Abstract
We have been investigating a long-term nonprogressor who was found to be human immunodeficiency virus type 1 (HIV-1) seropositive in 1985 and has survived with stable CD4+ T-cell counts (>1,000 CD4 cells/μl) without any AIDS-related illness. We have previously reported that repeated attempts to measure HIV-1 RNA in the peripheral mononuclear cells obtained from this subject have invariably failed. In the present study, we have analyzed the molecular nature of the HIV-1 quasispecies infecting this patient by PCR amplification of two proviral regions, the 5′ long terminal repeat (5′LTR)/gag leader and the nef gene, directly from fresh uncultured peripheral mononuclear cells, followed by length polymorphism analysis (with 1994, 1995, and 1996 samples) and sequencing (with a 1996 sample). Only proviral forms with nef deletions were revealed by length polymorphism analysis in samples from all three time points. Sequence analysis of the nef gene from the 1996 sample confirmed the presence of similar proviral quasispecies characterized by the presence of several deletions located in the nef-alone and the nef/U3 overlapping regions. Length polymorphism analysis of the 5′LTR/gag leader region suggested the existence of two major quasispecies populations, one characterized by the presence of forms carrying deletions in the U3 region and the other showing a completely intact, full-length 5′LTR. Evidence of the role of nef gene defects in long-term survival of HIV-1-infected patients has been provided so far in two independent investigations involving patients infected with HIV through blood transfusion. Here we show the existence of a similar condition in a subject who acquired HIV-1 seropositivity through the sexual route.
Investigations aimed at elucidating the biological basis of the peculiar condition commonly known as long-term nonprogressive human immunodeficiency virus type 1 (HIV-1) infection, a condition that affects only a small percentage of the total HIV-1-infected population (33), have rapidly increased in the past few years. For most of the long-term nonprogressor (LTNP) patients investigated, much of the attention has been devoted to the characterization of the specific antiviral host immune response and the genotypic properties of the infecting viral quasispecies (6, 30). The latter approach, generally based on the analysis of PCR-derived molecular clones of specific proviral regions, has generated interesting data. In one of the first studies of this kind, by Michael et al. (26), naturally occurring genotypes of the 5′ long terminal repeat (5′LTR)/gag leader region were characterized for four LTNP patients. A wide spectrum of intra- and interpatient sequence variability was observed, with a remarkable number of point mutations and length polymorphisms in cis- and trans-acting regulatory elements present in this region. A wide range of functional transcriptional activities was associated with these mutations in a reporter gene assay. In another study by the same group (27), the molecular characterization of the vif, vpr, vpu, tat1, and rev accessory genes, as well as of the nef gene proviral sequences, was carried out for one LTNP patient. This study revealed that 64% of the proviruses present in the peripheral blood mononuclear cells (PBMC) were grossly defective in either the vif, vpr, vpu, or tat1 gene, while a much lower percentage of the nef genes, 2 of 24 molecular clones (8.4%), were found to be defective, because of a frameshift and a nonsense mutation, respectively. Investigations of the nef gene in 10 LTNPs by the group of D. D. Ho revealed no gross deletions or obvious sequence abnormalities in the proviral genes of all of these subjects (17) and conserved wild-type nef gene biological function as determined by a single-cell infection assay (18). Evidence that gross viral defects in the nef gene can be responsible for, or contribute to, the absence of disease progression in a particular subpopulation of LTNPs has come from two independent investigations. In the first one, an LTNP subject affected by hemophilia was found to harbor proviruses containing only gross deletions in the nef gene (21). The second, more recent, investigation demonstrated that a group of LTNPs were infected by a single blood donor with a particularly attenuated HIV-1 strain, characterized by the presence of only genomes with gross nef deletions (8). These results are confirming for humans what was already known for rhesus monkeys infected with nef deletion variants of simian immunodeficiency virus (SIV) (19).
The nef gene is present only in primate lentiviruses. Although the function of the nef gene has been quite controversial (7), it has been documented that in primary blood lymphocytes and macrophages, Nef promotes HIV-1 replication (10, 28, 35). It was also shown that Nef stimulates HIV-1 proviral DNA synthesis (2). In in vitro experiments with T-cell lines, it has been shown that Nef protein downregulates the cell surface CD4 expression (3, 31). Interestingly, Nef also seems to play a role in signal transduction and cellular activation (11). A role for Nef in viral and cell activation has been described by Fujinaga et al. (12): they have demonstrated that extracellular Nef protein can activate HIV-1 from latent to productive infection both in infected T-cell lines and in PBMC from asymptomatic carriers even when these cells were cultivated without phytohemagglutinin (PHA) and interleukin 2 (IL-2).
We have been studying a cohort of 11 LTNP patients characterized not only by the absence of recoverable infectious virus but, most notably, by undetectable levels of HIV-1 RNA species in uncultured PBMC (13). When the PBMC derived from six of these subjects were put into culture and stimulated in vitro by PHA and phorbol 12-myristate 13-acetate (PMA), once again no detectable levels of specific HIV-1 RNA were found (14). Repeated attempts at in vitro stimulation of PBMC derived from one of these patients, termed SG1, over a period of 3 years have all failed. These results strongly suggest that the PBMC of SG1 may harbor replication-defective proviral quasispecies, and this, in turn, may explain the stability of the nonprogressive condition in this patient. Hoping to elucidate the molecular basis of this presumed defectivity, we have analyzed the 5′LTR/gag leader and nef gene proviral regions present in fresh uncultured PBMC. Strikingly, in both of these populations the nef gene was found to be defective due to the presence of inactivating deletions. This finding confirms and strengthens the evidence for the role of nef gene defects in the long-term survival from HIV-1 infection.
MATERIALS AND METHODS
Study patient.
The patient, referred to as patient SG1, belongs to a cohort of 11 HIV-1-positive subjects who, since 1993, have met all the criteria to be classified as LTNPs (30). Some virological and immunological features of this cohort have been previously reported (13, 14).
Sample preparation.
PBMC were prepared over Ficoll-Hypaque (Lymphoprep; Nycomed Pharma AS, Oslo, Norway) from EDTA-treated peripheral blood freshly drawn from SG1. Peripheral blood lymphocytes (PBL) were obtained by removing the macrophages for adherence to plastic surface as previously described (9). CD4 counts were cytofluorometrically determined with anti-Leu3a monoclonal antibody (Becton Dickinson, San Jose, Calif.). CD8-depleted PBMC were obtained by magnetic purging of PBMC with M-450 CD8 Dynabeads (Dynal, Great Neck, N.Y.) under standard conditions. The effectiveness of the depletion procedure was ascertained by fluorescence-activated cell sorter analysis with anti-Leu2a monoclonal antibody (Becton Dickinson). After purging, levels of residual CD8 cells lower than 0.1% were found.
Virus isolation procedures.
PBMC (5 × 106) from patient SG1 and mixed PMBC (5 × 106) from three seronegative donors were separately resuspended in 5 ml of RPMI 1640–10% fetal calf serum (RPMI) supplemented with PHA (2 μg/ml) and incubated at 37°C for 48 h in a humidified atmosphere containing 5% CO2. After this period, the cells were washed, mixed together, resuspended in 10 ml of RPMI supplemented with recombinant IL-2 (100 U/ml), and incubated at 37°C. Culture supernatants were collected at 7-day intervals over a period of 4 weeks and assayed for p24 production by a standard procedure (HIV-1 p24 Elisa Kit; Du Pont, Brussels, Belgium).
HIV-1 RNA induction by PHA-PMA.
CD8-depleted PBMC (107) were resuspended in 5 ml of RPMI supplemented with PMA (100 nM), PHA (2 μg/ml), and IL-2 (100 U/ml). Culture aliquots were collected at 7-day intervals over a period of 4 weeks and assayed for the presence of HIV-1 RNA.
PCR detection of HIV-1 DNA and HIV-1 RNA.
HIV-1 provirus in adherent macrophages and PBL was assayed by PCR analysis of the HIV-1 gag region by using SK38 and SK39 primers as previously described (14). The presence of extragenomic viral DNA was evaluated by the PCR positivity for two-LTR circular DNA molecules as described by Pang et al. (29). The HIV RNA in PBMC was assayed by reverse transcription-PCR analysis of the gag region as previously described (14).
Quantitative RNA analysis.
The HIV-1 RNA load in plasma was measured by the Amplicor HIV monitor test (Roche Diagnostic System, Inc., Branchburg, N.J.) according to the manufacturer’s instructions.
Quantitative assay of HIV-1 provirus in PBMC.
