Summary
Background
Recent studies have reported the predictive and prognostic value of novel transcriptional factor-based molecular subtypes in small-cell lung cancer (SCLC). We conducted an in-depth analysis pairing multi-omics data with immunohistochemistry (IHC) to elucidate the underlying characteristics associated with differences in clinical outcomes between subtypes.
Methods
IHC (n = 252), target exome sequencing (n = 422), and whole transcriptome sequencing (WTS, n = 189) data generated from 427 patients (86.4% males, 13.6% females) with SCLC were comprehensively analysed. The differences in the mutation profile, gene expression profile, and inflammed signatures were analysed according to the IHC-based molecular subtype.
Findings
IHC-based molecular subtyping, comprised of 90 limited-disease (35.7%) and 162 extensive-disease (64.3%), revealed a high incidence of ASCL1 subtype (IHC-A, 56.3%) followed by ASCL1/NEUROD1 co-expressed (IHC-AN, 17.9%), NEUROD1 (IHC-N, 12.3%), POU2F3 (IHC-P, 9.1%), triple-negative (IHC-TN, 4.4%) subtypes. IHC-based subtype showing high concordance with WTS-based subtyping and non-negative matrix factorization (NMF) clusterization method. IHC-AN subtype resembled IHC-A (rather than IHC-N) in terms of both gene expression profiles and clinical outcomes. Favourable median overall survival was observed in IHC-A (15.2 months) compared to IHC-N (8.0 months, adjusted HR 2.3, 95% CI 1.4–3.9, p = 0.002) and IHC-P (8.3 months, adjusted HR 1.7, 95% CI 0.9–3.2, p = 0.076). Inflamed tumours made up 25% of cases (including 53% of IHC-P, 26% of IHC-A, 17% of IHC-AN, but only 11% of IHC-N). Consistent with recent findings, inflamed tumours were more likely to benefit from first-line immunotherapy treatment than non-inflamed phenotype (p = 0.002).
Interpretation
This study provides fundamental data, including the incidence and basic demographics of molecular subtypes of SCLC using both IHC and WTS from a comparably large, real-world Asian/non-Western patient cohort, showing high concordance with the previous NMF-based SCLC model. In addition, we revealed underlying biological pathway activities, immunogenicity, and treatment outcomes based on molecular subtype, possibly related to the difference in clinical outcomes, including immunotherapy response.
Funding
This work was supported by AstraZeneca, Future Medicine 2030 Project of the Samsung Medical Center [grant number SMX1240011], the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) [grant number 2020R1C1C1010626] and the 7th AstraZeneca-KHIDI (Korea Health Industry Development Institute) oncology research program.
Keywords: Small cell lung cancer, Molecular subtype, ASCL1, NEUROD1, POU2F3
Research in context.
Evidence before this study
Small-cell lung cancer (SCLC) exhibits distinct molecular subtypes. While recent trials have highlighted immunotherapy advantages for specific patients, the precise link between these molecular subtypes and their responsiveness to immunotherapy remains uncertain.
Added value of this study
Conducted on a substantial scale, this study demonstrated the practicality of using immunohistochemistry to explore genomic, transcriptomic, and immune traits in newly identified SCLC subtypes (-A, -N, -P). The research unveiled survival differences among these subtypes, highlighting superior outcomes for A and AN compared to N and P. Additionally, our findings show a closer resemblance of subtype AN to A than to N in both genomic and transcriptomic profiles. A significant discovery was the presence of the inflamed phenotype across molecular subtypes with varying prevalence, indicating that the inflamed phenotype is not exclusive to transcriptional subtypes of SCLC.
Implications of all the available evidence
Each subtype is associated with distinct pathway altered and subsequent therapeutic vulnerabilities, opening new opportunities for personalized treatment. Recognizing the inflamed phenotype as an additional dimension in the proposed molecular classification of SCLC may optimize the current treatment paradigm of SCLC including immunotherapy.
Introduction
Small cell lung cancer (SCLC) is the most aggressive form of lung cancer, accounting for approximately 15% of all newly diagnosed lung cancers. The clinical features of SCLC are characterised by rapid proliferation, with clinical response to platinum-based chemotherapy combined with DNA topoisomerase inhibitor.1,2 Despite the rapid response to chemotherapy, the 1- and 2-year overall survival (OS) of extensive-disease (ED) SCLC remains approximately 29% and 7%, respectively.3 Introduction of immune checkpoint blockade (ICB) improved the median OS among patients with treatment-naïve ED-SCLC. However, the median OS remains dismal at around 12 months.4
To explore novel treatment options for SCLC, its characteristics have been investigated based on comprehensive genomic profiling using DNA sequencing which revealed almost universal bi-allelic inactivation of TP53 and RB1, and frequent inactivating mutations in NOTCH family genes.5,6 However, somatic alterations in SCLC demonstrated limited predictive and prognostic value, and targeted approaches based on such alterations were insufficient to prove clinical benefit.7,8 Beyond somatic alterations, the subtyping of SCLC has recently moved on to transcription factor-based molecular classification such as SCLC-A (ASCL1), SCLC-N (NEUROD1), SCLC-P (POU2F3) and SCLC-I defined as inflamed subtype with lack of significant expression in A/N/P.9 The consensus classification includes neuroendocrine (NE) subset which expresses ASCL1 and/or NEUROD1, and non-NE subset expressing POU2F3. Evidence supports the hypothesis that differences in gene expression based on certain subtypes might be related to the distinct cell of origin, such as POU2F3 subtype derived from chemosensory tuft cells.10 Moreover, each subtype is associated with distinct pathway altered and subsequent therapeutic vulnerabilities,9 opening new opportunities for personalized treatment.
There has been a subsequent observation that ASCL1 and NEUROD1 are co-expressed in a single tumour,11,12 and the question remains whether this accounts for a distinct subtype or a transitional state, as the plasticity between each subtype (ASCL1 to NEUROD1) was observed in pre-clinical models.13,14 In addition, emerging evidence suggests that treatment failure might be related to the intra-subtype heterogeneity of SCLC, which increases during disease progression.9,15 These findings underscore the complexity of molecular subtypes and indicates the clear need for in-depth analysis of the underlying characteristics of each molecular subtype.
More importantly, additional research into the clinical relevance of these subtypes is needed, including as it relates to limited vs. extensive stage disease and with different types of systemic treatment (chemotherapy vs. chemo-immunotherapy). For example, a certain subtype itself may be correlated with a poor prognosis to the standard treatment. Moreover, an increasing body of evidence suggests that the molecular classification could be correlated with clinical profile and potentially used as a biomarker for subtype-directed therapy.16 A prominent example has been shown in post hoc analysis from IMpower133 and CASPIAN study, which demonstrated that the clinical benefit of ICB was enhanced in inflamed SCLC subtype by non-negative matrix factorization (NMF) method, (NMF-I)—tumours with low expression of all three transcription factors.9,17 However, whether major transcription factors are mutually exclusive to inflamed gene signatures needs clarification.18,19 Furthermore, translating these observations into clinics awaits validation on large-scale data.
In this study, we carried out a comprehensive analysis of the molecular subtypes of SCLC. To expand its clinical utility, subtyping was primarily performed with IHC-based molecular subtyping and was corroborated by target exome sequencing and gene expression data to explore differences in the underlying pathway activity and immunogenicity. Matched with clinical outcomes, we uncovered the prognostic and predictive significance of the molecular subtypes.
