Abstract
Apoptosis is regulated cell death that depends on caspases. A specific initiator caspase is involved upstream of each apoptotic signaling pathway. Characterized in nematode, fly, and mammals, intrinsic apoptosis is considered to be ancestral, conserved among animals, and depends on shared initiators: caspase-9, Apaf-1 and Bcl-2. However, the biochemical role of mitochondria, the pivotal function of cytochrome c and the modality of caspase activation remain highly heterogeneous and hide profound molecular divergence among apoptotic pathways in animals. Uncovering the phylogenetic history of apoptotic actors, especially caspases, is crucial to shed light on the evolutionary history of intrinsic apoptosis. Here, we demonstrate with phylogenetic analyses that caspase-9, the fundamental key of intrinsic apoptosis, is deuterostome-specific, while caspase-2 is ancestral to bilaterians. Our analysis of Bcl-2 and Apaf-1 confirms heterogeneity in functional organization of apoptotic pathways in animals. Our results support emergence of distinct intrinsic apoptotic pathways during metazoan evolution.
Keywords: intrinsic apoptosis, initiator caspases, cell death evolution, phylogeny
Introduction
Apoptosis, regulated cell death defined by a set of morphological features and dependent on caspases, occurs during metazoan development, tissue homeostasis, and regeneration (Hipfner & Cohen, 2004; Jacobson et al., 1997; Jeffery & Gorički, 2021; Krasovec et al., 2019, 2021, 2022; Vriz et al., 2014). Pioneering works in Caenorhabditis elegans gave rise to a model of the molecular network of apoptosis decision, execution, engulfment-degradation, and more fundamentally the first description of an apoptotic regulation pathway at the molecular level (Ellis & Horvitz, 1986; Hengartner, 2000; Lettre & Hengartner, 2006). Investigations on apoptosis were next extended to Drosophila melanogaster and mammals, and recently to emerging models such as cnidarians, ascidians, mollusks, and others (Chambon et al., 2002; Cikala et al., 1999; Krasovec et al., 2021; Sokolova, 2009). Currently, two main apoptotic regulation pathways are described, recently named intrinsic (formerly known as mitochondrial pathway) and extrinsic apoptosis (previously named death receptor pathway) by the Nomenclature Committee on Cell Death (Galluzzi et al., 2018). Apoptotic pathways rely on a specific initiator caspase possessing a CARD (Caspase Recruitment Domain) or DED (Death Effector Domain) prodomain for intrinsic and extrinsic apoptosis, respectively. Each initiator caspase is activated into a protein complex specific to its pathway. Extrinsic apoptosis is triggered by activation of TNF receptors which recruit FADD, forming the DISC complex with initiator caspase-8 leading to its activation(Galluzzi et al., 2018). While extrinsic apoptosis is well-documented in mammals, it is absent in Caenorhabditis and Drosophila. However, intrinsic apoptosis was described in mammals as well as Caenorhabditis and Drosophila (Ellis & Horvitz, 1986; Hengartner, 2000; Lettre & Hengartner, 2006). Building on work in Caenorhabditis, investigation of apoptotic regulation pathways in Drosophila and mammals imposed the paradigm that the intrinsic apoptotic “molecular program” is conserved throughout animal evolution (Bender et al., 2012; Driscoll, 1996; Hengartner, 2000; Lettre & Hengartner, 2006; Steller, 2008). Intrinsic apoptosis depends on three components which are the key initiator caspase-9 (named Ced-3 and Dronc in Caenorhabditis and Drosophila, respectively), the activator Apaf-1 (named Ced-4 and Dark in Caenorhabditis and Drosophila, respectively) and the Bcl-2 multigenic family (Figure 1) (Hengartner, 2000; Steller, 2008). Initiator caspase-9 is activated by Apaf-1 and forms the apoptosome (Bao & Shi, 2007; Dorstyn et al., 2018; Shi, 2006). However, recent research has revealed evolutionary divergence between these models, with major functional diversifications (Dorstyn et al., 2018; Young et al., 2020). Notably, apoptosome platforms are different between Caenorhabditis, Drosophila, and mammals. In mammals, and conversely to ecdysozoans, the release of cytochrome c is mandatory for triggering intrinsic apoptosis and results from Mitochondrial Outer Membrane Permeabilization (MOMP) regulated by the Bcl-2 multigene family (Chipuk et al., 2010; Kalkavan & Green, 2018). Bcl-2 proteins, composed of BH domains, are categorized as either proapoptotic, such as Bcl-2 or Bcl-xl, and antiapoptotic, such as Bax, Bak, or Bok (Chipuk et al., 2010). The interaction and balance between Bcl-2 controls MOMP and subsequent cytochrome c release. While the Bcl-2 family is diverse in metazoans, it is poorly represented in Drosophila (two members; Buffy and Debcl) and Caenorhabditis (one member; Ced-9) in which their functions are secondary (Hengartner, 2000; Steller, 2008). Importantly, the initiation of apoptotic pathway in Drosophila depends on an increase of inhibition, controlled by three specific actors. Importantly, and independently of functional approaches, the phylogenetic history of intrinsic apoptosis actors, mostly the caspase family, is the cornerstone which sheds light on intrinsic apoptosis evolution. Caspases with a CARD prodomain (referred to as CARD-caspases), in particular, are diversified in several phyla, but their evolution remains enigmatic. Vertebrates, in addition to caspase-9, possess a set of inflammatory caspases (caspase-1, 4, and 5) which play a role in the immune system and the inflammatory response (T.-J. Fan et al., 2005). Caspase-2 is involved in various molecular pathways, including the DNA damage response and cell cycle regulation. Several CARD-caspases have been identified in mollusk genomes, such as in the oyster Crassostrea gigas, conversely to Caenorhabditis and Drosophila where Ced-3 and Dronc, respectively, are the only representative of CARD-caspases. In this study, we investigated the evolutionary history of the major actors of intrinsic apoptosis. We conducted separate phylogenetic analyses of the three fundamental actors of intrinsic apoptosis: the initiator CARD-caspases, the effector Bcl-2 family members, and the activator Apaf-1 proteins, across major animal phyla and describe their evolutionary patterns. Despite functional similarities, these actors, especially initiator caspases, are engaged in relatively independent evolutionary histories that involve distinct signaling pathways. Surprisingly, we discovered that Ced-3 and Dronc are not orthologous to mammalian caspase-9 but rather to caspase-2, challenging the prevailing vision of a conservation of intrinsic apoptosis actors across evolution. More fundamentally, caspase-9 is specific to Deuterostomia, while caspase-2 is ancestral and shared among bilaterian animals. Lophotrochozoans present a specific diversification of CARD-caspases, leading us to designate a CARD-caspase group specific to mollusks and annelids, caspase-Y.
Figure 1.
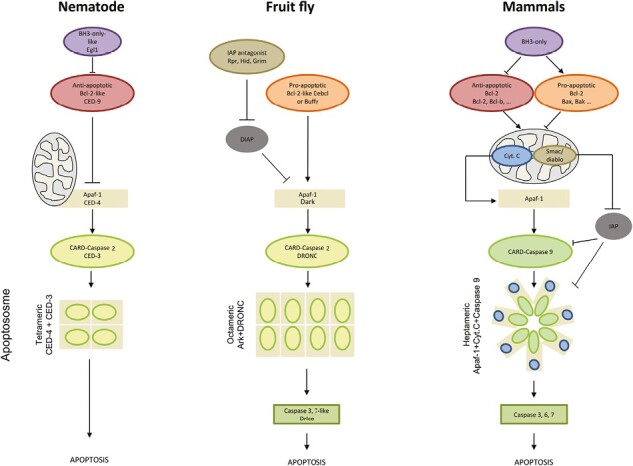
Schematic representations of intrinsic apoptosis in nematode, fruit fly, and mammals illustrate the dependence on three major components: the Bcl-2 family, a CARD-caspase, and Apaf-1, which forms the apoptosome platform (structurally different between species) with the initiator caspase. In mammals, the initiator caspase is caspase-9, while in ecdysozoans, it is caspase-2 (result of this study). In mammals, the release of cytochrome c forms the apoptosome after mitochondrial membrane permeabilization. The initiation of the fruit fly pathway involves the removal of inhibition by Rpr, Hid, and Grim. The function of mitochondria is minor in ecdysozoans and cytochrome c is not required to initiate apoptosis. Nematodes lack executioner caspases.
Results and discussion
Initiator caspases of intrinsic apoptotic pathways are not orthologous genes
Initiation and execution of apoptotic signaling pathways fundamentally depend on caspases which are widely distributed and diversified among all metazoan phyla (Bell & Megeney, 2017; Crawford et al., 2012; Shrestha et al., 2020; Uren et al., 2000).
Caspases are a class of proteases composed of three protein domains: the prodomain, the small P10, and the large P20 (Elmore, 2007; Thornberry & Lazebnik, 1998). Initiator caspases, specific to each apoptotic pathway, have a Caspase Recruiting Domain (CARD) prodomain or two DED prodomains. Intrinsic and extrinsic apoptosis are distinct and involve specific initiators, either caspase-9 (CARD prodomain) or caspase-8/-10 (DED prodomains), respectively. Both pathways trigger activation of common executioner caspases, leading to apoptosis execution (Galluzzi et al., 2018; Hengartner, 2000). First, we conducted a phylogenetic analysis on the whole caspase family at the metazoan scale with restricted species sampling, but representative of animal diversity. The topology we obtained is composed of several monophyletic groups globally corresponding to caspase types (Figure 2). CARD-caspases form a well-supported cluster, with only three divergent sequences excluded from this clade (Hydra caspase-X and Octopus caspase-Y1 and Y2). These positions likely indicate specific evolution in these species and do not affect deep nodes. The CARD-caspase clade maintains taxonomic coherence, with non-bilaterians positioned at the base of the group, and bilaterian sequences forming a monophyletic cluster. Bilaterian sequences are divided across two groups, first caspase-9 with deuterostomian caspases exclusively, and secondly [Caspase-2 + Inflammatory caspases + Caspase-Y] comprising of deuterostomian, lophotrochozoan, and ecdysozoan sequences. Due to the coherent CARD-caspase clustering, and given the crucial role of caspase-9 in initiating intrinsic apoptosis, we next choose to focus on CARD-caspases only. Using this strategy allowed us to considerably increase the species sampling for an in-depth analysis of their evolutionary history.
Figure 2.