The number of HIV-DNA copies present in the PBMC of patient SG1 was determined by semiquantitative PCR with procedures previously described in detail (13). Briefly, a sample containing 106 PBMC was treated with 100 μl of lysis buffer (10 mM Tris-HCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% Tween 20 and 200 μg of proteinase K per ml) and incubated at 56°C for 2 h and at 95°C for 10 min. PCR was performed with the HIV-1 gag primers (SK38 and SK39), using 10 μl of cell lysate. PCR products were electrophoresed on a 2% agarose gel and transferred to a Hybond N+ membrane (Amersham International PLC, Buckinghamshire, United Kingdom). The filters were subjected to hybridization with 32P-end-labelled SK19 probe (2 × 106 dpm/pmole) and autoradiographed (15-h exposure). The relative absorbance of the PCR signals was measured by laser densitometric analysis (LKB Ultrascan XL) of autoradiographic films. The number of HIV DNA copies in the PBMC samples was calculated by using the regression coefficient of the linear part of the calibration curve. The calibration curve was obtained by gag amplification of 10-fold-diluted (from 105 to 101 cells/ml) lysates of LAV-8E5 cells containing a single provirus per cell.
Exogenous HIV-1 infection of PBL from patient SG1.
PBL of patient SG1 were PHA stimulated for 3 days. A portion of these lymphocytes, containing 2 × 106 CD4+ cells, was depleted of CD8 lymphocytes. A second portion of lymphocytes containing the same number of CD4+ cells was not subjected to CD8-purging procedures. Both CD8-depleted and non-CD8-depleted cells were exposed to 5 × 105 50% tissue culture infective doses of HIV strain H9/IIIb for 4 h at 37°C and then washed, resuspended in 5 ml of RPMI supplemented with IL-2, and incubated at 37°C. Equivalent amounts of non-CD8-depleted and CD8-depleted PBL of SG1 were put in culture without exogenous virus infection. Two additional cultures, containing PHA-stimulated PBL of an HIV-seronegative individual depleted of CD8 lymphocytes and not depleted, were infected with H9/IIIb virus and incubated as described above. At various time intervals, the supernatants of all of the cultures were assayed for the amount of cell-released p24 antigen. Preparation and titration of the H9/IIIb infecting virus were performed as previously described (4).
Amplification, molecular cloning, and nucleotide sequencing of the proviral nef gene and 5′LTR/gag region (bp 128 to 784) from patient PBMC.
Cell lysates were obtained by treating each PBMC sample with 200 μg of proteinase K per ml in lysis buffer (10 mM Tris-HCl [pH 8.00], 5 mM EDTA, 0.1% sodium dodecyl sulfate [SDS]) overnight at 40°C. In order to amplify the HIV-1 proviral DNA present in low copy numbers, a nested-PCR strategy was employed. For the first round, 10 μl of PBL lysate (equivalent to 5 × 105 cells) was assembled in a 50-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 5 mM MgCl2 under a thin layer of solid paraffin (hot-start PCR). After being kept at 95°C for 10 min, each tube was overlaid with 50 μl of a mixture containing 50 pmol each of the outer primer pairs (LTR-1–LTR-2 or Nef-1–Nef-2), 0.4 mM each deoxynucleoside triphosphate (dNTP), 2 μl of Tween 20, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 10 U of Taq DNA polymerase. They were then cycled in a Perkin-Elmer 9600 DNA thermal cycler as follows: (i) 95°C (2 min); (ii) 94°C (1 min), 57°C (1 min), and 72°C (2.5 min) for 2 cycles; (iii) 94°C (45 s), 57°C (30 s), and 72°C (2.5 min) for 25 cycles; and (iv) 10 min at 72°C. Ten microliters of the first-round PCR mixture was overlaid on top of a thin solid-paraffin layer (hot-start PCR) separating the lower 90 μl of reaction mixture, containing 50 pmol each of the inner primer pairs (LTR-3–LTR-4 or Nef-3–Nef-4), 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each dNTP, and 10 U of Taq DNA polymerase, and amplified by using the same cycling conditions as for the first round. The outer primers were as follows: LTR-1, 5′ CACACAAGGCTAYTTCCCTGA 3′ (positions 59 to 79), and LTR-2, 5′ TCCYCYTGGCCTTAACCGAAT 3′ (positions 864 to 844), for the 5′LTR/gag leader region; Nef-1, 5′ GTAGCTGAAGGGACAGATAGGGTTAT 3′ (8678 to 8703), and Nef-2, 5′ GCACTCAAGGCAAGCTTTATTGAGGC 3′ (9622 to 9597), for the nef gene region. The inner primers were as follows: LTR-3, 5′ TGGATGGTGCTWCAAGYTAGT 3′ (128 to 148), and LTR-4, TCCTTCTAGCCTCCGCTAGTC 3′ (784 to 764), for the 5′ LTR/gag leader region; Nef-3, 5′ ACATACCTAGAAGAATAAGACAGG 3′ (8739 to 8762), and Nef-4, 5′ GTCCCCAGCGGAAAGTCCCTTGTA 3′ (9443 to 9420), for the nef gene region. (In these sequences, Y = C or T, W = A or T, and numbers in parentheses refer to NL4-3 sequence positions [1]). To improve sampling, for each of the two proviral regions, PCR products were generated from three independent amplifications and then pooled before cloning. PCR products were purified by spin-dialysis with Chromaspin C-100 columns (Clontech, Inc.) prior to cloning into the pGEM-T (Promega, Inc.) vector. Recombinant pGEM-T clones were identified by colony PCR, and plasmid DNA was prepared by rapid column purification (Qiagen, Inc.). Individual clones, 11 for the nef region (N clones) and 10 for the 5′LTR/gag leader region (L clones), were fully sequenced in both strands with the Sequenase version 2.0 DNA sequencing kit (U.S. Biochemicals) by the dideoxy chain termination method (32).
PCR-generated misincorporation errors were determined by control experiments using a defined template, with the same experimental conditions employed for the patient samples but in separate experiments to prevent contamination of patient samples. The error rate was found to be about four substitutions in 6,050 bases sequenced, or 0.066%. No length variants were observed in these control experiments. Great care was taken to avoid the possibility of sample cross-contamination: all of the manipulations involved in PBMC lysate preparation, PCR sample preparation, and PCR product analysis were performed in different laboratory spaces, and negative controls (no lysate and PBMC lysate from a seronegative donor) were included in each PCR experiment. In addition, PCRs with different proviral regions were never performed in the same experiment, and this precaution was also taken for the molecular cloning experiments. Nucleotide sequences were aligned with the Clustal program (16).
Colony PCR.
The colonies obtained from the subcloning experiments were grown overnight in standard Luria-Bertani medium containing ampicillin (100 μg/ml). The bacterial pellet obtained from about 100 μl of the overnight culture was resuspended in 20 μl of water, boiled for 10 min, and centrifuged to pellet cellular debris. The supernatant was subjected to single-round amplification with the inner primers. The amplified product was analyzed by agarose gel electrophoresis.
Amplification and characterization of the proviral 5′LTR termini.
The entire 5′LTR/gag leader region was investigated by PCR, using the LF-1 (5′ TGGAAGGGCKARTTYACTCC 3′ [K = G or T; R = G or A; Y = C or T; positions 1 to 20, referred to NL4-3]) and LTR-2 (see previous paragraph) oligonucleotides as sense and antisense primers, respectively. The sequence of the LF-1 oligonucleotide was chosen from the consensus sequence derived from the our nef clones. PCR was performed in a 50-μl volume containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 0.2 mM each dNTP, 50 pmol of each primer, and 1.5 U of AmpliTaq Gold (Perkin-Elmer Cetus). Before the amplification, the enzyme was activated by heating the reaction mixture at 95°C for 10 min. The amplification conditions were as follows: 2 cycles of 95°C for 2 min, 57°C for 1 min, and 72°C for 2.5 min; 25 cycles of 95°C for 45 s, 57°C for 30 s, and 72°C for 2.5 min; and an incubation at 72°C for 5 min. The PCR products generated after one round of amplification were identified by Southern blotting. The amplified bands were transferred from the agarose gel to a Hybond N+ membrane (Amersham) in 0.4 M NaOH under a slight vacuum and preincubated at 56°C for 2 h in 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–5× Denhardt’s solution–0.5% SDS–10 μg of denatured salmon sperm DNA per ml. The hybridization was carried out at 56°C overnight in the same mixture containing 10 pmol of the T4 polynucleotide kinase 32P-labelled L2-F (5′ GGGCCAGGGATCAGATATCC 3′ ; positions 99 to 118) or L3-R (5′ GGCGGGACTGGGGAGTGG 3′; positions 390 to 407) probe. (Positions refer to the NL4-3 clone.) After being washed with 2× SSC–1% SDS at room temperature, the membranes were autoradiographed.
