Abstract
Human foamy virus (HFV) is the prototype of the Spumavirus genus of retroviruses. These viruses have a genomic organization close to that of other complex retroviruses but have similarities to hepadnaviruses such as human hepatitis B virus (HBV). Both HFV and HBV express their Pol protein independently of their structural proteins. Retroviruses and hepadnaviruses differ in their requirements for particle assembly and genome packaging. Assembly of retroviral particles containing RNA genomes requires only the Gag structural protein. The Pol protein is not required for capsid assembly, and the Env surface glycoprotein is not required for release of virions from the cell. In contrast, assembly of extracellular HBV particles containing DNA requires core structural protein and polymerase (P protein) for assembly of nucleocapsids and requires surface glycoproteins for release from the cell. We investigated the requirements for synthesis of extracellular HFV particles by constructing mutants with either the pol or env gene deleted. We found that the Pol protein is dispensable for production of extracellular particles containing viral nucleic acid. In the absence of Env, intracellular particles are synthesized but few or no extracellular particles could be detected. Thus, foamy virus assembly is distinct from that of other reverse transcriptase-encoding mammalian viruses.
Human foamy virus (HFV) is the prototype of the Spumavirus genus of the Retroviridae family (35). It was originally isolated in 1971 from tissue culture cells derived from a human cancer patient (1), but, based on homology with other primate isolates, it is now thought to be of chimpanzee origin (19, 41). HFV is a complex retrovirus which contains several accessory genes in addition to the canonical retroviral gag, pol, and env genes (13). One of these genes, bel1 or tas, encodes a transactivator protein (22, 36) which is required for viral transcription and replication (29). HFV Pol is a 127-kDa polyprotein consisting of protease (PR), reverse transcriptase (RT), RNase H (RN), and integrase domains (IN) (31). Unlike other retroviruses, however, HFV expresses Pol independently of Gag from a spliced subgenomic RNA (11, 27, 47). Expression of Pol as a Gag-Pol fusion protein, in the case of other retroviruses, provides mechanisms for both expression of Pol and assembly of the viral enzymes into particles through Gag-Gag interactions (18, 21). In addition, for most retroviruses, Pol is completely dispensable for virus assembly and RNA packaging, with both processes depending entirely upon Gag (32, 42). While the Env protein determines tissue tropism for the mature virion, it is dispensable for assembly, packaging, and budding (42).
Pararetroviruses have evolved very different mechanisms for packaging viral RNA and for budding. Hepadnaviruses such as human hepatitis B virus (HBV) require their RT (P protein) for encapsidation of genomic RNA (3, 33, 34). Assembly is initiated by binding of the nascent P protein to a secondary structure in the viral RNA called epsilon (ɛ), after which capsid protein dimers are recruited to complete capsid formation (5). Capsids form in the cytoplasm and are thought to be transported to the rough endoplasmic reticulum, where they acquire the surface glycoproteins during budding into the lumen of the endoplasmic reticulum (ER). In contrast to retroviruses, HBV requires expression of the surface glycoproteins for release of extracellular particles (7).
While HFV shares features with both retroviruses and the other mammalian RT encoding-viruses such as hepadnaviruses, its replication pathway is distinct from both of these. As in the case of HBV, cell-free virions contain significant amounts of DNA (47, 49), indicating either that reverse transcription is initiated prior to assembly or that the Pol protein is highly active inside virions. However, unlike HBV, where a majority of virions contain nicked, circular double-stranded DNA (30), only about 10% of HFV particles contain full-length double-stranded DNA (47). While both HBV and HFV express their Pol proteins independently of their structural proteins, they initiate reverse transcription by different mechanisms. In HBV, reverse transcription is coupled to assembly which is initiated by Pol binding to HBV RNA (4, 34, 45). HFV RNA contains a primer binding site for tRNALys, and all evidence suggests that reverse transcription proceeds by a retroviral pathway (23). The roles of Gag, Pol, and Env in HFV assembly are not known. One recent study suggested that expression of Gag alone did not lead to particle formation (12).
In this study, we have examined the roles of Pol and Env in the assembly of HFV. We demonstrate that as with other retroviruses, the Pol protein is not required for particle assembly or encapsidation of genomic RNA. In contrast, we have found that like hepadnaviruses, the HFV Env protein is required for production of extracellular virions but not for intracellular particle formation.
MATERIALS AND METHODS
Recombinant plasmid DNAs.
HFV proviral deletion mutants ΔPol569 and ΔEnv190 were generated by linearizing the pHSRV13 (35) plasmid at unique PacI and BspEII restriction sites, respectively, digestion with Bal31 exonuclease, and religation after treatment with Klenow DNA polymerase. ΔPol569 and ΔEnv190 contain 569- and 190-bp deletions, respectively, which both result in translational frameshifting. The env mutant ΔMN (deleted from MroI [position 6957] to NdeI [position 8970]), was provided by Martin Lochelt.
The vector pSGC11 was used to make probes for RNase protection analysis of HFV nucleic acids. pSGC11 contains a 427-bp fragment of the HFV long terminal repeat which was amplified by PCR from the pHSRV13 proviral plasmid with primers containing BamHI and EcoRI restriction sites. These were used for cloning into pBluescript SKII+ (Stratagene). The oligonucleotide primers were U3BAMP11 (5′-ACTTGGATCCGATAATGTTTTAAGGAATACT-3′) and U5ECOP11 (5′-AGCTGAATTCTGTATATTGATTATCCTAAGG-3′).
Cells and culture.
FAB indicator cells (foamy virus activation of β-galactosidase), described elsewhere (48), were maintained in Dulbecco’s modified Eagle’s medium (DME) supplemented with 5% fetal bovine serum (FBS) and 0.4 mg of hygromycin per ml. Before transfection, the cells were trypsinized and placed in DME with 5% FBS only. Human embryonic lung fibroblasts (HEL) were maintained in DME with 10% FBS.
