Abstract
Little is known about the metabolic basis of life-history trade-offs but lipid stores seem to play a pivotal role. During reproduction, an energetically highly costly process, animals mobilize fat reserves. Conversely, reduced or curtailed reproduction promotes lipid storage in many animals. Systemic signals from the gonad seem to be involved: Caenorhabditis elegans lacking germline stem cells display endocrine changes, have increased fat stores and are long-lived. Similarly, germline-ablated Drosophila melanogaster exhibit major somatic physiological changes, but whether and how germline loss affects lipid metabolism remains largely unclear. Here we show that germline-ablated flies have profoundly altered energy metabolism at the transcriptional level and store excess fat as compared to fertile flies. Germline activity thus constrains or represses fat accumulation, and this effect is conserved between flies and worms. More broadly, our findings confirm that lipids represent a major energetic currency in which costs of reproduction are paid.
Keywords: germline, cost of reproduction, trade-offs, energy stores, fat reserves, lipid metabolism
…as Goethe expressed it, “in order to spend on one side, nature is forced to economise on the other side.” … natural selection is continually trying to economise in every part of the organisation. If under changed conditions of life a structure, before useful, becomes less useful, its diminution will be favoured, for it will profit the individual not to have its nutriment wasted in building up a useless structure.
(Darwin, 1859, p. 147–148)
It would be instructive to know not only by what physiological mechanisms a just apportionment is made between the nutriment devoted to the gonads and that devoted to the rest of the parental organism, but also what circumstances in the life-history and environment would render profitable the diversion of a greater or lesser share of the available resources towards reproduction.
(Fisher, 1930, p. 43–44)
Introduction
Understanding how trade-offs constrain adaptation is a central, long-standing problem in evolutionary biology (Bell & Koufopanou, 1986; Fisher, 1930; Roff, 2007; Roff & Fairbairn, 2007; Stearns, 1989). Despite much work on life-history trade-offs, however, still little is known about their biochemical, physiological or metabolic underpinnings (Barnes & Partridge, 2003; Calow, 1979; Flatt & Heyland, 2011; Harshman & Zera, 2007; Leroi, 2001; Rose & Bradley, 1998; Williams, 2005; Zera & Harshman, 2001).
Fat (i.e., lipids called triglycerides or triacylglycerides, TAG) represents a major form of energy storage that seems to play a central role in many life-history trade-offs (Gáliková & Klepsatel, 2018; Townsend & Calow, 1981; van der Horst et al., 2002; Zera, 2005; Zhao & Zera, 2002). In insects, where many trade-off studies have been performed, the abundance of total lipid or TAG is positively correlated with longevity, starvation survival, and/or energetically demanding activities such as fecundity, flight, or diapause (Chippindale et al., 1993, 1996; Dingle, 1996; Djawdan et al., 1998; Hansen et al., 2013; Leroi et al., 1994; Rion & Kawecki, 2007; Rose et al., 1992; Service, 1987; Service & Rose, 1985; Service et al., 1985; 1988; Zera & Larsen, 2001; Zhao & Zera, 2002; Zwaan et al., 1995).
In particular, fat is a major currency in which the energetically costly process of reproduction is paid: animals mobilize and spend down their fat reserves during reproduction (Bronson, 1989; Carey, 1996; Rose & Bradley, 1998; Townsend & Calow, 1981). Conversely, reduced or curtailed reproduction (e.g., gonadectomy; hypogonadism, a gonadal hormone deficiency) causes excess fat storage in many mammals (e.g., humans, monkeys, cats, dogs, rodents) (Corona et al., 2009; Hansen et al., 2013; McElroy & Wade, 1987; Stotsenburg, 1913; Wilson & Roehrborn, 1999).
Likewise, in many insects (e.g., fruit flies, blow flies, bugs, locusts, grasshoppers) ovariectomy causes an enlargement (hypertrophy) of the “fat body,” the insect equivalent of mammalian adipose and liver tissues (Hansen et al., 2013; Judd et al., 2011; Socha et al., 1991; Strong, 1967; Thomsen & Hamburger, 1955). Hypertrophy of the fat body has also been observed in female-sterile Drosophila mutants; remarkably, a normal-sized fat body can be restored by implanting wild-type ovaries into the mutants (Butterworth & Bodenstein, 1968; Doane, 1961). In C. elegans, germline-less and long-lived glp-1 mutants, as well as several other sterile mutants, have increased fat stores (Chaturbedi & Lee, 2023; McCormick et al., 2012; O’Rourke et al., 2009). Such “failure reveals design” (Frank, 2016): the above cases of “reproductive failure” suggest a common pattern whereby under normal conditions gonadal (or germline) activity constrains or represses the growth of adipose tissue and thus reduces lipid stores (Butterworth & Bodenstein, 1968; Chippindale et al., 1993; Leroi, 2001).
Similar to the observations in C. elegans and related nematodes (Arantes-Oliveira et al., 2002; Hsin & Kenyon, 1999; Rae et al., 2012), we have previously found that loss of germline stem cells in Drosophila alters insulin/insulin-like growth factor signaling (IIS) and carbohydrate metabolism, extends lifespan and promotes innate immunity (Flatt et al., 2008; Rodrigues et al., 2021). Yet, how loss of germline stem cell proliferation impacts lipid metabolism in the fly remains poorly understood (Parisi et al., 2010, 2011). More generally, the above findings hint at profound but poorly understood connections between the gonad, germline activity, metabolism and somatic maintenance that might be important for understanding the nature of physiological constraints upon life history (Flatt et al., 2008; Hansen et al., 2013; O’Rourke et al., 2009; Wang et al., 2008).
To further examine these fundamental issues, we sought to investigate “conflicts” (i.e., trade-offs) between reproduction and metabolism over patterns of gene expression (Stearns & Magwene, 2003) in adult female Drosophila melanogaster by analyzing transcriptome-wide changes with RNA sequencing (RNA-seq) in response to simultaneous manipulation of reproduction (germline ablated vs. fertile control flies) and diet (varying levels of dietary yeast). We were particularly interested in testing whether the effects of germline loss upon fat metabolism might be conserved between the nematode worm and the fruit fly.
Our results show that germline-less and fertile flies differ in the expression of numerous genes involved in energy metabolism, especially lipid metabolism. In support of these transcriptomic results, we find that germline-ablated flies possess excess fat accumulation, similar to previous findings in C. elegans. These findings suggest that the energetic trade-off between investment into reproduction (germline activity) vs. lipid storage (in support of somatic maintenance) is evolutionarily conserved.
Results and discussion
We analyzed transcriptome-wide patterns of gene expression in female flies in response to both reproductive and dietary manipulation. To directly manipulate costs of reproduction we used germline-ablated flies (hereafter referred to as “germline-less” or “sterile”) vs. fertile control flies. Germline ablation was achieved by driving overexpression of bag of marbles (UASp-bam+) with a germline-specific nanos (nos)-GAL4::VP16 driver, which causes loss of germ cells in the late third larval instar or early adult and hence abolishes egg production (see Figure 1; Chen & McKearin, 2003; Flatt et al., 2008; Rodrigues et al., 2021; see Materials and Methods for details). Because the metabolic demands of fecundity vs. somatic maintenance depend on nutritional input (Chippindale et al., 1993; Djawdan et al., 1996; Flatt, 2011; Lee et al., 2008; Min et al., 2007; Simmons & Bradley, 1997; Skorupa et al., 2008), we also manipulated dietary yeast levels (2, 4, 8, or 12% of total food volume; see Materials and Methods; cf. Tatar, 2007), the main protein source of flies (see Figure 1). Considering the joint effects of reproduction and diet is also relevant as diet and germline signals are known to interact in affecting C. elegans lifespan (Crawford et al., 2007). Figure 1 illustrates the effects on fecundity of our 2-way design; as is well known, increasing yeast levels promote egg production (Min et al., 2007; Simmons & Bradley, 1997; Skorupa et al., 2008), and transgenic germline ablation abolishes reproductive output (Flatt et al., 2008).
Figure 1.
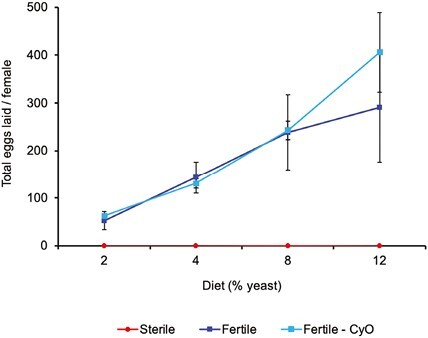
Effects of treatments (germline ablation; dietary yeast) on female fecundity. In our transcriptomic study we sought to manipulate reproductive physiology and metabolism of female D. melanogaster in two ways: by manipulating reproduction (germline ablation vs. fertile control flies) and by manipulating yeast levels in the fly food. The graph shows the average total number of eggs laid per female over a 20-day period as a function of the yeast level in the diet (2%, 4%, 8%, 12%). Red curve: germline-less (sterile) flies (yw; +/+; nanos-GAL4::VP16/nanos-GAL4::VP16); dark blue curve: fertile control genotype (y1,w1118); light blue curve: a second fertile control genotype (y,w; CyO/+; nanos-GAL4::VP16/+); error bars represent the standard error of the mean. As is well known, increasing dietary yeast levels promote female fecundity. By contrast, germline-ablated flies are unable to produce eggs. Data were analyzed with a fully factorial two-way fixed-effects type II ANOVA on rank-transformed egg counts, revealing the following effects: Reproduction (germline-less vs. fertile controls; F1,67 = 62.6, p < .0001); Diet (F3,67 = 4.4, p = .0068); and Reproduction × Diet (F3,67 = 1.4, p = .26). The two fertile controls were not statistically different from each other (F1,47 = 1.6, p = .21); we therefore pooled them for the above-mentioned analysis.
To study gene expression changes in response to these treatments and their interaction, we used RNA-seq. We examined expression changes in two tissues of key importance in endocrine physiology and energy metabolism, the fat body (the fly equivalent of mammalian adipose and liver tissues) and the head (as a proxy for the brain) (Baker & Thummel, 2007; Leopold and Perrimon 2007). Because age can have large effects on gene expression (Carnes et al., 2015; Pletcher et al., 2002), we examined transcriptional responses in young vs. old flies (10 vs. 38 days after eclosion; see Materials and Methods).
Previous work by Parisi et al. (2010) has also analyzed patterns of nongonadal gene expression using the germline-less Drosophila mutant tudor. A potential caveat of using such maternal-effect mutants is that they act during development (Boswell & Mahowald, 1985) and might thus exhibit confounding developmental effects (Flatt et al., 2008; Rodrigues et al., 2021). Here instead, we used an alternative method for germline ablation, enabling us to study the effects of germline loss in a manner that excludes potential developmental carry-over effects. While we analyzed expression in two tissues of female flies at two adult ages across 4 yeast levels, Parisi et al. (2010) examined expression in carcasses (the totality of nongonad tissue) of both females and males at 5–7 days of adult age on a single diet. Despite these major differences, the experiments of Parisi et al. (2010) and ours are complementary and provide a helpful comparison.
