Abstract
Human cytomegalovirus (HCMV) infection inhibits cell cycle progression and alters the expression of cyclins E, A, and B (F. M. Jault, J.-M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector, J. Virol. 69:6697–6704, 1995). In this study, we examined cell cycle progression, cyclin gene expression, and early viral events when the infection was initiated at different points in the cell cycle (G0, G1, and S). In all cases, infection led to cell cycle arrest. Cells infected in G0 or G1 phase also showed a complete or partial absence, respectively, of cellular DNA synthesis at a time when DNA synthesis occurred in the corresponding mock-infected cells. In contrast, when cells were infected near or during S phase, many cells were able to pass through S phase and undergo mitosis prior to cell cycle arrest. S-phase infection also produced a delay in the appearance of the viral cytopathic effect and in the synthesis of immediate-early and early proteins. Labeling of cells with bromodeoxyuridine immediately prior to HCMV infection in S phase revealed that viral protein expression occurred primarily in cells which were not engaged in DNA synthesis at the time of infection. The viral-mediated induction of cyclin E, maintenance of cyclin-B protein levels, and inhibitory effects on the accumulation of cyclin A were not significantly affected when infection occurred during different phases of the cell cycle (G0, G1, and S). However, there was a delay in the observed inhibition of cyclin A in cells infected during S phase. This finding was in accord with the pattern of cell cycle progression and delay in viral gene expression associated with S-phase infection. Analysis of the mRNA revealed that the effects of the virus on cyclin E and cyclin A, but not on cyclin B, were primarily at the transcriptional level.
Human cytomegalovirus (HCMV), a member of the betaherpesvirus family, is a ubiquitous human pathogen that is medically relevant due to the severe threat it poses to immunodeficient individuals and newborns (9). Entry of HCMV into the host cell has been shown to produce rapid cellular activation in a manner analogous to growth factor stimulation. For example, there is a transient induction of Fos, Jun, and Myc, as well as increased expression of ornithine decarboxylase, thymidine kinase, DNA polymerase α, and dihydrofolate reductase (2, 5). However, this cellular activation does not lead to cell division (1, 23). In a previous study, we found that in addition to inducing cell cycle arrest, HCMV infection also altered the steady-state levels of several proteins involved in cell cycle regulation. HCMV induced elevated levels of both cyclins E and B as well as their associated kinase activities (23). In contrast, the synthesis of cyclin A appeared to be inhibited, and only at late times in the infection was there any increase in the levels of the protein and its associated kinase activity. Also, we and others have noted that the infection leads to the accumulation of high levels of p53 and retinoblastoma protein (Rb) (23, 29, 38). These striking effects of HCMV on the cell cycle and regulatory proteins have now been confirmed and extended by several investigators (7, 8, 18, 26, 34).
The results of the above studies support the hypothesis that HCMV has adopted a strategy of early cellular activation which facilitates its own replication at the expense of host cellular DNA replication (7, 18, 26). In considering this model of virus-host interaction, however, it is important to take into account the relatively extended life cycle of the virus. By viral standards, there is a long lag between entry into the cell and the initiation of viral DNA replication. In addition, viral replication takes place over a relatively extended period of time. Although viral DNA replication can be detected by 24 h postinfection (p.i.), an additional 24 to 36 h are required for viral replication to reach high levels. As a result, release of detectable quantities of viral progeny begins only after 72 h p.i. (28). Therefore, a valid model of the HCMV life cycle and its effect on the host cell must include not only the prominent changes brought about in the first hours of the infection, but also the subsequent effects on the host which occur throughout the first 72 h p.i., ending with successful virion production.
Elucidation of the mechanisms by which HCMV regulates its replication, while altering the metabolism of the host cell, requires an examination of the host cell cycle. The cell cycle is divided into four phases—G1, S, G2, and M. An additional phase of G0, or quiescence, defines a state in which cells are metabolically active but not cycling and therefore not dividing (35). In tissue culture, the G0 state can be achieved by withdrawal of growth factors via serum starvation or by contact inhibition. Addition of serum or relief from contact inhibition, respectively, then enables cells to reenter the cell cycle in G1. The regulated progression through successive phases of the cell cycle is at least partly dependent on a specific class of protein kinases, called cyclin-dependent kinases (cdks). The activities and specificities of these kinases are in turn dependent upon phosphorylation and dephosphorylation of their catalytic subunits as well as their association with various regulatory proteins, which include cyclins and cdk inhibitors (19, 21, 32, 37). Cyclins are differentially expressed during the cell cycle, with regulation occurring at the level of both transcription and protein degradation (for a complete review, see reference 33).
Cyclins expressed during the G1 phase include the D-type cyclins and cyclin E. The D-type cyclins are synthesized in early G1 in response to cytokines, while cyclin E accumulates during the second half of G1. Cyclin E in association with cdk2 has been shown to be necessary for G1/S transition (4). A second class of cyclins includes cyclins A and B, which regulate the S and G2/M phases of the cycle. Cyclin A accumulates at the initiation of S phase and, in association with cdk2, appears to be required for DNA synthesis (33). Cyclin B begins to accumulate late in S through G2 phase and forms a complex with cdc2 to generate maturation-promoting factor. The binding of cyclin B to cdc2, however, is normally insufficient for maturation-promoting factor activity. Instead, this activity is both positively and negatively controlled by an intricate upstream network of kinases and phosphatases which target cdc2. Cyclin A, in complex with cdc2, also appears to cooperate with cyclin B-cdc2 to facilitate mitosis (24).
In this work, we have extended our studies on the virus-host interactions which lead to cell cycle dysregulation and the altered expression of cyclins E, A, and B. Cells were infected at three different points in the cell cycle (G0, G1, and S phases) and assayed for effects on cell cycle progression as well as cyclin protein expression. Additionally, we have determined how cyclin mRNA is affected when HCMV infection occurs in G0. We found that the effects of the infection on the expression of cyclins E and A are primarily mediated at the level of RNA transcription. However, mRNA levels of cyclin B were not significantly altered by the infection. This observation leads to the notion that maintenance of cyclin B in infected cells is primarily due to the absence of cell cycle-mediated degradation of the protein.
The viral effects on cyclin protein levels occurred regardless of the cell cycle phase at the beginning of the infection, although the observed steady-state levels were also dependent on the amount of cyclin protein present at the initial stages of the infection. Our studies also showed that cells infected in G0 phase and most cells infected in G1 phase did not initiate cellular DNA synthesis at a time corresponding to S phase in the mock-infected cells. In contrast, approximately 50% of cells infected in S phase were able to progress through G2/M before they underwent cell cycle arrest. This lag in cell cycle arrest was associated with a delay in both viral gene expression and the appearance of the cytopathic effect (CPE). By immunostaining it appeared that when cells were actively engaged in DNA replication at the time of the infection, virus was able to enter the cells but viral gene expression was preferentially delayed.
MATERIALS AND METHODS
Cell culture and virus.
All experiments described were performed with human foreskin fibroblasts (HFFs) prior to passage 20. HFFs were propagated in Earle’s minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS; Summit Biotechnology) (MEM–10% FBS), 45 μg of gentamicin sulfate (Gemini Bio-Products, Inc.)/ml, 1.4 μg of amphotericin B (Fungizone)/ml, 1.8 μM l-glutamine, 180 U of penicillin G/ml, and 180 μg of streptomycin sulfate/ml; tissue culture media and other listed supplements were obtained from the Core Cell Culture Facility of the University of California, San Diego. To synchronize cells, HFFs were grown to confluence and maintained at high density for 3 days prior to trypsinization and replating at 106 cells/75-cm2 flask.
HCMV Towne was obtained from the American Type Culture Collection and was stored at −80°C in MEM–10% FBS containing 1% dimethyl sulfoxide. For G0 infections, cells were trypsinized and infected at a multiplicity of infection (MOI) of 5 at the time of replating. In the case of G1- and S-phase infections, cells were infected at 12 and 24 h, respectively, after plating. Viral supernatants were always diluted to between 1:5 and 1:10 with MEM–10% FBS prior to infection. Viral supernatants were removed at 12 h p.i. and replaced with MEM–10% FBS. Mock infections were performed with medium conditioned on actively growing cells for 3 days. Conditioned medium was adjusted to 1% dimethyl sulfoxide and stored at −80°C until use.
Ab.
Cyclin B (GNS-1) and Cyclin E (HE12) mouse monoclonal antibodies (Ab) were obtained from PharMingen. The mouse monoclonal Ab directed against the immediate-early (IE) proteins IE1 72 and IE2 86 (CH16.O) was obtained from the Goodwin Institute (Plantation, Fla.). The rabbit polyclonal Ab directed against cyclin A was kindly provided by Tony Hunter (Salk Institute).
Rabbit antiserum generated against the common amino terminus of the 2.2-kb proteins (UL112-113) was prepared as follows. The plasmid pMY2, which contains a 692-bp DpnI fragment encoding the N terminus of the 2.2-kb early protein sequence (amino acids 3 to 233) fused to sequence encoding the C terminus of glutathione S-transferase (GST), was used to transform the DH5α strain of Escherichia coli (Gibco BRL) as directed by the manufacturer. Purification of the GST–2.2-kb protein was accomplished as described previously by Klucher et al. with the addition of a final dialysis step to eliminate the presence of protease inhibitors (25). Antiserum to the fusion protein was produced essentially as described previously by Cranmer et al. (11).
