Abstract
Replication of mixtures of two or more human immunodeficiency virus type 1 (HIV-1) variants would be expected to result in the eventual selection of the fittest virus due to Darwinian competition among the variants. The relative proportions of known HIV-1 variants (which may differ only by a single nucleotide from a standard “wild-type” virus, HIV-1HXB2) in mixed viral cultures were quantified by analysis of automated sequence signals of reverse transcriptase PCR products. With this method, the relative levels of replicative fitness of several zidovudine (3′-azidothymidine)-resistant HIV-1HXB2 variants were estimated under controlled in vitro conditions by measuring the rate of change in the proportions of viral variants as they replicated in cell cultures both in the presence and in the absence of drug selection pressure. These variants were engineered to contain commonly observed zidovudine resistance mutations in the HIV-1 reverse transcriptase (M41L, K70R, T215Y, and M41L+T215Y). In the absence of zidovudine, all variants tested displayed reduced replicative fitness compared to wild-type HIV-1HXB2. The order of relative fitness was wild type > K70R ≫ T215Y = M41L+T215Y > M41L. Mixed cultures in the presence of zidovudine showed a dose-dependent selection pressure against the wild-type virus which varied according to the resistance profile of each virus. The information gathered from this approach provides insight into competition among multiple HIV-1 variants, which likely occurs in vivo with drug selection pressure, and may be applicable in more complex mathematical models for predicting the emergence of HIV-1 variants after the initiation of antiretroviral therapy.
To date, all antiretroviral agents used as monotherapy to treat human immunodeficiency virus type 1 (HIV-1) infection appear to result in the selection of drug-resistant mutants, which ultimately limits the benefits of such therapy (for a review, see reference 16). HIV-1 variants resistant to zidovudine (3′-azidothymidine), commonly used for the treatment of HIV-1 infection, have been isolated from patients who have undergone long-term monotherapy or combination antiretroviral drug therapy (9, 12, 13, 18, 21, 24). These variants can contain in the HIV-1 reverse transcriptase (RT) multiple mutations which tend to emerge in a specific order (2). Typically, HIV-1 with a K70R substitution is selected after several months of zidovudine treatment; this variant is often later replaced by a variant with a T215Y substitution. Higher-level zidovudine resistance coincides with the emergence of variants containing both the M41L and the T215Y substitutions and/or other multiple substitutions (9, 19). Interestingly, these drug-resistant isolates appear to persist for some time after the cessation of zidovudine therapy (1, 3, 25) but are eventually replaced by virus with fewer mutations (6–8). The large population size and rapid replication of HIV-1 (10, 28) result in the rapid selection of the fittest HIV-1 variants in the presence of antiretroviral drugs, as even small differences in viral fitness quickly lead to the replacement of the less fit HIV-1 variants in a population (4). This phenomenon has allowed the estimation of the in vivo fitness of two zidovudine-resistant HIV-1 variants (6, 7).
Previously, it was not possible to assess the impact of single base changes on viral fitness by measuring differences in the growth kinetics of common zidovudine-resistant HIV-1 variants in cell cultures (or final virus titers), as these differences were too small to detect by standard culture methods (14).
The aim of this study was to assess the effect of mutations conferring zidovudine resistance on HIV-1 replicative fitness by measuring the change in the proportions of viruses with these mutations (in a constant genetic background) during several replication cycles in a well-defined culture system (9). These data were used to generate values for the relative levels of fitness in this environment of common zidovudine-resistant HIV-1 variants as a function of drug selection pressure. Relative viral fitness values derived in this way may aid in the understanding of HIV-1 evolution, particularly the emergence of drug-resistant HIV-1 in patients receiving antiretroviral drugs.
MATERIALS AND METHODS
Viral passage.
Cloned HIV-1 variants with specific mutations associated with zidovudine resistance (previously prepared by site-directed mutagenesis) were mixed together in known ratios. These mixed viral samples were used to infect 4 × 106 MT-4 cells at a multiplicity of infection of <0.01. After 4 to 6 days, 0.5 ml of the supernatant was removed and used to reinfect a fresh aliquot of 4 × 106 MT-4 cells, and this process was repeated. The relative proportions of wild-type and mutant viral strains were determined by automated sequencing after each viral passage (see below).
