Abstract
At present, safe and effective treatment drugs are urgently needed for diabetic kidney disease (DKD). Circulating protein biomarkers with causal genetic evidence represent promising drug targets, which provides an opportunity to identify new therapeutic targets. Summary data from two protein quantitative trait loci studies are presented, one involving 4,907 plasma proteins data from 35,559 individuals and the other encompassing 4,657 plasma proteins among 7,213 European Americans. Summary statistics for DKD were obtained from a large genome-wide association study (3,345 cases and 2,372 controls) and the FinnGen study (3,676 cases and 283,456 controls). Mendelian randomization (MR) analysis was conducted to examine the potential targets for DKD. The colocalization analysis was used to detect whether the potential proteins exist in the shared causal variants. To enhance the credibility of the results, external validation was conducted. Additionally, enrichment analysis, assessment of protein druggability, and the protein-protein interaction networks were used to further enrich the research findings. The proteome-wide MR analyses identified 21 blood proteins that may causally be associated with DKD. Colocalization analysis further supported a causal relationship between 12 proteins and DKD, with external validation confirming 4 of these proteins, and TGFBI was affirmed through two separate group data sets. These results indicate that targeting these four proteins could be a promising approach for treating DKD, and warrant further clinical investigations.
Article Highlights
We undertook this study to identify potential therapeutic targets for diabetic kidney disease (DKD).
We investigated which proteins are associated with DKD and which protein drug targets can treat DKD.
We identified and replicated four plasma proteins associated with DKD, namely, CBLN1, COL6A2, ITIH3, and TGFBI, which shared common causal variants.
The four proteins, CBLN1, COL6A2, ITIH3 and TGFBI, will provide guidance and new directions for targeted therapies for DKD.
Introduction
Diabetic kidney disease (DKD), a prevalent complication of both type 1 and type 2 diabetes, stands as the primary catalyst for end-stage renal disease (ESRD) (1). In 2021, approximately 536.6 million people between the ages of 20 and 79 years were estimated to have diabetes worldwide, and this number is projected to increase to 783.2 million people by 2045 (2). As the number of patients with diabetes increases, the incidence rate of DKD is also showing a worrying trend. This not only imposes substantial social and economic costs but also poses a grave risk to human health and well-being (3). Currently, the treatment options for DKD are limited, with a majority focused on managing symptoms, such as regulating blood pressure and glucose levels. Major drugs include inhibitors of the renin-angiotensin-aldosterone system, sodium-dependent glucose transporter 2 (SGLT-2) (4), and finerenone (5) (a nonsteroidal, selective mineralocorticoid receptor antagonist). Unfortunately, these medications cannot provide a complete cure for the condition and can only slow down its progression to some extent. DKD develops rapidly into ESRD, and patients with ESRD can only receive kidney replacement therapy through dialysis or kidney transplantation (6). Therefore, it is particularly urgent to carry out research and development of therapeutic drugs for DKD.
Proteins often have specific binding sites or regions that can be targeted by small molecules or biologics, allowing for the development of drugs that can interact with them in a precise and controlled manner (7). Recently, thousands of protein quantitative trait loci (pQTL) for plasma proteins have been identified through genome-wide association studies (GWAS) (8). These studies not only enable the testing of causal effects of plasma proteins on DKD but also hold promise in identifying potential biomarkers and assessing the risk and protective factors associated with DKD. Mendelian randomization (MR) is a statistical method that uses genetic variation as a tool to evaluate causal relationships between exposures and outcomes. These genetic variations are typically independent of confounding factors and remain unaffected by postnatal environmental, behavioral, psychological, or socioeconomic influences (9). Using MR to integrate GWAS and pQTL data can help determine drug targets in advance, reducing experimental bias and minimizing confounding factors. This approach makes the most efficient use of experimental resources and time, avoids redundant work, and accelerates the research and development process (10).
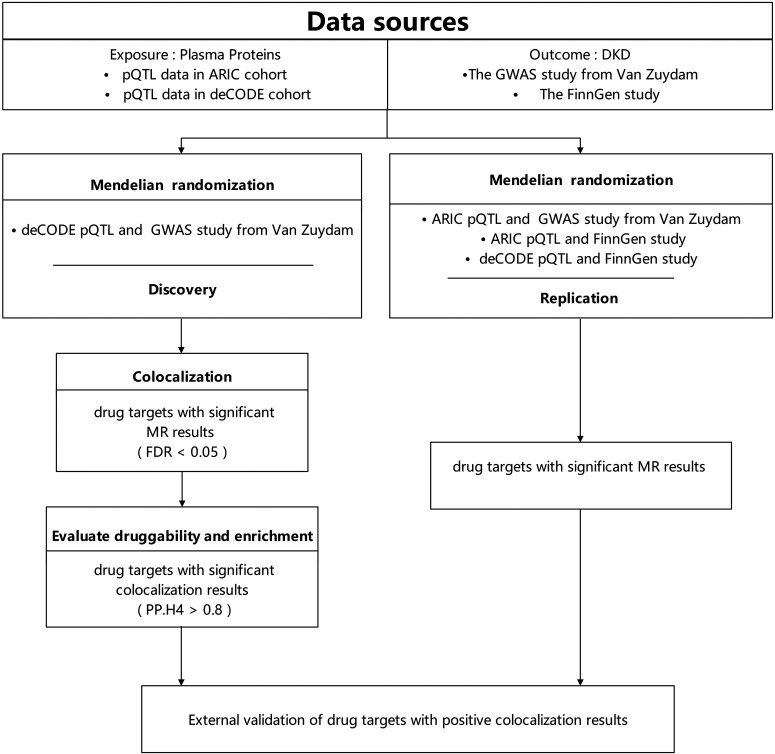
In this study, we selected two pQTL data sets as exposure variables while concurrently using two GWAS data sets as outcome variables. Initially, we used the MR method to analyze the GWAS data sourced from van Zuydam et al. (11), in conjunction with the deCODE pQTL data (12). Subsequently, we utilized a colocalization approach to further refine the results of MR analysis. These sequential steps constitute the primary research findings. To gain insights into the functions and interactions of biomolecules, enrichment analysis was conducted. Additionally, we assessed the druggability of these proteins and explored their relationships with the targets of current medications for DKD. To enhance the credibility of the primary protein results, additional validation was conducted using three sets of data: FinnGen study and deCODE pQTL data, the GWAS data from van Zuydam et al. (11) and Atherosclerosis Risk in Communities (ARIC) pQTL data, along with FinnGen study and ARIC pQTL data (13,14). This comprehensive analysis aims to seek the strongest evidence to further substantiate our research findings (Fig. 1).
