Abstract
The hepatitis A virus cellular receptor 1 (HAVcr-1) cDNA codes for a class I integral membrane glycoprotein, termed havcr-1, of unknown natural function which serves as an African green monkey kidney (AGMK) cell receptor for HAV. The extracellular domain of havcr-1 has an N-terminal Cys-rich region that displays homology with sequences of members of the immunoglobulin superfamily, followed by a Thr/Ser/Pro (TSP)-rich region characteristic of mucin-like O-glycosylated proteins. The havcr-1 glycoprotein contains four putative N-glycosylation sites, two in the Cys-rich region and two in the TSP-rich region. To characterize havcr-1 and define region(s) involved in HAV receptor function, we expressed the TSP-rich region in Escherichia coli fused to glutathione S-transferase and generated antibodies (Ab) in rabbits (anti-GST2 Ab). Western blot analysis with anti-GST2 Ab detected 62- and 65-kDa bands in AGMK cells and 59-, 62-, and 65-kDa bands in dog cells transfected with the HAVcr-1 cDNA (cr5 cells) but not in dog cells transfected with the vector alone (DR2 cells). Treatment of AGMK and cr5 cell extracts with peptide-N-glycosidase F resulted in the collapse of the havcr-1-specific bands into a single band of 56 kDa, which indicated that different N-glycosylated forms of havcr-1 were expressed in these cells. Treatment of AGMK and cr5 cells with tunicamycin reduced binding of protective monoclonal Ab (MAb) 190/4, which suggested that N-glycans are required for binding of MAb 190/4 to havcr-1. To test this hypothesis, havcr-1 mutants lacking the N-glycosylation motif at the first site (mut1), second site (mut2), and both (mut3) sites were constructed and transfected into dog cells. Binding of MAb 190/4 and HAV to mut1 and mut3 cells was highly reduced, while binding to mut2 cells was not affected and binding to dog cells expressing an havcr-1 construct containing a deletion of the Cys-rich region (d1− cells) was undetectable. HAV-infected cr5 and mut2 cells but not mut1, mut3, d1−, and DR2 cells developed the characteristic cytoplasmic granular fluorescence of HAV-infected cells. These results indicate that the Cys-rich region of havcr-1 and its first N-glycosylation site are required for binding of protective MAb 190/4 and HAV receptor function.
Viral hepatitis is a major public health problem, with estimated annual medical costs of billions of dollars. The Center for Disease Control and Prevention estimated that in the United States alone, hepatitis A virus (HAV), the causative agent of acute hepatitis in humans, produces substantial morbidity and mortality, with an estimated 125,000 to 200,000 infections occurring each year and approximately 100 deaths from fulminant hepatitis. HAV is the only member of the hepatovirus genus of the Picornaviridae, a family of small, nonenveloped, positive-strand RNA viruses that include human pathogens such as poliovirus (PV) and rhinovirus as well as animal pathogens such as foot-and-mouth disease virus and encephalomyocarditis virus. Hepatitis A is transmitted via the oral-fecal route and can be prevented by vaccination with cell culture-adapted formalin-inactivated HAV (6, 22). The HAV RNA genome of about 7,500 nucleotides (nt) is covalently linked to the small virus-encoded VPg protein at its 5′ end (21) and has a poly(A) tail at its 3′ end. The approximately 750-nt long 5′ nontranslated region of the HAV genome codes for a long and complex internal ribosome entry site which directs the cap-independent translation of the viral message (reference ;9; and references therein). The HAV mRNA contains a single long open reading frame, which is translated into a polyprotein from which the structural proteins VP0, VP3, and VP1 and nonstructural proteins are cleaved by 3Cpro, the only HAV-encoded protease (8, 17). Sixty copies of VP0, VP3, and VP1 assemble into viral capsids, which, in association with the HAV genome, form provirions that undergo a slow RNA-dependent maturation cleavage of VP0 into VP4 and VP2 (2). VP4 of HAV is a very small protein of 21 to 23 amino acids which, in contrast to VP4s of all other picornaviruses, has not yet been found in the viral capsid (5, 12, 19).
Although there have been major advances in our knowledge about human hepatitis viruses, very little is known about the mechanisms of their cell entry. Cellular receptors for human hepatitis viruses have been difficult to characterize due to poor in vitro viral growth, association of virions with serum and cell-derived materials which mask genuine virus-receptor interactions leading to cell entry, and attachment of virions to susceptible and nonsusceptible cells. We identified havcr-1 as an African green monkey kidney (AGMK) cellular receptor for HAV using protective monoclonal antibody (MAb) 190/4 as a probe (10). Ashida and Hamada recently identified a protein very similar to havcr-1 in S.la/Ve-1 cells, hybrid cells between marmoset liver and Vero cells, as an HAV receptor using the independently derived protective MAb 2H4 to screen a cDNA library (1). Nucleotide sequence analysis revealed that the HAVcr-1 cDNA codes for a novel mucin-like class I integral membrane glycoprotein, termed havcr-1, whose extracellular domain contains four putative N-glycosylation sites and two distinctive regions: an N-terminal Cys-rich region that displays homology to sequences of members of the immunoglobulin superfamily, and a mucin-like C-terminal region containing 27 repeats of the consensus PTTTTL. Our knowledge about the interaction of HAV with havcr-1 is currently limited, and the natural function(s) and ligand(s) of this receptor are unknown. In this work we characterize different species of havcr-1 migrating between 59 and 65 kDa as N-glycosylated forms of a 56-kDa band present after removal of N-glycans with peptide-N-glycosidase F (PNGase F). We also determined, using N-glycosylation and deletion mutants, that the havcr-1 Cys-rich region and its first N-glycosylation site are required for HAV receptor function. Further characterization of havcr-1 and the HAV–havcr-1 interaction will help us to understand the mechanism of cell entry of HAV and possibly to develop drugs which can prevent such interaction.
MATERIALS AND METHODS
Cells and viruses.
The continuous clone GL37 of AGMK cells was selected for supporting optimal growth of HAV (20). Fetal rhesus monkey kidney (FRhK-4) cells were obtained from S. Emerson, National Institute of Allergy and Infectious Diseases. Canine osteogenic sarcoma D-17 cells, obtained from the American Type Culture Collection (Rockville, Md.), were cotransfected with pCMVEBNA and pSV2neo, and G418-resistant cell clones with increased efficiencies of transfection for episomal plasmids containing an Epstein-Barr virus P1 origin of replication were selected (16, 18). A G418-resistant cell clone, Perro6D, which had 10- to 100-times-higher transfection efficiency with pDR2 (16, 18) than parental D-17 cells was isolated and used for transfection with HAVcr-1 cDNA constructs cloned into the pDR2 vector. Cell lines were grown in Eagle’s minimal essential medium (EMEM) containing 10% fetal bovine serum (FBS) at 37°C in a CO2 incubator.
Human tissue culture-adapted HAV strain HM175 of genotype 1B, which was derived from infectious cDNA (3), and tissue culture-adapted HAV strain KRM003 of genotype 3B (20) were grown in AGMK cells.
Antisera.
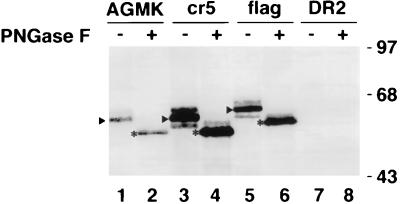
Anti-havcr-1 antiserum was obtained from rabbits immunized with recombinant protein GST2, which consists of the mucin-like region of havcr-1 fused to the C terminus of glutathione S-transferase (GST) expressed in the C600 strain of Escherichia coli harboring pGEXcrpt. To construct this plasmid, a 584-bp BstBI/HincII DNA fragment coding for nt 607 to 1291 of the HAVcr-1 cDNA was cut from pDR2GL37/5 (10), filled in with DNA polymerase I Klenow enzyme, and cloned into the SmaI site of the expression vector pGEX-3X (Pharmacia Biotech Inc.). The resulting plasmid, pGEXcrpt, encodes a fusion protein consisting of GST followed by 228 amino acids of the Thr/Ser/Pro (TSP)-rich region of havcr-1. To prepare antiserum against the fusion polypeptide, a 50-ml culture of E. coli containing pGEXcrpt was induced with 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 2 h at 37°C. Bacteria were pelleted and lysed, and the GST2 fusion protein was purified by binding to glutathione-agarose and elution with 5 mM reduced glutathione as recommended by the manufacturer (Pharmacia Biotech Inc.). The GST2 protein was emulsified in Freund’s complete adjuvant and inoculated into two female New Zealand White rabbits. After several booster injections, the resulting antiserum reacted in Western blot analysis with havcr-1-specific bands of 62 to 65 kDa in AGMK cells (see Fig. 2, lane 1) and of 59 to 65 kDa in dog cells transfected with the HAVcr-1 cDNA (see Fig. 2, lane 5) but not in dog cells transfected with the vector pDR2 (see Fig. 2, lane 7).