Cell lysate Preparation.
At the desired times p.i., HFFs were trypsinized, washed once with phosphate-buffered saline (PBS), pelleted, and lysed with reducing sample buffer (2% sodium dodecyl sulfate [SDS], 10% glycerol, 1% β-mercaptoethanol, 60 mM Tris [pH 6.8], 1 mM sodium metabisulfite, 1 mM benzamidine, 3 μg of aprotinin/ml, 10 mM sodium pyrophosphate, 50 mM sodium fluoride, 0.5 mM sodium orthovanadate, and 4 mM phenylmethylsulfonyl fluoride). Lysates were then sonicated briefly, boiled for 5 min, centrifuged at a relative centrifugal force of 16,000 for 10 min at 4°C, and stored at −80°C until use. Protein concentrations were determined by A280 by using a bovine serum albumin standard curve.
Gel electrophoresis and Western blotting.
One hundred micrograms of each lysate sample was boiled at 100°C for 5 min before separation by electrophoresis on SDS–10% polyacrylamide gels. Following electrophoresis, proteins were transferred to Immobilon-P membranes (Millipore) by electroblotting at 100 V for 1 h in transfer buffer (50 mM Tris, 380 mM glycine, 0.1% SDS, and 20% methanol). The transferred protein was visualized by staining with amido black. Blots were blocked for 1 h in Tris-buffered saline blocking solution containing 5% nonfat dry milk, 0.1% Tween 20, and 0.01% Antifoam A emulsion (Sigma). After a quick wash with a washing solution of 0.1% Tween 20 in Tris-buffered saline, blots were incubated with primary Ab in blocking solution for 1 to 2 h. Blots were washed once quickly, once for 15 min, and twice for 5 min (each time) before incubation with horseradish peroxidase-linked anti-mouse or anti-rabbit immunoglobulin G secondary Ab at a dilution of 1:1,700 for 1 h in blocking solution. This incubation was followed by one 15-min and three 5-min washes. Proteins were visualized by using enhanced-chemiluminescence reagents as instructed by the manufacturer (Amersham or Pierce) and exposure of the blot to film. When stripping of the blots was necessary, the membranes were incubated in stripping buffer (0.1 M β-mercaptoethanol, 2% SDS, and 62.5 mM Tris [pH 6.7]) at 50°C for 30 min, followed by one quick wash and two 10-min washes in washing solution before blocking for the next Ab.
Analysis of DNA content.
HFFs were cultured and infected as described above. At the time of harvest, cells were treated with Passage Ease (VEC TEC, Inc.), washed with PBS, counted, and stored at 4°C in PBS with 40% ethanol. DNA content was determined by flow cytometry following staining of the cells with propidium iodide as described previously (10, 12, 39).
Preparation of mRNA and Northern analysis.
HFFs were cultured and infected as described above. For each specific RNA sample, at least 6 × 107 cells were used. Total RNA was isolated with an Rneasy Midi Total RNA purification kit (Qiagen), and mRNA was then isolated from total RNA with the PolyAtract mRNA isolation system (Promega). mRNA was quantitated by A260, ethanol precipitated, and resuspended in RNase-free H2O for electrophoresis.
mRNA (1 μg) was separated on a 1% agarose-formaldehyde gel, transferred to Nytran membranes (Schleicher & Schuell) by overnight diffusion, and finally cross-linked to the membrane with a UV Stratalinker 1800 (Stratagene) by using standard protocols (3). Hybridization of the membranes was performed as directed by the manufacturer (Schleicher & Schuell). DNA probes for cyclins A, B1, and E were generously provided by Steve Reed (Scripps Institute), and the probe for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was acquired from Ambion, Inc. DNA probes (30 to 75 ng) were 32P labeled with a Prime-It II kit (Stratagene) and isolated through Nuctrap probe purification columns (Stratagene). Membranes were then washed and exposed to film with an intensifying screen at −80°C. Northern blots were also analyzed by PhosphorImager (Molecular Dynamics) to verify normalization and to determine fold differences in signal between the various samples.
Immunofluorescence.
Confluent HFFs were trypsinized and subsequently seeded onto coverslips in 10-cm-diameter plates at 8 × 105 cells/plate in MEM–10% FBS. After 2 h to allow for adherence of the cells, the plates were washed once with PBS, followed by addition of MEM–0.1% FBS to begin a 48-h serum starvation. Following serum starvation, those plates which were being used for S-phase infection were refed with MEM–10% FBS to initiate growth, while those plates designated for G0 infection were refed with MEM–0.1% FBS. After 26 h, cells were either infected at an MOI of 5 in MEM–10% FBS or mock infected with conditioned medium. At the desired times (12 or 36 h p.i.), plates were washed twice with PBS. Cells were then fixed for 10 min in a 3% paraformaldehyde-PBS solution, then permeabilized for 5 min in 1% Triton X-100 solution, and finally washed extensively with PBS. The coverslips were stored at 4°C in PBS prior to use for immunofluorescence. Coverslips were then incubated for 10 min with mouse monoclonal Ab directed against the IE proteins (CH16.0) at a dilution of 1:100 in PBS–0.2% gelatin, followed by extensive washing in PBS. They were then incubated for 10 min with fluorescein isothiocyanate-conjugated goat anti-mouse secondary Ab (Jackson Laboratory) diluted 1:400 in PBS–0.2% gelatin, followed by extensive washing. Finally, coverslips were incubated in Hoechst dye (Calbiochem) diluted 1:100 in PBS–0.2% gelatin for 2 min, followed by extensive washing, and were mounted onto glass slides with glycerol-paraphenylenediamine (an antiphotobleaching agent). Immunofluorescence analysis was performed on a Nikon EFD-3 fluorescence microscope. The microscope was equipped with Neofluor objectives and an MTI charge-coupled device 2 camera. Images were captured with NIH Image 1.59.
BrdU double-labeling experiment.
Confluent HFFs were trypsinized and plated onto glass coverslips at 106 cells/10-cm-diameter plate in MEM–10% FBS. After 23 h, the medium was removed and replaced with fresh MEM–10% FBS containing bromodeoxyuridine (BrdU) labeling mix (Boehringer Mannheim). Plates were incubated for 30 min, then washed three times with fresh medium, with the final wash lasting for 30 min. Following this incubation (at 24 h postplating), cells were infected at an MOI of 5 or were mock infected. After 2 h, cells were washed extensively in fresh medium. Coverslips were harvested at 12 and 28 h p.i. and prepared for immunofluorescence analysis as described above. Unless otherwise noted, Ab were diluted in a PBS–0.2% gelatin solution. Before incubation with specific antisera, all coverslips were preincubated with normal goat serum for 10 min. After a wash in PBS, cells were incubated with a rat Ab directed against BrdU (Accurate Scientific), diluted in a solution designed to expose the labeled residues, for 30 min (5 μl of 1 M MgCl2, 0.5 μl of DNase I, and 1 μl of Ab in 250 μl of PBS–0.2% gelatin). After extensive washing in PBS, coverslips were incubated for 10 min with a donkey anti-rat secondary Ab that had been cross-adsorbed for other species and conjugated with Cy3. After a wash, coverslips were incubated with either a mouse monoclonal Ab directed against pp65 (Goodwin Institute, Plantation, Fla.) or a combination of CH16.0 and a monoclonal Ab against IE1 72 (from William Britt) for 10 min. After more washes, the slips were finally incubated with a cross-adsorbed donkey anti-mouse secondary Ab conjugated with fluorescein isothiocyanate and with Hoechst dye to illuminate the DNA for 10 min. Coverslips were mounted and examined as described above, except that a Zeiss fluorescence microscope was used for analysis.
RESULTS
The pattern of HCMV-mediated cell cycle arrest is dependent on the phase of the cell cycle at the time of infection.
We previously demonstrated that infection of HFFs with HCMV immediately after release of cells from growth arrest led to an arrest in cell cycle progression. In these initial experiments, cells were growth arrested in G0 by incubation in medium containing 0.5% FBS for approximately 45 h prior to infection with virus in medium containing 10% FBS (23). We subsequently found that better synchrony of cells could be obtained when they were growth arrested by contact inhibition. Thus, for the experiments described below, HFFs were maintained at confluence for 3 days in medium containing 10% FBS before being replated at a lower density (also in medium with 10% FBS) and infected with HCMV. At defined times p.i., cells were harvested for analysis of DNA content by flow cytometry.