Methods
Patients and study design
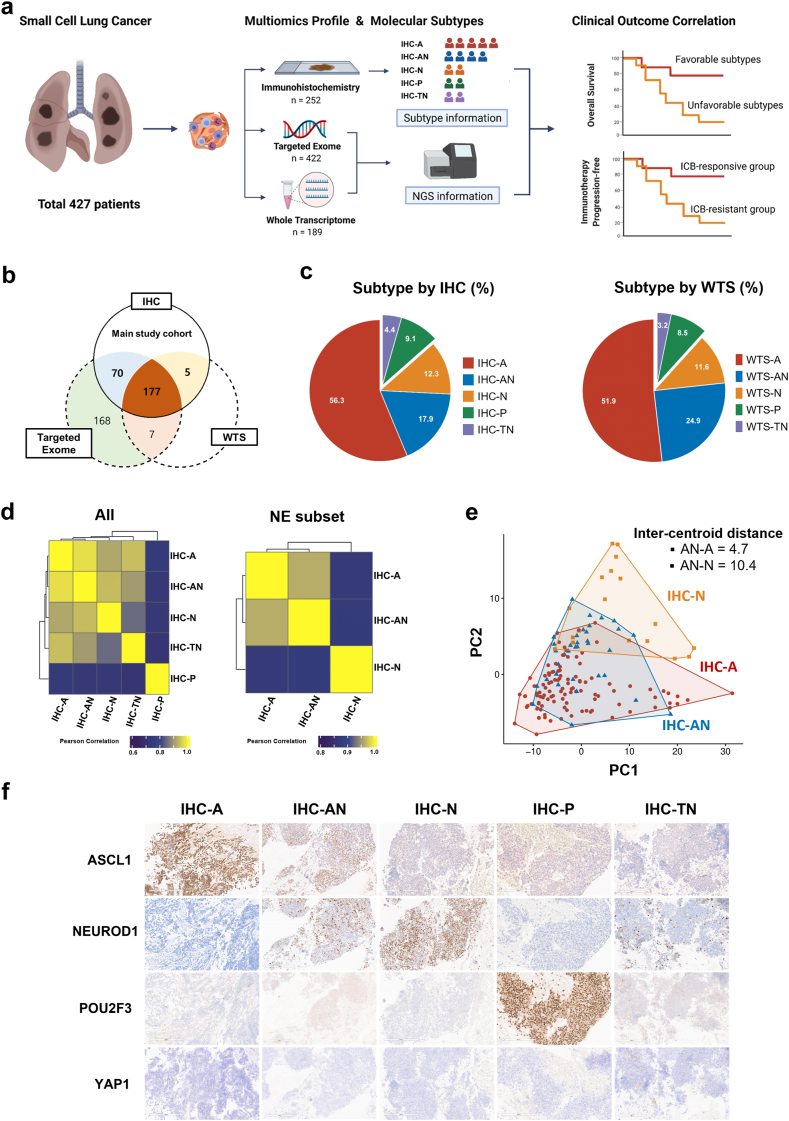
This study was designed as part of an exploratory analysis using prospectively collected samples from a multi-center, biomarker-driven, umbrella clinical trial called SUKSES (Small Cell Lung Cancer Umbrella Korea StudiES, NCT02688894),20 and patients from Samsung Medical Center were included for the current research. Formalin-fixed, paraffin-embedded (FFPE) samples from January 2013 to December 2020 were acquired from 427 patients with pathologically confirmed de novo SCLC who agreed to undergo molecular screening before treatment (Fig. 1a).
Fig. 1.
Transcription factor-based molecular subtyping of SCLC. (a) Study design. (b) Venn diagram showing the number of samples assessed by IHC (main study cohort), targeted exome, and WTS. (c) IHC subtypes (left) and WTS subtypes (right). (d) Differentially expressed genes among IHC subtypes (left) and among NE subset (right). (e) PCA of IHC-A, IHC-AN, and SCLC-N. (f) Representative H&E images of the five molecular subtypes.
Immunohistochemical staining (IHC, n = 252), targeted exome sequencing (TES, n = 422), and whole transcriptome sequencing (WTS, n = 182) were performed completely or partly depending on the tissue availability (Fig. 1b). The IHC cohort was primarily analysed to evaluate the clinical outcomes according to molecular subtypes of SCLC. Additionally, genomic and transcriptomic correlates of SCLC subtypes were explored at the intersection of the IHC and TES cohorts and the intersection of the IHC and WTS cohorts, respectively.
Ethics statement
All patients provided informed consent for sample acquisition for research purposes, and the study was approved by the institutional review board protocol (Samsung Medical Center IRB nos. 2013-10-112 and 2016-02-071). This approval ensures that the study complies with the ethical standards for research involving human participants, as outlined in the Declaration of Helsinki.
Clinical data acquisition and outcome definition
Clinical data were retrieved directly from the SUKSES trial registry, which was prospectively and regularly updated by a trained data manager, including treatment histories, responses, and survival outcomes. Survival information was updated in December 2021 through electronic medical records and the National Cancer Registry. The objective response was assessed by physicians using the Response Evaluation Criteria in Solid Tumours (RECIST) v1.1. Overall survival (OS) was defined as the duration from the date of diagnosis to the date of death from any cause or the last follow-up. Progression-free survival (PFS) was defined as the duration from the date of diagnosis until disease progression or death, whichever occurs first. The durable clinical benefit (DCB) to the combination ICB treatment was defined as a response maintained for more than six months, whereas non-durable benefit (NDB) was defined as not achieving a DCB.
Histology and immunohistochemical (IHC) staining
Tumour H&E slides were thoroughly evaluated by thoracic pathologists to determine the proportion of viable tumour cells and their representativeness. Consecutive 4-μm-thick tissue sections were cut from FFPE tissues and stained for ASCL1, NEUROD1, POU2F3, and YAP1. Following primary antibodies were applied: ASCL1 (Clone 24B72D11.1, BD Biosciences, Cat# 556604, RRID:AB_396479, 1:25) and YAP1 (Clone 2F12, Abcam, Cat# ab56701, RRID:AB_2219140, 1:200), both stained using the DAKO Autostainer Link 48. Antigen retrieval was performed with a low pH solution (DAKO) for 20 min, followed by primary antibody incubation of 10 min for ASCL1 and 60 min for YAP1. NEUROD1 (Clone EPR20766, Abcam, Cat# ab213725, RRID:AB_2801303, 1:250) and POU2F3 (Polyclonal, Novus Biologicals, Cat# NBP1-83966, RRID:AB_11024500, 1:200) were processed on the VENTANA BenchMark ULTRA system, using CC1 solution for antigen retrieval (64 min for NEUROD1 and 48 min for POU2F3), with a 60-min primary antibody incubation. FFPE cell lines pellets known to express ASCL1, NEUROD1, POU2F3, and YAP1 were used to establish optimal condition, sensitivity and specificity for each IHC.
Targeted exome sequencing (TES)
DNA sequencing was performed using CancerSCAN version 2, a tumour-only, targeted NGS panel with exonic regions of 381 cancer-related genes (1.07 Mb).21 A list of the genes covered in the CancerSCAN panel is provided in Table 1, Appendix p 10. Genomic DNA (200 ng) from the tumour was extracted from FFPE samples using an AllPrep DNA/RNA Mini kit (Qiagen, Hilden, Germany) and prepared as previously described.21 Paired-end sequencing was performed using the HiSeq 2500 platform (Illumina, San Diego, CA, USA). For the TES, 800 × was set as the mean target coverage. Sequenced reads were aligned to the hg19 reference genome using BWA-MEM (v0.7.5). SAMTOOLS (v0.1.18), Picard (v1.93), and GATK (v3.1-1) were used for manipulating the aligned sequenced data. Variants were detected using MuTect2 for single nucleotide variations (SNVs) and using Pindel (v0.2.4) for small indels. We applied several procedures to filter out the putative germline variants as previously described. ANNOVAR was used to functionally annotate variants. In addition to SNVs and indels, somatic copy number variations (CNV) and tumour purity were detected using custom algorithms as previously described. If the adjusted amplitude of the copy change is greater than 1 or less than 1 (in log scale), the region is called as amplification or deletion, respectively. Annotated SNV, indel, and CNV data was merged and converted to a MAF object using R package maftools. Mutated genes at the intersection of the CancerSCAN panel and the a priori oncogenic pathway templates were visualized in a heatmap using R package maftools. A pathway was considered altered in a tumour if ≥1 gene within the corresponding pathway template was altered.
Table 1.