Transient transfection.
FAB cells were transfected with proviral constructs by using Lipofectamine reagent (Gibco-BRL). Liposome complexes were generated by diluting 5 μg of plasmid DNA in 750 μl of DME lacking penicillin and streptomycin (P/S−) and adding 750 μl of DME P/S− containing 25 μl of Lipofectamine reagent. This mixture was allowed to stand for 30 min while the FAB cells were rinsed with DME P/S−. The mixture was added to a 10-cm plate with cells at 75% confluency (approximately 106 cells per plate) and rocked gently before the addition of another 6 ml of DME P/S−. Transfection mixtures were incubated for 2 to 3 h at 37°C under 6% CO2 and washed twice with isotonic buffer, and 8 to 10 ml of DME with P/S and 5% FBS were added to each plate.
Expression and purification of recombinant proteins.
The central domain of HFV Gag and the RNase H domain of Pol were cloned into pGM484 for overexpression in Escherichia coli. Nucleotide sequences 1494 to 2658 and 4635 to 5978 of HSRV13 were amplified by PCR with primers containing 5′ BamHI or 3′ EcoRI restriction sites and three in-frame CACCAT (six-His tag) repeats at the 5′ end of the target sequence. Protein expression was induced in E. coli JDBE3 by addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) during log-phase growth for 4 h. Both recombinant proteins were recovered from the insoluble fraction of the cells after lysis by sonication. Denatured proteins were partially purified by Ni+ column chromatography as recommended by the column manufacturer (Qiagen) and further purified by denaturing polyacrylamide gel electrophoresis.
Antibodies.
New Zealand White rabbits were immunized with recombinant proteins corresponding to the central region of the HFV Gag protein and the RNase H domain of the Pol protein for the generation of polyclonal monospecific antisera (see the previous section). Pure proteins were isolated by polyacrylamide gel electrophoresis and homogenized in Freund’s incomplete adjuvent for injection. Antiserum against this domain of Gag recognizes the 78-kDa Gag precursor and the 74-kDa cleavage product. Antiserum against the RNase H domain of Pol recognizes the 127-kDa Pol precursor and the 80-kDa product from which integrase has been cleaved by the viral protease.
RIPA.
Transiently transfected FAB cells were washed with DME 24 h posttransfection, and labeled for 12 h with [35S]methionine (NEN Research Products) at 50 μCi/ml in DME lacking Met and Cys and containing 5% dialyzed FBS. The supernatants were passed through 0.45-μm Nalgene syringe filters, and the virus was pelleted through 20% sucrose cushions. Virus pellets were resuspended directly in antibody buffer (20 mM Tris [pH 7.5], 50 mM NaCl, 0.5% Nonidet P-40 [NP-40], 0.5% sodium dodecyl sulfate [SDS], 0.5% deoxycholate [DOC], 0.5% aprotinin, ex tempora 10 mM iodacetamide) for the radioimmunoprecipitation assay (RIPA). Plates (10-cm) of cells were disrupted in 1 ml of Ab buffer, and chromosomal DNA was sheared by passing the extract through a 23-gauge needle. Cell debris were pelleted by microcentrifugation, and incorporation was measured by trichloroacetic acid precipitation. Lysates were incubated for 2 h at room temperature in the presence of protein A-Sepharose (Pharmacia) and 2 μl of rabbit antiserum (either anti-Gag or anti-Pol). Protein A beads were washed twice with high-stringency RIPA buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1% NP-40, 1% DOC, 0.1% SDS, 0.5% aprotinin), once with high-salt buffer (10 mM Tris [pH 7.4], 2M NaCl, 1% NP-40, 1% DOC), and then once with Tris-EDTA (TE). The beads were boiled for 5 min in 2× denaturing SDS-polyacrylamide gel electrophoresis Laemmli sample buffer, and samples were electrophoresed through 10% acrylamide gels. Quantitation was performed with a PhosphorImager and ImageQuant software.
RNase protection assay (RPA).
Total nucleic acids from concentrated virions or whole cells were detected with a Direct Protect kit (Ambion). Purified nucleic acids were treated with RNase-free DNase or DNase-free RNase and then diluted in lysis buffer for further analysis. Radiolabeled RNA probe was generated with pSGC11 (see above) for in vitro transcription with T7 RNA polymerase and [32P]UTP (Dupont; 3,000 Ci/mmol) after digestion with EagI. Quantitation was performed with a PhosphorImager and ImageQuant software.
Intracellular capsid analysis.
Transiently transfected FAB cells were washed with isotonic buffer 36 h posttransfection and lysed by two different methods. Half the plates were lysed gently with Triton X lysis buffer (0.25 M sucrose, 0.5% Triton X-100, 10 mM Tris [pH 7.5], 1 mM EDTA, 140 mM NaCl), and the other half were lysed completely in RIPA Ab buffer to solubilize all intracellular structures. Lysates were cleared of cellular debris by centrifugation at 2,000 rpm in an IEC HN-SII clinical centrifuge for 30 min and then placed on top of a 20% sucrose cushion and centrifuged at 24,000 rpm in an SW28 rotor (Beckman) for 3 h. Pellets were analyzed for the presence of intracellular particles by Western blotting (immunoblotting) with Gag antiserum. The RIPA Ab buffer-generated pellets were treated with DNase before SDS-polyacrylamide gel electrophoresis. Samples were electrophoresed through 10% acrylamide gels and transferred to Immobilon-P (polyvinylidene difluoride) membranes with an Ellard Instrumentation Inc. semi-dry transfer apparatus at 0.8 mA/cm2 for 1 h. Transfer buffer consisted of 15% methanol, 192 mM glycine, and 25 mM Tris-base. The membranes were blocked for 1 h with phosphate-buffered saline–5% nonfat dry milk and then probed with Gag antiserum at 1:2,000 in block–0.05% Tween for 2 h. The membranes were washed three times with block–Tween and the probed with donkey anti-rabbit immunoglobulin Ig conjugated with horseradish peroxidase (Amersham) at 1:7,000 in block+Tween for 2 h. After the membranes were washed in block-Tween and then straight phosphate-buffered saline for a total of 30 min, Gag/donkey anti-rabbit immunoglobulin-conjugated horseradish peroxidase-specific bands were detected by enhanced chemiluminescence as recommended by the manufacturer (Amersham).
