Abstract
Artificial intelligence (AI) now enables automated interpretation of medical images. However, AI’s potential use for interventional image analysis remains largely untapped. This is because the post hoc analysis of data collected during live procedures has fundamental and practical limitations, including ethical considerations, expense, scalability, data integrity and a lack of ground truth. Here we demonstrate that creating realistic simulated images from human models is a viable alternative and complement to large-scale in situ data collection. We show that training AI image analysis models on realistically synthesized data, combined with contemporary domain generalization techniques, results in machine learning models that on real data perform comparably to models trained on a precisely matched real data training set. We find that our model transfer paradigm for X-ray image analysis, which we refer to as SyntheX, can even outperform real-data-trained models due to the effectiveness of training on a larger dataset. SyntheX provides an opportunity to markedly accelerate the conception, design and evaluation of X-ray-based intelligent systems. In addition, SyntheX provides the opportunity to test novel instrumentation, design complementary surgical approaches, and envision novel techniques that improve outcomes, save time or mitigate human error, free from the ethical and practical considerations of live human data collection.
Advances in robotics and artificial intelligence (AI) are bringing autonomous surgical systems closer to reality. However, developing the AI backbones of such systems currently depends on collecting training data during routine surgeries. This remains one of the largest barriers to widespread use of AI systems in interventional clinical settings, versus triage or diagnostic settings, as the acquisition and annotation of interventional data is time intensive and costly. Furthermore, while this approach can contribute to the automation or streamlining of existing surgical workflows, robotic and autonomous systems promise even more substantial advances: novel and super-human techniques that improve outcomes, save time or mitigate human error. This is perhaps the most exciting frontier of computer-assisted intervention research.
Conventional approaches for curating data for AI development (that is, sourcing it retroactively from clinical practice) are insufficient for training AI models that benefit interventions that use novel instrumentation, different access points or more flexible imaging. This is because they are, by definition, incompatible with contemporary clinical practices and such data do not emerge from routine care. Furthermore, these novel systems are not readily approved, and thus not easily or quickly introduced into clinical practice. Ex vivo experimentation does not suffer the same ethical constraints; however, it is costly and requires mature prototypes, and therefore does not scale well.
A promising alternative to these strategies is simulation, that is, the in silico generation of synthetic interventional training data and imagery from human models. Simulation offers a rich environment for training both human and machine surgeons alike, and sidesteps ethical considerations that arise when exploring procedures outside the standard of care. Perhaps most importantly, in silico surgical sandboxes enable rapid prototyping during the research phase. Simulation paradigms are inexpensive, scalable and rich with information. While intra-operative data are generated in highly unstructured and uncontrolled environments, and require manual annotation, simulation can provide detailed ground-truth data for every element of the procedure, including tool and anatomy pose, which are invaluable for AI development.
However, simulations can fall short of real surgery in one key aspect: realism. The difference in characteristics between real and simulated data is commonly referred to as the ‘domain gap’. The ability of an AI model to perform on data from a different domain, that is, with a domain gap from the data it was trained on, is called ‘domain generalization’. Domain gaps are problematic because of the well-documented brittleness of AI systems1, which exhibit vastly deteriorated performance across domain gaps. This may happen even with simple differences, such as noise statistics, contrast level and other minutiae2–5. This unfortunate circumstance, which applies to all machine learning tasks, has motivated research in the AI field on simulation-to-reality (Sim2Real) transfer, the development of domain transfer methods.
In this Article, we present SyntheX, a framework for developing generalizable AI algorithms for X-ray image analysis solely based on synthetic data simulated from annotated computed tomography (CT). Using realistic simulation of X-ray image formation from CT and using domain randomization to train AI models, SyntheX creates AI models that retain their performance under domain shift, enabling evaluation and deployment on clinical X-rays acquired in the real world. The overall concept of SyntheX is illustrated in Fig. 1 and we demonstrate its utility and validity on three clinical applications: hip imaging, surgical robotic tool detection and coronavirus disease 2019 (COVID-19) lesion segmentation.
Fig. 1 |. Overall concept of SyntheX.
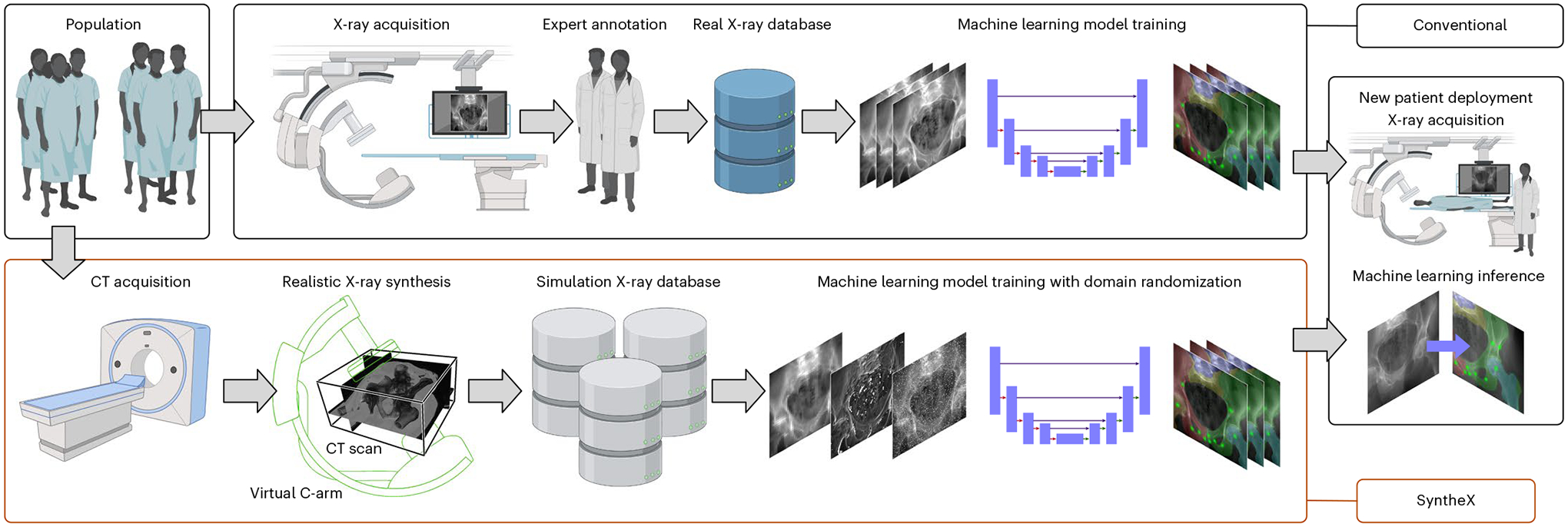
Top: conventional approach for learning-based tasks on medical imaging. Curating a relevant database of real X-ray samples requires real-data acquisition and costly annotation from domain experts. Bottom: SyntheX enables simplified and scaled-up data curation because data generation is synthetic and synthesized data can be annotated automatically through propagation from the 3D model, which can be CT scans or volumetric surgical tool models. SyntheX results in deep learning image analysis models that perform comparably to or better than real-data-trained models. Figure created with Biorender.com.
At the core of our report is an experiment on precisely controlled data from the hip-imaging task that isolates and quantifies the effect of domain shift for AI-based X-ray image analysis. Using CT images from human cadavers and corresponding C-arm X-ray images acquired from two different imaging systems during surgical exploration, we generated a hip-image dataset consisting of geometrically identical images across various synthetic and the real domains to train AI models for hip-image analysis. To our knowledge, no study so far has isolated the effect of domain generalization using precisely matched datasets across domains. This work also demonstrates a feasible and cost-effective way to train AI image analysis models for clinical intervention on synthetic data in a way that provides comparable performance to training on real clinical data in multiple applications. We also demonstrate that the model’s performance increases substantially as the number of synthetic training samples increases, which high-lights the key advantage of SyntheX: making available large amounts of well-annotated data for model training or pre-training.
Clinical tasks
We demonstrate the benefits of SyntheX on three X-ray image analysis downstream tasks: hip imaging, surgical robotic tool detection and COVID-19 lesion segmentation in chest X-ray (Fig. 2). All of the three tasks use deep neural networks to make clinically meaningful predictions on X-ray images. We introduce the clinical motivations for each task in the following sections. Details of the deep network and training/evaluation paradigm are described in ‘Model and evaluation paradigm’.
Fig. 2 |. Clinical tasks.
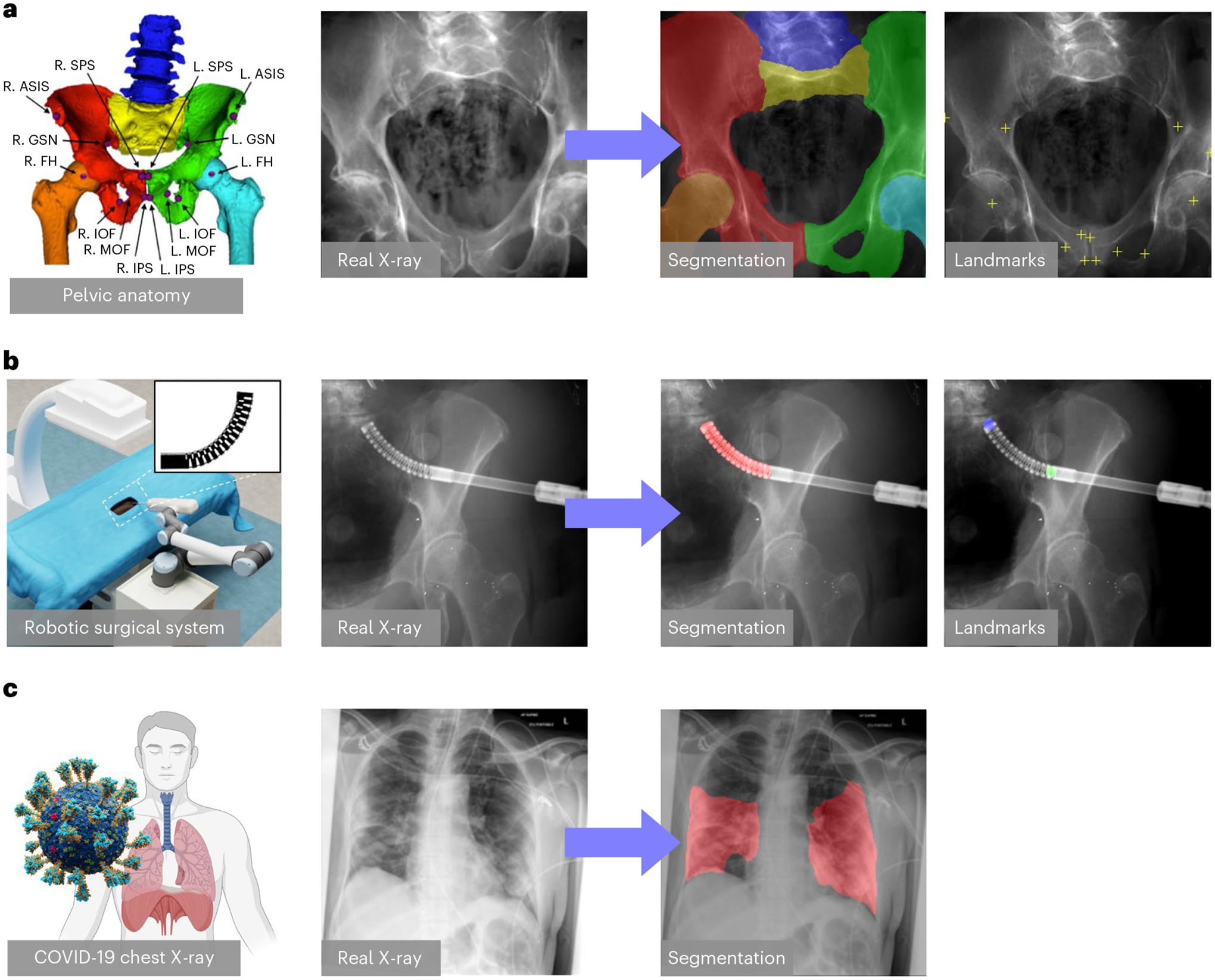
a, Hip imaging. The hip anatomical structures include left and right hemipelvis, lumbar vertebrae, upper sacrum, and left and right femurs, which are illustrated by different colours in the leftmost hip rendering. The anatomical landmarks consist of left (L.) and right (R.) anterior superior iliac spine (ASIS), centre of femoral head (FH), superior pubic symphysis (SPS), inferior pubic symphysis (IPS), medial obturator foramen (MOF), inferior obturator foramen (IOF) and the greater sciatic notch (GSN). These landmarks are useful in identifying the anterior pelvic plane and initializing the 2D/3D registration of both pelvis and femur81,82. b, Surgical robotic tool detection. An illustration of the image-guided robotic surgical system is shown on the left. A picture of the continuum manipulator (CM) is shown in the top right corner. An example real X-ray image and the corresponding segmentation and landmarks of the CM is shown on the right. c, COVID-19 CXR lesion segmentation. A real CXR image of COVID-19 infection is shown with its lesion segmentation mask.
Hip imaging
Computer-assisted surgical systems for X-ray-based image guidance have been developed for trauma surgery6, total hip arthroplasty7, knee surgery8, femoroplasty9, pelvis osteotomy10 and spine surgery11. The main challenge in these procedures is to facilitate intra-operative image-based navigation by continually recovering the spatial tool-to-tissue relationships from two-dimensional (2D) transmission X-ray images. One effective approach to achieving spatial alignment is the identification of known structures and landmarks in the 2D X-ray image, which then are used to infer poses12,13.
In the context of hip imaging, we define six anatomical structures and fourteen landmarks as the most relevant known structures. They are shown in Fig. 2a. We trained deep networks using SyntheX to make these detections on X-ray images. Synthetic images were generated using CT scans selected from the New Mexico Decedent Image Database14. The three-dimensional (3D) anatomical landmarks were manually annotated and the anatomical structures were segmented using the automatic method described in ref.15, which were then projected to 2D as labels following the simulation X-ray geometries. We evaluate the performance of our model on 366 real X-ray images collected from 6 cadaveric specimens using the Siemens CIOS Fusion imaging system and another 60 real X-ray images from a separate cadaveric specimen using the BrainLab LoopX imaging system. On real images, ground-truth target structures were annotated semi-automatically. This real dataset also serves as the basis for our precisely controlled experiments that isolate the effect of the domain gap. We provide substantially more details on the creation, annotation and synthetic duplication of this dataset in ‘Precisely matched hip dataset’.