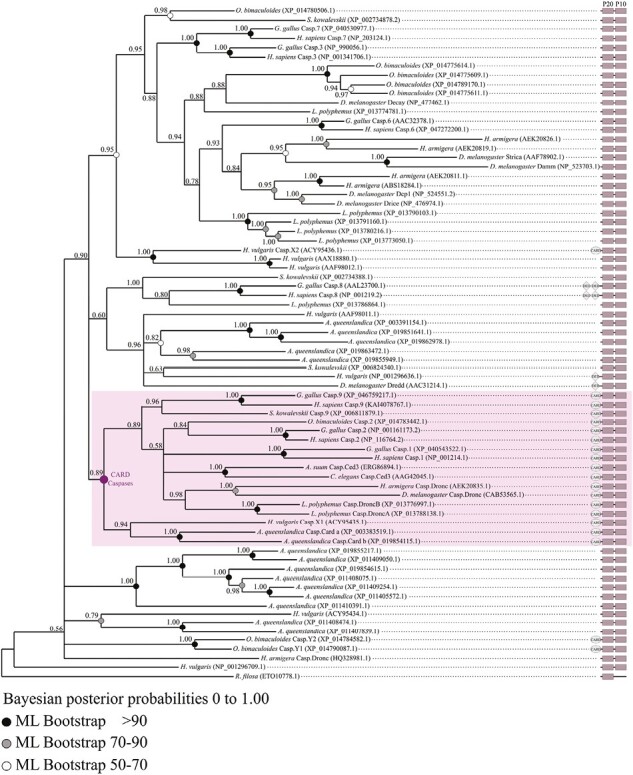
Topology of the metazoan caspase phylogeny obtained by Bayesian inference using full-length sequence alignments with a Reticulomyxa filosa sequence as out group. CARD-caspases cluster together, with only three sequences appearing outside of the group. These caspases belong to Hydra vulgaris and Octopus bimaculoides and are likely divergent when compared to other CARD-caspases. The distribution of CARD-caspases suggests a monophyly of this caspase type. The topology of the CARD-caspase cluster shows a similar species distribution as in the analysis made on CARD-caspases only (Figure 3). Non-bilaterian animals branch early, bilaterians are monophyletic and split across two main groups, the deuterostomian caspase-9 and the bilaterian caspase-2.
We confirmed distribution of CARD-caspases in most animal phyla and reconstructed their phylogenetic relationships (Figure 3, Supplementary Table S1). Both Bayesian inference and maximum likelihood methodologies gave consistent topologies, identifying four strongly supported monophyletic groups: caspase-9 (PP – Posterior Probability- = 1.00), caspase-2 (PP = 0.62), vertebrate inflammatory caspases (PP = 1.00), lophotrochozoan caspase-Y (PP = 0.98), with non-bilaterian caspase-X presenting a paraphyletic distribution. Monophyly of caspase-9 and caspase-2 families remains robust, grouping together with Inflammatory caspases and caspase-Y in a clade strictly diversified within bilaterian animals (PP = 0.81). The sequences of non-bilaterians [Cnidaria + Ctenophora + Placozoa] formed several groups, here named caspase-X (Figure 3). Switching between metazoan putative “basal” out groups (i.e., Amphimedon queenslandica (Porifera) or Pleurobrachia bachei (Ctenophora)) does not affect caspase topology (Supplementary Figure S1). All bilaterian sequences, including caspase-9, caspase-2, inflammatory caspases, and caspase-Y remained monophyletic (Supplementary Figure S1).
Figure 3.
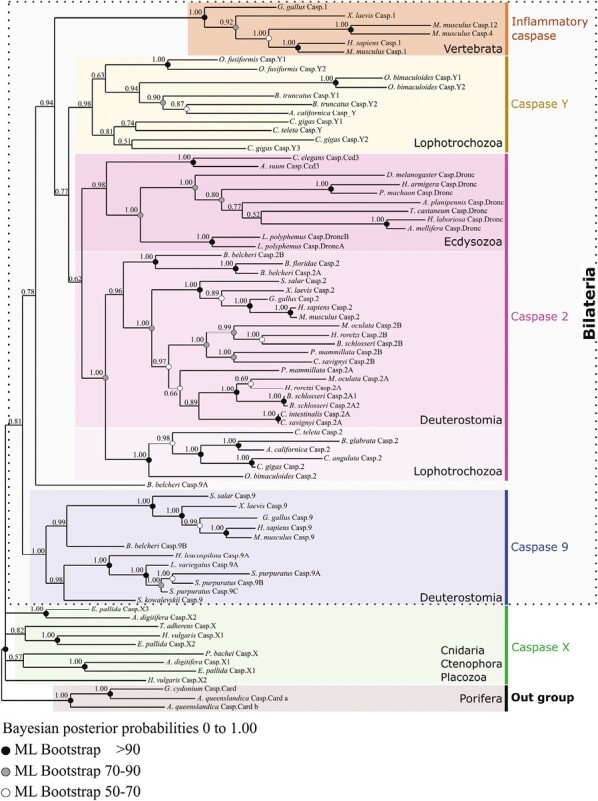
Topology of metazoan CARD-caspase phylogeny obtained by Bayesian inference using full-length sequence alignments. Four strongly supported monophyletic groups are identified: caspase-9, caspase-2, inflammatory caspases and caspase-Y. Together they form a clade strictly corresponding to bilaterian animals. Caspase-2 is widely distributed among bilaterians [Deuterostomia + Ecdysozoa + Lophotrochozoa/Spiralia] reflecting an ancestral origin. Conversely, caspase-9 is strictly restricted to deuterostomian animals. Inflammatory caspases are restricted to vertebrates, and caspase-Y are protostomian specific. Sequences of non-bilaterians (Cnidaria + Ctenophora + Placozoa) form divergent paraphyletic groups. The selected out group are the CARD-caspases of Porifera Amphimedon queenslandica.
Caspase-2 appears to be widely distributed among bilaterians and sequence evolution is congruent with the bilaterian groups [Deuterostomia + Ecdysozoa + Lophotrochozoa/Spiralia], reflecting its ancestral origin. Interestingly caspase-9 is restricted to deuterostome animals. This distribution could be interpreted either as the early emergence of caspase-9 in all bilaterians followed by a secondary loss in protostomes, or as a specific derived character of deuterostomes. The diversity of the genomes explored and the depth of taxonomic sampling unambiguously confirms this result. More specifically, the robustness of the deuterostome caspase-9 clade was explored (Supplementary Figure S2), and its specific diversification among [Vertebrata + Cephalochordata + Echinodermata + Hemichordata] was confirmed. However, the relative position of the cephalochordate Branchiostoma belcheri caspase-9A paralogous gene remains unstable. Consistent with previous studies on the ascidian Ciona intestinalis (Terajima et al., 2003), the five representatives of ascidian genomes studied are devoid of any caspase-9, likely due to a unique secondary loss in urochordates, the sister group of vertebrates (Delsuc et al., 2006).
Unexpectedly, Drosophila Dronc and Caenorhabditis Ced-3 are distinctly identified as orthologous (i.e., coming from speciation) to caspase-2 of vertebrates, not to caspase-9, as was previously reported and largely accepted (Bao & Shi, 2007; Fogarty et al., 2016; Jaroszewski et al., 2000; Lim et al., 2021; Napoletano et al., 2017; Lee et al., 2016; Fan & Bergmann, 2010; Fan et al., 2014). All Dronc/Ced-3 proteins of insects, horseshoe crab (Xiphosura), and nematodes form a strongly supported monophyletic clade (PP = 0.98) revealing single gene conservation without duplication (except horseshoe crab). Thus, the caspase-2 clade of ecdysozoans is the sister group of caspase-2 clade of [Lophotrochozoa + Deuterostomia]. The absence of identifiable caspase-2 in echinoderms and hemichordates (i.e., Ambulacraria) can probably be interpreted as a clade-specific derived loss. To test the robustness of the topology and identify potential artifacts that rapidly evolving sequences can introduce (long branches), we conducted additional analysis. First, we removed the divergent Branchiostoma belcheri caspase-9A (Supplementary Figure S3), and secondly the nematodes Ced-3 (Supplementary Figure S4). In both cases, the topology remains stable, conserving the evolutionary relationships between caspases which we established in the previous analysis. Importantly, the robustness of a number of nodes significantly increases, likely due to the elimination of fast evolving sequences. The robustness of the bilaterian clade increases from 0.81 PP to 0.93 PP and 0.89 PP when we deleted Branchiostoma Caspase-9A and both nematode Ced-3, respectively. The robustness of the caspase-2 clade also increases drastically to 0.82 PP and 0.98 PP, and long branches of arthropods Dronc remain sisters of both [Deuterostomia + Lophotrochozoa] caspase-2.
Diversification of CARD-caspases in lophotrochozoans led us to specifically name the group restricted to mollusks and annelids caspase-Y. This diversification poses interesting questions about their evolutionary origin, the uncertainty of which makes it challenging to propose an orthologous relationship with vertebrate genes. Three plausible scenarios could explain emergence of caspase-Y. First, a duplication of caspase-2 in lophotrochozoans, followed by fast evolution, resulting in a long branch and divergent sequences. Secondly, an ancestral acquisition in bilaterians, followed by losses in both ecdysozoan and deuterostomian animals. The position of caspase-Y as sister of the caspase-2 clade in the topology may suggest this scenario. Thirdly, a similar origin of both vertebrates’ inflammatory caspases and caspase-Y, implying a loss in ecdysozoans. The fast evolution of both inflammatory and caspase-Y sequences might currently obscure their common origin.
Taken globally, our analysis shows that the traditionally considered conserved initiator caspases of intrinsic apoptosis are not orthologous, but are divergent genes which may have originated from ancestral duplication (i.e., paralogous genes) among bilaterians. This finding challenges the conventional understanding of the evolutionary relationships among these caspases.
The caspase activation regulator apoptosome is structurally divergent in metazoans
In intrinsic apoptosis, initiator caspase activation depends on recruitment by a pivotal, shared component, the apoptosome platform (Dorstyn et al., 2018; Hengartner, 2000). Apoptosome formation results from assembly between Caenorhabditis Ced-4, Drosophila Dark and human Apaf-1 with their respective initiator caspases Ced-3, Dronc and procaspase-9 (Dorstyn et al., 2018). CARD and other domains (NB-ARC) are highly conserved in Apaf-1, Dark and Ced-4 proteins (Zmasek & Godzik, 2013; Zmasek et al., 2007; Zou et al., 1997). However, with the exception of the majority of nematode species (Young et al., 2020), Apaf-1 possesses WD40 repeats at its C-terminus which bind to cytochrome c. This binding is required in mammals for Apaf-1 oligomerisation and apoptosome formation (Zou et al., 1997, 1999), while Ced-4 and Dark do not require cytochrome c for their general assembly into an apoptosome (Dorstyn et al., 2002, 2018). In addition, the regulation of apoptosome structure assembly is specific to each species with an octameric Dark, a tetrameric Ced-4, and the heptameric Apaf-1 in Drosophila, Caenorhabditis and mammals, respectively. These major differences likely reveal evolutionary divergence between animal apoptosome formation and procaspase activation mechanisms, probably differentially modulating the cell death execution pathway (Cheng et al., 2016; Dorstyn et al., 2018; Shi, 2006) (Figure 1).