The shorter PCR product (about 680 bp) was excised from the gel. The DNA was purified with a QIAquick gel extraction kit (Quiagen, Inc) and cloned into the pGEM-T vector. Five individual clones (5′L clones) were sequenced as described above.
LD-PCR, restriction analysis, and sequencing.
Almost-full-length and 3′-half HIV-1 proviruses were obtained by employing a nested long-distance PCR (LD-PCR). The primers used were as follows: 626s, 5′ TCTCTAGCAGTGGCGCCCGAACAGGG; 691s, 5′ GCAGGACTCGGCTTGCTGAAGC; 5048s, 5′ ACAGATGGCAGGTGATGATTGTGT; 9614a, 5′ GGCAAGCTTTATTGAGGCTTAAG; and 9680a, 5′ GGTCTGAGGGATCTCTAGTTACCAGAGTC. Each primer is designated by a number that reflects the position of the first 5′ nucleotide with respect to NL4-3 coordinates and a letter, either s or a, that indicates, respectively, sense or antisense strand. PCR amplifications were performed with a long-distance mixture of the thermostable DNA polymerases Taq (5 U/μl) (Perkin-Elmer Cetus) and Pfu (5 U/μl) (Stratagene) at a ratio of 8:1 (vol/vol). First- and second-round PCRs were conducted in 50-μl volumes containing 20 mM Tris-HCl (pH 8.7), 10 mM KCl, 2 mM MgSO4, 10 mM (NH4)2SO4, 0.1 mg of bovine serum albumin per ml, 200 μM each dNTP, 4 pmol of each primer, and 0.5 μl of long-distance enzyme mixture. All of the reaction mixtures were overlaid with a drop of liquid paraffin and run on a Perkin-Elmer Thermal Cycler 9600 (2 min of denaturation at 92°C followed by 40 cycles of 92°C for 30 s, 60°C for 30 s, and 68°C for 10 min, plus a final extension at 68°C for 10 min).
Genomic DNA equivalent to about 2 × 105 PBMC was used as the template in each first-round PCR, and 5 μl of the first-round PCR product was used as the template for the second-round PCR. The sensitivity of the PCR was such that a fragment of the expected size was detected starting from about 90 copies of pNL4-3 (data not shown).
Restriction enzyme digests were obtained by treating 10 μl of PCR products with 5 U of the appropriate restriction enzyme, along with the specific buffer, for 2 to 3 h at 37°C.
Partial sequencing of the nef region was carried out on the gel-purified PCR product (Geneclean; BIO 101, Inc.) by cycle sequencing with the fluorescence-labelled primer 5′TCTTGAAGTACTCCGGATGC (positions 9399 to 93799) and the Sequitherm EXCEL Long-Read DNA Sequencing Kit (Epicentre Technologies Corporation, Madison, Wis.) by the procedures specified by the manufacturer.
Nucleotide sequence accession numbers.
The sequences described here have been deposited in GenBank under accession no. U89846 through U89855 for the nef gene and U89856 through U89866 for the 5′LTR region.
RESULTS
Characteristics of the patient.
Patient SG1 is a 35-year-old man who has not experienced HIV-associated clinical signs or symptoms in the past 11 years since infection. He was found to be HIV-1 seropositive by enzyme-linked immunosorbent assay and Western blotting in November 1985. He has continued to be HIV-1 seropositive. In 1995, this patient showed weak positivity for Treponema antibodies which disappeared in 1997. He has always been negative for hepatitis B virus and hepatitis C virus markers. PBL analysis in November 1992 showed 1,023 CD4 T cells per μl, with a CD4/CD8 ratio of 1.39. CD4 counts have fluctuated around 1,106.2 ± 68.3 (mean ± standard deviation) cells per μl over the past 4 years, with an overall CD4 slope of 1,346 ± 1,109 (mean ± standard error). Patient SG1 has never received antiretroviral treatment.
Stability of the virological parameters.
The virological parameters of patient SG1 since October 1993 are summarized in Table 1. The proviral DNA burden ranged between 93 and 98 copies per 105 PBMC over a 3-year period. Searches for extragenomic HIV-1 DNA in PBL and proviral DNA in macrophages were always negative. The most remarkable virological feature of this subject has been the inability not only to isolate the virus from PBMC but, most notably, to detect virus-specific RNA forms, in both PBMC and plasma. Furthermore, PMA-PHA-stimulated lymphocytes were consistently p24 and HIV RNA negative.
TABLE 1.
Virological features of patient SG1
| Time point | Viral isolation from PBMC | HIV-1 DNA in macrophages | HIV-1 DNA copies/105 PBMC (mean ± SD)a | Extragenomic HIV-1 DNA | HIV-1 RNA in PHA-PMA-stimulated lymphocytes | HIV-1 RNA copies/ml of plasma |
|---|---|---|---|---|---|---|
| October 1993 | Negative | UDb | 98 ± 12 | UD | Absent | |
| May 1994 | Negative | UD | 99 ± 20 | UD | Absent | UD |
| May 1995 | Negative | UD | ||||
| June 1996 | Negative | UD | UD | Absent | UD | |
| November 1996 | Negative | 93 ± 18 | UD | Absent | UD |
Values for triplicate experiments.
UD, undetectable.
HIV-1 replication in PBL from patient SG1 with or without exogenous HIV-1 infection.
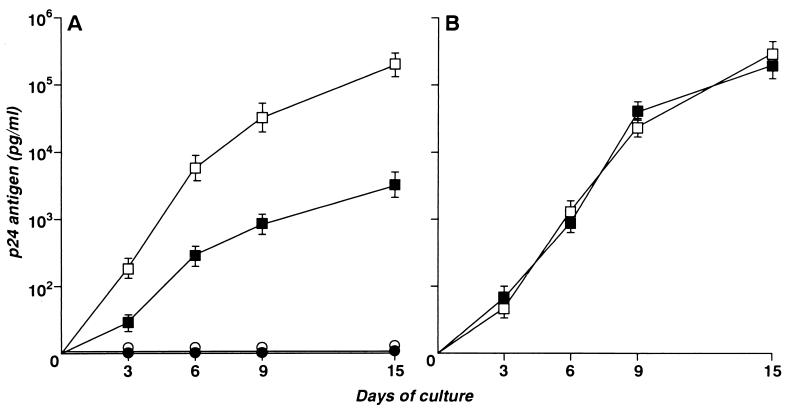
To determine how lymphocytes from patient SG1 responded to HIV-1 reinfection in vitro, we performed the experiment with the results shown in Fig. 1. CD8-depleted and non-CD8-depleted PBL obtained from patient SG1 were PHA stimulated, exogenously infected with HIV-1, and expanded in the presence of IL-2, and p24 antigen levels in the culture supernatants were determined. A parallel experiment was performed without exogenous infection (Fig. 1A). Two additional cultures, containing PBL of an HIV-1-seronegative subject depleted of CD8 cells or not depleted, were infected with HIV-1 and investigated in parallel (Fig. 1B). Without exogenous HIV infection, non-CD8-depleted and CD8-depleted PBL from SG1 did not generate detectable amounts of p24 antigen. In contrast, after HIV-1 superinfection, consistent levels of p24 antigen were readily released in the culture medium, with an absolute amount of p24 in CD8-depleted PBL that was about 50 times higher than that in non-CD8-depleted PBL. The experiment performed with PBL from the seronegative donor showed an high level of p24, of the same magnitude in the supernatants of both CD8-depleted and non-CD8-depleted cultures. This experiment demonstrates that the CD4 lymphocytes from patient SG1 were susceptible to in vitro HIV-1 superinfection. Moreover, there is clear evidence that CD8 lymphocytes from this subject had a strong anti-HIV activity.
FIG. 1.
(A) Kinetics of p24 antigen accumulation in the supernatants of undepleted (closed symbols) and CD8-depleted (open symbols) PBMC cultures from patient SG1 with (squares) and without (circles) exogenous infection with HIV-1. (B) Kinetics of p24 antigen accumulation in the supernatants of HIV-infected undepleted (▪) and CD8-depleted (□) PBMC cultures from an HIV-seronegative individual. Error bars indicate standard deviations.