FIG. 2.
Western blot analysis of the expression of HAVcr-1 in dog cell transfectants. The expression of HAVcr-1 in AGMK cells (lanes 1 and 2) was compared with the expression in dog cells transfected with wt HAVcr-1 cDNA (cr5 cells [lanes 3 and 4]), HAVcr-1 cDNA tagged with a FLAG peptide (flag cells [lanes 5 and 6]), and the vector pDR2 alone (DR2 cells [lanes 7 and 8]). Cytoplasmic extracts prepared in RSB–1% NP-40 containing 20 to 25 μg of total protein were treated with 500 U of PNGase F (+ [lanes 2, 4, 6, and 8]) or mock-treated (− [lanes 1, 3, 5, and 7]), separated by SDS–12.5% polyacrylamide gel electrophoresis, transferred to PVDF membranes, and probed with rabbit anti-GST2 Ab directed against the TSP-rich region of havcr-1. Arrowheads point to the most abundant N-glycosylated form of havcr-1, which in AGMK and cr5 cells corresponds to a 62-kDa band and in flag cells corresponds to a 64-kDa band due to the FLAG insertion. Asterisks mark the most abundant havcr-1-specific band observed after treatment of cell extracts with PNGase F. The positions of prestained molecular mass markers and their sizes in kilodaltons are shown on the right.
Murine MAbs 190/4 directed against havcr-1 (10) and M2 directed against the FLAG peptide DTKDDDDK (Eastman Kodak Co.) were purified through protein A-agarose columns. Unlabeled and 125I-labeled human anti-HAV polyclonal antisera were obtained from an HAVAB kit (Abbott Laboratories). Fluorescein isothiocyanate (FITC)-labeled goat anti-human Ab (Accurate Inc.) was used to detect HAV by indirect immunofluorescence analysis. Alkaline phosphatase-labeled goat anti-rabbit and peroxidase-labeled goat anti-mouse Abs were used as suggested by the manufacturer (Kirkegaard & Perry Laboratories, Inc.).
Plasmid constructs.
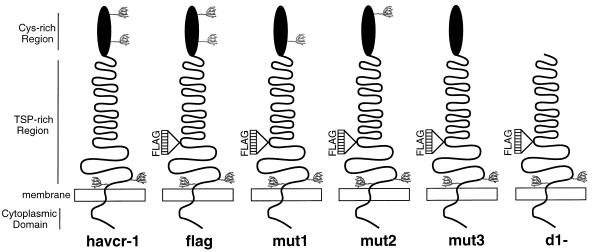
Recombinant DNA manipulations were done by standard methods (15). Constructions were verified by automatic nucleotide sequencing in an ABI Prism model 377 automatic sequencer with an ABI PRISM dye terminator cycle-sequencing ready-reaction kit (Perkin-Elmer Cetus, Inc.). All plasmids were grown in E. coli DH5α and purified by chromatography with plasmid preparation kits as recommended by the manufacturer (Qiagen, Inc.). The nucleotide and deduced amino acid positions of the HAVcr-1 cDNA are according to the previously published sequence (10). Figure 1 shows a schematic drawing of the receptors encoded by the following HAVcr-1 cDNA constructs used in this study.
FIG. 1.
Schematic drawing of havcr-1 mutants. The extracellular domain of havcr-1 is composed of an N-terminal Cys-rich region that has homology to members of the immunoglobulin superfamily and a TSP-rich C-terminal region that contains 27 repeats of the consensus PTTTTL, which resembles mucin-like molecules. There are four putative N-glycosylation sites in havcr-1 (tree-like structures): two in the Cys-rich region and two in the TSP-rich region. Synthetic oligonucleotides coding for a FLAG peptide were inserted into the unique NcoI site of the coding region of the HAVcr-1 cDNA in the TSP-rich region, between the end of the hexameric repeats and the transmembrane domain, and the receptor was termed flag. The N-glycosylation mutant receptors mut1, mut2, and mut3 differ from flag in their numbers and positions of N-glycosylation sites in the Cys-rich region. The Cys-rich region of flag was deleted, and the resulting receptor was termed d1−.
(i) pDR2HAVcrFlag.
To introduce a tag epitope into the extracellular domain of havcr-1, the 1,520-bp DNA fragment coding for nt 175 to 1695 of the HAVcr-1 cDNA was cut from pDR2GL37/5 (10) with BstYI and AseI and filled in with DNA polymerase I Klenow enzyme. This cDNA fragment was subcloned into pBlueBacIII (Invitrogen Corp.), cut with BamHI and HindIII, and filled in with Klenow enzyme, and the resulting construct was named pBBHAVcr-1. Annealed synthetic oligonucleotides HAVcr-flag1+ and HAVcr-1flag1− (Table 1), which code for a FLAG peptide and an SmaI restriction site, were inserted into the unique NcoI site of pBBHAVcr-1 at nt 1037 of the HAVcr-1 cDNA. The nucleotide sequences of several clones were analyzed, and a clone containing the correct orientation of the inserted oligonucleotides was selected and termed pBBHAVcrFlag. This plasmid was linearized with HindIII, filled in with Klenow enzyme, and cut with AflII, and the resulting 1,377-bp cDNA fragment containing nt 218 to 1695 of the HAVcr-1 cDNA was subcloned into the AflII and XbaI filled-in sites of pDR2GL37/5. The resulting construct, pDR2HAVcrFlag, codes for a FLAG-tagged receptor termed flag (Fig. 1).
TABLE 1.
Oligonucleotides used in this study
| Namea | Sequenceb |
|---|---|
| cr196-218+ | ATG CAT CTT CAA GTG GTC ATC TT |
| mut1+ | AGC TTT TGG GGA ACC TTT CAC GCA GGG ATG TCT CTT TGA CTA TAG CAA ATA CAG |
| mut1− | CTG TAT TTG CTA TAG TCA AAG AGA CAT CCC TGC GTG AAA GGT TCC CCA AA |
| mut2− | ACT GTC AGA CAC AGC TGT ATT TGC TAT AGT CAA AGA GAC ATC CCT GCG TGA AAG GACPvuII CCC CAA AAG CTT ATA GCG TGT CTC CTT CCG ATA GGT GAC GTG GGT TCC ATT GGT CCA HindIII GAC AAT GCC ATC TGG GCA TGA GAA AAC AGA ACA TGT GCC TC |
| mut3− | GAC CCC CAA AAG CTT ATA GCG TGT CTC CTT CCG ATA GGT GAC GTG GGT TCC TTG GGT HindIII CCA GAC AAT GCC AT |
| crfD1+ | TGT CAG TTC TTA AGC CTC ATC CTA CAT CTG GCA GAT TCT GTA GCC GAT TCT GTA AAT AflII GTT GAT GGA GTG GCA GGT ACC CCA CCC |
| crfD1− | GCT AGT TCG AAC AGT TCT GAC AAT TGG AGT AGT GAC TCT GGG TGG GGT ACC TGC CAC TCC BstBI |
| HAVcr-flag1+ | CAT GGA CTA CAA GGA CGA CGA TGA CAA GCC CGG GTT SmaI |
| HAVcr-flag1− | CAT GAA CCC GGG CTT GTC ATC GTC GTC CTT GTA GTC SmaI |
The last character of the oligonucleotide name indicates its polarity. −, negative sense; +, positive sense.
Mutations are in boldface type. Restriction sites are underlined and named below the line.
(ii) pDR2HAVcrD1−.
The BamHI/XbaI HAVcr-1 cDNA fragment of pDR2HAVcrFlag coding for the flag receptor was subcloned into the BamHI- and XbaI-cut pFastBac1 (Gibco BRL, Inc.), and the resulting construct was named pFBHAVcrFlag. Synthetic oligonucleotides crfD1+ and crfD1− (Table 1) were annealed, filled in with Taq DNA polymerase (Perkin-Elmer Cetus, Inc.), cut with AflII and BstBI, and cloned into AflII- and BstBI-cut pFBHAVcrFlag. The resulting construct, pFastBacHAVcrD1−, contains a deletion of 288 nt of the Cys-rich region of HAVcr-1 cDNA (from nt 283 to 570) which encompasses all the Cys residues present in the extracellular domain. The BamHI/XbaI DNA fragment of pFastBacHAVcrD1− was subcloned into BamHI- and XbaI-cut pDR2. The resulting construct, pDR2HAVcrD1−, codes for a receptor named d1−, which contains a deletion of almost all the Cys-rich region of havcr-1 (Fig. 1).
(iii) pDR2HAVcrmut2.