PCR amplification and sequencing.
RT-PCR amplification of the RT region of HIV-1 was carried out as previously described (9, 20). PCR products were subsequently sequenced with Sequenase dye terminator chemistry and an automated fluorescence sequencer (Applied Biosystems). The data were imported into the software packages Factura and Sequence Navigator for further analysis of relative peak heights as previously described (9).
Plasma sample preparation.
HIV-1 RNA from infected plasma samples (or mixtures of plasma) from two HIV-1-positive patients was extracted with 1.8 ml of guanidine thiocyanate (5 M), incubated at 65°C for 10 min, and precipitated with isopropanol. The resulting pellet was washed once with ethanol. These plasma samples had been previously quantified for HIV-1 RNA with the Roche Amplicor assay (22, 23). The 5′ RT region of the virus from these patients was amplified by nested RT-PCR and sequenced on an ABI 373 automated sequencer as previously described (19).
Calculation of fitness.
The relative levels of fitness of two viruses can be approximated from the equation p/q = [p(0)/q(0)] × (fitness)T, where p is the proportion of less fit virus, q is the proportion of more fit virus, 0 indicates time zero, and T is the time in viral generations (6). In a single cycle of viral replication (in the absence of drug), for every 100 viable progeny produced from a wild-type virus, a virus which is 10% less fit will produce 90 viable progeny in each replicative cycle. For competition among more than two viral variants, relative fitness values can be estimated in a similar fashion (6).
The number of plaques (N) produced in a typical experiment done to determine the dose-response curve for the inhibition of viral replication (50% inhibitory concentration [IC50] curve) can be approximated by the equation N = N(0)/[1 + ([drug]/IC50x)], where x is a factor related to the steepness of the dose-response curve. For a drug-resistant isolate (with a higher ICr50), the relative number of plaques arising from the resistant strain (Nr) at a particular drug concentration can be approximated from the equation Nr = f(0)/[1 + ([drug]/ICr50x)], where f(0) is the relative viral fitness in the absence of drug. As a function of drug concentration, the relative number of viable progeny (drug-resistant virus/wild-type virus) produced can be expected to be approximated by the ratio of the dose-response curves (N/Nr).
RESULTS
Reproducibility of RT-PCR and sequence peak height determinations.
We first determined the feasibility of using RT-PCR and automated sequence analysis to quantify relative and absolute HIV-1 copy numbers in plasma samples. HIV-1 RNA in plasma samples from two HIV-1-infected patients (with slightly different RT sequences) was quantified with the Roche Amplicor assay. The samples were mixed together to produce equal copy numbers from each patient. Seven independent RT-PCR and automated sequencing runs were performed. The amount of HIV-1 RNA in each sample was estimated from the relative peak heights on the electropherograms from the first nine bases which differed between the two patient samples (Table 1). When all nine positions were used to calculate the relative amounts of the two variants, the average ratio was 59.5% (range, 47 to 65%; coefficient of variation, 10%), indicating that the method is both consistent when equal proportions are used and reasonably reproducible. Note that the relative peak heights were somewhat base dependent. For example, the relative signal at base 158 was consistently lower than that at base 170 (Table 1), indicating slightly different kinetics of base incorporation during the sequencing reactions (19). The values obtained, however, were consistent across all seven replicates (average coefficient of variation, <25%). Similar results were obtained when other bases were examined or when a different sequencing primer was used (data not shown).
TABLE 1.