Characteristics of the main study cohort.
| Variables | Overall (n = 252) | VALSG stage |
|
|---|---|---|---|
| Limited disease |
Extensive disease |
||
| (n = 90) | (n = 162) | ||
| Age, year | 66 [60, 72] | 64 [59, 70] | 67 [61, 72] |
| Sex, male | 216 (86) | 77 (86) | 139 (86) |
| Smoker | 227 (90) | 82 (91) | 145 (89) |
| ECOG performance status | |||
| 0 | 11 (4) | 7 (8) | 4 (2) |
| 1 | 228 (91) | 79 (88) | 149 (92) |
| ≥2 | 13 (5) | 4 (4) | 9 (6) |
| Sites of biopsy | |||
| Lymph nodes | 168 (67) | 60 (67) | 108 (67) |
| Lunga | 72 (29) | 27 (30)∗ | 45 (28) |
| Brain | 6 (2) | 0 (0) | 6 (4) |
| Other sites | 6 (2) | 3 (3) | 3 (2) |
| TNM stage (7th) | |||
| I | 8 (3) | 8 (9) | 0 (0) |
| II | 9 (4) | 9 (10) | 0 (0) |
| IIIA | 34 (14) | 34 (38) | 0 (0) |
| IIIB | 39 (16) | 39 (43) | 0 (0) |
| IV (M1a) | 20 (8) | 0 (0) | 20 (12) |
| IV (M1b) | 142 (56) | 0 (0) | 142 (88) |
| Metastatic sites, ED | |||
| Brain | 26 (10) | 0 (0) | 26 (16) |
| Liver | 22 (9) | 0 (0) | 22 (14) |
| Bone | 37 (15) | 0 (0) | 37 (23) |
| Adrenal gland | 13 (5) | 0 (0) | 13 (8) |
| Other organs | 44 (18) | 0 (0) | 44 (27) |
| First-line treatment, LD | |||
| Concurrent chemoradiation | 80 (32) | 80 (89) | 0 (0) |
| Surgery + adjuvant chemotherapy | 10 (4) | 10 (11) | 0 (0) |
| First-line treatment, ED | |||
| Chemotherapy + immunotherapy | 41 (16) | 0 (0) | 41 (25) |
| Chemotherapy only | |||
| Etoposide + platinum | 118 (47) | 0 (0) | 118 (73) |
| Irinotecan + platinum | 1 (0) | 0 (0) | 1 (1) |
| Other regimens | 2 (1) | 0 (0) | 2 (1) |
Data are shown as number (%) or median (interquartile range). TNM stage was presented in accordance with the 7th edition of the American Joint Committee for Cancer Staging Manual.
IASLC, International Association of Study for Lung Cancer; ASA, American Society of Anesthesiologists classification; PET, Positron Emission Tomography; MRI, Magnetic resonance imaging; LVI, lymphovascular invasion; PNI, perineural invasion; EGFR, epidermal growth factor receptor.
Samples from nine cases of limited disease were obtained through curative resection, while the remaining samples were acquired from biopsies.
Whole transcriptome sequencing (WTS)
RNA (1 μg) was extracted from formalin-fixed paraffin-embedded (FFPE) samples containing more than 70% tumour proportion on the corresponding H&E slides. The AllPrep DNA/RNA Mini Kit (Qiagen) and the TruSeq RNA Access Library Prep Kit v2 (Illumina) were used for RNA extraction and preparation. RNA concentration and integrity were measured using a NanoDrop 8000 UV-Vis spectrometer (NanoDrop Technologies Inc., Wilmington, DE, USA). cDNA libraries were qualified with the Agilent 2100 BioAnalyser (Agilent Technologies, Santa Clara, CA, USA). Paired-end sequencing was performed using the HiSeq 2500 platform (Illumina). The reads from FASTQ files were mapped against hg19 using the 2-pass mode of STAR (v2.4.0). RNA-SeQC was conducted to control the quality of the BAM files. Raw read counts mapped to genes were analysed for transcript abundance using RSEM (v1.2.18), and poorly expressed samples were eliminated using the cutoff of total read count less than 1 million. Normalised read counts were transformed to log2 counts per million (log2 CPM) with the R package edgeR and used for downstream analyses.
Differentially expressed genes (DEGs) were calculated and visualized using the R package edgeR and EnhancedVolcacno with default parameters. To compare similarities of DEGs across SCLC subtypes, the overlap correlation of DEG was calculated using a binarized matrix method (whether a DEG belonged to the subtype or not) and visualized via hierarchical clustering. Gene set enrichment analysis (GSEA) for MSigDB Hallmark pathways was performed using the R package fgsea. the web-based module (GenePattern, Broad Institute). Gene set scoring for individual sample was performed using the R package singscore with default parameters. The gene sets used for single sample scoring in this study are summarized in Table 2, Appendix p 11.
Table 2.
Adjusted hazard ratios for overall survival.
| Molecular subtypes | Events/Patients | Overalla |
Limited diseaseb |
Extensive diseaseb |
|||
|---|---|---|---|---|---|---|---|
| Adjusted HR (95% CI) | p | Adjusted HR (95% CI) | p | Adjusted HR (95% CI) | p | ||
| IHC-A | 116/142 | 1.00 (Reference) | – | 1.00 (Reference) | – | 1.00 (Reference) | – |
| IHC-AN | 36/45 | 0.88 (0.60–1.30) | 0.53 | 0.86 (0.44–1.68) | 0.67 | 0.91 (0.55–1.50) | 0.70 |
| IHC-N | 28/31 | 1.89 (1.24–2.88) | 0.0031 | 1.14 (0.50–2.61) | 0.75 | 2.32 (1.38–3.92) | 0.0022 |
| IHC-P | 21/23 | 1.00 (0.61–1.64) | 0.99 | 0.54 (0.22–1.34) | 0.18 | 1.74 (0.94–3.19) | 0.076 |
| IHC-TN | 9/11 | 1.28 (0.64–2.56) | 0.49 | 1.40 (0.32–6.21) | 0.66 | 1.30 (0.56–2.98) | 0.54 |
Adjusted for age at diagnosis, sex, ECOG, and VALSG stage.
Adjusted for age at diagnosis, sex, ECOG, TNM stage, and lines of therapy received. HR, hazard ratio; CI, confidence interval; IHC, immunohistochemistry.
Assessment of IHC molecular subtype
The molecular subtypes were determined using predefined IHC criteria (Fig. 1a, Appendix p 1). Briefly, the H-score was derived as the sum of the proportion multiplied by its corresponding intensity (range: 0–300). First, samples with high POU2F3 expression (H-score ≥200) were assigned as IHC-P, as POU2F3 expression of this level were highly exclusive to other subtype markers, while lower expression was associated with the expression of ASCL1 or NEUROD1. Next, samples with non-significant expression (H-score <50) of both ASCL1 and NEUROD1 were assigned as IHC-TN (triple-negative). In the remaining samples, a significant overlap between ASCL1 and NEUROD1 expression was observed, as previously reported.11 Therefore, we assigned subtypes based on dominant marker (i.e., more than 2-fold of the comparator, Fig. 1b, Appendix p 1), resulting in the following three subtypes: IHC-A (ASCL1-dominant), IHC-N (NEUROD1-dominant), and IHC-AN (double-positive without dominance on either side). All interpretations were conducted by two independent thoracic pathologists with no access to clinical outcomes.
Assessment of WTS molecular subtype
The WTS-based molecular subtypes were evaluated using a supervised method (WTS subtypes: WTS-A, WTS-AN, WTS-N, WTS-P, and WTS-TN, Fig. 1c, Appendix p 1). For WTS-based molecular subtyping, we used z-normalised signature scores of ASCL1, NEUROD1, and POU2F3 based on gene sets representative of each subtype. Signature gene sets unique to ASCL1 and NEUROD1 were obtained from the publication by Borromeo et al.22 Tuft cell-like signature was obtained from the publication by Huang et al. and used as a POU2F3 subtype gene set.10 Thresholds of z-value for determining each subtype were manually selected on the basis of the score distribution (Fig. 1d, Appendix p 1). Thereafter, samples were annotated with WTS subtype of either WTS-A, WTS-AN, WTS-N, WTS-P, or WTS-TN. To assess the robustness of IHC-based subtype determination, the calling accuracy between two methods, the standard IHC and WTS, was evaluated. We further correlated our IHC subtype with the most recent consensus classification method, using NMF-identified subtypes, NMF-A, NMF-N, NMF-P, NMF-I as previously reported.9
Assessment of inflamed phenotype and tumour microenvironment (TME) subtype
Unsupervised hierarchical clustering (k = 2) was performed in the WTS cohort to annotate “Inflamed” and “Non-inflamed” types. Following three signatures of effector cells (T-cell and NK-cell) were used for clustering: T-cell inflamed signature, cytolytic signature, and NK-cell signature. TME subtype was determined using the algorithm molecular functional portrait (MFP), a transcriptomic-based TME classification platform developed by BostonGene (Waltham, MA, USA). Using an expression matrix of the WTS cohort as input, TME subtypes were classified into four subtypes as follows: IE/F (immune enriched/fibrotic), IE (immune enriched, non-fibrotic), F (fibrotic), and D (depleted). In addition to TME subtype information, signature scores of diverse immune subsets related to ICB response were also calculated using the MFP algorithm. We also utilised MFP algorithm to calculate scores for cancer-associated fibroblasts (CAF), matrix remodeling, and epithelial–mesenchymal transition (EMT) based on single-sample Gene Set Enrichment Analysis (ssGSEA). The gene sets for each category were as follows: For CAFs, the gene set included: COL1A1, COL1A2, COL5A1, ACTA2, FGF2, FAP, LRP1, CD248, COL6A1, COL6A2, COL6A3, CXCL12, FBLN1, LUM, MFAP5, MMP3, MMP2, PDGFRB, and PDGFRA. For the matrix component, the gene set comprised: FN1, COL1A1, COL1A2, COL4A1, COL3A1, VTN, LGALS7, LGALS9, LAMA3, LAMB3, LAMC2, TNC, ELN, COL5A1, and COL11A1. For EMT, the gene set included: SNAI1, SNAI2, TWIST1, TWIST2, ZEB1, ZEB2, and CDH2.