The second putative N-glycosylation site of the Cys-rich region of havcr-1 was mutated from NLS to VLS by PCR technology. Briefly, a 299-bp cDNA fragment from nt 196 to 495 was amplified by PCR with oligonucleotides cr196-218+ and mut2− (Table 1) as the primers and pDR2HavcrFlag as the template. The amplified cDNA fragment, which codes for the N-to-V change at amino acid 82 and a silent A-to-T mutation at nt 432 that creates a unique HindIII site, was digested at nt 218 with AflII and at nt 482 with PvuII and cloned into AflII- and PvuII-cut pFBHAVcrFlag, and the resulting plasmid was named pFBHAVcrmut2. The BamHI/XbaI HAVcr-1 cDNA fragment of pFBHAVcrFlagmut2 was subcloned into BamHI- and XbaI-cut pDR2. The resulting construct, pDR2HAVcrmut2, codes for the mut2 receptor (Fig. 1), which contains only the first N-glycosylation site in the Cys-rich region.
(iv) pDR2HAVcrmut3.
The first and second putative N-glycosylation sites of the Cys-rich region of havcr-1 were mutated from NGT and NLS to QGT and VLS, respectively, by PCR technology. To do so, a 210-bp HAVcr-1 PCR fragment coding for nt 218 to 428 of the HAVcr-1 cDNA was amplified with pDR2HAVcrflag as the template and synthetic oligonucleotides cr196-218+ and mut3− (Table 1) as the primers. This PCR fragment, which contains an N-to-Q change at amino acid 65, was cut with AflII and HindIII and cloned into AflII- and HindIII-cut pFBHAVcrmut2, and the resulting construct was named pFBHAVcrmut3. The BamHI/XbaI HAVcr-1 cDNA fragment of pFBHAVcrmut3 was subcloned into BamHI- and XbaI-cut pDR2. The resulting construct, pDR2HAVcrmut3, codes for the mut3 receptor (Fig. 1) with no N-glycosylation sites in the Cys-rich region.
(v) pDR2HAVcrmut1.
An HAVcr-1 cDNA construct containing only the first N-glycosylation site of the Cys-rich region was obtained by mutating back to wild type (wt) the second N-glycosylation site of mut3. To do so, synthetic oligonucleotides mut1+ and mut1− (Table 1) were annealed and cloned into HindIII- and PvuII-cut pFBHAVcrmut3. The resulting construct, pFBHAVcrmut1, codes for a QGT mutated site at amino acids 65 to 67 and an NLS wt N-glycosylation site at amino acids 82 to 84 of the Cys-rich region of havcr-1. The BamHI/XbaI HAVcr-1 cDNA fragment of pFBHAVcrmut1 was subcloned into BamHI- and XbaI-cut pDR2. The resulting construct, pDR2HAVcrmut1, codes for an mut1 receptor (Fig. 1) containing only the second N-glycosylation site in the Cys-rich region.
Transfection and selection of dog cells expressing the HAVcr-1 cDNA constructs.
Perro6D cell monolayers grown in 25-cm2 flasks were transfected with 1 μg of plasmid and 10 μl of DOSPER as recommended by the manufacturer (Boehringer Mannheim, Inc.) in a final volume of 3 ml of OptiMEM (Gibco BRL, Inc.). After overnight incubation at 37°C under 5% CO2, 3 ml of EMEM containing 10% FBS was added to each 25-cm2 flask. After 24 h of incubation, the transfection medium was changed to selection medium consisting of EMEM–10% FBS containing 250 μg of hygromycin per ml and cells were selected for 1 week. After 7 days of treatment with hygromycin, approximately 20 to 30% of the transfected cells survived whereas none of the mock-transfected cells resisted the antibiotic selection. The expression of the transfected receptors remained constant in the hygromycin-resistant dog cells passaged for at least 6 months with selection medium (EMEM–10% FBS–250 μg of hygromycin per ml).
Cell surface ELISA.
Expression of HAVcr-1 in AGMK cells and dog cell transfectants was analyzed by a cell surface enzyme-linked immunosorbent assay (ELISA) as described previously (10). Briefly, duplicate wells of unfixed cells grown in 96-well plates were treated with twofold dilutions of 190/4 or M2 MAbs for 1 h at room temperature (RT), washed extensively, and treated with a 1:1,000 dilution of affinity-purified peroxidase-labeled anti-mouse antibody for 1 h at RT. After being washed extensively, 100 μl of the one-component tetramethylbenzidine (TMB) substrate (Kirkegaard & Perry Laboratories, Inc.) was added per well. The reaction was stopped with 1% H2SO4, and absorbance was read at 450 nm. The difference in the optical densities at 450 nm (OD450) of the duplicate wells was within the experimental error of 5 to 10%, average values were highly reproducible, and backgrounds were below 0.1 OD450 unit. The mean OD450 of duplicate wells was calculated and graphed versus the log10 of the antibody dilution.
Western blot analysis.
Confluent monolayers of dog cell transfectants or AGMK cells grown in 25-cm2 flasks were scraped into 1 ml of phosphate-buffered saline (PBS), pelleted, resuspended in 0.2 ml of reticulocyte standard buffer (RSB; 10 mM NaCl, 10 mM Tris-HCl [pH 7.2]) containing 1% Nonidet P-40 (NP-40), and incubated for 2 min at RT. After the nuclei were removed by centrifugation at 12,000 × g, the total amount of protein in the supernatant was determined by the Bradford method with a protein assay kit (Bio-Rad Laboratories); these cytoplasmic extracts were used immediately or stored at −70°C. Approximately one-third of the cytoplasmic extract obtained from a cell monolayer grown in a 25-cm2 flask, corresponding to 20 to 25 μg of total protein, was loaded per well and fractionated in sodium dodecyl sulfate (SDS)–polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride (PVDF; Immobilon P; Millipore, Inc.) membranes, probed with a 1:1,000 dilution of rabbit anti-GST2 Ab, and stained with a 1:5,000 dilution of alkaline phosphatase-labeled goat anti-rabbit antibody. The substrate 5-bromo-4-chloro-3-indolylphosphate (BCIP)–nitroblue tetrazolium was used as recommended by the manufacturer (Kirkegaard & Perry Laboratories).
Tunicamycin treatment.
AGMK cells and dog cell transfectants grown in 96-well plates were treated with 1 μg of tunicamycin per ml in EMEM–10% FBS for 3, 2, or 1 day (or were untreated) and incubated at 37°C under 5% CO2. After treatment, expression of the 190/4 and M2 epitopes at cell surfaces was determined by ELISA as described above.
In vitro removal of N-linked glycans.
For complete deglycosylation, cytoplasmic extracts (20 to 25 μg) of AGMK cells and dog cell transfectants were denatured in 0.5% SDS–1% β-mercaptoethanol at 100°C for 10 min and treated with 500 U of PNGase F at 37°C for 1 h in 50 mM Na2PO4–1% NP-40 (pH 7.5) as recommended by the manufacturer (New England Biolabs). For partial deglycosylation, cytoplasmic extracts were denatured and treated with twofold dilutions of PNGase F (1:20 to 1:640) in 50 mM Na2PO4–1% NP-40 (pH 7.5) at 37°C for 1 h. After PNGase F treatment, cell extracts were analyzed by Western blot analysis with rabbit anti-GST2 Ab as described above.
HAV binding assay.
Binding of HAV HM175 to AGMK cells and dog cell transfectants was quantitated by radioimmunoassay as reported previously (10) but with minor modifications. HAV was purified from 20 15-cm2 dishes containing confluent monolayers of AGMK cells infected with HAV HM175 at a multiplicity of infection (MOI) of 1 50% tissue culture infectious dose (TCID50)/cell for one week at 35°C in a CO2 incubator. Cytoplasmic extracts were prepared as described above in 5 ml of RSB–1% NP-40 and treated with 1% SDS and 1% Sarkosyl overnight at RT, and HAV was pelleted through a 4-ml 40% sucrose-NTE (150 mM NaCl, 10 mM Tris-HCl [pH 7.4], 1 mM EDTA [pH 8.0]) cushion by centrifugation at 40,000 rpm for 4 h at 4°C in a Beckman SW40 rotor. The pelleted virus was resuspended in 2 ml of NTE, aliquoted, and stored at −70°C. Purified HAV HM175 had a titer of approximately 1010 TCID50s/ml as assessed with 96-well plates containing confluent monolayers of FRhK-4 cells (4). The binding assay was done with 80%-confluent monolayers of AGMK cells and dog cell transfectants grown in 96-well plates. Duplicate wells were treated with 50 μl of a 1:20 dilution of purified HAV HM175 in EMEM–10% FBS for 1 h at 35°C in a CO2 incubator. Monolayers were washed four times with EMEM–10% FBS, fixed with 80% methanol, blocked with 5% bovine serum albumin in PBS, incubated with 50 μl of 125I-labeled human anti-HAV Ab, washed four times with PBS, and exposed to X-ray film (XAR-2; Kodak) with an intensifying screen for 24 to 96 h. After exposure, the 96-well plates were stained with 1% crystal violet and absorbance at 595 nm was determined in an ELISA plate reader (Bio-Rad Laboratories), which indicated that similar numbers of cells (within 5 to 10% similar) were present in each well. Densitometric analysis of the autoradiography was performed on a Macintosh Quadra950 computer with the public domain NIH Image program (written by Wayne Rasband at the National Institutes of Health).
