Abstract
The high-throughput genomic and proteomic scanning approaches allow investigators to measure the quantification of genome-wide genes (or gene products) for certain disease conditions, which plays an essential role in promoting the discovery of disease mechanisms. The high-throughput approaches often generate a large gene list of interest (GOIs), such as differentially expressed genes/proteins. However, researchers have to perform manual triage and validation to explore the most promising, biologically plausible linkages between the known disease genes and GOIs (disease signals) for further study. Here, to address this challenge, we proposed a network-based strategy DDK-Linker to facilitate the exploration of disease signals hidden in omics data by linking GOIs to disease knowns genes. Specifically, it reconstructed gene distances in the protein–protein interaction (PPI) network through six network methods (random walk with restart, Deepwalk, Node2Vec, LINE, HOPE, Laplacian) to discover disease signals in omics data that have shorter distances to disease genes. Furthermore, benefiting from the establishment of knowledge base we established, the abundant bioinformatics annotations were provided for each candidate disease signal. To assist in omics data interpretation and facilitate the usage, we have developed this strategy into an application that users can access through a website or download the R package. We believe DDK-Linker will accelerate the exploring of disease genes and drug targets in a variety of omics data, such as genomics, transcriptomics and proteomics data, and provide clues for complex disease mechanism and pharmacological research. DDK-Linker is freely accessible at http://ddklinker.ncpsb.org.cn/.
Keywords: network diffusion, network embedding, disease signal, omics data analysis, genes of interest, gene distance
INTRODUCTION
Human complex diseases such as cancers are caused by multiple genetic and environmental factors and involve considerably complex pathological processes [1]. High-throughput experimental approaches are allowing researchers to study biological systems from a global perspective, generating a large gene list of interest (GOIs), such as SNP sites from genome-wide association study (GWAS) analysis, differentially expressed mRNA from transcriptome data and differentially expressed proteins from proteome data [2, 3]. The GOIs contain deep insights into the complex nature of diseases, paradoxically, the growing landscape of diverse and interconnected information between genes often hinders the elucidation of their biologic meaning and further translation studies [4, 5]. Routine bioinformatics analysis often stops after statistical analysis (such as differential gene expression analysis) and simple bioinformatics annotation [such as Gene Ontology (GO) analysis] [6]. Researchers have to manually review the in-depth domain knowledge in the massive volume of databases and publications to find disease signals, which are the most promising, biologically plausible linkages between the GOIs and known disease genes [7]. It is usually time-consuming and often laborious to perform these personalized data analyses for human diseases [8]. Interpreting the biology of large interesting gene list (ranging from hundreds to thousands of genes) is still a challenging and formidable task [9].
In fact, the disease proteins (the products of disease genes) are not randomly dispersed within the interactome, but tend to interact with each other [10]. On the other hand, despite two proteins can be involved in the same biological pathway without a physical interaction, it has been noted that proteins associated with identical or similar diseases are more likely to share the same topological structure or have similar neighbors in protein interaction networks [11]. Protein–protein interaction (PPI) network is considered to represent a platform through which researchers have the opportunity to systematically identify disease-related genes based on the relationships between genes with similar functions [12].
Many network-based algorithms have been successfully used to discover candidate disease gene products [11, 13–17]. Among them, network diffusion-based and network embedding-based algorithms can utilize network features to discover nodes with strong interactional tendency or similar topological structure [18, 19]. As a popular network diffusion method, random walk with restart (RWR) simulates random walks starting from seed nodes (such as known disease genes) [13, 20]. Networking embedding methods offer avenues to learn the latent features or embeddings for network nodes, including matrix factorization based (e.g. Laplacian and HOPE), random walk–based (e.g. Deepwalk and Node2Vec) and neural network (e.g. LINE and SDNE)–based strategies [21]. Matrix factorization embedding techniques such as Laplacian eigenmap have shown promising results for a variety biomedical graph analysis task [22, 23]. HOPE considers the high-order proximity of the network to preserve the graph structure [24]. Deepwalk and Node2Vec are Embedding techniques using random walks to generate sequences of nodes and then feed the sequences into the Skip-gram mode to learn node representations [25]. As a neural network-based algorithm, LINE directly models node embedding vectors by approximating the first-order proximity and second-order proximity of nodes, which can be seen as a single-layer MLP (multilayer perceptron) model [22]. These popular algorithms can quantify the similarity between genes in biological networks (such as the PPI network) and have been successfully utilized to discover candidate disease gene products, but they can’t construct the linkages to known disease genes. Therefore, we aim to employ these network-based algorithms to establish linkages between omics data and disease knowledge, enabling the exploration of disease signals.
Also to alleviate the bottleneck in interpreting omics data, we proposed the DDK-Linker strategy to explore and annotate disease signals by linking GOIs from omics data with known disease genes. DDK-Linker is designed to analyze GOIs from multi-omics data (such as genome, transcriptome and proteome data). It employed six network-based methods to reconstruct gene distance within the PPI network, aiming to identify disease signals in omics data with shorter distances to disease genes. Moreover, with the support of our established disease knowledge base, in-depth bioinformatics annotations were supplied for each identified disease signal. We further utilized the R Shiny framework to develop an interactive interface that integrates six network-based algorithms and a high-confidence disease knowledge base for disease signals recommendation and annotation. The proposed strategy, together with an automated GOIs linkage workflow and an interactive interface, significantly reduces disease omics data interpretation time from months to mere minutes.