In total, we identified 8,644 differentially expressed genes (DEG). To explore overall patterns of expression changes we used principal components analysis (PCA) (Figure 2).
Figure 2.
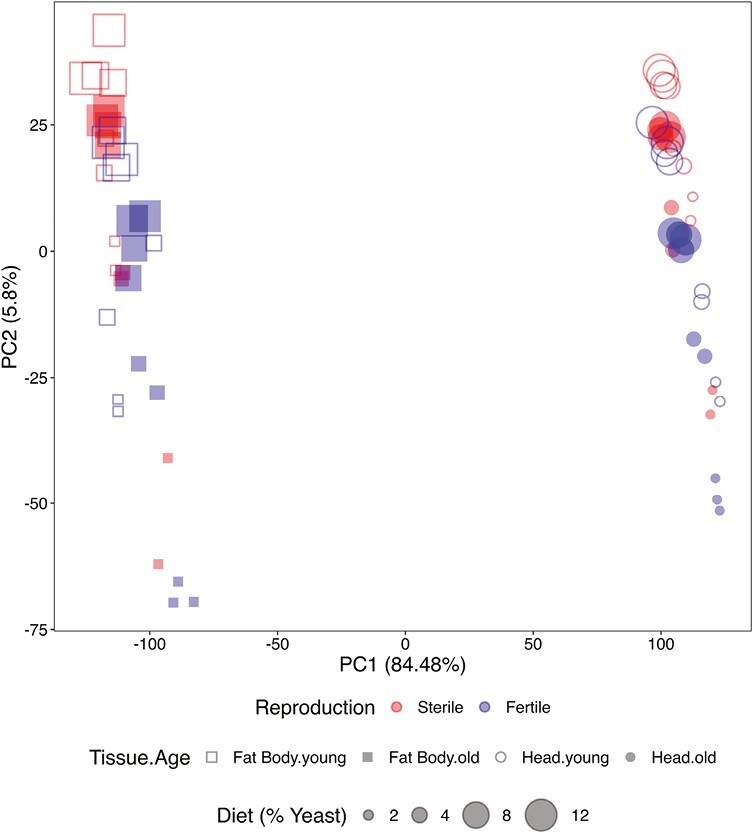
PCA of differentially expressed genes (DEG). PC1 separates the expression data by tissue (fat body vs. head), whereas PC2 separates the data by yeast levels and, more weakly, by reproductive status (germline-less vs. fertile flies). PCA plots based on normalized reads. Red symbols: germline-less (sterile) flies; blue symbols: fertile flies; squares: fat body; circles: heads; open symbols: young flies; filled symbols: old flies. Different symbol sizes represent the different yeast concentrations, ranging from smallest (2%) to largest (12%).
The first principal component (PC1) clearly separated fat body and head, explaining 84.48% of the variance in the data (Figure 2). PC2 separated the different yeast diets and, to a lesser extent, the two reproductive states (fertile vs. germline-less), but only explained 5.8% of the variance (Figure 2). Increasing yeast from to 2 to 8% had large effects on expression, while increasing yeast from 8 to 12% had marginal effects, as indicated by the clustering of the two high yeast levels in the PCA plot (Figure 2).
To facilitate further analyses, we divided our data by tissue and age into four subsets: (a) fat body, young; (b) fat body, old; (c) head, young; and (d) head, old. PCAs on these subsets resulted in a clear separation of reproductive states (Figure 3).
Figure 3.
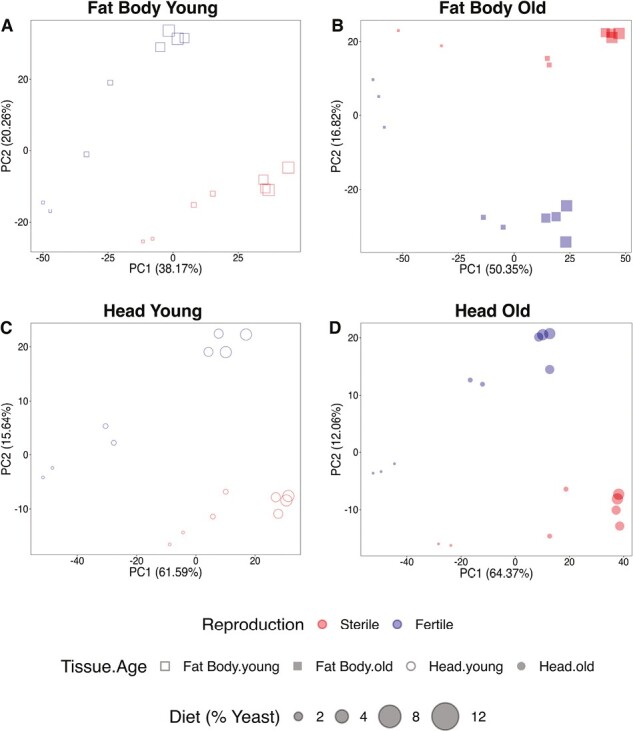
PCAs of differentially expressed genes (DEG) by tissue and age. (A–D) represent separate PCAs for each data sub-dataset: (A) fat body, young; (B) fat body, old; (C) head, young; and (D) head, old. In all four subsets, the two reproductive states (germline-less vs. fertile) are separated into distinct expression groups. PCA plots based on normalized reads. Red symbols: germline-less (sterile) flies; blue symbols: fertile flies; squares: fat body; circles: heads; open symbols: young flies; filled symbols: old flies. Different symbol sizes represent the different yeast concentrations, ranging from smallest (2%) to largest (12%).
Reproduction has major effects on expression of energy metabolic pathways
We next used linear models in limma-voom (Ritchie et al., 2015; see Materials and Methods) to identify expression effects on individual transcripts of (a) reproduction (R; germline-less vs. fertile), (b) diet (D; 2%, 4%, 8%, 12% yeast), and (c) the interaction between reproduction and diet (R X D) in each data subset. We only considered DEG with an absolute fold change (FC) ≥ 1.5 (log2 [FC] ≤ −0.58 or log2 [FC] ≥ 0.58) as candidates for analyses, resulting in a “universe” of 8644 DEG (Supplementary Table S1; see Materials and Methods).
Reproduction (germline ablated vs. fertile flies) affected the expression of 1,390 genes (16% of all DEG; Figure 4; Supplementary Table S1). Expression changes due to differences in reproduction were more prevalent in the fat body than in the head: 802 genes were differentially expressed in fat bodies of young flies and 749 DEG in fat bodies of old flies (Figure 4A and B, Supplementary Table S1). In contrast, we only found 472 and 416 DEG in the heads of young and old flies, respectively (Figure 4C and D, Supplementary Table S1).
Figure 4.
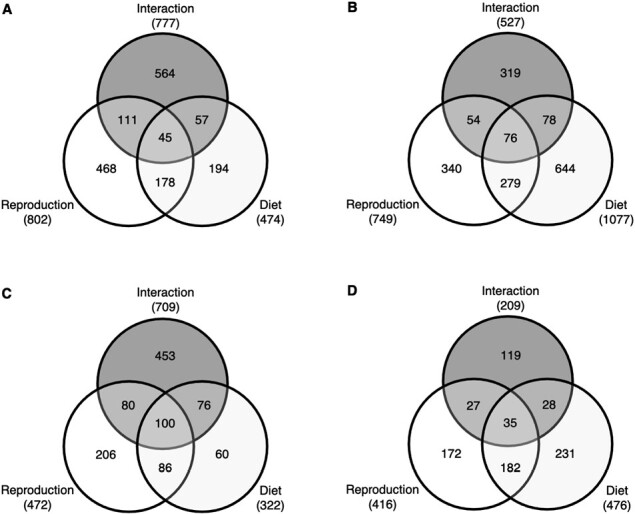
Number of significantly differentially expressed genes (DEG) in response to reproduction, diet and their interaction. Venn diagrams present the number of significantly DEG for the main effects of reproduction and diet and their interaction in the different data subsets: (A) fat body in young flies; (B) fat body in old flies; (C) head in young flies; and (D) head in old flies. See main text and Materials and Methods for further details.
DEG affected by reproduction were significantly enriched for lipid, carbohydrate and protein metabolism, as well as for signal transduction, immunity (also cf. Rodrigues et al., 2021) and neuronal physiology (Supplementary Tables S2 and S3). Notably, pathways and GO terms related to lipid metabolism were consistently and significantly enriched in all four data subsets, with a relatively large number of hits (Supplementary Tables S2 and S3).
Our results on the effects of germline removal vs. fertility on gene expression in fat body and head agree well with those of Parisi et al. (2010) who found that germline-less tudor mutants express many genes involved in energy capture and utilization (but also see Parisi et al., 2011). Such energy-related expression changes in distant tissues outside the gonad thus likely reflect the metabolic demands of reproduction.
Our above results are also consistent with previous work showing that germline removal in Drosophila has systemic effects on carbohydrate stores, insulin signaling, and immunity (Flatt et al., 2008; Rodrigues et al., 2021). Similarly, evidence suggests the existence of an endocrine feedback loop between germline stem cells in the gonad and the brain (Flatt et al., 2008; Hsu & Drummond-Barbosa, 2009; Hsu et al., 2008; LaFever & Drummond-Barbosa, 2005; Narbonne & Roy, 2006). As discussed by Parisi et al. (2010), such long-range effects of germline removal on gene expression and metabolism indicate that Drosophila has a germline/soma hormonal axis that is similar to the well known hypothalamic–pituitary–gonadal axis of mammals.
Dietary yeast levels and the interaction between reproduction and diet also had major effects on expression (Figure 4). Yeast levels affected the expression of 1,346 genes (15.6% of all DEG; Supplementary Table S1), with the highest number of DEG (1077) found in the fat body of old flies (Figure 4B, Supplementary Table S1). Genes whose expression was affected by diet were enriched for carbohydrate, amino acid and nucleotide metabolism, as well as for immunity and pathways related to molecule transport (Supplementary Tables S4 and S5). In particular, we identified a large number of pathway and GO-term enrichment hits for lipid metabolism, especially in old flies (Supplementary Tables S4 and S5). The reproduction by diet interaction also affected the expression of many genes (1,787 = 20.7% of all DEG; Figure 4, Supplementary Table S1), which were mostly enriched for RNA and protein metabolism (Supplementary Tables S6 and S7). Again, we found enrichment of pathways and GO terms related to lipid metabolism in all four subsets, with a particularly strong signature in the heads of young flies (Supplementary Tables S6 and S7).
Many transcriptional effects of reproduction, diet and their interaction thus seem to converge on the regulation of lipid metabolism. This is in strong agreement with the facts that (a) gamete production relies on mobilizing energy from fat; (b) curtailed reproduction causes excess fat storage; and (c) dietary yeast promotes fecundity but suppresses fat accumulation (see Introduction; cf. Simmons & Bradley, 1997; Skorupa et al., 2008). Given these compelling connections, we focus our discussion below on lipid metabolism (a discussion of other transcriptome-wide expression changes is beyond the scope of this paper; also see our related experiments and analyses in Rodrigues et al., 2021).