Figure 1.
Overview of the study design in our MR and colocalization study.
Research Design and Methods
Proteomic Data Source
The proteomic data come from a study conducted by the deCODE Genetics team. In the study, the researchers used Illumina single nucleotide polymorphism (SNP) chips to perform whole-genome sequencing on 49,708 individuals of Icelandic descent, generating their genotype and phenotype information. Based on these data, they estimated the genomes of 166,281 Icelanders. Among these genotyped individuals, 35,559 individuals simultaneously underwent protein measurement using a multianalyte, modified aptamer binding assay called SOMAscan version 4, and 4,907 plasma proteins were detected (12). We acquired additional whole-blood pQTL data from the ARIC study, which encompassed 4,657 plasma proteins derived from a cohort of 7,213 European Americans (13). For each plasma protein from two pQTL data sets, SNPs with a minor allele frequency of at least 1% and that are genome-wide significant (with P < 5.0 × 10−8) are retained. Additionally, SNPs that are in high linkage disequilibrium (LD) with each other (with an LD R2 value greater than 0.1 in the 1000 Genomes Project from the European population) are considered to be redundant and are removed from the analysis (Supplementary Table 1).
Outcome Data Sources
The main outcome data used in this study were sourced from the GWAS by van Zuydam et al. (11) in 2018, and included patients with type 1 and type 2 diabetes. To accurately assess the severity of kidney disease, the article defined seven binary phenotypes related to DKD using albumin-to-creatinine ratio, albumin excretion rate, and estimated glomerular filtration rate indicators for classification. We chose the main phenotype “all DKD,” which includes 3,345 patients with type 2 diabetes with DKD ranging from microalbuminuria to ESRD as cases, and 2,372 patients with type 2 diabetes with normal urinary albumin as controls. The GWAS summary data for another DKD study came from the Finnish Biobank Alliance (FinnGen) version 8 data (https://r8.finngen.fi/), which include 287,132 Finnish adult participants (3,676 cases and 283,456 controls) (14). The above GWAS data used in the study came from two independent, nonoverlapping samples of European ancestry (Supplementary Table 1).
MR Analysis
MR is a powerful method for inferring causal relationships between exposures and outcomes using genetic variants as instrumental variables (15). In this study, we used MR with the “TwoSampleMR” method to investigate the relationship between plasma proteins and DKD and rigorously followed the guidelines outlined in STROBE-MR (Strengthening the Reporting of Observational Studies in Epidemiology-Mendelian Randomization) (16,17). Based on three core assumptions, we applied several filtering rules to select the genetic instruments: 1) a significance threshold of P < 5 × 10−8, 2) one strength standard being F-statistic >10, 3) a minimum physical distance of 1,000 kb between any two genetic variants, and 4) an LD threshold of r2 <0.1 between any two genes (based on the 1000 Genomes European reference panel). In the subsequent analysis, for proteins instrumented by only one SNP, we used the Wald ratio method. The inverse variance weighted method was used for proteins with more than one SNP as instrumental variables. We used false discovery rate (FDR) correction to control the FDR when conducting multiple comparisons, and set the FDR <0.05. In external validation, we used a nominal P value threshold of less than 0.05 to determine statistical significance. The GWAS data for DKD provide a valuable resource for identifying and replicating genetic associations, which is a significant advantage in MR studies. Cochran Q tests are used to assess the heterogeneity of individual causal effects. The MR-Egger intercept term is also used to evaluate horizontal pleiotropy. When P values are less than 0.05 in these tests, it usually indicates the presence of heterogeneity or pleiotropy (18).
Colocalization Analysis
Colocalization is a further analysis that strengthens the results of genetic studies by searching for evidence of the same genetic variation being associated with both exposure and outcome. This helps to confirm that genetic variation is truly causally related to the outcome, rather than being a result of LD or other confounding factors. We used the coloc R package (19) for colocalization analysis to determine whether the identified association between known proteins and DKD is driven by LD. The Bayesian analysis assesses support for five mutually exclusive hypotheses: H0, whether a genetic variation is not associated with any trait; H1, associated only with one trait; H2, associated only with another trait; H3, associated with both traits but with different causal variations; and H4, associated with both traits and sharing the same causal variation (19). We calculated the posterior probabilities (PP) for each hypothesis and determined the presence of colocalization evidence for a protein based on a protein-based PP.H4 > 0.8.
Pathway and Functional Enrichment Analysis
We performed gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis using KEGG Orthology-Based Annotation System (KOBAS) (20) to gain a better understanding of the biological functions and metabolic pathways of similar expressed proteins. GO is an analytical method used to study the commonalities of genes in biological processes (BP), molecular functions (MF), and cellular components (CC). It determines the enrichment of the genes in each GO annotation by comparing the differences between the analyzed genes and the reference genome, and generates enrichment analysis results. The main focus of KEGG enrichment analysis results is the enrichment of genes in metabolic pathways (21). Enrichment analysis was conducted on a group of proteins with positive colocalization results, and the results were filtered to include only those with a significance level of corrected P value < 0.05.
