Abstract
We describe humans with rare biallelic loss-of-function PTCRA variants impairing pre-TCRα expression. Low circulating naïve αβ T cell counts at birth persisted over time, with normal memory αβ and high γδ T cell counts. Their TCRα repertoire was biased, suggesting that noncanonical thymic differentiation pathways can rescue αβ T cell development. Only a minority of these individuals were sick, with infection, lymphoproliferation, and/or autoimmunity. We also report that 1 in 4000 individuals from the Middle East and South Asia are homozygous for a common hypomorphic PTCRA variant. They had normal circulating naïve αβ T cell counts but high γδ T cell counts. Although residual pre-TCRα expression drove the differentiation of more αβ T cells, autoimmune conditions were more frequent in these patients than in the general population.
One-Sentence Summary:
Functional αβ T cells and late-onset immunological conditions in humans with rare or common inherited pre-TCRα deficiency.
Introduction
αβ and γδ T lymphocytes constitute two of the three cellular lineages of adaptive immunity in jawed vertebrates. In a process clarified in mice, they are generated from progenitor stem cells by differentiation in the thymus (1). Double-negative (DN) thymocytes, which lack both CD4 and CD8, are the most immature cells. They differentiate into mature TCRαβ– or TCRγδ–expressing T cells. Cells branch off into these two lineages during early thymopoiesis, which occurs at the same time as TRD, TRG, and TRB locus rearrangements (2-4). Productive TRD and TRG rearrangements then lead to TCRγδ expression on the cell surface, promoting maturation into γδ T cells. Alternatively, following productive TRB locus rearrangement, a TCRβ chain may dimerize with a pre-TCRα protein to generate a pre-TCR. This heterodimer is expressed on the cell surface and, during a process known as β-selection, it promotes a burst of proliferation and differentiation into CD4+CD8+ double-positive (DP) thymocytes. The TRA loci on the DP thymocytes then undergo successive waves of rearrangement (5-8), leading to the expression of TCRαβ heterodimers on the cell surface, and downregulation of the pre-TCRα chain (8, 9). After undergoing negative and positive selection, TCRαβ+ thymocytes eventually differentiate into CD4+ or CD8+ single-positive (SP) mature T cells and migrate to the periphery (10, 11). In four-week-old mice, pre-TCRα loss is associated with a >95% decrease in DP thymocyte counts (12). Although peripheral T cells have not been extensively studied in these mice, only a few TCRαβ cells are detected in lymph nodes (LNs) (5% normal levels), with the cells displaying normal TCR diversity (12, 13). In these studies, the mice remained healthy in pathogen-free conditions but were not challenged with pathogens. They did not develop overt phenotypes but, to our knowledge, no data have been published for Ptcra−/− mice beyond the age of 2 months (12-14). The consequences of pre-TCRα deficiency in humans remain unknown. We therefore searched for patients with biallelic germline PTCRA variants likely to cause pre-TCRα deficiency.
Identification of rare biallelic predicted loss-of-function PTCRA variants in seven kindreds
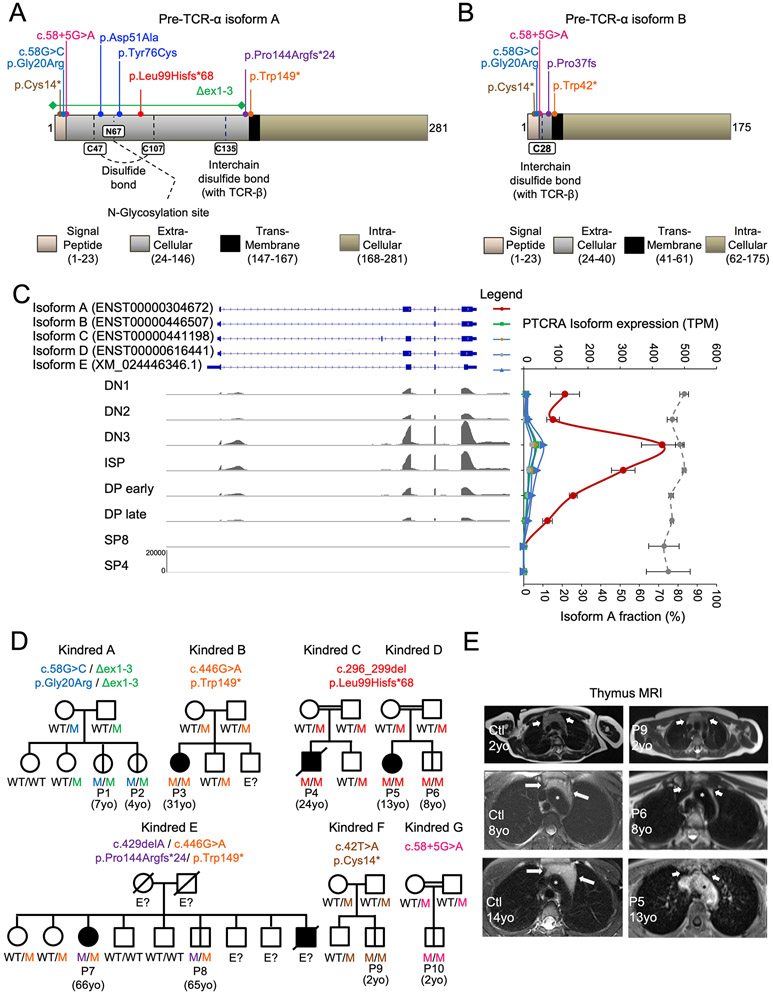
PTCRA encodes two functional isoforms in humans and mice (15). Isoform B is 106 amino acids shorter than isoform A and lacks part of the extracellular domain (Fig. 1, A and B). We reanalyzed a public RNAseq dataset corresponding to eight sorted thymocyte subsets from healthy controls (Fig. 1C) (16) and found that isoform A was the principal pre-TCRα isoform in all human thymocyte subsets (Supplementary Text). Unless otherwise specified, we refer below to isoform A. We searched for biallelic predicted loss-of-function (pLOF) variants of the PTCRA isoform A including large deletions, frameshift insertions or deletions, premature stop codons, and variants affecting essential splice sites or the start codon. No biallelic pLOF variants meeting these criteria have ever been reported in public databases (17-19). In our in-house database containing data for >25,000 patients, including four other unrelated patients identified by newborn screening (P1, P2, P9, and P10), we identified 10 patients from seven kindreds, all carrying biallelic pLOF variants (Fig. 1D; fig. S1, A to C; and Supplementary Text). The seven pLOF variants in these individuals were present in the homozygous state in five kindreds and in the compound heterozygous state in two kindreds. Five variants were private to the kindreds identified, and two were reported in major public databases, but only in the heterozygous state, with a MAF <10−4 (17-19). Two variants were predicted to affect a splice site, two were small frameshift deletions, two led to premature stop codons, and one was a large deletion. The c.58G>C substitution was a missense variant (p.Gly20Arg) but was considered to be pLOF because it was predicted to impair splicing between exons 1 and 2. Apart from the seven pLOF variants identified, only 15 biallelic coding variants, all missense and not predicted to be LOF, were found in public databases or in the HGID in-house database (fig. S1A).
Fig. 1. Autosomal recessive pre-TCRα deficiency.
(A to B) Schematic representations of the isoform A (A) and B (B) proteins encoded by PTCRA. (C) Abundance of the indicated pre-TCRα isoforms in transcripts per million (TPM) across thymocyte developmental stages (DN1, DN2, DN3, ISP, DP early, DP late, and single-positive SP8 and SP4). The proportion of total PTCRA transcripts corresponding to isoform A in each thymocyte subset is indicated on the graph (dashed gray line). (D) Pedigree of the seven unrelated families displaying familial segregation of the mutant PTCRA alleles. The indicated mutant alleles, each with a unique color code, are labeled “M" in the pedigree. Individuals of unknown genotype are labeled “E?”. Asymptomatic individuals are annotated with a vertical bar. (E) MRI on axial sections at the level of the aortic arch: T1-weighted sequences after gadolinium injection (P5) and T2-weighted sequences (P6, P9, and controls), for P5, P6, P9, and age- and sex-matched controls. In patients, the thymic lodge, located between the sternum and the aortic arch (asterisk), appears empty (P5 and P6) or small (P9). By contrast, the thymus is clearly visible in controls, even after the onset of puberty (14-year-old girl).
Clinical features of patients with biallelic pLOF PTCRA variants
These 10 patients came from seven unrelated families and were of four different ethnicities (Fig. 1D; data S1; and Supplementary Text). Six of 10 patients with predicted pre-TCRα deficiency, including the four identified by neonatal screening, were clinically asymptomatic at their most recent evaluation (at the ages of 2, 2, 4, 7, 8, and 65 years). The other four patients (13, 24, 31, and 66 years of age) displayed infection, lymphoproliferation, and/or autoimmunity with an onset during their teens or in adulthood (age at onset: 13, 13, 15, and 25 years, respectively). One of these patients died from SARS-CoV-2 pneumonia at the age of 24 years. P9 had a small thymus on MRI at the age of 2 years, whereas P5 and P6 had no visible thymus at the ages of 13 and 8 years, respectively (Fig. 1E). Three of the nine patients with pLOF PTCRA variants tested were found to produce autoantibodies, several of which were associated with clinical manifestations (Fig. S2, A to E, and Supplementary Text). Anti-thyroid autoantibodies and/or clinically overt thyroiditis were found in three of the nine patients. P7, who suffered from recurrent herpes infections, had autoantibodies against type I IFNs (Fig. S2A). All known genetic etiologies of these antibodies disrupt T cell tolerance, due to mutations affecting thymocyte development or medullary thymic epithelial cells (20-24).
Patient alleles cause mRNA decay or premature translational termination
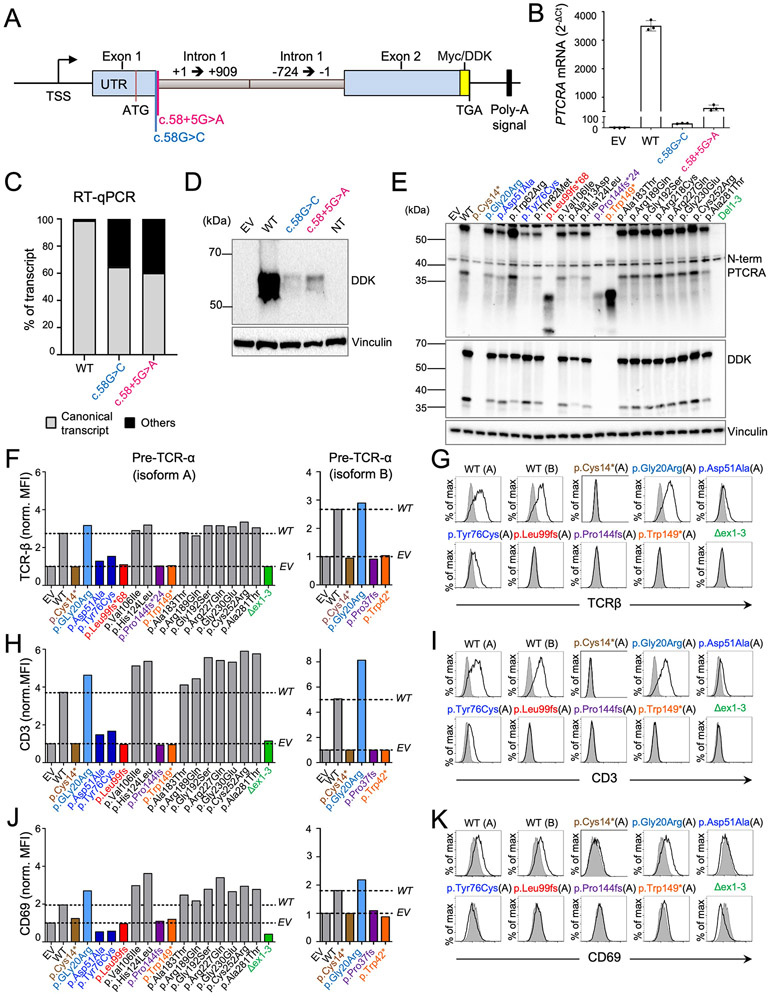
We then investigated the impact of the pLOF variants on PTCRA mRNA and protein. We were unable to test primary cells from the patients because pre-TCRα is expressed only in the thymus. First, using an artificial construct containing the gDNA sequence of PTCRA from the 5′ UTR to the end of exon 2 (Fig. 2A), we demonstrated that two of the seven pLOF variants (c.58G>C and c.58+5G>A) severely impaired pre-TCRα expression in vitro by mRNA decay (Fig. 2, B to D; fig. S3A; and Supplementary Text). Second, we transfected HEK293T cells with C-terminally DDK-tagged complementary DNAs (cDNAs) encoding the wild-type (WT) pre-TCRα, one of the six coding pLOF variants identified in the patients, or one of the 15 non-pLOF missense variants identified in the homozygous state in public databases or in our in-house cohort (fig. S1A). Cell extracts were subjected to SDS-PAGE followed by immunoblotting and immunodetection with a monoclonal antibody against DDK- or the N-terminus of pre-TCRα (Fig. 2E; fig. S3B; and Supplementary Text). All variants found in the homozygous state in public databases or in our in-house HGID cohort were normally expressed in this system. By contrast, cDNAs encoding pLOF variants yielded a truncated protein or no protein at all, except for the p.Gly20Arg (c.58G>C) variant, which produced normal amounts of protein in this cDNA overexpression system (Fig. 2E), but was subject to mRNA decay in our artificial gene system (Fig. 2, B to D). Thus, the pLOF variants identified in the patients impair PTCRA expression by mRNA decay or premature translation termination.
Fig. 2. Patient mutations and two mutants from gnomAD are loss-of-function or severely hypomorphic.
(A) Schematic representation of the artificial gene created to study the splicing between exons 1 and 2 of PTCRA. The two mutations tested are depicted. TSS: transcription start site. (B to D) HEK293T cells were transfected with an empty vector (EV) or with plasmids encoding the artificial gene with the WT or mutant PTCRA sequence described in A. (B) The RNA was subjected to RT-qPCR for PTCRA with a probe spanning the splice junction between exons 1 and 2. Data are displayed as 2−ΔCt values after normalization relative to an endogenous control (ΔCt). The bar graphs show the mean ± SEM of three technical replicates, and are representative of three independent experiments. (C) Exon trapping. Bar graph showing the proportion of canonical or noncanonical PTCRA transcripts in the transfected HEK293T cells. (D) Total protein extracts were subjected to immunoblotting with an antibody against the DDK tag or GAPDH. Data representative of three independent experiments. (E) HEK293T cells were transfected with an empty plasmid or with a plasmid carrying a C-terminal DDK-tagged cDNA encoding the WT or the indicated variants of PTCRA isoform A. Total protein extracts were subjected to immunoblotting with an antibody against the DDK tag, pre-TCRα, or Vinculin. Data representative of four independent experiments. (F to K) TCRα–deficient Jurkat cells were transduced with an EV or with a plasmid encoding the WT isoform A, the WT isoform B or the indicated variant of pre-TCRα. The expression of TCRβ (F and G), CD3ζ (H and I) or CD69 (J and K) at the cell surface was evaluated by flow cytometry on the transduced cell lines. Data representative of three independent experiments. (F, H, and J) Histograms showing the mean fluorescence intensity (MFI) of cells transduced with the indicated PTCRA allele normalized against the MFI for EV. (G, I, and K) Representative flow cytometry histogram plot for the indicated PTCRA alleles. Cells transduced with the PTCRA alleles (black line, unshaded area) are compared to the cells transduced with the EV (shaded).
Patient variants are loss of function and two variants from public databases are severely hypomorphic
We assessed the ability of the pre-TCRα variants to stabilize TCRβ and CD3 at the cell surface in the TCRα–deficient JR3.11 Jurkat cell line (25, 26). The transduction of these cells with the WT isoform A of pre-TCRα restored the expression of TCRβ and CD3 at the cell surface (Fig. 2, F to I, and fig. S3, C to F). As expected, none of the cDNAs encoding variants from the patients except the cDNA encoding the p.Gly20Arg (c.58G>C) variant restored the cell-surface expression of TCRβ and CD3. By contrast, 13 of the 15 biallelic variants reported in public or in-house databases restored the expression of TCRβ and CD3. The p.Asp51Ala and p.Tyr76Cys variants induced only very low levels of TCRβ and CD3 expression (Fig. 2, F to I, and fig. S3, C to F). Pre-TCR can signal autonomously when expressed at the cell surface. Its successful expression is, therefore, associated with cell-surface expression of the CD69 activation marker (25). Accordingly, all the pre-TCRα-encoding constructs that restored the expression of TCRβ and CD3 at the cell surface also induced weak CD69 expression (Fig. 2, J and K, and fig. S3, G and H). Neither the pLOF variants from the patients nor the p.Asp51Ala and p.Tyr76Cys variants from the public Genome Aggregation Database (gnomAD) V2.1.1 induced CD69 expression on JR3.11 Jurkat cells. Similar findings were obtained when the deleterious variants were tested for their impact on isoform B (Fig. 2, F to K, and fig. S3, C, D, and G). Thus, the seven alleles from the patients are biochemically LOF and the patients are predicted to have an autosomal recessive, complete form of pre-TCRα deficiency. Moreover, two missense variants (p.Asp51Ala and p.Tyr76Cys) found in the homozygous state in the general population are highly deleterious for pre-TCRα function.
