Summary
We evaluated the diagnostic performance of a multimodal deep-learning (DL) model for ovarian mass differential diagnosis. This single-center retrospective study included 1,054 ultrasound (US)-detected ovarian tumors (699 benign and 355 malignant). Patients were randomly divided into training (n = 675), validation (n = 169), and testing (n = 210) sets. The model was developed using ResNet-50. Three DL-based models were proposed for benign-malignant classification of these lesions: single-modality model that only utilized US images; dual-modality model that used US images and menopausal status as inputs; and multi-modality model that integrated US images, menopausal status, and serum indicators. After 5-fold cross-validation, 210 lesions were tested. We evaluated the three models using the area under the curve (AUC), accuracy, sensitivity, and specificity. The multimodal model outperformed the single- and dual-modality models with 93.80% accuracy and 0.983 AUC. The Multimodal ResNet-50 DL model outperformed the single- and dual-modality models in identifying benign and malignant ovarian tumors.
Subject areas: diagnostics, cancer, Artificial intelligence
Graphical abstract
Highlights
-
•
Multimodal deep-learning model evaluated for ovarian tumor diagnosis
-
•
Single, dual, and multi-modality models compared for diagnostic performance
-
•
Multimodal DL model achieves 0.983 AUC in ovarian tumor diagnosis
Diagnostics; Cancer; Artificial Intelligence
Introduction
Ovarian cancer is the most lethal gynecological malignancy, with a low five-year survival rate of 46%.1 Due to the lack of effective screening, 70% of patients with ovarian cancer are already in advanced stages when diagnosed, with only a 28% five-year survival rate. However, early detection of ovarian cancer can improve the survival rate.2,3 The treatment strategies and prognoses of benign and malignant ovarian tumors vary significantly. For benign lesions, follow-up programs are recommended to minimize stress and ovarian function damage caused by invasive biopsies and surgeries.4,5 For malignant masses, gynecological oncologists must perform a comprehensive assessment of tumor staging and design reasonable treatment strategies.6,7,8 Therefore, preoperative differentiation between benign and malignant ovarian tumors is essential for effective treatment.
Ultrasonography is a safe and cost-effective method for identifying ovarian tumors. This is the preferred choice for identifying adnexal masses.9 However, its accuracy can be affected by the ovarian tissue complexity and the doctor subjectivity, leading to misdiagnoses.10,11 According to current research, the effective diagnosis of ovarian cancer remains a challenge.2 Artificial Intelligence (AI) has shown significant potential in the medical field, assisting in the diagnosis of various cancers, including ovarian cancer.12,13 Deep learning (DL) is a branch of AI that can automatically learn mid- and high-level abstract features from raw data such as ultrasound (US), mammograms, and magnetic resonance imaging, thereby replacing the traditional time-consuming manual extraction method.14 Convolutional neural network (CNN)-based architectures have been successfully implemented for automatic medical image analysis.15 Studies have shown AI’s diagnostic performance in ovarian tumor detection is comparable to that of human experts.16,17,18
Previous studies have shown that the use of molecular features can improve the diagnostic accuracy for ovarian cancer. However, these studies mainly focused on single sources of information.19,20,21 In clinical practice, ovarian cancer diagnosis should include imaging methods, serum tumor markers, and patient age. For example, the United States Food and Drug Administration recommends using CA125 to assess treatment response and monitor residual disease or recurrent hazards after first-line treatment. A link between the CA125 clinical stage and survival rate has been reported in several studies.22,23,24 HE4 is a promising ovarian cancer biomarker with higher specificity for diagnosing ovarian cancer than CA125.25,26,27 Thus, these markers could be used to screen for ovarian cancer.3
Therefore, improving the US diagnostic accuracy for ovarian masses is necessary. Our study aimed to build a multimodal model to improve AI diagnostics for benign and malignant ovarian tumors.
Results
Study population
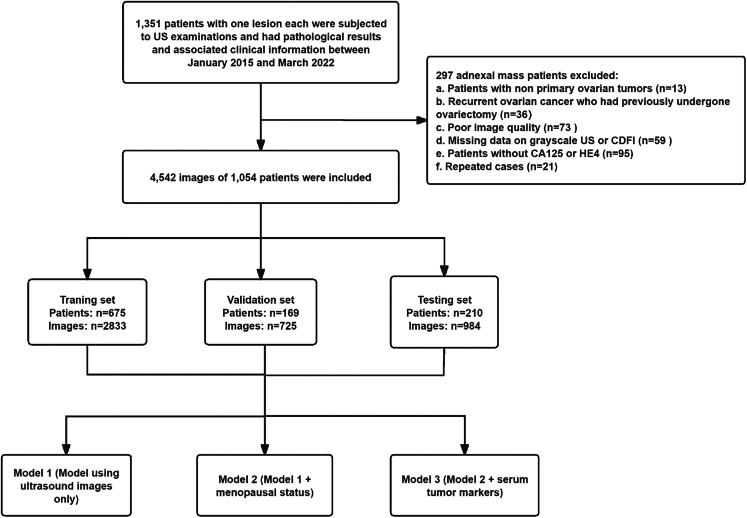
As shown in Table 1 and Figure 1, we obtained 4,542 US images and clinical data from 1,054 patients with primary ovarian tumors screened between January 2015 and March 2022 at the Shenzhen People’s Hospital. Of the 1,054 tumors evaluated, 699 (2,611 images) were benign and 355 (1,931 images) were malignant, according to the pathological results. The average patient age was 40.8 years (range, 15–81 years) for the training set, 41.8 years (range, 15–79) years for the validation set, and 42.4 years (range, 14–86 years) for the test set. No statistically significant differences in baseline information were observed among the training, validation, and test sets (p > 0.05). The model architecture is shown in Figure 2. The histopathological subtypes of all tumors are detailed in Table S1.
Table 1.