Druggable Proteins Identification and Protein-Protein Interaction Network
In order to assess the druggability of colocalized positive proteins, our methodology begins with an initial query of the DrugBank database (https://go.drugbank.com/, accessed on 16 September 2022) (22). This database provides comprehensive documentation of these proteins, including details regarding associated drugs and their developmental pathways. This methodology facilitates the discernment of drug repurposing opportunities, where drugs initially formulated for different medical indications demonstrate potential efficacy in the treatment of DKD. Subsequently, we investigate the targets of medications that are currently approved for the treatment of DKD using the Open Targets database, with the search term “Diabetic Kidney Disease” (https://platform.opentargets.org/, accessed on 16 September 2023) (23). Following this, we use version 12.0 of the Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/) to explore the protein-protein interaction (PPI) network connecting the identified targets with the approved drug targets for DKD (24).
Data and Resource Availability
The pQTL summary data from deCODE can be found at https://www.decode.com/summarydata/, and those of the ARIC study can be found at http://nilanjanchatterjeelab.org/pwas/.
GWAS summary statistics are available, by application, from https://www.finngen.fi/en/access_results and https://www.ebi.ac.uk/gwas/.
R package “TwoSampleMR” (version 0.5.6) is available at https://github.com/MRCIEU/TwoSampleMR.
R package “Coloc” (version 5.5.2) is available at https://cran.r-project.org/web/packages/coloc/.
All the data and code are accessible in public databases and open for public access. Further inquiries can be directed to the corresponding author.
Results
MR Reveals 21 Plasma Proteins Causally Associated With DKD, Using pQTL
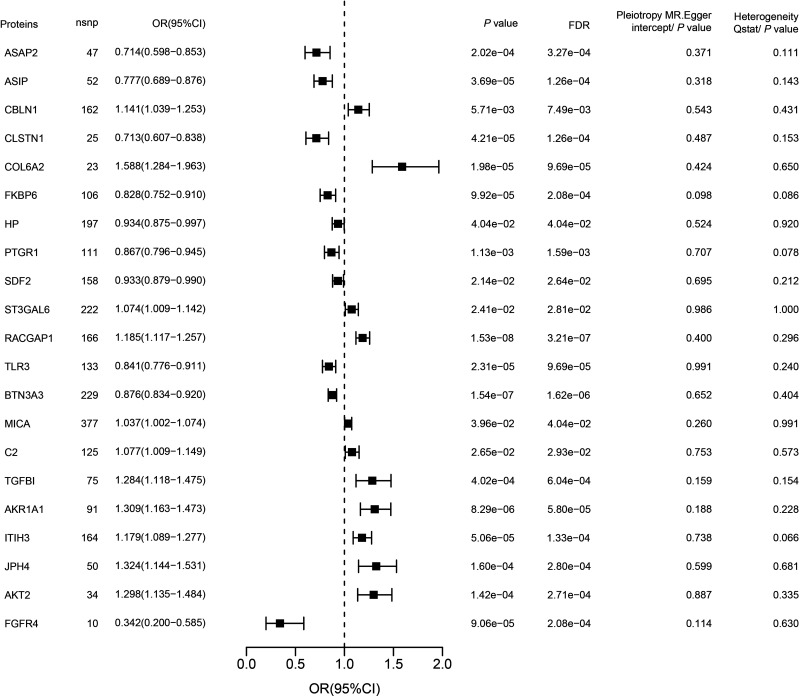
In this study, we used the GWAS data from van Zuydam et al. (11) and deCODE pQTL data as our discovery cohorts. Through MR analysis, we successfully identified 21 plasma proteins that have a causal relationship with DKD (Supplementary Table 2 and Fig. 2). Among these proteins, 11 were identified as risk proteins, potentially exacerbating the progression of DKD. Notably, RACGAP1 exhibited the most significant MR result (odds ratio [OR] = 1.185 [1.117, 1.257], FDR = 3.21E-07). Conversely, we also identified 10 protective proteins that may help mitigate the risk of DKD. Among them, BTN3A3 displayed the most significant MR result (OR = 0.876 [0.834, 0.920], FDR = 1.62E-06). It is important to emphasize that, in our preliminary analysis, we did not find significant heterogeneity and pleiotropy among the analyzed proteins. Detailed results are illustrated in Fig. 2. Throughout the entire analysis process, we did not delete any instrumental variables of protein due to noncompliance with the F-value criterion (Supplementary Table 3).
Figure 2.
The 21 significant potential drug targets in two independent DKD case-control cohorts. The y axis shows the protein name. The x axis shows the OR. The error bars represent the DKD OR per 1 SD increase in protein expression, calculated using the Wald ratio (if 1 SNP) or inverse variance weighted method (if >1 SNP) and corrected for the number of genes tested. nsnp, number of SNP.
Candidate Therapeutic Targets for DKD Identified by Colocalization
We conducted colocalization analysis on the 21 proteins from MR results to further determine the possibility of shared causal genetic variation associated with DKD and pQTL. The results showed that 12 proteins may share causal variation within DKD (PP.H4 > 0.8) and could be candidate drug target proteins (Supplementary Table 4 and Supplementary Fig. 1). Among these proteins, TGFBI, CLSTN1, PTGR1, and CBLN1 are emphasized as the strongest candidates for DKD risk (PP.H4 = 1.0). Additionally, FKBP6, ASAP2, COL6A2, and ASIP are also considered to be closely associated with DKD risk (PP.H4 > 0.95).