Statistical methods
Descriptive statistics were used to summarize the patient characteristics. Fisher's exact test was used to assess the association between two categorical variables, which was necessitated by more than 20% of the cells having expected counts below five. Differences in continuous variables between the groups were tested using the Wilcoxon rank-sum test (also known as Mann-Whiney U test). Concordance between subtype calling methods was evaluated using balanced accuracy and Cohen's unweighted kappa value calculated from the a two- or three-class confusion matrix. Pearson's correlation (coefficient r) was used to analyse the strength and direction of the linear relationship between two continuous variables. The survival time was measured from the date of diagnosis, which serves as both the origin and the start time for our analysis. Kaplan–Meier estimates and log-rank tests were used for survival analysis, and median survival time was calculated using the Brookmeyer and Crowley method. The Cox proportional hazards regression model was used to calculate the adjusted hazard ratio (HR) with confidence interval (CI). The proportional hazards assumption was assessed using Schoenfeld residuals, and the linearity assumption was checked through a graphical method using Martingale residuals from the Cox model. Variables were selected through a univariable approach, which were then refined using backward elimination in the multivariable model. In the overall patients, the models were adjusted for age at diagnosis, sex, ECOG, VALSG stage, and lines of therapy received. In the stratified analysis of limited-stage and extensive-stage SCLC, the model was adjusted for age at diagnosis, sex, ECOG, TNM stage, and lines of therapy received. Statistical significance was set at p < 0.05.
Software and reproducibility
Data statistics and bioinformatic analyses were performed using R (https://www.r-project.org/) and Bioconductor packages (https://www.bioconductor.org/). All figures and graphs were generated using the R package ggplot2. The script used for the data processing and figure generation is available at https://github.com/SeHoonLab/molecular-subtype-SCLC, including targeted sequencing data and transcriptome data. Clinical and IHC staining data is provided as supplementary information in Table S3, Appendix p 13.
Role of funders
The funders were not involved in the study design, data collection, data analysis, interpretation or writing of the manuscript.
Results
Study population and IHC-based molecular subtyping
The main IHC cohort (n = 252) comprised 90 patients with limited-disease (LD, 35.7%) and 162 patients with extensive-disease (ED, 64.3%). The median age was 66 years, with a sex breakdown of 86% and 14% for males and females, respectively. Among the ED patients, forty-one (25%) received combination ICB as the first-line treatment. Detailed patient and sample characteristics of the main study cohort are summarized in Table 1.
A breakdown of the molecular subtypes by IHC revealed that IHC-A was the dominant subtype in 56.3% (n = 142) of the tumours, followed by IHC-AN, IHC-N, IHC-P, and IHC-TN at 17.9% (n = 45), 12.3% (n = 31), 9.1% (n = 23), and 4.4% (n = 11), respectively (Fig. 1c). The proportion of molecular subtypes did not significantly vary based on clinical factors, including smoking status, biopsy sites, and initial VALSG stage (Fig. 2, Appendix p 2).
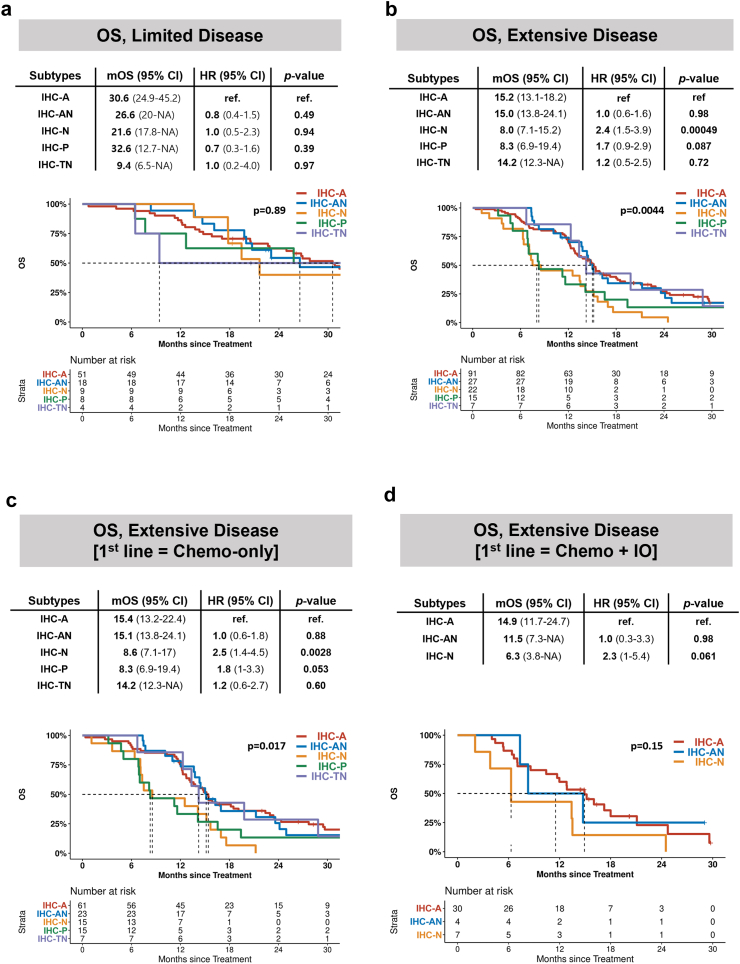
Fig. 2.
Survival analysis of IHC subtype. OS in patients with (a) limited disease and (b) extensive disease. OS in extensive disease patients treated with (c) front-line chemotherapy and (d) front-line chemotherapy plus immunotherapy.
IHC provides a clinically feasible approach for classifying SCLC subtypes
Marked differences in scores of a priori signature gene sets were observed across molecular subtypes, which supported the validity of IHC-based subtyping (Fig. 2a, Appendix p 2). Clustering of differentially expressed genes (DEGs) identified IHC-P as the most unique subtype in terms of expression profiles (Fig. 1d, left). Among neuroendocrine (NE) subsets, which includes IHC-A, AN, and N, we found that the DEG profile of IHC-AN correlated more strongly with IHC-A than with IHC-N (Pearson's coefficient, 0.96 vs. 0.87, Fig. 1d, right). Same results were reproduced using WTS subtype (Fig. 2b, Appendix p 2). Additionally, principal component analysis revealed that IHC-AN was clustered more closely with IHC-A than with IHC-N (inter-centroid distance, 4.7 vs. 10.4, Fig. 1e).
The IHC subtype was highly concordant with the WTS subtype in discriminating IHC-P from the other subtypes (balanced accuracy = 98%, Fig. 2B, Appendix p 2). When discriminating between IHC-A, IHC-AN, and IHC-N, combining IHC-AN with IHC-A improved the agreement between IHC and WTS-based subtyping (balanced accuracy = 92%; Fig. 2c, Appendix p 2), which suggests a closer similarity of the IHC-AN subtype to IHC-A than to IHC-N. NMF classification was constructed with different axis compared to our IHC-based analysis, NMF-I instead of IHC-AN and IHC-TN (Fig. 3a, Appendix p 3). Despite the difference in the method of categorization, we observed very high concordant in discriminating IHC-P from other subtypes (balanced accuracy = 100%) and IHC-AN combined with IHC-A (balanced accuracy = 91.2%), which is very similar to the analysis using WTS subtype (Fig. 3b and c, Appendix p 3). In addition, NMF-I was constructed with 14.7% of IHC-A, 20.8% of IHC-AN, 8.3% of IHC-N but none from IHC-P and IHC-TN. Collectively, we defined the molecular subtypes of SCLC according to the IHC-based criteria and conducted further analyses. Representative images of IHC-based molecular subtypes are shown in Fig. 1f.