Demographic and clinical variables of patients in the training, validation, and test sets
| Characteristics | Training cohort (n = 675) | Validation cohort (n = 169) | Test cohort (n = 210) |
|---|---|---|---|
| Age (years)a | 40.8 (15–81) | 41.8 (15–79) | 42.4 (14–86) |
| Histologic tumor type | |||
| No. of benign | 447 (66.2%) | 112 (66.3%) | 140 (66.7%) |
| No. of malignant | 228 (33.7%) | 57 (33.7%) | 70 (33.3%) |
| CA-125 median, range (U/mL)a | 186.55 (3.6–5832) | 283.07 (5.9–3419) | 239.20 (3.6–4837) |
| CA-125 (U/mL) | |||
| ≤35 | 380 (56.2%) | 89(52.6%) | 119 (56.7%) |
| >35 | 295 (43.7%) | 80(47.3%) | 91 (43.3%) |
| HE4 median, range (pmol/L)a | 188.39 (9.86–4492) | 212.40 (11.78–1500) | 167.34 (11.34–4609) |
| HE4 (pmol/L) | |||
| ≤140 | 510 (75.6%) | 123 (72.8%) | 162 (77.1%) |
| >140 | 165 (24.4%) | 46 (27.2%) | 48 (22.9%) |
| Menopause | |||
| Pre | 493 (73.0%) | 117 (69.2%) | 147 (70%) |
| Post | 182 (27.0%) | 52 (30.8%) | 63 (30%) |
Unless otherwise specified, data in parentheses are percentages.
CA-125, carbohydrate antigen 125; HE4, human epididymal protein 4.
Numbers in parentheses are the range.
Figure 1.
Flowchart of eligibility criteria and the procedure for deep learning
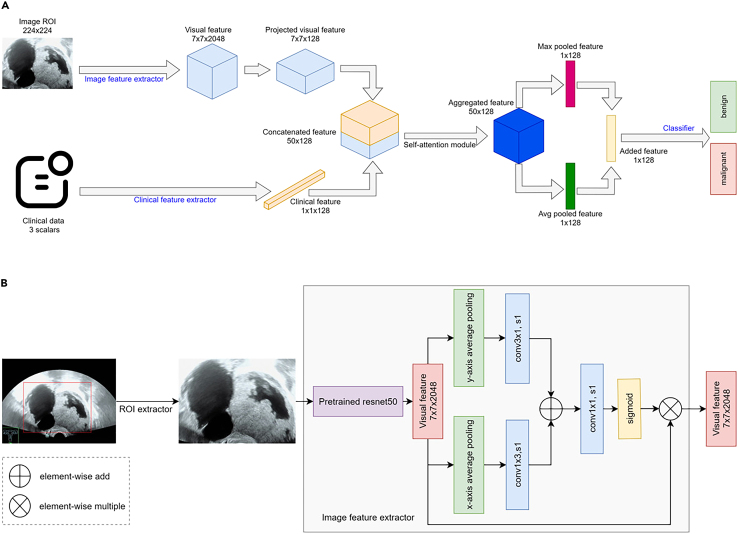
Figure 2.
Architecture of models
(A) Flowchart of the multimodal input and overall workflow for deep learning-based methods.
(B) Image feature extractor network structure.
Performance of the three ResNet-50 models in the benign-malignant classification
As shown in Table 2, a 5-fold cross-validation was performed on the training set. Each fold reported the AUCs of the models as well as the mean AUC and standard deviation. Model 3 had the highest AUC for each fold, best mean AUC, and narrowest standard deviation. This was followed by Model 2, which outperformed the single US image-based model in all evaluation metrics.
Table 2.
Performance of 5-fold cross-validation on training set with different modal inputs
| AUC | Modality | Fold-1 | Fold-2 | Fold-3 | Fold-4 | Fold-5 | Mean ± 1 std.dev |
|---|---|---|---|---|---|---|---|
| Model 1 | US | 0.909 | 0.959 | 0.962 | 0.923 | 0.938 | 0.937 ± 0.021 |
| Model 2 | US + menopausal status | 0.943 | 0.977 | 0.983 | 0.952 | 0.957 | 0.960 ± 0.015 |
| Model 3 | US + menopausal status+serum tumor markers | 0.964 | 0.993 | 0.986 | 0.954 | 0.977 | 0.971 ± 0.014 |
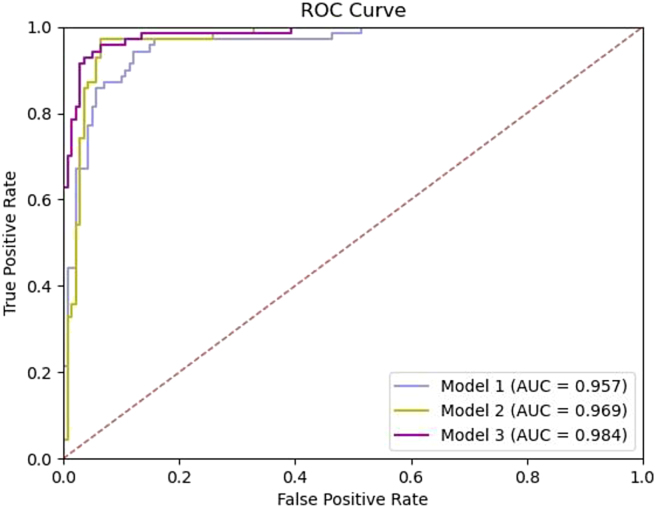
These results are consistent with those of the 5-fold cross-validation of the training set. Compared with the single US imaging modality and menopausal status-embedded models, Model 3 showed the best performance in predicting lesion malignancy. Model 1 achieved an AUC of 0.957 (95% confidence interval [CI] = 0.930–0.984). Model 2 achieved an AUC of 0.969 (95% CI = 0.946–0.992). With the additional serum tumor marker data, Model 3 attained a significantly better AUC of 0.984 (95% CI = 0.969–0.998) with p < 0.05 (Figure 3).
