Abstract
The herpes simplex virus type 1 (HSV-1) origin of DNA replication, oriS, contains three binding sites for the viral origin binding protein (OBP) flanked by transcriptional regulatory elements of the immediate-early genes encoding ICP4 and ICP22/47. To assess the role of flanking sequences in oriS function, plasmids containing oriS and either wild-type or mutant flanking sequences were tested in transient DNA replication assays. Although the ICP4 and ICP22/47 regulatory regions were shown to enhance oriS function, most individual elements in these regions, including the VP16-responsive TAATGARAT elements, were found to be dispensable for oriS function. In contrast, two oriS core-adjacent regulatory (Oscar) elements, OscarL and OscarR, at the base of the oriS palindrome were shown to enhance oriS function significantly and additively. Specifically, mutational disruption of either element reduced oriS-dependent DNA replication by 60 to 70%, and disruption of both elements reduced replication by 90%. The properties of protein-DNA complexes formed in gel mobility shift assays using uninfected and HSV-1-infected Vero cell nuclear extracts demonstrated that both OscarL and OscarR are binding sites for cellular proteins. Whereas OscarR does not correspond to the consensus binding site of any known transcription factor, OscarL contains a consensus binding site for the transcription factor Sp1. Gel mobility shift and supershift experiments using antibodies directed against members of the Sp1 family of transcription factors demonstrated the presence of Sp1 and Sp3, but not Sp2 or Sp4, in the protein-DNA complexes formed at OscarL. The abilities of OscarL and OscarR to bind their respective cellular proteins correlated directly with the efficiency of oriS-dependent DNA replication. Cooperative interactions between the Oscar-binding factors and proteins binding to adjacent OBP binding sites were not observed. Notably, Oscar element mutations that impaired oriS-dependent DNA replication had no detectable effect on either basal or induced levels of transcription from the ICP4 and ICP22/47 promoters, as determined by RNase protection assays. The Oscar elements thus appear to provide binding sites for cellular proteins that facilitate oriS-dependent DNA replication but have no effect on transcription of oriS-flanking genes.
Characterization of viral origins of DNA replication in eukaryotic cells has demonstrated that most viral origins consist of two components: an essential core sequence that recruits the origin recognition proteins and specifies the initiation site, and auxiliary regions that modulate the efficiency of replication initiated at the core (for reviews, see references 17 to 19). The auxiliary regions of viral replication origins also serve as promoter-enhancer regions for genes flanking the origins. The interaction of cellular and viral transcription factors with auxiliary regions can directly influence the efficiency of origin-dependent DNA replication as well as transcription of origin-flanking genes. This functional organization is evident in the origins of polyomavirus (20, 31), simian virus 40 (SV40) (10, 16, 34, 36, 46, 47), bovine papillomavirus (50, 67), adenovirus (61, 79), Epstein-Barr virus (28, 60, 82), and herpes simplex virus (HSV) (80).
The HSV type 1 (HSV-1) genome contains three origins of DNA replication: a single copy of oriL, located in the middle of the unique long segment of the genome, and two copies of oriS, located in the inverted-repeat sequences flanking the unique short segment of the genome. Replication of the virus requires at least one intact origin and the expression of seven viral genes (35, 55, 81). Both oriS and oriL can support plasmid replication in transient DNA replication assays when the essential viral proteins are provided in trans by superinfection with HSV-1 or by cotransfection with plasmids expressing these proteins (9, 33, 66, 68, 70, 75, 77, 81).
Like the origins of other DNA-containing viruses, the core origins of HSV-1 DNA replication are located in the promoter-regulatory regions of divergently transcribed genes (70, 77). oriL lies between the transcriptional start sites of two early genes specifying the DNA polymerase and the single-stranded-DNA binding protein, ICP8. Similarly, oriS lies between the start sites of the immediate-early genes encoding ICP4 and ICP22/47. The core component of oriS is 80 bp in length and contains a 45-bp imperfect palindrome with an AT-rich sequence at its apex and three binding sites for the viral origin binding proteins (OBP and OBPC) which partially overlap the binding site of a cellular factor, OF-1 (3, 4, 12, 13, 23, 24, 49, 51, 54, 70, 76, 80). Within the 822-bp intergenic region containing the oriS core are numerous binding sites for transcription factors that play critical roles in the expression of ICP4 and ICP22/47 (1, 7, 38, 56–58, 72).
The proximity of the oriS core and transcriptional regulatory elements suggests the possibility of interplay between factors affecting transcription and initiation of DNA replication. Deletion analysis of the shared promoter-regulatory region of the genes encoding ICP4 and ICP22/47 has shown that this region enhances the efficiency of oriS function in transient plasmid DNA replication assays (80). Six recurring elements with the consensus CCCGTTGG, distributed between the transcription start site of ICP4 and the oriS core (see Fig. 1), were suggested to play a role in the observed stimulation of oriS efficiency but were not tested for this function (80). In addition, the promoter-regulatory region containing oriS also contains four TAATGARAT elements responsive to the HSV-1 transcriptional activator, VP16 (see Fig. 1). The acidic transactivation domain of VP16 is capable of binding protein A, a cellular DNA replication protein (32, 48). In a fusion protein construct containing a GAL4 DNA-binding domain VP16 was shown to stimulate bovine papillomavirus DNA replication in vitro (48) and polyomavirus DNA replication in transient assays (32). The roles that VP16 and TAATGARAT elements may play in HSV-1 origin-dependent DNA replication have not been investigated.
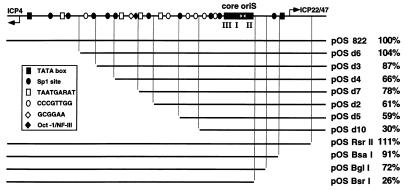
FIG. 1.
Deletion analysis of the promoter-regulatory region flanking oriS. At the top of the figure is a diagram of the 822-bp intergenic region between the divergently transcribed genes encoding ICP4 and ICP22/47. This region contains oriS and the promoter-regulatory elements of these two immediate-early genes. The long black rectangle represents the 80-bp minimum oriS sequence that consists of an AT-rich stretch (white bow tie) at the center of an imperfect palindrome and three binding sites (I, II, and III) for OBP and OBPC. Other symbols designate individual cis-acting elements in the oriS-flanking region. The extent of oriS-flanking sequences in the wild-type plasmid pOS 822 and the remaining sequences in deletion constructs are represented as black lines. The replication efficiency of each plasmid in transient DNA replication assays is shown to the right of the name of the plasmid. The results from at least three experiments per plasmid were averaged and normalized against the replication efficiency of pOS 822.
To test the roles of CCCGTTGG and TAATGARAT elements in oriS-dependent DNA replication as well as to identify additional oriS auxiliary elements, we conducted a systematic analysis of the contribution of the sequences flanking the oriS core to oriS function in transient plasmid DNA replication assays. During the course of these studies, we demonstrated the interaction of cellular proteins, including the transcription factors, Sp1 and Sp3, with two newly identified regulatory elements immediately adjacent to the oriS core that facilitate oriS-dependent DNA replication but have no detectable effect on transcription of oriS-flanking genes.
MATERIALS AND METHODS
Virus and cell line.
The KOS strain of HSV-1 was used as the wild-type virus, and all viral DNA sequences were derived from KOS DNA. The virus was grown and assayed in Vero cells, which were propagated as described previously (15).
Plasmids.
pOS 822 and pOS 80 have been described previously (80). Plasmids in which oriS-flanking sequences were deleted from the ICP4 transcription start site toward the core origin (Fig. 1) were constructed from pOS 822, using the Erase-a-Base System (catalog no. E5850) essentially as described by the manufacturer (Promega, Madison, Wis.). Briefly, pOS 822 was digested to completion with HindIII and KpnI. The linearized plasmids were incubated with exonuclease III at room temperature for various lengths of time. The exonuclease III-treated DNAs were then incubated with S1 nuclease to remove single-stranded tails and then with the Klenow fragment of DNA polymerase to generate blunt ends, which were subsequently ligated to recircularize the deletion-containing plasmids. The deletion end points were determined by DNA sequencing by the dideoxynucleotide method using Sequenase (United States Biochemicals, Cleveland, Ohio).
Plasmids in which oriS-flanking sequences were deleted from the ICP22/47 transcription start site toward the core origin (Fig. 1) were constructed as follows. pOS 822 was digested with XhoI and BsrI. A 664-bp fragment was isolated and treated with the Klenow fragment of DNA polymerase to generate blunt ends. SacI linkers were added to the ends. Finally, the fragment was inserted into pGEM7Zf+ to generate pOS BsrI. In addition, pOS 822 was partially digested with BglI, partially digested with BsaI, or completely digested with RsrII. The full-length, linearized plasmid DNAs were isolated and treated with the Klenow fragment of DNA polymerase to generate blunt ends. SacI linkers were added to the ends. Finally, the linearized plasmid DNAs were recircularized to generate pOS BglI, pOS BsaI, and pOS RsrII. The deletion end points of these four plasmids were confirmed by DNA sequencing.
Mutations in TAATGARAT elements 1 and 2 (counting from the ICP4 transcriptional start site) and in the GCGGAA sequence (72) were transferred from plasmids generously provided by Steven J. Triezenberg (Michigan State University, East Lansing) into the pOS 822 background by standard restriction enzyme digestions and ligations (Fig. 2). Oligonucleotide-directed mutagenesis was performed on TAATGARAT elements 3 and 4 and on the CCCGTTGG elements by the method of Kunkel (45) as described previously (2). Specifically, the TAATGARAT elements were changed to TCCGTARAT (72) and the CCCGTTGG elements were changed to CCCTGGTG (specific nucleotide changes are indicated by boldface type). All mutations were confirmed by DNA sequencing. The internal deletions present in pOS 16 and pOS 20 were detected during repeated attempts to perform oligonucleotide-directed mutagenesis on the four CCCGTTGG elements closest to the core oriS. The end points of the deletions were determined by DNA sequencing.