Surgical robotic tool detection
Automatic detection of the surgical tool from intra-operative images is an important step for robot-assisted surgery as it enables vision-based control16. Because training a detection model requires sufficient image data with ground-truth labels, developing such models is possible only after the surgical robot is mature and deployed clinically. We demonstrate AI model development for custom and pre-clinical surgical robotic tools.
We consider a continuum manipulator (CM) as the target object. CMs have been investigated in minimally invasive robot-assisted orthopaedic procedures because of their substantial dexterity and stiffness17,18, but they are not currently used clinically nor easily manufactured for extensive cadaveric testing. Using SyntheX, we address CM detection, which consists of segmenting the CM body and predicting distinct landmarks in the X-ray images. The semantic segmentation mask covers the 27 alternating notches that discern the CM from the other surgical tools; the landmarks are defined as the start and end points of the CM centreline19. Synthetic images were generated using CT scans selected from the New Mexico Decedent Image Database14 and a computer-aided design model of the CM. Three-dimensional CM segmentations and landmark locations were determined through forward kinematics and then projected to 2D as training labels using the X-ray geometry. The performance was evaluated on 264 real X-ray images of the CM during pre-clinical cadaveric testing. These images were acquired at different scenarios, including different cadaver specimens, with or without drilling tool inserted, positions of the tool, and multiple scanner acquisition settings. We present example simulation and real X-ray images in Extended Data Fig. 1. On real images, ground-truth segmentation masks and landmark locations were annotated manually.
COVID-19 lesion segmentation
Chest X-ray (CXR) has emerged as a major tool to assist in COVID-19 diagnosis and guide treatment. Numerous studies have proposed the use of AI models for COVID-19 diagnosis from CXR and efforts to collect and annotate large amounts of CXR images are underway. Annotating these images in 2D is expensive and fundamentally limited in its accuracy due to the integrative nature of X-ray transmission imaging. While localizing COVID-19 presence is possible, deriving quantitative CXR analysis solely from CXR images is impossible. Given the availability of CT scans of patients suffering from COVID-19, we demonstrate lung-imaging applications using SyntheX.
Specifically, we consider the task of COVID-19 lesion segmentation, which is possible also from CXR to enable comparison. We used the open-source COVID-19 CT dataset released by ImagEng lab20 and the CT scans released by the University of Electronic Science and Technology of China (UESTC)21 to generate synthetic CXR images. A 3D infection mask was created for each CT using the automatic lesion segmentation method COPLE-Net21. We followed the same realistic X-ray synthesis pipeline and generated synthetic images and labels using the paired CT scan and segmentation mask from various geometries. The lesion labels were projected following the same geometries. The segmentation performance was tested on the benchmark dataset QaTa-COV1922, which contains 2,951 real COVID-19 CXR samples. Ground-truth segmentation masks for the COVID-19 lesions in these CXR images are supplied with the benchmark, and were created in a human–machine collaborative approach.
Precisely controlled investigations on hip imaging
Beyond presenting SyntheX for various clinical tasks, we present experiments on a unique dataset for hip imaging that enables the isolation of the effect that the domain gap has on Sim2Real AI model transfer. On the task of anatomical landmark detection and anatomy segmentation in hip X-ray, we study the most commonly used domain generalization techniques, namely, domain randomization and domain adaptation, and further consider different X-ray simulators, image resolution and training dataset size. We introduce details on these experiments next.
Precisely matched hip dataset
We created an accurately annotated dataset of 366 real hip fluoroscopic images and corresponding high-resolution CT scans of six lower torso cadaveric specimens with manual label annotations12, which constitutes the basis of our unique dataset that enables precisely controlled benchmarking of domain shift. For each of the real X-ray images, the X-ray camera pose was accurately estimated using a comprehensive 2D/3D image registration pipeline12. We then generated synthetic X-ray images (digitally reconstructed radiographs (DRRs)) that precisely recreate the spatial configurations and anatomy of the real X-ray images and differ in only the realism of the simulation (Fig. 3a). Because synthetic images precisely match the real dataset, all labels in 2D and 3D apply equally. Details of the dataset creation are introduced in ‘Benchmark hip-imaging investigation’.
Fig. 3 |. Precisely controlled hip-imaging X-ray database.
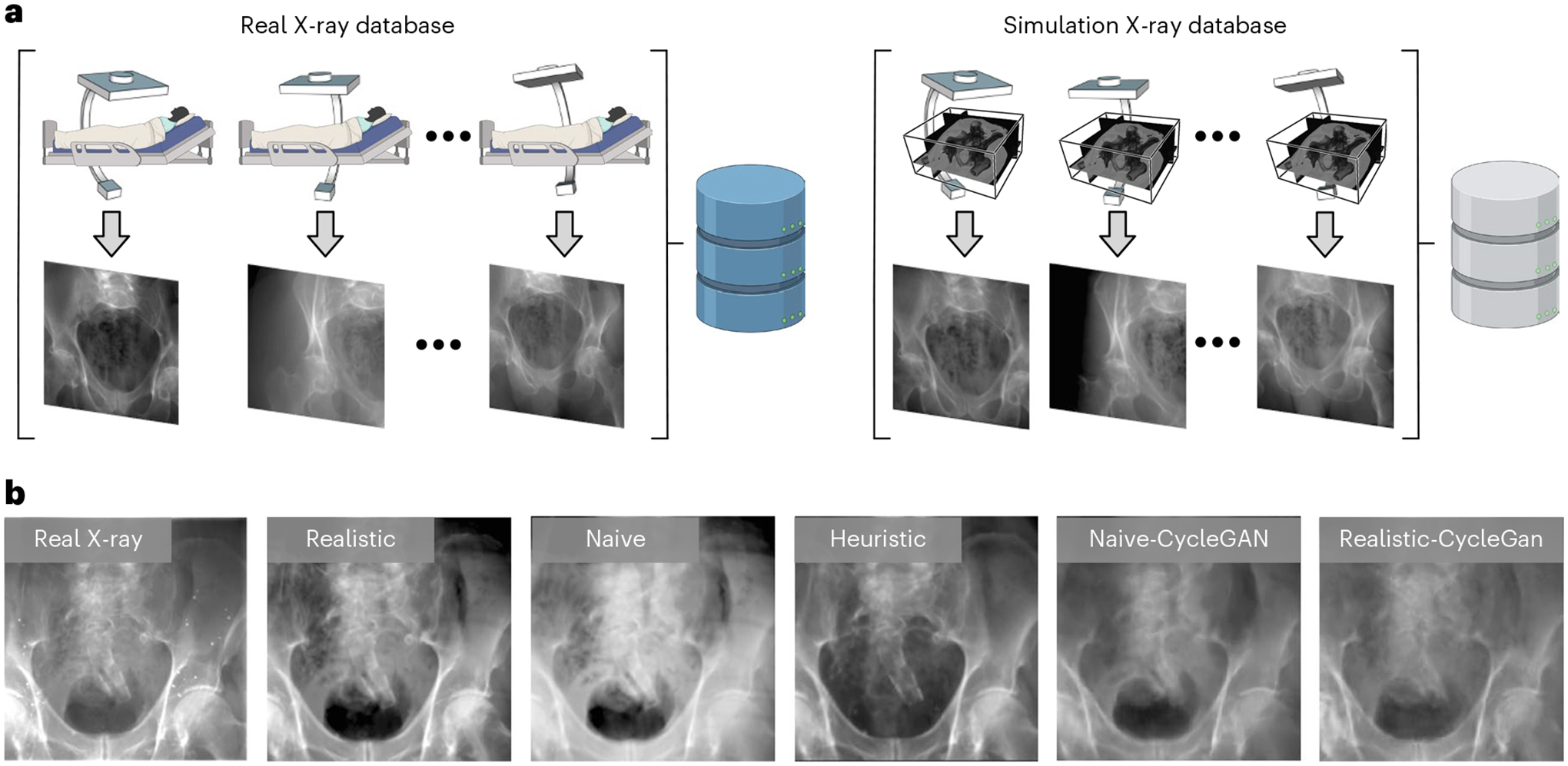
a, Generation of precisely matched synthetic and real X-ray database. Real X-rays and CT scans are acquired from cadaveric specimens and registered to obtain the relative camera poses. Using these poses, synthetic X-rays can be generated from the CT scans that precisely match the real X-ray data in all aspects but appearance. b, Changes in (synthetic) X-ray appearance based on simulation paradigm.
We studied three different X-ray image simulation techniques: naive DRR generation, xreg DRR10 and DeepDRR23,24, which we refer to as naive, heuristic and realistic simulations. They differ in the considerations of modelling realistic X-ray imaging physical effects. Figure 3b shows a comparison of image appearance between the different simulators and a corresponding real X-ray image.
We have collected data on an additional lower torso cadaveric specimen using the Brainlab LoopX imaging system, which is different from the Siemens CIOS Fusion C-arm system for collecting the 366 controlled study data. High-resolution CT scans of the specimen were acquired. We collected 60 X-ray images of the cadaveric specimen to test our model’s generalization performance. These data differ from all images previously used in the controlled investigations for training and testing in regards to anatomy, acquisition protocol and X-ray scanner characteristics. We performed the same 2D/3D image registration pipeline and generated 2D segmentation and landmark labels.
Domain randomization and adaptation
Domain randomization is a domain generalization technique that inflicts marked changes on the appearance of the input images. This produces training samples with markedly altered appearance, which forces the network to discover more robust associations between input image features and desired target. These more robust associations have been demonstrated to improve the generalization of machine learning models when transferred from one domain to another (here, from simulated to real X-ray images, respectively). We implemented two levels of domain randomization effects, namely, regular domain randomization and strong domain randomization. Details are described in ‘Domain randomization’.
Other than domain randomization, which does not assume knowledge or sampling of the target domain at training time, domain adaptation techniques attempt to mitigate the domain gap’s detrimental effect by aligning features across the source (training domain; here, simulated data) and target domain (deployment domain; here, real X-ray images). As such, domain adaptation techniques require samples from the target domain at training time. Recent domain adaptation techniques have increased the suitability of the approach for Sim2Real transfer because they now allow for the use of unlabelled data in the target domain. We conducted experiments using two common domain adaptation methods: CycleGAN, a generative adversarial network with cycle consistency25 and adversarial discriminative domain adaptation (ADDA)26. The two methods are similar in that they attempt to align properties of real and synthetic domains, and differ based on what properties they seek to align. While CycleGAN operates directly on the images, ADDA seeks to align higher-level feature representations, that is, image features after multiple convolutional neural network layers. Example CycleGAN generated images are shown in Fig. 3b. More details of CycleGAN and ADDA training are provided in ‘Domain adaptation’.
Model and evaluation paradigm
As the focus of our experiments is to demonstrate convincing Sim2Real performance, we rely on a well-established backbone network architecture, namely, TransUNet27, for all tasks. TransUNet is a state-of-the-art medical image segmentation framework, which has shown convincing performance across various tasks27. Segmentation networks for all clinical applications are trained to minimize the Dice loss (Lseg)28, which evaluates the overlap between predicted and ground-truth segmentation labels. For hip-image analysis and surgical tool detection, we adjust the TransUNet architecture as shown in Extended Data Fig. 2 to concurrently estimate landmark locations. Reference landmark locations are represented as symmetric Gaussian distributions centred on the true landmark locations (zero when the landmark is invisible). This additional prediction target is penalized using (Lld), the mean squared error between network prediction and reference landmark heatmap.
For evaluation purposes, we report the landmark accuracy as the l2 distance between predicted and ground-truth landmark positions. Further, we use the Dice score to quantitatively assess segmentation quality for hip imaging and surgical tool detection. The COVID-19 lesion segmentation performance is reported using confusion matrix metrics to enable comparison with previous work21.
For all three tasks, we report both Sim2Real and Real2Real (reality-to-reality) performances. The Sim2Real performance was computed on all testing real X-ray data. The Real2Real experiments were conducted using k-fold cross-validation, and we report the performance as an average of all testing folds. For the hip-imaging benchmark studies, we further carefully designed the evaluation paradigm in a leave-one-specimen-out fashion. For each experiment, the training and validation data consisted of all labelled images from all but one specimen while all labelled images from the remaining specimen were used as test data. The same data split was strictly preserved also for training of domain adaptation methods to avoid leakage and optimistic bias. On the scaled-up dataset, we used all synthetic images for training and evaluated on all real data in the benchmark dataset.
A specially designed assessment curvature plot is used for reporting pelvic landmark detection performance. This way of measuring landmark detection performance provides detailed information on the two desirable attributes of such an algorithm: (1) completeness and (2) precision of detected landmarks. The direct network output for each landmark prediction is a heatmap intensity image (I). To distinguish the landmark prediction confidence, we compute a normalized cross-correlation between I and the Gaussian landmark heatmap Igauss, ncc(I, Igauss)12. Landmarks are considered valid (activated) if ncc(I, Igauss) is higher than a confidence threshold, ϕ, (ncc(I, Igauss) > ϕ). The kth predicted landmark location is reported using the image coordinate of the maximum intensity pixel. Given the ground-truth location , the mean landmark detection error (eld) is reported as the average l2 distance error over all activated landmarks: , where K is the total number of activated landmarks. The ratio (p) of the activated landmarks over all landmarks is a function of ϕ. Thus, we created plots to demonstrate the relationship between eld and p, which shows the change of the error as we lower the threshold to activate more landmarks. Ideally, we would like a model to have a 0.0 mm error with a 100% activation percentage, corresponding to a measurement in the bottom right corner of the plots in Fig. 4. Following the convention in previous work12, we selected a threshold of 0.9 (ncc(I, Igauss) > 0.9) to report the numeric results for all ablation study methods in Table 1. This threshold selects the network’s confident predictions for evaluation.