RESULTS
Overview of DDK-Linker
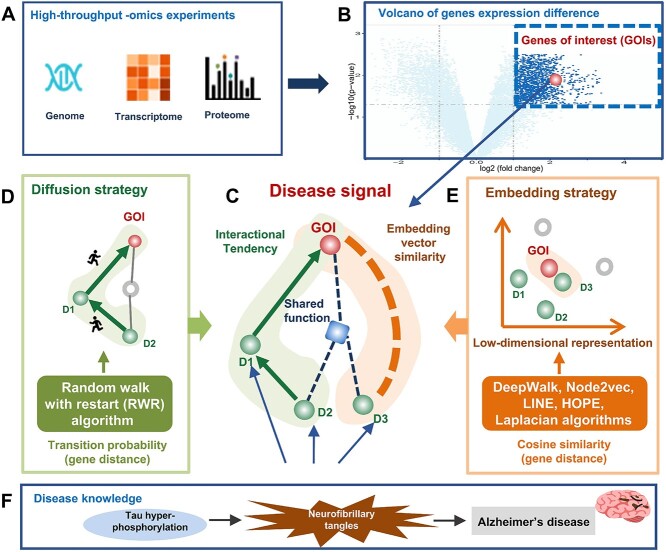
We proposed the strategy DDK-Linker to satisfy the urgent needs of the disease-specific signals identification from high-throughput datasets (Figure 1A). Specifically, DDK-Linker focus on establishing the linkage between GOIs from disease omics data and disease genes (Figure 1B). To realize the pipeline interpretation of omics high-throughput data, DDK-Linker first maps the GOIs and known disease genes in a specific disease to the PPI network (Figure 1C). Then, the six network-based methods (Random Walk with Restart [26], Deepwalk [27], Node2Vec [28], LINE [29], HOPE [24], Laplacian [23]) are employed to measure the distance between GOIs and disease genes, thereby unveiling disease signals characterized by shorter gene distances (Figure S1). In network diffusion, transfer probability was used to measure the gene distance in the network (Figure 1D), while in network embedding, cosine similarity was used to quantify this distance (Figure 1E). Importantly, DDK-Linker not only presents the high confidence disease signals, but also provides GO, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and other biological items to interpret each disease signal, which provide important clues for uncovering disease mechanisms (Figure 1F).
Figure 1.
Illustration for the identification of disease signal from high-throughput omics experiments. (A) High-throughput experimental approaches such as gene expression microarrays or quantitative proteomics generate GOIs (such as DEGs/proteins). (B) The volcano plot illustrates the distribution of GOIs. (C) Disease signals are the most promising, biologically plausible linkages between the known disease genes and GOIs. (D) Diffusion strategy was developed to reconstruct the gene distance in the PPI network by transition probability. (E) Embedding strategy was established to reconstruct the gene distance in the PPI network by embedding vector cosine similarity. (F) The underlying diseases knowledge base.
Evaluation of the gene distance to identify the disease signals based on six different network analysis methods
Here, we introduce gene distance to represent the strength of linkages between genes. For network diffusion algorithm, transfer probability is employed to represent the effective connectivity between genes, and for network embedding algorithms, we use cosine similarity to represent the topological similarity between genes.
To examine whether transfer probabilities and cosine similarity are suitable gene distance index for discovering disease signals, we measured differences in gene distances generated by these six network methods between disease genes and non-disease genes. Following Nguyen et al.’s methodology, we chose genes with scores exceeding 0.3 in the DisGeNET [30] database as known disease genes [31–33]. As for the non-disease genes, we followed the same procedure outlined in previous literature [34, 35]. We randomly selected an equivalent number of genes from outside the DisGeNET database to the known disease genes, forming the negative dataset. We ultimately selected 465 diseases with more than 15 disease genes.
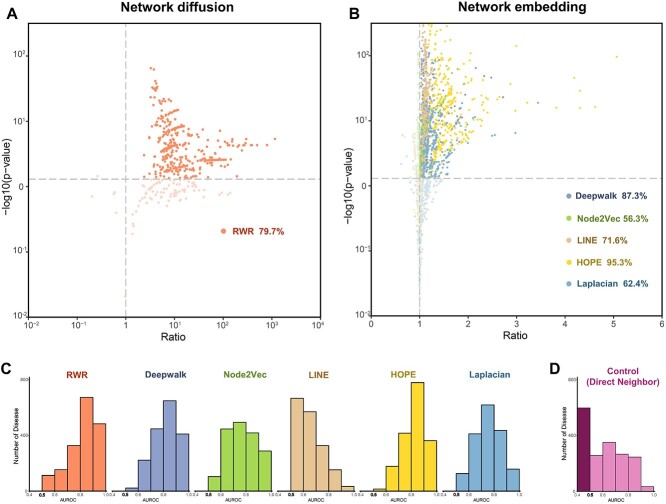
Secondly, we computed the respective gene–disease gene distance (DDD) and disease gene–non-disease gene distance (DND) for each disease. Finally, the differences between DDD and DND for one disease were assessed through t-tests and ratio values [mean (DDD)/mean (DND)]. The results reveal that the gene distance (transfer probability) obtained from the RWR algorithm shows DDD is smaller than DND in 79.7% of diseases (with P-value <0.05 and ratio > 1, Figure 2A). Furthermore, gene distances computed by other five network embedding algorithms (Deepwalk, Node2Vec, LINE, HOPE, Laplacian) also exhibit significant differences between DDD and DND in the majority of diseases (Figure 2B). Notably, the HOPE algorithm demonstrates excellent performance in over 95% of diseases. This indicates that the cosine similarity between genes obtained through network embedding method can represent the distance characteristics between disease genes.
Figure 2.
Evaluation of the gene distance for disease signals based on six different network linkage algorithms. (A) For the RWR algorithm, transfer probability was used to measure the distance between genes. The scatter plot depicts the distance difference between DDD and DND among diseases, with each point corresponding to one disease. DDD: distance between disease gene and disease gene. DND: distance between non disease gene and disease gene. Ratio = mean (DDD)/mean (DND). P-value was obtained by t-test. (B) For the five embedding algorithms, cosine similarity was used to measure the distance between genes. It reflects the distance differences between DDD and DND in various diseases, with each point representing one disease. (C) AUC for gene distances from six algorithms to distinguish disease gene among 1750 diseases. Histograms were used to describe the AUC distribution. (D) AUC for control method (guilt by association) to distinguish disease gene among 1750 diseases.