Length polymorphism analysis of the nef and 5′LTR/gag leader regions.
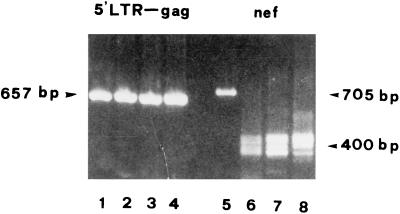
PBMC lysates obtained from three time points spanning a 2-year period (May 1994, October 1995, and June 1996) were subjected to two separate nested PCRs for the nef and 5′LTR/gag leader regions. As a control for reference size, we used the NL4-3 clone of HIV-1. The agarose gel analysis shown in Fig. 2 represents the typical, highly reproducible pattern obtained from a series of multiple PCR experiments. The 5′LTR/gag leader region amplification performed with the LTR-1–LTR-3 and LTR-2–LTR-4 primers yielded only a single, full-length DNA fragment in all of the sequential samples, which was identical in size to that obtained from the NL4-3 control (expected size, 657 bp). In contrast, all of the nef region amplifications yielded DNA fragments that migrated much faster than the 705-bp fragment obtained from the NL4-3 control, clearly indicating the presence of large deletions. Samples from all three time points studied showed a cluster of multiple DNA fragments by gel analysis, with very similar patterns. Although at least four bands in each sample were detectable, these were very closely scattered around a length of approximately 420 to 380 nucleotides. The absence of full-length nef gene fragments was further confirmed by Southern blot analysis (data not shown).
FIG. 2.
Analysis of length polymorphism of 5′LTR/gag leader (bp 128 to 784) and nef gene regions in PBMC obtained from patient SG1 in May 1994 (lanes 2 and 6), October 1995 (lanes 3 and 7), and June 1996 (lanes 4 and 8). DNA fragments from the second PCR amplification were separated by electrophoresis through a 2% agarose gel and visualized by ethidium bromide staining. The positive control (lanes 1 and 5) consists of plasmid pNL4-3, which yields two full-length bands of 657 bp (5′LTR/gag leader) and 705 bp (nef gene).
5′LTR/gag leader region sequence analysis shows full-length conserved sequences with low sequence divergence.
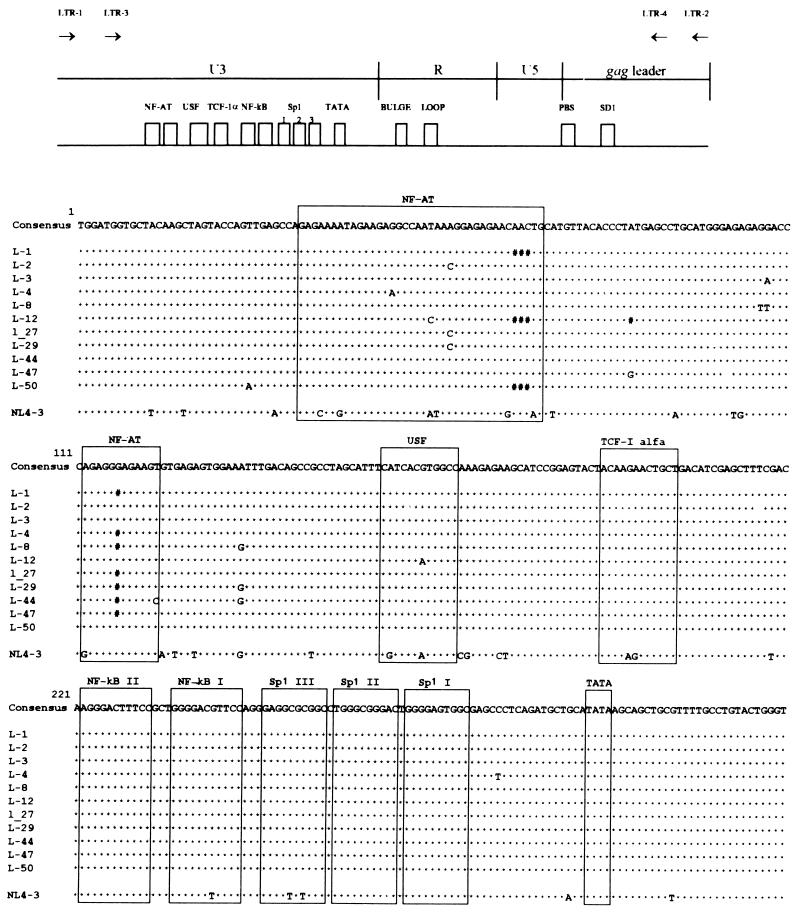
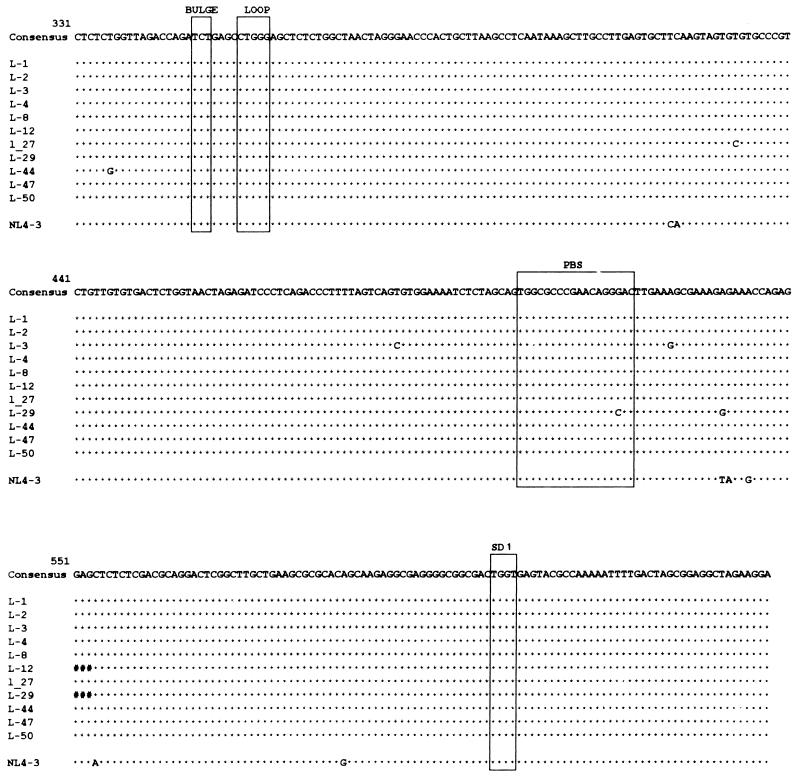
The apparent lack of viral RNA in patient SG1 could be due to a transcriptionally silent 5′LTR. Consequently, we looked for possible abnormalities in the 5′LTR region. The 5′LTR/gag leader region was amplified from a PBMC sample, obtained in June 1996, by a nested PCR. This amplicon encompasses NL4-3 sequence positions 128 to 784 and contains almost all of the cis-acting sequences, from the AP-1 binding site through the PSI site, characterized in the prototype LTRs (36). The amplification products were cloned, and the complete sequences of 11 molecular clones were determined. A complete alignment of all of the sequences obtained is shown in Fig. 3. Consistent with the agarose gel analysis (see above), all of the clones were about the same size, with a minimal difference from the longest clone, L-3 (657 bp [same size as NL4-3]) to the shortest clone, L-12 (650 bp). This slight length variation was due to two 3-bp deletions, in the first NF-AT element and the region following the primer binding site, and a 1-bp deletion in the second NF-AT element. Two other clones (L-1 and L-50) had the same 3-bp deletion in the first NF-AT element. All of the clones represented distinct sequence variants, with sequence divergence ranging from 0% (clone L-1) to 0.6% (clone L-29). Most notably, the sequence variation among the clones, with respect to the consensus sequence, was very limited. A G-to-A substitution in clone L-4, a T-to-C mutation in clone L-12, and an A-to-C change in clones L-2, L-27, and L-29 were located in the first NF-AT element. A T-to-C substitution in clone L-44 was the only change in the second NF-AT element. A G-to-A mutation in the USF element was found in clone L-12. A comparison of the cis-acting motif consensus, with respect to the reference NL4-3 sequence, showed a limited number of base changes. Most notably, a G-to-T transversion occurred in the first NF-κB element, and two C-to-T transversions occurred in the third Sp1 element.
FIG. 3.