Fig. 4 |. Plots of average landmark detection error with respect to activated landmark percentage.
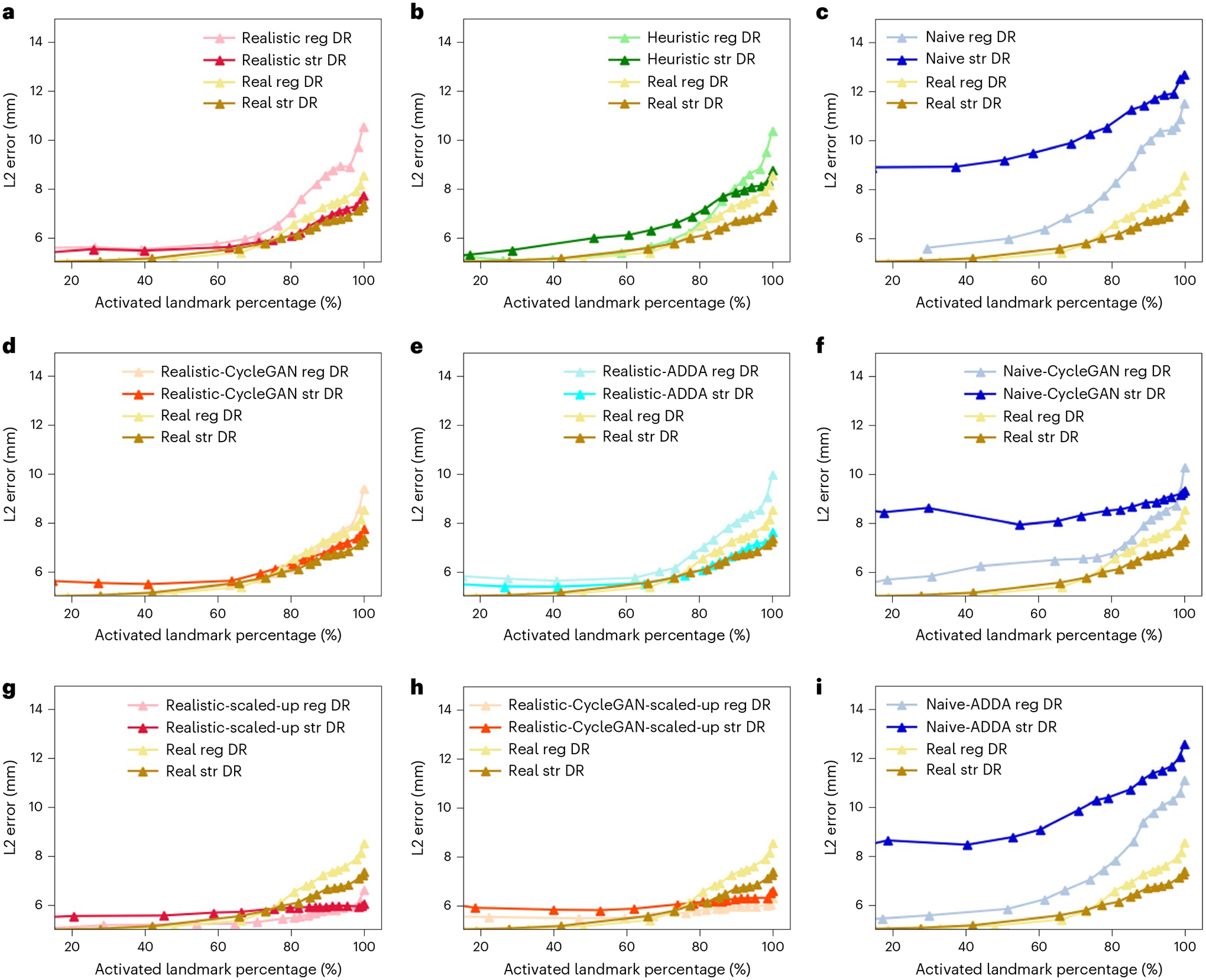
The Real2Real performance on the controlled dataset is shown in gold. An ideal curve should approach the bottom right corner: all landmarks detected with perfect localization. Each plot compares the baseline Real2Real performance curve to various Sim2Real methods that are evaluated on the same real data test set. The Sim2Real technique of the specific method is identified in the top legend of each plot. We use real, realistic, heuristic and naive to refer to the image domains with decreasing level of realism, which are defined in ‘Benchmark hip-imaging investigation’. Domain names followed by ‘CycleGAN’ mean the training data are generated using CycleGAN trained between the specific image domain and the real image domain; ‘reg DR’ and ‘str DR’ refer to regular domain randomization and strong domain randomization, respectively. a–c Performance comparison of methods trained on precisely matched datasets. d–f,i, Evaluation of the added effect of using domain adaptation techniques again using precisely matched datasets. g,h, Improvements in Sim2Real performance on the same real data test set when a larger, scaled-up synthetic training set is used. All the results correspond to input image size of 360 × 360 px.
Table 1 |.
Hip-imaging landmark detection errors and segmentation Dice scores
| Training domain | Landmark detection errors (mm) | Dice score | ||||||
|---|---|---|---|---|---|---|---|---|
| Regular DR | Strong DR | Regular DR | Strong DR | |||||
| Mean | CI | Mean | CI | Mean | CI | Mean | CI | |
| RealXray (Real2Real) | 6.90 ± 10.69 | 0.39 | 6.46 ± 8.21 | 0.30 | 0.80 ± 0.24 | 0.01 | 0.79 ± 0.25 | 0.01 |
| Realistic | 7.59 ± 13.80 | 0.51 | 6.44 ± 7.05 | 0.26 | 0.78 ± 0.25 | 0.01 | 0.80 ± 0.23 | 0.01 |
| Heuristic | 6.83 ± 9.39 | 0.35 | 7.18 ± 7.93 | 0.29 | 0.76 ± 0.27 | 0.01 | 0.79 ± 0.24 | 0.01 |
| Naive | 8.23 ± 14.18 | 0.53 | 10.50 ± 12.34 | 0.47 | 0.69 ± 0.29 | 0.01 | 0.73 ± 0.26 | 0.01 |
| Realistic-Cyc | 6.57 ± 8.22 | 0.30 | 6.62 ± 6.82 | 0.25 | 0.79 ± 0.25 | 0.01 | 0.80 ± 0.23 | 0.01 |
| Naive-Cyc | 7.35 ± 12.10 | 0.44 | 8.66 ± 13.16 | 0.48 | 0.78 ± 0.25 | 0.01 | 0.79 ± 0.23 | 0.01 |
| Realistic-ADDA | 7.33 ± 13.21 | 0.48 | 6.41 ± 6.27 | 0.23 | 0.79 ± 0.24 | 0.01 | 0.80 ± 0.23 | 0.01 |
| Naive-ADDA | 7.82 ± 13.25 | 0.49 | 10.38 ± 13.50 | 0.51 | 0.70 ± 0.29 | 0.01 | 0.73 ± 0.26 | 0.01 |
| Realistic-Scaled | 5.71 ± 4.31 | 0.16 | 5.95 ± 3.52 | 0.13 | 0.85 ± 0.23 | 0.01 | 0.86 ± 0.21 | 0.01 |
| Realistic-Cyc-Scaled | 5.88 ± 3.73 | 0.13 | 6.20 ± 3.56 | 0.13 | 0.84 ± 0.23 | 0.01 | 0.85 ± 0.21 | 0.01 |
| Realistic-Scaled (HD) | 5.19 ± 3.95 | 0.14 | 5.48 ± 3.37 | 0.12 | 0.84 ± 0.23 | 0.01 | 0.87 ± 0.20 | 0.01 |
The Landmark errors are reported at a heatmap threshold of 0.9. ALL errors are reported as a mean of sixfold individual testing on 366 real hip X-ray images. The Lower Landmark errors correspond to better performance. The Dice score ranges from 0 to 1, with larger values corresponding to better segmentation performance The best performance result is bolded. Real2Real refers to training and testing both in real domain datasets. CI refers to confidence intervals. They are computed using the 2-tailed z-test with a critical value for a 95% level of confidence (p < 0.05) DR, domain randomization; Cyc, CycleGAN; ‘-Scaled’, training on scaled-up dataset; HD, higher image resolution of 480×480 px.
Results
Primary findings
We find that across all three clinical tasks, namely, hip imaging, surgical robotic tool detection and COVID-19 lesion segmentation, models trained using the SyntheX Sim2Real model transfer paradigm when evaluated on real data perform comparably to or even better than models trained directly on real data. This finding suggests that SyntheX, that is, the realistic simulation of X-ray images from CT combined with domain randomization, is a feasible cost- and time-effective, and valuable approach to the development of learning-based X-ray image analysis algorithms that preserve performance during deployment on real data.
Hip imaging
We present the multi-task detection results of hip imaging on images with 360 × 360 px in Extended Data Tables 1 and 2. Both landmark detection and anatomical structure segmentation performance achieved using SyntheX Sim2Real model transfer are superior to those of Real2Real when considering averaged metrics. The Sim2Real predictions are more stable with respect to their standard deviation: landmark error of 3.52 mm, Dice score of 0.21, compared with 8.21 mm and 0.25, respectively, for Real2Real. We attribute this improvement to the flexibility of the SyntheX approach, providing the possibility of simulating a richer spectrum of image appearances from more hip CT samples and varied X-ray geometries compared with the limited data sourced from complex real-world experiments.
Our Sim2Real model’s performance on the 60 real X-ray images acquired by the BrainLab LoopX imaging system achieves a mean landmark detection error of 6.16 ± 5.15 mm and a Dice score of 0.84 ± 0.12, which is similar to the performance reported on the 366 Siemens real X-rays. This result suggests the strong generalization ability of the SyntheX-trained model across different imaging acquisition systems.
Considering individual anatomical landmark and structure, we have noticed that the Sim2Real detection accuracies of most landmarks are superior or comparable to Real2Real accuracies, expect for superior pubic symphysis and inferior pubic symphysis. This is potentially because the left and right positions of superior pubic symphysis and inferior pubic symphysis are very close, and thus their local geometric features are ambiguous during simulation. The Sim2Real segmentation performance is consistently better than Real2Real in all six structures. The detection accuracy of landmark ASIS and the segmentation accuracy of structure Sacrum are the worst in both Sim2Real and Real2Real, which is because the feature appearances change more drastically in varying projection geometries than the other landmarks and structures.
In addition, we particularly studied the Sim2Real performance change with respect to the number of generated training data samples. In the hip-imaging task, we generated an increasing number of scaled-up simulation images as training data using CT scans from the New Mexico Decedent Image Database14. We generated 500 synthetic X-ray images for every CT scan following the same randomized geometry distribution, and created four training datasets that contain 1,000, 2,000, 5,000 and 10,000 images. We trained the same network model using the same hyperparameters on these four datasets until convergence and reported testing performance on the 366 real hip X-ray images. The landmark performance curves are presented in Extended Data Fig. 3. Numeric results are present in Extended Data Table 3. We can clearly observe that the Sim2Real performances consistently improve as the number of training data increases.
Surgical robotic tool detection
The results of the surgical tool detection task are summarized in Extended Data Tables 4 and 5. The landmark detection errors of Sim2Real and Real2Real are comparable to a mean localization accuracy of 1.10 mm and 1.19 mm, respectively. However, the standard deviation of the Sim2Real error is substantially smaller: 0.88 mm versus 2.49 mm. Further, with respect to segmentation Dice score, Sim2Real outperforms Real2Real by a large margin achieving a mean Dice score of 0.92 ± 0.07 compared with 0.41 ± 0.23, respectively. Overall, the results suggest that SyntheX is a viable approach to developing deep neural networks for this task, especially when the robotic hardware is in the prototypic stages.
COVID-19 lesion segmentation
The results of COVID-19 lesion segmentation are presented in Extended Data Table 6. The overall mean accuracy of SyntheX training reaches 85.03% compared with 93.95% for the real data training. The Sim2Real performance is similar to Real2Real in terms of sensitivity and specificity, but falls short in the other metrics. As the 3D CT scans for training X-ray image generation were from different patients compared with the real X-rays and the lesion annotations were performed by different expert clinicians, there is an inconsistency in the lesion appearance between training data and real X-ray data, which potentially causes the performance deterioration. Similar effects have previously been reported for related tasks, such as lung nodule detection29 and thoracic disease classification30. The results suggest that SyntheX is capable of handling soft tissue-based tasks, such as COVID-19 lesion segmentation.
Sim2Real benchmark findings
On the basis of our precisely controlled hip-imaging ablation studies, including comparisons of (1) simulation environment, (2) domain randomization and domain adaptation effects, and (3) image resolution, we observed that training using realistic simulation with strong domain randomization performs on a par with models trained on real data or models trained on synthetic data but with domain adaptation, yet, does not require any real data at training time. Training using realistic simulation consistently outperformed naive or heuristic simulations. The above findings can be observed in Fig. 4 and Table 1, where the model trained with realistic simulation achieved a mean landmark detection error of 6.44 ± 7.05 mm, and a mean Dice score of 0.80 ± 0.23. The mean landmark and segmentation results of the Real2Real and realistic-CycleGAN models were 6.46 ± 8.21 mm, 0.79 ± 0.25, and 6.62 ± 6.82 mm, 0.80 ± 0.23, respectively. The mean landmark errors of heuristic and naive models were all above 7 mm, and their mean Dice scores were all below 0.80. Training using scaled-up realistic simulation data with domain randomization achieved the best performance on this task, even outperforming real data-trained models due to the effectiveness of larger training data. The best performance results are highlighted in Table 1. Thus, realistic simulation of X-ray images from CT combined with domain randomization, which we refer to as the SyntheX model transfer concept, is a most promising approach to catalyse learning-based X-ray image analysis. The specially designed landmark detection error metric plot, which summarizes the results across all ablations on images with 360 × 360 px, is shown in Fig. 4. We plotted the Real2Real performance using gold curves as a baseline comparison with all the other ablation methods.
