Abstract
Purpose:
Recent advances in computer vision and machine learning have resulted in endoscopic video-based solutions for dense reconstruction of the anatomy. To effectively use these systems in surgical navigation, a reliable image-based technique is required to constantly track the endoscopic camera’s position within the anatomy, despite frequent removal and re-insertion. In this work, we investigate the use of recent learning-based keypoint descriptors for six degree-of-freedom camera pose estimation in intraoperative endoscopic sequences and under changes in anatomy due to surgical resection.
Methods:
Our method employs a dense structure from motion (SfM) reconstruction of the preoperative anatomy, obtained with a state-of-the-art patient-specific learning-based descriptor. During the reconstruction step, each estimated 3D point is associated with a descriptor. This information is employed in the intraoperative sequences to establish 2D-3D correspondences for Perspective-n-Point (PnP) camera pose estimation. We evaluate this method in six intraoperative sequences that include anatomical modifications obtained from two cadaveric subjects.
Results
show that this approach led to translation and rotation errors of 3.9 mm and 0.2 radians, respectively, with 21.86% of localized cameras averaged over the six sequences. In comparison to an additional learning-based descriptor (HardNet++), the selected descriptor can achieve a better percentage of localized cameras with similar pose estimation performance. We further discussed potential error causes and limitations of the proposed approach.
Conclusion:
Patient-specific learning-based descriptors can relocalize images that are well distributed across the inspected anatomy, even where the anatomy is modified. However, camera relocalization in endoscopic sequences remains a persistently challenging problem, and future research is necessary to increase the robustness and accuracy of this technique.
Keywords: Camera relocalization, Sinus surgery navigation, Learning-based descriptors, Anatomical landmark recognition
1. Introduction
Background:
The adoption of minimally invasive surgery and digital endoscopy has brought benefits to patients, including reduced recovery and hospitalization times [1, 2]. However, the use of the body’s natural cavities as pathways for endoscopic intervention entails intricate and occluded workspace environments, constrained fields of vision, and an inherent distance between target anatomy locations and surgeons’ direct visual reach. In this respect, it is critical to provide surgeons with useful information through improved visualization and spatial understanding of instruments-anatomy interactions. To achieve this, numerous algorithms and computer vision applications have been developed that leverage the image information provided by high-resolution endoscopy [3–5].
Standard approaches for endoscopic navigation use optical or electromagnetic tracking to estimate the position of the endoscope camera with respect to a reference coordinate frame. However, this practice has additional constraints like resource needs, time, and ease-of-use, which can negatively affect overall workflow, thus reducing adoption rates when deployed in clinical settings [6]. In this regard, an endoscope-centered solution is critical to enable navigation, while circumventing the downsides of current solutions.
Longitudinal endoscopic assessments generally involve multiple scopings of the anatomy of interest to follow the progression of a surgery, disease or treatment. In particular, during surgical procedures, the endoscope camera is inserted and removed from the anatomy multiple times, while documenting the anatomical variations brought by the surgeon. In this setting, a reliable navigation system must be able to recover the endoscope’s position across multiple intraoperative scopings. This task is referred to as camera relocalization, and is closely associated with anatomical reconstruction from surgical video, which can provide the model and reference frame for camera localization. Previous works have explored this association through Structure from Motion (SfM) [7] and Simultaneous Localization and Mapping (SLAM)-based approaches [5], but are limited to single scopings, where all frames are connected by at least some keypoint correspondences in successive frames. However, endoscope retraction violates this assumption and entails a loss of spatial calibration to the corresponding anatomical model. This motivates the development of a method that establishes correspondences between an anatomical model and the endoscopic frames from different scopings.
Related work:
Camera pose estimation methods have been widely explored in applications in autonomous navigation, robotics, and augmented reality [8]. End-to-end pose regression based on deep learning have been proposed, however, it has been shown in [9] that this family of methods is outperformed by strategies that handle pose estimation via 3D structure. Structure-based approaches utilize SfM in order to construct 3D representations (e.g. point cloud) of the environment based on images showing the same scene but from different views [7]. This algorithm can be used to generate an initial reconstruction for more complex applications such as dense reconstructions [4] and neural-based surface representations [10]. SfM allows for explicit association of points in the 3D structure to keypoints in the 2D image [11], and the resulting correspondences can be employed to solve for the 6-DoF camera pose using methods like Perspective-n-Point (PnP) [12].