Figure 3.
Comparison of diagnostic efficacy in the testing set
Model 1 (using US images only), Model 2 (Model 1+ menopausal status), Model 3 (Model 2+ serum tumor makers). ROC, receiver operating characteristic; AUC, area under the curve.
Table 3 shows the accuracy, sensitivity, and specificity of each ResNet-50 model for the test set. The accuracy, sensitivity, and specificity of Model 1 were 90.00%, 94.28%, and 87.62%, respectively. The model’s performance increased stepwise with the integration of menopausal status and serum tumor markers. The accuracy, sensitivity, and specificity of Model 3 were 94.76%, 94.28%, and 95.00%, respectively.
Table 3.
Comparison of the efficacy of AI model in the testing set
| Modality | AUC (95%CI) | Cut-off | Sensitivity (%) | Specificity (%) | Accuracy (%) | p value |
|---|---|---|---|---|---|---|
| Model 1 | 0.957 (0.930–0.984) | 0.341 | 94.28 | 87.62 | 90.00 | <0.001 |
| Model 2 | 0.969 (0.946–0.992) | 0.397 | 97.14 | 93.57 | 94.76 | <0.001 |
| Model 3 | 0.984 (0.969–0.998) | 0.590 | 94.28 | 95.00 | 94.76 | <0.001 |
| Model 1 | 0.957 (0.930–0.984) | 0.385 | 90.00 | 88.57 | 89.04 | <0.001 |
| Model 2 | 0.969 (0.946–0.992) | 0.479 | 94.28 | 92.91 | <0.001 | |
| Model 3 | 0.984 (0.969–0.998) | 0.802 | 97.14 | 94.76 | <0.001 |
We compared the sensitivity and specificity of the three models with different cutoff values to evaluate the benefits of embedding clinical information into AI systems. At the sensitivity level of Models 1 and 3 (94.28%), Model 3 had significantly higher specificity (95.00% for Model 3 and 87.62% for Model 1). To improve the detection rate, we set a default sensitivity level of 90.00% with specificities of 88.57%, 94.28%, and 97.14% for Models 1, 2, and 3, respectively.
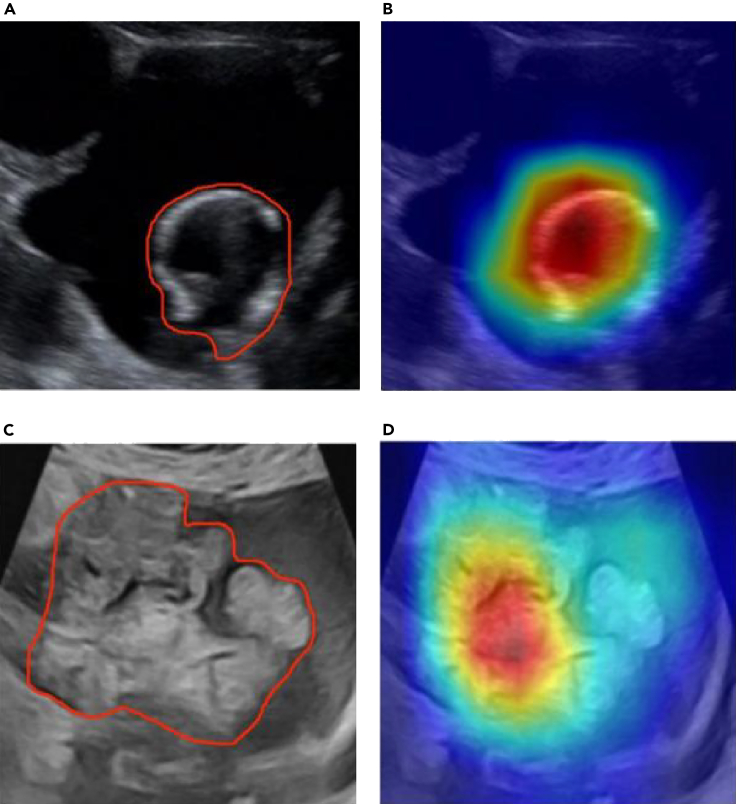
To analyze the classification regions of interest of the network model on ovarian US images, we used gradient-weighted class activation maps to visualize the classification decisions of the model.28 The feature-attention heat maps for US images are shown in Figure 4. Color-scale maps were obtained by overlapping the attention of the network on different features in the original images. By comparing the attention map of the lesion area traced by a senior sonographer with that of a DL network, it was found that, through network learning, the generated attention was similar to that of a senior sonographer all around the lesion and its surrounding area.
Figure 4.
US images and heat maps of two cases with ovarian lesions
(A and B) Images of a 36-year-old female with mucinous cystadenoma. US image (A) showed a 6.8-cm cystic anechoic mass with irregular separation inside. The overlaid heatmap (B) shows that the attention is on the cystic anechoic part, which is consistent with the focus of the senior sonographers.
(C and D) Images of a 55-year-old female with serous cystadenocarcinoma. US image (C) shows a 9.0-cm cystic solid mass with irregular solid masses. Overlaid heatmap (D) shows the irregular real space occupying part of the attention in the package, which is consistent with the concerns of senior sonographers.
Discussion
Using ResNet-50 DL neural networks, we successfully developed a multimodal model to differentiate between benign and malignant ovarian tumors. Model 3 produced the most accurate predictions on the test set, with an AUC of 0.984, a sensitivity of 94.28%, and a specificity of 95.00%. Our results demonstrate that it outperform ResNet-50 at predicting benign and malignant ovarian tumors. This study used a multimodal DL model that incorporated US images, menopausal status, and tumor markers to predict and analyze benign and malignant ovarian lesions.