The effect of domain randomization
Across all experiments, we observed that networks trained with strong domain randomization consistently achieved better performance than those with regular domain randomization. This is expected because strong domain randomization introduces more drastic augmentations, which samples a much wider spectrum of possible image appearance and promotes the discovery of more robust features that are less prone to overfitting. The only exception is the training on naively simulated images, where training with strong domain randomization results in much worse performance compared with regular domain randomization. We attribute this to the fact that the contrast of bony structures, which are most informative for the task considered here, are already much less pronounced in naive simulations. Strong domain randomization then further increases problem complexity, to the point where performance deteriorates.
From Fig. 4a–c, we see that realistic simulation (DeepDRR) outperforms all other X-ray simulation paradigms in both regular domain randomization and strong domain randomization settings. Realistic simulation trained using strong domain randomization even outperforms Real2Real with regular domain randomization. As our experiments were precisely controlled and the only difference between the two scenarios is the image appearance due to varied simulation paradigm in the training set, this result supports the hypothesis that realistic simulation of X-rays using DeepDRR performs best for model transfer to real data. The strong domain randomization scheme includes a rich collection of image augmentation methods. The Sim2Real testing results on real X-ray images acquired from a different acquisition system, the BrainLab LoopX system, have shown similar performance. This suggests that models trained with SyntheX generalize to images across acquisition settings.
The effect of domain adaptation
From Fig. 4d,f, we observe that both realistic-CycleGAN and naive-CycleGAN achieve comparable performance to Real2Real. This means that images generated from synthetic images via CycleGAN have similar appearance, despite the synthetic training domains being different. The improvements over training purely on the respective synthetic domains (Fig. 4a,c) confirms that CycleGAN is useful for domain generalization. ADDA training also improves the performance over non-adapted transfer, but does not perform at the level of CycleGAN models. Interestingly, ADDA with strong domain randomization shows deteriorated performance compared with regular domain randomization (Fig. 4e,i). This is because the marked and random appearance changes due to domain randomization complicate domain discrimination, which in turn has adverse effects on overall model performance.
Scaling up the training data
We selected the best performing methods from the above domain randomization and domain adaptation ablations on the controlled dataset. These methods were realistic simulation with domain randomization and CycleGAN training based on realistic simulation, respectively, and trained on the scaled-up dataset, which contains a much larger variety of anatomical shape and imaging geometry, that is, synthetic C-arm poses.
With more training data and geometric variety, we found that all scaled-up experiments outperform the Real2Real baseline on the benchmark dataset (Fig. 4g,h). The model trained with strong domain randomization on realistically synthesized but large data (SyntheX, as reported above) achieved a mean landmark distance error of 5.95 ± 3.52 mm, and a mean Dice score of 0.86 ± 0.21. For segmentation performance, SyntheX is substantially better than the Real2Real baseline (P = 2.3 × 10−5 using a one-tailed t-test). Landmark detection also performed better, but the improvement was not significant at the P = 0.05 confidence level (P = 0.14 using a one-tailed t-test), suggesting that our real dataset was adequate to train landmark detection models. Figure 5 presents a collection of qualitative visualizations of the detection performance of this synthetic-data-trained model when applied to real data. This result suggests that training with strong domain randomization and/or adaptation on large-scale, realistically synthesized data is a feasible alternative to training on real data. Training on large-scale data processed by CycleGAN achieved comparable performance (6.20 ± 3.56 mm) as pure realistic simulation with domain randomization, but comes with the disadvantage that real data with sufficient variability must be available at training time to enable CycleGAN training.
Fig. 5 |. Qualitative results of the segmentation and landmark detection.
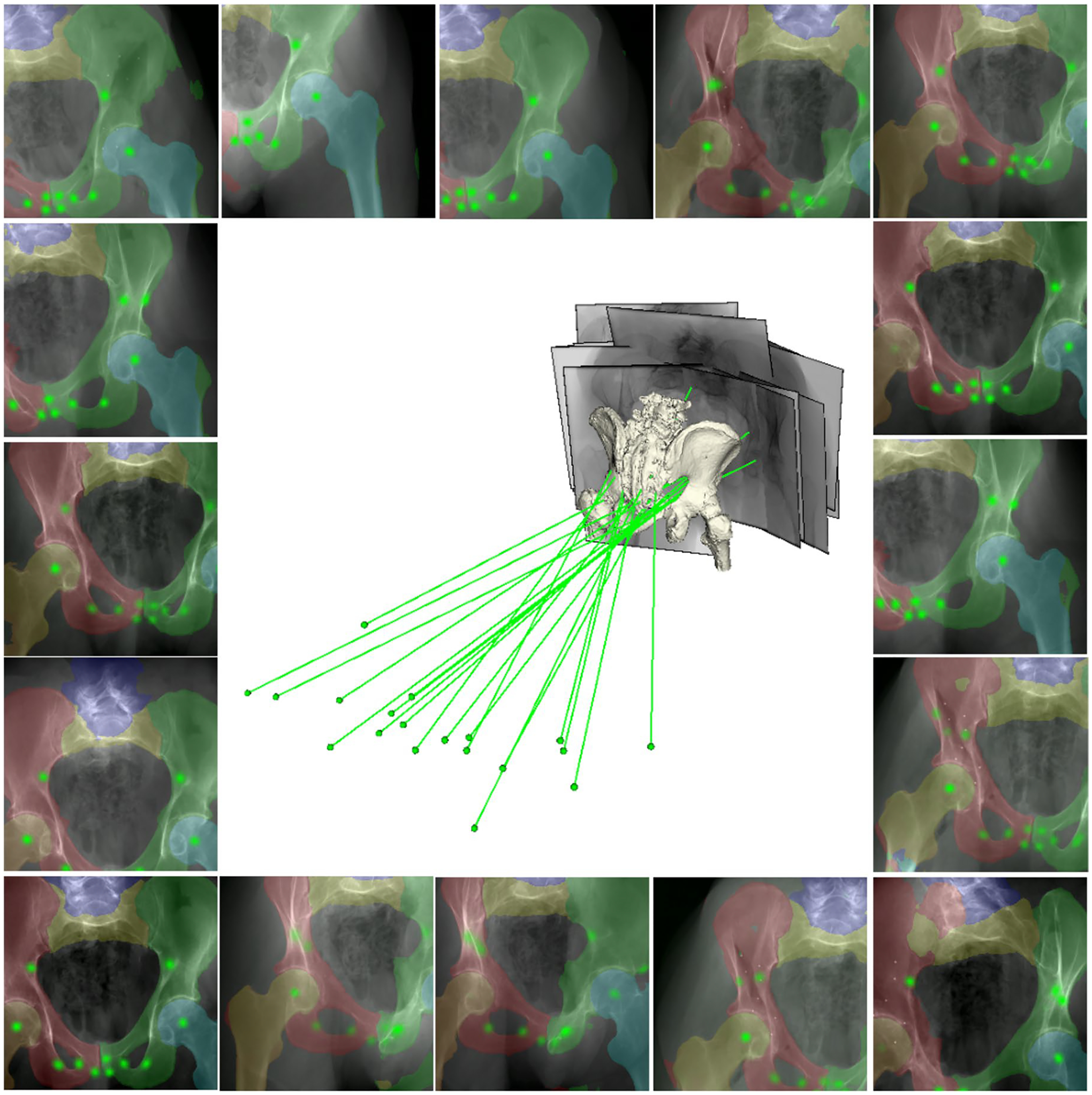
The results are presented as overlays on testing data using the model trained with scaled-up SyntheX data. Anatomical segmentation structures are blended with various colours. Landmark heatmap responses are visualized in green. The projection geometries corresponding to the images relative to a 3D bone mesh model of the anatomy are presented in the centre. The X-ray sources are shown as green dots and the principal rays are shown as green lines.
Discussion
We present general use cases of SyntheX for various scenarios, including purely bony anatomy (the hip), a metallic artificial surgical tool and soft tissue (lung COVID-19 lesion). Our experiments on three varied clinical tasks demonstrate that the performance of models trained using SyntheX–on real data–meets or exceeds the performance of real-data-trained models. We show that generating realistic synthetic data is a viable resource for developing machine learning models and is comparable to collecting largely annotated real clinical data.
Using synthetic data to train machine learning algorithms is receiving increasing attention. In general computer vision, the Sim2Real problem has been explored extensively for self-driving perception31–36 and robotic manipulation37–42. In diagnostic medical image analysis, GAN-based synthesis of novel samples has been used to augment available training data for magnetic resonance imaging43–48, CT46,49, ultrasound50, retinal51–53, skin lesion54,55 and CXR56 images. In computer-assisted interventions, early successes on the Sim2Real problem include analysis on endoscopic images3,57–59 and intra-operative X-ray60–62. The controlled study here validates this approach in the X-ray domain by showing that Sim2Real compares favourably to Real2Real training.
The hip-imaging ablation experiments reliably quantify the effect of the domain gap on real data performance for varied Sim2Real model transfer approaches. This is because all aleatoric factors that usually confound such experiments are precisely controlled for, with alterations to image appearances due to the varied image simulation paradigms being the only source of mismatch. The aleatoric factors that we controlled include anatomy, imaging geometries, ground-truth labels, network architectures and hyperparameters. The number of training samples is the same for all experiments. Use of domain randomization and adaptation techniques does not create additional samples but merely changes the appearance of samples on the pixel level. In particular, the viewpoints and 3D scene recreated in the simulation were identical to the real images, which to our knowledge has not yet been achieved. From these results, we draw the following conclusions.
Physics-based, realistic simulation of training data using the DeepDRR framework results in models that generalize better to the real data domain compared with models trained on less realistic, that is, naive or heuristic, simulation paradigms. This suggests, not surprisingly, that matching the real image domain as closely as possible directly benefits generalization performance.
Realistic simulation combined with strong domain randomization (SyntheX) performs on a par with both the best domain adaptation method (CycleGAN with domain randomization) and real-data training when models are trained on matched datasets. However, because SyntheX does not require any real data at training time, this paradigm has clear advantages over domain adaptation. Specifically, it saves the effort of acquiring real data early in development or designing additional machine learning architectures that perform adaptation. This makes SyntheX particularly appealing for the development of novel instruments or robotic components, real images of which can simply not be acquired early during conceptualization.
Realistic simulation using DeepDRR is as computationally efficient as naive simulation, both of which are orders of magnitude faster than Monte Carlo simulation23. Further, realistic simulation using DeepDRR brings substantial benefits in regards to Sim2Real performance and self-contained data generation and training. These findings are encouraging and strongly support the hypothesis that training on synthetic radiographs simulated from 3D CT is a viable alternative to real data training, or at a minimum, a strong candidate for pre-training.
Compared with acquiring real patient data, generating large-scale simulation data is more flexible, time efficient, low-cost and avoids privacy concerns. For the hip-image analysis use case, we performed experiments based on 10,000 synthetic images from 20 hip CT scans. Training with realistic simulation and strong domain randomization outperformed Real2Real training at the 90% activation level but generally improved performance as seen by a flattened activation versus error curve (Fig. 4g). The performance of training with CycleGAN with larger datasets was similar. These findings suggest that scaling-up data for training is an effective strategy to improve performance both inside and outside of the training domain. Scaling up training data is costly or impossible in real settings, but in comparison is easily possible using data synthesis. Having access to more varied data samples during training helps the network parameter optimization find a more stable solution that also transfers better.
We have found that Sim2Real model transfer performs best for scenarios where real data and corresponding annotations are particularly hard to obtain. This is evidenced by the change in the performance gap between Real2Real and Sim2Real training, where Sim2Real performs particularly well for scenarios where little real data are available, such as for hip imaging and robot tool detection, and hardly matches Real2Real performance for use cases where abundant real data exist, such as COVID-19 lesion segmentation. The value of SyntheX thus primarily derives from the possibility of generating large synthetic training datasets for innovative applications, for example, including custom-designed hardware19,63 or novel robotic imaging paradigms64,65, the data for which could not otherwise be obtained. Second, SyntheX can complement real datasets by providing synthetic samples that exhibit increased variability in anatomies, imaging geometries or scene composition. Finally, the SyntheX simulation paradigm enables generation of precise annotations, for example, the lesion volume in the COVID-19 use case, that could not be derived otherwise.
Interestingly, although domain adaptation techniques (CycleGAN and ADDA) have access to data in the real domain, these methods outperformed domain generalization techniques (here, domain randomization) by only a small margin in the controlled study. The performance of ADDA training heavily depends on the choices of additional hyperparameters, such as the design of the discriminator, number of training cycles between task and discriminator network updates, and learning rates, among others. Thus, it is non-trivial to find the best training settings, and these settings are unlikely to apply to other tasks. Because CycleGAN performs image-to-image translation, a complicated task, it requires sufficient and sufficiently diverse data in the real domain to avoid overfitting. Further, using CycleGAN requires an additional training step of a large model, which is memory intensive and generally requires long training time. In certain cases, CycleGAN models could also introduce undesired effects. A previous study found that the performance of CycleGAN is highly dependent on the dataset, potentially resulting in unrealistic images with less information content than the original images66. Moreover, although ref.67 showed that image-to-image translation may closer approximate real X-rays according to image similarity metrics, our study shows that the advantage over domain randomization in terms of downstream task performance is marginal. Finally, because real domain data are being used in both domain adaptation paradigms, adjustments to the real-data target domain, for example, use of a different C-arm X-ray imaging device or design changes to surgical hardware, may require de novo acquisition of real data and re-training of the models. In contrast, SyntheX resembles a plug-and-play module, to be integrated into any learning-based medical imaging tasks, which is easy to set up and use. Similar to multiscale modelling68 and in silico virtual clinical trials69,70, SyntheX has the potential to envision, implement and virtually deploy solutions for image-guided procedures and evaluate their potential utility and adequacy. This makes SyntheX a promising tool that may replicate traditional development workflows solely using computational tools.
Our scaled-up hip-imaging experiments using SyntheX achieved a mean landmark detection error of 5.95 ± 3.52 mm. A pelvic landmark detection error of 5–6 mm is frequently reported in the literature: ref.12 reported a mean error of 5.0 mm12 and and ref.5 reported a mean error of 5.6 ± 4.5 mm. This accuracy was tested to be effective in initializing the 2D/3D pelvis registration and achieving less than 1° error for 86% of the images12. We consider this detection accuracy to be sufficient for related hip-imaging tasks. Extended Data Fig. 4 shows histograms of the C-arm geometry variations in the real hip-imaging dataset. The C-arm geometry is reported as the rotation difference of each view’s pelvis registration pose with respect to the standard anterior/posterior pose. We have observed that most of the C-arm geometries are within 30°. This range of C-arm geometry distribution is typical for pelvic procedures, such as osteotomy10.