We conducted exhaustive studies by reciprocal BLAST and phylogenetic analyses of Apaf-1 homologs and confirmed their conservation in the majority of metazoan phyla (Figure 4, Supplementary Table S2) (Zmasek & Godzik, 2013; Zmasek et al., 2007). Our analysis includes sequences from a previous study (Zmasek et al., 2007) and is supplemented with sequences we have specifically identified (Supplementary Tables S2 and S4). We distinguished Apaf-1 genes with CARD and NB-ARC domains, likely involved in apoptosis regulation, from Apaf-1-like, which lack CARD domains but possess a range of unconventional domains. The differentiation of Apaf-1 and Apaf-1-like genes based on domain composition likely adds a layer of complexity to the functional roles of these homologs.
Figure 4.
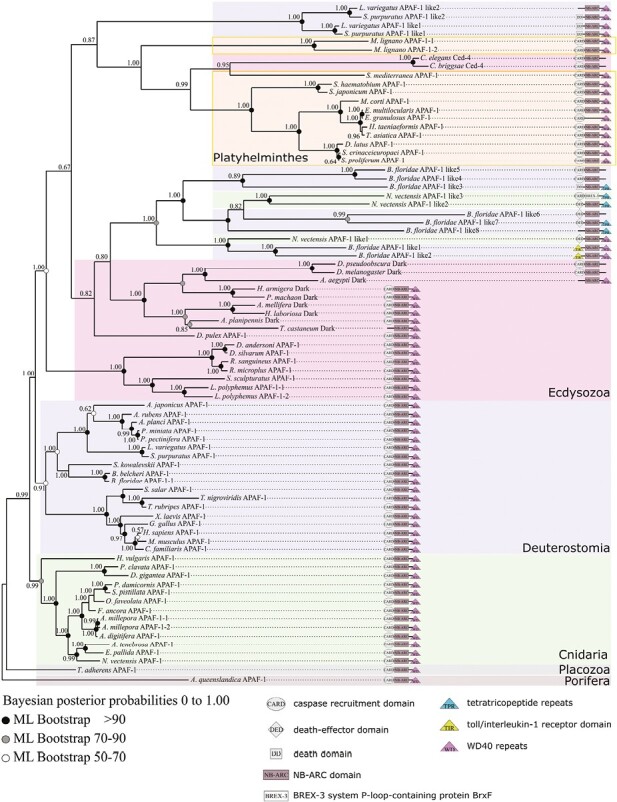
Phylogeny of Apaf-1 at the metazoan scale made with the full-length sequence alignment. Several monophyletic groups are identified but with inconsistent clustering. Apaf-1 is characterized by the presence of CARD and NB-ARC domain, while Apaf-1 like exhibit various unusual domains. Cnidarians are monophyletic, with the exception of three divergent Apaf-1 like of Nematostella. Deuterostomes are grouped in a coherent cluster of Apaf-1, but variations in domain composition of Apaf-1 like in two echinoderms, Strongylocentrotus purpuratus and Lytechinus variegatus and in the cephalochordate Branchiostoma floridae are observed. These Apaf-1 like are grouped inside protostomians, likely due to high divergence and long branch attraction. Protostomians are composed of ecdysozoans and platyhelminths. Ctenophores, mollusks, annelids, and urochordates do not have any Apaf-1 genes. Among metazoans, Apaf-1 is characterized by high sequence divergence and fast independent evolution, making homology hypotheses difficult to establish. The selected out group is Apaf-1 of Porifera Amphimedon queenslandica.
Our topology presents several clades that are common between Bayesian inference method and maximum likelihood estimation, shedding light on the ancient acquisition of Apaf-1 in Metazoa (Figure 4). Furthermore, the topology shows an accumulation of divergent paralogs (Apaf-1-like) in specific species, likely a consequence of loss or reorganization of domains. Consistently with metazoan relationships, non-bilaterian animals branch close to the root with the first branching Placozoa. We did not find Apaf-1 in the Pleurobrachia pileus genome (Ctenophora). Consequently, we extend our search to all ctenophore genomes and we were unable to find any candidates. Cnidarian Apaf-1 forms a monophyletic group and is the sister group of bilaterians. However, the three extensively modified Apaf-1-like sequences from the cnidarian Nematostella vectensis, which lack a CARD domain, branch within bilaterian sequences. Despite these exceptions, bilaterians are monophyletic and divided into two major and relatively well-supported clades. The highly conserved sequences of most deuterostomes form a well-supported monophyletic group, subdivided into the four major groups which are echinoderms, hemichordates, cephalochordates, and vertebrates. Previous works have suggested the absence of Apaf-1 in the ascidian Ciona (Terajima et al., 2003). We looked for Apaf-1 across currently available ascidian genomes, and did not detect any candidates, indicating a loss of Apaf-1 throughout urochordates, similar to the case of caspase-9. Although most Apaf-1 sequences in deuterostomes are well-conserved, variations in domain composition can be observed in two echinoderms, Strongylocentrotus purpuratus and Lytechinus variegatus (where the CARD domain is replaced by a DEAD domain or lost), and in the cephalochordate Branchiostoma floridae (where, in addition to Apaf-1, numerous highly divergent paralogues are identified). Likely due to highly divergent sequences, these Apaf-1 like are grouped within protostomians.
Within protostomians, the evolution of Apaf-1 appears more complex, and the topology is altered by the presence of numerous divergent sequences, likely associated with long branch attraction. Thus, the Apaf-1 sequences of ecdysozoans have a paraphyletic organization, but within this group, those of insects and chelicerates remain monophyletic. Some insect sequences, referred to as Dark, indeed exhibit significant modifications (loss of the CARD domain in Tribolium castaneum and Aedes aegypti, loss of the WD40 domain in Drosophila), which could lead to identifying these Apaf-1 paralogs as Dark-like.
Regarding the lophotrochozoans, we were not able to detect any Apaf-1 in mollusks and annelids, despite an extensive search in all available genomes. This is consistent with a previous study in which Apaf-1 was not found in the mollusk Crassostrea (Li et al., 2017). However, most species of flatworms (platyhelminths) present a classical organization of the Apaf-1 domain, and form a monophyletic clade in our topology, except for the insertion of two nematode (Caenorhabditis) sequences (Ced-4). This insertion likely results from long branch attraction, probably associated with the loss or divergence of the WD40 domain. We note that, despite the loss of the NB-ARC domain in Echinococcus granulosus, Apaf-1 has phylogenetically diverged very little from the sequences that are related to it.
Our results suggest an ancient acquisition of Apaf-1, but also show specific accumulation of divergent paralogs in some species. Theoretically, differences in the quality of the genome assembly across species could account for some of the putative losses. However, the fact that we did not detected Apaf-1 in any urochordates, mollusks or ctenophores suggests independent losses in these groups. Consequently, Apaf-1 does not present a linear evolution, our results suggest a complex, independent and species-specific evolution from a common ancestor of metazoans.
Importantly, a previous analysis of several genomes (cnidarian, nematode, fly, amphioxus, sea urchin, human) identified independent clades of Apaf-1 genes among metazoans, inconsistent with species phylogeny (Zmasek et al., 2007). This study highlighted that Ced-4, Dark, and Apaf-1 are not orthologous genes between ecdysozoans and vertebrates. Therefore, despite its fundamental role in apoptosis regulation (Ferraro et al., 2011; Shakeri et al., 2017; Zermati et al., 2007), Apaf-1 was independently lost in diverse metazoan phyla, and significantly diverges in the cnidarian Nematostella vectensis, the cephalochordate Branchiostoma floridae, and echinoderms (Zmasek et al., 2007). Consequently, given the pivotal role of Apaf-1 with a CARD domain in apoptosis and the resulting selection pressure, we hypothesize a relaxation of functional constraints on atypical Apaf-1 molecules (deprived of the CARD domain and likely not involved in oligomerisation processes). This relaxation could drive the diversification of Apaf-1-like paralogous genes in certain species. These findings suggest distinct evolutionary trajectories among species and convergence in the modality of apoptosome formation. This could explain functional analyses showing variations in apoptosome formation and subsequent caspase activation among taxa (Young et al., 2020).
The regulation of apoptosis by the Bcl-2 family is variable among metazoans
Intrinsic apoptosis is ultimately regulated by the Bcl-2 proteins, composed of several Bcl-2 homologous (BH) domains (Banjara et al., 2020; E. F. Lee et al., 2011). In mammals, the balance between prosurvival (four BH1–BH4 domains) and proapoptotic proteins (Bax/Bak/Bok and BH-3-only) of Bcl-2 controls initiation of intrinsic apoptosis. Conformational changes of the three-dimensional structures and interactions between Bcl-2 actors enable the assembly of pore-like structures controlling MOMP (Kalkavan & Green, 2018).
Multiple sequence alignments of metazoan Bcl-2 family proteins (Figure 5, Supplementary Table S3) (but with overrepresented chordates reflective of greater availability of vertebrate genomes) confirm the widespread distribution and early origin of Bcl-2 in Metazoa (Banjara et al., 2020; Dunn et al., 2006; Young et al., 2020; Zmasek & Godzik, 2013). Proteins clustered into five monophyletic groups, consistent with classical Bcl-2 functional classification (i.e., three “pro-apoptotic” clades: Bok, Bak, Bax, and two less supported prosurvival – “anti-apoptotic” – groups: Bcl-2/W/XL and a more complex Bcl-B/Mcl-1/Bfl-1 clade) (Figure 5). However, relationships among the five well-supported groups were not conclusively resolved despite the use of various methodologies and out group selections (Figure 5A and B). Consequently, each of the five Bcl-2 clades seems conserved across evolution, and homology (orthology) relationships inside each clade can be established. However, due to the challenge of addressing the relationship between these five monophyletic groups (Bok, Bak, Bax, Bcl-2/W/XL, Bcl-B/Mcl-1/Bfl-1), it seems difficult to hypothesize their evolutionary pattern, history, and determine a potential ancestral group. In addition, very divergent sequences from mollusks such as Biomphalaria glabrata (Bcl-like2, Bcl-like3), the cnidarian Hydra vulgaris (Bcl-like1) or from the urochordate Ciona intestinalis (Bcl-like1) were not assigned to a particular class.
Figure 5.