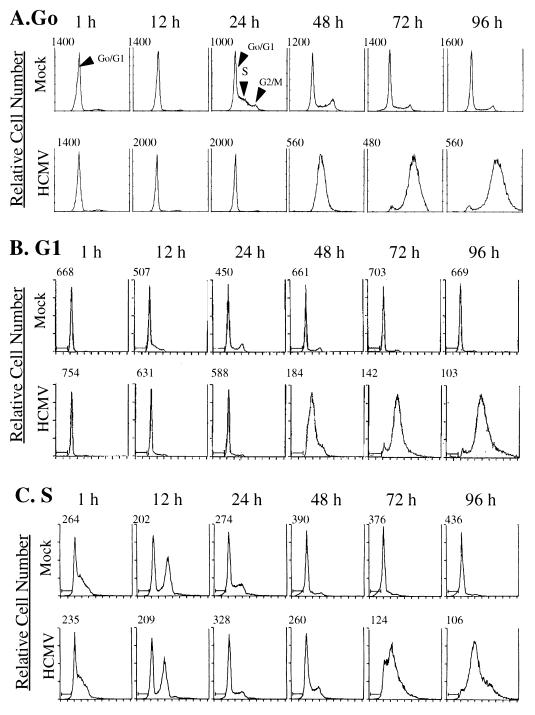
Figure 1A shows a fluorescence-activated cell sorter (FACS) analysis from an experiment referred to as a G0-phase infection. Cells were replated at a lower density, which released them from growth arrest, and were simultaneously infected with HCMV. At 1 through 12 h p.i., all cells in mock-infected and HCMV-infected cultures appeared to be in the G0/G1 phase of the cell cycle. As expected, approximately 50% of mock-infected cells were in S phase by 24 h postplating. Furthermore, mock-infected cells continued to cycle until reaching confluence at approximately 96 h p.i. In contrast, on the basis of DNA content, none of the infected cells appeared to enter S phase at the 24-h time point. This delay was similar to what was previously observed when HFFs were infected following release from serum starvation (23). It was not until 48 h p.i., when viral DNA replication was well under way, that an apparent increase in the DNA content of infected cells was observed.
FIG. 1.
Flow cytometry analysis of HFFs infected at various points in the cell cycle. HFFs were synchronized by contact inhibition and stimulated to cycle by replating at lower density. Cells were infected at the time of replating (G0) (A), 12 h postplating (G1) (B), or 24 h post-plating (S phase) (C). Samples of mock- and HCMV-infected cells were harvested at defined times p.i. (indicated above the histograms) and analyzed for DNA content by flow cytometry following staining of the DNA with propidium iodide. The number above each histogram on the left side indicates relative cell number.
To determine if the effects of HCMV infection on the cell cycle were dependent on the phase of the cycle at the time of infection, we varied the experiment so that cells were infected with HCMV either 12 h (G1-phase infection) or 24 h (S-phase infection) after being replated at a lower density. As described above, the cells were then harvested for analysis of DNA content by flow cytometry at defined times p.i. The results of the G1 infection (Fig. 1B) were similar to those observed with the G0 infection. As expected, mock-infected cells began to enter S phase approximately 12 h after mock infection and proceeded to cycle normally. During this initial 12-h period, the percentage of cells in G1 decreased from 91 to 64%, while the percentage of cells in S increased from 9 to 34%. In contrast, DNA replication in the HCMV-infected cells seemed to be partially inhibited, as evidenced by the fact that during the same initial 12-h period, a smaller percentage of cells had progressed from G1 to S. In this case, the percentage of cells in G1 decreased only to 79%, while the percentage in S increased to 18%. The cells in the infected culture which did undergo an increase in DNA content likely represented a population that had progressed sufficiently through the G1 phase to begin DNA replication despite viral infection (see below). We believe that it is unlikely that these are uninfected cells, since all infections were performed at high MOIs and all cells displayed staining for the pp65 tegument protein at 12 h p.i. (see below) and distinct CPEs by 36 h p.i. As in the G0 infection, the G1-infected cells did display an apparent increase in DNA content at 48 h p.i.
Figure 1C shows the flow cytometry results when cells were infected 24 h after being replated at a lower density (S-phase infection). At 1 h p.i., 56% of the mock-infected cells were in S phase, and by 8 h, a major fraction (37%) were in G2/M. These cells continued to cycle until they reached confluence at approximately 72 h p.i. We found that HCMV-infected cells displayed a DNA content very similar to that of mock-infected cells for the first 12 h of the infection. HCMV-infected cells appeared to advance into G2/M by 12 h p.i. as efficiently as the mock-infected cells. Between 12 and 24 h p.i., both mock-infected and infected cultures also showed a decrease in the percentage of cells in G2/M and a concomitant increase in the percentage in G1. However, at 24 h p.i., it was clear that a significantly greater percentage of the mock-infected population had again entered S phase compared to the infected cultures, which indicated that cellular DNA replication was inhibited (or delayed) by the infection. As was observed for the infections initiated at other points in the cell cycle, there was a major accumulation of DNA in the S-phase-infected cells after 48 h p.i. These results suggest that HCMV infection in S phase does not immediately inhibit cellular DNA replication until after cells have passed through G2/M.
The phase of the cell cycle at the beginning of the infection influences the progression of the CPE.
In the course of the experiments described above, we observed a quantitative and qualitative difference in CPE when cells were infected at different times in the cell cycle. To determine whether these differences were real or simply the result of inherent variability in the experiments performed at different times, we performed an experiment whereby cells at different phases of the cell cycle were simultaneously infected with a single stock of virus. To accomplish this, contact-inhibited cell cultures were replated to a lower density at staggered times so that at the time of infection, there were sets of flasks containing cells either in G0, G1, or S phase. The results of this experiment confirmed our initial observations.
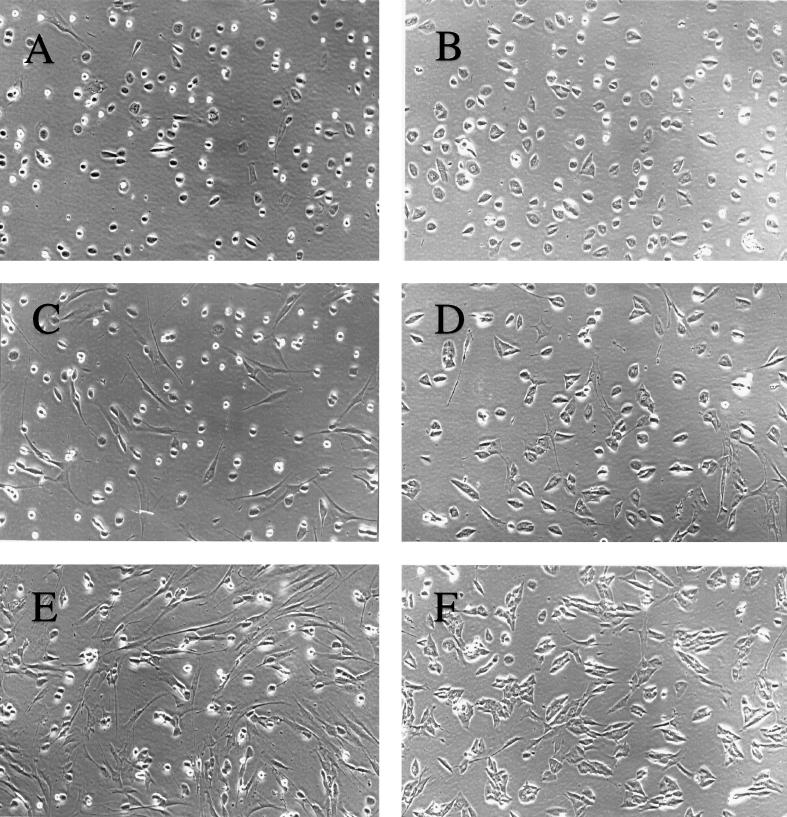
Figure 2A shows that almost all the cells infected in G0 phase were tightly rounded by 12 h p.i. For the most part, this was also true of G1-phase infections, although a slightly higher percentage of cells had no observable CPE at this time point (Fig. 2C). In contrast, we observed that approximately 50% of cells infected in S phase displayed no CPE at 12 h p.i. (Fig. 2E). Although one interpretation of this result is that only a fraction of the cells were actually infected, this was found not to be the case.
FIG. 2.
Differential CPE as a result of infection of cells at different phases of the cell cycle. HFFs were synchronized by contact inhibition and stimulated to cycle by replating at lower density. To control for the possibility of variability in the infections, the times of replating were staggered so that all infections (G0, G1, and S phase) could be performed concurrently with the same stock of virus. For each type of infection (G0, G1, and S phase), one representative flask of four was used for photography. Cells were photographed in situ with a 35-mm camera attached to a phase microscope. All photographs depict cells infected by HCMV at an MOI of 5. The cell cycle phases at the time of infection and the times p.i. when cells were photographed are as follows: G0, 12 (A) and 36 (B) h; G1, 12 (C) and 36 (D) h; S, 12 (E) and 36 (F) h. Magnification, ×93.
At 12 h p.i., the viral inoculum was removed from the cells and replaced with fresh medium. The cells were examined at various times thereafter, and by 36 h p.i., nearly 100% of the cells in each of the cultures showed CPE (Fig. 2B, D, and F). At this time p.i., viral replication in the G0-phase-infected cells had progressed such that most of the cells had initiated the cellular enlargement characteristic of the infection (Fig. 2B). The phenotype of the G1-infected cells was similar, except that the stage of cytomegaly appeared to be more heterogeneous (Fig. 2D). Interestingly, virtually the entire population of the S-phase-infected cells at 36 h p.i. showed cytomegaly, although the morphology of the cells differed somewhat from that of those infected in G0 or G1 (Fig. 2F).
The observation that all of the S-phase-infected cells displayed CPE by 36 h p.i. indicated that all of the cells had been infected by the 12-h time point, but the progress of the infection likely differed from that in the G0-phase- and G1-phase-infected cells. Moreover, since infectious virus is not produced at detectable levels until approximately 72 h p.i., we considered it unlikely that the observed CPE in the S-phase-infected cells was the result of secondary infection. Similar results were also obtained when cells were released from growth arrest induced by serum starvation and then infected at the time of addition of high levels of serum (G0-phase infection) or 26 h later (S-phase infection). This ruled out the possibility that an artifact may have been introduced by release of the cells from growth arrest with trypsin and replating of the cells at a lower density (data not shown). Therefore, it appears that the differences in the kinetics of the CPE are directly related to the phase of the cell cycle at the time of infection.