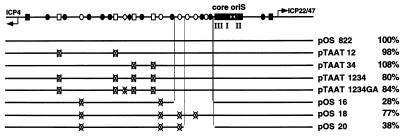
FIG. 2.
Effect of TAATGARAT and CCCGTTGG elements on oriS function. At the top of the figure is a diagram of the 822-bp oriS as described in the legend to Fig. 1. The extent of oriS-flanking sequences in the wild-type plasmid, pOS 822, and substitution and deletion constructs thereof are represented as black lines. Also indicated are the TAATGARAT (white rectangle), GCGGAA (white diamond), and CCCGTTGG (white oval) elements. An X in place of a cis-acting element indicates the disruption of that element by substitution mutagenesis. The replication efficiency of each plasmid in transient DNA replication assays was normalized against the replication efficiency of pOS 822 and is shown to the right of the name of the plasmid. The test results for the TAATGARAT mutant plasmids from two experiments were averaged, and the results of the CCCGTTGG mutant plasmids from four experiments were averaged.
pOS 80L, pOS 80R, and pOS 80LR were constructed by adding back oriS-flanking sequences to pOS 80 (Fig. 3). DNA fragments containing the left (ICP4) or the right (ICP22/47) side of oriS-flanking sequences were generated by PCR with pOS 822 as the template and the following primer sets: 5′-GGGGGTACCGGATCCGTGTCGGCAGC-3′ and 5′-CCGAAGCTTCGGGGGAACCCCGCCAG-3′ for the left-sided fragment and 5′-GCCAAGCTTACTCCGCGCCGGCCCC-3′ and 5′-CCCCTCGAGCCTAGGCTACGGCTGCG-3′ for the right-sided fragment. The left-sided PCR product was digested with KpnI and BstBI and inserted into pOS 80 to generate pOS 80L. The right-sided PCR product was digested with HindIII and SacI and inserted into pOS 80 to generate pOS 80R and into pOS 80L to generate pOS 80LR. The primer sets used in the PCRs had been designed so that the reconstituted flanking sequences retained wild-type spacing and orientation with respect to the oriS core. Consequently, plasmid sequences connecting the PCR products to pOS 80 became linker substitution mutations replacing equal-length wild-type sequences that normally flank the oriS core. These properties were confirmed by DNA sequencing.
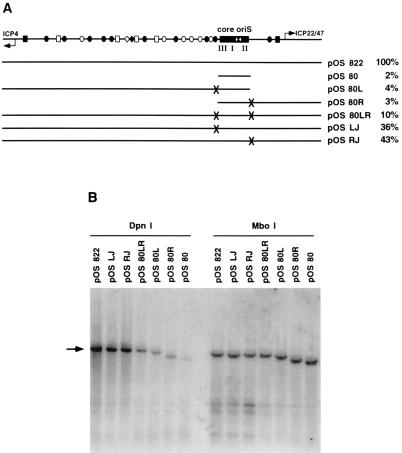
FIG. 3.
Identification of the Oscar elements. (A) Diagram of oriS and the extent of its flanking sequences in individual plasmids. X’s mark the locations of plasmid linker sequences that replace equal-length segments of HSV DNA adjacent to the minimum oriS. Specifically, on the ICP4 side of oriS, sequence GGGGCCGC was replaced by TTCGAAAT (the left junction or LJ mutation), and on the ICP22/47 side of oriS, wild-type sequence CCGTGCCC was replaced by ATAAGTTT (the right junction or RJ mutation). The replication efficiency of each plasmid in transient DNA replication assays is shown to the right of the name of the plasmid. The results of at least three experiments were normalized against the replication efficiency of pOS 822 and then averaged. (B) Southern blot of a transient plasmid DNA replication assay. Vero cells were transfected with the indicated plasmids. At 18 h posttransfection, cells were superinfected with wild-type HSV-1 strain KOS at an MOI of 10 PFU/cell. Infected cells were harvested at 18 h p.i. Total cellular DNA was extracted and digested with restriction enzymes to linearize the plasmids and to distinguish between input DNA (MboI resistant) and newly replicated DNA (DpnI resistant). Southern blots of digested extracts were probed with 32P-labeled plasmid vector sequences. The black arrow indicates the positions of linearized plasmids. Faster-migrating bands are the cleavage products of DpnI or MboI digestions. The intensities of unit-length bands were quantified by using a PhosphorImager. The ratio of the DpnI signal to the MboI signal for each plasmid represents the fold replication over input and serves as an internally controlled measure of replication efficiency.
pOS80 LJ was the product of a three-way ligation involving a 641-bp XhoI-BsrI fragment from pOS 80LR, a 208-bp BsrI-SacI fragment from pOS 822, and the pGEM7Zf+ vector digested with XhoI and SacI (Fig. 3). Similarly, pOS 80RJ was the product of a three-way ligation involving a 664-bp XhoI-BsrI fragment from pOS 822, a 205-bp BsrI-SacI fragment from pOS 80LR, and the pGEM7Zf+ vector digested with XhoI and SacI. The identities of both pOS 80LJ and pOS 80RJ were verified by restriction enzyme digestion.
pOS L10, pOS L11, and pOS R7 (see Fig. 8) were constructed as follows. A 284-bp BssHII-BsaI fragment was isolated from pOS 822 and inserted into vector pNEB 193, which had been digested with XmaI and AscI, to generate pOS 284. The relatively small size of the oriS insert in pOS 284 facilitated mutagenesis of the oriS core-adjacent regulatory (Oscar) elements and subsequent verification by DNA sequencing. Directed mutagenesis by PCR to introduce restriction sites was performed on pOS 284 as described elsewhere (2). M13 forward and reverse sequencing primers were used as flanking primers together with the following mutagenic primers (the nucleotide changes are shown in boldface): L10 top, 5′-AAGAATTCGGGGCCGCCGGGTAAA-3′; L10 bottom, 5′-CGGCGGCGAATTCCCCTTGGGGCG-3′; L11 top, 5′-AAGGGGAATTCGCCGCCGGGTAAA-3′; L11 bottom, 5′-CGGCGGCCCCGAATTCTTGGGGCG-3′; R7 top, 5′-CACTGGCGCCGTTAACGACTCCGC-3′; and R7 bottom, 5′-GCGGAGTCGTTAACGGCGCCAGTG-3′. The L10 and L11 mutations introduced an EcoRI restriction site. The R7 mutation introduced an HpaI restriction site. Six PCRs were performed, and the products were digested by restriction enzymes as listed in Table 1. The digested products of reactions 1 and 2 (Table 1) were ligated together and inserted into pNEB 193 to generate pOS 284 L10. The digested products of reactions 3 and 4 (Table 1) were ligated together and inserted into pNEB 193 to generate pOS 284 L11. The digested products of reactions 5 and 6 (Table 1) were ligated together and inserted into pNEB 193 to generate pOS 284 R7. In turn, pOS 284 L10 and pOS 284 L11 were used as templates for directed mutagenesis by PCR, as described above for reactions 5 and 6, to generate pOS 284 L10R7 and pOS 284 L11R7. The oriS inserts in the pOS 284 series of plasmids were sequenced completely to verify that the desired substitutions in the Oscar elements had been achieved without spurious mutations occurring elsewhere. Finally, members of the pOS 284 series of plasmids were digested with BssHII and BsaI, and the oriS inserts containing mutations in the Oscar elements were used to replace the equivalent wild-type sequences in pOS 822 to generate the pOS 822 series of plasmids with mutations in the Oscar elements.
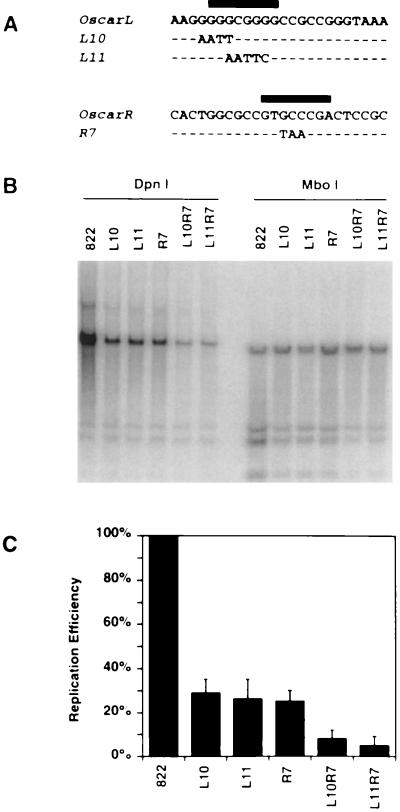
FIG. 8.
Replication efficiencies of wild-type and mutant oriS-containing plasmids. (A) Nucleotide sequences of OscarL and OscarR and the mutated forms of OscarL (L10 and L11) and OscarR (R7) with substitutions in their respective binding sites (indicated with black bars). (B) Southern blot analysis of a transient DNA replication assay testing the replication efficiencies of oriS plasmids carrying the Oscar mutations. The plasmids L10R7 and L11R7 carry the indicated mutations in both OscarL and OscarR. (C) Graph of replication efficiencies of wild-type and Oscar mutant plasmids. The replication efficiency of each plasmid was normalized to that of the wild-type plasmid pOS 822. The graph shows the means and standard deviations of data from three experiments.
