SUMMARY
Although social interactions are known to drive pathogen transmission, the contributions of socially transmissible host-associated mutualists and commensals to host health and disease remain poorly explored. We use the concept of the social microbiome—the microbial metacommunity of a social network of hosts—to analyze the implications of social microbial transmission for host health and disease. We investigate the contributions of socially transmissible microbes to both eco-evolutionary microbiome community processes (colonization resistance, the evolution of virulence, and reactions to ecological disturbance) and microbial transmission-based processes (transmission of microbes with metabolic and immune effects, inter-specific transmission, transmission of antibiotic-resistant microbes, and transmission of viruses). We consider the implications of social microbial transmission for communicable and non-communicable diseases and evaluate the importance of a socially transmissible component underlying canonically non-communicable diseases. The social transmission of mutualists and commensals may play a significant, under-appreciated role in the social determinants of health and may act as a hidden force in social evolution.
INTRODUCTION
Eukaryotic life originated from prokaryotic life, evolved amidst microbiomes, and now harbors distinct host-associated microbiomes.1 These microbes (collectively, the microbiota) shape the phenotypes of their hosts, influencing energy metabolism,2,3 immunity,4,5 and even psychological development and behavior, including social behavior.6,7,8 Furthermore, the host’s social context, interactions, and relationships influence the composition of its microbiome, and several exciting discoveries have revealed that endogenous microbes are readily transmissible between hosts through social interactions.9–17 In this regard, socially transmissible microbes may be an under-appreciated aspect of the social determinants of health11,18 and may contribute to both the causes and consequences of variation in host sociality and health. Much research has focused on the costs of enhanced pathogen dispersal in social networks and the rather more aggressive transmission strategies of pathogens.19 However, comparatively less is known about the social transmission of mutualistic and commensal microbes and whether social animals derive any significant benefits from such social microbial transmission. Indeed, although social evolution may have driven the emergence of pathogen avoidance and control behaviors,19–21 it has also been suggested that social behaviors and social structures supporting the transmission of commensal and mutualistic microbes could have emerged over the course of social evolution.22–26 In other words, given that both pathogenic and non-pathogenic microbes exert substantial effects on host physiology and are socially transmissible, we believe that it is time to move beyond the focus on pathogen transmission. Here, we examine the implications of the social transmission of commensals and mutualists for host health and disease and also consider the role of such transmission in social evolution.
We first provide a synthesis of the social transmission of microbes through the lens of the social microbiome concept27 (i.e., the microbial metacommunity of an animal social network, together with its genes and gene products; Figure 1A; Table 1). We focus primarily on the gut microbiome because its associations with host health are better characterized, but we also discuss the microbiomes of other body sites. Throughout this Perspective, we refer to five levels of social-ecological forces that shape the social microbiome (Figure 1A), including microbial exchanges occurring at the inter-host level (level 1), the network level (level 2), the inter-group level (level 3), the species level (level 4), and the inter-species level (level 5). We then define two general dimensions under which various relationships between social microbial transmission and host health and disease can be analyzed (Figure 1B). One dimension can be conceptualized as a set of broader eco-evolutionary processes occurring at the level of complex, whole-microbiome communities and entails processes such as (1) colonization resistance, (2) the evolution of virulence and microbial transmissibility, and (3) the reactions of the microbiome to ecological disturbance. The second dimension can be conceptualized as the dispersal of specific microbes between hosts and entails processes such as (1) the transmission of microbes with appreciable metabolic and immunological effects, (2) inter-specific transmission and zoonotic spillovers, (3) the transmission of antibiotic-resistant microbes and microbial genes, and (4) the transmission of viruses from the host virome. We describe a range of effects, outcomes, and predictions pertaining to these categories (Figure 1B) as well as empirical approaches to test those predictions. Finally, we analyze the role of the social transmission of microbes in relation to communicable diseases (infectious diseases caused by pathogenic microorganisms) and non-communicable diseases (chronic diseases typically attributed to host factors, such as cardiovascular diseases, autoimmune diseases, metabolic diseases, atopic diseases, neurological conditions, and cancers). We evaluate the possibility that non-communicable diseases entail a communicable component by virtue of the social transmission of microbes.28 Depending on the nature of the host-microbe interactions and other host factors, this socially transmissible component could either mitigate or exacerbate disease risk and severity.
Figure 1. Social-ecological forces shaping the social microbiome and the implications of socially transmitted microbes for host health and disease.
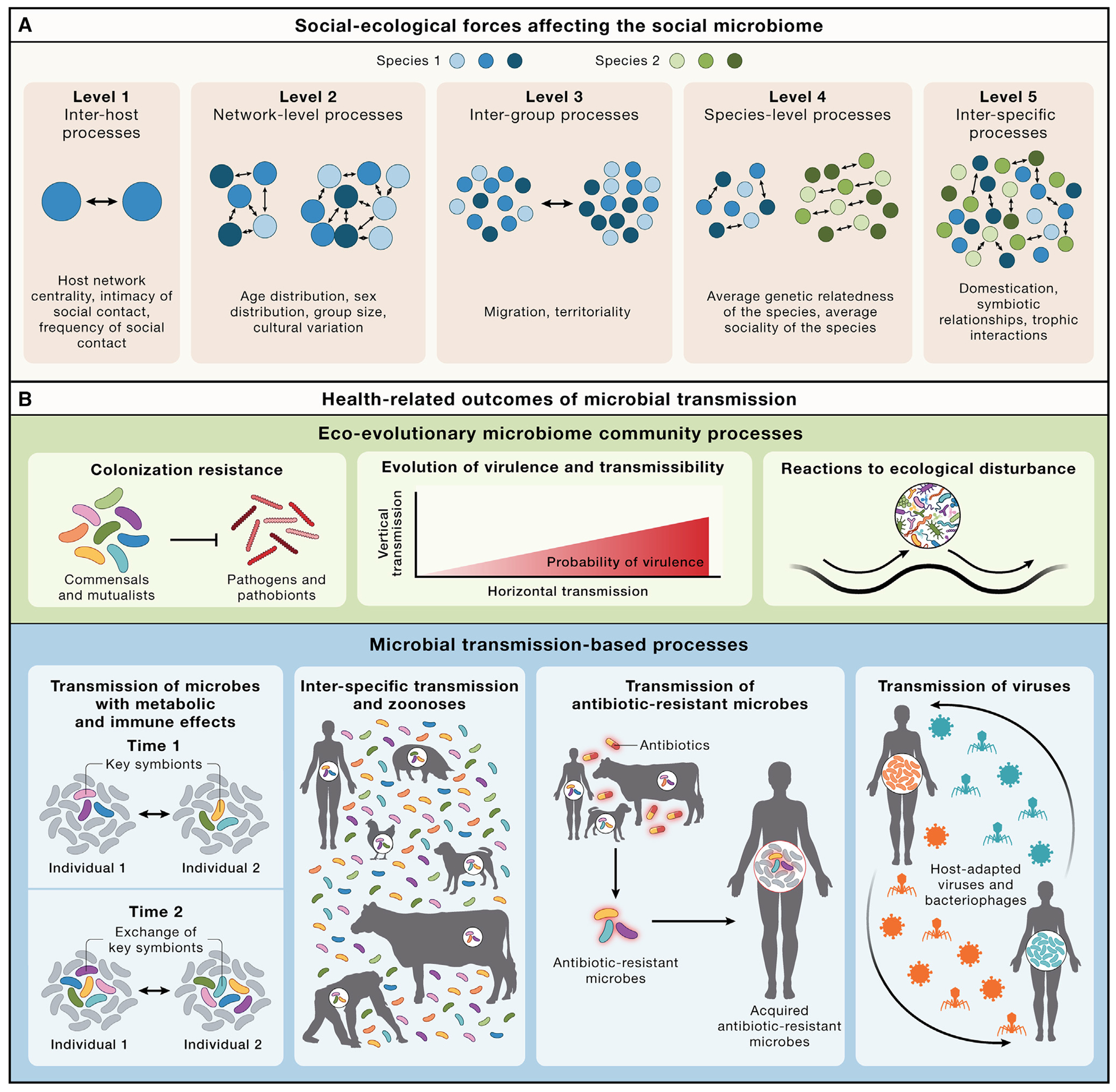
(A) Processes at different social-ecological scales influence the social microbiome. Blue and green circles denote unique host individuals. These processes need not be mutually exclusive.
(B) Health-relevant processes predicted to be affected by the social transmission of microbes. These can be categorized under two broad dimensions: eco-evolutionary microbiome community processes, and microbial transmission-based processes. The visualizations of these processes are simplified for convenience, and greater nuance is provided in the text. These processes need not be mutually exclusive.
Table 1.
Processes at different scales influencing the social microbiome
| Levela | Factors | Host taxa | Parameters | General effects | Examples |
|---|---|---|---|---|---|
| Level 1: Inter-host processes | Dyadic factors that affect transmission between two hosts | Non-human animals | Social associations | Gut microbial similarity is highest amongst pairs of hosts with the strongest social associations. | Studies in mice and non-human primates show that the strength of social ties predicts the extent to which individuals share microbes.12–14 |
| Humans | Intimacy and frequency of social contact and host network position | Microbial exchange between social partners increases with the strength of the social relationships. | The extent of social transmission of microbes between social partners depends on the type of relationship (e.g., close partners share more microbes than friends) and the hos’s centrality in the social network.10,15,17,29 More socially central individuals have microbiomes that more closely resemble the social microbiome.29 | ||
| Social norms | Cultural and social norms pertaining to pairwise interactions amongst hosts can influence microbial transmission. | Cultural differences in the types of social greetings, the level of acceptable physical proximity to another individual, customs such as co-sleeping with infants, and permissible forms of sexual behavior should affect rates of social microbial transmission. Many ritual practices likely shape the microbiome. In Fiji, attendees at community meetings routinely drink Kava from the same bowl.30 Such practices likely transfer oral microbes amongst individuals. |
|||
|
| |||||
| Level 2: Network-level processes | Network-level factors that affect microbial transmission between multiple interacting hosts within a group | Non-human animals | Age and sex distributions of group members | The degree of modularity in the microbial transmission landscape within host populations can be shaped by network assortment (i.e., the fact that animals exhibit different social behaviors depending on factors such as age and sex). | Composition of the microbiome co-varies with host age and sex.31 Age- or sex-based differences in the microbiome may result in part from age- or sex-biased modularity in the social microbiome. |
| Humans | Cultural practices | Cultural practices can influence microbial composition and transmission in human social networks in ways that differ systematically between cultures. | The Tsimane forager-horticulturalists of Bolivia feed infants a specialized fermented drink inoculated with oral microbes, and infants fed this drink show distinct microbiome compositions compared with infants not fed this drink.32 Circumcision reduces male susceptibility to sexually transmitted diseases and the likelihood of transmitting disease to sexual partners.33 Comparisons of the penile microbiota for circumcised and uncircumcised males suggest that circumcision substantially alters local microbial ecology.34 | ||
|
| |||||
| Level 3: Inter-group processes | Host population-level factors that influence microbial dispersal across distantly connected components of the host social network (e.g., distinct social groups) | Non-human animals | Frequency and intimacy of social contact between groups | More frequent contact between social groups (e.g., through home range overlap or migration) will facilitate the spread of microbes between those groups. For instance, microbial exchange should be more likely between bonobo social groups (which have overlapping territories) than between chimpanzee social groups (which generally do not have overlapping territories). | The microbiomes of male baboons migrating from one group to another undergo gradual remodeling to resemble the social microbiome of the receiving group while still retaining a signature of the natal group.35 |
| Humans | Travel and immigration contribute to the sharing of microbes between groups. | Travel facilitates the spread of microbes, including pathogens such as SARS-CoV-2 variants.36 Individuals residing in different villages show some degree of microbiome strain sharing across villages.29 |
|||
|
| |||||
| Level 4: Species-level processes | Species-level factors that influence the social transmission of microbes | Humans and non-human animalsb | Average sociality of the species, the nature of the breeding system, and the average genetic relatedness amongst group members | Highly social species (e.g., spotted hyaenas, lions, chimpanzees, and honeybees) are expected to experience higher rates of microbial transmission. | Honeybees—which are eusocial—acquire gut microbes through social interactions,37 whereas Drosophila, which are non-eusocial, appear to acquire gut microbes through diet and other environmental sources.38 |
|
| |||||
| Level 5: Inter-specific processes | Between-species factors that influence microbial transmission amongst sympatric host species living in the same area | Humans and non-human animalsc | Trophic interactions, symbiotic interactions and inter-specific social behaviors | Antagonistic interactions (e.g., predation, herbivory, and parasitism) spread pathogenic, commensal, and mutualistic microbes.39 | In North American mammals, gut microbiome similarity amongst predator and prey species is elevated relative to the similarity expected based on host evolutionary divergence.40 |
| Sympatric host species exchange microbes through sharing environments and exploiting overlapping niches. | Sympatric populations of chimpanzees and gorillas have been shown to harbor convergent gut microbes relative to allopatric populations, sharing >50% more bacterial lineages on average.41 | ||||
| Factors affecting interactions between humans and domesticated animals | Domestication provides opportunities for microbial transmission between human and domesticated animal microbiomes. | Humans and dogs exchange microbes with one another,42,43 and humans exchange microbes with a range of agricultural animals, including pigs and poultry.44,45 | |||
The five levels need not be mutually exclusive, and some processes can be considered at multiple levels.
Humans and non-human animals are grouped together because level 4 captures factors that intrinsically distinguish host species from one another.
Humans and non-human animals are grouped together because the kinds of interactions described for humans, by definition of level 5, require microbial exchanges with non-human animals.
MICROBIAL TRANSMISSION IN THE SOCIAL MICROBIOME
Animal gut microbiomes are highly dynamic ecosystems that display considerable variation within and between hosts over time.46 Microbial composition is shaped by environmental influences, such as diet and the dispersal of microbes from external sources, as well as factors intrinsic to hosts such as physiology and genetics2,46 (Figure 2; Table 2). Metacommunity theory supplies a useful framework for understanding these dynamics.47–50 Under this framework, each host’s microbiome is an “island,” a distinct community shaped by ecological processes operating both within hosts (including microbe-microbe interactions and host-mediated selection on microbes) and between hosts (including social transmission and selection imposed by the external environment) (Figure 2; Table 2). In this regard, the social microbiome refers to the microbial metacommunity of an animal social network (as well as its genes and gene products), wherein networks of host islands can form distinct biological archipelagos27 (Figure 1A; Table 1). Moreover, different social groups of the same host species inhabiting similar ecologies often have distinct social microbiomes, a phenomenon that has been observed across the animal kingdom, including humans.27 Each microbiome is embedded in a social-ecological network and is connected to other microbiomes by microbial transmission. The social transmission of microbes can be analyzed at five distinct, but not mutually exclusive, levels of increasing ecological scale. These range from inter-host to inter-specific interactions that can influence the nature and frequency of microbial exchange between hosts27 (Figure 1A; Table 1). Importantly, the social transmission of microbes has been shown to covary with and reflect host social networks.9,10,13–15,17,29,51 Indeed, socially transmitted microbes appear to be detectable even for second-order interactions in humans.29 For example, if A interacts with B, and B interacts with C, then C bears a microbial trace of the commensals and mutualists from A that C acquired via interactions with B.29 This phenomenon has previously been observed for pathogens such as Mycobacterium tuberculosis.52 However, if such patterns also characterize the transmission of commensals and mutualists, then it would suggest that an individual’s extended social network affects microbiome composition through intermediating social partners that serve as reservoirs of microbes from other parts of the social network.
Figure 2. A metacommunity framework for microbiome assembly and microbial transmission in the social microbiome.