Typically, hand-crafted features such as SIFT [13] are used to establish 2D-3D correspondences, which has shown to perform well in reconstructing scenes from natural images [7]. However, these features do not perform well in challenging conditions such as illumination changes and lack of texture present in endoscopic images. Liu et. al [3, 4] demonstrate that these problems can be mitigated with learning-based descriptors that allow for reliable reconstruction of the structure, while computing an expressive feature representation for the estimated 3D points in the reconstruction space.
Contributions:
This work presents and evaluates a structure-based endoscope relocalization method in sinus endoscopy. We generate an initial anatomical 3D reconstruction utilizing the methods of [3] and [4]. Our relocalization pipeline then performs feature matching between the 3D reconstruction and 2D images of unseen endoscopy video. The 2D-3D correspondences were then used for for pose estimation of the video frames relative to the initial model. We compare the performance of different keypoint descriptors proposed for point matching and correspondence generation.
The method is evaluated under the challenging environment presented by the sinus anatomy with monocular endoscopic sequences obtained on cadaver data. Our evaluation environment consists of an initial video sequence generated by scoping the undisturbed anatomy. Relocalization is then evaluated on a repeated endoscope pass on the undisturbed anatomy as well as two subsequent scopings of the same anatomy after surgical manipulation, i. e., resection of structures, which alters the 3D anatomy visible in those videos. Our experimental setup is further described in Section 3.1. We aim to provide an analysis of potential limitations and benefits of learning-based descriptors for camera relocalization in a surgical environment.
2. Methods
We define the relocalization problem as the task of finding the 3 × 3 rotation matrix R and translation vector t ∈ R3 of a query image I in an endoscopic sequence relative to a previously established anatomy coordinate frame. The image I is indexed by its 2D pixel position x, and hence, we use the notation I(x) to indicate the value of the image I (or of any array) at position x. To generate the initial model w.r.t. to which images will be relocalized, we rely on SfM combined with a learning-based keypoint descriptor dx = F(x) to generate an initial point cloud of the anatomy, with Xi ∈ ℝ3. Furthermore, each 3D point in C is linked to a descriptor d, that was used to establish correspondences for the generating 2D points. We will represent this association as dX = P(X). The paired descriptors are later used in the relocalization process to find a set S of 2D-3D correspondences between C and the query image I. These correspondences are finally used to estimate the corresponding camera pose. Figure 1 shows an overview of our method, organized into three steps: 1) 3D anatomy model reconstruction, 2) keypoint relocalization, and 3) endoscope image pose estimation.
Fig. 1. Endoscope relocalization pipeline.
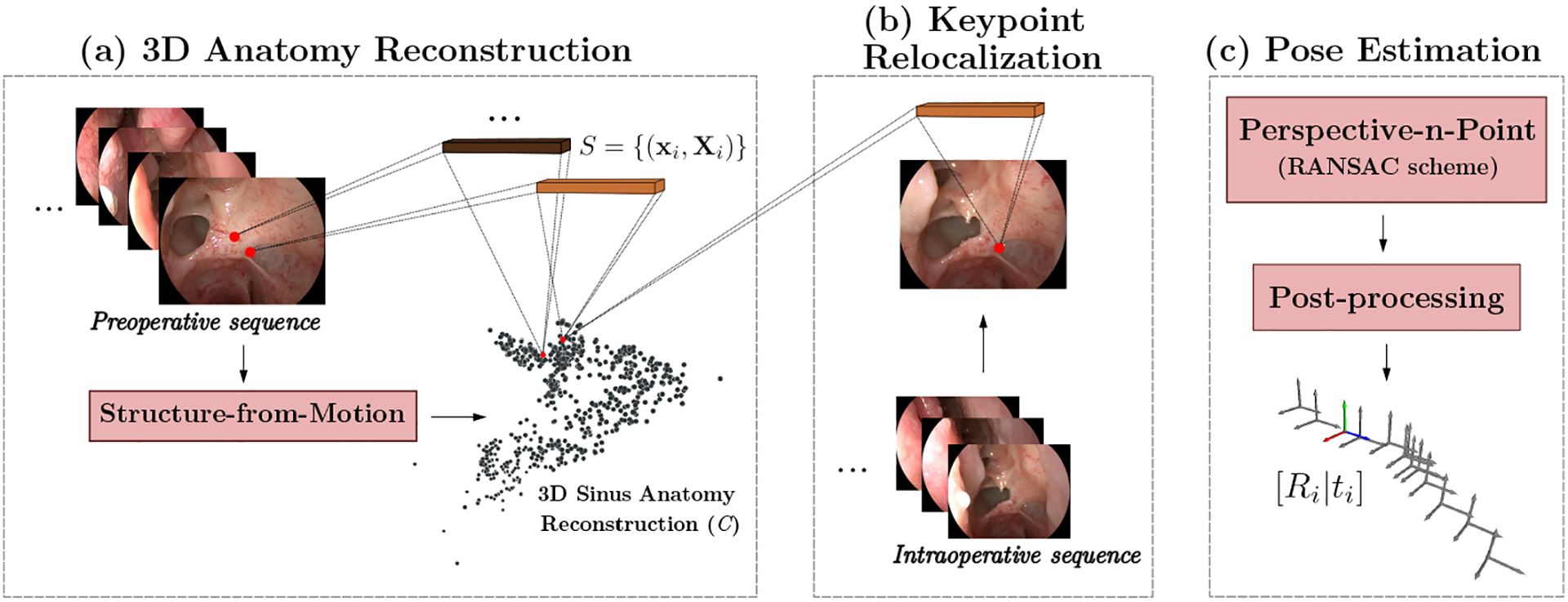
The relocalization process comprises three main stages: a) Using preoperative data, we generate a dense point cloud C of the anatomy using a learning-based descriptor extraction and matching process [3] along with SfM. We relate every point in C with a numerical descriptor (depicted as brown blocks). b) The relocalization of a query image Iq occurs by extracting its corresponding descriptor representation, and finding matches with the point cloud descriptors to generate a query 2D-3D correspondence set S. c) The set S is used by a standard PnP solver that integrates RANSAC for correspondence outlier rejection. Best viewed in color.
2.1. 3D Anatomy Reconstruction
To generate the corresponding point cloud C, we rely on a SfM algorithm [7] on the frames of an initial endoscope sequence. We generated keypoint matches by employing a learning-based descriptor utilizing the dense descriptor extraction and matching method described in [3]. The input for the network is a pair of images Is, It, as source and target respectively, and the output is a set of 2D-2D correspondences between the two inputs.