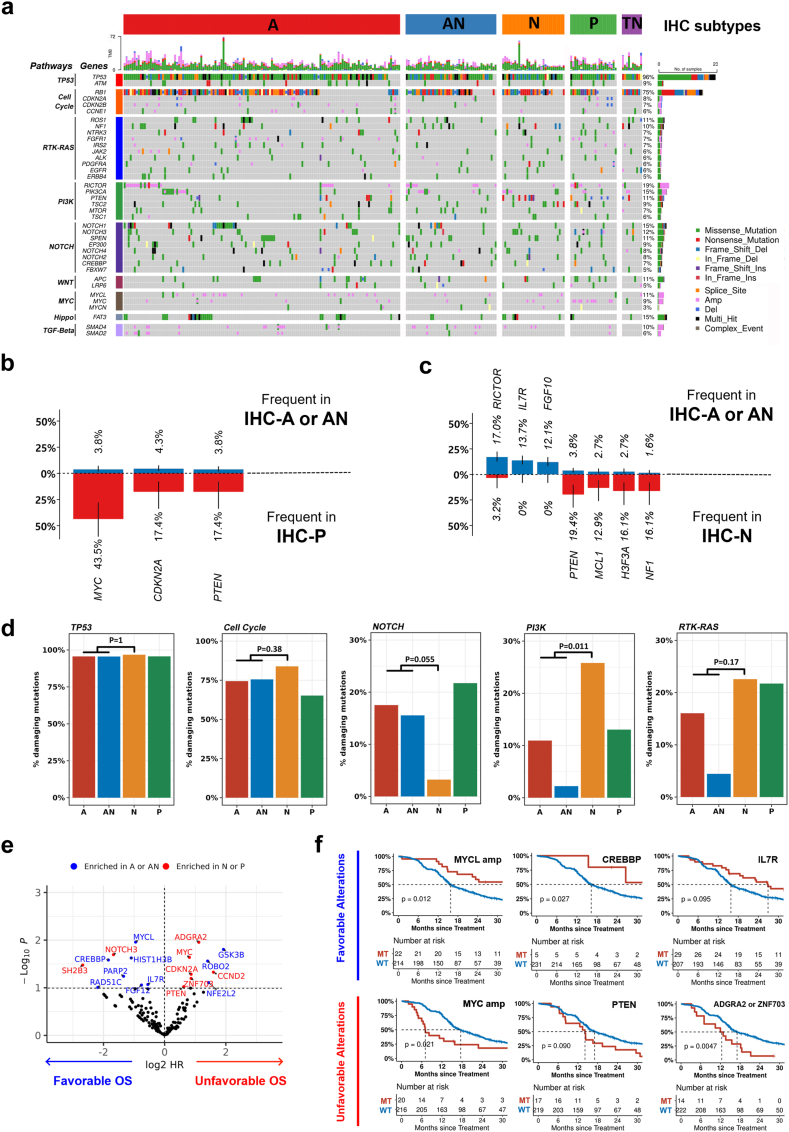
Fig. 3.
Characteristic genomic alterations among IHC subtypes. (a) Genomic alterations across SCLC subtypes. (b) Frequent genomic alterations found in IHC-P compared to A/AN. (c) Frequent genomic alterations found in IHC-N compared to A/AN. (d) Comparison of different pathway alterations among NE subset. (e) Genetic alterations associated with prognosis. Genetic alterations enriched in IHC-A or AN and IHC-N or P are indicated. (f) OS according to genomic alterations with favourable (top) or unfavourable (bottom) prognostic effect.
IHC-based subtypes are also concordant with NMF-based subtypes
NMF classification was constructed along different axes compared to our IHC-based analysis, using NMF-I instead of IHC-AN and IHC-TN (Fig. 3a, Appendix p 3). Despite the methodological differences in categorization, a very high level of concordance was observed when discriminating IHC-P subtype from other subtypes (balanced accuracy = 100%). Additionally, IHC-AN combined with IHC-A was well discriminated from IHC-N (balanced accuracy = 91.2%), which aligns with the analysis using WTS subtype (Fig. 3b and c, Appendix p 3). These findings underscore the practicality and validity of the IHC method as a reliable approach to molecular subtyping.
Survival outcomes vary across molecular subtypes
Clinical outcomes were analysed according to molecular subtype with stratification of the initial stage. In the overall cohort, the median follow-up time was 16.5 (IQR, 11.6–28.7) months. No difference in OS between the molecular subtypes in patients with LD were observed (Fig. 2a). However, among ED patients, significant differences in OS between molecular subtypes were observed, showing that IHC-A and IHC-AN had median OS of 15.2 months (95% CI, 13.1–18.2) and 15.0 months (95% CI, 13.8–24.1), respectively (Fig. 2b). In contrast, IHC-N had a significantly decreased median OS of 8.0 months (95% CI, 7.1–15.2, p = 0.00049), and IHC-P showed a decreased median OS of 8.3 months (95% CI, 6.9–19.4, p = 0.087). Subgroup analysis also supported the poor prognostic nature of IHC-N, regardless of whether immunotherapy was combined as first-line therapy or not (Fig. 2c and d).
After adjustment for age, sex, ECOG performance status, TNM stage, and total lines of therapy received, adjusted HRs (aHR) for OS comparing IHC-trans and IHC-N to IHC-A were 0.91 (95% CI, 0.55–1.50, p = 0.699) and 2.32 (95% CI, 1.38–3.92, p = 0.002) in patients with ED, respectively (Table 2). IHC-P was also associated with an increased risk of death among patients with ED, although it did not reach statistical significance (aHR, 1.74, 95% CI, 0.94–3.19, p = 0.076). Survival analysis based on PFS also showed shorter survival in IHC-N (p = 0.001) than in IHC-A (Fig. 4a–c, Appendix p 4).
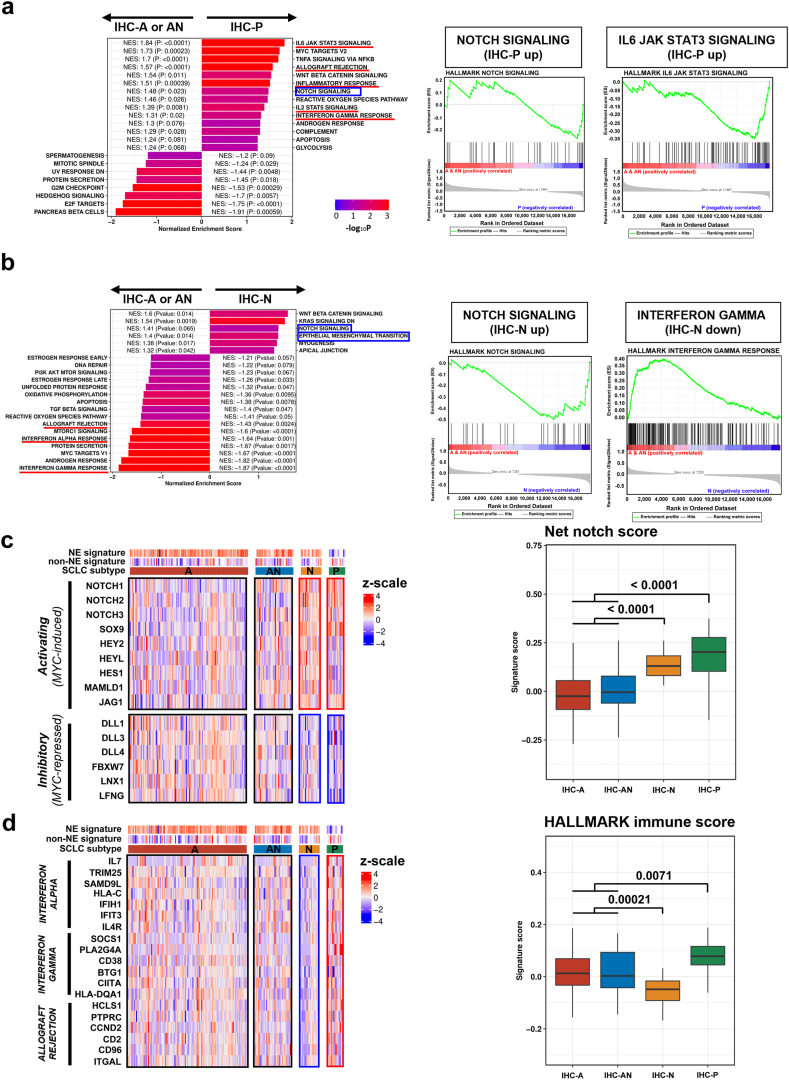
Fig. 4.
Difference in notch and immune pathway activities across IHC subtypes. (a) Gene set enrichment analysis of IHC-P compared to IHC-A/AN. (b) Gene set enrichment analysis of IHC-N compared to IHC-A/AN. (c) Differences in notch activating (MYC-induced) and notch inhibitory (MYC-repressed) gene expression across IHC subtypes. (d) Differences in HALLMARK immune gene expression across IHC subtypes.