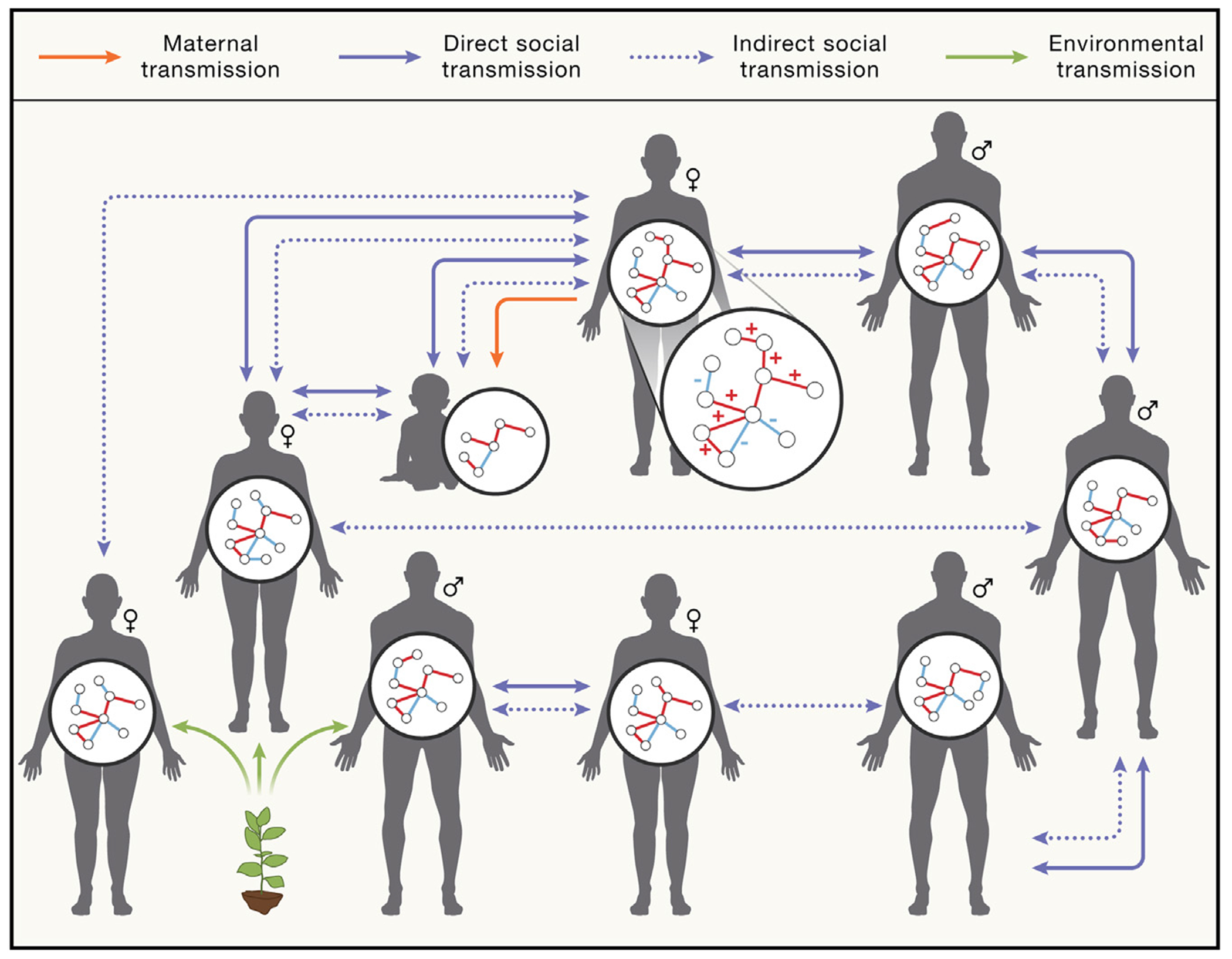
The microbiome is assembled in a host through microbial intra-community interactions (microbe-microbe interactions) occurring in a selective context mediated by host physiology and genetics (host-microbe interactions) and ultimately defined by transmission. Microbe-microbe interactions are visualized in the microbiome inset, where “+” indicates cooperative interactions (e.g., cross-feeding) and “−” indicates antagonistic interactions (e.g., competition). Social transmission can occur via independent pathways that create distinct ecological landscapes for microbes across hosts: direct social transmission (solid purple arrows) and indirect social transmission (dashed purple arrows). Direct social transmission involves microbial exchanges between microbiotas enabled by social contact. Indirect social transmission increases microbiome similarity between hosts that overlap in geographical space, though the hosts themselves may not come into direct contact. Maternal transmission (orange arrow) from body sites including the vagina, gut, and skin is an important form of social transmission that drives early microbiome assembly in infants. The infant microbiome shown here has fewer nodes and edges, representing that it is simpler and less diverse at this developmental stage. The infant microbiome is strongly shaped by maternal transmission, but also receives both direct and indirect social microbial transmission. In addition to social transmission, some members of the microbiome can be acquired directly from the environment independent of social transmission (green arrows), as exemplified by microbial transmission from the soil microbiome to the gut.53 Environmentally acquired microbes are typically generalists and are not strictly adapted to living within animal hosts. They comprise a minority of host-associated microbes in mammals and many other animal species.
Table 2.
Social transmission modes and their effects in humans and other animals
| Transmission mode | Host taxa | Transmission effects on microbiome composition and host health |
|---|---|---|
| Parental transmission | Non-human animals | Multicellular organisms acquire microorganisms beginning in early life.50 In many host taxa, these initial microbes are acquired from the mother. For example, egg-laying animals such as toads coat their eggs with probiotic secretions that serve as both protective layers and initial microbial inocula for the offspring,54 whereas many insects transmit their endosymbionts directly through their eggs.55 Young birds assemble their initial microbiomes in part from their parents through close contact such as regurgitation.56 In most mammals, offspring are initially colonized by microbes from the maternal vagina and distal gut during parturition.57,58 |
| Humans | Humans acquire their initial microbial populations via maternal exposure during parturition. Infants born vaginally are initially colonized by microbes from the maternal vaginal and gastrointestinal tracts, and infants born via caesarean section are first colonized by microbes from human skin and hospital surfaces.57,58 Early colonizers acquired from the mother may influence the trajectory of subsequent microbiome assembly and host development. For example, the founding microbial communities facilitate maturation of the immune system.59 Disruption of maternal transmission (e.g., via caesarean delivery) may contribute to disease risk,60 although the long-term consequences of birth mode for human health remain debated. | |
|
| ||
| Direct social transmission (mediated by social contact) | Non-human animals | In mammals, the signature of maternal transmission gradually yields to non-maternal direct social transmission. For instance, in wild mice, young (but weaned) individuals initially harbor microbiomes similar to their mothers, but these maternal signatures are gradually replaced by microbial transmission from other individuals in the social network as the animal matures.61 In non-human primates, grooming intensity predicts the degree of sharing of microbes.14 |
| Humans | Direct social interactions function as transmission routes for a range of microbes, especially anaerobic gut microbes. In general, greater social intimacy predicts increased microbe-sharing between humans.10,15–17,29 | |
|
| ||
| Indirect social transmission (mediated by the environment) | Non-human animals | Microbes can undergo indirect social transmission between hosts through the external environment.15,51 In laboratory mice, aerobic bacteria are more likely than anaerobic bacteria to adopt environmental transmission modes,62 a pattern that has also been observed in wild rodents.61 Recent comparative studies have shown that aerobic microbial lineages are also more likely than anaerobes to display distributions across mammalian species consistent with indirect social transmission.63 In a recent meta-analysis, the degree of bias for social transmission that bacterial genera displayed within laboratory mice60 was significantly and positively associated with the degree to which strains within the genera were shared between mammalian species.61 These results suggest that the traits that affect transmission within host species may scale up to affect transmission between host species. |
| Humans | In humans, bacteria displaying broad geographic distributions also tend to be enriched in sporulation genes,64 further supporting a role for aerotolerance in indirect social transmission. Many of the most common human pathogens and pathobionts, such as Escherichia coli and other Proteobacteria as well as Clostridioides difficile, are known to persist in environmental reservoirs that enable their transmission between hosts in the absence of direct social contact.11 These observations are congruent with evolutionary theory, which predicts that uncoupling of host and microbial fitness through frequent indirect social transmission can favor the evolution of increased virulence relative to strict maternal transmission.62 | |
The social transmission of microbes can be considered across three broad forms11,27,51,62 (Figure 2; Table 2): (1) parental transmission that occurs in early life and is sufficiently influential to warrant independent consideration as a form of social microbial transmission, (2) direct social transmission in which animals acquire microbes horizontally via social interactions, and (3) indirect social transmission in shared environments in which microbes are transmitted between hosts via incidental contact with fecal matter or other host-associated microbes with endurance mechanisms that enable persistence in the extra-host environment. Overall, social environments can therefore exert significant effects on the composition and function of animal microbiomes (Figures 1 and 2; Tables 1 and 2). In this Perspective, we focus on direct and indirect social transmission in the context of the social microbiome.
Microbiome composition is influenced by pairwise associations within social networks,12–14,29,65 and the effects of social interactions on microbial composition can extend from birth into adulthood61,64,66 (Figure 2; Table 2). Recent human examples illustrate the dynamic and nested nature of social effects on microbiome composition. Within households, co-habitation leads to enhanced microbial strain sharing between mothers and offspring,16 between siblings, and between non-kin.15,64 Individuals within the same household typically share 12% of their gut microbial strains, whereas strain sharing between individuals in the same village is 4%–8%.15,29 In addition to strain sharing between individuals, network-level characteristics of the household can affect the microbial composition of the inhabitants. For instance, the relative abundances of certain bacterial genera within infants, including Lactobacillus, Clostridium, Enterobacter, and Klebsiella, have been shown to be associated with the size of the household and the number of siblings.67 The gut microbiome becomes more stable and displays more adult-like features at approximately three years of age.68 Following this, the quantity of shared strains between pairs does not depend on kinship status (mother-infant, father-infant, partner-partner, or sibling-sibling), but rather on social context.15 Moreover, the influence of co-habitation appears to be stronger than age in strain sharing patterns among adult twins.15 This suggests that the strain sharing patterns observed in adults are more dependent on social relationships than on the maternally derived microbiome.
Signatures of the social transmission of microbes have been observed across a range of host body sites, including the gut,10,15,17,29 skin,69 and mouth.10,15 Moreover, the microbes at a particular body site may migrate to new sites. For instance, recent work has shown that there is extensive transmission from the oral microbiome to the gut microbiome within individual humans,70 though in some cases such transmission is associated with pathologies such as rheumatoid arthritis71 and inflammatory bowel disease.72 The specific taxa that are socially transmissible and the degree of social structuring of the microbiome can vary across body sites. For instance, one study found that individuals who display evidence of social transmission of gut microbes do not always display evidence of social transmission of oral microbes.10 In contrast, other work has found higher transmissibility of generally aerotolerant oral microbes compared to the mostly anaerobic gut microbes, with the latter being less likely to persist for sufficiently long in the oxygen-rich external environment to colonize new hosts.15 Indeed, the longer the duration of co-habitation (e.g., partners or parents with their offspring), the greater the similarity of the oral microbiomes of the individuals.15 Similarly, skin microbes of dogs and their owners show stronger evidence of inter-specific transmission than gut microbes.42 Overall, the effects of direct and indirect social transmission on microbiomes vary amongst body sites: aerotolerant skin microbes may be more readily transmissible between hosts through shared environments (indirect social transmission), whereas anaerobic gut microbes may require more intimate social contact to undergo transmission (direct social transmission). Within body sites, specific bacterial taxa may be primed or better suited to social transmission. For instance, in baboons, gut bacteria belonging to the Bifidobacterium and Fusobacterium genera show stronger evidence of social transmission than other bacterial taxa.14 In contrast, the social transmission of bacteria appears to be independent of bacterial taxonomy in humans.10 This suggests that most microbial taxa in humans may be socially transmissible—at least in principle. Regardless of the variation in the degree of social transmission of microbes across body sites, across microbial taxa, and across host populations, the social transmission of microbes appears to be a widespread and robust determinant of microbiome composition in humans and non-human animals.
HOST HEALTH AND DISEASE IN THE CONTEXT OF THE SOCIAL MICROBIOME
Group living and differentiated social bonds confer numerous fitness advantages upon individuals, including protection from predation, enhanced access to mates, and assistance in acquiring and defending resources. The social determinants of health framework examines the connections between sociality and both health and evolutionary fitness.18 Furthermore, social context and social relationships, including social rank and connectedness, exert major consequences upon individual health and wellbeing.18 It is therefore not surprising that various aspects of the social environment—including social rank, social integration, and early-life adversity—are amongst the strongest and most consistent predictors of individual morbidity and mortality.18 The strength of these links has drawn attention in both the social and natural sciences that share common interests in the biological processes that connect the social environment to animal health, disease outcomes, and mortality risk. Often, research focuses on genetic, epigenetic, immune, and endocrine processes through which the social environment interacts with individual physiological processes to affect health and evolutionary fitness.18 Researchers are now beginning to highlight the potential role of the microbiome in mediating the relationship between social interactions and host health status. We develop here the concept that socially transmissible microbes and social-behavioral drivers of microbiome composition may contribute to these effects.73
Within the social determinants of health framework, one of the consequences of sociality on health is the exposure to transmissible microbes. This includes the effects of both pathogens and the rather more overlooked commensals and mutualists. With respect to pathogens, a most venerable field of enquiry has long investigated the connections between pathogen transmission and host sociality. For example, individuals living in larger groups, with higher rates of social contact, operating in specific network positions or structures, or engaging in longer and more intimate contact with conspecifics, face higher communicable disease risk than isolated individuals, and as such, hosts may have evolved various social behaviors to avoid or control pathogens.19–21,74 The transmission strategies of commensals and mutualists are currently under-appreciated11, but if they were to differ from the transmission strategies of pathogens, this could potentially select for the evolution of various social behaviors that benefit host health through microbial transmission. A difficulty with this proposal is that pathogenic and non-pathogenic gut microbes often employ the same, or similar, transmission strategies.11 Thus, although a wide range of social behaviors—including grooming, co-feeding, mouth-mouth interactions, nursing, and coprophagy—have been hypothesized to facilitate the transmission of bacteria that confer metabolic and immunological benefits,22–26 it is unclear whether the transmission strategies of commensals and mutualists are sufficiently distinct from those of pathogens, or sufficiently beneficial, to favor the emergence of social behaviors that facilitate such transmission. Indeed, there are alternative evolutionary explanations for many of these behaviors independent of their effects on microbial transmission. Future research and modeling efforts on the differences in the transmission strategies of mutualists, commensals, and pathogens may be able to shed light on the relationship between social microbial transmission and the evolution of sociality. A central question in this vein is whether there is sufficient variation in the transmission strategies of mutualists, commensals, and pathogens for natural selection to favor the emergence of social behaviors that facilitate the transmission of beneficial microbes but not harmful ones. Of course, the evolution of social behaviors favoring transmission would also depend on the relative benefits of commensals and mutualists versus the detriments of pathogen exposure for the host, not only differentiation among routes of transmission.
CONSEQUENCES OF THE SOCIAL MICROBIOME FOR HOST HEALTH AND DISEASE: ECO-EVOLUTIONARY MICROBIOME COMMUNITY PROCESSES
Several of the effects of the social transmission of microbes occur at the whole-microbiome community level, including colonization resistance, the evolution of virulence and transmissibility, and reactions to ecological disturbance (Figure 1B).
Colonization resistance
Colonization resistance refers to the intrinsic capacity of an individual’s microbiome to thwart invasive pathogen colonization and proliferation.75,76 Several common members of the microbiome such as Clostridioides difficile (formerly classified as Clostridium difficile) are pathobionts (i.e., opportunistically pathogenic), rendering invasion and pathogenesis a matter of ecological context in many cases. Here, we consider a typical or healthy microbiome as one that offers little opportunity for microbes to invade and disproportionately colonize ecological niches. We predict that the social microbiome will influence host colonization resistance.
Commensal and mutualistic microbes contribute to colonization resistance via various mechanisms. These include directly competing with each other and with pathogens for space and nutrients, secreting antimicrobial molecules, altering the biochemical properties of the gut environment, and training the immune system to distinguish between harmless and potentially dangerous microbes11,75,76 (Figure 3).
Figure 3. Interactions between socially transmissible gut bacteria and the host immune system.