Alignment of 11 HIV-1 5′LTR/gag leader sequences in PBMC derived from patient SG1 in June 1996, along with the alignment of the consensus for these sequences and the alignment of the NL4-3 sequences with respect to the consensus. Asterisks indicate identity to the consensus sequence. A number symbol indicates the absence of a base pair. The positions of the motifs for the NF-AT, USF, TCF-1 alpha, NF-κB, and Sp1 sites, TATAA box, bulge and loop elements of the TAR, primer binding site (PBS), and SD 1 (major 5′ splice donor) are boxed. A schematic drawing of the LTR/gag leader region is shown above the alignment, and the positions of the nested-PCR primers are indicated by arrows at the top.
nef region sequence analysis reveals two large deletions located in the nef/U3 nonoverlapping region and in the nef/U3 overlapping region.
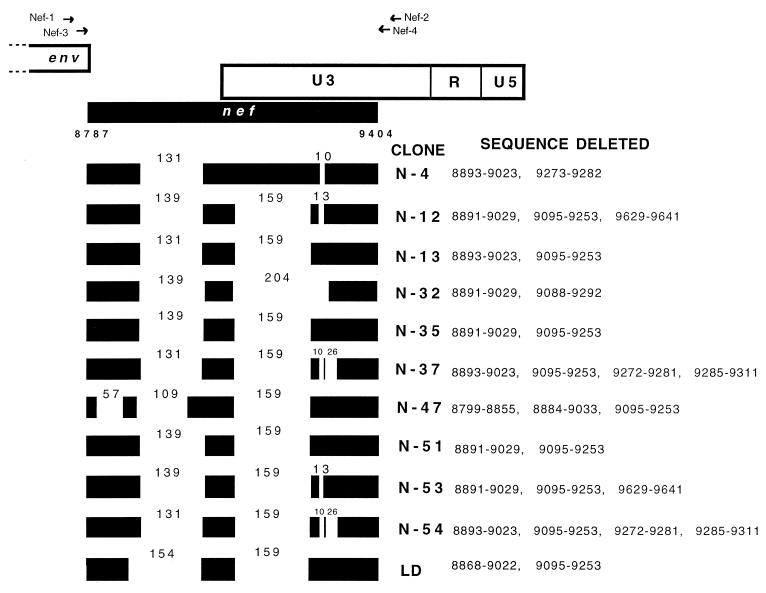
The PCR length polymorphism analysis of the nef gene region revealed that patient SG1 harbored only deleted forms of this gene. In order to characterize these deletions at the sequence level, we molecularly cloned the PCR products obtained from the June 1996 sample. Fifty recombinant bacterial clones were picked and screened by colony PCR, all of them containing nef gene deletion variants. The majority of the clones exhibited similar sizes of approximately 400 bp by gel analysis; only clone N-4 was significantly larger (about 550 bp). Clone N-47 appeared to be slightly smaller than the average. The complete nucleotide sequences of 10 clones (8 randomly chosen clones plus clones N-4 and N-47) were determined. In Fig. 4, an alignment of the deletions present in all of the clones sequenced is shown. Two main large deletions characterized 9 of the 10 molecular clones investigated; they were slightly variable in size and boundary but were located in approximately the same regions. The first main deletion encompassed 109 to 139 nucleotides in the nef-alone region, and the second main deletion mapped to the nef/U3 overlapping region. An identical nef/U3 deletion of 159 bp was present in 8 of 10 clones, with only N-4 and N-32 being different. One or two additional small deletions (10 to 26 bp) may follow the main one in some of the clones (Fig. 4). Clone N-47 had an additional 56-bp deletion located upstream of the main nef-alone deletion. Of note, while the main nef-alone deletion removes both the highly conserved acidic domain and the highly conserved (Pxx)4 motif from Nef, all of the downstream deletions located in the nef/U3 region leave the polypurine tract, TATAA box, and NF-κB and Sp-1 binding sites intact.
FIG. 4.
Locations of the deletions present within the nef gene in PBMC derived from patient SG1 in June 1996. A schematic drawing of the genomic structure of the NL4-3 HIV-1 clone (1) for the same region is shown at the top. The positions of the nested-PCR primers are indicated by arrows. Black boxes represent normal sequence, while blank spaces represent deletions. Nucleotide numbering refers to NL4-3 sequence positions. The numbers above the blank regions represent the sizes of the deletions. The deletions shown in the LD row are referred to the sequencing of the LD-PCR product.
Identification of proviruses with partially deleted 5′LTR termini.
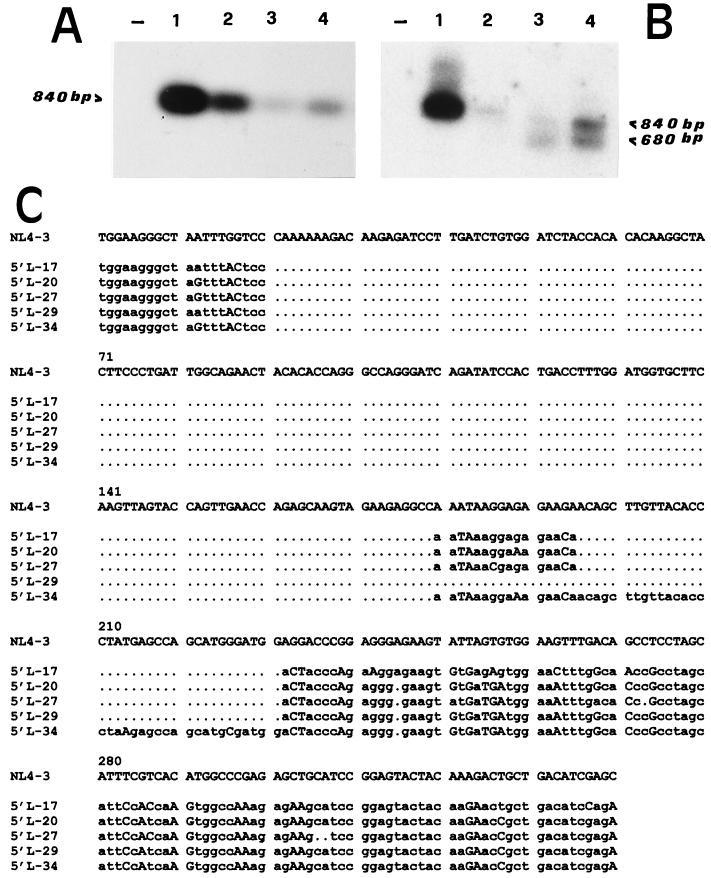
Sequence analysis of the nef region revealed that the upstream outer-inner primer pair (LTR-1 and LTR-3), designed for the 5′LTR/gag leader region amplification, mapped exactly inside the nef/U3 overlapping downstream deletion. This fact makes it impossible to recover similarly deleted 5′LTR/gag leader proviral region variants that may be represented in the proviral population harbored in SG1 PBMC. Thus, a new PCR assay was performed on SG1 samples from two time points, using a new sense oligonucleotide (LF-1) spanning the beginning of the U3 immediately before the nef/U3 deletions. The LTR-2 primer was used as an antisense oligonucleotide. Products were analyzed by Southern blotting with two 32P-labelled probes able to recognize, respectively, the inside (L2-F) and the outside (L3-R) of the nef/U3 deletions present in the majority of the nef clones sequenced. The results are shown in Fig. 5. In the SG1 DNA samples as well as in the NL4-3 DNA (used as a control), the L2-F probe recognized only an 840-bp band corresponding to the expected full-length size for this amplification (Fig. 5A). The L3-R probe recognized an additional band of about 680 bp in the SG1 samples but not in the control NL4-3 (Fig. 5B). The 680-bp product obtained from the SG1 sample of June 1996 was cloned and sequenced (Fig. 5C). All clones showed large deletions in the U3 region. The total sizes of the deletions were 170 bp (5′L-34 clone), 206 bp (5′L-17, 5′L-20, and 5′L-27 clones), and 221 bp (5′L-29 clone).
FIG. 5.
Identification of proviruses with partially deleted 5′LTR termini: Southern blotting and sequence analysis of the 5′LTRs of proviruses in SG1 PBMC. Hybridization with L2-F (A) and L3-R (B) 32P-labelled probes is shown. Lanes −, negative controls (DNA extracted from 105 PBMC of a healthy blood donor). Lanes 1 and 2, positive controls (1 pg [lane 1] and 1 fg [lane 2] of pNL4-3 DNA mixed with DNA extracted from 105 PBMC of a healthy donor). The other lanes show PCR products of DNA extracted from 105 PBMC collected from patient SG1 at the May 1995 and June 1996 time points. (C) Alignment of partial sequences relative to the 5′LTR regions of clones obtained from the 680-bp PCR product.