In this classification task, we used transfer learning methods. Specifically, in the backbone section of ResNet-50 described in the STAR methods section, we transferred the pre-trained weights to ImageNet. This approach offers advantages such as alleviating data scarcity (the annotation cost of medical images is high), reducing overfitting, and accelerating model convergence. In addition, because of the domain differences between natural and medical images, we did not freeze the transferred weights post-transfer to alleviate the potential decrease in classification performance caused by these differences; hence, all parameters of the entire network model participated in training.
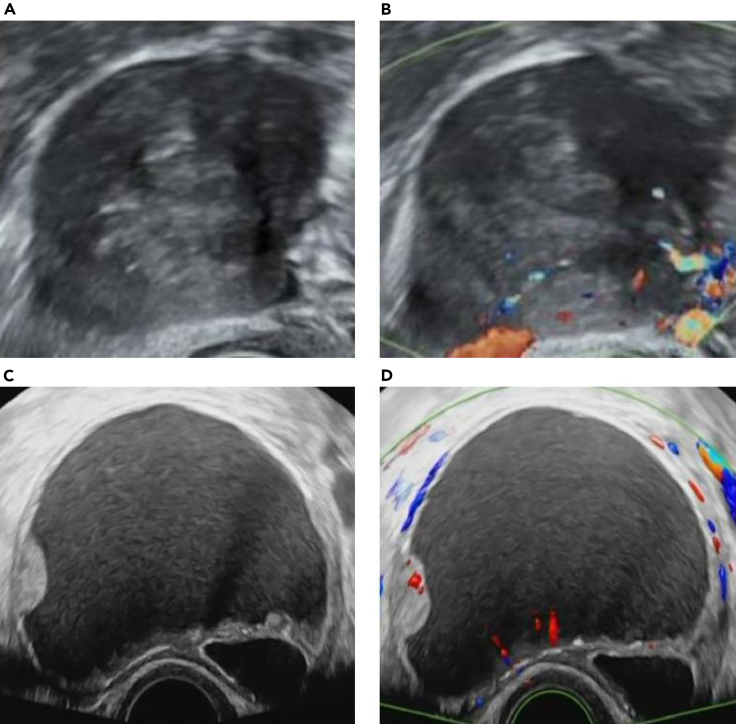
In our study, Model 1 used only US images, with an AUC of 0.957. After adding menopausal status, we found that the diagnostic efficiency was slightly higher, and the AUC reached 0.969, which may be essential for assessing the risk of complicated ovarian tumors. As shown in Figures 5A and 5B, this patient was a premenopausal woman; the masses were solid with clear boundaries, and blood flow signals were apparent. Although the images demonstrated malignant characteristics, the patient was pathologically diagnosed as having a theca cell tumor. Model 2 correctly classified it as benign, whereas Model 1 generated an inaccurate malignant classification. Therefore, the addition of menopausal status can improve the efficiency of the model and assist in the diagnosis of benign and malignant lesions. This is consistent with previous findings that most ovarian cancers are prone to occur in postmenopausal women, while the incidence rate of ovarian cancer is low in premenopausal women.29
Figure 5.
US images of two cases with ovarian lesions
(A and B) Images of a 44-year-old woman with a solid mass in the right ovary. (A) Grayscale US image showing the maximal size plane of the mass.
(B) Color Doppler US image. The pathological diagnosis was cell tumor. This was categorized correctly by Model 2, but not by Model 1.
(C and D) Images of a 38-year-old woman with a unilocular cyst and solid component in the left ovary.
(C) Grayscale US image showing the maximal plane of the mass, and (D) color Doppler US image of the solid component. A pathological diagnosis of mucinous cystadenoma was established. This was misdiagnosed using Models 1 and 2, whereas Model 3 was correct.
Model 3 may assist in addressing the main disadvantages of US in clinical use, which are its high rate of false positives and low specificity. With the aid of additional clinical indicators, Models 2 and 3 obtained higher specificities than single-modality Model 1. When we reviewed the cases correctly predicted by Model 3 but misdiagnosed by Models 1 and 2, we discovered that only 7 of the 18 cases were benign. This result highlights that CA125 and HE4 can help the AI model better diagnose ovarian masses, improve diagnostic specificity, and reduce unnecessary surgery and patient burden. With more clinical information, AI systems can perform better.
As shown in Figures 5C and 5D, Model 3 classified the case as benign, but Models 1 and 2 classified it as malignant. The pathological result was mucinous cystadenoma, which is difficult to diagnose even by experienced US physicians because the misleading solid components in the cyst demonstrate suspicious malignant characteristics. In this study, we combined clinical indicators to build a multimodal DL model that can be used as an auxiliary diagnostic tool for sonographers. Before obtaining histopathological results, sonographers can use the model’s probabilities to better define the nature of the tumor.