The network consists of a feature extraction model F = f(I) to compute the dense descriptor maps Fs, Ft on both the source and target images. A set of 2D keypoints locations is sampled on the source image Is. For each keypoint location, the transposed descriptor is employed as a convolutional kernel and is applied to the whole target descriptor map Ft to compute the response map to the keypoint defined as:
| (1) |
The position of the correspondence in the target image for the source keypoint is given by the pixel coordinate that maximizes the response map Due to the learning-based description of point features that considers both local and global context, this method is able to generate a dense set of correspondences between a pair of images. The output matches are used by an SfM algorithm to generate a dense point cloud C of the sinus anatomy. In addition, we obtain an estimate of the preoperative camera trajectory with respect to C, employed for the later refinement step.
2.2. 3D Point Descriptors
Using the results of SfM, it is possible to establish a correspondence between the estimated 3D positions and the 2D keypoints that generated these positions. In general, each X ∈ C can be projected into a 2D keypoint x* in a particular image Iref. Considering this relationship, we define the descriptor for each Xi ∈ C as:
| (2) |
Since Xi can have a projection in multiple images (and hence multiple 2D locations), we select the descriptor for the first projection found. This selection follows the definition of point descriptors, whereby any descriptor will cause a maximum response when convolved with a true correspondence in a new image. In Online Resource 1 - Figure 1, we provide a visual example to complement on the projection similarity of unique 3D point descriptors on 2D images.
2.3. Keypoint Relocalization
The relocalization process relies on the information generated during the reconstruction step. Considering an input image Iq from the initial query image sequence, in our case an endoscopic video sequence of the undisturbed anatomy, we first employ a dense feature extraction model f to generate the query descriptor map Fq = f(Iq). Note that f is fixed and used for feature extraction of all test sequences (pre- and intraoperative).
In order to find the 2D-3D correspondence set S, we follow the same strategy described in Section 2.1, but this time to establish correspondences between a point Xi ∈ C and a location x′ ∈ Iq. Considering the descriptor di = P(Xi), we use Equation (1) to compute the response map . To select valid matches, we establish a response threshold τq. In this manner, if the maximum value of the response map is bigger than τq, we set its location to be a correspondence for Xi. Otherwise, we consider that Xi does not have a match in Iq. With this in mind, the matching process can be formulated as:
| (3) |
The empty value ∅ indicates no correspondence was found. We run this process over the whole point cloud to find a set correspondences between C and Iq. Moreover, to avoid setting a fixed response threshold τ for all input images and sequences, we design a τq selection method that adapts to the 2D-3D correspondences found for each query frame Iq. In this regard, 2D-3D correspondences are filtered progressively, decreasing the response threshold in fixed steps until a minimum number of correspondences is obtained. This procedure allows for a tailored selection of the highest-confidence correspondences for each query frame Iq, and ensures a satisfactory and correct pose estimation in the subsequent PnP stage. This design choice is supported by an additional experiment, which maintained the response threshold fixed for all query frames, and showed a considerable decrease in the localization rate of our method. Additional details are reported in Online Resource 1 - Figure 2.