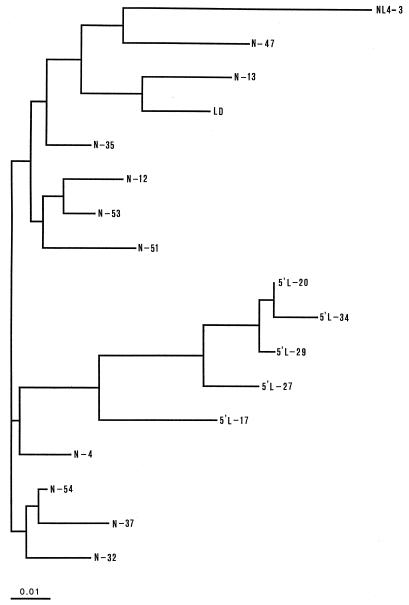
These results suggest that the SG1 DNA samples contained a mixture of two types of proviruses, one carrying the undeleted full-length 5′LTR and the other missing an upstream portion of 5′-U3, similar in size and location to those characterized in the nef/U3 deletion clones. Phylogenetic analysis by the neighbor-joining method further validates the relationship among the clones from different origins containing the U3 deletions (Fig. 6).
FIG. 6.
Phylogenetic analysis of HIV-1 nef/U3 quasispecies in patient SG1, showing the relationship between the sequences derived from different sets of experiments. The tree was constructed by the neighbor-joining method, using Clustal (16). One thousand bootstrap replications were performed. NL4-3 was chosen as the prototype sequence.
Either almost the entire HIV-1 provirus or its 3′ proviral half can be amplified from PBMC by LD-PCR.
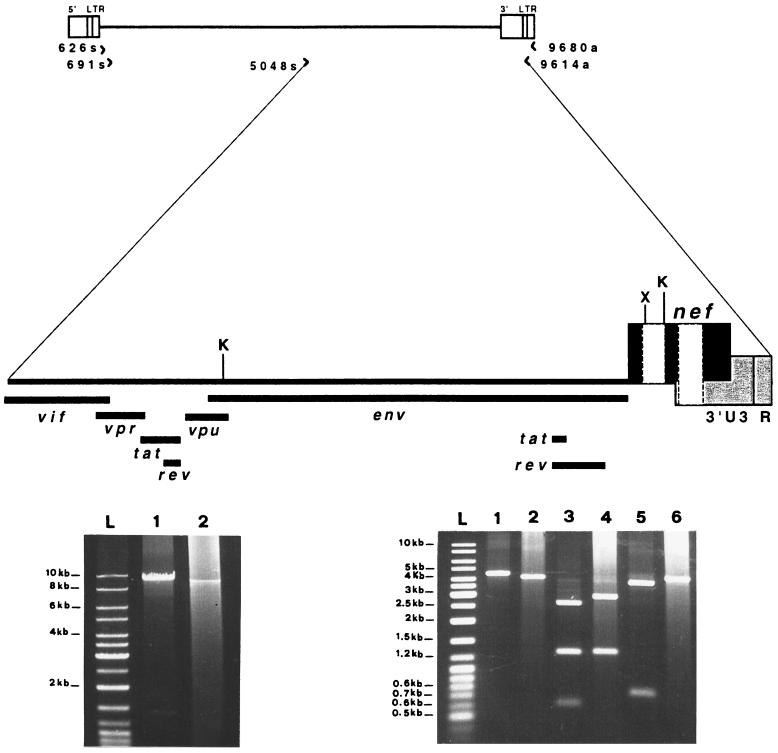
The possible presence of additional deletions in other regulatory and/or accessory genes of the provirus was investigated by LD-PCR. A nested LD-PCR strategy was designed in order to recover an HIV-1 provirus corresponding to almost the full length (positions 691 to 9614 of the NL4-3 map). This strategy allowed the recovery of a distinct DNA fragment of about 9 kb, as detected by ethidium bromide staining after electrophoretic separation of the PCR product obtained from the 1996 PBMC sample (Fig. 7, bottom left, lane 2). Interestingly, this fragment seemed to be slightly shorter than the fragment obtained from the plasmid NL4-3 amplification run in parallel (Fig. 7, bottom left, lane 1). This result was obtained with genomic DNA equivalent to about 2 × 105 PBMC. A smaller amount of PBMC genomic DNA failed to give any detectable band. The yield of the reaction was low and was not improved by increasing the input of genomic DNA (data not shown). Replacing the upstream inner primer (691s) with another primer located at the beginning of the second half of the provirus (5048s) in the second round of the PCR increased the yield of the PCR product considerably (Fig. 7, bottom right, lane 2), allowing further characterizations to be carried out. The ca. 4.5-kb 3′ half of the proviral genome so obtained contains all of the accessory and regulatory genes as well as almost the entire 3′LTR. Most noticeably, the fragment obtained was clearly shorter than that obtained from the control plasmid NL4-3 amplification (Fig. 7, bottom right, lane 1), indicating the presence of a deletion(s) in the SG1 product. The difference in size from the NL4-3 fragment was consistent with the presence of a deletion similar in size to those present in the nef clones. In order to verify this possibility, aliquots of the LD-PCR product were digested with the restriction enzymes XhoI and KpnI, which have recognition sites at the boundary of (XhoI) or inside (KpnI) the NL4-3 region (Fig. 7, top) missing in the nef clones. The cutting patterns generated by the two enzymes are shown in Fig. 7, bottom right, lanes 3 to 6. The digestion of the SG1 product with XhoI did not recognize any site leaving the fragment intact (Fig. 7, bottom right, lane 6). Also, the KpnI site located inside the nef deletion was not recognized, as indicated by the specific pattern of digestion produced (Fig. 7, bottom right, lane 4). In fact, the cutting pattern of the SG1 product digested with this enzyme showed two bands. The first one (about 1.3 kb) was identical in size to the one produced by the NL4-3 fragment cut in parallel (Fig. 7, bottom right, lane 3), indicating the conservation of the upstream recognition site for this enzyme. The second fragment (about 3.0 kb) was about 200 to 300 bp shorter than would be expected from a simple sequence change of the second KpnI site, thus indicating the presence of a deletion. Finally, partial sequencing of the terminal part of the 4.5-kb fragment demonstrated the presence of two deletions (Fig. 4, LD). The upstream deletion (positions 8868 to 9022) was 154 bp long and located in the nef-alone region. The downstream deletion (positions 9095 to 9253) corresponded to the largest U3 deletion most frequently found in the nef analysis.
FIG. 7.
LD-PCR analysis of nearly full-length HIV-1 provirus (about 9 kb) and restriction enzyme analysis of the 3′ half of the viral genome (about 4.5 kb). (Top) Schematic illustration of HIV-1 proviral DNA showing the positions and the orientations of the nested PCR primers used to generate either the 9- or 4.5-kb fragment. The magnified 3′ half of the provirus shows the locations of the regulatory and accessory genes, the restriction sites (K, KpnI; X, XhoI), and two nef deletions (white boxes). The reference is the HIV-1 NL4-3 map. (Bottom left) Agarose gel electrophoresis (0.8%) of the second-round PCR products obtained from the 9-kb amplification (nested primer pairs, 626s-9680a for first round and 691s-9614a for second round). Lane 1, pNL4-3 (10 fg of plasmid diluted in DNA extracted from PBMC of a seronegative subject). Lane 2, SG1 (DNA extracted from 2 × 105 PBMC). Lane L, molecular size ladder (MBI, Fermentas). (Bottom right) Restriction site analysis of the 3′ half of the viral genome amplified by the nested primer pairs 626s-9680a (first round) and 5048s-9614a (second round). Lane 1, undigested pNL4-3; lane 2, undigested SG1; lanes 3 and 5, pNL4-3 digested with KpnI and XhoI, respectively; lanes 4 and 6, SG1 digested with KpnI and XhoI, respectively.
Taken together, these results both confirm the presence of nef deletions and strongly suggest the absence of length polymorphism in the other parts of the provirus.
The short direct repeat CCAA is present at the large U3 deletion junction in most of the sequences recovered from SG1.
The alignments of the nef clones and the 5′LTR deletion clones identified the presence of the direct repeat CCAA at the deletion junction in almost all of the sequences (Fig. 8). The partial sequence obtained from the LD-PCR-derived provirus also confirmed the presence of this characteristic.
FIG. 8.