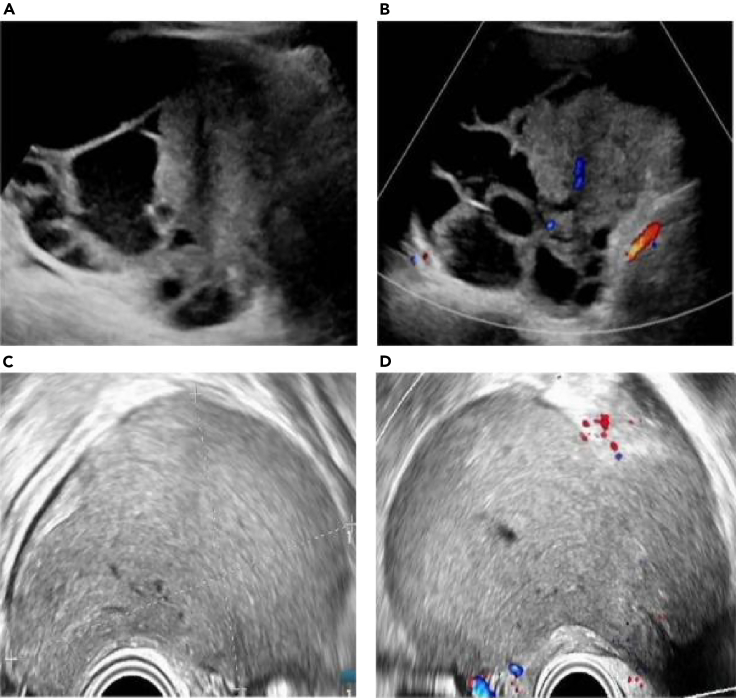
Model 3 also showed a few false negatives and false positives. As shown in Figures 6A and 6B, Model 3 classified this case as benign, whereas the pathological result was a serous cystadenocarcinoma, and the US image showed suspicious malignant features, such as a solid mass with blood flow signals. However, HE4 in this case was negative (56.3 pmol/L), CA125 was positive (63.2 U/mL), and the patient was premenopausal (43 years old). The reason for the missed diagnosis may be that the HE4 and menopausal conditions of the patient affected the judgment of the model. By contrast, as shown in Figures 6C and 6D, Model 3 classified this case as malignant; however, the pathological result was an ovarian endometriosis cyst. The US image shows a cystic mass that appears to be a solid mass, but in fact, it has dense dotted echoes inside. The HE4 of this case was negative (13.7 pmol/L), the CA125 was positive (108.3 U/mL), and the patient was premenopausal (23 years old). The probable cause of the misdiagnosis may have been the static image and CA125 value of this case, resulting in the incorrect judgment of the model. The presented cases illustrates that, in practical applications, we can combine models with dynamic images and clinical experience to improve diagnostic efficiency.
Figure 6.
US images of two cases with ovarian lesions
(A and B) Images of a 43-year-old woman with a solid cyst in the left ovary. Model 3 classified this case as benign, whereas the pathological result was serous cystadenocarcinoma, and the US image showed suspicious malignant features, such as a solid mass with blood flow signals. The HE4 in this case was negative (56.3 pmol/L) and CA125 was positive (63.2 U/mL), and the patient was premenopausal.
(C and D) Images of a 23-year-old woman with cystic right ovary. Model 3 classified this case as malignant; however, the pathological diagnosis was an ovarian endometriotic cyst. The US image shows a cystic mass without a blood flow signal that appears as a solid mass; however, it has dense dotted echoes. The patient’s HE4 was negative (13.7 pmol/L), the CA125 was positive (108.3 U/mL), and they were premenopausal.
In conclusion, the multimodality model outperformed the single-modality model by integrating menopausal status and serum tumor markers with US images. We anticipate that this multimodality model will be an effective tool for improving US performance in identifying benign and malignant ovarian tumors in clinical practice.
Limitations of the study
Our study has some limitations. First, this was a retrospective study, and the results were influenced by limited data. However, large-scale prospective studies are required for further improvements. We will continue to conduct prospective research in the next step to avoid selection bias. Second, this was a single-center study. With more participating centers, we can further improve and validate our model using a more diverse population. Third, the experiment did not include a comparison with the experts’ subjective evaluations, potentially limiting the generalizability of the diagnostic performance evaluated using our multimodality model. However, literature15,17 reports the consistency of expert subjective and DL model evaluations, alleviating this concern. Fourth, this study did not evaluate the importance of imaging versus demographic and laboratory factors in the Model 3 prediction.
Fifth, due to the malignant tendency of borderline tumors, follow-up is not recommended. Misclassification as benign may cause physicians to misjudge and affect treatment. The existing 38 cases of borderline ovarian tumors are not sufficient to illustrate the problem. In future research, we will attempt to focus on borderline ovarian tumors by expanding the sample size.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| Python | van Rossum,30 | https://www.python.org/ |
| Tensorflow | Abadi et al.31 | https://www.tensorflow.org/ |
| NumPy | Harris et al.32 | https://numpy.org/ |
| Code for this paper | Yang,33 | https://doi.org/10.17632/f5p4h32p46.2 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Fajin Dong (dongfajin@szhospital.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Data are not publicly shared but is available upon reasonable request from the lead contact.
-
•
All original code has been deposited at Mendeley Data and is publicly available as of the date of publication. DOIs are listed in the key resources table (https://doi.org/10.17632/f5p4h32p46.2).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Method details
Ethics
This study was approved by the ethics committee of Shenzhen People’s Hospital, and the requirement for informed consent was waived due to the retrospective data collection. This study was conducted in accordance with the principles of the Declaration of Helsinki.
Patients and datasets
The data flow is shown in Figure 1. We retrospectively enrolled 1351 patients who underwent US examinations within two weeks prior to surgery, biopsy, or other antineoplastic treatments and had pathological results and associated clinical information at Shenzhen People’s Hospital from January 2015 to March 2022. The inclusion criteria were as follows: (a) patients with ovarian tumors who had undergone surgical resection or biopsy; (b) patients who had undergone grayscale and color Doppler US within two weeks before surgery, biopsy, or other antineoplastic treatments; and (c) the examination of serum tumor markers was completed before treatment and surgery. The exclusion criteria were as follows: (a) patients with non-primary ovarian tumors, (b) patients with recurrent ovarian cancer who had previously undergone ovariectomy, (c) patients with poor US image quality, (d) missing grayscale US or CDFI data, (e) patients without CA125 or HE4, and (f) repeated cases. Finally, 1054 patients were included and randomly divided into training (675), validation (169), and testing (210) sets. Clinical data, including age, menopausal status, pathological findings, serum tumor markers (CA125 and HE4), and US diagnosis reports, were collected from the picture archiving and communication system (PACS) of Shenzhen People’s Hospital.
Ultrasound image collection protocol
Two physicians with more than five years of experience in gynecological ultrasonography reviewed and filtered the US images based on the inclusion and exclusion criteria. All images were annotated by the two doctors to manually segment the lesion region from the input US images using an open-source, commonly used labeling tool (LabelMe; http://labelme.csail.mit.edu/Release3.0/).