Electron microscopy.
Thin-section transmission electron microscopy (TEM) was performed on FAB cells, which were transiently transfected with ΔPol569, ΔEnv190, and wild-type HFV (HSRV13). At 36 h posttransfection, the cells were fixed in Karnofsky’s reagent for sectioning and further staining. The cells were harvested 36 h posttransfection and prepared for sectioning and staining as previously described (2). The sections were analyzed under a JEOL 100SX transmission electron microscope at 80 kV.
RESULTS
The Env glycoprotein is required for efficient budding from the cell but not for particle formation.
We were interested in the role of the Env glycoprotein in the HFV assembly pathway. A deletion mutant (ΔEnv190) was used to analyze virus assembly and RNA packaging. We predicted that such a mutant would assemble and bud normally from the cell, since it was wild type for Gag and Pol proteins and therefore would contain RNA as is the case of murine leukemia virus and other retroviruses. Transient expression was used to compare the assembly of virus particles and RNA packaging in the presence or absence of Env. Transfections were performed in FAB cells which contain an HFV LTR driving β-galactosidase expression (48); thus, only cells containing the Bel 1 (Tas) transactivator protein will turn blue after being stained with the chromogenic substrate X-Gal (5-bromo-4-chloro-3-indolyl-5-β-d-galactopyranoside). This allowed comparison of transfection efficiency prior to virus particle and protein analysis. Equivalent transfection efficiencies were observed with wild-type and mutant proviruses, usually in the range of 10 to 20%.
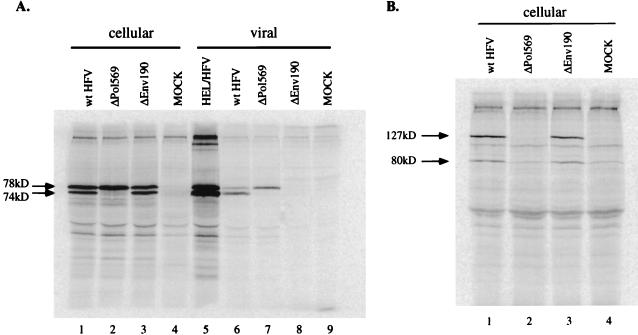
To investigate intra- and extracellular viral protein expression, RIPA was performed with Gag antiserum. The major products detected with this serum are of 74 and 78 kDa. Unlike other retroviruses, no cleavage to mature Gag proteins is detected (17). Intracellular expression levels of Gag (Fig. 1A, lanes 1 and 3) were similar for wild-type and ΔEnv190 as measured by RIPA. However, no detectable Gag protein was present in the supernatant of ΔEnv190-transfected cells (lane 8), indicating that extracellular virus was not produced in the absence of the Env glycoprotein. Quantitative analysis of the wild-type bands corresponding to extracellular Gag indicated a 20-fold difference between wild-type and ΔEnv190 at 36 h posttransfection (lanes 6 and 8), which was the limit of the sensitivity of the assay. We also looked at Pol protein expression. The major proteins detected by our RNase H antiserum are a 127-kDa precursor and an 80-kDa Pro-Pol (PR-RT-RN) product from which the 40-kDa integrase domain has been cleaved (31). We found no difference in intracellular Pol expression between HFV wild type (Fig. 1B, lane 1) and ΔEnv190 (lane 3). Since no particles were detected in the supernatant, it was not surprising that no RNA could be detected (Fig. 2B, lane 5). A different env mutant, ΔMN (provided by Martin Löchelt, Heidelberg, Germany), was also tested for comparison. This mutant, whose deletion spans a much larger portion of the env gene, yielded the same result (data not shown). The ΔMN construct had also been previously characterized for correct expression of the other major HFV genes (28) and does not interfere with the essential internal promoter for Bel 1 transcription (28). At very late times after transfection of these mutants, a very small amount of viral Gag could be detected in the culture supernatant (data not shown). However, some of this signal could be Gag released from dying cells.
FIG. 1.
RIPA of cellular and viral Gag and Pol proteins. Assay conditions and antibodies are described in Materials and Methods. (A) RIPA with anti-Gag antiserum. Lanes 1 to 4 contain cellular Gag proteins from transfected cells. Lanes: 1, wild-type (wt) HFV; 2, ΔPol569; 3, ΔEnv190; 4, mock-transfected FAB cells. Lane 5 contains virus from the supernatant of labelled, acutely infected HEL cells. Lanes 6 to 9 contain extracellular viral Gag proteins from the supernatants of the transfections. Lanes: 6, wild-type HFV; 7, ΔPol569; 8, ΔEnv190; 9, mock-transfected cells. Lanes 6 and 7 were used to normalize the number of wild-type HFV and ΔPol569 particles for the packaging experiments. (B) RIPA with anti-Pol antiserum. Cellular lysates for this figure were identical to those used with the anti-Gag antiserum. Lanes: 1, wild-type HFV; 2, ΔPol569; 3, ΔEnv190; 4, mock-transfected cells.