TABLE 1.
Primer pairs and restriction enzymes used for PCRs
| Reaction | Primer set | Restriction enzymes |
|---|---|---|
| 1 | L10 top-M13 forward | KpnI-EcoRI |
| 2 | L10 bottom-M13 reverse | EcoRI-BamHI |
| 3 | L11 top-M13 forward | KpnI-EcoRI |
| 4 | L11 bottom-M13 reverse | EcoRI-BamHI |
| 5 | R7 top-M13 forward | KpnI-HpaI |
| 6 | R7 bottom-M13 reverse | HpaI-BamHI |
ICP4-CAT and ICP22/47-CAT promoter-reporter plasmids (see Fig. 10) were constructed as follows. The 822-bp BamHI insert of pOS 822, which contains oriS, its flanking transcriptional regulatory regions, and the transcriptional start sites of ICP4 and ICP22/47, was inserted into a chloramphenical acetyltransferase (CAT) reporter plasmid called pAN3 in both of the two possible orientations. One orientation generated pAN6, in which the ICP4 promoter drives CAT gene transcription. The other orientation generated pAN7, in which the ICP22/47 promoter drives CAT gene transcription. pAN3 was chosen as the CAT reporter vector because it contains three tandem copies of SV40 polyadenylation signals in an 810-bp fragment inserted immediately 5′ of the polylinker cloning sites in front of the CAT gene. The presence of this trimer cassette minimizes the production of read-through transcripts originating from cryptic promoters in plasmid vector sequences (52).
FIG. 10.
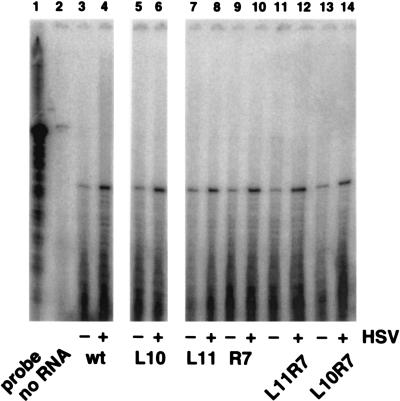
Effects of mutations in the Oscar elements on transcription from the ICP4 promoter. Vero cells were transfected with ICP4 promoter-CAT reporter plasmids. After 18 h, the transfected cells were either mock infected or infected with HSV-1 strain KOS at an MOI of 10 PFU/cell to provide viral regulatory proteins in trans. At 4 h p.i., total RNA was extracted from the cells and the levels of CAT RNA message were measured by RNase protection assay. Lanes: 1, full-length CAT antisense riboprobe; 2, no RNA extract; 3 to 14, protected fragments indicating the levels of CAT message transcribed from the ICP4 promoter carrying wild-type or mutant Oscar elements in mock-infected cells (−) or in HSV-infected cells (+).
Transient plasmid replication assay.
Vero cells (2 × 106) were plated in 100-mm-diameter dishes and incubated at 37°C overnight. Three hours before transfection, the medium was replaced with fresh medium. The cells were transfected with 10 μg of test plasmid and 10 μg of salmon sperm DNA by the calcium phosphate–N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid (BES) method (11) as described elsewhere (2). At 18 to 24 h posttransfection, cells were superinfected with HSV-1 strain KOS at a multiplicity of infection (MOI) of 10 PFU/cell. Except where otherwise indicated, cells were harvested 18 h after infection, and total cellular DNA was extracted as described previously (80). Ten micrograms of DNA was digested with HindIII to linearize the test plasmid and with either DpnI or MboI to distinguish between replicated plasmid (DpnI-resistant) and input plasmid (MboI-resistant) DNA. The digested DNA was analyzed by Southern blot hybridization with 32P-labeled nick-translated pGEM7Zf+ vector sequences as the probe. The resulting bands were quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). The ratio of replicated plasmid to input plasmid represented the replication efficiency of the test plasmid.
Oligonucleotides.
Single-stranded oligonucleotides were synthesized at either the Molecular Biology Core Facility, Dana-Farber Cancer Institute, or the Nucleic Acid Facility, Cancer Research Center, University of Pennsylvania. Complementary oligonucleotides were annealed by heating them to 95°C for 3 min and then slowly cooling them to room temperature. Double-stranded probes were separated from unannealed single-stranded oligonucleotides by polyacrylamide gel electrophoresis and eluted in elution buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA [pH 8.0], and 0.1% sodium dodecyl sulfate) (62). Gel-purified double-stranded probes were radioactively labeled by the use of T4 polynucleotide kinase and [γ-32P]ATP (6,000 Ci/mmol; Dupont NEN, Boston, Mass.) to a specific activity of ∼108 cpm/μg. Unlabeled double-stranded probes were used as specific and nonspecific competitors in gel mobility shift assays.
Nuclear extracts.
All centrifugation steps in the preparation of nuclear extracts were performed at 4°C. All buffers were ice cold. Except for phosphate-buffered saline (PBS), all buffers contained the following protease inhibitors, added just before use: 0.5 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 μg of pepstatin per ml, 100 μg of Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK) per ml, and 100 μg of N-tosyl-l-phenylalanine chloromethyl ketone (TPCK) per ml. The method used for the preparation of nuclear extracts was a modification of a procedure described previously (25). Vero cells (107 to 108) were washed once in PBS, harvested in PBS, collected by centrifugation (for 5 min at 2,000 rpm in a Sorvall RC3 centrifuge), and resuspended in 1 ml of hypotonic buffer (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2). The cells were collected by centrifugation (5 min at 750 × g in an Eppendorf microcentrifuge), resuspended gently in 1 ml of hypotonic buffer containing 0.5% Nonidet P-40, and incubated on ice for 5 min. Nuclei were recovered by centrifugation (5 min at 750 × g), resuspended gently in 1 ml of hypotonic buffer, and collected again by centrifugation (5 min at ∼750 × g). The nuclei were resuspended gently in 100 to 300 μl of high-salt extraction buffer (20 mM HEPES [pH 7.9], 25% glycerol, 0.42 M KCl, 1.5 mM MgCl2, 0.2 mM EDTA) and incubated at 4°C for 30 min with gentle rocking. Following this incubation, the mixture was centrifuged for 10 min at 12,000 × g (maximum speed) in a microcentrifuge. The supernatant extract was dialyzed at 4°C twice for 15 min each time in dialysis buffer (20 mM HEPES [pH 7.9], 20% glycerol, 100 mM KCl, 0.2 mM EDTA). After dialysis, the extract was centrifuged again for 10 min at 12,000 × g to remove any precipitated material. The supernatant was recovered, aliquoted, and frozen quickly in liquid nitrogen. Frozen extracts were stored at −70°C. The protein concentrations of the extracts were determined by the method of Bradford (6), using the Bio-Rad (Melville, N.Y.) protein assay reagent and bovine serum albumin as a standard.
Gel mobility shift assays.
For each binding reaction, 5 μg of extract was diluted to 5 μl with dialysis buffer and preincubated with 1 μl (4 μg) of poly(dI-dC) (Pharmacia) on ice for 5 min. A 4-μl mixture containing 250 pg of labeled oligonucleotide probe, competitor oligonucleotide (if any), 1 μl of 10% polyvinyl alcohol, and 1 μl of 2.5 mM DTT was prepared separately at room temperature. The binding reaction was initiated by adding the prepared extract to the probe mixture. The final binding reaction mixture contained the following: 5 μg of extract, 250 pg of labeled oligonucleotide probe, 10 mM HEPES [pH 7.9], 50 mM KCl, 10% glycerol, 1% polyvinyl alcohol, 0.5 M DTT, and 0.1 mM EDTA. Binding reaction mixtures were incubated at room temperature for 40 min. Protein-DNA complexes were separated by electrophoresis in 6% polyacrylamide (37.5:1 acrylamide-bisacrylamide) gels prepared and run in 0.5× Tris-borate-EDTA buffer at room temperature (62).
Polyclonal antibodies against Sp1, Sp2, Sp3, and Sp4 were obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.). In gel mobility shift-supershift experiments, 1 μl of antibodies was added to the binding reaction after 40 min of incubation and the incubation was continued for an additional 20 min.
Transient expression assay.
Vero cells were transfected with various CAT expression plasmids as described above for the transient DNA replication assays. At 18 to 24 h posttransfection, cells were mock infected or superinfected with HSV-1 strain KOS at an MOI of 10 PFU/cell. At 4 h postinfection, cells were harvested and cytoplasmic RNA was prepared as described previously (39). RNA samples were stored in ethanol at −70°C. [α-32P]CTP-labeled RNA probe was prepared from pTRI-CAT (Ambion) template, using SP6 RNA polymerase (Promega). RNase protection assays were performed as described previously (39).
RESULTS
Systematic analysis of oriS-flanking sequences.
We set out to test the hypothesis that the interaction of cellular and viral transcription factors with auxiliary sequences flanking the oriS core enhances the efficiency of oriS-dependent DNA replication. To assess the roles that the promoter-regulatory sequences flanking the oriS core play in oriS-dependent DNA replication, plasmid constructs containing the full-length 822-bp oriS core and auxiliary sequences were tested in transient plasmid DNA replication assays, as were plasmids with progressive deletions in oriS-flanking sequences.