MAb 190/4-mediated inhibition of binding of purified HAV HM175 to AGMK cells and dog cell transfectants was also quantitated by radioimmunoassay as described previously (10). Briefly, 80%-confluent cell monolayers grown in 96-well plates were treated with 50 μl of 20-μg/ml anti-havcr-1 MAb 190/4 per well or left untreated for 1 h at 35°C. Cells were infected with 50 μl of a 1:10 dilution of purified HAV HM175 in EMEM–10% FBS per well for 1 h at 35°C in a CO2 incubator. Monolayers were washed four times with PBS–2% FBS, fixed with 80% methanol, blocked with 5% bovine serum albumin in PBS, incubated with 50 μl of 125I-labeled human anti-HAV Ab per well, washed four times with PBS, and exposed to X-ray film (XAR-2; Kodak) with an intensifying screen for 24 to 96 h. Densitometric analysis of the autoradiography was performed as described above. The percentage of inhibition of binding of HAV to the cells was calculated by comparing the integrated OD of wells treated with MAb 190/4 and those left untreated.
HAV infectivity assay.
Dog cell transfectants grown in eight-well permanox culture slides (Nunc, Inc.) were infected with 107 to 108 TCID50s of HAV KRM003 purified as described above or mock infected for 6 h at 35°C under 5% CO2. After being washed three times with EMEM–10% FBS, cells were incubated for 3 days at 35°C under 5% CO2. Cell monolayers were fixed with cold acetone and stained with a 1:1,000 dilution of human anti-HAV Ab and then with a 1:300 dilution of FITC-labeled goat anti-human Ab. Immunofluorescent micrographs were taken with a Zeiss Axioscope microscope at ×1,000 with an oil immersion objective.
RESULTS
Characterization of havcr-1.
To characterize havcr-1, we raised antibodies against an E. coli-expressed GST2 recombinant protein which consisted of the TSP-rich region of havcr-1 fused to the C terminus of GST. Western blot analysis (Fig. 2) of AGMK cell extracts showed that the anti-GST2 Ab reacted with two bands (lane 1): a faint band of approximately 65 kDa and a major band of approximately 62 kDa (lane 1). Treatment of AGMK cell extracts with PNGase F resulted in the collapse of the two bands into a band of 56 kDa (lane 2), which suggested that the 62- and 65-kDa bands were different N-glycosylated forms of havcr-1. To further analyze this observation, we transfected Perro6D cells with pDR2GL37/5 or the vector pDR2 and selected hygromycin-resistant cells, which were termed cr5 or DR2 cells, respectively. A large number of hygromycin-resistant cells, between 10 and 30% of the monolayer, were selected in 3 to 7 days. Western blot analysis of cr5 cells showed that the anti-GST2 Ab reacted with three bands (lane 3): a 65-kDa band that comigrated with the faint band observed in AGMK cells, a prominent 62-kDa band that comigrated with the 62-kDa band of AGMK cells, and a 59-kDa band that was not detected in AGMK cells. Treatment of cr5 cell extracts with PNGase F (lane 4) resulted in the collapse of the three bands into a major band of approximately 56 kDa (lane 4) which comigrated with the band observed in AGMK cells extracts treated with PNGase F (compare lanes 2 and 4). No bands were detected in DR2 cell extracts that were left untreated (lane 7) or treated with PNGase F (lane 8), which indicated that the anti-GST2 Ab reacted specifically against havcr-1. Similar amounts of total protein were loaded in each well; therefore, the higher levels of havcr-1 detected in the dog cell transfectants indicated that dog cells expressed more havcr-1 than AGMK cells (compare lanes 3 and 4 with lanes 1 and 2). Removal of N-glycans from havcr-1 resulted in a 56-kDa protein (lanes 2 and 4), which is larger than the 46 kDa predicted for the protein backbone. This 10-kDa difference may be due to O-linked glycans attached to the mucin-like region (10), phosphorylation, or other posttranslational modification of havcr-1.
Expression of havcr-1 glycosylation mutants.
To further understand the pattern of glycosylation of havcr-1, we introduced mutations in the HAVcr-1 cDNA and transfected the constructs into Perro6D cells. The anti-GST2 antisera did not react with havcr-1 expressed at the cell surfaces of AGMK and cr5 cells probably due to the O glycosylation of the mucin-like region. Therefore, we introduced a tag epitope into the mucin-like region to monitor the expression of havcr-1 at the cell surfaces of the dog cell transfectants. To do so, we cloned a 36-bp synthetic DNA fragment coding for the FLAG octapeptide and four additional amino acids into the NcoI site at nt 1037 of the HAVcr-1 cDNA. Perro6D cells transfected with the resulting construct, pDR2HAVcrFlag, were selected with hygromycin and termed flag cells. A cell surface ELISA using the anti-FLAG MAb M2 showed that the FLAG epitope was expressed at the cell surfaces of flag cells. Western blot analysis of flag cells (Fig. 2) showed that the anti-GST2 Ab reacted with three bands: two minor bands of 67 and 61 kDa and a major band of 64 kDa (lane 5) which migrated slightly higher than similar bands observed in cr5 cells (compare lane 5 with lane 3) due to the 12-amino-acid insertion. After treatment of flag cell extracts with PNGase F (lane 6), the three bands collapsed into a major 58-kDa band observed in AGMK and cr5 cells (compare lane 6 with lanes 2 and 4). These results indicated that the insertion of the FLAG epitope into havcr-1 did not affect the expression of the receptor as assessed by Western blot analysis.
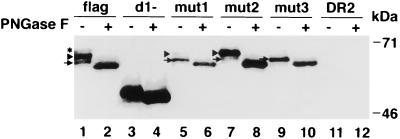
To determine whether the TSP-rich region of havcr-1 was N glycosylated, we deleted almost all the Cys-rich region of the HAVcr-1 cDNA. The resulting construct, pDR2HAVcrD1−, was transfected into Perro6D cells, and hygromycin-resistant cells were selected and termed d1− cells. Western blot analysis of d1− cells (Fig. 3) showed that the anti-GST2 Ab reacted with a 51-kDa band (lane 3) that was converted to a 49-kDa band upon PNGase F treatment (lane 4), which suggested that only one of the two N-glycosylation sites of the TSP-rich region was glycosylated. It should be pointed out that untreated and PNGase F-treated DR2 cell extracts did not react with the anti-GST2 Ab (lanes 11 and 12), which indicated that the bands observed in Fig. 3 were havcr-1 specific.
FIG. 3.
Western blot analysis of havcr-1 mutants expressed in dog cells. Cytoplasmic extracts of flag (lanes 1 and 2), d1− (lanes 3 and 4), mut1 (lanes 5 and 6), mut2 (lanes 7 and 8), and mut3 (lanes 9 and 10) cells or of control DR2 cells (lanes 11 and 12) were prepared in RSB–1% NP-40. Cell extracts containing 20 to 25 μg of total protein were treated with 500 U of PNGase F (+ [lanes 2, 4, 6, 8, 10, and 12]) or mock treated (− [lanes 1, 3, 5, 7, 9, and 11]), separated by SDS–12.5% polyacrylamide gel electrophoresis, transferred to PVDF membranes, and probed with rabbit anti-GST2 Ab directed against the TSP-rich region of havcr-1. The asterisk marks the 67-kDa band corresponding to a form of havcr-1 containing three N-glycans. Arrowheads point to 64-kDa havcr-1 forms containing two N-glycans. Arrows point to 62-kDa havcr-1 forms containing only one N-glycan. The positions of prestained molecular mass markers and their sizes are shown on the right.