For sensitivity analysis, survival outcomes were evaluated using the WTS-based definitions of subtypes. Compared to the WTS-A type, the WTS-N type (p = 0.016) and WTS-P type (p = 0.065) showed decreased OS (Fig. 4d, Appendix p 4). A similar trend was observed using NMF classification (Fig. 4F, Appendix p 4).
Mutational landscape analysis indicates different genetic backgrounds across molecular subtypes
We explored the genomic features potentially associated with molecular subtypes (Fig. 3a). MYC amplification, CDKN2A deletion, and PTEN loss-of-function alterations were significantly enriched in IHC-P compared to IHC-A or IHC-AN (Fig. 3b). IHC-N showed no RICTOR, IL7R, or FGF10 amplification (all located in chromosomal 5p.13) but had more frequent alterations in PTEN, MCL1, H3F3A, and NF1 genes than IHC-A or IHC-AN (Fig. 3c).
In terms of oncogenic pathways, we observed that TP53 and cell cycle pathways were universally altered without subtype-specific enrichment (Fig. 3d). However, NOTCH pathway alterations were more frequent in IHC-A/AN than in IHC-N (18% vs. 3%, p = 0.082). In contrast, the PIK3 pathway was altered more frequently in IHC-N than in IHC-A/AN (26% vs. 6%, p = 0.043). The RTK-RAS pathway was also more frequently altered in IHC-N, although the difference was not statistically significant (22% vs. 11%, p = 0.173). Details of alterations with subtype differences are summarized in Fig. 5a–d, Appendix p 5.
Fig. 5.
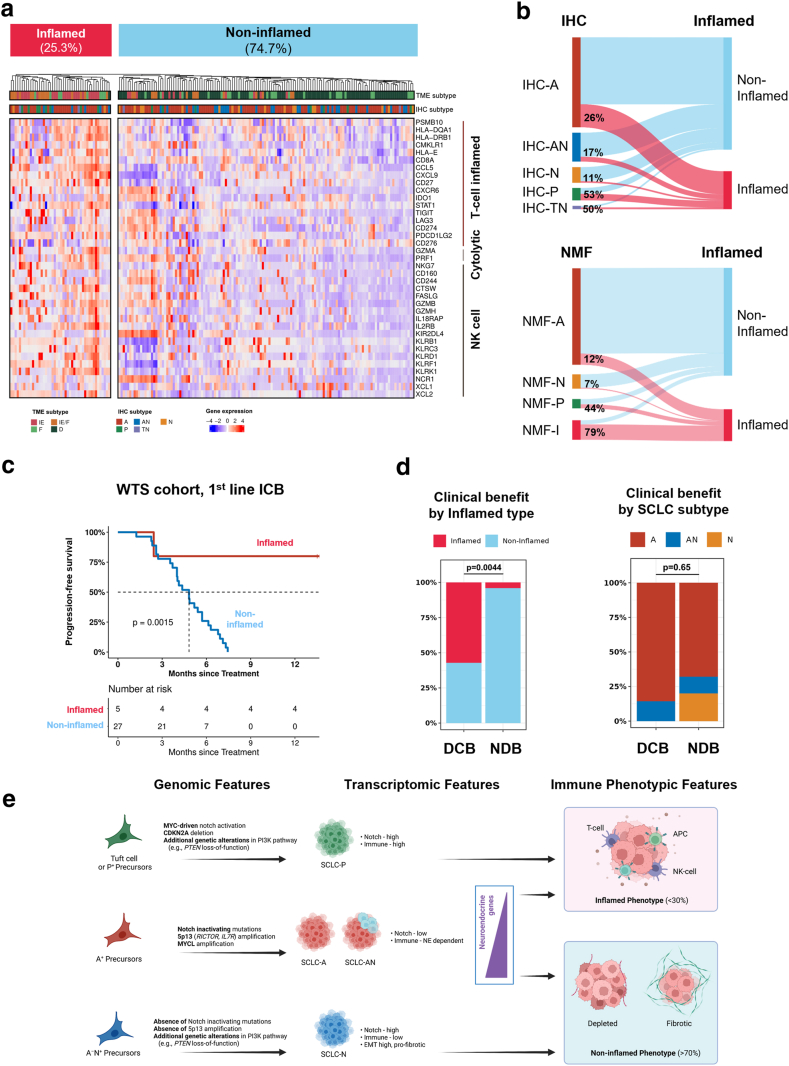
Inflamed phenotypes and their association with IHC subtype. (a) Inflamed phenotypes defined by hierarchical clustering (k = 2) using T cell, NK cell, and cytolytic activities in the WTS cohort. (b) Distribution of inflamed phenotype across IHC and NMF subtypes. (c) Progression-free survival by inflamed phenotype among patients treated with first-line immunotherapy. (d) Clinical benefit from front-line immunotherapy stratified by inflamed phenotype (left) and IHC subtype (right). (e) Summary of findings from this study.
We evaluated the association of genomic alterations with molecular subtypes and patient survival to evaluate the genomic features underlying tumour characteristics and the prognostic nature of molecular subtypes. Eighteen candidate genes were identified to have favourable (n = 9) or unfavourable (n = 9) effects on survival (log-rank p < 10−1), and were classified into two categories: A/AN-enriched or N/P-enriched (Fig. 3e). We observed that seven out of the nine (78%) unfavourable genetic alterations were mapped to N/P-enriched, including MYC amplification (HR = 1.4, p = 0.023), PTEN loss-of-function mutations (HR = 1.3, p = 0.095), or ADGRA2/ZNF703 amplification (HR = 1.6, p = 0.005, Fig. 3F).
Transcriptomic analysis revealed different activities of Notch and immune-related signaling pathway across molecular subtypes
We leveraged the transcriptomic data of 182 patients to identify the features of gene expression and/or pathway activities that may be associated with molecular subtypes. DEGs of IHC-P and IHC-N compared to IHC-A/AN are presented in Fig. S6a and b, Appendix p 6 respectively. Compared with IHC-A/AN, IHC-P exhibited significant enrichment in several HALLMARK immune-related pathways (normalised enrichment score [NES] >1), and also had an enriched HALLMARK notch signaling pathway (Fig. 4a). Unlike IHC-P, IHC-N showed significant enrichment in the HALLMARK notch signaling (NES = 1.41) and the HALLMARK epithelial–mesenchymal transition (EMT, NES = 1.40), but negative enrichment in HALLMARK immune-related pathways (Fig. 4b).
Considering the significance of crosstalk between MYC and notch pathways in SCLC tumour biology, we used a net notch score that reflected the difference in expression levels of MYC-induced targets (e.g., NOTCH1, NOTCH2, NOTCH3) and MYC-repressed targets (e.g., DLL3, FBXW7) of the notch pathway. Interestingly, both IHC-N and IHC-P showed significantly increased net notch scores compared to IHC-A/AN (p < 0.0001, Fig. 4c). Although IHC-P had the highest HALLMARK immune score among the subtypes, IHC-N had the lowest HALLMARK immune score (p < 0.0001, Fig. 4d). IHC-A and IHC-AN showed a broad spectrum of immune scores (Fig. 4d), indicating that some proportions of IHC-A and IHC-AN might have high immune activities. Collectively, while IHC-P may be characterised by high–notch activity along with high–immune activity, IHC-N might be characterised by high–notch activity but low–immune activity.
Inflamed phenotype, predictive of ICB response in SCLC, is not exclusive to specific subtype but displays varying prevalence among molecular subtypes
Using hierarchical clustering (k = 2) of gene expression derived from effector immune cells, all SCLCs were classified as inflamed or non-inflamed phenotype (Fig. 5a). Proportion of inflamed phenotype based on the IHC subtypes is shown in Figs. S7 and 8, Appendix p 7 and 8. On average, the inflamed type accounted for 25.3% of all SCLCs. However, its proportion differed substantially across molecular subtypes (Fig. 5b). Inflamed phenotype accounted for the largest proportion (53%) of IHC-P, followed by 50% of IHC-TN, 17% of IHC-AN, and 26% of IHC-A. IHC-N rarely (11%) had an inflamed phenotype. Using the NMF-based classification, similar results were reproduced, showing enrichment in inflamed phenotype in NMF-I (79%), NMF-P (44%), and NMF-N (7%). In parallel, we correlated molecular subtypes with their tumour microenvironment (Fig. S9a, Appendix p 9). The IE environment was found in a subset of IHC-P (66.7%) and IHC-A (33.9%), but rare among IHC-N (15.8%). Higher cancer-associated fibroblast score (p = 0.031), intercellular matrix score (p = 0.009), and EMT score (p = 0.113) were observed in IHC-N compared to IHC-A/AN (Fig. S9b, Appendix p 9).