Serial deletion of left-hand flanking sequence from the transcriptional start site of ICP4 to the oriS core progressively reduced the oriS replication efficiency (Fig. 1). Similar results were obtained by serial deletion of the right-hand sequence from the transcription start site of ICP22/47 to the oriS core. In both deletion series, reductions in oriS efficiency were gradual. Thus, 59% of oriS function remained after 439 bp of the 546-bp left-hand flanking sequence had been deleted (pOS d5), and 72% of oriS function remained after 166 bp of the 195-bp right-hand flanking sequence had been deleted (pOS BglI). Only after nearly all of the left-hand flanking sequence or all of the right-hand flanking sequence had been removed did oriS function show major reductions. Thus, with only 51 bp of the left-hand flanking sequence, oriS function was 30% (pOS d10), and in the total absence of the right-hand flanking sequence, oriS function was 26% (pOS BsrI). These results are consistent with the existence of multiple redundant cis-acting elements spaced throughout left- and right-hand flanking sequences which together facilitate maximum oriS-dependent DNA replication.
Interestingly, deletion of the TATA box and transcription start site of either ICP4 (pOS d6) or ICP22/47 (pOS RsrII) had no effect on the stimulatory activity of oriS-flanking sequences (Fig. 1), suggesting that transcription and the assembly of the transcription initiation complex on the ICP4 and ICP22/47 promoters are not critical for oriS-dependent DNA replication.
Effect of TAATGARAT and CCCGTTGG elements on oriS function.
Since the HSV-1 transcriptional activator VP16 can interact with cellular DNA replication protein A (32, 48) and can stimulate bovine papillomavirus DNA replication in vitro (48) and polyomavirus DNA replication in transient assays (32), we investigated the requirement for the VP16-responsive cis-acting elements flanking oriS in oriS-dependent DNA replication (Fig. 2). These elements, which include four TAATGARAT elements and a GCGGAA motif located between the ICP4 transcriptional start site and the oriS core, have been shown to mediate VP16 transactivation of the ICP4 promoter (72). Plasmids containing substitution mutations known to disrupt VP16 transactivation of the ICP4 promoter at these elements were tested in transient DNA replication assays. The results of these tests demonstrate that the TAATGARAT elements and the GCGGAA motif play no major roles in modulating the efficiency of oriS-dependent DNA replication (Fig. 2).
Since six sequence motifs with the consensus CCCGTTGG in the oriS left-hand flanking region have been suggested to enhance oriS function (80), we examined the roles of these elements in oriS-dependent DNA replication by site-directed mutagenesis and transient DNA replication assays. Although substitution mutagenesis of four of the six CCCGTTGG elements (pOS 18) was not sufficient to significantly affect oriS function, elimination of all six CCCGTTGG elements by a combination of substitution and deletion mutagenesis (pOS 16 and pOS 20) led to a ∼70% reduction in oriS function (Fig. 2). These findings could be taken to suggest a cumulative effect of these motifs on oriS function, implicating a role for redundancy in the origin-enhancing activity of the CCCGTTGG elements. However, in retrospect, a more likely explanation is that novel cis-acting elements immediately adjacent to oriS that enhance oriS function were affected by the deletions as described below.
Identification of the Oscar elements.
As an alternative means of identifying specific flanking sequences that can enhance the function of the minimum core origin contained in pOS 80 (80), oriS-flanking sequences were reconstituted by ligating them to pOS 80 (Fig. 3A). The plasmid pOS 80LR, which contains both the left and the right wild-type oriS-flanking regions, replicated five times more efficiently than pOS 80 but only 10% as efficiently as pOS 822 (Fig. 3B). pOS 80LR differs from pOS 822 in that the former plasmid contains two linker substitutions (denoted by the X’s in Fig. 3A) adjoining the minimum oriS. This observation indicated that although the cis-acting elements distal to the linkers enhance oriS function, the linkers themselves appeared to mark the sites of two novel cis-acting elements in oriS that are critical for optimal origin function. Consistent with this interpretation, pOS LJ and pOS RJ, which contain linker substitutions only in the left- or the right-hand site, respectively, replicated to intermediate levels (36 and 43%, respectively), between the efficiencies of pOS 80LR (10%) and pOS 822 (100%) (Fig. 3B). These novel cis-acting elements, OscarL and OscarR, are regulatory elements on the left and right sides of the minimum oriS, respectively.
Identification of cellular proteins that bind to the Oscar elements.
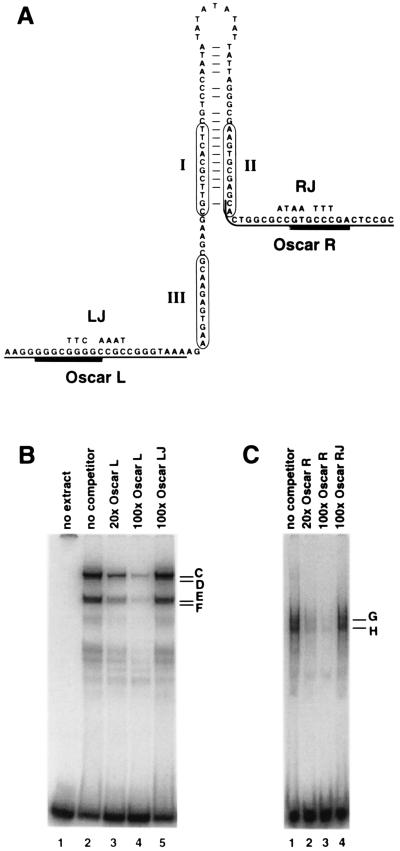
To determine whether protein-DNA complexes form at the Oscar elements, double-stranded DNA oligonucleotides containing OscarL or OscarR sequences (Fig. 4A) were used as probes in gel mobility shift assays. Proteins in HSV-1-infected Vero cell nuclear extracts formed four discernible complexes with OscarL (Fig. 4B, lane 2) and two with OscarR (Fig. 4C, lane 1). The formation of these complexes was shown to be specific for Oscar elements since they were subject to competition from excess unlabeled wild-type probes but not from mutant probes containing linker substitution mutations (Fig. 4B, lanes 3 to 5; Fig. 4C, lanes 2 to 4).
FIG. 4.
Protein-DNA complexes that form at OscarL and OscarR. (A) The minimum oriS and the Oscar elements. The nucleotide sequences are displayed as a hypothetical stem-loop structure to illustrate the palindromic nature of the sequences. The three binding sites for OBP are circled. The nucleotide changes introduced by the junction substitutions, LJ and RJ, are noted above the wild-type sequences. The underlined sequences indicate the OscarL and OscarR probes used in gel mobility shift assays. Heavy underlining indicates the actual protein binding sites in OscarL and OscarR as determined in Fig. 6. (B) Gel mobility shift assays using the OscarL probe and HSV-infected Vero cell nuclear extract. Lanes: 1, no extract; 2, no specific competitor; 3 and 4, 20- and 100-fold molar ratios of unlabeled OscarL competitor, respectively; 5, 100-fold molar ratio of a mutant oligonucleotide competitor containing the LJ mutation. (C) Gel mobility shift assays of OscarR probe and HSV-infected Vero cell nuclear extract. Lanes: 1, no specific competitor; 2 and 3, 20- and 100-fold molar ratios of unlabeled OscarR competitor, respectively; 4, 100-fold molar ratio of a mutant oligonucleotide competitor containing the RJ mutation.
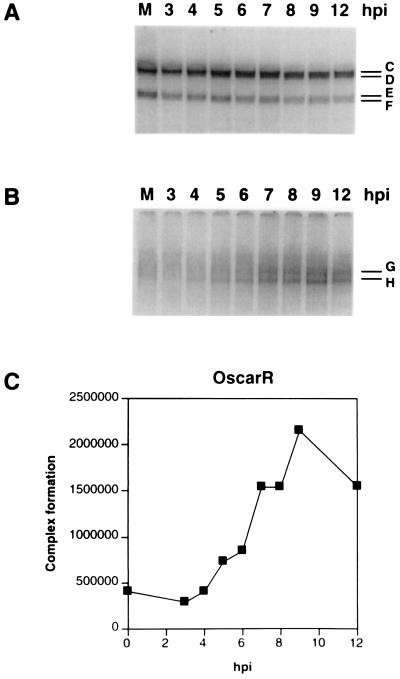
Whereas the OscarL complexes that formed with uninfected-cell extracts (Fig. 5A, lane M) remained essentially unchanged in number, mobility, and intensity during the course of HSV-1 infection (Fig. 5A), the OscarR complexes, which also formed with uninfected-cell extracts (Fig. 5B, lane M, and data not shown), exhibited a significant increase in intensity, but not in number or mobility, between 3 and 9 h postinfection (p.i.) (Fig. 5B). This enhanced intensity was evident through 12 h p.i. (Fig. 5B) and 18 h p.i. (data not shown). The formation of identical complexes by proteins from mock-infected and infected Vero cell nuclear extracts indicates that the OscarL and OscarR complexes are likely composed of cellular proteins (Fig. 5A and B, lanes M).
FIG. 5.
Kinetics of OscarL and OscarR complex formation. Gel mobility shift assays were performed with OscarL (A) or OscarR (B), and nuclear extracts were prepared from Vero cells that were mock infected (lane M) or infected with HSV-1 and harvested at selected times postinfection (3, 4, 5, 6, 7, 8, 9, and 12 h p.i.). In other experiments, cells were also harvested at 18 h p.i. The letters correspond to specific complexes. (C) Quantities of OscarR-specific complexes observed during the time course, expressed in PhosphorImager units.
Mapping of protein binding sites in OscarL and OscarR.