The motif (Asn-X-Thr/Ser) of the first and second N-glycosylation sites of the Cys-rich region of havcr-1 were altered by site-directed mutagenesis to prevent glycosylation. The electrophoretic mobilities of the N-glycosylation mutants and the flag receptor were compared by Western blot analysis with the anti-GST2 Ab (Fig. 3). The first N-glycosylation site of havcr-1 located at amino acid positions 65 to 67 of the Cys-rich region was mutated from NGT to QGT. The resulting construct, pDR2HAVcrmut1, was transfected into Perro6D cells, and hygromycin-resistant cells were selected and named mut1 cells. Western blot analysis of mut1 cells (lane 5) showed that the anti-GST2 Ab reacted with a faint 64-kDa band and a strong 61-kDa band which comigrated with similar bands present in flag cells (compare lanes 1 and 5). A similar analysis was done with the second N-glycosylation site of the Cys-rich region located at amino acid 82 to 84 of havcr-1, which was mutated from NLS to VLS. The resulting construct, pDR2HAVcrmut2, was transfected into Perro6D cells, and hygromycin-resistant cells were selected and named mut2 cells. Western blot analysis of mut2 cells (lane 7) showed that the anti-GST2 Ab reacted with two bands: a minor band of 61 kDa and a major band of 64 kDa. These mut2 bands comigrated with similar bands detected in flag and mut1 cells (compare lanes 1, 5, and 7). Perro6D cells were also transfected with pDR2HAVcrmut3, a construct in which the first and second N-glycosylation sites of the Cys-rich region of havcr-1 were mutated. Hygromycin-resistant cells were selected and termed mut3 cells. Western blot analysis of mut3 cells showed that the anti-GST2 Ab reacted only with a 61-kDa band (lane 9), which comigrated with similar 61 kDa bands found in flag, mut1, and mut2 cells (lanes 1, 5, 7, and 9). Treatment of cell extracts of flag, mut1, mut2, and mut3 cells with PNGase F resulted in the collapse of all havcr-1-specific bands into comigrating 58-kDa bands (lanes 2, 6, 8, and 10). This result indicated that the 67- to 61-kDa bands that reacted with the anti-GST2 Ab were different N-glycosylation forms of havcr-1. The anti-GST2 Ab recognized a 67-kDa band only in flag cells (lane 1) and not in mut1, mut2, and mut3 cells, which expressed havcr-1 mutants lacking one or two N-glycosylation sites. Therefore, our results suggested that the fully N-glycosylated form of havcr-1 contained three sites occupied by N-glycans.
Partial N-deglycosylation analysis of the havcr-1 mutants.
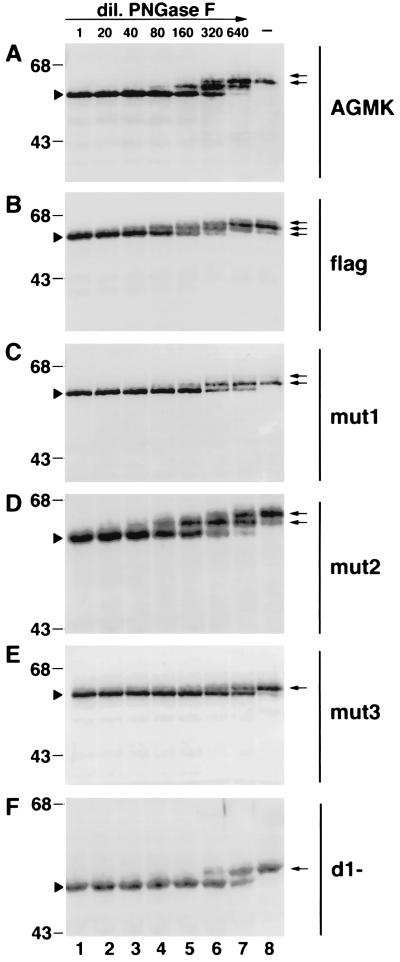
To confirm our observation that three of the four putative N-glycosylation sites of havcr-1, two in the Cys-rich region and one in the TSP-rich region, were glycosylated (Fig. 3), we performed partial PNGase F digestion of cell extracts of AGMK cells and dog cell transfectants. Western blot analysis of AGMK cell extracts (Fig. 4A) treated with undiluted PNGase F (lane 1) resulted in a single 56-kDa band (lane 1) corresponding to a completely N-deglycosylated form of havcr-1. A second havcr-1 discrete band of approximately 59 kDa, containing N-glycans attached to only one site, appeared at the 1/20 dilution of PNGase F and became the strongest band at the 1/320 dilution of PNGase F (lane 6). A third band of approximately 62 kDa, representing an havcr-1 form containing two N-glycans, appeared very faintly at the 1/160 dilution of PNGase F (lane 5) and became the most prominent band at the 1/640 dilution of PNGase F (lane 7). A faint 65-kDa band observed at the 1/640 dilution of PNGase F (lane 7) corresponded to a minor form of havcr-1 containing three N-glycans. Comparison of the mobilities of the discrete bands produced by the partial PNGase F digestion (lanes 1 to 7) with the mobilities of the havcr-1 bands present in untreated extracts indicated that AGMK cells expressed two N-glycosylated forms of havcr-1 (lane 8): a major 62-kDa form containing two N-glycans and a minor 65-kDa form containing three N-glycans. A similar analysis indicated that flag cells expressed three havcr-1 N-glycosylation forms (Fig. 4B) of 61, 64, and 67 kDa (lane 8) that contained one to three N-glycans, respectively, which migrated a little bit higher than similar bands observed in AGMK and cr5 cells due to the insertion of the 12 amino acid residues containing the FLAG epitope. The presence in flag and cr5 cells but not in AGMK cells of a form of havcr-1 containing only one N-glycan may be due to differences in the glycosylation machinery of the dog and monkey cells. The partial PNGase F deglycosylation analysis revealed that mut1 (Fig. 4C) and mut2 (Fig. 4D) cells expressed receptors of 61 and 64 kDa containing one and two N-glycans, respectively, which migrated slower than the 58-kDa band corresponding to the fully N-deglycosylated form of the same receptor. Partial PNGase F deglycosylation analysis of mut3 cells (Fig. 4E) clearly showed the existence of only one N-glycosylated form of havcr-1 of 61 kDa (lane 8), which migrated higher than the 58-kDa fully N-deglycosylated form of mut3 (lane 1). This result suggested that one of the N-glycosylation sites of the TSP-rich region was glycosylated. Further confirmation came from the partial PNGase F digestion of cytoplasmic extracts of d1− cells (Fig. 4F), which showed only one N-glycosylated form of the d1− receptor migrating as a 51-kDa band (lane 8) higher than the fully N-deglycosylated 49-kDa form (lane 1). Considered together, the partial N-deglycosylation results further suggested that havcr-1 contained three N-glycosylated sites, two in the Cys-rich region and one in the TSP-rich region. Direct biochemical confirmation of the existence of these three N-glycosylation sites will require radiolabeling of havcr-1 with sugars and determination of the number of sugar-labeled tryptic peptides.
FIG. 4.
Partial PNGase F digestion of havcr-1 mutants. Cytoplasmic extracts of AGMK (A), flag (B), mut1 (C), mut2 (D), mut3 (E), and d1− (F) cells were prepared in RSB–1% NP-40. Aliquots containing 20 to 25 μg of total protein of each cytoplasmic extract were treated with undiluted PNGase F (500 U) (lanes 1), treated with twofold dilutions of PNGase F from 1:20 to 1:640 (lanes 2 to 7), or mock treated (lanes 8). After treatment, proteins were separated by SDS–12.5% polyacrylamide gel electrophoresis, transferred to PVDF membranes, and probed with rabbit anti-GST2 Ab directed against the TSP-rich region of havcr-1. Arrowheads point to completely N-deglycosylated forms of the receptors. Arrows point to N-glycosylated forms of the receptors expressed in the different cell lines. The positions of prestained molecular mass markers and their sizes in kilodaltons are shown on the left. dil., dilution.
N-glycosylation-dependent binding of MAb 190/4 to havcr-1.