For each lesion, three to seven images were selected, including grayscale US images (one image), images with calipers enclosed at the edge of the lesion (one image), and images with color Doppler (one image). Other images with specific US characteristics were also selected, including those with solid components, irregular walls, and papillary projections in the tumor (one to four images, if any). These US images were recorded by trained physicians in real clinical workflow using the following commercially available units (GE Voluson E10 and E9, GE Healthcare; Philips EPIQ7, Philips Healthcare; and Mindray Resona7, Mindray) equipped with 5.0–9.0 MHz, 4.0–8.0 MHz, and 3.0–10.0 MHz transvaginal probes, and 1.0–5.0 MHz transabdominal probes. In total, 4,542 US images of the enrolled patients were used to train, validate, and test the proposed DL model.
Image processing
Object detection was performed for each input US image to extract the region of interest using the YOLO model.34 Padding and resizing were performed on the ROIs to reshape them to 224 × 224 pixels. The reshaped ROIs were the inputs for the ResNet-50-based models.
Architecture of models
ResNet-50 is a DL model proposed by Microsoft Research in 2015. This model is characterized by introducing "Residual Blocks" in each layer, where new transformations are added based on the previous layer while preserving the original input. This enables the network to learn the differences between the inputs and outputs better, thus improving the model’s performance. Compared with other models, ResNet-50 introduces the idea of residual learning, which addresses the issue of vanishing gradients in DL. By design, Residual Blocks allow for better model training. Additionally, the model’s depth should be adapted to the scale of the data; in our case, a depth of 50 layers achieved the best results.
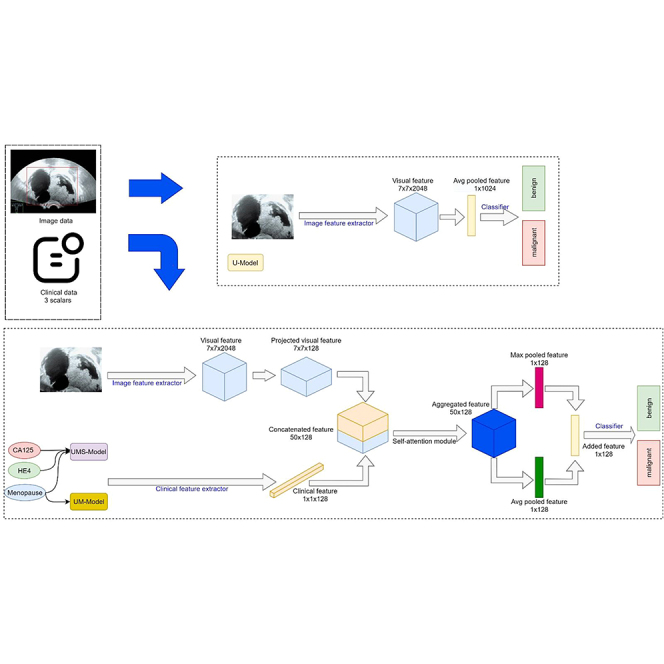
Figure 2A depicts the architecture of our models. In the top branch, we used ResNet-50 to extract features from the images. It contains four stages and downsamples the image five times to create a final image feature matrix with a shape of 7 × 7×2048. As shown in Figure 2B, strip pooling followed by nonanisotropic convolution was used to convolve the feature in both the height and width dimensions to capture the long-range context. The results of the non-anisotropic convolution are added together, followed by a sigmoid function to generate a spatial attention map. The attention map is then used to scale the original image feature matrix. Clinical information on the lower branch, such as menopausal status and serum tumor markers (CA125 and HE4), are used to assist in prediction. A multilayer perceptron extracts information from the clinical data and projects it onto a feature vector of 128 sizes.
To fuse the information from the images and clinical data, we performed a 1 × 1 convolution to project the feature matrix of the image onto the clinical space. The image feature matrix, whose shape was 7 × 7×2048, became 7 × 7×128 after a 1 × 1 convolution layer, and we combined the first two dimensions; thus, its final shape was 49 × 128. The reshaped image features (49 × 128) and clinical data features (1 × 128) were concatenated in the channel dimensions, resulting in a merged feature with a shape of 50 × 128. The concatenated features are passed to the multihead self-attention module, where three heads are used to conduct interactive feature fusion. Subsequently, max pooling and average pooling were utilized on the fused features, and the results were added to create a 128-size feature vector for classification. Finally, a fully connected layer is used as the classifier for the final prediction.
Models
Three models were constructed using diverse input modalities. Model 1 used only US images, with no clinical data for classification. The lower branch in Figure 2B was removed from Model 1. Menopausal status is utilized as additional clinical information for model 2 to assist diagnosis with US images. In Model 3, menopausal status and serum tumor markers (CA125 and HE4) were used in the clinical data branch to share information with the image branch.
Quantification and statistical analysis
Sensitivity, specificity, and accuracy are the primary metrics of the models based on different modalities. The receiver operating characteristic curves of the models were obtained to assess their ability to classify benign malignancies. The AUC and binomial-accurate 95% CI were reported to quantitatively describe the performance of the models. Statistical differences between AUCs were compared using a Delong test,35 with p values <0.05 considered significant. All statistical analyses were performed using R 3.4.3 software (https://cran.r-project.org/) and Python v3.9. Borderline ovarian tumors were considered malignant based on statistical analyses.36
Acknowledgments
We thank all authors for their assistance in the preparation of this manuscript. The authors state that this work has not received any funding.