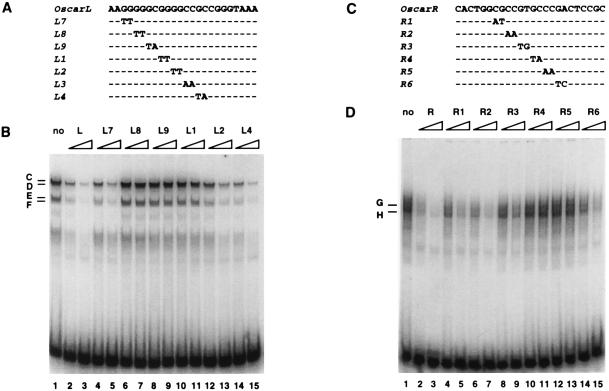
To fine map the binding sites of protein complexes that form at OscarR, a panel of mutant oligonucleotides containing 2-bp substitutions in OscarR was used as the competitor in gel mobility shift competition assays (Fig. 6C). Whereas oligonucleotides R1 and R2 competed for OscarR binding almost as effectively as the wild-type probe, R3 and R6 competed less effectively, and R4 and R5 did not compete at all (Fig. 6D). Notably, competition was equally efficient for both complexes G and H in these tests. The binding site of OscarR complexes G and H was deduced from these tests to consist of the nucleotides affected by mutations in R3 through R6, namely GTGCCCGA. A search of the TransFac transcription factor database revealed that the OscarR protein binding sequence does not correspond with any known transcription factor binding site.
FIG. 6.
Mapping the binding sites at OscarL and OscarR. Mutant oligonucleotides containing 2-bp substitutions spanning the sequences of OscarL (A) or OscarR (C) were used as competitors against the OscarL (B) or OscarR (D) wild-type probe, respectively, in gel mobility shift assays. Dashes in the sequences of the mutant competitors signify nucleotides that are identical to the wild-type sequence. Gel mobility shift assays were performed in the absence of competitor (lane 1), in the presence of excess unlabeled wild-type probe (lanes 2 and 3), or in the presence of unlabeled mutant oligonucleotides (lanes 4 through 15). The triangular ramps signify 20-fold (left) and 100-fold (right) molar excesses of each competitor. The letters on the left indicate specific complexes.
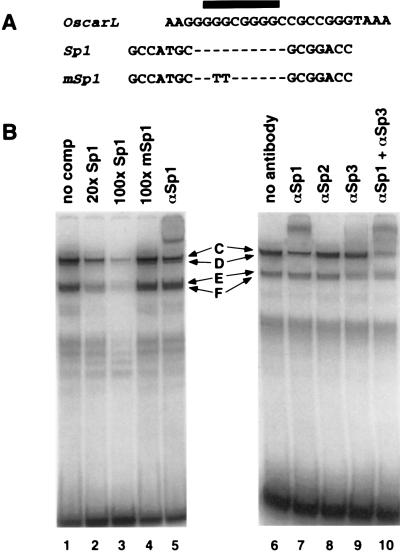
Using a similar approach, the common binding site of OscarL complexes C to F was shown to be GGGCGGGG (Fig. 6A and B), which is a perfect match for the consensus binding site of the cellular transcription factor Sp1.
Members of the Sp1 family of transcription factors bind to OscarL.
Consistent with the identity of the binding site of OscarL complexes C to F and the consensus Sp1 binding site, an oligonucleotide probe containing the consensus Sp1 binding site competed for the formation of all four OscarL-specific complexes in gel mobility shift assays, whereas a mutant probe (mSp1) did not compete for any of the OscarL complexes (Fig. 7B, lanes 1 to 4). In gel mobility shift-supershift assays, an antibody against Sp1 supershifted complex D but did not affect the remaining complexes (Fig. 7B, lanes 5 and 7). Antibodies against Sp2 (Fig. 7B, lane 8) or Sp4 (data not shown) did not affect any of the OscarL-specific complexes. Addition of antibody against Sp3 led to a slight reduction in complexes E and F as well as a reduction in complex C formation (Fig. 7B; compare lane 6 with lane 9 and lane 7 with lane 10). Collectively, these results indicate that cellular transcription factors Sp1 and Sp3 bind to OscarL. Interestingly, although competition with the Sp1 consensus probe affected all four OscarL complexes equally, some of these complexes, especially E and F, were resistant to supershifting even with large excesses of the four antibodies. This suggests that other cellular proteins that share Sp1 DNA binding specificity besides those tested here also bind to OscarL.
FIG. 7.
Members of the Sp1 family of transcription factors bind to OscarL. (A) The nucleotide sequences of OscarL, a probe containing an Sp1 binding site, and a probe containing a mutant Sp1 site unable to bind Sp1. The black bar denotes the OscarL-specific binding site determined in Fig. 6. Dashes in the sequences of the Sp1 wild-type and mutant probes represent nucleotides that are identical to the OscarL sequence. (B) Gel mobility shift assays using the OscarL probe and HSV-infected Vero cell nuclear extract. In competition experiments (lanes 1 to 4), the gel mobility shift assays were performed in the absence of a specific oligonucleotide competitor (lane 1), in the presence of a 20-fold (lane 2) or 100-fold (lane 3) molar excess of an Sp1 competitor, or in the presence of a 100-fold molar excess of mSp1 competitor (lane 4). In gel mobility shift-supershift experiments (lanes 5 to 10), after OscarL and Vero nuclear extract had been incubated for 40 min, the indicated antibodies (prefixed with α) were added, and the reaction mixtures were incubated for an additional 20 min. Specific complexes formed are indicated by letters.
Correlation between protein-DNA complex formation at the Oscar elements and efficiency of oriS-dependent DNA replication.
Based on the binding site mapping data shown in Fig. 6, specific mutations designed to abrogate complex formation at OscarL and OscarR were introduced into pOS 822. Plasmids carrying these mutations, designated L10, L11, and R7 (Fig. 8A), were tested in transient DNA replication assays, and the corresponding mutant oligonucleotides were used in gel shift assays to assess protein-DNA complex formation. Consistent with the results of competition experiments (Fig. 6), specific protein-DNA complexes did not form with the mutant OscarL probes, L10 and L11, or with the mutant OscarR probe, R7, when either mock-infected or HSV-infected Vero cell nuclear extracts were used as the source of protein (data not shown). In transient DNA replication assays, disruption of either OscarL or OscarR alone (pOS L10, pOS L11, or pOS R7) reduced the efficiency of DNA replication by 70%, and disruption of both (pOS L10R7 or pOS L11R7) reduced the efficiency of DNA replication by 90% (Fig. 8B and C). These findings demonstrate a direct correlation between protein-DNA complex formation at OscarL and OscarR and the efficiency of oriS-dependent DNA replication.
Absence of cooperative interaction between OscarR-binding proteins and OBP/OBPC.
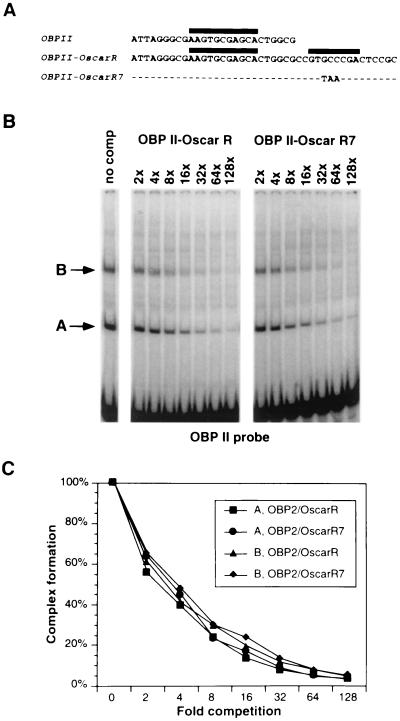
The protein binding site at OscarL is 14 bp from a recognized binding site (III) for the virus-specified origin binding proteins OBP and OBPC, and the protein binding site of OscarR is 8 bp from binding site II (Fig. 4A). To determine whether formation of protein complexes at OscarR alters the affinity of OBP or OBPC for binding site II, gel mobility shift competition assays were performed with an oligonucleotide containing OBP binding site II (OBP II) as a probe and competitor oligonucleotides OBP II-OscarR and OBP II-OscarR7 (Fig. 9A). Both competitors contain wild-type OBP II. OBP II-OscarR contains wild-type OscarR sequences, whereas OBP II-OscarR7 contains substitutions in OscarR that render it unable to form complexes G and H.
FIG. 9.
Absence of cooperative binding activity between the OscarR-binding factors and OBP or OBPC. (A) Nucleotide sequences of OBP binding site II in oriS (OBP II), which was used as the labeled probe in gel mobility shift assays. Two competitor oligonucleotides were used, one containing the wild-type OscarR adjacent to OBP II (OBPII-OscarR) and the other containing a nonfunctional OscarR that could no longer bind OscarR-specific cellular proteins (OBPII-OscarR7). (B) Gel mobility shift competition assays using OBP II probe and HSV-infected Vero cell nuclear extract. The binding reactions were performed in the absence of a specific oligonucleotide competitor (no comp) or in the presence of increasing molar ratios of specific competitors as indicated. Complexes A and B contain the viral proteins OBPC and OBP, respectively. (C) The intensities of complexes A and B were quantified with a PhosphorImager and plotted against the molar ratios of oligonucleotide competitors.