2.4. Pose Estimation
Given the set of 2D-3D correspondences between Iq and C, we use a standard PnP approach to compute the pose associated to Iq within the preoperative 3D model. It defines a equation system that is solved for R and t:
| (4) |
To reduce the impact of potential outliers found during the described 2D-3D correspondence search, we employ a PnP solver that incorporates a RANSAC scheme to solve for camera pose. By setting a maximum allowed distance of 12.0 pixels between observed and computed point projections during pose estimation, this implementation assists in the selection of the most adequate point correspondences from the computed set S.
Trajectory Post-processing.
We incorporate additional measures to discard large deviations in the estimated endoscopic camera poses, and confine possible solutions to the 3D anatomical space. First, we obtain the tightest bounding cube around the preoperative point cloud and reject any pose estimation located outside of its limits. Second, a temporal-based refinement of the translation component of accepted pose estimates is achieved through a median filter. Finally, we use the preoperative camera trajectory estimated by SfM as a reference for likely endoscopic camera poses. To do so, first, we compute the average difference μsfm between consecutive cameras in the SfM poses. Then each computed intraoperative pose is paired with its closest preoperative counterpart. Considering the space restrictions of the anatomy, the intraoperative pose is accepted if its distance with the nearest preoperative camera is smaller than 2μsfm and its rotation difference is lower than 45 degrees.
2.5. Pose Evaluation
We obtain ground-truth camera poses from optical tracking. Given that pose estimations are computed with respect to the coordinate frame generated with SfM, we require a transformation between this and the optical tracker space. To do so, we register the SfM trajectory of the initial sequence to its measured counterpart in the coordinate frame of the optical tracker, enabling direct evaluation of camera pose in the optical tracker space.
Our quantitative evaluation considers independent error measures for the translation and rotation components of valid pose estimates. We evaluate translation vectors using the L2 distance between the reference and estimated counterparts. To evaluate the rotation matrices, we compute the residual rotation between the predicted and ground truth poses’ rotation as . Note that for a perfect estimation, RΔ equals the identity matrix I3×3. Then, we compute the angle around the rotation axis in radians, as defined below:
| (5) |
where tr(·) expresses the trace of the residual rotation matrix [14].
3. Experiments and Results
We evaluate our pipeline with six endoscopic sequences acquired during a simulated sinus surgery setting on two cadaveric subjects. The scoping sequences were obtained with a rigid endoscope connected to a Storz Image1 HD camera head, and performed by an experienced sinus surgeon under an IRB-approved protocol. Two sequences were acquired with an undisturbed anatomy, while the remaining sequences were captured after surgical intervention that removed parts (cadaver Subject # 1) or the complete lamella of the uncinate (cadaver Subject # 2). All the videos were pre-processed to remove the sections outside the subject and sinus anatomy. Regarding optical tracking, an NDI Polaris Hybrid Position Sensor was employed to obtain pose information of the endoscope relative to the patient anatomy utilizing rigidly attached marker spheres. We performed a hand-eye calibration with the system described in [15] to obtain the position of the camera to determine the ground-truth camera pose relative to the anatomy. The tracking information is time-paired with the endoscopic video to obtain a one-to-one correspondence between video frames and measured poses.
3.1. Experimental Setup
For both subjects, we use one of the sequences with an undisturbed anatomy as initial input to generate the reconstruction and 3D descriptors following the method described in Section 2.1. The second undisturbed scoping, together with the two sequences displaying the surgical progression, were employed as intraoperative sequences to evaluate the 2D-3D matching process and camera pose estimation. Henceforth, these sequences are referred to as Undisturbed Anatomy, Progression Step # 1 and Progression Step # 2. For cadaveric Subject # 1, these sequences contain respectively 930, 746, and 656 query images, while for cadaveric Subject # 2 they contain 1710, 241, and 565 query images. Due to the closeness of the raw frames in the temporal domain, we uniformly sample 310, 249, and 219 (Subject # 1) and 570, 241, and 283 (Subject # 2) from the raw sequences to perform the experiments, ensuring a rich distribution of cameras along the sequences and avoiding content redundancy.