Population genetics of the Asp51Ala and Tyr76Cys variants
The p.Asp51Ala and p.Tyr76Cys variants identified in gnomAD affect residues interacting with TCRβ (fig. S4A) (27). The p.Asp51Ala variant affects a charged residue in the extracellular domain. In the mouse, knock-in mutations of such residues impair the interaction between pre-TCRα and TCRβ, leading to a decrease in the count of DP thymocytes and an increase in γδ T cell counts (28, 29). The p.Asp51Ala and p.Tyr76Cys variants may, therefore, impair dimerization between pre-TCRα and TCRβ. In gnomAD v2.1.1 and the Centogene Biodatabank, the pTyr76Cys variant was most frequent in sub-Saharan Africans, with a MAF of ~0.0037 versus 0.0003 in the global population from gnomAD V2.1.1 (fig. S4B and tables S1 to S3). Thus, ~0.001% of Africans would be expected to have a partial deficiency of pre-TCRα (~1/73,000 individuals). In various databases, the p.Asp51Ala variant is more frequent in individuals from South Asia and the Middle East, whose MAF is ~0.01. By contrast, the MAF for the global population from gnomAD V2.1.1 is about 0.002 (fig. S4C and tables S1 to S3). In these populations—which together account for almost two billion individuals—the p.Asp51Ala allele can be regarded as “common” (MAF >1%). Thus, 1/1000 to 1/10,000 Middle Eastern and South Asian individuals would be predicted to have a partial form of recessive pre-TCRα deficiency. We analyzed the exomes of two Iranian kindreds carrying the homozygous p.Asp51Ala variant and estimated that the most recent common ancestor carrying the variant lived about 8000 years ago (95% confidence interval: 2511-29,430 years). This finding suggests that there is no strong depletion of individuals homozygous for PTCRA in these populations. Thus, considering only the p.Asp51Ala and p.Tyr76Cys alleles, ~1/180,000 individuals worldwide may have a partial form of pre-TCRα deficiency. In particular, the p.Asp51Ala variant is found in the homozygous state in 1/1000 to 1/10,000 individuals in the populations of South Asia and the Middle East.
Homozygosity for the Asp51Ala allele is a risk factor for autoimmunity
We investigated the impact of the p.Asp51Ala variant on immunological phenotypes by analyzing the reported phenotypes of individuals homozygous and heterozygous for this variants among the South Asians included in the UK Biobank. The frequencies of autoimmunity (~20%) and hypothyroidism (~10%) codes were similar in individuals heterozygous for p.Asp51Ala and controls (table S4). By contrast, three (75%) homozygous carriers had autoimmunity-related codes and one (25%) had a hypothyroidism-related code. Homozygote 1 suffered from hypothyroidism and lichen planus at the ages of 48 and 52 years, respectively. Homozygote 2 presented thrombocytopenia and Henoch–Schönlein purpura at the age of 50 years, and Homozygote 3 suffered from rheumatoid arthritis at the age of 50 years. No autoimmunity was reported in Homozygote 4, but he suffered from hypoxemic COVID-19 pneumonia at age 61. No lymphoproliferation was reported in any of the four homozygotes. We also analyzed the phenotype of the homozygotes identified in other cohorts (table S1). One of the two homozygotes in the Qatar Biobank was asymptomatic at the age of 46 years, and the other suffered from hypothyroidism with autoantibodies against thyroid peroxidase (TPO) at the age of 31 years. Two homozygotes were identified in a Saudi database and clinical data were available for only one, an otherwise healthy 38-year-old man with vitiligo. His 40-year-old sister was shown, by Sanger sequencing, to be homozygous for the variant, but was asymptomatic. In an Iranian database of individuals recruited on the basis of neurological phenotypes and Sanger sequencing data for the relatives of the proband, we identified three homozygous carriers of the p.Asp51Ala variant (P11, P12, and P13) (fig. S4D and Supplementary Text). None of these individuals presented unusual susceptibility to infection. However, two of the three children suffered from hypothyroidism. The thymic compartment of P11 (9 years old) contained tissue with abnormal properties on MRI, suggesting that the content of the thymus was abnormal (fig. S4E). Thus, evidence of autoimmunity was obtained for seven of the 11 (64%) homozygotes for whom clinical information was available. Finally, using the Centogene cohort, we identified 51 additional individuals homozygous for p.Asp51Ala (tables S5 and S6 and Supplementary Text). In this cohort, the association between autoimmunity and homozygosity for the p.Asp51Ala variant was confirmed, with an odds ratio (OR) of 5.02 relative to heterozygotes and WT subjects (95% CI=1.750054 to 11.816898, adjusted P=0.009965). Thus, homozygosity for the p.Asp51Ala variant appears to be a significant risk factor for the development of autoimmune disease in individuals of Middle Eastern and South Asian origin.
Low CD3+ T cell counts in newborns with complete pre-TCRα deficiency
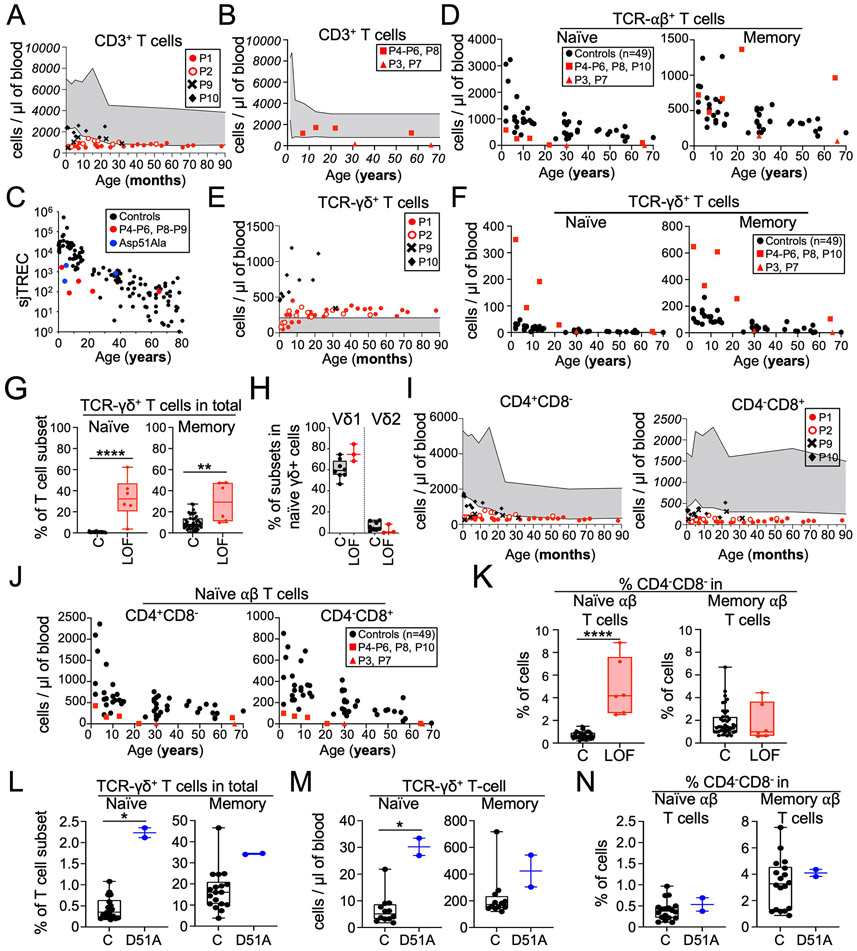
The asymptomatic pre-TCRα–deficient patients (P1, P2, P6, P8, and P9)—like the patient with a mild clinical presentation (P5)—had normal or near-normal distributions of leukocyte subsets other than T cells, and normal antibody responses to antigens (data S1; fig. S5; and Supplementary Text). By contrast, patients with clinical autoimmunity (P3, P4, and P7) were diagnosed with CVID and presented progressive cytopenia for multiple cell types. Pre-TCRα deficiency affects thymocyte differentiation in mice. We consequently investigated the blood T cell compartment of the patients. Except for P10, all patients (P1, P2, and P9) followed from birth displayed T cell lymphopenia early in life (Fig. 3A). Their total T cell counts remained stable over time, reaching counts at the low end of the normal range by the age of 3 years, when a physiological decline of CD3+ T cell counts is observed in normal individuals. Relative to age-matched controls, all patients other than P3 and P7 (aged 31 and 66 years respectively, both displaying progressive pancytopenia) had normal or near-normal blood counts of total CD3+ T cells at their most recent follow-up visit (Fig. 3, A and B). Moreover, all patients under the age of 30 years had low proportions of single-joint TRECs (sjTRECs) among PBMCs, suggesting poor thymic output (Fig. 3C). These data suggested that pre-TCRα deficiency impairs T cell development, resulting in low T cell counts in infancy, facilitating detection by newborn TREC level screening. However, the total T cell counts of the patients gradually increases, eventually reaching the normal range for age-matched controls (Fig. 3A). Moreover, these T cells proliferated normally upon mitogen stimulation in vitro (data S1)
Fig. 3. T cell immunophenotyping for patients with pre-TCRα deficiency.
(A and B) CD3+ T cell counts as a function of age. The control range is represented by the gray area. (C) Thymic function was assessed in pre-TCRα–deficient patients (red and blue dots) and healthy local controls (black dots; n=101). The concentration of sjTRECs in the blood (sjTREC/105 PBMCs) is presented as a function of age. (D) TCRαβ+ T cell counts as a function of age for naïve and memory T cells. (E) TCRγδ+ T cell counts as a function of age. The control range is represented by the gray area. (F) TCRγδ+ T cell counts as a function of age for naïve and memory T cells. (G) Frequency of TCRγδ+ T cells among total naïve (CD3+CD45RA+CCR7+) and memory (defined as non-naïve CD3+) T cells from patients (P3-P6 and P8) and controls (n=46). (H) Frequency of TCRγδ1+ and TCRγδ2+ T cells among naïve TCRγδ+ T cells from patients (P4, P8, and P9) and controls (n=8). (I) Cell counts as a function of age, for CD4+CD8− T cells and CD4−CD8+ T cells. The control range is represented by the gray area. (J) Naïve αβ T cell counts as a function of age, for CD4+CD8− T cells and CD4−CD8+ T cells. αβ T cells are defined here as CD3+TCRγδ− cells. (K) Frequency of CD4−CD8− cells in αβ (defined here as CD3+TCRγδ−) naïve (right) and memory (left) T cells from patients (P3-P6 and P8) and controls. (L to N) Phenotyping of individuals homozygous for the p.Asp51Ala mutation (D51A) and controls (n=12-18). (L) Frequency of TCRγδ+ T cells among total naïve and memory T cells. (M) TCRγδ+ T cell counts for naïve and memory T cells. (N) Frequency of CD4−CD8− cells among αβ (defined here as CD3+TCRγδ−) naïve (right) and memory (left) T cells. (B, D, F, and J) P3 suffered from severe enteropathy and was on rituximab treatment. P7 received chemotherapy for lymphoma. These two patients are therefore depicted with triangles. (G, H, and K to N) C: controls; LOF: patients homozygous for LOF variants; and D51A: individuals homozygous for the p.Asp51Ala mutation.
Patients with complete pre-TCRα deficiency have high naïve γδ and low naïve αβ T cell numbers
The mouse pre-TCRα is essential for αβ T cell development, but is dispensable for γδ T cell development (12). We therefore studied the impact of complete human pre-TCRα deficiency on the two major T cell lineages. Patients had lower blood counts of naïve αβ T cells than age-matched controls, but normal counts of memory αβ T cells (Fig. 3D). Total γδ T cell counts were high from early childhood (Fig. 3E). In children and adults, both naïve and memory γδ T cell counts remained normal to high (Fig. 3F). Accordingly, the proportion of γδ T cells among naïve T cells was higher in patients (median=32.3; range 3.7-62.3) than in controls (median=0.6; range 0.1-2.2) (Fig. 3G). However, the proportion of γδ T cells among memory T cells was above the upper limit of the control range only in P4, P5 and P10. The proportions of δ1+ and δ2+ γδ T cells among naïve T cells were normal in patients with pre-TCRα deficiency (Fig. 3H). Thus, pre-TCRα deficiency has different impacts on the thymic outputs of αβ and γδ T cells, impairing the production of αβ T cells and favoring the production of γδ T cells. Nevertheless, most circulating T cells (including naïve T cells) were TCRαβ+.
Patients with complete pre-TCRα deficiency have normal memory αβ T cell counts and low MAIT cell counts
We then investigated the αβ T cell compartment in more depth. The infants had low total CD4−CD8+ and CD4+CD8− T cell counts (Fig. 3I), which normalized between childhood and adulthood (fig. S6A). However, as expected from their low naïve αβ T cell counts, pre-TCRα–deficient children and adults had low counts of naïve CD4+ and CD8+ T cells (Fig. 3J). The low naïve T cell counts of the patients were accompanied by a higher proportion of both CD4 and CD8 effector memory T cells (fig. S6, B and C). The proportion of regulatory T cells (Tregs) among CD4+ T cells was in the range of controls for all patients (fig. S6D). Within the memory CD4+ T cell compartment, the frequencies of T helper subsets were within or near the control range (fig. S6E). Accordingly, comparisons with controls revealed no major differences in the production of T helper (Th)1 (IFN-γ), Th2 (IL-13), and Th17 (IL-17A) cytokines by the patients’ memory CD4+ T cells following stimulation (fig. S6F). In addition, pre-TCRα–deficient patients had lower levels of CD161+TCRVα7.2+ mucosal-associated invariant T (MAIT) cells than controls and normal frequencies of invariant natural killer T cells (iNKT) among T cells (fig. S6G). The frequency of TCRVα7.2+ cells was low among memory αβ T cells, but normal among naïve αβ T cells (fig. S6H), suggesting that the low frequency of MAIT cells was not due to impaired V(D)J rearrangement. Thus, patients with complete pre-TCRα deficiency have low total naïve αβ T cell counts, normal αβ T memory-cell counts from childhood onward, and a low frequency of MAIT cells.
Patients with complete pre-TCRα deficiency have a high proportion of CD4−CD8− DN αβ T cells among naïve T cells
Blood γδ T cells typically have a CD4−CD8−/lo phenotype, defining a T cell lineage that does not pass through the CD4+CD8+ DP stage in the thymus. In TCR-transgenic mice expressing TCRαβ at the DN stage in the thymus, a small abnormal population of TCRαβ cells with a CD4−CD8− phenotype is observed in the periphery (4, 30-32). Fate mapping has shown that these cells do not pass through the CD4+CD8+ DP stage. Instead, they are thought to use the γδ differentiation pathway despite their expression of a TCRαβ (33). We therefore investigated whether a fraction of TCRαβ+ cells in the periphery in pre-TCRα deficient patients harbored the same phenotype. We found no difference in the frequency of CD4−CD8− cells among memory T cells from controls and patients (Fig. 3K). However, the frequency of CD4−CD8− DN cells among naïve TCRαβ+ T cells from pre-TCRα–deficient patients was higher (median=4.2%; range: 2.5 to 8.9%) than that in age-matched controls (median=0.6%; range 0.2 to 1.5%) (Fig. 3K). These CD4−CD8− DN cells did not have high levels of HLA-DR or CD38 expression and were, therefore, probably not chronically activated (fig. S6I) (34). Moreover, only small proportions of naïve CD4−CD8−TCRαβ+ T cells from the pre-TCRα–deficient patients expressed MAIT (CD161+TCRVα7.2+), iNKT (TCRVα24-Jα18+), Treg (CD127−CD25+), or intraepithelial lymphocyte (IEL) markers (CLA, CD103, NKG2C, and NKG2A), suggesting that most DN αβ T cells from the patients do not belong to an unconventional αβ T cell subset (fig. S6J). Thus, Pre-TCRα deficiency is associated with an approximately eightfold increase in the proportion of CD4−CD8− T cells in the naïve TCRαβ+ T cell compartment. As in mice (4), DN αβ T cells in humans may therefore develop via an alternative T cell differentiation pathway.
Low TREC levels and a high proportion of γδ T cells among the naïve T cells of p.Asp51Ala homozygotes
We tested the hypothesis that homozygosity for the hypomorphic p.Asp51Ala variant affects T cell differentiation. We determined the sjTREC levels of three patients (P11-P13). These levels were low in the two youngest patients (Fig. 3C). We also performed extensive immunophenotyping on these two children, which showed their counts and proportions of myeloid, B, and NK cells to be normal (fig. S7, A to D). The T cell counts of these patients were within the normal range for age-matched controls, as were the proportions of naïve and memory T cell subsets, and other T helper subsets, Tregs, iNKT, and MAIT cells (fig. S7, E to I). Nevertheless, the counts and proportions (among naïve T cells) of blood γδ T cells were higher in the p.Asp51Ala homozygotes than in controls (Fig. 3, L and M). By contrast to the findings for patients with complete pre-TCRα deficiency (Fig. 3K), the proportion of CD4−CD8− cells in the naïve αβ T cell compartment was normal (Fig. 3N). Compared to patients with complete pre-TCRα deficiency, p.Asp51Ala homozygotes generally had a narrower but still distinctive immunological phenotype, with higher proportions of γδ T cells among naïve T cells. This was reminiscent of mice with mutations that affect similar charged residues of pre-TCRα (28, 29).