As shown in Fig. 9B and C, both OBPII-OscarR and OBPII-OscarR7 competed equally well with OBP II for formation of complex A (containing OBPC) (3, 4) and complex B (containing OBP) (13), indicating that in this experimental system, formation of complexes G and H does not alter the affinity of OBP or OBPC for binding site II.
Similar gel mobility shift competition experiments were performed to determine whether formation of protein complexes at OscarL alters the affinity of OBP for binding site III. Unlike binding sites I and II, OBP has an extremely low affinity for binding site III and OBPC has none (13, 23). Thus, oligonucleotides containing binding site III did not form detectable complexes A and B (data not shown). Consequently, competition for their formation could not be measured.
Oscar elements and transcription of oriS-flanking genes.
Because the Oscar elements are located in the transcriptional regulatory region shared by the divergently transcribed immediate-early genes encoding ICP4 and ICP22/47, and since members of the Sp1 family of transcription factors interact with OscarL, it was of interest to determine whether the Oscar elements are involved in the transcriptional regulation of these genes.
Vero cells were transfected with plasmids carrying the CAT reporter gene under the control of the ICP4 promoter. Twenty-four hours after transfection, cells were either mock infected, to measure basal transcription levels, or superinfected, to provide viral transactivators. After 4 h, total RNA was extracted and the levels of CAT message were measured by RNase protection assay. As shown in Fig. 10, the mutations in L10, L11, and R7, which disrupt binding of proteins to the Oscar elements and impair oriS function, had no detectable effect on either basal or induced transcription of the ICP4 gene flanking oriS. Similar results were obtained with the ICP22/47 promoter CAT reporter gene (data not shown).
DISCUSSION
Auxiliary function of the transcriptional regulatory region flanking oriS in origin-dependent DNA replication.
In the presence of the seven essential HSV-1 DNA replication proteins, the 80-bp oriS core, which contains the three OBP binding sites and the AT-rich sequence, is sufficient to serve as the template for the initiation of DNA replication. This minimum 80-bp origin functions inefficiently, however, in the absence of auxiliary flanking sequences. In addition to their roles in origin function, the oriS-flanking sequences also serve as the promoter-regulatory regions of the genes encoding ICP4 and ICP22/47. Since deletion of the TATA box and transcriptional start site of either gene had no effect on oriS function, it appears that although the efficiency of oriS function is significantly enhanced by flanking promoter-regulatory sequences, oriS function is not dependent on the process of transcription or even on the assembly of the transcription initiation complex in the ICP4 or ICP22/47 promoters—at least in transient assays in Vero cells.
The results of the systematic deletion analysis reported here confirm and extend the observations of Wong and Schaffer (80) by demonstrating that progressive loss of oriS-flanking sequences on either side of the core origin leads to a progressive reduction in the efficiency of oriS-dependent DNA replication. In this previous study (80), an oriS-containing plasmid was used as an internal standard in transient DNA replication assays, raising concern that there might have been competition between the test plasmids and the internal standard for viral and cellular factors. The present study did not utilize a replication-competent control plasmid but used instead the amount of MboI-resistant input test plasmid DNA as the internal control for each sample. That both studies yielded similar results regarding the overall contribution of oriS-flanking sequences confirms the initial observation. In addition, the systematic deletion analysis conducted in this study demonstrated that the sequence between the oriS core and the ICP22/47 transcriptional start site is as important for oriS-dependent DNA replication as the sequence between the oriS core and the ICP4 start site.
Although incremental deletion of sequences on either side of oriS toward the core origin led to a progressive reduction in oriS function, major reductions in DNA replication efficiency did not occur until the deletion end points were within 50 bp of the core origin. These observations suggest that elements in oriS-flanking sequences that enhance oriS function might be redundant and that sequential deletion of these elements results in a cumulative reduction in oriS functional efficiency. Moreover, they indicate that elements most proximal to the core origin likely have the greatest effect on origin function.
Consistent with the above-stated hypothesis, site-directed mutagenesis of more-distal elements, such as TAATGARAT and CCCGTTGG elements, did not have major effects on the efficiency of oriS-dependent DNA replication. Mutagenesis of the TAATGARAT elements also demonstrated that, unlike the enhancing effect of VP16 on bovine papilloma- and polyomaviruses (32, 48), VP16 does not affect the initiation of oriS-dependent DNA replication in HSV-1 (at least in Vero cells). Likewise, mutagenesis of four of the six CCCGTTGG elements, which were initially suggested to enhance oriS-dependent DNA replication (80), reduced oriS function by only 23%.
Auxiliary function of Oscar elements in oriS-dependent DNA replication.
In contrast to site-specific mutations in distal oriS-flanking sequences, linker substitutions affecting elements immediately adjacent to the oriS core reduced oriS function significantly even when all other oriS-flanking sequences were wild type. Thus, OscarL and OscarR were identified as specific and novel auxiliary components of oriS. Each Oscar element on its own is responsible for a significant enhancement of oriS function, and together their stimulatory effects on oriS-dependent DNA replication are additive.
In HSV-2, two tandem-duplicated copies of oriS are present in each repeat sequence flanking the unique short segment of the genome (78). Notably, the duplicated oriS sequences contain not only the core elements (AT-rich sequence and OBP binding sites) but also the OscarL and OscarR elements. The HSV-2 oriS OscarL element has the same nucleotide sequence as its HSV-1 counterpart, and the HSV-2 oriS OscarR differs from its HSV-1 counterpart by two conservative nucleotide changes in the 8-bp protein binding site. In HSV-2 oriS, a 51-bp region to the left and a 15-bp region to the right of the core origin, containing OscarL and OscarR, respectively, can each enhance oriS function approximately 10-fold (49). These observations indicate that the roles of the Oscar elements have been conserved among the herpes simplex viruses. In HSV-1 and HSV-2, OscarR is present at oriS but not at oriL. Inspection of origin sequences in other alphaherpesviruses, including varicella-zoster virus (14, 69), equine herpesvirus (5, 71), Marek’s disease virus (8), pseudorabies virus (44), and bovine herpesvirus (63), did not reveal the presence of obvious OscarR elements near these origins.
Although it is possible that OscarL and OscarR enhance oriS-dependent DNA replication through cis-acting effects on DNA conformation, their effects on oriS function correlate directly with their ability to serve as binding sites for specific proteins. The cellular transcription factors Sp1 and Sp3 form complexes with OscarL, and the formation of these protein complexes at OscarL correlates directly with the efficiency of oriS function. In addition, OscarR forms complexes with an unidentified cellular factor(s), and the ability of this factor(s) to associate with OscarR is also required for maximum oriS function.
In competition experiments (80), cotransfection of increasing molar excesses of a competitor plasmid carrying most of the oriS-flanking region without the oriS core reduced the replication efficiency of a wild-type oriS test plasmid significantly, presumably by competing for trans-acting factors that bind to flanking sequences. Although the competitor plasmid used in that study did not contain OscarL, it contained several Sp1 binding sites and OscarR. Thus, the basis of the previously observed inhibition might be competition with the wild-type oriS plasmid for binding of Sp1, Sp3, and the OscarR-binding factor(s).
Although our data indicate that protein-DNA complexes that form at both Oscar elements contain only cellular proteins, we cannot rule out the possibility that viral infection may alter the composition or posttranslational modification of the proteins in these complexes.
OscarR-binding factor(s).
Mutational disruption of OscarR did not affect the ability of the adjacent OBP binding site II to compete for binding of OBP and OBPC in gel mobility shift assays. This result implies that the OscarR mutation does not have deleterious effects on the local DNA conformation that might affect binding of OBP and OBPC to site II. It also argues against a stabilizing interaction between an OscarR-binding factor and OBP or OBPC bound at site II. In the context of viral infection, however, both DNA conformational changes and protein-protein interactions (including interactions with OBP or OBPC) remain possible mechanisms by which OscarR might enhance oriS function.
The OscarR-binding cellular factor(s) is likely to be a novel protein with interesting properties. It enhances oriS function through interaction with a specific DNA sequence which is unique in the transcription factor binding site database and does not appear to interact cooperatively with OBP or OBPC, and its DNA binding activity (or expression level) is enhanced by HSV-1 infection. Purification and characterization of the OscarR-binding factor(s) should further elucidate its function within the cell as well as its role in the HSV-1 life cycle.
Interaction of the Sp1 family of transcription factors with OscarL.
The OscarL-binding proteins in Vero cell nuclear extracts include Sp1 and Sp3 but not Sp2 or Sp4. Originally identified as a cellular transcription factor required for SV40 gene expression, Sp1 stimulates transcription by binding to GC-rich elements in a wide variety of cellular and viral promoters (21, 22, 38, 40–42). Sp1 is constitutively expressed, and its activity can be modified by phosphorylation (37) or interaction with cofactors (53, 73, 74). Sp3 is an inhibitory member of the Sp1 family of transcription factors and shares the same preference and affinity for the Sp1 consensus DNA binding site (26, 27, 43). Both Sp1 and Sp3 are ubiquitously expressed (26, 43) and can interact functionally with the retinoblastoma protein to regulate the transcription of a variety of growth-control genes, including c-fos, c-myc, and transforming growth factor β (73, 74).
Since Sp2 is the member of the Sp1 family with the lowest affinity for the Sp1 consensus binding site (26, 43), and since the expression of Sp4 is restricted to certain neuronal cell types (26), it is not surprising that Sp2 and Sp4 were not detected in gel mobility shift assays using Vero cell nuclear extracts. At this point it remains unclear whether the transcriptional activator, Sp1, the transcriptional repressor, Sp3, or both modulate oriS function or whether in neuronal cells Sp4 is also a member of the OscarL protein-DNA complex.