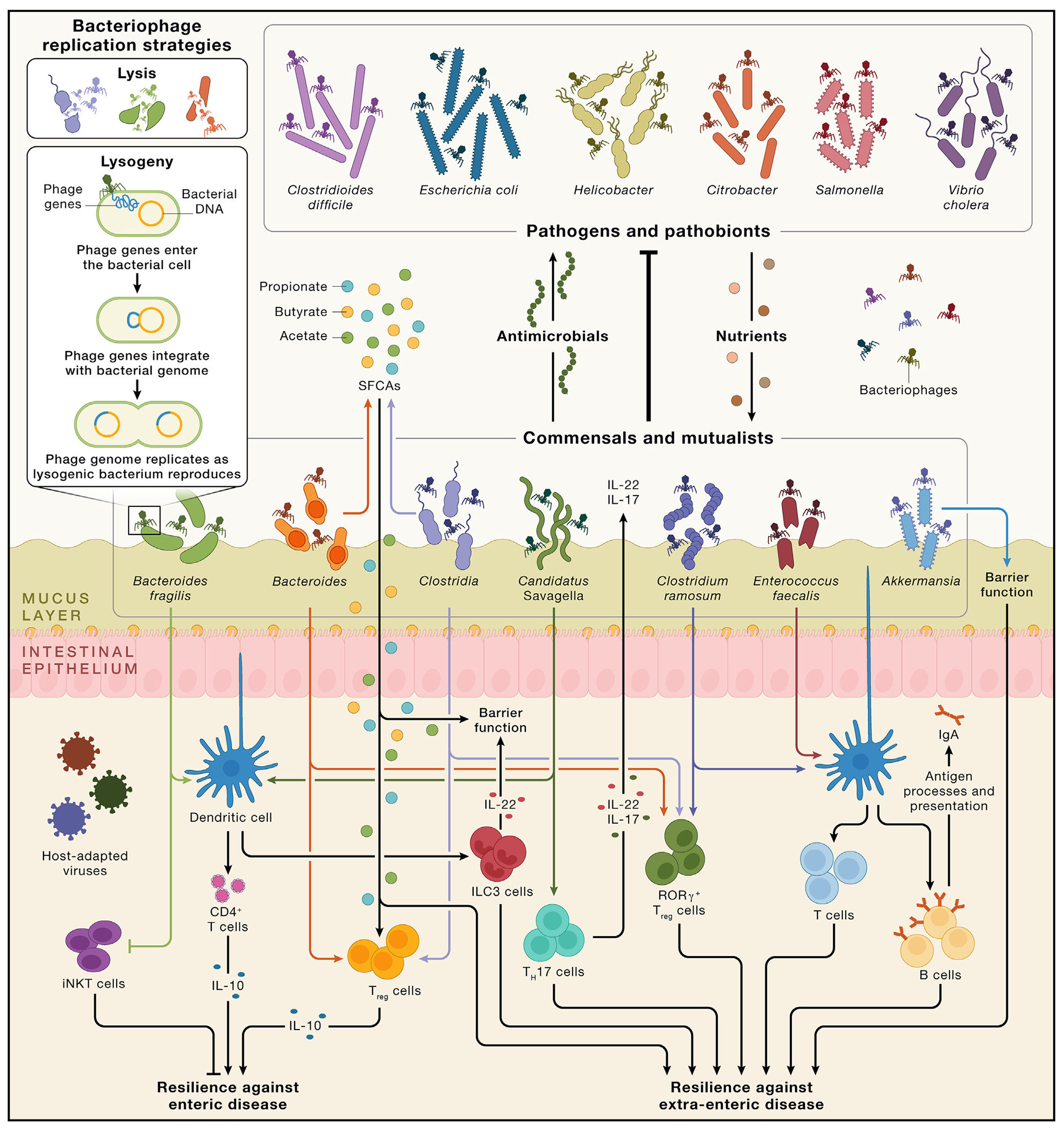
The upper half of the figure visualizes colonization resistance conferred by commensal and mutualistic gut bacteria through physical space occupation, secretion of antimicrobial molecules (shown here by the upward movement of antimicrobials) and nutrient absorption (shown here by the downward movement of nutrients such as fibers, proteins, and organic acids to represent the competition that pathogens and pathobionts face from the commensals and mutualists). Though we list bacteria as either mutualists and commensals or pathogens, these features are matters of ecological context. For instance, Clostridioides difficile is a pathobiont, commensal under most circumstances but capable of pathogenesis under circumstances of low ecological competition. Similarly, there are also both commensal and pathogenic strains of Escherichia coli (the latter is shown here). All bacterial communities are regulated by bacteriophages, which themselves should be socially transmissible when the bacteria they infect are transmitted. Bacteriophages regulate bacteria via lysis (wherein viruses replicate in bacterial cells, ultimately killing the cell and releasing new virions) and lysogeny (wherein the viral genome integrates with the bacterial genome and replicates alongside the bacterium). In other words, bacterial effects on the host are tri-partite functions of bacteria-host interactions, bacteria-bacteria interactions, and bacteria-bacteriophage interactions. Many bacteriophages are highly adapted to specific bacteria, shown here by the matching of colors between bacteriophages and bacteria. The lower half of the figure illustrates examples of how various socially transmissible bacterial taxa can affect multiple immune processes, including cell types and molecules. Similarly, a given immune process can be sensitive to the actions of various bacteria known to be socially transmissible. For example, dendritic cells, which can extend into the lumen and sample the local environment to trigger subsequent immunological effects,77 can be affected by Bacteroides fragilis, Candidatus Savagella (segmented filamentous bacteria), Clostridium ramosum, and Enterococcus faecalis. These kinds of interactions exert downstream effects on host health and confer resilience against enteric and extra-enteric disease. Bacteria can influence multiple immune cells. For example, Bacteroides fragilis also inhibits iNKT cells, which can exacerbate colitis. Bacteria also produce short-chain fatty acids (SCFAs) such as acetate, propionate, and butyrate, which serve as energetic substrates in the gut and in distal tissues and interact with various immune processes such as the induction of Treg cells. SCFAs are important not only for colonic energy salvage, with butyrate alone meeting 60-70% of the energy demands of the colonic epithelium,78 but also for gut barrier integrity. The host is also colonized by several eukaryotic-adapted viruses, including pegiviruses and allenoviruses.
In mammals, some of the most common gut bacterial taxa are involved in maintaining host colonization resistance75,76 and are also socially transmissible.14,15 Socially transmissible microbes can affect colonization resistance through processes involving specific taxa, as well as emergent community properties of the whole microbiome. We consider four key attributes of the microbiome that influence colonization resistance that can be affected by social transmission processes48,79: (1) the presence of specific microbial taxa important for colonization resistance, (2) microbiome diversity, (3) microbiome stability, and (4) microbiome similarity amongst hosts.
First, several host-associated microbes can be beneficial for host colonization resistance via consuming resources necessary for pathogen survival or pathobiont overgrowth (i.e., competitive exclusion). For instance, commensal strains of Escherichia coli consume many of the same organic acids, amino acids, and other nutrients required for the growth of pathogenic strains of enterohaemorrhagic Escherichia coli.80,81 Such competition from the commensal strains inhibit the growth of the pathogenic strains. Microbial taxa can also contribute to colonization resistance in context-dependent ways. For example, commensal strains of Escherichia coli exert little effect on the growth of the pathogens Klebsiella pneumoniae and Salmonella enterica in a simple co-culture.82 However, they are crucial as part of a more diverse microbial community where they contribute to the capacity of other microbes to out-compete these pathogens through nutrient depletion.82 As another example, depletion of dietary amino acids by commensal microbes in the mouse gut supports colonization resistance against the highly transmissible pathogen Citrobacter rodentium (used in murine models to mimic pathogenic Escherichia coli), which also depends on these amino acids.83 Moreover, specific gut microbes may also protect the host against pathogen colonization by altering the ecological conditions in the gut, creating hostile environments for potential pathogens. For instance, Bifidobacterium spp. can prevent pathogenic Escherichia coli from colonizing the gut by lowering the pH of the local environment.84
Second, the diversity of the microbiome may enhance its capacity to use all available niche space, and thus resist colonization.85 This hypothesis is predicated on the ecological theory that biodiversity is negatively associated with a community’s invasibility (i.e., the vulnerability of a community to invasions).86 Consistent with this proposal, a recent study found that the diversity of the gut microbiomes of gnotobiotic mice linearly increased the microbiomes’ capacities to resist pathogen invasion.82 Diverse microbiomes harbor many competing microbes, which help stabilize the community against perturbations48 and occupy ecological niches that could otherwise be exploited by invaders.85 The ecological niche space that most common bacterial pathogens might exploit can be saturated in a high-diversity microbiome, with commensal and mutualistic gut microbes utilizing most available nutrients, thereby holding pathogens and pathobionts at low abundances and limiting invasions.76 Paralleling these theories, evidence from antibiotic treatment of humans and mice supports the hypothesis that extreme reductions in microbiome diversity can render hosts more vulnerable to pathogen invasion.87
Considerable debate persists over the consistency and linearity of the correlation between microbiome diversity and colonization resistance88 and between microbiome diversity and host health. Indeed, although high microbiome diversity is commonly associated with better host health,89–93 several studies have also found that high microbiome diversity is related to poor health outcomes90,91,94 or is unrelated to health. Importantly, microbiome diversity can be positively associated with some pathogens but negatively associated with others.95 Similarly, the effects of social transmission on microbiome diversity are more complex than a simple positive sociality-diversity association. Although social interactions may increase diversity,12 extensive social interactions and large social groups may also reduce diversity in some cases.27 For instance, a negative association between the degree of social interactions and the average microbial diversity within individual hosts has recently been demonstrated in free-living populations of red-bellied lemurs65 and yellow-bellied marmots.96 This negative relationship can occur if, for instance, a particular microbial lineage possesses a competitive advantage within hosts over other lineages. In socially fragmented populations, such a microbe may only come to dominate the microbiome of a few hosts, whereas in socially connected populations, the microbe is likely to spread to, and proliferate within, most or all hosts. These variations hint at a complex relationship between microbiome diversity and colonization resistance. Rather than a uniformly positive relationship between colonization resistance and microbiome diversity, there may instead exist a “tipping point” of diversity reduction that can unbalance the microbiome, creating ecological niche space conducive to pathogen invasions.97
Third, colonization resistance is inherently linked to the stability of the microbiome, an emergent community property that may be influenced by social microbial transmission. Generally, stable communities are expected to be more resistant to invasion than unstable communities, because instability in community composition can create ecological niche space, thereby providing opportunities for invasion.79,86 Instability is considered an aspect of a dysbiotic microbiome state in humans,98 and instability-associated perturbations may lead to pathogenic overgrowth of some taxa.99 Indeed, opportunistic pathogenesis of typically commensal microbes through overgrowth can be a causal mechanism underlying diarrhea.99 For example, traveler’s diarrhea often appears without an obvious enteric pathogen, and instead seems to be attributable to commensal microbe overgrowth associated with dysbiosis.100 These instability-driven invasions illustrate how pathogenesis may occur due to sudden availability of niche space rather than the invasive tendencies of a pathogen per se. The extent to which microbiome stability is influenced by social microbial transmission is an understudied area. For instance, whether an individual’s position in a social network shapes microbiome turnover (a common measure of stability) has not yet been thoroughly explored,101 likely due to a paucity of detailed longitudinal data from natural host-microbiome systems. Some evidence suggests that social network instability may be associated with gut microbiome instability. For example, amongst wild Verreaux’s sifakas, individuals with more unstable social ties show higher gut microbiome turnover rates.102 Unstable social ties could affect microbiome composition and stability, with social stress contributing to the association between social instability and microbiome instability. Unstable social relationships trigger hormonal stress responses which in turn may lead to compositional changes7 and may in turn cause reductions in the stability of the microbiome. Future research could experimentally manipulate social rank in model animals and examine how social network position interacts with factors such as stress and microbiome composition to affect host phenotypes, including colonization resistance.
Fourth, colonization resistance can be influenced by microbiome similarity between socially interacting hosts. This is because social interactions increase the similarity of microbiomes between hosts.10,15,29 Enhanced similarity of microbial communities across hosts could theoretically both enhance or diminish colonization resistance, and we discuss each possibility in turn. First, with respect to enhancing colonization resistance, a host may display higher resistance to colonization by familiar microbes due to pre-acclimation of the host’s immune system to those microbes. For example, many microbes that are typically considered commensal or mutualistic can become pathogenic under certain conditions, with Clostridioides difficile as a canonical example. The shift to pathogenesis may partly depend on how acclimated the host is to a given microbe. Human studies suggest that host-microbe interactions train host adaptive immunity, reducing pathogenesis caused by familiar microbes.103 In contrast, unfamiliar microbes may be more likely to become pathogenic.104 The degree of microbiome similarity amongst social partners may thus affect the likelihood that microbes become pathogenic in the new host following transmission. For example, imagine that a host interacts with a novel social partner whose immune system is unaccustomed to the host’s commensals and mutualists. Such an interaction may be more likely to lead to pathogenesis relative to interactions amongst hosts with a history of social interactions and exchange of microbes. This is because familiar hosts are expected to have more similar microbiomes and immune training. Second, high degrees of microbiome similarity could also diminish colonization resistance. Specifically, because social interactions enhance the similarity between microbiomes,10,15,29 this microbial similarity may also confer advantages to pathogens that have developed mechanisms to overcome or subvert colonization resistance. Individuals with microbiomes that closely resemble the social microbiome may thus also be the most susceptible to invasions by pathogens that have previously succeeded in invading similar microbial communities. Studying pathogen transmission through social networks as a function of the degree of similarity between an individual’s microbiome and the social microbiome should yield insights into the rate at which pathogens spread in the social microbiome. This would enable assessments of whether transmission is positively associated with the degree of microbiome similarity between hosts in the social network.
All social transmission effects on colonization resistance can be influenced by the various social-ecological forces that contextualize the social microbiome (Figure 1A). For example, larger and more heterogeneous groups should provide the maximum number of colonization opportunities (level 2). Similarly, host species that are on average more social (level 4) may experience higher rates of potentially invasive transmission events. However, they might also possess greater intrinsic colonization resistance owing to the greater number of opportunities they have for the transmission of non-pathogenic microorganisms. Because interacting with others possessing dissimilar microbiomes can also enhance diversity by introducing new microbes to a host, there is an inherent trade-off between “safe” sharing of commensal and mutualistic microbes and acquiring more diverse (but potentially more dangerous) microbes. Primate research suggests that distributing a set of familiar microbes amongst social partners might help maintain diversity, as any microbe lost to local extinction in any host can be reacquired through social contacts. Maintaining diversity could also reduce the risk of acquiring completely unfamiliar microbes—which might possess greater potential for pathogenesis—through social interactions.65,105,106
Evolution of virulence and transmissibility
Social transmission of gut microbes is expected to affect the evolution of virulence among members of the social microbiome. Strict transmission of gut microbes within host genealogies (i.e., vertical transmission) creates a situation in which the long-term fitness of microbial lineages is dependent on the host.107 Under this scenario, strains that severely decrease host fitness may decrease their own fitness and suffer an evolutionary disadvantage relative to less pathogenic strains,108 unless impairing the host is central to the fitness strategy of the microbe. One example of this phenomenon is the parasitic fungus Ophiocordyceps unilateralis that controls and ultimately kills its ant hosts to enhance the distribution of its spores.6 The extraordinarily virulent rabies virus and the protozoan parasite Toxoplasma gondii are prominent examples of microbe-mediated impairment of the host. Overall, microbial control of host fitness is expected to evolve only rarely and under very precise circumstances.6,109 Therefore a high degree of microbial dependence on the host should typically favor reduced virulence. However, the possibility of social transmission of microbes, especially amongst non-kin, may partially decouple microbial fitness from host fitness. Opportunities to colonize multiple unrelated hosts could potentially increase the long-term fitness of microbial lineages that exert deleterious effects, which might otherwise have been disfavored by selection in microbial lineages that display high fidelity to host lineages. Under this scenario, virulence could evolve if the negative effects of severe host illness or death on microbial fitness are outweighed by the positive effects that virulence yields for within-host microbial fitness.
Although increasing the opportunities for horizontal (social) transmission of microbes may promote the evolution of virulence, evolutionary theory also predicts that increasing opportunities for social transmission may in some cases select for reduced virulence in microbes that are both vertically and socially transmissible.110 For instance, high rates of social transmission can increase the prevalence of a microbe in a host population, thereby reducing further opportunities for social transmission and decreasing the fitness of virulent strains that rely on social transmission.110 Thus, the effect of social transmission on the evolution of virulence in members of the social microbiome is certainly more complex than a positive linear relationship between opportunities for social transmission and virulence.111 Transmission opportunities for many microbes (pathogens, commensals, and mutualists alike) can be conceptualized in terms of varying social-ecological forces affecting the social microbiome (Figure 1A). For example, transmission opportunities are likely maximized by frequent and intimate social contacts and in more centrally networked individuals (level 1). Transmission opportunities are also likely to be increased amongst groups of larger size (level 2), between groups that have greater numbers of migrating individuals (level 3), amongst more social species, species with greater average within-group genetic relatedness, and in seasons and environments that promote close social interactions (level 4), and under circumstances of greater inter-species contact (level 5). Notably, some of these interactions may be indirect, resulting from multi-partite connections between individuals and populations, as recently observed in bats.112 In these ‘cryptic’ connections, microbial lineages are socially transmitted between hosts that never directly interact (i.e., indirect social transmission; Figure 2; Table 2).
The social microbiome may also affect the evolution of traits critical for microbial transmissibility. Long-term pathogen fitness is a function of the number of new hosts that the pathogen can infect, and the same is likely true for gut-adapted commensals and mutualists. A social network in which hosts are closely connected reduces the spatial and temporal distance between potential hosts, and allows host-adapted microbes to transmit across the social network with greater success than amongst more solitary hosts. This is especially relevant for members of the gut microbiome, many of which are obligate anaerobic bacteria that do not possess adequate endurance mechanisms for significant persistence in the oxygen-rich external environment.11 Thus, a dense social network with many proximal hosts should increase colonization opportunities for anaerobic gut bacteria. Indeed, where direct social transmission of microbes has been studied, the bacterial taxa that are most socially transmissible are also the least likely to persist in aerobic external environments, possessing fewer mechanisms supporting extra-host survival.14
Endurance mechanisms such as sporulation facilitate bacterial survival in extra-host environments.11 Sporulating bacteria are significantly more aerotolerant than non-sporulating bacteria.113 Unlike obligate anaerobes, spore-forming bacteria can readily disperse across individuals independent of direct social contact. Concordantly, gut microbes transmitted through direct social contact between wild mice are mostly anaerobic, whereas gut bacteria transmitted indirectly through shared environments are enriched in aerobic spore-forming taxa.51 Genera containing sporulating bacteria appear to represent up to 30% of the microbial abundance in the gut and are found across several prevalent bacterial families, including Lachnospiraceae, Ruminococcaceae, and Clostridiaceae.11,113 Notably, the pathobiont Clostridioides difficile produces metabolically dormant and highly resistant spores that facilitate both persistence within the host during hostile conditions and indirect social transmission through shared environments.113
The evolution of microbial endurance mechanisms, such as the capacity to form spores, may be shaped by the degree of sociality of the host species. Regarding the relationship between host sociality and the strength of selection for endurance mechanisms, one may postulate two competing hypotheses. First, endurance mechanisms such as sporulation may be selected against in microbes confined to host species that are solitary or have few social partners. This is because social contact may be so sparse that opportunities for colonizing another host are too limited to support the evolution of endurance mechanisms that enable efficient indirect social transmission to new hosts. Second, evolutionary pressures may select for enhanced endurance mechanisms in more solitary host species relative to social host species, enabling microbes to persist in the environment to reach new, infrequent hosts via indirect social transmission. Of course, acquisition of microbes from other sympatric species (level 5; Figure 1A; Table 1) could also affect these kinds of outcomes. These hypotheses could be empirically or meta-analytically tested by comparing the presence or absence of endurance mechanisms amongst microbial lineages associated with host species that vary in their degree of sociality while accounting for interactions with other sympatric species.