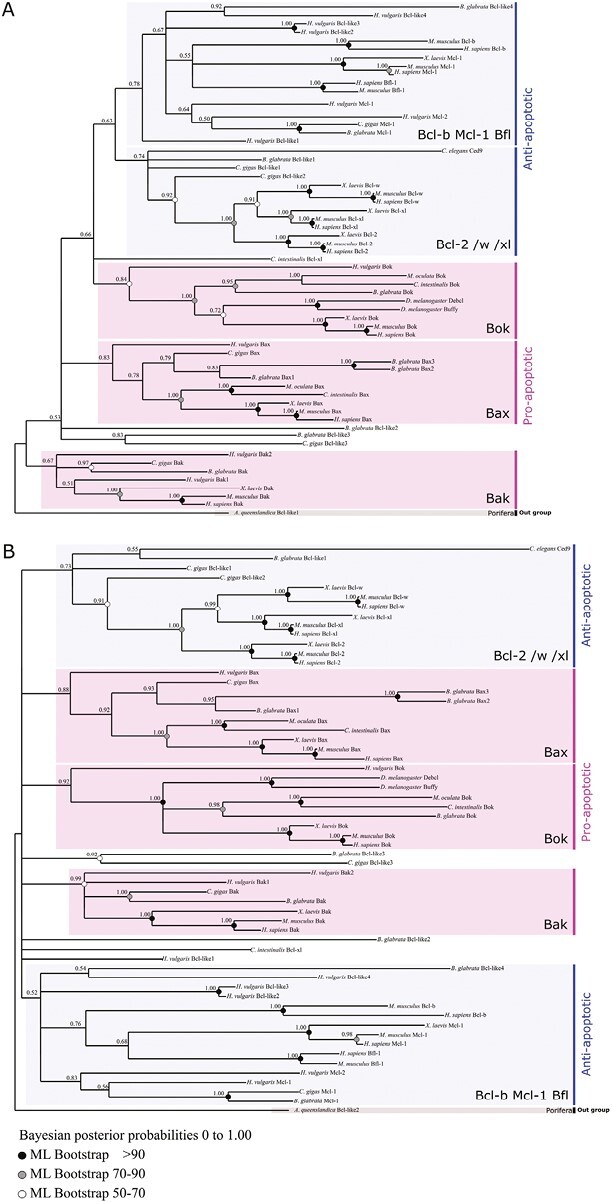
Topology of Bcl-2 family phylogeny from Bayesian inference and maximum likelihood at the metazoan scale using out groups Amphimedon queenslandica Bcl-like 1 (XP_003383425.1) (A) and Bcl-like 2 (XP_003387574.1) (B). Bcl-2 proteins strictly clustered into five monophyletic groups: three “pro-apoptotic” clades (Bok, Bak, Bax) and two less supported “anti-apoptotic” groups: the Bcl-2 clade (Bcl-2/W/XL) and a more complex (Bcl-B/Mcl-1/Bfl-1) clade. Each group includes bilaterian as well as non-bilaterian sequences which suggests a deep origin of this complex multigenic family. Clustering into these five groups is consistent between analyses, while relationships among them is not well resolved.
Caenorhabditis lacks proapoptotic Bcl-2 and possesses only the prosurvival Ced-9 (homolog to vertebrates Bcl-2/w/xl), and two BH-3 only proteins (Egl-1 and Ced-13). Conversely, only two proapoptotic Bok-like paralogues (Debcl and Buffy) were present in Drosophila (Figure 5), but their functions remain unclear (Clavier et al., 2016). Coexistence of both anti- and pro-apoptotic Bcl-2 in mollusks underlies the divergence observed within protostomes. Contrarily, similarities in Bcl-2 family composition are observed within some deuterostomes (mammals and echinoderms).
Finally, despite the conservation of almost all homologous Bcl-2 genes since early metazoan evolution, the accumulation of key differences may result in divergent initiation mechanisms within intrinsic apoptosis signaling pathways among animals.
Apoptotic mitochondrial pathways are divergent among metazoans
Functional evidence shows that caspase-2 members play a critical role in various cell death processes and are also independently involved in a range of nonapoptotic functions, including cell cycle regulation, DNA repair, and tumor suppression (Krumschnabel, Sohm & et al., 2009; Lassus et al., 2002; Olsson et al., 2015; Zhivotovsky & Orrenius, 2005). This implication of caspase-2 in myriad signaling pathways, and its ability to interact with a range of molecules, highlights its functional versatility (Braga et al., 2008; Krumschnabel, Manzl & et al., 2009; Krumschnabel, Sohm & et al., 2009; Lavrik et al., 2006; Olsson et al., 2009, 2015). As previously reported, our phylogenetic analyses support the wide distribution of caspase-2 in bilaterians, suggesting a probable ancestral multifunctionality.
The major functional similarities, leading to an initial misinterpretation of phylogenetic position of Ced-3 and Dronc, suggest a common evolutionary origin of caspase-2 and -9 genes. We hypothesized here that caspase-9 originates from bilaterian-specific duplication of a caspase-like ancestor gene, followed by a loss of caspase-9 in protostomians (Figure 6). Among deuterostomes the two families of paralogs have been preserved in vertebrates and probably in cephalochordates (Figures 3 and 6). Caspase-2 retains multifunctional activity, and in mammals can interact with the PIDDosome platform containing P53, adapter molecule RAIDD, and signaling complex DISC, activating both extrinsic and intrinsic apoptosis, and DNA damage pathways (Duan & Dixit, 1997; Haupt et al., 2003; Lavrik et al., 2006; Tinel & Tschopp, 2004). Conversely, the caspase-9 gene likely underwent a functional divergence with specialization in allosteric interactions with the apoptosome.
Figure 6.
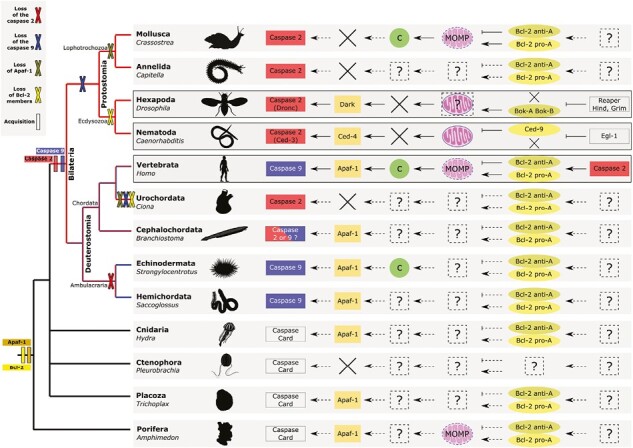
Reconstruction of convergent hypothetical intrinsic apoptotic pathways among metazoans according to molecular actors detected and identified in their genomes. Variability of intrinsic apoptotic pathways among animals emerged from convergences and recruitment of distinct actors with independent evolutionary history. Caspase-2 is bilaterian-specific and the initiator of ecdysozoans intrinsic apoptosis. Caspase-9 is restricted to deuterostomes and the specific initiator of mammalian intrinsic apoptosis. Deuterostomes exhibit several losses (i.e., caspase-2 in cephalochordates, caspase-9 in urochordates) or duplication (i.e., caspase-9 in echinoderms), highlighting a putative evolutionary flexibility in apoptotic pathway establishment. Mitochondrial functions diverge among phyla and cytochrome c (circle) release thanks to mitochondrial outer membrane permeabilization (MOMP) is specific only to mammals and possibly echinoderms. Cross indicate absence/loss. The convergent evolutionary histories reflect a probable phylum-specific adaptive process leading to parallel evolution of mitochondrial apoptotic pathways observed among animals. Animal draws come from PhyloPic (https://www.phylopic.org/).
Due to its pivotal role as a mediator of genomic stability through involvement in cell proliferation, oxidative stress, aging and cell death, the molecular divergence of the caspase-2 gene could be highly constrained during evolution, likely due to the potential initiation of tumorigenesis in case of signaling cascade destabilization. However, during the radiation of deuterostomes, both presence of caspase-2 and -9 likely induced redundancy and could have facilitated the loss of one or the other of them (Conant & Wolfe, 2008; Fares et al., 2013; Force et al., 1999). Notably, a caspase-2 duplication cooccurred with the loss of caspase-9 in urochordates, which may suggest a clade-specific relaxation of purifying selection on the caspase-2 gene (Figures 3 and 6). Similarly, caspase-2 has been lost, or strongly diverged, while caspase-9 presents a de novo relative expansion in Strongylocentrotus purpuratus (Echinodermata) and Saccoglossus kowalevskii (Hemichordata).
Typically, gene losses in Caenorhabditis and Drosophila have led to the conclusion that apoptotic pathways in ecdysozoans are simpler than those in vertebrates (Elmore, 2007; Meier et al., 2000). However, compared to mammals, ecdysozoans have pathways organized around different paralogous genes in addition to the smaller number of canonical apoptotic actors. The absence of orthologous gene relationships results in a very different structural organization of apoptosome platforms but also generates important functional divergences (i.e., mechanisms of regulating assembly, CARD-CARD interactions with procaspases) (Dorstyn et al., 2018).
Consistent with mammalian caspase-2 functions, Ced-3 has both initiator as well as executioner caspase activities (Degterev et al., 2003; Fuchs & Steller, 2015; Lavrik et al., 2006; Mancini et al., 2000). Dronc is involved in various processes such as compensatory cell proliferation, inhibition of cell migration or spermatid differentiation, similarly to the functionalities of mammalian caspase-2 (Huh et al., 2004; Ouyang et al., 2011). Due to its interaction with Dark (Apaf-1 paralog), the only CARD-caspase in Drosophila (Dronc) has been erroneously classified as caspase-9 (Kumar & Doumanis, 2000; Steller, 2008). Unlike the organization of the mammal apoptosome, both Caenorhabditis and Drosophila present neither MOMP nor the release and necessity of cytochrome c to activate Ced-3 and Dronc via Ced-4 and Dark, respectively (Figures 1 and 6) (Lettre & Hengartner, 2006; Meier et al., 2000).
Like other protostomes, mollusks are devoid of caspase-9, but caspase-2 has been identified in bivalves and is suspected to function in “a caspase-9-like manner” (Vogeler et al., 2021). Hence, caspase-2 seems involved in apoptotic processes in mollusks. Surprisingly, despite the absence of caspase-9 and Apaf-1 (and consequently a mammalian-like apoptosome), this peculiar pathway is associated with cytochrome c release (Estévez-Calvar et al., 2013; Li et al., 2017; Pirger et al., 2009; Romero et al., 2011; Yang et al., 2015; Zhang et al., 2011). The complexity of intrinsic apoptosis in mollusks seems significant, but divergent from what was observed in ecdysozoans or vertebrates, with a putative expansion of caspases that participate both in immunity, stress responses, and apoptosis (Figure 6) (Estévez-Calvar et al., 2013; Piquet et al., 2019; Plachetzki et al., 2020; Sokolova, 2009; Sokolova et al., 2004; Vogeler et al., 2021).