Exploring the Biological Significance by Enrichment Analysis
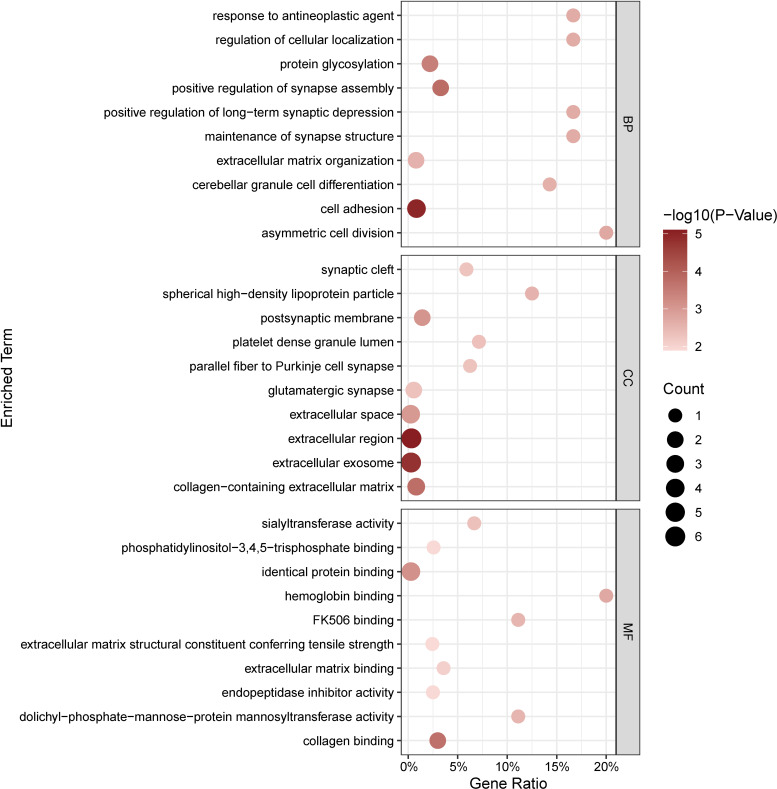
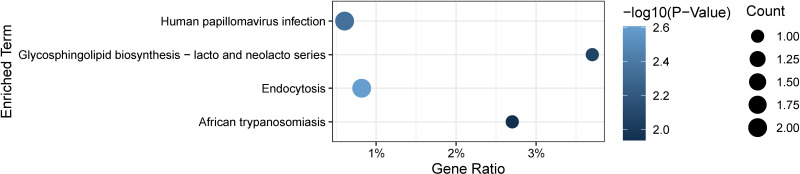
Through GO and KEGG analysis, we can gain a comprehensive understanding of the biological functions, metabolic pathways, and interactions of similar expressed proteins. GO annotation results of the top 10 proteins showed that asymmetric cell division, response to antineoplastic agent, and regulation of cellular localization were chiefly enriched in BP, while CC included spherical HDL particles, platelet dense granule lumens, and parallel fiber to Purkinje cell synapses. In addition, MF of the top 10 proteins mainly involved hemoglobin binding, FK506 binding, and dolichyl-phosphate-mannose-protein mannosyltransferase activity (Fig. 3). On performing KEGG enrichment analysis, we found that the proteins were mainly involved in endocytosis, human papillomavirus infection, and glycosphingolipid biosynthesis-lacto and neolacto series (Fig. 4).
Figure 3.
GO enrichment analysis of 12 identified proteins for treatment of DKD. Significantly enriched GO terms of similar expressed proteins in DKD.
Figure 4.
Key signaling pathways of 12 identified proteins associated of DKD. The x axis represents gene ratio, and the y axis represents different biological pathways. The size of the circle represents protein count. Different colors of circles represent different P values.
Proteins’ Druggability and Association With Current DKD Medications
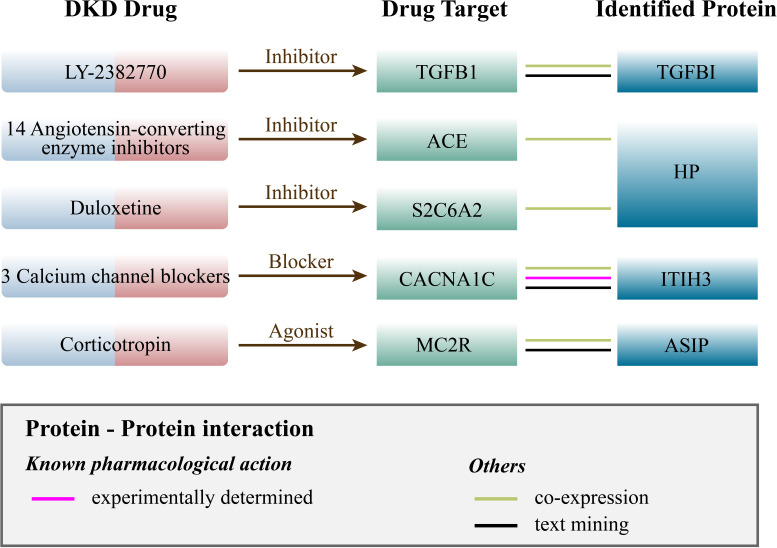
Based on results from the Opentarget database, we identified 79 proteins related to current medications (Supplementary Table 5). Through PPI network analysis, we discovered interactions between four proteins (ITIH3, HP, TGFBI, and ASIP) and the targets of five medications currently used for DKD treatment. Specifically, ITIH3 is associated with CACNA1C, which is the target of amlodipine, nisoldipine, and nifedipine (calcium channel blockers). HP is associated with two target proteins: SLC6A2, corresponding to the medication duloxetine, and ACE, which is the target of 14 angiotensin-converting enzyme inhibitors drugs. Additionally, ASIP is associated with MC2R (targeted by corticotropin), and TGFBI is related to TGFB1 (targeted by LY-2382770) (Fig. 5 and Supplementary Fig. 2). We also conducted a search for identified potential causal proteins in the DrugBank database. Among these proteins, SDF2 and PTGR1 have compounds as their drug targets, while ITIH3 and HP are targeted by zinc and related supplements (Supplementary Table 6).