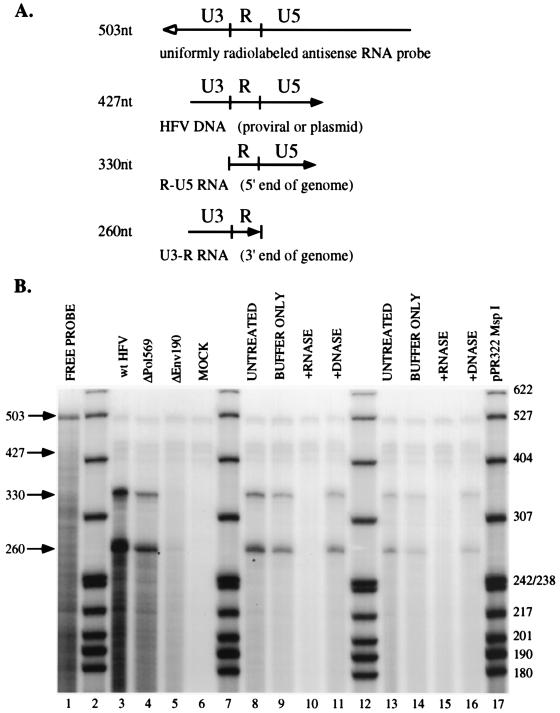
FIG. 2.
RPA of wild-type and mutant HFV nucleic acids. (A) The pSGC11 RNA probe is depicted, along with the predicted fragments protected by viral nucleic acids. The open arrow depicts the antisense probe, and the solid arrow represents the direction of viral transcription. (B) Results of RPA. Lanes 2, 7, 12, and 17 contain pBR322 MspI-digested, radiolabelled marker DNA. Other lanes: 1, 1% of total undigested free probe; 3, untreated wild-type (wt) HFV; 4, untreated ΔPol569; 5, untreated ΔEnv190; 6, mock-transfected cell supernatant; 8, untreated wild-type HFV; 9, mock-digested wild-type HFV; 10, RNase-treated wild-type HFV; 11, DNase-treated wild-type HFV; 13, untreated ΔPol569; 14, mock-digested ΔPol569; 15, RNase-treated ΔPol569; 16, DNase-treated ΔPol569. Lanes 3 to 6 were used for quantitation.
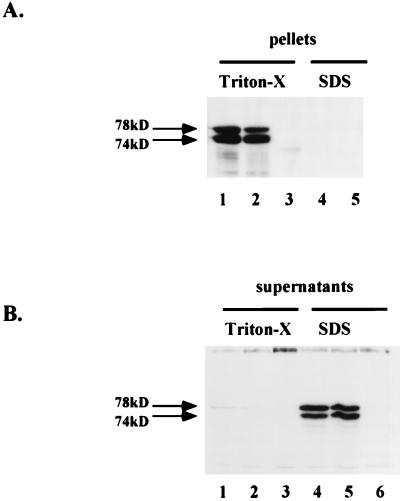
We next examined the effect of the Env deletion on intracellular particle formation. The strategy was to lyse wild-type- or ΔEnv190-transfected cells in different detergent-containing buffers. Lysis buffer containing Triton-X should lyse cell membranes but not disrupt intracellular capsids, whereas lysis with SDS should disrupt the integrity of virions as well as membranes. Methods similar to this have been used to quantitate intracellular retroviral type D capsids (Mason-Pfizer monkey virus) as well as HBV nucleocapsids (26, 37). Figure 3A indicates the amount of viral Gag present in pellets after ultracentrifugation of both Triton X and SDS lysates. We found similar levels of pelletable Gag from Triton X lysates for both wild-type-transfected (lane 1) and ΔEnv190-transfected (Lane 2) cells. No Gag was recovered in pellets after complete lysis in SDS-containing Ab buffer (lanes 4 and 5) or from mock-transfected cells (lane 3). Conversely, little or no Gag protein was detectable in the ultracentrifugation supernatants of Triton X-lysed cells (Fig. 3B, lanes 1 and 2), whereas wild-type and ΔEnv190 Gag were detected at similar levels in the SDS lysis supernatants, in which capsid structures are disrupted (lanes 4 and 5). These results indicate that the intracellular Gag seen in ΔEnv190-transfected cells, like wild-type-transfected cells, is present in assembled particles and not as free protein. Thus, we conclude that Env is not required for the assembly of intracellular capsids but is required for the release of virions from the cell.
FIG. 3.
Comparison of intracellular particle formation by wild-type and ΔEnv190 by using Western blot analysis. Cell lysates were prepared as discussed in Materials and Methods. (A) Pellets. Lanes: 1 to 3, Triton X-lysed cells; 4 and 5, SDS-lysed cells. Lanes 1 and 4 contain wild-type HFV; lanes 2 and 5 contain ΔEnv190; lane 3 contains mock-transfected cells. (B) Supernatants. Lanes: 1 to 3, Triton X-lysed cells; 4 to 6, SDS-lysed cells. Lanes 1 and 4 contain wild-type HPV; lanes 2 and 5 contain ΔEnv190; lanes 3 and 6 contain mock-transfected cells.