Reactions to ecological disturbance
Ecological disturbances refer to transient environmental events that precipitate significant ecosystem and ecological change (e.g., floods, forest fires, hurricanes).114 One definition of ecological resilience is the extent to which a disturbed ecosystem recovers and returns to or resembles its pre-disturbance state.115 Principles from disturbance ecology and ecological resilience in macroecological systems can also be fruitfully applied toward understanding microecological processes, including host-microbe interactions.50 Disturbances to the ecology of the microbiome, including exposure to a new diet, antibiotic exposure, illness, or infection, can result in the loss of endogenous microbial populations and their replacement with other microbial populations (Figure 4). For instance, antibiotic-induced disturbances and subsequent microbial losses allow ecological niches in the gut to become available for colonization, leaving the host vulnerable to invasion by foreign and potentially pathogenic microbes or to the unrestrained growth of pathobionts such as Clostridioides difficile.116
Figure 4. Stability landscape of the gut microbiome in the social microbiome.
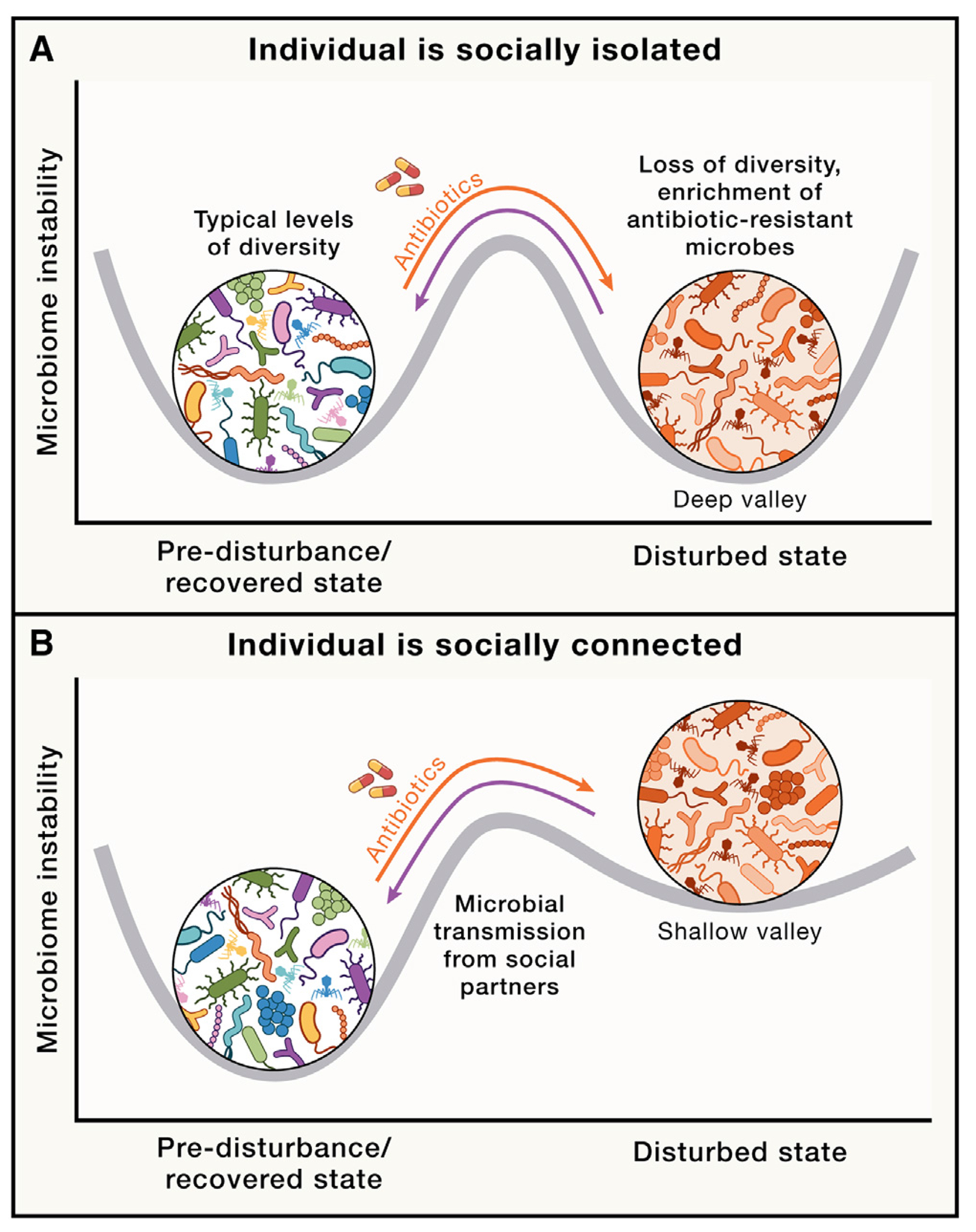
Stability landscapes provide useful views of how microbiomes react to, and recover from, ecological disturbances such as antibiotic exposure.23 Curved gray lines indicate possible stability landscapes of an individual’s microbiome resulting from the combined effects of within-host dynamics and inter-host microbial transmission. Deeper valleys represent higher stability (i.e., lower instability). Undisturbed, recovered, and antibiotic-disturbed states are shown. Orange and purple arrows represent transitions between undisturbed (pre-disturbance) and antibiotic-disturbed states and between antibiotic-disturbed and recovered states, respectively. Fewer opportunities for social interaction may be hypothesized to result in higher transition peaks between disturbed and recovered states, corresponding to greater difficulty in moving from disturbed to recovered states. Conversely, increased social interactions may provide a greater number of opportunities for microbes from the social microbiome to recolonize the host, resulting in shallower valleys and transition peaks, indicating greater ease in moving from disturbed to recovered states. When hosts are socially isolated, disturbed microbiome states may be as stable as undisturbed states due to lack of transmission from individuals with undisturbed microbiomes, as shown in (A). If the host with a disturbed microbiome is socially connected to many healthy hosts, the undisturbed state is expected to be more stable than the disturbed state, as shown in (B), given the effects of social transmission of gut microbiota from healthy hosts.
Exposure to a social network of conspecifics may enhance the microbiome’s resilience by providing a metacommunity of socially available microbes from proximal hosts to facilitate post-disturbance recolonization (Figure 4). For example, in humans, even short courses of antibiotics precipitate substantial reductions in bacterial diversity,117–121 and the microbiome can remain perturbed for months or years after antibiotic exposure.118,119,121 Although early-life antibiotic-induced disruption of the gut microbiome can exert lifelong consequences,122 antibiotic-mediated perturbations are often mild amongst adults, with the microbiome tending to return to stable, largely preexposure states fairly quickly following the cessation of antibiotic treatment.57,120,121,123 The capacity of the adult gut microbiome to return to baseline states following perturbation probably reflects mechanisms of host control but may also be facilitated by the dense social networks that provide numerous opportunities for social microbial transmission. Exposure to human-associated microbes from the surrounding social network and environment may compensate for losses in commensal and mutualistic microbial populations following antibiotic treatment. However, such a pattern may be more likely to hold for familiar social partners. In contrast, exposure to unfamiliar social partners after antibiotic treatment may result in pathogenesis, owing to the potential transmission of new microbes for which the individual lacks immune tolerance. These dynamics might be tested in experimentally tractable social species such as mice and bees in which antibiotic-treated animals are exposed to familiar or novel social partners after antibiotic exposure.
Various social-ecological forces from across the five levels of the social microbiome (Figure 1A) could influence the probability of successful recovery from disturbance. Microbial transmission from conspecifics may covary positively with the extent to which the individual is networked in a social group and the frequency and intimacy of social contacts (level 1), the size of the social network (level 2), and the number of interactions across more distant components of social networks (level 3). On average, more social species (level 4) may be expected to experience more rapid post-disturbance recolonization.
Nevertheless, the microbiome is generally resilient to perturbations.48,79 An important area of research, therefore, is to understand the relative contribution of social transmission to microbiome resilience compared to other biological mechanisms (e.g., host immunity). This question could be empirically examined in model organisms by exposing hosts to standardized antibiotic treatment while manipulating host social structure (e.g., housing animals individually or in groups of varying size). Supporting the importance of social transmission in microbiome resilience, antibiotic-induced ablation of the honeybee microbiome increased mortality, but seven days of exposure to other hosts from the hive partially restored bacterial composition in antibiotic-treated bees.124 In contrast, bees housed individually remained depleted of bacteria relative to antibiotic-free control bees.124 Similarly, recovery of the mouse gut microbiome after antibiotic treatment was accelerated when antibiotic-treated mice were co-housed with untreated mice, which served as microbial reservoirs.125,126 Hence, social partners may contribute to the resilience of the gut microbiome following antibiotic-induced disturbance. Colony models of rodent social networks could be used to examine microbiome recovery from disturbance in settings that more closely mimic natural social environments. It may be that more central individuals who have more social interactions are able to recover more rapidly than more peripheral individuals by virtue of enhanced microbial transmission and acquisition (Figure 4). Nevertheless, because individuals who are more integrated within social networks tend to have better health in general,18 careful treatment will be required to experimentally disentangle the two processes.
CONSEQUENCES OF THE SOCIAL MICROBIOME FOR HOST HEALTH AND DISEASE: MICROBIAL TRANSMISSION-BASED PROCESSES
Although several host health-related effects of social microbial transmission are based on processes at the whole-microbiome community level, sociality also drives the transmission of specific microbes that affect host health and disease, including the transmission of microbes with metabolic and immune effects, inter-specific transmission, the transmission of antibiotic-resistant microbes, and the transmission of viruses (Figure 1B).
Transmission of microbes with metabolic and immune effects
Social interactions may promote the transmission of specific microbes that exert appreciable effects on host metabolism and immunity. For example, a study in baboons tested the transmission of microbes through host social and grooming networks.35 “Core” microbial taxa often contribute toward generating crucial ecosystem services for their hosts, including the digestion of complex carbohydrates, the synthesis of vitamins, and the production of short-chain fatty acids (SCFAs) such as butyrate, the primary energetic substrate for colonocytes.78,127–129 Given their significant contributions to the host, it was thought that the abundance of core microbes within hosts would be too important for host health to depend on host social interactions.35 Contrary to this expectation, the presence and abundance of these core microbes (including the mutualistic genera Bifidobacterium and Faecalibacterium) were predicted by social group membership and social behavior. Socially transmissible gut microbes may also contribute to the host’s capacity to exploit a particular niche. For instance, desert woodrats consume tannin-rich plants that are metabolized by gut microbes such as Enterococcus faecalis,130 a socially transmissible taxon.131 These microbes are necessary for the appropriate degradation of tannins, and their absence predicts the body mass reduction and liver damage typically associated with tannin consumption.130 This is also one route through which the social transmission of microbes may facilitate host adaptation to novel dietary resources. This proposal could be experimentally investigated by cohousing rodents that lack microbes capable of degrading diet-derived xenobiotics with rodents that possess such bacteria. We predict that rodents lacking these bacteria will acquire them via direct and indirect social transmission, and will be better able to tolerate the xenobiotics. Such a phenomenon has important implications for understanding host range expansions. Socially transmissible microbes that enable hosts to exploit new resources and niches could eventually facilitate the dispersal of hosts into new ecologies.
The microbiome also plays a crucial role in shaping and regulating host immunity,4,5 and specific socially transmissible microbial taxa may affect the general immune status of the host (Figure 3). For instance, a recent study of wild macaques found that host sociability was positively correlated with the presence of mutualistic gut bacterial genera such as Faecalibacterium, which confer anti-inflammatory and other beneficial effects on health.132 However, less sociable macaques displayed increased levels of the genus Streptococcus, whose members include several pathogens and pathobionts.132 Through SCFA-mediated signaling, effects on barrier function, and other mechanisms, gut microbes can modulate a broad range of host immune cell populations (Figure 3; Table 3), and microbiome-immune interactions can exacerbate or protect the host from both enteric diseases and various extra-enteric diseases including cancer, autoimmune diseases, and viral infections including, potentially, SARS-CoV-2.4,133,134 Common microbial metabolites such as SCFAs (which are also produced by socially transmissible microbes) can exert significant effects on host immunity. For instance, SCFAs induce regulatory T (Treg) cells in the colon, conferring resilience against colitis in mouse models.135,136 Individual bacterial species also affect the frequencies of diverse immune cell types.137 Several of these microbes also interact with drugs and can alter drug metabolism, with consequences for host drug responses and treatment outcomes (Table 3). Crucially, microbes that exert these effects have also been observed to be socially transmissible10,14–16,35,51 (Figure 3; Table 3). These lines of evidence point to the possibility of immune effects of socially transmissible microbes.
Table 3.
Implications of socially transmitted microbes for host immunity
| Domain of interaction | Type | Association | Socially transmissible bacterial families, genera, or speciesb | Characterized bacteria × immune interactions that may be sensitive to social structuringe | Hostg |
|---|---|---|---|---|---|
| Bacterial interactions with host immune processes a | Host immune cell types and cytokines | Gut bacteria affect immune cell populations throughout the colon, small intestine, and mesenteric lymph nodes, as well as in extra-enteric tissue.137 | Akkermansia 3,10 | Correlates strongly with blood concentrations of neutrophils, lymphocytes, and monocytes.138 | Human |
| Faecalibacterium 10,139 | Correlates strongly with blood concentrations of neutrophils, lymphocytes, and monocytes.138 | Human | |||
| Ruminococcus 10 | Correlates strongly with blood concentrations of neutrophils, lymphocytes, and monocytes.138 | Human | |||
| Clostridium 3,10,14 | Induces Treg cells.140–142 | Mouse | |||
| Bacteroides 3,10,15,16 | Induces Treg cells.140–142 | Mouse | |||
| Bacteroides fragilis 15 | Induces CD4+ T cells to release interleukin-10 (IL-10).140,143 | Mouse | |||
| Bacteroides fragilis 15 | Produces sphingolipids that inhibit invariant natural killer T (iNKT) cell numbers during neonatal development, a reduction that persists into adulthood and protects the host from oxazolone-induced colitis.144 | Mouse | |||
| Candidatus Savagella (segmented filamentous bacteria)145 | Stimulates T helper 17 (TH17) cells to release IL-17 and IL-22, conferring resilience against the enteric pathogen Citrobacter rodentium146 | Mouse | |||
| Gut integrity and homeostasis | Commensal and mutualistic gut microbes of typically colonized mice induce the expression of antimicrobials (e.g., RegIIIγ) by epithelial cells to promote physical separation between the gut microbiome and the epithelial layer.147 | Segmented filamentous bacteria145 | Stimulates type 3 innate lymphoid cells (ILC3s) to release IL-22 to enhance gut barrier function.148 | Mouse | |
| Enterococcus faecalis 131 | Interacts with dendritic cells to induce luminal IgA via B cells, which binds to bacterial cells and inhibits their capacity to translocate across the gut epithelium.149 | Mouse | |||
| Enterococcus faecium 3 | Stimulates RegIIIγ secretion to promote tolerance to Salmonella infection.150,151 | Mouse | |||
| Responses to colitis | Gut bacteria regulate responses to colitis. | Clostridium ramosum 3 | Increases the relative number of RORγ+Treg cells, which enhance resilience against colitis induced by 2,4,6-trinitrobenzenesulfonic acid (TNBS).152 | Mouse | |
| Bacteroides thetaiotaomicron 3 | Increases the relative number of RORγ+Treg cells, which enhance resilience against TNBS-induced colitis.152 | Mouse | |||
| Bacteroides fragilis 15 | Induces the production of IL-10, which enhances protection against Helicobacter hepaticus and TNBS-induced colitis.140 | Mouse | |||
| Clostridium 3,10,14 | Induces Treg cells to protect from colitis and ovalbumin-induced allergic diarrhea141,f. | Mouse | |||
|
| |||||
| Bacterial interactions with drugs | Immune checkpoint inhibitor cancer therapy | Gut microbial composition predicts the efficacy of immune checkpoint inhibitor therapy for cancer, and various bacteria-immune interactions facilitate response to immune checkpoint inhibitors (e.g., anti-PD-1 therapy and anti-CTLA4 therapy). | Akkermansia muciniphila 3,10 | More abundant in patients with epithelial tumors who responded to anti-PD-1 therapy.153 | Human |
| Bifidobacteriaceae 10,16 | Overrepresented in responders relative to non-responders to anti-PD-1 therapy.154 | Human | |||
| Faecalibacterium 10,139 | Overrepresented in responders relative to non-responders to anti-PD-1 therapy.155 | Human | |||
| Enterococcus faecium 3 | Secretes a hydrolase which generates muropeptides to promote response to anti-PD-L1 therapy in a NOD2-dependent manner.156 | Mouse | |||
| Akkermansia muciniphila 3,10 | Promotes release of STING agonists to stimulate an anti-tumor response that is dependent on type 1 interferon signaling.157 | Mouse | |||
| Coprobacillus cateniformis 139,158 | Suppresses expression of PD-L2 on dendritic cells to enhance the efficacy of anti-PD-L1 therapy.159 | Mouse | |||
| Bifidobacterium pseudolongum 10,16,c | Secretes inosine which signals through the adenosine 2A receptor on T cells to promote responses to anti-CTLA4 therapy.160 | Mouse | |||
| Vaccines | The gut microbiome modulates host responses to vaccination. | Streptococcus bovis 10,d | Positive correlation with enhanced responses to oral rotavirus vaccines in infants.161 | Human | |
| Bacteroides 3,10,16 | Negative correlation with enhanced responses to oral rotavirus vaccines in infants.161 | Human | |||
| Prevotella 10,14,139 | Negative correlation with enhanced responses to oral rotavirus vaccines in infants.161 | Human | |||
| Prescription drugs for non-communicable diseases | The gut microbiome alters drug metabolism. | Enterococcus faecalis 131 | Transforms the prodrug Levodopa (used to treat Parkinson’s Disease) into dopamine in the gut via a decarboxylation reaction, reducing the fraction crossing the blood-brain barrier and being converted in situ to dopamine for host benefit.162 |
Mouse | |
| Eggerthella lenta 158 | Transforms the drug digoxin (used to treat atrial fibrillation and heart failure) to inactive dihydrodigoxin, reducing bioavailable digoxin for host use.163 | Mouse | |||
| Bacteroides uniformis 3,139 | Sequesters the drug duloxetine (used to treat depression), reducing bioavailable duloxetine for host benefit.164 | In vitro | |||
Several of these processes are visualized in Figure 3.