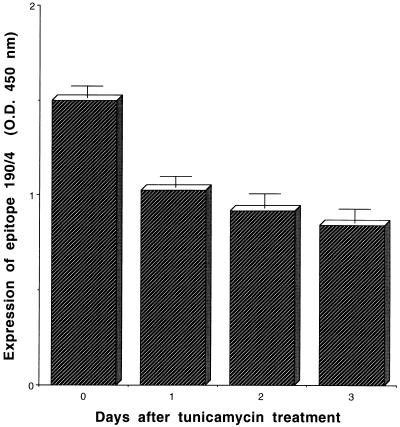
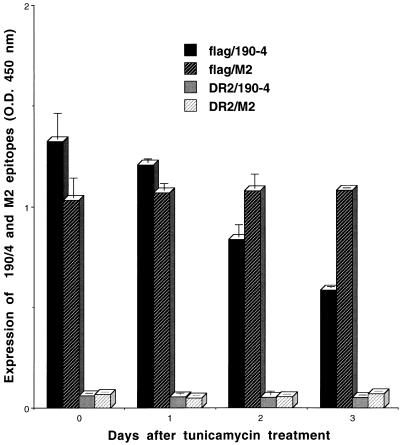
To analyze whether the expression of protective epitope 190/4 was glycosylation dependent, we treated AGMK cells with 10 μg of tunicamycin per ml for 12 to 72 h and analyzed the expression of epitope 190/4 at cell surfaces by ELISA (Fig. 5). The expression level of the 190/4 epitope at the cell surfaces of AGMK cells was reduced 31.6, 38.8, and 43.7% after 12, 48, and 72 h of treatment with tunicamycin, respectively. These data suggested that the attachment of N-glycans to havcr-1 was important for the expression of the 190/4 epitope but did not address the possibility that cell surface expression of havcr-1 was affected by the tunicamycin treatment. Therefore, we used flag cells to monitor the expression of FLAG-tagged havcr-1 at cell surfaces using MAb M2. An ELISA of cell surface expression (Fig. 6) showed that while expression of the M2 epitope remained constant in flag cells, expression of the 190/4 epitope was reduced 8.76, 36.96, and 55.78% after treatment with tunicamycin for 1, 2, and 3 days, respectively. Since the M2 and 190/4 epitopes are in the same molecule, our data indicated that expression of the 190/4 epitope was indeed affected by tunicamycin treatment and suggested that N-glycans are required for the expression of the 190/4 epitope. It should be pointed out that, regardless of the tunicamycin treatment, DR2 cells did not express the M2 and 190/4 epitopes at their cell surfaces (Fig. 6).
FIG. 5.
Effect of tunicamycin treatment on the expression of protective epitope 190/4 in AGMK cells. AGMK cells were treated with 1 μg of tunicamycin per ml for 0, 1, 2, and 3 days. After treatment, expression of the 190/4 epitope was quantitated by cell surface ELISA with a saturating concentration of MAb 190/4 (200 μg/ml), peroxidase-labeled goat anti-mouse Ab, and TMB substrate. Absorbance was read at 450 nm. Data are means of values from quadruplicate wells ± standard errors of the means.
FIG. 6.
Effect of tunicamycin treatment on the expression of the protective 190/4 epitope in dog cell transfectants. Dog cells expressing the flag receptor or control DR2 cells transfected with the vector alone were treated with 1 μg of tunicamycin per ml for 0, 1, 2, and 3 days. After treatment, expression of the 190/4 (flag/190-4 and DR2/190-4) and M2 (flag/M2 and DR2/M2) epitopes was determined by cell surface ELISA with a saturating concentration of 190/4 and M2 MAbs (200 μg/ml), peroxidase-labeled goat anti-mouse Ab, and TMB substrate. Absorbance was read at 450 nm. Data are means of values from duplicate wells ± standard errors of the means.
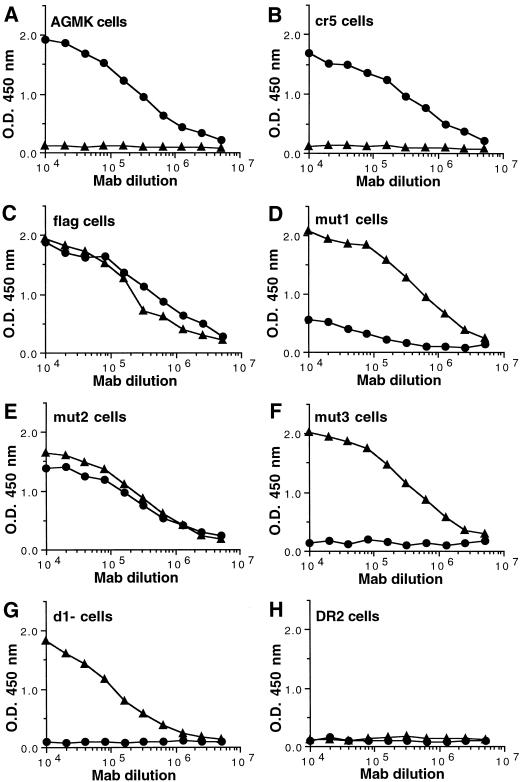
To further confirm the importance of N-glycans in the expression of the 190/4 epitope, we analyzed binding of MAb 190/4 to the cell surfaces of the dog cell transfectants. Due to the FLAG tag inserted into havcr-1 mutants, we were able to compare the levels of expression of the M2 and 190/4 epitopes at cell surfaces by ELISA (Fig. 7). Control AGMK and cr5 cells expressed similar levels of the 190/4 epitope (Fig. 7A and B) and did not express the M2 epitope (Fig. 7A and B), whereas control DR2 cells reacted with neither 190/4 nor M2 MAbs (Fig. 7H). On the other hand, flag cells expressed both epitopes at their surfaces (Fig. 7C). Similar levels of the M2 epitope were expressed in flag and mut1 cells (Fig. 7C and D), but the mutation of the first N-glycosylation site in mut1 resulted in an at least fivefold decrease in the expression of the 190/4 epitope (Fig. 7D) compared to that in flag cells. As expected, the mutation of the second N-glycosylation site in mut2 did not affect the expression of the 190/4 epitope (compare Fig. 7E and C). The double mutation of the first and second N-glycosylation sites in mut3 (Fig. 7F) did not affect the expression of the M2 epitope, but it drastically reduced the expression of the 190/4 epitope to undetectable levels. These data indicated that the expression of the 190/4 epitope was affected by the lack of glycans attached to the first N-glycosylation site in mut1 and completely abrogated by the lack of glycans at both N-glycosylation sites in mut3. However, the lack of glycans at the second N-glycosylation site by itself (mut2) did not affect the expression of the 190/4 epitope but had a synergistic effect in combination with the mutation at the first N-glycosylation site (in mut3) which further reduced the expression of the 190/4 epitope. These data also suggested that the lack of glycans at the first N-glycosylation site affected the conformation of the Cys-rich region, which was further deformed by the lack of glycans at the second N-glycosylation site.
FIG. 7.
Expression of the protective epitope 190/4 at the cell surfaces of dog cell transfectants. Expression of the 190/4 and M2 epitopes at the cell surfaces of AGMK (A), cr5 (B), flag (C), mut1 (D), mut2 (E), mut3 (F), d1− (G), and DR2 (H) cells was determined by ELISA with twofold dilutions of MAb 190/4 (circles) or M2 (triangles). Absorbance at 450 nm (y axis) was ploted versus the log10 of the MAb dilution (x axis). Data are means of results for duplicate wells; duplicate values varied by less than 10%. The results are from one experiment which was repeated at least two times, with an experimental error of approximately 5 to 10%.
Since our data indicated that N-glycans of the Cys-rich domain of havcr-1 are important for binding of MAb 190/4, it was of interest to determine whether the Cys-rich domain itself is required for binding. ELISA of the cell surfaces of d1− cells (Fig. 7H) showed that the deletion of the Cys-rich region of havcr-1 completely abrogated the expression of the 190/4 epitope but did not affect the expression of the M2 epitope, which hinted that MAb 190/4 may bind to the Cys-rich region of havcr-1.
Binding of HAV to dog cells expressing havcr-1 mutants.
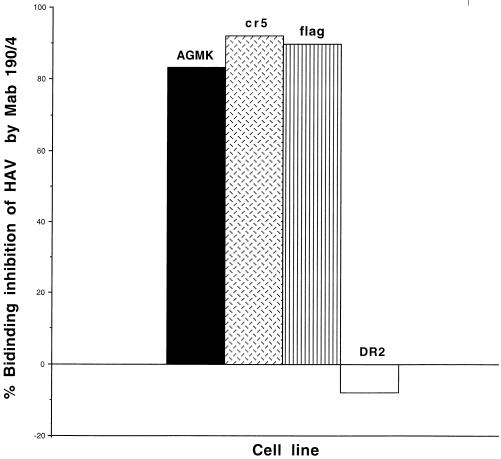
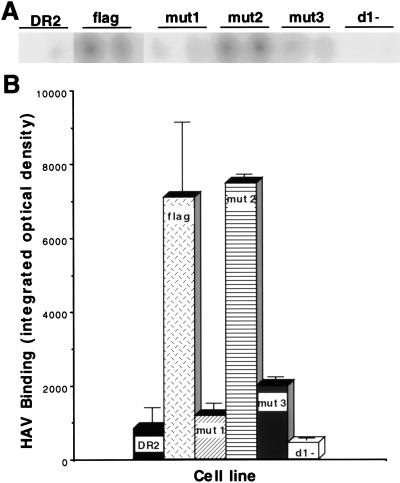
To determine whether HAV binds specifically to havcr-1 expressed at the cell surfaces of dog cell transfectants, AGMK, cr5, flag, and DR2 cells grown in 96-well plates were treated with protective MAb 190/4 or mock treated for 1 h and infected with purified HAV HM175 for 1 h at 35°C. After being washed extensively, monolayers were fixed and bound HAV was detected with 125I-labeled human anti-HAV antibody. The resulting autoradiograph was scanned, and binding of HAV HM175 to MAb 190/4- and mock-treated cells was quantitated with the NIH Image program (Fig. 8). Treatment with MAb 190/4 inhibited binding of HAV HM175 to AGMK, cr5, and flag cells by 83.4, 91.1, and 89.9%, respectively, whereas the same treatment of DR2 cells produced no observable inhibition. These data indicated that HAV bound specifically to havcr-1 expressed on the dog cell transfectants and that the FLAG peptide inserted into the TSP-rich region of havcr-1 did not interfere with binding of HAV to the flag receptor. Therefore, we used the dog cell transfectants to analyze the importance of the N-glycans and Cys-rich region of havcr-1 for binding of HAV (Fig. 9). Binding of HAV HM175 to mut1 cells was approximately seven times lower than binding to flag cells, which indicated that the mutation of the first N-glycosylation site of havcr-1 had a significant effect on virus binding. Similar levels of binding of HAV HM175 to mut2 and flag cells indicated that the mutation of the second N-glycosylation site did not affect virus binding. Binding of HAV HM175 to mut3 cells was also reduced significantly mainly due to the lack of glycans attached to the first rather than to the second N-glycosylation site. Interestingly, binding of HAV HM175 to d1− cells was reduced approximately 16 times compared to binding to flag cells, which clearly indicated that the Cys-rich region of havcr-1 was required for binding of HAV.