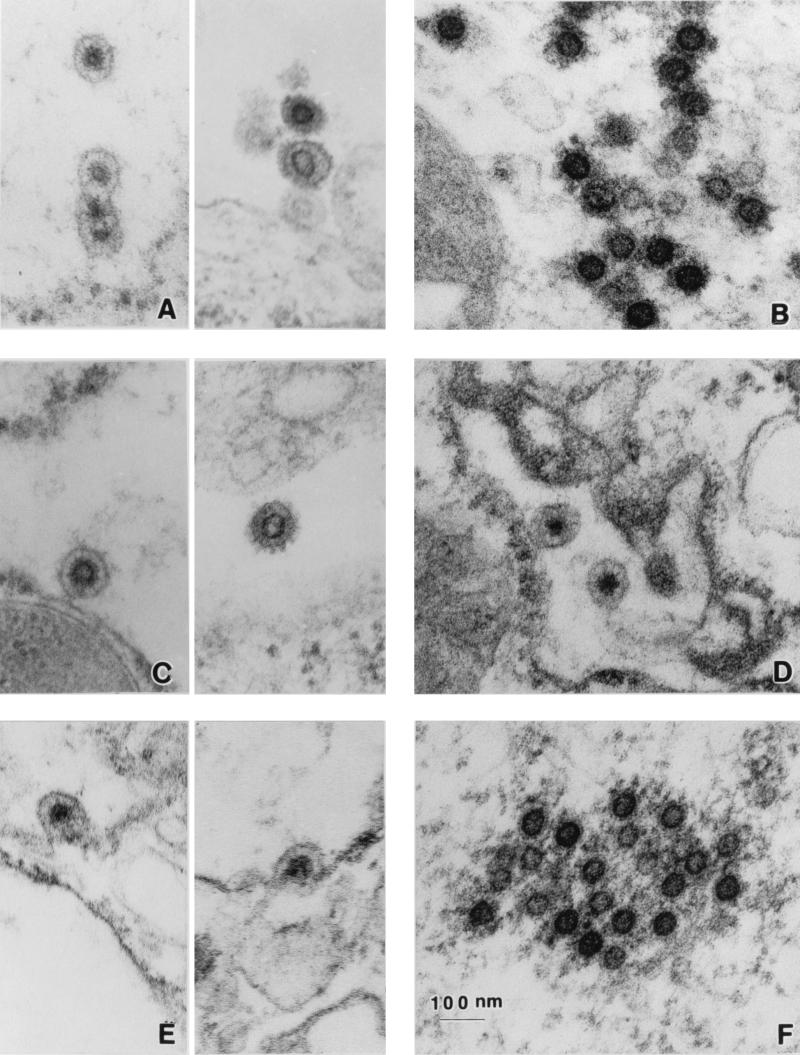
TEM was performed to verify that particles assembled in the absence of Env resemble the wild type. We were able to detect intracellular particles in FAB cells transiently transfected with both wild-type and ΔEnv190 DNA. ΔEnv190 particles were seen in intracellular compartments (Fig. 4D); they resemble wild-type particles (Fig. 4A) and were occasionally found budding from intracellular membranes (Fig. 4E). No particles were detected at or near the cell surface, and none were found budding from the plasma membrane. Preformed cytoplasmic capsids could also be found in both wild-type-transfected (Fig. 4B) and ΔEnv190-transfected (Fig. 4F) cells.
FIG. 4.
TEM analysis of wild-type and mutant HFV particles. (A and B) Representative examples of wild-type HFV-transfected cells. (C) ΔPol569-transfected cells. (D to F) ΔEnv190-transfected cells. For wild-type (A) and ΔPol569 (C) enveloped particles, intracellular virions are shown on the left and extracellular virions are shown on the right. Cytoplasmic capsids are shown only for wild-type (B) and ΔEnv190 (F) capsids. Descriptions for fixing and staining of transfected FAB cells can be found in Materials and Methods.
The Pol protein is not required for RNA packaging.
Unlike other retroviruses, HFV Pol is expressed independently of Gag. We were interested to find whether the HFV assembly pathway is dependent on the Pol protein, as is the case for HBV. A pol deletion mutant (ΔPol569) was used to evaluate whether the Pol protein was required for RNA packaging. The viral protease, encoded within the Pol precursor, cleaves the 78-kDa Gag precursor near the C terminus to release 74- and 4-kDa products. We used RIPA to analyze Gag and Pol protein expression in HFV wild-type- and ΔPol569-transfected cells. As expected, we did not see any cleavage of intracellular Gag (Fig. 1A, lane 2) or virion-associated Gag from supernatants (lane 7) of ΔPol569-transfected cells. Also as expected, no Pol protein could be detected in ΔPol569-transfected cells (Fig. 1B, lane 2).
Viral RNA was quantitated by RPA. The probe (pSGC11) used in these experiments distinguishes RNA from DNA, since it spans part of U3, all of R, and part of U5 (Fig. 2A). DNA (either endogenous viral DNA or plasmid from the transfection) protects an RNA probe fragment corresponding to 427 nucleotides (nt). Genomic viral RNA protects two fragments of the riboprobe: the 5′ end of the genome protects a 330-nt fragment of the probe corresponding to R-U5, and the 3′ end of the genome protects a 260-nt fragment corresponding to U3-R. Under the conditions used for hybridization in this assay, RNA-RNA hybrids are detected more readily than RNA-DNA hybrids (data not shown). The signal in lanes 3 and 4 of Fig. 2B is from RNA, as determined by comparison to the molecular weight markers, although virion DNA is easily detected in virus isolated from the media of acutely infected cells (data not shown). Virus used for RPA assays was isolated as soon as it was detectable by RIPA with anti-Gag antiserum in the supernatants (36 h posttransfection) in an effort to compare assembly prior to spread of wild-type virus. We found that similar levels of RNA were detected in wild-type HFV and ΔPol569 particles (Fig. 2B, lanes 3 and 4). To verify the finding that genomic RNA is packaged by the Pol mutant, nucleic acids were purified from virus particles and subjected to treatment with RNase-free DNase (lanes 11 and 16) or DNase-free RNase (lanes 10 and 15) prior to RPA. Both wild-type and ΔPol569 signals disappeared after treatment with RNase but not DNase. This demonstrates that the probe is discriminating between RNA and DNA in the RPA reactions and that RNA is packaged by the ΔPol569 mutant.
As a control in these studies, we compared the ΔPol569 mutant to a protease point mutant (D/A) since it was shown that it assembles particles and expresses full-length Pol protein but is not replication competent (24). We considered that if the Pol protein was required for packaging, this mutant would serve as a positive control in the absence of viral spread. We found that the assembly, RNA packaging, and budding of D/A particles were identical to those for ΔPol569 (data not shown).