Reproducibility of HIV-1 quantification by sequencing
| Replicate | Resulta for base:
|
Avg for run (± SD) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 17 | 50 | 116 | 137 | 152 | 158 | 160 | 161 | 170 | ||
| 1 | 84.6 | 61.5 | 62.5 | 50.0 | 31.3 | 34.4 | 83.9 | 80.8 | 86.6 | 64 ± 22 |
| 2 | 73.9 | 53.9 | 33.3 | 60.7 | 29.7 | 41.6 | 61.3 | 63.0 | 75.5 | 55 ± 16 |
| 3 | 78.9 | 79.4 | 14.3 | 67.5 | 53.3 | 41.7 | 78.6 | 71.4 | 85.4 | 63 ± 23 |
| 4 | 65.7 | 60.4 | 44.4 | 42.4 | 25.3 | 18.4 | 49.2 | 50.0 | 67.4 | 47 ± 17 |
| 5 | 73.8 | 69.2 | 63.1 | 54.8 | 30.0 | 28.9 | 80.0 | 63.2 | 91.0 | 62 ± 21 |
| 6 | 70.7 | 78.8 | 61.5 | 66.7 | 42.5 | 64.0 | 72.2 | 57.8 | 77.8 | 65 ± 11 |
| 7 | 66.7 | 80.0 | 36.8 | 59.3 | 68.8 | 15.4 | 71.7 | 65.0 | 79.0 | 60 ± 21 |
| Avg at each codon (± SD) | 73 ± 7 | 69 ± 11 | 45 ± 18 | 57 ± 9 | 40 ± 16 | 35 ± 16 | 79 ± 12 | 64 ± 10 | 80 ± 8 | |
The relative percentage of automated DNA signal strengths from two viral strains differing at the indicated bases is given for each of seven replicate runs of nested RT-PCR and automated sequencing.
Fitness of resistant variants in the absence of drug.
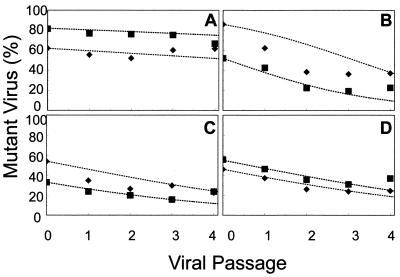
The relative ratios of wild-type HIV-1HXB2 and zidovudine-resistant HIV-1HXB2 variants in cultures containing predetermined mixtures of viruses were measured by population-based sequencing in the absence and presence of zidovudine in order to establish the effects of mutations on viral fitness under controlled conditions. Four HIV-1HXB2 variants previously prepared by site-directed mutagenesis were used in this study (13, 18); they had amino acid substitutions at RT codons K70R, M41L, T215Y, and M41L+T215Y. Each mutant was mixed with wild-type HIV-1HXB2 to produce mixtures of two or three different initial ratios of viruses ranging from about 30% mutant to 70% wild type to about 80% mutant to 20% wild type. The mixtures were cultured over 5 to 6 days, with approximately two replication cycles per viral passage, assuming a viral replication cycle of 2 to 3 days (5, 23). The ratio of mutant to wild-type HIV-1HXB2 was quantified after every passage for each mixture (Fig. 1). The data were used to predict relative levels of HIV-1 variant fitness by use of the equations given in Materials and Methods over the first four viral passages in order to minimize the impact of any possible competing mutants (Fig. 1). The average relative levels of fitness of the K70R, M41L, T215Y, and M41L+T215Y variants compared with that of the wild type were estimated from the curves to be 97, 80, 85, and 85%, respectively (Fig. 1), although the best-fit line did not match the data well in all cases (see Fig. 1B for an example). The order of fitness of the wild type and zidovudine-resistant variants in this environment from this set of experiments therefore appeared to be wild type > K70R ≫ T215Y = M41L+T215Y > M41L.
FIG. 1.
Replication competition between wild-type HIV-1HXB2 and HIV-1HXB2-based zidovudine-resistant clones. The relative proportions of DNA signal contributed by various zidovudine-resistant mutant viruses compared to wild-type virus over several passages in the absence of zidovudine are indicated for each of two (▪ and ⧫) independent initial starting ratios of mutant to wild type. Mutant viruses had the mutation K70R (A), M41L (B), T215Y (C), or M41L+T215Y (D). Dotted lines show the best fit.
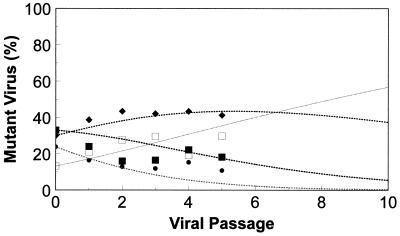
Because of the discrepancy between the data and the predicted values observed in some cases, a mixed infection of these four variants was examined in the absence of zidovudine in order to confirm the order of viral fitness determined above. Under these conditions, it was expected that the wild type—being the fittest—would quickly predominate; the K70R variant (only slightly less fit) would remain relatively abundant; and the T215Y and M41L variants (which were the least fit) would quickly be outgrown and decline to very low levels. These predictions were confirmed by mixing the four HIV-1HXB2 variants together in approximately equal quantities and allowing them to replicate in competition for five passages (Fig. 2). Predicted values based on the relative fitness values calculated above for the wild type and the HIV-1HXB2 variants (Fig. 2) were consistent with the observed data points.