Figure 5.
Interaction between current DKD medications targets and identified potential drug targets.
External Validation of Potential Drug Targets for DKD
The most crucial aspect for reliable effect estimation in our study was the validation process, which involved the use of different data types and independent replication cohorts. To further bolster the evidence for DKD risk, we created three distinct validation queues by combining various data sets. Specifically, we incorporated the GWAS data from van Zuydam et al. (11) and ARIC pQTL data to identify COL6A2 and TGFBI. CBLN1 was discovered in the FinnGen study and ARIC pQTL data, while TGFBI and ITIH3 were found in the FinnGen study and deCODE pQTL data (Supplementary Tables 7–9).
Discussion
When searching for new drug targets to treat DKD, the human proteome becomes a critically important field of study. In this manuscript, following the MR-STROBE guidelines (Supplementary Table 10), we initially used MR methodology to preliminarily identify 21 potential protein targets, followed by colocalization analysis, resulting in the selection of 12 proteins. Enrichment analysis, protein druggability, and PPI network have deepened our understanding of the mechanisms and drug utilization related to these proteins. Finally, through external validation, we successfully confirmed the potential drug target properties of four proteins, including COL6A2, CBLN1, TGFBI, and ITIH3. In particular, TGFBI, which underwent two rounds of external validation, provides the most robust evidence in this context (Table 1).
Table 1.
Evidence supporting potential proteins for which expression was significantly associated with DKD
| Protein | DKD outcome | Colocalization | Duplication | MR-Egger intercept test | Cochran Q test | Druggable proteins identification | PPI network |
|---|---|---|---|---|---|---|---|
| FKBP6 | Protection | √ | √ | √ | |||
| ASAP2 | Protection | √ | √ | √ | |||
| SDF2 | Protection | √ | √ | √ | √ | ||
| CLSTN1 | Protection | √ | √ | √ | |||
| COL6A2 | Risk | √ | √ | √ | √ | ||
| PTGR1 | Protection | √ | √ | √ | √ | ||
| TGFBI | Risk | √ | √ | √ | √ | √ | |
| ASIP | Protection | √ | √ | √ | √ | ||
| ST3GAL6 | Risk | √ | √ | √ | |||
| ITIH3 | Risk | √ | √ | √ | √ | √ | √ |
| HP | Protection | √ | √ | √ | √ | √ | |
| CBLN1 | Risk | √ | √ | √ | √ |
A check mark denotes pass; a blank denotes fail.
TGFBI, originally named βig-h3 and also known as BIGH3, is a gene that encodes a protein in the extracellular matrix (ECM) of corneal epithelial cells. TGFBI is generally expressed in the renal proximal tubules, and, under physiological conditions, it can mediate the adhesion, extension, and migration of tubular epithelial cells through interaction with α3β1 integrin-interaction motifs (25). This is consistent with the results of our enrichment analysis. During kidney injury repair, TGFBI promotes the migration of primary renal proximal tubular epithelial cells to facilitate the reconstruction of the proximal tubule (26). However, under diabetic conditions, TGFBI is a key factor in the development and progression of DKD. Transforming growth factor-β (TGF-β) is known to play a major role in the development of renal hypertrophy and ECM accumulation in diabetes, and its expression of βig-h3 is highly induced. Thus, research suggests that βig-h3 can be used as a marker to measure the biological activity of TGF-β in the kidney (27). Animal experiments have found that TGF-β or glucose significantly increases the production of βig-h3 in vitro in a dose-dependent manner (28). Moritz et al. (29) posited that the damage caused by BIGH3 to the kidney is mainly due to the activation of various signaling pathways by macrophages under high glucose conditions, which induces the production of TGF-β 1(TGFB1, belonging to the TGF-β family) and stimulates the expression of the TGFBI gene encoding the ECM protein BIGH3, leading to renal cell apoptosis. The increase in the number of apoptotic cells further promotes the infiltration of phagocytic cells, forming a vicious cycle (29). While TGFB1 is the primary target of LY-2382770 in DKD treatment, it may also impact TGFBI. This discovery provides a direction for further research to delve deeper into the multiple mechanisms of action of LY-2382770 and to validate whether it can comprehensively intervene in the pathogenesis of DKD.
ITIH3 is a plasma protein that belongs to the inter-α-trypsin inhibitors (ITIs) family and is also a component of serine protease inhibitors (30). ITIH3 and hyaluronic acid (HA) can come together to form a complex that can bolster the adhesion and durability of HA (31). Under normal conditions, renomedullary interstitial cells are the main cells responsible for producing HA in the kidney, which helps to regulate fluid balance throughout the body and maintain normal glomerular filtration and endothelial stability (32,33). When DKD occurs, high blood glucose and high insulin levels can cause kidney cells to synthesize and secrete excessive HA, which can accelerate renal function decline and lead to renal fibrosis (34). The main pathogenesis includes activation of the PKC/TGF-β1 pathway, increased prostaglandin production, ischemia and hypoxia, and inflammatory infiltration (33). Based on our enrichment results, ITIH3 was found to be primarily involved in HA metabolism. Therefore, we dare to speculate that reducing the levels of ITIH3 protein or preventing its binding with HA could potentially be a therapeutic approach for controlling the synthesis and accumulation of HA in DKD (35). STRING analysis indicated physical interaction between ITIH3 and CACNA1C (the treatment targets for amlodipine, nisoldipine, and nifedipine), but there is currently no literature explaining their mechanism, implying that these two proteins are closely associated, though not necessarily in direct contact. Moreover, according to DrugBank, zinc is listed as the drug corresponding to ITIH3, which suggests the need for further exploration.