The descriptor model was initialized with the pre-trained weights provided by the authors of [3]. Further training was carried out for a fixed number of 52 epochs on the initial sequence in a self-supervised fashion, to perform patient-specific fine-tuning. The model with the highest accuracy on the validation set was then used for our relocalization method. We maintained the default architecture and training parameters, as used in the original work [3]. We used the learned descriptors in a general purpose Structure-from-Motion pipeline [7] to generate a 3D reconstruction of the sinus anatomy. After keypoint relocalization and 2D-3D correspondence search, we made use of the OpenCV PnP-RANSAC implementation, using the EPnP solver [12, 16], to solve the PnP problem. All experiments were conducted on one NVIDIA RTX3090 GPU, in a 128GB RAM memory PC.
Additional Descriptors.
We evaluated the performance of SIFT and an alternative learning-based descriptor, HardNet++ [17], in the relocalization problem. This model was trained until convergence, initialized with the weights provided by the authors and aiming for fine-tuning over the same preoperative endoscopic sequence as before. Further, to ensure a fair evaluation, we fixed the SfM reconstruction and vary the descriptor paired with the 3D points of P.
3.2. Pose Estimation
We evaluated the localization performance of the proposed pipeline, using [3] as a keypoint descriptor. Average translation and rotation errors are reported in millimeters and radians in Table 1. We also report the percentage of localized cameras, denoting the number of valid solutions computed by the PnP algorithm out of the original number of frames. Invalid cases included frames where not enough correspondences were found or the relocalized camera was excluded from the estimated trajectory in the post-processing stage.
Table 1.
Performance comparison for estimated trajectories employing the proposed method. Results are reported as mean error ± standard deviation.
| Subject # | Sequence | Translation error (mm) | Orientation error (rad) | % of Localized Cameras |
|---|---|---|---|---|
| 1 | Undisturbed Anatomy | 3.33 ± 2.33 | 0.2 ± 0.14 | 27.42 |
| Progression Step # 1 | 2.28 ± 1.21 | 0.22 ± 0.14 | 19.68 | |
| Progression Step # 2 | 2.6 ± 1.43 | 0.26 ± 0.16 | 6.85 | |
| 2 | Undisturbed Anatomy | 4.76 ± 1.69 | 0.17 ± 0.07 | 14.04 |
| Progression Step # 1 | 3.87 ± 0.81 | 0.31 ± 0.12 | 13.69 | |
| Progression Step # 2 | 6.4 ± 3.08 | 0.28 ± 0.16 | 49.47 |
Overall, performance is comparable across the six test sequences. Interestingly, in cadaver Subject # 1, the translation error is larger for the Undisturbed Anatomy sequence. However, the percentage of correctly localized cameras is ~8% bigger than the corresponding disturbed sequences, increasing the possibility of outliers that lie inside the anatomy. Even though the employed descriptor can localize cameras with similar performance along all sequences, it is clear that the number of cameras decreases for Progression Step # 1 and Progression Step # 2 sequences in Subject # 1. Subject #2 follows a similar pattern for the Undisturbed Anatomy and Progression Step # 1 sequences. However, Progression Step # 2 deviates from the behavior observed in Subject # 1. The percentage of localized cameras is more prominent, but the error increases by around 2 mm compared with the previous steps. Similarly, the standard deviation has an increment of up to 2.2 mm compared to the previous progressions. This situation indicates that more correspondences are found, but the level of the noise in the predictions increases. We attribute this increase to the removal of the entire lamella in this particular sequence.
Furthermore, Table 2 presents the results obtained when modifying the nature and source of the 3D point descriptors, between hand-crafted [13] and the selected learning-based descriptors [3, 17]. With respect to SIFT [13], the learning-based nature of these descriptors proves beneficial to derive expressive features from images of the sinus anatomy, and can more suitably localize keypoints despite their photometric inconsistencies and smooth appearances across a sequence. The learning-based descriptors exhibited consistent performance based on comparable translation and rotation errors. While the Dense descriptor was able to relocalize a higher percentage of cameras than HardNet++ for each of the sequences, these percentages are still ultimately low indicating that sinus anatomy provides a challenging environment for their generalizability, and overall capacity to recognize keypoints in endoscopic images. The complexity of the camera relocalization problem in endoscopic surgery is evidenced by overall low camera relocalization rates, and high camera pose estimation errors compared to natural scenes.
Table 2.