Loss of germline proliferation causes upregulation of lipid metabolic genes
In support of the above enrichment results (Supplementary Tables S2–S7), analyses of individual DEG using linear models revealed (a) upregulation of many genes involved in lipid metabolism in germline-less relative to fertile flies (Figure 5); (b) a positive relation between the expression levels of many of these transcripts and increasing dietary yeast levels (Figure 6); and (c) for several lipid metabolic genes significant interactions between the effects of reproduction and diet (Figure 7).
Figure 5.
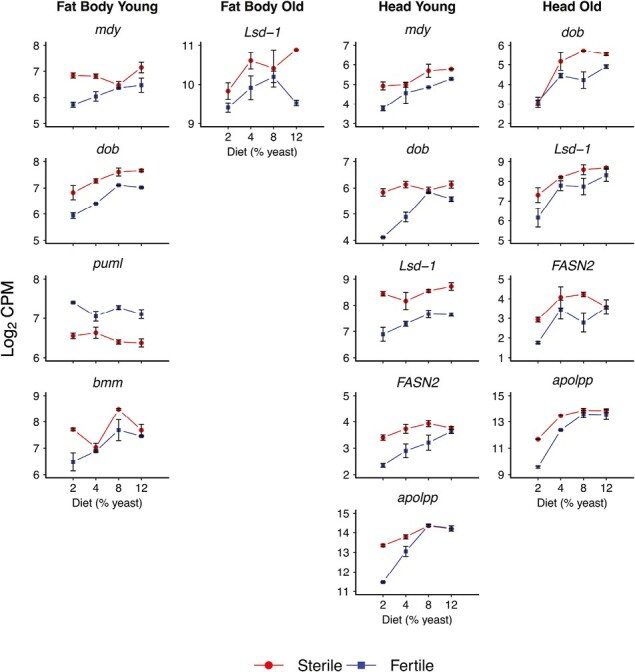
Effects of germline ablation vs. fertility on expression of genes in lipid metabolism. The figure shows a selection of genes involved in lipid metabolism whose expression is significantly affected by reproduction, i.e., germline removal vs. fertility (also see Supplementary Table S1). Germline removal seems to cause the upregulation of genes involved in both lipid anabolism and catabolism. Columns represent the four expression data subsets (fat body, young; fat body, old; head, young; and head, old). The x-axes display percentage dietary yeast (2%, 4%, 8%, 12%), and y-axes indicate expression values (log2 of the counts per million, CPM). Error bars represent standard errors of the mean. Red curves (“reaction norms”) depict expression in germline-less (sterile) flies, whereas blue curves or reaction norms represent fertile control flies. Note that the results for mdy, dob, Lsd-1, and apolpp are also displayed in Figures 6 and 7 because these transcripts were also significantly affected by diet and the reproduction × diet interaction.
Figure 6.
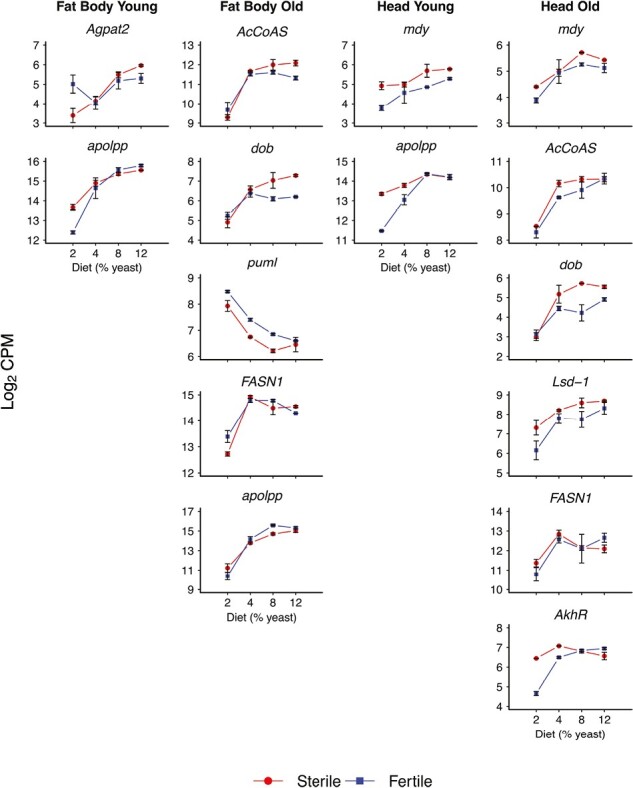
Effects of diet on expression of genes in lipid metabolism. The figure shows a selection of genes involved in lipid metabolism whose expression is significantly affected by dietary yeast concentration (also see Supplementary Table S1). Increasing yeast levels tend to lead to higher expression of genes involved in both lipid anabolism and catabolism. Columns represent the four expression data subsets (fat body, young; fat body, old; head, young; and head, old). The x-axes display percentage dietary yeast (2%, 4%, 8%, 12%), and y-axes indicate expression values (log2 of the counts per million, CPM). Error bars represent standard errors of the mean. Red curves (“reaction norms”) depict expression in germline-less (sterile) flies, whereas blue curves or reaction norms represent fertile control flies. Note that expression levels for mdy, AcCoAS, Agpat2, dob, FASN1, apolpp, and AkhR are also displayed in Figures 5 and 7 because these transcripts were also significantly affected by reproduction and the reproduction by diet interaction.
Figure 7.
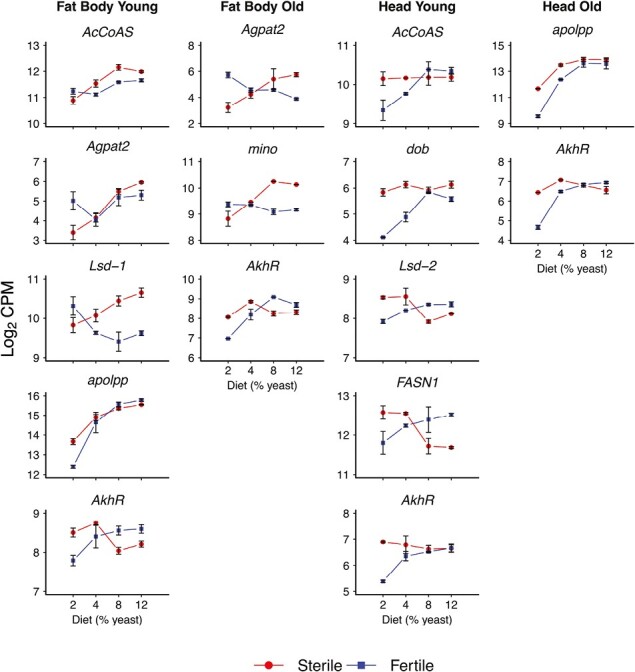
Effects of the reproduction by diet interaction on expression of genes in lipid metabolism. The figure shows genes involved in lipid metabolism whose expression is significantly affected by the interaction between reproduction and yeast concentration (see Supplementary Table S1). Columns represent the four expression data subsets (fat body, young; fat body, old; head, young; and head, old). The x-axes display percentage dietary yeast (2%, 4%, 8%, 12%), and y-axes indicate expression values (log2 of the counts per million, CPM). Error bars represent standard errors of the mean. Red curves (“reaction norms”) depict expression in germline-less (sterile) flies, whereas blue curves or reaction norms represent fertile control flies. Note that AcCoAS, Agpat2, dob, Lsd-1, FASN1, apolpp, and AkhR are also displayed in Figures 5 and 6 because their expression was also significantly affected by the main effects of reproduction and diet.
Figures 5–7 illustrate the expression effects of reproduction (Figure 5), dietary yeast (Figure 6), and the interaction between reproduction and diet (Figure 7) in the form of “reaction norm” plots: these plots depict expression levels of germline-less vs. fertile flies as a function of dietary yeast levels, plotted separately for the different tissues and age classes. Before discussing these results, we provide a brief overview of the key processes involved in lipid metabolism.
Brief overview of triacylglycerol/ lipid metabolism
Lipid metabolism is defined as the synthesis and breakdown of triacylglycerol (TAG) lipids, which represent the most important energy store for supporting metabolic homeostasis, reproduction and survival (Hansen et al., 2013; Heier & Kühnlein, 2018; Lehmann, 2018). The physiological (especially nutritional) state of the organism determines the different functions of lipid metabolism. Upon feeding under optimal diet conditions, ingested fat is processed and stored as TAG in lipid droplets in the fat body. Conversely, under poor diet conditions, or when reproduction demands it, fat reserves are mobilized and TAG are processed into smaller molecules to produce the energy necessary to support vital processes (Heier & Kühnlein, 2018; also cf. Introduction). At the same time, the fat reserves need to be replenished by the synthesis of new TAG molecules (Heier & Kühnlein, 2018).
Major endocrine signaling pathways, such as the IIS, target of rapamycin (TOR) and adipokinetic hormone (AKH) pathways, are the principal coordinators of lipid metabolism in response to organismal demands upon metabolism. The IIS/TOR pathways are important for initiating and regulating TAG synthesis (Heier & Kühnlein, 2018; Lehmann, 2018; Nässel & Vanden Broeck, 2016; Teleman, 2009), whereas AKH maintains homeostasis by mobilizing lipids in response to a negative energy balance, for example under starvation (Baumbach et al., 2014; Bharucha et al., 2008; Gáliková et al., 2015; Grönke et al., 2007; Isabel et al., 2005; Lee & Park, 2004; Liao et al., 2021; Mochanová et al., 2018).
Below we discuss the results in Figures 5–7 in terms of lipid anabolism vs. catabolism: TAG synthesis to support fat storage vs. mobilization of TAG stores and lipid transport to “fuel” energetically costly processes such as reproduction.
Lipid anabolism: TAG synthesis
Major genes implicated in lipid anabolism, including minotaur (mino, FBgn0027579, CG5508), midway (mdy, FBgn0004797, CG31991), 1-Acylglycerol-3-phosphate O-acyltransferase 2 (Agpat2, FBgn0026718, CG17608), and Fatty acid synthase genes 1 and 2 (FASN1, FBgn0283427, CG3523; FASN2, FBgn0042627, CG3524), were differentially expressed in response to reproduction, diet and their interaction (Figures 5–7, Supplementary Table S1).