We investigated several molecular parameters that could be used to delineate inflamed phenotype in IHC-A/AN, the largest subtype (Fig. S9c, Appendix p 9). The neuroendocrine (NE) score had a moderately negative correlation with the inflamed score (r = −0.32, p < 0.0001). Net notch score had a weakly positive correlation with the inflamed score (r = 0.15, p = 0.070). However, IHC-N had low inflamed scores regardless of the NE score (r = 0.19, p = 0.425) and net notch score (r = −0.18, p = 0.453).
Among patients who received combined first-line ICB treatment (n = 32), the inflamed phenotype showed a significantly longer median PFS compared with the non-inflamed phenotype (mPFS, Not reached vs. 4.82 months, p = 0.002; Fig. 5c). The inflamed phenotype (p = 0.004, Fig. 5d) and TME subtypes (p = 0.010, Fig. S9d, Appendix p 9) significantly predicted durable clinical benefit from first-line ICB combination. Compared to IHC-A/AN, IHC-N showed a shorter median PFS (4.0 months vs. 5.2 months, p = 0.112) to first-line ICB treatment (Fig. S9e, Appendix p 9).
Discussion
SCLC has long been considered a homogeneous disease based on its relatively consistent histology, until the recent proposal of distinct transcriptomic subtypes.23 Although potential prognostic and therapeutic implications have been suggested, the clinical relevance of these molecular subtypes remains obscure.9,14 Here, using IHC-based molecular subtyping to expand its clinical utility, our comprehensive analysis corroborated by targeted exome sequencing and gene expression profiling provides the prognostic and predictive significance of the molecular subtypes. Importantly, we found that IHC-N exhibits a distinct high-notch, low–immune profile distinct from IHC-A/AN and correlates with poor outcomes for both chemotherapy and immunotherapy. In addition, we showed a high-notch, high–immune profile of IHC-P, which is intrinsically aggressive but may be salvaged through immunotherapy. Furthermore, we found that the major SCLC subtypes, IHC-A and SCLC-AN, are not entirely immune-cold, and that low-NE/high–immune profiles among them may identify candidates for immunotherapy. Together, these findings may have real-world implications for subtype-based treatment approaches and can ultimately help improve clinical outcomes (Fig. 5e).
Since the initial proposal of SCLC subtypes based on the expression of representative transcriptional factor,23 there has been a suggestion in the method and nomenclature of SCLC subtypes.9 One of the representative methods was using NMF based approach, which was able to segment the potential responder of ICB (SCLC-I or NMF-I from this article). Based on the advantages of clinical utility using IHC-based approach, we matched IHC-based classification with the NMF-based method, which showed high concordance, especially in IHC-P type. However, due to the reason the NMF classification categorizes NMF-I as mutually exclusive to the other -A, -N, -P types, NMF-I patients were inevitably constructed with IHC-A (68.4%), IHC-N (5.3%) or IHC-AN (26.3%) (Fig. 3a, Appendix p 3). Further analysis using hierarchical clusterization using effector gene-related signature also included 79% of NMF-I type, which support that NMF classification has benefit in identifying inflamed phenotype in patients who is available for the WTS result. Notably, we identified that both IHC-P (53%) and NMF-P (44%) include a high proportion of inflamed phenotype, as previously reported.18,19 These findings support the necessity of further improvement in the clinically feasible classification method in segmenting inflamed phenotypes admitting that there are limitations in the IHC-based classification method.
We demonstrated that IHC-A and IHC-AN are similar in terms of genetic, transcriptomic profile. Given that ASCL1-expressing SCLC can switch to NEUROD1-expressing SCLCs, this suggests that ASCL1/NEUROD1 co-expressing tumours may represent a transitional state that might be closely related to IHC-A.13 In contrast, pure IHC-N showed genetic and transcriptomic features that are apart from those of IHC-A and IHC-AN. Strikingly, we found that NOTCH mutations are rarely found in IHC-N. Furthermore, our data suggest that IHC-N differs from IHC-A and IHC-AN in terms of notch pathway activation, which is supported by transcriptomic analysis. As growing evidence indicates that different types of SCLCs may arise from the different cell of origin and the type-specific genetic alterations, a distinct pathway involved in tumourigenesis of IHC-N may warrant further investigation.24
Furthermore, we found that IHC-N was associated with poor prognosis. There are several hypotheses to explain aggressive behavior of IHC-N. First, a higher incidence of PTEN alteration in IHC-N, related to accelerated tumour progression and metastasis, could result in poor prognosis, which is also observed in the PTEN-inactivated (Rb/p53/pten) mouse model.25 Second, the poor prognosis of IHC-N may be related to the high incidence of PI3K pathway alterations, which are recently found to be associated with chemoresistance.26 Finally, the low immune score observed in IHC-N (Fig. 4d) might be related to the comparably poor treatment outcome, which will be emphasized as immunotherapy becomes the standard treatment for SCLC. Our study identified that the molecular subtypes were significantly associated with inflamed/non-inflamed phenotypes, which may correlate with the clinical response to immunotherapy. Consistent with previous reports,9,18 we found that IHC-P and IHC-TN, the two most low-NE subtypes of SCLC, represent the most inflamed type (more than 50% inflamed), while IHC-N represents the coldest subtype (about 11% inflamed).
Notably, a recent study suggested notch signaling as a determinant of response to ICB in SCLC.27 In our study, IHC-N was a notch activated subtype but was not correlated with inflamed microenvironment. Therefore, our data supports that the role of notch signaling in immune infiltrations may not be universally applied to all SCLC subtypes, and that IHC-N may have an additional mechanism of immune evasion such as pro-fibrotic or high EMT features (Fig. S9b, Appendix p 9). Furthermore, compared to notch signaling, the loss of NE gene expression was more strongly correlated with inflamed phenotype (Fig. S9c, Appendix p 9). This may be attributed to a notch-independent mechanism,24 and further investigations on determinants of immune environment of SCLC are warranted.
Our findings related to the different clinical meaning of notch pathway activation based on molecular subtypes may help in designing future immunotherapeutic strategies for SCLC. As the survival benefit of immunotherapy is only modest in SCLC,4 novel strategies are focused on provoking the immunogenicity of SCLC. A promising example is LSD1 inhibition, which can induce antigen presentation and enhance the immunogenicity of SCLC.28,29 Intriguingly, a recent study reported that NEUROD1-positive SCLC was associated with a lack of immunogenic response to LSD1 inhibition compared with ASCL1-positive SCLC.30,31 LSD1 inhibition suppresses ASCL1 and upregulates notch pathway,25 which in turn make SCLC non-NE and immunogenic.24 Once again, the finding that LSD1 inhibition did not induce an immunogenic response in IHC-N may also support the hypothesis that the role of notch pathway activation in immunogenicity of IHC-N is not significant.
The limitations of this study are related to its single-institutional nature, which can be potentially biased. We also acknowledge that unmeasured confounding remains a concern that could impact our results, necessitating a cautious interpretation of the reported associations. Additionally, we concede that the hazard ratios employed in our analysis may carry a built-in selection bias. We were unable to conduct internal validation by splitting our cohort into training and validation sets due to the relatively small sample size and the rarity of the disease. The reporting of the standard error of zero for a kappa estimate of 100% reflects an upward bias, potentially misleading interpretations about the kappa statistic's precision and reliability. To overcome those methodological limitations, further research is required on a larger sample size and independent validation, especially in patients exposed to first-line immunotherapy.
Considering the intratumoural subtype heterogeneity of SCLC and the inherent limitations in assessing subtypes in clinical samples, our findings, especially those regarding the A, AN, and N types, should be interpreted with caution. In addition, the lack of serial sampling data leaves the question of whether IHC-based subtypes are interchangeable, as some evidence suggests that SCLC subtypes could represent different stages of tumour progression.15 Moreover, we recognize that the current study lacks functional data. Incorporating such data in future research is crucial for validating our current findings and enhancing their application in a clinical context. Last but not least, overall survival based on the molecular subtypes in limited disease shows no significant difference. However, as the concomitant or maintenance treatment to concurrent chemoradiotherapy is actively investigated, our result including demographics of molecular subtypes could assist the further development of combination strategies in limited disease.
In conclusion, our comprehensive analysis using transcription factor based molecular subtypes provides the prognostic and predictive significance of the molecular subtypes. This may inform subtype-directed approaches and help improve outcomes of small-cell lung cancer.
Contributors
Study conception, design: Sehhoon Park, Tae Hee Hong, Soohyun Hwang, Se-Hoon Lee, Keunchil Park.
