16,176 bp. The gene synteny, codon usage analysis, and amino acid frequency are similar to other Fannia species reported previously from the Palearctic region. Phylogenetic analysis supports the monophyly of the genus and its close relationship with the Muscidae family. Further taxonomic sampling is required to delve deep into the phylogenetic hypothesis of the proposed species-groups and subgroups based only on morphology and to study the mitochondrial genome evolution in the Fannia genus, particularly in the Neotropics where there is a diversity concentration of Fannia compared to other biogeographic regions.
Data availability
The complete Fannia pusio mitochondrial genome and Illumina raw sequence reads have been deposited in GenBank (OQ692989) and the Sequence Read Archive (BioProject ID: PRJNA1003469, SRR25570796), respectively. Mitogenomes from other Fannia species and others were downloaded from GenBank, with accession numbers and references listed in Table 1.
Additional information
No additional information is available for this paper.
CRediT authorship contribution statement
Yesica S. Durango-Manrique: Writing – review & editing, Investigation, Data curation, Conceptualization. Andrés López-Rubio: Writing – review & editing, Resources, Investigation, Formal analysis. Lina A. Gutiérrez: Writing – review & editing, Resources, Methodology, Investigation, Funding acquisition, Conceptualization. Juan P. Isaza: Writing – review & editing, Resources, Methodology, Investigation, Formal analysis, Data curation. Giovan F. Gómez: Writing – review & editing, Writing – original draft, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Comité para el Desarrollo de la Investigación -CODEI-, Tecnológico de Antioquia (Grant 206001206); Dirección de Investigación y Transferencia -CIDI, Universidad Pontificia Bolivariana; and Universidad Nacional de Colombia. The funders had no role in study design, data collection and analysis, the decision to publish, or manuscript preparation. We thank the Bioforense research group members who performed field collections and the Colección Entomológica Tecnológico de Antioquia (CETdeA).
Contributor Information
Yesica S. Durango-Manrique, Email: durangom.yesica@gmail.com.
Andrés López-Rubio, Email: alopezru@tdea.edu.co.
Lina A. Gutiérrez, Email: lina.gutierrezb@upb.edu.co.
Juan P. Isaza, Email: juan.isazaa@upb.edu.co.
Giovan F. Gómez, Email: gfgomezg@unal.edu.co.
References
- 1.Cameron S.L. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu. Rev. Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- 2.Allio R., Donega S., Galtier N., Nabholz B. Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Mol. Biol. Evol. 2017;34:2762–2772. doi: 10.1093/molbev/msx197. [DOI] [PubMed] [Google Scholar]
- 3.Wachi N., Matsubayashi K.W., Maeto K. Application of next-generation sequencing to the study of non-model insects. Entomol. Sci. 2018;21:3–11. doi: 10.1111/ens.12281. [DOI] [Google Scholar]
- 4.Cameron S.L., Lambkin C.L., Barker S.C., Whiting M.F. A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst. Entomol. 2007;32:40–59. doi: 10.1111/j.1365-3113.2006.00355.x. [DOI] [Google Scholar]
- 5.Durango-Manrique Y., López-Rubio A., Gómez G.F. vol. 15. Boletín del Museo Entomológico Francisco Luis Gallego; 2023. pp. 9–31. (Una revisión del género Fannia Robineau-Desvoidy, 1830 en el Neotrópico). [Google Scholar]
- 6.Albuquerque D. de O., Pamplona D., De Carvalho C.J.B. Arquivos Do Museu Nacional; 1981. Contribuição ao conhecimento dos Fannia R. D., 1830 da região neotropical. (Diptera, Fanniidae) pp. 9–34. [Google Scholar]
- 7.Domínguez M.C., Roig-Juñent S.A. A phylogeny of the family Fanniidae Schnabl (Insecta: Diptera: Calyptratae) based on adult morphological characters, with special reference to the Austral species of the genus Fannia. Invertebr Syst. 2008;22:563–587. doi: 10.1071/IS08003. [DOI] [Google Scholar]
- 8.Grzywacz A., Prado Castro C. New records of Fannia Robineau-Desvoidy (Diptera: Fanniidae) collected on pig carrion in Portugal with additional data on the distribution of F. conspecta Rudzinski. Entomol Fenn. 2003;23(2012):169–176. doi: 10.33338/ef.84582. [DOI] [Google Scholar]