Pre-TCRα deficiency impairs the generation of TCRαβ+ but not TCRγδ+ T cells in vitro
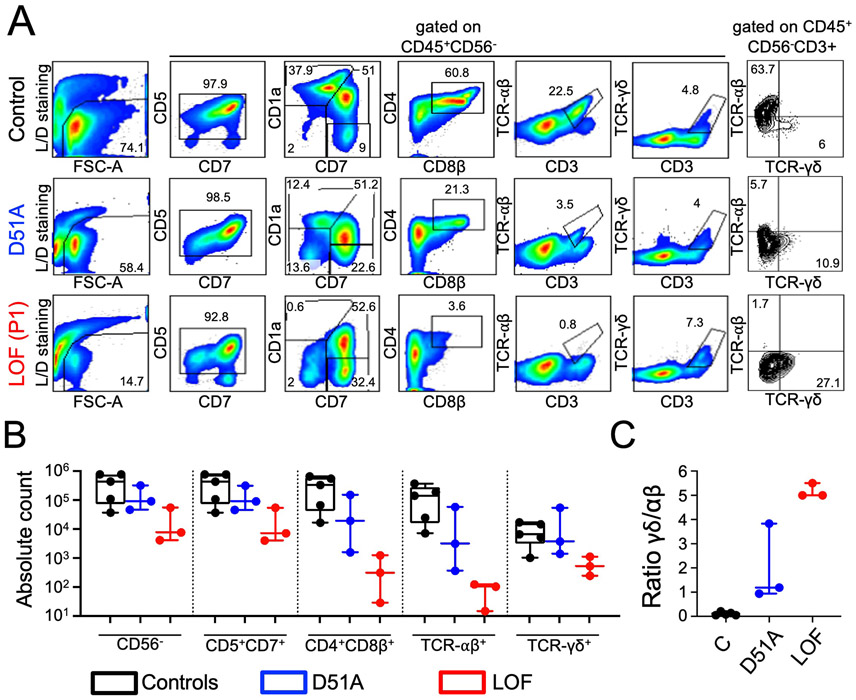
We assessed the impact of the patients’ PTCRA genotype on the early stages of T cell differentiation by isolating blood CD34+ cells from three pre-TCRα–deficient patients and five healthy controls, and inducing their differentiation in vitro in an artificial thymic organoid (ATO) system (Fig. 4) (35). After 5 weeks of culture, control CD34+ cells remained highly viable (around 74%) (Fig. 4A). Efficient differentiation into CD4+CD8+ DP cells (mean ~52% of CD45+CD56− cells), TCRαβ+CD3+ SP cells (~23%), and TCRγδ+CD3+ cells (~1.8%) was observed. By contrast, after 5 weeks of culture, viability was much lower for the CD34+ cells isolated from all three pre-TCRα–deficient patients (7 to 39%). The differentiation of these cells into T cells was impaired, with a block at the CD7+CD1a+CD4−CD8β− DN stage and an almost total absence of CD4+CD8+ DP cells (mean ~2% of CD45+CD56− cells) and TCRαβ+CD3+ SP cells (~0.5%). However, a significant fraction of the cells were TCRγδ+CD3+ (~3.5%). A determination of absolute counts per ATO of cells at various stages of differentiation confirmed the deficit of CD4+CD8β+ DP and TCRαβ+CD3+ cells and the presence of a significant number of TCRγδ+ cells in the three pre-TCRα–deficient patients studied, relative to controls (Fig. 4, A and B). In particular, the ratio of TCRγδ+ cells to TCRαβ+ cells was markedly higher in the pre-TCRα–deficient patients (~5) than in controls (~0.1) (Fig. 4C). Finally, ATOs generated with CD34+ cells from the three patients homozygous for the p.Asp51Ala variant had a phenotype intermediate between those of the controls and pre-TCRα–deficient patients (Fig. 4). Thus, complete pre-TCRα deficiency almost completely abolishes human αβ T cell differentiation in vitro, whereas partial deficiency due to p.Asp51Ala homozygosity has a milder impact.
Fig. 4. Impaired generation of TCRαβ+ T cells in pre-TCRα–deficient ATOs.
In vitro T cell differentiation from positively selected peripheral blood CD34+ cells obtained from a healthy control, three patients with the p.Asp51Ala variant (D51A) and three patients with LOF PTCRA mutations (P1, P5, P6) after 5 weeks of culture in the ATO system. (A) Flow cytometry plots showing the expression of early and late T cell differentiation markers (CD7, CD5, CD1a, CD4, CD8β, TCRαβ, TCRγδ, and CD3) following gating on LIVE/DEAD−CD45+CD56− cells. The data shown correspond to one control, one p.Asp51Ala patient and P1. (B) Plots of absolute counts/ATO for the various stages of T cell differentiation, for the cells isolated from the ATOs. (C) Bar graphs showing the ratio of absolute counts of TCRγδ+ cells to absolute counts of TCRαβ+ cells per ATO. C: controls, LOF: patients homozygous for LOF variants, D51A: individuals homozygous for the p.Asp51Ala variant.
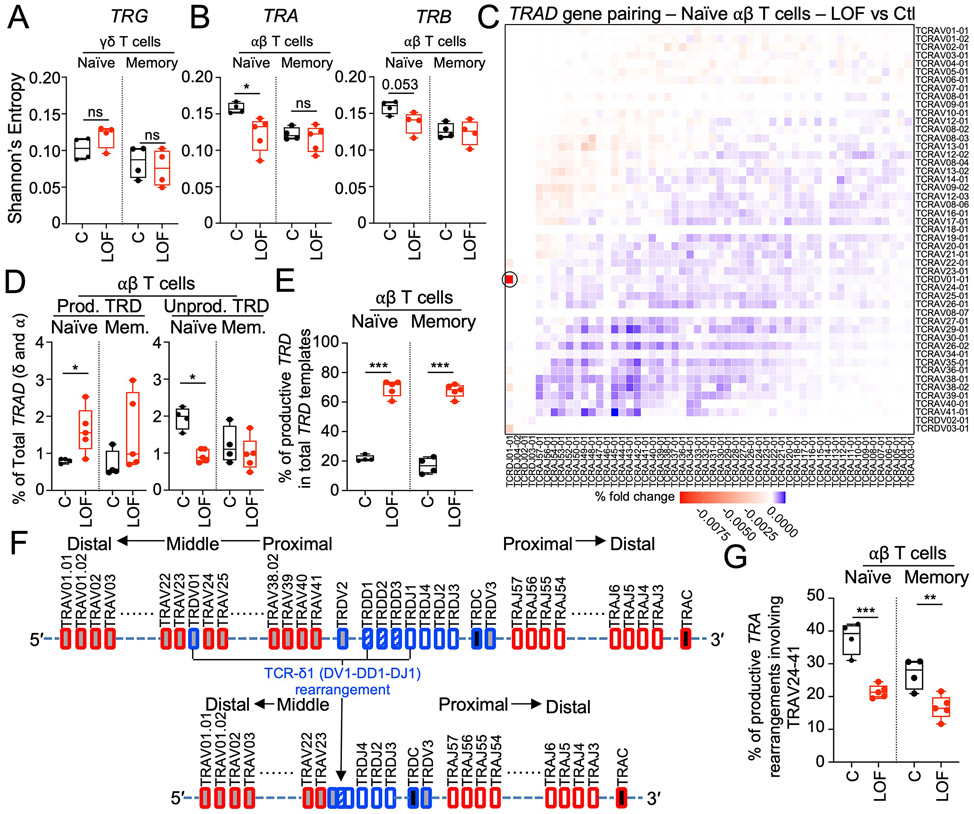
Sequencing of the TRAD locus in αβ T cells reveals an enrichment in proximal TCRδ1 and a depletion of distal MAIT cell rearrangements
The unexpectedly modest impact of pre-TCRα deficiency on αβ T cell development in vivo raised the question of how these cells developed in the absence of a major TCR component during the β-selection process. In patients with complete pre-TCRα deficiency, the circulating αβ and γδ TCR repertoire diversities were slightly low and normal, respectively, (Fig. 5, A and B; fig. S8A; table S7; and Supplementary Text). We then investigated whether the patients displayed preferential usage of productive V-J rearrangements at the TRAD locus in genomic DNA from purified naïve and memory αβ T cells. The most common productive V-J recombination at the TRAD locus in the naïve and memory αβ T cells of the patients was TRDV01:TRDJ01 (i.e., TCRδ1) (Fig. 5C and fig. S8B). The percentages of productive and nonproductive TRD rearrangements among total TRAD rearrangements were significantly higher and lower, respectively, in the patients’ naïve αβ T cells than in those of the controls (Fig. 5D). Productive TRD rearrangements (involving any TRDV) consequently accounted for ~70% of the total TRD rearrangements detected in the αβ T cells of patients with complete pre-TCRα deficiency but only ~20% of those in healthy controls (P<0.0001) (Fig. 5E and fig. S8C). An analysis of the TRAD locus from purified naïve and memory αβ T cells showed a depletion of the TRAV genes removed during TCRδ1 rearrangement (TRAV24-TRAV41) in the productive TRAD rearrangements in the αβ T cells of patients relative to controls (Fig. 5, C, F and G, and fig. S8B). As a result, TRAV genes distal to TRDV01 (TRAV01-TRAV23) were enriched in the patients’ αβ T cells (Fig. 5C and fig. S8B). TCRδ1 accounted for ~70% of total naïve γδ T cells (Fig. 3H), so such a pattern would be expected for TCRα repertoires preferentially rearranged from a TCRδ1 template, with TRAV23 becoming the most proximal TRAV gene following successful TCRδ1 (TRDV01:TRDJ01) rearrangement (fig. S8D). Thus, our TRAD repertoire analysis suggests that, in absence of pre-TCRα, TCRα rearrangements preferentially occur from a productive TCRδ1 template.
Fig. 5. Biases in the TRAD rearrangement repertoire indicate that TCRα chains are mostly generated by rearrangement of a TCRδ1 template in pre-TCRα–deficient individuals.
(A) Shannon’s entropy for TCRγ rearrangements in naïve and memory γδ T cells from controls (black; n=4) and pre-TCRα–deficient individuals (red; P1, P2, P4, and P8). (B) Shannon’s entropy for TCRα and TCRβ rearrangements in naïve and memory αβ T cells from controls (black; n=4) and pre-TCRα–deficient individuals (red; P1, P2, P4, P8, and P9). (C) Heatmap of paired gene rearrangements at the TRAD locus for naïve αβ T cells from four controls compared with five pre-TCRα–deficient individuals (P1, P2, P4, P8, and P9). The red color highlights V-J gene pairings overused in patients and the blue color highlights V-J gene pairings overused in controls. The TCRδ1 (TRDV1:TRDJ1) rearrangement is indicated with a black circle. (D) Fraction of TCRδ rearrangements in total productive TRAD rearrangements from sorted naïve and memory αβ T cells from controls (black; n=4) and pre-TCRα–deficient individuals (red; P1, P2, P4, P8, and P9). (E) Fraction of productive TCRδ rearrangements among total TCRδ rearrangements in naïve and memory αβ T cells from controls (black; n=4) and pre-TCRα–deficient individuals (red; P1, P2, P4, P8, and P9) (red). (F) Schematic representation of the TRAD locus before and after TCRδ1 rearrangement. (G) Percentage of productive TRA rearrangements involving TRAV24-41 in sorted naïve and memory αβ T cells from controls (black; n=4) and pre-TCRα–deficient individuals (red; P1, P2, P4, P8, P9). Unpaired t tests were used for comparisons in panels A, B, D, and E.
TCRδ1 is not a surrogate for pre-TCRα
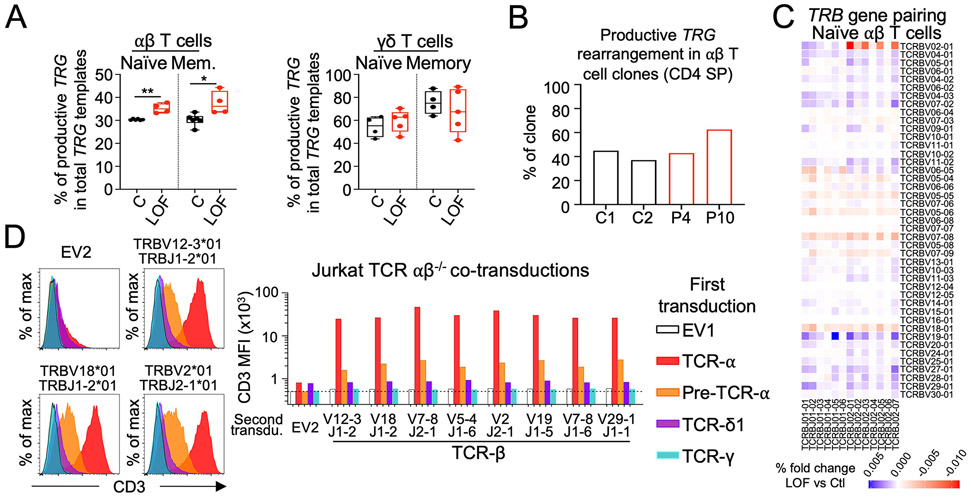
Having excluded the possibility that most αβ T cells preferentially differentiate from γδ+ thymocytes in the absence of pre-TCRα (Fig. 6,A and B; tables S8 and S9; and Supplementary Text), we hypothesized that TCRδ might act as a surrogate for pre-TCRα in the formation of a pre-TCR complex with specific TCRβ rearrangements and CD3. Consistent with this hypothesis, we found that the TCRβ repertoire was biased in patients with pre-TCRα deficiency, with an enrichment in rearrangements involving the middle TRBV genes and any TRBJ gene or involving the distal TRBV02-1 gene and any TRBJ02 gene (Fig. 6C and fig S8, E and F). We transduced TCRαβ–deficient Jurkat cells with TCRδ1, pre-TCRα, TCRα, or TCRγ cDNA. These stable cell lines were cotransduced with an empty vector or one of eight selected TCRβ chains, and CD3 stabilization at the cell surface was assessed by flow cytometry. TCRδ1 and TCRα alone stabilized low amounts of CD3 on the cell surface, whereas neither pre-TCRα, TCRβ, nor TCRγ alone could stabilize CD3 expression at detectable levels on the cell surface. Relative to transduction with single chains, we observed no enhancement of CD3 stabilization following cotransduction with TCRδ1 or TCRγ together with any of the tested TCRβ chains, including the TCRβ chain with the rearrangement frequently found in pre-TCRα–deficient patients and a TCRβ chain previously suggested to stabilize TCRδ1 expression (36). By contrast, pre-TCRα or TCRα stabilized CD3 expression at high levels on the cell surface following cotransduction with any TCRβ construct (Fig. 6D). Thus, in this system, TCRδ1 and TCRγ are unable to replace the pre-TCRα to stabilize TCRβ expression at the cell surface.
Fig. 6. γδ+ thymocytes do not preferentially differentiate into αβ T cells in the absence of pre-TCRα, and TCRδ1 cannot act as a surrogate for pre-TCRα.
(A) Fraction of productive TCRγ among total TCRγ templates in sorted naïve and memory αβ (left) or γδ (right) T cells from controls (black, n=4) and pre-TCRα–deficient individuals (red, P1, P2, P4, P9). Unpaired t tests were used for all comparisons. (B) Fraction of expanded αβ T cell clones from two controls and two pre-TCRα–deficient individuals with a productive TRG rearrangement at the gDNA level. Of note ~90% of these clones were CD4+CD8−. (C) Heatmap of paired gene rearrangements of the TRB locus for naïve αβ T cells from controls compared with five pre-TCRα–deficient individuals (P1, P2, P4, P8). The red color highlights V-J gene pairings overused in patients and the blue color highlights V-J gene pairings overused in controls. (D) TCRαβ–deficient Jurkat cells were stably transduced with an empty plasmid or with a plasmid encoding TCRα, pre-TCRα, TCRδ1 or TCRγ. Each of the resulting cell lines was then cotransduced with another empty plasmid or with a plasmid encoding one of eight selected TCRβ chains. The expression of CD3 at the cell surface was evaluated by flow cytometry. Representative flow cytometry histogram plot for three independent experiments. Representative flow cytometry data are shown on the left. A recapitulative bar graph of the MFI for CD3 for each cell line is shown on the right.