To access the effectiveness of gene distances calculated based on six network algorithms for discerning disease genes, we hypothesized a strong association between the probability of candidate genes becoming disease genes and their distances from known genes. Therefore, we evaluated the area under the curve (AUC) using leave-one-out cross-validation (LOOCV) to estimate the performance of six gene distances in disease gene discovery. The AUC serves as an indicator of the effectiveness of the corresponding assessment system. An ideal test with perfect discrimination (100% sensitivity, 100% specificity) has an AUC of 1.0, while a non-informative prediction holds an area of 0.5, suggesting that it may be achieved by sheer guess. The closer a test’s AUC is to 1.0, the greater its overall effectiveness. The results indicate that the AUC for all six algorithms exceeded 0.5 across all diseases (Figure 2C). Among them, the HOPE algorithm shows the highest average AUC across the 1750 diseases, while the LINE algorithm has the lowest average AUC. Notably, for the five algorithms excluding LINE, most diseases show AUC values concentrated between 0.7 and 0.9. These strongly suggest that gene distance calculated using network-based algorithms can identify disease genes. Therefore, gene distance serves as a valuable metric to measure the connections between genes, facilitating the discovery of disease signals.
The principle of ‘guilt-by-association’ (GBA) [36], which states the biological entity’s function is inferred by examining its direct neighbors, has been foundational for network-based inference algorithms, including network diffusion [37]. A straightforward analysis using the direct neighbors’ approach can provide the examination of the relationship between potential disease genes and known disease genes through raw network distance. In a PPI network, if a gene is connected to a greater number of disease genes, it is considered to have a stronger association with this disease. We also examined the effectiveness of the direct neighbor method in discovering disease genes. Our findings reveal that direct neighbors’ approach performs poorly in some diseases as it only considers adjacent genes, with an AUC less than 0.5 in 598 out of 1750 diseases (Figure 2D). Furthermore, we conducted pairwise t-tests to compare the performance between the direct neighbor algorithm and the six network-based algorithms across different diseases. We found significant differences (P-value <0.01) between the direct neighbor algorithm and network-based algorithms. This demonstrates that the six network-based methods have an advantage in describing gene distance and identifying disease signals compared to the direct neighbor method.
The efficiency varies among different gene distance in identifying disease signals
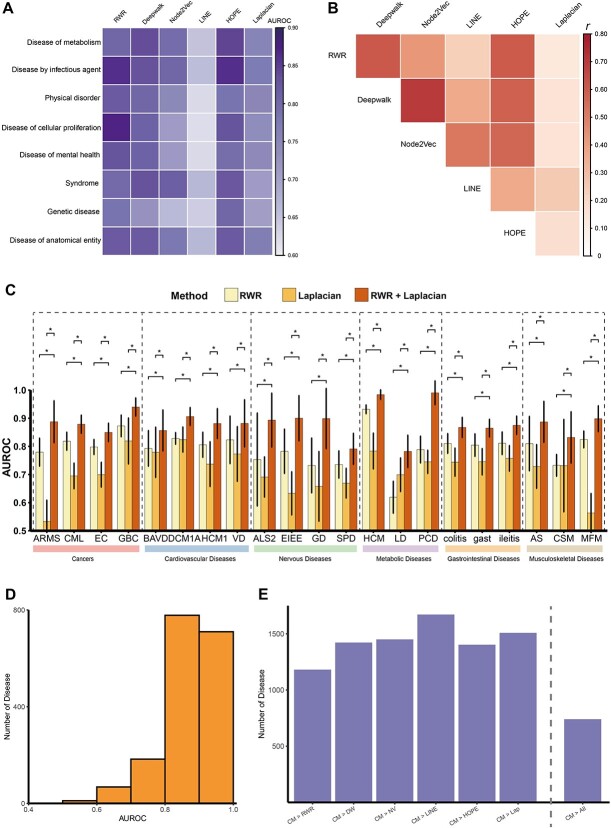
Considering the distinct molecular mechanisms and pathways involved in various disease classes, we classified all diseases into eight disease categories based on disease ontology. We compared the performance of the six gene distance in various disease types by calculating the average AUC for each disease type. The results indicate that, in metabolic diseases, the Deepwalk and HOPE algorithms outperform the others. For infectious diseases, RWR and HOPE show better performance. RWR also demonstrates excellent performance in cell proliferation–related diseases and neurological diseases (Figure 3A). Additionally, genetic diseases perform poorly across multiple algorithms (RWR, Deepwalk, Node2Vec and Laplacian), possibly due to the stochastic nature of genetic mutations in genetic diseases.
Figure 3.
The efficiency varied among different gene distance in identifying disease signals. (A) Difference in the disease signal detection efficiency of six algorithms across eight disease categories. (B) Pairwise correlation between the efficiency of different methods for exploring disease signals. r: Pearson correlation coefficient. (C) Combining of algorithms can improve the effectiveness of exploring disease signals for certain diseases. AUC comparison for various methods (RWR, Laplacian, combined RWR with Laplacian) through different type of diseases. ARMS: alveolar rhabdomyosarcoma, CML: chronic myeloid leukemia, EC: esophageal cancer, GBC: gallbladder cancer, BAVD: bicuspid aortic valve disease, DCM1A: dilated cardiomyopathy 1A, HCM1: hypertrophic cardiomyopathy 1, VD: vascular disease, ALS2: amyotrophic lateral sclerosis type 2, EIEE: early infantile epileptic encephalopathy, GD: generalized dystonia, SPD: secondary Parkinson disease, HCM: hypercalcemia, LD: Leigh disease, PCD: pyruvate carboxylase deficiency disease, gast: gastroenteritis, AS: ankylosing spondylitis, CSM: congenital structural myopathy, MFM: myofibrillar myopathy, *: P-value <0.05. (D) AUROC for the combined method based on consensus score to distinguish disease gene among 1750 diseases. Histograms were used to describe the AUROC distribution. (E) Number of the diseases where consensus score outperforms individual score or any single scores. CM: Combined method based on consensus score, RWR: random walk with restart, DW: Deepwalk, NV: Node2Vec, Lap: Laplacian methods, CM > ALL: the consensus score exceeds any single scores.