FIG. 2.
Replication competition of a mixture of several isolates in the absence of zidovudine. Nearly equal mixtures of wild-type virus (□) and three zidovudine-resistant variants, K70R (⧫), M41L (•), and T215Y (▪), were allowed to replicate in the absence of zidovudine. Observed data are indicated by the data points, while data calculated based upon the relative fitness values from Fig. 1 and the equations given in Materials and Methods are indicated by the dotted lines.
The T215Y and M41L+T215Y variants appeared to have nearly equal relative levels of fitness when grown in competition with the wild type (Fig. 1C and D). This was also the case when they were allowed to replicate in competition with each other in the absence of drug (Fig. 3A). Thus, individual reductions in fitness resulting from the substitutions at codons 41 and 215 may not necessarily be additive.
FIG. 3.
Effect of zidovudine selection pressure on growth of mutant viruses with equal initial levels of fitness. (A) Virus with mutation M41L+T215Y was grown in competition with virus with mutation T215Y at nearly equal starting quantities in the absence of zidovudine. (B and C) Mixtures of wild-type and mutant viruses at an initial ratio of 9:1 were allowed to replicate in the presence of 0.2 (▪) or 2 (⧫) μM zidovudine. Mutant virus in the mixture was virus M41L+T215Y (B) or the less zidovudine-resistant virus T215Y (C). Data points represent observed values, and dotted lines represent values calculated based upon the fitness values from Fig. 1, the known IC50 values, and the equations given in Materials and Methods.
Fitness of resistant variants in the presence of drug.
The addition of zidovudine to cultures with mixed populations of HIV-1 variants would be expected to create selection pressure favoring the growth of drug-resistant mutants in a manner related to the zidovudine concentration and the sensitivity of the variant to the drug. To test this prediction, mixtures of 90% wild-type HIV-1HXB2 plus 10% T215Y variant and 90% wild-type HIV-1HXB2 plus 10% M41L+T215Y variant were grown in the presence of 0.2 or 2 μM zidovudine for four passages (Fig. 3B and C). The data showed that the rate of increase in the proportion of the mutant was higher for both the T215Y variant and the M41L+T215Y variant at 2 μM zidovudine than at 0.2 μM zidovudine, indicating that the selection pressure for these zidovudine-resistant mutants was related to the drug concentration. In addition, the M41L+T215Y variant was selected more rapidly than the T215Y variant at both drug concentrations, indicating that fitness in the presence of drug was dependent on the resistance profiles of these viruses (in the absence of zidovudine, the T215Y and M41L+T215Y variants were approximately equally fit [Fig. 3A]).
Titration of selection pressure.
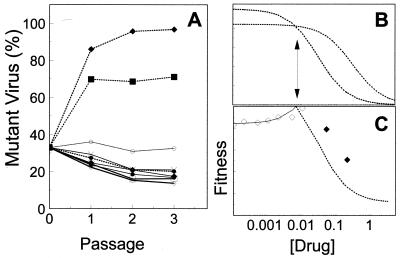
To further examine the effects of the concentration of zidovudine on the rate of selection of HIV-1 variants, a mixture of 70% wild-type HIV-1HXB2 and 30% T215Y variant was grown in the presence of a range of zidovudine concentrations (0 to 0.2 μM) for three passages (Fig. 4A). At low concentrations of zidovudine, the relative proportion of the T215Y variant in the culture decreased and the wild type predominated. However, at concentrations higher than about 0.01 μM zidovudine, the relative amount of the T215Y variant increased. These data were used to calculate the relative fitness of the predominant virus at each drug concentration (Fig. 4B). Below 0.01 μM zidovudine, the T215Y variant was approximately 10 to 20% less fit than the wild type, as indicated above. At a concentration close to 0.01 μM zidovudine, the relative fitness equaled 1.0, indicating that the wild type and the T215Y variant replicated at nearly constant rates and that their relative proportions did not change. Above 0.01 μM zidovudine, the wild type was less fit than the T215Y variant, and the resistant virus outgrew the wild type. Dose-response curves for wild-type HIV-1HXB2 and the T215Y variant are shown in Fig. 4C; these were determined with an initial fitness of the T215Y variant of 85%. The ratios of the dose-response curves coincided with the relative fitness values derived experimentally (Fig. 4B). Thus, the IC50 of a drug for a particular HIV-1 variant and the relative fitness of the variant in the absence of the drug can be used to approximate the characteristics of an evolving viral population at different drug concentrations.