Author contributions
J.X.: conceptualization, methodology, investigation. Q.L.: conceptualization, methodology, investigation, supervision. F.D.: conceptualization, methodology, investigation, analyzed the data, supervision. Z.W.: resources, methodology, writing – original draft, writing – review & editing, analyzed the data. S.L.: resources, analyzed the data, methodology. J.C.: analyzed the data, methodology. Yitao Jiang: analyzed the data, resources. S.S.: software, supervision, analyzed the data. C.C.: software, supervision, analyzed the data, funding acquisition. Y.Y., J.Z., Yang Jiao, Y.Z., and F.X.: analyzed the data, resources.
Declaration of interests
The authors declare no competing interests.
Published: March 4, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109403.
Contributor Information
Jinfeng Xu, Email: xujinfeng@yahoo.com.
Qi Lin, Email: linqik@sina.com.
Fajin Dong, Email: dongfajin@szhospital.com.
Supplemental information
References
- 1.Lheureux S., Braunstein M., Oza A.M. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA A Cancer J. Clin. 2019;69:280–304. doi: 10.3322/caac.21559. [DOI] [PubMed] [Google Scholar]
- 2.Henderson J.T., Webber E.M., Sawaya G.F. Screening for ovarian cancer: updated evidence report and systematic review for the US Preventive Services Task Force. JAMA. 2018;319:595–606. doi: 10.1001/jama.2017.21421. [DOI] [PubMed] [Google Scholar]
- 3.Hurwitz L.M., Pinsky P.F., Trabert B. General population screening for ovarian cancer. Lancet. 2021;397:2128–2130. doi: 10.1016/S0140-6736(21)01061-8. [DOI] [PubMed] [Google Scholar]
- 4.Froyman W., Landolfo C., De Cock B., Wynants L., Sladkevicius P., Testa A.C., Van Holsbeke C., Domali E., Fruscio R., Epstein E., et al. Risk of complications in patients with conservatively managed ovarian tumours (IOTA5): a 2-year interim analysis of a multicentre, prospective, cohort study. Lancet Oncol. 2019;20:448–458. doi: 10.1016/S1470-2045(18)30837-4. [DOI] [PubMed] [Google Scholar]
- 5.Wang R., Cai Y., Lee I.K., Hu R., Purkayastha S., Pan I., Yi T., Tran T.M.L., Lu S., Liu T., et al. Evaluation of a convolutional neural network for ovarian tumor differentiation based on magnetic resonance imaging. Eur. Radiol. 2021;31:4960–4971. doi: 10.1007/s00330-020-07266-x. [DOI] [PubMed] [Google Scholar]
- 6.Abramowicz J.S., Timmerman D. Ovarian mass-differentiating benign from malignant: the value of the International Ovarian Tumor Analysis ultrasound rules. Am. J. Obstet. Gynecol. 2017;217:652–660. doi: 10.1016/j.ajog.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong D.K., Alvarez R.D., Bakkum-Gamez J.N., Barroilhet L., Behbakht K., Berchuck A., Chen L.M., Cristea M., DeRosa M., Eisenhauer E.L., et al. Ovarian Cancer. version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021;19:191–226. doi: 10.6004/jnccn.2021.0007. [DOI] [PubMed] [Google Scholar]
- 8.Chua K.J.C., Patel R.D., Trivedi R., Greenberg P., Beiter K., Magliaro T., Patel U., Varughese J. Accuracy in referrals to gynecologic oncologists based on clinical presentation for ovarian Mass. Diagnostics. 2020;10 doi: 10.3390/diagnostics10020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischerova D., Zikan M., Semeradova I., Slama J., Kocian R., Dundr P., Nemejcova K., Burgetova A., Dusek L., Cibula D. Ultrasound in preoperative assessment of pelvic and abdominal spread in patients with ovarian cancer: a prospective study. Ultrasound Obstet. 2017;49:263–274. doi: 10.1002/uog.15942. [DOI] [PubMed] [Google Scholar]
- 10.Smith-Bindman R., Poder L., Johnson E., Miglioretti D.L. Risk of malignant ovarian cancer based on ultrasonography findings in a large unselected population. JAMA Intern. Med. 2019;179:71–77. doi: 10.1001/jamainternmed.2018.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePriest P.D., DeSimone C.P. Ultrasound screening for the early detection of ovarian cancer. J. Clin. Oncol. 2003;21:194s. doi: 10.1200/JCO.2003.02.054. [DOI] [PubMed] [Google Scholar]
- 12.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 13.Koch A.H., Jeelof L.S., Muntinga C.L.P., Gootzen T.A., van de Kruis N.M.A., Nederend J., Boers T., van der Sommen F., Piek J.M.J. Analysis of computer-aided diagnostics in the preoperative diagnosis of ovarian cancer: a systematic review. Insights into imaging. 2023;14:34. doi: 10.1186/s13244-022-01345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y., Lei Y., Wang T., Curran W.J., Liu T., Yang X. Deep learning in medical image registration: a review. Phys. Med. Biol. 2020;65:20TR01. doi: 10.1088/1361-6560/ab843e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H., Yang B.W., Qian L., Meng Y.S., Bai X.H., Hong X.W., He X., Jiang M.J., Yuan F., Du Q.W., et al. Deep learning prediction of ovarian malignancy at US compared with O-RADS and expert assessment. Radiology. 2022;304:106–113. doi: 10.1148/radiol.211367. [DOI] [PubMed] [Google Scholar]
- 16.Wang H., Liu C., Zhao Z., Zhang C., Wang X., Li H., Wu H., Liu X., Li C., Qi L., et al. Application of deep convolutional neural networks for discriminating benign, borderline, and malignant serous ovarian tumors from ultrasound images. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.770683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christiansen F., Epstein E.L., Smedberg E., Åkerlund M., Smith K., Epstein E. Ultrasound image analysis using deep neural networks for discriminating between benign and malignant ovarian tumors: comparison with expert subjective assessment. Ultrasound Obstet. Gynecol. 2021;57:155–163. doi: 10.1002/uog.23530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y., Zeng S., Xu X., Li H., Yao S., Song K., Li X., Chen L., Tang J., Xing H., et al. Deep learning-enabled pelvic ultrasound images for accurate diagnosis of ovarian cancer in China: a retrospective, multicentre, diagnostic study. Lancet Digit. Health. 2022;4:e179. doi: 10.1016/S2589-7500(21)00278-8. [DOI] [PubMed] [Google Scholar]