Unexpectedly, mammals present a unique case (possibly with the cephalochordates) in which both caspase-2 and caspase-9 are conserved and involved in apoptosis. This putative functional redundancy (i.e., recruitment, autoactivation, or transactivation, homodimerization and subsequent interchain proteolytic cleavage) likely led to the functional specialization observed for caspase-9. This may result from the specificity of the mammalian mitochondrial pathway and the nonapoptotic function of caspase-9 (An et al., 2020; Hollville & Deshmukh, 2018; Madadi et al., 2019; Tran et al., 2017). Finally, echinoderms seem to uniquely have intrinsic apoptosis similar to mammals, with a caspase-9, Bcl-2, Apaf-1, and a MOMP with cytochrome c release (Figure 6) (Bender et al., 2012; R. Tamura et al., 2018).
While we can envisage a weak parsimonious scenario suggesting a common ancestral apoptotic pathway in deuterostomes (implying independent secondary losses in hemichordates, cephalochordates, and urochordates), the similarities observed between echinoderms and mammals more probably reflect “functional convergences” based on independent recruitment of apoptotic actors.
Conclusion
The apoptotic networks of Caenorhabditis and Drosophila do not reflect ancestral conditions from which mammalian-grade apoptotic complexity emerged. On the contrary, as recently suggested, they represent a derived condition specific to ecdysozoans among animals (Belyi et al., 2010; Lu et al., 2009; Plachetzki et al., 2020; Zmasek & Godzik, 2013).
The core components of intrinsic apoptotic pathways, especially initiator caspases and the apoptosome platform, are not ancestral in metazoans. Our phylogenetic analyses highlight an unexpected evolutionary history: while the bilaterian caspase-2-mediated apoptotic toolkit emerged ancestrally and remains multifunctional, the caspase-9 mediator of the mammalian apoptosome is specific to deuterostomes.
The major functional divergences in mitochondrial apoptotic pathways observed in animals (Berthelet & Dubrez, 2013; Fuchs & Steller, 2015; Ribeiro Lopes et al., 2019, 2020) may primarily originate in the recruitment of paralogous actors from the same multigenic families. The evolution of these diverse pathways, underlined by nonorthologous molecular actors, may reflect putative adaptive processes or taxon-specific constraints which ultimately result in divergent evolutionary histories. Notably, the abundance of the apoptotic genetic repertoire has been suggested to be linked to the persistence of stem cells in adults across different phyla (Aubrey, Janic & et al., 2018; Aubrey, Kelly & et al., 2018).
Finally, mitochondria-mediated apoptosis, like other programmed cell deaths, may have evolved before and during metazoans diversification to shape developmental processes, modulate immune response, and adapt the cellular environment to environmental constraints.
Material and methods
Sequence dataset construction
Putative metazoan caspases with a CARD prodomain (CARD-caspases) were identified using tBLASTn and BLASTp searches with human caspases, Ced-3, and Dronc as queries on NCBI, ANISEED (ascidians), EchinoBase (Strongylocentrotus purpuratus), and Neurobase (Pleurobrachia bachei) databases, and followed by reciprocal BLAST. After identification of CARD-caspases in target species, sequences were added as queries to conduct BLAST searches in close relatives (i.e., identified CARD-caspases of Crassostrea gigas were used as queries to search in other mollusks). Sequences with an e-value less than 1e-10 were retained. All identified sequences were analyzed with ScanProsite (ExPaSy) (Gattiker et al., 2002) and InterProScan (EMBL-EBI) (Quevillon et al., 2005) to verify the presence of specific caspase domains. Additionally, the proposed caspase-2 from Crassostrea angulata was added (Yang et al., 2015). Caspase family proteins are short (containing the large common P20 and the small P10 domains) with a high number of genes per species which rapidly limits the relevance of the phylogenetic analyses. To reduce artifact branching and unreadable topologies, and to maximize phylogenetic diversity across metazoans, the dataset was built using CARD-caspase gene repertoires of selected species. A full list of all caspase sequences is provided in Supplementary Table S1. Taxon and genes were selected to have a wide diversity of major metazoan phyla and to have an equilibrium inside and between each group. We chose representative species for each phylum and took care not to unreasonably increase the number of sequences by redundant choice. We chose species where we detected caspases with CARD domains, DED domains, and executioner caspases, suggesting a high quality of sequences allowing identification of all members of the caspase family. For each taxon, all caspases with a CARD prodomain have been included in the analysis.
Identification of metazoan Apaf-1 was made using tBLASTn and BLASTp using human Apaf-1, nematode Ced-4, and fly Dark as queries on NCBI, ANISEED (ascidians), and neurobase.rc.ufl.edu (Pleurobrachia bachei) databases, and followed by reciprocal BLAST. Potential resulting sequences were analyzed with ScanProsite and also InterProScan. A full list of all Apaf-1 sequences is provided in Supplementary Table S2. Apaf-1 data set comprises of sequences from a previous study (Zmasek et al., 2007) in addition to our new sequences.
Metazoan Bcl-2 was identified by using tBLASTn and BLASTp searches with human Bcl-2 as the query sequence on NCBI, and then again on downloaded genomes and transcriptomes, followed by reciprocal BLAST. All identified sequences were analyzed with ScanProsite (ExPaSy) and InterProScan (EMBL-EBI) to verify the presence of BH domains. Due to short sequence length, BH-3-only were not considered. A full list of all Bcl-2 sequences is provided in Supplementary Table S3.
Multiple alignments of protein sequences were generated using the MAFFT software version 7 (Katoh & Standley, 2013) with default parameters and also Clustal Omega (Sievers et al., 2011) to verify the congruence of the different alignments. All sequences were then manually checked in BioEdit 7.2 software (HALL, 1999) to verify the presence of specific domains previously identified. Gblocks version 0.91b (Castresana, 2000) was used to remove vacancies and blur sites. Final alignments comprise of 224, 230, 235, 147, and 1,201 amino acids for all types of metazoan caspases alignment, metazoan CARD-caspases alignment, deuterostomian CARD-caspases alignment, metazoan Bcl-2 alignment, and Apaf-1 alignment, respectively. Apaf-1 alignment was done on the full sequence alignment (all domains).
Taxon sampling strategy
Proteins in multigene families like caspases and Bcl-2 are short, making it challenging to conduct phylogenetic analyses with a large number of species without sacrificing reliability. The initial focus was on CARD-caspases, aiming to include at least one representative genome from major metazoan phyla: vertebrates, urochordates, cephalochordates, hemichordates, echinoderms, ecdysozoans (insects, chelicerates, nematodes), mollusks (bivalves, cephalopods), annelids, cnidarians, placozoans, ctenophores, sponges, totaling 42 species (Supplementary Table S4). In certain phyla, particularly nematodes, the rate of evolution is high. However, the variations in sequences are specific to the terminal nodes and have little influence on the deep nodes, which are the primary focus of the analyses. Selection criteria included a blast search for caspases in all genomes, choosing species with identified caspases (i.e., Caenorhabditis, Drosophila, Ciona, Crassostrea, Mus, Xenopus, Hydra) and good genome quality. Model animals and taxa with functional or well-studied genomic data were prioritized. Due to the short length of caspases, species sampling was reduced in order to add all caspases of the family when conducting the phylogeny on all types of caspases. Caspase studies maintain consistency as all species included in all type of caspases phylogeny are also represented in the CARD-caspases analysis (Supplementary Table S4).
The sampling for the Apaf-1 phylogeny aimed to improve representation of deuterostomians and cnidarians to test the paraphyly of Apaf-1 from these group reported in a previous study (Supplementary Table S4). Platyhelminths were added to investigate the impact of lophotrochozoan sequences, due to the absence of Apaf-1 in mollusks and annelids. Consistency was maintained across analyses by including all species from the CARD-caspases analysis in Apaf-1 analysis and sequences provided in the paper by Zmasek et al (2007). Species not represented in the Apaf-1 dataset but which are in the CARD-caspases analysis lacked Apaf-1 due to specific losses (i.e., mollusks, annelids, ascidians, ctenophores) or undetected despite theoretical presence (i.e., nematode Ascaris suun).
Bcl-2 alignment is very short, leading to challenges in the analysis. To maintain readability and reliability, the sampling was minimized while ensuring representation of metazoan diversity. Species for which Bcl-2 were already known or studied were prioritized. The study maintains overall consistency in the analyses by ensuring that species included in Bcl-2 analysis are all represented in the CARD-caspases analysis (Supplementary Table S4).
Phylogenetic analysis
Phylogenetic analyses were carried out from the amino-acid alignment with Maximum Likelihood (ML) method in PhyML 3.1 (Guindon et al., 2010), a combined ML tree search with 1,000 bootstrap replicates was produced and then visualized using Seaview (Gouy et al., 2010). The best amino-acid evolution models to conduct analyses were determined using MEGA11 (Tamura et al., 2021) and determined to be Blossum32 for all types of caspases alignment, WAG for sequences of CARD-caspases alignments (CARD, P20, P10), and LG for both Bcl-2 and Apaf-1 alignments.
Bayesian analyses were performed using MrBayes (v3.2.6) (Ronquist & Huelsenbeck, 2003) under mixed model. For each analysis, one-fourth of the topologies were discarded as burn-in values, while the remaining ones were used to calculate posterior probability. The run for metazoan caspases (all types, Figure 2) alignment was carried out for 5,000,000 generations with 15 randomly started simultaneous Markov chains (1 cold chain, 14 heated chains) and sampled every 100 generations. The run for metazoan CARD-caspases alignment (Figure 3, Supplementary Figure S1-S4) was carried out for 2,500,000 generations with 15 randomly started simultaneous Markov chains (1 cold chain, 14 heated chains) and sampled every 100 generations. The run for deuterostomian CARD-caspases alignment (Supplementary Figure S2) was carried out for 500,000 generations with 5 randomly started simultaneous Markov chains (1 cold chain, 4 heated chains) and sampled every 100 generations. The run for metazoan Apaf-1 alignment (Figure 4) was carried out for 5,000,000 generations with 5 randomly started simultaneous Markov chains (1 cold chain, 4 heated chains) and sampled every 100 generations. The run for metazoan Bcl-2 alignment (Figure 5) was carried out for 5,000,000 generations with 20 randomly started simultaneous Markov chains (1 cold chain, 19 heated chains) and sampled every 100 generations. ML bootstrap values higher than 50% and Bayesian posterior probabilities are indicated on the Bayesian tree.