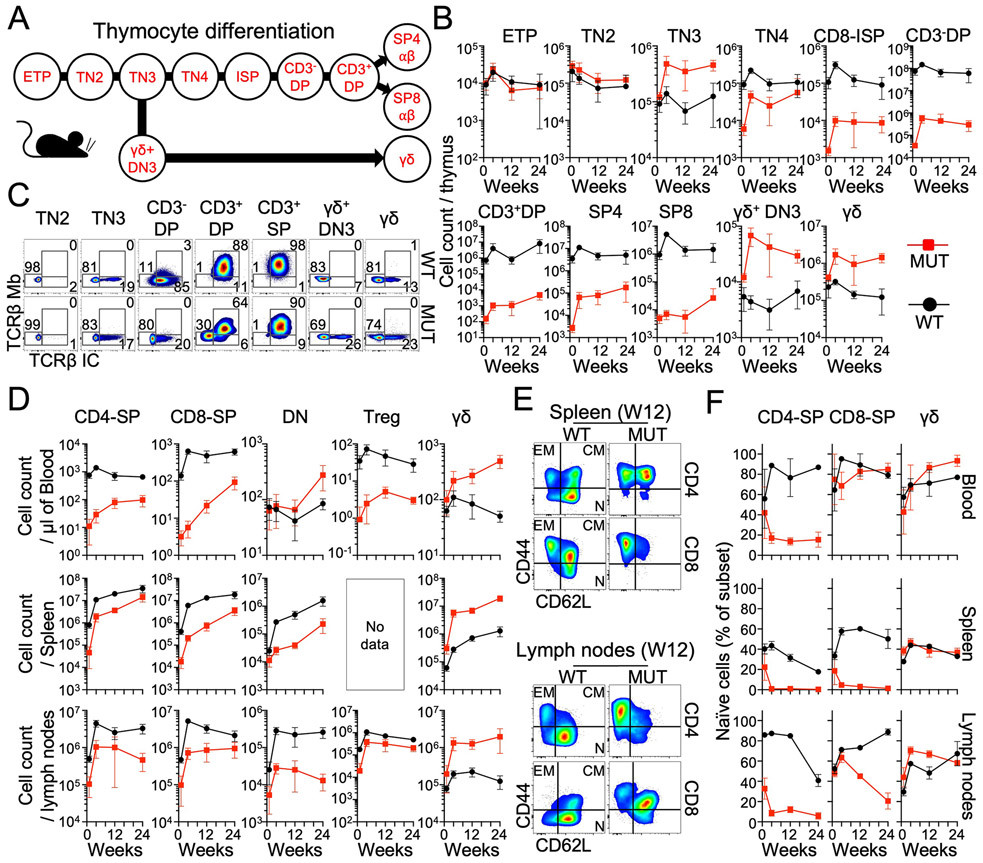
Longitudinal study of peripheral T cells in 1-to-24-week-old Ptcra−/− mice
We found that the peripheral αβ T cell counts of pre-TCRα–deficient humans normalized with age due to the physiological decrease in T cell counts with age in healthy individuals and an accumulation of memory αβ T cells in the patients. Four-week-old Ptcra−/− mice have been reported to have 5% normal T cells in the LNs (12). However, T cell dynamics in the thymus and periphery have not been studied during aging. We therefore sought to reassess and extend mouse immunophenotyping longitudinally by studying the thymus, blood, spleen, and LNs of 1-, 4-, 12- and 24-week-old Ptcra−/− and control mice. The skewed thymocyte differentiation observed in pre-TCRα–deficient mice remained stable in the aging thymus between the ages of 1 month and at least 6 months (Fig. 7, A to C, and Supplementary Text). Contrasting with the previous report of 5% normal T cells, we found that the CD4+ and CD8+ αβ T cell counts in the LNs of 4-week-old mice corresponded to 23% and 14% the normal level, respectively (Fig. 7D) (12). CD4+ αβ T cell counts were 39% and 26% the normal values in 12- and 24-week-old Ptcra−/− mice, respectively, whereas CD8+ αβ T cell counts were 14% and 45% the normal values in 12- and 24-week-old Ptcra−/− mice, respectively. Circulating CD4+ αβ T cell counts in Ptcra−/− mice were 2%, 11%, and 15% the normal level, whereas circulating CD8+ αβ T cell counts were 1%, 5%, and 15% normal levels in 4-, 12- and 24-week-old mice, respectively (Fig. 7D). Splenic CD4+ αβ T cell counts in Ptcra−/− mice were 17%, 18%, and 40% the normal values, whereas splenic CD8+ αβ T cell counts were 3%, 5%, and 19% the normal values in 4-, 12- and 24-week-old mice, respectively (Fig. 7D). Treg counts mirrored the counts of total CD4+ αβ T cells in the LNs and the blood (Fig. 7D). The increase in CD4+ and CD8+ αβ T cell counts in the periphery in aging Ptcra−/− mice was driven by an accumulation of memory T cells, except for blood CD8+ T cells, which remained mostly naïve in 24-week-old mice (Fig. 7, E and F). The γδ T cell counts of Ptcra−/− mice were higher than those of control mice at all timepoints with a greater increase in the spleen (10-tos20-fold between weeks 4 and 24 (W4 and W24)) and LNs (7-16-fold between W4 and W24) than in the blood (2-to-10-fold increase between W4 and W24) (Fig. 7D). As in humans, circulating αβ CD4−CD8− DN T cells were more abundant in Ptcra−/− compared with WT mice (Fig. 7D). Splenic and LN αβ CD4−CD8− DN T cell counts did not significantly differ between Ptcra−/− and mice, however. Thus, CD4+ and CD8+ αβ T cell counts increase in aging Ptcra−/− mice mostly due to an expansion of the memory T cell compartment. These findings are consistent with our pre-TCRα–deficient patient data, highlighting the importance of studying aging animals when exploring thymic phenotypes.
Fig. 7. Longitudinal studies of Ptcra−/− mice.
(A) Schematic representation of thymocyte differentiation stages in mice. (B) Cell counts per thymus in Ptcra−/− mice and WT mice aged 0-24 weeks, for the various thymocyte developmental stages. (C) Intracellular (IC) and membrane (Mb) expression of TCRβ for the indicated thymocyte subsets from Ptcra−/− mice (bottom panel) and WT mice (upper panel). Cytometry data representative of 6 Ptcra−/− mice and 6 WT mice are shown. (D) Counts of cells per microliter of blood or per spleen or per LN for Ptcra−/− mice (red) and WT mice (black) aged 0-24 weeks for the indicated T cell subsets, including CD4-SP (CD3+TCRγδ−CD4+CD8−), CD8-SP (CD3+TCRγδ−CD4−CD8+), DN (CD3+TCR−γδ−CD4−CD8−), Treg (CD3+TCRγδ−CD4+CD8−CD25+), and γδ (CD3+TCRγδ−CD4+CD8−). (E) Representative flow cytometry plots of naïve and memory cell staining for CD4 and CD8 αβ T cells from the spleen and LNs of 12-week-old WT (black) and Ptcra−/− mice (red). EM: effector memory; CM: central memory; N: Naïve. (F) Frequency of naïve cells among CD4-SP and CD8-SP αβ T cells, and of γδ T cells in the indicated tissue of Ptcra−/− mice (red) and WT mice (black) from 0-24 weeks of age. (B, D and F) The data shown are the mean and standard deviation of 4 to 6 animals at each age.
Discussion
The mouse and human PTCRA genes were discovered in 1994 and 1995, respectively, and the first pre-TCRα–deficient mice were described in 1995 (12, 37, 38). We report here 10 humans from seven kindreds and three distant ancestries with AR complete pre-TCRα deficiency. Given the severity of αβ T cell deficiency in pre-TCRα–deficient mice (12), it was expected that pre-TCRα–deficient humans would suffer from life-threatening infections in infancy. To our surprise, this was not the case, and six of our 10 patients, aged 2 to 65 years, have remained healthy. The remaining four patients have exhibited severe infection, lymphoproliferation, or autoimmunity beginning between the ages of 13 to 25 years. This relatively mild clinical phenotype is likely due to an age-dependent accumulation of normal numbers of diverse, functional memory αβ T cells. With hindsight, these findings do not conflict with the reported role of pre-TCRα in mice. Indeed, the impact of the Ptcra−/− genotype on thymocytes and peripheral T cells have not been studied in aging mice and young mice, which survived in pathogen-free conditions, and have not been challenged with pathogens. As in humans, we observed a progressive increase in mouse blood αβ T cell counts with age, driven by memory cell accumulation. These findings highlight the need for caution when extrapolating phenotypes from mutant mice, which are often studied at a young age and in a narrow range of experimental conditions, to humans (39, 40). They also suggest that it can be productive to revisit mouse phenotypes based on human studies. In both mice and humans, αβ T cells can develop in the absence of pre-TCRα.
These findings raise questions about how diverse naïve αβ T cells develop in the absence of pre-TCRα. Our first hypothesis was that early productive proximal TRA rearrangements may permit αβ T cell development (41). However, we observed a depletion of productive TRAD rearrangements involving proximal TRAV genes (TRAV24-TRAV41) and an enrichment in rearrangements involving distal TRAV genes (TRAV1-TRAV23) in the patients’ naïve αβ T cells. Moreover, an abnormal enrichment in the productive TCRδ1 (TRDV01:01-TRDJ01:01) rearrangement was observed in αβ T cells from patients. As TRAV segments preferentially recombine with symmetric TRAJ segments (proximal V with proximal J, distal V with distal J) (42, 43), the TCRα repertoire observed in the absence of pre-TCRα—with a depletion of rearrangements involving proximal TRAV and an enrichment in rearrangements involving distal TRAV—suggests that these TCRα rearrangements occurred preferentially with a TCRδ1 template (fig. S8D). We therefore tested the hypothesis that TCRδ permits αβ T cell development. However, we found that TCRδ1 was unable to act as a surrogate for pre-TCRα in the formation of a pre-TCR. Moreover, similar to controls, most CD4+ SP αβ T cell clones from the patients did not carry a productive TRG rearrangement, suggesting that most of the patients’ T cells were unlikely to have differentiated directly from γδ+ thymocytes. These findings call for alternative hypotheses that may account for αβ T cell differentiation in the absence of pre-TCRα, which are consistent with the associated rearrangement bias observed at the TRAD locus.
We also identified two alleles affecting residues interacting with TCRβ responsible for partial pre-TCRα deficiency in homozygotes. Homozygosity for the p.Tyr76Cys variant affects about 1/76,000 individuals in sub-Saharan Africa. Homozygosity for p.Asp51Ala is more common, affecting between 1/1000 and 1/10,000 individuals in South Asian and Middle Eastern countries. The p.Asp51 residue was previously identified as potentially crucial for pre-TCRα function due to its conservation and negative charge (28). We show that this amino acid plays a crucial role in pre-TCRα dimerization with TCRβ. Humans homozygous for p.Asp51Ala have a partial form of pre-TCRα deficiency. They have normal cell counts of blood αβ T cell subsets, but high counts of naïve γδ T cells and low sjTREC levels. This phenotype is reminiscent of that of transgenic mice with substitutions in the extracellular domain of pre-TCRα, which have a phenotype intermediate between those of WT and Ptcra−/− mice (29). The clinical phenotype of individuals with partial pre-TCRα deficiency is milder than that of individuals with complete pre-TCRα deficiency. The patients are asymptomatic or display isolated autoimmune manifestations. Homozygosity for hypomorphic mutations of PTCRA should be considered in patients with isolated autoimmunity, particularly the p.Asp51Ala substitution in individuals of South Asian or Middle Eastern origin. Collectively, our findings demonstrate that human pre-TCRα is largely redundant for αβ T cell development, but that its complete or partial deficiency can result in late-onset clinical manifestations (including autoimmunity in particular) with incomplete penetrance.
Materials and Methods
Informed consent
Written informed consent was obtained from legally authorized representatives in accordance with the Declaration of Helsinki. The study was approved by the ethics committee of INSERM (RCB 2010-A00634-35 et 2008-A01078-47), the UZ/KU Leuven ethical committee for research (reference number s58466), the Children’s Hospital of Philadelphia Institutional Review Board, and Tokyo Medical and Dental University (G2000-103). For studies of in vitro T cell differentiation and high-throughput sequencing of TCR repertoires, patients gave consent to participate in protocol 18-I-0128 approved by the NIH IRB and registered at www.clinicaltrials.gov as protocol NCT03610802.
Whole-exome sequencing, whole-genome sequencing, and Sanger sequencing
P1, P9, and P10 underwent whole-exome sequencing as part of clinical care after a presumptive positive result for neonatal SCID screening. As a sibling of P1, P2 underwent neonatal SCID screening (presumptive positive result) and confirmatory testing for the familial mutations identified in P1. Commercial whole-genome sequencing (Macrogen) was performed for P3, P4, P5, and P7, due to their clinical history of immunodeficiency or autoimmunity. P6 and P8 were identified by regular Sanger sequencing for familial segregation analysis. The frequency of the p.Asp51Ala (Chr6(GRCh37):g.42890858A>C)) variant was evaluated in different cohorts including the South Asians from the UK biobank (44), 800 Qataris from the Qatar Genome Program (QGP) and Qatar Biobank (QBB) longitudinal study (45), Kuwaitis (46), Iranians (neurological phenotypes only), and Saudis (patients with suspected Mendelian diseases and their parents).
Founder effect analysis for the p.Leu99Hisfs*68 and p.Asp51Ala variants
The occurrence of homozygosity for the p.Leu99Hisfs*68 and p.Asp51Ala variants in different kindreds suggested a founder effect. An analysis of the whole-exome sequencing data revealed that P4 (Kindred C) and P5 (Kindred D), both of whom are homozygous for p.Leu99Hisfs68*, have a homozygous haplotype around PTCRA and encompassing a 1.38 Mb region corresponding to 73 SNVs in common. The ESTIAGE method estimated the age of the most recent common ancestor (MRCA) of the two patients at 60 generations (95% CI (19-258 generations)) (47). Assuming a generation time of 27 years (48), the MRCA of these patients with the p.Leu99Hisfs68* mutation would have lived about 1600 (513-6966) years ago. Similarly, an analysis of the whole-exome sequencing data of P11 (Kindred H) and P13 (Kindred I), both homozygous for p.Asp51Ala, showed that they had a homozygous haplotype around PTCRA encompassing a 580 kb region corresponding to 39 SNPs in common. The ESTIAGE method estimated the age of the most recent common ancestor (MRCA) of these two patients at 301 generations (95% CI (93-1090 generations)) (47). Assuming a generation time of 27 years (48), the MRCA of these patients with the p.Asp51Ala mutation would have lived about 8000 (2511-29,430) years ago.
Analysis of the UK Biobank data
We used the UK Biobank plink-formatted population-level exome original quality functional equivalent exome files for n=454,713 individuals (field 23155, with genotypes set to missing when read depth was <7 for single-nucleotide variations). For the UK Biobank cohort, the participants were aged 50 to 87 years as of 2021, and 55% were female. We further restricted our analysis to participants with South Asian ancestry, using field 21000. We considered a participant to be of South Asian ancestry if they had one of the following data codes for field 21000: 3001 (Indian), 3002 (Pakistani), 3 (Asian or Asian British), 3003 (Bangladeshi), or 3004 (any other Asian background). ICD10 codes and associated dates were collected from inpatient data (category 2000), cancer registries (category 100092) and first occurrences (category 1712), defined as the earliest occurrences of ICDs in the general practice, inpatient and death data, at three-digit resolution. The ICD10 codes used for autoimmune diseases were as follows: E03 (hypothyroidism), E10 (type 1 diabetes), M35 (Sjogren’s disease), G73.7 (Addison’s disease), K90.0 (celiac disease), M33 (dermatomyositis), M34 (systemic sclerosis), E05.0 (Graves’ disease), G35 (multiple sclerosis), M06 (rheumatoid arthritis), G70 (myasthenia gravis), D51.0 (pernicious anemia), M32 (systemic lupus erythematosus), D69.6 (autoimmune cytopenia), L80 (vitiligo), and L40 (psoriasis).
Analysis of the Centogene Biodatabank
Exome and genome analyses performed at Centogene up to April 21, 2023 were analyzed (Centogene started exome and genome sequencing in 2014 and 2016, respectively). Only variants with a base coverage ≥10, read frequency ≥30, and variant quality ≥220 (only for exome sequencing) were retained for further analyses. We then selected all related genetic information for the variants NM_138296.3:c.152A>C (PTCRA p.(Asp51Ala)) and NM_138296.3:c.227A>G (PTCRA p.(Tyr76Cys)). We extracted information concerning the patient’s year of birth, sex, country of origin, family relationship, reported genetic test results of ES/GS-tested individuals, and HPO-encoded clinical information from our database when available. All the available de-identified data were aggregated at individual level.
We calculated p.(Asp51Ala) allele frequencies and their binomial confidence intervals by country and geographic region. For the phenotypic analysis, we stratified the cohort by PTCRA p.(Asp51Ala) genotype (homozygotes (HOM), heterozygotes (HET), and wild-type (WT)) and counted the number of occurrences per individual of any HPO term from a predefined list of 24 autoimmunity-related terms (table S5). We analyzed the difference in the proportions of individuals with a matching autoimmune-related phenotype (having at least one of 24 predefined HPO terms) by PTCRA p.(Asp51Ala) genotype. We tested the hypothesis of an association between the individual’s PTCRA p.(Asp51Ala) genotype and the presence of an autoimmune-related phenotype by enrichment analysis. Briefly, we calculated the odds ratio of having a positive match to the autoimmunity-related HPO terms from the predefined list, comparing all PTCRA p.(Asp51Ala) genotype groups in Fisher’s exact test. Statistical analyses and figures were produced with RStudio (version 2023.03.1 Build 446, Posit Software, rstudio.com), using R Statistical Software (version 4.3.0, R Core Team 2023, R-project.org) and the tidyverse package (version 2.0.0, Posit Software, tidyverse.org).
RNA-seq analysis of sorted human thymocyte subsets
Data for biological triplicates of RNA-seq performed on sorted primary human thymic T cell subsets (Thy1/DN1, Thy2/DN2, Thy3/DN3, ISP4/ISP, DP early, DP late, and single-positive SP8 and SP4) were downloaded with the SRA toolkit and the fastq-dump v2.9.6 tool (dataset accession number BioProject: PRJNA741323) (16). The sequence reads were aligned with the human hg38 reference genome assembly with HISAT2 v2.2.1, using the -k 1 function (49). The principal pre-TCRα isoforms were detected with a combination of cufflinks v2.2.1 de novo transcript assembly (50) and the manual curation of transcript databases: Ensembl hg38 v96 and NCBI Refseq genes v110. Expression was estimated for five pre-TCRα isoforms, along with the full gene list in Ensembl v96, with kallisto v0.46.1, for each sample (51). Estimated normalized isoform expression, expressed in transcripts per million (TPM), was used to compare expression levels across thymocyte developmental stages and to calculate the abundance of isoform A relative to the other isoforms. The aligned reads were converted to BAM format with samtools v1.14 and the triplicates were combined with the merge function and loaded onto the Integrated Genome Viewer for figure preparation (52, 53).