To quantitate the amount of RNA packaged per virion, particles were normalized to viral Gag protein by RIPA (Fig. 1A, lanes 6 and 7). PhosphorImager analysis was used to determine the efficiency of RNA packaging for ΔPol569 relative to the wild type. Ratios of RNA (Fig. 2B, lanes 3 and 4) to Gag (Fig. 1A, lanes 6 and 7) indicate that the ΔPol569 mutant packages genomic RNA with wild-type efficiency. This experiment was repeated four times, each time determining the Gag/RNA ratio for both wild-type HFV and ΔPol569 and then calculating the efficiency of packaging by comparing wild-type HFV(Gag/RNA) and ΔPol569(Gag/RNA) values. The average packaging efficiency of wild-type HFV with respect to ΔPol569 was 0.96 with a standard deviation of ±0.31.
The Pol protein is not required for virus assembly or budding.
TEM was used to analyze particle formation in the absence of the Pol protein. Pol mutant particles were detected by TEM in both intracellular compartments as well as at the plasma membrane (Fig. 4C). The intracellular (left) and extracellular (right) particles formed by the ΔPol569 mutant (Fig. 4C) are very similar to those of the wild type (Fig. 4A). The pol mutant particles are detected in the extracellular media at wild-type levels, indicating that budding is not impaired by the lack of Pol protein. This data is consistent with results obtained with a protease active-site mutant of HFV (D/A) and indicates that cleavage or partial cleavage of the Gag protein is not required for virus assembly or budding. These results demonstrate that virus assembly occurs in the absence of the Pol protein, as in other retroviruses.
DISCUSSION
While assembly of conventional retroviruses occurs without either the Pol protein or the Env protein, we demonstrate here that HFV assembly is unique. Transient expression of proviral HFV pol and env mutants in FAB cells has permitted us to evaluate the roles of Pol and Env during the late stages of HFV replication. We have found that like all other retroviruses studied, HFV Pol is not required for assembly, RNA packaging, or budding. Particles produced by a mutant which is lacking the Pol protein resemble wild-type HFV (Fig. 4). This finding confirms previous data that the activity of the viral protease is not required for particle formation or release from the cell. However, it is in contrast to a previous report in which it was suggested that a protease point mutant (D/A) assembles aberrantly and that cleavage of the Gag precursor is required for normal virion morphology (24). Although it is possible that our mutant (ΔPol569), which retains a portion of the protease open reading frame, has some protease activity, no Gag cleavage could be detected with anti-Gag antiserum (Fig. 1A, lanes 2 and 7). ΔPol569 packages wild-type levels of RNA as measured by RNase protection analysis of viral nucleic acids isolated from extracellular particles.
Analysis of the env deletion mutant gave an unexpected result. Although Gag and Pol expression from the env mutant was equivalent to that from the wild type, extracellular particles were undetectable by RIPA (Fig. 1A, lane 8) or RNase protection (Fig. 2B, lane 5). Intracellular particles were, however, detected by TEM. Preformed capsids were found in the cytoplasm (Fig. 4F), some budding was seen (Fig. 4E), and some particles appeared within membrane-bound cytoplasmic structures such as the lumen of the rough endoplasmic reticulum, the Golgi complex, or secretory vesicles (Fig. 4D). Comparison of intracellular particle levels demonstrated that Env is not required for wild-type levels of capsid assembly and that this function is mediated by Gag alone (Fig. 3). While the role of Gag in assembly seems to be retrovirus-like, intracellular particle retention in the absence of Env is atypical for most retroviruses, which do not require the Env protein for budding or particle release from the cell (42). However, this finding is reminiscent of the hepadnavirus assembly pathway in which there is a strict requirement of surface glycoproteins for particle release but not nucleocapsid formation (7).
One possible explanation for this observation is that a signal in the Env protein interacts with the secretory machinery and actively induces the transport of enveloped virions to the cell surface. In the absence of Env, virions could be retained in cytoplasmic secretory vesicles. Most cells use a constitutive secretory pathway for the transport of membrane components and glycoproteins of the extracellular matrix. More specialized secretory cells can use an additional pathway, which is regulated. These cells are able to store soluble proteins and other substances in secretory vesicles for regulated release (reviewed in reference 20). If this were the case, we might have expected to see a greater accumulation of env mutant virions within intracellular compartments. Also, this type of mechanism would be highly cell type specific and could have implications for both cytopathicity and virus production. As yet, we have not analyzed our mutants in cells other than fibroblasts.
Additionally, as with other retroviruses such as human immunodeficiency virus type 1 (HIV-1), there may be a definable interaction between the cytoplasmic tail of the Env protein and the N terminus of Gag (8–10, 14). It may be that this interaction enhances Gag multimerization and hence particle assembly. If the interaction between Gag and Env is required for efficient particle formation, it is also possible that expression, modification, and localization of mature Env protein are the rate-limiting steps in mature-particle formation. In HIV-2, the cytoplasmic tail of the Env glycoprotein is important in enhancing particle release (6, 39). However, there was only about a 10-fold effect, comparable to the enhancement of particle release by the Vpu protein of HIV-1 (43, 44). The foamy virus env mutant phenotype is therefore more similar to that of HBV envelope protein mutants, which are not released in the absence of their surface glycoproteins (7).