FIG. 4.
Titration of zidovudine selection pressure. Nine mixtures of wild-type and T215Y viruses were allowed to replicate for three passages in the presence of increasing micromolar concentrations of zidovudine: 0 (—○—), 0.0001 (—•—), 0.0002 (—□—), 0.0005 (—⋄—), 0.001 (—♦—), 0.002 (—×—), 0.005 (··•··), 0.01 (··○··), 0.05 (··▪··), and 0.2 (··♦··). (A) Relative proportions of mutant viruses. (B) IC50 curves determined with a fitness value of 0.85 in the absence of zidovudine for the mutant and wild-type viruses. (C) Observed relative replicative fitness values for the two strains at each concentration (data points) and values determined by the ratio of the curves in panel B (dotted lines). Note that at concentrations higher than 0.01 μM zidovudine, the wild-type virus (⧫) had a lower relative fitness than the mutant virus, while at drug concentrations lower than this value, the wild-type virus was more fit (◊).
Selection pressure acting upon mixed infections.
Zidovudine therapy in vivo acts upon a number of competing zidovudine-resistant variants. Therefore, replication competition between wild-type virus and the K70R, T215Y, and M41L variants was studied as a simplified model of the in vivo situation. The four viruses were mixed in approximately equal proportions and cultured with 0.2 or 2 μM zidovudine for nine passages (Fig. 5). At 0.2 μM zidovudine, drug-resistant HIV-1 variants had a competitive advantage over the wild type, which quickly decreased to undetectable levels. Virus with the T215Y substitution was strongly selected and rapidly predominated, such that virtually 100% of the viral population consisted of the T215Y variant after about six passages, closely approximating the predicted values. In contrast, virus with the K70R mutation remained at about 30% of the viral population, while virus with the M41L mutation was initially competed out of the culture, dropping to about 5% of the population after three passages. Subsequently, the proportion of this variant appeared to increase to 30% of the viral population after nine passages (Fig. 5A). These selection trends became more exaggerated when the viruses were grown with 2 μM zidovudine due to the greater drug-related selection pressure. Wild-type HIV-1 levels declined, and the variant with the T215Y substitution took over almost immediately. The proportion of the variant with the K70R mutation remained relatively constant, and the variant with the M41L mutation quickly rebounded after a single passage (Fig. 5B). The overlapping proportions of viruses with specific resistance mutations (percentages added up to far greater than 100% after one or two passages) indicated that variants containing multiple mutations accumulated as the population evolved. In particular, since the M41L+T215Y variant had a competitive advantage over the T215Y variant in the presence of this concentration of zidovudine (Fig. 3), once this combination of mutations was generated, it was rapidly selected.
FIG. 5.
Replication of mixtures of viruses in the presence of zidovudine. Mixtures of wild-type HIV-1HXB2 (□) and three zidovudine-resistant variants, K70R (⧫), M41L (•), and T215Y (▪), similar to those shown in Fig. 2 were allowed to replicate in the presence of 0.2 (A) or 2 (B) μM zidovudine. Data points represent observed values, and dotted lines represent calculated values obtained with the values and methods indicated earlier. Extensive growth of the M41L+T215Y virus is indicated by the solid line.