It should be noted that it is difficult to achieve complete synchronization of primary fibroblasts. Therefore, only approximately 50% of the cell population in the S-phase-infected cells were clearly in S phase at the time of infection, as measured by flow cytometry (see Fig. 1C). As a result, approximately 50% of cells still appeared to be in G1, based on analysis of DNA content. These percentages correspond well with our observation that approximately 50% of S-phase-infected cells displayed CPE at 12 h p.i., although we cannot determine from these data whether the subsets were, in fact, the same cell population. However, since the majority of the cells in cultures that were infected prior to S phase (G0 and G1 infections) showed clear CPE by 12 h p.i., it seemed most likely that infection of cells during DNA synthesis resulted in a delay in the appearance of the CPE.
Cells infected in S phase show a delay in the expression of HCMV IE and early proteins.
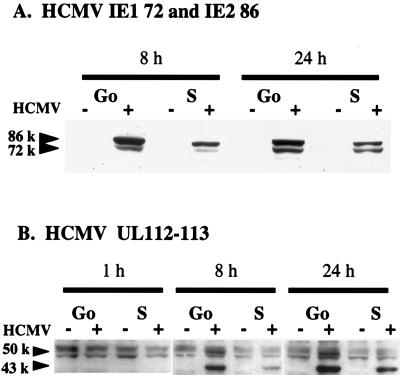
In view of the above results, it was important to determine whether the delay in the appearance of the CPE was associated with a delay in HCMV protein expression. To address this question, cells in G0 and S phase were simultaneously infected with the same stock of virus, using the procedure described above, and at various times p.i., cell lysates were prepared and analyzed by Western blotting for HCMV viral proteins. As probes, we used an Ab that recognizes the domain which is common to the IE proteins IE1 72 and IE2 86 (although the Ab apparently has a higher affinity for IE2 86) and an Ab to the 2.2-kb family of HCMV early viral proteins (UL112-113).
IE protein could not be detected in either G0- or S-phase-infected cells at 1 h p.i. (data not shown), and as expected, there was no viral protein detected in mock-infected cells at any time. However, by 8 h p.i., IE1 72 and IE2 86 were present in both the G0- and S-phase-infected cells, although it was clear that the levels in S-phase-infected cells were lower (Fig. 3A). At 24 h p.i., there was still more IE protein in the G0-phase-infected cells than in cells infected at S phase, but the difference between the two was diminished. This difference continued to decrease, and at later time points, the patterns of IE protein expression in the two cultures were comparable (reference 27 and data not shown).
FIG. 3.
Effect of cell cycle phase at the time of HCMV infection on viral protein expression. HFFs were synchronized by contact inhibition and stimulated to cycle by replating at lower density. As described for Fig. 2, cells were concurrently infected at the time of replating (G0) or 24 h postplating (S phase). At defined times p.i., cultures were harvested for Western analysis. Cell lysates (100 μg) from each time point were subjected to electrophoresis and subsequent immunoblotting analysis with Ab against the HCMV IE proteins IE1 72 and IE2 86 (A) or 2.2-kb early proteins UL112-113 (B).
The kinetics of the appearance of the 2.2-kb family of early proteins were also slower in the S-phase-infected cells (Fig. 3B). Two bands specific for the 2.2-kb proteins (a major band of 43 kDa and a minor band of 50 kDa) were detected at 8 h p.i. in both the G0- and S-phase-infected cells, with the levels in S-phase-infected cells being significantly lower. It should be noted that two nonspecific bands, one above and one below the specific 50-kDa band, were detected with our Ab. The difference in protein levels was somewhat less at 24 h p.i., and as was the case for the IE proteins, the levels of expression of the 2.2-kb proteins in the G0- and S-phase-infected cells were similar at later times in the infection (reference 27 and data not shown).
Taken together, the kinetics of viral protein expression and the appearance of the CPE point to a general delay in the viral life cycle when cells are infected during S phase relative to that observed when cells are infected immediately after release from G0. This delay was most apparent during the first 24 h p.i., after which the steady-state levels of viral proteins and the CPE displayed minimal differences between cells infected at different times.
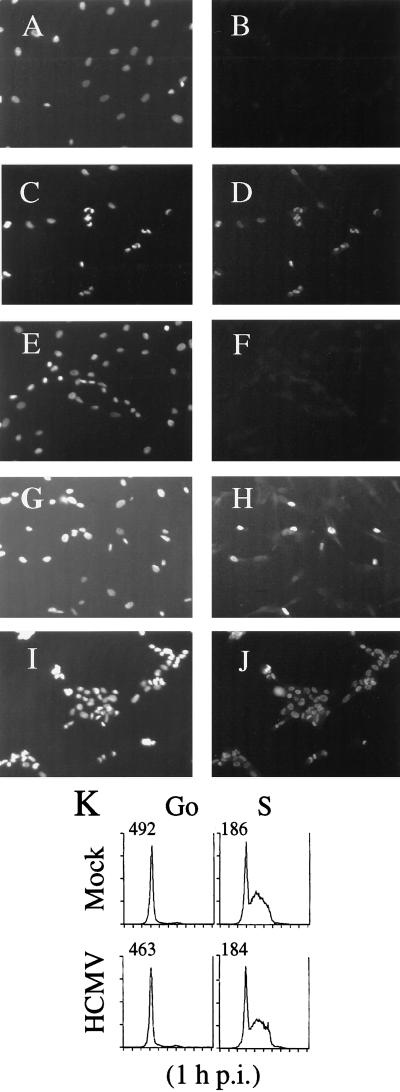
The results of the above experiments raised the question of whether there was an inhibition of viral protein expression in only a fraction of S-phase-infected cells or whether the accumulation of the IE and early viral proteins simply proceeded at a slower rate in all of the cells. To distinguish between these possibilities, we assayed for the presence of the IE proteins by indirect immunofluorescence (Fig. 4). For these experiments, cells were arrested in G0 by serum starvation for 2 days. Serum was then added to one population of cells, which were allowed to progress to S phase (26 h following serum stimulation as determined by flow cytometry [Fig. 4K]). After 26 h, serum was added to a second group of cells to release them from G0. The cells just released from G0 and the cells in S phase were then both infected simultaneously with the same stock of HCMV. At the desired times p.i., the cells were fixed, permeabilized, and stained with Ab against the IE proteins.
FIG. 4.
Effect of cell cycle phase at the time of HCMV infection on IE protein expression in individual cells as determined by immunofluorescence. HFFs, grown on coverslips, were serum starved, followed by serum stimulation to generate plates containing cells either in G0 or in S phase, thus allowing concurrent infection (see Materials and Methods). At defined times p.i., coverslips were then processed for immunofluorescence. Cells were stained with Ab to the IE viral proteins (B, D, F, H, and J), and Hoechst dye was used to illuminate the DNA in the cells (A, C, E, G, and I). Shown are G0-phase mock-infected (A and B) and HCMV-infected (C and D) cells at 12 h p.i., S-phase mock-infected cells at 12 h p.i. (E and F), and S-phase HCMV-infected cells at 12 (G and H) and 36 (I and J) h p.i. Magnification, ×108. (K) Samples of mock- and HCMV-infected cells (from G0- and S-phase infections) were harvested at 1 h p.i. and analyzed for DNA content by flow cytometry following staining of the DNA with propidium iodide.
Most of the cells infected in G0 displayed IE expression and showed distinct cell rounding at 12 h p.i. (Fig. 4C and D). In contrast, at this time point, IE protein was detected in less than 50% of the cells in the S-phase-infected cultures (Fig. 4G and H). However, by 36 h p.i., almost all S-phase-infected cells contained IE protein and showed CPE (Fig. 4I and J). These results were consistent with the above data, which demonstrated significant CPE and IE protein expression in S-phase-infected cells by 36 h p.i. We conclude from these experiments that HCMV infection of cells in or near S phase leads to a defined delay in the initiation of characteristic early events in the viral life cycle.
Viral gene expression and cellular DNA synthesis do not occur simultaneously in the majority of HCMV-infected cells.