For example, germline ablation caused upregulation of mdy and FASN2 relative to fertile flies, most prominently in young flies (Figure 5; Supplementary Table S1). mdy is involved in the last steps of TAG synthesis, specifically in the formation of long-chain fatty acids, whereas genes of the FASN family are implicated in de novo fatty acid formation (Barber et al., 2005; Baumbach et al., 2014; Beller et al., 2010; Heier & Kühnlein, 2018; Smith et al., 2003). Mutations in these genes are known to cause reduced TAG levels (Barber et al., 2005; Baumbach et al., 2014; Heier & Kühnlein, 2018; Schüpbach & Wieschaus, 1991; Wicker-Thomas et al., 2015). Interestingly, mdy is implicated in reproduction: it was first identified in a genetic screen for female sterility (Schüpbach & Wieschaus, 1991), and mdy mutants exhibit reduced oocyte lipid stores and egg chamber degeneration during mid-oogenesis (Buszczak et al., 2002). Loss of germline proliferation in Drosophila thus seems to increase the expression of several components of TAG synthesis, maybe consistent with previous results from other organisms showing that curtailed reproduction causes accumulation of excess fat (see Introduction).
Diet also affected the expression of genes in lipid anabolism. Depending on the tissue and age, the expression of mdy, FASN1, FASN2, and Agpat2 was positively affected by increasing yeast levels, typically followed by a plateau at higher yeast concentrations (Figure 6; Supplementary Table S1). At first glance, these findings are a bit puzzling, given that high yeast levels suppress fat accumulation in Drosophila (Simmons & Bradley, 1997; Skorupa et al., 2008). An important caveat is that transcript levels are unlikely to bear a 1:1 relation to realized levels of lipid store, and the effects of expression changes on fat stores will depend on the balance of anabolic vs. catabolic effects, as also discussed below.
Lipid anabolic genes such as mino (involved in the conversion of fatty acids into more complex lipids; Vagin et al., 2013), Agpat2, and FASN1 were also affected by the interaction between reproduction and diet, revealing several intriguing patterns (Figure 7; Supplementary Table S1). For instance, the expression levels of both mino and Agpat2 were higher in fertile than germline-less flies at the lowest yeast concentration (2%), but this pattern was reversed for higher yeast concentrations, with expression being higher in germline-less than fertile flies (Figure 7). On the other hand, FASN1 showed exactly the opposite interaction pattern as mino and Agpat2 (Figure 7).
These and other “interaction” results in Figure 7 indicate that the effects of germline proliferation vs. fertility on the expression of specific lipid metabolic genes depend critically on diet levels. Given the current state of our knowledge, it is very difficult to interpret such complex patterns beyond documenting their existence. Nonetheless, the presence of such interaction effects in response to well-defined experimental manipulations suggests that they may well be functionally relevant. This seems especially likely for reaction norms that cross over (e.g., as seen for mino and other transcripts; Figure 7): such crossing curves involve the reversal of a given effect at some crossing point and thus represent a very strong form of interaction.
Lipid catabolism: mobilization and transport of lipids
In terms of TAG catabolism, we found several genes involved in lipid mobilization to be differentially expressed in our dataset. For example, one of the main genes responsible for TAG breakdown (lipolysis), brummer (bmm, FBgn0036449, CG5295; see Grönke et al., 2005), was differentially expressed in response to germline ablation in the fat body of young flies (Figure 5, Supplementary Table S1).
In addition to bmm, we also observed differential expression of doppelgänger von brummer (dob, FBgn0030607, CG5560) and pummelig (puml, FBgn0033226, CG1882), both of which are induced by starvation and thought to have similar lipase functions as bmm (Grönke et al., 2005; Birner-Gruenberger et al., 2012; R. Kühnlein, pers. comm.; Figures 5 and 6, Supplementary Table S1). Expression of bmm and dob tended to be higher in germline-less flies, perhaps consistent with increased lipolysis upon germline ablation, but the opposite trend was seen for puml (Figures 5 and 6).
Expression of another component of lipid catabolism, adipokinetic hormone receptor (AkhR, FBgn0025595, CG11325), a member of the adipokinetic hormone (Akh) pathway, was affected by diet and especially by the reproduction by diet interaction (Figures 6 and 7, Supplementary Table S1). Notably, Akh signaling is a main regulator of bmm expression under poor diet conditions; similar to bmm, AkhR is involved in accumulation and mobilization of fat stores (Grönke et al., 2007). Mutants of Akhr and of Akh, the gene encoding the hormone ligand, are obese as adults and highly starvation resistant; although they down-spend their fat reserves during starvation at a similar rate as control flies, they maintain constitutively greater fat stores than control flies (Gáliková et al., 2015). Double mutants of bmm and AkhR are extremely obese and unable to mobilize body fat even when fully starved (Grönke et al., 2007).
Under low yeast conditions AkhR was upregulated in germline-less flies relative to fertile flies, but at higher yeast levels this pattern was reversed (Figure 7). Despite this interaction, AkhR expression tended to be more constant across yeast levels in germline-ablated flies, maybe suggesting that AKH signaling responds less to yeast levels in germline-less than fertile flies.
Two genes called Lipid storage droplet 1 and 2 (Lsd-1, FBgn0039114, CG10374; Lsd-2, FBgn0030608, CG9057), which are involved in regulating lipase activity on lipid droplets (Kimmel et al., 2010), were also differentially expressed in response to the reproduction, diet or their interaction (Figures 5–7, Supplementary Table S1). Lsd-1 was affected by reproduction and diet: expression tended to be higher in germline-less flies and increased with higher yeast concentration (Figures 5 and 6); and Lsd-2 expression was affected by the interaction between reproduction and diet in the heads of young flies (Figure 7).
Once mobilized, TAG must be transported from the fat body to other tissues. An important gene involved in this process is apolipophorin (apolpp, FBgn0087002, CG11064), which encodes the precursor of lipophorin (Lpp), the major lipoprotein responsible for carrying lipids through the hemolymph (Sundermeyer et al., 1996). Expression of apolpp was affected by reproduction, diet and their interaction across tissues and age classes (Figures 5–7, Supplementary Table S1). Overall, expression of apolpp tended to increase with higher yeast concentration (Figure 6). While apolpp was upregulated in germline-less flies at a low yeast level, its expression was very similar between germline-less and fertile flies at higher yeast concentrations (Figure 7).
Qualitative patterns in the dataset
Overall, two qualitative patterns emerge from the above analyses with regard to the effects of reproduction on the expression of genes involved in lipid metabolism (see Figures 5–7; see statistics in Supplementary Table S1).
First, germline-less flies often tend to exhibit significantly higher expression levels than fertile flies for both lipid anabolic and catabolic genes, suggesting that lipid metabolism might be increased in germline-less flies as compared to fertile control flies. Germline loss might thus possibly cause both higher lipid synthesis and higher breakdown.
Second, while expression levels of anabolic and catabolic genes typically increased with increasing yeast levels for both germline-less and fertile flies, the reaction norms of major lipid catabolism genes often tended to be flatter across yeast levels for germline-less as compared to fertile flies. If true, this pattern suggests that germline-less flies might break down lipids at a lower rate than fertile flies and that germline loss tilts the “metabolic balance” towards increased anabolism, thereby potentially causing increased fat storage.
Germline loss in the gonad causes excess fat storage in the soma
To test the above prediction, we determined the fat content of germline-less vs. fertile control female flies using a TAG assay (see Materials and Methods). As expected, we found that germline-less females exhibit excess fat storage as compared to fertile females (Figure 8 [data of MAR, CDV, TF]; this result is also supported by several independent previous experiments by our group [unpublished data of M. Gáliková; J. Steger {Steger, 2010}; S. Carvalho; data not shown]).
Figure 8.
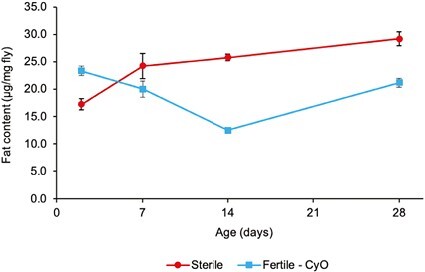
TAG content of germline-less vs. fertile control flies. The graph shows the TAG content (μg fat/ mg fly) of germline-ablated (sterile) female flies (red curve) vs. that of fertile control female flies (blue curve) as a function of adult age. Error bars represent standard errors of the mean. Starting at around 7 days of adult age, germline-less flies begin to harbor significantly higher levels of TAG levels than fertile flies. Analysis using a fully factorial two-way fixed-effects type II ANOVA revealed significant effects of Reproduction (germline-less vs. fertile control; F1,116 = 28.5); Age (F3,116 = 7.6); and Age × Reproduction (F3,116 = 24.8); in all three cases, p < .001.
The fact that germline-ablated D. melanogaster show increased fat accumulation mirrors observations in gonadectomized insects and germline-ablated C. elegans (e.g., Butterworth & Bodenstein, 1968; Judd et al., 2011; O’Rourke et al., 2009; Chaturbedi & Lee, 2023; also cf. Hansen et al., 2013) and suggests that the effects of germline loss upon lipid storage are evolutionarily conserved.
While the proximate causes of increased fat storage upon loss of germline stem cells in the fly await mechanistic study, it is noteworthy that germline-less flies are characterized by increased whole-body expression of the translation inhibitor 4E-BP whose activity is controlled by the IIS/TOR pathways (Flatt et al., 2008); interestingly, systemic activation of 4E-BP causes a net increase in fat accumulation in adipose tissue (Teleman et al., 2005). Similarly, germline-less flies exhibit upregulation of the insulin binding protein Imp-L2 (Flatt et al., 2008); and overexpression of Imp-L2 has been found to cause upregulation of 4E-BP, improve oxidative stress resistance and extend lifespan, reduce fecundity and—notably—to increase lipid storage (Alic et al., 2011). It is thus an interesting possibility that the effects of germline ablation on lipid storage might be mediated by 4E-BP (and/or by Imp-L2).
Summary and conclusions
Here we have used a transcriptomic approach to identify potential “conflicts” (i.e., trade-offs) between reproduction and metabolism over patterns of gene expression in Drosophila. Our main findings and conclusions can be summarized as follows:
(1) Increasing dietary yeast levels tend to increase the expression of genes involved in both lipid anabolism and catabolism. Since fecundity increases with higher yeast levels, this might indicate an increased turnover of lipid pools (i.e., higher synthesis and breakdown) in order to meet the high energetic demands of egg manufacture. Yet, since high yeast levels repress fat accumulation despite promoting fecundity (e.g., Simmons & Bradley, 1997; Skorupa et al., 2008), this implies that under high yeast conditions “expenditure” might exceed “income.”
(2) Germline-less flies tend to exhibit higher expression of both lipid anabolic and catabolic genes as compared to fertile control flies; lack of germline activity in the gonad has systemic effects on the expression of genes involved in lipid metabolism in tissues outside of the gonad, i.e., in the fat body and head.
(3) Despite this increased expression of both anabolic and catabolic genes, the reaction norms for several major catabolic genes are shallower in germline-less as compared to fertile flies, consistent with the idea that breakdown of lipids in germline-ablated flies might be reduced. In support of this hypothesis, germline-less flies exhibit significantly increased fat storage as compared to fertile flies, similar to previous observations in C. elegans (e.g., Chaturbedi & Lee, 2023; O’Rourke et al., 2009).