The out group for all types of caspases phylogeny made at the metazoans scale is a caspase-like of Reticulomyxa filosa (ETO10778.1) (Klim et al., 2018). The out group for the metazoan CARD-caspase phylogeny are the only two caspases with a CARD prodomain of the Porifera Amphimedon queenslandica (XP_003383519 and XP_019854115.1) (Figure 3, Figure S3-S4) or the only one of the ctenophore Pleurobrachia bachei (Sb 2658116) (Supplementary Figure S1). Analyses of CARD-caspases were made independently at the deuterostome scale with four different out groups to test their effect on the stability of the topology: (a) CARD-caspase-Y of the annelid Capitella teleta (ELT97848.1), (b) CARD-caspase-2 of the mollusk Aplysia californica (XP_005113266), (c) CARD-caspase-X2 of cnidarian Hydra vulgaris (NP_001274285.1), and (d) CARD-caspase Ced-3 of the ecdysozoan Caenorhabditis elegans (AAG42045.1) (Supplementary Figure S2). For the metazoan Apaf-1 phylogenies, the selected out group is Apaf-1 (XP_019855714.1) of Porifera Amphimedon queenslandica (Figure 4). For the metazoan Bcl-2 phylogenies, out groups used to test their effect on the stability of the topology are: (a) Bcl-2-like1 (XP_003383425.1) and (b) Bcl-2-like2 (XP_003387574.1) of Porifera Amphimedon queenslandica (Figure 5).
Supplementary Material
Acknowledgments
Authors thank Sébastien Darras (Sorbonne Université, Banyuls-sur-Mer), Christine Vesque (Sorbonne Université, Paris), Jérôme Gros (Institut Pasteur, Paris), Sabine Hennequin (Sorbonne Université, Paris), Miguel Arenas (University of Vigo, Spain), the Gazave team (Institut Jacques Monod, Paris) and Uri Frank (University of Galway, Ireland) for helpful comments.
Contributor Information
Gabriel Krasovec, ISYEB - UMR 7205, Institut de Systématique, Evolution et Biodiversité, Sorbonne Université, CNRS, MNHN, Paris, France.
Helen R Horkan, Centre for Chromosome Biology, School of Biological and Chemical Sciences, University of Galway, Galway, Ireland.
Éric Quéinnec, ISYEB - UMR 7205, Institut de Systématique, Evolution et Biodiversité, Sorbonne Université, CNRS, MNHN, Paris, France.
Jean-Philippe Chambon, Centre de Recherche en Biologie cellulaire de Montpellier (CRBM), Université de Montpellier, CNRS, Montpellier, France.
Data and code availability
All data needed to evaluate the conclusions in this study are present in the paper and the Supplementary Materials. Any requests can be addressed to the corresponding author GK. Alignments used to conduct phylogenetic analyses are available at Figshare data base (DOI: doi.org/10.6084/m9.figshare.24441217).
Author contributions
J.P. and E.Q. managed the project. G.K. made analyses and generated the data, with participation of H.H. G.K. and E.Q. wrote the manuscript. All authors approved the manuscript.
Funding
The work of G.K was supported by a Ph.D. fellowship from the French Ministry of Education, Research and Innovation. G.K is currently funded by a postdoctoral fellowship from the Fondation ARC pour la recherche sur le cancer (project ARCPOST-DOC2022070005318). H.R.H. is a doctoral student in the Science Foundation Ireland Centre for Research Training in Genomics Data Science (grant no. 18/CRT/6214).
Conflict of interest: The authors declare no conflict of interest.
References
- An, H. -K., Chung, K. M., Park, H., Hong, J., Gim, J. -E., Choi, H., Lee, Y. W., Choi, J., Mun, J. Y., & Yu, S. -W. (2020). CASP9 (caspase 9) is essential for autophagosome maturation through regulation of mitochondrial homeostasis. Autophagy, 16(9), 1598–1617. 10.1080/15548627.2019.1695398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubrey, B. J., Janic, A., Chen, Y., Chang, C., Lieschke, E. C., Diepstraten, S. T., Kueh, A. J., Bernardini, J. P., Dewson, G., O’Reilly, L. A., Whitehead, L., Voss, A. K., Smyth, G. K., Strasser, A., & Kelly, G. L. (2018). Mutant TRP53 exerts a target gene-selective dominant-negative effect to drive tumor development. Genes & Development, 32(21-22), 1420–1429. 10.1101/gad.314286.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubrey, B. J., Kelly, G. L., Janic, A., Herold, M. J., & Strasser, A. (2018). How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death and Differentiation, 25(1), 104–113. 10.1038/cdd.2017.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banjara, S., Suraweera, C. D., Hinds, M. G., & Kvansakul, M. (2020). The Bcl-2 family: Ancient origins, conserved structures, and divergent mechanisms. Biomolecules, 10(1), 128. 10.3390/biom10010128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, Q., & Shi, Y. (2007). Apoptosome: A platform for the activation of initiator caspases. Cell Death and Differentiation, 14(1), 56–65. 10.1038/sj.cdd.4402028 [DOI] [PubMed] [Google Scholar]
- Bell, R. A. V., & Megeney, L. A. (2017). Evolution of caspase-mediated cell death and differentiation: Twins separated at birth. Cell Death & Differentiation, 24(8), 1359–1368. 10.1038/cdd.2017.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi, V. A., Ak, P., Markert, E., Wang, H., Hu, W., Puzio-Kuter, A., & Levine, A. J. (2010). The origins and evolution of the p53 family of genes. Cold Spring Harbor Perspectives in Biology, 2(6), a001198. 10.1101/cshperspect.a001198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, C. E., Fitzgerald, P., Tait, S. W. G., Llambi, F., McStay, G. P., Tupper, D. O., Pellettieri, J., Sánchez Alvarado, A., Salvesen, G. S., & Green, D. R. (2012). Mitochondrial pathway of apoptosis is ancestral in metazoans. Proceedings of the National Academy of Sciences of the United States of America, 109(13), 4904–4909. 10.1073/pnas.1120680109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelet, J., & Dubrez, L. (2013). Regulation of apoptosis by Inhibitors of Apoptosis (IAPs). Cells, 2(1), 163–187. 10.3390/cells2010163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga, M., Sinha Hikim, A. P., Datta, S., Ferrini, M. G., Brown, D., Kovacheva, E. L., Gonzalez-Cadavid, N. F., & Sinha-Hikim, I. (2008). Involvement of oxidative stress and caspase 2-mediated intrinsic pathway signaling in age-related increase in muscle cell apoptosis in mice. Apoptosis, 13(6), 822–832. 10.1007/s10495-008-0216-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 17(4), 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chambon, J. -P., Soule, J., Pomies, P., Fort, P., Sahuquet, A., Alexandre, D., Mangeat, P. -H., & Baghdiguian, S. (2002). Tail regression in Ciona intestinalis (Prochordate) involves a Caspase-dependent apoptosis event associated with ERK activation. Development (Cambridge, England), 129(13), 3105–3114. 10.1242/dev.129.13.3105 [DOI] [PubMed] [Google Scholar]
- Cheng, T. C., Hong, C., Akey, I. V., Yuan, S., & Akey, C. W. (2016). A near atomic structure of the active human apoptosome. eLife, 5, e17755. 10.7554/eLife.17755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk, J. E., Moldoveanu, T., Llambi, F., Parsons, M. J., & Green, D. R. (2010). The BCL-2 family reunion. Molecular Cell, 37(3), 299–310. 10.1016/j.molcel.2010.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cikala, M., Wilm, B., Hobmayer, E., Böttger, A., & David, C. N. (1999). Identification of caspases and apoptosis in the simple metazoan Hydra. Current Biology, 9(17), 959–962. 10.1016/s0960-9822(99)80423-0 [DOI] [PubMed] [Google Scholar]
- Clavier, A., Rincheval-Arnold, A., Colin, J., Mignotte, B., & Guénal, I. (2016). Apoptosis in Drosophila: Which role for mitochondria? Apoptosis, 21(3), 239–251. 10.1007/s10495-015-1209-y [DOI] [PubMed] [Google Scholar]
- Conant, G. C., & Wolfe, K. H. (2008). Turning a hobby into a job: How duplicated genes find new functions. Nature Reviews. Genetics, 9(12), 938–950. 10.1038/nrg2482 [DOI] [PubMed] [Google Scholar]
- Crawford, E. D., Seaman, J. E., Barber, A. E., David, D. C., Babbitt, P. C., Burlingame, A. L., & Wells, J. A. (2012). Conservation of caspase substrates across metazoans suggests hierarchical importance of signaling pathways over specific targets and cleavage site motifs in apoptosis. Cell Death & Differentiation, 19(12), 2040–2048. 10.1038/cdd.2012.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev, A., Boyce, M., & Yuan, J. (2003). A decade of caspases. Oncogene, 22(53), 8543–8567. 10.1038/sj.onc.1207107 [DOI] [PubMed] [Google Scholar]
- Delsuc, F., Brinkmann, H., Chourrout, D., & Philippe, H. (2006). Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature, 439(7079), 965–968. 10.1038/nature04336 [DOI] [PubMed] [Google Scholar]
- Dorstyn, L., Akey, C. W., & Kumar, S. (2018). New insights into apoptosome structure and function. Cell Death and Differentiation, 25(7), 1194–1208. 10.1038/s41418-017-0025-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn, L., Read, S., Cakouros, D., Huh, J. R., Hay, B. A., & Kumar, S. (2002). The role of cytochrome c in caspase activation in Drosophila melanogaster cells. The Journal of Cell Biology, 156(6), 1089–1098. 10.1083/jcb.200111107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll, M. (1996). Cell death in C elegans: Molecular insights into mechanisms conserved between nematodes and mammals. Brain Pathology (Zurich, Switzerland), 6(4), 411–425. 10.1111/j.1750-3639.1996.tb00873.x [DOI] [PubMed] [Google Scholar]
- Duan, H., & Dixit, V. M. (1997). RAIDD is a new « death » adaptor molecule. Nature, 385(6611), 86–89. 10.1038/385086a0 [DOI] [PubMed] [Google Scholar]
- Dunn, S. R., Phillips, W. S., Spatafora, J. W., Green, D. R., & Weis, V. M. (2006). Highly conserved caspase and Bcl-2 homologues from the sea anemone Aiptasia pallida: Lower metazoans as models for the study of apoptosis evolution. Journal of Molecular Evolution, 63(1), 95–107. 10.1007/s00239-005-0236-7 [DOI] [PubMed] [Google Scholar]
- Ellis, H. M., & Horvitz, H. R. (1986). Genetic control of programmed cell death in the nematode C elegans. Cell, 44(6), 817–829. 10.1016/0092-8674(86)90004-8 [DOI] [PubMed] [Google Scholar]