- 9.Grzywacz A., Trzeciak P., Wiegmann B.M., Cassel B.K., Pape T., Walczak K., Bystrowski C., Nelson L., Piwczyński M. Towards a new classification of Muscidae (Diptera): a comparison of hypotheses based on multiple molecular phylogenetic approaches. Syst. Entomol. 2021;46:508–525. doi: 10.1111/syen.12473. [DOI] [Google Scholar]
- 10.Domínguez M.C. Description of a new species of Fannia (Diptera: Fanniidae) from the argentinean patagonia. Rev. Soc. Entomol. Argent. 2023;82:22–27. [Google Scholar]
- 11.Grzywacz A., Jarmusz M., Walczak K., Skowronek R., Johnston N.P., Szpila K. DNA barcoding identifies unknown females and larvae of Fannia R.-D. (Diptera: Fanniidae) from carrion succession experiment and case report. Insects. 2021;12:381. doi: 10.3390/insects12050381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boore J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. www.biology.lsa [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun H., Zhou K., Song D. Mitochondrial genome and phylogenetic reconstruction of arthropods. Zool. Res. 2003;24 [Google Scholar]
- 14.Shao S., Yang L., Hu G., Li L., Wang Y., Tao L. Application of omics techniques in forensic entomology research. Acta Trop. 2023;246 doi: 10.1016/j.actatropica.2023.106985. [DOI] [PubMed] [Google Scholar]
- 15.Ren L., Zhang X., Li Y., Shang Y., Chen S., Wang S., Qu Y., Cai J., Guo Y. Comparative analysis of mitochondrial genomes among the subfamily Sarcophaginae (Diptera: sarcophagidae) and phylogenetic implications. Int. J. Biol. Macromol. 2020;161:214–222. doi: 10.1016/j.ijbiomac.2020.06.043. [DOI] [PubMed] [Google Scholar]
- 16.Ge M., Wang D., Liang H., Zhu J., Shi X., Tian J. The complete mitochondrial genome of Fannia canicularis (Diptera: Fanniidae) Mitochondrial DNA B Resour. 2022;7:1841–1842. doi: 10.1080/23802359.2022.2134744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasconcelos S.D., Costa D.L., Oliveira D.L. Entomological evidence in a case of a suicide victim by hanging: first collaboration between entomologists and forensic police in north-eastern Brazil. Aust. J. Forensic Sci. 2019;51:231–239. doi: 10.1080/00450618.2017.1356870. [DOI] [Google Scholar]
- 18.Carvalho L.M.L., Thyssen P.J., Linhares A.X., Palhares F. A checklist of arthropods associated with pig carrion and human corpses in Southeastern Brazil. Mem. Inst. Oswaldo Cruz. 2000;95:135–138. doi: 10.1590/S0074-02762000000100023. [DOI] [PubMed] [Google Scholar]
- 19.Rezende L. do C., de Oliveira T.M., Teixeira C.M., Santos M.A. de S., Cunha L.M., Silva M.X., Martins N.R. da S. Synanthropic diptera affecting layer poultry farms: a review. Arq Inst Biol (São Paulo). 2019;86 doi: 10.1590/1808-1657000922017. [DOI] [Google Scholar]
- 20.De Carvalho C., Pont A., Couri M., Pamplona D. A catalogue of the Fanniidae (Diptera) of the neotropical region. Zootaxa. 2003;219:1–32. doi: 10.5281/zenodo.156183. [DOI] [Google Scholar]
- 21.Grisales D., de Carvalho C.J.B. Highland biodiversity of Fanniidae (insecta, Diptera): fourteen new species from the andes and Central America. Zootaxa. 2019:330–360. doi: 10.11646/zootaxa.4551.3.4. [DOI] [PubMed] [Google Scholar]
- 22.Durango Y., Ramírez-Mora M. Fannia Robineau-Desvoidy (Diptera: Fanniidae) of Colombia: new species, identification key and updated checklist. Zootaxa. 2019;4604:301–325. doi: 10.11646/zootaxa.4604.2.4. [DOI] [PubMed] [Google Scholar]
- 23.Barros de Carvalho C.J., de Mello-Patiu C.A. Key to the adults of the most common forensic species of Diptera in South America. Rev. Bras. Entomol. 2008;52:390–406. doi: 10.1590/S0085-56262008000300012. [DOI] [Google Scholar]