(4) These results confirm that germline activity trades off with fat storage and that developing oocytes represent a major energetic sink for lipids (see Introduction; cf. Van Antwerpen et al., 2005). Removal of this sink in the gonad, for example, through ablation of proliferating germline stem cells, has systemic effects that cause fat accumulation in the soma (cf. Butterworth & Bodenstein, 1968).
(5) Germline loss is a sufficient but not a necessary cause for increased lipid storage as gonadectomy, hypogonadism, or other forms of sterility also cause excess fat storage (see Introduction; cf. Chaturbedi & Lee, 2023; Hansen et al., 2013).
(6) The effects of germline activity (or reproduction more broadly) on lipid metabolism represent a highly regulated, evolutionarily conserved process that involves a feedback loop between the gonad and the soma (cf. Butterworth & Bodenstein, 1968; Doane, 1961), not a passive process of energy allocation between reproduction and somatic maintenance.
It will clearly be of considerable interest to learn more about the signals that coordinate such metabolic costs of reproduction and how they constrain life-history evolution (cf. Fisher, 1930).
Materials and methods
Drosophila strains and maintenance
To obtain germline-less flies we used the binary GAL4 > UAS system, by crossing a nanos-GAL4::VP16 driver line (full genotype: y,w; +/+; nanos-GAL4::VP16/nanos-GAL4::VP16; Van Doren et al., 1998) to a UASp-bag of marbles (bam; full genotype: y,w; UAS-bam/CyO; +/+; Chen & McKearin, 2003) responder line (for details see Flatt et al., 2008). Ectopic overexpression of bam under the control of the nanos-GAL4::VP16 driver leads to the loss of germline stem cells at the late L3 pupal stage or in early adulthood (Chen & McKearin, 2003; Flatt et al., 2008). To obtain adult flies for the experiments, we employed the following procedure. Using light CO2 anesthesia, we collected virgin females and unmated males from the nanos-GAL4 and UAS-bam stocks within 2 hr of eclosion and kept them separate for 3 days. After 3 days, 15 females from one strain and 15 males from the other were placed in a bottle containing 25 ml of medium, allowing flies to mate and females to lay eggs during 24 hr. Crosses were set up reciprocally in both directions (cross 1: UAS-bam females × nanos-GAL4 males; cross 2: nanos-GAL4 females × UAS-bam males). These crosses yielded 50% germline-less (sterile) progeny (y,w; nanos-GAL4::VP16/+; UAS-bam/+; called “Sterile”) and 50% fertile progeny (y,w; CyO/+; nanos-GAL4::VP16/+; called “Fertile - CyO”). Adult F1 flies from these crosses were collected within 2 hr of eclosion and sexed under light CO2 anesthesia; the dominant CyO mutation on the second chromosome of the UAS strain was used as a marker to distinguish between germline-less and fertile females. Prior assays revealed no differences between the two cross directions in terms of fat content: cross directionality had neither a significant effect on the fat content (μg fat/ mg fly) of germline-less F1 flies (type II ANOVA, effect of cross direction: F1,14 = 1.4, p = .26) nor on that of fertile F1 flies (F1,16 = 0.39, p = .54) (unpublished data of CDV and TF). For our experiments here, we therefore pooled F1 females from both crosses in equal proportions. From these crosses, we reared F1 progeny to adulthood.
As fertile control genotypes we used, depending on the experiment, (i) fertile females derived from the above-mentioned crosses (i.e., y,w; CyO/+; nanos-GAL4::VP16/+ = “Fertile - CyO”) and/or (ii) fertile y1, w1118 mutant females (y1,w1118; +/+; +/+; called “Fertile”), i.e., the strain that provided the genetic background for the UAS and GAL4 strains (obtained through multi-generation backcrossing; Flatt et al., 2008). Control (ii) was handled as described above for transgenic crosses. Fecundity assays involved both control genotypes (i) and (ii); fat assays employed control (i); and RNA-seq experiments used control (ii).
Note that multiple (or different) control genotypes are commonly used (and useful) when employing the binary GAL4 > UAS transgenic system (cf. Flatt et al., 2008). This is because there is typically no single, “perfect” control genotype: even with repeated backcrossing and isogenization of backgrounds, the binary GAL4 > UAS system (which involves crossing two distinct strains) does not necessarily guarantee complete isogenicity. Importantly, both control genotypes, (i) and (ii), yielded identical results in terms of fecundity (see analysis in Figure 1), suggesting that both controls can be viewed as being equivalent.
Stocks were maintained and crosses and experiments performed at 25 °C and 60% relative air humidity on a 12 hr:12 hr light:dark cycle, using controlled larval densities to avoid overcrowding. Stocks were reared on a standard laboratory diet consisting of agar (7 g/L), sugar (50 g/L, sucrose), yeast (50 g/L), cornmeal (50 g/L), 20% nipagin (10 ml/L), and propionic acid (6 ml/L).
Fecundity assay
In the fecundity assay, we compared germline-less flies with both controls (i) and (ii), as mentioned above. Upon eclosion, one female and one male from the same genotype were put into a vial containing one of the four diet treatments (2%, 4%, 8%, and 12% yeast in the total food volume) (see Supplementary Table S8).
The range of yeast levels used in our experiments (also see the section on RNA-seq below) was chosen based on a large body of prior work on dietary manipulation in Drosophila (e.g., reviewed in Min & Tatar, 2006; Min et al., 2007; Tatar, 2007 and references therein; also cf. ). The majority of such experiments has used yeast levels falling into the range of 1–16% yeast, with levels of approximately 2–5% yeast typically maximizing lifespan (“dietary restriction” effect) at the expense of reduced fecundity and with lower (malnutrition) or higher yeast levels (overfeeding) causing shortened lifespan (cf. Tatar, 2007).
For each genotype and diet, we used seven replicate vials. Flies were transferred daily to a new vial with fresh food, and daily per-capita fecundity was quantified by counting eggs in the old vial. Fecundity was measured over a 20-day period; Figure 1 shows the average number of eggs laid per female over the 20-day period. Egg count data were analyzed with a fully factorial two-way fixed-effects type II analysis of variance (ANOVA) on rank-transformed counts.
Fat assay
To measure and compare the fat content of germline-less flies vs. fertile control (i) flies we used a triglyceride assay. Genotypes were selected and separated upon eclosion; flies were kept in vials in mixed-sex groups consisting of approximately 10–15 adults and allowed to mate freely. Flies were transferred every 2–3 days into new vials with fresh food. Measures of fat content were performed on female flies of four different age classes: (a) 1–2 day-old, not yet reproductively fully mature/active females; (b) 5–8 day-old females at (or close to) peak fecundity; (c) 12–14 day-old females whose fecundity levels start to decline; and (d) 28–40 day-old females whose fecundity is relatively low to very low. For each age class, groups of 10–20 females from each genotype were collected, weighed, snap-frozen, and stored at −20 °C. For fat measurements, we separated females into groups of two flies per sample and measured fat content (µg/ mg fly) with the Serum Triglyceride Determination kit from Sigma, using triolein for establishing standard curves. The data were analyzed using two-way fixed-effects type II ANOVA with factors genotype and age and their interaction.
RNA-seq and transcriptomic analyses
Experimental flies, RNA extraction and sequencing
Germline-less and control genotype (ii) flies were collected upon eclosion and transferred to 1-L demography cages (Tatar et al., 2001) within a 24-h period. Cages with germline-less flies were set up with 65 females and 35 males from each cross direction (see above), thus giving a total of 130 females and 70 males per cage. For control flies, the same total number of females (130) and males (70) was transferred to each cage. Four replicate cages were set up per genotype, with one cage per diet treatment (i.e., either 2%, 4%, 8%, or 12% yeast) (Supplementary Table S8) until the end of the experiment (see above for the choice of yeast levels). Fresh food was supplied every second day, and dead flies were removed from cages
Adult females from each cage were sampled for RNA extractions at two different ages: 10 day-old females (“young” group) and 38 day-old females (“old” group). For each cage and time point, four groups of 5 females were sampled (20 females in total), providing two replicates for fat body and head tissue extractions. Each sample consisted of a pool of tissue from five females.
For fat body samples, groups of five female flies were cold-anesthetized at 4 °C for 2–3 min. Fat bodies were dissected in ice-cold 1× PBS and collected attached to the cuticle to ensure that the entire fat body tissue was collected. Such samples are often referred to as “fat body enriched” samples (DiAngelo & Birnbaum, 2009). Once detached from the other organs and separated from the thorax, fat body enriched tissue was transferred to sterile tubes with 200 µl of homogenization buffer from the RNA isolation kit (MagMAX-96 Total RNA Isolation Kit [ThermoFisher Scientific, Waltham, MA, USA]). Tissue samples were homogenized using a pestle rotor until no visible tissue could be recognized in the solution. Samples were stored at −80 °C until RNA extraction.
For head samples, RNA from entire heads was sampled, with the samples thus including the brain, the head capsule and the head (pericerebral) fat body, but excluding the retrocerebral complex (which contains the corpus allatum and the corpora cardiaca). Groups of five females were transferred into sterile tubes and snap-frozen using liquid nitrogen. Tubes were shaken manually in order to separate heads from bodies. Heads were transferred to new sterile tubes containing 200 µl of homogenization buffer from the RNA isolation kit. As described above, tissues were homogenized in the solution and kept at −80 °C until RNA extraction.
Total RNA was extracted from 66 samples (2 genotypes × 2 tissues × 2 age classes × 4 diets × 2 (or 3) replicates; see Supplementary Table S9) using the MagMAX-96 Total RNA Isolation Kit (ThermoFisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol on a MagMAX Express Magnetic Particle Processor (ThermoFisher Scientific, Waltham, MA, USA). Prior to sequencing RNA quality was measured using Fragment Analyzer (Advanced Analytical). Total RNA from each sample was sequenced using the Illumina HiSeq 4000 platform at BGI (Hong Kong, China), with the following parameters: paired-end, 100bp length, and approximately 200× coverage. Library preparation was performed by BGI using the TruSeq RNA kit (Illumina, Ca. USA), following the BGI in-house protocol.
Analysis of RNA-seq data
Reads were cleaned by BGI using “SOAPnuke” (https://github.com/BGI-flexlab/SOAPnuke) using the following parameters: -n 0.05 -l 20 -q 0.2 -p 1 -i -Q 2 -G --seqType 1. Upon receipt of the reads, we performed quality assessment using FastQC (v.0.11.7; Andrews, 2010) and assessed that a second cleaning step was necessary. After the cleaning step, reads were trimmed with Q-score below 35 using Cutadapt (v.1.15; Martin, 2011). Trimmed reads were aligned to the D. melanogaster transcriptome (release 6.17) using Kallisto (v.0.43.0; Bray et al., 2016), and a quantification list of transcript abundances was generated. To identify differentially expressed genes we used the Bioconductor package edgeR (v.3.20.8; Robinson et al., 2009) in R (v.3.5.0; http://www.R-project.org). Genes with less than two counts per million in at least 12 samples were excluded. The final number of differentially expressed genes in this experiment was 8,644 (i.e., 62% of all genes).