Despite the promising outlook, our study has several limitations. First, while the real X-ray and CT datasets of cadaveric specimens used for the hip imaging and robotic tool segmentation task are of a respectable size for this type of application, it is small compared with some dataset sizes in general computer vision applications. However, the effort, facilities, time and, therefore, costs required to acquire and annotate a dataset of even this size are substantial due to the nature of the data. Further, we note that using a few hundred images, as we do for the hip-imaging X-ray tasks, is a typical size in the literature5,12,71–76, and most of the existing work on developing machine learning solutions for intra-operative X-ray analysis tasks, such as 2D/3D registration, do not develop nor test their methods on any real data13. In summary, while datasets of the size reported here may not accurately reflect all of the variability one may expect during image-based surgery, the models trained on our datasets performed well on held-out data, using both leave-one-subject-out cross-validations and an independent test set, and performed comparably to previous studies on larger datasets5,77.
Second, the performance we report is limited by the quality of the CT and annotations. The spatial resolution of CT scans (between 0.5 mm and 1.0 mm in hip imaging and surgical robot tool segmentation; between 1.0 mm and 2.0 mm in COVID-19 lesion segmentation, isotropic) imposed a limitation on the resolution that can be achieved in 2D simulation. Pixel sizes of conventional detectors are as small as 0.2 mm, smaller than the highest-resolution scenario considered here. However, contemporary computer vision algorithms for image analysis tasks have considered only downsampled images in the ranges described here. Another issue arises from annotation mismatch, especially when annotations are generated using different processes for SyntheX training and evaluation on real 2D X-ray images. This challenge arose specifically in the COVID-19 lesion segmentation task, where 3D lesion labels generated from the pre-trained lesion segmentation network and used for SyntheX training are not consistent with the annotations on real 2D X-ray data. This is primarily for two reasons. First, because CT scans and CXR images were not from the same patients, COVID-19 disease stages and extent of lesions were varied; second, because real CXRs were annotated in 2D, smaller or more opaque parts of COVID-19 lesions may have been missed due to the projective and integrative nature of X-ray imaging. This mismatch in ground-truth definition is unobserved but establishes an upper bound on the possible Sim2Real performance. Further, realism of simulation can be improved with higher-quality CT scans, super-resolution techniques and advanced modelling techniques to more realistically represent anatomy at higher resolutions.
Third, SyntheX performs X-ray image synthesis from existing human models, which does not manipulate pathologies/lesions within healthy patient scans. For example, in the application of COVID-19 lesion segmentation, the CT scans were acquired from patients that were infected by COVID-19 and contained lesions naturally. Our X-ray synthesis model followed the same routine to generate images from the CT recordings, which then present lesions in the 2D domain as well. Future work will consider expanding on our current work by researching possibilities to advance human modelling.
Conclusion
In this paper, we demonstrated that realistic simulation of image formation from human models combined with domain generalization or adaptation techniques is a viable alternative to large-scale real-data collection. We demonstrate its utility on three variant clinical tasks, namely hip imaging, surgical robotic tool detection and COVID-19 lesion segmentation. On the basis of controlled experiments on a pelvic X-ray dataset, which is precisely reproduced in varied synthetic domains, we quantified the effect of simulation realism and domain adaptation and generalization techniques on Sim2Real transfer performance. We found promising Sim2Real performance of all models that were trained on realistically simulated data. The specific combination of training on realistic synthesis and strong domain randomization, which we refer to as SyntheX, is particularly promising. SyntheX-trained models perform on a par with real-data-trained models, making realistic simulation of X-ray-based clinical workflows and procedures a viable alternative or complement to real-data acquisition. Because SyntheX does not require real data at training time, it is particularly promising for the development of machine learning models for novel clinical workflows or devices, including surgical robotics, before these solutions exist physically.
Methods
We introduce further details on the domain randomization and domain adaptation methods applied in our studies. We then provide additional information on experimental set-up and network training details of the clinical tasks and benchmark investigations.
Domain randomization
Domain randomization effects were applied to the input images during network training. We studied two domain randomization levels: regular and strong domain randomization. Regular domain randomization included the most frequently used data augmentation schemes. For strong domain randomization, we included more drastic effects and combined them together. We use x to denote a training image sample. The domain randomization techniques we introduced are as follows.
Regular domain randomization included the following. (1) Gaussian noise injection: x + N(0, σ), where N is normal distribution and σ was uniformly chosen from the interval (0.005, 0.1) multiplied by the image intensity range. (2) Gamma transform: norm(x)γ, where x was normalized by its maximum and minimum value and γ was uniformly selected from the interval (0.7, 1.3). (3) Random crop: x was cropped at random locations using a square shape, which has the dimension of 90% x size. Regular domain randomization methods were applied to every training iteration.
Strong domain randomization included the following. (1) Inverting: max(x) – x, where the maximum intensity value was subtracted from all image pixels. (2) Impulse/pepper/salt noise injection: 10% of pixels in x were replaced with one type of noise including impulse, pepper and salt. (3) Affine transform: a random 2D affine warp including translation, rotation, shear and scale factors was applied. (4) Contrast: x was processed with one type of the contrast manipulations including linear contrast, log contrast and sigmoid contrast. (5) Blurring: x was processed with a blurring method including Gaussian blur N(μ = 0, σ = 3.0), where μ is the mean of normal distribution, and average blur (kernel size between 2 × 2 and 7 × 7). (6) Box corruption: a random number of box regions were corrupted with large noise. (7) Dropout: either randomly dropped 1–10% of pixels in x to 0, or dropped them in a rectangular region with 2–5% of the image size. (8) Sharpening and embossing: sharpen x blended the original image with a sharpened version with an alpha between 0 and 1 (no and full sharpening effect). Embossing added the sharpened version rather than blending it. (9) One of the pooling methods was applied to x: average pooling, max pooling, min pooling and median pooling. All of the pooling kernel sizes were between 2 × 2 and 4 × 4. (10) Multiply: either changed brightness or multiplied x element wise with 50–150% of the original value. (11) Distort: distorted local areas of x with a random piece-wise affine transformation. For each image, we still applied basic domain randomization but only randomly concatenated up to two strong domain randomization methods during each training iteration to avoid too heavy augmentation.
Domain adaptation
We select the two most frequently used domain adaptation approaches for our comparison study, which are CycleGAN25 and ADDA26. CycleGAN was trained using unpaired synthetic and real images before task network training. All synthetic images were then processed with trained CycleGAN generators, to alter their appearance to match real data. We strictly enforced the data split used during task-model training so that images from the test set were excluded during both CycleGAN and task network training. ADDA introduced an adversarial discriminator branch as an additional loss to discriminate between features derived from synthetic and real images. We followed the design of ref.26 to build the discriminator for ADDA training on the task of semantic segmentation. Both CycleGAN and ADDA models were tested using realistic and naive simulation images.
CycleGAN.
CycleGAN was applied to learn mapping functions between two image domains X and Y given training samples where xi ∈ X and where yj ∈ Y. Letters i and j indicate the sample index of the total sample number N and M, respectively. The model includes two mapping functions G: X → Y and F: Y → X, and two adversarial discriminators DX and DY. The objective contains two terms: adversarial loss to match the distribution between generated and target image domain; and cycle-consistency loss to ensure learned mapping functions are cycle-consistent. For one mapping function G: X → Y with its discriminator DY, the first term, adversarial loss, can be expressed as:
| (1) |
where G generates images G(x) with an appearance similar to images from domain Y, while DY tries to distinguish between translated samples G(x) and real samples y. Overall, G aims to minimize this objective against an adversary D that tries to maximize it. Similarly, there is an adversarial loss for the mapping function F: Y → X with its discriminator DX.
The second term, cycle-consistency loss, can be expressed as:
| (2) |
where for each image x from domain X, x should be recovered after one translation cycle, that is, x → G(x) → F(G(x)) ≈ x. Similarly, each image y from domain Y should be recovered as well. A previous study25 argued that learned mapping functions should be cycle-consistent to further reduce the space of possible mapping functions. The above formulation using domain discrimination and cycle consistency enables unpaired image translation, that is, learning the mappings G(x) and F(y) without corresponding images.
The overall objective for CycleGAN training is expressed as:
| (3) |
where λ controls the relative importance of cycle-consistency loss, aiming to solve:
| (4) |
For the generator network, 6 blocks for 128 × 128 images and 9 blocks for 256 × 256 and higher-resolution training images were used with instance normalization. For the discriminator network, a 70 × 70 PatchGAN was used.
Adversarial discriminative domain adaptation.
We applied the idea of ref.26 on our pelvis segmentation and landmark localization task. The architecture consists of three components: segmentation and localization network, decoder, and discriminator. The input to the segmentation and localization network is image (x) and the output prediction feature is z. The loss is Lseg and Lld as introduced in ‘Clinical tasks’. The decoder shared the same TransUNet architecture, takes z as input and the output is the reconstruction R(z). The reconstruction loss, Lrecons, is the mean squared error between x and z. The discriminator was trained using an adversarial loss:
| (5) |
where H and W are the dimensions of the discriminator output, s = 0 when D takes target domain prediction (Yt) as input, and s = 1 when taking source domain prediction (Ys) as input. The discriminator contributed an adversarial loss during training to bring in domain transfer knowledge. The adversarial loss is defined as:
| (6) |
where t refers to the target domain. Thus, the total training loss can be written as:
| (7) |
where λadv and λrecons are weight hyperparameters, which are empirically chosen to be 0.001 and 0.01, as suggested by ref.26.
Clinical tasks experimental details
The SyntheX simulation environment was set up to approximate a Siemens CIOS Fusion C-Arm, which has image dimensions of 1,536 × 1,536, isotropic pixel spacing of 0.194 mm per pixel, a source-to-detector distance of 1,020 mm and a principal point at the centre of the image.
Hip imaging.
Synthetic hip X-rays were created using 20 CT scans from the New Mexico Decedent Image Database14. During simulation, we uniformly sampled the CT volume rotation in [−45°, 45°], and translation left/right in [−50 mm, 50 mm], interior/superior in [−20 mm, 20 mm], and anterior/posterior in [−100 mm, 100 mm]. We generated 18,000 images for training and 2,000 images for validation. Ground-truth segmentation and landmark labels were projected from 3D using the projection geometry.
We consistently trained the model for 20 epochs and selected the final converged model for evaluation. Strong domain randomization was applied at training time (see ‘Domain randomization’). During evaluation, a threshold of 0.5 was used for segmentation and the landmark prediction was selected using the highest heatmap response location.
Robotic surgical tool detection.
We created 100 voxelized models of the CM in various configurations by sampling its curvature control point angles form a Gaussian distribution N(μ = 0, σ = 2.5°). The CM base pose was uniformly sampled left and right anterior oblique views (LAO/RAO) in [−30°, 30°], cranio and caudal views (CRAN/CAUD) in [−10°, 10°], source-to-isocentre distance in [600 mm, 900 mm], and translation in x, y axes following a Gaussian distribution N(μ = 0 mm, σ = 10 mm). We created DeepDRR synthetic images by projecting randomly selected hip CT scans from the 20 New Mexico Decedent Image Database CT scans used for hip imaging together with the CM model, which include 28,000 for training and 2,800 for validation. Ground-truth segmentation and landmark labels were projected following each simulation geometry.
The network training details are in ‘Network training details’, and strong domain randomization was applied (see ‘Domain randomization’). The network was trained for ten epochs and the final converged model was selected for evaluation. The performance was evaluated on 264 real CM X-ray images with manual ground-truth label annotations. During evaluation, a threshold of 0.5 was used for segmentation and the landmark prediction was selected using the highest heatmap response location. The network was trained for 50 epochs for the fivefold Real2Real experiments. The testing and evaluation routines are the same.
COVID-19 lesion segmentation.
We used 81 high-quality CT scans from ImagEng lab20 and 62 CT scans with resolution less than 2 mm from UESTC21, all diagnosed as COVID-19 cases, to generate synthetic CXR data. The 3D lesion segmentations of CTs from ImagEng lab were generated using the pre-trained COPLE-Net21. During DeepDRR simulation, we uniformly sampled the view pose of CT scans, rotation from [−5°, 5°] in all three axes and source-to-isocentre distance in [350 mm, 650 mm], resulting in 18,000 training images and 1,800 validation images with a resolution of 224 × 224 px. A random shearing transformation from [−30°, 30°] was applied on the CT scan and segmentations were obtained with a threshold of 0.5 on the predicted response. The corresponding lesion mask was projected from the 3D segmentation using the simulation projection geometry.
The network training set-ups follow the descriptions in ‘Network training details’. Strong domain randomization was applied during training time (see ‘Domain randomization’). We trained the network for 20 epochs and selected the final converged model for testing. The performance was evaluated on a 2,951 real COVID-19 benchmark dataset22. During evaluation, the network segmentation mask was created using a threshold value of 0.5 on the original prediction. The network was trained for 50 epochs for the fivefold Real2Real experiments. The testing and evaluation routines are the same.
Benchmark hip-imaging investigation.
For every X-ray image, ground-truth X-ray camera poses relative to the CT scan were estimated using an automatic intensity-based 2D/3D registration of the pelvis and both femurs12. Every CT scan was annotated with segmentation of anatomical structures and anatomical landmark locations defined in Fig. 2a. Two-dimensional labels for every X-ray image were then generated automatically by forward projecting the reference 3D annotations using the corresponding ground-truth C-arm pose.