DISCUSSION
The large population size and rapid rate of replication of HIV-1 in vivo (10, 28) imply that it can be considered an ideal Darwinian population which evolves on the basis of selection of the fittest strain (4, 11). For example, if a viral population has achieved a steady state, then the frequency of a given mutant can be approximated as the mutation frequency (estimated to be 10−4 to 10−5 per replicative cycle) divided by the negative coefficient of selection (4). One method of assessing the effect of a given mutation on HIV-1 fitness in a controlled environment is to perform replication competition assays with viruses having defined mutations and directly measure the change in the proportion of mixed bases at different positions in the RT gene over time from electropherograms (population-based sequencing) (9). Here we show that population-based sequencing of PCR products is a reasonably reproducible technique that can be used for determining relative (or absolute) amounts of specific variants of HIV-1 and that this information may be usefully applied in simple models of viral fitness and drug selection pressure.
Population-based sequencing could be used to determine absolute levels of virus by including a known amount of an internal standard differing from the material to be amplified at a few nucleic acid positions, as was done here (Table 1). This approach has several potential advantages over other methods of quantitative PCR. Viral quantification can be combined with experiments designed to examine biologically or clinically relevant regions of HIV-1, such as regions which confer drug resistance. For example, a double mutation at RT codon 215 which confers 16-fold resistance to zidovudine was detected in one of the HIV-1 samples examined here. It is also relatively easy to generate an internal standard, and the presence of PCR contamination can be directly checked simply by analyzing the sequences of the PCR products formed. Finally, in theory, this technique could be applicable to any virus of interest. While the repeatability of population-based sequencing appears to be acceptable, drawbacks of this approach include the relatively low throughput of the method compared to that of commercial methods of HIV-1 quantification (23) and the requirement for nested PCR steps to produce sufficient material for sequencing. However, advances in sequencing technology which increase throughput and sensitivity and decrease cost may make this a useful approach to viral quantification in the future.
In this study, population-based sequencing was used to determine the relative proportions of HIV-1 variants under well-controlled in vitro conditions during several cycles of viral replication, allowing viral evolution to be monitored at the level of the individual base. It should be noted, however, that even though HIV-1 was initially added as cloned material, it rapidly mutated and recombined into a population of related variants (quasispecies). Therefore, these experiments actually compared populations of variants with or without a given mutation. This constant variation and stochastic error likely account for the majority of the difference between the experimental data and the predicted curves.
Resistance to zidovudine is typically conferred by the stepwise accumulation of mutations at RT codon K70R, followed or replaced by T215F or T215Y, M41L, and others (2, 14). The results described here provide a rationale for the ordered appearance of these mutations as a function of the initial proportion of a given variant (determined by its fitness) and the drug selection pressure (determined by the level of drug resistance). In the absence of zidovudine, direct competition of wild-type HIV-1HXB2 with individual HXB2-based zidovudine-resistant variants led to the predominance of the wild type and a reduction in drug-resistant variants at a rate dependent upon their relative levels of fitness. In the case of the K70R variant (relative fitness, about 97%), the rate of reduction was relatively low. This result suggests that this variant would be the most prevalent single mutant in an equilibrium viral population (4) and is consistent with the observation that K70R is generally the first substitution selected upon the initiation of zidovudine therapy (2).
It was particularly interesting to observe the growth of both the M41L and the T215Y mutants in the presence of high concentrations of zidovudine (Fig. 5B and, to a lesser extent, Fig. 5A). Although the M41L and T215Y variants were added to the culture as separate clones, the high frequencies of both mutations imply that they must have become linked within individual variants (essentially all of the viruses appeared to have the codon 215 substitution). Once linkage occurred, viruses with the M41L+T215Y combination rapidly outgrew those with single M41L or T215Y mutations, due to the much higher degree of zidovudine resistance (64-fold) conferred by the combination versus the 16-fold or 4-fold resistance conferred by the T215Y or M41L single mutation, respectively (17) (Fig. 3). The M41L+T215Y mutation might have occurred as a result of sequential mutation of either variant or as a result of viral recombination between the M41L variant and the T215Y variant. This latter possibility seems more likely, as rapid recombination between codons has been demonstrated to occur under conditions similar to those described here (15). Regardless of the mechanism, these results demonstrate a stepwise accumulation of zidovudine resistance mutations in a manner similar to that observed in vivo.