- Elmore, S. (2007). Apoptosis: A review of programmed cell death. Toxicologic Pathology, 35(4), 495–516. 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez-Calvar, N., Romero, A., Figueras, A., & Novoa, B. (2013). Genes of the mitochondrial apoptotic pathway in Mytilus galloprovincialis. PLoS One, 8(4), e61502. 10.1371/journal.pone.0061502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, T. -J., Han, L. -H., Cong, R. -S., & Liang, J. (2005). Caspase family proteases and apoptosis. Acta Biochimica et Biophysica Sinica, 37(11), 719–727. 10.1111/j.1745-7270.2005.00108.x [DOI] [PubMed] [Google Scholar]
- Fan, Y., & Bergmann, A. (2010). The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death and Differentiation, 17(3), 534–539. 10.1038/cdd.2009.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Y., Wang, S., Hernandez, J., Yenigun, V. B., Hertlein, G., Fogarty, C. E., Lindblad, J. L., & Bergmann, A. (2014). Genetic models of apoptosis-induced proliferation decipher activation of JNK and identify a requirement of EGFR signaling for tissue regenerative responses in Drosophila. PLoS Genetics, 10(1), e1004131. 10.1371/journal.pgen.1004131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares, M. A., Keane, O. M., Toft, C., Carretero-Paulet, L., & Jones, G. W. (2013). The roles of whole-genome and small-scale duplications in the functional specialization of Saccharomyces cerevisiae genes. PLoS Genetics, 9(1), e1003176. 10.1371/journal.pgen.1003176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro, E., Pesaresi, M. G., De Zio, D., Cencioni, M. T., Gortat, A., Cozzolino, M., Berghella, L., Salvatore, A. M., Oettinghaus, B., Scorrano, L., Pérez-Payà, E., & Cecconi, F. (2011). Apaf1 plays a pro-survival role by regulating centrosome morphology and function. Journal of Cell Science, 124(Pt 20), 3450–3463. 10.1242/jcs.086298 [DOI] [PubMed] [Google Scholar]
- Fogarty, C. E., Diwanji, N., Lindblad, J. L., Tare, M., Amcheslavsky, A., Makhijani, K., Brückner, K., Fan, Y., & Bergmann, A. (2016). Extracellular reactive oxygen species drive apoptosis-induced proliferation via drosophila macrophages. Current Biology, 26(5), 575–584. 10.1016/j.cub.2015.12.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L., & Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics, 151(4), 1531–1545. 10.1093/genetics/151.4.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, Y., & Steller, H. (2015). Live to die another way: Modes of programmed cell death and the signals emanating from dying cells. Nature Reviews. Molecular Cell biology, 16(6), 329–344. 10.1038/nrm3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi, L., Vitale, I., Aaronson, S. A., Abrams, J. M., Adam, D., Agostinis, P., Alnemri, E. S., Altucci, L., Amelio, I., Andrews, D. W., Annicchiarico-Petruzzelli, M., Antonov, A. V., Arama, E., Baehrecke, E. H., Barlev, N. A., Bazan, N. G., Bernassola, F., Bertrand, M. J. M., Bianchi, K., & Kroemer, G. (2018). Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death and Differentiation, 25(3), 486–541. 10.1038/s41418-017-0012-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattiker, A., Gasteiger, E., & Bairoch, A. (2002). ScanProsite: A reference implementation of a PROSITE scanning tool. Applied Bioinformatics, 1(2), 107–108. [PubMed] [Google Scholar]
- Gouy, M., Guindon, S., & Gascuel, O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution, 27(2), 221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- Guindon, S., Dufayard, J. -F., Lefort, V., Anisimova, M., Hordijk, W., & Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 30. Systematic Biology, 59(3), 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98. [Google Scholar]
- Haupt, S., Berger, M., Goldberg, Z., & Haupt, Y. (2003). Apoptosis—The p53 network. Journal of Cell Science, 116(Pt 20), 4077–4085. 10.1242/jcs.00739 [DOI] [PubMed] [Google Scholar]
- Hengartner, M. O. (2000). The biochemistry of apoptosis. Nature, 407(6805), 770–776. 10.1038/35037710 [DOI] [PubMed] [Google Scholar]
- Hipfner, D. R., & Cohen, S. M. (2004). Connecting proliferation and apoptosis in development and disease. Nature Reviews. Molecular cell biology, 5(10), 805–815. 10.1038/nrm1491 [DOI] [PubMed] [Google Scholar]
- Hollville, E., & Deshmukh, M. (2018). Physiological functions of non-apoptotic caspase activity in the nervous system. Seminars in Cell & Developmental Biology, 82, 127–136. 10.1016/j.semcdb.2017.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, J. R., Guo, M., & Hay, B. A. (2004). Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Current Biology: CB, 14(14), 1262–1266. 10.1016/j.cub.2004.06.015 [DOI] [PubMed] [Google Scholar]
- Jacobson, M. D., Weil, M., & Raff, M. C. (1997). Programmed cell death in animal development. Cell, 88(3), 347–354. 10.1016/s0092-8674(00)81873-5 [DOI] [PubMed] [Google Scholar]
- Jaroszewski, L., Rychlewski, L., Reed, J. C., & Godzik, A. (2000). ATP-activated oligomerization as a mechanism for apoptosis regulation: Fold and mechanism prediction for CED-4. Proteins, 39(3), 197–203. [DOI] [PubMed] [Google Scholar]
- Jeffery, W. R., & Gorički, S. (2021). Apoptosis is a generator of Wnt-dependent regeneration and homeostatic cell renewal in the ascidian Ciona. Biology Open, 10(4), bio058526. 10.1242/bio.058526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkavan, H., & Green, D. R. (2018). MOMP, cell suicide as a BCL-2 family business. Cell Death and Differentiation, 25(1), 46–55. 10.1038/cdd.2017.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K., & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klim, J., Gładki, A., Kucharczyk, R., Zielenkiewicz, U., & Kaczanowski, S. (2018). Ancestral state reconstruction of the apoptosis machinery in the common ancestor of eukaryotes. G3: Genes, Genomes, Genetics, 8(6), 2121–2134. 10.1534/g3.118.200295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasovec, G., Horkan, H. R., Quéinnec, E., & Chambon, J. -P. (2022). The constructive function of apoptosis: More than a dead-end job. Frontiers in Cell and Developmental Biology, 10, 1033645. 10.3389/fcell.2022.1033645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasovec, G., Pottin, K., Rosello, M., Quéinnec, E., & Chambon, J. -P. (2021). Apoptosis and cell proliferation during metamorphosis of the planula larva of Clytia hemisphaerica (Hydrozoa, Cnidaria). Developmental Dynamics, 250(12), 1739–1758. 10.1002/dvdy.376 [DOI] [PubMed] [Google Scholar]
- Krasovec, G., Robine, K., Quéinnec, E., Karaiskou, A., & Chambon, J. P. (2019). Ci-hox12 tail gradient precedes and participates in the control of the apoptotic-dependent tail regression during Ciona larva metamorphosis. Developmental Biology, 448(2), 237–246. 10.1016/j.ydbio.2018.12.010 [DOI] [PubMed] [Google Scholar]
- Krumschnabel, G., Manzl, C., & Villunger, A. (2009). Caspase-2: Killer, savior and safeguard—Emerging versatile roles for an ill-defined caspase. Oncogene, 28(35), 3093–3096. 10.1038/onc.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumschnabel, G., Sohm, B., Bock, F., Manzl, C., & Villunger, A. (2009). The enigma of caspase-2: The laymen’s view. Cell Death & Differentiation, 16(2), 195–207. 10.1038/cdd.2008.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., & Doumanis, J. (2000). The fly caspases. Cell Death and Differentiation, 7(11), 1039–1044. 10.1038/sj.cdd.4400756 [DOI] [PubMed] [Google Scholar]
- Lassus, P., Opitz-Araya, X., & Lazebnik, Y. (2002). Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science (New York, N.Y.), 297(5585), 1352–1354. 10.1126/science.1074721 [DOI] [PubMed] [Google Scholar]
- Lavrik, I. N., Golks, A., Baumann, S., & Krammer, P. H. (2006). Caspase-2 is activated at the CD95 death-inducing signaling complex in the course of CD95-induced apoptosis. Blood, 108(2), 559–565. 10.1182/blood-2005-07-007096 [DOI] [PubMed] [Google Scholar]
- Lee, E. F., Clarke, O. B., Evangelista, M., Feng, Z., Speed, T. P., Tchoubrieva, E. B., Strasser, A., Kalinna, B. H., Colman, P. M., & Fairlie, W. D. (2011). Discovery and molecular characterization of a Bcl-2-regulated cell death pathway in schistosomes. Proceedings of the National Academy of Sciences of the United States of America, 108(17), 6999–7003. 10.1073/pnas.1100652108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T. V., Kamber Kaya, H. E., Simin, R., Baehrecke, E. H., & Bergmann, A. (2016). The initiator caspase Dronc is subject of enhanced autophagy upon proteasome impairment in Drosophila. Cell Death and Differentiation, 23(9), 1555–1564. 10.1038/cdd.2016.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre, G., & Hengartner, M. O. (2006). Developmental apoptosis in C elegans: A complex CEDnario. Nature Reviews. Molecular cell biology, 7(2), 97–108. 10.1038/nrm1836 [DOI] [PubMed] [Google Scholar]
- Li, Y., Zhang, L., Qu, T., Tang, X., Li, L., & Zhang, G. (2017). Conservation and divergence of mitochondrial apoptosis pathway in the Pacific oyster, Crassostrea gigas. Cell Death & Disease, 8(7), e2915–e2915. 10.1038/cddis.2017.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, Y., Dorstyn, L., & Kumar, S. (2021). The p53-caspase-2 axis in the cell cycle and DNA damage response. Experimental & Molecular Medicine, 53(4), 517–527. 10.1038/s12276-021-00590-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W. -J., Amatruda, J. F., & Abrams, J. M. (2009). p53 ancestry: Gazing through an evolutionary lens. Nature Reviews. Cancer, 9(10), 758–762. 10.1038/nrc2732 [DOI] [PubMed] [Google Scholar]
- Madadi, Z., Akbari-Birgani, S., Monfared, P. D., & Mohammadi, S. (2019). The non-apoptotic role of caspase-9 promotes differentiation in leukemic cells. Biochimica et Biophysica Acta, Molecular Cell Research, 1866(12), 118524. 10.1016/j.bbamcr.2019.118524 [DOI] [PubMed] [Google Scholar]