- 19.Landolfo C., Achten E.T.L., Ceusters J., Baert T., Froyman W., Heremans R., Vanderstichele A., Thirion G., Van Hoylandt A., Claes S., et al. Assessment of protein biomarkers for preoperative differential diagnosis between benign and malignant ovarian tumors. Gynecol. Oncol. 2020;159:811–819. doi: 10.1016/j.ygyno.2020.09.025. [DOI] [PubMed] [Google Scholar]
- 20.Gahlawat A.W., Witte T., Haarhuis L., Schott S. A novel circulating miRNA panel for non-invasive ovarian cancer diagnosis and prognosis. Br. J. Cancer. 2022;127:1550–1556. doi: 10.1038/s41416-022-01925-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L., Ma R., Luo C., Xie Q., Ning X., Sun K., Meng F., Zhou M., Sun J. Noninvasive early differential diagnosis and progression monitoring of ovarian cancer using the copy number alterations of plasma cell-free DNA. Transl. Res. : J. Lab. Clin. Med. 2023;262:12–24. doi: 10.1016/j.trsl.2023.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Goff B.A., Agnew K., Neradilek M.B., Gray H.J., Liao J.B., Urban R.R. Combining a symptom index, CA125 and HE4 (triple screen) to detect ovarian cancer in women with a pelvic mass. Gynecol. Oncol. 2017;147:291–295. doi: 10.1016/j.ygyno.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Zhang M., Cheng S., Jin Y., Zhao Y., Wang Y. Roles of CA125 in diagnosis, prediction, and oncogenesis of ovarian cancer. Biochim. Biophys. Acta Rev. Canc. 2021;1875 doi: 10.1016/j.bbcan.2021.188503. [DOI] [PubMed] [Google Scholar]
- 24.Paramasivam S., Tripcony L., Crandon A., Quinn M., Hammond I., Marsden D., Proietto A., Davy M., Carter J., Nicklin J., et al. Prognostic importance of preoperative CA-125 in International Federation of Gynecology and Obstetrics stage I epithelial ovarian cancer: an Australian multicenter study. J. Clin. Oncol. 2005;23:5938–5942. doi: 10.1200/JCO.2005.08.151. [DOI] [PubMed] [Google Scholar]
- 25.Lycke M., Kristjansdottir B., Sundfeldt K. A multicenter clinical trial validating the performance of HE4, CA125, risk of ovarian malignancy algorithm and risk of malignancy index. Gynecol. Oncol. 2018;151:159–165. doi: 10.1016/j.ygyno.2018.08.025. [DOI] [PubMed] [Google Scholar]
- 26.Matsuo K., Tanabe K., Hayashi M., Ikeda M., Yasaka M., Machida H., Shida M., Sato K., Yoshida H., Hirasawa T., et al. Utility of comprehensive serum glycopeptide spectra analysis (CSGSA) for the detection of early stage epithelial ovarian cancer. Cancers. 2020;12 doi: 10.3390/cancers12092374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dochez V., Caillon H., Vaucel E., Dimet J., Winer N., Ducarme G. Biomarkers and algorithms for diagnosis of ovarian cancer: CA125, HE4, RMI and ROMA, a review. J. Ovarian Res. 2019;12:28. doi: 10.1186/s13048-019-0503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. IEEE International Conference on Computer Vision (ICCV); 22-29. 2017. Grad-CAM: visual explanations from deep networks via gradient-based localization; pp. 618–626. [DOI] [Google Scholar]
- 29.Pfeiffer R.M., Park Y., Kreimer A.R., Lacey J.V., Jr., Pee D., Greenlee R.T., Buys S.S., Hollenbeck A., Rosner B., Gail M.H., et al. Risk prediction for breast, endometrial, and ovarian cancer in white women aged 50 y or older: derivation and validation from population-based cohort studies. PLoS Med. 2013;10 doi: 10.1371/journal.pmed.1001492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Rossum G. Department of Computer Science [CS]; 1995. Python Reference Manual. [Google Scholar]
- 31.Abadi M., Barham P., Chen J., Chen Z., Davis A., Dean J., Devin M., Ghemawat S., Irving G., Isard M., Kudlur M. {TensorFlow}: a system for {Large-Scale} machine learning. In 12th USENIX symposium on operating systems design and implementation. OSDI. 2016;16:265–283. [Google Scholar]
- 32.Harris C.R., Millman K.J., van der Walt S.J., Gommers R., Virtanen P., Cournapeau D., Wieser E., Taylor J., Berg S., Smith N.J., et al. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y. 2023. Code for OvaryNet(Mendeley Data) [DOI] [Google Scholar]
- 34.Hnewa M., Radha H. IEEE Transactions on Image Processing: a publication of the IEEE Signal Processing Society. 2023. Integrated Multiscale Domain Adaptive YOLO. [DOI] [PubMed] [Google Scholar]
- 35.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 36.Piovano E., Cavallero C., Fuso L., Viora E., Ferrero A., Gregori G., Grillo C., Macchi C., Mengozzi G., Mitidieri M., et al. Diagnostic accuracy and cost-effectiveness of different strategies to triage women with adnexal masses: a prospective study. Ultrasound Obstet. Gynecol. 2017;50:395–403. doi: 10.1002/uog.17320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data are not publicly shared but is available upon reasonable request from the lead contact.
-
•
All original code has been deposited at Mendeley Data and is publicly available as of the date of publication. DOIs are listed in the key resources table (https://doi.org/10.17632/f5p4h32p46.2).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.