To reduce the complexity and dimensionality of our dataset prior to further analyses beyond PCA (see Figure 2), we divided the dataset by tissue (fat body vs. head tissues), according to PC1, and by age class, thus resulting in four data subsets: (a) fat body, young; (b) fat body, old; (c) head, young; and (d) head, old.
To identify gene expression changes in response to reproductive manipulation (sterile vs. fertile), dietary manipulation (different yeast levels), and their interaction we performed factorial analyses with the Bioconductor package “limma-voom” (v.3.34.7; Ritchie et al., 2015) and using the “makeConstrast” function. To calculate a global F-test across pairwise comparisons we employed the “eBayes” function and applied the Benjamini-Hochberg procedure to all p-values in order to account for multiple testing (Benjamini & Hochberg, 1995). Next, we selected differentially expressed candidate genes based on a significance threshold of <5% (adjusted F-test p < .05). Because some differentially expressed genes exhibited rather small fold-changes (FC), we used an additional FC-based cutoff and only considered genes with an absolute FC equal to or greater than 1.5 as candidates (log2 [FC] ≤ -0.58 or log2 [FC] ≥ 0.58) between groups (Supplementary Table S1).
To perform pathway enrichment analyses based on candidate genes, we used the Bioconductor package “ReactomePA” (v.1.28.0; Yu & He, 2016). p-values were corrected for multiple testing using the Benjamini-Hochberg method (Benjamini & Hochberg, 1995). As a complementary approach, we additionally performed gene ontology (GO) term enrichment analyses using the Bioconductor package “topGO” (v.2.16.0; Alexa & Rahnenführer, 2010), with a minimum node size of 5. In order to to be more conservative and stringent, we applied a lower p-value filter (adjusted p-value < .005) in the GO-term analysis.
Supplementary Material
Acknowledgments
We are grateful to two anonymous reviewers for valuable comments on a previous version of our paper. We thank our colleagues H. Aguilaniu, L. Falquet, J. Korb, R. Kühnlein, R. Rohr, and B.J. Zwaan, as well as former members of the Flatt lab, in particular M. Gálikova, J. Steger, S. Carvalho, and D. Martynow, for support and discussion over the years. This paper was written as part of the research carried out by the DFG Collaborative Research Unit (RU) “Sociality and the Reversal of the Fecundity–Longevity Trade-off” (DFG FOR 2281).
Contributor Information
Marisa A Rodrigues, Department of Ecology and Evolution, University of Lausanne, Lausanne, Switzerland; Department of Biology, University of Fribourg, Fribourg, Switzerland.
Chantal Dauphin-Villemant, Department of Ecology and Evolution, University of Lausanne, Lausanne, Switzerland.
Margot Paris, Department of Biology, University of Fribourg, Fribourg, Switzerland.
Martin Kapun, Department of Ecology and Evolution, University of Lausanne, Lausanne, Switzerland; Department of Biology, University of Fribourg, Fribourg, Switzerland; Central Research Laboratories, Natural History Museum Vienna, Vienna, Austria; Division of Cell and Developmental Biology, Medical University of Vienna, Vienna, Austria.
Esra Durmaz Mitchell, Department of Ecology and Evolution, University of Lausanne, Lausanne, Switzerland; Department of Biology, University of Fribourg, Fribourg, Switzerland; Department of Biochemistry and Molecular Biology, University of Southern Denmark, Odense, Denmark.
Envel Kerdaffrec, Department of Biology, University of Fribourg, Fribourg, Switzerland.
Thomas Flatt, Department of Ecology and Evolution, University of Lausanne, Lausanne, Switzerland; Department of Biology, University of Fribourg, Fribourg, Switzerland.
Data and code availability
The RNA-seq data are available from the Short Read Archive (SRA) under SRA accession PRJNA672962 (http://www.ncbi.nlm.nih.gov/bioproject/672962). The raw data in Figures 2 and 8 are available at Dryad: https://doi.org/10.5061/dryad.8cz8w9gxt. The scripts and code used for data processing and analyses are available in the Supplementary Materials and Methods file (pdf) associated with this paper.
Author contributions
Definitions according to CRediT (https://casrai.org/credit/): M.A.R.: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing—original draft, Writing—review & editing; C.D.V.: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Software, Writing—review & editing; M.P.: Software, Writing—review & editing; M.K.: Methodology, Software; E.D.M.: Methodology, Writing—review & editing; E.K.: Software, Writing—review & editing; T.F.: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing—review & editing.
Funding
Over the years, our research on this project was funded by the Swiss National Science Foundation (SNSF grants 310030E-164207 and 31003A_182262 to T.F.), the Novartis Foundation for Medical-Biological Research (grants 13C154 and 19B149 to T.F.), the DFG Collaborative Research Unit (RU) “Sociality and the Reversal of the Fecundity–Longevity Trade-off” (DFG FOR 2281), the Austrian Science Foundation (FWF grant P21498-B11 to T.F.), and the European Molecular Biology Organization (EMBO long-term fellowship ALT 248-2018 to E.K.)
Conflict of interest: The authors declare no conflict of interest.
References
- Alexa, A., & Rahnenführer, J. (2010). topGO: Enrichment analysis for gene ontology. https://bioconductor.org/packages/release/bioc/html/topGO.html
- Alic, N., Hoddinott, M. P., Vinti, G., & Partridge, L. (2011). Lifespan extension by increased expression of the Drosophila homologue of the IGFBP7 tumour suppressor. Aging Cell, 10(1), 137–147. 10.1111/j.1474-9726.2010.00653.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Arantes-Oliveira, N., Apfeld, J., Dillin, A., & Kenyon, C. (2002). Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science, 295(5554), 502–505. 10.1126/science.1065768 [DOI] [PubMed] [Google Scholar]
- Baker, K. D., & Thummel, C. S. (2007). Diabetic larvae and obese flies—Emerging studies of metabolism in Drosophila. Cell Metabolism, 6(4), 257–266. 10.1016/j.cmet.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber, M. C., Price, N. T., & Travers, M. T. (2005). Structure and regulation of acetyl-CoA carboxylase genes of metazoa. Biochimica et Biophysica Acta, 1733(1), 1–28. 10.1016/j.bbalip.2004.12.001 [DOI] [PubMed] [Google Scholar]
- Barnes, A., & Partridge, L. (2003). Costing reproduction. Animal Behaviour, 66, 199–204. [Google Scholar]
- Baumbach, J., Hummel, P., Bickmeyer, I., Kowalczyk, K. M., Frank, M., Knorr, K., Hildebrandt, A., Riedel, D., Jäckle, H., & Kühnlein, R. P. (2014). A Drosophila in vivo screen identifies store-operated calcium entry as a key regulator of adiposity. Cell Metabolism, 19(2), 331–343. 10.1016/j.cmet.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Bell, G., & Koufopanou, V. (1986). The cost of reproduction. In Dawkins R. & Ridley M. (Eds.), Oxford surveys in evolutionary biology (pp. 83–131). Oxford University Press. [Google Scholar]
- Beller, M., Bulankina, A. V., Hsiao, H. -H., Urlaub, H., Jäckle, H., & Kühnlein, R. P. (2010). PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metabolism, 12(5), 521–532. 10.1016/j.cmet.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57(1), 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bharucha, K. N., Tarr, P., & Zipursky, S. L. (2008). A glucagon-like endocrine pathway in Drosophila modulates both lipid and carbohydrate homeostasis. The Journal of Experimental Biology, 211(Pt 19), 3103–3110. 10.1242/jeb.016451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner-Gruenberger, R., Bickmeyer, I., Lange, J., Hehlert, P., Hermetter, A., Kollroser, M., Rechberger, G. N., & Kühnlein, R. P. (2012). Functional fat body proteomics and gene targeting reveal in vivo functions of Drosophila melanogaster α-Esterase-7. Insect Biochemistry and Molecular Biology, 42(3), 220–229. 10.1016/j.ibmb.2011.12.004 [DOI] [PubMed] [Google Scholar]
- Boswell, R., & Mahowald, A. (1985). tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell, 43(1), 97–104. [DOI] [PubMed] [Google Scholar]
- Bray, N. L., Pimentel, H., Melsted, P., & Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology, 34(5), 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Bronson, F. H. (1989). Mammalian reproductive biology. University of Chicago Press. [Google Scholar]
- Buszczak, M., Lu, X., Segraves, W. A., Chang, T. Y., & Cooley, L. (2002). Mutations in the midway gene disrupt a Drosophila acyl coenzyme A: Diacylglycerol acyltransferase. Genetics, 160(4), 1511–1518. 10.1093/genetics/160.4.1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth, F. M., & Bodenstein, D. (1968). Adipose tissue of Drosophila melanogaster 3: The effect of the ovary on cell growth and the storage of lipid and glycogen in the adult tissue. The Journal of Experimental Zoology, 167(2), 207–217. 10.1002/jez.1401670209 [DOI] [PubMed] [Google Scholar]
- Calow, P. (1979). The cost of reproduction—A physiological approach. Biological Reviews, 54(1), 23–40. 10.1111/j.1469-185x.1979.tb00866.x [DOI] [PubMed] [Google Scholar]
- Carey, C. (1996). Avian energetics and nutritional ecology. Chapman & Hall. [Google Scholar]
- Carnes, M. U., Campbell, T., Huang, W., Butler, D. G., Carbone, M. A., Duncan, L. H., Harbajan, S. V., King, E. M., Peterson, K. R., Weitzel, A., Zhou, S., & Mackay, T. F. C. (2015). The genomic basis of postponed senescence in Drosophila melanogaster. PLoS One, 10(9), e0138569. 10.1371/journal.pone.0138569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturbedi, A., & Lee, S. S. (2023). Different gametogenesis states uniquely impact longevity in Caenorhabditis elegans. Preprint, bioRxiv: 2023.2006.2013.544885. [Google Scholar]
- Chen, D., & McKearin, D. M. (2003). A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development, 130(6), 1159–1170. 10.1242/dev.00325 [DOI] [PubMed] [Google Scholar]
- Chippindale, A. K., Chu, T. J. F., & Rose, M. R. (1996). Complex trade-offs and the evolution of starvation resistance in Drosophila melanogaster. Evolution, 50(2), 753–766. 10.1111/j.1558-5646.1996.tb03885.x [DOI] [PubMed] [Google Scholar]
- Chippindale, A. K., Leroi, A. M., Kim, S. B., & Rose, M. R. (1993). Phenotypic plasticity and selection in Drosophila life-history evolution 1 Nutrition and the cost of reproduction. Journal of Evolutionary Biology, 6(2), 171–193. 10.1046/j.1420-9101.1993.6020171.x [DOI] [Google Scholar]