Several objections to the approach described here for determining viral fitness can be made. The method does not distinguish between sequences derived from viable and nonviable viruses and therefore may not reflect the fitness of the viable population (26), and the viral background used in these experiments (HXB2) is a defective virus lacking a functional Nef protein (26). The first criticism does not appear to be valid since, by definition, nonviable viruses do not contribute progeny and therefore could not contribute to the changes in viral sequence observed. HIV-1HXB2 has a defective Nef region; however, the experiments described here were designed to determine the relative effects of specific mutations on viral replication in an isogenic background. HXB2 has been used in a similar way to examine the effects of specific mutations on drug resistance, and this method appears to have in vivo relevance (2, 9, 13–15, 19). In addition, no specific interactions between nef and one of the RT mutations examined have been demonstrated. However, it must be emphasized that the environment of the viral background, T-cell line, and culture conditions used can be extrapolated to other situations only with great caution. Indeed, no single viral background or cell type can be considered an ideal system.
Only a limited amount of in vivo fitness data is available to compare to the results reported here (6, 7). In these studies, the T215Y and M41L+T215Y variants were replaced (at a range of rates) under conditions in which zidovudine selection pressure was absent, implying a reduced fitness (10 to 25%) of these isolates compared with the isolates which replaced them (6, 7). These results are broadly similar to the value for the relative fitness of T215Y compared to the wild type obtained here. However, the sets of experiments are not directly comparable. The in vivo experiments examined fitness after the removal of zidovudine selection pressure, and other compensatory changes which stabilized the mutations were likely to have occurred during the period of zidovudine therapy. A further complication is that the typical “wild-type” virus may not be the variant which arises upon the removal of drug selection pressure (6, 7). For example, following the removal of zidovudine selection pressure, the bases at codon 215 (TAC) were replaced by a mixture of GAC and TCC rather than the more commonly observed ACC (6, 7). In fact, as one of the more drug-susceptible viruses, the wild type would be expected to be present only at extremely low levels following long-term antiretroviral therapy.
The variant which predominates upon cessation of therapy would represent contributions from its level of drug resistance (which helps to determine the relative prevalence of the mutant during therapy) and its inherent fitness in the absence of drug (which helps to determine the rate at which it could be expected to take over the population from the resistant mutant). These considerations may help to explain the apparent continued development of new mutations after the cessation of treatment which has been reported on several occasions. The M41L mutation appears to be relatively stable in vivo in the absence of selection (6), but the fact that the M41L mutation is rarely observed in untreated patients (6) implies a low level of fitness in a zidovudine-free environment. In our study, the proportion of the M41L variant declined very rapidly compared to that of the wild type (relative fitness, about 80%).
Taken together, these results suggest that the approach of Goudsmit et al. (6, 7) is a more useful tool for predicting the behavior of viral populations after cessation of therapy than the in vitro values calculated here. The approach used here may be particularly useful for models of the behavior of multiple strains of drug-resistant variants after the initiation of antiretroviral therapy and for defining the effects of known mutations on viral replication in a fixed environment.
REFERENCES
- 1.Albert J, Wahlberg J, Lundeberg J, Cox S, Sandstrom E, Wahren B, Uhlen M. Persistence of azidothymidine-resistant human immunodeficiency virus type 1 RNA genotypes in posttreatment sera. J Virol. 1992;66:5627–5630. doi: 10.1128/jvi.66.9.5627-5630.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher C A, O’Sullivan E, Mulder J W, Ramautarsing C, Kellam P, Darby G, Lange J M, Goudsmit J, Larder B A. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J Infect Dis. 1992;165:105–110. doi: 10.1093/infdis/165.1.105. [DOI] [PubMed] [Google Scholar]