DNA sequence at the deletion junction. Sequences of nef clones, 5′LTR deletion clones (consensus) and the LD-PCR product (LD) are compared with the corresponding sequence of HIV-1 NL4-3. The direct repeats CCAA are underlined. The numbers underneath the nef and 5′LTR deletion clones refer to the frequency of the repeats among them. nt, nucleotides.
DISCUSSION
The apparent absence of viral replication seems to be the hallmark of patient SG1, the LTNP subject of this study. This conclusion has been supported, during a long follow-up, by the constant inability to isolate the virus and to detect HIV-1 RNA species in both plasma and PBMC derived from this patient. Several other studies have documented an apparent lack of viral replication in some LTNP individuals. Most notably, in one of the first reports on seropositive nonprogressors, Lifson et al. found that 22 of 23 nonprogressors lacked viral RNA (23). Other authors (6, 15) described LTNP patients with very low or undetectable levels of viremia associated with the inability to isolate HIV-1 by use of sensitive culture techniques. Among these reports, only Greenough et al. (15) undertook a molecular analysis of the regulatory genes, finding no significant abnormalities. All of these data led us to believe that patient SG1 may harbor a population of defective HIV-1 quasispecies. Viral defectivity may also help explain the stability of the nonprogressive condition in this patient. To test this hypothesis, we first analyzed the 5′LTR/gag leader proviral region, because gross defects in this region would readily hamper the ability of virus to express RNA and to replicate. Contrary to this expectation, the PCR analysis, using primers encompassing the region from the 5′-U3 (position 59 of HIV-1 NL4-3) to the beginning of the gag leader (position 844), revealed only full-length proviral forms at all the three time points examined. Also, the sequences obtained at the most recent time point (1996 sample) showed a prototype 5′LTR region quite similar to the corresponding region of HIV-1 NL4-3. Clearly, this result seemed to rule out the possibility that LTR defects are responsible for the observed phenotype, and we directed our attention to other proviral regions.
Recently, alterations of the nef accessory gene have been associated with the LTNP phenotype. The apparently pivotal role of nef in in vivo pathogenesis has already been demonstrated in the SIV model of AIDS infection. Monkeys infected with SIV containing a nef deletion have low viral loads, a normal CD4 count, and no signs of disease progression (19), a phenotype which closely mimics the characteristics present in LTNP subjects. This finding spurred a series of investigations concerning the state of the nef gene in these patients. One study of 10 LTNP subjects, however, revealed only full-length functional forms of the nef gene (17, 18). In other investigations, mixed populations of defective forms of the nef gene were found to coexist with functional ones within the same LTNP subjects (24, 27). It may be argued that it would be difficult to evaluate the precise role played by nef-defective virus variants in these cases, since complementation events with the nef-functional virus variants could take place. In fact, a report by McNeary et al. suggested that Nef-functional viruses could serve as helpers for Nef-defective viruses in a group of HIV-1-infected people (25). Kirchoff et al. (21), in contrast, described for the first time an LTNP hemophiliac patient apparently bearing only deleted forms of the nef gene in the PBMC. Indeed, a similar result has been shown more recently by Deacon et al. (8) for a group of Australian transfusion recipients infected by the same blood donor. The last two reports differed in that while Deacon et al. were able to cultivate virus from many of the LTNP subjects studied, Kirchoff et al. were unable to recover virus from the hemophiliac subject even after repeated culture attempts. Thus, there is strong evidence that, at least in a limited number of HIV-1-infected LTNP humans, the presence of only nef deletion viruses can indeed be responsible for the stability of the nonprogressive condition.
These results prompted us to investigate the structure of the nef gene in patient SG1. Length polymorphism analysis (with 1994, 1995, and 1996 samples) and sequence analysis (with a 1996 sample) revealed the presence of only grossly deleted nef gene variants. All of the molecular clones sequenced were characterized by a similarly sized large deletion, spanning a very similar region of the nef-alone part of the gene. Additional deletions were also present in the different clones; in particular, a large deletion of 159 bp was present in almost all of the clones, spanning the first portion of the nef/U3 overlapping region. Only one molecular clone, N-4, was unique in that in addition to the large deletion in the nef-alone region, it had only a small 10-bp deletion in the U3 nef coding region. This kind of underrepresentation may well be the result of lower efficiency of amplification compared to that for the smaller molecules. In fact, when the nef region of plasmid pNL4-3 was coamplified with increasing amounts of a plasmid containing a nef deletion of 290 bp (N-13 clone), a ratio of 1:20 (undeleted to deleted) was enough to eliminate the detection of the undeleted form (data not shown). This could also help explain why we did not observe clones containing completely intact U3 sequences, whose existance was predicted by the 5′LTR/gag leader analysis performed with the LTR-1 and LTR-2 primers.
Another apparently contrasting point is that the nef gene analysis appeared to be in disagreement with the previous LTR analysis, which did not show deletions in the U3 part of the LTR. To clarify this point, we reamplified the entire 5′LTR/gag leader region, replacing the previous sense primer with another one, positioned exactly at the beginning of the 5′-U3, which is able to prime the short stretch of sequence which is conserved in almost all of the nef gene molecular clones sequenced. Southern blot analysis of the amplified products clearly showed the presence of a smaller fragment (about 680 bp) as well as a full-length fragment (840 bp). Two observations indicate that the 680-bp fragment represents the equivalent of the nef/U3 overlapping-region molecular clones. First, an oligonucleotide recognizing the nef/U3 deletion region failed to hybridize with the smaller fragment. Second, all of the clones obtained by the 680-bp product had large deletions in the U3 region.
The data obtained from the present investigation, taken together, seem to indicate that patient SG1 is currently harboring proviral nef-defective quasispecies, consisting of two distinguishable major molecular forms. These two forms differ from each other with respect to the presence or absence of U3 deletions in the LTRs.
In this study, the analysis of the nef gene has been limited to the last 2 years because of the lack of availability of older blood samples from this subject. Therefore, we can only speculate on the evolution of these deletions over time. In particular, the finding of only nef deletion HIV-1 quasispecies in this LTNP subject appears to be very rare and difficult to interpret. In fact, a number of different investigations have demonstrated that Nef-functional viruses have a strong selective in vivo advantage over Nef-defective variants (5, 19, 34). Therefore, if a subject is infected for the first time with an initial inoculum containing Nef-functional HIV-1 quasispecies, it is unlikely that the selective process would favor the emergence and progressive accumulation of nef deletion variants over time, with the complete elimination of full-length nef forms. Results obtained from rhesus macaques infected with a particular nef deletion SIV strain (20) and from one LTNP human patient (21) seem to suggest that once a single nef deletion has occurred, additional deletions are likely to accumulate with time.
Based on these considerations, we suggest that patient SG1 was initially infected with a particularly attenuated HIV-1 strain and that this strain already carried at least one deletion in the nef coding region. The apparent coexistence of two major proviral quasispecies, different only in the complexity of the nef deletions, indicates that some level of viral replication may have occurred during the first year of infection. During this period, a particularly effective anti-HIV cellular and humoral response could have been established and could progressively have cleared all the replication-competent quasispecies harbored in patient SG1. It has been firmly established that the noncytotoxic anti-HIV activity of CD8 cells may play a critical role in preventing progression to disease (22). In this regard, it is worth noting that patient SG1 demonstrated a strong CD8-dependent anti-HIV-1 activity even after years of apparent absence of viral replication. CD8-dependent anti-HIV activity together with a very effective immunological response could have spared only a population of virus genomes characterized by the inability to replicate.
We are still unable to fully explain the molecular mechanism(s) responsible for the apparent absence of viral RNA in the PBMC of patient SG1. In fact, the most critical cis-acting elements of the LTRs, specifically the NF-κB and Sp1 binding sites, as well as the TATA box, were found to be remarkably intact in all of the molecular clones examined. It is also unlikely that host cellular factors could adversely affect viral transcription, since the lymphocytes of this subject were susceptible to infection with a common HIV-1 strain. In addition, the LD-PCR analysis failed to reveal the presence of gross deletions in other important regions of the provirus.
In conclusion, this investigation strengthens the association between nef gene defectivity and LTNP status, supporting the idea that a nef defect may be sufficient to determine the nonprogressive condition in a restricted number of HIV-1-infected subjects.