To explore potential associations between the effectiveness of different algorithms in diseases, simple linear regression was employed to calculate the correlations between the AUC of each network algorithm. The analysis revealed that Deepwalk exhibits the strongest correlation with Node2Vec (correlation coefficient of 0.71) and a relatively high correlation coefficient of 0.61 with RWR (Figure 3B). Additionally, the HOPE algorithm shows correlation coefficients exceeding 0.5 with RWR, Deepwalk and Node2Vec. On the other hand, Laplacian demonstrates low correlations with other algorithms, with the lowest correlation coefficient with RWR being only 0.13. This may be attributed to the embedding principle of the Laplacian algorithm, which differs from other methods as it utilizes matrix factorization for node embedding [23]. These findings underscore the associations and distinctions among different algorithms, suggesting that, in practical analyses, algorithms with strong correlations can potentially be interchangeable.
In the correlation analysis of different algorithms, we observed the lowest correlation between RWR and Laplacian. Hence, considering the potential complementarity of the two algorithms in exploring disease signals, we combined the Laplacian with RWR to explore disease signals and calculated the AUC for this integrated method. Interestingly, across various disease categories, we found a significant improvement in the AUC when combining the two methods compared to using a single algorithm (Figure 3C). This enhancement may be attributed to the differences in how network embedding (Laplacian) and network diffusion (RWR) methods correlate nodes. The integration of these two algorithms improves prediction accuracy in some diseases, highlighting the capability of the integration to compensate for the limitations of individual methods.
To integrate six network-based algorithms, we built a consensus score that groups together all six independent scores. First, we ranked genome-wide candidate disease genes according to each independent score. Then, we used the average rank from these six independent scores to establish the consensus score. Leave-one-out cross-validation and the ROC curve were used to assess the efficacy of the combined method based on consensus score in identifying disease genes across 1750 diseases. The results indicate that the consensus score generally outperforms all single scores, with 85% of diseases achieving an AUC value exceeding 0.8 (Figure 3D). Furthermore, we found that for each independent score, more than half of the diseases performed worse than the consensus score in identifying disease genes (Figure 3E). Specifically, in 740 diseases (42.3%), the AUC of the consensus score exceeded that of any single score, showing satisfactory predictive performance.
Interactive application was established to mine disease signals from disease omics data
To accommodate diversity of opinions and to enable researchers to conveniently explore disease signals in omics data, we developed an interactive application, DDK-Linker, which can be accessed without requiring login credentials and support mainstream web browsers, including Microsoft Edge, Chrome, Firefox and Safari. We also provided DDK-Linker’s corresponding R package for convenient local usage.
Input and customize parameters
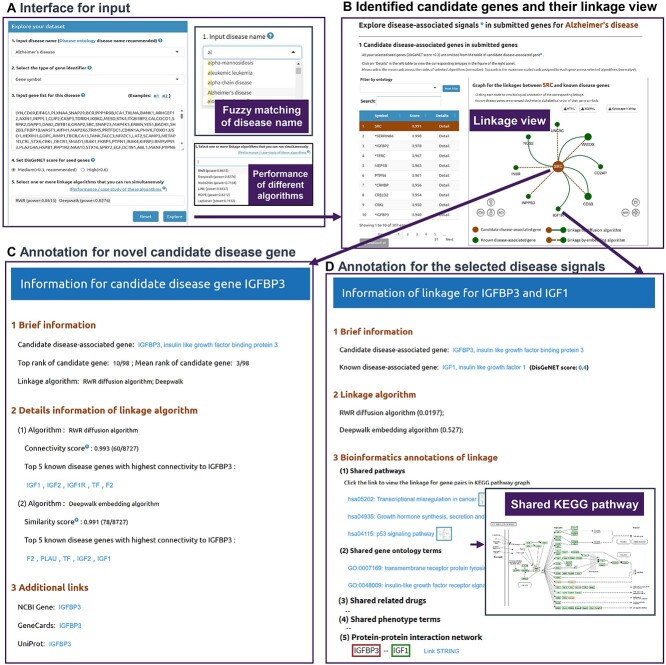
Benefiting from the establish of high confidence disease knowledge base, DDK-Linker realizes analysis in specific disease (Figure 4A). Users can designate one disease by providing its name in disease ontology. Currently, DDK-Linker contains 1750 diseases. An embedded plug-in can help users select the disease name through approximate string matching. Users could input a disease name directly, and DDK-Linker can help users match the standard disease name.
Figure 4.
DDK-Linker interactive interface allows users to uncover disease signals. (A) The input interface accepts the disease name designated by users and the related gene list of interest from a high-throughput omics dataset. The user can customize parameters, such as disease gene confidence score and network linkage algorithms. DDK-Linker can present power scores for each disease under different algorithms. (B) Network view for disease signals shows the linkages between candidate disease–associated genes and known disease–associated genes. The central node represents the candidate disease gene from GOIs and the surrounding nodes are the known disease genes. There are two types of linkages representing different link algorithms: the solid line represents the association from network diffusion strategy, and the dashed line represents the association from network embedding strategy. Users can click each node/edge to view detailed annotations of the corresponding linkage (C, D). (C) Bioinformatics annotations for the candidate disease–associated gene. (D) Detailed information for certain disease signal between candidate disease–associated gene and known disease–associated gene. The inset graph shows the graph to illustrate this linkage in KEGG pathway.
The next step is for users to input GOIs derived from high-throughput omics. Users can do this by pasting the GOIs into the submission box according to the specified format. To support different types of gene lists derived from multi-omics data, DDK-Linker accepts five types of gene identifiers: Entrez Gene Symbol and Entrez Gene ID for genes, RefSeq RNA ID for mRNA and Uniprot and Refseq protein AC for proteins. For the mapping of gene, mRNA and protein identifiers, we utilized the id mapping file (version 20240118) from the UniProt database.