- Corona, G., Mannucci, E., Forti, G., & Maggi, M. (2009). Hypogonadism, ED, metabolic syndrome and obesity: A pathological link supporting cardiovascular diseases. International Journal of Andrology, 32(6), 587–598. 10.1111/j.1365-2605.2008.00951.x [DOI] [PubMed] [Google Scholar]
- Crawford, D., Libina, N., & Kenyon, C. (2007). Caenorhabditis elegans integrates food and reproductive signals in lifespan determination. Aging Cell, 6(5), 715–721. 10.1111/j.1474-9726.2007.00327.x [DOI] [PubMed] [Google Scholar]
- Darwin, C. (1859). On the origins of species by means of natural selection. John Murray, London. [Google Scholar]
- DiAngelo, J. R., & Birnbaum M. J. (2009). Regulation of fat cell mass by insulin in Drosophila melanogaster. Molecular and Cellular Biology, 29(24), 6341–6352. 10.1128/MCB.00675-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle, H. (1996). Migration: The biology of life on the move. Oxford University Press. [Google Scholar]
- Djawdan, M., Chippindale, A. K., Rose, M. R., & Bradley, T. J. (1998). Metabolic reserves and evolved stress resistance in Drosophila melanogaster. Physiological Zoology, 71(5), 584–594. 10.1086/515963 [DOI] [PubMed] [Google Scholar]
- Djawdan, M., Sugiyama, T. T., Schlaeger, L. K., Bradley, T. J., & Rose, M. R. (1996). Metabolic aspects of the trade-off between fecundity and longevity in Drosophila melanogaster. Physiological Zoology, 69(5), 1176–1195. 10.1086/physzool.69.5.30164252 [DOI] [Google Scholar]
- Doane, W. W. (1961). Developmental physiology of the mutant female sterile(2)adipose of Drosophila melanogaster III Corpus allatum-complex and ovarian transplantations. The Journal of experimental zoology, 146, 275–298. 10.1002/jez.1401460307 [DOI] [PubMed] [Google Scholar]
- Fisher, R. A. (1930). The genetical theory of natural selection. Oxford at the Clarendon Press. [Google Scholar]
- Flatt, T. (2011). Survival costs of reproduction in Drosophila. Experimental Gerontology, 46(5), 369–375. 10.1016/j.exger.2010.10.008 [DOI] [PubMed] [Google Scholar]
- Flatt, T., & Heyland, A. (Eds.). (2011). Mechanisms of life history evolution. The genetics and physiology of life history traits and trade-offs. Oxford University Press. [Google Scholar]
- Flatt, T., Min, K. J., D’Alterio, C., Villa-Cuesta, E., Cumbers, J., Lehmann, R., Jones, D. L., & Tatar, M. (2008). Drosophila germ-line modulation of insulin signaling and lifespan. Proceedings of the National Academy of Sciences of the United States of America, 105(17), 6368–6373. 10.1073/pnas.0709128105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, S. A. (2016). Puzzles in modern biology I Male sterility, failure reveals design. F1000Research, 5, 2533. 10.12688/f1000research.9789.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gáliková, M., Diesner, M., Klepsatel, P., Hehlert, P., Xu, Y., Bickmeyer, I., Predel, R., & Kühnlein, R. P. (2015). Energy homeostasis control in Drosophila Adipokinetic hormone mutants. Genetics, 201(2), 665–683. 10.1534/genetics.115.178897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gáliková, M., & Klepsatel, P. (2018). Obesity and aging in the Drosophila Model. International Journal of Molecular Sciences, 19(7), 1896. 10.3390/ijms19071896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke, S., Mildner, A., Fellert, S., Tennagels, N., Petry, S., Müller, G., Jäckle, H., & Kühnlein, R. P. (2005). Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metabolism, 1(5), 323–330. 10.1016/j.cmet.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Grönke, S., Muller, G., Hirsch, J., Fellert, S., Andreou, A., Haase, T., Jackle, H., & Kuhnlein, R. (2007). Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biology, 5, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, M., Flatt, T., & Aguilaniu, H. (2013). Reproduction, fat metabolism, and life span: What is the connection? Cell Metabolism, 17(1), 10–19. 10.1016/j.cmet.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman, L., & Zera, A. (2007). The cost of reproduction: The devil in the details. Trends in Ecology and Evolution, 22, 80–86. [DOI] [PubMed] [Google Scholar]
- Heier, C., & Kühnlein, R. P. (2018). Triacylglycerol metabolism in Drosophila melanogaster. Genetics, 210(4), 1163–1184. 10.1534/genetics.118.301583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin, H., & Kenyon, C. (1999). Signals from the reproductive system regulate the lifespan of C elegans. Nature, 399(6734), 362–366. 10.1038/20694 [DOI] [PubMed] [Google Scholar]
- Hsu, H. J., & Drummond-Barbosa, D. (2009). Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proceedings of the National Academy of Sciences of the United States of America, 106(4), 1117–1121. 10.1073/pnas.0809144106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, H. J., LaFever, L., & Drummond-Barbosa, D. (2008). Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Developmental Biology, 313(2), 700–712. 10.1016/j.ydbio.2007.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel, G., Martin, J. R., Chidami, S., Veenstra, J. A., & Rosay, P. (2005). AKH-producing neuroendocrine cell ablation decreases trehalose and induces behavioral changes in Drosophila. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 288(2), R531–R538. 10.1152/ajpregu.00158.2004 [DOI] [PubMed] [Google Scholar]
- Judd, E. T., Wessels, F. J., Drewry, M. D., Grove, M., Wright, K., Hahn, D. A., & Hatle, J. D. (2011). Ovariectomy in grasshoppers increases somatic storage, but proportional allocation of ingested nutrients to somatic tissues is unchanged. Aging Cell, 10(6), 972–979. 10.1111/j.1474-9726.2011.00737.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel, A. R., Brasaemle, D. L., McAndrews-Hill, M., Sztalryd, C., & Londos, C. (2010). Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. Journal of Lipid Research, 51(3), 468–471. 10.1194/jlr.R000034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFever, L., & Drummond-Barbosa, D. (2005). Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science, 309(5737), 1071–1073. 10.1126/science.1111410 [DOI] [PubMed] [Google Scholar]
- Lee, G., & Park, J. H. (2004). Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the Adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics, 167(1), 311–323. 10.1534/genetics.167.1.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. P., Simpson, S. J., Clissold, F. J., Brooks, R., Ballard, J. W. O., Taylor, P. W., Soran, N., & Raubenheimer, D. (2008). Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proceedings of the National Academy of Sciences of the United States of America, 105(7), 2498–2503. 10.1073/pnas.0710787105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, M. (2018). Endocrine and physiological regulation of neutral fat storage in Drosophila. Molecular and Cellular Endocrinology, 461, 165–177. 10.1016/j.mce.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold, P., & Perrimon, N. (2007). Drosophila and the genetics of the internal milieu. Nature, 450(7167), 186–188. 10.1038/nature06286 [DOI] [PubMed] [Google Scholar]
- Leroi, A. (2001). Molecular signals versus the Loi de Balancement. Trends in Ecology and Evolution, 16(1), 24–29. 10.1016/s0169-5347(00)02032-2 [DOI] [PubMed] [Google Scholar]
- Leroi, A., Kim, S. N., & Rose, M. R. (1994). The evolution of phenotypic life-history trade-offs: An experimental study using Drosophila melanogaster. American Naturalist, 144, 661–676. [Google Scholar]
- Liao, S., Amcoff, M., & Nässel, D. R. (2021). Impact of high-fat diet on lifespan, metabolism, fecundity and behavioral senescence in Drosophila. Insect Biochemisry and Molecular Biology, 133, 103495. 10.1016/j.ibmb.2020.103495 [DOI] [PubMed] [Google Scholar]
- Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal, 17, 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- McCormick, M., Chen, K., Ramaswamy, P., & Kenyon, C. (2012). New genes that extend Caenorhabditis elegans’ lifespan in response to reproductive signals. Aging Cell, 11(2), 192–202. 10.1111/j.1474-9726.2011.00768.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy, J. F., & Wade, G. N. (1987). Short- and long-term effects of ovariectomy on food intake, body weight, carcass composition, and brown adipose tissue in rats. Physiology & Behavior, 39(3), 361–365. 10.1016/0031-9384(87)90235-6 [DOI] [PubMed] [Google Scholar]
- Min, K. J., Flatt, T., Kulaots, I., & Tatar, M. (2007). Counting calories in Drosophila diet restriction. Experimental Gerontology, 42(3), 247–251. 10.1016/j.exger.2006.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, K. J., & Tatar, M. (2006). Drosophila diet restriction in practice: Do flies consume fewer nutrients? Mechanisms of Ageing and Development, 127(1), 93–96. 10.1016/j.mad.2005.09.004 [DOI] [PubMed] [Google Scholar]
- Mochanová, M., Tomčala, A., Svobodová, Z., & Kodrík, D. (2018). Role of adipokinetic hormone during starvation in Drosophila. Comparative biochemistry and physiology. Part B, Biochemistry & molecular biology, 226, 26–35. 10.1016/j.cbpb.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Narbonne, P., & Roy, R. (2006). Regulation of germline stem cell proliferation downstream of nutrient sensing. Cell Division, 1, 29. 10.1186/1747-1028-1-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel, D. R., & Vanden Broeck, J. (2016). Insulin/IGF signaling in Drosophila and other insects: factors that regulate production, release and post-release action of the insulin-like peptides. Cellular and molecular life sciences : CMLS, 73(2), 271–290. 10.1007/s00018-015-2063-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke, E. J., Soukas, A. A., Carr, C. E., & Ruvkun, G. (2009). C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metabolism, 10(5), 430–435. 10.1016/j.cmet.2009.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M., Li, R., & Oliver, B. (2011). Lipid profiles of female and male Drosophila. BMC Research Notes, 4, 198. 10.1186/1756-0500-4-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M. J., Gupta, V., Sturgill, D., Warren, J. T., Jallon, J. -M., Malone, J. H., Zhang, Y., Gilbert, L. I., & Oliver, B. (2010). Germline-dependent gene expression in distant non-gonadal somatic tissues of Drosophila. BMC Genomics, 11, 346. 10.1186/1471-2164-11-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher, S. D., Macdonald, S. J., Marguerie, R., Certa, U., Stearns, S. C., Goldstein, D. B., & Partridge, L. (2002). Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Current Biology: CB, 12(9), 712–723. 10.1016/s0960-9822(02)00808-4 [DOI] [PubMed] [Google Scholar]