Performance comparison for estimated trajectories using different 3D point descriptors.
| Descriptor type | Sequence | Translation error (mm) | Orientation error (rad) | % of Localized Cameras |
|---|---|---|---|---|
| SIFT [13] | Undisturbed Anatomy | 1.65 ± 0.26 | 0.2 ± 0.004 | 1.94 |
| Progression Step # 1 | - | - | 0 | |
| Progression Step # 2 | - | - | 0 | |
| HardNet++ [17] | Undisturbed Anatomy | 3.28 ± 1.63 | 0.19 ± 0.11 | 19.35 |
| Progression Step # 1 | 1.73 ± 1.65 | 0.18 ± 0.11 | 8.43 | |
| Progression Step # 2 | 4.17 ± 2.33 | 0.23 ± 0.15 | 3.2 | |
| Dense descriptor [3] | Undisturbed Anatomy | 3.33 ± 2.33 | 0.2 ± 0.14 | 27.42 |
| Progression Step # 1 | 2.28 ± 1.21 | 0.22 ± 0.14 | 19.68 | |
| Progression Step # 2 | 2.6 ± 1.43 | 0.26 ± 0.14 | 6.85 |
4. Analysis and Discussion
Spatial Distribution of Localized Cameras.
The relocalization strategy employs additional restrictions based on the preoperative SfM results, allowing us to filter out potentially mislocalized cameras and establish a criterion for considering a camera as “correctly localized”. Consequently, the cameras that meet this criterion have a lower error, reducing the number of valid cameras to an average of 21.86% (Table 1). An important aspect to consider is the ability of the descriptor to find proper correspondences across the entire sequence. In this regard, we verify that the localized cameras do not collapse into a single section of the sequence but instead distribute over the entire endoscope trajectory. In Figure 2, we report a depiction of two exemplary estimated trajectories with respect to their corresponding ground-truth trajectories and anatomical model used as reference for the localization process.
Fig. 2.
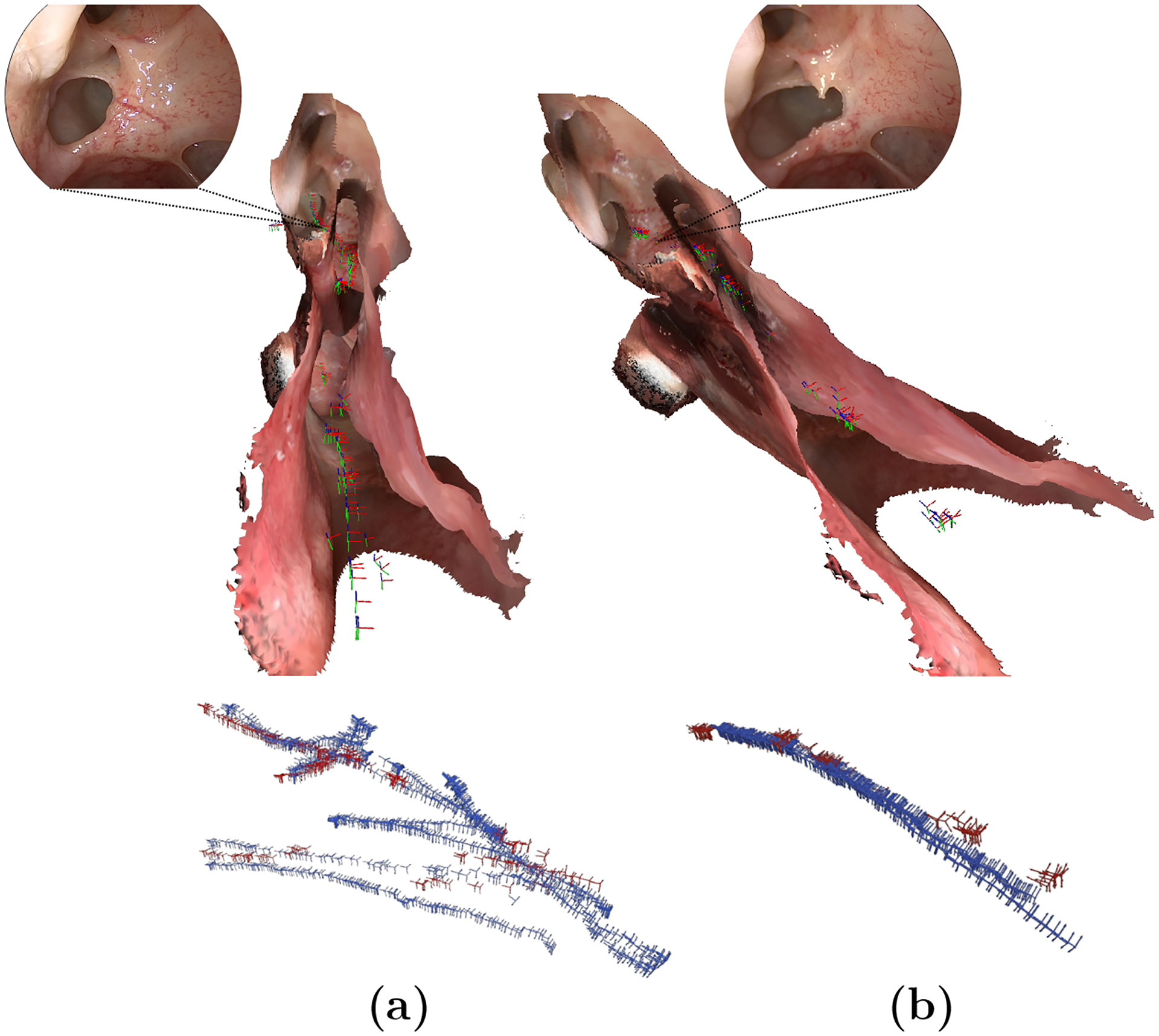
Spatial distribution of valid cameras (red) w.r.t its ground-truth trajectory (blue) and 3D anatomical model. (a) and (b) correspond to sequences of Undisturbed Anatomy and Progression Step # 1 of Subject # 1, respectively. A comparative frame of the operated region is also presented. Best viewed in color.
Overall, we observe that the estimations are interspersed and cover suitably the progression of the different intraoperative sequences. As shown in Figure 2, this distribution is reflected in the spatial domain, as the localized cameras attend to several regions in the anatomy, and particularly cover the operated region near the lamella of the uncinate. This observation refers back to the appropriate relocalization of salient features, pertaining to anatomical landmarks along the intraoperative scopings. To complement these observations, the localized cameras were also shown to be well-distributed in the temporal domain of the ground-truth trajectory (Online Resource 1 - Figure 3).
Effect of Noisy 2D-3D Correspondences in PnP Localization.