We generated synthetic data using three DRR simulators: naive DRR, xreg DRR and DeepDRR. Naive DRR generation amounts to simple ray-casting and does not consider any imaging physics. This amounts to the assumption of a mono-energetic source, single material objects and no image corruption, for example, due to noise or scattering. Heuristic simulation performs a linear thresholding of the CT Hounsfield units to differentiate materials between air and anatomy before ray-casting. While this results in a more realistic appearance of the resulting DRRs, in that the tissue contrast is increased, the effect does not model imaging physical effects. Realistic simulation (DeepDRR) simulates imaging physics by considering the full spectrum of the X-ray source, and relies on machine learning for material decomposition and scatter estimation. It also considers both signal-dependent noise as well as readout noise together with detector saturation.
Network training details
We used Pytorch for all implementations and trained the networks from the pre-trained vision transformer model78. The use of pre-trained model is suggested in the TransUNet paper27. The networks were trained using stochastic gradient descent with an initial learning rate of 0.1, Nesterov momentum of 0.9, weight decay of 0.00001 and a constant batch size of 5 images. The learning rate was decayed with a gamma of 0.5 for every 10 epochs during training. The multi-task network training loss is equally weighted between landmark detection loss and segmentation loss. All experiments were conducted on an Nvidia GeForce RTX 3090 Graphics Card with 24 GB memory. It takes around 2 h to generate 10,000 synthetic hip-imaging images. The average network training time for 10,000 data is about 5 h until convergence.
Extended Data
Extended Data Fig. 1 |. X-ray Images of the continuum manipulator.
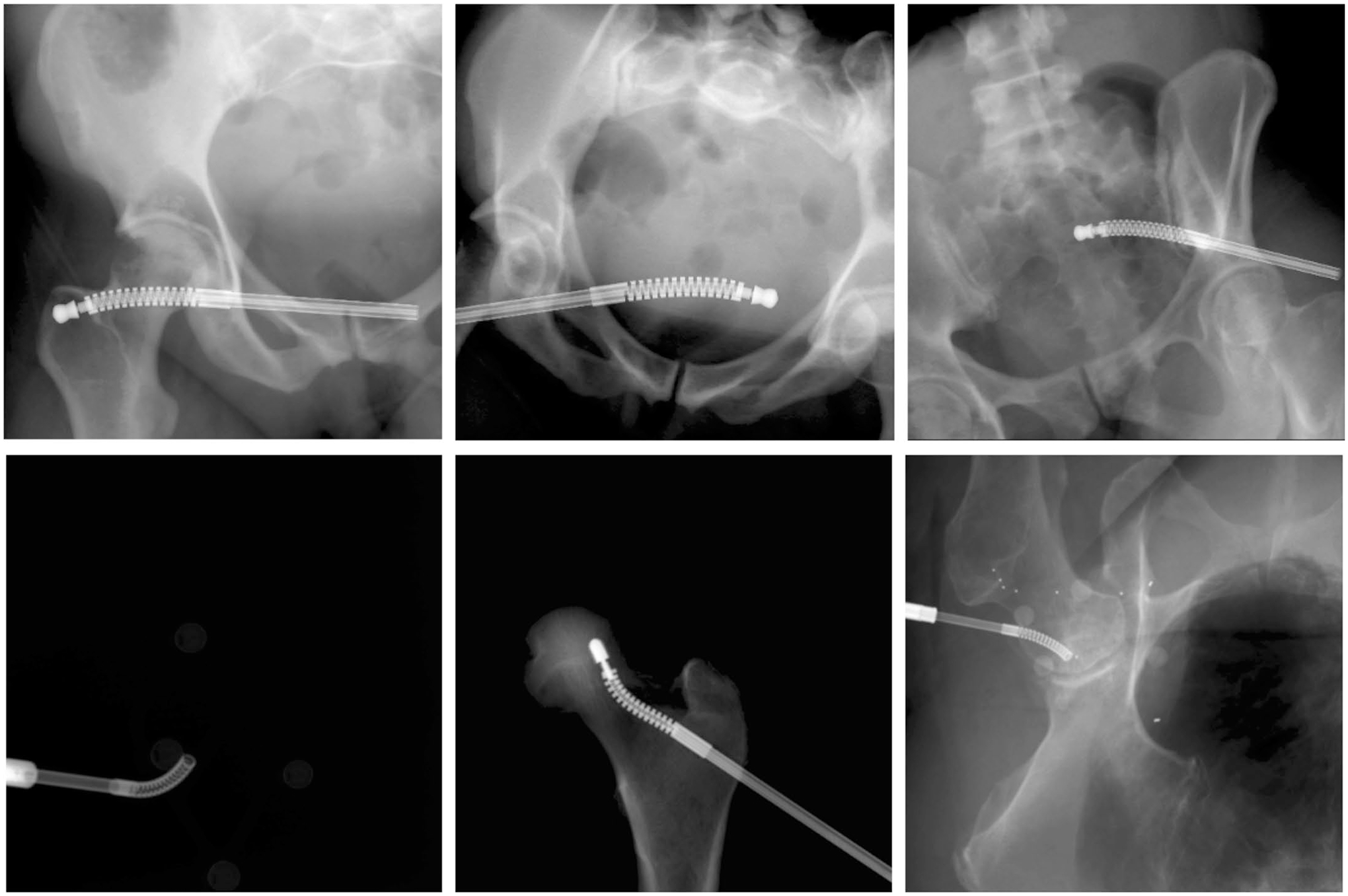
Upper Row: Example synthetic X-ray images of the continuum manipulator. Lower Row: Example real X-ray images of the continuum manipulator.
Extended Data Fig. 2 |. Multi-task network architecture.
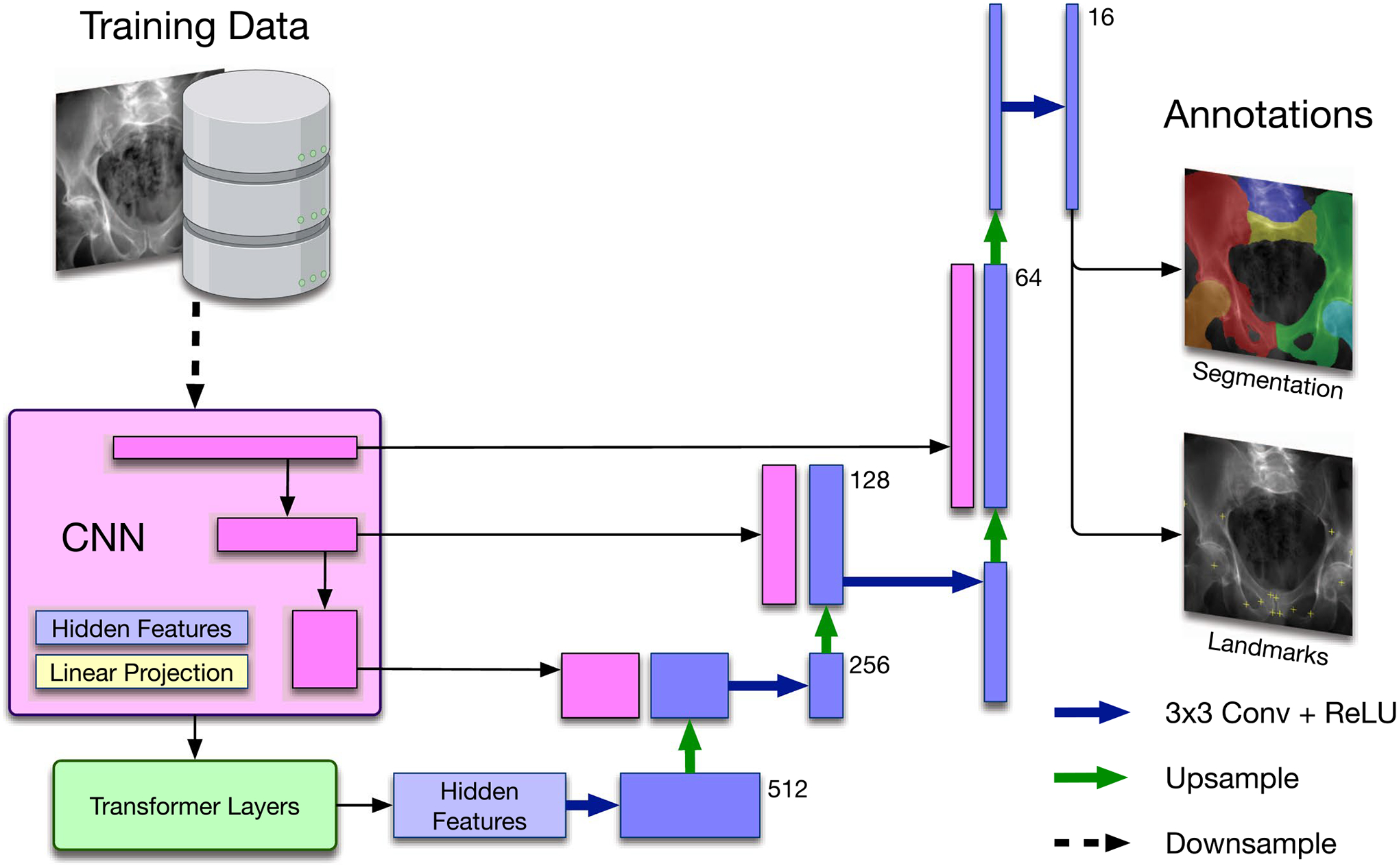
TransUNet based concurrent segmentation and landmark detection network architecture for multi-task learning.
Extended Data Fig. 3 |. Scaled-up dataset landmark detection performance comparison.
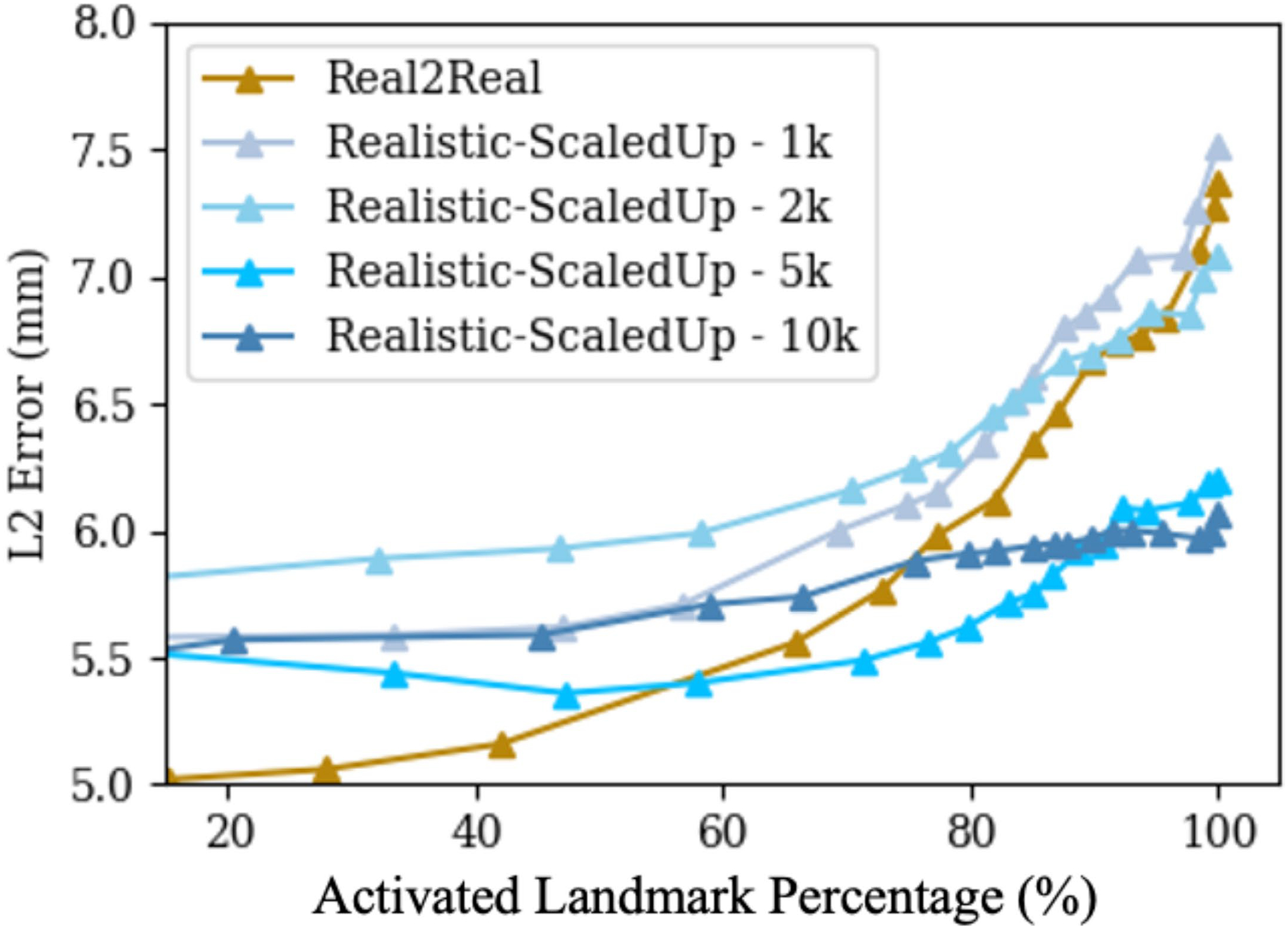
Real2Real performance curve is present in dark gold colour, and the Sim2Real performance curves corresponding to increasing scaled-up training data sizes are present in different levels of blue colours.
Extended Data Fig. 4 |.