Transcription and DNA replication at oriS.
Although the efficiency of oriS-dependent DNA replication is augmented by auxiliary sequences in the transcriptional regulatory region flanking oriS, initiation of DNA replication at oriS does not appear to depend on the process of transcription or on the assembly of the transcription initiation complex on the ICP4 or ICP22/47 promoter in transient assays. Moreover, DNA replication and transcription in the oriS region appear to be enhanced by distinct auxiliary elements. Consistent with this observation, the TAATGARAT elements, which are known to play important roles in the transcriptional regulation of these genes, do not affect the efficiency of oriS-dependent DNA replication. In addition to the Sp1 site in OscarL, nine other Sp1 binding sites are present in oriS-flanking sequences. Deletion of those sites that have been shown to be important for the transcription of oriS-flanking genes (38, 56) did not affect oriS-dependent DNA replication. In contrast, the Sp1 site in OscarL, which is important for oriS-dependent DNA replication, is apparently not important for transcription. Similarly, the OscarR element is important for DNA replication but not transcription. These experimental observations suggest, but do not prove, that transcription and DNA replication are uncoupled at oriS in vivo.
Cellular proteins and HSV-1 DNA replication.
In summary, we have identified and characterized two novel cis-acting elements flanking the oriS core—OscarL and OscarR—that interact with cellular transcription factors and other cellular proteins to enhance oriS-dependent DNA replication. Sp1, Sp3, and possibly the OscarR-binding factors join a growing list of cellular proteins that have been implicated in HSV-1 origin-dependent DNA replication. A cellular protein designated origin factor 1 has been found to interact with the core sequences of both HSV-1 origins (oriS and oriL) and to enhance OBP binding to binding site I in oriS (12, 29). Recently, a glucocorticoid receptor-like protein was shown to interact with the glucocorticoid response element identified in the core sequences of oriL, and dexamethasone treatment was shown to enhance oriL function while repressing oriS function in PC12 cells differentiated by nerve growth factor (30). Collectively, these observations suggest that cellular proteins are intimately involved in initiation of HSV-1 DNA replication.
Although rolling circle DNA replication has been accomplished in vitro with protein extracts from mammalian cells infected with HSV-1 (59, 65) or insect cells infected with recombinant baculoviruses expressing HSV-1 DNA replication proteins (64), such replication is origin and OBP independent and is actually inhibited by OBP when the template contains oriS (65). To gain a better understanding of HSV-1 origin-dependent initiation, it would be worthwhile to investigate specifically how the cellular proteins that enhance oriS-dependent DNA replication in transient assays might function in cell-free systems.
ACKNOWLEDGMENTS
We thank Robert Jordan, Luis Schang, Elli Mendoza, Bill Halford, Amy Francis, and Jennifer Isler for helpful discussions and Paige Hasling for preparation of the manuscript.
These studies were supported by research grant R01 AI28537 from the National Institute of Allergy and Infectious Diseases. A. Nguyen-Huynh was the recipient of a Howard Hughes Predoctoral Fellowship.
REFERENCES
- 1.apRhys C M J, Ciufo D M, O’Neill E A, Kelly T J, Hayward G S. Overlapping octamer and TAATGARAT motifs in the VF65-response elements in herpes simplex virus immediate-early promoters represent independent binding sites for cellular nuclear factor III. J Virol. 1989;63:2798–2812. doi: 10.1128/jvi.63.6.2798-2812.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 3.Baradaran K, Dabrowski C E, Schaffer P A. Transcriptional analysis of the region of the herpes simplex virus type 1 genome containing the UL8, UL9, and UL10 genes and identification of a novel delayed-early gene product, OBPC. J Virol. 1994;68:4251–4261. doi: 10.1128/jvi.68.7.4251-4261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baradaran K, Hardwicke M A, Dabrowski C E, Schaffer P A. Properties of the novel herpes simplex virus type 1 origin binding protein, OBPC. J Virol. 1996;70:5673–5679. doi: 10.1128/jvi.70.8.5673-5679.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumann R P, Yalamanchili V R R, O’Callaghan D J. Functional mapping and DNA sequence of an equine herpesvirus 1 origin of replication. J Virol. 1989;63:1275–1283. doi: 10.1128/jvi.63.3.1275-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford M M. Rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Bzik K J, Preston C M. Analysis of DNA sequences which regulate the transcription of herpes simplex virus immediate early gene 3: DNA sequences required for enhancer-like activity and response to transactivation by a virion polypeptide. Nucleic Acids Res. 1986;14:929–943. doi: 10.1093/nar/14.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camp H S, Coussens P M, Silva R F. Cloning, sequencing, and functional analysis of a Marek’s disease virus origin of DNA replication. J Virol. 1991;65:6320–6324. doi: 10.1128/jvi.65.11.6320-6324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Challberg M D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci USA. 1986;83:9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandrasekharappa S C, Subramanian K N. Effects of position and orientation of the 72-base-pair-repeat transcriptional enhancer on replication from the simian virus 40 core origin. J Virol. 1987;61:2973–2980. doi: 10.1128/jvi.61.10.2973-2980.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dabrowski C E, Carmillo P J, Schaffer P A. Cellular protein interactions with herpes simplex virus type 1 oriS. Mol Cell Biol. 1994;14:2545–2555. doi: 10.1128/mcb.14.4.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dabrowski C E, Schaffer P A. Herpes simplex virus type 1 origin-specific binding protein: oriS-binding properties and effects of cellular proteins. J Virol. 1991;65:3140–3150. doi: 10.1128/jvi.65.6.3140-3150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davison A J, Scott J E. DNA sequence of the major inverted repeat in the varicella-zoster virus genome. J Gen Virol. 1985;66:207–220. doi: 10.1099/0022-1317-66-2-207. [DOI] [PubMed] [Google Scholar]
- 15.DeLuca N A, Schaffer P A. Activation of immediate-early, early, and late promoters by temperature-sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Mol Cell Biol. 1985;5:1997–2008. doi: 10.1128/mcb.5.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLucia A L, Deb S, Partin K, Tegtmeyer P. Functional interactions of the simian virus 40 core origin of replication with flanking regulatory sequences. J Virol. 1986;57:138–144. doi: 10.1128/jvi.57.1.138-144.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DePamphilis M L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- 18.DePamphilis M L. Eukaryotic replication origins. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 45–86. [Google Scholar]
- 19.DePamphilis M L. Transcriptional elements as components of eukaryotic origin of DNA replication. Cell. 1988;52:635–638. doi: 10.1016/0092-8674(88)90398-4. [DOI] [PubMed] [Google Scholar]
- 20.de Villiers J, Schaffner W, Tyndall C, Lupton S, Kamen R. Polyoma virus DNA replication requires an enhancer. Nature. 1984;312:242–246. doi: 10.1038/312242a0. [DOI] [PubMed] [Google Scholar]
- 21.Dynan W S, Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983;32:669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- 22.Dynan W S, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 23.Elias P, Gustafsson C L, Hammarsten O. The origin binding protein of herpes simplex virus 1 binds cooperatively to the viral origin of replication oriS. J Biol Chem. 1990;265:17167–17173. [PubMed] [Google Scholar]
- 24.Elias P, Lehman I R. Interaction of origin binding protein with an origin of replication of herpes simplex virus 1. Proc Natl Acad Sci USA. 1988;85:2959–2963. doi: 10.1073/pnas.85.9.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frazier D P, Cox D, Godshalk E M, Schaffer P A. Identification of cis-acting sequences in the promoter of the herpes simplex virus type 1 latency-associated transcripts required for activation by nerve growth factor and sodium butyrate in PC12 cells. J Virol. 1996;70:7433–7444. doi: 10.1128/jvi.70.11.7433-7444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagen G, Muller S, Beato M, Suske G. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 1992;20:5519–5525. doi: 10.1093/nar/20.21.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 29.Hardwicke M A, Schaffer P A. Cloning and characterization of herpes simplex virus type 1 oriL: comparison of replication and protein-DNA complex formation by oriL and oriS. J Virol. 1995;69:1377–1388. doi: 10.1128/jvi.69.3.1377-1388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardwicke M A, Schaffer P A. Differential effects of nerve growth factor and dexamethasone on herpes simplex virus type 1 oriL- and oriS-dependent DNA replication in PC12 cells. J Virol. 1997;71:3580–3587. doi: 10.1128/jvi.71.5.3580-3587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassell J A, Muller W J, Mueller C R. The dual role of the polyomavirus enhancer in transcription and DNA replication. Cancer Cells. 1986;6:561–569. [Google Scholar]
- 32.He Z, Brinton B T, Greenblatt J, Hassell J A, Ingles C J. The transactivator proteins VP16 and GAL4 bind replication factor A. Cell. 1993;73:1223–1232. doi: 10.1016/0092-8674(93)90650-f. [DOI] [PubMed] [Google Scholar]