To understand potential error sources, we evaluate the robustness of PnP against noisy inputs. We apply different levels of additive Gaussian noise to the 2D-3D correspondences to simulate a mismatch between the 3D points and the predicted 2D location over a subset of 106 images uniformly sampled from Subject # 1 - Progression Step # 1. We find that the PnP presents a certain level of robustness to noise in the estimated 2D keypoint location. However, noise in the 3D component leads to higher errors (above 10 cm in translation, and 2 radians for rotations) in the estimated pose. These findings suggest the presence of false positive 3D matches in the query images as one of the error sources for the camera pose estimation. Furthermore, it is possible that the training strategies employed with the descriptors generate a high-sensitivity matching process. This allows strong correspondences to be found in a local vicinity (favorable for SfM) but may present a low specificity in long-range matches, contributing to errors in the relocalization process. Additional details and visualizations are given in Online Resource 1 (Figures 4 and 5).
Overall Performance Suggests Need for Further Research.
The use of the same descriptor for SfM reconstruction and 2D-3D matching brings advantages, as only one single model is required for both tasks. Nevertheless, to apply these models to the relocalization task, it is necessary to understand and address their limitations. Even though patient-specific descriptors are advantageous over other descriptor models, results show that extending these descriptors to problems beyond their primary SfM task requires additional effort. Moreover, we identify the inclusion of incorrect 3D locations in the 2D-3D correspondence sets as a strong contribution to the errors in the estimation of the endoscopic camera poses. In light of this, the employed feature model’s specificity is a critical point that needs to be attended to. However, additional aspects also need to be considered, including the influence of the noise in the pseudo-ground truth used to train the model given the self-supervised nature of the descriptor employed.
5. Conclusion
This work presents an application and evaluation of patient-specific learning-based descriptors for the endoscope camera relocalization problem. The main objective of the descriptor is to define 2D-3D correspondences between a preoperative point cloud and the individual frames of a new endoscopic sequence. Patient-specific learning-based descriptors can relocalize endoscopic images that are well distributed across the inspected anatomy, even in cases where the anatomy is modified. However, camera relocalization in endoscopic sequences remains a challenging problem, and future research is necessary to increase both the robustness and the accuracy of these techniques, before optimizing them for clinical use.
Supplementary Material
Acknowledgments.
Isabela Hernández acknowledges the support of the 2021 Uniandes-DeepMind Scholarship.
Funding:
This work was funded in part by Johns Hopkins University internal funds and in part by NIH R01EB030511. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplementary information. This article has an accompanying supplementary file (Online Resource 1). We kindly direct the reader to this material for additional details and visualizations.