Multimerization of Gag is likely to be concentration dependent (46), and if interaction with Env at a specific membrane occurs, it could enhance particle formation at that membrane. It has been demonstrated that the matrix (MA) domain of Gag can target the structural polyproteins to specific locations in the cell and direct the site of assembly. In one case, a single point mutation in MA changed a type D retrovirus to type C, which was able to bud from the plasma membrane without preforming capsids (38). Recently it has been reported that nonbudding, immature intracellular A-type particles can be directed to the plasma membrane for assembly and release by altering the endoplasmic reticulum targeting signal within Gag (46). Although nuclear localization of HFV Gag has been reported, it does not seem to be required for replication (40), and no other targeted localization has been described.
In addition, it has been observed that both HFV and HBV surface glycoproteins contain ER retrieval signals (16, 25) while other retroviral Env proteins do not. The biological relevance of this observation for HFV remains to be determined, although it has been reported that the HFV ER retrieval signal is not required for intracellular budding or budding from the plasma membrane. Mutants with mutations in the ER retrieval signal bud more often from the plasma membrane but release fewer particles with than does the wild type (15). These data are consistent with the idea that colocalization of Gag and Env may occur more efficiently at the membranes of the ER, enhanced by the ER retrieval signal in Env, but they do not explain why particles which still form intracellularly without Env are not released from the cell.
There appears to be an evolutionary relationship between foamy viruses and other retroviruses, as well as hepadnaviruses such as HBV. Several features of HFV replication are of particular interest in this regard. Like HBV, HFV expresses RT independently of the structural proteins. HFV lacks extensive proteolytic processing of the Gag structural protein (17) and contains GR-box nucleic acid binding motifs instead of the canonical retroviral Cys-His boxes (13). In addition, infectious double-stranded DNA can be extracted from a small percentage of extracellular HFV virions (49). However, the HBV assembly pathway is initiated and completed very differently from that of retroviruses. The P protein (RT and RNase H) initiates the assembly process and is required for genome encapsidation (3, 34), and surface glycoproteins are required for the release of virus from the cell (7). Thus, while HFV may represent an evolutionary bridge between retroviruses and hepadnaviruses, its replication is distinct from both.
ACKNOWLEDGMENTS
D.N.B. was supported by Public Health Service National Research Service Award T32 GM07270, from the National Institute of General Medical Sciences. This investigation was also supported by grant CA18282 from the National Cancer Institute to M.L.L.
We thank Michael Emerman for critical comments on and review of the manuscript, Martin Löchelt for stimulating discussion regarding this research during his sabbatical at FHCRC, and Scott Eastman for assistance with rabbit antisera.
REFERENCES
- 1.Achong B G, Mansell W A, Epstein M A, Clifford P. An unusual virus in cultures from a human nasopharyngeal carcinoma. J Natl Cancer Inst. 1971;46:299–307. [PubMed] [Google Scholar]
- 2.Aronoff R, Hajjar A M, Linial M L. Avian retroviral RNA encapsidation: reexamination of functional 5′ RNA sequences and the role of nucleocapsid Cys-His motifs. J Virol. 1993;67:178–188. doi: 10.1128/jvi.67.1.178-188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Junker-Niepmann M, Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager R, Schaller H. Hepadnavirus assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnbaum F, Nassal M. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bour S, Schubert U, Peden K, Strebel K. The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: a Vpu-like factor? J Virol. 1996;70:820–829. doi: 10.1128/jvi.70.2.820-829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bugelski P J, Maleeff B E, Klinkner A M, Ventre J, Hart T K. Ultrastructural evidence of an interaction between Env and Gag proteins during assembly of HIV type 1. AIDS Res Hum Retroviruses. 1995;11:55–64. doi: 10.1089/aid.1995.11.55. [DOI] [PubMed] [Google Scholar]
- 9.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 10.Dorfman T, Mammano F, Haseltine W A, Gottlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enssle J, Jordan I, Mauer B, Rethwilm A. Foamy virus reverse transcriptase is expressed independently from the Gag protein. Proc Natl Acad Sci USA. 1996;93:4137–4141. doi: 10.1073/pnas.93.9.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer N, Enssle J, Muller J, Rethwilm A, Niewiesk S. Characterization of human foamy virus proteins expressed by recombinant vaccinia viruses. AIDS Res Hum Retroviruses. 1997;13:517–521. doi: 10.1089/aid.1997.13.517. [DOI] [PubMed] [Google Scholar]
- 13.Flugel R M. Spumaviruses: a group of complex retroviruses. J Acquired Immune Defic Syndr. 1992;4:739–759. [PubMed] [Google Scholar]
- 14.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goepfert, P. A. and M. J. Mulligan. Personal communication.