- 24.Ratnasingham S., Hebert P.D.N. BOLD: the barcode of life data system. Mol. Ecol. Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. http://www.barcodinglife.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., Karsch-Mizrachi I., GenBank Nucleic Acids Res. 2020;48:D84. doi: 10.1093/nar/gkz956. –D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prjibelski A., Antipov D., Meleshko D., Lapidus A., Korobeynikov A. Using SPAdes de novo assembler. Current Protocols in Bioinformatics. 2020;70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 28.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milne I., Stephen G., Bayer M., Pritchard L., Cardle L., Shawand P.D., Marshall D. Using tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 31.Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., Pütz J., Middendorf M., Stadler P.F. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 32.Rombel I.T., Sykes K.F., Rayner S., Johnston S.A., Orf-Finder A vector for high-throughput gene identification. Gene. 2002;282:33–41. doi: 10.1016/S0378-1119(01)00819-8. [DOI] [PubMed] [Google Scholar]
- 33.Chan P.P., Lin B.Y., Mak A.J., Lowe T.M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021;49:9077–9096. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perna N.T., Kocher T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995;41:353–358. doi: 10.1007/BF00186547. [DOI] [PubMed] [Google Scholar]
- 35.Cucini C., Leo C., Iannotti N., Boschi S., Brunetti C., Pons J., Fanciulli P.P., Frati F., Carapelli A., Nardi F. EZmito: a simple and fast tool for multiple mitogenome analyses. Mitochondrial DNA B Resour. 2021;6:1101–1109. doi: 10.1080/23802359.2021.1899865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rozas J., Ferrer-Mata A., Sanchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sanchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 38.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by Maximum Likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 39.Darriba D., Taboada G.L., Doallo R., Posada D. JModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q., Iq-Tree A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lartillot N., Lepage T., Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 42.Song F., Li H., Jiang P., Zhou X., Liu J., Sun C., Vogler A.P., Cai W. Capturing the phylogeny of holometabola with mitochondrial genome data and Bayesian site-heterogeneous mixture models. Genome Biol Evol. 2016;8:1411–1426. doi: 10.1093/gbe/evw086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller M.A., Pfeiffer W., Schwartz T. 2010 Gateway Computing Environments Workshop. GCE); 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. [Google Scholar]
- 44.Ciccarelli F.D., Doerks T., Von Mering C., Creevey C.J., Snel B., Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311(1979):1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 45.Sun Z., Wan D.G., Murphy R.W., Ma L., Zhang X.S., Huang D.W. Comparison of base composition and codon usage in insect mitochondrial genomes. Genes Genom. 2009;31:65–72. doi: 10.1007/BF03191139. [DOI] [Google Scholar]
- 46.Chandra S.B.C., Vlk J.L., Kapatral V. Comparative insect mitochondrial genomes: differences despite conserved genome synteny. Afr. J. Biotechnol. 2006;5:1308–1318. http://www.academicjournals.org/AJB [Google Scholar]
- 47.Xie H., Sun L., Huang J., Wang H., Jiao Y., Yan J., Guan Y. The complete mitochondrial genome of Fannia scalaris (Diptera: Muscidae) Mitochondrial DNA B Resour. 2021;6:1757–1758. doi: 10.1080/23802359.2020.1820391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis O.L., Farr C.L., Kaguni L.S. Drosophila melanogaster mitochondrial DNA: completion of the nucleotide sequence and evolutionary comparisons. Insect Mol. Biol. 1995;4:263–278. doi: 10.1111/j.1365-2583.1995.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 49.Staton J.L., Daehler L.L., Brown W.M. Mitochondrial gene arrangement of the horseshoe crab Limulus polyphemus L.: conservation of major features among arthropod classes. Mol. Biol. Evol. 1997;14:867–874. doi: 10.1093/oxfordjournals.molbev.a025828. [DOI] [PubMed] [Google Scholar]