- 33.Heilbronn R, zur Hausen H. A subset of herpes simplex virus replication genes induces DNA amplification within the host cell genome. J Virol. 1989;63:3683–3692. doi: 10.1128/jvi.63.9.3683-3692.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hertz G Z, Mertz J E. Bidirectional promoter elements of simian virus 40 are required for efficient replication of the viral DNA. Mol Cell Biol. 1986;6:3513–3522. doi: 10.1128/mcb.6.10.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Igarashi K, Fawl R, Roller R J, Roizman B. Construction and properties of a recombinant herpes simplex virus 1 lacking both S-component origins of DNA synthesis. J Virol. 1993;67:2123–2132. doi: 10.1128/jvi.67.4.2123-2132.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Innis J W, Scott W A. DNA replication and chromatin structure of simian virus 40 insertion mutants. Mol Cell Biol. 1984;4:1499–1507. doi: 10.1128/mcb.4.8.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson S P, MacDonald J J, Lees-Miller S, Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell. 1990;63:155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- 38.Jones K A, Tjian R. Sp1 binds to promoter sequences and activates herpes simplex virus immediate-early gene transcription in vitro. Nature. 1985;317:179–182. doi: 10.1038/317179a0. [DOI] [PubMed] [Google Scholar]
- 39.Jordan R, Schaffer P A. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J Virol. 1997;71:6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadonaga J T, Carner K R, Masiarz F R, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA-binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 41.Kadonaga J T, Courey A J, Ladika J, Tjian R. Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science. 1988;242:1566. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
- 42.Kadonaga J T, Jones K A, Tjian R. Promoter-specific activation of RNA polymerase II transcription by Sp1. Trends Biochem Sci. 1986;11:20–23. [Google Scholar]
- 43.Kingsley C, Winoto A. Cloning of GT box-binding proteins: a novel Sp1 multigene family regulating T-cell receptor gene expression. Mol Cell Biol. 1992;12:4251–4261. doi: 10.1128/mcb.12.10.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klupp B G, Kern H, Mettenleiter T C. The virulence-determining genomic BamHI fragment 4 of pseudorabies virus contains genes corresponding to the UL15 (partial), UL18, UL19, UL20, and UL21 genes of herpes simplex virus and a putative origin of replication. Virology. 1992;191:900–908. doi: 10.1016/0042-6822(92)90265-q. [DOI] [PubMed] [Google Scholar]
- 45.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee-Chen G-J, Woodworth-Gutai M. Simian virus 40 DNA replication: functional organization of regulatory elements. Mol Cell Biol. 1986;6:3086–3093. doi: 10.1128/mcb.6.9.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J J, Peden K W C, Dixon R A F, Kelly T. Functional organization of the simian virus 40 origin of DNA replication. Mol Cell Biol. 1986;6:1117–1128. doi: 10.1128/mcb.6.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li R, Botchan M R. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 49.Lockshon D, Galloway D A. Sequence and structural requirements of a herpes simplex viral DNA replication origin. Mol Cell Biol. 1988;8:4018–4027. doi: 10.1128/mcb.8.10.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lusky M, Botchan M R. Transient replication of bovine papilloma virus type 1 plasmids: cis and trans requirements. Proc Natl Acad Sci USA. 1986;83:3609–3613. doi: 10.1073/pnas.83.11.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin D W, Deb S P, Klauer J S, Deb S. Analysis of the herpes simplex virus type 1 OriS sequence: mapping of functional domains. J Virol. 1991;65:4359–4369. doi: 10.1128/jvi.65.8.4359-4369.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maxwell I H, Harrison G S, Wood W M, Maxwell F. A DNA cassette containing a trimerized SV40 polyadenylation signal which efficiently blocks spurious plasmid-initiated transcription. BioTechniques. 1989;7:276–280. [PubMed] [Google Scholar]
- 53.Murata Y, Kim H G, Rogers K T, Udvadia A J, Horowitz J M. Negative regulation of Sp1 trans-activation is correlated with the binding of cellular proteins to the amino terminus of the Sp1 trans-activation domain. J Biol Chem. 1994;269:20674–20681. [PubMed] [Google Scholar]
- 54.Olivo P D, Nelson N J, Challberg M D. Herpes simplex virus DNA replication: the UL9 gene encodes an origin-binding protein. Proc Natl Acad Sci USA. 1988;85:5414–5418. doi: 10.1073/pnas.85.15.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Polvino-Bodnar M, Orberg P K, Schaffer P A. Herpes simplex virus type 1 oriL is not required for virus replication or for the establishment and reactivation of latent infection in mice. J Virol. 1987;61:3528–3535. doi: 10.1128/jvi.61.11.3528-3535.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Preston C M, Cordingley M G, Stow N D. Analysis of DNA sequences which regulate the transcription of a herpes simplex virus immediate early gene. J Virol. 1984;50:708–716. doi: 10.1128/jvi.50.3.708-716.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Preston C M, Frame M C, Campbell M E M. A complex formed between cell components and an HSV structural polypeptide binds to a viral immediate early gene regulatory sequence. Cell. 1988;52:425–434. doi: 10.1016/s0092-8674(88)80035-7. [DOI] [PubMed] [Google Scholar]
- 58.Preston C M, Tannahill D. Effects of orientation and position on the activity of a herpes simplex virus immediate early gene far-upstream region. Virology. 1984;137:439–444. doi: 10.1016/0042-6822(84)90238-1. [DOI] [PubMed] [Google Scholar]
- 59.Rabkin S D, Hanlon B. Herpes simplex virus DNA synthesis at a preformed replication fork in vitro. J Virol. 1990;64:4957–4967. doi: 10.1128/jvi.64.10.4957-4967.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosenfeld P J, O’Neill E A, Wides R J, Kelly T J. Sequence-specific interactions between cellular DNA-binding proteins and the adenovirus origin of DNA replication. Mol Cell Biol. 1987;7:875–886. doi: 10.1128/mcb.7.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 63.Schwyzer M, Wirth U V, Vogt B, Fraefel C. BICP22 of bovine herpes virus 1 is encoded by a spliced 1.7 kb RNA which exhibits immediate early and late transcription kinetics. J Gen Virol. 1994;75:1703–1711. doi: 10.1099/0022-1317-75-7-1703. [DOI] [PubMed] [Google Scholar]
- 64.Skaliter R, Lehman I R. Rolling circle DNA replication in vitro by a complex of herpes simplex virus type 1-encoded enzymes. Proc Natl Acad Sci USA. 1994;91:10665–10669. doi: 10.1073/pnas.91.22.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skaliter R, Makhov A M, Griffith J D, Lehman I R. Rolling circle DNA replication by extracts of herpes simplex virus type 1-infected human cells. J Virol. 1996;70:1132–1136. doi: 10.1128/jvi.70.2.1132-1136.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spaete R R, Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982;30:295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- 67.Stenlund A, Bream G L, Botchan M R. A promoter with an internal regulatory domain is part of the origin of replication in BPV-1. Science. 1987;236:1666–1671. doi: 10.1126/science.3037693. [DOI] [PubMed] [Google Scholar]
- 68.Stow N D. Localization of an origin of DNA replication within the TRs/IRs repeated region of the herpes simplex virus type 1 genome. EMBO J. 1982;1:863–867. doi: 10.1002/j.1460-2075.1982.tb01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stow N D, Davison A J. Identification of a varicella-zoster virus origin of DNA replication and its activation by herpes simplex virus type 1 gene products. J Gen Virol. 1986;67:1613–1623. doi: 10.1099/0022-1317-67-8-1613. [DOI] [PubMed] [Google Scholar]
- 70.Stow N D, McMonagle E C. Characterization of the TRs/IRs origin of DNA replication of herpes simplex virus type 1. Virology. 1983;130:427–438. doi: 10.1016/0042-6822(83)90097-1. [DOI] [PubMed] [Google Scholar]
- 71.Telford E A R, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus 1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 72.Triezenberg S J, LaMarco K L, McKnight S L. Evidence of DNA: protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes Dev. 1988;2:730–742. doi: 10.1101/gad.2.6.730. [DOI] [PubMed] [Google Scholar]
- 73.Udvadia A J, Rogers K T, Higgins P D, Murata Y, Martin K H, Humphrey P A, Horowitz J M. Sp-1 binds promoter elements regulated by the RB protein and Sp-1-mediated transcription is stimulated by RB coexpression. Proc Natl Acad Sci USA. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Udvadia A J, Templeton D J, Horowitz J M. Functional interactions between the retinoblastoma (Rb) protein and Sp-family members: superactivation by Rb requires amino acids necessary for growth suppression. Proc Natl Acad Sci USA. 1995;92:3953–3957. doi: 10.1073/pnas.92.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vlazny D A, Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genome. Proc Natl Acad Sci USA. 1981;78:742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weir H M, Calder J M, Stow N D. Binding of the herpes simplex virus type 1 UL9 gene product to an origin of viral DNA replication. Nucleic Acids Res. 1989;17:1409–1426. doi: 10.1093/nar/17.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weller S K, Spadaro A, Schaffer J E, Murray A W, Maxam A M, Schaffer P A. Cloning, sequencing, and functional analysis of oriL, a herpes simplex virus type 1 origin of DNA synthesis. Mol Cell Biol. 1985;5:930–942. doi: 10.1128/mcb.5.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whitton J L, Clements J B. Replication origins and a sequence involved in coordinate induction of the immediate-early gene family are conserved in an intergenic region of herpes simplex virus. Nucleic Acids Res. 1984;12:2061–2079. doi: 10.1093/nar/12.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wides R J, Challberg M D, Rawlins D R, Kelly T J. Adenovirus origin of DNA replication: sequence requirements for replication in vitro. Mol Cell Biol. 1987;7:864–874. doi: 10.1128/mcb.7.2.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong S W, Schaffer P A. Elements in the transcriptional regulatory region flanking herpes simplex virus type 1 oriS stimulate origin function. J Virol. 1991;65:2601–2611. doi: 10.1128/jvi.65.5.2601-2611.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu C A, Nelson N J, McGeoch D J, Challberg M D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wysokenski D A, Yates J L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989;63:2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]