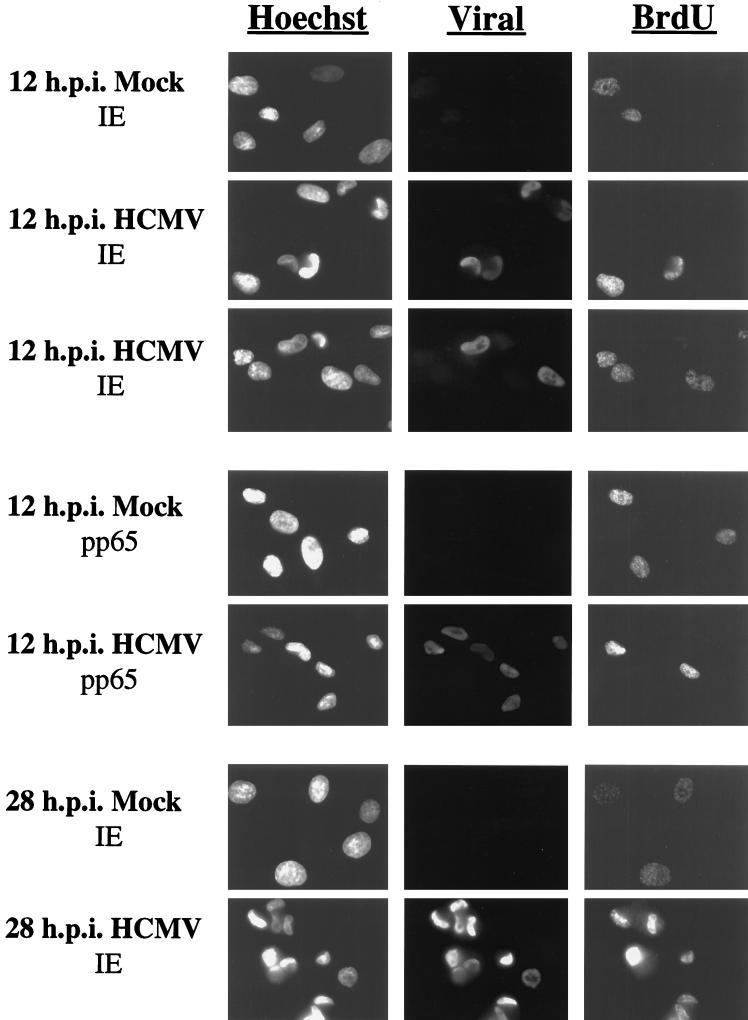
The partial (50%) infection observed in S-phase-infected cells at 12 h p.i. raised the question of how cells which expressed IE protein at 12 h.p.i. differed from those cells in which the infection was apparently delayed. One possible explanation was that the status of those cells which were actually in S phase as measured by FACS analysis was not favorable for the initiation of the HCMV infection. This hypothesis would predict that cells which expressed IE protein by 12 h p.i. in the S-phase-infected population represented those cells which had not initiated cellular DNA synthesis, since FACS analysis indicated that approximately 50% of cells in the population maintained an apparent G0/G1 status at 12 h p.i. Alternatively, cells may have become refractory to HCMV infection as a result of some other factor associated with cell cycle progression. In this case, the efficiency of infection would likely decrease to approximately 50% in a stochastic manner regardless of whether cells had initiated DNA synthesis at the time of infection. To differentiate between these possibilities, cells were labeled with BrdU immediately prior to infection to allow detection of cells actively engaged in DNA synthesis. In this experiment, the viral inoculum was removed from the cells after 2 h to avoid the possibility that adsorption and penetration would be asynchronous. At 12 h p.i., cells were then fixed and double stained with Ab against BrdU and HCMV IE proteins. At least 200 cells were analyzed to determine if there was a direct relationship between cellular DNA synthesis and viral gene expression.
When cells were labeled with BrdU immediately prior to mock or HCMV infection in S phase, approximately 50% of the cells were positive for BrdU when fixed and stained at 12 h p.i. (Fig. 5). This correlated with the percentage of cells observed to be in S phase by FACS analysis. In addition, approximately 32% of the cells were expressing viral IE proteins. However, only 5% of all cells were positive for both BrdU and HCMV IE protein. This double-positive population represented only 14% of those cells positive for IE gene expression. Therefore, the vast majority of cells which were positive for IE gene expression at 12 h p.i. were not engaged in cell DNA synthesis at the time of infection. Finally, approximately 18% of the cells were double negative. Therefore, IE gene expression was also delayed in some cells that were not actively synthesizing DNA at the time of infection.
FIG. 5.
Cells infected in S phase display a preferential delay in viral protein expression during cellular DNA synthesis. HFFs were synchronized by contact inhibition and stimulated to cycle by replating at lower density onto coverslips. At 23 h postplating, cells were labeled with BrdU for 30 min, followed by infection at 24 h postplating (S phase) with HCMV. At 12 and 28 h p.i., coverslips were processed for immunofluorescence. Cells were double stained with Ab against BrdU and HCMV IE protein or against BrdU and HCMV pp65 tegument protein. In both cases, cells were stained with Hoechst dye to illuminate DNA. Two fields of HCMV-infected cells double-stained with Ab against BrdU and IE protein are shown. Magnification, ×333.
To exclude the possibility that the restriction to infection was occurring at the point of viral entry, the cells were also stained with Ab to the pp65 (UL83) viral tegument protein (Fig. 5). The majority (>95%) of the cells in the population showed the clear presence of this viral protein in the nucleus at 12 h p.i., which indicated that viral entry had occurred regardless of whether the cells were actively synthesizing DNA at the time of infection. In addition, as seen previously, >95% of the cells were expressing the HCMV IE protein at 28 h p.i. (Fig. 5). These results further support our conclusion that progression of the infection is delayed but not inhibited in S-phase-infected cells. Taken together, our results show that while HCMV successfully enters cells infected in S phase, a delay in viral protein expression is more likely to occur if the cell has initiated DNA synthesis. However, this delay is temporary, and viral gene expression does eventually occur, most likely after the cells have progressed through G2/M.
HCMV infection at different points in the cell cycle has little effect on virus-mediated changes in the expression of cell cycle-regulatory proteins.
Previously, we showed that HCMV is able to alter markedly the expression of cyclins A, B, and E when cells are infected at the time of release from G0 induced by serum starvation (23). Since these cyclins are differentially expressed during the normal cell cycle, we decided to determine whether the observed HCMV-mediated effects on these cyclins were dependent upon the phase of the cell cycle at the time of infection. To address this question, we harvested cells that were infected and mock infected as described above (in G0, G1, and S phases), and we analyzed the levels of the cyclins at specific times p.i. by Western blotting.
Cyclin A.
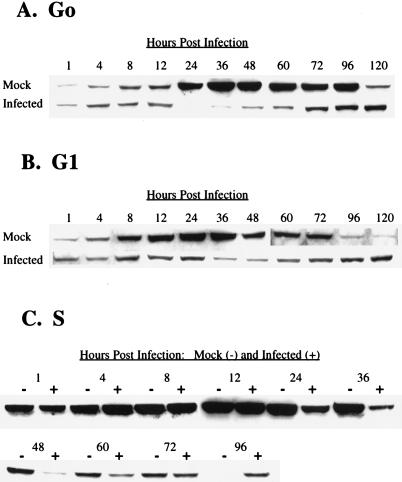
The results of the G0-phase infection for cyclin A were comparable to those reported previously (23). During the first 12 h of the experiment, both mock- and HCMV-infected cells contained only small amounts of cyclin A, which likely represented protein that was present in the cells when they were replated (Fig. 6A). In mock-infected cells, cyclin A accumulated to high levels by 24 h postplating, a time which coincided with the observed entry into S phase as determined by flow cytometry (see Fig. 1A). In contrast, cyclin-A protein did not accumulate in HCMV-infected cells, and at 24 h p.i., the levels declined below the limits of detection. In mock-infected cells, the amount of cyclin A remained high until 96 h p.i., consistent with the observed cycling of the cells. At 120 h, however, we detected a significant drop in cyclin-A levels, which was likely a consequence of the mock-infected cells reaching confluence. In the HCMV-infected cells, a small amount of cyclin A was again detected at 36 h p.i., and levels continued to increase slowly throughout the infection. Therefore, HCMV infection in G0 is able to efficiently inhibit an increase in the steady-state levels of cyclin A. Only at later times in the infection is there a gradual accumulation of a modest amount of this cyclin.
FIG. 6.
Effect of HCMV infection on the steady-state kinetics of cyclin A. HFFs were synchronized by contact inhibition and stimulated to cycle by replating at lower density. Cells were infected at the time of replating (G0) (A), 12 h postplating (G1) (B), or 24 h postplating (S phase) (C). At defined times p.i., cells were harvested for Western analysis. Cell lysates (100 μg) from each time point were subjected to electrophoresis and subsequent immunoblotting analysis with anti-cyclin A Ab.
The results of Western blot analysis of cyclin A when cells were infected 12 h after replating (G1 phase) are shown in Fig. 6B. The pattern of cyclin-A accumulation in the mock-infected cells was as expected. The cells accumulated high levels of this cyclin by 12 h p.i., which correlated with entry into S phase, as measured by flow cytometry (see Fig. 1B). Cyclin-A levels remained elevated in mock-infected cultures until approximately 96 h.p.i., when cells reached confluence. In HCMV-infected cells, there was a modest increase of cyclin A between 4 and 8 h p.i., but the levels then gradually decreased and did not begin to show any increase again until 60 h p.i. However, HCMV-infected cells never achieved the peak cyclin-A levels reached in mock-infected cultures, even at late times in the infection, when the amount of cyclin A in the infected cells reached a maximum. These results suggest that when cells are infected in G1, cyclin-A accumulation is also inhibited at a relatively early point in the infection. However, there is less of a differential between mock- and HCMV-infected cells than is observed during a G0 infection, due to the synthesis of some cyclin A prior to the onset of the HCMV-mediated inhibition. Nevertheless, the overall pattern of decline and rise of cyclin-A levels is comparable for cells infected in G0 and G1.