Sanger sequencing and TA cloning
Genomic DNA was obtained from whole blood from the patients. The PTCRA mutations identified by WES were checked by amplifying the corresponding gDNA regions with a recombinant Taq polymerase (Thermo Fisher Scientific). PCR products were purified by centrifugation through Sephadex G-50 Superfine resin (Merck) before and after the sequencing reaction, which was performed with the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) and the primers previously used to amplify the region of interest. Purified sequencing products were analyzed with an ABI Prism 3500 apparatus (Applied Biosystems) and aligned with the genomic sequence of PTCRA (Ensembl) with Serial Cloner 2.6 software. We checked that the compound mutations found in P7 and P8 really were in two different alleles by cloning the PCR amplicons from the gDNA of these patients with the TOPO TA cloning kit (Thermo Fisher Scientific) and using them for the one-shot transformation of TOP10 chemically competent Escherichia coli cells (Thermo Fisher Scientific). PCR with the M13 primers supplied with the TA cloning kit was performed on individual colonies before sequencing.
Cell culture
HEK293T cells were cultured in DMEM (#61965059, Gibco) supplemented with 10% fetal bovine serum (Sigma-Aldrich). TCRα–deficient JR3.11 Jurkat cells and TCRαβ–deficient J76 Jurkat cells were cultured in RPMI (# 61870044, Gibco) supplemented with 10% fetal bovine serum (25). All cell lines were cultured at 37°C under an atmosphere containing 5% CO2. For transfection, HEK293T cells were plated at a density of 8×105 cells per well in six-well plates. PBMCs were isolated from whole-blood samples by Ficoll-Hypaque centrifugation (Amer-sham-Pharmacia-Biotech).
Plasmids and transient transfection
The C-terminally Myc/DDK-tagged pCMV6 empty vector and the human PTCRA expression vectors were purchased from Origene (NM_138296; #RC215794). Constructs carrying mutant alleles were generated by direct mutagenesis with the CloneAmp Hifi premix and polymerase (#639298, Takara). The resulting PCR products were digested with DpnI (#R0176L, New England Biolabs) for 1 hour at 37°C, amplified in competent E. coli cells (#C3019H, New England Biolabs), and purified with a Maxiprep kit (#12663, Qiagen). Isoform B of pre-TCRα and the mutant allele lacking exons 1 to 3 were obtained by opening the Isoform A WT vector by PCR with primers flanking the region to be deleted and then using the Quick Blunting™ Kit (#E1201L, New England Biolabs) and the Quick Ligation™ Kit (#M2200S, New England Biolabs) as recommended by the manufacturer. Lentiviral plasmids carrying the various PTCRA variants were generated by inserting the cDNA from the pCMV6 plasmids into an empty pTrip-SFFV-ΔNGFR vector (modified pTRIP-SFFV-mtagBFP-2A; addgene, plasmid #102585). This was achieved by digesting the empty pTrip-SFFV-ΔNGFR vector with XhoI and BamHI for 1 hour at 37°C. The cDNA of interest was amplified by PCR and inserted into the vector by homologous recombination with the In-Fusion® HD Cloning Kit according to the manufacturer’s instructions (#638911, Takara). Lentiviral plasmids pTrip-SFFV-ΔNGFR encoding TCRα, TCRδ, TCRγ, or TCRγδ and pTrip-SFFV-GFP encoding various forms of TCRβ were synthesized by TwistBioscience, after onboarding our empty vector plasmid. HEK293T cells were transiently transfected in the presence of the X-tremeGENE 9 DNA Transfection Reagent (#6365787001, Roche), in accordance with the manufacturer’s instructions.
Choice of TCRα, TCRβ, TCRγ and TCRδ1 cDNA sequences
The TCRα sequence was obtained from addgene plasmid #128544 (after removal of the intron). The choice of TCRβ was based on a comparison of the TRB repertoire of naïve αβ cells from patients and controls. The TRBV2*01∣TRBJ2-1*01, TRBV7-8*01∣TRBJ2-1*01, TRBV5-4*01∣TRBJ1-6*01 and TRBV7-8*01∣TRBJ1-6*01 rearrangements were selected because they were found to be overused in patients’ cells. Conversely, the TRBV19*01∣TRBJ1-5*01 and TRBV29-1*01∣TRBJ1-1*01 rearrangements were selected because they were less frequently detected in the patients’ cells than in control cells. The TRBV12-3*01∣TRBJ1-2*01 and TRBV18*01∣TRBJ1-2*01 TCRβ chains were selected because they are the chains expressed in the Jurkat and DN-D41 cell lines, respectively (54). These two cell lines were used as controls. It has been suggested that DN-D41 expresses the TCRδ1/TCRβ heterodimer at the cell surface (36). The full-length TCRδ1, TCRγ and TCRβ sequences were assembled with stitchR software (55). The TCRγ sequence used was the example data provided by stitchR. The CDR3 of the TCRδ1 and TCRβ sequences were randomly picked from the most frequent αβ or γδ T cell clones of the patients with the V-J of interest according to our TCR bulk sequencing data. The full sequences of all the TCRs used in this study are provided in the supplementary materials.
Artificial gene and exon trapping for the c.58G>C and c.58+5G>A alleles
We cloned PTCRA gDNA from a control, as described in Fig. 2A. The inserted PTCRA gDNA sequence, extending from the 5′ UTR to the end of exon 2, is represented in blue (exons) and gray (intron 1). Briefly, PTCRA gDNA containing exon 1 and the first 909 nucleotides of intron 1 was amplified with CloneAmp Hifi premix (Takara), the forward primer 5′- GAGATCTGCCGCCGCGTAGAAGGCAGTCTTGTGGGTGC-3′, and the reverse primer 5′-AAGGAACTCAGTTCCTCCAGGACTCAACCTCCAGA-3′. Similarly, PTCRA gDNA containing the last 724 nucleotides of intron 1 and exon 2 was amplified with the forward primer 5′-GGAACTGAGTTCCTTGAGAGCAGGGACAATGACTTAC-3′ and the reverse primer 5′-CTCGAGCGGCCGCGTACGCGTTGACAGATGCATGGGCTGTGTAC-3′. The Infusion Cloning kit (Clontech) was used to insert both PCR products between the ASIS1 and Mlu1 cloning sites of the pCMV6 entry vector (Origene) by homologous recombination. The c.58+G>C or c.58+5G>A mutation was generated by mutagenesis. We extracted mRNA from HEK293T cells after 24 hours of transfection with the WT, c.58G>C or c.58+5G>A exon-trapping vectors. The cDNA for the PTCRA transcript was amplified with a recombinant Taq polymerase (Thermo Fisher Scientific), a forward primer 5′-TAGAAGGCAGTCTTGTGGGTGC-3′ binding to the 5′ UTR of PTCRA, and a reverse primer 5′-CATTTGCTGCCAGATCCTCTT-3′ binding to the in-frame C-terminal Myc/DDK tag. The PCR products were then cloned with the TOPO TA cloning kit (Thermo Fisher Scientific) and used for the one-shot transformation of TOP10 chemically competent E. coli cells (Thermo Fisher Scientific). Splice variants of PTCRA from individual colonies were amplified with the M13 primers supplied with the TA cloning kit before sequencing and alignment with the PTCRA cDNA (NM_138296.2), with SnapGene software used to identify alternative splicing variants. We screened 82 colonies for the WT PTCRA exon-trapping vector, 65 for the c.58G>C PTCRA exon-trapping vector, and 83 for the c.58+5G>A PTCRA exon-trapping vector.
mRNA purification, and RT-qPCR
Total RNA was extracted from the indicated cells with the RNeasy Extraction Kit (Qiagen). RNA was reverse-transcribed with the SuperScript II reverse transcriptase (Thermo Fisher Scientific) and oligo-dT primers (Thermo Fisher Scientific). We then performed qPCR with the Applied Biosystems Assays-on-Demand probes/primers specific for PTCRA-FAM (Hs00300125_m1) on 100 ng cDNA. The data were normalized relative to the expression (ΔCt) of GUS (13-glucuronidase-VIC, 4326320E) and are expressed as 2−ΔCt values.
Cell lysis and immunoblotting
The exon-trapping primers were designed to retain the translation frame of the Myc/DDK tag from the pCMV6 plasmid (Fig. 2A). The end of exon 2 was, therefore, fused to a Myc/DDK tag for detection of the artificial fusion protein. Total protein was extracted from HEK293T cells after 24 hours of transfection with the WT, c.58G>C or c.58+5G>A exon-trapping vectors. We assessed the expression of PTCRA variants by extracting total protein from HEK293T cells 48 hours after transfection with the various pCMV6 plasmids encoding the PTCRA variants. Total protein extracts were obtained by incubating cells with lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 2 mM EDTA, and 0.5% Triton X-100). A mixture of protease and phosphatase inhibitors was added to the buffers: aprotinin (10 μg/ml; Sigma-Aldrich), PMSF (1 mM; Sigma-Aldrich), leupeptin (10 μg/ml; Sigma-Aldrich), protease inhibitor cocktail (Sigma-Aldrich). After 30 min of lysis at 4°C, the cells were centrifuged for 10 min at 16,000g, and the supernatant was collected for immunoblotting. For each variant, we separated 20 μg of total protein by SDS-PAGE and immunoblotting was performed with antibodies against the DDK Tag (1:3000, HRP-coupled, M2, #A8592, Sigma-Aldrich), PTCRA (1:3000, PA5-95578, Invitrogen), GAPDH (1:5000, FL335, #sc47724 HRP, Santa Cruz Biotechnology) or vinculin (1:5000, EPR8185, #ab129002, Abcam). Staining was detected with the Clarity Western ECL substrate (Biorad, #1705061) or SuperSignal West Femto (ThermoScientific, #34096) with ChemiDoc MP (Biorad).
Lentivirus production and transduction
The lentiviruses used for the transduction of TCRα–deficient JR3.11 Jurkat cells and TCRαβ–deficient J76 Jurkat cells were produced by transfecting HEK293T cells with pCMV-VSV-G (0.2 μg) (56), pHXB2 env (0.2 μg; NIH-AIDS Reagent Program; #1069), psPAX2 (1 μg; gift from D. Trono; Addgene plasmid #12260) and a vector containing the sequence for transduction. The vectors containing the sequences for transduction were pTrip-SFFV-ΔNGFR (empty vector), pTrip-SFFV-GFP (empty vector), pTrip-SFFV-ΔNGFR-PTCRA-WT, the other pTrip-SFFV-ΔNGFR vectors containing the PTCRA variants studied, pTrip-SFFV-ΔNGFR-TCRα, pTrip-SFFV-ΔNGFR-TCRδ, pTrip-SFFV-ΔNGFR-TCRγ, pTrip-SFFV-ΔNGFR-TCRγδ and the pTrip-SFFV-GFP-TCRβ vectors. HEK293T cells were transfected in six-well plates and the medium was replaced after 6 hours of incubation. The virus-containing supernatant was collected and passed through a 0.2-μm filter 24 hours after the medium was changed. Protamine sulfate (8 μg/ml) was added to the virus-containing supernatant, which was then added to Jurkat cells (immediately after seeding), which were spinoculated for 2 hours at 1200g and 25°C. The cells were then cultured for 48 hours at 37°C under an atmosphere containing 5% CO2, without shaking. Transduction efficiency was then checked by flow cytometry with the GFP tag or an anti-CD271 antibody (#557196, BD, 1:500). Transduced cells were sorted with a magnetic MACS® Column and the CD271 MicroBead Kit (#130-099-023, Miltenyi Biotec), as recommended by the manufacturer.
Flow cytometry analysis of JR3.11 Jurkat cells and J76 Jurkat cells
Transduced JR3.11 Jurkat cells and J76 Jurkat cells were stained with antibodies against CD271 (#557196, BD, 1:500), CD3 (#555333, BD, 2:50), C1β TCR (#565776, BD, 2:50), or CD69 (#310912, Biolegend, 1:400) and incubated with the Aqua Live/Dead Cell Stain Kit (Thermo Fisher Scientific) for 1 hour at room temperature before analysis with a Gallios (Beckman Coulter) flow cytometer. All data were analyzed with FlowJo 10.5.3 software.
Intracellular cytokine staining
Intracellular Cytokine Staining (ICS) was performed as previously described (57). Briefly, PBMCs were thawed in cRPMI (RPMI 1640 1X supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1% L-glutamine and 1% Pen-Strep) and centrifuged before being resuspended at a concentration of 2 × 106 cells/ml in cRPMI. Cell pellets were resuspended in surface antibody cocktail (table S10) and incubated for 20 min. Cells were then permeabilized with permeabilization reagent (Invitrogen) and incubated for 20 min followed by a wash with PERM Buffer. Cell suspensions were centrifuged, and the pellets stained with intracellular antibody cocktail (table S10) for 60 minutes. Lastly, cells were washed with permeabilization buffer before being resuspended in 1.6% PFA to fix. The fixed cells were stored overnight at 4°C and analyzed on Aurora Spectral flow cytometer (Cytek).
Mass cytometry (CyTOF)
CyTOF was performed with various strategies. One involved the use of whole blood in the Maxpar Direct Immune Profiling Assay, 30 Markers (Standard Biotools, ref: 201334), according to the manufacturer’s instructions. All the samples for whole-blood staining were processed within 24 hours of collection. P10, P12, and P13 were phenotyped by the same protocol but with a customized antibody panel (table S11). We investigated the T cell subsets, including IEL markers, with another CyTOF staining panel for cryopreserved samples (IEL panel, table S12). PBMCs were thawed and 4×106 cells were immediately stained according to the Standard Biotools protocol. The antibodies against TCR Vδ1 and TCR Vδ2 were added after 10 min of staining with the other antibodies to prevent interference with the binding of the TCRγδ antibody. For both whole blood and IEL panels, cells were frozen at −80°C after iridium staining and stored at the same temperature until acquisition on a Helios machine (Standard Biotools). In addition to whole-blood immunophenotyping, we also performed immunophenotyping on cryopreserved PBMCs for some patients (table S13). Single-cell suspensions were centrifuged to obtain a cell pellet, which was then incubated with 20 μM lanthanum-139 (Trace Sciences)–loaded maleimido-mono-amine-DOTA (Macrocyclics) in PBS for 10 min at room temperature for live–dead discrimination (LD). Cells were washed in staining buffer and resuspended in surface antibody cocktail, incubated for 30 min at room temperature, washed twice in staining buffer, fixed, permeabilized with the FoxP3 staining buffer set (eBioscience), and subjected to intracellular staining for 60 min at room temperature. Cells were washed twice and then fixed by overnight incubation in 1.6% PFA (Electron Microscopy Sciences) solution supplemented with 125 nM iridium at 4°C. Before data acquisition on a CyTOF Helios flow cytometer (Standard Biotools), cells were washed twice in PBS and once in dH2O. Custom conjugation to isotope-loaded polymers was performed with the MAXPAR kit (Standard Biotools). The data were analyzed with OMIQ software.
Luciferase reporter assays for autoantibodies against IFNs
The blocking activity of anti-IFN-α, anti-IFN-β, and anti-IFN-ω autoantibodies was assessed in a luciferase reporter assay, as described elsewhere (58).
Protein array for assessing autoantibodies
Protein arrays (HuProt™ v4.0 from CDI laboratories) for assessing autoantibodies were performed as previously described (24).