COL6A2 is the gene responsible for encoding Collagen VI, a vital structural protein. Collagen VI are present in the ECM of nearly all tissues, where their primary function is to provide support and structural integrity to these tissues, aligning with our enrichment results (36). However, within the context of DKD, several critical pathological mechanisms come into play. DKD is characterized by chronic inflammation and metabolic disturbances, leading to the accumulation of ECM components in renal tissues. This accumulation results in a range of detrimental effects, including glomerulosclerosis, vascular constriction, and damage to renal tubules (37). A prominent feature of late-stage DKD is the formation of nodular lesions, and Collagen VI is implicated in this process. Immunohistochemical studies have revealed a strong and uniform distribution of type VI collagen throughout these nodules, potentially causing irreversible harm to the glomerular matrix (38,39). Moreover, α3(VI) cleaved C5 domain, also known as endotrophin, is considered a pivotal driver of fibrosis in DKD. It triggers renal tissue fibrosis through multiple mechanisms, including inflammation, apoptosis, angiogenesis, and the accumulation of myofibroblasts. The fibrotic process significantly contributes to the progression of DKD to ESRD (40). Additionally, a prospective study involving 198 early-stage DKD patients demonstrated that the PRO-C6 in the serum (S-PRO-C6), generated as a result of Collagen VI activity, can independently predict the risk of cardiovascular events, overall mortality, and declining kidney function (41).
CBLN1 is a member of the C1q family’s CBLN subfamily, primarily expressed in cerebellar granule cells. Its function involves regulating the synaptic connections between Purkinje cells and parallel fibers to maintain cerebellar coordination (42). In vivo and in vitro experiments in mice have revealed a striking phenomenon—the high expression of CBLN1 in the pancreas, indicating its potential role in the development of diabetes. This suggests its potential role in the development of diabetes. Notably, cerebellin (CER), CBLN1’s derivative, has been found to inhibit insulin activity, with its mechanism involving negative regulation of cAMP and calcium-dependent pathways (43). Additionally, the impact of CER extends beyond insulin secretion, leading to a significant increase in the secretion of hormones such as epinephrine, aldosterone, and cortisol/corticosterone. This effect further elevates blood glucose levels, creating unfavorable conditions for the progression of diabetes (44). Although CBLN1 is typically known for its expression in the cerebellum and its neural functions, as a member of the C1q family, it may have undiscovered roles in the field of immunology. Therefore, we propose a bold hypothesis: CBLN1 influences the development of diabetes and its complications by mediating immune responses (45). While we have recognized the potential significance of CBLN1 in diabetes research, further experiments are needed to delve deeper into its direct relationship with DKD.
In addition to the four proteins that underwent external validation, the remaining eight proteins identified through colocalization analysis continued to demonstrate strong potential, further expanding our pool of potential drug targets. ASAP2, as a protein associated with iron metabolism and insulin signaling, may play a crucial regulatory role in DKD. Qin’s discovery suggests that ASAP2 induces the expression of SLC7A11 by regulating SOX2 through miR-770-5p, which, in turn, reduces inflammation and oxidative stress while alleviating ferroptosis. This mechanism offers protection against DKD (46). Additionally, research from Zhao et al. (47) underscores the significance of ASAP2 in DKD, as it reveals that RL15 can form a complex with ASAP2, thereby mediating insulin signaling. SDF2 associates with endoplasmic reticulum (ER) stress, with functions that include participating in the quality control of newly synthesized proteins. Studies indicate that ER stress and the unfolded protein response are activated, with SDF2 playing a regulatory role in this process and contributing to protective mechanisms (48). ER stress is a critical mechanism in the development of DKD, leading to damage in various kidney cell types (49). Based on these research findings, it can be inferred that SDF2 mitigates kidney damage in DKD patients by modulating the ER stress process. ASIP is typically expressed in human adipose tissue and has been shown to be involved in lipid metabolism (50). However, when ASIP is expressed in the pancreas, it stimulates calcium signaling in pancreatic β-cells, leading to increased insulin secretion (51). HP is an acute-phase protein. When diabetes leads to increased oxidative stress, HP binds to free hemoglobin and serves multiple functions: protecting hemoglobin from oxidative damage and aiding macrophages in clearing fragmented hemoglobin. Additionally, the formation of large complex molecules through this binding prevents free hemoglobin from obstructing renal tubules, thus maintaining kidney function (52,53). Moreover, HP has demonstrated an expansion in drug-related research, which is certainly worth our attention. ST3GAL6, PTGR1, FKBP6, and CLSTN1 have limited research on their association with diabetes and DKD. It is necessary for us to conduct more comprehensive studies to fill the gaps in our understanding of their involvement in these conditions.
There are certain limitations that need to be acknowledged. First, the GWAS data we used in this study were derived exclusively from European populations, which may limit the generalizability of our findings to other ethnic groups. Second, although we used the largest GWAS data set for DKD currently available, the sample size is still relatively small, which may increase the risk of bias and limit the statistical power of the study. Third, we need to further elucidate the biological mechanisms underlying our in vivo and in vitro experimental results to better understand the therapeutic efficacy of the protein targeted in our research. Finally, despite the limitation of the original GWAS data lacking detailed gender information to the extent that we could not differentiate by gender in target proteins, it is crucial to emphasize that this constraint does not compromise the validity of our results. The study’s findings remain robust and insightful.
In summary, we have successfully extended the current biomarkers of DKD and gained a deeper understanding of its pathogenesis. Through MR and colocalization analysis, we identified 12 plasma proteins associated with DKD, among which ITIH3, COL6A2, CBLN1, and TGFBI had the most common causal variants. These findings provide guidance and new directions for targeted therapies. We believe that these research results will contribute to the prevention and treatment of DKD, and may also help reduce the personal and societal burden of kidney diseases.
This article contains supplementary material online at https://doi.org/10.2337/figshare.24964149.