The kinetics of cyclin-A expression in cells infected 24 h after replating (S phase) are shown in Fig. 6C. In accord with flow cytometry data showing passage of both mock- and HCMV-infected cells from S to G2/M during the first 12 h p.i. (see Fig. 1C), cyclin-A levels were correspondingly high at 1 h p.i., with a continuous increase in both infected and mock-infected cells during the first 12 h p.i. The mock-infected cells maintained high levels of cyclin A through 36 h p.i., and then these levels began to decline as cells reached confluence. At 24 h p.i. there was a clear decrease in the amount of this protein in HCMV-infected cells, and this decline continued through 48 h p.i. As was observed when cells were infected with HCMV in G0 or G1, cyclin-A levels in S-phase-infected cells began to increase slowly after 60 h p.i. Taken together, the results of these experiments suggest that HCMV-mediated inhibition of cyclin-A expression does not occur immediately after infection of the cell, but rather the infection must progress for approximately 8 to 12 h. Therefore, if cells are infected at a point in the cell cycle when they are actively synthesizing cyclin A or if there is a delay in the initiation of viral-gene expression, there will be a parallel increase in cyclin-A levels in both the mock- and HCMV-infected cells during the first 8 to 12 h p.i. Cyclin-A levels will then begin to decline until late times in the infection, when there appears to be some accumulation in the infected cells.
Cyclin E.
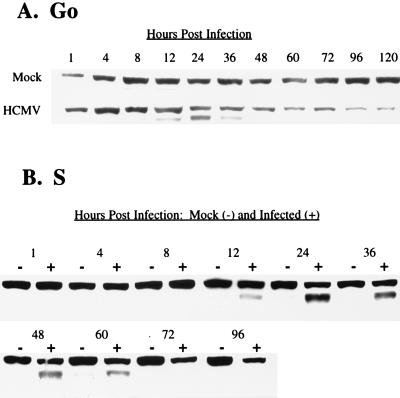
The steady-state levels of cyclin E during the courses of G0- and S-phase infections are shown in Fig. 7A and B, respectively. In the G0 infection, there was an overall increase in the levels of cyclin E in infected cells beginning at 12 h p.i. This increase was primarily due to the induction of a second, smaller isoform of cyclin E, which is barely detectable in the mock-infected cells. At 48 h p.i., this smaller species of cyclin E was no longer seen in infected cells. As the infection progressed, the overall level of cyclin E gradually declined, falling below the levels seen in mock-infected cells at the corresponding time points. Similar results were obtained for both mock- and HCMV-infected cells during a G1 infection (data not shown).
FIG. 7.
Effect of HCMV infection on steady-state kinetics of cyclin E. HFFs were synchronized by contact inhibition and stimulated to cycle by replating at lower density. Cells were infected at the time of replating (G0) (A) or 24 h postplating (S phase) (B). At defined times p.i., cells were harvested for Western analysis with anti-cyclin E Ab.
Infection of cells in S phase had little effect on the overall pattern of cyclin-E accumulation in infected cells, except that induction of the smaller species appeared to remain at detectable levels for a longer period. It is likely that these prolonged kinetics are due to the observed delay in the initial stages of the infection in a fraction of the cells when cultures are infected in S phase. This is in contrast to infection of cells in G0 or G1, where there is a rapid initiation of viral-gene expression and a relatively synchronous pattern of viral replication.
Cyclin B.
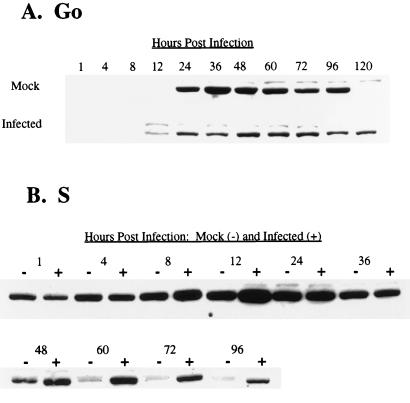
Figure 8A shows the effect of HCMV on the steady-state levels of cyclin B during a G0-phase infection. Expression of cyclin B was detected in mock-infected cells by 24 h postplating, and the levels were sustained until after 96 h p.i., when cells reached confluence. The detection of cyclin B at 24 h p.i. correlates with the expected high-level expression occurring late in S phase and in the G2 phase of the cell cycle in preparation for mitosis (35). In the experiment shown, we did not detect a phase-specific decline of cyclin-B levels. However, such a fluctuation has been observed in other experiments when mock-infected cells were harvested as they traversed the short interval between M and G1, when cyclin B is degraded (data not shown). As previously reported, HCMV infection of cells in G0 did not delay the time of induction of cyclin B relative to that in mock-infected cells, in direct contrast to the effects of the virus on cyclin A (23). The major difference between mock- and HCMV-infected cells was the maintenance of cyclin-B levels in infected cells, which contrasted with a decline in cyclin-B levels at a corresponding time in mock-infected cells. Also, in our experiments we observed no periodic fluctuations of cyclin-B levels in infected cells. Similar results were obtained when cells were infected in G1 phase (data not shown).
FIG. 8.
Effect of HCMV infection on steady-state kinetics of cyclin B. HFFs were synchronized by contact inhibition and stimulated to cycle by replating at lower density. Cells were infected at the time of replating (G0) (A) or 24 h postplating (S phase) (B). At defined times p.i., cells were harvested for Western analysis with anti-cyclin B Ab.
Infection of cells in S phase again had little effect on the pattern of cyclin-B accumulation, relative to mock cells, at early times (Fig. 8B). As expected, there were moderately high levels of cyclin B at 1 h p.i. in both mock- and HCMV-infected cells. These levels continued to increase during the first 12 h as cells moved from S to G2/M, with slightly higher levels accumulating in infected cells than in mock-infected cells. After this time point, the amount of cyclin B in mock- and HCMV-infected cells was similar until after 48 h p.i., when the levels of cyclin B notably declined in mock-infected cells as they became confluent. In contrast, HCMV-infected cells maintained significant levels of cyclin B through 96 h p.i.
The effects of HCMV infection on the expression of cyclins are mediated in part at the transcriptional level.
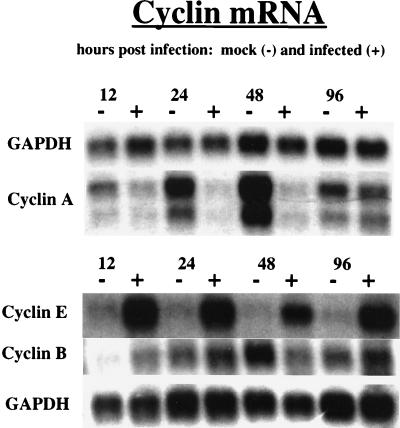
To examine further the molecular basis for the effects of HCMV infection on expression of cyclins E, A, and B, we proceeded to measure the levels of their corresponding mRNAs. For these experiments, the cells were infected in the G0 phase, and at various times p.i. the cells were harvested. Poly(A)-containing RNA was then isolated and analyzed on Northern blots with probes specific for the individual cyclin genes (Fig. 9). To control for the amount of RNA in each lane, blots were also hybridized with a probe for GAPDH RNA, since GAPDH RNA levels are unaffected by the infection.
FIG. 9.
Effects of HCMV infection on mRNA levels of cyclins A, B, and E. HFFs were synchronized by contact inhibition and stimulated to cycle by replating at lower density. The cells were infected with HCMV or mock infected at the time of replating (G0). At defined times p.i., 6 × 107 cells were harvested from both mock- and HCMV-infected cell populations for the isolation of mRNA. The mRNA was then used for Northern analysis as described in Materials and Methods.
The pattern of cyclin-A mRNA accumulation in both HCMV- and mock-infected cells correlated well with the protein levels described above. We observed that the amount of cyclin-A mRNA in HCMV-infected cells at 24 h p.i. was greatly reduced relative to that in mock-infected cells, and it remained at this low level until after 48 h p.i. In contrast, the levels in mock-infected cells continued to increase, resulting in a significant difference between infected and mock-infected cells at 48 h p.i. However, by 96 h p.i., an increase in cyclin-A mRNA levels was detected in infected cells, which coincided with a significant decrease in mock-infected cells. The net effect at 96 h p.i. was that the levels of cyclin-A mRNA were similar. Thus, analogous to the kinetics of cyclin-A protein accumulation, cyclin-A mRNA levels in HCMV-infected cells remained low until late in the infection. Although these data indicate that the effect of HCMV infection on cyclin-A expression is at the level of transcription, we cannot determine from these experiments whether the mRNA kinetics are a result of inhibition of RNA synthesis or enhanced RNA degradation.
Northern analysis also revealed a clear effect of the infection on cyclin-E mRNA levels. However, in this case the effect was the opposite of that observed for cyclin A. By 12 h p.i., HCMV-infected cells underwent a significant induction of cyclin-E mRNA that was sustained through 96 h p.i. The magnitude of the cyclin-E mRNA induction in infected cells was somewhat surprising in view of the Western blot analysis described above, which showed a relatively smaller increase in the levels of cyclin-E protein at 24 h p.i. and a decline in those levels after 48 h p.i. (Fig. 7). Collectively, these results indicate that during the infection there may be differential regulation of both cyclin-E mRNA accumulation and the synthesis and degradation of the protein.