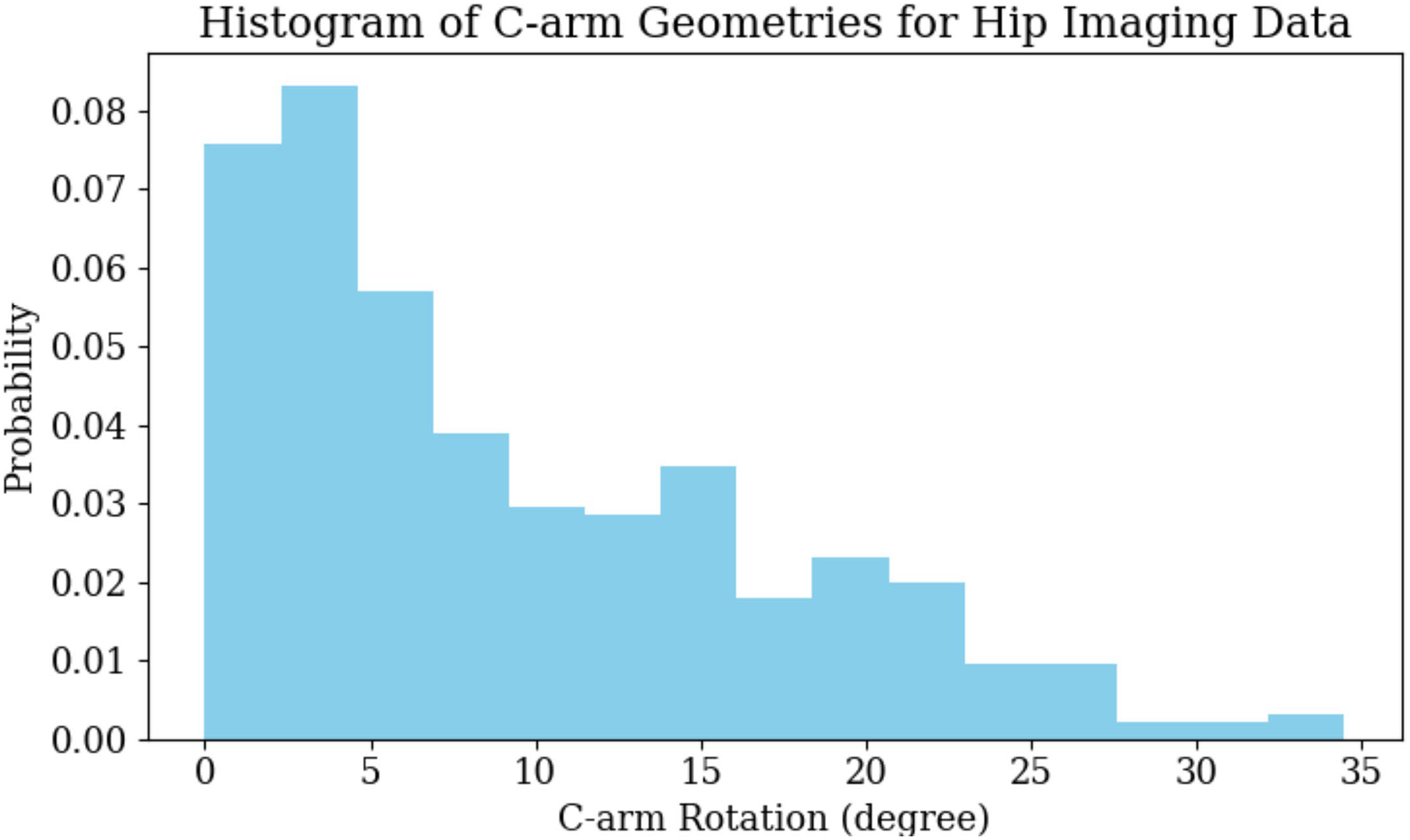
Histogram of C-arm Geometries for Hip Imaging Data.
Extended Data Table 1 |.
Individual Landmark Error (mm) for hip imaging as a mean of 6-fold individual testing on 366 real hip X-ray images
| Sim2Real | Real2Real | |||
|---|---|---|---|---|
| Mean | CI | Mean | CI | |
| L.FH | 5.20 ± 1.66 | 0.23 | 3.51 ± 1.53 | 0.21 |
| R.FH | 6.14 ± 4.12 | 0.60 | 8.48 ± 4.41 | 0.64 |
| L.GSN | 6.08 ± 2.90 | 0.38 | 6.68 ± 4.43 | 0.58 |
| R.GSN | 5.48 ± 2.58 | 0.35 | 8.20 ± 3.93 | 0.55 |
| L.IOF | 5.15 ± 2.49 | 0.34 | 6.41 ± 8.42 | 1.20 |
| R.IOF | 3.62 ± 3.26 | 0.45 | 4.68 ± 4.76 | 0.67 |
| L.MOF | 3.75 ± 2.37 | 0.30 | 5.42 ± 3.57 | 0.46 |
| R.MOF | 4.85 ± 2.71 | 0.35 | 8.60 ± 4.01 | 0.51 |
| L.SPS | 9.48 ± 2.99 | 0.35 | 5.32 ± 4.70 | 0.55 |
| R.SPS | 7.21 ± 2.68 | 0.34 | 5.87 ± 5.20 | 0.62 |
| L.IPS | 6.32 ± 2.64 | 0.34 | 4.11 ± 2.45 | 0.31 |
| R.IPS | 4.48 ± 1.63 | 0.21 | 3.56 ± 1.87 | 0.23 |
| L.ASIS | 6.85 ± 5.02 | 0.76 | 12.62 ± 9.47 | 1.62 |
| R.ASIS | 9.05 ± 5.40 | 0.88 | 13.50 ± 30.46 | 5.34 |
| All | 5.95 ± 3.52 | 0.13 | 6.46 ± 8.21 | 0.3 |
CI refers to confidence intervals. They are computed using the 2-tailed z-test with a critical value for a 95% level of confidence (p<0.05). Real2Real refers to training and testing both in real domain datasets. Sim2Real means training in simulation dataset and testing in real dataset.
Extended Data Table 2 |.
Individual Dice Score for hip imaging as a mean of 6-fold individual testing on 366 real hip X-ray images
| Sim2Real | Real2Real | |||
|---|---|---|---|---|
| Mean | CI | Mean | CI | |
| L.Pel | 0.89 ± 0.21 | 0.02 | 0.86 ± 0.19 | 0.02 |
| R.Pel | 0.89 ± 0.18 | 0.02 | 0.88 ± 0.15 | 0.02 |
| Verteb | 0.79 ± 0.19 | 0.02 | 0.69 ± 0.27 | 0.03 |
| Sacrum | 0.74 ± 0.13 | 0.01 | 0.55 ± 0.19 | 0.02 |
| L.Fem | 0.95 ± 0.14 | 0.01 | 0.91 ± 0.21 | 0.02 |
| R.Fem | 0.88 ± 0.27 | 0.03 | 0.86 ± 0.28 | 0.03 |
| All | 0.86 ± 0.21 | 0.01 | 0.79 ± 0.25 | 0.01 |
CI refers to confidence intervals. They are computed using the 2-tailed z-test with a critical value for a 95% level of confidence (p<0.05). Real2Real refers to training and testing both in real domain datasets. Sim2Real means training in simulation dataset and testing in real dataset.
Extended Data Table 3 |.
Average Landmark Error (mm) for hip imaging as a mean of 6-fold individual testing on 366 real hip X-ray images
| Landmark Error (mm) | Dice Score | |||
|---|---|---|---|---|
| Mean | CI | Mean | CI | |
| Real | 6.46 ± 8.21 | 0.13 | 0.79 ± 0.25 | 0.01 |
| Sim 1k | 6.61 ± 7.27 | 0.26 | 0.82 ± 0.23 | 0.01 |
| Sim 2k | 6.56 ± 4.78 | 0.17 | 0.81 ± 0.24 | 0.01 |
| Sim 5k | 5.82 ± 4.52 | 0.16 | 0.82 ± 0.24 | 0.01 |
| Sim 10k | 5.95 ± 3.52 | 0.13 | 0.86 ± 0.21 | 0.01 |
CI refers to confidence intervals. They are computed using the 2-tailed z-test with a critical value for a 95% level of confidence (p<0.05). The Sim numbers from 1k to 10k in the left most column refer to the size of scaled-up simulation dataset.
Extended Data Table 4 |.
Average landmark error (mm) for surgical tool detection as a mean of 5-fold individual testing on 264 real X-ray images of the continuum manipulator
| Sim2Real | Real2Real | |||
|---|---|---|---|---|
| Mean | CI | Mean | CI | |
| Base | 1.09 ± 0.69 | 0.09 | 1.09 ± 0.89 | 0.11 |
| Tip | 1.12 ± 1.04 | 0.13 | 1.29 ± 3.40 | 0.43 |
| All | 1.10 ± 0.88 | 0.08 | 1.19 ± 2.49 | 0.22 |
CI refers to confidence intervals. They are computed using the 2-tailed z-test with a critical value for a 95% level of confidence (p<0.05). Real2Real refers to training and testing both in real domain datasets. Sim2Real means training in simulation dataset and testing in real dataset.
Extended Data Table 5 |.
Average Dice Score for surgical tool detection as a mean of 5-fold individual testing on 264 real X-ray images of the continuum manipulator
| Sim2Real | Real2Real | ||
|---|---|---|---|
| Mean | CI | Mean | CI |
| 0.92 ± 0.07 | 0.01 | 0.41 ± 0.23 | 0.03 |
CI refers to confidence intervals. They are computed using the 2-tailed z-test with a critical value for a 95% level of confidence (p<0.05). Real2Real refers to training and testing both in real domain datasets. Sim2Real means training in simulation dataset and testing in real dataset.
Extended Data Table 6 |.
Average performance metrics (%) for COVID-19 infected region segmentation as a mean of 5-fold individual testing on 2,951 real COVID-19 real chest X-ray images
| Sensitivity | Specifcity | Precision | F1-Score | F2-Score | Accuracy | |
|---|---|---|---|---|---|---|
| Sim2Real | 80.28 ± 15.74 | 87.41 ± 6.78 | 48.67 ± 27.23 | 54.69 ± 23.06 | 63.81 ± 25.51 | 85.22 ± 5.89 |
| Real2Real | 79.83 ± 17.37 | 96.92 ± 3.51 | 75.16 ± 25.71 | 73.54 ± 20.35 | 76.09 ± 25.45 | 94.05 ± 4.54 |
Real2Real refers to training and testing both in real domain datasets. Sim2Real means training in simulation dataset and testing in real dataset.
Acknowledgements
We gratefully acknowledge financial support from NIH NIBIB Trailblazer R21 EB028505, NIH R01 EB023939, NIH R01 EB016703 and Johns Hopkins University internal funds.
Footnotes
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
The codes developed for this study are available in the SyntheX GitHub repository available at https://github.com/arcadelab/SyntheX (ref.80). An updated repository for DeepDRR is available at https://github.com/arcadelab/deepdrr. The xReg registration software module is at https://github.com/rg2/xreg. We used the open-source software 3D Slicer 4.10.2 for processing the CT scans (https://www.slicer.org/). We used the open-source software labelme v5.0.0 for annotating the 2D segmentation masks of X-ray data (https://github.com/wkentaro/labelme). We used the open-source software ImageJ Version 2.3.0/1.53q to overlay the 2D image data and labels (https://imagej.nih.gov/ij/).
Competing interests
The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s42256-023-00629-1.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s42256-023-00629-1.
Data availability
We provide access web links for public data used in our study. The DOI link to the dataset is https://doi.org/10.7281/T1/2PGJQU (ref.79). The hip-imaging CT scans are selected from the New Mexico Decedent Image Database at https://nmdid.unm.edu/resources/data-information. The hip-imaging real cadaveric CT scans and X-rays can be accessed at https://github.com/rg2/DeepFluoroLabeling-IPCAI2020. The COVID-19 lung CT scans can be accessed at https://www.imagenglab.com/news-ite/covid-19/. The COVID-19 real CXR data can be accessed at https://www.kaggle.com/datasets/aysendegerli/qatacov19-dataset. The COVID-19 3D lesion segmentation pre-trained network module and associated CT scans can be accessed upon third-party restriction at https://github.com/HiLab-git/COPLE-Net.
References
- 1.Drenkow N, Sani N, Shpitser I & Unberath M Robustness in deep learning for computer vision: mind the gap? Preprint at https://arxiv.org/abs/2112.00639 (2021).
- 2.Taori R et al. Measuring robustness to natural distribution shifts in image classification. Preprint at https://arxiv.org/abs/2007.00644 (2020).