- Mancini, M., Machamer, C. E., Roy, S., Nicholson, D. W., Thornberry, N. A., Casciola-Rosen, L. A., & Rosen, A. (2000). Caspase-2 is localized at the Golgi complex and cleaves Golgin-160 during apoptosis. The Journal of Cell Biology, 149(3), 603–612. 10.1083/jcb.149.3.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, P., Finch, A., & Evan, G. (2000). Apoptosis in development. Nature, 407(6805), 796–801. 10.1038/35037734 [DOI] [PubMed] [Google Scholar]
- Napoletano, F., Gibert, B., Yacobi-Sharon, K., Vincent, S., Favrot, C., Mehlen, P., Girard, V., Teil, M., Chatelain, G., Walter, L., Arama, E., & Mollereau, B. (2017). P53-dependent programmed necrosis controls germ cell homeostasis during spermatogenesis. PLoS Genetics, 13(9), e1007024. 10.1371/journal.pgen.1007024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson, M., Forsberg, J., & Zhivotovsky, B. (2015). Caspase-2: The reinvented enzyme. Oncogene, 34(15), 1877–1882. 10.1038/onc.2014.139 [DOI] [PubMed] [Google Scholar]
- Olsson, M., Vakifahmetoglu, H., Abruzzo, P. M., Högstrand, K., Grandien, A., & Zhivotovsky, B. (2009). DISC-mediated activation of caspase-2 in DNA damage-induced apoptosis. Oncogene, 28(18), 1949–1959. 10.1038/onc.2009.36 [DOI] [PubMed] [Google Scholar]
- Ouyang, Y., Petritsch, C., Wen, H., Jan, L., Jan, Y. N., & Lu, B. (2011). Dronc caspase exerts a non-apoptotic function to restrain Phospho-Numb-induced ectopic Neuroblast formation in Drosophila. Development (Cambridge, England), 138(11), 2185–2196. 10.1242/dev.058347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquet, B., Shillito, B., Lallier, F. H., Duperron, S., & Andersen, A. C. (2019). High rates of apoptosis visualized in the symbiont-bearing gills of deep-sea Bathymodiolus mussels. PLoS One, 14(2), e0211499. 10.1371/journal.pone.0211499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirger, Z., Rácz, B., & Kiss, T. (2009). Dopamine-induced programmed cell death is associated with cytochrome c release and caspase-3 activation in snail salivary gland cells. Biology of the Cell, 101(2), 105–116. 10.1042/BC20070168 [DOI] [PubMed] [Google Scholar]
- Plachetzki, D. C., Pankey, M. S., MacManes, M. D., Lesser, M. P., & Walker, C. W. (2020). The genome of the softshell clam Mya Arenaria and the evolution of apoptosis. Genome Biology and Evolution, 12(10), 1681–1693. 10.1093/gbe/evaa143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., & Lopez, R. (2005). InterProScan: Protein domains identifier. Nucleic Acids Research, 33(Web Server issue), W116–W120. 10.1093/nar/gki442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro Lopes, M., Parisot, N., Callaerts, P., & Calevro, F. (2019). Genetic diversity of the apoptotic pathway in insects. In Pontarotti P. (Éd.), Evolution, origin of life, concepts and methods (pp. 253–285). Springer International Publishing. 10.1007/978-3-030-30363-1_13 [DOI] [Google Scholar]
- Ribeiro Lopes, M., Parisot, N., Gaget, K., Huygens, C., Peignier, S., Duport, G., Orlans, J., Charles, H., Baatsen, P., Jousselin, E., Da Silva, P., Hens, K., Callaerts, P., & Calevro, F. (2020). Evolutionary novelty in the apoptotic pathway of aphids. Proceedings of the National Academy of Sciences of the United States of America, 117(51), 32545–32556. 10.1073/pnas.2013847117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, A., Estévez-Calvar, N., Dios, S., Figueras, A., & Novoa, B. (2011). New insights into the apoptotic process in mollusks: Characterization of caspase genes in Mytilus galloprovincialis. PLoS One, 6(2), e17003. 10.1371/journal.pone.0017003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist, F., & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxford, England), 19(12), 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Shakeri, R., Kheirollahi, A., & Davoodi, J. (2017). Apaf-1: Regulation and function in cell death. Biochimie, 135, 111–125. 10.1016/j.biochi.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Shi, Y. (2006). Mechanical aspects of apoptosome assembly. Current Opinion in Cell Biology, 18(6), 677–684. 10.1016/j.ceb.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Shrestha, S., Tung, J., Grinshpon, R. D., Swartz, P., Hamilton, P. T., Dimos, B., Mydlarz, L., & Clark, A. C. (2020). Caspases from scleractinian coral show unique regulatory features. The Journal of Biological Chemistry, 295(43), 14578–14591. 10.1074/jbc.RA120.014345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J., Thompson, J. D., & Higgins, D. G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology, 7, 539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova, I. M. (2009). Apoptosis in molluscan immune defense. Invertebrate Survival Journal, 6(1), 49–58. [Google Scholar]
- Sokolova, I. M., Evans, S., & Hughes, F. M. (2004). Cadmium-induced apoptosis in oyster Hemocytes involves disturbance of cellular energy balance but no mitochondrial permeability transition. The Journal of Experimental Biology, 207(Pt 19), 3369–3380. 10.1242/jeb.01152 [DOI] [PubMed] [Google Scholar]
- Steller, H. (2008). Regulation of apoptosis in Drosophila. Cell Death and Differentiation, 15(7), 1132–1138. 10.1038/cdd.2008.50 [DOI] [PubMed] [Google Scholar]
- Tamura, K., Stecher, G., & Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 38(7), 3022–3027. 10.1093/molbev/msab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, R., Takada, M., Sakaue, M., Yoshida, A., Ohi, S., Hirano, K., Hayakawa, T., Hirohashi, N., Yura, K., & Chiba, K. (2018). Starfish Apaf-1 activates effector caspase-3/9 upon apoptosis of aged eggs. Scientific Reports, 8(1), 1611. 10.1038/s41598-018-19845-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terajima, D., Shida, K., Takada, N., Kasuya, A., Rokhsar, D., Satoh, N., Satake, M., & Wang, H. -G. (2003). Identification of candidate genes encoding the core components of the cell death machinery in the Ciona intestinalis genome. Cell Death and Differentiation, 10(6), 749–753. 10.1038/sj.cdd.4401223 [DOI] [PubMed] [Google Scholar]
- Thornberry, N. A., & Lazebnik, Y. (1998). Caspases: Enemies within. Science (New York, N.Y.), 281(5381), 1312–1316. 10.1126/science.281.5381.1312 [DOI] [PubMed] [Google Scholar]
- Tinel, A., & Tschopp, J. (2004). The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science (New York, N.Y.), 304(5672), 843–846. 10.1126/science.1095432 [DOI] [PubMed] [Google Scholar]
- Tran, H. T., Fransen, M., Dimitrakopoulou, D., Van Imschoot, G., Willemarck, N., & Vleminckx, K. (2017). Caspase-9 has a nonapoptotic function in Xenopus embryonic primitive blood formation. Journal of Cell Science, 130(14), 2371–2381. 10.1242/jcs.186411 [DOI] [PubMed] [Google Scholar]
- Uren, A. G., O’Rourke, K., Aravind, L., Pisabarro, M. T., Seshagiri, S., Koonin, E. V., & Dixit, V. M. (2000). Identification of Paracaspases and Metacaspases: Two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Molecular Cell, 6(4), 961–967. 10.1016/S1097-2765(05)00086-9 [DOI] [PubMed] [Google Scholar]
- Vogeler, S., Carboni, S., Li, X., & Joyce, A. (2021). Phylogenetic analysis of the caspase family in bivalves: Implications for programmed cell death, immune response and development. BMC Genomics, 22(1), 80. 10.1186/s12864-021-07380-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriz, S., Reiter, S., & Galliot, B. (2014). Cell death: A program to regenerate. Current Topics in Developmental Biology, 108, 121–151. 10.1016/B978-0-12-391498-9.00002-4 [DOI] [PubMed] [Google Scholar]
- Yang, B., Li, L., Pu, F., You, W., Huang, H., & Ke, C. (2015). Molecular cloning of two molluscan caspases and gene functional analysis during Crassostrea angulata (Fujian oyster) larval metamorphosis. Molecular Biology Reports, 42(5), 963–975. 10.1007/s11033-014-3833-y [DOI] [PubMed] [Google Scholar]
- Young, N. D., Harris, T. J., Evangelista, M., Tran, S., Wouters, M. A., Soares da Costa, T. P., Kershaw, N. J., Gasser, R. B., Smith, B. J., Lee, E. F., & Fairlie, W. D. (2020). Diversity in the intrinsic apoptosis pathway of nematodes. Communications Biology, 3(1), 478. 10.1038/s42003-020-01208-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zermati, Y., Mouhamad, S., Stergiou, L., Besse, B., Galluzzi, L., Boehrer, S., Pauleau, A. -L., Rosselli, F., D’Amelio, M., Amendola, R., Castedo, M., Hengartner, M., Soria, J. -C., Cecconi, F., & Kroemer, G. (2007). Nonapoptotic role for Apaf-1 in the DNA damage checkpoint. Molecular Cell, 28(4), 624–637. 10.1016/j.molcel.2007.09.030 [DOI] [PubMed] [Google Scholar]
- Zhang, L., Li, L., & Zhang, G. (2011). Gene discovery, comparative analysis and expression profile reveal the complexity of the Crassostrea gigas apoptosis system. Developmental and Comparative Immunology, 35(5), 603–610. 10.1016/j.dci.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Zhivotovsky, B., & Orrenius, S. (2005). Caspase-2 function in response to DNA damage. Biochemical and Biophysical Research Communications, 331(3), 859–867. 10.1016/j.bbrc.2005.03.191 [DOI] [PubMed] [Google Scholar]
- Zmasek, C. M., & Godzik, A. (2013). Evolution of the animal apoptosis network. Cold Spring Harbor Perspectives in Biology, 5(3), a008649. 10.1101/cshperspect.a008649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek, C. M., Zhang, Q., Ye, Y., & Godzik, A. (2007). Surprising complexity of the ancestral apoptosis network. Genome Biology, 8(10), R226. 10.1186/gb-2007-8-10-r226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, H., Henzel, W. J., Liu, X., Lutschg, A., & Wang, X. (1997). Apaf-1, a human protein homologous to C elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell, 90(3), 405–413. 10.1016/s0092-8674(00)80501-2 [DOI] [PubMed] [Google Scholar]
- Zou, H., Li, Y., Liu, X., & Wang, X. (1999). An APAF-1cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. The Journal of Biological Chemistry, 274(17), 11549–11556. 10.1074/jbc.274.17.11549 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in this study are present in the paper and the Supplementary Materials. Any requests can be addressed to the corresponding author GK. Alignments used to conduct phylogenetic analyses are available at Figshare data base (DOI: doi.org/10.6084/m9.figshare.24441217).