Cyclin-B mRNA levels were found to be the least affected by HCMV infection. At 12 h p.i., there was slightly more cyclin-B mRNA in HCMV-infected cells than in mock-infected cells. By 24 h p.i., this difference was no longer apparent, and at 48 h p.i. there was a modest increase in cyclin-B mRNA levels in mock-infected cells. At 96 h p.i., mRNA levels were again similar. These results indicate that HCMV infection does not lead to a significant change in cyclin-B mRNA levels. However, there does appear to be a slightly earlier induction of mRNA in infected cells following release from cell cycle arrest. In accord with this, we have observed in some experiments slightly higher amounts of cyclin-B protein in infected cells at early time points. Interestingly, the cyclin-B mRNA is maintained at a similar level in both mock- and HCMV-infected cells at later time points, when the amount of cyclin-B protein in mock-infected cells declines. It is likely that the observed difference in cyclin-B protein levels at these later times is due to the degradation of cyclin B in mock-infected cells as a result of mitosis and a corresponding lack of new synthesis as cells become confluent. In contrast, this phase-specific degradation does not appear to occur in HCMV-infected cells which have undergone cell cycle arrest. These results suggest that the sustained levels of cyclin-B protein in infected cells are primarily the result of effects on the rate of protein degradation rather than transcription.
DISCUSSION
Viruses have evolved a variety of mechanisms to alter cellular regulatory functions to enhance their own replication. In the case of HCMV, the virus brings about changes in cellular metabolism that are usually associated with the signaling pathways leading to cell proliferation (2, 5). However, in activating the host cell to allow optimal viral replication, HCMV infection alters the expression of key cell cycle-regulatory proteins, which ultimately leads to cell cycle arrest (7, 18, 23, 26, 34). In this report, we present data which provide insight into the nature of the virus-host interactions that culminate in cell cycle arrest as well as the molecular mechanisms underlying the marked effects on the cyclins.
The current view of normal cell cycle progression is that cellular DNA synthesis requires cyclin-A expression, which is regulated at the G1/S boundary at the level of transcription (22). Our findings support this model. In G0-infected cells, DNA replication could not be detected during the first 48 h p.i., and this lack of cellular DNA synthesis correlated with the absence of cyclin-A mRNA and protein. Interestingly, we observed that there was a modest increase in cyclin-A mRNA and protein levels in the infected cells after 48 h p.i., independent of the cell cycle phase in which the infection occurred. Previously, we showed that cyclin-A-associated kinase activity also undergoes a similar increase only after 48 h p.i. (23). Since the peak of viral DNA replication occurs after 48 h p.i., it is possible that these two events are linked, and studies are in progress to address this question. The prolonged nature of the HCMV life cycle may make the virus vulnerable to competition from normal cellular DNA synthesis, which would likely occur before HCMV is able to initiate its own replication. By delaying host DNA synthesis, HCMV may be forcing the cell to compete for replication machinery and nucleotides later in the infection, when the levels of viral gene products are optimal for viral DNA replication. This may also account for the delay in the initiation of viral gene expression when cells are already engaged in DNA replication at the time of infection.
In direct contrast to the effects on cyclin A, we previously showed that HCMV infection induces cyclin E and its associated kinase activity to high levels at 24 h p.i. (23). Bresnahan et al. have also reported that translocation of cdk2 into the nucleus and the induction of cyclin E require viral gene expression, with inhibition of cellular cdk2 activity causing inhibition of HCMV replication (6–8). In this study, we demonstrate that cells infected in G0, G1, or S phase produce high levels of a second, smaller isoform of cyclin E. The significance of the second isoform is unknown, although it is clear that mock-infected cells also express this protein, albeit at significantly lower levels. Moreover, Ohtsubo et al. have presented evidence that both forms of cyclin E, which differ in length only by 15 amino acids at the amino terminus, are functional (31). Taken together, these data support the hypothesis that the up-regulation of cyclin E is an important mechanism by which the virus facilitates its replication.
The induction of the cyclin-E gene by HCMV is most striking at the mRNA level. By 12 h p.i., the amount of cyclin-E mRNA was considerably greater in infected cells than in mock-infected cells, and this large difference between mock-infected and infected cells was maintained throughout the entire infection. These results suggest that induction of cyclin-E transcription occurs relatively early in the infection. However, induction of cyclin-E mRNA is only partially reflected at the protein level. In fact, after 36 h p.i., the induced smaller isoform of cyclin-E protein is barely detectable in both infected and mock-infected cells. In addition, the larger isoform decreases in infected cells to levels below those seen in mock-infected cells. Cyclin-E-associated kinase activity also begins to decrease after its maximum is reached at 24 h p.i. (23). The underlying regulation which creates the observed differential between cyclin-E mRNA and protein expression is currently under investigation. Nevertheless, it would appear that the virus requires cyclin-E function primarily during the first 24 to 36 h p.i., when the cyclin-E protein level and associated kinase activity are at their highest. The high levels of cyclin-E-associated kinase activity, coupled with previously observed hyperphosphorylation of Rb, support the notion that the virus has promoted many of the molecular events associated with the G1/S transition in a normally cycling cell (4, 6–8, 23). Therefore, the virus likely requires these events to prime the cellular environment to a state most advantageous for the ensuing viral replication.
A key question that emerges from these studies is how the HCMV infection imposes such striking and diametrically opposed effects on the cyclin-E and cyclin-A mRNAs. In uninfected cells, the induction of transcription of cyclin-E and cyclin-A mRNAs are cell cycle dependent, with activation of the cyclin-E promoter occurring during G1 and activation of the cyclin-A promoter occurring as the cells enter S phase. Both promoters contain several transcription factor binding sites, but it appears that a sequence with similarity to the binding site for the transcription factor E2F most likely is responsible for the cell cycle-dependent regulation of both genes (13, 20, 22, 30, 36). In the case of the cyclin-E promoter, there are 5 E2F sites and 12 Sp-1 sites located between nucleotides −380 and +30 (20, 30). The cyclin-A promoter appears to contain a variant E2F site (referred to as the cell cycle-dependent element, or CDE), as well as a 5-bp region (cell cycle genes homology region, or CHR) located 6 nucleotides 3′ to the CDE, which are both important for the cell cycle-dependent regulation (40). The cyclin-A promoter also contains several other upstream transcription factor binding sites, including NF-Y, Sp-1, and ATF/CREB, that may contribute to its regulation (17, 22). At present, we do not know whether the HCMV infection mediates its effect on the cyclin-E and cyclin-A mRNA levels through any of these sites. The issue can be critically addressed only through a careful analysis of the effects of the infection on specific transcription factors and their interaction with the endogenous promoters in the context of host cell chromatin, and these studies are currently in progress.
In contrast to the clear effects that the HCMV infection has on the synthesis of cyclin-E and cyclin-A mRNAs, only modest changes in the levels of cyclin-B mRNA were observed. As previously reported, HCMV infection did not significantly alter the kinetics of cyclin-B protein expression (23). Moderate levels of cyclin B were maintained throughout the infection, even at late time points, when the cyclin was no longer detectable in mock-infected cells. This pattern of sustained cyclin-B protein expression was observed regardless of the phase of the cell cycle in which the infection occurred. Taken together, these data indicate that HCMV infection has only a small effect on cyclin-B transcription, although there may be a slightly earlier induction of transcription when cells are infected immediately after release from growth arrest. We propose that the maintenance of cyclin-B protein in HCMV-infected cells is likely due to the arrest of the cell cycle and the lack of the periodic degradation of cyclin B by the proteasome, which normally occurs during mitosis.
Our finding that HCMV infection imposes an inhibitory effect on cyclin-A mRNA and protein expression but allows cyclin B to be expressed at apparently normal levels is somewhat surprising, considering that during the normal cell cycle, cyclin B is strictly expressed after cyclin A. Cyclin B begins to accumulate late in S phase and forms an inactive complex with cdc2 kinase due to the presence of inhibitory phosphates on cdc2. At the G2/M transition, a sophisticated network of kinases and phosphatases converge to activate the cyclin B-cdc2 complex, which in turn phosphorylates the substrates that presumably are required for rapid entry of the cell into mitosis (24). The fact that cyclin B is in an active kinase complex during the HCMV infection indicates that many of the upstream components of this regulatory pathway normally associated with the S and G2 phases of the cell cycle are also functioning (23, 24). Thus, to optimize its own replication, it appears that the virus has selectively usurped a number of activities associated with different phases of the normal cell cycle regardless of the expense for the host. It is clear that the net result of this dysregulation is the loss of cell cycle progression and cell division. However, the above observations highlight the difficulty in defining the point of HCMV-mediated cell cycle arrest in terms of the conventional cell cycle phases associated with normally dividing cells.
An interesting result of this study was the observation that progression of the HCMV-induced CPE is dependent on the cell cycle phase at the beginning of the infection. We found that a significantly lower percentage of cells infected in S phase were rounded by 12 h p.i. compared with G0- or even G1-infected cells. It was clear that this phenomenon was not due to a variable number of infectious particles in the inoculum, since by 36 h p.i., virtually all cells displayed characteristic CPE regardless of the timing of infection. Furthermore, even when the infecting inoculum was removed from S-phase cells after 2 h of incubation, practically all of the cells showed the presence of the HCMV pp65 tegument protein in the nucleus at 12 h p.i., indicating that viral entry was not affected. Therefore, it appeared that following viral entry, the CPE represented at least one early event in the viral life cycle which was delayed but not inhibited by S-phase infection.