- 16.Goepfert P A, Shaw K L, Ritter G D, Mulligan M J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J Virol. 1997;71:778–784. doi: 10.1128/jvi.71.1.778-784.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahn H, Baunach G, Brautigam S, Mergia A, Neumann-Haefelin D, Daniel M D, McClure M O, Rethwilm A. Reactivity of primate sera to foamy virus Gag and Bet proteins. J Gen Virol. 1994;75:2635–2644. doi: 10.1099/0022-1317-75-10-2635. [DOI] [PubMed] [Google Scholar]
- 18.Hatfield D L, Levin J G, Rein A, Oroszlan S. Translation suppression in retroviral gene expression. Adv Virus Res. 1992;41:193–239. doi: 10.1016/S0065-3527(08)60037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herchenroder O, Renne R, Loncar D, Cobb E K, Murthy K K, Schneider J, Mergia A, Luciw P A. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV) Virology. 1994;201:187–199. doi: 10.1006/viro.1994.1285. [DOI] [PubMed] [Google Scholar]
- 20.Hong W, Tang B L. Protein trafficking along the exocytotic pathway. Bioessays. 1993;15:231–8. doi: 10.1002/bies.950150403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. In: Swanstrom R, Vogt P K, editors. Retroviruses: strategies of replication. Berlin, Germany: Springer-Verlag KG; 1990. pp. 93–124. [DOI] [PubMed] [Google Scholar]
- 22.Keller A, Partin K M, Lochelt M, Bannert H, Flugel R M, Cullen B R. Characterization of the transcriptional trans activator of human foamy retrovirus. J Virol. 1991;65:2589–2594. doi: 10.1128/jvi.65.5.2589-2594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kogel D, Aboud M, Flugel R M. Molecular biological characterization of the human foamy virus reverse transcriptase and ribonuclease H domains. Virology. 1995;213:97–108. doi: 10.1006/viro.1995.1550. [DOI] [PubMed] [Google Scholar]
- 24.Konvalinka J, Lochelt M, Zentgraf H, Flugel R M, Krausslich H-G. Active spumavirus proteinase is essential for virus infectivity but not for formation of the Pol polyprotein. J Virol. 1995;69:7264–7268. doi: 10.1128/jvi.69.11.7264-7268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroki K, Russnak R, Ganem D. Novel N-terminal amino acid sequence required for retention of a hepatitis B virus glycoprotein in the endoplasmic reticulum. Mol Cell Biol. 1989;9:4459–4466. doi: 10.1128/mcb.9.10.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavine J, Hirsch R, Ganem D. A system for studying the selective encapsidation of hepadnavirus RNA. J Virol. 1989;63:4257–4263. doi: 10.1128/jvi.63.10.4257-4263.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lochelt M, Flugel R M. The human foamy virus pol gene is expressed as a Pro-Pol polyprotein and not as a Gag-Pol fusion protein. J Virol. 1996;70:1033–1040. doi: 10.1128/jvi.70.2.1033-1040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lochelt M, Yu S F, Linial M L, Flugel R M. The human foamy virus internal promoter is required for efficient gene expression and infectivity. Virology. 1995;206:601–610. doi: 10.1016/s0042-6822(95)80077-8. [DOI] [PubMed] [Google Scholar]
- 29.Lochelt M, Zentgraf H, Flugel R M. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel 1 gene. Virology. 1991;184:43–54. doi: 10.1016/0042-6822(91)90820-2. [DOI] [PubMed] [Google Scholar]
- 30.Nassal M, Schaller H. Hepatitis B virus replication. Trends Microbiol. 1993;1:221–228. doi: 10.1016/0966-842x(93)90136-f. [DOI] [PubMed] [Google Scholar]
- 31.Netzer K, Schliephake A, Maurer B, Watanabe R, Aguzzi A, Rethwilm A. Identification of pol-related gene products of human foamy virus. Virology. 1993;192:336–338. doi: 10.1006/viro.1993.1039. [DOI] [PubMed] [Google Scholar]
- 32.Oertle S, Spahr P F. Role of the Gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J Virol. 1990;64:5757–5763. doi: 10.1128/jvi.64.12.5757-5763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollack J R, Ganem D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol. 1993;67:3254–3263. doi: 10.1128/jvi.67.6.3254-3263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollack J R, Ganem D. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J Virol. 1994;68:5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rethwilm A, Darai G, Rosen A, Maurer B, Flugel R M. Molecular cloning of the genome of human spumaretrovirus. Gene. 1987;59:19–28. doi: 10.1016/0378-1119(87)90262-9. [DOI] [PubMed] [Google Scholar]
- 36.Rethwilm A, Erlwein O, Baunach G, Maurer B, ter Meulen V. The transcriptional transactivator of human foamy virus maps to the bel 1 genomic region. Proc Natl Acad Sci USA. 1991;88:941–945. doi: 10.1073/pnas.88.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhee S S, Hunter E. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J Virol. 1987;61:1045–1053. doi: 10.1128/jvi.61.4.1045-1053.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhee S S, Hunter E. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 39.Ritter G D, Jr, Yamshchikov G, Cohen S J, Mulligan M J. Human immunodeficiency virus type 2 glycoprotein enhancement of particle budding: role of the cytoplasmic domain. J Virol. 1996;70:2669–2673. doi: 10.1128/jvi.70.4.2669-2673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schliephake A W, Rethwilm A. Nuclear localization of foamy virus Gag precursor protein. J Virol. 1994;68:4946–4954. doi: 10.1128/jvi.68.8.4946-4954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweizer M, Neumann-Haefelin D. Phylogenetic analysis of primate foamy viruses by comparison of pol sequences. Virology. 1995;207:577–582. doi: 10.1006/viro.1995.1120. [DOI] [PubMed] [Google Scholar]
- 42.Shields A, Witte W N, Rothenberg E, Baltimore D. High frequency of aberrant expression of Moloney murine leukemia virus in clonal infections. Cell. 1978;14:601–609. doi: 10.1016/0092-8674(78)90245-3. [DOI] [PubMed] [Google Scholar]
- 43.Strebel K, Klimkait T, Maldarelli F, Martin M A. Molecular and biochemical analyses of human immunodeficiency virus type 1 vpu protein. J Virol. 1989;63:3784–3791. doi: 10.1128/jvi.63.9.3784-3791.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terwilliger E F, Cohen E A, Lu Y C, Sodroski J G, Haseltine W A. Functional role of human immunodeficiency virus type 1 vpu. Proc Natl Acad Sci USA. 1989;86:5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang G H, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welker R, Janetzko A, Krausslich H G. Plasma membrane targeting of chimeric intracisternal A-type particle polyproteins leads to particle release and specific activation of the viral proteinase. J Virol. 1997;71:5209–5217. doi: 10.1128/jvi.71.7.5209-5217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu S F, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. Human foamy virus replication—a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]
- 48.Yu S F, Linial M L. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J Virol. 1993;67:6618–6624. doi: 10.1128/jvi.67.11.6618-6624.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, S. F., M. D. Sullivan, and M. L. Linial. Evidence that the human foamy virus genome is DNA. Submitted for publication. [DOI] [PMC free article] [PubMed]