Single-cell RNA sequencing (5′ transcriptomics, αβ and γδ TCR)
CD3+ T cells were sorted by flow cytometry from the PBMCs of P1, P2 (two independent samples analyzed, collected at the ages of 12 and 18 months), P3, P4, and healthy controls matched for age. Cryopreserved peripheral blood mononuclear cells (PBMC) in R10 medium (RPMI 1640, 10% FBS, 2 mM L-glutamine, 100 U/ml of penicillin, and 100 μg/ml of streptomycin) were thawed and immediately centrifuged to obtain a cell pellet. The cells were then incubated for 15 min at 37°C under an atmosphere containing 5% CO2 in the presence of Benzonase (Millipore Sigma, cat. 70664) diluted 1:1000 in R10 medium. The cells were then washed once in R10 and once in FACS buffer (2% FBS in PBS). For staining, cells were resuspended in 50 μl of a mixture of LIVE/DEAD Fixable Blue Dead Cell Stain (cat. L34962) diluted 1:200 in PBS and anti-CCR7 APC-Cy7 antibody (Biolegend, cat. 353212) and incubated for 10 min at 37°C under an atmosphere containing 5% CO2. Cells were then labeled with 1 μl of oligonucleotide-linked hashing antibody (Totalseq-C, Biolegend) and stained by incubation with 50 μl of antibody cocktail diluted in Brilliant Stain Buffer (BD Biosciences, cat. 566349) for 20 min at room temperature. The antibody cocktail contained the following antibodies: anti-CD14 BV510 (Biolegend, cat. 301842), anti-CD19 BV510 (Biolegend, cat. 302242), anti-CD56 BV510 (Biolegend, cat. 362534), anti-CD8 BV785 (Biolegend, cat. 301046), anti-CD4 PECy7 (Biolegend, cat. 300512), anti-CD95 AlexaFluor 700 (Biolegend, cat. 305648), anti-PD-1 BV750 (Biolegend, cat. 329966), anti-CD69 FITC (Biolegend, cat. 310904), anti-CD40L BV421 (Biolegend, cat. 310824), anti-CD3 BUV805 (BD Biosciences, cat. 741999), anti-CD45RA PECF594 (BD Biosciences, cat. 562298), anti-CD25 BUV661 (BD Biosciences, cat. 741685), anti-CXCR3 BV711 (BD Biosciences, cat. 563156), anti-HLA-DR PECy5.5 (Invitrogen, cat. MHLDR18), anti-CXCR5 APC (Invitrogen, cat. 17-9185-42). The cells were washed twice with FACS buffer and resuspended in R10 for sorting. We sorted 12,000 T cells (CD3+CD14−CD19−CD56−Live/Dead−) from each sample with a BD FACSymphony S6 Cell Sorter instrument (BD Biosciences) running BD FACSDiva Software version 9.5.1 (BD Biosciences). Sorted cells were pooled four by four, and each pool was loaded in a different lane of the 10x Genomics Chromium Chip for sequencing. For the sequencing of single-cell V(D)J repertoires for sorted T cells, the cell suspension was loaded on the 10x Genomics Chromium Instrument according to the manufacturer’s protocol for the Next GEM Single-Cell 5′ Kit v1.1 (10x Genomics PN-1000165) to generate gel bead-in-emulsions and for GEM-RT and the amplification of total cDNA. Following purification with SPRIselect beads (Beckman Coulter), specific TCR targets were amplified from the cDNA with the PTCR1 primer (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTC-3′) and constant region primers: TRB (5′-TGCTTCTGATGGCTCAAACACAGCGACCT-3′), TRA (5′-TCTCAGCTGGTACACGGCAGGGTCAGGGT-3′), TRG (5′-GAAGGAAGAAAAATAGTGGGCTTGGGGGAAAC-3′), or TRD (5′-CACCAGACAAGCGACATTTGTTCC-3′) with a barcode and the P7 sequence added to the constant region primers. The Illumina-ready libraries were sequenced by paired-end MiSeq with 2×300 base-pair reads to obtain VDJ sequences.
Evaluation of TCR entropy and TCR chain combinations in scRNA-seq data
We processed 10x single-cell transcriptome libraries with Cellranger (v6.1.1) and Seurat (v4.0.4). The TCRαβ and TCRγδ libraries were demultiplexed and cell barcodes were assigned with Minnn (v10.1). TCR libraries were annotated with MiXCR (v3.0.13) and then separated by subject. The numbers of αβ or γδ TCRs for the patients and controls were calculated by counting the numbers of cells expressing both TRA and TRB V-J genes and both TRG and TRD V-J genes. The final counts corresponded to the intersection of cells expressing combinations of TRA, TRB, TRG or TRD genes, accounting for 10,365, 21,755, 2233, and 1440 cells, respectively, for the controls (n=11) and 5963, 11,416, 2095, and 529 cells, respectively, for the patients (n=5). The diversity of α, β, γ, and δ TCRs was estimated by calculating Shannon’s entropy (H) index. Entropy was calculated by summing the frequencies of each clone (CDR3 amino-acid sequence) and multiplying by the base 2 logarithm of the same frequency over all cells expressing TRA, TRB, TRG or TRD V-J genes. Higher H-index values indicate a more diverse distribution of CDR3 clones (59).
VirScan - phage immunoprecipitation-sequencing (PhIP-Seq)
Antibody profiling by phage immunoprecipitation-sequencing (PhIP-Seq) was performed on plasma samples from patients and controls as previously described (60).
In vitro T cell differentiation in the artificial thymic organoid (ATO) system
In vitro T cell differentiation was studied by coculturing peripheral blood CD34+ cells with a DLL4-expressing stromal cell line (MS5-hDLL4) in the ATO system, as previously described (61), but with minor modifications. Briefly, CD34+ peripheral blood cells from five normal donors, three patients with partial pre-TCRα deficiency (P11-P13), and three patients with complete pre-TCRα deficiency (P1, P5 and P6) were positively selected with the CD34 MicroBead UltraPure kit (Miltenyi Biotec) on an AutoMACS Pro Separator. We mixed 1000-1500 CD34+ cells with 150,000 MS5-hDLL4 cells per ATO. Each ATO (5 μl) was then plated in a 0.4 μM Millicell Transwell insert and placed in one well of a six-well plate in 1 ml complete RB27 medium supplemented with rhIL-7 (5 ng/ml), rhFlt3-L (5 ng/ml), and 30 μM L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate. Each insert contained a maximum of two ATOs. For the first 3 weeks of culture, the medium was also supplemented with 10 ng/ml of rhSCF. After 5 weeks in culture, MACS buffer (PBS supplemented with 0.5% BSA and 2 mM EDTA) was added to each well and ATOs were dissociated by manual pipetting. The cells were then collected into a pellet by centrifugation, resuspended in FACS buffer (2% FBS in PBS), counted, and stained with the antibody cocktail described in table S14. Events were acquired on a BD LSR II Fortessa flow cytometer (BD Biosciences, San Jose, CA) and analyzed with FlowJo software version 10.6.1 (FlowJo, LLC, Ashland, OR).
Mouse experiments
Mice were bred under specific pathogen-free conditions in CIPHE animal facilities (agreement number: B1301407) and handled in accordance with institutional committee and European guidelines for animal care. C57BL/6 mice were purchased from Janvier Laboratories. Ptcratm1(icre)Hjf KO mice have been described elsewhere (62) and were rederived from the INFRAFRONTIER/EMMA archive (EM:08347). Multiparametric immunophenotyping was performed at the CIPHE-PHENOMIN (INSERM, US012) flow cytometry facility. Peripheral blood (PB) was collected by submandibular puncture into Microvette® 500 K3 EDTA tubes (Sarstedt). Hematological analysis was performed on a Procyte Dx (IDDEX) machine, in accordance with the manufacturer’s recommendations. Peripheral blood leukocytes were analyzed with a Lyse No Wash protocol and 1X FACS Lysing Solution (BD Biosciences). Leukocytes from the spleen and thymus were extracted according to the protocol of the International Mouse Phenotyping Consortium (IMPC_IMM_002). Red blood cells were not lysed for thymic leukocyte preparations. Lymph node (LN) T cells were isolated from pooled inguinal, brachial, axillary, and submandibular LNs. Briefly, organs were disrupted with the OctoGentleMACS system (Miltenyi Biotec), using 600 Mandl units of collagenase D (Roche Life Science) and 30 μg of DNAse I (Sigma), for 20 min at room temperature. The cell suspension was filtered and the cells were counted. Red blood cells were lysed by incubation for 1 min at room temperature with ammonium-chloride-potassium (ACK) lysis solution (eBioscience). Before staining, the cells were incubated for 10 min on ice with an anti-CD16/32 (2.4G2) antibody to block Fc receptors. In all experiments, DAPI (Invitrogen) staining was used to exclude dead cells from the analysis. Multiparameter FACS acquisition was performed on a Fortessa LSRII SORP or Canto 10C system (BD Biosciences). The analysis was performed with FACSDiva 9.01 (BD Biosciences) software. Doublets were systematically excluded on the basis of side scatter (SSC) and forward scatter (FSC) parameters. The antibodies used for immunophenotyping are listed in table S15. The thymocyte subsets were defined as ETP (CD4−CD8a−CD3e−CD44+CD25−ckit+), TN2 (CD4−CD8a−CD3e−CD44+CD25+ckit+), TN3 (CD4−CD8a−CD3e−CD44−CD25+γδ−), TN4 (CD4−CD8a−CD3e−CD44−CD25−γδ−), ISP (CD4−CD8a+CD3e−CD44−CD25−γδ−), iDP (CD4+CD8a+CD3e−CD44−CD25−γδ−), mDP (CD4+CD8a+CD3e+CD44−CD25−γδ−), SP4 (CD4+CD8a−CD3e+CD44−γδ−), SP8 (CD4−CD8a+CD3e+CD44−γδ−), γδ TN3 (CD3e+CD25+γδ+) or γδ (CD3e+CD25−γδ+).
High-throughput sequencing (HTS) of the human TCR repertoire from the gDNA of sorted naïve and memory T cells
PBMCs were stained with antibodies against CD3 (#565491, BD, 1:50), CD45RA (#130-092-247, Miltenyi Biotec, 2:50), CCR7 (#130-120-600, Miltenyi Biotec, 2:50), TCRγδ (#331218, Biolegend, 1:50), or TCRαβ (#555548, BD, 2:50) and incubated with the Aqua Live/Dead Cell Stain Kit (Thermo Fisher Scientific) for 30 min at room temperature. Naïve and memory αβ and γδ T cells were sorted with a FACSAria cell sorter (Becton Dickinson, San Jose, CA) on the basis of CD45RA and CCR7 expression. DNA extraction was performed with the DNeasy Blood & Tissue Kit (#69504; Qiagen). The rearranged TRAD, TRB, and TRG genomic loci were sequenced by Adaptive Biotechnologies (Seattle, WA) as a commercial service. The data were then analyzed with ImmunoSeq online tools (Adaptive Biotechnologies) and custom R scripts. The frequencies of productive and nonproductive TRD, TRG, TRB, and TRA rearrangements were analyzed for both unique and total TRD, TRG, TRB, or TRA sequences obtained from the sorted αβ and γδ T cell subsets. The frequency distributions for individual clonotypes (including TRBV-to-TRBJ pairing and TRAV-to-TRAJ pairing) were analyzed within unique sequences. Diversity indices were calculated and heat-map representations of the frequencies of individual TRAV/TRDV to TRAJ/TRD gene pairs and TRBV-to-TRBJ gene pairs were produced with R software version 4.2.0 (2022-04-22 ucrt) and the R packages Tidyverse (1.3.2) and Immunarch (0.6.9).
High-throughput sequencing (HTS) of the human TRG locus from the gDNA of clonally expanded T cells
T cell clones were obtained by sorting naïve T cells (CD3+CD45RA+CCR7+TCRαβ+TCRγδ−) with a BD FACSAria III SORP cell sorter (Becton Dickinson, San Jose, CA) and DIVA 9.1 software. Cells were sorted, one cell per well, in 96-well plates containing 50 μl of ImmunoCult™-XF T Cell Expansion Medium (StemCell Technologies, REF #10981) supplemented with IL-2 (1 ng/ml) and ImmunoCult™ Human CD3/CD28/CD2 T cell Activator (StemCell Technologies ,REF #10990,1:40) per well. Every 2 days, fresh medium with IL-2 (1 ng/ml) was added to the cells. Clones were visible under a microscope 1 week after sorting. Clones were reactivated every three weeks with ImmunoCult™ Human CD3/CD28/CD2 T cell Activator (StemCell Technologies, #10990,1:80). DNA was extracted from clones with the DNeasy Blood & Tissue Kit (#69504 ; Qiagen). The TRG gene repertoire was investigated by next-generation sequencing (NGS). For library preparation, PCR was performed on 100 ng of genomic DNA with a published protocol (63), but with adaptation of the primers for a NGS version of the assay (table S16). Dual barcoding of the primers made the simultaneous multiplexing of samples possible. After library purification, sequencing was performed on an Illumina™ MiSeq platform. Sequencing data analysis, including demultiplexing, quality control and clonotype assignment, was performed with the Vidjil pipeline (https://www.vidjil.org). IMGT V-QUEST (https://www.imgt.org/IMGT_vquest/analysis) was used for further TRGV and TRGJ annotation and for CDR3 characterization.
TREC levels
Single-joint T cell receptor excision circles (sjTRECs) were quantified by nested quantitative polymerase chain reaction (qPCR), with the primers and standard curve plasmid described by Dion et al. (64). The qPCR protocol was adapted as described in Roux et al. (65) using approximately 500 ng of purified gDNA for each quantification.
Statistics
Analyses were performed with GraphPad Prism V10.1.1 software. Two-tailed Mann–Whitney tests or unpaired t tests were used for single comparisons of independent groups. In the corresponding figures, n.s. indicates not significant; ****P<0.0001, ***P<0.001; **P<0.01; and *P<0.05.
Supplementary Material
Acknowledgments:
We thank the patients and their families for participating in the study. We thank the members of the HGID laboratory for providing excellent comments on the paper. We also wish to thank L. Hadjem from the CIPHE facility, and the members of the CYPS mass cytometry core facility team (Pitié Salpêtrière Hospital) for providing outstanding technical assistance. We thank A. Liston, S. Humblet-Baron, and M. Willemsen (Laboratory of Adaptive Immunology, KU Leuven). We thank the National Facility for Autoimmunity and Serology Profiling at SciLifeLab for their excellent technical support with the protein microarray studies. We thank Qatar Genome and the Qatar Biobank (QBB) management and staff for allowing us to access and analyze QBB/QGP samples and data and the Integrated Genomics Services team of Sidra Medicine for generating and processing WGS data for QBB study participants. We also thank S. Elledge (Brigham and Women’s Hospital and Harvard Medical School) for providing the VirScan phage library used in this study. This research was performed with the UK Biobank resource under application number 40436.
Funding:
This study was supported in part by a grant from the St. Giles Foundation, The Rockefeller University, Institut National de la Santé et de la Recherche Médicale (INSERM), Paris Cité University, the National Center for Research Resources, the National Center for Advancing Sciences of the National Institutes of Health (UL1TR001866), the French National Research Agency (ANR) under the “Investments for the Future” program (ANR-10-IAHU-01), the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID), ANR CARMIL2 (ANR-21-CE15-0034), ANR-RHU program (ANR-21-RHUS-08-COVIFERON), the HORIZON-HLTH-2021-DISEASE-04 program under grant agreement 01057100 (UNDINE), the French Foundation for Medical Research (EQU201903007798), ITMO Cancer of Aviesan and INCa within the framework of the 2021–2030 Cancer Control Strategy (funds administered by Institut National de la Santé et de la Recherche Médicale), the Square Foundation, William E. Ford, General Atlantic’s Chairman and Chief Executive Officer, Gabriel Caillaux, General Atlantic’s Co-President, Managing Director and Head of business in EMEA, and the General Atlantic Foundation, the Qatar National Research Fund (PPM1-1220-150017), Sidra Medicine (SDR400048), the SCOR Corporate Foundation for Science, Institut National de la Santé et de la Recherche Médicale, and the University of Paris. Open Access funding was provided by Rockefeller University. D.L. was supported by a Fonds de Recherche du Québec – Santé Chercheur-Boursier Junior 1 award. T.L.V. and P.B. were supported by the Bettencourt-Schueller Foundation and the MD–PhD program of the Imagine Institute. A.H., T.A.T., and F.A.-M. are supported by institutional funding from the Kuwait Foundation for the Advancement of Sciences. M.Mo. is supported by the ANRS. P.B. was supported by the FRM (EA20170638020). The work by A.Ce., D.E., and N.L. was funded by grants from the Swedish Research Council (2021-03118) and the Göran Gustafsson Foundation (2141 and 2227) to N.L. L.D.N. is supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, MD, USA (grant ZIA AI001222). F.S. (11B5520N, fellow), V.S. (1804523N), I.M. (1805821N), and R.S. (1805518N, 1805523N, senior clinical investigator fellow) are supported by the Fonds Wetenschappelijk Onderzoek - Vlaanderen National Fund for Scientific Research (FWO). E.H. holds the Bank of Montreal Chair for Pediatric Immunology. F.A.Q. was supported by the Ibn Rushd Fellowship Award – King Abdullah University of Science and Technology and King Abdulaziz City for Science and Technology. D.E. was supported by Clas Groschinsky Memorial Foundation (M21116). This study was supported by the VIB Grand Challenge program (Translational science initiative on PID, GC01-C01 for I.M. and R.S.). I.M. and R.S. are members of the European Reference Network for Rare Immunodeficiency, Autoinflammatory and Autoimmune Diseases (Project ID No 739543). I.M. is funded by the FWO Vlaanderen G0B5120N and by C16/18/007 KU Leuven and by the Jeffrey Modell Foundation and is a senior clinical investigator at FWO Vlaanderen. S.E.H. is supported by a K08AI135091 grant, the Burroughs Wellcome Fund, and the CHOP Research Institute.
Footnotes
Competing interests: I.M. received consultancy fees from Boehringer-Ingelheim and research grant from CSL Behring, outside this work and paid to KU Leuven. S.H. declares that she is on the Advisory Board for Horizon Therapeutics without relation to this work. The other authors have no competing interests to declare.
Availability of data and materials: The materials and reagents used are commercially available and nonproprietary. All raw and processed data and biological materials, including immortalized cell lines from patients are available from the the corresponding author via a material transfer agreement with INSERM. The RNA-seq data for sorted primary human thymic T cell subsets have already been published in the BioProject repository under the accession number PRJNA741323 (16). Single-cell RNA sequencing data are available in the MIAME compliant gene expression omnibus database (GEO: GSE243927). Raw data for the immunoblots and qPCR are available from Dryad (66). The entire TCR sequencing dataset is accessible via the adaptive biotechnologies website (67).