Article Information
Acknowledgments. The authors thank all investigators and participants for providing the GWAS data mentioned in the article.
Funding. This work was supported by the National Natural Science Foundation of China (nos. 82004316 and 81973799).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. W.Z. and L.M. researched data. W.Z., L.M., and H.X. wrote the manuscript. Q.Z., T.G., X.Z., and H.X. contributed to discussion and reviewed and edited the manuscript. H.X. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Funding Statement
This work was supported by the National Natural Science Foundation of China (nos. 82004316 and 81973799).
Footnotes
W.Z. and L.M. contributed equally to this work and share first authorship.
References
- 1. Yuan CM, Nee R, Ceckowski KA, Knight KR, Abbott KC.. Diabetic nephropathy as the cause of end-stage kidney disease reported on the medical evidence form CMS2728 at a single center. Clin Kidney J 2017;10:257–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sun H, Saeedi P, Karuranga S, et al. IDF Diabetes Atlas: global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res Clin Pract 2022;183:109119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. GBD Chronic Kidney Disease Collaboration . Global, regional, and national burden of chronic kidney disease, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020;395:709–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cervantes CE, Hanouneh M, Jaar BG. From screening to treatment: the new landscape of diabetic kidney disease. BMC Med 2022;20(1):329 [DOI] [PMC free article] [PubMed]
- 5. Bakris GL, Agarwal R, Anker SD, et al. ; FIDELIO-DKD Investigators . Effect of finerenone on chronic kidney disease outcomes in type 2 diabetes. N Engl J Med 2020;383:2219–2229 [DOI] [PubMed] [Google Scholar]
- 6. Kofod DH, Carlson N, Ballegaard EF, et al. Cardiovascular mortality in patients with advanced chronic kidney disease with and without diabetes: a nationwide cohort study. Cardiovasc Diabetol 2023;22:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng J, Haberland V, Baird D, et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat Genet 2020;52:1122–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Emilsson V, Ilkov M, Lamb JR, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018;361:769–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emdin CA, Khera AV, Kathiresan S.. Mendelian randomization. JAMA 2017;318:1925–1926 [DOI] [PubMed] [Google Scholar]
- 10. Reay WR, Cairns MJ.. Advancing the use of genome-wide association studies for drug repurposing. Nat Rev Genet 2021;22:658–671 [DOI] [PubMed] [Google Scholar]
- 11. van Zuydam NR, Ahlqvist E, Sandholm N, et al. ; Finnish Diabetic Nephropathy Study (FinnDiane) ; Hong Kong Diabetes Registry Theme-based Research Scheme Project Group; Warren 3 and Genetics of Kidneys in Diabetes (GoKinD) Study Group; GENIE (GEnetics of Nephropathy an International Effort) Consortium; Diabetes Control and Complications Trial (DCCT)/Epidemiology of Diabetes Interventions and Complications (EDIC) Research Group; SUrrogate markers for Micro- and Macrovascular hard endpoints for Innovative diabetes Tools (SUMMIT) Consortium. A genome-wide association study of diabetic kidney disease in subjects with type 2 diabetes. Diabetes 2018;67:1414–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferkingstad E, Sulem P, Atlason BA, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet 2021;53:1712–1721 [DOI] [PubMed] [Google Scholar]
- 13. Zhang J, Dutta D, Köttgen A, et al. ; CKDGen Consortium . Plasma proteome analyses in individuals of European and African ancestry identify cis-pQTLs and models for proteome-wide association studies. Nat Genet 2022;54:593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kurki MI, Karjalainen J, Palta P, et al. ; FinnGen . FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 2023;613:508–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davey Smith G, Hemani G.. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23(R1):R89–R98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Skrivankova VW, Richmond RC, Woolf BAR, et al. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization: The STROBE-MR statement. JAMA 2021;326:1614–1621 [DOI] [PubMed] [Google Scholar]
- 17. Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burgess S, Thompson SG.. Erratum to: Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol 2017;32:391–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 2014;10:e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bu D, Luo H, Huo P, et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res 2021;49(W1):W317–W325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie LM, Huang YF, Liu YL, et al. Identification of the hub genes and the signaling pathways in human iPSC-cardiomyocytes infected by SARS-CoV-2. Biochem Genet 2022;60:2052–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Finan C, Gaulton A, Kruger FA, et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med 2017;9:eaag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ochoa D, Hercules A, Carmona M, et al. The next-generation Open Targets Platform: reimagined, redesigned, rebuilt. Nucleic Acids Res 2023;51(D1):D1353–D1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023;51(D1):D638–D646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Runager K, Enghild JJ, Klintworth GK.. Focus on molecules: transforming growth factor beta induced protein (TGFBIp). Exp Eye Res 2008;87:298–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park SW, Bae JS, Kim KS, et al. Beta ig-h3 promotes renal proximal tubular epithelial cell adhesion, migration and proliferation through the interaction with α3β1 integrin. Exp Mol Med 2004;36:211–219 [DOI] [PubMed] [Google Scholar]