Users will then select a few parameters that will be used to perform linkage analysis and generate the results. First, users can select one or more network-based methods (DDK-Linker provided six algorithms) to discover disease signals according to their needs. Besides, to meet different requirements, users can customize a cutoff value to select genes as known disease genes that are used in the linkage strategies. In addition, DDK-Linker presents the performance (AUC) of six algorithms in various diseases in the drop-down box for algorithm selection. When users designate one disease, users can choose the corresponding method based on the AUC scores of different algorithms.
Results pages
After submitting, DDK-Linker performs the linkage analysis to discover disease-associated signals, presents the list of candidate disease–associated genes in submitted genes and illustrates their linkages with known disease genes (network view, right panel). For candidate disease–associated genes, we provided annotations of gene molecular functions and biological pathways. We have developed gene filtering functions based on GO functions and KEGG pathways, and users can use this function to further select their interested gene sets from top-ranked candidate genes. Additionally, in the candidate disease–associated genes table panel, we provide a score indicator. This score is calculated as the average of the rankings assigned by the user selection algorithm. This score could serve as an indication of the similarity between candidate disease genes and known disease genes, and the genes with higher rankings are more likely to be closely related to the disease and have higher priority for experimental verification. Considering that DDK-Linker aims to mine novel disease–associated genes from omics data, known disease genes were excluded from result list to avoid potential misleading. For known disease genes in submitted list, the corresponding supporting literature information is presented in the table below. Users can also download all analysis results of DDK-Linker by clicking on download buttons.
Disease signals identified
For each candidate disease–associated gene, a network view was designed to present its related disease signals (the linkage between candidate disease–associated genes and known disease genes) (Figure 4B). The central node is the candidate disease genes, and the surrounding nodes are the known disease genes detected through linkage analysis. There are two types of linkages representing different link methods: a solid green line represents the linkage from network diffusion algorithm. A brown dashed line represents the linkage from network embedding algorithm. If users want to obtain the detailed scores of the linkage methods, they can click the central node in the network view, which will lead to the page of detailed information for the candidate disease gene (Figure 4C). The disease signal annotation page can also be presented by clicking the surrounding known disease gene nodes (Figure 4D). To facilitate further analysis by users, we provide three network view files: HTML, XGMML and Cytoscape style xml. Users can download these files and make personalized modifications in Cytoscape locally.
Detailed information to interpret disease signals
First, users can further find the score of the selected algorithm to see which algorithms are involved in this disease signals (Figure 4D). To help users understand biological mechanism and generate hypotheses, we also collected abundant bioinformatic annotations for each disease signal, including the GO term [38], KEGG pathway [39], drug [40], phenotype term [41] and PPI networks [42]. Generally speaking, the biological function of the living cell is a result of numerous interacting molecules; it cannot be ascribed to just a single molecule. The shared KEGG terms indicate that the candidate and known disease genes participate in the same biological function, while the shared GO terms reveal the biological processes, cellular components and molecular functions implicated in this disease. We aim to construct the relationship between candidate disease genes and drugs to find disease-related drugs through drug annotation. Serving as crucial bridges between medical experimental discoveries and clinical practices, phenotypes contribute significantly to translational medicine. The shared phenotype terms suggest that the candidate disease genes may act on this phenotype together with known genes. PPI networks have the potential to elucidate the complex relationships between candidate disease genes and known disease-associated genes. Users could obtain more details about the interaction by clicking on STRING hyperlinks in this detailed information page (Figure 4D).
Local installation of DDK-Linker
For the utilization of DDK-Linker on a local computer, the entire pipeline for disease signal discovery has been implemented in R using Shiny (http://shiny.rstudio.com) to facilitate its usage. Currently, there is no explicit limit to the number of simultaneous users. Users can install the DDK-Linker R package (π-DDK-Linker) along with R itself on their local computer (R is freely downloadable from http://r-project.org/ for all major operating systems). Once installed locally, the complete analysis workflow of π-DDK-Linker can be executed on the local computer.
USE CASE: DDK-LINKER UNCOVERS THE KEY DISEASE SIGNALS FROM PROTEOMICS AND GWAS DATA IN ALZHEIMER’S DISEASE
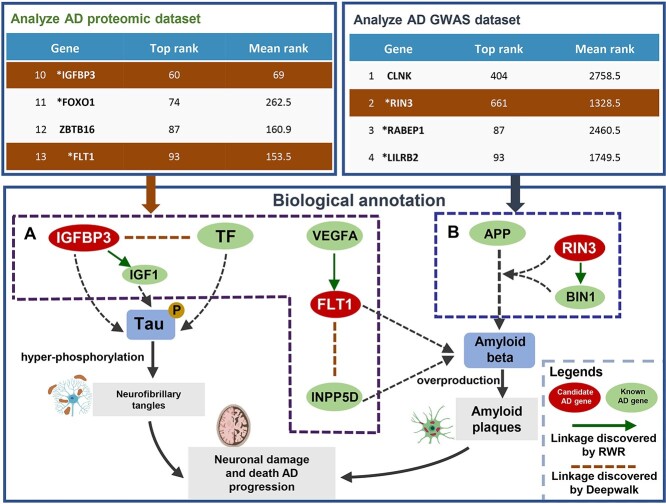
Today, the presence of these amyloid plaques and neurofibrillary tangles are still essential for the pathological diagnosis of Alzheimer’s disease (AD) [43]. In recent years, various omics studies, including genomics, transcriptomics and proteomics, have revealed the mechanisms leading to neuronal death and identified biomolecular markers associated with AD [44]. Here, we used DDK-Linker to uncover the disease signals from plasma proteome and GWAS AD gene lists (Figure 5).
Figure 5.