- Rae, R., Sinha, A., & Sommer, R. J. (2012). Genome-wide analysis of germline signaling genes regulating longevity and innate immunity in the nematode Pristionchus pacificus. PLoS Pathogens, 8(8), e1002864. 10.1371/journal.ppat.1002864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rion, S., & Kawecki, T. J. (2007). Evolutionary biology of starvation resistance: What we have learned from Drosophila. Journal of Evolutionary Biology, 20(5), 1655–1664. 10.1111/j.1420-9101.2007.01405.x [DOI] [PubMed] [Google Scholar]
- Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., & Smyth, G. K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research, 43(7), e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M. D., McCarthy, D. J., & Smyth, G. K. (2009). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1), 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, M. A., Merckelbach, A., Durmaz, E., Kerdaffrec, E., & Flatt, T. (2021). Transcriptomic evidence for a trade-off between germline proliferation and immunity in Drosophila. Evolution Letters, 5(6), 644–656. 10.1002/evl3.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff, D. A. (2007). Contributions of genomics to life-history theory. Nature Reviews Genetics, 8(2), 116–125. 10.1038/nrg2040 [DOI] [PubMed] [Google Scholar]
- Roff, D. A., & Fairbairn, D. J. (2007). The evolution of trade-offs: Where are we? Journal of Evolutionary Biology, 20(2), 433–447. 10.1111/j.1420-9101.2006.01255.x [DOI] [PubMed] [Google Scholar]
- Rose, M. R., & Bradley, T. J. (1998). Evolutionary physiology of the cost of reproduction. Oikos, 83(3), 443–451. 10.2307/3546672 [DOI] [Google Scholar]
- Rose, M. R., Vu, L. N., Park, S. U., & Graves, J. L.Jr (1992). Selection on stress resistance increases longevity in Drosophila melanogaster. Experimental Gerontology, 27(2), 241–250. 10.1016/0531-5565(92)90048-5 [DOI] [PubMed] [Google Scholar]
- Schüpbach, T., & Wieschaus, E. (1991). Female sterile mutations on the second chromosome of Drosophila melanogaster II Mutations blocking oogenesis or altering egg morphology. Genetics, 129(4), 1119–1136. 10.1093/genetics/129.4.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service, P. M. (1987). Physiological mechanisms of increased stress resistance in Drosophila melanogaster selected for postponed senescence. Physiological Zoology, 60(3), 321–326. 10.1086/physzool.60.3.30162285 [DOI] [Google Scholar]
- Service, P. M., Hutchinson, E. W., MacKinley, M. D., & Rose, M. R. (1985). Resistance to environmental stress in Drosophila melanogaster selected for postponed senescence. Physiological Zoology, 58, 380–389. [Google Scholar]
- Service, P. M., Hutchinson, E. W., & Rose, M. R. (1988). Multiple genetic mechanisms for the evolution of senescence in Drosophila melanogaster. Evolution, 42(4), 708–716. 10.1111/j.1558-5646.1988.tb02489.x [DOI] [PubMed] [Google Scholar]
- Service, P. M., & Rose, M. R. (1985). Genetic covariation among life-history components: The effects of novel environments. Evolution, 39(4), 943–945. 10.1111/j.1558-5646.1985.tb00436.x [DOI] [PubMed] [Google Scholar]
- Simmons, F. H., & Bradley, T. J. (1997). An analysis of resource allocation in response to dietary yeast in Drosophila melanogaster. Journal of Insect Physiology, 43(8), 779–788. 10.1016/s0022-1910(97)00037-1 [DOI] [PubMed] [Google Scholar]
- Skorupa, D. A., Dervisefendic, A., Zwiener, J., & Pletcher, S. D. (2008). Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell, 7(4), 478–490. 10.1111/j.1474-9726.2008.00400.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S., Witkowski, A., & Joshi, A. K. (2003). Structural and functional organization of the animal fatty acid synthase. Progress in Lipid Research, 42(4), 289–317. 10.1016/s0163-7827(02)00067-x [DOI] [PubMed] [Google Scholar]
- Socha, R., Sula, J., Kodrik, D., & Gelbic, I. (1991). Hormonal control of vitellogenin synthesis in Pyrrhocoris apterus (l) (Heteroptera). Journal of Insect Physiology, 37(11), 805–816. 10.1016/0022-1910(91)90077-d [DOI] [Google Scholar]
- Stearns, S. C. (1989). Trade-offs in life-history evolution. Functional Ecology, 3(3), 259–268. 10.2307/2389364 [DOI] [Google Scholar]
- Stearns, S. C., & Magwene, P.; American Society of Naturalists. (2003). The naturalist in a world of genomics. The American Naturalist, 161(2), 171–180. 10.1086/367983 [DOI] [PubMed] [Google Scholar]
- Steger, J. (2010). Effects of germline ablation on fat metabolism in Drosophila melanogaster [Unpublished Thesis for the Degree of Bachelor of Science (Biomedicine & Biotechnology)]. Institute of Population Genetics. [Google Scholar]
- Stotsenburg, J. M. (1913). The effect of spaying and semi-spaying young albino rats (Mus norvegicus albinus) on the growth in body weight and body length. The Anatomical Record, 7(6), 183–194. 10.1002/ar.1090070602 [DOI] [Google Scholar]
- Strong, L. (1967). Feeding activity, sexual maturation, hormones, and water balance in the female African migratory locust. Journal of Insect Physiology, 13(4), 495–507. 10.1016/0022-1910(67)90061-3 [DOI] [PubMed] [Google Scholar]
- Sundermeyer, K., Hendricks, J. K., Prasad, S. V., & Wells, M. A. (1996). The precursor protein of the structural apolipoproteins of lipophorin: cDNA and deduced amino acid sequence. Insect Biochemistry and Molecular Biology, 26(8-9), 735–738. 10.1016/s0965-1748(96)00060-4 [DOI] [PubMed] [Google Scholar]
- Tatar, M. (2007). Diet restriction in Drosophila melanogaster design and analysis. Interdisciplinary Topics in Gerontology, 35, 115–136. 10.1159/000096559 [DOI] [PubMed] [Google Scholar]
- Tatar, M., Chien, S. A., & Priest, N. K. (2001). Negligible senescence during reproductive dormancy in Drosophila melanogaster. The American Naturalist, 158(3), 248–258. 10.1086/321320 [DOI] [PubMed] [Google Scholar]
- Teleman, A., Chen, Y., & Cohen, S. (2005). 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes & Development, 19(16), 1844–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman, A. A. (2009). Molecular mechanisms of metabolic regulation by insulin in Drosophila. The Biochemical Journal, 425(1), 13–26. 10.1042/BJ20091181 [DOI] [PubMed] [Google Scholar]
- Thomsen, E., & Hamburger, K. (1955). Oxygen consumption of castrated females of the blow-fly, Calliphora erythrocephala Meig. Journal of Experimental Biology, 32(4), 692–699. 10.1242/jeb.32.4.692 [DOI] [Google Scholar]
- Townsend, C. R., & Calow, P. (1981). Physiological ecology: An evolutionary approach to resource use. Blackwell; 4. [Google Scholar]
- Vagin, V. V., Yu, Y., Jankowska, A., Luo, Y., Wasik, K. A., Malone, C. D., Harrison, E., Rosebrock, A., Wakimoto, B. T., Fagegaltier, D., Muerdter, F., & Hannon, G. J. (2013). Minotaur is critical for primary piRNA biogenesis. RNA, 19(8), 1064–1077. 10.1261/rna.039669.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Antwerpen, R., Daphne, Q.-D. P., & Ziegler, R. (2005). Accumulation of lipids in insect oocytes. in Reproductive Biology of Invertebrates, Vol.12, Part B (pp. 281–304). CRC Press. [Google Scholar]
- van der Horst, D. J., van Hoof, D., van Marrewijk, W. J., & Rodenburg, K. W. (2002). Alternative lipid mobilization: The insect shuttle system. Molecular and Cellular Biochemistry, 239(1-2), 113–119. [PubMed] [Google Scholar]
- Van Doren, M., Williamson, A. L., & Lehmann, R. (1998). Regulation of zygotic gene expression in Drosophila primordial germ cells. Current Biology: CB, 8(4), 243–246. 10.1016/s0960-9822(98)70091-0 [DOI] [PubMed] [Google Scholar]
- Wang, M., O’Rourke, E., & Ruvkun, G. (2008). Fat metabolism links germline stem cells and longevity in C elegans. Science, 322, 957–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker-Thomas, C., Garrido, D., Bontonou, G., Napal, L., Mazuras, N., Denis, B., Rubin, T., Parvy, J. P., & Montagne, J. (2015). Flexible origin of hydrocarbon/pheromone precursors in Drosophila melanogaster. Journal of Lipid Research, 56(11), 2094–2101. 10.1194/jlr.M060368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, T. D. (2005). Mechanisms underlying the costs of egg production. Bioscience, 55(1), 39–48. 10.1641/0006-3568(2005)055[0039:mutcoe]2.0.co;2 [DOI] [Google Scholar]
- Wilson, J. D., & Roehrborn, C. (1999). Long-term consequences of castration in men: lessons from the Skoptzy and the eunuchs of the Chinese and Ottoman courts. The Journal of Clinical Endocrinology and Metabolism, 84(12), 4324–4331. 10.1210/jcem.84.12.6206 [DOI] [PubMed] [Google Scholar]
- Yu, G., & He, Q. Y. (2016). ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Molecular Biosystems, 12(2), 477–479. 10.1039/c5mb00663e [DOI] [PubMed] [Google Scholar]
- Zera, A. J. (2005). Intermediary metabolism and life history trade-offs: Lipid metabolism in lines of the wing-polymorphic cricket, Gryllus firmus, selected for flight capability vs early age reproduction. Integrative and Comparative Biology, 45(3), 511–524. 10.1093/icb/45.3.511 [DOI] [PubMed] [Google Scholar]
- Zera, A. J., & Harshman, L. G. (2001). The physiology of life history trade-offs in animals. Annual Review of Ecology and Systematics, 32(1), 95–126. 10.1146/annurev.ecolsys.32.081501.114006 [DOI] [Google Scholar]
- Zera, A. J., & Larsen, A. (2001). The metabolic basis of life history variation: Genetic and phenotypic differences in lipid reserves among life history morphs of the wing-polymorphic cricket, Gryllus firmus. Journal of Insect Physiology, 47(10), 1147–1160. 10.1016/s0022-1910(01)00096-8 [DOI] [PubMed] [Google Scholar]
- Zhao, Z., & Zera, A. J. (2002). Differential lipid biosynthesis underlies a tradeoff between reproduction and flight capability in a wing-polymorphic cricket. Proceedings of the National Academy of Sciences of the United States of America, 99(26), 16829–16834. 10.1073/pnas.262533999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaan, B., Bijlsma, R., & Hoekstra, R. F. (1995). Direct selection on life span in Drosophila melanogaster. Evolution, 49(4), 649–659. 10.1111/j.1558-5646.1995.tb02301.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data are available from the Short Read Archive (SRA) under SRA accession PRJNA672962 (http://www.ncbi.nlm.nih.gov/bioproject/672962). The raw data in Figures 2 and 8 are available at Dryad: https://doi.org/10.5061/dryad.8cz8w9gxt. The scripts and code used for data processing and analyses are available in the Supplementary Materials and Methods file (pdf) associated with this paper.