References and Notes
- 1.Yui MA, Rothenberg EV, Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol 14, 529–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rast JP, Anderson MK, Strong SJ, Luer C, Litman RT, Litman GW, α, β, γ, and δ T Cell Antigen Receptor Genes Arose Early in Vertebrate Phylogeny. Immunity 6, 1–11 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Krangel MS, Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol 21, 133–139 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreslavsky T, Gleimer M, von Boehmer H, αβ versus γδ lineage choice at the first TCR-controlled checkpoint. Curr Opin Immunol 22, 185–192 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malissen M, Trucy J, Letourneur F, Rebai N, Dunn DE, Fitch FW, Hood L, Malissen B, A T cell clone expresses two T cell receptor α genes but uses one αβ heterodimer for allorecognition and self MHC-restricted antigen recognition. Cell 55, 49–59 (1988). [DOI] [PubMed] [Google Scholar]
- 6.Casanova JL, Romero P, Widmann C, Kourilsky P, Maryanski JL, T cell receptor genes in a series of class I major histocompatibility complex-restricted cytotoxic T lymphocyte clones specific for a Plasmodium berghei nonapeptide: implications for T cell allelic exclusion and antigen-specific repertoire. J Exp Med 174, 1371–83 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borgulya P, Kishi H, Uematsu Y, von Boehmer H, Exclusion and inclusion of α and β T cell receptor alleles. Cell 69, 529–537 (1992). [DOI] [PubMed] [Google Scholar]
- 8.Brady BL, Steinel NC, Bassing CH, Antigen Receptor Allelic Exclusion: An Update and Reappraisal. J Immunol 185, 3801–3808 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Boehmer H, Melchers F, Checkpoints in lymphocyte development and autoimmune disease. Nat Immunol 11, 14–20 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Xing Y, Hogquist KA, T-Cell Tolerance: Central and Peripheral. Cold Spring Harb Perspect Biol 4, a006957 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein L, Kyewski B, Allen PM, Hogquist KA, Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol 14, 377–391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H, Crucial role of the pre-T-cell receptor α gene in development of ap but not γδ T cells. Nature 375, 795–798 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Mancini S, Candéias SM, Fehling HJ, von Boehmer H, Jouvin-Marche E, Marche PN, TCR α-Chain Repertoire in pTα-Deficient Mice Is Diverse and Developmentally Regulated: Implications for Pre-TCR Functions and TCRA Gene Rearrangement. J Immunol 163, 6053–6059 (1999). [PubMed] [Google Scholar]
- 14.Buer J, Aifantis I, DiSanto JP, Fehling HJ, von Boehmer H, Role of different T cell receptors in the development of pre-T cells. J Exp Med 185, 1541–7 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saint-Ruf C, Lechner O, Feinberg J, von Boehmer H, Genomic structure of the human pre-T cell receptor α chain and expression of two mRNA isoforms. European Journal of Immunology 28, 3824–3831 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Sun V, Sharpley M, Kaczor-Urbanowicz KE, Chang P, Montel-Hagen A, Lopez S, Zampieri A, Zhu Y, de Barros SC, Parekh C, Casero D, Banerjee U, Crooks GM, The Metabolic Landscape of Thymic T Cell Development In Vivo and In Vitro. Frontiers in Immunology 12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren Z, Povysil G, Hostyk JA, Cui H, Bhardwaj N, Goldstein DB, ATAV: a comprehensive platform for population-scale genomic analyses. BMC Bioinformatics 22, 149 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott EM, Halees A, Itan Y, Spencer EG, He Y, Azab MA, Gabriel SB, Belkadi A, Boisson B, Abel L, Clark AG, Alkuraya FS, Casanova J-L, Gleeson JG, Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet 48, 1071–1076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O’Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Neale BM, Daly MJ, MacArthur DG, The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D, Projection of an Immunological Self Shadow Within the Thymus by the Aire Protein. Science 298, 1395–1401 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Walter JE, Rosen LB, Csomos K, Rosenberg JM, Mathew D, Keszei M, Ujhazi B, Chen K, Lee YN, Tirosh I, Dobbs K, Al-Herz W, Cowan MJ, Puck J, Bleesing JJ, Grimley MS, Malech H, Ravin SSD, Gennery AR, Abraham RS, Joshi AY, Boyce TG, Butte MJ, Nadeau KC, Balboni I, Sullivan KE, Akhter J, Adeli M, El-Feky RA, El-Ghoneimy DH, Dbaibo G, Wakim R, Azzari C, Palma P, Cancrini C, Capuder K, Condino-Neto A, Costa-Carvalho BT, Oliveira JB, Roifman C, Buchbinder D, Kumanovics A, Franco JL, Niehues T, Schuetz C, Kuijpers T, Yee C, Chou J, Masaad MJ, Geha R, Uzel G, Gelman R, Holland SM, Recher M, Utz PJ, Browne SK, Notarangelo LD, Broad-spectrum antibodies against self-antigens and cytokines in RAG deficiency. J Clin Invest 125, 4135–4148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg JM, Maccari ME, Barzaghi F, Allenspach EJ, Pignata C, Weber G, Torgerson TR, Utz PJ, Bacchetta R, Neutralizing Anti-Cytokine Autoantibodies Against Interferon-α in Immunodysregulation Polyendocrinopathy Enteropathy X-Linked. Frontiers in Immunology 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bastard P, Orlova E, Sozaeva L, Lévy R, James A, Schmitt MM, Ochoa S, Kareva M, Rodina Y, Gervais A, Le Voyer T, Rosain J, Philippot Q, Neehus A-L, Shaw E, Migaud M, Bizien L, Ekwall O, Berg S, Beccuti G, Ghizzoni L, Thiriez G, Pavot A, Goujard C, Frémond M-L, Carter E, Rothenbuhler A, Linglart A, Mignot B, Comte A, Cheikh N, Hermine O, Breivik L, Husebye ES, Humbert S, Rohrlich P, Coaquette A, Vuoto F, Faure K, Mahlaoui N, Kotnik P, Battelino T, Trebušak Podkrajšek K, Kisand K, Ferré EMN, DiMaggio T, Rosen LB, Burbelo PD, McIntyre M, Kann NY, Shcherbina A, Pavlova M, Kolodkina A, Holland SM, Zhang S-Y, Crow YJ, Notarangelo LD, Su HC, Abel L, Anderson MS, Jouanguy E, Neven B, Puel A, Casanova J-L, Lionakis MS, Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J Exp Med 218, e20210554 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Voyer T, Parent AV, Liu X, Cederholm A, Gervais A, Rosain J, Nguyen T, Perez Lorenzo M, Rackaityte E, Rinchai D, Zhang P, Bizien L, Hancioglu G, Ghillani-Dalbin P, Charuel J-L, Philippot Q, Gueye MS, Maglorius Renkilaraj MRL, Ogishi M, Soudée C, Migaud M, Rozenberg F, Momenilandi M, Riller Q, Imberti L, Delmonte OM, Müller G, Keller B, Orrego J, Franco Gallego WA, Rubin T, Emiroglu M, Parvaneh N, Eriksson D, Aranda-Guillen M, Berrios DI, Vong L, Katelaris CH, Mustillo P, Raedler J, Bohlen J, Bengi Celik J, Astudillo C, Winter S, McLean C, Guffroy A, DeRisi JL, Yu D, Miller C, Feng Y, Guichard A, Béziat V, Bustamante J, Pan-Hammarström Q, Zhang Y, Rosen LB, Holland SM, Bosticardo M, Kenney H, Castagnoli R, Slade CA, Boztuğ K, Mahlaoui N, Latour S, Abraham RS, Lougaris V, Hauck F, Sediva A, Atschekzei F, Sogkas G, Poli MC, Slatter MA, Palterer B, Keller MD, Pinzon-Charry A, Sullivan A, Droney L, Suan D, Wong M, Kane A, Hu H, Ma C, Grombiříková H, Ciznar P, Dalal I, Aladjidi N, Hie M, Lazaro E, Franco J, Keles S, Malphettes M, Pasquet M, Maccari ME, Meinhardt A, Ikinciogullari A, Shahrooei M, Celmeli F, Frosk P, Goodnow CC, Gray PE, Belot A, Kuehn HS, Rosenzweig SD, Miyara M, Licciardi F, Servettaz A, Barlogis V, Le Guenno G, Herrmann V-M, Kuijpers T, Ducoux G, Sarrot-Reynauld F, Schuetz C, Cunningham-Rundles C, Rieux-Laucat F, Tangye SG, Sobacchi C, Doffinger R, Warnatz K, Grimbacher B, Fieschi C, Berteloot L, Bryant VL, Trouillet Assant S, Su H, Neven B, Abel L, Zhang Q, Boisson B, Cobat A, Jouanguy E, Kampe O, Bastard P, Roifman CM, Landegren N, Notarangelo LD, Anderson MS, Casanova J-L, Puel A, Autoantibodies against type I IFNs in humans with alternative NF-κB pathway deficiency. Nature 623, 803–813 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramiro AR, Navarro MN, Carreira A, Carrasco YR, de Yébenes VG, Carrillo G, Millán JLS, Rubin B, Toribio ML, Differential Developmental Regulation and Functional Effects on Pre-TCR Surface Expression of Human pTαa and pTαb Spliced Isoforms. The Journal of Immunology 167, 5106–5114 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Arnaud J, Huchenq A, Vernhes MC, Caspar-Bauguil S, Lenfant F, Sancho J, Terhorst C, Rubin B, The interchain disulfide bond between TCR alpha beta heterodimers on human T cells is not required for TCR-CD3 membrane expression and signal transduction. International Immunology 9, 615–626 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Pang SS, Berry R, Chen Z, Kjer-Nielsen L, Perugini MA, King GF, Wang C, Chew SH, La Gruta NL, Williams NK, Beddoe T, Tiganis T, Cowieson NP, Godfrey DI, Purcell AW, Wilce MCJ, McCluskey J, Rossjohn J, The structural basis for autonomous dimerization of the pre-T-cell antigen receptor. Nature 467, 844–848 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Yamasaki S, Ishikawa E, Sakuma M, Ogata K, Sakata-Sogawa K, Hiroshima M, Wiest DL, Tokunaga M, Saito T, Mechanistic basis of pre–T cell receptor–mediated autonomous signaling critical for thymocyte development. Nat Immunol 7, 67–75 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Ishikawa E, Miyake Y, Hara H, Saito T, Yamasaki S, Germ-line elimination of electric charge on pre–T-cell receptor (TCR) impairs autonomous signaling for β-selection and TCR repertoire formation. PNAS 107, 19979–19984 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buer J, Aifantis I, DiSanto JP, Fehling HJ, von Boehmer H, T-cell development in the absence of the pre-T-cell receptor. Immunology Letters 57, 5–8 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Terrence K, Pavlovich CP, Matechak EO, Fowlkes BJ, Premature Expression of T Cell Receptor (Tcr)αβ Suppresses Tcrγδ Gene Rearrangement but Permits Development of γδ Lineage T Cells. J Exp Med 192, 537–548 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruno L, Fehling HJ, von Boehmer H, The αβ T Cell Receptor Can Replace the γδ Receptor in the Development of γδ Lineage Cells. Immunity 5, 343–352 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Aifantis I, Bassing CH, Garbe AI, Sawai K, Alt FW, von Boehmer H, The E delta enhancer controls the generation of CD4- CD8- alphabetaTCR-expressing T cells that can give rise to different lineages of alphabeta T cells. J Exp Med 203, 1543–1550 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ndhlovu ZM, Kamya P, Mewalal N, Kløverpris HN, Nkosi T, Pretorius K, Laher F, Ogunshola F, Chopera D, Shekhar K, Ghebremichael M, Ismail N, Moodley A, Malik A, Leslie A, Goulder PJR, Buus S, Chakraborty A, Dong K, Ndung’u T, Walker BD, Magnitude and Kinetics of CD8+ T Cell Activation during Hyperacute HIV Infection Impact Viral Set Point. Immunity 43, 591–604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosticardo M, Pala F, Calzoni E, Delmonte OM, Dobbs K, Gardner CL, Sacchetti N, Kawai T, Garabedian EK, Draper D, Bergerson JRE, DeRavin SS, Freeman AF, Güngör T, Hartog N, Holland SM, Kohn DB, Malech HL, Markert ML, Weinacht KG, Villa A, Seet CS, Montel-Hagen A, Crooks GM, Notarangelo LD, Artificial thymic organoids represent a reliable tool to study T-cell differentiation in patients with severe T-cell lymphopenia. Blood Advances 4, 2611–2616 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hochstenbach F, Brenner MB, T-cell receptor δ-chain can substitute for α to form a βδ heterodimer. Nature 340, 562–565 (1989). [DOI] [PubMed] [Google Scholar]
- 37.Porto PD, Bruno L, Mattei MG, von Boehmer H, Saint-Ruf C, Cloning and comparative analysis of the human pre-T-cell receptor alpha-chain gene. PNAS 92, 12105–12109 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saint-Ruf C, Ungewiss K, Groettrup M, Bruno L, Fehling HJ, von Boehmer H, Analysis and Expression of a Cloned Pre-T Cell Receptor Gene. Science 266, 1208–1212 (1994). [DOI] [PubMed] [Google Scholar]
- 39.Gros P, Casanova J-L, Reconciling Mouse and Human Immunology at the Altar of Genetics. Annu Rev Immunol, doi: 10.1146/annurev-immunol-101721-065201 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Medetgul-Ernar K, Davis MM, Standing on the shoulders of mice. Immunity 55, 1343–1353 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mancini SJC, Candéias SM, Di Santo JP, Ferrier P, Marche PN, Jouvin-Marche E, TCRA Gene Rearrangement in Immature Thymocytes in Absence of CD3, Pre-TCR, and TCR Signaling1. J Immunol 167, 4485–4493 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Aude-Garcia C, Gallagher M, Marche PN, Jouvin-Marche E, Preferential ADV-AJ association during recombination in the mouse T-cell receptor alpha/delta locus. Immunogenetics 52, 224–230 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Pasqual N, Gallagher M, Aude-Garcia C, Loiodice M, Thuderoz F, Demongeot J, Ceredig R, Marche PN, Jouvin-Marche E, Quantitative and Qualitative Changes in V-J α Rearrangements During Mouse Thymocytes Differentiation : Implication For a Limited T Cell Receptor α Chain Repertoire. Journal of Experimental Medicine 196, 1163–1174 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, Cortes A, Welsh S, Young A, Effingham M, McVean G, Leslie S, Allen N, Donnelly P, Marchini J, The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan T, Rahman M, Ahmed I, Al Ali F, Jithesh PV, Marr N, Human leukocyte antigen class II gene diversity tunes antibody repertoires to common pathogens. Frontiers in Immunology 13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hebbar P, Abubaker JA, Abu-Farha M, Alsmadi O, Elkum N, Alkayal F, John SE, Channanath A, Iqbal R, Pitkaniemi J, Tuomilehto J, Sladek R, Al-Mulla F, Thanaraj TA, Genome-wide landscape establishes novel association signals for metabolic traits in the Arab population. Hum Genet 140, 505–528 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Genin E, Tullio-Pelet A, Begeot F, Lyonnet S, Abel L, Estimating the age of rare disease mutations: the example of Triple-A syndrome. J Med Genet 41, 445–449 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang RJ, Al-Saffar SI, Rogers J, Hahn MW, Human generation times across the past 250,000 years. Science Advances 9, eabm7047 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL, Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H, Twelve years of SAMtools and BCFtools. GigaScience 10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorvaldsdóttir H, Robinson JT, Mesirov JP, Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan K-T, Ding L-W, Sun Q-Y, Lao Z-T, Chien W, Ren X, Xiao J-F, Loh XY, Xu L, Lill M, Mayakonda A, Lin D-C, Yang H, Koeffler HP, Profiling the B/T cell receptor repertoire of lymphocyte derived cell lines. BMC Cancer 18, 940 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heather JM, Spindler MJ, Alonso MH, Shui YI, Millar DG, Johnson DS, Cobbold M, Hata AN, Stitchr: stitching coding TCR nucleotide sequences from V/J/CDR3 information. Nucleic Acids Research 50, e68 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen ISY, Hahn WC, Sharp PA, Weinberg RA, Novina CD, Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conrey PE, Denu L, O’Boyle KC, Rozich I, Green J, Maslanka J, Lubin J-B, Duranova T, Haltzman BL, Gianchetti L, Oldridge DA, De Luna N, Vella LA, Allman D, Spergel JM, Tanes C, Bittinger K, Henrickson SE, Silverman MA, IgA deficiency destabilizes homeostasis toward intestinal microbes and increases systemic immune dysregulation. Science Immunology 8, eade2335 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bastard P, Gervais A, Voyer TL, Rosain J, Philippot Q, Manry J, Michailidis E, Hoffmann H-H, Eto S, Garcia-Prat M, Bizien L, Parra-Martínez A, Yang R, Haljasmägi L, Migaud M, Särekannu K, Maslovskaja J, de Prost N, Tandjaoui-Lambiotte Y, Luyt C-E, Amador-Borrero B, Gaudet A, Poissy J, Morel P, Richard P, Cognasse F, Troya J, Trouillet-Assant S, Belot A, Saker K, Garçon P, Rivière JG, Lagier J-C, Gentile S, Rosen LB, Shaw E, Morio T, Tanaka J, Dalmau D, Tharaux P-L, Sene D, Stepanian A, Megarbane B, Triantafyllia V, Fekkar A, Heath JR, Franco JL, Anaya J-M, Solé-Violán J, Imberti L, Biondi A, Bonfanti P, Castagnoli R, Delmonte OM, Zhang Y, Snow AL, Holland SM, Biggs C, Moncada-Vélez M, Arias AA, Lorenzo L, Boucherit S, Coulibaly B, Anglicheau D, Planas AM, Haerynck F, Duvlis S, Nussbaum RL, Ozcelik T, Keles S, Bousfiha AA, Bakkouri JE, Ramirez-Santana C, Paul S, Pan-Hammarström Q, Hammarström L, Dupont A, Kurolap A, Metz CN, Aiuti A, Casari G, Lampasona V, Ciceri F, Barreiros LA, Dominguez-Garrido E, Vidigal M, Zatz M, van de Beek D, Sahanic S, Tancevski I, Stepanovskyy Y, Boyarchuk O, Nukui Y, Tsumura M, Vidaur L, Tangye SG, Burrel S, Duffy D, Quintana-Murci L, Klocperk A, Kann NY, Shcherbina A, Lau Y-L, Leung D, Coulongeat M, Marlet J, Koning R, Reyes LF, Chauvineau-Grenier A, Venet F, Monneret G, Nussenzweig MC, Arrestier R, Boudhabhay I, Baris-Feldman H, Hagin D, Wauters J, Meyts I, Dyer AH, Kennelly SP, Bourke NM, Halwani R, Sharif-Askari NS, Dorgham K, Sallette J, Sedkaoui SM, AlKhater S, Rigo-Bonnin R, Morandeira F, Roussel L, Vinh DC, Ostrowski SR, Condino-Neto A, Prando C, Bonradenko A, Spaan AN, Gilardin L, Fellay J, Lyonnet S, Bilguvar K, Lifton RP, Mane S, H. Lab§, C. Clinicians§, C.