Exploration of GWAS and proteomic data for disease signals of AD based on reconstructed gene distances. (A) Proteome (the dataset from ‘Alzheimers Dement. 88:102’): DDK-Linker identified FLT1 and IGFBP3 as candidate genes. FLT1 and IGFBP3 interact with VEGFA and IGF1, respectively. Additionally, TF and INPP5D exhibit similarity associations with IGFBP3 and FLT1, respectively. These identified genes can be confirmed by the manual curation. Both IGFBP3 and FLT1 have been reported to be involved in the pathological process of amyloid plaques and tau phosphorylation in AD. (B) GWAS (the dataset from ‘Nat Genet. 53:1722’): the linkage between RIN3 (from GWAS datasets) and BIN1 as potential AD signals by DDK-Linker. In fact, RIN3 was validated to promote amyloid-β deposition.
Analysis of AD proteomics dataset
Plasma proteins are increasingly recognized as potential biomarkers for AD. A study systematically analyzed the plasma proteome to discover novel AD blood biomarkers and established high-performance diagnostics for AD [45]. Here, we use the dataset that includes 429 human plasma proteins derived from the differentially expressed genes (DEGs) analysis of patients with AD versus healthy controls. We used the combination linkage algorithm (‘RWR’ and ‘Deepwalk’, which have demonstrated the best performance in AD) to identify the disease signals. The known AD genes (such as TF, IGF1, VEGFA and INPP5D, indicated in green in Figure 5) sourced from the DisGeNET database (DisGeNET score > 0.3) were used as seed genes in algorithms. After submission, DDK-Linker performed the linkage analysis to discover disease-associated signals (307 candidate disease–associated genes) in submitted genes. Among the top 20 high-confidence candidate genes, two most promising and biologically plausible genes were identified: FLT1 and IGFBP3 (Figure 5A). FLT1 has exhibited disease signal with the known disease gene VEGFA (both RWR and Deepwalk algorithms) and INPP5D (Deepwalk algorithm) in DDK-Linker. Therefore, it is speculated that FLT1 participates in similar disease processes. In fact, FLT1 was found to be the receptor of VEGFA, associating with amyloid plaque [46]. INPP5D has also been reported to be related with amyloid-β (Aβ) deposition in AD patients, just like FLT1 [47]. We also found the disease signal IGFBP3-IGF1 and IGFBP3-TF, candidate disease gene IGFBP3 is linked to known disease genes IGF1 and TF (both RWR and Deepwalk algorithms). By the bioinformatics annotation page, we noted all these three genes (IGFBP3, IGF1 and TF) share the same GO term of ‘phosphorylation’. The most common primary characteristics of AD is that neurofibrillary tangles are largely composed of hyper-phosphorylated tau [48]. Both IGF1 and TF have been reported to be related with neurofibrillary tangles [49, 50]. Thus, we inferred that IGFBP3 may be related to tau phosphorylation. These linkages provided us crucial clues for further experiments. Interesting, IGFBP3 has also been found to affect the tau phosphorylation by binding to IGF1 [50]. Furthermore, DDK-Linker also identified some genes that could potentially become novel biomarkers for AD. The gene PTPN6 ranked sixth in candidate AD-associated genes, and recently, Kiratikanon et al. pointed out that PTPN6 is a promising biomarker for AD [51].
Analysis of AD GWAS dataset
Wightman et al. identified 38 independent risk loci by GWAS, and we found candidate disease–associated genes in this gene list [52]. DDK-Linker presents the disease signals between the candidate disease gene RIN3 and known disease gene BIN1 is also noteworthy (Figure 5B). In fact, it has been demonstrated that BIN1 directly interacts with RIN3 to initiate endocytosis, a process essential for the cleavage of β-amyloid precursor protein to generate Aβ, which is the critical component of senile plaques in AD [53].
Here, we analyzed two gene lists from plasma proteome and GWAS to explore the AD disease signals through DDK-Linker. Further biological annotations of the top disease signals indicate involvement in the formation of amyloid plaques and neurofibrillary tangles, providing plenty of clues for further mechanism study.
CONCLUSION AND DISCUSSION
In this study, we proposed the strategy DDK-Linker to address the urgent need for the disease-specific signals identification from multiple omics datasets, such as genomics, transcriptomics and proteomics. We conducted an extensive literature review and found six algorithms (Random Walk with Restart, Deepwalk, Node2Vec, LINE, HOPE, Laplacian), which have been widely used in disease gene prediction (see Supplementary Tables S1 and S2 for more details on the application of the six algorithms). Utilizing these six network-based algorithms, we recalculated the distances between genes within the network, determining the connectivity strength between GOIs and disease genes. This enabled the identification of disease signals most closely correlated with the respective diseases.
DDK-Linker demonstrates satisfactory performance on DisGeNET dataset. To further assess its robustness and effectiveness, we also used another training set, DISEASE, to train and test our DDK-Linker system. Using the same protocol as that used for DisGeNET, we found that the results of DDK-Linker on DISEASE were consistent through leave-one-out cross-validation and ROC analysis. Among the 810 diseases in DISEASE, all six algorithms show satisfactory performance in predicting disease genes (Figure S2).
To help users choose a suitable algorithm for their GOIs, we presented the predictive efficacy (AUC) of six algorithms on 1750 diseases in the algorithm selection drop-down box of the DDK-Linker input panel. When users designate one disease, they can choose the corresponding method based on the AUC scores of different algorithms. In addition, we also assessed the predictive efficacy of six algorithms in different types of diseases. We first grouped the 1750 diseases into eight major categories according to the Disease Ontology and selected five major categories (cell proliferation diseases, mental health diseases, anatomical entity diseases, syndromes and metabolic diseases), each of which included over 50 diseases, for subsequent analysis. Then we calculated the proportion of the most suitable algorithm (highest AUC) for each type of disease. We found that there is certain correlation between the algorithm and the type of disease. For example, RWR performs better in diseases of cellular proliferation and mental diseases, while HOPE algorithm performs best in metabolic diseases (Figure S3). All these instructions on regarding which diseases are more suitable for DDK-Linker will help users use DDK-Linker more effectively.