References in this column indicate published studies supporting the social transmission of these bacterial families, genera, or species.
Research indicates that Bifidobacteriaceae and Bifidobacterium species are socially transmissible.
Research indicates that Streptococcus species are socially transmissible.
This column summarizes immune effects and associations of the bacterial taxa listed in the preceding column, although it remains to be investigated whether social transmission of these bacteria induces these effects in new hosts.
Researchers administered a cocktail of Clostridia species, of which several did not belong to the Clostridium genus.
Host species in which the bacteria × immune association described in the preceding column was observed.
Inter-specific transmission and zoonoses
Zoonoses and zoonotic spillovers are long-standing concerns across biomedicine and public health, and featured prominently in debates concerning the origins of SARS-CoV-2. Microbial transmission between animals of different species is readily interpretable as a level 5 process of the social-ecological forces that contextualize the social microbiome (Figure 1;Table 1). Inter-specific microbial transmission occurs in a range of settings. For example, anole lizards, coyotes, and sparrows residing in urban environments harbor gut bacterial lineages that are typically found in humans.165 Other cases include microbial transmission across predator-prey networks,40 and microbial transmission via interactions with domesticated animals.42 Such interactions between host species have well-recognized potential for inter-host pathogen transfer. For instance, spending time in pig farms increased the abundance of harmful microbes in human visitors.44
More generally, host species living in close contact with one another and potentially sharing microbes166 may allow for a decoupling of host and gut microbial fitness. Such decoupling could potentiate the emergence of virulence in members of the social microbiome. Many of the most virulent human diseases such as Ebola and acquired immune deficiency syndrome (AIDS) are the result of zoonotic infections from other host species.167 Many kinds of interactions can result in such infections. For instance, hunting and consuming bushmeat increases the risk of acquiring zoonotic viruses, which introduces pathogens into human social networks.167 Similarly, several human viral, bacterial, and eukaryotic pathogens can infect and cause disease in other species, including various great apes.168
Although research in this area has focused on zoonoses and the negative consequences of inter-specific microbial transmission, there may also be some benefits. For instance, amongst sympatric species living in close proximity, such interspecific transmission may enhance the microbial diversity of individuals and social groups.166 Similarly, interacting with livestock may contribute to increased microbial diversity of the human gut microbiome.169 As discussed earlier, diverse social microbiomes represent ecological communities that may be able to better resist potential pathogens via colonization resistance75,76 and also via direct and indirect suppression of pathogens such as viruses in the host.134 In some cases, these inter-specific interactions may also exert beneficial effects on immune physiology. For instance, exposing mice to dog-associated house dust enriched beneficial Lactobacillus johnsonii, dampened biomarkers of inflammation, and protected the mice against subsequent respiratory infection and pathology.43 These kinds of proposals could be investigated by colonizing germ-free mice with a mixed or more diverse microbiome comprising microbes from a few other host species and testing resistance to pathogen colonization compared to mice colonized with microbes from a single host species (the former group may show greater resistance to an experimentally introduced pathogen). We could similarly measure pathogen resistance in germ-free mice colonized with “naturally” mixed host microbiota samples from farms, watering holes, or other contexts in which several animals exchange microbes. Such experiments would help us better understand whether there are beneficial effects of microbiomes that include microbes from diverse host species. Overall, although most research on the inter-specific transmission of microbes focuses on disease risk, there is simultaneous potential for inter-specific microbial transmission to yield hidden benefits for hosts that warrants further investigation.
Transmission of antibiotic resistance
The development of antibiotic resistance is an ancient adaptation that enables bacteria to compete with one another.170 However, the widespread exploitation of antibiotics for medical and agricultural purposes is driving the emergence of antibiotic resistance to levels that pose serious public health risks. Much research on antibiotic resistance has focused on either intrahost development of antibiotic resistance following exposure to antibiotics or acquisition of antibiotic-resistant bacterial genes via lateral gene transfer.170
Here, we discuss how the different social-ecological forces shaping the social microbiome (Figure 1A; Table 1) can be exploited to examine the dispersal of antibiotic-resistant microbes at various levels in novel ways. For instance, at level 1, individuals sharing a household are predicted to acquire antibiotic-resistant microbes from co-habitants undergoing antibiotic treatment. This may become exacerbated under longer courses of antibiotics that last for many months or years. Two observations support such a proposal. First, bacteria belonging to the Bacteroidales order are amongst the most transmissible within households.16 Second, Bacteroidales members act as reservoirs for antibiotic resistance genes.171 Thus, some of the most transmissible species and strains may also be highly effective at spreading antibiotic resistance. At level 2, cultures, societies, and countries differ in the extent to which they use antibiotics, creating culture-dependent transmission landscapes for antibiotic-resistant microbes and genes. At level 3, humans traveling over long distances can experience different degrees of exposure to antibiotic-resistant microbes and genes. At level 5, the transfer of antibiotic-resistant microbes and genes has been observed between humans and other animals sharing environments.172 For instance, companion animals are a potential source of antibiotic-resistant microbes and genes.173 Furthermore, individuals working with antibiotic-exposed agricultural animals or in environments inhabited by these animals show evidence of microbiome remodeling and the acquisition of antibiotic-resistant microbes and microbial genes.44,45 There are at least three primary concerns over such acquisitions174: First, the antibiotic-resistant bacteria may directly infect humans. Second, the adaptation of resistant strains following initial infections from livestock may lead to sustained transmission in humans. Third, antibiotic resistance genes emerging in livestock may be acquired by human pathogens via lateral gene transfer between bacteria. All such routes may contribute to a hidden, socially transmissible layer of antibiotic resistance. The implication, of course, is that such transmission may render future antibiotic treatment less effective for an individual’s social contacts and caregivers. The possibility and magnitude of such an effect demand further empirical enquiry.
Transmission of viruses and the social virome
Nearly all organisms are hosts to multiple viruses. Apart from pathogenic viruses such as SARS-CoV-2 and human immunodeficiency virus that cause acute disease, multicellular organisms also harbor intrinsic viral populations—viromes—that consist of two distinct components: host-adapted commensal or conditionally pathogenic viruses that replicate in host cells, and a much larger contingent of bacterial viruses (bacteriophages, or phages) and archaeal viruses that infect the prokaryotic members of the host microbiome (Figure 3). These viral members are also a part of the individual microbiome, and the social virome forms a part of the social microbiome.27
In humans, common members of the host-adapted virome include endogenous retroviruses (i.e., components of viral genomes that are integrated with the human genome), anelloviruses, pegiviruses, polyomaviruses and papillomaviruses, parvoviruses, and herpesviruses.175 Although some of these viruses (papillomaviruses and herpesviruses, in particular) are associated with pathology and cause sporadic disease, including several types of cancer,176 most do not appear to be associated with any pathology. For instance, anelloviruses – an enigmatic group of small viruses with single-stranded DNA genomes that are nearly ubiquitous in humans177 – have not yet been shown to contribute to any disease. Some non-pathogenic or conditionally pathogenic viruses could even be considered mutualistic in specific contexts because of certain benefits they confer upon the host. For example, coinfection with human pegiviruses is associated with less virulent AIDS178 and Ebola infection.179 Generally, host-adapted viruses may be involved in shaping host immunity. In this vein, anelloviruses are thought to contribute to training the human immune system during development.180 Furthermore, infecting germ-free mice with murine norovirus facilitated typical immune development and promoted homeostasis in germ-free mice, while uninfected mice displayed aberrant developmental patterns characteristic of germ-free status.181 Thus, some host-adapted viruses can mimic the developmental effects of bacterial colonization of the host. The spread of these host-adapted viruses can occur via multiple routes and should be affected by social interactions. Some of these viruses, such as anelloviruses, can be transmitted via blood,177 and therefore their transmission should be subject to several of the same processes discussed in levels 1–5 of the social-ecological forces shaping the social microbiome (Figure 1A; Table 1).
The bulk of the host virome consists of viruses targeting the microbiome. Bacterial populations are universally controlled by the bacteriophages that infect them, and the host microbiome is no exception.182,183 Although the viral component of the microbiome remains incompletely characterized, advances in metagenomics have led to the discovery of numerous previously unknown groups of bacteriophages, some of which are highly abundant in the human gut. For instance, Crassvirales viruses infect members of the Bacteroidetes phylum, a major component of the human gut microbiome comprising bacteria that are difficult to cultivate.184,185 The challenges of cultivating Bacteroidetes members mean their associated bacteriophage communities remain relatively poorly characterized.
Phage-prokaryote associations are highly specific, with phage lineages usually infecting a very narrow range of prokaryotic hosts. To some extent, all microbiome-immune interactions (Figure 3) could reflect various bacteriophage-bacteria interactions. In other words, any bacterium is engaged not only in interacting with the host and competing with other bacteria, but also with adapting to bacteriophage presence and coordinating antiphage defenses.186 Thus, all or most bacterial effects on the host should be considered in the context of a tri-partite system of interactions comprising bacteria-host interactions, bacteria-bacteria interactions, and bacteria-bacteriophage interactions.187 There can be little doubt that bacteriophages and archaeal viruses exert profound bottom-up control upon the microbial populations they infect, alongside the top-down control exerted by the host immune system. Explorations of host-bacteria-bacteriophage interactions and their role in host health and disease are important avenues for research.
Because bacteriophages most likely accompany any bacterial transmission, bacteriophages should be subject to the same social-ecological processes across levels 1–5 that shape the social microbiome (Figure 1A; Table 1). Several recent studies have found evidence of such phenomena. For instance, pursuant to findings of socially structured gut bacterial communities in baboons,14 host social groups can also be distinguished on the basis of their bacteriophage communities.183 Paralleling results for bacterial composition, grooming intensity between baboons predicts bacteriophage community similarity, even after controlling for genetic relatedness183 (level 1; Figure 1A; Table 1). Such parallels between bacterial and bacteriophage dynamics have also been observed in human studies. For instance, in studies of adult monozygotic twins, bacteriophage diversity is closely correlated with bacterial diversity.188 Furthermore, as with bacteria, the infant virome bears a signature of birth mode, with vaginally delivered infants displaying more diverse viromes than infants delivered via caesarean section.189 Finally, as with the bacterial members of the gut microbiome,31 bacteriophage composition also changes during senescence.190 These findings highlight the parallels between bacteriophage and bacterial transmission and within-host dynamics in the social microbiome.
Bacteriophages can beneficially affect host health. For instance, amongst individuals with Vibrio cholera infection, those harboring bacteriophages that infect Vibrio cholera experience less virulent disease.191 These beneficial effects may also extend to psychological domains such as cognitive performance. For example, some bacteriophages infect bacteria that can impair host cognition, leading to enhanced host cognitive performance.192 This pattern has been observed in humans and experimentally demonstrated in mice and flies.192 Conversely, bacteriophages could harm host health by infecting bacteria that are beneficial for the host, or by infecting commensal and mutualistic bacteria that compete with pathogens and pathobionts.193 Overall, bacteriophage transmission likely comprises an under-investigated component of health and disease. Indeed, because bacteriophages are seldom quantified or intentionally controlled in studies, it is worth considering the possibility that many of the physiological effects currently attributed to the bacterial component of the microbiome arise in part from the myriad interactions between bacteria and their associated bacteriophages.
A more complete understanding of how microbial transmission affects host health requires explicit incorporation of the virome. For instance, the inclusion of viral persistence and replication strategies such as lysis-lysogeny switching and broader ecological processes such as predator-prey dynamics could further enhance our understanding of microbial transmission, assembly, succession, resilience, and functional effects in hosts. Overall, the types of social interactions that drive the transmission of microbes between hosts are expected to entail the concomitant transmission of the phages that infect those microbes. The transmission of viruses means that the social microbiome also comprises an inextricable social virome, with implications for host health that offer many opportunities for discovery.
IMPLICATIONS OF THE SOCIAL MICROBIOME FOR COMMUNICABLE AND NON-COMMUNICABLE DISEASES
The potential effects of socially transmissible microbes may also be relevant in the context of communicable diseases and non-communicable diseases. While communicable diseases have declined during the epidemiological transition, non-communicable diseases have become more prevalent and now account for ~75% of all deaths globally.194,195 Using the themes and principles developed in this Perspective, we propose that the social transmission of microbes may enhance or reduce host susceptibility to both communicable and non-communicable diseases (Figure 5), and thus that non-communicable diseases may harbor an under-appreciated communicable component.
Figure 5. Social transmission of microbes and disease risk.
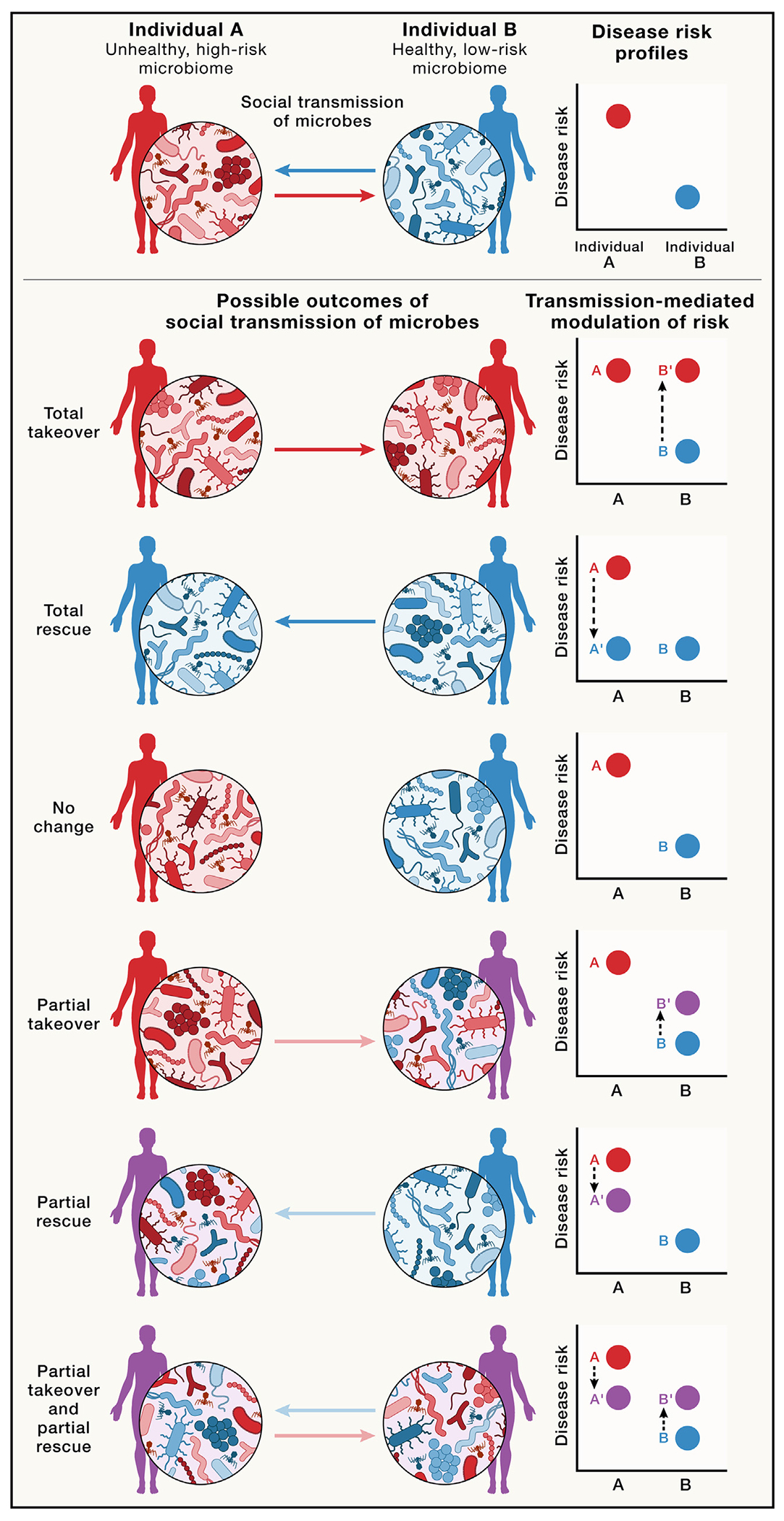
Social microbial transmission between two individuals—a relatively unhealthy individual with a high-risk microbiome (A; red) and a relatively healthy individual with a low-risk microbiome (B; blue)—can result in a range of outcomes. The first three scenarios (i.e., total takeover, total rescue, and no change, indicated by the darker arrows) can be useful to consider as hypothetical extremes, but the latter three transmission scenarios (partial take-over,28 partial rescue, and partial takeover and rescue, indicated by the lighter arrows) are more likely. The purple silhouettes illustrate the intermediate health status conferred by hybrid microbiomes resulting from social transmission. The degree to which socially acquired microbes alter gut microbiome structure and function can then modulate the degree of disease risk. On the right-hand side, we highlight the health outcomes associated with each kind of microbial change within individuals A and B. Interactions between the gut microbiome and various non-microbial factors (e.g., genetics, lifestyle, current health status) shape the expected risk of developing disease phenotypes, and responses of individuals to social microbial transmission (movement from A to A′ and B to B′) are expected to vary based on the microbiome profiles of the individual’s social contacts. We only show a limited number of scenarios here. There are of course multiple other possibilities, including varying degrees of “takeover” and “rescue” and varying degrees to which A′ and B′ return to their previous states (A and B) over time. The magnitude of these hypothesized effects must be determined empirically.