- 50.Talavera G., Vila R. What is the phylogenetic signal limit from mitogenomes? the reconciliation between mitochondrial and nuclear data in the Insecta class phylogeny. BMC Evol. Biol. 2011;11 doi: 10.1186/1471-2148-11-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shang Y., Ren L., Zhang X., Li Y., Zhang C., Guo Y. Characterization and comparative analysis of mitochondrial genomes among the Calliphoridae (Insecta: Diptera: oestroidea) and phylogenetic implications. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.799203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hassanin A. Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol. Phylogenet. Evol. 2006;38:100–116. doi: 10.1016/j.ympev.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 53.GilArriortua M., Saloña Bordas M.I., Kohnemann S., Pfeiffer H., de Pancorbo M.M. Molecular differentiation of Central European blowfly species (Diptera, Calliphoridae) using mitochondrial and nuclear genetic markers. Forensic Sci. Int. 2014;242:274–282. doi: 10.1016/j.forsciint.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 54.Singh D., Achint R. Molecular identification of some Indian Muscid flies (Diptera: Muscidae) based on mitochondrial gene COII. Int J Zool Stud. 2017;2:101–105. [Google Scholar]
- 55.Zajac B.K., Martin-Vega D., Feddern N., Fremdt H., e Castro C.P., Szpila K., Reckel F., Schütt S., Verhoff M.A., Amendt J., Zehner R. Molecular identification and phylogenetic analysis of the forensically important family Piophilidae (Diptera) from different European locations. Forensic Sci. Int. 2016;259:77–84. doi: 10.1016/j.forsciint.2015.12.024. [DOI] [PubMed] [Google Scholar]
- 56.Zajac B.K., Sontigun N., Wannasan A., Verhoff M.A., Sukontason K., Amendt J., Zehner R. Application of DNA barcoding for identifying forensically relevant Diptera from northern Thailand. Parasitol. Res. 2016;115:2307–2320. doi: 10.1007/s00436-016-4977-6. [DOI] [PubMed] [Google Scholar]
- 57.Bergsten J., Bilton D.T., Fujisawa T., Elliott M., Monaghan M.T., Balke M., Hendrich L., Geijer J., Herrmann J., Foster G.N., Ribera I., Nilsson A.N., Barraclough T.G., Vogler A.P. The effect of geographical scale of sampling on DNA barcoding. Syst. Biol. 2012;61:851–869. doi: 10.1093/sysbio/sys037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phillips J.D., Gillis D.J., Hanner R.H. Incomplete estimates of genetic diversity within species: implications for DNA barcoding. Ecol. Evol. 2019;9:2996–3010. doi: 10.1002/ece3.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maia B.M.C., Madeira-Ott T., Thyssen P.J. Status and challenges for the molecular characterization of Fannia (insecta, Diptera, Fanniidae) in Brazil. Revista Dos Trabalhos de Iniciação Científica Da UNICAMP. 2018 doi: 10.20396/revpibic262018186. [DOI] [Google Scholar]
- 60.Song N., Liang A. The complete mitochondrial genome sequence of Geisha distinctissima (Hemiptera: flatidae) and comparison with other hemipteran insects. Acta Biochim. Biophys. Sin. 2009;41:206–216. doi: 10.1093/abbs/gmp003. [DOI] [PubMed] [Google Scholar]
- 61.Wang S., Lei Z., Wang H., Dong B., Ren B. The complete mitochondrial genome of the leafminer Liriomyza trifolii (Diptera: agromyzidae) Mol. Biol. Rep. 2011;38:687–692. doi: 10.1007/s11033-010-0155-6. [DOI] [PubMed] [Google Scholar]
- 62.Cha S.Y., Yoon H.J., Lee E.M., Yoon M.H., Hwang J.S., Jin B.R., Han Y.S., Kim I. The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignitus (Hymenoptera: apidae) Gene. 2007;392:206–220. doi: 10.1016/j.gene.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 63.Song W., Ye B., Cao X., Yin H., Zhang D. The complete mitochondrial genome of Phlaeoba tenebrosa (Orthoptera: acridoidea: Acrididae) Mitochondrial DNA Part A. 2016;27:409–410. doi: 10.3109/19401736.2014.898281. [DOI] [PubMed] [Google Scholar]