In the future, we will continue to update DDK-Linker to alleviate the bottleneck in the interpretation of omics data. There are two main directions. First, we will develop new methods for calculating gene distance. The current software only uses PPI networks to calculate gene distance. We plan to apply gene knowledge graphs to this process, which contain more protein feature information beyond protein interactions, such as function annotations and family classification information. We expect that these new embedding methods based on knowledge graphs can provide a more accurate description of gene distance. Second, we will expand the use of gene distance for discoveries beyond disease gene associations. Currently, DDK-Linker mainly uses gene distance to establish associations between GOIs and known disease genes. However, the framework of DDK-Linker can be expanded for the discovery of gene–drug/phenotype associations. The current version of DDK-Linker does not include known drug/phenotype-related genes, which could serve as seed genes to identify novel gene–drug/phenotype associations. We plan to incorporate this information into DDK-Linker.
Key Points
We developed a network-based strategy DDK-Linker to link high-throughput omics genes (GOIs) with disease genes by reconstructing gene distances in a PPI network.
We assessed the performance for discovering disease signal of six network algorithms across 1750 diseases.
DDK-Linker is designed to efficiently discover and annotate disease signals hidden in high-throughput data.
DDK-Linker is supposed to greatly accelerate the speed of specific disease omics data interpretation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the bioinformatics platform at Phoenix Center for the strong and stable IT support.
Author Biographies
Xiangren Kong is a master’s student at State Key Laboratory of Medical Proteomics, National Center for Protein Sciences (Beijing).
Lihong Diao is a PhD student at the School of Traditional Chinese Medicine, Beijing University of Chinese Medicine.
Peng Jiang is a master’s student at State Key Laboratory of Medical Proteomics, National Center for Protein Sciences (Beijing).
Shiyan Nie is a master’s student at State Key Laboratory of Medical Proteomics, National Center for Protein Sciences (Beijing).
Shuzhen Guo is a professor at the School of Traditional Chinese Medicine, Beijing University of Chinese Medicine.
Dong Li is a professor at State Key Laboratory of Medical Proteomics, National Center for Protein Sciences (Beijing).
Contributor Information
Xiangren Kong, State Key Laboratory of Medical Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China.
Lihong Diao, State Key Laboratory of Medical Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China; School of Traditional Chinese Medicine, Beijing University of Chinese Medicine, Beijing 100029, China.
Peng Jiang, State Key Laboratory of Medical Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China.
Shiyan Nie, State Key Laboratory of Medical Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China.
Shuzhen Guo, School of Traditional Chinese Medicine, Beijing University of Chinese Medicine, Beijing 100029, China.
Dong Li, State Key Laboratory of Medical Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China.
FUNDING
This work was supported by National Key Research and Development Program of China [2021YFA1301603, 2023YFF1204600] and National Natural Science Foundation of China [32088101, 32271518].
DATA AVAILABILITY
DDK-Linker is available at http://ddklinker.ncpsb.org.cn/. See Supplementary Materials and Methods for more details of materials and methods.
References
- 1. Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet 2002;3:391–7. [DOI] [PubMed] [Google Scholar]
- 2. O'Brien MA, Costin BN, Miles MF. Using genome-wide expression profiling to define gene networks relevant to the study of complex traits: from RNA integrity to network topology. Int Rev Neurobiol 2012;104:91–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reimand J, Isserlin R, Voisin V, et al. Pathway enrichment analysis and visualization of omics data using g:profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc 2019;14:482–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hassani-Pak K, Singh A, Brandizi M, et al. KnetMiner: a comprehensive approach for supporting evidence-based gene discovery and complex trait analysis across species. Plant Biotechnol J 2021;19:1670–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yan J, Risacher SL, Shen L, Saykin AJ. Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Brief Bioinform 2018;19:1370–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang H, Ao M, Boja A, et al. OmicsOne: associate omics data with phenotypes in one-click. Clin Proteomics 2021;18:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blatti C, Emad A, Berry MJ, et al. Knowledge-guided analysis of "omics" data using the KnowEnG cloud platform. PLoS Biol 2020;18:e3000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li R, Li L, Xu Y, Yang J. Machine learning meets omics: applications and perspectives. Brief Bioinform 2022;23:23. [DOI] [PubMed] [Google Scholar]
- 9. Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Menche J, Sharma A, Kitsak M, et al. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 2015;347:1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu H, Guan J, Li H, et al. Predicting the disease genes of multiple sclerosis based on network representation learning. Front Genet 2020;11:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Safari-Alighiarloo N, Taghizadeh M, Rezaei-Tavirani M, et al. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol Hepatol Bed Bench 2014;7:17–31. [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J, Suo Y, Liu M, Xu X. Identification of genes related to proliferative diabetic retinopathy through RWR algorithm based on protein-protein interaction network. Biochim Biophys Acta Mol Basis Dis 2018;1864:2369–75. [DOI] [PubMed] [Google Scholar]
- 14. Li G, Luo J, Xiao Q, et al. Predicting MicroRNA-disease associations using network topological similarity based on DeepWalk. IEEE Access 2017;5:24032–9. [Google Scholar]
- 15. Peng J, Guan J, Shang X. Predicting Parkinson's disease genes based on Node2vec and autoencoder. Front Genet 2019;10:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou J-R, You Z-H, Cheng L, Ji BY. Prediction of lncRNA-disease associations via an embedding learning HOPE in heterogeneous information networks. Mol Ther Nucleic Acids 2021;23:277–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gong Y, Niu Y, Zhang W, Li X. A network embedding-based multiple information integration method for the MiRNA-disease association prediction. BMC Bioinformatics 2019;20:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shi M, Tang Y, Zhu X. Topology and content co-alignment graph convolutional learning. IEEE Trans Neural Netw Learn Syst 2022;33:7899–907. [DOI] [PubMed] [Google Scholar]