Social transmission of microbes and communicable diseases
Socially transmissible microbes may modulate the risks and effects of communicable diseases. For example, microbes can facilitate viral infection of the host, including via enhancing the stability of virions as they bind to host cells or suppressing host antiviral responses.134 To the extent that such microbes are transmissible, host responses to viral infection may be at least partially a function of socially acquired microbes. Furthermore, because gut microbes that are transmissible interact with and modulate host vaccine responses (Table 3), the host’s vaccine-mediated resistance to various bacterial and viral pathogens could be at least partly influenced by socially acquired microbes.
The social transmission of antibiotic-resistant microbes and microbial genes could also enhance the recipient’s resistance to antibiotic treatment during future bacterial infection. Indeed, recent in vitro evidence suggests that even non-antibiotic drugs such as antidepressants may drive the emergence of antibiotic resistance at clinically relevant concentrations.196 This could potentially create further opportunities for the social transmission of antibiotic-resistant microbes. For instance, we can imagine a hypothetical scenario in which one household member is under treatment for major depressive disorder. Not only is the patient at risk of developing antidepressant-mediated antibiotic resistance, there is also the possibility that these antibiotic-resistant microbes will be transmitted to other household members. This creates a route via which the treatment of a non-communicable disease may affect the outcomes of treatments for communicable diseases in the patient’s social network.
The social transmission of microbes could also contribute to the mitigation of communicable diseases. Socially acquired microbes could contribute to colonization resistance, thereby protecting the host against various bacterial pathogens and reducing the risk and severity of disease (Figure 3). The protective effects of socially acquired microbes against communicable diseases are well known in insects. For instance, in bumblebees, hosts gain gut bacteria via contact with the feces of nestmates after pupal eclosion, and these bacteria provide protection against parasitic infection by the virulent trypanosomatid Crithidia bombi.197 This principle was recently extended to the design of synthetic probiotics to protect hosts. Synthetic symbionts administered to honeybees enhanced resilience to Nosema ceranae, a microsporidian parasite frequently associated with colony collapse disorder.198 Crucially, the synthetic probiotics spread among co-housed bees, a phenomenon with implications for hive-wide protection.198
Among mammals, protection from disease has been shown to be a transmissible phenotype: when gut microbes sourced from humans showing differential susceptibility to enteric infections were transferred into germ-free mice, the recipient mice recapitulated their donor’s resistance traits when exposed to Citrobacter rodentium infection.139 Specifically, when the most susceptible mice (i.e., mice colonized with microbes from the most susceptible humans) were co-housed with the most resistant mice (i.e., mice colonized with microbes from the most resistant humans), the susceptible mice became more resistant but the resistant mice did not become more susceptible to Citrobacter rodentium infection139 (visualized in the “rescue” scenarios in Figure 5). In addition, endogenous microbial populations interact with pathogenic eukaryotic-adapted viruses in ways that may suppress viral infection – for example, by interfering with virions as they attempt to attach to host cells.134 The transmission of bacteriophages could also reduce the virulence of certain infectious diseases as exemplified by the beneficial effects of bacteriophages on Vibrio cholera infection in humans.191 Moreover, some research suggests that bacteria from the socially transmissible bacterial genera Bifidobacterium, Lactobacillus, and Streptococcus (Table 3) may be associated with protection against malaria.199 As in insects, the social transmission of microbes may thus protect mammalian hosts against infections or limit their severity via multiple mechanisms. Furthermore, the design of synthetic, transmissible probiotics for bees198 also has implications for similar approaches targeting humans, and designing transmissible probiotics and treatments might be feasible for human communicable diseases. For example, certain probiotics can drive Staphylococcus aureus decolonization, with beneficial effects on human health.200 If probiotics administered to targeted hosts can colonize the hosts and are transmissible across hosts, this could present opportunities as well as novel ethical dilemmas for the management of communicable diseases.
Socially transmissible microbes and non-communicable diseases
Non-communicable diseases have canonically been considered to arise from non-transmissible risk factors, including genetic and lifestyle features. However, non-communicable diseases may also possess a communicable component arising from the social transmission of microbes,28 and this could be involved in both exacerbating and mitigating disease risk.
Numerous conditions originally classified as non-communicable are now undergoing evaluation for their microbial correlates and causes, including metabolic conditions,3 atherosclerosis,201 various cancers,202 and brain-related syndromes and conditions.145,203,204 Transplanting feces from patients to germ-free animals demonstrates that donor microbes can drive the emergence of a range of clinically relevant phenotypes in naïve recipients. However, many of these studies involve transferring whole gut microbial communities from donor to recipient rather than the transmission of selected members of the microbiota via social interactions. Moreover, several microbes that are socially transmissible can also interfere with the metabolism of drugs intended for the treatment of cancer and various other non-communicable diseases including Parkinson’s Disease, cardiovascular disease, and depression (Table 3). Thus, the next step is to test whether microbes that are associated with disease or which intefere with drug metabolism are socially transmissible.
Empirical support is growing for the general hypothesis that many non-communicable diseases may possess communicable components because the microbes that shape host susceptibilities, outcomes, and responses to treatment are socially transmissible. For example, in a mouse model of autism, the social transmission of segmented filamentous bacteria during pregnancy was shown to interact with maternal immune activation to induce maternal TH17 cell populations that in turn drove social behavioral deficits in offspring.145
Socially transmitted microbes may also confer some protection against non-communicable diseases. For instance, household size, which is expected to correlate positively with within-group microbial transmission (level 2, Figure 1A), was negatively associated with incidence of inflammatory bowel disease.205 A form of this epidemiological pattern was also observed in mouse studies wherein mice living in larger groups displayed enhanced resilience to colitis induced by dextran sulphate sodium compared to mice living in smaller groups.205 This protection was associated with shifts in microbial composition attributable to increased group size.205
With respect to metabolic conditions, co-housing mice harboring gut microbes from human twins discordant for obesity led to biased transfer of lean twin-derived microbes, which protected the mice with the obese twin’s microbes from gaining weight3 (visualized in the “rescue” scenarios in Figure 5). However, these protective effects were diet-dependent, manifesting only when recipient mice were fed chow or diets relatively low in fat and high in fruits and vegetables. This suggests that social transmission of microbes and lifestyle factors such as diet may interact to shape host health outcomes. Similarly, co-housing also improved outcomes for immune checkpoint blockade therapy in melanoma: when therapy-resistant mice were co-housed with therapy-responsive mice, the resistant mice displayed improved treatment outcomes that were attributable to social acquisition of beneficial microbes from the responsive mice.206 Notably, the responsive mice did not become unresponsive to therapy,206 suggesting social transmission of benefits without obvious costs in this case. Furthermore, host responses to cancer treatments are associated with microbial composition, and microbes important to host responsiveness are socially transmissible (Table 3). Thus, the treatment of non-communicable conditions such as cancer may be influenced at least in part by the actions of socially transmissible microbes.
Our argument is not, of course, that conditions traditionally considered non-communicable should be reclassified as communicable based on the possibility of disease-mediating social transmission of microbes, or that microbes from a less healthy individual will necessarily predispose their social contacts to developing disease. Indeed, from ecological theory and the empirical pattern that health-associated microbiomes tend to be more diverse and resilient,3,139,206 the effects of transmission may be more likely to occur in the direction of “rescue” than “takeover” (Figure 5). Of course, beneficial transmission does not always occur,145 and such possibilities warrant further investigation.28 Whether social transmission of non-pathogenic microbes tends to exacerbate or diminish non-communicable-disease risk remains an open question. Likewise, the relevance to humans remains to be established, as most studies to date have been performed in laboratory mice exhibiting coprophagic behaviors that can be expected to facilitate high-fidelity social transmission. Overall, social exposure to, and acquisition of, disease-associated or protective microbes could enrich or diminish the probability of developing clinical phenotypes, respectively, with the degree of modulation likely interacting with host-specific features such as genetics and lifestyle (Figure 5). It is of course possible that the magnitude of the effects of socially transmissible microbes on host health and disease may be no more than small - at this stage we must learn more. Nonetheless, these effects are worth investigating as contributors to the social determinants of health and possibly for their role in social evolution.
CONCLUSIONS: SOCIAL TRANSMISSION OF MICROBES AND HUMAN HEALTH
In this Perspective, we have applied the social microbiome concept (Figure 1A; Table 1) to examine the implications of the social transmission of microbes for host health, disease, and social evolution. Socially transmitted microbes can affect a broad range of processes relevant to host health that can be categorized in terms of eco-evolutionary microbiome community processes and microbial transmission-based processes (Figure 1B). Crucially, socially transmissible commensals and mutualists may modify disease risk for both communicable and non-communicable diseases (Figure 5). If non-communicable disease risks and outcomes can indeed be affected by socially transmissible microbes, we must consider the possibility that non-communicable diseases possess a communicable component.28 Investigating the relevance to humans and the mechanisms via which socially transmissible microbes alter the risk of developing communicable and non-communicable diseases may facilitate the emergence of both knowledge and therapies concerning these diseases.
Understanding the transmission ecologies of pathogenic and non-pathogenic microbes will be an important area of research11: if pathogens, mutualists, and commensals exploit at least somewhat different transmission strategies, or differentially change host behavior (e.g., pathogens triggering social isolation and sickness behavior), then particular social structures could exert distinct effects on the transmission of pathogenic versus mutualistic or commensal microbes. This presents differential targets for natural selection with implications for social evolution. Moreover, the microbes that disperse over host social networks include not just bacterial and viral components – as we have discussed here – but also archaeal, fungal, and various eukaryotic microbiome members. In other words, the social microbiome comprises not only a bacterial component and a social virome, but also a social archaeome, a social mycobiome, and a social eukaryome.
Furthermore, just as human cultural evolution has generated behaviors and practices that restrain the spread of pathogens,207 cultural evolution may also favor the emergence of behavioral patterns and practices to facilitate the transmission of commensal and mutualistic microbes between humans. All organisms and the environment are connected to some degree via microbial transmission, which is also a basic premise of the One Health view of the health of humans, other animals, and the environment. Elucidating these microbial connections may thus also be useful beyond human disease contexts and aid in the management of global health challenges. Ultimately, viewing social microbial transmission through a broader lens – one accommodating commensals and mutualists as well as pathogens – can help us better understand microbial signals influencing the social determinants of health and the pleiotropic roles of transmissible microbes in social evolution.
ACKNOWLEDGMENTS
The authors thank Cary Allen-Blevins, Melanie Colvin, Arjun Dutta, Mira-Rose Kingsbury Lee, Timothy Kistner, Grace Rubin, Laura Schell, Emily Venable, and Nikolas Weyland for feedback on various aspects of the manuscript. A.S., C.J.A.M., S.H., N.G.O.I., K.V.-A.J., and J.H. declare no relevant funding. A.R. reports funding from the Kone Foundation (202007064). M.V.-C. reports funding from a Beatriz Galindo Junior Fellowship from the Spanish Ministry of Universities (BG22/00172) and a Knowledge Generation Project grant from the Spanish Ministry of Science and Innovation (PID2022-139328OA-I00). I.L.B. receives support from the NOMIS Foundation (GR108454-CON-80002144), the David and Lucile Packard Foundation, and from the Pew Charitable Trusts. E.A.A. reports funding from the National Science Foundation (DEB-1840223) and the National Institutes of Health (R01 AG071684). L.B.B. reports funding from the National Institutes of Health (R01 GM134376 and P30 DK042086). B.B.F. reports funding from the Canadian Institute for Advanced Research (FL-001238/Appt 2410; additional sponsor reference number: FS22-058/Appt 2410) and the Canadian Institutes of Health Research (FDN-159935). E.V.K. reports funding from the Intramural Research Program of the National Institutes of Health of the United States of America (National Library of Medicine). F.S.G. reports funding from the National Cancer Institute (1 K22 CA258960-01). R.N.C. reports funding from the National Science Foundation (BCS-1919892 and BCS-2142073), the William F. Milton Fund, and the Harvard University Dean’s Competitive Fund for Promising Scholarship. A.H.M. reports funding from the National Institute of General Medical Sciences (R35 GM138284). Figures 4 and 5 were originally prepared using BioRender.com
Footnotes
DECLARATION OF INTERESTS
A.S., C.J.A.M., A.R., N.G.O.I., K.V.-A.J., M.V.-C., I.L.B., J.H., E.A.A., L.B.B., B.B.F., R.N.C., and A.H.M. declare no competing interests. S.H. is a current employee of SilverCloud Health and SilverCloud Health is a subsidiary of Amwell. S.H. holds shares in Amwell. F.S.G. is listed as an inventor on patent applications pertaining to the treatment of cancer or the study of the human microbiome. E.V.K. is listed as an inventor on patents and patent applications pertaining to CRISPR enzymes and systems or protein regulation.