- 27. Gilbert RE, Wilkinson-Berka JL, Johnson DW, et al. Renal expression of transforming growth factor-β inducible gene-h3 (βig-h3) in normal and diabetic rats. Kidney Int 1998;54:1052–1062 [DOI] [PubMed] [Google Scholar]
- 28. Lee SH, Bae JS, Park SH, et al. Expression of TGF-β–induced matrix protein βig-h3 is up-regulated in the diabetic rat kidney and human proximal tubular epithelial cells treated with high glucose. Kidney Int 2003;64:1012–1021 [DOI] [PubMed] [Google Scholar]
- 29. Moritz RJ, LeBaron RG, Phelix CF, et al. Macrophage TGF-β1 and the proapoptotic extracellular matrix protein BIGH3 induce renal cell apoptosis in prediabetic and diabetic conditions. Int J Clin Med 2016;7:496–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lord MS, Melrose J, Day AJ, Whitelock JM.. The inter-α-trypsin inhibitor family: versatile molecules in biology and pathology. J Histochem Cytochem 2020;68:907–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fries E, Kaczmarczyk A.. Inter-alpha-inhibitor, hyaluronan and inflammation. Acta Biochim Pol 2003;50:735–742 [PubMed] [Google Scholar]
- 32. van den Berg BM, Wang G, Boels MGS, et al. Glomerular function and structural integrity depend on hyaluronan synthesis by glomerular endothelium. J Am Soc Nephrol 2019;30:1886–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stridh S, Palm F, Hansell P.. Renal interstitial hyaluronan: functional aspects during normal and pathological conditions. Am J Physiol Regul Integr Comp Physiol 2012;302:R1235–R1249 [DOI] [PubMed] [Google Scholar]
- 34. Takeda M, Babazono T, Nitta K, Iwamoto Y.. High glucose stimulates hyaluronan production by renal interstitial fibroblasts through the protein kinase C and transforming growth factor-beta cascade. Metabolism 2001;50:789–794 [DOI] [PubMed] [Google Scholar]
- 35. Selman G, Martinez L, Lightle A, et al. A hyaluronan synthesis inhibitor delays the progression of diabetic kidney disease in a mouse experimental model. Kidney360 2021;2:809–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lamandé SR, Bateman JF.. Collagen VI disorders: insights on form and function in the extracellular matrix and beyond. Matrix Biol 2018;71-72:348–367 [DOI] [PubMed] [Google Scholar]
- 37. Kolset SO, Reinholt FP, Jenssen T.. Diabetic nephropathy and extracellular matrix. J Histochem Cytochem 2012;60:976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mise K, Ueno T, Hoshino J, et al. Nodular lesions in diabetic nephropathy: collagen staining and renal prognosis. Diabetes Res Clin Pract 2017;127:187–197 [DOI] [PubMed] [Google Scholar]
- 39. Nerlich AG, Schleicher ED, Wiest I, Specks U, Timpl R.. Immunohistochemical localization of collagen VI in diabetic glomeruli. Kidney Int 1994;45:1648–1656 [DOI] [PubMed] [Google Scholar]
- 40. Williams L, Layton T, Yang N, Feldmann M, Nanchahal J.. Collagen VI as a driver and disease biomarker in human fibrosis. FEBS J 2022;289:3603–3629 [DOI] [PubMed] [Google Scholar]
- 41. Rasmussen DGK, Hansen TW, von Scholten BJ, et al. Higher collagen VI formation is associated with all-cause mortality in patients with type 2 diabetes and microalbuminuria. Diabetes Care 2018;41:1493–1500 [DOI] [PubMed] [Google Scholar]
- 42. Yuzaki M. Cbln1 and its family proteins in synapse formation and maintenance. Curr Opin Neurobiol 2011;21:215–220 [DOI] [PubMed] [Google Scholar]
- 43. Albertin G, Malendowicz LK, Macchi C, Markowska A, Nussdorfer GG.. Cerebellin stimulates the secretory activity of the rat adrenal gland: in vitro and in vivo studies. Neuropeptides 2000;34:7–11 [DOI] [PubMed] [Google Scholar]
- 44. Strowski MZ, Kaczmarek P, Mergler S, et al. Insulinostatic activity of cerebellin—evidence from in vivo and in vitro studies in rats. Regul Pept 2009;157:19–24 [DOI] [PubMed] [Google Scholar]
- 45. Yuzaki M. Synapse formation and maintenance by C1q family proteins: a new class of secreted synapse organizers. Eur J Neurosci 2010;32:191–197 [DOI] [PubMed] [Google Scholar]
- 46. Li Q, Meng X, Hua Q.. Circ ASAP2 decreased inflammation and ferroptosis in diabetic nephropathy through SOX2/SLC7A11 by miR-770-5p. Acta Diabetol 2023;60:29–42 [DOI] [PubMed] [Google Scholar]
- 47. Zhao J, Wang M, Deng W, et al. ADP-ribosylation factor-like GTPase 15 enhances insulin-induced AKT phosphorylation in the IR/IRS1/AKT pathway by interacting with ASAP2 and regulating PDPK1 activity. Biochem Biophys Res Commun 2017;486:865–871 [DOI] [PubMed] [Google Scholar]
- 48. Yang M, Liu C, Jiang N, et al. Endoplasmic reticulum homeostasis: a potential target for diabetic nephropathy. Front Endocrinol (Lausanne) 2023;14:1182848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lorenzon AR, Moreli JB, de Macedo Melo R, et al. Stromal cell-derived factor (SDF) 2 and the endoplasmic reticulum stress response of trophoblast cells in gestational diabetes mellitus and in vitro hyperglycaemic condition. Curr Vasc Pharmacol 2021;19:201–209 [DOI] [PubMed] [Google Scholar]
- 50. Claycombe KJ, Wang Y, Jones BH, et al. Transcriptional regulation of the adipocyte fatty acid synthase gene by agouti: interaction with insulin. Physiol Genomics 2000;3:157–162 [DOI] [PubMed] [Google Scholar]
- 51. Kempf E, Landgraf K, Stein R, et al. Aberrant expression of agouti signaling protein (ASIP) as a cause of monogenic severe childhood obesity. Nat Metab 2022;4:1697–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Amiri AA, Hashemi-Soteh MB, Haghshenas MR, Daneshvar F, Rastegar A, Farazmand T.. Haptoglobin polymorphism in individuals with type 2 diabetic microangiopathy. N Am J Med Sci 2013;5:529–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wan BN, Zhou SG, Wang M, Zhang X, Ji G.. Progress on haptoglobin and metabolic diseases. World J Diabetes 2021;12:206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]