- 3.Mahmood F & Durr NJ Deep learning and conditional random fields-based depth estimation and topographical reconstruction from conventional endoscopy. Med. Image Anal 48, 230–243 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Gu W, Gao C, Grupp R, Fotouhi J & Unberath M Extended capture range of rigid 2D/3D registration by estimating riemannian pose gradients. In International Workshop on Machine Learning in Medical Imaging 281–291 (Springer, 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bier B et al. Learning to detect anatomical landmarks of the pelvis in X-rays from arbitrary views. Int. J. Comput. Assist. Radiol. Surg 14, 1463–1473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung K, Tang N, Cheung L & Ng E Image-guided navigation in orthopaedic trauma. J. Bone Joint Surg. Br 92, 1332–1337 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Kelley TC & Swank ML Role of navigation in total hip arthroplasty. J. Bone Joint Surg. Am 91, 153–158 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Kordon F et al. Multi-task localization and segmentation for X-ray guided planning in knee surgery. In International Conference on Medical Image Computing and Computer-Assisted Intervention 622–630 (Springer, 2019). [Google Scholar]
- 9.Gao C, Grupp RB, Unberath M, Taylor RH & Armand M Fiducial-free 2D/3D registration of the proximal femur for robot-assisted femoroplasty. In Medical Imaging2020: Image-Guided Procedures, Robotic Interventions, and Modeling Vol. 11315, 113151C (International Society for Optics and Photonics, 2020). [Google Scholar]
- 10.Grupp RB et al. Pose estimation of periacetabular osteotomy fragments with intraoperative X-ray navigation. IEEE Trans. Biomed. Eng 67, 441–452 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nolte L-P et al. A new approach to computer-aided spine surgery: fluoroscopy-based surgical navigation. Eur. Spine J 9, S078–S088 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grupp RB et al. Automatic annotation of hip anatomy in fluoroscopy for robust and efficient 2D/3D registration. Int. J. Comput. Assist. Radiol. Surg 15, 759–769 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Unberath M et al. The impact of machine learning on 2D/3D registration for image-guided interventions: a systematic review and perspective Preprint at https://arxiv.org/abs/2108.02238 (2021). [DOI] [PMC free article] [PubMed]
- 14.Edgar H et al. New Mexico Decedent Image Database (Office of the Medical Investigator, University of New Mexico, 2020); 10.25827/5s8c-n515 [DOI] [Google Scholar]
- 15.Krčah M, Székely G & Blanc R Fully automatic and fast segmentation of the femur bone from 3D-CT images with no shape prior. In 2011 IEEE International Symposium on Biomedical Imaging: from Nano to Macro 2087–2090 (IEEE, 2011). [Google Scholar]
- 16.Bouget D, Allan M, Stoyanov D & Jannin P Vision-based and marker-less surgical tool detection and tracking: a review of the literature. Med. Image Anal 35, 633–654 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Alambeigi F et al. A curved-drilling approach in core decompression of the femoral head osteonecrosis using a continuum manipulator. IEEE Robot. Autom. Lett 2, 1480–1487 (2017). [Google Scholar]
- 18.Bakhtiarinejad M et al. A biomechanical study on the use of curved drilling technique for treatment of osteonecrosis of femoral head. In Computational Biomechanics for Medicine (eds Nash M, Nielsen P et al.) 87–97 (Springer, 2020). [Google Scholar]
- 19.Gao C et al. Fluoroscopic navigation for a surgical robotic system including a continuum manipulator. IEEE Trans. Biomed. Eng 69, 453–464 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaffino P et al. An open-source COVID-19 CT dataset with automatic lung tissue classification for radiomics. Bioengineering 8, 26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G et al. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging 39, 2653–2663 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Degerli A et al. COVID-19 infection map generation and detection from chest X-ray images. Health Inf. Sci. Syst 9, 15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unberath M et al. DeepDRR—a catalyst for machine learning in fluoroscopy-guided procedures. In International Conference on Medical Image Computing and Computer-Assisted Intervention 98–106 (Springer, 2018). [Google Scholar]
- 24.Unberath M et al. Enabling machine learning in X-ray-based procedures via realistic simulation of image formation. Int. J. Comput. Assist. Radiol. Surg 14, 1517–1528 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J-Y, Park T, Isola P & Efros AA Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proc. IEEE International Conference on Computer Vision 2223–2232 (2017). [Google Scholar]
- 26.Haq MM & Huang J Adversarial domain adaptation for cell segmentation. In Proceedings of the Third Conference on Medical Imaging with Deep Learning (eds Arbel T, Ben Ayed I et al.) 277–287 (PMLR, 2020). [Google Scholar]
- 27.Chen J et al. Transunet: transformers make strong encoders for medical image segmentation Preprint at https://arxiv.org/abs/2102.04306 (2021).
- 28.Milletari F, Navab N & Ahmadi S-A V-Net: fully convolutional neural networks for volumetric medical image segmentation. In 2016 Fourth International Conference on 3D Vision 565–571 (IEEE, 2016). [Google Scholar]
- 29.Schultheiss M et al. Lung nodule detection in chest X-rays using synthetic ground-truth data comparing cnn-based diagnosis to human performance. Sci. Rep 11, 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H et al. SDFN: segmentation-based deep fusion network for thoracic disease classification in chest X-ray images. Comput. Med. Imaging Graph 75, 66–73 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganin Y et al. Domain-adversarial training of neural networks. J. Mach. Learn. Res 17, 2096–2030 (2016). [Google Scholar]
- 32.Zhang Y, David P & Gong B Curriculum domain adaptation for semantic segmentation of urban scenes. In Proc. IEEE International Conference on Computer Vision. 2039–2049 (2017). [Google Scholar]
- 33.Hoffman J et al. CyCADA: cycle-consistent adversarial domain adaptation. In Proc. 35th International Conference on Machine Learning, Proc. Machine Learning Research Vol. 80 (eds Dy J & Krause A) 1989–1998 (PMLR, 2018); https://proceedings.mlr.press/v80/hoffman18a.html [Google Scholar]
- 34.Zhang Q, Zhang J, Liu W & Tao D Category anchor-guided unsupervised domain adaptation for semantic segmentation. Adv. Neural Inf. Process. Syst 32, 435–445 (2019). [Google Scholar]
- 35.Hoyer L, Dai D & Van Gool L DAFormer: improving network architectures and training strategies for domain-adaptive semantic segmentation. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition 9924–9935 (IEEE, 2022). [Google Scholar]
- 36.Wen L-H & Jo K-H Deep learning-based perception systems for autonomous driving: a comprehensive survey. Neurocomputing 489, 255–270 (2022). [Google Scholar]
- 37.Tobin J et al. Domain randomization for transferring deep neural networks from simulation to the real world. In IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS) 23–30 (IEEE, 2017). [Google Scholar]
- 38.Bousmalis K et al. Using simulation and domain adaptation to improve efficiency of deep robotic grasping. In ICRA 4243–4250 (IEEE, 2018). [Google Scholar]
- 39.Zhang F, Leitner J, Milford M & Corke P Modular deep Q networks for sim-to-real transfer of visuo-motor policies In ACRA (2017); http://arxiv.org/abs/1610.06781
- 40.Hundt A et al. “Good robot!”: Efficient reinforcement learning for multi-step visual tasks with sim to real transfer. IEEE Rob. Autom. Lett 5, 6724–6731 (2020). [Google Scholar]
- 41.Zhao W, Queralta JP & Westerlund T Sim-to-real transfer in deep reinforcement learning for robotics: a survey. In 2020 IEEE Symposium Series on Computational Intelligence 737–744 (IEEE, 2020). [Google Scholar]
- 42.Hu H et al. A sim-to-real pipeline for deep reinforcement learning for autonomous robot navigation in cluttered rough terrain. IEEE Rob. Autom. Lett 6, 6569–6576 (2021). [Google Scholar]
- 43.Han C et al. GAN-based synthetic brain MR image generation. In 2018 IEEE 15th International Symposium on Biomedical Imaging 734–738 (IEEE, 2018). [Google Scholar]
- 44.Shin H-C et al. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In Simulation and Synthesis in Medical Imaging 1–11 (Springer, 2018). [Google Scholar]
- 45.Han C et al. Combining noise-to-image and image-to-image GANs: brain MR image augmentation for tumor detection. IEEE Access 7, 156966–156977 (2019). [Google Scholar]
- 46.Uzunova H, Ehrhardt J & Handels H Memory-efficient GAN-based domain translation of high resolution 3D medical images. Comput. Med. Imaging Graph 86, 101801 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Zia T, Murtaza S, Bashir N, Windridge D & Nisar Z VANT-GAN: adversarial learning for discrepancy-based visual attribution in medical imaging. Pattern Recognit. Lett 156, 112–118 (2022). [Google Scholar]
- 48.Fernandez-Quilez A, Parvez O, Eftestøl T, Kjosavik SR & Oppedal K Improving prostate cancer triage with GAN-based synthetically generated prostate ADC MRI. In Proc. Medical Imaging 2022: Computer-Aided Diagnosis Vol. 12033, 436–441 (SPIE, 2022). [Google Scholar]
- 49.Frid-Adar M, Klang E, Amitai M, Goldberger J & Greenspan H Synthetic data augmentation using GAN for improved liver lesion classification. In 2018 IEEE 15th International Symposium on Biomedical Imaging 289–293 (IEEE, 2018). [Google Scholar]
- 50.Pang T, Wong JHD, Ng WL & Chan CS Semi-supervised GAN-based radiomics model for data augmentation in breast ultrasound mass classification. Comput. Methods Programs Biomed 203, 106018 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Iqbal T & Ali H Generative adversarial network for medical images (MI-GAN). J. Med. Syst 42, 231–11 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Wang S et al. Diabetic retinopathy diagnosis using multichannel generative adversarial network with semisupervision. IEEE Trans. Autom. Sci. Eng 18, 574–585 (2020). [Google Scholar]
- 53.You A, Kim JK, Ryu IH & Yoo TK Application of generative adversarial networks (GAN) for ophthalmology image domains: a survey. Eye Vis 9, 1–19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahmad B et al. Improving skin cancer classification using heavy-tailed Student t-distribution in generative adversarial networks (TED-GAN). Diagnostics 11, 2147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghorbani A, Natarajan V, Coz D & Liu Y DermGAN: synthetic generation of clinical skin images with pathology. In Machine Learning for Health Workshop 155–170 (PMLR, 2020); https://proceedings.mlr.press/v116/ghorbani20a.html [Google Scholar]
- 56.Waheed A et al. CovidGAN: data augmentation using auxiliary classifier GAN for improved COVID-19 detection. IEEE Access 8, 91916–91923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trovato G et al. Development of a colon endoscope robot that adjusts its locomotion through the use of reinforcement learning. Int. J. Comput. Assist. Radiol. Surg 5, 317–325 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Su Y-H et al. Local style preservation in improved GAN-driven synthetic image generation for endoscopic tool segmentation. Sensors 21, 5163 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cartucho J, Tukra S, Li Y, Elson S, D. & Giannarou, S. VisionBlender: a tool to efficiently generate computer vision datasets for robotic surgery. Comput. Methods Biomech. Biomed. Eng. Imaging Vis 9, 331–338 (2021). [Google Scholar]
- 60.Kausch L et al. Toward automatic C-arm positioning for standard projections in orthopedic surgery. Int. J. Comput. Assist. Radiol. Surg 15, 1095–1105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kausch L et al. C-arm positioning for spinal standard projections in different intra-operative settings. In International Conference on Medical Image Computing and Computer-Assisted Intervention 352–362 (Springer, 2021). [Google Scholar]
- 62.Van Houtte J, Audenaert E, Zheng G & Sijbers J Deep learning-based 2D/3D registration of an atlas to biplanar X-ray images. Int. J. Comput. Assist. Radiol. Surg 17, 1333–1342 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao C, Unberath M, Taylor R & Armand M Localizing dexterous surgical tools in X-ray for image-based navigation Preprint at https://arxiv.org/abs/1901.06672 (2019).
- 64.Zaech J-N et al. Learning to avoid poor images: towards task-aware C-arm cone-beam CT trajectories. In International Conference on Medical Image Computing and Computer-Assisted Intervention 11–19 (Springer, 2019). [Google Scholar]
- 65.Thies M et al. A learning-based method for online adjustment of C-arm cone-beam CT source trajectories for artifact avoidance. Int. J. Comput. Assist. Radiol. Surg 15, 1787–1796 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park J, Han DK & Ko H Adaptive weighted multi-discriminator cyclegan for underwater image enhancement. J. Mar. Sci. Eng 7, 200 (2019). [Google Scholar]
- 67.Dhont J, Verellen D, Mollaert I, Vanreusel V & Vandemeulebroucke J RealDRR—rendering of realistic digitally reconstructed radiographs using locally trained image-to-image translation. Radiother. Oncol 153, 213–219 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Alber M et al. Integrating machine learning and multiscale modeling—perspectives, challenges, and opportunities in the biological, biomedical, and behavioral sciences. NPJ Digit. Med 2, 115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Viceconti M, Henney A & Morley-Fletcher E In silico clinical trials: how computer simulation will transform the biomedical industry. Int. J. Clin. Trials 3, 37–46 (2016). [Google Scholar]
- 70.Badano A et al. Evaluation of digital breast tomosynthesis as replacement of full-field digital mammography using an in silico imaging trial. JAMA Netw. Open 1, e185474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grimm M, Esteban J, Unberath M & Navab N Pose-dependent weights and domain randomization for fully automatic X-ray to CT registration Preprint at https://arxiv.org/abs/2011.07294 (2020). [DOI] [PubMed]
- 72.Miao S, Wang ZJ & Liao R A CNN regression approach for real-time 2D/3D registration. IEEE Trans. Med. Imaging 35, 1352–1363 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Miao S et al. Dilated FCN for multi-agent 2D/3D medical image registration. In Proc. AAAI Conference on Artificial Intelligence Vol. 32, No. 1 (AAAI, 2018). [Google Scholar]
- 74.Zhang Y, Miao S, Mansi T & Liao R Task driven generative modeling for unsupervised domain adaptation: application to X-ray image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention 599–607 (Springer, 2018). [Google Scholar]
- 75.Zhang Y, Miao S, Mansi T & Liao R Unsupervised X-ray image segmentation with task driven generative adversarial networks. Med. Image Anal 62, 101664 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Shiode R et al. 2D–3D reconstruction of distal forearm bone from actual X-ray images of the wrist using convolutional neural networks. Sci. Rep 11, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bier B et al. X-ray-transform invariant anatomical landmark detection for pelvic trauma surgery. In International Conference on Medical Image Computing and Computer-Assisted Intervention 55–63 (Springer, 2018). [Google Scholar]
- 78.Dosovitskiy A et al. An image is worth 16×16 words: transformers for image recognition at scale Preprint at 10.48550/arXiv.2010.11929 (2021). [DOI]
- 79.Gao C et al. Data associated with the publication: Synthetic data accelerates the development of generalizable learning-based algorithms for X-ray image analysis 10.7281/T1/2PGJQU, Johns Hopkins Research Data Repository, V1 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao C. arcadelab/synthex: synthex. Zenodo 10.5281/zenodo.7537597 (2023). [DOI] [Google Scholar]
- 81.Gao C et al. Fiducial-free 2D/3D registration for robot-assisted femoroplasty. IEEE Trans. Med. Robot. Bionics 2, 437–446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nikou C, Jaramaz B, DiGioia AM & Levison TJ Description of anatomic coordinate systems and rationale for use in an image-guided total hip replacement system. In International Conference on Medical Image Computing and Computer-Assisted Intervention 1188–1194 (Springer, 2000). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We provide access web links for public data used in our study. The DOI link to the dataset is https://doi.org/10.7281/T1/2PGJQU (ref.79). The hip-imaging CT scans are selected from the New Mexico Decedent Image Database at https://nmdid.unm.edu/resources/data-information. The hip-imaging real cadaveric CT scans and X-rays can be accessed at https://github.com/rg2/DeepFluoroLabeling-IPCAI2020. The COVID-19 lung CT scans can be accessed at https://www.imagenglab.com/news-ite/covid-19/. The COVID-19 real CXR data can be accessed at https://www.kaggle.com/datasets/aysendegerli/qatacov19-dataset. The COVID-19 3D lesion segmentation pre-trained network module and associated CT scans can be accessed upon third-party restriction at https://github.com/HiLab-git/COPLE-Net.