FIG. 8.
MAb 190/4-mediated binding inhibition of HAV to dog cell transfectants. Monolayers of AGMK, cr5, flag, and DR2 cells grown in 96-well plates were treated with 20 μg of MAb 190/4 per ml or mock treated, infected with purified HAV HM175 for 1 h, and washed extensively. HAV bound to the cells was detected with 125I-labeled human anti-HAV Ab and autoradiography. The binding inhibition was calculated by densitometric analysis of the autoradiography results as percentages calculated from the integrated OD remaining after monolayers were treated with protective MAb 190/4 compared to those of untreated monolayers. The results are from one experiment which was repeated three times, with an experimental error of approximately 5 to 10%.
FIG. 9.
Binding of HAV to dog cells transfected with the HAVcr-1 mutants. Dog cell transfectants expressing flag, mut1, mut2, mut3, and d1− receptors or control DR2 cells transfected with the vector alone were grown in 96-well plates and infected with purified HAV HM175 for 1 h at 35°C. After extensive washing, monolayers were fixed and HAV bound to the cells was detected with 125I-labeled human anti-HAV Ab. (A) Direct autoradiography of the 96-well plate showing duplicate wells of each cell line. (B) HAV binding to each cell line was determined by densitometric analysis of the autoradiograph shown in panel A. Data are means of results from duplicate wells ± standard errors of the means.
Susceptibility of dog cell transfectants to HAV infection.
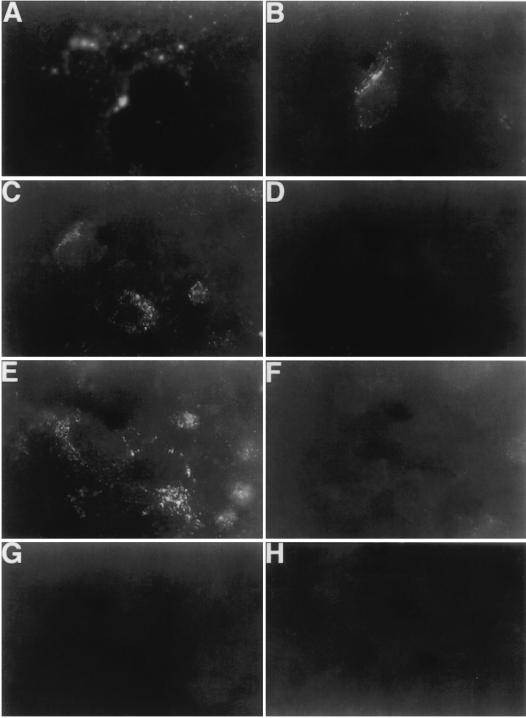
Dog cells are not susceptible to HAV infection or transfection of infectious HAV RNA, which indicated that these cells contain an internal block(s) to HAV replication (4). However, infection of cr5 cells with HAV KRM003 resulted in the characteristic granular cytoplasmic fluorescence of HAV-infected cells, which lasted for several weeks but became undetectable after 1 month postinfection. This limited level of susceptibility of the dog cell transfectants to HAV infection is similar to that previously reported for Ltk− cells transfected with the HAVcr-1 cDNA (10). Since receptor-minus but otherwise fully HAV-susceptible cell lines have not yet been identified, we used the dog cell transfectants to analyze the importance of the N-glycans and Cys-rich region in limited HAV infection of the dog cells. To do so, we infected 80%-confluent monolayers of dog cell transfectants with an MOI of 100 to 1,000 TCID50s of HAV KRM003 per cell and studied the cells by indirect immunofluorescence analysis after staining them with human anti-HAV antibody (Table 2). At 3 days postinfection, HAV-specific immunofluorescence was detected in 100% of the AGMK cells and approximately 60% of cr5, flag, and mut2 cells but not in mut1, mut3, d1−, and DR2 cells. Figure 10 shows the characteristic granular cytoplasmic fluorescence of HAV-infected cells observed in HAV-infected AGMK, cr5, flag, and mut2 cells (Fig. 10A to C and E) but not in HAV-infected mut1, mut3, d1−, and DR2 cells (Fig. 10D and F to H), which indicated that the havcr-1 Cys-rich region and its first N-glycosylation site are required for the limited susceptibility of dog cell transfectants to HAV infection. These results correlated well with our data on binding of protective MAb 190/4 and HAV (Fig. 7 and 9) to the dog cell transfectants, which indicated that binding of HAV to havcr-1 is relevant to the susceptibility of the dog cell transfectants to HAV infection.
TABLE 2.
Indirect immunofluorescence analysis of dog cell transfectants infected with HAV
| Cells | Level of IFa |
|---|---|
| AGMK | ++++ |
| cr5 | +++ |
| flag | +++ |
| mut1 | − |
| mut2 | +++ |
| mut3 | − |
| d1− | − |
| DR2 | − |
Percentage of cells showing the characteristic cytoplasmic granular immunofluorescence (IF) of HAV-infected cells, as follows: ++++, 76 to 100%; +++, 51 to 75%; and −, not detectable.
FIG. 10.
Detection of HAV antigen in the cytoplasms of dog cell transfectants by indirect immunofluorescence analysis. Monolayers of AGMK (A), cr5 (B), flag (C), mut1 (D), mut2 (E), mut3 (F), d1− (G), and DR2 cells grown in eight-well slides were infected with an MOI of 100 to 1,000 TCID50s of purified HAV KRM003 per cell for 6 h, washed extensively, and incubated at 35°C for 3 days. Cells were fixed with cold acetone and stained with human anti-HAV Ab and FITC-labeled goat anti-human Ab. Mock-infected cells did not immunofluoresce (data not shown).
DISCUSSION
Very little is known about the interaction of HAV with its cellular receptor havcr-1. Since picornaviruses such as PV and human rhinovirus interact with the first immunoglobulin domain of their cellular receptors, the homology of the Cys-rich region of havcr-1 with members of the immunoglobulin superfamily suggested that HAV might interact with the N-terminal Cys-rich region of havcr-1 (10). The first domain of the PV receptor, which is necessary and sufficient for PV binding and uptake (for a review, see reference 23), contains two N-glycans that are not required for PV binding. Therefore, it was of interest to study the pattern of N-glycosylation of havcr-1 and determine the role of the N-glycans in HAV binding. Our results indicated that one of the two N-glycosylation sites of the TSP-rich region and the first N-glycosylation site of the Cys-rich region are preferentially glycosylated, resulting in the accumulation of a prominent form of havcr-1 containing two N-glycans (Fig. 2 to 4). Our data also suggested that the second N-glycosylation site of the Cys-rich region of havcr-1 is also glycosylated but to a much lower extent than the first site (Fig. 3). In d1− and mut3 cells, we detected only one N-glycosylation form of havcr-1 (Fig. 3), which suggested that the putative N-glycosylation site of the TSP-rich region adjacent to the transmembrane domain is not occupied by N-glycans due to its proximity to the plasma membrane. Consequently, we never detected an havcr-1 form containing N-glycans attached to all four putative N-glycosylation sites. Treatment of AGMK cells with tunicamycin, which inhibits N glycosylation, showed that N-glycans are required for the expression of protective epitope 190/4 (Fig. 5 and 6). Unfortunately, the tunicamycin treatment inhibited HAV replication in AGMK cells (data not shown) and we were unable to determine the receptor function of N-deglycosylated forms of havcr-1 in AGMK cells. Therefore, we mutated the first site (mut1), second site (mut2), or both (mut3) N-glycosylation sites of the Cys-rich region of havcr-1 (Fig. 1) and showed that binding of protective MAb 190/4 (Fig. 7) and HAV (Fig. 9), as well as the limited susceptibility of dog cells to HAV infection (Table 2), was drastically reduced in mut1 and mut3 but that binding was not affected in mut2 cells. These data suggested that N-glycans are needed to maintain the conformation of havcr-1 required for binding and uptake of HAV or that the first N-glycan itself forms part of the binding site. This N-glycosylation requirement for receptor function is not unique to the HAV–havcr-1 interaction and has also been observed for the interaction of measles virus with CD46 (13, 14).