- 3.Boucher C A, van Leeuwen R, Kellam P, Schipper P, Tijnagel J, Lange J M, Larder B A. Effects of discontinuation of zidovudine treatment on zidovudine sensitivity of human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1525–1530. doi: 10.1128/aac.37.7.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 5.Dimitrov D S, Willey R L, Sato H, Chang L J, Blumenthal R, Martin M A. Quantitation of human immunodeficiency virus type 1 infection kinetics. J Virol. 1993;67:2182–2190. doi: 10.1128/jvi.67.4.2182-2190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goudsmit J, de Ronde A, de Rooij E, de Boer R. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J Virol. 1997;71:4479–4484. doi: 10.1128/jvi.71.6.4479-4484.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goudsmit J, de Ronde A, Ho D D, Perelson A S. Human immunodeficiency virus fitness in vivo: calculations based on a single zidovudine resistance mutation at codon 215 of reverse transcriptase. J Virol. 1996;70:5662–5666. doi: 10.1128/jvi.70.8.5662-5664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gurusinghe A D, Land S A, Birch C, McGavin C, Hooker D J, Tachedjian G, Doherty R, Deacon N J. Reverse transcriptase mutations in sequential HIV-1 isolates in a patient with AIDS. J Med Virol. 1995;46:238–243. doi: 10.1002/jmv.1890460312. [DOI] [PubMed] [Google Scholar]
- 9.Harrigan P R, Kinghorn I, Bloor S, Kemp S D, Najera I, Kohli A, Larder B A. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J Virol. 1996;70:5930–5934. doi: 10.1128/jvi.70.9.5930-5934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 11.Holland J J, de la Torre J C, Clarke D K, Duarte E. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J Virol. 1991;65:2960–2967. doi: 10.1128/jvi.65.6.2960-2967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hooker D J, Tachedjian G, Solomon A E, Gurusinghe A D, Land S, Birch C, Anderson J L, Roy B M, Arnold E, Deacon N J. An in vivo mutation from leucine to tryptophan at position 210 in human immunodeficiency virus type 1 reverse transcriptase contributes to high-level resistance to 3′-azido-3′-deoxythymidine. J Virol. 1996;70:8010–8018. doi: 10.1128/jvi.70.11.8010-8018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellam P, Boucher C A, Larder B A. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc Natl Acad Sci USA. 1992;89:1934–1938. doi: 10.1073/pnas.89.5.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellam P, Boucher C A, Tijnagel J M, Larder B A. Zidovudine treatment results in the selection of human immunodeficiency virus type 1 variants whose genotypes confer increasing levels of drug resistance. J Gen Virol. 1994;75:341–351. doi: 10.1099/0022-1317-75-2-341. [DOI] [PubMed] [Google Scholar]
- 15.Kellam P, Larder B A. Retroviral recombination can lead to linkage of reverse transcriptase mutations that confer increased zidovudine resistance. J Virol. 1995;69:669–674. doi: 10.1128/jvi.69.2.669-674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuritzkes, D. R. 1996. Clinical significance of drug resistance in HIV-1 infection. AIDS 10(Suppl. 5):S27–S31. [DOI] [PubMed]
- 17.Larder, B. A. 1995. Viral resistance and the selection of antiretroviral combinations. J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 10(Suppl. 1):S28–S33. [PubMed]
- 18.Larder B A, Darby G, Richman D D. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 19.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 20.Larder B A, Kohli A, Kellam P, Kemp S D, Kronick M, Henfrey R D. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature. 1993;365:671–673. doi: 10.1038/365671a0. [DOI] [PubMed] [Google Scholar]
- 21.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 22.McDonald C K, Kuritzkes D R. Human immunodeficiency virus type 1 protease inhibitors. Arch Intern Med. 1997;157:951–959. [PubMed] [Google Scholar]
- 23.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 25.Rooke R, Tremblay M, Soudeyns H, DeStephano L, Yao X J, Fanning M, Montaner J S, O’Shaughnessy M, Gelmon K, Tsoukas C the Canadian Zidovudine Multi-Centre Study Group. Isolation of drug-resistant variants of HIV-1 from patients on long-term zidovudine therapy. AIDS. 1989;3:411–415. doi: 10.1097/00002030-198907000-00001. [DOI] [PubMed] [Google Scholar]
- 26.Smith M S, Koerber K L, Pagano J S. Long-term persistence of zidovudine resistance mutations in plasma isolates of human immunodeficiency virus type 1 of dideoxyinosine-treated patients removed from zidovudine therapy. J Infect Dis. 1994;169:184–188. doi: 10.1093/infdis/169.1.184. [DOI] [PubMed] [Google Scholar]
- 27.Wainberg M A. Reverse transcriptase fidelity and HIV-1 variation. Science. 1997;275:229–230. [PubMed] [Google Scholar]
- 28.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]