Conflict of interest/Competing interests: The authors have no competing interests to declare that are relevant to the content of this article.
Code availability:
The source code is available at https://github.com/arcadelab/camera-relocalization.
Consent to participate: This study was performed under the approved IRB00267324 protocol on non-living subjects, for which informed consent was not required.
References
- [1].Mirota DJ, Masaru I, Hager GD: Vision-Based Navigation in Image-Guided Interventions. Annu Rev Biomed Eng (2011) [DOI] [PubMed] [Google Scholar]
- [2].Yeung BPM, Gourlay T: A technical review of flexible endoscopic multitasking platforms. International Journal of Surgery (2012) [DOI] [PubMed] [Google Scholar]
- [3].Liu X, Zheng Y, Killeen B, Ishii M, Hager GD, Taylor RH, Unberath M: Extremely Dense Point Correspondences using a Learned Feature Descriptor. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 4847–4856 (2020) [Google Scholar]
- [4].Liu X, Stiber M, Huang J, Ishii M, Hager GD, Taylor RH, Unberath M: Reconstructing Sinus Anatomy from Endoscopic Video – Towards a Radiation-Free Approach for Quantitative Longitudinal Assessment. In: Martel AL, Abolmaesumi P, Stoyanov D, Mateus D, Zuluaga MA, Zhou SK, Racoceanu D, Joskowicz L (eds.) Medical Image Computing and Computer Assisted Intervention – MICCAI 2020, pp. 3–13. Springer, Cham: (2020) [Google Scholar]
- [5].Liu X, Li Z, Ishii M, Hager GD, Taylor RH, Unberath M: SAGE: SLAM with Appearance and Geometry Prior for Endoscopy. In: ICRA; (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Waelkens P, Van Oosterom M, Van den Berg N, Navab N, Leeuwen FWB: Surgical Navigation: An Overview of the State-of-the-Art Clinical Applications, (2016)
- [7].Schonberger JL, Frahm J-M: Structure-From-Motion Revisited. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4104–4113 (2016) [Google Scholar]
- [8].Kendall A, Grimes M, Cipolla R: PoseNet: A Convolutional Network for Real-Time 6-DOF Camera Relocalization. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 2938–2946 (2015) [Google Scholar]
- [9].Sattler T, Zhou Q, Pollefeys M, Leal-Taixe L: Understanding the Limitations of CNN-based Absolute Camera Pose Regression. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 3302–3312 (2019) [Google Scholar]
- [10].Mildenhall B, Srinivasan PP, Tancik M, Barron JT, Ramamoorthi R, Ng R: NeRF: Representing Scenes as Neural Radiance Fields for View Synthesis. Communications of the ACM 65(1), 99–106 (2021) [Google Scholar]
- [11].Sattler T, Leibe B, Kobbelt L: Fast image-based localization using direct 2D-to-3D matching. In: 2011 International Conference on Computer Vision, pp. 667–674 (2011). IEEE [Google Scholar]
- [12].Lepetit V, Moreno-Noguer F, Fua P: EPnP: An Accurate O(n) Solution to the PnP Problem. International Journal Of Computer Vision (2009) [Google Scholar]
- [13].Lowe DG: Distinctive Image Features from Scale-Invariant Keypoints. Int. J. Comput. Vision 60(2), 91–110 (2004). 10.1023/B:VISI.0000029664.99615.94 [DOI] [Google Scholar]
- [14].Strobl K, Hirzinger G: Optimal Hand-Eye Calibration, pp. 4647–4653 (2006). 10.1109/IROS.2006.282250 [DOI] [Google Scholar]
- [15].Vagdargi P, Uneri A, Jones C, Wu P, Han R, Luciano M, Anderson W, Hager G, Siewerdsen J: Robot-assisted ventriculoscopic 3D reconstruction for guidance of deep-brain stimulation surgery. In: Medical Imaging 2021: Image-Guided Procedures, Robotic Interventions, and Modeling, vol. 11598, pp. 47–54 (2021). SPIE [Google Scholar]
- [16].Moreno-Noguer F, Lepetit V, Fua P: Accurate Non-Iterative O(n) Solution to the PnP Problem. In: 11th IEEE International Conference on Computer Vision (2007) [Google Scholar]
- [17].Mishchuk A, Mishkin D, Radenovic F, Matas J: Working hard to know your neighbor’s margins: Local descriptor learning loss. Advances in Neural Information Processing Systems 30 (2017) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