Several viruses, such as PV, mouse hepatitis virus, and human immunodeficiency virus, bind to the first domain of receptors that are members of the immunoglobulin superfamily (for a review, see reference 23). Our results showed that the N-terminal Cys-rich region of havcr-1 is required for binding of protective MAb 190/4 and HAV (Fig. 7 and 9), which suggested that HAV binds to this region. Further experiments involving binding of HAV to chimeric receptors containing only the Cys-rich domain of havcr-1 will be required to determine whether this N-terminal region of havcr-1 is sufficient for virus binding and uptake. The role of the TSP-rich mucin-like region of havcr-1 in binding and uptake of HAV is currently unknown. However, it is possible that the only function of the TSP-rich region is to extend the Cys-rich region above the cell surface as in the “lollipop on a stick” model (7). Further research will be required to elucidate the function of the TSP-rich region in the HAV–havcr-1 interaction.
The lack of a receptor-minus but otherwise fully susceptible cell line for HAV replication has been problematic. Although we devoted considerable effort to finding one, we have not been able to identify such a cell line (4). In this paper we showed that dog cells transfected with HAVcr-1 cDNA gain limited susceptibility to HAV infection (Table 2 and Fig. 10), similar to what was previously shown for Ltk− transfectants (10). A similar phenomenon of poor picornavirus replication has recently been shown for enterovirus 70 in NIH 3T3 cells expressing decay-accelerating factor (CD55), the enterovirus 70 cellular receptor (11). We are currently trying to adapt HAV to grow in the dog cell transfectants by multiple rounds of infection and serial passaging of the infected cells for several months. It is possible that inhibitors or the lack of factors required for the efficient replication of HAV is responsible for the limited susceptibility of Ltk− and dog cells to HAV infection. If this is the case, the Ltk− and dog cell transfectants expressing havcr-1 could be used as tools to identify factors required for efficient HAV replication.
ACKNOWLEDGMENTS
We thank Stephen Feinstone, Dino Feigelstock, Sara Gagneten, Barry Falgout, and Hira Nakhasi for comments on the manuscript.
REFERENCES
- 1.Ashida M, Hamada C. Molecular cloning of the hepatitis A virus receptor from a simian cell line. J Gen Virol. 1997;78:1565–1569. doi: 10.1099/0022-1317-78-7-1565. [DOI] [PubMed] [Google Scholar]
- 2.Bishop N E, Anderson D A. RNA-dependent cleavage of VP0 capsid protein in provirions of hepatitis A virus. Virology. 1993;197:616–623. doi: 10.1006/viro.1993.1636. [DOI] [PubMed] [Google Scholar]
- 3.Cohen J I, Ticehurst J R, Feinstone S M, Rosenblum B, Purcell R H. Hepatitis A virus cDNA and its RNA transcripts are infectious in cell culture. J Virol. 1987;61:3035–3039. doi: 10.1128/jvi.61.10.3035-3039.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dotzauer A, Feinstone S M, Kaplan G. Susceptibility of nonprimate cell lines to hepatitis A virus infection. J Virol. 1994;68:6064–6068. doi: 10.1128/jvi.68.9.6064-6068.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gauss-Muller V, Lottspeich F, Deinhardt F. Characterization of hepatitis A virus structural proteins. Virology. 1986;155:732–736. doi: 10.1016/0042-6822(86)90234-5. [DOI] [PubMed] [Google Scholar]
- 6.Innis B L, Snitbhan R, Kunasol P, Laorakpongse T, Poopatanakool W, Kozik C A, Suntayakorn S, Suknuntapong T, Safary A, Tang D B, et al. Protection against hepatitis A by an inactivated vaccine. JAMA. 1994;271:1328–1334. [PubMed] [Google Scholar]
- 7.Jentoft N. Why are proteins O-glysylated? Trends Biochem Sci. 1990;15:291–294. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 8.Jia X Y, Summers D F, Ehrenfeld E. Primary cleavage of the HAV capsid protein precursor in the middle of the proposed 2A coding region. Virology. 1993;193:515–519. doi: 10.1006/viro.1993.1157. [DOI] [PubMed] [Google Scholar]
- 9.Jia X Y, Tesar M, Summers D F, Ehrenfeld E. Replication of hepatitis A viruses with chimeric 5′ nontranslated regions. J Virol. 1996;70:2861–2868. doi: 10.1128/jvi.70.5.2861-2868.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan G, Totsuka A, Thompson P, Akatsuka T, Moritsugu Y, Feinstone S M. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J. 1996;15:4282–4296. [PMC free article] [PubMed] [Google Scholar]
- 11.Karnauchow T M, Tolson D L, Harrison B A, Altman E, Lublin D M, Dimock K. The HeLa cell receptor for enterovirus 70 is decay-accelerating factor (CD55) J Virol. 1996;70:5143–5152. doi: 10.1128/jvi.70.8.5143-5152.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kusov Y, Kazachkov Y A, Dzagurov G K, Khozinskaya G A, Balayan M S, Gauss-Muller V. Identification of precursors of structural proteins VP1 and VP2 of hepatitis A virus. J Med Virol. 1992;37:220–227. doi: 10.1002/jmv.1890370313. [DOI] [PubMed] [Google Scholar]
- 13.Maisner A, Alvarez J, Liszewski M K, Atkinson D J, Atkinson J P, Herrler G. The N-glycan of the SCR 2 region is essential for membrane cofactor protein (CD46) to function as a measles virus receptor. J Virol. 1996;70:4973–4977. doi: 10.1128/jvi.70.8.4973-4977.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maisner A, Scheneider-Schaulies J, Liszewski M K, Atkinson J P, Herrler G. Binding of measles virus to membrane cofactor protein (CD46): importance of disulfide bonds and N-glycans for the receptor function. J Virol. 1994;68:6299–6304. doi: 10.1128/jvi.68.10.6299-6304.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 16.Murphy A J M, Kung A L, Swirski R A, Schimke R T. cDNA expression cloning in human cells using the pλDR2 episomal vector system. Methods (Orlando) 1992;4:111–131. [Google Scholar]
- 17.Schultheiss T, Sommergruber W, Kusov Y, Gauss-Muller V. Cleavage specificity of purified recombinant hepatitis A virus 3C proteinase on natural substrates. J Virol. 1995;69:1727–1733. doi: 10.1128/jvi.69.3.1727-1733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swirski R A, Van Den Berg D, Murphy A J M, Lambert C M, Friedberg E C, Schimke R T. Improvements in the Epstein-Barr-based shuttle vector system for direct cloning in human tissue culture cells. Methods (Orlando) 1992;4:133–142. [Google Scholar]
- 19.Tesar M, Jia X Y, Summers D F, Ehrenfeld E. Analysis of a potential myristoylation site in hepatitis A virus capsid protein VP4. Virology. 1993;194:616–626. doi: 10.1006/viro.1993.1301. [DOI] [PubMed] [Google Scholar]
- 20.Totsuka A, Moritsugu Y. Hepatitis A vaccine development in Japan. In: Nishioka K, Suzuki H, Mishiro S, Oda T, editors. Viral hepatitis and liver disease. Tokyo, Japan: Springer-Verlag; 1994. pp. 509–513. [Google Scholar]
- 21.Weitz M, Baroudy B M, Maloy W L, Ticehurst J R, Purcell R H. Detection of a genome-linked protein (VPg) of hepatitis A virus and its comparison with other picornaviral VPgs. J Virol. 1986;60:124–130. doi: 10.1128/jvi.60.1.124-130.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werzberger A, Mensch B, Kuter B, Brown L, Lewis J, Sitrin R, Miller W, Shouval D, Wiens B, Calandra G, et al. A controlled trial of a formalin-inactivated hepatitis A vaccine in healthy children. N Engl J Med. 1992;327:453–457. doi: 10.1056/NEJM199208133270702. [DOI] [PubMed] [Google Scholar]
- 23.Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]