In accord with the above observations, we found that viral IE and early proteins accumulated slowly during the first 24 to 36 h p.i. in S-phase-infected cells. Particularly striking were the immunofluorescence data, which indicated that fewer cells infected in S phase contain IE protein at 12 h p.i. compared to G0-infected cells. Double staining of cells with Ab against BrdU and the HCMV IE protein demonstrated that viral protein expression in cells which have initiated DNA synthesis at the time of infection is preferentially delayed at 12 h p.i. In addition, approximately 20% of cells were observed to be double negative for BrdU and IE protein expression at 12 h.p.i. Since viral entry was not affected, we propose that these double-negative cells were either in G2/M phase or far enough into the G1 phase of the cell cycle and past a putative restriction point at the time of infection to produce a delay in viral gene expression. However, it is possible that these double-negative cells may have delayed the infection by a mechanism independent of the cells’ proximity to S phase. Nevertheless, by 36 h p.i., all of the cells infected in S phase were clearly expressing viral gene products and showing CPE. This indicates that the S-phase-dependent delays in viral protein production and CPE do not translate into a significant change for the long-term progression of the infection. In addition, viral titers of supernatants from G0-, G1-, and S-phase infections displayed comparable growth curves (data not shown).
Collectively, the above results demonstrate that when cells are infected in S phase, there is a lag in the progression of the viral life cycle that is associated with a significant fraction of the cells progressing through S and G2/M. It is possible that the intracellular environment during active DNA synthesis is not favorable for the early stages of viral replication. As a result, a delay of the infection may be required until cells complete S phase and reach a point in the cell cycle where conditions may be more favorable for HCMV gene expression. In fact, early studies by DeMarchi and Kaplan led them to propose that (i) cellular DNA replication and efficient synthesis of viral proteins were mutually exclusive events and (ii) an early gene product expressed in cells fully permissive for the infection was required for this apparent inhibition of cell DNA synthesis (14–16). Our data are also consistent with the hypothesis that viral proteins expressed early in the infection may need to reach a certain threshold for cellular DNA synthesis to be delayed. Alternatively, cells which are infected beyond a certain point in the cell cycle may remain committed to DNA replication and continue to progress through the cycle for some period of time before arresting. In this scenario, such progression may be associated with a delay in viral gene expression. In accord with this, Lu and Shenk have recently reported that in an asynchronous population of growing cells, there was not an immediate halt in the cell cycle, and cell cycle progression was inhibited at multiple points (26).
The studies presented here, coupled with previous work from our lab and others, demonstrate that HCMV brings about dramatic and temporally ordered changes in the expression of host cell regulatory proteins, leading to arrest of the cell cycle. We now must determine the relationship between these alterations in cell cycle proteins and viral replication, as well as the mechanisms governing the delay in viral gene expression when cells are infected in S phase. Our hope is that the knowledge gained from these studies not only will contribute to our understanding of the processes by which HCMV causes disease but also will provide additional insight into the complex pathways involved in cell cycle regulation.
ACKNOWLEDGMENTS
We thank Maziar Younessian for the production of the Ab against the HCMV 2.2-kb proteins (UL112-113). In addition, we thank Chuck Clark and Irene Smith, whose assistance was instrumental in the timely completion of this work. Finally, we thank Anita McElroy, Christopher Morello, and Steve Rodems for helpful discussions during the course of this research as well as the critical reading of the manuscript.
This work was supported by NIH grant CA34729 and a grant from the University of California Cancer Research coordinating committee. B.S.S. was supported by NIH training grant T32 GMO7240-22.
REFERENCES
- 1.AbuBakar S, Au W W, Legator M S, Albrecht T. Induction of chromosome aberrations and mitotic arrest by cytomegalovirus in human cells. Environ Mol Mutagen. 1988;12:409–420. doi: 10.1002/em.2860120409. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht T, Boldogh I, Fons M, Lee C H, AbuBakar S, Russell J M, Au W W. Cell-activation responses to cytomegalovirus infection: relationship to the phasing of CMV replication and to the induction of cellular damage. Subcell Biochem. 1989;15:157–202. [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 4.Beijersbergen R L, Bernards R. Cell cycle regulation by the retinoblastoma family of growth inhibitory proteins. Biochim Biophys Acta. 1996;1287:103–120. doi: 10.1016/0304-419x(96)00002-9. [DOI] [PubMed] [Google Scholar]
- 5.Boldogh I, AbuBakar S, Deng C Z, Albrecht T. Transcriptional activation of cellular oncogenes fos, jun, and myc by human cytomegalovirus. J Virol. 1991;65:1568–1571. doi: 10.1128/jvi.65.3.1568-1571.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnahan W A, Boldogh I, Chi P, Thompson E A, Albrecht T. Inhibition of cellular Cdk2 activity blocks human cytomegalovirus replication. Virology. 1997;231:239–247. doi: 10.1006/viro.1997.8489. [DOI] [PubMed] [Google Scholar]
- 7.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 8.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Human cytomegalovirus infection results in altered cdk2 subcellular localization. J Gen Virol. 1997;78:1993–1997. doi: 10.1099/0022-1317-78-8-1993. [DOI] [PubMed] [Google Scholar]
- 9.Britt W, Alford C. Cytomegalovirus. In: Fields B N, et al., editors. Fields Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523. [Google Scholar]
- 10.Corbeil J, Richman D D. Productive infection and subsequent interaction of CD4-gp120 at cellular membrane required for HIV-induced apoptosis of CD4+ T cells. J Gen Virol. 1995;76:681–690. doi: 10.1099/0022-1317-76-3-681. [DOI] [PubMed] [Google Scholar]
- 11.Cranmer L D, Clark C, Spector D H. Cloning, characterization, and expression of the murine cytomegalovirus homologue of the human cytomegalovirus 28-kDa matrix phosphoprotein (UL99) Virology. 1994;205:417–429. doi: 10.1006/viro.1994.1662. [DOI] [PubMed] [Google Scholar]
- 12.Darzynkiewicz Z, Bruno S, Bino G D, Gorczyca W, Hotz M A, Lassota P, Traganos F. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13:795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- 13.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeMarchi J M. Correlation between stimulation of host cell DNA synthesis by human cytomegalovirus and lack of expression of a subset of early virus gene. Virology. 1983;129:274–286. doi: 10.1016/0042-6822(83)90167-8. [DOI] [PubMed] [Google Scholar]
- 15.DeMarchi J M, Kaplan A S. Replication of human cytomegalovirus DNA: lack of dependence on cell DNA synthesis. J Virol. 1976;18:1063–1070. doi: 10.1128/jvi.18.3.1063-1070.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeMarchi J M, Kaplan A S. Physiological state of human embryonic lung cells affects their response to human cytomegalovirus. J Virol. 1977;23:126–132. doi: 10.1128/jvi.23.1.126-132.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desdouets C, Matesic G, Molina C A, Foulkes N S, Sassone-Corsi P, Brechot C, Sobszak-Thepot J. Cell cycle regulation of cyclin A gene expression by the cyclic AMP transcription factors CREB and CREM. Mol Cell Biol. 1995;15:3301–3309. doi: 10.1128/mcb.15.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enoch T, Nurse P. Coupling M phase and S phase: controls maintaining the dependence of mitosis on chromosome replication. Cell. 1991;65:921–923. doi: 10.1016/0092-8674(91)90542-7. [DOI] [PubMed] [Google Scholar]
- 20.Geng Y, Eaton E N, Picon M, Roberts J M, Lundberg A S, Gifford A, Sardet C, Weinberg R A. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- 21.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 22.Henglein B, Chenivesse X, Wang J, Eick D, Brechot C. Structure and cell cycle-regulated transcription of the human cyclin A gene. Proc Natl Acad Sci USA. 1994;91:5490–5494. doi: 10.1073/pnas.91.12.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jault F M, Jault J-M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated RB, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King R W, Jackson P K, Kirschner M W. Mitosis in transition. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 25.Klucher K M, Sommer M, Kadonaga J T, Spector D H. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol Cell Biol. 1993;13:1238–1250. doi: 10.1128/mcb.13.2.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu M, Shenk T. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McElroy, A., and D. H. Spector. 1997. Unpublished data.
- 28.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, et al., editors. Fields Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 29.Muganda P, Mendoza O, Hernandez J, Qian Q. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J Virol. 1994;68:8028–8034. doi: 10.1128/jvi.68.12.8028-8034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohtani K, Degregori J, Nevins J R. Regulation of the cyclin E promoter by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pines J. Cyclins and cyclin-dependent kinases: take your partners. Trends Biochem Sci. 1993;18:195–197. doi: 10.1016/0968-0004(93)90185-p. [DOI] [PubMed] [Google Scholar]
- 33.Pines J. Cyclins and cyclin-dependent kinases: theme and variations. Adv Cancer Res. 1995;66:181–212. doi: 10.1016/s0065-230x(08)60254-7. [DOI] [PubMed] [Google Scholar]
- 34.Poma E E, Kowalik T F, Zhu L, Sinclair J H, Huang E-S. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J Virol. 1996;70:7867–7877. doi: 10.1128/jvi.70.11.7867-7877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy G P. Cell cycle: regulatory events in G1→S transition of mammalian cells. J Cell Biochem. 1994;54:379–386. doi: 10.1002/jcb.240540404. [DOI] [PubMed] [Google Scholar]
- 36.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherr C J. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 38.Speir E, Modali R, Huang E-S, Leon M B, Shawl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 39.Telford W G, King L E, Fraker P J. Comparative evaluation of several DNA binding dyes in the detection of apoptosis-associated chromatin degradation by flow cytometry. Cytometry. 1992;13:137–143. doi: 10.1002/cyto.990130205. [DOI] [PubMed] [Google Scholar]
- 40.Zwicker J, Lucibello F C, Wolfraim L A, Gross C, Truss M, Engeland K, Muller R. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]