- 64.Hurst L.D. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18:486–487. doi: 10.1016/S0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- 65.Sun S., Li Q., Kong L., Yu H. Complete mitochondrial genomes of Trisidos kiyoni and Potiarca pilula: varied mitochondrial genome size and highly rearranged gene order in Arcidae. Sci. Rep. 2016;6 doi: 10.1038/srep33794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu W., Lin S., Liu H. Mitochondrial genomes of five Hyphessobrycon tetras and their phylogenetic implications. Ecol. Evol. 2021;11:12754–12764. doi: 10.1002/ece3.8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xing Z.P., Liang X., Wang X., Hu H.Y., Huang Y.X. 1994: New Gene Order in Encyrtidae (Hymenoptera, Chalcidoidea), Zookeys. 2022. 2022. Novel gene rearrangement pattern in mitochondrial genome of Ooencyrtus plautus Huang & Noyes; pp. 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nedbal M.A., Flynn J.J. Do the combined effects of the asymmetric process of replication and DNA damage from oxygen radicals produce a mutation-rate signature in the mitochondrial genome? Mol. Biol. Evol. 1998;15:219–223. doi: 10.1093/oxfordjournals.molbev.a025917. [DOI] [PubMed] [Google Scholar]
- 69.Wang D., Liu F., Wang L., Huang S., Yu J. Nonsynonymous substitution rate (Ka) is a relatively consistent parameter for defining fast-evolving and slow-evolving protein-coding genes. Biol. Direct. 2011;6:13. doi: 10.1186/1745-6150-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu M., Wu T., Ju H., Ma X., Fang Z., Chang Q. Phylogenetic analysis of mitochondrial genome of Tabanidae (Diptera: Tabanidae) reveals the present status of Tabanidae classification. Insects. 2022;13 doi: 10.3390/insects13080695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan L., Liu H., Ge X., Yang G., Xie G., Yang Y. A mitochondrial genome phylogeny of Cleridae (Coleoptera, Cleroidea) Insects. 2022;13 doi: 10.3390/insects13020118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Breinholt J.W., Kawahara A.Y. Phylotranscriptomics: saturated third codon positions radically influence the estimation of trees based on next-gen data. Genome Biol Evol. 2013;5:2082–2092. doi: 10.1093/gbe/evt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kutty S.N., Pape T., Pont A., Wiegmann B.M., Meier R. The Muscoidea (Diptera: Calyptratae) are paraphyletic: evidence from four mitochondrial and four nuclear genes. Mol. Phylogenet. Evol. 2008;49:639–652. doi: 10.1016/j.ympev.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 74.Chillcott J.G. A revision of the nearctic species of Fanniinae (Diptera: Muscidae) Can. Entomol. 1960;92:5–295. doi: 10.4039/entm9214fv. [DOI] [Google Scholar]
- 75.Hennig W. Vorarbeiten zu einem phylogenetischen system der Muscidae (Diptera: cyclorrhapha) Stuttgarter Beitrage Zur Naturkunde. 1965;141:1–100. [Google Scholar]
- 76.Pont A. A revision of Australian Fanniidae (Diptera : calyptrata) Aust. J. Zool. Suppl. Ser. 1977;25:1. doi: 10.1071/ajzs051. [DOI] [Google Scholar]
- 77.Rozkošný R., Gregor F., Pont A.C. The European Fanniidae (Diptera) Acta Scientarum Naturalium Acadeniae Scientiarum Bohemicae (Brno) 1997;31:1–80. [Google Scholar]
- 78.Domínguez M.C., Pont A.C. Fauna N Z; 2014. Fanniidae (Insecta: Diptera) pp. 1–91. [DOI] [Google Scholar]
- 79.Domínguez M.C., Roig-Juñent S.A. Historical biogeographic analysis of the family Fanniidae (Diptera: Calyptratae), with special reference to the austral species of the genus Fannia (Diptera: Fanniidae) using dispersal-vicariance analysis. Rev. Chil. Hist. Nat. 2011;84:65–82. [Google Scholar]
- 80.Kutty S.N., Pape T., Wiegmann B.M., Meier R. Molecular phylogeny of the Calyptratae (Diptera: cyclorrhapha) with an emphasis on the superfamily Oestroidea and the position of Mystacinobiidae and McAlpine's fly. Syst. Entomol. 2010;35:614–635. doi: 10.1111/j.1365-3113.2010.00536.x. [DOI] [Google Scholar]
- 81.Sorokina V.S., Ovtshinnikova O.G. The phylogenetic relationships of the Fanniidae within the Muscoid grade (Diptera: calyptrata) based on the musculature of the male terminalia. Insects. 2022;13 doi: 10.3390/insects13020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete Fannia pusio mitochondrial genome and Illumina raw sequence reads have been deposited in GenBank (OQ692989) and the Sequence Read Archive (BioProject ID: PRJNA1003469, SRR25570796), respectively. Mitogenomes from other Fannia species and others were downloaded from GenBank, with accession numbers and references listed in Table 1.