-S. Clinicians§, N. I. R. to C. Group§, N.-C. S. Group§, D. Chge§, D. B. D. Study§, S. J. Hospital, S. C. I. Group§, F. C. C. S. Group§, I. COVID-Group§, T. M. I. Consortium§, C.-C. Cohort§, A. U. Covid-19, B. Investigators§, C. H. G. Effort§, C. Cohort§, 3C-Dijon Study§, C. Health-Care§, E. du S. study Group§, Anderson MS, Boisson B, Béziat V, Zhang S-Y, Vandreakos E, Hermine O, Pujol A, Peterson P, Mogensen TH, Rowen L, Mond J, Debette S, de Lamballerie X, Duval X, Mentré F, Zins M, Soler-Palacin P, Colobran R, Gorochov G, Solanich X, Susen S, Martinez-Picado J, Raoult D, Vasse M, Gregersen PK, Piemonti L, Rodríguez-Gallego C, Notarangelo LD, Su HC, Kisand K, Okada S, Puel A, Jouanguy E, Rice CM, Tiberghien P, Zhang Q, Cobat A, Abel L, Casanova J-L, Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Science Immunology 6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shannon CE, The mathematical theory of communication. 1963. MD Comput 14, 306–317 (1997). [PubMed] [Google Scholar]
- 60.Béziat V, Rapaport F, Hu J, Titeux M, Bonnet des Claustres M, Bourgey M, Griffin H, Bandet É, Ma CS, Sherkat R, Rokni-Zadeh H, Louis DM, Changi-Ashtiani M, Delmonte OM, Fukushima T, Habib T, Guennoun A, Khan T, Bender N, Rahman M, About F, Yang R, Rao G, Rouzaud C, Li J, Shearer D, Balogh K, Al Ali F, Ata M, Dabiri S, Momenilandi M, Nammour J, Alyanakian M-A, Leruez-Ville M, Guenat D, Materna M, Marcot L, Vladikine N, Soret C, Vahidnezhad H, Youssefian L, Saeidian AH, Uitto J, Catherinot É, Navabi SS, Zarhrate M, Woodley DT, Jeljeli M, Abraham T, Belkaya S, Lorenzo L, Rosain J, Bayat M, Lanternier F, Lortholary O, Zakavi F, Gros P, Orth G, Abel L, Prétet J-L, Fraitag S, Jouanguy E, Davis MM, Tangye SG, Notarangelo LD, Marr N, Waterboer T, Langlais D, Doorbar J, Hovnanian A, Christensen N, Bossuyt X, Shahrooei M, Casanova J-L, Humans with inherited T cell CD28 deficiency are susceptible to skin papillomaviruses but are otherwise healthy. Cell 184, 3812–3828.e30 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seet CS, He C, Bethune MT, Li S, Chick B, Gschweng EH, Zhu Y, Kim K, Kohn DB, Baltimore D, Crooks GM, Montel-Hagen A, Generation of mature T cells from human hematopoietic stem and progenitor cells in artificial thymic organoids. Nat Methods 14, 521–530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luche H, Nageswara Rao T, Kumar S, Tasdogan A, Beckel F, Blum C, Martins VC, Rodewald H-R, Fehling HJ, In vivo fate mapping identifies pre-TCRα expression as an intra- and extrathymic, but not prethymic, marker of T lymphopoiesis. J Exp Med 210, 699–714 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Armand M, Derrieux C, Beldjord K, Wabeke T, Lenze D, Boone E, Bruggemann M, Evans PAS, Gameiro P, Hummel M, Villarese P, Groenen PJTA, Langerak AW, Macintyre EA, Davi F, A New and Simple TRG Multiplex PCR Assay for Assessment of T-cell Clonality: A Comparative Study from the EuroClonality Consortium. Hemasphere 3, e255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dion M-L, Sékaly R-P, Cheynier R, “Estimating Thymic Function Through Quantification of T-Cell Receptor Excision Circles” in Immunological Tolerance: Methods and Protocols, Fairchild PJ, Ed. (Humana Press, Totowa, NJ, 2007; 10.1007/978-1-59745-395-0_12)Methods in Molecular Biology™, pp. 197–213. [DOI] [PubMed] [Google Scholar]
- 65.Roux HM, Marouf A, Dutrieux J, Charmeteau-De Muylder B, Figueiredo-Morgado S, Avettand-Fenoel V, Cuvelier P, Naudin C, Bouaziz F, Geri G, Couëdel-Courteille A, Squara P, Marullo S, Cheynier R, Genetically determined thymic function affects strength and duration of immune response in COVID patients with pneumonia. Science Advances 9, eadh7969 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Materna M, Delmonte OM, Bosticardo M, Momenilandi M, Conrey P, Charmeteau-De Muylder B, Bravetti C, Bellworthy R, Cederholm A, Staels F, Ganoza CA, Darko S, Sayed S, Le Floc’h C, Ogishi M, Rinchai D, Guenoun A, Bolze A, Khan T, Gervais A, Krüger R, Völler M, Palterer B, Sadeghi-Shabestari M, Langlois de Septenville A, Schramm CA, Shah S, Tello-Cajiao J, Pala F, Amini K, Campos JS, Santana Lima N, Eriksson D, Lévy R, Seeleuthner Y, Jyonouchi S, Ata M, Al Ali F, Deswarte C, Pereira A, Mégret J, Le Voyer T, Bastard P, Berteloot L, Dussiot M, Vladikine N, Cardenas PP, Jouanguy E, Alqahtani M, Hasan A, Thanaraj TA, Rosain J, Al Qureshah F, Sabato V, Alyanakian MA, Leruez-Ville M, Rosenberg F, Haddad E, Regueiro JR, Toribio ML, Kelsen JR, Salehi M, Nassiry S, Torabizadeh M, Rokni-Zadeh H, Changi-Ashtiani M, Vatandoost N, Moravej H, Akrami SM, Mazloomrezaei M, Cobat A, Meyts I, Etsushi T, Nishimura M, Moriya K, Mizukami T, Imai K, Abel L, Malissen B, Al-Mulla F, Alkuraya FS, Parvaneh N, von Bernuth H, Beetz C, Davi F, Douek DC, Cheynier R, Langlais D, Landegren N, Marr N, Morio T, Shahrooei M, Schrijvers R, Henrickson SE, Luche H, Notarangelo LD, Casanova J-L, Béziat V, Data from: From rare to common PTCRA variants: Immunopathological range of human pre-TCRα deficiencies, Dryad Digital Repository (2024); 10.5061/dryad.9zw3r22m8. [DOI] [Google Scholar]
- 67.Materna M, Delmonte OM, Bosticardo M, Momenilandi M, Conrey P, Charmeteau-De Muylder B, Bravetti C, Bellworthy R, Cederholm A, Staels F, Ganoza CA, Darko S, Sayed S, Le Floc’h C, Ogishi M, Rinchai D, Guenoun A, Bolze A, Khan T, Gervais A, Krüger R, Völler M, Palterer B, Sadeghi-Shabestari M, Langlois de Septenville A, Schramm CA, Shah S, Tello-Cajiao J, Pala F, Amini K, Campos JS, Santana Lima N, Eriksson D, Lévy R, Seeleuthner Y, Jyonouchi S, Ata M, Al Ali F, Deswarte C, Pereira A, Mégret J, Le Voyer T, Bastard P, Berteloot L, Dussiot M, Vladikine N, Cardenas PP, Jouanguy E, Alqahtani M, Hasan A, Thanaraj TA, Rosain J, Al Qureshah F, Sabato V, Alyanakian MA, Leruez-Ville M, Rosenberg F, Haddad E, Regueiro JR, Toribio ML, Kelsen JR, Salehi M, Nassiry S, Torabizadeh M, Rokni-Zadeh H, Changi-Ashtiani M, Vatandoost N, Moravej H, Akrami SM, Mazloomrezaei M, Cobat A, Meyts I, Etsushi T, Nishimura M, Moriya K, Mizukami T, Imai K, Abel L, Malissen B, Al-Mulla F, Alkuraya FS, Parvaneh N, von Bernuth H, Beetz C, Davi F, Douek DC, Cheynier R, Langlais D, Landegren N, Marr N, Morio T, Shahrooei M, Schrijvers R, Henrickson SE, Luche H, Notarangelo LD, Casanova J-L, Béziat V, Data from: From rare to common PTCRA variants: Immunopathological range of human pre-TCRα deficiencies, Adaptive Biotechnologies (2024); 10.21417/MM2024S. [DOI] [Google Scholar]
- 68.Zhang P, Bigio B, Rapaport F, Zhang S-Y, Casanova J-L, Abel L, Boisson B, Itan Y, PopViz: a webserver for visualizing minor allele frequencies and damage prediction scores of human genetic variations. Bioinformatics 34, 4307–4309 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Itan Y, Shang L, Boisson B, Patin E, Bolze A, Moncada-Vélez M, Scott E, Ciancanelli MJ, Lafaille FG, Markle JG, Martinez-Barricarte R, de Jong SJ, Kong X-F, Nitschke P, Belkadi A, Bustamante J, Puel A, Boisson-Dupuis S, Stenson PD, Gleeson JG, Cooper DN, Quintana-Murci L, Claverie J-M, Zhang S-Y, Abel L, Casanova J-L, The human gene damage index as a gene-level approach to prioritizing exome variants. PNAS 112, 13615–13620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rapaport F, Boisson B, Gregor A, Béziat V, Boisson-Dupuis S, Bustamante J, Jouanguy E, Puel A, Rosain J, Zhang Q, Zhang S-Y, Gleeson JG, Quintana-Murci L, Casanova J-L, Abel L, Patin E, Negative selection on human genes underlying inborn errors depends on disease outcome and both the mode and mechanism of inheritance. PNAS 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Desmet F-O, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C, Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37, e67–e67 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Itan Y, Shang L, Boisson B, Ciancanelli MJ, Markle JG, Martinez-Barricarte R, Scott E, Shah I, Stenson PD, Gleeson J, Cooper DN, Quintana-Murci L, Zhang S-Y, Abel L, Casanova J-L, The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Meth 13, 109–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J, A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann H-H, Zhang Y, Dorgham K, Philippot Q, Rosain J, Béziat V, Manry J, Shaw E, Haljasmägi L, Peterson P, Lorenzo L, Bizien L, Trouillet-Assant S, Dobbs K, de Jesus AA, Belot A, Kallaste A, Catherinot E, Tandjaoui-Lambiotte Y, Pen JL, Kerner G, Bigio B, Seeleuthner Y, Yang R, Bolze A, Spaan AN, Delmonte OM, Abers MS, Aiuti A, Casari G, Lampasona V, Piemonti L, Ciceri F, Bilguvar K, Lifton RP, Vasse M, Smadja DM, Migaud M, Hadjadj J, Terrier B, Duffy D, Quintana-Murci L, van de Beek D, Roussel L, Vinh DC, Tangye SG, Haerynck F, Dalmau D, Martinez-Picado J, Brodin P, Nussenzweig MC, Boisson-Dupuis S, Rodríguez-Gallego C, Vogt G, Mogensen TH, Oler AJ, Gu J, Burbelo PD, Cohen JI, Biondi A, Bettini LR, D’Angio M, Bonfanti P, Rossignol P, Mayaux J, Rieux-Laucat F, Husebye ES, Fusco F, Ursini MV, Imberti L, Sottini A, Paghera S, Quiros-Roldan E, Rossi C, Castagnoli R, Montagna D, Licari A, Marseglia GL, Duval X, Ghosn J, H. Lab§, N.-U. I. R. to C. Group§, C. Clinicians§, C.-S. Clinicians§, I. C. Group§, F. C. C. S. Group§, T. M. I. Consortium§, C.-C. Cohort§, A. U. C.-19 Biobank§, C. H. G. Effort§, Tsang JS, Goldbach-Mansky R, Kisand K, Lionakis MS, Puel A, Zhang S-Y, Holland SM, Gorochov G, Jouanguy E, Rice CM, Cobat A, Notarangelo LD, Abel L, Su HC, Casanova J-L, Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pozzetto B, Mogensen KE, Tovey MG, Gresser I, Characteristics of Autoantibodies to Human Interferon in a Patient with Varicella-Zoster Disease. The Journal of Infectious Diseases 150, 707–713 (1984). [DOI] [PubMed] [Google Scholar]
- 76.Busnadiego I, Abela IA, Frey PM, Hofmaenner DA, Scheier TC, Schuepbach RA, Buehler PK, Brugger SD, Hale BG, Critically ill COVID-19 patients with neutralizing autoantibodies against type I interferons have increased risk of herpesvirus disease. PLOS Biology 20, e3001709 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mathian A, Breillat P, Dorgham K, Bastard P, Charre C, Lhote R, Quentric P, Moyon Q, Mariaggi A-A, Mouries-Martin S, Mellot C, Anna F, Haroche J, Cohen-Aubart F, Sterlin D, Zahr N, Gervais A, Voyer TL, Bizien L, Amiot Q, Pha M, Hié M, Chasset F, Yssel H, Miyara M, Charneau P, Ghillani-Dalbin P, Casanova J-L, Rozenberg F, Amoura Z, Gorochov G, Lower disease activity but higher risk of severe COVID-19 and herpes zoster in patients with systemic lupus erythematosus with pre-existing autoantibodies neutralising IFN-α. Annals of the Rheumatic Diseases 81, 1695–1703 (2022). [DOI] [PubMed] [Google Scholar]
- 78.Kaji K, Fertig N, Medsger TA Jr., Satoh T, Hoshino K, Hamaguchi Y, Hasegawa M, Lucas M, Schnure A, Ogawa F, Sato S, Takehara K, Fujimoto M, Kuwana M, Autoantibodies to RuvBL1 and RuvBL2: A Novel Systemic Sclerosis–Related Antibody Associated With Diffuse Cutaneous and Skeletal Muscle Involvement. Arthritis Care & Research 66, 575–584 (2014). [DOI] [PubMed] [Google Scholar]
- 79.Franceschini F, Cavazzana I, Anti-Ro/SSA and La/SSB antibodies. Autoimmunity 38, 55–63 (2005). [DOI] [PubMed] [Google Scholar]
- 80.Kasimiotis H, Myers MA, Argentaro A, Mertin S, Fida S, Ferraro T, Olsson J, Rowley MJ, Harley VR, Sex-determining region Y-related protein SOX13 is a diabetes autoantigen expressed in pancreatic islets. Diabetes 49, 555–561 (2000). [DOI] [PubMed] [Google Scholar]
- 81.Kreslavsky T, Garbe AI, Krueger A, von Boehmer H, T cell receptor–instructed αβ versus γδ lineage commitment revealed by single-cell analysis. J Exp Med 205, 1173–1186 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van Coppernolle S, Vanhee S, Verstichel G, Snauwaert S, van der Spek A, Velghe I, Sinnesael M, Heemskerk MH, Taghon T, Leclercq G, Plum J, Langerak AW, Kerre T, Vandekerckhove B, Notch induces human T-cell receptor γδ+ thymocytes to differentiate along a parallel, highly proliferative and bipotent CD4 CD8 double-positive pathway. Leukemia 26, 127–138 (2012). [DOI] [PubMed] [Google Scholar]
- 83.Garman RD, Doherty PJ, Raulet DH, Diversity, rearrangement, and expression of murine T cell gamma genes. Cell 45, 733–742 (1986). [DOI] [PubMed] [Google Scholar]
- 84.Ishida I, Verbeek S, Bonneville M, Itohara S, Berns A, Tonegawa S, T-cell receptor gamma delta and gamma transgenic mice suggest a role of a gamma gene silencer in the generation of alpha beta T cells. Proceedings of the National Academy of Sciences 87, 3067–3071 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kang J, Fehling HJ, Laplace C, Malissen M, Cado D, Raulet DH, T Cell Receptor γ Gene Regulatory Sequences Prevent the Function of a Novel TCRγ/pTα Pre–T Cell Receptor. Immunity 8, 713–721 (1998). [DOI] [PubMed] [Google Scholar]
- 86.John SE, Antony D, Eaaswarkhanth M, Hebbar P, Channanath AM, Thomas D, Devarajan S, Tuomilehto J, Al-Mulla F, Alsmadi O, Thanaraj TA, Assessment of coding region variants in Kuwaiti population: implications for medical genetics and population genomics. Sci Rep 8, 16583 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.