ACKNOWLEDGMENT
This work was supported by the Italian AIDS Project, grant 9402-02, from the Italian Ministry of Health.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken C, Trono D. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol. 1995;69:5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiken C, Konner J, Landau N R, Lenburg M E, Trono D. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 4.Benedetto A, Garbuglia A R, Di Caro A, Lo Presti E, Alfani E, Delfini C. Virus free survival and down-regulation of CD4 in C8166 cells infected with human immunodeficiency virus type 1 at low density. J Gen Virol. 1993;74:2595–2601. doi: 10.1099/0022-1317-74-12-2595. [DOI] [PubMed] [Google Scholar]
- 5.Blumberg B M, Epstein L G, Saito Y, Chen D, Sharer L R, Anand R. Human immunodeficiency virus type 1 Nef quasispecies in pathological tissue. J Virol. 1992;66:5256–5264. doi: 10.1128/jvi.66.9.5256-5264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao Y, Qim L, Zhang L, Safrit G, Ho D D. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N Engl J Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 7.Cullen B R. The role of Nef in the replication cycles of the human and simian immunodeficiency viruses. Virology. 1994;205:1–6. doi: 10.1006/viro.1994.1613. [DOI] [PubMed] [Google Scholar]
- 8.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, Lawson V A, Crowe S, Maers A, Sonza S, Laermont J, Sullivan J S, Cunningham A, Dwyer D, Dowton D, Mills J. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 9.Delfini C, Garbuglia A R, Alfani E, Di Caro A, Sette P, Benedetto A. Heroin addicts infected by HBV and HIV have a low prevalence of HBV DNA in peripheral blood mononuclear cells. J Med Virol. 1993;41:114–119. doi: 10.1002/jmv.1890410206. [DOI] [PubMed] [Google Scholar]
- 10.De Ronde A, Klaver B, Keulen W, Smith L, Goudsmit J. Natural HIV-1 Nef accelerates virus replication in primary human lymphocytes. Virology. 1992;188:391–395. doi: 10.1016/0042-6822(92)90772-h. [DOI] [PubMed] [Google Scholar]
- 11.Du Z, Lang S M, Sasseville V G, Lackner A A, Ilynskii P O, Daniel M D, Jung J U, Desrosiers R C. Identification of nef allele that causes lymphocyte activation and acute disease in macaque monkeys. Cell. 1995;82:665–674. doi: 10.1016/0092-8674(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 12.Fujinaga K, Zhong Q, Nakaya T, Kameoka M, Meguro T, Yamada K, Ikuta K. Extracellular Nef protein regulates productive HIV-1 infection from latency. J Immunol. 1995;155:5289–5298. [PubMed] [Google Scholar]
- 13.Garbuglia A R, Salvi R, Di Caro A, Montella F, Di Sora F, Recchia O, Delfini C, Benedetto A. Peripheral lymphocytes of clinically non progressor patients harbor inactive and uninducible HIV proviruses. J Med Virol. 1995;46:116–121. doi: 10.1002/jmv.1890460206. [DOI] [PubMed] [Google Scholar]
- 14.Garbuglia A R, Salvi R, Di Caro A, Cappiello G, Montella F, Di Sora F, Recchia O, Lauria F, Benedetto A. In vitro activation of HIV RNA expression in peripheral blood lymphocytes as a marker to predict the stability of non progressive status in long-term survivors. AIDS. 1996;10:17–21. doi: 10.1097/00002030-199601000-00003. [DOI] [PubMed] [Google Scholar]
- 15.Greenough T C, Somasundaran M, Brettler D B, Hesselton R M, Alimenti A, Kirchhoff F, Panicali D, Sullivan J L. Normal immune function and inability to isolate virus in culture in an individual with long-term human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 1994;10:395–403. doi: 10.1089/aid.1994.10.395. [DOI] [PubMed] [Google Scholar]
- 16.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y, Zhang L, Ho D D. Characterization of nef sequences in long-term survivors of human immunodeficiency virus type 1 infection. J Virol. 1995;69:93–100. doi: 10.1128/jvi.69.1.93-100.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Zhang L, Ho D D. Biological characterization of nef in long-term survivors of human immunodeficiency virus type 1 infection. J Virol. 1995;69:8142–8146. doi: 10.1128/jvi.69.12.8142-8146.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kestler H W, III, Ringler D J, Mori K, Daniel D L, Desrosiers R C. Importance of the nef gene for maintenance of high virus load and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 20.Kirchhoff F, Kestler III H W, Desrosiers R C. Upstream U3 sequences in simian immunodeficiency virus are selectively deleted in vivo in the absence of an intact nef gene. J Virol. 1994;68:2031–2037. doi: 10.1128/jvi.68.3.2031-2037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. Brief report: absence of intact nef sequences in a long-term survivor with non progressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 22.Levi J A, Mackewicz C E, Barker E. Controlling HIV pathogenesis: the role of the noncytotoxic anti-HIV response of CD8+ T cells. Immunol Today. 1996;17:217–224. doi: 10.1016/0167-5699(96)10011-6. [DOI] [PubMed] [Google Scholar]
- 23.Lifson A R, Buchbinder S P, Sheppard H W, Mawle A C, Wilber J C, Stanley M, Hart C E, Hessol N A, Holmberg S D. Long-term human immunodeficiency virus infection in asymptomatic homosexual and bisexual men with normal CD4+ lymphocyte counts: immunologic and virologic characteristics. J Infect Dis. 1991;163:959–965. doi: 10.1093/infdis/163.5.959. [DOI] [PubMed] [Google Scholar]
- 24.Mariani R, Kirchhoff F, Greenough T C, Sullivan J L, Desrosiers R C, Skowronski J. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J Virol. 1996;70:7752–7764. doi: 10.1128/jvi.70.11.7752-7764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNearney T, Hornickova Z, Templeton A, Birdwell A, Arens M, Markham R, Saah A, Ratner L. Nef and LTR sequence variation from sequentially derived human immunodeficiency virus type 1 isolates. Virology. 1995;208:388–398. doi: 10.1006/viro.1995.1166. [DOI] [PubMed] [Google Scholar]
- 26.Michael N L, D’Arcy L, Ehremberg P K, Redfield R R. Naturally occurring genotypes of the human immunodeficiency virus type 1 long terminal repeat display a wide range of basal and Tat-induced transcriptional activities. J Virol. 1994;68:3163–3174. doi: 10.1128/jvi.68.5.3163-3174.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michael N L, Chang G, D’Arcy L A, Ehremberg P K, Mariani R, Busch M P, Birx D L, Schwartz D H. Defective accessory genes in a human immunodeficiency virus type 1-infected long-term survivor lacking recoverable virus. J Virol. 1995;69:4228–4236. doi: 10.1128/jvi.69.7.4228-4236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller M D, Warmendam M T, Gaston I, Greene W C, Feimberg M B. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med. 1994;179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang S, Konyanagi Y, Miles S, Wiley C, Vinters H V, Chen I. High levels of unintegrated HIV-1 DNA in brain tissue of AIDS dementia complex. Nature. 1990;343:85–89. doi: 10.1038/343085a0. [DOI] [PubMed] [Google Scholar]
- 30.Pantaleo G, Menzo S, Vaccarezza M, Graziosi C, Cohen O J, Demarest J F, Montefiori D, Orenstein J M, Fox C, Schrager L K, Margolick J B, Buchbinder S, Giorgi J V, Fauci A S. Studies in subjects with long-term non progressive human immunodeficiency virus infection. N Engl J Med. 1995;332:209–216. doi: 10.1056/NEJM199501263320402. [DOI] [PubMed] [Google Scholar]
- 31.Rhee S S, Marsh J W. Human immunodeficiency virus type 1 NEF-induced down-modulation of CD4 is due to rapid internalization and degradation of surface CD4. J Virol. 1994;68:5156–5163. doi: 10.1128/jvi.68.8.5156-5163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrager, L. K., J. M. Young, M. G. Fouler, B. J. Mathieson, and S. H. Vermund. 1994. Long-term survivors of HIV-1 infection: definition and research challenges. AIDS 8(Suppl. 1):S95–S108.
- 34.Shugars D C, Smith M S, Glueck D H, Nantermet P V, Seillier-Moiseiwitsch F, Swanstrom R. Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo. J Virol. 1993;67:4639–4650. doi: 10.1128/jvi.67.8.4639-4650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spina C A, Kwoh T J, Chowers M Y, Guatelli J C, Richman D D. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescient CD4 lymphocytes. J Exp Med. 1994;179:115–123. doi: 10.1084/jem.179.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Starcich B, Ratner L, Josephs S F, Okamoto T, Gallo R C, Wong-Staal F. Characterization of long terminal repeat sequences of HTLV-III. Science. 1985;277:538–540. doi: 10.1126/science.2981438. [DOI] [PubMed] [Google Scholar]