- 19. Sumathipala M, Maiorino E, Weiss ST, Sharma A. Network diffusion approach to predict LncRNA disease associations using multi-type biological networks: LION. Front Physiol 2019;10:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ata SK, Wu M, Fang Y, et al. Recent advances in network-based methods for disease gene prediction. Brief Bioinform 2021;22:22. [DOI] [PubMed] [Google Scholar]
- 21. Makarov I, Kiselev D, Nikitinsky N, Subelj L. Survey on graph embeddings and their applications to machine learning problems on graphs. PeerJ Computer Science 2021;7:e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yue X, Wang Z, Huang J, et al. Graph embedding on biomedical networks: methods, applications and evaluations. Bioinformatics (Oxford, England) 2020;36:1241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Belkin M, Niyogi P. Laplacian Eigenmaps for dimensionality reduction and data representation. Neural Comput 2003;15:1373–96. [Google Scholar]
- 24. Ou M, Cui P, Pei Jet al. Asymmetric transitivity preserving graph embedding. In Krishnapuram B, Shah M, Smola AJ, Aggarwal CC, Shen D, Rastogi R, (eds), In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. San Francisco, California, USA: Association for Computing Machinery, 2016, 1105–14. [Google Scholar]
- 25. Zhang Y, Wang Z, Wang S, Shang J. Comparative analysis of unsupervised protein similarity prediction based on graph embedding. Front Genet 2021;12:744334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Köhler S, Bauer S, Horn D, Robinson PN. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet 2008;82:949–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perozzi B, Al-Rfou R, Skiena S. DeepWalk: online learning of social representations. In Macskassy SA, Perlich C, Leskovec J, Wang W, Ghani R, (eds), In: Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York, New York, USA: Association for Computing Machinery, 2014, 701–10. [Google Scholar]
- 28. Grover A, Leskovec J. node2vec: Scalable Feature Learning for Networks. In Krishnapuram B, Shah M, Smola AJ, Aggarwal CC, Shen D, Rastogi R, (eds), In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. San Francisco, California, USA: Association for Computing Machinery, 2016, 855–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang J, Qu M, Wang Met al. LINE: Large-scale Information Network Embedding. In Gangemi A, Leonardi S, Panconesi A, (eds), In: Proceedings of the 24th International Conference on World Wide Web. Florence, Italy: International World Wide Web Conferences Steering Committee, 2015, 1067–77.
- 30. Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res 2020;48:D845–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nguyen HCT, Baik B, Yoon S, et al. Benchmarking integration of single-cell differential expression. Nat Commun 2023;14:1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jané P, Xu X, Taelman V, et al. The Imageable genome. Nat Commun 2023;14:7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lazo de la Vega L, Yu W, Machini K, et al. A framework for automated gene selection in genomic applications, genetics in medicine. Genet Med 2021;23:1993–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Binder J, Ursu O, Bologa C, et al. Machine learning prediction and tau-based screening identifies potential Alzheimer's disease genes relevant to immunity. Commun Biol 2022;5:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shu J, Li Y, Wang S, et al. Disease gene prediction with privileged information and heteroscedastic dropout. Bioinformatics 2021;37:i410–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oliver S. Guilt-by-association goes global. Nature 2000;403:601–2. [DOI] [PubMed] [Google Scholar]
- 37. Cowen L, Ideker T, Raphael BJ, Sharan R. Network propagation: a universal amplifier of genetic associations. Nat Rev Genet 2017;18:551–62. [DOI] [PubMed] [Google Scholar]
- 38. The gene ontology resource: enriching a GOld mine. Nucleic Acids Res 2021;49:D325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kanehisa M, Furumichi M, Sato Y, et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res 2023;51:D587–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Davis AP, Grondin CJ, Johnson RJ, et al. Comparative Toxicogenomics database (CTD): update 2021. Nucleic Acids Res 2021;49:D1138–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Köhler S, Gargano M, Matentzoglu N, et al. The human phenotype ontology in 2021. Nucleic Acids Res 2021;49:D1207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Keshava Prasad TS, Goel R, Kandasamy K, et al. Human protein reference database--2009 update. Nucleic Acids Res 2009;37:D767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. DeTure MA, Dickson DW. The neuropathological diagnosis of Alzheimer's disease. Mol Neurodegener 2019;14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tan MS, Cheah P-L, Chin A-V, et al. A review on omics-based biomarkers discovery for Alzheimer's disease from the bioinformatics perspectives: statistical approach vs machine learning approach. Comput Biol Med 2021;139:104947. [DOI] [PubMed] [Google Scholar]
- 45. Jiang Y, Zhou X, Ip FC, et al. Large-scale plasma proteomic profiling identifies a high-performance biomarker panel for Alzheimer's disease screening and staging. Alzheimers Dement 2022;18:88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mahoney ER, Dumitrescu L, Moore AM, et al. Brain expression of the vascular endothelial growth factor gene family in cognitive aging and Alzheimer's disease. Mol Psychiatry 2021;26:888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsai AP, Lin PB-C, Dong C, et al. INPP5D expression is associated with risk for Alzheimer's disease and induced by plaque-associated microglia. Neurobiol Dis 2021;153:105303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boutajangout A, Sigurdsson EM, Krishnamurthy PK. Tau as a therapeutic target for Alzheimer's disease. Curr Alzheimer Res 2011;8:666–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hoshi K, Ito H, Abe E, et al. Transferrin biosynthesized in the brain is a novel biomarker for Alzheimer's disease. Metabolites 2021;11(9):616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Watanabe K, Uemura K, Asada M, et al. The participation of insulin-like growth factor-binding protein 3 released by astrocytes in the pathology of Alzheimer's disease. Mol Brain 2015;8:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kiratikanon S, Chattipakorn SC, Chattipakorn N, Kumfu S. The regulatory effects of PTPN6 on inflammatory process: reports from mice to men. Arch Biochem Biophys 2022;721:109189. [DOI] [PubMed] [Google Scholar]
- 52. Wightman DP, Jansen IE, Savage JE, et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer's disease. Nat Genet 2021;53:1276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bhattacharyya R, Teves CAF, Long A, et al. The neuronal-specific isoform of BIN1 regulates β-secretase cleavage of APP and Aβ generation in a RIN3-dependent manner. Sci Rep 2022;12:3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DDK-Linker is available at http://ddklinker.ncpsb.org.cn/. See Supplementary Materials and Methods for more details of materials and methods.