REFERENCES
- 1.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA 110, 3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmody RN, and Bisanz JE (2023). Roles of the gut microbiome in weight management. Nat. Rev. Microbiol 21, 535–550. [DOI] [PubMed] [Google Scholar]
- 3.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng D, Liwinski T, and Elinav E (2020). Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belkaid Y, and Hand TW (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson KV-A, and Foster KR (2018). Why does the microbiome affect behaviour? Nat. Rev. Microbiol 16, 647–655. [DOI] [PubMed] [Google Scholar]
- 7.Sarkar A, Harty S, Johnson KV-A, Moeller AH, Carmody RN, Lehto SM, Erdman SE, Dunbar RIM, and Burnet PWJ (2020). The role of the microbiome in the neurobiology of social behaviour. Biol. Rev 95, 1131–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarkar A, Harty S, Lehto SM, Moeller AH, Dinan TG, Dunbar RIM, Cryan JF, and Burnet PWJ (2018). The microbiome in psychology and cognitive neuroscience. Trends Cogn. Sci 22, 611–636. [DOI] [PubMed] [Google Scholar]
- 9.Antwis RE, Lea JMD, Unwin B, and Shultz S (2018). Gut microbiome composition is associated with spatial structuring and social interactions in semi-feral Welsh Mountain ponies. Microbiome 6, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brito IL, Gurry T, Zhao S, Huang K, Young SK, Shea TP, Naisilisili W, Jenkins AP, Jupiter SD, Gevers D, and Alm EJ (2019). Transmission of human-associated microbiota along family and social networks. Nat. Microbiol 4, 964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Browne HP, Neville BA, Forster SC, and Lawley TD (2017).Transmission of the gut microbiota: Spreading of health. Nat. Rev. Microbiol 15, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moeller AH, Foerster S, Wilson ML, Pusey AE, Hahn BH, and Ochman H (2016). Social behavior shapes the chimpanzee pan-microbiome. Sci. Adv 2, e1500997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raulo A, Allen BE, Troitsky T, Husby A, Firth JA, Coulson T, and Knowles SCL (2021). Social networks strongly predict the gut microbiota of wild mice. ISME J. 15, 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tung J, Barreiro LB, Burns MB, Grenier J-C, Lynch J, Grieneisen LE, Altmann J, Alberts SC, Blekhman R, and Archie EA (2015). Social networks predict gut microbiome composition in wild baboons. eLife 4, e05224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valles-Colomer M, Blanco-Míguez A, Manghi P, Asnicar F, Dubois L, Golzato D, Armanini F, Cumbo F, Huang KD, Manara S, et al. (2023). The person-to-person transmission landscape of the gut and oral microbiomes. Nature 614, 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valles-Colomer M, Bacigalupe R, Vieira-Silva S, Suzuki S, Darzi Y, Tito RY, Yamada T, Segata N, Raes J, and Falony G (2022). Variation and transmission of the human gut microbiota across multiple familial generations. Nat. Microbiol 7, 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter MM, Olm MR, Merrill BD, Dahan D, Tripathi S, Spencer SP, Yu FB, Jain S, Neff N, Jha AR, et al. (2023). Ultra-deep sequencing of Hadza hunter-gatherers recovers vanishing gut microbes. Cell 186, 3111–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snyder-Mackler N, Burger JR, Gaydosh L, Belsky DW, Noppert GA, Campos FA, Bartolomucci A, Yang YC, Aiello AE, O’Rand A, et al. (2020). Social determinants of health and survival in humans and other animals. Science 368, eaax9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altizer S, Nunn CL, Thrall PH, Gittleman JL, Antonovics J, Cunningham AA, Dobson AP, Ezenwa V, Jones KE, Pedersen AB, et al. (2003). Social organization and parasite risk in mammals: Integrating theory and empirical studies. Annu. Rev. Ecol. Evol. Syst 34, 517–547. [Google Scholar]
- 20.Cremer S, Pull CD, and Fürst MA (2018). Social immunity: Emergence and evolution of colony-level disease protection. Annu. Rev. Entomol 63, 105–123. [DOI] [PubMed] [Google Scholar]
- 21.Loehle C (1995). Social barriers to pathogen transmission in wild animal populations. Ecology 76, 326–335. [Google Scholar]
- 22.Dunn RR, Amato KR, Archie EA, Arandjelovic M, Crittenden AN, and Nichols LM (2020). The internal, external and extended microbiomes of hominins. Front. Ecol. Evol 8, 25. [Google Scholar]
- 23.Archie EA, and Theis KR (2011). Animal behaviour meets microbial ecology. Anim. Behav 82, 425–436. [Google Scholar]
- 24.Lombardo MP (2008). Access to mutualistic endosymbiotic microbes: An underappreciated benefit of group living. Behav. Ecol. Sociobiol 62, 479–497. [Google Scholar]
- 25.Montiel-Castro AJ, González-Cervantes RM, Bravo-Ruiseco G, and Pacheco-López G (2013). The microbiota-gut-brain axis: Neurobehavioral correlates, health and sociality. Front. Integr. Neurosci 7, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Troyer K (1984). Microbes, herbivory and the evolution of social behavior. J. Theor. Biol 106, 157–169. [Google Scholar]
- 27.Sarkar A, Harty S, Johnson KV-A, Moeller AH, Archie EA, Schell LD, Carmody RN, Clutton-Brock TH, Dunbar RIM, and Burnet PWJ (2020). Microbial transmission in animal social networks and the social microbiome. Nat. Ecol. Evol 4, 1020–1035. [DOI] [PubMed] [Google Scholar]
- 28.Finlay BB; CIFAR Humans and the Microbiome (2020). Are noncommunicable diseases communicable? Science 367, 250–251. [DOI] [PubMed] [Google Scholar]
- 29.Pullman J, Beghini F, Alexander M, Shridhar SV, Prinster D, Brito IL, and Christakis NA (2023). Detailed social network interactions and gut microbiome strain-sharing within isolated Honduras villages. Preprint at bioRxiv. 10.1101/2023.04.06.535875. [DOI] [Google Scholar]
- 30.Toren C (1990). Making sense of hierarchy: Cognition as social process in Fiji (The Athlone Press; ). [Google Scholar]
- 31.Kundu P, Blacher E, Elinav E, and Pettersson S (2017). Our gut microbiome: The evolving inner self. Cell 171, 1481–1493. [DOI] [PubMed] [Google Scholar]
- 32.Sprockett DD, Martin M, Costello EK, Burns AR, Holmes SP, Gurven MD, and Relman DA (2020). Microbiota assembly, structure, and dynamics among Tsimane horticulturalists of the Bolivian Amazon. Nat. Commun 11, 3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farley TM, Samuelson J, Grabowski MK, Ameyan W, Gray RH, and Baggaley R (2020). Impact of male circumcision on risk of HIV infection in men in a changing epidemic context – systematic review and meta-analysis. J. Int. AIDS Soc 23, e25490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu CM, Hungate BA, Tobian AAR, Serwadda D, Ravel J, Lester R, Kigozi G, Aziz M, Galiwango RM, Nalugoda F, et al. (2013). Male circumcision significantly reduces prevalence and load of genital anaerobic bacteria. mBio 4, e00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grieneisen LE, Livermore J, Alberts S, Tung J, and Archie EA (2017). Group living and male dispersal predict the core gut microbiome in wild baboons. Integr. Comp. Biol 57, 770–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tegally H, Wilkinson E, Tsui JL-H, Moir M, Martin D, Brito AF, Giovanetti M, Khan K, Huber C, Bogoch II, et al. (2023). Dispersal patterns and influence of air travel during the global expansion of SARS-CoV-2 variants of concern. Cell 186, 3277–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powell JE, Martinson VG, Urban-Mead K, and Moran NA (2014). Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol 80, 7378–7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandler JA, Lang JM, Bhatnagar S, Eisen JA, and Kopp A (2011). Bacterial communities of diverse Drosophila species: Ecological context of a host–microbe model system. PLoS Genet. 7, e1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grupstra CGB, Lemoine NP, Cook C, and Correa AMS (2022). Thank you for biting: Dispersal of beneficial microbiota through “antagonistic” interactions. Trends Microbiol. 30, 930–939. [DOI] [PubMed] [Google Scholar]
- 40.Moeller AH, Suzuki TA, Lin D, Lacey EA, Wasser SK, and Nachman MW (2017). Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proc. Natl. Acad. Sci. USA 114, 13768–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moeller AH, Peeters M, Ndjango J-B, Li Y, Hahn BH, and Ochman H (2013). Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Res. 23, 1715–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Caporaso JG, Knights D, Clemente JC, Nakielny S, et al. (2013). Cohabiting family members share microbiota with one another and with their dogs. eLife 2, e00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, Boushey HA, Zoratti E, Ownby D, Lukacs NW, and Lynch SV (2014). House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc. Natl. Acad. Sci. USA 111, 805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun J, Liao X-P, D’Souza AW, Boolchandani M, Li S-H, Cheng K, Luis Martínez J, Li L, Feng Y-J, Fang L-X, et al. (2020). Environmental remodeling of human gut microbiota and antibiotic resistome in livestock farms. Nat. Commun 11, 1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X-R, Lian X-L, Su T-T, Long T-F, Li M-Y, Feng X-Y, Sun R-Y, Cui Z-H, Tang T, Xia J, et al. (2021). Duck wastes as a potential reservoir of novel antibiotic resistance genes. Sci. Total Environ 771, 144828. [DOI] [PubMed] [Google Scholar]
- 46.Grieneisen L, Dasari M, Gould TJ, Björk JR, Grenier J-C, Yotova V, Jansen D, Gottel N, Gordon JB, Learn NH, et al. (2021). Gut microbiome heritability is nearly universal but environmentally contingent. Science 373, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, Holt RD, Shurin JB, Law R, Tilman D, et al. (2004). The metacommunity concept: A framework for multi-scale community ecology. Ecol. Lett 7, 601–613. [Google Scholar]
- 48.Coyte KZ, Schluter J, and Foster KR (2015). The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. [DOI] [PubMed] [Google Scholar]
- 49.Miller ET, Svanbäck R, and Bohannan BJM (2018). Microbiomes as metacommunities: Understanding host-associated microbes through metacommunity ecology. Trends Ecol. Evol 33, 926–935. [DOI] [PubMed] [Google Scholar]
- 50.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, and Relman DA (2012). The application of ecological theory toward an understanding of the human microbiome. Science 336, 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raulo A, Bürkner P, Dale J, English H, Finerty G, Lamberth C, Firth JA, Coulson T, and Knowles SC (2023). Social and environmental transmission spread different sets of gut microbes in wild mice. Preprint at bioRxiv. 10.1101/2023.07.20.549849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gardy JL, Johnston JC, Ho Sui SJ, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, et al. (2011). Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N. Engl. J. Med 364, 730–739. [DOI] [PubMed] [Google Scholar]
- 53.Banerjee S, and van der Heijden MGA (2023). Soil microbiomes and one health. Nat. Rev. Microbiol 21, 6–20. [DOI] [PubMed] [Google Scholar]
- 54.Kamalakkannan V, Salim AA, and Capon RJ (2017). Microbiome-mediated biotransformation of cane toad bufagenins. J. Nat. Prod 80, 2012–2017. [DOI] [PubMed] [Google Scholar]
- 55.Koga R, Meng X-Y, Tsuchida T, and Fukatsu T (2012). Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte–embryo interface. Proc. Natl. Acad. Sci. USA 109, E1230–E1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matheen MIA, Gillings MR, and Dudaniec RY (2022). Dominant factors shaping the gut microbiota of wild birds. Emu - Austral Ornithology 122, 255–268. [Google Scholar]
- 57.Yassour M, Vatanen T, Siljander H, Hämäläinen AM, Härkönen T, Ryhänen SJ, Franzosa EA, Vlamakis H, Huttenhower C, Gevers D, et al. (2016). Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med 8, 343ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, and Knight R (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 107, 11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lubin J-B, Green J, Maddux S, Denu L, Duranova T, Lanza M, Wynosky-Dolfi M, Flores JN, Grimes LP, Brodsky IE, et al. (2023). Arresting microbiome development limits immune system maturation and resistance to infection in mice. Cell Host Microbe 31, 554–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mueller NT, Bakacs E, Combellick J, Grigoryan Z, and Dominguez-Bello MG (2015). The infant microbiome development: Mom matters. Trends Mol. Med 21, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wanelik KM, Raulo A, Troitsky T, Husby A, and Knowles SCL (2023). Maternal transmission gives way to social transmission during gut microbiota assembly in wild mice. Anim. Microbiome 5, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moeller AH, Suzuki TA, Phifer-Rixey M, and Nachman MW (2018). Transmission modes of the mammalian gut microbiota. Science 362, 453–457. [DOI] [PubMed] [Google Scholar]
- 63.Mazel F, Guisan A, and Parfrey LW (2023). Transmission mode and dispersal traits correlate with host specificity in mammalian gut microbes. Mol. Ecol 10.1111/mec.16862. [DOI] [PubMed] [Google Scholar]
- 64.Hildebrand F, Gossmann TI, Frioux C, Özkurt E, Myers PN, Ferretti P, Kuhn M, Bahram M, Nielsen HB, and Bork P (2021). Dispersal strategies shape persistence and evolution of human gut bacteria. Cell Host Microbe 29, 1167–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raulo A, Ruokolainen L, Lane A, Amato K, Knight R, Leigh S, Stumpf R, White B, Nelson KE, Baden AL, and Tecot SR (2018). Social behaviour and gut microbiota in red-bellied lemurs (Eulemur rubriventer): In search of the role of immunity in the evolution of sociality. J. Anim. Ecol 87, 388–399. [DOI] [PubMed] [Google Scholar]
- 66.Johnson KV-A (2020). Gut microbiome composition and diversity are related to human personality traits. Hum. Microb. J 15, 100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lane AA, McGuire MK, McGuire MA, Williams JE, Lackey KA, Hagen EH, Kaul A, Gindola D, Gebeyehu D, Flores KE, et al. (2019). Household composition and the infant fecal microbiome: The INSPIRE study. Am. J. Phys. Anthropol 169, 526–539. [DOI] [PubMed] [Google Scholar]
- 68.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni H, Metcalf GA, et al. (2018). Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, et al. (2014). Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, Wirbel J, Maistrenko OM, Alves RJ, Bergsten E, et al. (2019). Extensive transmission of microbes along the gastrointestinal tract. eLife 8, e42693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al. (2015). The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med 21, 895–905. [DOI] [PubMed] [Google Scholar]
- 72.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, VanTreuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Amato KR, Arrieta M-C, Azad MB, Bailey MT, Broussard JL, Bruggeling CE, Claud EC, Costello EK, Davenport ER, Dutilh BE, et al. (2021). The human gut microbiome and health inequities. Proc. Natl. Acad. Sci. USA 118, e2017947118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rook G, Bäckhed F, Levin BR, McFall-Ngai MJ, and McLean AR (2017). Evolution, human-microbe interactions, and life history plasticity. Lancet 390, 521–530. [DOI] [PubMed] [Google Scholar]
- 75.Buffie CG, and Pamer EG (2013). Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol 13, 790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caballero-Flores G, Pickard JM, and Núñez G (2023). Microbiota-mediated colonization resistance: mechanisms and regulation. Nat. Rev. Microbiol 21, 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hashimoto D, Miller J, and Merad M (2011). Dendritic cell and macrophage heterogeneity in vivo. Immunity 35, 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cummings JH, and Macfarlane GT (1997). Role of intestinal bacteria in nutrient metabolism. J. Parenter. Enteral Nutr 21, 357–365. [DOI] [PubMed] [Google Scholar]
- 79.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, and Knight R (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Momose Y, Hirayama K, and Itoh K (2008). Effect of organic acids on inhibition of Escherichia coli O157:H7 colonization in gnotobiotic mice associated with infant intestinal microbiota. Antonie Leeuwenhoek 93, 141–149. [DOI] [PubMed] [Google Scholar]
- 81.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, et al. (2008). Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun 76, 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spragge F, Bakkeren E, Jahn MT, Araujo EBN, Pearson CF, Wang X, Pankhurst L, Cunrath O, and Foster KR (2023). Microbiome diversity protects against pathogens by nutrient blocking. Science 382, eadj3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caballero-Flores G, Pickard JM, Fukuda S, Inohara N, and Núñez G (2020). An enteric pathogen subverts colonization resistance by evading competition for amino acids in the gut. Cell Host Microbe 28, 526–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. (2011). Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547. [DOI] [PubMed] [Google Scholar]
- 85.Lawley TD, and Walker AW (2013). Intestinal colonization resistance. Immunology 138, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Levine JM, and D’Antonio CM (1999). Elton revisited: A review of evidence linking diversity and invasibility. Oikos 87, 15–26. [Google Scholar]
- 87.Kim S, Covington A, and Pamer EG (2017). The intestinal microbiota: Antibiotics, colonization resistance, and enteric pathogens. Immunol. Rev 279, 90–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reese AT, and Dunn RR (2018). Drivers of microbiome biodiversity: A review of general rules, feces, and ignorance. mBio 9, e01294–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HMB, Coakley M, Lakshminarayanan B, O’Sullivan O, et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184. [DOI] [PubMed] [Google Scholar]
- 90.Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, Knight R, Fontenot AP, and Palmer BE (2013). Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 14, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnson KV-A, and Burnet PWJ (2016). Microbiome: should we diversify from diversity? Gut Microb. 7, 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thaiss CA, Itav S, Rothschild D, Meijer MT, Levy M, Moresi C, Dohnalová L, Braverman S, Rozin S, Malitsky S, et al. (2016). Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 540, 544–551. [DOI] [PubMed] [Google Scholar]
- 93.Smith P, Willemsen D, Popkes M, Metge F, Gandiwa E, Reichard M, and Valenzano DR (2017). Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. eLife 6, e27014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, Wang W, Tang W, Tan Z, Shi J, et al. (2015). Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun 48, 186–194. [DOI] [PubMed] [Google Scholar]
- 95.Weldon L, Abolins S, Lenzi L, Bourne C, Riley EM, and Viney M (2015). The gut microbiota of wild mice. PLoS One 10, e0134643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pfau M, Degregori S, Johnson G, Tennenbaum SR, Barber PH, Philson CS, and Blumstein DT (2023). The social microbiome: Gut microbiome diversity and abundance are negatively associated with sociality in a wild mammal. R. Soc. Open Sci 10, 231305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lahti L, Salojärvi J, Salonen A, Scheffer M, and de Vos WM (2014). Tipping elements in the human intestinal ecosystem. Nat. Commun 5, 4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zaneveld JR, McMinds R, and Vega Thurber R (2017). Stress and stability: Applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol 2, 17121. [DOI] [PubMed] [Google Scholar]
- 99.Dey P, and Ray Chaudhuri S (2023). The opportunistic nature of gut commensal microbiota. Crit. Rev. Microbiol 49, 739–763. [DOI] [PubMed] [Google Scholar]
- 100.Youmans BP, Ajami NJ, Jiang Z-D, Campbell F, Wadsworth WD, Petrosino JF, DuPont HL, and Highlander SK (2015). Characterization of the human gut microbiome during travelers’ diarrhea. Gut Microb. 6, 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Björk JR, Dasari M, Grieneisen L, and Archie EA (2019). Primate microbiomes over time: Longitudinal answers to standing questions in microbiome research. Am. J. Primatol 81, e22970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Perofsky AC, Ancel Meyers L, Abondano LA, Di Fiore A, and Lewis RJ (2021). Social groups constrain the spatiotemporal dynamics of wild sifaka gut microbiomes. Mol. Ecol 30, 6759–6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao Q, and Elson CO (2018). Adaptive immune education by gut microbiota antigens. Immunology 154, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Feng T, and Elson CO (2011). Adaptive immunity in the host–microbiota dialog. Mucosal Immunol. 4, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Amato KR (2016). Incorporating the gut microbiota into models of human and non-human primate ecology and evolution. Am. J. Phys. Anthropol 159, 196–215. [DOI] [PubMed] [Google Scholar]