Provision of study materials or patients: Sehhoon Park, Se-Hoon Lee, Jong Ho Cho, Yong Soo Choi, Jhingook Kim, Young Mog Shim, Hong Kwan Kim, Hyun-Ae Jung, Jong-Mu Sun, Jin Seok Ahn, Myung-Ju Ahn, Keunchil Park.
Collection and assembly of data: Sehhoon Park, Tae Hee Hong, Soohyun Hwang, Se-Hoon Lee.
Data verification: Sehhoon Park, Tae Hee Hong, Soohyun Hwang.
Data analysis and interpretation: Tae Hee Hong, Soohyun Hwang, Jiyeon Kim, Simon Heeke, Carl M. Gay, Lauren Averett Byers.
Writing of manuscript: Sehhoon Park, Tae Hee Hong, Soohyun Hwang, Simon Heeke, Carl M. Gay, Lauren Averett Byers, John V. Heymach, Yoon-La Choi, Se-Hoon Lee, Keunchil Park.
Final approval of manuscript: All authors.
Accountable for all aspects of the work: All authors.
Data sharing statement
The targeted sequencing data and transcriptome data used in this study are publicly available from https://github.com/SeHoonLab/molecular-subtype-SCLC. Clinical and IHC staining data is provided as supplementary information in Table S3, Appendix p 13.
Declaration of interests
The authors have declared that no conflict of interest exists.
Acknowledgements
This work was supported by AstraZeneca, Future Medicine 2030 Project of the Samsung Medical Center [grant number SMX1240011], the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) [grant number 2020R1C1C1010626] and the 7th AstraZeneca-KHIDI (Korea Health Industry Development Institute) oncology research program.
Prior presentation
Presented at 2023 AACR annual meeting.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105062.
Contributor Information
Yoon-La Choi, Email: ylachoi@skku.edu.
Se-Hoon Lee, Email: shlee119@skku.edu.
Keunchil Park, Email: KPark3@mdanderson.org.
Appendix A. Supplementary data
References
- 1.Socinski M.A., Smit E.F., Lorigan P., et al. Phase III study of pemetrexed plus carboplatin compared with etoposide plus carboplatin in chemotherapy-naive patients with extensive-stage small-cell lung cancer. J Clin Oncol. 2009;27:4787–4792. doi: 10.1200/JCO.2009.23.1548. [DOI] [PubMed] [Google Scholar]
- 2.Noda K., Nishiwaki Y., Kawahara M., et al. Irinotecan plus cisplatin compared with etoposide plus cisplatin for extensive small-cell lung cancer. N Engl J Med. 2002;346:85–91. doi: 10.1056/NEJMoa003034. [DOI] [PubMed] [Google Scholar]
- 3.Amarasena I.U., Chatterjee S., Walters J.A., Wood-Baker R., Fong K.M. Platinum versus non-platinum chemotherapy regimens for small cell lung cancer. Cochrane Database Syst Rev. 2015;2015:CD006849. doi: 10.1002/14651858.CD006849.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horn L., Mansfield A.S., Szczesna A., et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med. 2018;379:2220–2229. doi: 10.1056/NEJMoa1809064. [DOI] [PubMed] [Google Scholar]
- 5.George J., Lim J.S., Jang S.J., et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peifer M., Fernandez-Cuesta L., Sos M.L., et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park S., Lee H., Lee B., et al. DNA damage response and repair pathway alteration and its association with tumor mutation burden and platinum-based chemotherapy in SCLC. J Thorac Oncol. 2019;14:1640–1650. doi: 10.1016/j.jtho.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Park S., Shim J., Mortimer P.G.S., et al. Biomarker-driven phase 2 umbrella trial study for patients with recurrent small cell lung cancer failing platinum-based chemotherapy. Cancer. 2020;126:4002–4012. doi: 10.1002/cncr.33048. [DOI] [PubMed] [Google Scholar]
- 9.Gay C.M., Stewart C.A., Park E.M., et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell. 2021;39:346–360.e7. doi: 10.1016/j.ccell.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y.H., Klingbeil O., He X.Y., et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev. 2018;32:915–928. doi: 10.1101/gad.314815.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baine M.K., Hsieh M.S., Lai W.V., et al. SCLC subtypes defined by ASCL1, NEUROD1, POU2F3, and YAP1: a comprehensive immunohistochemical and histopathologic characterisation. J Thorac Oncol. 2020;15:1823–1835. doi: 10.1016/j.jtho.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Megyesfalvi Z., Barany N., Lantos A., et al. Expression patterns and prognostic relevance of subtype-specific transcription factors in surgically resected small-cell lung cancer: an international multicenter study. J Pathol. 2022;257:674–686. doi: 10.1002/path.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ireland A.S., Micinski A.M., Kastner D.W., et al. MYC drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell. 2020;38:60–78.e12. doi: 10.1016/j.ccell.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mollaoglu G., Guthrie M.R., Bohm S., et al. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell. 2017;31:270–285. doi: 10.1016/j.ccell.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart C.A., Gay C.M., Xi Y., et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat Cancer. 2020;1:423–436. doi: 10.1038/s43018-019-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirier J.T., George J., Owonikoko T.K., et al. New approaches to SCLC therapy: from the laboratory to the clinic. J Thorac Oncol. 2020;15:520–540. doi: 10.1016/j.jtho.2020.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie M., Chugh P., Broadhurst H. Abstract CT024: Durvalumab (D)+ platinum-etoposide (EP) in 1L extensive-stage small-cell lung cancer (ES-SCLC): Exploratory analysis of SCLC molecular subtypes in CASPIAN. Cancer Res. 2022;82 [Google Scholar]
- 18.Best S.A., Hess J.B., Souza-Fonseca-Guimaraes F., et al. Harnessing natural killer immunity in metastatic SCLC. J Thorac Oncol. 2020;15:1507–1521. doi: 10.1016/j.jtho.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Sutherland K.D., Ireland A.S., Oliver T.G. Killing SCLC: insights into how to target a shapeshifting tumor. Genes Dev. 2022;36:241–258. doi: 10.1101/gad.349359.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The molecular screening study for the umbrella trial (SUKSES) in small cell lung cancer patients ( NCT02688894) 2016. https://ClinicalTrials.gov/show/NCT02688894 [updated April 29 2022]. Available from: [Google Scholar]
- 21.Shin H.T., Choi Y.L., Yun J.W., et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun. 2017;8:1377. doi: 10.1038/s41467-017-01470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borromeo M.D., Savage T.K., Kollipara R.K., et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 2016;16:1259–1272. doi: 10.1016/j.celrep.2016.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudin C.M., Poirier J.T., Byers L.A., et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer. 2019;19:289–297. doi: 10.1038/s41568-019-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferone G., Lee M.C., Sage J., Berns A. Cells of origin of lung cancers: lessons from mouse studies. Genes Dev. 2020;34:1017–1032. doi: 10.1101/gad.338228.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui M., Augert A., Rongione M., et al. PTEN is a potent suppressor of small cell lung cancer. Mol Cancer Res. 2014;12:654–659. doi: 10.1158/1541-7786.MCR-13-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Y., Chen Y., Tang H., et al. Activation of PI3K/AKT pathway is a potential mechanism of treatment resistance in small cell lung cancer. Clin Cancer Res. 2022;28:526–539. doi: 10.1158/1078-0432.CCR-21-1943. [DOI] [PubMed] [Google Scholar]
- 27.Roper N., Velez M.J., Chiappori A., et al. Notch signaling and efficacy of PD-1/PD-L1 blockade in relapsed small cell lung cancer. Nat Commun. 2021;12:3880. doi: 10.1038/s41467-021-24164-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiatt J.B., Sandborg H., Garrison S.M., et al. Inhibition of LSD1 with bomedemstat sensitizes small cell lung cancer to immune checkpoint blockade and T cell killing. Clin Cancer Res. 2022;28:4551–4564. doi: 10.1158/1078-0432.CCR-22-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen E.M., Taniguchi H., Chan J.M., et al. Targeting lysine-specific demethylase 1 rescues major histocompatibility complex class I antigen presentation and overcomes programmed death-ligand 1 blockade resistance in SCLC. J Thorac Oncol. 2022;17:1014–1031. doi: 10.1016/j.jtho.2022.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H.-Y., Durmaz Y.T., Li Y., et al. Regulation of neuroendocrine plasticity by the RNA-binding protein ZFP36L1. Nat Commun. 2022;13:4998. doi: 10.1038/s41467-022-31998-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Augert A., Eastwood E., Ibrahim A.H., et al. Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci Signal. 2019;12 doi: 10.1126/scisignal.aau2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.