- 106.Grieneisen LE, Charpentier MJE, Alberts SC, Blekhman R, Brad-burd G, Tung J, and Archie EA (2019). Genes, geology and germs: Gut microbiota across a primate hybrid zone are explained by site soil properties, not host species. Proc. Biol. Sci 286, 20190431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ewald PW (1987). Transmission modes and evolution of the parasitism-mutualism continuum. Ann. N. Y. Acad. Sci 503, 295–306. [DOI] [PubMed] [Google Scholar]
- 108.Stewart AD, Logsdon JM, and Kelley SE (2005). An empirical study of the evolution of virulence under both horizontal and vertical transmission. Evolution 59, 730–739. [PubMed] [Google Scholar]
- 109.Foster KR, Schluter J, Coyte KZ, and Rakoff-Nahoum S (2017). The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lipsitch M, Siller S, and Nowak MA (1996). The evolution of virulence in pathogens with vertical and horizontal transmission. Evolution 50, 1729–1741. [DOI] [PubMed] [Google Scholar]
- 111.Ebert D, and Bull JJ (2003). Challenging the trade-off model for the evolution of virulence: Is virulence management feasible? Trends Microbiol. 11, 15–20. [DOI] [PubMed] [Google Scholar]
- 112.Hoyt JR, Langwig KE, White JP, Kaarakka HM, Redell JA, Kurta A, DePue JE, Scullon WH, Parise KL, Foster JT, et al. (2018). Cryptic connections illuminate pathogen transmission within community networks. Nature 563, 710–713. [DOI] [PubMed] [Google Scholar]
- 113.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, and Lawley TD (2016). Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dale VH, Joyce LA, McNulty S, Neilson RP, Ayres MP, Flannigan MD, Hanson PJ, Irland LC, Lugo AE, Peterson CJ, et al. (2001). Climate change and forest disturbances: climate change can affect forests by altering the frequency, intensity, duration, and timing of fire, drought, introduced species, insect and pathogen outbreaks, hurricanes, windstorms, ice storms, or landslides. Bioscience 51, 723–734. [Google Scholar]
- 115.Gunderson LH (2000). Ecological resilience—in theory and application. Annu. Rev. Ecol. Syst 31, 425–439. [Google Scholar]
- 116.de Nies L, Kobras CM, and Stracy M (2023). Antibiotic-induced collateral damage to the microbiota and associated infections. Nat. Rev. Microbiol 21, 789–804. [DOI] [PubMed] [Google Scholar]
- 117.Hagan T, Cortese M, Rouphael N, Boudreau C, Linde C, Maddur MS, Das J, Wang H, Guthmiller J, Zheng N-Y, et al. (2019). Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell 178, 1313–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jernberg C, Löfmark S, Edlund C, and Jansson JK (2007). Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1, 56–66. [DOI] [PubMed] [Google Scholar]
- 119.Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, and Engstrand L (2010). Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One 5, e9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dethlefsen L, and Relman DA (2011). Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA 108, 4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Palleja A, Mikkelsen KH, Forslund SK, Kashani A, Allin KH, Nielsen T, Hansen TH, Liang S, Feng Q, Zhang C, et al. (2018). Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol 3, 1255–1265. [DOI] [PubMed] [Google Scholar]
- 122.Kimura I, Miyamoto J, Ohue-Kitano R, Watanabe K, Yamada T, Onuki M, Aoki R, Isobe Y, Kashihara D, Inoue D, et al. (2020). Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science 367, eaaw8429. [DOI] [PubMed] [Google Scholar]
- 123.Dethlefsen L, Huse S, Sogin ML, and Relman DA (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6, e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Raymann K, Shaffer Z, and Moran NA (2017). Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 15, e2001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Reese AT, Cho EH, Klitzman B, Nichols SP, Wisniewski NA, Villa MM, Durand HK, Jiang S, Midani FS, Nimmagadda SN, et al. (2018). Antibiotic-induced changes in the microbiota disrupt redox dynamics in the gut. eLife 7, e35987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ng KM, Aranda-Díaz A, Tropini C, Frankel MR, Van Treuren W, O’Loughlin CT, Merrill BD, Yu FB, Pruss KM, Oliveira RA, et al. (2019). Recovery of the gut microbiota after antibiotics depends on host diet, community context, and environmental reservoirs. Cell Host Microbe 26, 650–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Clavel T, Lepage P, and Charrier C (2014). The family Coriobacteriaceae. In The prokaryotes: Actinobacteria, Rosenberg E, DeLong EF, Lory S, Stackebrandt E, and Thompson F, eds. (Springer; ), pp. 201–238. [Google Scholar]
- 128.Martín R, Rios-Covian D, Huillet E, Auger S, Khazaal S, Bermúdez-Humarán LG, Sokol H, Chatel J-M, and Langella P (2023). Faecalibacterium: A bacterial genus with promising human health applications. FEMS Microbiol. Rev 47, fuad039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.O’Callaghan A, and van Sinderen D (2016). Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol 7, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kohl KD, Stengel A, and Dearing MD (2016). Inoculation of tannin-degrading bacteria into novel hosts increases performance on tannin-rich diets. Environ. Microbiol 18, 1720–1729. [DOI] [PubMed] [Google Scholar]
- 131.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, et al. (2014). Multistate point-prevalence survey of health care–associated infections. N. Engl. J. Med 370, 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Johnson KV-A, Watson KK, Dunbar RIM, and Burnet PWJ (2022). Sociability in a non-captive macaque population is associated with beneficial gut bacteria. Front. Microbiol 13, 1032495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sarkar A, Harty S, Moeller AH, Klein SL, Erdman SE, Friston KJ, and Carmody RN (2021). The gut microbiome as a biomarker of differential susceptibility to SARS-CoV-2. Trends Mol. Med 27, 1115–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li N, Ma W-T, Pang M, Fan Q-L, and Hua J-L (2019). The commensal microbiota and viral infection: A comprehensive review. Front. Immunol 10, 1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. [DOI] [PubMed] [Google Scholar]
- 136.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, and Garrett WS (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, et al. (2017). Mining the human gut microbiota for immunomodulatory organisms. Cell 168, 928–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schluter J, Peled JU, Taylor BP, Markey KA, Smith M, Taur Y, Niehus R, Staffas A, Dai A, Fontana E, et al. (2020). The gut microbiota is associated with immune cell dynamics in humans. Nature 588, 303–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Porras AM, Shi Q, Zhou H, Callahan R, Montenegro-Bethancourt G, Solomons N, and Brito IL (2021). Geographic differences in gut microbiota composition impact susceptibility to enteric infection. Cell Rep. 36, 109457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mazmanian SK, Round JL, and Kasper DL (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625. [DOI] [PubMed] [Google Scholar]
- 141.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. [DOI] [PubMed] [Google Scholar]
- 142.Faith JJ, Ahern PP, Ridaura VK, Cheng J, and Gordon JI (2014). Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci. Transl. Med 6, 220ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dasgupta S, Erturk-Hasdemir D, Ochoa-Reparaz J, Reinecker H-C, and Kasper DL (2014). Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe 15, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, and Kasper DL (2014). Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kim S, Kim H, Yim YS, Ha S, Atarashi K, Tan TG, Longman RS, Honda K, Littman DR, Choi GB, and Huh JR (2017). Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature 549, 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, and Hooper LV (2011). The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee J-Y, Ziel JW, Miraldi ER, Domingos AI, et al. (2015). An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 163, 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Macpherson AJ, and Uhr T (2004). Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303, 1662–1665. [DOI] [PubMed] [Google Scholar]
- 150.Pedicord VA, Lockhart AAK, Rangan KJ, Craig JW, Loschko J, Rogoz A, Hang HC, and Mucida D (2016). Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance. Sci. Immunol 1, eaai7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rangan KJ, Pedicord VA, Wang Y-C, Kim B, Lu Y, Shaham S, Mucida D, and Hang HC (2016). A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science 353, 1434–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, et al. (2015). Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Dail-lère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. [DOI] [PubMed] [Google Scholar]
- 154.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, Luke JJ, and Gajewski TF (2018). The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Griffin ME, Espinosa J, Becker JL, Luo J-D, Carroll TS, Jha JK, Fanger GR, and Hang HC (2021). Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science 373, 1040–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lam KC, Araya RE, Huang A, Chen Q, Di Modica M, Rodrigues RR, Lopès A, Johnson SB, Schwarz B, Bohrnsen E, et al. (2021). Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell 184, 5338–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, et al. (2016). Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351, aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Park JS, Gazzaniga FS, Wu M, Luthens AK, Gillis J, Zheng W, LaFleur MW, Johnson SB, Morad G, Park EM, et al. (2023). Targeting PD-L2-RGMb overcomes microbiome-related immunotherapy resistance. Nature 617, 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, Paik S, Stagg J, Groves RA, Gallo M, et al. (2020). Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 369, 1481–1489. [DOI] [PubMed] [Google Scholar]
- 161.Harris VC, Armah G, Fuentes S, Korpela KE, Parashar U, Victor JC, Tate J, de Weerth C, Giaquinto C, Wiersinga WJ, et al. (2017). Significant correlation between the infant gut microbiome and rotavirus vaccine response in rural Ghana. J. Infect. Dis 215, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Maini Rekdal V, Bess EN, Bisanz JE, Turnbaugh PJ, and Balskus EP (2019). Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science 364, eaau6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, and Turnbaugh PJ (2013). Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Klünemann M, Andrejev S, Blasche S, Mateus A, Phapale P, Devendran S, Vappiani J, Simon B, Scott TA, Kafkia E, et al. (2021). Bioaccumulation of therapeutic drugs by human gut bacteria. Nature 597, 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dillard BA, Chung AK, Gunderson AR, Campbell-Staton SC, and Moeller AH (2022). Humanization of wildlife gut microbiota in urban environments. eLife 11, e76381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ellis RJ, Bruce KD, Jenkins C, Stothard JR, Ajarova L, Mugisha L, and Viney ME (2013). Comparison of the distal gut microbiota from people and animals in Africa. PLoS One 8, e54783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wolfe ND, Dunavan CP, and Diamond J (2007). Origins of major human infectious diseases. Nature 447, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dunay E, Apakupakul K, Leard S, Palmer JL, and Deem SL (2018). Pathogen transmission from humans to great apes is a growing threat to primate conservation. EcoHealth 15, 148–162. [DOI] [PubMed] [Google Scholar]
- 169.Mosites E, Sammons M, Otiang E, Eng A, Noecker C, Manor O, Hilton S, Thumbi SM, Onyango C, Garland-Lewis G, et al. (2017). Microbiome sharing between children, livestock and household surfaces in western Kenya. PLoS One 12, e0171017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Smith WPJ, Wucher BR, Nadell CD, and Foster KR (2023). Bacterial defences: mechanisms, evolution and antimicrobial resistance. Nat. Rev. Microbiol 21, 519–534. [DOI] [PubMed] [Google Scholar]
- 171.Yan W, Hall AB, and Jiang X (2022). Bacteroidales species in the human gut are a reservoir of antibiotic resistance genes regulated by invertible promoters. npj Biofilms Microbiomes 8, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Maciel-Guerra A, Baker M, Hu Y, Wang W, Zhang X, Rong J, Zhang Y, Zhang J, Kaler J, Renney D, et al. (2023). Dissecting microbial communities and resistomes for interconnected humans, soil, and livestock. ISME J. 17, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pomba C, Rantala M, Greko C, Baptiste KE, Catry B, van Duijkeren E, Mateus A, Moreno MA, Pyörälä S, Ružauskas M, et al. (2017). Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother 72, 957–968. [DOI] [PubMed] [Google Scholar]
- 174.Chang Q, Wang W, Regev-Yochay G, Lipsitch M, and Hanage WP (2015). Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl 8, 240–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Koonin EV, Dolja VV, and Krupovic M (2021). The healthy human vi-rome: From virus-host symbiosis to disease. Curr. Opin. Virol 47, 86–94. [DOI] [PubMed] [Google Scholar]
- 176.Ghoreshi Z-A-S, Molaei HR, and Arefinia N (2023). The role of DNA viruses in human cancer. Cancer Inform. 22, 11769351231154186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Arze CA, Springer S, Dudas G, Patel S, Bhattacharyya A, Swami-nathan H, Brugnara C, Delagrave S, Ong T, Kahvejian A, et al. (2021). Global genome analysis reveals a vast and dynamic anellovirus landscape within the human virome. Cell Host Microbe 29, 1305–1315. [DOI] [PubMed] [Google Scholar]
- 178.Singh S, and Blackard JT (2017). Human pegivirus (HPgV) infection in sub-Saharan Africa—a call for a renewed research agenda. Rev. Med. Virol 27, e1951. [DOI] [PubMed] [Google Scholar]
- 179.Lauck M, Bailey AL, Andersen KG, Goldberg TL, Sabeti PC, and O’Connor DH (2015). GB virus C coinfections in west African Ebola patients. J. Virol 89, 2425–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Freer G, Maggi F, Pifferi M, Di Cicco ME, Peroni DG, and Pistello M (2018). The virome and its major component, anellovirus, a convoluted system molding human immune defenses and possibly affecting the development of asthma and respiratory diseases in childhood. Front. Microbiol 9, 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kernbauer E, Ding Y, and Cadwell K (2014). An enteric virus can replace the beneficial function of commensal bacteria. Nature 516, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Virgin HW (2014). The virome in mammalian physiology and disease. Cell 157, 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gogarten JF, Röhlemann M, Archie E, Tung J, Akoua-Koffi C, Bang C, Deschner T, Muyembe-Tamfun J-J, Robbins MM, Schubert G, et al. (2021). Primate phageomes are structured by superhost phylogeny and environment. Proc. Natl. Acad. Sci. USA 118, e2013535118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Koonin EV, and Yutin N (2020). The crAss-like phage group: How metagenomics reshaped the human virome. Trends Microbiol. 28, 349–359. [DOI] [PubMed] [Google Scholar]
- 185.Yutin N, Makarova KS, Gussow AB, Krupovic M, Segall A, Edwards RA, and Koonin EV (2018). Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat. Microbiol 3, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Georjon H, and Bernheim A (2023). The highly diverse antiphage defence systems of bacteria. Nat. Rev. Microbiol 21, 686–700. [DOI] [PubMed] [Google Scholar]
- 187.Mirzaei MK, and Maurice CF (2017). Ménage à trois in the human gut: Interactions between host, bacteria and phages. Nat. Rev. Microbiol 15, 397–408. [DOI] [PubMed] [Google Scholar]
- 188.Moreno-Gallego JL, Chou S-P, Di Rienzi SC, Goodrich JK, Spector TD, Bell JT, Youngblut ND, Hewson I, Reyes A, and Ley RE (2019). Virome diversity correlates with intestinal microbiome diversity in adult monozygotic twins. Cell Host Microbe 25, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Draper LA, Ryan FJ, Smith MK, Jalanka J, Mattila E, Arkkila PA, Ross RP, Satokari R, and Hill C (2018). Long-term colonization with donor bacteriophages following successful faecal microbial transplantation. Microbiome 6, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Johansen J, Atarashi K, Arai Y, Hirose N, Sørensen SJ, Vatanen T, Knip M, Honda K, Xavier RJ, Rasmussen S, and Plichta DR (2023). Centenarians have a diverse gut virome with the potential to modulate metabolism and promote healthy lifespan. Nat. Microbiol 8, 1064–1078. [DOI] [PubMed] [Google Scholar]
- 191.Seed KD, Yen M, Shapiro BJ, Hilaire IJ, Charles RC, Teng JE, Ivers LC, Boncy J, Harris JB, and Camilli A (2014). Evolutionary consequences of intra-patient phage predation on microbial populations. eLife 3, e03497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mayneris-Perxachs J, Castells-Nobau A, Arnoriaga-Rodríguez M, Garre-Olmo J, Puig J, Ramos R, Martínez-Hernández F, Burokas A, Coll C, Moreno-Navarrete JM, et al. (2022). Caudovirales bacteriophages are associated with improved executive function and memory in flies, mice, and humans. Cell Host Microbe 30, 340–356. [DOI] [PubMed] [Google Scholar]
- 193.Khan Mirzaei M, Khan MAA, Ghosh P, Taranu ZE, Taguer M, Ru J, Chowdhury R, Kabir MM, Deng L, Mondal D, and Maurice CF (2020). Bacteriophages isolated from stunted children can regulate gut bacterial communities in an age-specific manner. Cell Host Microbe 27, 199–212.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Omran A,R. (1971). The epidemiological transition: A theory of the epidemiology of population change. Milbank Mem. Fund. Q 49, 509–538. [PubMed] [Google Scholar]
- 195.Lea AJ, Clark AG, Dahl AW, Devinsky O, Garcia AR, Golden CD, Kamau J, Kraft TS, Lim YAL, Martins DJ, et al. (2023). Applying an evolutionary mismatch framework to understand disease susceptibility. PLoS Biol. 21, e3002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang Y, Yu Z, Ding P, Lu J, Mao L, Ngiam L, Yuan Z, Engelstädter J, Schembri MA, and Guo J (2023). Antidepressants can induce mutation and enhance persistence toward multiple antibiotics. Proc. Natl. Acad. Sci. USA 120, e2208344120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Koch H, and Schmid-Hempel P (2011). Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. USA 108, 19288–19292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Huang Q, Lariviere PJ, Powell JE, and Moran NA (2023). Engineered gut symbiont inhibits microsporidian parasite and improves honey bee survival. Proc. Natl. Acad. Sci. USA 120, e2220922120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ippolito MM, Denny JE, Langelier C, Sears CL, and Schmidt NW (2018). Malaria and the microbiome: A systematic review. Clin. Infect. Dis 67, 1831–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Piewngam P, Khongthong S, Roekngam N, Theapparat Y, Sunpaweravong S, Faroongsarng D, and Otto M (2023). Probiotic for pathogen-specific Staphylococcus aureus decolonisation in Thailand: a phase 2, double-blind, randomised, placebo-controlled trial. Lancet. Microbe 4, e75–e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, Gu X, Huang Y, Zamanian-Daryoush M, Culley MK, et al. (2015). Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Garrett WS (2015). Cancer and the microbiota. Science 348, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kelly JR, Borre Y, O’ Brien C, Patterson E, El Aidy S, Deane J, Kennedy PJ, Beers S, Scott K, Moloney G, et al. (2016). Transferring the blues: Depression-associated gut microbiota induces neurobe-havioural changes in the rat. J. Psychiatr. Res 82, 109–118. [DOI] [PubMed] [Google Scholar]
- 204.Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, II-han ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, et al. (2016). Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cho S, Stroup BM, Britto SL, Ruan W, Schady D, Hoffman KL, and Kellermayer R (2023). Increased number of children in households may protect against inflammatory bowel disease. Pediatr. Res 93, 535–540. [DOI] [PubMed] [Google Scholar]
- 206.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre M-L, et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science 350, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Henrich J (2015). The secret of our success: How culture is driving human evolution, domesticating our species, and making us smarter (Princeton University Press; ). [Google Scholar]


